Abstract
The dorsal lingual epithelium, which is composed of taste buds and keratinocytes differentiated from K14+ basal cells, discriminates taste compounds and maintains the epithelial barrier. N6-methyladenosine (m6A) is the most abundant mRNA modification in eukaryotic cells. How METTL3-mediated m6A modification regulates K14+ basal cell fate during dorsal lingual epithelium formation and regeneration remains unclear. Here we show knockout of Mettl3 in K14+ cells reduced the taste buds and enhanced keratinocytes. Deletion of Mettl3 led to increased basal cell proliferation and decreased cell division in taste buds. Conditional Mettl3 knock-in mice showed little impact on taste buds or keratinization, but displayed increased proliferation of cells around taste buds in a protective manner during post-irradiation recovery. Mechanically, we revealed that the most frequent m6A modifications were enriched in Hippo and Wnt signaling, and specific peaks were observed near the stop codons of Lats1 and FZD7. Our study elucidates that METTL3 is essential for taste bud formation and could promote the quantity recovery of taste bud after radiation.
Subject terms: Stem-cell niche, Genetics research
Introduction
The lingual epithelium consists of non-taste epithelium and taste epithelium. The non-taste epithelium covers a large proportion of the tongue’s surface. In mice, fungiform papillae (FFP), surrounded by mechanosensory filiform papillae (FLP), are distributed in the front part of the dorsal lingual epithelium1. The single circumvallate papilla (CVP) in mice, containing numerous taste buds, is located in the midline of the posterior lingual epithelium, whereas singular FFP houses only one taste bud2.
The taste system is mediated by the taste buds and innervated sensory neurons. Murine taste buds contain 50–100 elongated epithelial cells, which can be categorized into several types (types I, type II, and type III)3. Through different specific receptors, taste buds can detect five taste qualities: bitter, salt, sweet, sour, and umami (savory)4–6. Dysgeusia is common in patients undergoing head and neck radiotherapy7–9. Some patients may recover from taste dysfunction after some months or years, but a small minority of patients may suffer from permanent taste loss10,11. Loss of taste buds after radiation is caused by natural taste cell death and the interruption of taste cell replenishment8.
Previous studies using lineage tracing indicate that basal cells expressing cytokeratin 5 (K5 or Krt5) and K14 are progenitors of both non-taste epithelium and taste epithelium in mice12. Taste buds undergo continuous turnover, with an average life span of 10–14 days, while the non-taste epithelium takes 5–7 days to be renewed13,14. In non-taste epithelium, K5/K14+ basal progenitors differentiate into K13+ (KRT13) keratinocytes, which make up the suprabasal epithelial layers of the FLP and FFP15,16. K5/K14+ basal progenitors can also generate new cells into taste buds, subsequently developing into mature taste cell types1. Type I cells resemble glia and are the most abundant cells present in taste buds. However, the specific function of type I cells remains elusive17,18. Some researchers regard type I cells as salt detector4. Type II cells can transduce different signals by detecting bitter, sweet, and umami tasting stimuli, whereas type III cells can transduce sour flavors19–21. These three different cell types can be identified via distinctive markers: type I cells express ecto-ATPase, NTPdase2; type II cells express α-gustducin, phospholipase Cβ2; and type III cells express NCAM and SNAP-2522–24.
As the most abundant mRNA modification, N6-methyladenosine (m6A) regulates mRNA fate, including stability, splicing, transport, localization, and translation25–28. Importantly, the m6A modification is a reversible and dynamic process. Recent studies have demonstrated that the m6A modification can be catalyzed by an RNA methyltransferase complex, which consists of methyltransferase-like 3 (METTL3), Wilms tumor 1-associated protein (WTAP), METTL14, and other proteins. This modification can be removed by m6A eraser proteins, including alkylation repair homolog protein 5 (ALKBH5) and fat mass and obesity-associated protein (FTO)29–33. Over the past few years, METTL3, the most important component of the RNA methyltransferase complex, has been reported to play critical functions in embryonic development, stem cell differentiation, and tumor progression28,34–39. However, the role of m6A modification in lingual epithelial homeostasis remains elusive.
Here, our group generated an epidermis-specific Mettl3 knockout mouse model and found that METTL3 was an essential RNA methyltransferase that regulated lingual epithelium progenitor differentiation and was crucial for taste bud development. Moreover, overexpression of Mettl3 promoted taste bud recovery from radiation injury. We also discovered a mechanistic pathway by identifying downstream target genes and signals.
Results
Deletion of Mettl3 in epidermal progenitors leads to taste bud defects
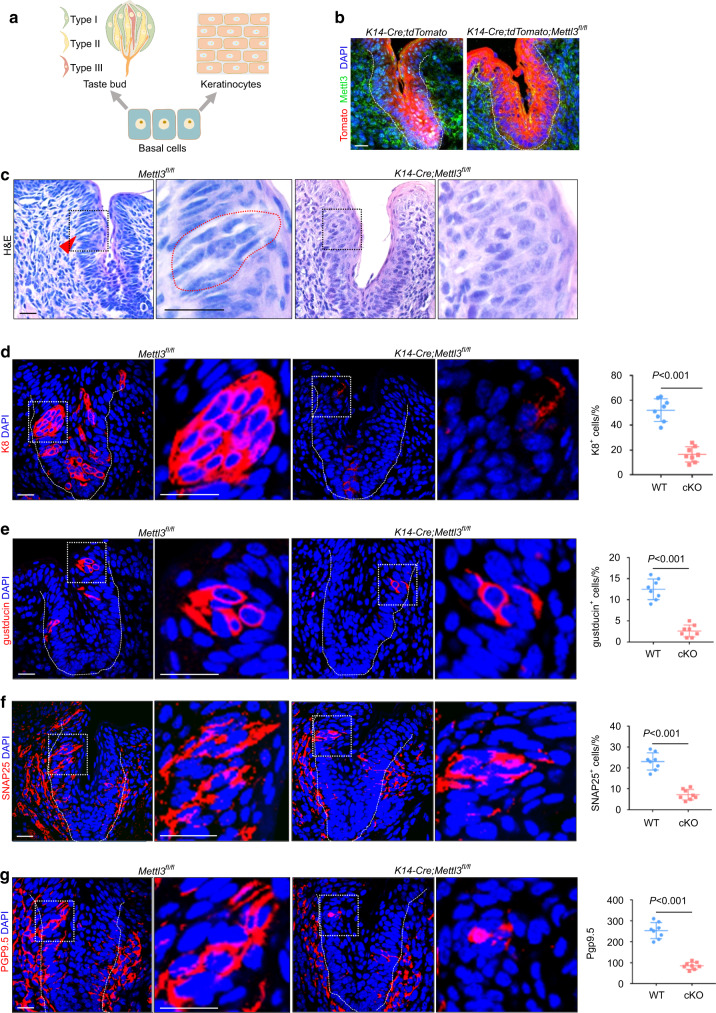
As previously described, basal progenitors develop into taste cells (K8) and differentiated keratinocytes (K13) (Fig. 1a)15,16,40. To explore whether deletion of Mettl3 affected taste bud development, we crossed Mettl3fl/fl mice28 with K14-Cre transgenic mice to conditionally delete Mettl3 from epithelium basal progenitors. K14-Cre;Mettl3fl/fl mice were viable and born at Mendelian’s ratio. Most of them survived by postnatal day 4 (P4), and few could survive by P7. We also generated the K14-Cre;tdTomato;Mettl3fl/fl mice, in which K14+ cells and their daughter cells were labeled with tdTomato fluorescence, and confirmed that METTL3 was largely abolished within Tomato+ cells in the CVP (Fig. 1b).
