Abstract
Peanut protein concentrates (PPCs) were subjected to hydrolysis by crude protease extract (CPE) obtained from three fungi viz; Rhizopus oligosporus, Trichoderma reesei, and Aspergillus oryzae and the effect on structural, functional and in-vitro protein digestibility (IVPD) properties were studied. Particle size was found significantly (p ≤ 0.05) lower in hydrolyzed samples than un-treated samples. Fourier transform infrared spectroscopy (FTIR) spectrum of hydrolyzed samples displayed intense absorbance peaks in the wavelength ranging from 1500 to 2600 cm−1. Peanut protein concentrates hydrolyzed by CPE from R. oligosporus showed higher surface hydrophobicity (564.18). Total sulfhydryl content was found lower in all the hydrolyzed samples whereas, reverse trend was observed for exposed sulfhydryl content. The structural changes simultaneously affected the functional and IVPD attributes of hydrolyzed PPCs. In comparison to the PPCs hydrolysed using crude extracts from T. reesei and R. oligosporus, PPCs hydrolysed by A, oryzae showed higher solubility, water and oil binding capacity, foaming capacity and foam stability. Higher IVPD values of 86.70% was also found in PPCs hydrolyzed with CPE of A. oryzae. The study established that CPE hydrolysis of PPCs has potential for scale-up studies and may serve as a cost effective alternative to protein hydrolysis with pure enzymes.
Keywords: Peanut protein concentrate, Hydrolysis, Crude protease extract, Conformational changes, Solubility
Introduction
Peanut (Arachis hypogea L.), a major source of edible oil and protein is considered valuable for animal and human nutrition. Defatted peanut meal containing about 30–35% protein with higher bioavailability and lower anti-nutritional factors is underutilized product of the peanut industry (Yadav et al 2012). Extraction of protein from defatted peanut meal and their applications in formulation of protein beverages can be of great significance (Malik et al 2016). Plant based protein drinks could also serve as an alternative to animal protein drinks due to some dietary restrictions and religious concerns (Jain et al 2019). Peanut protein has promising nutritional profile, however, its uses in food formulations are limited owing to lower solubility and poor emulsifying properties.
Different approaches like high-intensity ultrasound (HIUS) (Mir et al 2019); heat treatment (Mir et al 2020) and irradiation (Malik and Saini 2017) have been studied for modification of native protein structure in order to enhance the functional properties. Enzymatic hydrolysis with pure enzymes is also used for enhancing functional properties of the protein and usually preferred from the food safety point of view. Yu et al (2007) and Quist et al (2009) reported that enzymatic hydrolysis can generate lower molecular weight polypeptides and free amino acids. The lower molecular weight polypeptides are easily absorbed by the intestine and also possess lower allergenic effects. In addition, the biological functions like antihypertension, opioid agonists, orantagonists, immunomodulatory, antithrombotic, antioxidant, anti-cancer, and antimicrobial activities of hydrolyzed peanut proteins have also been reported by Elias et al (2008).
Though, modification with pure enzyme gives promising results with respect to protein functionality, but its uses are limited due to higher cost of pure enzymes which in turn increases the overall cost of the process and final product. To overcome these limitations, crude protease extracts obtained from different fungi can be explored and utilized for hydrolyzing different kinds of proteins. Oliveira et al (2010) reported that proteases obtained from fungi have tremendous importance due to their potential of catalyzing the hydrolysis reactions. In addition, crude protease extracts obtained from different microbial sources are preferred over pure enzymes obtained from plant and animal sources as they possess almost all the characteristics required for biotechnological applications (Gupta et al 2005). Among different microbial sources, fungal proteases exhibit higher activity and have the potential for commercial applications. Therefore, the present research was carried out with the major objectives: (1) Extraction of crude protease extracts from R. oligosporus, T. reesei, and A. oryzae by solid state fermentation, (2) Isolation of protein concentrate from defatted peanut flour and its enzymatic hydrolysis by crude protease extracts, and (3) Effect of enzymatic hydrolysis on the structural and functional characteristics of peanut protein concentrates.
Materials and methods
Materials
Commercially available de-oiled peanut cake (protein content, 36.5%, db) were procured from the local market of Ludhiana, Punjab, India from a local supplier. The cake was pulverized in a laboratory grinder in order to reduce particle size and passed through 60 mesh (0.250 mm) standard sieve. Defatted flour was dried in hot air oven for 2 h at 40–45 °C to remove any residual solvent, thereafter used for the preparation of protein concentrate. All chemicals used in the experiments were of analytical grade and purchased from Sigma Aldrich, MO, USA. Papain was obtained from Hi Media Laboratories Pvt Ltd, Mumbai, India. Cultures of T. reesei, A. oryzae and R. oligosporus were obtained from Bio-process Engineering Laboratory, ICAR- Central Institute of Post-Harvest Engineering and Technology (ICAR-CIPHET), Ludhiana, India.
Methods
Preparation of peanut protein concentrate
Peanut protein concentrate was prepared according to the method of Yu et al (2007) using alkali extraction and isoelectric precipitation method. Defatted peanut flour was mixed with water in the ratio of 1/10 (w/v), and pH of the mixture was adjusted to 10.0 with 1.0 N sodium hydroxide (NaOH). The suspension was stirred at room temperature (30 ± 2 °C) for 2 h and centrifuged at 3000 g for 15 min. Supernatant was collected and the pH was adjusted to 4.5 (iso-electric point) with 1.0 N hydrochloric acid (HCl). The precipitates obtained were centrifuged at 3000 g for15 min. The supernatant was discarded and the precipitate was neutralized, freeze dried and stored under refrigerated conditions for further use.
