Abstract
Food-borne pathogens are a severe threat to human illness and death world-wide. Researchers have reported more than 250 food-borne diseases. Most of these are infections caused by a wide variety of bacteria, viruses, and parasites. It has a significant economic impact also. Detection of pathogenic microbes is thus essential for food safety. Such identification techniques could meet the following parameters viz., the accuracy of detection techniques that are quick, efficient, economical, highly sensitive, specific, and non-labor intensive. The various available methods for detecting food pathogens are classified into different groups, each having its advantages and disadvantages. The conventional methods are usually the first choice of detection even though they are laborious. Modern techniques such as biosensors, immunological assays, and macromolecule-based (nucleic acid) methods are being developed and refined to overcome traditional methods’ limitations. Early detection of pathogens and secure food safety at each stage of food processing to storage, utilizing improved methodologies are mandatory. This review summarizes the deadly food pathogens leading to significant outbreaks and discusses the importance of early detection methods and advanced detection methods in comparison.
Keywords: Food-borne disease, Microbe, Conventional method, Modern technique, Early detection, Significant outbreak
Introduction
In the food industry, selecting particular identification methods and screening of food-borne pathogens must ensure food safety. In each phase, from crop harvesting to food packaging and trading, food contamination is highly desirable. Among all conventional techniques in practice, there are still methods and strategies required to determine the minimal quantity of emerging pathogens in food and meat processing industries. In turn, pathogenic microbes that emerge into superbugs could pose a threat to human consumers in upcoming decades, and need significant concern, especially in food safety management. Food-borne pathogens are a significant threat to consumers that contaminate edible products, leading to several diseases and outbreaks. Though there are quite a lot of food-borne diseases prevailing globally, a major cause being the Food-borne zoonoses. The World Health Organization (WHO) supporting the previous statement, reports that more than two million people die per year from diarrheal diseases induced mostly by consuming infected food (Westrell et al. 2009). Patients are reported with severe nausea, vomiting, diarrhea, and abdominal cramps to mortal diseased conditions (Abebe et al. 2020). Improper food product handling and storage can affect one country’s public health and economic sectors through restricted export and food trading worldwide (Kangethe et al. 2019). Various techniques are applied for the detection of these pathogens. Microbial and associated contaminant detection methods are classified into conventional and modern methods based on their principles, advantages, and disadvantages. The conventional methods involve homogenizing food samples, enrichment of viable pathogens, identification of targets by growing on a selective medium which makes this method quite challenging when applied to food and water-borne microbial identification. The standard microbiological detection methods have their inadequacies, such that they are laborious and time-consuming; whereas, the modern methods vary from being fast, accurate, sensitive, specific, and non-labor intensive (Priyanka et al. 2016).
Thus, to overcome these inadequacies, different ways with high specificity and sensitivity are still targeted (Adzitey et al. 2012). To estimate micro to nano-sized food contaminants, multiple technologies are proposed to improve rapidity, accuracy, time-efficient, and labor-saving pathogen detection methods to qualify food products before reaching the consumers’ hands. Those techniques must also ensure an in-depth analysis of any food pathogen with reliability, efficiency in providing quick results, and selection. (Zhang et al. 2020). Measurement of nano-sized food pathogens and contaminants, Nanotechnology, and nanomaterials are being used in biosensors such as Quantum dots, Gold nanoparticles, silver nanoparticles, etc. The other technique is High Throughput Sequencing that can generate thousands to millions of sequence reads (Sai Anand et al. 2019). The latest development in identifying pathogenic bacteria using advanced nanostructures, high throughput sequencing for detecting food-borne pathogens (Sekse et al. 2017).
Therefore, the objectives of this review paper are to:
To mention available microbial and associated food contaminants and details of outbreak, food-borne pathogen detection methods with their characteristic properties.
An overview of conventional, modern, and advanced methods and their comparison benefits on detecting food-borne pathogens.
Outbreak and pathogens- food contaminants
Food safety management is focused on eliminating food-borne pathogens or reducing the pathogenic load in food contents. It is a must to detect pathogens in the early stages of microbial growth to reduce food-borne outbreaks (Pinu 2016). Antibiotic treatment of animal diseases during farming started in the 1930s, and such practice to date. This practice impacts the microbial load of animals that could gain antibiotic-resistant genes (Allen 2014). It resulted in new emerging food-borne pathogens in 2000. Such food-borne pathogens, Campylobacter, Noroviruses, Yersinia enterocolitica, Salmonella Enteritidis, Listeria monocytogenes, Vibrio cholerae O1, V. parahaemolyticus, V. vulnificus, VTEC E. coli, Cyclospora, and prions follows the new transmission mode. After the first report in 1894, Staphylococcal associated food poisoning, food intoxication syndrome, a continuous threat to the milk and meat processing industry (Jakobsen et al. 2011). In 2015, the European Union (EU) reported 4362 food-borne outbreaks. Bacterial pathogens mediated those outbreaks. Salmonella sp. (21.8%) and Campylobacter sp. (8.9%) accounted for 33.7% of all reported outbreaks. Bacterial toxins caused 19.5% in 2014; other factors such as viruses accounted for 9.2%, parasites contributed less than 3% of the total outbreak in 2015 (Fig. 1).
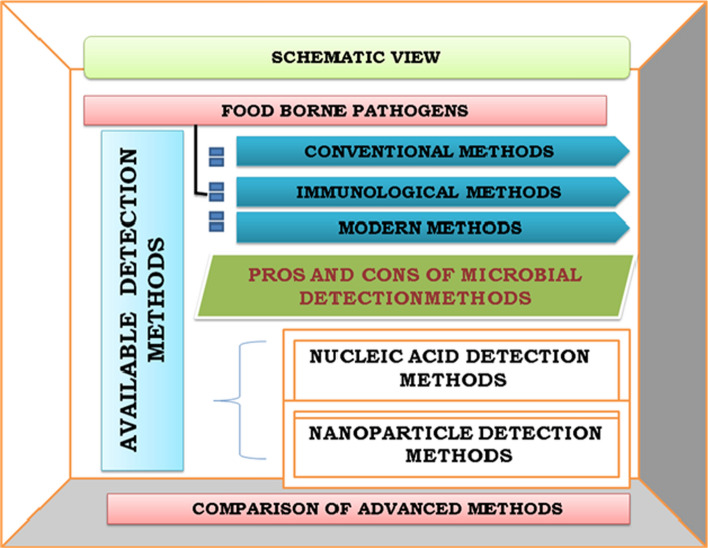
Fig. 1.
Schematic view of available detection methods
Bacterial foodborne pathogens
Campylobacter
It is a curved gram-negative rod, microaerophilic, cytochrome oxidase-positive unveiling corkscrew motility (Guerry 2007). It is a commensal in the intestine of many wild and domestic animals, particularly avian species, including poultry. Intestinal colonization results in healthy animals as carriers. There are 17 species and 6 subspecies belonging to the Campylobacter genus, of which C. jejuni (subspecies jejuni) and C. coli are most frequently encountered in human diseases (Guerry 2007). A few other species reported, such as C. lari, and C. upsaliensis have also been isolated from diarrhoeal disease patients but are reported less often. The C. coli flagellum comprises two strongly homologous flagellins, the major one, FlaA, and the minor one, FlaB. (Guerry 2007). The flaA gene is involved in adherence, gastrointestinal tract colonization, and invasion of the host cells (Jain et al. 2008), consequently arresting the immune response. C.jejuni consists of polar flagellum composed of O-linked glycosylated flagellin; for the regulation of Campylobacter flagellum, a two-component system composed of the FlgS sensor and the FlgR response regulator is vital (Dastia et al. 2010). Campylobacter spp. produce toxins such as CDT holotoxin, which consists of three subunits encoded by the genes cdtA, cdtB, and cdtC, triggering the arrest of eukaryotic cells, stopping them from entering mitosis in the G2/M phase of the cell cycle and thus contributing to cell death (Yamasaki et al. 2006; Ge et al. 2008; Zilbauer et al. 2008). The disease caused by Campylobacter infection is Campylobacteriosis. The risk of food-borne pathogens, including Food-borne illnesses, especially for young children, can be severe. Food is generally reported to be the primary source of transmission through undercooked meat and meat products, as well as raw or contaminated milk. The UK Government has raised the focus of the "Campylobacter Innovation Strategy" from 2010 to 2015 as Campylobacter is considered the most severe cause of food poisoning and accounted for an estimated 321,000 cases in England and Wales 2008, including more than 15,000 hospitalizations and 76 deaths (Portner et al. 2007). These microorganisms can develop viable but non-culturable cells under adverse growth conditions (Portner et al. 2007). Under laboratory conditions, Cappelier (1997) found that Campylobacter strains isolated from soil around the broiler house may have been transformed into viable but non-cultivable forms and may have been cultivable after passing through the intestinal tract of the chickens (Davies et al. 2020). The Nordic region was highly prone to Campylobacter outbreaks. Almost 36 outbreaks have occurred and affected nearly 7000 people in this region (Davies et al. 2020).In 2017 Campylobacter jejuni cross-contamination caused an outbreak of infection in Seoul. Contamination occurred due to chicken served in a particular region.The outbreak count was calculated to be 20.4% from the overall out surge (Kang et al. 2019). Another primary disorder caused by Campylobacter is Guillain-Barré Syndrome and the outbreak associated with the pathogen occurred in Peru in 2019. The outbreak related to Campylobacter had a different seasonal pattern between April and July (Ramos et al. 2021).In 2013 a large outbreak was reported in Australia due to infection caused by Campylobacter jejuni from the food (chicken) served. 56 cases were identified in the primary investigation associated with the outbreak of gastroenteritis caused by C.jejuni. ST991 sequence type was isolated from the food sample and identified and related with the out surge (Moffatt et al. 2016).
E.coli O157:H7
It is a gram-negative, rod-shaped bacterium and releases shiga toxin, which causes an outbreak. Shiga toxin-producing Escherichia coli (STEC) causes 5–10% of Hemolytic Uremic Syndrome (HUS)(Byrne et al. 2020).The epidemic outbreak caused by this bacterial species is associated with the toxin that contaminates food products. Nausea, fever, diarrhea, chills are the primary symptoms due to the infection caused by E. coli O157: H7. It affects infants, adults, and older people through infection severity, toxins produced by the pathogen in an infected host cause chronic illness (Byrne et al. 2020). In particular, strain O157:H7 63,000 illnesses and 20 deaths, 2100 hospitalization each year (Byrne et al. 2020). Two consecutive outbreaks were recorded in 1982 associated with fast food contamination of this strain. During 1993, an outbreak was recorded with hamburgers contamination that concluded that beef processing and handling defects in retail and restaurants. In 2011 around 4000 cases and 50 deaths were recorded within two months’ duration. Recently in 2015, 5901 cases were reported. Due to pathogenic traits carried out from enteroaggregative E.Coli and its capacity of Shiga toxin production shed an unusual impact on affected adults with HUS (Coulombe et al. 2020). Those patients were found with neurological symptoms too. The caseation in Canada regarding E. coli O157: H7 from the year 2008–2018 is based on the contamination of green leafy vegetables. Groundwater contamination caused by the release of Shiga toxin was the primary source of identification associated with the outbreak due to E. coli O157: H7. The outbreak caused a severe production loss of the food products based on these vegetables. The consumption caused a high level of food-borne illness for the people who consumed it (Byrne et al. 2020). E. coli O157: H7 outbreak occurred in raw meat produced in England in 2017; an epidemiological investigation is done to identify the rate of severity caused by the bacterium. Raw meat was the prime source for the origin of an outbreak in the region, causing gastrointestinal illness for the population. Raw milk, cheese, meat from the cross-contaminated source was the ideal mode of transmission through direct and non-direct contact in humans (Treacy et al. 2019). Infection can occur through the Carcass contamination of the slaughter and processing method. The production of Shiga toxin by the bacterium in infected humans causes several severe symptoms such as abdominal pain, vomiting, a uraemic syndrome associated with acute kidney failure. Pre-harvest and post-harvest strategies need to be updated to prevent the upsurge of the bacterium causing an epidemic (Diancourt et al. 2019).
The California Department of Public Health (CDPH) and the U.S. Food and Drug Administration (FDA) evaluated the list of ingredients of the recalled salads and identified that romaine lettuce from a farm in California was a likely vehicle for the outbreak. In 2015, a Shiga toxin-producing Escherichia coli O157:H7, linked to commercial chicken salad, caused 19 illnesses in 7 states. 5 illnesses, and two developed hemolytic-uremic syndrome (HUS), a type of kidney failure. No deaths were reported there (Mikhail et al. 2018).