Fig. 1.
Deletion of Mettl3 in epidermal progenitors leads to taste bud defects. a A schematic of basal progenitor cell differentiation. b METTL3 staining in circumvallate papillae from mice pups at P4. c H&E staining of taste buds (red arrow) in circumvallate papillae of P4 mice. d Staining and quantification of K8 + cells in circumvallate papillae (n = 8 biological replicates, P < 0.001 by unpaired two-tailed Student’s t-test). e, f Expression of GNAT3 and SNAP25 and the indicated quantification (n = 8 biological replicates, P < 0.001 by unpaired two-tailed Student’s t-test). g Detection of PGP9.5 in circumvallate papillae. Integrated density of PGP9.5 per trench profile (n = 8 biological replicates, P < 0.001 by unpaired two-tailed Student’s t-test). Dotted lines in b, d, e, f, and g indicate basal lamina. Scale bar: 20 μmol·L−1
Because of their short lifespan, we sacrificed K14-Cre; Mettl3fl/fl mice at P4 to explore whether taste bud differentiation was affected. Compared to control CVPs, the taste buds in mutant CVPs could not be recognized by hematoxylin and eosin (H&E) staining (Fig. 1c). K8 is a marker of differentiated taste bud cells40. Immunofluorescence staining revealed that the number of K8+ cells in the CVP was significantly decreased (Fig. 1d). We also observed a reduced number of type II cells (marked by gustducin) and type III cells (marked by SNAP25) in mutant CVP (Fig. 1e, f). Consistent with the taste bud loss phenotype, the innervated areas (marked by PGP9.5) of mutant CVP were remarkably reduced (Fig. 1g).
Deletion of Mettl3 leads to abnormal keratinization of lingual epithelium
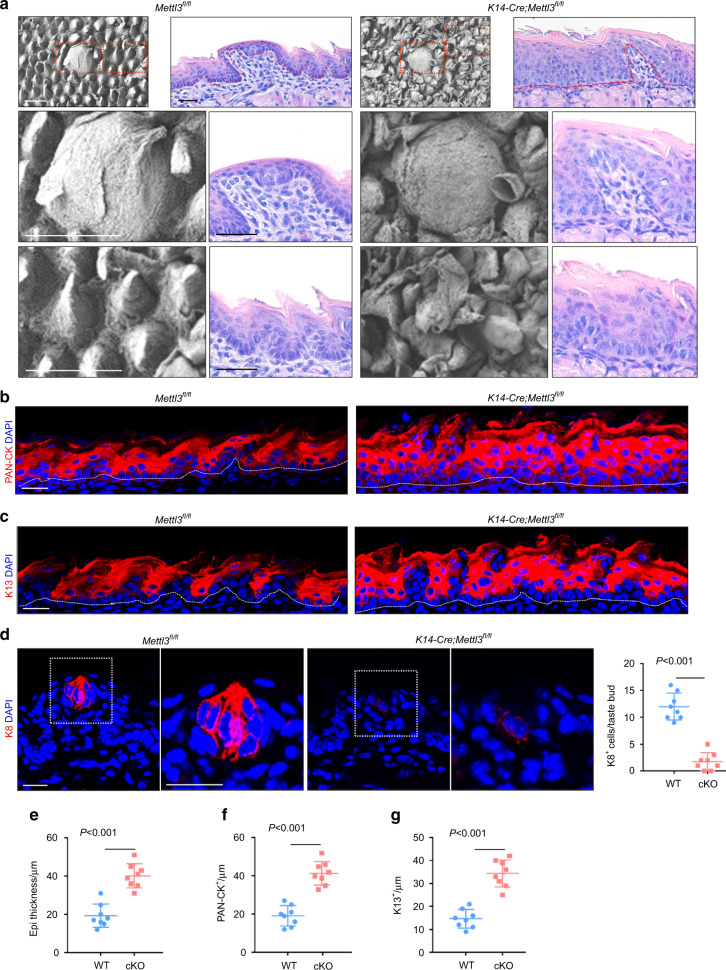
We then examined whether METTL3 regulated keratinization of the lingual epithelium. Scanning electron microscopy (SEM) showed that epithelial-specific Mettl3 deletion caused morphological abnormalities in FLP at P4 (Fig. 2a). Excessive keratinized fragments were observed on the surface of the tongue epithelium in mutant mice (Fig. 2a). H&E staining showed that the thickness of the epithelium was increased and the cell alignment was irregular compared to that of control mice (Fig. 2a). Quantitative analysis of epithelium thickness showed that the entire epithelium thickness of the mutants was almost double that of the controls (Fig. 2e). This observation was confirmed by immunofluorescence staining of PAN-CK and K13 cells (Fig. 2b, c, f, g). Although there was no obvious difference in the appearance of FFP by SEM, the number of K8+ taste cells in FFP decreased in the mutant mice (Fig. 2d).
Fig. 2.
