Abstract
Aim: Head and neck squamous cell carcinoma (HNSCC) is the sixth most commonly diagnosed malignancy worldwide. Overexpressed of microRNA-21-5p (miR-21-5p) has been reported to be involved in the development of HNSCC. However, the role of miR-21-5p in HNSCC is still not fully elucidated. The purpose of this study was to explore the underlying molecular mechanisms of miR-21-5p in HNSCC. Methods: RT-qPCR was used to determine the differential expression levels of miR-21-5p in tissue samples of HNSCC patients. Meta-analysis was performed based on miRNA expression data collected from the Gene Expression Omnibus (GEO) database, The Cancer Genome Atlas (TCGA), and published articles to evaluate the expression of miR-21-5p in HNSCC. We investigated the biological function of miR-21-5P by gene ontology enrichment and target prediction analysis. Furthermore, RT-qPCR and IHC were conducted to verify the expression of target genes. Finally, Kaplan–Meier survival analysis was performed to assessed the prognostic value of the putative miR-21-5p target genes. Results: MiR-21-5p was significantly overexpressed in HNSCC compared to healthy tissues (P < .05) and showed potent predictive power with a summary receiver operating characteristic of 0.90. Meanwhile, the expression of miR-21-5p was significantly correlated with tumor stage, T stage and smoking in HNSCC (P < .05). A total of 71 down-regulated genes, both HNSCC-related and miR-21-p5-related, were obtained from the analytical integration. Two predicted genes (ADH7, RDH12) were down-regulated in HNSCC, and significantly negatively correlated with miR-21-5p. IHC and RT-qPCR demonstrated that the expression of ADH7 and RDH12 in HNSCC samples was significantly lower than control. And high expression of ADH7 was associated with better DFS of HNSCC patients. Conclusions: miR-21-5p may target at ADH7, RDH12 and participate in regulation of retinol metabolism, which might affect the prognosis of HNSCC. High expression of ADH7 may indicate better prognosis in HNSCC patients.
Keywords: HNSCC, MicroRNA-21-5p, RT-qPCR, IHC, TCGA, bioinformatics
Introduction
Head and neck squamous cell carcinoma (HNSCC) is the sixth most common malignancy in the world and encompasses tumors arising in oral cavity, nasal cavity, nasopharynx, oropharynx, larynx, hypopharynx, paranasal sinus, and salivary glands, mainly associated with tobacco and alcohol consumption. 1 Currently, HNSCC has a global incidence of around 880 000 new cases worldwide annually and around 450 000 deaths. 1 However, because the early symptoms of HNSCC are not obvious, Over half of the patients are diagnosed at an advanced stage with an approximate 50% 5-year survival rate.2–4 The optimal treatment for HNSCC consists of surgical resection, radiotherapy, chemotherapy, immunotherapy, and other targeted therapies. Despite the therapeutic progress made in recent years, the clinical outcome of HNSCC patients has improved only marginally, mainly due to lymph node metastasis and local recurrences. As to today, the molecular mechanism of HNSCC has not been fully explored, and we still lack relevant biomarkers to guide therapeutic options and evaluate the prognosis of patients with HNSCC. Therefore, to improve prognostic prediction and treatment outcomes, there is a dire need to identify novel molecular therapeutic targets and robust biomarkers for HNSCC.
MicroRNA (miRNA) is a non-coding small RNA, with a length of approximately 21 to 23 nucleotides. 5 MiRNAs modulate the expression of numerous genes by inhibiting translation or destabilizing the mRNA. 6 Aberrant miRNA expression has been associated with multiple pernicious tumors in humans, reflecting their critical biological roles. Recent studies showed that microRNA-21-5p (miR-21-5p) downregulates various tumor suppressor genes and promotes the invasion and proliferation of cancer cells. 7 It is overexpressed in numerous malignancies, such as cervical cancer, 8 lung cancer, 9 laryngeal squamous cell carcinoma, 10 and hepatocellular carcinoma, 11 indicating its potential as a prognostic and therapeutic marker. MiR-21-5p dysregulation also correlates with various HNSCC subtypes and has been proposed as a novel biomarker for diagnostic and prognostic. 12 Previous study 13 showed that miR-21-5p was a suitable marker to distinguish between HNSCC and non-cancerous tissues. Upregulated miR-21-5p has been detected in HNSCC tissues, although the underlying molecular mechanism is not fully revealed.