Listeria monocytogenes
Listeria monocytogenes is a gram-positive, non-sporing bacteria species that are highly motile. The food contaminant disease caused by L.monocytogenes is known as listeriosis. Water and soil are the two central sources associated with the transmission of L.monocytogenes and, followed by favorable and optimum temperature, provide a suitable environment for transmission and infection outbreaks. Aerobic atmospheric conditions also play a prime role in the transmission process. Raw vegetables, dairy products, and undercooked meat are the target sources for the surge of infection. The symptoms include severe muscular pains, a rise in temperature, and diarrhea (Ricci et al. 2018).
Listeria sp. contamination in the food industry, a silent threat, could contaminate food products even after many years. This strain’s infection had a high mortality rate of 20–30%; even it can multiply in cold storages and refrigerators (Teixeira et al. 2020). However, these food manufacturers and exporters would need expeditious techniques to detect and put out the result simultaneously. Recent scientific advancements brought about fast-track techniques that employ nucleic-acid-based or immunological assays; nevertheless, the power of each strain and the differences in their ability to cause disease remains a puzzle (Jemmi et al.2006). In 2011, an active outbreak of Listeria monocytogenes was linked to the consumption of contaminated whole cantaloupes. The cantaloupes were grown on a farm in Colorado, but it caused illnesses in 28 states in total. Even though the cantaloupes were recalled, nearly 150 people became ill, 143 were hospitalized, and 33 died. A threatening outbreak of listeriosis bubbled up out of Michigan in late February involving Enoki mushrooms grown in South Korea. In its latest report on April 8, the centers for disease control and prevention centers for disease control and prevention (CDC) reported that 36 confirmed cases in 17 states involve 30 hospitalizations and 4 deaths. In 2012, 44 persons were affected by this strain by consuming contaminated chicken liver from Sweden (Lahti et al. 2017). In 2015, 270 deaths were reported due to listeriosis. L.monocytogenes are efficiently killed by pasteurization and cooking. Mostly, Ready to Eat (RTE) food types pose a risk for listeriosis. Campylobacter spp. was also associated with RTE associated spread.
In 2012, 44 persons were affected by this strain by consuming contaminated chicken liver from Sweden (Lahti et al. 2017). A listeriosis outbreak that arose in the United States in 2015 is closely linked with the contamination of ice cream from a production line. The investigation process from the contaminated ice cream samples indicated the infection infectivity range among the selected samples. The indication level ranged nearly 99% in the tested samples (Chen et al. 2016). Further, similar cases emerged in South Africa in 2017. The listeriosis outbreak was closely linked and related to the processed meat contamination by L. monocytogenes. 937 cases were recognized because of the condition caused by the surge. Epidemiological Investigation reports figured that low middle-income countries and regions were highly prone to the listeriosis outbreak (Juno Thomas.2020).
Staphylococcus aureus
It is a rod-shaped and gram-positive bacterium. Various strains of Staphylococcus cause illness by affecting the respiratory tract and the epithelial layer. Consuming processed food contaminated by the pathogen could be the main reason for the infectious pathogen outbreak (Guidi et al. 2018). The contamination can happen on account of improper hygiene and maintenance methods followed in preparing the food. Due to infection and upsurge of the virulent pathogen, symptoms are emesis, nausea, and abdominal cramps after intaking contaminated food within eight hours. The general and seasonal outbreak of Staphylococcus aureus occurs in Europe and across other western provinces. An outbreak due to Staphylococcus aureus occurred in Italy in 2017 (Nasheri et al. 2019). The upsurge happened due to the intake of contaminated food by the workers. Staphylococcus food poisoning investigation took place to understand the internal mechanism of infection by the pathogen and to identify the mode of transmission that occurred through it. The effect of illness analyzed through the food samples collected and recorded from the restaurant served the workers’ food. The end report provided after the investigation connected with the outbreak identified the intense severity of the pathogen (Guidi et al. 2018).
Clostridium perfringens
It is a spore-forming, gram-positive bacterium which produces toxins. It causes food poisoning by infecting the host and by causing an outbreak through it. The investigation reports summarize that the initial source for the bacterium is red meat, dairy products, nearly contaminated food, and the consumption of contaminated food distributions all over the regions caused an outbreak in England during 2018. The disease caused by the bacteria includes Necrotic enteritis and enterotoxaemia. Four toxins are produced; by the pathogen that consists of alpha, beta, epsilon, and iota. Strains of the bacteria are associated with the classification and identification of different types of strains in Clostridium perfringens. The provisional report provided that toxins produced by the Clostridium perfringens were the main cause for the food-borne illness outbreak (Bhattacharya et al. 2020). The spores from the contaminated soil could be the reason for the spread of the bacterium. Raw foods, non-processed, and optimum temperature maintenance for storage are the ideal reasons for the transmission of the bacteria directly and indirectly (Stelzer et al. 2019).
Salmonella
It is a gram-negative bacterium and commonly found in the intestinal tract of the animal and human beings. There are 2500 serotypes of Salmonella, but only a few, such as Salmonella typhimurium, Salmonella enteritidis, Salmonella subsp. enterica serovar enteritidis, and typhimurium, cause food-borne illnesses. The second predominant food-borne pathogen causes gastroenteritis bacteria. The primary salmonellosis vehicle is contaminated food and water, poultry, eggs, meat, and milk products. These organisms have a more remarkable ability to multiply in foodstuff and survive in it for several years. Salmonella typhimurium causes severe diarrhea, food poisoning, and it is associated with a significant risk of causing cardiovascular, bone, and joint infections in humans. (Afzal et al. 2015).
A Salmonella poona outbreak caused 907 illnesses which were linked to cucumbers from Mexico. This dramatic outbreak affected 40 states, 204 hospitalizations, and it caused 4 deaths (Laughlin et al. 2019). A Salmonella outbreak in 2008 related to contaminated peanut butter caused illnesses in 714 people in 46 states. Nearly 171 people were under hospitalizations, and the infections may have contributed to 9 deaths (Chang et al. 2013).
Viruses, fungi, parasites and prions-food borne pathogens
Virus
Some viruses, such as Norovirus, hepatitis E, hepatitis A, and rotavirus, always threaten food safety next to the bacterial mediated outbreak. Norovirus comes under the family Caliciviridae and genus Norovirus (NoV). NoV virus structure is a positive sense, non-enveloped, and single-stranded RNA virus and into seven genogroups in which GI, GII, GIV groups distinctly infect humans. Viral infection and its spread occur due to direct contact with highly contaminated surface soil in the agricultural lands; polluted water is a significant source, specifically SE in upsurge. Around 50% of the outbreak is related to food-borne illness caused by Norovirus (Zhou et al. 2019). NoV outbreak is a prime concern in western countries regarding sickness caused by this pathogen and its impact on food production and consumption (Zhou et al. 2019; Nasheri et al. 2019). North America and European countries were affected by the NoV epidemic; the predominant spread occurred from the frozen fruits and vegetables. Nearly 80% of the NoV outbreak is associated and related to contaminated fruits and vegetables. Specifically, the epidemic origin occurred from the infected raspberries strawberries. Five thousand cases are recorded due to illness caused by the contaminated frozen strawberries and 3000 cases from frozen raspberries. The outbreak has been recorded from the year 2008–2018 through the GPHIN system. Multiple infections are identified and recorded in India, China, Japan recently through the GPHIN method related to NoV (Meghnath et al. 2019). The hard encoded capsid protein provides stability to the virus from the external environment and completely protects the cellular machinery and genome (Compaoré et al. 2020). Thus, the pathogen’s virulence and severity are much higher and significantly impact food processing industries. Hepatitis E virus (HEV) is through water or food, especially raw shellfish, by accidental sewage contamination. It had been reported as an emerging zoonosis with higher chances of an endemic outbreak of human infections.
Fungi and molds
Fungi are eukaryotic, non-motile, spore-releasing organisms. Fungi are classified based on their phenotypic characteristics ranging from unicellular to multicellular level, based on the mode of nutrition, by genotype characteristics and metabolic activities. Molds are the subtypes of fungi that are differentiated by the filamentous structure known as hyphae. Food contamination caused by fungi species is due to mycotoxins released. The toxin spread by the fungal species is highly linked with the food-borne outbreak. Some fungi and molds could cause food contamination, such as Aspergillus sp and aflatoxins from Aspergillus flavus, Fusarium, Alternaria, Mucormycetes Candida sp. The food-borne outbreak occurred in Tanzania in 2016 due to the intake of food products with a high level of aflatoxins. 68 cases were reported during the epidemiological investigation, and the source for the outbreak was contaminated maize with a high ratio of aflatoxins. 68 cases were reported during the epidemiological investigation, and the source for the outbreak was contaminated maize with a high ratio of aflatoxins (Kamala et al. 2018). A similar food-borne outbreak took place in provinces of rural South-Africa regions. Aflatoxin contamination was identified in the homegrown maizes, and test samples of maizes were collected and assessed to determine the ratio of mycotoxin level. Both aflatoxins and fumonisins ratio level was higher than the indicated level (Mngqawa et al. 2016). Fruit samples collected from a supermarket in Washington DC were assessed to recognize the contamination level of fungi and their types. A total of 38 percent of the samples evaluated contained Candida lambica, Candida pulcherrima, Rhodotorula spp., present in a higher number in the salad sample collected (Tournas et al. 2006). In Canada, one pregnant woman was affected by Candida kefyr through frequent organic dairy products, leading to bloodstream infection of this strain and transmitted to her premature twin infants through the placenta (Pineda et al. 2012).
Parasites
Clinically proven food-borne transmission of parasites to cause human diseases are the other category of food-borne pathogens. Currently, about 300 parasitic worms and 70 protozoan species are known to infect humans and animals. Toxoplasma gondii is a protozoan organism that causes a disease called toxoplasmosis. The disease’s transmission and outbreak occur through oocysts from the contaminated soil, water, and food. The bradyzoites specify the food-borne zoonosis. Oocyst and cyst from the protozoan infect and contaminate the raw meat, which ideally leads to the pathogen’s spread and outbreak (Shapiro et al. 2019).
Toxoplasma gondii
Toxoplasma gondii is a protozoan organism that causes a disease called toxoplasmosis. The transmission and outbreak of the disease occur through oocysts from the contaminated soil, water, and food. The bradyzoites specify the food-borne zoonosis. Oocyst and cyst from the protozoan infect and contaminate the raw meat, which ideally leads to the spread and outbreak of the pathogen (Stelzer et al. 2019). Toxoplasma gondii outbreaks are caused due to raw uncooked foods and fresh foods, including green vegetables. T.gondii oocyst tends to resist and present from days to months. Toxoplasma gondii outbreak consists of raw uncooked foods and fresh foods, including green vegetables (Shapiro et al. 2019). T.gondii oocyst tends to resist and present from days to months. The unprocessed products produced in the farms from the contaminated soil and irrigation water are fundamental reasons for causing transmission and an outbreak. Polluting seawater contributes to the transmission of the oocysts and gives rise to a vulnerable threat to the sea fishes and other organisms. Overall transmission is caused by the contaminated soil, livestock and poultry products, and seafood as a direct one or intermediate transference. Fever, appetite-loss are the symptoms due to protozoan attacks in the infected host acting halfway and directly. Other factors accountable for the outbreak involve optimum temperature, environmental conditions. The severity caused by the pathogen is much higher when compared with other microbes.
The severity caused by the pathogen is much higher when compared with other microbes. A Toxoplasmosis outbreak occurred in brazil in august 2016. The transmission is associated with the cheese prepared from the contaminated raw milk from the farm. Almost 250 people from the locality tested to identify the severity of the outbreak caused by Toxoplasma gondii (da Costa et al. 2020a, b). Acute toxoplasmosis surge occurred among deer hunters through the intake of undercooked deer meat in the United States. Almost ten members were affected by toxoplasmosis and identified by the primary epidemiological investigation. In 2016, a toxoplasmosis outbreak happened in an institutional restaurant affecting 20 people (Gaulin et al. 2020). The transmission was associated with raw vegetables served in the restaurant and affecting the individuals. From the cases mentioned; the prime reason for transmission and outbreak of Toxoplasma gondii is due to the unhygienic storage of raw vegetables and raw meat. Oocyst transmission causes the illness among the individuals affected by toxoplasmosis (Pinto-Ferreira et al. 2019).