Deletion of Mettl3 leads to abnormal keratinization of lingual epithelium. a P4 K14-cre; Mettl3fl/fl mice displayed abnormal filiform papilla and keratinized fragments (red arrow), assayed by SEM. Scale bar: 50 μmol·L−1. Deletion of Mettl3 caused the epithelium thickening, as revealed by H&E staining. Scale bar: 20 μmol·L−1. b The detection of PAN-CK indicates epithelial thickening in mutant mice. Scale bar: 20 μmol·L−1. c Increased expression of keratinocyte markers K13 in K14-cre; Mettl3fl/fl mice. Scale bar: 20 μmol·L−1. d Decreased expression of taste bud markers K8 and quantification of K8 + cells in K14-cre; Mettl3fl/fl fungiform papillae section (for each condition n = 8 biological replicates, P < 0.001 by unpaired two-tailed Student’s t-test). Scale bar: 20 μmol·L−1. Dotted lines in a, b, and c indicate the basal lamina. e Quantitative analysis of entire epithelium thickness, P < 0.001 by unpaired two-tailed Student’s t-test. f Quantitative analysis of PAN-CK+ epithelium thickness, P < 0.001 by unpaired two-tailed Student’s t-test. g Quantitative analysis of K13+ epithelium thickness, P < 0.001 by unpaired two-tailed Student’s t-test)
Deletion of Mettl3 increases basal cell proliferation but decreases cell proliferation around taste buds
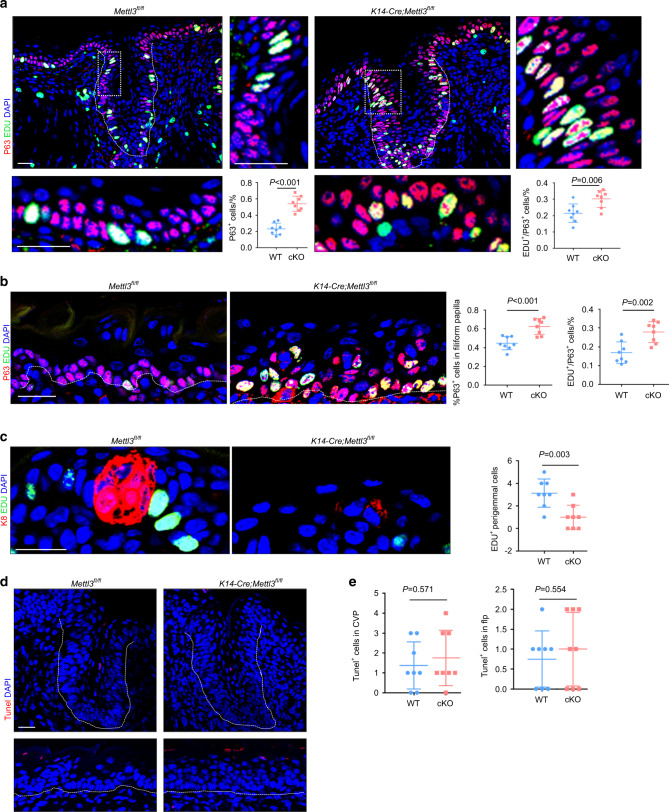
Transcription factor p63 is a crucial regulator of epidermal development and marks basal stem cells41. We noted that the number of P63+ cells in the CVP increased in the mutants (Fig. 3a). According to immunofluorescence results for P63 and 5-ethynyl-2′-deoxyuridine (EdU), the proliferation of P63+ cells in mutant CVP was more active than in control mice (Fig. 3a). Consistent results were observed in the FLP analysis (Fig. 3b). Deletion of Mettl3 led to an increase in the number of P63+ cells and promoted the proliferation of basal cells (Fig. 3b). In contrast, cell proliferation around taste buds reduced in Mettl3 knockout mice (Fig. 3c). Deletion of Mettl3 did not affect apoptosis of taste or non-taste cells (Fig. 3d, e).
Fig. 3.
Deletion of Mettl3 increases basal cell proliferation but decreases cell proliferation around taste buds. a Immunofluorescence for P63 and EdU in (a) Immunofluorescence for P63 and EdU in circumvallate papillae. Scale bar: 20 μmol·L−1. Quantification results are shown (for each condition n = 8 biological replicates, P < 0.001 and P = 0.006 by unpaired two-tailed Student’s t-test). b Immunofluorescence staining for P63 and EdU in filiform papillae. P63 + cells/total cells and EdU+ cells/P63 + cells are counted (for each condition n = 8 biological replicates, P < 0.001 and P = 0.002 by unpaired two-tailed Student’s t-test). c Immunofluorescence staining for K8 and EdU of perigemmal cells in fungiform papillae. Scale bar: 20 μmol·L−1. EDU + perigemmal cells are counted in fungiform papillae (for each condition n = 8 biological replicates, P = 0.003 by unpaired two-tailed Student’s t-test). d There is no difference in apoptosis, as assayed by TUNEL immunofluorescence. Scale bar: 20 μmol·L−1. e Quantitative analysis of TUNEL+ cells in CVP, P = 0.571 by unpaired two-tailed Student’s t-test). Quantitative analysis of TUNEL+ cells in FLP, P = 0.554 by unpaired two-tailed Student’s t-test)
Overexpression of Mettl3 promotes taste bud recovery after irradiation
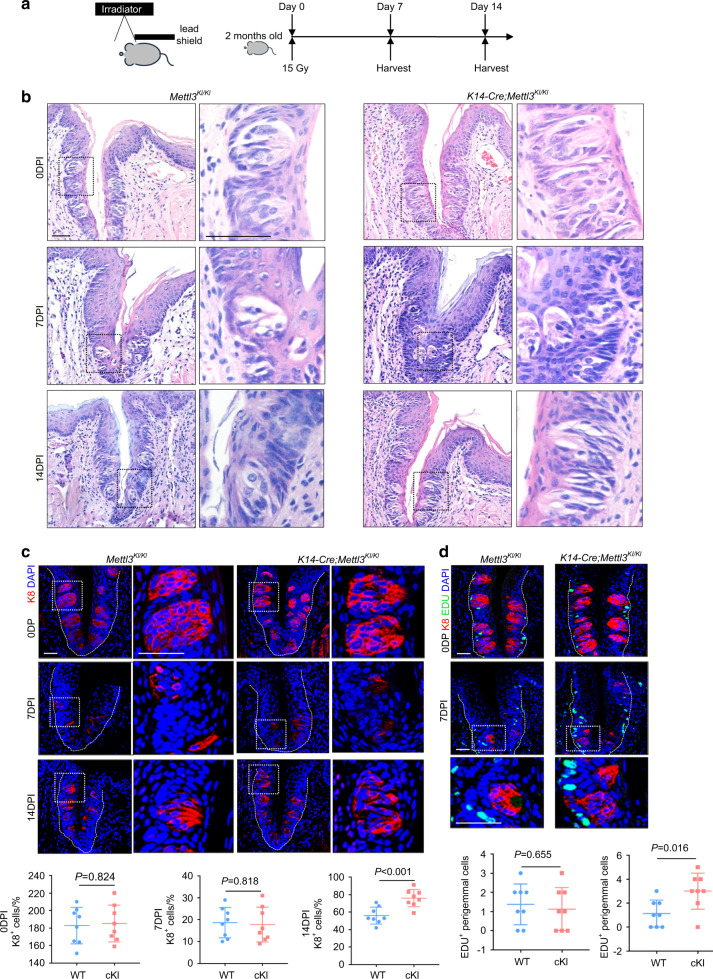
Next, we investigated whether Mettl3 overexpression could prevent lingual epithelial disorders. To this end, we generated K14-Cre driven Mettl3-tdTomato knock-in mice (K14-Cre;Mettl3KI/KI) to conditionally overexpress Mettl3 in epidermal progenitor cells28. As K14-Cre;Mettl3KI/KI mice did not exhibit a significant change in taste bud and epithelium development, we challenged them with 15 Gy irradiation (Fig. 4a). Both knock-in and control mice exhibited a severe loss of taste buds at 7 days post-irradiation (dpi) (Fig. 4b, c), indicating that overexpression of Mettl3 could not protect mice from epithelial injury due to irradiation. Notably, at 14 dpi, there were more recovered taste buds in the knock-in mice (Fig. 4b, c). Furthermore, overexpression of Mettl3 increased the proliferation of cells around taste buds at 7 dpi (Fig. 4d).