Determination of chemical characteristics of peanut protein concentrate
Chemical characteristics of native PPCs, such as moisture, protein, ash, fat and crude fiber content were determined according to the standard methods of AOAC (2006). The carbohydrate content was estimated by subtracting the sum of percentage of moisture, crude fat, crude protein and ash from 100.
Preparation of crude protease extract using different fungal cultures
Crude protease extract (CPE) from R. oligosporus, A. oryzae and T. reesei was prepared by the method described by Su et al (2011) with some modifications. Wheat bran (20 g), defatted peanut flour (8 g), calcium chloride (0.028 g) and deionized water (28 mL) were mixed and autoclaved at 121 °C for 30 min in a conical flask. Spores from each of the isolates were harvested following inoculation on PDA plates and incubation at 30 °C for 72 h. The plates were removed from the incubator followed by harvesting of spores using sterile water. The spores count was determined with a hemocytometer and the spore concentration of 1 × 108 spores/mL was used for enzyme production (Sandhu et al 2011). The sterilized mixture was cooled, inoculated individually with 2 mL spore suspension of R. oligosporus, A. oryzae and T. reesei and incubated at 30 °C for 68 h. After 3 days of incubation, 100 mL of 0.2 M phosphate buffer (pH 7.0) was added to each of the flasks and vortexed for 30 min. The mixture was centrifuged at 10,000 g for 30 min at 4 °C. The obtained supernatant (CPE) was stored for further use.
Determination of protease activity
The proteolytic activities of CPE and papain were determined by the procedure as described by Su et al (2011). Crude protease extracts showed protease activity of 15,478–17,522 USP units/mg in comparison to papain (30,000 USP units/mg).
Enzymatic hydrolysis of PPC with crude fungal protease extracts
Hydrolysis of PPC was carried out as per the method described by Silvestre et al (2012) with minor modifications. The PPCs were dissolved in distilled water to make 12.5% (w/v) solution and pH was adjusted to 6.5 using 2% citric acid solution for optimum activity of CPE. The solution was allowed to hydrate at 30 °C for 2 h. Thereafter, the temperature of the solution was increased to 50 °C. Crude protease extract (0.75%, w/w of protein) from each of the fungal source was added separately and the solution was continuously stirred for 1 h. The pH was maintained at 6.5 using 1 N NaOH solution during hydrolysis process. At the end of hydrolysis, the enzyme was inactivated by heating the mixture in a water bath at 85 °C for 10 min. Hydrolysates obtained were centrifuged at 5000 g for 20 min at 20 °C and pellets were collected, freeze dried and stored for further analysis. Papain (0.37%, w/w of protein) was used in place of CPE for hydrolyzing peanut protein concentrates. Peanut protein concentrates (PPCs) hydrolyzed with crude protease extracts of R. oligosporus, A. oryzae and T. reesei were designated as HPPCR, HPPCA and HPPCT. Peanut protein concentrates hydrolyzed with papain were designated as HPPCPs, while native peanut protein concentrates were designated as NPPC.
Determination of degree of hydrolysis
The degree of hydrolysis (DH), defined as the percent ratio of the number of peptide bonds broken (h) to the total number of peptide bonds in the substrate (htot), was determined with the o-pthaldialdehyde (OPA) as described by Nielsen et al (2001). The assay was carried out by mixing 0.4 mL of a 0.45 μ filtered sample (or control) and 0.3 mL OPA reagent. After vortex for 5 s and incubation at room temperature (30 ± 2 °C) for exactly 2 min, the absorbance of the mixture was measured in UV spectrophotometer (Model: UV-2700, Shimadzu, Tokyo, Japan) at 340 nm. L-serine (0.9516 meq/L) was used as positive control and distilled water as negative control. Calibration curve of L-serine was prepared in distilled water. The DH was calculated by the following equation:
| 1 |
where serine-NH2 is meqv serine in NH2/g protein; S is sample volume in litre; D is dilution volume; P is protein content in sample volume; V is the sample volume in assay; α, β and htot constants for casein are 1.039. 0.383 and 8.2, respectively.
| 2 |
where h = (Serine-NH2 –β) / α meq/g protein.
Determination of Particle size distribution (PSD)
Particle size analyzer based on laser light scattering ((Model Horiba Partica LA-950V2, Japan) was used for the determination of particle size and surface area of native and hydrolyzed samples. The particle size distributions (PSDs), i.e., particle size at 10% (Dv10), median diameter and 90% (Dv90) of the volume distribution were calculated from the graph. The specific surface area (cm2/cm3) was also calculated.
Fourier transform infrared (FTIR) spectroscopy
Effect of enzymatic hydrolysis of peanut protein concentrate on FTIR spectra was recorded on ALPHA, BRUKER (Laser class 1, Germany) using the compressed potassium bromide pellets. Moisture free protein concentrate samples were mixed with dry potassium bromide in the ratio of 1:100, and compressed in a hydraulic press to form transparent pellets. The spectra were recorded in the transmittance mode at 4000–400 cm−1 region.
Total and exposed sulfhydryl contents
Total and exposed sulfhydryl groups of native and hydrolyzed peanut protein concentrates were determined according to the method of Yin et al (2009).
Surface hydrophobicity (H0)
Fluorescent probe (8- anilinonaphthalene-1-sulfonic acid, ANS) was used for the determination of surface hydrophobicity of native and hydrolyzed PPCs using the established procedure of Wagner et al (2000). The protein surface hydrophobicity was obtained by calculating the initial slope of the fluorescence index versus the protein concentration plot.