Prion
Prion diseases spread through affected mammal meat trading and subsequent transmission to humans. They are a group of chronic fatal neurodegenerative disorders affecting several animals, scrapie in sheep and goats, bovine spongiform encephalopathy (BSE) or mad cow disease, chronic wasting disease (CWD) in deer and elk, in cattle, and variant Creutzfeldt-Jakob disease (vCJD) in humans (Doherr. 2007). A total of 229 vCJD patients have been identified in 12 countries since it was first discovered in the UK in 1996. Only by strict amendment of food control followed from 1992 to 2000, the number of BSE and vCJD epidemic cases reduced.
It is essential to devoid such pathogens from food manufacturing to consumption. For food processing, packaging, and storage in each stage, microbial pathogens must be identified and eliminated to ensure food safety. This review discussed a set of modern methods to assess micro and nano-level detection of microbial pathogens and components of food products and available conventional methods.
Available detection methods
It is essential to identify bacterial pathogens, toxins, and spore formations in food, which prevents food-borne disease outbreaks. The detection of microorganisms is based on the molecular, biochemical, immunological, or genetic means, enrichment (enumerations, counting of bacterial growth on plates). Detection methods are primarily used in food products and environmental monitoring for pathogenic organisms, indicators, and spoilers. Cultural enrichment methods, which are widely used, consume time and are deficient for rapid food pathogen detection. The presence of microorganisms in food and water is essential to be detected (Law et al. 2015).
Microbial detection methods
Conventional microbial methods tend to be labor-intensive and time-consuming. Traditional approaches focus on phenotypic detection through staining, culture, and basic biochemical studies (Baraketi et al. 2018). Modern methods are more powerful, rapid, and the novel provides more sensitive, precise reproducible results. Some current techniques frequently used to detect microorganisms are Immunological, nucleic acid, biochemical analytical methods.
Conventional methods
Many conventional approaches are available to detect pathogens and endotoxin, depending on culturing the microorganisms on agar plates. A routine microbiological technique involves homogenizing food samples, enrichment of viable pathogens if a load of pathogens is minimal, and further identifies the target pathogen by growing it on a selective medium accompanied by biochemical tests. To confirm the existence of the target, more subtyping studies are required to establish precise targets (Sharma and Mutharasan 2013). Enrichment plays a vital role in reviving injured cells (due to heat, cold, acid, or osmotic shocks during food processing), increasing target pathogens, and minimizing inhibitory compounds in processed food products (Gracias et al. 2004). Most conventional methods are labor-intensive and require 2–3 days to produce initial outcomes, and it takes up to one week to identify a particular pathogen. The performance ratio of this detection system is high and cost-effective. The culture of E.coli O157: H7 on Sorbitol MacConkey agar (SMAC) based on the concept of sorbitol fermentation is one of the best-known examples, which demonstrates a high success rate and also suggests that the method is very cost-effective. However, the main downside to this technique is the slow turnaround and false-positive findings due to the production of sorbitol fermentation serotypes, not O157 and O157 STEC. The main drawback of the culture-based process is slow growth in which the spare time lapses to achieve the final result and is assumed to be lethal. CHROMagar is more potent than SMAC, while CHROMagar is not susceptible to all strains (Priyanka et al. 2016). Cefsulodin Irgasan Novobiocin (CIN) agar is a selective medium known for the differentiation of bacterial species used to distinguish between Yersinia enterocolitica and non-Y. enterocolitica. Y. enterocolitica chromogenic medium is used where agar has fermented sugar cellobiose, these chromogenic substrates and selective inhibitors that kill competitive bacteria. Purple or blue colonies are produced on CIN agar, the primary food pathogens that cause Yersiniosis. In biochemical experiments, the bacteriophage is used instead of antibodies that have been an essential biological tool (Denis et al. 2011).
Immunological based methods
Immunoassay is based on an antigen–antibody interaction in which the specific antibody will bind to its particular specific antigen. The sensitivity and specificity are determined by the binding strength of antigen and antibody. Immunoassay involves the use of monoclonal, polyclonal, and antibodies. ELISA and lateral flow immunoassays are mainly used for the detection of immunoassays. ELISA allows the detection of bacterial toxins and can handle a large number of samples. This method is helpful in detecting Shiga toxin in E.coli O157 and non-O157 STEC (Baraketi et al. 2018). The polyclonal antibody was used as a conjugate for detection with HPR. Monoclonal antibodies are better than polyclonal antibodies because they have monovalence. In monoclonal antibodies, the antibodies produced are against one specific antigen. Specific growth conditions raise the analyzed time and complexity as ELISA is not much possible for toxin pathogens. Lateral flow assay is of low cost, dependable, precise, sensitive, and easy to operate, but still requires labeling antibody and antigen. The membrane strip and the lateral flow provide a quick and straightforward form of detection assay. The IFI methodology indicates more false-positive rates than ELISA and PCR. (Law et al. 2015) (Table 1).
Table 1.
Different immunological assays to detect the food-borne pathogens
| Pathogens | Methods | Food matrix | References |
|---|---|---|---|
| Listeria, Salmonella typhi Shiga toxins producing E.coli (STEC), E.coli O157: H7 and verotoxin | Lateral flow assay | Raw ground beef, beef trims, boneless beef, raw ground chicken, chicken carcass, sliced cooked turkey, liquid eggs, peanut butter, and tomato samples | Baraketi et al. 2018 |
| Listeria, salmonella spp., E.coli O157: H7 | Enzyme-linked immunosorbent assay | Meat, poultry, dairy products, nuts, fruit, all foods and animal feeds | Law et al. 2015 |
Modern methods
The DNA microarray is commonly used for the identification of microbes in food. Magnetic nanoparticles strengthen oligonucleotide microarray assay was developed for quick and delicate identification of Escherichia coli O157: H7, Salmonella enteric in food. Microarray-based microbial identification is primarily based on the hybridization of pre-amplified microbial DNA sequences to mask species-specific oligonucleotide probes. Each probe contains a specified dye that is fluorescent after hybridization, denatured to produce a single-stranded fragment. The fluorescence intensity corresponds to the concentration of each nucleic acid fragment labeled. This approach is gaining importance and becoming a useful gizmo. The detection of food-borne pathogens microarray is meant to target the Internal Spacer (ITS) sequence. ITS region of 5 Bacillus spp, B. anthracis, B.cereus, B. thuringiensis, B.mycoides, and B. weihenstephanensis were inspected as these have a peak homologous DNA level, which makes it hard to differentiate. DNA microarray is one of the best solutions to the present problem. DNA oligonucleotide microarray allows concurrent identification of multiple food-borne pathogens. (Priyanka et al. 2016).
PCR (polymerase chain reaction) is one of the most commonly used molecular methods for microbial detections of food-borne pathogens. The PCR theory is that the genes of various microbes can be amplified and further analyzed. Unique primers advance with every single gene, such as Salmonella fimA and pathogenic E. coli AFA for amplification. The size of the amplified product was 120 bp relative to the DNA marker. Identification uses agarose gel electrophoresis, fluorescent probes, unique shading, eventual staining with ethidium bromide, and molecular beacon. The multiplex PCR assay is developed to detect and recognize indicator microbial organisms of bacteria like Escherichia coli, Staphylococcus aureus, and Salmonella in a single reaction. These indicator bacteria are determined for the sensitivity and specificity of each primer. This process provides the exact observation of nucleic acid detection with a low concentration of starting amount (Wang et al. 2015).
Microbial spore detection
Spores are often produced in Bacillus and Clostridium species. Identification and enumeration of spores produced by the pathogens allow the recognition of potentially problematic species, whether from a consistency, quantity, hygiene, or pathogenic perspective. Protein-based, DNA-based approaches are used to identify spore-forming microbes (McHugh et al. 2017).
Protein-based methods
This assay was developed by manipulating DNA that encodes the proteins of interest and various positions of immunization. ELISA is used for the observation of entire cells of B. cereus. The subtractive screen ensures monoclonal antibodies (mAbs) are unique against B. cereus, and this approach has a lower detection limit of 0.9*103 cells/ml in phosphate-buffered saline. This assay is used to detect food samples penetrated with various pathogens, whereas culturing is not needed. This method effectively identifies B. cereus cells in the mixed samples with no intervention (McHugh et al. 2017). ELISA is not clear for the recognition of spores, as well as vegetative bacteria. It can be differentiated between the live and dead cells of B. cereus of surface antigen. The protein-based method is particularly true for spore formation in pathogens, whose presence is concerned but is currently in a dormant state during the sample testing.
DNA based methods
Random amplified polymorphic DNA PCR uses sort primers to amplify several DNA segments visualized on the agarose gel. Fingerprint analysis makes it easier to distinguish between species and strains. This approach was applied to the colonies of Geobacillus stearothermophilus, Anoxybacillus flavithermus, Bacillus licheniformis as they are the contaminant of WMPs and SMPs and in buttermilk and goat milk. The sequence of pro-printing synthesizes several polymorphic loci. The sequence is digitized and can be compared using a Pearson correlation to classify strains. Pyroprinting is developed and used for source monitoring. This process is used mainly for the identification of endospore-forming bacilli in raw milk. A culture-based test has been developed for the identification of spore-forming bacteria. Isolation of B.cereus has been developed by the baccara and mannitol egg yolk polymyxin agar (MYP). MYP agar is not as selective as baccara agar, which potentially leads to false-positive test results. Some Sulfite Reducing Clostridia (SRCs) have the capacity to reduce sulfite under anaerobic condition. The culture-based method isolates and specifies the cultural species (McHugh et al. 2017).
Biosensor
The biosensor is an analytical instrument composed of two main parts: a bioreceptor and a transducer. The target gene is recognized by the bioreceptor. A transducer is a detectable electrical signal of biological interaction (Alahi et al. 2017). A DNA-based pencil graphite electrode biosensor is being progressed to observe the toxin gene present in B. cereus, in which the toxin gene primers are immobilized on gold nanoparticles. The positive outcome is measured by the rise of charge resistance on the biosensor to hybridize target DNA to the toxin (Law et al. 2015) (Table 2).
Table 2.
The different biosensors used for pathogen detection
| Pathogens | Methods | Food matrix | References |
|---|---|---|---|
| Salmonella choleraesuis serotype Typhimurium, Listeria monocytogenes, Campylobacter jejuni, and Escherichia coli O157: H7 | Optical biosensor | Apple juice, cucumber, broiler meat, and ground beef | Zhao et al. 2014 |
| Escherichia Coli O157: H7 and Salmonella | Electrochemical biosensor | Milk, ground beef, chicken extract | Baraketi et al. 2018 |
| Salmonella and Campylobacter | Interferometric biosensor | Poultry products | Alahi et al. 2017 |
| Staphylococcal enterotoxin B and Botulinum toxin A | Fluorescent immunoassay biosensor | Tomatoes, beans, sweet corn, and mushrooms | Zhao et al. 2014 |
| Escherichia Coli O157, Listeria, Salmonella, and Campylobacter | Electro immunoassay biosensor | Chicken breast | Alahi et al. 2017 |
Microbial detection methods
Microorganisms such as bacteria produce toxins that cause infection and illness, directly affect the host tissues, and disable the immune system. New approaches are being developed for better isolation and understanding of these toxins. Detection techniques for pathogens and toxins can differ in their cost-effectiveness, scale, reaction, and reliability (Zhao et al. 2014). Some toxin-producing bacteria like E. coli, Salmonella enterica, Vibrio cholera, Bacillus cereus, Staphylococcus aureus, Campylobacter jejuni, Listeria monocytogenes, Salmonella spp. causes food-borne disease. Nucleic acid, biosensors, and immunologically based approaches are the most widely used methods (Wang et al. 2016).
Nucleic acid methods
These methods identify organisms, strains, or particular DNA or RNA sequences in target pathogens and hybridize the target nucleic acid sequence to the synthetic oligonucleotides complementing the target sequence of food-borne pathogens such as C. botulinum, V.cholerae, S. aureus, E. coli are capable of producing toxins (Law et al. 2015). Toxin producing genes can be evaluated using these techniques. These methods are used to detect particular genes in target pathogens, which are unclear or mistakenly interpreted. These techniques are more valuable because they are time-efficient and not labor-intensive. Nucleic acid approaches are PCR-based and non-PCR-based identification methods (Baraketi et al. 2018).