Fig. 4.
Overexpression of Mettl3 promotes taste bud recovery after irradiation. a Injury model for irradiation. b H&E-stained circumvallate papillae sections reveal that overexpression of Mettl3 does not impact taste bud development in normal physiology (0 dpi), nor protect taste buds from irradiation (7 dpi). Taste buds in mutants are significantly increased by 14 dpi. Scale bar: 20 μmol·L−1. c Immunofluorescence of K8 shows taste buds at 0 dpi, 7 dpi and 14 dpi. Scale bar: 20 μmol·L−1. K8 + cells in circumvallate papillae are counted (for each condition n = 8 biological replicates, P = 0.824 (0 dpi), P = 0.818 (7 dpi), P < 0.001 (14 dpi) by unpaired two-tailed Student’s t-test). d Immunofluorescence staining for K8 and EdU reveals increased proliferation of perigemmal cells in circumvallate papillae. Scale bar: 20 μmol·L−1. EDU + perigemmal cells are counted in circumvallate papillae (for each condition n = 8 biological replicates, P = 0.665 (0 dpi), P = 0.016 (7 dpi) by unpaired two-tailed Student’s t-test)
METTL3-mediated m6A RNA methylation regulates Hippo and Wnt pathways
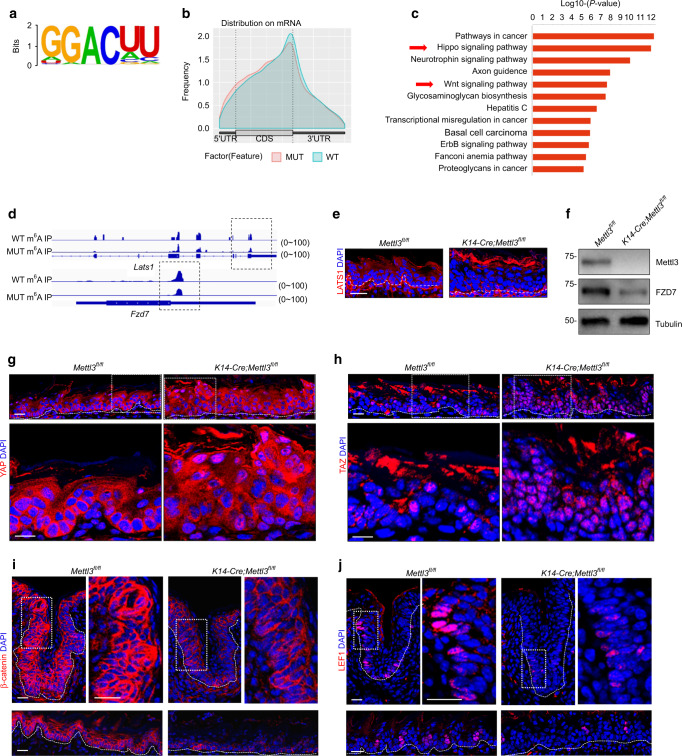
To explore the underlying mechanisms, we performed m6A RNA immunoprecipitation sequencing (m6ARIP-seq) of the lingual epithelium in K14-Cre; Mettl3fl/fl mice and their Mettl3fl/fl littermates at P4. Consistent with previous reports28,42, m6A peaks shared a common GGACU motif (Fig. 5a) and were enriched around the stop codon (Fig. 5b). Gene pathway analysis revealed that the most frequent changes involving m6A modifications after Mettl3 deletion were enriched in the Hippo and Wnt signaling pathways (Fig. 5c).
Fig. 5.
METTL3-mediated m6A RNA methylation regulates Hippo and Wnt pathways. a Consensus motif of m6A sites in Mettl3fl/fl mice. b Enrichment of m6ARIP-seq peaks in Mettl3fl/fl mice and K14-cre; Mettl3fl/fl mice at P4. c KEGG pathway enrichment for highly modified mRNAs. d The read density from m6ARIP-seq experiments shows the m6A peaks in Mettl3fl/fl mice and decreased peaks in K14-cre; Mettl3fl/fl mice. e Decreased expression of LATS1 in conditional Mettl3 knock-out mice at P4. Scale bar: 20 μmol·L−1. f Western blot analysis of FZD7 in Mettl3fl/fl mice and K14-cre; Mettl3fl/fl mice. g, h Immunofluorescence assays are used to detect the expression and location of YAP and TAZ. Scale bar: 20 μmol·L−1. i, j Immunofluorescence assays are used to detect the expression and localization of β-catenin and LEF1. Scale bar: 20 μmol·L−1
Specific peaks were observed near the stop codons of the large tumor suppressor kinase 1 (Lats1) and Frizzled class receptor 7 (Fzd7) (Fig. 5d). The deletion of Mettl3 significantly decreased the abundance of m6A modifications (Fig. 5d). Immunofluorescence staining and western blot analysis confirmed reduced LATS1 and FZD7 protein levels, respectively (Fig. 5e, f). Loss of METTL3 led to increased nuclear localization of YAP and TAZ in the basal epithelium (Fig. 5g, h). FZD7, a Wnt receptor, transduces signals and activates the Wnt signaling pathway43,44. Immunofluorescence staining for β-catenin indicated that nuclear β-catenin expression by K14-Cre; Mettl3fl/fl mice at P4 was significantly decreased in terms of both taste and non-taste epithelia (Fig. 5i). In addition, LEF1 expression was decreased in K14-Cre; Mettl3fl/fl mice (Fig. 5j).