Determination of functional characteristics of peanut protein concentrates
Solubility of native and hydrolyzed PPCs was determined according to the method described by Achouri et al (2012) at 30 ± 2 °C. Water binding capacity was determined using the method described by Beuchat (1977). The method of Chakraborty (1986) was adopted for the determination of oil binding capacity.
Foaming capacity and foam stability
The method described by Fekria et al (2012) was used for measuring foaming capacity (FC) and foam stability (FS) of protein samples with some modifications. Dispersions were prepared by dissolving sample (2.0 g) in water (100 mL). The solutions were whipped with a mixer blender for 5 min at high speed. After blending, the content was shifted into a cylinder and the foam volume was recorded. At different time intervals (10, 20, and 30 min), changes in volume was recorded. Percentage FC and FS were calculated according to the following equations:
| 3 |
where Vw is the volume of whipped sample and Vuw is the volume of un-whipped sample.
| 4 |
where Vt is the change in volume of foam with time and Vi is the initial volume of foam.
Determination of in-vitro protein digestibility (IVPD)
The method of Hsu et al (1977) was followed for the determination of in-vitro protein digestibility of native and hydrolyzed peanut protein concentrates with some modifications. Water suspensions of 25 mL containing peanut protein concentrate (6.25 mg/mL) were prepared and the pH was maintained at 8.0 using 0.1 N NaOH. Multi-enzyme solution (cocktail) was prepared by mixing 1.6 mg/mL Trypsin, 3.1 mg/mL Chymotrypsin, and 1.3 mg/mL Protease. The cocktail was maintained in an ice bath and the pH was adjusted to 8 using 0.1 N NaOH. From this cocktail, 2.5 mL was taken and added to protein suspension already maintained at 37 ºC in a water bath shaker. The pH drop after 10 min was measured by a calibrated digital pH meter (Model: Five Easy FEP20, Mettler-Toledo, Switzerland). Casein protein was used as a standard. The IVPD was calculated according to the following equation:
| 5 |
where Xf is the pH value of the solution after 10 min of enzymatic digestion.
Statistical analysis
The data presented in tables and figures are the mean of three replicates. Error bars represent deviation. Data were statistically analyzed using one-way analysis of variance (ANOVA). Duncan's multiple range tests (p ≤ 0.05) was used to determine significant differences among the mean values using Statistica software version 7.0.
Results and discussion
Chemical composition
Moisture content of protein concentrate was 5.09%, which is important from the microbiological point of view. Protein content was 72.08%. Total ash and insoluble ash content were 10.07 and 0.28%, respectively. Higher ash content depicts the presence of minerals in protein concentrate, while the insoluble ash generally refers to silica content. Crude fibre and fat content were 0.68 and 0.83%, respectively. Carbohydrate obtained by the difference method was found to be 10.97%.
Degree of hydrolysis (DH)
Peanut protein concentrates was subjected to CPE hydrolysis for 1 h. Degree of hydrolysis increased with increase in time (Fig. 1a). At 60 min, the highest DH value of 16.4% was found in PPCs hydrolysed with CPE from A. oryzae followed by CPE from R. oligosporus (15.6%) and CPE of T. reesei (15.0%). The DH value for samples hydrolysed with papain was 15.9% and comparable with other CPE hydrolysed samples. This indicates that the CPE from all three fungal sources contained protease enzymes, which hydrolysed the protein samples. Higher values (43.4%) of DH in defatted peanut meal hydrolysed with CPE from Aspergillus has also been reported by Su et al (2011). Hydrolysis through CPE might be associated with the presence of cellulolytic enzymes that could possibly decreased the viscosity of the reaction mixture and enhanced the protein dissolution and finally DH. The Aspergillus oryzae strain used in this study produces a consortium of enzymes comprising of cellulases, xylanases and pectinases (Sandhu et al 2011). Higher DH value observed in case of CPE of A. oryzae in comparison with CPE of R. oligosporous and T. reesei could also be due to the presence of larger expansion of hydrolytic genes in A. oryzae as reported by Vishwanatha et al (2009).
Fig. 1.
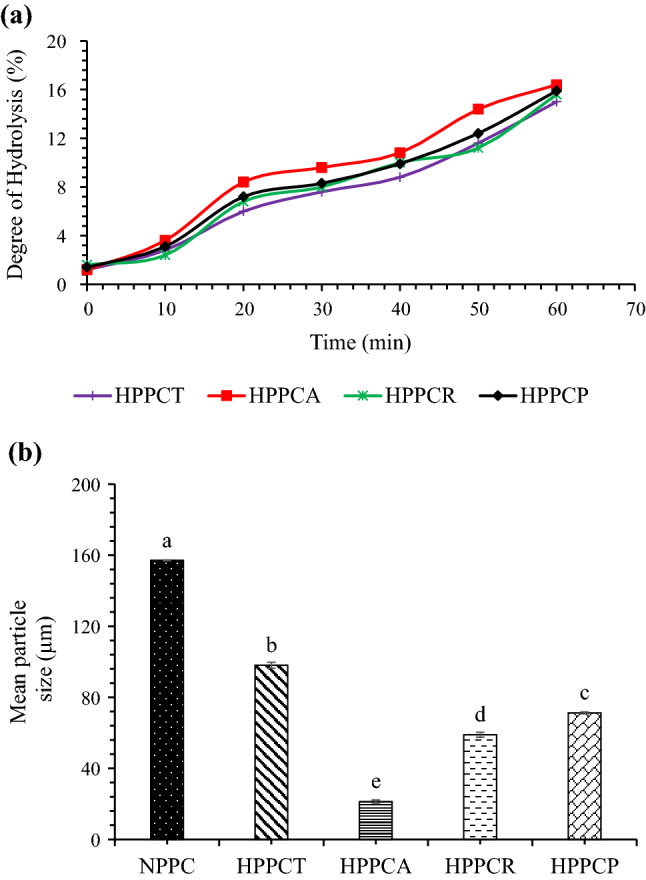
Effect of crude protease extract enzymatic hydrolysis on degree of hydrolysis (a) and particle size (b) of peanut protein concentrate. [HPPCT = hydrolyzed peanut protein concentrate with crude protease extract of Trichoderma reesei, HPPCA = hydrolyzed peanut protein concentrate with crude protease extract of Aspergillus oryzae, HPPCR = hydrolyzed peanut protein concentrate with crude protease extract of Rhizopus oligosporus and HPPCP = hydrolyzed peanut protein concentrate with papain]
Particle size