The PCR-based approach is one of the most common techniques used to identify food pathogens and toxins. The PCR approach is designed for DNA amplification In Vitro and other microbes. PCR methods have been improved for quicker identification with the development of real-time or quantitative PCR, multiplex PCR, oligonucleotide DNA microarrays, which detect five or more pathogens at the same time, as these methods are developed for monitoring PCR and amplification of PCR. Food materials are homogeneous and have undergone a centrifugation process before DNA extraction. PCR has yielded the identification limits of centrifugation for individual food products. Multiplex PCR is a traditional PCR approach in which multiple targets can be identified. This multiplex PCR method can identify genes, gene mutations, or genomic markers using a multiple set of primers, where a simple PCR uses a single set of primers. A multiplex PCR method is used to detect S. aureus, Cronobacter sakazakii, E. coli O156: H7, B. cereus, V. parahaemolyticus, and Salmonella spp. The detection limit of these pathogens ranges from 102 to 104 CFU /ml (Law et al. 2015).
The quantitative real-time method is the quantity, and this monitors the formation of the amplified DNA products. The amplified DNA is detected using the fluorescence-labeled moiety and is detected by the thermocycler. One of the qPCR methods used SYBR green, an intercalating dye that binds to double-stranded DNA’s minor groove. The amplified DNA is quantified by measuring the fluorescence intensity in the intercalating dye. Other qPCR are developed using probes. Probe-based assays are reproducible and provide a higher degree of sensitivity than SYBR Green assays. (Zhao et al. 2014).
Loop-mediated isothermal amplification is a non-PCR-based nucleic acid detection method. The targets of the LAMP are assays, stx1, stx2, and EAEes. In the LAMP reaction, 4–6 primers target a particular sequence, and a strand displaced but DNA polymerase to amplify target DNA copies under isothermal conditions. The result obtained in this process is considered to be quicker, more adaptive, and more accurate to the identification of STEC strains than the qPCR findings. One of the benefits of this LAMP is the lack of false-positive or false-negative effects. Primer dependent technology amplifies nucleic acids in a single reaction at isothermal conditions is Nucleic Acid Sequence-Based Amplification (NASBA). This method is used to detect an in vitro sequence of RNAs. The amplified products can be visualized using an electrophoresis gel or an enzyme-linked gel assay. Whereas NASBA product identification is also known to be labor-intensive. (Law et al. 2015).
DNA microarray is a small device that consists of short single-stranded DNA oligonucleotides. The target DNA is extracted and is labeled by this microarray using a fluorescent dye. The single-stranded DNA molecules bind to their complementary probes on the array. Double-stranded DNA is formed and is visualized using the fluorescence signal. Oligonucleotide microarray technology is susceptible and specific to the target sequence, detects multiple pathogenic microbial organisms, and is non-labor intensive (Table 3).
Table 3.
Different nucleic acid methods used to detect food-borne pathogens
| Pathogens | Methods | Food matrix | References |
|---|---|---|---|
| Staphylococcus, Salmonella spp., Listeria spp. (except L. grayil) | PCR– Polymerase chain reaction | Ground beef, poultry, dairy, vegetables, bakery products, meat, seafood, milk powder | Alahi et al. 2017 |
|
STEC O26, O103, O111, O145 Sorbitol fermenting O157 and non-sorbitol fermenting O157, E. coliO154:H7, Salmonella entertidis |
Multiplex PCR | Raw ground beef, beef trim, fresh pork sausage, sprouted seed (soy, alfafa, and leek), raw-milk cheese, minas cheese, carcasses, minced beef | Baraketi et al. 2018 |
| Salmonella spp., Stx and ear genes STEC screening, E. coli O45, O103, O145, Listeria species, Listeria monocytogenes | Real-time PCR | Meat, poultry, dairy, fruits, vegetables, beef trim, dairy, ready-to-eat meat, seafood, bakery products, pet food, raw ground beef | Zhao et al. 2014 |
| Escherichia Coli, Salmonella Enteritidis, Listeria monocytogenes, Bacillus circulans, Salmonella enteritidis, and Salmonella typhimurium, Bacillus amyloliquefaciens, Bacillus cereus | NASBA | Drinking water, fresh meat, ready to eat salads, cooked ham and smoked salmon slices, milk | Law et al. 2015 |
| Vibrio vulnificus, Salmonella spp. and Shigella spp., Vibrio parahaemolyticus | LAMP | Oysters, milk, spinach, lettuce and sprouts, fish, shrimp, mussels | Baraketi et al. 2018 |
| Salmonella entrica, Listeria monocytogenes, Campylobacter jejuni, Enterococcus faecalis, Yersinia enterocolitica, and Vibrio parahaemolyticus, proteus spp. | Oligonucleotide DNA microarray | Lettuce, spinach, sprouts, pork, meat, chicken, fish, beef, turkey, milk | Law et al. 2015 |
Pros and cons of food-borne pathogen detection methods
It is essential to protect food that harbors bacteria and fungi cells and their toxins, spores. Most of the microbes are harmless to humans; hence existing pathogens in less number are a potent threat to living human’s health and protection. Continual outbreaks and incidents may weaken our socio-economic and health care system of one country. As per the WHO report 2015, around 2.2 million deaths per annum due to food and water-borne disorder among them 1.9 million are children. Food and lifestyle modification in recent years directed to encourage consumption of ready to eat food consumption. In turn, it increased the chance of vulnerability of pathogens viz., E. coli, Salmonella, Listeria, and Campylobacter jejuni and adulterants of meat, vegetables, fruits and milk products.
Conventional detection could serve in the front line to identify and characterize pathogens based on cultivation procedures. Specific features and biochemical properties of microbes while cultivating in enrichment broths and selective media are quite helpful in microbial pathogen confirmation. (Zhao et al. 2014). These methods included microbiological and biochemical examination that are accurate, low operational cost, but time-consuming, not suitable to incorporate for on-site analysis. This type of method can identify one type of pathogen at a time. Also, manual error and experienced professionals are other limitations of culture-based methods. Such low sensitivity and false-negative results due to Viable Non-Culturable (VBNC) pathogens may increase food-borne disease risk (Ramirez-Castillo et al. 2015).
Nucleic acid-based detection methods
Identification procedures should be revised to meet the LOD of nucleic acid-based identification methods. Polymerase chain reaction (PCR), multiplex polymerase chain reaction, real-time polymerase chain reaction, and DNA microarray are commonly used. In recent years, owing to high reactivity and high identification limit (LOD) of food-borne pathogens and convenient protocols, such approaches have been thoroughly implemented. Except formalin stored and preserved samples, others can be processed through PCR to avoid DNA spoilage.
Polymerase chain reaction (PCR) and variants
Apart from universal primers, each time needs prior DNA sequence for primer designing in PCR method. PCR variants such as nested PCR could detect even 0.05 viral copies per cell. Multiplex PCR is the concurrent identification of targets in a single reaction well with a separate pair of primers for each target. This technique requires two or more probes that can be separated from each other and detected simultaneously. It provides the identification of false-negative samples at a short time interval (Lauri and Mariani 2009). Around the same time, five separate strains of bacteria have been detected, including Salmonella enteritidis, Staphylococcus aureus, Streptococcus pyogenes, Listeria monocytogenes, and Escherichia coli using 16S rDNA amplification with respective primer sets by multiplex PCR. When increasing primer numbers, some interference could result in the amplification process.
Real-time or quantitative PCR (qPCR) is a method used to track the progress of PCR reactions in real-time. Simultaneously, a minimal volume of PCR product (DNA, cDNA, or RNA) may be detected. This PCR is distinct from primary PCR; it does not require agarose gel electrophoresis to detect PCR products. During the activity, the PCR product can be quantified in the entire reaction by measuring the fluorescent signal generated with unique intercalating dyes. The quantitative PCR is the confirmation of the analytes through the melting curve analysis.
Besides, we can calculate how many amplicons are produced and how many non-specific or primer-dimers are created during the PCR reaction by conducting a melting curve analysis. Besides, qPCR has contributed to numerous commercial qPCR kits to identify food-borne pathogens such as Salmonella, Listeria monocytogenes, Escherichia coli, and Campylobacter. For example, commercial qPCR kit for Salmonella identification is used for shrimp and meat samples (Lauri and Mariani 2009). Minimal sample handling and low-level cross-contamination, High throughput analysis, electrophoresis independent amplicon detection are benefits, and only drawbacks exist in operation costs while handling more samples.
Nucleic acid sequence-based amplification (NASBA)
Nucleic acid sequence-based amplification, generally referred to as NASBA, is a molecular biology process used to amplify RNAs. Loop-mediated isothermal amplification is another method for isothermal amplification. More than 109 copies of the nucleic acid sequence can be generated in just 30 min by three enzyme actions. Costly thermocycling equipment is not needed as the reaction occurs isothermally at 41 °C. It helps to answer better to RT-PCR, as it gives faster elaboration kinetics. This technique is primarily used to validate the replication of the DNA virus by detecting late mRNA expression. It also facilitates the identification of human mRNA sequences without the possibility of DNA contamination.
There are also some drawbacks of NASBA due to RNA integrity and other amplification procedures (Law et al. 2015). Due to the operating temperature being isothermal at 41 °C, a single melting step is needed before the amplification reaction to facilitate the annealing of the primers to the target. Furthermore, since the specificity of the reactions depends on thermolabile enzymes, the reaction temperature cannot reach 42 °C without compromising it. Finally, the amplified RNA target sequence length should be in the range of 120–250 nucleotides, whereas shorter or longer sequences being amplified less effectively (Fakruddin et al. 2012).
Loop-mediated isothermal amplification (LAMP)
LAMP technology is primarily used to detect multiple parasitic infections such as malaria and leishmaniasis. The LAMP technique offers many benefits, including non-target DNA amplification, RNA conversion by Reverse Transcriptase enzyme, inexpensive techniques, and independent electrophoresis technique, ideal for creating a lab-on-a-chip microfluidic device. There is no internal control in PCR; hence ensuring amplification reaction needs to be replicated. Other steps of primer designing are carried out with existing software. Unlike PCR, the end product is a complement fragment, not suitable for cloning and extended molecular biology techniques.
The clarity of the method combined with PCR’s sense and clarity puts LAMP technology in a perfect place for future growth. Furthermore, LAMP has wide applications in clinical diagnosis to perform cost effective procedures in growing countries. This technique was developed to detect Schistosoma mansoni infections from fecal and sera samples of infected mice. Through the investigation, LAMP was also used to distinguish various types of Taenia (T. solium, T. Asiatica) from eggs in stool samples. For, e.g., the identification of Nicator American infections from human stool samples successfully using a LAMP microfluidic chip (LOC-LAMP) instrument to detect the presence of Schistosoma mansoni in infected mice hosts (Blais et al. 1997).
Nanoparticle based detection methods
The biotechnological advancement in the recent past has delivered diverge methods that provide new opportunities to detect food pathogens. Though the traditional methods still represent the first choice for detection, new techniques are being developed due to its limited identification capability and meet the growing demand to design ultrasensitive, rapid, and selective pathogen detection methods. The nanomaterial-based biosensor technology and High Throughput Sequencing (HTS) have emerged as an alternative to the time-consuming conventional methods (Ornerova et al. 2011).
Several regulatory agencies world-wide define the safety of nanomaterials, provide suggestions and guidance concerning their application in commercial products. The European Food Safety Authority (EFSA) Scientific Committee released a guideline recommending characterizing usage of engineered nanomaterials. According to the current regulation, there are no requirements for "nano" and the industries are solely responsible for meeting the safety standards set by The Food and Drug Administration (FDA). FDA supports and approves the innovation of new edge-cutting technologies and they promote the safe use of nanotechnology. Overall, regulatory bodies conduct research studies and provide transparent regulations for the use of nanomaterials and also inform the general public about the perks and risks involved in employing nanotechnology for food pathogen detection (Mustafa and Andreescu 2020).
Biosensors with advanced nanostructures
With advances in nanotechnology and nanoscience, nanomaterial-based biosensors have demonstrated the considerable potential to enhance microbial identification in food due to their unique physical, biological, mechanical, optical, and magnetic properties. Several sophisticated nanomaterials are now continually being used in the construction of biosensors as it has increased sensitivity and can lower the detection limits down to even individual molecules. Nanomaterials prove promising, that not only provides an enhanced, high specific surface area and high chemical reactivity, but also enables the possibility to immobilize substantial quantities of the bioreceptor units at reduced volumes. (Holzinger et al. 2014).