Discussion
In recent years, many studies have reported a role for METTL3 in stem cell differentiation. METTL3-mediated m6A modification regulates the development of the hematopoietic system, spermatogenesis, and other organs45,46. Our research group also found that m6A modification mediated by METTL3 plays a key role in regulating the fate of bone marrow mesenchymal stem cells28. In this study, we uncovered an essential role for METTL3 in lingual epithelial homeostasis. METTL3 was widely expressed in the lingual epithelium. Deletion of Mettl3 led to severe defects in taste bud development and abnormal epithelial thickening. In addition, overexpression of Mettl3 promoted taste bud recovery from radiation damage. Furthermore, the Wnt and Hippo signaling pathways may be responsible for the striking phenotypic defects in taste buds and keratinizing epithelium.
The Wnt/β-catenin pathway is required for taste bud development and taste cell renewal15,47,48. Basal cells with high expression of β-catenin can give rise to taste cells, whereas lower levels of β-catenin expression promote keratinocyte fate1,15. Conditional β-catenin knockout in progenitor cells causes a decrease in taste cells48. Here, we showed that the deletion of Mettl3 resulted in the downregulation of β-catenin and LEF1, which led to defects in taste cell development.
However, a study demonstrated that conditional knockout of Ctnnb1 in the epidermis led to FLP and FFP developmental defects and thinner epithelium, which did not match our phenotype49. In our study, β-catenin expression was extremely low after Mettl3 knockout, whereas the lingual epithelium was thickened. This finding reminded us that different mechanisms regulate the non-taste epithelium. METTL3-mediated m6A modification has been identified as the most abundant mRNA modification that regulates biological processes in mRNA27,50. Deletion of Mettl3 reduces the m6A peaks of many mRNAs, which changes their fate choices. The m6ARIP-seq analysis of the lingual epithelium showed that m6A modifications were mainly enriched in the Hippo and Wnt signaling pathways (Fig. 5c). LATS1 is a crucial kinase that phosphorylates and inactivates the transcriptional coactivators YAP and TAZ51–53. In our study, we found that loss of METTL3 reduced the m6A modification of LATS1 and further inhibited the Hippo pathway, resulting in abnormal proliferation of keratinizing epithelium.
Another area for investigation is how METTL3 regulates cellular physiological functions and pathological progression through m6A modification. A number of studies have elaborated on the role of RNA m6A modifications in alternative splicing42,45. A recent study also pointed out that m6A methylation regulates an array of chromosome-associated regulatory RNAs (carRNAs) to globally tune chromatin state and gene transcription54. In some studies, METTL3 and m6A modifications have been shown to enhance mRNA stability to promote cell proliferation37,55. m6A has been shown to promote mRNA translation in certain cell types56,57. In our previous study, the deletion of Mettl3 slowed down the translation of target proteins and further inhibited downstream signaling28. m6A modifications are important and function depending on the cellular context. Thus, we did not further investigate mRNA metabolism in this study.
Dysgeusia is common and significant in patients after receiving head and neck radiotherapy. A previous study found that radiation interrupts the renewal of taste bud cells by inhibiting the proliferation and differentiation of basal progenitor cells, resulting in taste bud injury in mice8. In subsequent studies, they found that activation of the Wnt/β-catenin signaling pathway promotes the recovery of taste cells from radiation58. To determine the function of METTL3 in radiation-induced gustation dysfunction, conditional Mettl3 knock-in mice were exposed to radiation and analyzed for taste bud maintenance and recovery. We found that overexpression of Mettl3 promoted taste bud recovery from radiation damage by increasing the proliferation of taste bud progenitor cells. Interestingly, overexpression of Mettl3 did not protect taste buds from radiation injury.
In conclusion, we elucidated that METTL3 was an essential regulator of lingual epithelial homeostasis by regulating m6A modification. Deletion of Mettl3 in the epidermis reduced the expression of LATS1 and FZD7 and further blocked downstream pathways, which led to taste bud defects and epithelial thickening. In addition, overexpression of Mettl3 promoted taste bud recovery from radiation damage by increasing the proliferation of taste bud progenitor cells.
Materials and methods
Mice
Mettl3fl/+ and Mettl3KI/+ mice were generated using the CRISPR-Cas9 system, as described previously28. K14-Cre transgenic mice were kindly provided by Dr. Demeng Chen from Sun Yat-sen University (Guangzhou, China). Rosa26-tdTomato mice were purchased from the Jackson Laboratory (Cat No:007905, Pennsylvania, USA). Mettl3fl/fl and Mettl3KI/KI mice were crossed with K14-Cre transgenic mice to generate K14-Cre; Mettl3fl/fl mice and K14-Cre; Mettl3KI/KI mice. By mating Mettl3fl/fl mice with K14-Cre; tdTomato mice, we obtained K14-Cre; tdTomato; Mettl3fl/fl mice. All mice had a C57BL6/J background. The genotypes of transgenic mice were identified as previously described28.
All mice were housed in specific pathogen-free (SPF) facilities with a 12-hour light-dark illumination cycle. All studies performed on mice were approved by the Subcommittee on Research and Animal Care (SRAC) at Sichuan University.
Tissue preparation
After anesthesia with xylazine (10 mg·kg−1) and ketamine (80 mg·kg−1), mice were perfused transcardially with normal saline and 4% paraformaldehyde (PFA) in 0.1 mol·L−1 phosphate buffer. Tongues were dissected from the mandible and fixed in 4% PFA overnight at 4 °C. For frozen sections, tissues were transferred to 20% sucrose in 0.1 mol·L−1 phosphate buffer overnight at 4 °C. The samples were embedded in OCT compound (Sakura Finetek, Torrance, USA) and cryosectioned to 12 µm. For paraffin sections, the samples were dehydrated in graded ethanol and xylene and then embedded in 5-µm-thick paraffin sections using a microtome (Leica, RM2255, Wetzlar, German).
Histology and immunofluorescence
For hematoxylin and eosin (H&E) staining, the paraffin sections were de-waxed using graded xylene solutions. Staining was performed according to the manufacturer’s instructions (Solarbio Science and Technology, Beijing, China).
For immunohistochemistry, paraffin sections were prepared by the above procedures, microwaved in sodium citrate buffer, and incubated with primary and secondary antibodies (Boster Biological Technology, Wuhan, China). Finally, the sections were developed using the AEC (3-amino-9-ethylcarbazole) Staining Kit (Boster Biological Technology, Wuhan, China).