Particle size of native and hydrolyzed peanut protein concentrates is shown in Fig. 1b. It is clear from the figure that enzymatic hydrolysis caused significant changes in the particle size of PPCs. Native PPCs has particle size of about 157.12 µm which was significantly (p ≤ 0.05) higher than the particle size of hydrolyzed samples. Among hydrolyzed PPCs, the lowest particle size of 21.35 µm was observed in the samples hydrolyzed with CPE produced by A. oryzae, and the highest particle size of 98.04 µm was observed in the samples hydrolyzed with CPE obtained from T. reesei. Papain hydrolyzed samples showed higher particle size than samples hydrolyzed with CPE from A. oryzae and R. oligosporus but the values were lower than the samples hydrolyzed with CPE from T. reesei. The possible reasons for the decline in particle size could be owing to enzymatic hydrolysis, which might have induced strong electrostatic repulsion between the newly created particles. The newly formed particles may have less chances to aggregate which in turn is responsible for the lower values of particle size for hydrolyzed samples. Tang et al (2019) also observed reduction in particle size of egg yolk powder after enzymatic hydrolysis. Moreover, the results of particle size also justify the results of foaming capacity, showing higher values for PPCs with lower particle size (Table 1).
Table 1.
Effect of crude protease extract enzymatic hydrolysis on surface hydrophobicity, SH groups, foaming properties and in-vitro protein digestibility of peanut protein concentrate
| Sample | Surface hydrophobicity | Total SH (µmol/mg) |
Exposed SH (µmol/mg) |
Foaming capacity (%) | Foam stability (%) | In-vitro protein digestibility (%) | ||
|---|---|---|---|---|---|---|---|---|
| 10 min | 20 min | 30 min | ||||||
| NPPC | 213.51 ± 1.96e | 8.78 ± 0.01a | 1.11 ± 0.02c | 22.33 ± 1.24e | 52.92 ± 0.91e | 45.40 ± 1.23e | 30.18 ± 0.60e | 70.11 ± 1.83c |
| HPPCT | 442.07 ± 2.26b | 6.54 ± 0.02c | 3.35 ± 0.01a | 33.66 ± 2.05d | 67.70 ± 0.92d | 53.73 ± 2.06d | 48.07 ± 0.82d | 83.65 ± 2.08b |
| HPPCA | 340.64 ± 1.44d | 7.33 ± 0.02b | 2.56 ± 0.01b | 58.70 ± 1.68a | 88.28 ± 0.93a | 83.46 ± 1.42a | 77.48 ± 1.44a | 86.69 ± 0.78b |
| HPPCR | 564.18 ± 2.46a | 8.01 ± 0.02a | 1.88 ± 0.01c | 47.31 ± 1.25b | 76.25 ± 0.54b | 73.28 ± 0.93b | 67.13 ± 1.98b | 84.33 ± 0.82b |
| HPPCP | 392.23 ± 2.94c | 5.45 ± 0.29d | 3.56 ± 0.17a | 41.33 ± 1.24c | 73.07 ± 1.00c | 61.52 ± 0.86c | 57.66 ± 0.89c | 94.62 ± 0.59a |
n = 3, results are expressed as mean ± standard deviation, samples with different superscript letter in the same column differ significantly (p ≤ 0.05) on a particular property. [NPPC = native peanut protein concentrate, HPPCT = hydrolyzed peanut protein concentrate with crude protease extract of Trichoderma reesei, HPPCA = hydrolyzed peanut protein concentrate with crude protease extract of Aspergillus oryzae, HPPCR = hydrolyzed peanut protein concentrate with crude protease extract of Rhizopus oligosporus and HPPCP = hydrolyzed peanut protein concentrate with papain]
Fourier transform infra-red spectroscopy (FTIR) properties
Changes in secondary structure due to crude protease enzymatic hydrolysis of peanut protein concentrates was recorded by FTIR spectroscopy (Fig. 2). The results depict that small absorbance peaks were visible in the wavelength ranging from 500 to 100 cm−1, which corresponds to N–H, C–N and C = O amide bonds. The intensity of absorbance peaks slightly increased from 800 to 1200 cm−1, which corresponds to the deformation in the NH3 groups of peanut protein concentrates probably due to the free amino group of lysine and the presence of three asymmetrical CH3 bending, symmetrical CO3−1 stretching, and ring vibrations attributed to free amino acids i.e. valine, glutamic acid and aromatic amino acid i.e. phenylalanine (Pavia et al 2008). The zone ranging from 1600 to 1800 cm−1 known as Amide-I (C = O bond) region is the most vulnerable zone which depicts the changes in the secondary structure of protein molecules. In our results, we observed a sharp decline in the peaks for all the samples, however the peaks were more intense in case of hydrolyzed samples compared to the native. This clearly indicates that changes in the secondary structure was due to crude protease hydrolysis. Pavia et al (2008) reported that structural changes corresponding to this region mainly arises due to changes in β-sheet and α-helix of protein, which is mainly caused due to C-H stretching. Other intense peaks were also observed in the wavelength ranging from 2000 to 2600 cm−1, which further confirms the dominance of structural changes in hydrolyzed samples than unhydrolyzed. Some minor absorbance peaks were also observed in the wavelengths ranging from 2700 to 3500 cm−1, which correspond to hydrogen bonding, N–H stretching and C-H bending of amide bonds. Similar peaks were observed for the samples hydrolyzed with papain. The validity of FTIR is further corroborated by the results of surface hydrophobicity and SH-groups showing the presence of structural changes owing to hydrolysis. These structural changes profoundly affected different functional characteristics like solubility, WBC and OBC and foaming capabilities (Table 1).