Nanomaterials are of a size that ranges between 1 and 100 nm. Nanomaterials have distinct physicochemical properties due to their small size. As a result, most of the constituent atoms and molecules are present on the nanomaterial surface. Nanomaterials are classified based on chemical composition. They are distinguished into three types:
Carbon-based nanomaterials, consisting of carbon atoms such as carbon dots, nanotubes, etc.
Organic nanomaterials, consisting of polymeric nanomaterials such as nanofilms.
Inorganic nanomaterials are made up of either metallic components such as gold, silver, etc. or non-metallic nanomaterials that include magnetic nanoparticles, quantum dots, etc.
The nanomaterials exhibit a low melting point due to less binding energy as the number of atoms on the surface of nanoparticles is much higher. It increases the surface area per unit mass, thus ultimately showing an increase in chemical reactivity. Some nanoparticles can also act as artificial atoms because of their electronic behavior, as the spatial arrangement of an electron at nanoscales generates an energy spectrum that is quantized. If there are multiple unpaired electron spins, nanoparticles possess magnetic properties, showing magnetic properties like super magnetism, and are suited for contrast agents in MRI scanning images. Their different intriguing properties are being applied in various fields of study that include the food sector, as they help detect the food-borne pathogen (Pirzada et al. 2019).
Semiconductors nanocrystals
With its salient properties, the semiconductor nanocrystals can be used as biomarkers that help detect food-borne pathogens by applying analytical techniques. One of the essential detection techniques used is Quantum dots.
Quantum dots
Quantum dots are used as a fluorescence marker. They are fluorescent, semiconductor nanoparticles with diameters of about 2–10 nm. They can produce different colors that are governed by their caliber. They will emit the same symmetric, narrow-spectrum regardless of the excitation wavelength that is generally shorter than the emission peak wavelength and is tunable with particle size and composition. This property of theirs allows the possible detection of different emission peaks simultaneously when other quantum dots are stimulated with a single wavelength (Wang et al. 2020).
They can detect cellular toxicity that depends on the nanoparticle’s surface properties, aspect ratio, and exposure time. They represent the II–VI, III–V, and IV–VI group semiconductor materials, their binary, alloyed, and core-shell systems in three dimensions. One of the best quantum dot fluorophores is Cadmium selenide (CdSe) cores covered with a layer of Zinc sulfide (ZnS). In these nanostructures, the ZnS layer protects the CdSe from oxidation and prevents leaching into the surrounding medium, and, in turn, produces an enhancement in photoluminescence yield (Zhuo Zhao et al. 2016).
Magnetic nanoparticle
They form another class of nanomaterials that are utilized in biosensors by changing its magnetic field. These nanoparticles form a cluster of magnetic beads that vary in diameters ranging from 50 to 500 nm. They do not require pre-treatment of the sample and produce immediate results in robust magnetic and visual signals within 20 min. They have also risen as necessary fabricating materials as they have penetrable colors and can distinguish the target material from the complex matrix (Sai Anand et al. 2019).
Metal-based nanomaterials
Gold nanoparticles
The synthesis of gold nanoparticles (AuNPs) occurs in an organic solvent with a stabilizing mediator called a surfactant. The surfactant is loaded; different gold nanoparticles’ properties are adjusted by choosing Surfactants. The stability is accomplished by including surfactants that induce chemical binding or take up the gold nanoparticles’ appropriate mediator. When utilized in association with the redox enzyme, it oxidizes or reduces the analyte as the reaction’s substrate. The immobilized catalyst that binds to the analyte builds the current signal and confirms the pathogen’s detection. As used in conjugation with single stranded DNA (ssDNA) complementary to the microbial DNA (Kumar et al. 2020), the gold nanoparticle strengthens its bonding to the DNA-gold nanoparticles present on the transducer surface.
Silver nanoparticles
Silver, due to its intriguing properties and being one of the noble metals, has found extensive applications in different sectors like the food, pharmaceutical, and health sector. The silver nanoparticles are widely utilized on Surface-Enhanced Raman Spectroscopy (SERS)-based biosensors that show a significantly higher extinction coefficient than the gold nanoparticles and undergo electrochemical oxidation more easily. Silver nanoparticles have an excellent electrical conductivity that makes them ideal nanoparticles for electrochemical biosensors (Sai Anand et al. 2019). These nanoparticles are made ready for use through various procedures. They have successfully detected many human pathogens such as Klebsiella Pneumoniae, Staphylococcus Aureus, and many pathogenic bacteria such as Escherichia coli, Staphylococcus epidermidis, Listeria innocua.
Carbon-based nanomaterials
Carbon-based nanomaterials such as graphene, carbon dots are utilized for bacterial detection as they have remarkable surface modification and aspect ratio. The carbon dots developed from natural biomass acting as fluorescent biomarkers are inexpensive as they do not require additional magnetic or chemical treatments. The bacterial detection depends on the affinity of the carbon dots towards the cell surface of the bacteria. They also show the varying intensity and the spectral positions in the fluorescence spectra for various bacteria and distinguish among the different bacterial strains. The graphene nanomaterials are aptamers utilized as transducers of biosensors. It is one of the most commonly used nanomaterials in differently designed biosensors that could convert target and receptor molecules into a detectable measurement. They have different transduction modes based on their large surface area, electrical conductivity, and high electron transmission rate that helps in immobilizing various types of molecules. The graphene can be in the form of reduced graphene oxide, graphene oxide, or graphene. There will be a difference in bacterial detection by the biosensors based on graphene with different oxidation states and functional groups. The detection limit is also affected by the graphene orientation. This sensing platform is quite selective and enhanced by a coating of appropriate material. For example, graphene, when coated with antimicrobial peptides, a biorecognition molecule, was shown to have a stronger binding affinity to a bacterium present in the food analyzed (Kumar et al. 2020). At times, carbon electrodes are coated with graphene for bacterial detection. The other carbon-based nanomaterials include carbon tubes and carbon materials. Carbon nanotubes are immune-sensing platforms of nanoparticles like gold, antibodies, etc. There occurs an interaction between the immobilized antibodies on the sensing probe and the bacterium. This interaction causes an increase or decrease in the electron transfer resistance that is measured through spectroscopy. Different types of carbon nanotubes are single-walled carbon nanotube and multi-walled nanotube. When combined with fluorescence dyes such as cyanine through stacking interactions, the carbon soot nanoparticles can further enhance bacterial detection. The Transmission Electron Microscopy (TEM) images and confocal laser microscope images see the carbon soot nanoparticles and the bacterial binding to aptamers used for analysis.
Conducting polymers
The fabrication of biosensors can also utilize polymers because of their conducting properties. They are good conductors as they possess both semiconductor and metallic characteristics. The different types of nano polymers used for various applications are polythiophene, polyaniline, and polypyrrole. They also act as immobilizing agents to convey better signal transduction, high affectability, biocompatibility (Kumar et al. 2020) (Table 4).
Table 4.
Food pathogen detection by using different nanomaterials
| Nanomaterials | Sub-classification | Pathogens | Limit of detection | References |
|---|---|---|---|---|
| Semiconductor Nanomaterial | Quantum dots | E. coli | 30 CFU/mL | Kumar et al. 2020 |
| Inorganic metal oxide | Salmonella | 1 × 10–8 RIU | ||
| Magnetic nanomaterial | – | Bacillus anthracis | 2 × 105 spores/g of starch 5 × 105 spores/g of baking soda | Wang et al.2020 |
| E. coli O157: H7 | 6 × 104 spores/g of milk powder | |||
| 12 CFU mL−1 of broth and 30–300 CFU mL−1 | ||||
| Metal-based nanomaterial | Gold nanoparticles |
Norovirus L. monocytogenes |
60 copies mL−1 2 log CFU/g in spiked blueberries | Huang et al. 2015 |
| Silver nanoparticles | E. Coli O157: H7 | 10 CFU/mL | ||
| Carbon-based Nanomaterial |
Graphene Graphene (G) |
E. coli O157: H7 | 10–100 cells mL−1 | Sai-Anand (2019) |
| Graphene oxide (GO) | Methicillin-resistant Staphylococcus aureus | 10 CFU mL | ||
| Reduced graphene oxide | AFB1 | 0.12 ng mL−1 | ||
| Carbon Tubes | ||||
| Single-walled carbon tubes | Salmonella | 1 × 10–9 mol/L DNA | ||
| Multi walled carbon tubes | Salmonella enteritidis | 5.5 × 101 CFU mL−1 | ||
| Salmonella typhimurium | ||||
| Carbon Nanoparticles | V. parahaemolyticus, S. aureus, and S. Typhimurium | 6.7 × 101 CFU mL−1 | ||
| 25–50 CFU mL−1 | ||||
| Conducting Polymers | – | S. typhimurium | 3 CFU mL−1 | Kumar (2020) |
| L. monocytogenes | 4.1 PG mL−1 | |||
| Others | Silicon Nanomaterials | E. coli |
101 cells 102 cells |
Zhao. (2016) |
| Dendrimers | AFB1 | 0.40 ± 0.03 nM |
High throughput sequencing (HTS)
HTS is a prominent tool used for the screening of food pathogens. The HTS shows a wide range of inter laboratory variations. The lack of consistency, the lack of universally accepted definition of HTS, and the lack of protocols for their use have limited the integration of HTS into regulatory policies. (Barrick et al. 2017). Sequencing is an appealing approach for detecting food pathogens because nearly all pathogens contain DNA or RNA (Gu et al. 2019). This method aims to amplify specific DNA areas to focus on a DNA sequence at a targeted site, subsequently used as a molecular marker.
HTS can generate thousands to millions of sequence reads and up to a hundred billion base pairs (Sekse et al. 2017). The most recently developed techniques are Next Generation Sequencing (NGS), a versatile technology. Several NGS platforms are now available. The frequently used is Illumina. Illumina sequencing offers the highest-fidelity as it provides humongous data sets for a relatively short read where the error percentage is less than 1%. It also provides a range of choices that generate various numbers and lengths of DNA sequences. Illumina iSeq 100 can create a limit of 1.2 billion bases of succession for every run, while the HiSeq X Ten can deliver up to 1.8 trillion bases (Haynes et al. 2019).
A nanopore sequencing method is an alternative approach to next-generation sequencing. It relies on the threading of individual DNA or RNA molecules into engineered protein nanopores and the constant electrical current measurement through each pore.
Steps involved in the transformation of raw sequence data into a piece of information are as follows:
Assembly of shorter fragments into a complete sequence by mapping them against a known reference genome.
Compare the genomes assembled with the reference strains.
Comparing the assembled genome with reference strains allows a range of interferences, such as pathogen identification, high-resolution strain typing, and phenotypic characteristics. It provides for a well-cured and up-to-date reference database as pathogens proliferate. Each transformation method uses a range of bioinformatics techniques of the utmost importance since the generation of vast sequences involves a sophisticated operating system for analysis. Multiple software options are present where some are freely available. Examples of the software are CLC Genomics Workbench (Qiagen), Bionumerics (Applied Maths), and SeqSphere + (Ridom) (Haynes et al. 2019).
The fusion of nanotechnology allows the building and assembly of devices with much smaller sizes, making it easier for them to handle. Each nanomaterial has its own set of advantages and detection techniques. For example, some semi conductive nanoparticles show optical properties, while some of the magnetic nanoparticles use the NMR method to ease the pathogen’s detection (Sai Anand et al. 2019).
Comparison of advanced methods
Recently, modern techniques are widely applied in food pathogen detection to overcome the limitations of conventional methods. Even though conventional methods still represent the first line for pathogen detection. New techniques are being developed to extend the identification capability and meet the growing demand to attain food safety. Such methods should be ultrasensitive, rapid, and accurate in pathogen detections with ease of operation and cost-effective to globalization.
Conventional methods are adapted widely because of their accuracy, directionality in culturing pathogens from contaminated food samples. Enumeration of microbial load of food and water is required sequentially to avoid outbreaks and disease occurrence (Law et al. 2015). These techniques are labor-intensive; concerns are mainly given to identify phenotypic and biochemical characteristics of microbial pathogens (Baraketi et al. 2018). When compared with available modern methods as followed, these techniques have limitations in detecting pathogens indeed. Such a qualitative method could only determine the presence of a microbe in a tested food sample. The other prominent factor, limit of detection (LOD) determines the detectable microbe count in 25 g of food sample tested. LOD expected value for the culture plating method is 1 (CFU/g or colony-forming unit per gram).