To further investigate the diagnostic and prognostic value of miR-21-5P in HNSCC, patients tissue with HNSCC were used to identify the expression of miR-21-5p. Moreover, a meta-analysis of the miR-21-5p expression was performed with data collected from The Cancer Genome Atlas (TCGA), the Gene Expression Omnibus (GEO) database, and published literature, revealing a high expression, as well as with potential prognostic value in HNSCC.
The present study will reveal part of the underlying molecular mechanisms in HNSCC, which might contribute to the Clinical therapy and the prognosis to the HNSCC. Our findings indicate that miR-21-5p might mediate the development and progression of HNSCC and may be a potential diagnostic and prognostic marker.
Materials and Methods
Ethics
Ethical approval for this study was received from the Ethics Committee of Guangxi Medical University, the approval numbers were 2020(KY-E-181) and 2021(K-Y-E153). Written informed consents were obtained from all the patients for the publication.
RT-qPCR
In order to verify the expression level of miR-21-5p in HNSCC, 30 patients with HNSCC and 40 non-tumor controls were collected. Meanwhile, 25 patients with HNSCC and 25 non-tumor controls were included to examined the expression level of putative target genes. Tissue samples were obtained from HNSCC patients under treatment at the Department of Pathology of The First Affiliated Hospital of Guangxi Medical University, Nanning, China. RT-qPCR was performed on an ABI 7500 Cycler (Applied Biosystems) using SYBR-Green Master Mix (Takara). We performed the 2−ΔΔCt calculation method and U6 was used as the internal control. And for detection, we used the TaKaRa liaoning dalian (China) synthetic miR-21-5p and U6 primers, which had the following sequences: for Mir-21-5p the forward primer was: 5′- GCCTAGCTTATCAGACTGATGTTGA-3′ mRQ, and the reverse primer was: 5′ mRQ 3′. For U6 the following primers were used: forward 5′-GGAACGATACAGAGAGATTAGC-3′ and reverse: 5′-TGGAACGCTTCACGAATTTGC-3′. For the ADH7, the forward primer was: 5′-GTTCCACTTGCGTCGTCT-3′, the reverse primer was: 5′-CTGATGGGTTTGGTAGAGT-3′. And for RDH12, the forward primer was: 5′-CAATGCGGGAGTAATGAT-3′, the reverse primer was: 5′-AGGAGCAGGTAGGTGAGG-3′. All the experiments were replicated more than 3 times independently.
Literature Search Strategy
A systematic literature search was performed on the Wiley Online Library, the Cochrane Library, the Web of Science, EMBASE, Science Direct, PubMed, the China National Knowledge Infrastructure Database (CNKI), the Technology Journal Database (VIP), the China Biomedical Literature Database (CBM), the Wanfang Database, and the China Science databases for all relevant studies published before February 15, 2021. The keywords used were as follows: (“HNSCC” OR “SCC” OR Rsquamous cell carcinoma” OR Rsquamous cell cancer”) AND (“head and neck” OR “oral cavity” OR “oropharynx” OR “oropharyngeal” OR “oral” OR “hypopharynx” OR Rlarynx” OR “laryngopharyngeal” OR “laryngopharynx” OR “laryngeal” OR “laryngeal” OR “pharyngeal” OR “nose” OR “nasopharynx” OR “nasal sinus” OR “paranasal sinuses” OR “nasal cavity” OR “Rtonsil” OR “tonsillar” OR “tongue” OR “cheek” OR “buccal” OR “palatal” OR “lip”) AND (“microRNA-21” OR “microRNA21” OR “miRNA-21” OR “miRNA 21” OR “miRNA21” OR “miR-21” OR RmiR 21” OR “miR21”).