Fig. 2.
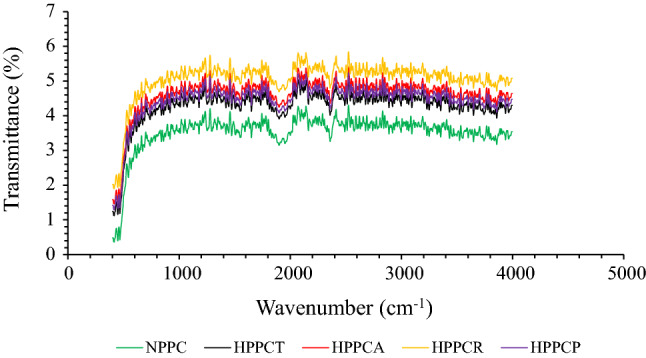
Effect of crude protease extract enzymatic hydrolysis on FTIR spectra of peanut protein concentrate. [NPPC = native peanut protein concentrate, HPPCT = hydrolyzed peanut protein concentrate with crude protease extract of Trichoderma reesei, HPPCA = hydrolyzed peanut protein concentrate with crude protease extract of Aspergillus oryzae, HPPCR = hydrolyzed peanut protein concentrate with crude protease extract of Rhizopus oligosporus and HPPCP = hydrolyzed peanut protein concentrate with papain]
Total and exposed SH content
The data showing the effect of CPE hydrolysis on the total and exposed SH content of PPCs are presented in Table 1. Higher total SH content (8.78) was found in native PPCs than hydrolyzed PPCs whereas, the lowest total SH content (6.54) was found in PPCs hydrolyzed with CPE obtained from T. reesei. Higher values for exposed SH content were observed in the hydrolyzed samples than native one. The values for total and exposed SH content of the samples hydrolyzed with papain were 5.45 ± 0.29 and 3.56 ± 0.17, respectively. Native PPCs showed exposed SH values of 1.11, whereas PPCs hydrolyzed with CPE of T. reesei, A. oryzae and R. oligosporus showed exposed SH values of 3.35, 2.56 and 1.88, respectively. SH groups are important functional parameters, which indicates about the strength of secondary and tertiary structure, and also reflect the extent of denaturation. Decline in the total SH content might be owing to the oxidation of disulfide bonds occurred due to the structural modifications. The structural changes occurred in PPCs due to hydrolysis were evident from the results of surface hydrophobicity (Table 1). These structural changes are associated with the improvement in other functional characteristics like solubility, water and oil binding capacity and foaming properties. Liu and Kuo (2016) reported that majority of -SH groups of native proteins are hidden inside and poorly accessible regions of the polypeptide chain, and usually do not come in contact with Ellman's reagent (DTNB). They proposed that changes in SH groups may be considered as an indicator of degree of unfolding in proteins. Enzymatic hydrolysis might have resulted in the unfolding of protein structure thereby exposing SH groups which ultimately increased the accessibility of DTNB to reaction sites.
Surface hydrophobicity (H0)
The results for the surface hydrophobicity of native and hydrolyzed PPCs are presented in Table 1. The data shows that surface hydrophobicity of hydrolyzed PPCs was significantly (p ≤ 0.05) higher than that of native PPCs. Among hydrolyzed PPCs, the highest surface hydrophobicity value of 564.18 was found in PPCs hydrolyzed with CPE obtained from R. oligosporus, and the lowest value of 340.64 was found in PPCs hydrolyzed with CPE obtained from A. oryzae. Peanut protein concentrates hydrolyzed with papain showed surface hydrophobicity value of 392.23. Zhao et al (2012) reported that enzymatic hydrolysis can unfold the structure of protein molecules, and thus exposes hidden hydrophobic groups. However, it must be noted that higher surface hydrophobicity of protein is detrimental for other functional characteristics, such as solubility. Once unfolding takes place, hydrophobic groups are exposed to a larger extent and the exposed hydrophobic groups can self-aggregate resulting in the decline in solubility of protein molecules. In the present study, we observed higher surface hydrophobicity of PPCs hydrolyzed with CPE of R. oligosporus. The solubility of this sample was also found significantly lower than other hydrolyzed samples, which may be due to the aggregation of already exposed hydrophobic groups.
Solubility
The results in Fig. 3a indicate that in comparison with native PPC, the solubility of hydrolysed PPCs was significantly (p ≤ 0.05) higher. Among hydrolysed samples, PPCs hydrolysed with CPE obtained from A. oryzae displayed higher solubility. Peanut protein concentrates hydrolysed with papain showed solubility value of 61% and did not differ significantly (p ≤ 0.05) with the samples hydrolysed with CPE from T. reesei. Improved solubility of hydrolysed proteins may be owing to the release of soluble peptides from insoluble aggregates or precipitates which favour the ionization of carboxylic and amino groups of amino acids. Zhao et al (2011) also observed higher solubility in peanut protein isolates hydrolyzed with commercial Alcalase enzyme. This may be associated with the fact that enzymatic hydrolysis results in the unfolding of secondary and tertiary structure of protein and increases the exposure of hydrophilic groups which ultimately improves hydration of protein molecules and thus solubility. Enzymatic hydrolysis might have also promoted the interaction between hydrophilic groups and water molecules due to the reduction in the peptide size, which increased the solubility.