For immunofluorescence, cryosections were thawed at room temperature (26 °C), rehydrated in 0.1 mol·L−1 phosphate-buffered saline and microwaved in sodium citrate buffer. After incubation with the primary antibody, the sections were incubated with Alexa Fluor-labeled secondary antibodies (Jackson Laboratory, Pennsylvania, USA).
Antibodies
The following antibodies were used: rabbit anti-METTL3 (1:200; Abcam, Cat No: ab195352), rabbit anti-KRT8 (1:200; Abcam, Cat No: ab53280), goat anti- gustducin (1:100; Aviva Systems Biology, Cat No: OAEB00418), rabbit anti-PGP9.5 (1:500, Thermo Fisher, Cat No: 480012), rabbit anti-SNAP25 (1:100; Proteintech, Cat No: 14903-1-AP), mouse anti- PAN-CK(1:100, Thermo Fisher, Cat No: MA1-82041), mouse anti-KRT13 (1:200; Abcam, Cat No: ab16112), rabbit anti-p63 (1:300; Abcam, Cat No: ab53039), mouse anti-LATS1 (1:100; Santa Cruz Biotechnology, Cat No: sc-398560), mouse anti-YAP (1:200; Santa Cruz Biotechnology, Cat No: sc-101199), mouse anti-TAZ (1:100; Santa Cruz, Cat No: sc-293183), rabbit anti-FZD7 (1:1 000 for western blot; Abcam, Cat No: ab64636), rabbit anti-β-catenin (1:250; Proteintech, Cat No: 51067-2-AP), rabbit anti-LEF1 (1:200; Abcam, Cat No: ab137872).
Irradiation
After anesthesia, the mice were covered with a custom-made lead shield, leaving the head and neck exposed. The mice were irradiated with 15 Gy in an X-ray irradiator (~1.25 Gy per min, Accela, X-RAD 160). Irradiated mice were returned to their cages for recovery.
TUNEL assay
To assess cell death, the TUNEL assay was performed using the In Situ Cell Death Detection Kit TMR Red (Boster Biological Technology, MK 1012-100, Wuhan, China). The sections were digested with proteinase K for 5 min and then washed in tris-HCl buffered saline three times. Sections were incubated with labeling buffer for 2 h at 37 °C prior to TUNEL reactions. Labeling buffer was prepared according to the manufacturer’s instructions. After two washes, the sections were incubated in blocking solution at room temperature for 30 min. The blocking solution was then removed. The fluorescence probes were used to detect cell death. Sections were counterstained with DAPI and imaged using laser scanning confocal microscopy (LSCM; Olympus FV3000, Tokyo, Japan).
EdU labeling
For EdU labeling, mice were injected with 25 µg of EdU per gram of body weight and euthanized after 1 h. Tongues were fixed overnight in 4% PFA and embedded in paraffin. After de-waxing, paraffin sections were incubated with the Click-iT EdU Imaging Kit (Invitrogen, CA, USA).
SEM
Tongue samples were fixed in 4% PFA overnight and dehydrated in a graded series of ethanol concentrations. Dehydrated samples were then incubated in 50% hexamethyldisilazane (Sigma-Aldrich, St. Louis, USA) for 20 min, followed by three solvent changes to 100% hexamethyldisilazane. After air-drying overnight, the samples were sputter-coated with gold-palladium. Specimens were examined and photographed using a SEM49.
Western blot
The lingual epithelium was collected from four mice in each group following previously described methods13. The tissues were lysed in RIPA buffer (Pierce, Rockford, IL, USA). The sample was then mixed with sample buffer containing 2% SDS and 1% 2-mercaptoethanol and heated at 95 °C for 5 min. The samples were separated on 10% SDS–polyacrylamide gels and transferred to polyvinylidene fluoride (PVDF) membranes using a semi-dry transfer apparatus (Bio-Rad, CA, USA)59. Incubation with rabbit anti-METTL3 (1:2 000; Abcam, Cat No: ab195352), rabbit anti-FZD7 (1:1 000; Abcam, Cat No: ab64636), rabbit anti-α-tubulin (1:2 000; Proteintech, Cat No: 11224-1-AP) at 4 °C was conducted overnight, followed by incubation with a horseradish peroxidase (HRP)-conjugated secondary antibody (1:5 000, Cell Signaling Technology, Cat No:7074). After washing, the blots were analyzed with Immobilon Reagents (Millipore, Burlington, USA) using a gel imaging system (Bio-Rad, CA, USA).
m6A MeRIP-Seq (m6A RNA immunoprecipitation sequencing)
Total RNA from the lingual epithelium was isolated using TRIzol reagent13. mRNA was enriched from total RNA using Immobilon Reagents (Millipore, Burlington, USA) and gel imaging systems (Bio-Rad, CA, USA). The mRNA was then fragmented with ZnCl2 buffer. Immunoprecipitation was performed using an anti-m6A antibody (1:100; Synaptic Systems, Cat No: 202003). Immunoprecipitated RNA was analyzed by high-throughput sequencing or RT-qPCR, as previously described28,60.
Statistical analysis
All data are presented as means ± standard error. For comparison between two independent groups, statistical differences were analyzed using unpaired two-tailed Student’s t-test. Statistical significance was set at p < 0.05.
Acknowledgements
We thank Professor Demeng Chen’s laboratory, Sun Yat-Sen University, for all instruction.
Author contributions
Conceptualization: R.X., Q.Y. Data curation: Q.X., X.Z., K.L., X.Z., R.X., Q.Y. Formal analysis: Q.X., X.Z., K.L., X.Z., Q.W., Methodology: Q.X., X.Z., K.L., X.Z., W.L., Investigation: Q.W., W.L., R.X., Q.Y. Resources: K.L., Q.W., W.L., R.X., R.T. Software: Q.X., C.L., X.Z., R.X., Validation: Q.X., X.Z., R.X., Q.Y. Visualization: Q.X., X.Z., R.X., Project administration: QY Supervision: R.X., Q.Y. Funding acquisition: Q.Y. Writing—original draft: Q.X., X.Z., K.L., X.Z., Q.W., W.L., R.X., Q.Y. Writing—review and editing: Q.X., X.Z., K.L., X.Z., Q.W., W.L., R.X., Q.Y.
Funding
This work was supported by the National Natural Science Foundation of China (81970913 and 82125006).
Data availability
All data are available in the main text.
Competing interests
The authors declare no competing interests.