Apart from confirming pathogen presence and strain identification, the culturing method was not suitable for emerging pathogens and its serotype of the same genus. Emerging serotypes are found with their phenotypic and genotypic diversity through adaptation to antibiotics. Hence there are false-positive results; error rate could occur with traditional methods. Thus, new microbial serotype and its biochemical and fermentation properties may also vary. At this junction, modern methods were applied further to test their efficacy in food pathogen detection.
Second, immunoassay techniques are used to identify pathogens with monoclonal and polyclonal antibody applications. The enzyme-linked immunosorbent assay (ELISA) and lateral flow immunosorbent assays are primarily used in food-borne detection methods. ELISA could be used to detect bacterial toxins and can accommodate a considerable number of samples. For, e.g., this approach is useful for detecting Shiga toxins in E. coli O157 and not O157 Shiga toxin producing Escherichia coli (STEC) (Baraketi et al. 2018). ELISA is not evidential to recognize spores and vegetative forms of bacteria.
Nucleic acid detection methods mainly depend on PCR (Polymerase Chain Reaction), an essential molecular technique applied for microbial pathogen detection. Multiplex PCR, this variant of PCR, could detect more pathogens in a single tube PCR reaction, less time consuming, and could detect 10–2–10–4 CFU/ml detection limit (Law et al. 2015). Quantitative real-time PCR provides more advantages than any PCR variant by live monitoring pathogen presence with accuracy during food processing stages of industrial sectors. The further probe-based assay could be of higher sensitivity than other reporter-based assays (Zhao et al. 2014). Associated RAPD (Random amplified polymorphic DNA) and DNA fingerprinting analysis could provide accurate differentiation at species and strain level of food pathogens.
The biosensor is an electronic instrument with two main parts, namely a bioreceptor and a transducer. Bioreceptor sensitizes target gene and identification; transducer further confirms electrical signal by biological interaction and product measurement. These data produced by the positive result due to an increase in the biosensor load resistance directly denote the hybridization of the target DNA to the toxin. Such signal identification confirms organisms or strains containing a particular genetic sequence of target pathogens (Law et al. 2015). Hybridization of DNA or RNA nucleic acid sequence to synthetic oligonucleotides does reflect the target sequence of food-borne pathogens viz.,Clostridium botulinum, Vibrio cholera, Staphylococcus aureus, Escherichia coli and toxins, and so on.
The field of food pathogen detection is evolving quite rapidly, which is evident from the significant emphasis on experimentation and analysis of better, efficient materials with high sensitivity, high magnitudes of detection levels, longer life, easy operation, and generating massive amounts of data in less time. Concerning this, nanomaterials have gained importance in the recent past owing to their distinctive properties such as fast reaction rate, relative ease for the sensing and discernment of specific pathogens, and selective nature.
Implementing nanomaterials does offer sophisticated tools for food pathogen detections. On the other side, nanomaterials are highly expensive and proven to be expensive to commercial-scale processes. Another disadvantage is that their particle size may cross-react, or expose pathogenic contents of microbial cells (toxins) that could contaminate food particles along with food pathogens. Therefore, these materials are not widely implied in food quality and safety detections (Mira Miralles et al. 2019).
Advantageous HTS operation cost may be reduced with changes in sample processing steps and more reliable than time-consuming procedures of biosensor applications. The output data from computational analysis must be compared with existing food pathogen species, or subspecies reported earlier. Further this field needs more upgradation and also repository information and databases must be reported world-wide to support and make HTS as a universal tool in identifying food pathogens (Gwinn et al. 2019; Yang et al. 2020).
Future scope of the mentioned detection techniques
In view from previous sections most valid points are taken to meet our future perspective of food borne pathogen detection. Conventional and Modern methods giving us a gist of the various detection techniques employed to detect the contamination of food by the associated pathogens. To overcome limitations of known methods, to make the detection of micro and even nano-sized organism contents easier, cheaper, less time-consuming, than in the past. Under conventional method proper updation in media to highlight pathogen detection, universal sampling methods must be followed. Combinations of immunological and molecular techniques would be quite helpful in exceeding LOD. As an example, the Immunological method in combination with RT-PCR is reported in the evaluation of Norovirus. Likewise, microscopic paramagnetic beads adapted with nuclease enzyme activity are beneficial in Listeria monocytogenes detection from milk and milk products, also for Campylobacter jejuni in both food and water samples. The main aim of quality management is to decrease the time taken for the detection of these food pathogen contaminations; improve the product quality, safety, and hygiene as well (Adley 2014). A blend of two or more of the above-stated techniques would help create new techniques which might minimize the present consequences of the detection methods (Hameed et al. 2018).
PCR and its variants shed impact on the detection and diagnostic methods of the pathogen in clinical, pharmaceutical, food, and as well as industrial sectors. Along with PCR, next-generation methods like High Throughput Sequencing (HTS) could determine pathogens at the molecular level precisely. Targeted protein expression through species-specific primer-based assays and detailing on species might add to the value of HTS (Murray et al. 2011). Also, the theory of Surface Plasmon Resonance (SPR) has been employed in optical biosensors for HTS (Bhunia et al. 2014). Another method is the use of Lamp (Loop-Mediated Isothermal Amplification) system in HTS which seemed like a quicker alternative to the usual methods, especially for foods that need extra care such as infant foods. The compatibility of the detection system is another boon to detection with accuracy in addition to being less time-consuming (Jin et al. 2020). Though these two methods are highly preferred, they still do not have a standardized protocol, hence, this procedure should be handled with utmost care for accurate results (Palomino-Camargo et al. 2014); Further up-gradation is needed to improve in automation, sensitivity, and specificity, satisfying the multiplex targets of the future times.
The next molecular technique trending at present times is DNA-based Biosensors. This particular technique became trending due to its better characteristics such as easy programmability etc. (Huo et al. 2020). There is a better preference for this technique due to its faster, simpler, and cost-effective technology. There are several subtypes of these sensors each employing its own respective technology (Abu-Salah et al. 2015). This technique is employed to detect whole-cell bacteria, which when made with eco-friendly material for its construction will give an additional benefit, miniaturizing its size compatible for movement and storage. The ubiquitous usage of these whole bacterial cell biosensors will be a major breakthrough in all fields of diagnostics (Ahmed et al. 2014). This technique is further foreseen to become available in the next decade and push up further other fields of diagnostics along with it (Abu-Salah et al. 2015).
Another class of modern methods of detection is the Nanoparticles-based detection methods, exhibiting its wide variety which puts into use specific technologies compatible to the biomaterial used for construction which shows a high range of sensitivity and specificity along with detecting paramagnetic properties provides a promising scope in the future (Sahoo et al. 2021) but again, the major con with this method is the high initial cost incurred Verma et al. 2015). Quantum Dots technique and application of this technique in Food Science is often common these days, where their photoluminescent properties have been put to use to monitor even in media such as water. But, usage of natural minerals which does not cause ecological harm as well would save any further contamination too. This method would place its strong foothold by enhancing its selectivity and robustness (Nsibande et al. 2016). Another innovative approach towards an eco-friendly and economically benefiting technique is the usage of Carbon Quantum Dots (CQDs) procured from food waste having superior optical and physical properties too (Fan et al. 2020) and the cheapest among them being the Graphene Quantum Dots (GQDs) which is novel on its side and has long term stability indeed. With this, there was a promising note that CQDs don’t need any further equipment too (Hossein Safardoust-Hojaghan et al. 2017). Additionally, one another is the Chitosan modified Cadmium Sulfide (CdS) quantum dots which are higher-up in all fields such as speed, biocompatibility, sensitivity, affinity to the substrate, etc. Proving its superiority in all the above-mentioned fields, this Quantum Dots is an excellent and novel biosensor that can be brought to use in the forthcoming years (Abdelhamid et al. 2013).
This review has elaborated on existing food-borne pathogen detection methods, along with the pros and cons of each technique and comparative perspective. The main focus was given to shed an impact on detection methods and suggestions to overcome the LOD of food microbial pathogens. The futuristic approach may complete with two or more techniques in a combined approach to meet out the limitations of each detection method. It would help in finding a solution for the limit of detection of food-borne pathogens.
Conclusion
In this review, we have covered and highlighted the various pathogens responsible for food-borne diseases leading to an outbreak among the public, the current and the advanced methods for detection of food pathogens, their characteristics, the perks and setbacks of each technique, and the scenario of early detection of these novel food pathogens. From this, the evolution of the field of food pathogen detection can be validated. Despite the exceptional effort to develop the different detection techniques, only a few are economically viable and commercially available. The ideal detection methods for identifying the pathogens in the food matrix reasonably are not simple. It involves many additional prerequisite steps for preparation and assembly before the actual detection.
The future prospects of the above-stated techniques are quite promising provided the protocol is properly followed and the procedure is done in a sterilized and favorable environment of that specific technique. Moreover, the development of any device for detecting pathogens also depends upon the type of food and the component nutrients present in food such as proteins, fats, carbohydrates, and fibers. Hence, the specific analytical tools and sampling methods are essential for detection in each food product. The development of a new technique or advancement of an existing system is possible only by conducting extensive research. Most importantly, it should be economically viable, applied, and utilized on an industrial scale, and not limited to, research purposes alone.
Acknowledgements
We thank SRM Institute of Science and Technology, Kattankulathur, Chennai for providing support for writing this review article.
Authors’ contributions
All the authors GV, HKVB, PS, SR, PS, AS, SS, SS have contributed equally and have read and approved the MS; and, are aware of its submission to JFST.
Funding
None.