Fig. 3.
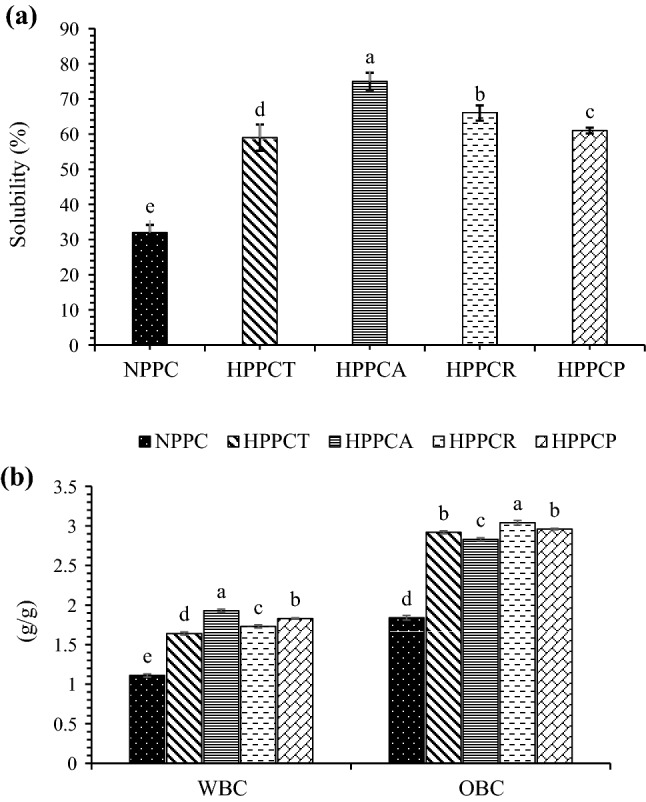
Effect of crude protease extract enzymatic hydrolysis on solubility (a), water binding and oil binding capacity (b) of peanut protein concentrate. [NPPC = native peanut protein concentrate, HPPCT = hydrolyzed peanut protein concentrate with crude protease extract of Trichoderma reesei, HPPCA = hydrolyzed peanut protein concentrate with crude protease extract of Aspergillus oryzae, HPPCR = hydrolyzed peanut protein concentrate with crude protease extract of Rhizopus oligosporus and HPPCP = hydrolyzed peanut protein concentrate with papain]
Water binding capacity
The effect of CPE hydrolysis on the water binding capacity of PPCs is shown in Fig. 3b. The native PPCs has water binding capacity value of 1.11 g/g only. Significant (p ≤ 0.05) increase in the water binding capacity of the samples was observed after hydrolysis. Higher water binding capacity value of 1.93 g/g was observed for the samples hydrolyzed with CPE obtained from A. oryzae (Fig. 3b), which was higher than the value (1.83 g/g) for samples hydrolyzed with papain. Improvement in water binding capacity of hydrolyzed PPCs is closely related to solubility. The structural changes which were evident from the results of surface hydrophobicity and SH groups may be the main contributors for the improvement in functional characteristics. The structural changes were owing to limited amount of unfolding, which occurred during hydrolysis. Once unfolding took place, the buried hydrophilic groups might have been exposed to the polar environment, which ultimately increased the solubility. Solubility is the pre-requisite for other functional properties, a small increment in solubility would affect other functional properties.
Oil binding capacity
Effect of CPE hydrolysis on the oil binding capacity of PPCs is shown in Fig. 3b. Similar trend was observed for oil binding capacity as for water binding capacity of hydrolyzed samples. Native PPCs showed lower oil binding capacity value (1.84 g/g), whereas hydrolyzed PPCs showed comparatively higher oil binding capacity. Higher oil binding capacity value of 3.04 g/g was found in PPCs hydrolyzed with CPE of Rhizopus oligosporus followed by PPCs hydrolyzed with A. oryzae and T. reesei (Fig. 3b). Papain hydrolysed samples had oil binding capacity value of 2.96 g/g, which was comparable with the samples hydrolyzed with CPEs obtained from all three fungal sources. The possible reasons for the improvement in oil binding capacity could be attributed to the changes in the secondary structure of protein molecules because of enzymatic hydrolysis. The non-polar side chains of proteins might have been exposed during hydrolysis, which bounded fat molecules. Higher surface area of the protein particles and the exposure of hydrophobic groups due to unfolding of chains might be the main reasons for improvement of oil binding capacity.
Foaming capacity and foam stability
The data for the effect of crude protease enzymatic hydrolysis on the foaming capacity and foam stability of PPCs are presented in Table 1. Native PPCs showed foaming capacity value of 22.33% only, which was significantly (p ≤ 0.05) lower than the foaming capacity of hydrolyzed PPCs. Among hydrolyzed PPCs, the highest foaming capacity value of 58.70% was observed in samples hydrolyzed with CPE obtained from A. oryzae followed by R. oligosporus (47.31%), papain (41.33%) and T. reesei (33.66%). Foams were more stable in A. oryzae hydrolyzed samples followed by R. oligosporus, papain and T. reesei. Improvement in foaming capacity and foam stability might be owing to lower particle size and higher solubility (Fig. 3a). This reconfirms the exposure of hydrophobic groups after hydrolysis. Higher foam stability may be attributed to lower particle size of hydrolyzed PPCs as mentioned previously in this manuscript. Panyam and Kilara (1996) observed that enzymatic hydrolysis of protein results in lower molecular weight peptides as well as brings some structural modifications. The structural modifications expose the hidden hydrophobic groups, which improve the surface properties of protein.