Contributor Information
Ruoshi Xu, Email: xurs@scu.edu.cn.
Quan Yuan, Email: yuanquan@scu.edu.cn.
References
- 1.Barlow LA. Progress and renewal in gustation: new insights into taste bud development. Development. 2015;142:3620–3629. doi: 10.1242/dev.120394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arvidson K, Friberg U. Human taste: response and taste bud number in fungiform papillae. Science. 1980;209:807–808. doi: 10.1126/science.7403846. [DOI] [PubMed] [Google Scholar]
- 3.Qin Y, Sukumaran SK, Margolskee RF. Nkx2-2 expressing taste cells in endoderm-derived taste papillae are committed to the type III lineage. Dev. Biol. 2021;477:232–40. doi: 10.1016/j.ydbio.2021.05.020. [DOI] [PubMed] [Google Scholar]
- 4.Liman ER, Zhang YV, Montell C. Peripheral coding of taste. Neuron. 2014;81:984–1000. doi: 10.1016/j.neuron.2014.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Simon SA, de Araujo IE, Gutierrez R, Nicolelis MA. The neural mechanisms of gustation: a distributed processing code. Nat. Rev. Neurosci. 2006;7:890–901. doi: 10.1038/nrn2006. [DOI] [PubMed] [Google Scholar]
- 6.Norris DM. Transduction mechanism in olfaction and gustation. Nature. 1969;222:1263–1264. doi: 10.1038/2221263a0. [DOI] [PubMed] [Google Scholar]
- 7.Stieb S, et al. Development and validation of a contouring guideline for the taste bud bearing tongue mucosa. Radiother. Oncol. 2021;157:63–69. doi: 10.1016/j.radonc.2020.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nguyen HM, Reyland ME, Barlow LA. Mechanisms of taste bud cell loss after head and neck irradiation. J. Neurosci. 2012;32:3474–3484. doi: 10.1523/JNEUROSCI.4167-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhu J, et al. LiCl Promotes Recovery of Radiation-Induced Oral Mucositis and Dysgeusia. J. Dent. Res. 2021;100:754–63. doi: 10.1177/0022034521994756. [DOI] [PubMed] [Google Scholar]
- 10.Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J. Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 11.Alfaro R., Crowder S., Sarma K. P., Arthur A. E., & Pepino M. Y. Taste and Smell Function in Head and Neck Cancer Survivors. Chem. Senses. 46, bjab026 (2021). [DOI] [PMC free article] [PubMed]
- 12.Golden E. J. et al. Onset of taste bud cell renewal starts at birth and coincides with a shift in SHH function. Elife. 10, e64013 (2021). [DOI] [PMC free article] [PubMed]
- 13.Castillo-Azofeifa D, et al. Sonic hedgehog from both nerves and epithelium is a key trophic factor for taste bud maintenance. Development. 2017;144:3054–65. doi: 10.1242/dev.150342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bloomquist RF, et al. Developmental plasticity of epithelial stem cells in tooth and taste bud renewal. Proc. Natl Acad. Sci. USA. 2019;116:17858–66. doi: 10.1073/pnas.1821202116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gaillard D, Xu M, Liu F, Millar SE, Barlow LA. beta-Catenin Signaling Biases Multipotent Lingual Epithelial Progenitors to Differentiate and Acquire Specific Taste Cell Fates. PLoS Genet. 2015;11:e1005208. doi: 10.1371/journal.pgen.1005208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Okubo T, Pevny LH, Hogan BL. Sox2 is required for development of taste bud sensory cells. Genes Dev. 2006;20:2654–2659. doi: 10.1101/gad.1457106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rodriguez YA, et al. “Tripartite Synapses” in Taste Buds: A Role for Type I Glial-like Taste Cells. J. Neurosci. 2021;41:9860–71. doi: 10.1523/JNEUROSCI.1444-21.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baumer-Harrison C, et al. Optogenetic Stimulation of Type I GAD65(+) Cells in Taste Buds Activates Gustatory Neurons and Drives Appetitive Licking Behavior in Sodium-Depleted Mice. J. Neurosci. 2020;40:7795–810. doi: 10.1523/JNEUROSCI.0597-20.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang AL, et al. The cells and logic for mammalian sour taste detection. Nature. 2006;442:934–938. doi: 10.1038/nature05084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang L, et al. The coding of valence and identity in the mammalian taste system. Nature. 2018;558:127–31. doi: 10.1038/s41586-018-0165-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Taruno A, et al. CALHM1 ion channel mediates purinergic neurotransmission of sweet, bitter and umami tastes. Nature. 2013;495:223–226. doi: 10.1038/nature11906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vandenbeuch A, et al. Role of the ectonucleotidase NTPDase2 in taste bud function. Proc. Natl Acad. Sci. USA. 2013;110:14789–14794. doi: 10.1073/pnas.1309468110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guarascio DM, Gonzalez-Velandia KY, Hernandez-Clavijo A, Menini A, Pifferi S. Functional expression of TMEM16A in taste bud cells. J. Physiol. 2021;599:3697–3714. doi: 10.1113/JP281645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Larson E. D., Vandenbeuch A., Anderson C. B., & Kinnamon S. C. GAD65Cre Drives Reporter Expression in Multiple Taste Cell Types. Chem Senses. 46, bjab033 (2021). [DOI] [PMC free article] [PubMed]
- 25.Wang T, Kong S, Tao M, Ju S. The potential role of RNA N6-methyladenosine in Cancer progression. Mol. Cancer. 2020;19:88. doi: 10.1186/s12943-020-01204-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.He L, et al. Functions of N6-methyladenosine and its role in cancer. Mol. Cancer. 2019;18:176. doi: 10.1186/s12943-019-1109-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao BS, Roundtree IA, He C. Post-transcriptional gene regulation by mRNA modifications. Nat. Rev. Mol. Cell Biol. 2017;18:31–42. doi: 10.1038/nrm.2016.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu Y, et al. Mettl3-mediated m(6)A RNA methylation regulates the fate of bone marrow mesenchymal stem cells and osteoporosis. Nat. Commun. 2018;9:4772. doi: 10.1038/s41467-018-06898-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zheng G, et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell. 2013;49:18–29. doi: 10.1016/j.molcel.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jia G, et al. Oxidative demethylation of 3-methylthymine and 3-methyluracil in single-stranded DNA and RNA by mouse and human FTO. FEBS Lett. 2008;582:3313–3319. doi: 10.1016/j.febslet.2008.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sledz P., Jinek M. Structural insights into the molecular mechanism of the m(6)A writer complex. Elife. 14, e18434 (2016). [DOI] [PMC free article] [PubMed]