Declaration
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Abdelhamid HN, Wu HF. Probing the interactions of chitosan capped CdS quantum dots with pathogenic bacteria and their biosensing application. J Mater Chem B Biomater. 2013;1(44):6094–6106. doi: 10.1039/c3tb21020k. [DOI] [PubMed] [Google Scholar]
- Abebe E, Gugsa G, Ahmed M. Review on major food-borne zoonotic bacterial pathogens. J Trop Med. 2020;2020:1. doi: 10.1155/2020/4674235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abu-Salah KM, Zourob MM, Mouffouk F, Alrokayan SA, Alaamery MA, Ansari AA. DNA-based nanobiosensors as an emerging platform for detection of disease. Sensors. 2015;15(6):14539–14568. doi: 10.3390/s150614539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adley CC. Past, present and future of sensors in food production. Foods. 2014;3(3):491–510. doi: 10.3390/foods3030491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adzitey F, Rusul G, Huda N, Cogan T, Corry J. Prevalence, antibiotic resistance and RAPD typing of Campylobacter species isolated from ducks, their rearing and processing environments in Penang. Malays Int J Food Microbiol. 2012;154(3):197–205. doi: 10.1016/j.ijfoodmicro.2012.01.006. [DOI] [PubMed] [Google Scholar]
- Afzal A, Hussain A, Irfan M, Malik KA. Molecular diagnostics for foodborne pathogen (Salmonella spp.) from poultry. Adv Life Sci. 2015;2(2):91–97. [Google Scholar]
- Ahmed A, Rushworth JV, Hirst NA, Millner PA. Biosensors for whole-cell bacterial detection. Clin Microbiol Rev. 2014;27(3):631–646. doi: 10.1128/CMR.00120-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alahi ME, Mukhopadhyay SC. Detection methodologies for pathogen and toxins: a review. Sensors. 2017;17(8):1885–1905. doi: 10.3390/s17081885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen HK. Antibiotic resistance gene discovery in food-producing animals. Curr Opin Microbiol. 2014;19:25–29. doi: 10.1016/j.mib.2014.06.001. [DOI] [PubMed] [Google Scholar]
- Baraketi A, Salmieri S, Lacroix M. Foodborne pathogens detection: persevering worldwide challenge. Biosens Technol Detect Pathog A Prospect Rapid Anal. 2018;53:1–20. [Google Scholar]
- Barrick A, Châtel A, Bruneau M, Mouneyrac C. The role of high-throughput screening in ecotoxicology and engineered nanomaterials. Environ Toxicol Chem. 2017;36(7):1704–1714. doi: 10.1002/etc.3811. [DOI] [PubMed] [Google Scholar]
- Bhattacharya A, Shantikumar S, Beaufoy D, Allman A, Fenelon D, Reynolds K, Normington A, Afza M, Todkill D. Outbreak of Clostridium perfringens food poisoning linked to leeks in cheese sauce: an unusual source. Epidemiol Infect. 2020;148:1–7. doi: 10.1017/S095026882000031X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhunia AK. One day to one hour: how quickly can foodborne pathogens be detected? Future Microbiol. 2014;9(8):935–946. doi: 10.2217/fmb.14.61. [DOI] [PubMed] [Google Scholar]
- Blais BW, Turner G, Sooknanan R, Malek LT. A nucleic acid sequence-based amplification system for detection of Listeria monocytogenes hlyA sequences. Appl Environ Microbiol. 1997;63(310–313):1–4. doi: 10.1128/aem.63.1.310-313.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne L, Kaindama L, Bentley M, Jenkins C, Aird H, Oliver I, Paranthaman K. Investigation into a national outbreak of STEC O157: H7 associated with frozen beef burgers, UK, 2017. Epidemiol Infect. 2020;148:1–23. doi: 10.1017/S0950268820001582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang AS, Sreedharan A, Schneider KR. Peanut and peanut products: a food safety perspective. Food Control. 2013;32(1):296–303. doi: 10.1016/j.foodcont.2012.12.007. [DOI] [Google Scholar]
- Chen YI, Burall LS, Macarisin D, Pouillot R, Strain E, De Jesus AJ, Laasri A, Wang H, Ali L, Tatavarthy A, Zhang G, Datta AR. Prevalence and level of Listeria monocytogenes in ice cream linked to a listeriosis outbreak in the United States. J Food Prot. 2016;79(11):1828–1832. doi: 10.4315/0362-028X.JFP-16-208. [DOI] [PubMed] [Google Scholar]
- Compaoré MK, Yougbaré VM, Dembélé RE, Nikièma F, Elie KE, Barro N. Retrospective study of the contamination of exported sesame by Salmonella species from 2007 to 2017 in Burkina Faso. Afr J Agric Res. 2020;16(8):1141–1147. doi: 10.5897/AJAR2020.14917. [DOI] [Google Scholar]
- Costa SP, Dias NM, Melo LD, Azeredo J, Santos SB, Carvalho CM. A novel flow cytometry assay based on bacteriophage-derived proteins for Staphylococcus detection in blood. Sci Rep. 2020;10(1):1–13. doi: 10.1038/s41598-019-56847-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulombe G, Catford A, Martinez-Perez A, Buenaventura E. Outbreaks of Escherichia coli O157: H7 infections linked to Romaine lettuce in Canada from 2008 to 2018: an analysis of food safety context. Journal of Food Protection. 2020;83(8):1444–1462. doi: 10.4315/JFP-20-029. [DOI] [PubMed] [Google Scholar]
- da Costa MA, Pinto-Ferreira F, de Almeida RPA, Martins FDC, Pires AL, Mareze M, Mitsuka-Breganó R, Freire RL, da Rocha Moreira RV, Borges JM, Navarro IT. Artisan fresh cheese from raw cow’s milk as a possible route of transmission in a toxoplasmosis outbreak. Br Zoonoses Public Health. 2020;67(2):122–129. doi: 10.1111/zph.12660. [DOI] [PubMed] [Google Scholar]
- Dastia JI, Tareen AM, Lugert R, Zautner AE, Groß U. Campylobacter jejuni: a brief overview on pathogenicity-associated factors and disease-mediating mechanisms. Int J Med Microbiol. 2010;300(4):205–211. doi: 10.1016/j.ijmm.2009.07.002. [DOI] [PubMed] [Google Scholar]
- Davies E, Ebbesen M, Johansson C, Kaden R, Rautelin H. Genomic and phenotypic Characterisation of Campylobacter jejuni isolates from a waterborne outbreak. Front Cell Infect Microbiol. 2020;10:1–7. doi: 10.3389/fcimb.2020.594856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denis M, Houard E, Labbé A, Fondrevez M, Salvat G. A selective chromogenic plate, YECA, for the detection of pathogenic Yersinia enterocolitica: specificity, sensitivity, and capacity to detect pathogenic Y. enterocolitica from pig tonsils. J Pathogens. 2011;2011:1–9. doi: 10.4061/2011/296275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diancourt L, Sautereau J, Criscuolo A, Popoff MR. Two Clostridium perfringens type E isolates in France. Toxins. 2019;11(3):138–148. doi: 10.3390/toxins11030138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherr MG. Brief review on the epidemiology of transmissible spongiform encephalopathies (TSE) Vaccine. 2007;25(30):5619–5624. doi: 10.1016/j.vaccine.2006.10.059. [DOI] [PubMed] [Google Scholar]
- Fakruddin MD, Mazumdar RM, Chowdhury A, Mannan KB. Nucleic acid sequence based amplification (NASBA)-prospects and applications. Int J Life Sci Pharma Res. 2012;2(1):106–121. [Google Scholar]
- Fan H, Zhang M, Bhandari B, Yang CH. Food waste as a carbon source in carbon quantum dots technology and their applications in food safety detection. Trends Food Sci Technol. 2020;95:86–96. doi: 10.1016/j.tifs.2019.11.008. [DOI] [Google Scholar]
- Gaulin C, Ramsay D, Thivierge K, Tataryn J, Courville A, Martin C, Cunningham P, Désilets J, Morin D, Dion R. Acute toxoplasmosis among Canadian deer hunters associated with consumption of undercooked deer meat hunted in the United States. Emerg Infect Dis. 2020;26(2):199–205. doi: 10.3201/eid2602.191218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge Z, Schauer DB, Fox JG. In vivo virulence properties of bacterial cytolethal-distending toxin. Cell Microbiol. 2008;10(8):1599–1607. doi: 10.1111/j.1462-5822.2008.01173.x. [DOI] [PubMed] [Google Scholar]
- Gracias KS, McKillip JL. A review of conventional detection and enumeration methods for pathogenic bacteria in food. Can J Microbiol. 2004;50(11):883–890. doi: 10.1139/w04-080. [DOI] [PubMed] [Google Scholar]
- Guerry P. Campylobacter flagella: not just for motility. Trends Microbiol. 2007;15(10):456–461. doi: 10.1016/j.tim.2007.09.006. [DOI] [PubMed] [Google Scholar]
- Guidi F, Duranti A, Gallina S, Nia Y, Petruzzelli A, Romano A, Travaglini V, Olivastri A, Calvaresi V, Decastelli L, Blasi G. Characterization of a staphylococcal food poisoning outbreak in a workplace canteen during the post-earthquake reconstruction of Central Italy. Toxins. 2018;10(12):523–534. doi: 10.3390/toxins10120523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gwinn M, MacCannell D, Armstrong GL. Next-generation sequencing of infectious pathogens. Jama. 2019;321(9):893–894. doi: 10.1001/jama.2018.21669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hameed S, Xie L, Ying Y. Conventional and emerging detection techniques for pathogenic bacteria in food science: a review. Trends Food Sci Technol. 2018;81(61–73):41. [Google Scholar]
- Haynes E, Jimenez E, Pardo MA, Helyar SJ. The future of NGS (Next Generation Sequencing) analysis in testing food authenticity. Food Control. 2019;101:134–143. doi: 10.1016/j.foodcont.2019.02.010. [DOI] [Google Scholar]
- Holzinger M, Le Goff A, Cosnier S. Nanomaterials for biosensing applications: a review. Front Chem. 2014;2:63–73. doi: 10.3389/fchem.2014.00063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Xu Z, Mao Y, Ji Y, Xu H, Xiong Y, Li Y. Gold nanoparticle-based dynamic light scattering immunoassay for ultrasensitive detection of Listeria monocytogenes in lettuces. Biosensor Bioelectron. 2015;15(66):184–90. doi: 10.1016/j.bios.2014.11.016. [DOI] [PubMed] [Google Scholar]
- Huo B, Hu Y, Gao Z, Li G. Recent advances on functional nucleic acid-based biosensors for detection of food contaminants. Talanta. 2020;222:121565–121577. doi: 10.1016/j.talanta.2020.121565. [DOI] [PubMed] [Google Scholar]
- Jain D, Prasad KN, Sinha S, Husain N. Differences in virulence attributes between cytolethal distending toxin positive and negative Campylobacter jejuni strains. J Med Microbiol. 2008;57(3):267–272. doi: 10.1099/jmm.0.47317-0. [DOI] [PubMed] [Google Scholar]
- Jakobsen RA, Heggebø R, Sunde EB, Skjervheim M. Staphylococcus aureus and Listeria monocytogenes in Norwegian raw milk cheese production. Food Microbiol. 2011;28(3):492–496. doi: 10.1016/j.fm.2010.10.017. [DOI] [PubMed] [Google Scholar]
- Jemmi T, Stephan R. Listeria monocytogenes: food-borne pathogen and hygiene indicator. Rev Sci Tech. 2006;25(2):571–580. doi: 10.20506/rst.25.2.1681. [DOI] [PubMed] [Google Scholar]
- Jin Z, Ding G, Li G, Yang G, Han Y, Hao N, Deng J, Zhang Y, Zhang W, Li W. Rapid detection of foodborne bacterial pathogens using visual high-throughput microfluidic chips. J Chem Technol Biotechnol. 2020;95(5):1460–1466. doi: 10.1002/jctb.6331. [DOI] [Google Scholar]
- Kamala A, Kimanya M, De Meulenaer B, Kolsteren P, Jacxsens L, Haesaert G, Kilango K, Magoha H, Tiisekwa B, Lachat C. Post-harvest interventions decrease aflatoxin and fumonisin contamination in maize and subsequent dietary exposure in Tanzanian infants: a cluster randomised-controlled trial. World Mycotoxin J. 2018;11(3):447–458. doi: 10.3920/WMJ2017.2234. [DOI] [Google Scholar]
- Kang CR, Bang JH, Cho SI. Campylobacter jejuni foodborne infection associated with cross-contamination: outbreak in Seoul in 2017. Infect Chemother. 2019;51(1):21–27. doi: 10.3947/ic.2019.51.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang’ethe E, Muriuki S, Karugia JT, Guthiga PM, Kirui L (2019) Scoping study report on national sood safety architecture of the horticulture value hain, Kenya, pp 16–18
- Kumar H, Kuča K, Bhatia SK, Saini K, Kaushal A, Verma R, Bhalla TC, Kumar D. Applications of nanotechnology in biosensor-based. Sensors. 2020;20(7):6–10. doi: 10.3390/s20071966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahti E, Löfdahl M, Ågren J, Hansson I, Olsson Engvall E. Confirmation of a campylobacteriosis outbreak associated with chicken liver pâté using PFGE and WGS. Zoonoses Public Health. 2017;64(1):14–20. doi: 10.1111/zph.12272. [DOI] [PubMed] [Google Scholar]
- Lancova K, Dip R, Antignac JP, Le Bizec B, Elliott CT, Naegeli H. Detection of hazardous food contaminants by transcriptomics fingerprinting. TrAC Trends Anal Chem. 2011;30(2):181–191. doi: 10.1016/j.trac.2010.10.013. [DOI] [Google Scholar]
- Laughlin M, Bottichio L, Weiss J, Higa J, McDonald E, Sowadsky R, Fejes D, Saupe A, Provo G, Seelman S, Concepción-Acevedo J. Multistate outbreak of Salmonella Poona infections associated with imported cucumbers, 2015–2016. Epidemiol Infect 147-153 [DOI] [PMC free article] [PubMed]