In-vitro protein digestibility (IVPD)
The data pertaining to the effect of enzymatic hydrolysis on the in-vitro protein digestibility are presented in Table 1. It can be observed that the IVPD of native PPC was comparatively lower (70.11%) than IVPD of hydrolyzed PPCs. Papain hydrolyzed samples showed IVPD value of 94.62%. Among CPE hydrolyzed samples, the higher IVPD value of 86.70% was found in samples hydrolyzed with CPE obtained from A. oryzae followed by R. oligosporus and T. reesei. Vishwanatha et al (2009) reported that A. oryzae has the higher expansion of hydrolytic genes (135 proteinase genes) and also possesses more secretory proteinase genes that function in acidic pH too. This could largely be responsible for significantly higher IVPD of PPC hydrolyzed with CPE obtained from A. oryzae, compared to PPCs hydrolysed by CPEs from the other two fungi. Microbial crude protease obtained from A. oryzae are non-toxic due to its long history of industrial uses and also due to the absence of expressed sequence tags for the genes responsible for aflatoxin production. It’s worth mentioning here that A. oryzae is used in the preparation of different fermented foods and has been accorded GRAS status by USFDA. Improvement in IVPD of hydrolyzed samples is due to the fact that proteases catalyze the hydrolysis of peptide bonds, which in turn converts proteins into peptides of smaller size and free amino acids. Lower molecular weight peptides and free amino acids can be easily digested by the small intestine and also possess lower allergenic effects. Su et al (2011) also observed that CPE obtained from A. oryzae possesses comparatively higher nutritional quality than other commercially available pure proteases.
Conclusion
It is evident from the study that hydrolysis using crude protease extracts is an effective approach for improving the functional as well as in-vitro digestibility of peanut protein concentrate. The CPE obtained from three fungal sources positively affected the different structural and functional characteristics of PPCs. Crude protease extracts obtained from A. oryzae was more effective in improving the functional and digestibility profile of PPCs in comparison to other two fungal CPEs. Therefore, this study has set a platform for conducting more comprehensive studies on the use of CPEs from A. oryzae for hydrolysis of complex protein isolates and concentrates from a variety of sources in order to improve the functional and nutritional characteristics of such proteins besides offering a cost effective solution.
Acknowledgements
The authors are highly thankful to National Agricultural Science Fund (NASF), New Delhi, India for providing funds for the study. Authors also acknowledge Bio-process Engineering Laboratory, ICAR-CIPHET Ludhiana, India for providing microbial cultures used in the study.
Declarations
Conflict of interest
The authors declare that there is no conflict of interest in the present study.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Achouri A, Nail V, Boye JI. Sesame protein isolate: fractionation, secondary structure and functional properties. Food Res Int. 2012;46:360–369. doi: 10.1016/j.foodres.2012.01.001. [DOI] [Google Scholar]
- AOAC . Official methods of analysis. 18. Arlington, VA: Association of Official Analytical Chemists, Washington, DC; 2006. [Google Scholar]
- Beuchat LR. Functional and electrophoretic characteristics of succinylated peanut flour proteins. J Agrl Food Chem. 1977;25:258. doi: 10.1021/jf60210a044. [DOI] [Google Scholar]
- Chakraborty P. Coconut protein isolate by ultrafiltration. In: Le Meguer M, Jelen P, editors. Food Engineering and Process Applications 2. New York: Elsevier Applied Science Publishers; 1986. pp. 308–315. [Google Scholar]
- Elias RJ, Kellerby SS, Decker EA. Antioxidant activity of proteins and peptides. Crit Rev Food Sci Nutr. 2008;48:430–441. doi: 10.1080/10408390701425615. [DOI] [PubMed] [Google Scholar]
- Fekria AM, Isam AMA, Suha OA, Elfadil EB. Nutritional and functional characterization of defatted seed cake of two sudanese groundnut (Arachis Hypogaea) cultivars. Int Food Res J. 2012;19:629–637. [Google Scholar]
- Gupta A, Roy I, Khare SK, Gupta MN. Purification and characterization of a solvent stable protease from pseudomonas aeruginosa. J Chrom A. 2005;1069(2):155–161. doi: 10.1016/j.chroma.2005.01.080. [DOI] [PubMed] [Google Scholar]
- Hsu HW, Vavak DL, Satterlee L, Miller GA. A multi-enzyme technique for estimating protein digestibility. J Food Sci. 1977;42(5):1269–1273. doi: 10.1111/j.1365-2621.1977.tb14476.x. [DOI] [Google Scholar]
- Jain A, Subramanian R, Manohar B, Radha C. Preparation, characterization and functional properties of Moringa oleifera seed protein isolate. J Food Sci Technol. 2019;56(4):2093–2104. doi: 10.1007/s13197-019-03690-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu HH, Kuo MI. Ultra high pressure homogenization effect on the proteins in soy flour. Food Hydrocol. 2016;52:741–748. doi: 10.1016/j.foodhyd.2015.08.018. [DOI] [Google Scholar]