- 32.Ping XL, et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 2014;24:177–189. doi: 10.1038/cr.2014.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang X, et al. Structural basis of N(6)-adenosine methylation by the METTL3-METTL14 complex. Nature. 2016;534:575–578. doi: 10.1038/nature18298. [DOI] [PubMed] [Google Scholar]
- 34.Geula S, et al. Stem cells. m6A mRNA methylation facilitates resolution of naive pluripotency toward differentiation. Science. 2015;347:1002–1006. doi: 10.1126/science.1261417. [DOI] [PubMed] [Google Scholar]
- 35.Chen T, et al. m(6)A RNA methylation is regulated by microRNAs and promotes reprogramming to pluripotency. Cell. Stem Cell. 2015;16:289–301. doi: 10.1016/j.stem.2015.01.016. [DOI] [PubMed] [Google Scholar]
- 36.Chen M, et al. RNA N6-methyladenosine methyltransferase-like 3 promotes liver cancer progression through YTHDF2-dependent posttranscriptional silencing of SOCS2. Hepatology. 2018;67:2254–70. doi: 10.1002/hep.29683. [DOI] [PubMed] [Google Scholar]
- 37.Li T, et al. METTL3 facilitates tumor progression via an m(6)A-IGF2BP2-dependent mechanism in colorectal carcinoma. Mol. Cancer. 2019;18:112. doi: 10.1186/s12943-019-1038-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lin W, Xu H, Wu Y, Wang J, Yuan Q. In silico genome-wide identification of m6A-associated SNPs as potential functional variants for periodontitis. J. Cell Physiol. 2020;235:900–908. doi: 10.1002/jcp.29005. [DOI] [PubMed] [Google Scholar]
- 39.Sheng R, et al. METTL3-Mediated m(6) A mRNA Methylation Modulates Tooth Root Formation by Affecting NFIC Translation. J. Bone Min. Res. 2021;36:412–23. doi: 10.1002/jbmr.4180. [DOI] [PubMed] [Google Scholar]
- 40.Knapp L, Lawton A, Oakley B, Wong L, Zhang C. Keratins as markers of differentiated taste cells of the rat. Differentiation. 1995;58:341–349. doi: 10.1046/j.1432-0436.1995.5850341.x. [DOI] [PubMed] [Google Scholar]
- 41.Yang A, et al. p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature. 1999;398:714–718. doi: 10.1038/19539. [DOI] [PubMed] [Google Scholar]
- 42.Dominissini D, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485:201–206. doi: 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- 43.Fernandez A, et al. The WNT receptor FZD7 is required for maintenance of the pluripotent state in human embryonic stem cells. Proc. Natl Acad. Sci. USA. 2014;111:1409–14. doi: 10.1073/pnas.1323697111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li G, et al. Frizzled7 Promotes Epithelial-to-mesenchymal Transition and Stemness Via Activating Canonical Wnt/beta-catenin Pathway in Gastric Cancer. Int J. Biol. Sci. 2018;14:280–93. doi: 10.7150/ijbs.23756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu K, et al. Mettl3-mediated m(6)A regulates spermatogonial differentiation and meiosis initiation. Cell Res. 2017;27:1100–1114. doi: 10.1038/cr.2017.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vu LP, et al. The N(6)-methyladenosine (m(6)A)-forming enzyme METTL3 controls myeloid differentiation of normal hematopoietic and leukemia cells. Nat. Med. 2017;23:1369–76. doi: 10.1038/nm.4416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu F, et al. Wnt-beta-catenin signaling initiates taste papilla development. Nat. Genet. 2007;39:106–12. doi: 10.1038/ng1932. [DOI] [PubMed] [Google Scholar]
- 48.Gaillard D, et al. beta-catenin is required for taste bud cell renewal and behavioral taste perception in adult mice. PLoS Genet. 2017;13:e1006990. doi: 10.1371/journal.pgen.1006990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xu M, et al. WNT10A mutation causes ectodermal dysplasia by impairing progenitor cell proliferation and KLF4-mediated differentiation. Nat. Commun. 2017;8:15397. doi: 10.1038/ncomms15397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Meyer KD, Jaffrey SR. The dynamic epitranscriptome: N6-methyladenosine and gene expression control. Nat. Rev. Mol. Cell Biol. 2014;15:313–326. doi: 10.1038/nrm3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pan D. The hippo signaling pathway in development and cancer. Dev. Cell. 2010;19:491–505. doi: 10.1016/j.devcel.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Halder G, Camargo FD. The hippo tumor suppressor network: from organ size control to stem cells and cancer. Cancer Res. 2013;73:6389–6392. doi: 10.1158/0008-5472.CAN-13-2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mo JS, Park HW, Guan KL. The Hippo signaling pathway in stem cell biology and cancer. EMBO Rep. 2014;15:642–56. doi: 10.15252/embr.201438638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu J, et al. N (6)-methyladenosine of chromosome-associated regulatory RNA regulates chromatin state and transcription. Science. 2020;367:580–586. doi: 10.1126/science.aay6018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhao W, et al. METTL3 Facilitates Oral Squamous Cell Carcinoma Tumorigenesis by Enhancing c-Myc Stability via YTHDF1-Mediated m(6)A Modification. Mol. Ther. Nucleic Acids. 2020;20:1–12. doi: 10.1016/j.omtn.2020.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu L, et al. METTL3 Promotes Tumorigenesis and Metastasis through BMI1 m(6)A Methylation in Oral Squamous Cell Carcinoma. Mol. Ther. 2020;28:2177–2190. doi: 10.1016/j.ymthe.2020.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jin H, et al. N(6)-methyladenosine modification of ITGA6 mRNA promotes the development and progression of bladder cancer. EBioMedicine. 2019;47:195–207. doi: 10.1016/j.ebiom.2019.07.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gaillard D, Shechtman LA, Millar SE, Barlow LA. Fractionated head and neck irradiation impacts taste progenitors, differentiated taste cells, and Wnt/beta-catenin signaling in adult mice. Sci. Rep. 2019;9:17934. doi: 10.1038/s41598-019-54216-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu Y, et al. DNA demethylase ALKBH1 promotes adipogenic differentiation via regulation of HIF-1 signaling. J. Biol. Chem. 2022;298:101499. doi: 10.1016/j.jbc.2021.101499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lin W, et al. Mapping the immune microenvironment for mandibular alveolar bone homeostasis at single-cell resolution. Bone Res. 2021;9:17. doi: 10.1038/s41413-021-00141-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data are available in the main text.