- Lauri A, Mariani PO. Potentials and limitations of molecular diagnostic methods in food safety. Genes Nutr. 2009;4:1–12. doi: 10.1007/s12263-008-0106-1p1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law JW, Ab Mutalib NS, Chan KG, Lee LH. Rapid methods for the detection of foodborne bacterial pathogens: principles, applications, advantages and limitations. Front Microbiol. 2015;5:770–789. doi: 10.3389/fmicb.2014.00770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHugh AJ, Feehily C, Hill C, Cotter PD. Detection and enumeration of spore-forming bacteria in powdered dairy products. Front Microbiol. 2017;8:109. doi: 10.3389/fmicb.2017.00109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meghnath K, Hasselback P, McCormick R, Prystajecky N, Taylor M, McIntyre L, amp; Bitzikos, O. Outbreaks of Norovirus and acute gastroenteritis associated with British Columbia Oysters, 2016–2017. Food and Environmental Virology. 2019;11(2):138–148. doi: 10.1007/s12560-019-09374-4. [DOI] [PubMed] [Google Scholar]
- Mikhail AFW, Jenkins C, Dallman TJ, Inns T, Douglas A, Martín AIC, Hawker J. An outbreak of Shiga toxin-producing Escherichia coli O157: H7 associated with contaminated salad leaves: epidemiological, genomic and food trace back investigations. Epidemiol Infect. 2018;146(2):187–196. doi: 10.1017/S0950268817002874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mira Miralles M, Maestre-Carballa L, Lluesma-Gomez M, Martinez-Garcia M. High-throughput 16S rRNA sequencing to assess potentially active bacteria and foodborne pathogens: a case example in ready-to-eat food. Foods. 2019;8(10):480–494. doi: 10.3390/foods8100480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mngqawa P, Shephard GS, Green IR, Ngobeni SH, De Rijk TC, Katerere DR. Mycotoxin contamination of home-grown maize in rural northern South Africa (Limpopo and Mpumalanga Provinces) Food Addit Contam Part B. 2016;9(1):38–45. doi: 10.1080/19393210.2015.1121928. [DOI] [PubMed] [Google Scholar]
- Moffatt CRM, Greig A, Valcanis M, Gao W, Seemann T, Howden BP, Kirk MD. A large outbreak of Campylobacter jejuni infection in a university college caused by chicken liver pâté, Australia. Epidemiol Infect. 2016;144(14):2971–2978. doi: 10.1017/S0950268816001187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray DC, Bunce M, Cannell BL, Oliver R, Houston J, White NE, Barrero RA, Bellgard MI, Haile J. DNA-based faecal dietary analysis: a comparison of qPCR and high throughput sequencing approaches. PLoS One. 2011;6(10):e25776–86. doi: 10.1371/journal.pone.0025776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustafa F, Andreescu S. Nanotechnology-based approaches for food sensing and packaging applications. RSC Adv. 2020;10(33):19309–19336. doi: 10.1039/D0RA01084G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasheri N, Vester A, Petronella N. Food-borne viral outbreaks are associated with frozen produce. Epidemiol Infect. 2019;147:1–8. doi: 10.1017/S0950268819001791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nsibande SA, Forbes PBC. Fluorescence detection of pesticides using quantum dot materials–review. Anal Chim Acta. 2016;945(9–22):3–9. doi: 10.1016/j.aca.2016.10.002. [DOI] [PubMed] [Google Scholar]
- Palomino-Camargo C, González-Muñoz Y. Molecular techniques for detection and identification of pathogens in food: advantages and limitations. Revis Peru Med Exp Salud Publ. 2014;31(3):535–546. [PubMed] [Google Scholar]
- Pineda C, Kaushik A, Kest H, Wickes B, Zauk A. Maternal sepsis, chorioamnionitis, and congenital Candida kefyr infection in premature twins. Pediatr Infect Dis J. 2012;31(3):320–322. doi: 10.1097/INF.0b013e31823eee1a. [DOI] [PubMed] [Google Scholar]
- Pinto-Ferreira F, Caldart ET, Pasquali AKS, Mitsuka-Breganó R, Freire RL, Navarro IT. Patterns of transmission and sources of infection in outbreaks of human toxoplasmosis. Emerg Infect Dis. 2019;25(12):2177–2182. doi: 10.3201/eid2512.181565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinu FR. Early detection of food pathogens and food spoilage microorganisms: application of metabolomics. Trends Food Sci Technol. 2016;54:213–215. doi: 10.1016/j.tifs.2016.05.018. [DOI] [Google Scholar]
- Pirzada M, Altintas Z. Nanomaterials for healthcare biosensing applications. Sensors. 2019;19(23):5311–5366. doi: 10.3390/s19235311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portner DC, Leuschner RG, Murray BS. Optimising the viability during storage of freeze-dried cell preparations of Campylobacter jejuni. Cryobiology. 2007;54(3):265–270. doi: 10.1016/j.cryobiol.2007.03.002. [DOI] [PubMed] [Google Scholar]
- Priyanka B, Patil RK, Dwarakanath S. A review on detection methods used for food-borne pathogens. Indian J Med Res. 2016;144(3):327–338. doi: 10.4103/0971-5916.198677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez-Castillo FY, Loera-Muro A, Jacques M, Garneau P, Avelar-González FJ, Harel J, Guerrero-Barrera AL. Waterborne pathogens: detection methods and challenges. Pathogens. 2015;4(2):307–334. doi: 10.3390/pathogens4020307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos AP, Leonhard SE, Halstead SK, Cuba MA, Castañeda CC, Dioses JA, Tipismana MA, Abanto JT, Llanos A, Gourlay D, Grogl M. Guillain-Barré syndrome outbreak in Peru 2019 associated with Campylobacter jejuni infection. Neurol Neuroimmunol Neuroinflammation. 2021;8(2):e952–975. doi: 10.1212/NXI.0000000000000952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricci A, Allende A, Bolton D, Chemaly M, Davies R, Fernández Escámez PS, Girones R, Herman L, Koutsoumanis K, Nørrung B. EFSA Panel on Biological Hazards (BIOHAZ), Listeria monocytogenes contamination of ready-to-eat foods and the risk for human health in the EU. EFSA J. 2018;16(1):e05134–5306. doi: 10.2903/j.efsa.2018.5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safardoust-Hojaghan H, Salavati-Niasari M, Amiri O, Hassanpour M. Preparation of highly luminescent nitrogen doped graphene quantum dots and their application as a probe for detection of Staphylococcus aureus and E coli. J Mol Liq. 2017;241(1114–1119):4–6. [Google Scholar]
- Sahoo M, Vishwakarma S, Panigrahi C, Kumar J. Nanotechnology: current applications and future scope in food. Food Frontiers. 2021;2(1):3–22. doi: 10.1002/fft2.58. [DOI] [Google Scholar]
- Sai-Anand G, Sivanesan A, Benzigar MR, Singh G, Gopalan AI, Baskar AV, Ilbeygi H, Ramadass K, Kambala V, Vinu A. Recent progress on the sensing of pathogenic bacteria using advanced nanostructures. Bull Chem Soc Jpn. 2019;92(1):216–244. doi: 10.1246/bcsj.20180280. [DOI] [Google Scholar]
- Sekse C, Holst-Jensen A, Dobrindt U, Johannessen GS, Li W, Spilsberg B, Shi J. High throughput sequencing for the detection of foodborne pathogens. Front Microbiol. 2017;8:2–6. doi: 10.3389/fmicb.2017.02029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro K, Bahia-Oliveira L, Dixon B, Dumètre A, de Wit LA, VanWormer E, Villena I. Environmental transmission of Toxoplasma gondii: oocysts in water, soil and food. Food Waterborne Parasitol. 2019;15:e49–67. doi: 10.1016/j.fawpar.2019.e00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma H, Mutharasan R. Review of biosensors for foodborne pathogens and toxins. Sens Actuators B Chem. 2013;183:535–549. doi: 10.1016/j.snb.2013.03.137. [DOI] [Google Scholar]
- Stelzer S, Basso W, Silván JB, Ortega-Mora LM, Maksimov P, Gethmann J, Conraths FJ, Schares G. Toxoplasma gondii infection and toxoplasmosis in farm animals: risk factors and economic impact. Food Waterborne Parasitol. 2019;15:e37–69. doi: 10.1016/j.fawpar.2019.e00037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira LA, Carvalho FT, Vallim DC, Pereira RC, Cunha Neto A, Vieira BS, Carvalho RC, Figueiredo EE. Listeria monocytogenes in export-approved beef from mato grosso, Brazil: prevalence, molecular characterization and resistance to antibiotics and disinfectants. Microorganisms. 2020;8(1):18–29. doi: 10.3390/microorganisms8010018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas J, Govender N, McCarthy KM, Erasmus LK, Doyle TJ, Allam M, Blumberg LH. Outbreak of listeriosis in South Africa associated with processed meat. N Engl J Med. 2020;382(7):632–643. doi: 10.1056/NEJMoa1907462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tournas VH, Heeres J, Burgess L. Moulds and yeasts in fruit salads and fruit juices. Food Microbiol. 2006;23(7):684–688. doi: 10.1016/j.fm.2006.01.003. [DOI] [PubMed] [Google Scholar]
- Treacy J, Jenkins C, Paranthaman K, Jorgensen F, Mueller-Doblies D, Anjum M, Kaindama L, Hartman H, Kirchner M, Carson T, Kar-Purkayastha I. Outbreak of Shiga toxin-producing Escherichia coli O157: H7 linked to raw drinking milk resolved by rapid application of advanced pathogen characterization methods, England, August to October 2017. EuroSurveillance. 2019;24(16):1800191–1800197. doi: 10.2807/1560-7917.ES.2019.24.16.1800191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma MS, Rogowski JL, Jones L, Gu FX. Colorimetric biosensing of pathogens using gold nanoparticles. Biotechnol Adv. 2015;33(6):666–680. doi: 10.1016/j.biotechadv.2015.03.003. [DOI] [PubMed] [Google Scholar]
- Wang Y, Salazar JK. Culture-independent rapid detection methods for bacterial pathogens and toxins in food matrices. Compr Rev Food Sci Food Safety. 2016;15(1):183–205. doi: 10.1111/1541-4337.12175. [DOI] [PubMed] [Google Scholar]
- Wang L, Ye C, Xu H, Aguilar ZP, Xiong Y, Lai W, Wei H. Development of an SD-PMA-mPCR assay with internal amplification control for rapid and sensitive detection of viable Salmonella spp., Shigella spp. and Staphylococcus aureus in food products. Food Control. 2015;57:314–320. doi: 10.1016/j.foodcont.2015.04.016. [DOI] [Google Scholar]
- Wang D, Lian F, Yao S, Liu Y, Wang J, Song X, Ge L, Wang Y, Zhao Y, Zhang J, Zhao C. Simultaneous detection of three foodborne pathogens based on immunomagnetic nanoparticles and fluorescent quantum dots. Am Chem Soc Omega. 2020;5(36):23070–23080. doi: 10.1021/acsomega.0c02833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westrell T, Ciampa N, Boelaert F, Helwigh B, Korsgaard H, Chríel M, Ammon A, Mäkelä P. Zoonotic infections in Europe in 2007: a summary of the EFSA-ECDC annual report. Eurosurveillance. 2009;14(3):19100–19103. doi: 10.2807/ese.14.03.19100-en. [DOI] [PubMed] [Google Scholar]
- Yamasaki S, Asakura M, Tsukamoto T, Faruque SM, Deb R, Ramamurthy T. Cytolethal distending toxin (CDT): genetic diversity, structure and role in diarrheal disease. Toxin Rev. 2006;25(1):61–88. doi: 10.1080/15569540500320938. [DOI] [Google Scholar]
- Yang M, Cousineau A, Liu X, Luo Y, Sun D, Li S, Gu T, Sun L, Dillow H, Lepine J, Xu M. Direct metatranscriptome RNA-seq and multiplex RT-PCR amplicon sequencing on Nanopore MinION–promising strategies for multiplex identification of viable pathogens in food. Front Microbiol. 2020;11:514–528. doi: 10.3389/fmicb.2020.00514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Belwal T, Li L, Lin X, Xu Y, Luo Z. Nanomaterial-based biosensors for sensing key food-borne pathogens: advances from recent decades. Compr Rev Food Sci Food Safety. 2020;19(4):1465–1487. doi: 10.1111/1541-4337.12576. [DOI] [PubMed] [Google Scholar]
- Zhao Z (2016) A portable and automatic biosensing instrument for detection of foodborne pathogenic bacteria in food samples 7–19
- Zhao X, Lin CW, Wang J, Oh DH. Advances in rapid detection methods for foodborne pathogens. J Microbiol Biotechnol. 2014;24(3):297–312. doi: 10.4014/jmb.1310.10013. [DOI] [PubMed] [Google Scholar]
- Zhou X, Kong DG, Li J, Pang BB, Zhao Y, Zhou JB, Zhang T, Xu JQ, Kobayashi N, Wang YH. An outbreak of gastroenteritis associated with GII. 17 Norovirus-contaminated secondary water supply system in Wuhan, China, 2017. Food Environ Virol. 2019;11(2):126–137. doi: 10.1007/s12560-019-09371-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilbauer M, Dorrell N, Wren BW, Bajaj-Elliott M. Campylobacter jejuni-mediated disease pathogenesis: an update. Trans R Soc Trop Med Hyg. 2008;102(2):123–129. doi: 10.1016/j.trstmh.2007.09.019. [DOI] [PubMed] [Google Scholar]