- Malik MA, Saini CS. Gamma irradiation of alkali extracted protein isolate from dephenolized sunflower meal. LWT-Food Sci Tech. 2017;84:204–211. doi: 10.1016/j.lwt.2017.05.067. [DOI] [Google Scholar]
- Malik MA, Sharma HK, Saini CS. Effect of removal of phenolic compounds on structural and thermal properties of sunflower protein isolate. J Food Sci Technol. 2016;53(9):3455–3464. doi: 10.1007/s13197-016-2320-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mir NA, Riar CS, Singh S. Structural modification of quinoa seed protein isolates (QPIs) by variable time sonification for improving its physicochemical and functional characteristics. Ultrason Sonochem. 2019;58:104700. doi: 10.1016/j.ultsonch.2019.104700. [DOI] [PubMed] [Google Scholar]
- Mir NA, Riar CS, Singh S. Structural modification in album (chenopodium album) protein isolates due to controlled thermal modification and its relationship with protein digestibility and functionality. Food Hydrocol. 2020;103:105708. doi: 10.1016/j.foodhyd.2020.105708. [DOI] [Google Scholar]
- Nielsen PM, Petersen D, Dambmann C. Improved method for determining food protein degree of hydrolysis. J Food Sci. 2001;66:642–646. doi: 10.1111/j.1365-2621.2001.tb04614.x. [DOI] [Google Scholar]
- Oliveira AND, Oliveira LAD, Andrade JS. Production and some properties of crude alkaline proteases of indigenous central Amazonian rhizobia strains. Braz Arch Bio Tech. 2010;53(5):1185–1195. doi: 10.1590/S1516-89132010000500024. [DOI] [Google Scholar]
- Panyam D, Kilara A. Enhancing the functionality of food proteins by enzymatic modification. Trends Food Sci Tech. 1996;7:120–125. doi: 10.1016/0924-2244(96)10012-1. [DOI] [Google Scholar]
- Pavia DL, Lampman GM, Kriz GS, Vyvyan JA. Introduction to Spectroscopy. Cengage Learning; 2008. [Google Scholar]
- Quist EE, Phillip RD, Saalia FK. The effect of enzyme systems and processing on the hydrolysis of peanut (Arachis hypogaea L.) protein. LWT-Food Sci Tech. 2009;42:1717–1721. doi: 10.1016/j.lwt.2009.04.005. [DOI] [Google Scholar]
- Sandhu SK, Oberoi HS, Dhaliwal SS, Babbar N, Kaur U, Nanda D, Kumar D. Ethanol production from Kinnow mandarin (Citrus reticulata) peels via simultaneous saccharification and fermentation using crude enzyme produced by Aspergillus oryzae and the thermos-tolerant Pichia kudriavzevii strain. Annals Microb. 2011;62:655–666. doi: 10.1007/s13213-011-0302-x. [DOI] [Google Scholar]
- Silvestre MP, Silva MR, Silva VD, Souza MW, Junior L, de Oliveira C, Afonso WD. Analysis of whey protein hydrolysates: peptide profile and ACE inhibitory activity. Braz J Pharm Sci. 2012;48:747–757. doi: 10.1590/S1984-82502012000400019. [DOI] [Google Scholar]
- Su G, Ren J, Yang B, Cui C, Zhao M. Comparison of hydrolysis characteristics on defatted peanut meal proteins between a protease extract from Aspergillus oryzae and commercial proteases. Food Chem. 2011;126:1306–1311. doi: 10.1016/j.foodchem.2010.11.083. [DOI] [Google Scholar]
- Tang S, Zhou X, Gouda M, Cai Z, Jin Y. Effect of enzymatic hydrolysis on the solubility of egg yolk powder from the changes in structure and functional properties. LWT-Food Sci Technol. 2019;110:214–222. doi: 10.1016/j.lwt.2019.04.070. [DOI] [Google Scholar]
- Vishwanatha KS, Rao AA, Singh SA. Characterization of acid protease expressed from Aspergillus oryzae MTCC 5341. Food Chem. 2009;114:402–407. doi: 10.1016/j.foodchem.2008.09.070. [DOI] [Google Scholar]
- Wagner JR, Sorgentini DA, Anon MC. Relation between solubility and surface hydrophobicity as an indicator of modifications during preparation processes of commercial and laboratory-prepared soy protein isolates. J Agril Food Chem. 2000;48(8):3159–3165. doi: 10.1021/jf990823b. [DOI] [PubMed] [Google Scholar]
- Yadav DN, Thakur N, Sunooj KV. Effect of partially de-oiled peanut meal flour (DPMF) on the nutritional, textural, organoleptic and physico chemical properties of biscuits. Food Nutr Sci. 2012;3:471–476. doi: 10.4236/fns.2012.34067. [DOI] [Google Scholar]
- Yin SW, Tang CH, Wen QB, Yang XQ. Functional and conformational properties of phaseolin (Phaseolus vulgris L.) and kidney bean protein isolate: a comparative study. J Sci Food Agri. 2009;90:599–607. doi: 10.1002/jsfa.3856. [DOI] [PubMed] [Google Scholar]
- Yu JM, Ahmedna M, Goktepe I. Peanut protein concentrate: production and functional properties as affected by processing. Food Chem. 2007;103(1):121–129. doi: 10.1016/j.foodchem.2006.08.012. [DOI] [Google Scholar]
- Zhao G, Liu Y, Zhao M, Ren J, Yang B. Enzymatic hydrolysis and their effects on conformational and functional properties of peanut protein isolate. Food Chem. 2011;127(4):1438–1443. doi: 10.1016/j.foodchem.2011.01.046. [DOI] [Google Scholar]
- Zhao Q, Xiong H, Selomulya C, Chen XD, Zhong H, Wang S, Sun W, Zhou Q. Enzymatic hydrolysis of rice dreg protein: effects of enzyme type on the functional properties and antioxidant activities of recovered proteins. Food Chem. 2012;134:1360–1367. doi: 10.1016/j.foodchem.2012.03.033. [DOI] [PubMed] [Google Scholar]