Selection Standard and Data Screening
Two researchers conducted the database searches in duplicate independently to minimize any bias. Publications were included if following standards were meet: (1) expression analysis conducted on HNSCC and non-cancerous tissue; (2) expression levels of miR-21-5p were assessed by reverse transcription quantitative PCR (RT-qPCR) and; (3) availability of sufficient data to determine relevant descriptive statistics, including mean and standard deviation (SD) and 95% confidence intervals (95% CI). Studies were excluded based on these criteria: (1) overlapping or duplicate publications; (2) other languages than English or Chinese; (3) unqualified data were included; (4) letters, conference reports, editorials, comments, reviews, meeting abstracts, or case reports; (5) the expression of miR-21-5p was not evaluated on human specimens. Each selected study was evaluated for data quality independently by reviewers, and the first author name, published year, country of publication, sample origin, sample size, and analysis strategies were extracted. We contacted the authors to acquire the initial dataset if the data was incomplete.
Microarray Data Collection From GEO
HNSCC microarray datasets uploaded before February 15, 2021, were collected from NCBI-GEO database (https://www.ncbi.nlm.nih.gov/gds/). 14 The keywords used for the search were as follows: (“HNSCC” OR “SCC” OR “squamous cell carcinoma” OR Rsquamous cell cancer”) AND (“head and neck” OR “oropharynx” OR “oropharyngeal” OR “oral” OR “oral cavity” OR “hypopharynx” OR “larynx” OR “laryngopharyngeal” OR “laryngopharynx” OR “laryngeal” OR “laryngeal” OR “pharyngeal” OR “nose” OR “nasopharynx” OR “nasal sinus” OR “paranasal sinuses” OR “nasal cavity” OR “tonsil” OR “tonsillar” OR “tongue” OR “cheek” OR “buccal” OR “palatal” OR “lip”) AND (“microRNA” OR “miRNA” OR “miRNA” OR “miR “ OR “miR”). The inclusion criteria are outlined as follows: (1) data comprised HNSCC and healthy tissue; (2) the association of miR-21-5p expression levels and clinical outcomes were evaluated; (3) availability of adequate data to determine relevant descriptive statistics, including mean, SD, and 95% confidence intervals (95% CI). The exclusion criteria were: (1) irrelevant to the research focus; (2) ineligible data; (3) not published in English or Chinese.
RNA Sequencing Data Extraction From TCGA
The expression levels and the clinicopathological parameters for miR-21-5p were downloaded and filtered from the OncoLnc website (http://www.oncolnc.org/; TCGA data version 28.0 February 2, 2021). 15
Target Gene Prediction
The differentially expressed genes (DEGs) in HNSCC tissues were retrieved from TGCA using GEPIA (http://gepia2.cancer-pku.cn), and downregulated genes with ∣log2FC∣>2 were selected. MiRWalk 3.0 16 (http://zmf.umm.uni-heidelberg.de/apps/zmf/mirwalk2/) was applied to identify potential target genes of miR-21-5p. Finally, overlapping genes were identified by VENNY (https://omictools. com/venny-tool).
Bioinformatics Functional Analysis
Gene Ontology (GO) 17 and the Kyoto Encyclopedia of Genes and Genomes (KEGG) 18 pathway enrichment analyses were performed by DAVID 6.8 (https://david.ncifcrf.gov/summery.jsp), and visualized by OmicShare tools (http://www.omicshare.com/tools). The online database STRING (https://string-db.org/) was utilized to develop a protein–protein interaction (PPI) network. UALCOULD 19 (http://ualcan.path.uab.edu/index.html) was used to evaluate the expression levels of the miR-21-5p potential target genes in HNSCC and non-cancerous tissues. We explored the correlation between miR-21-5p expression and underlying target genes using LinkedOmics’ 20 (http://www.linkedomics.org/) Spearman's analysis.
Protein Expression of Target Genes by Immunohistochemistry
IHC was performed to evaluate the protein expression of hub genes in HNSCC samples and non-tumor controls. Tissue samples were obtained from 15 HNSCC patients and 15 normal control at the First Affiliated Hospital of Guangxi Medical University. All of the tissue specimens were fixed in 4% formalin and embedded in paraffin. The immunohistochemistry (IHC) procedure was performed according to the manufacturer's protocols. Briefly, after antigen extraction, tissue sections were incubated with a polyclonal antibody at 4 °C overnight, and then incubation with a biotinylated secondary antibody and an avidin-biotin peroxidase complex (Sanying Biotechnology). Then, the immune reactions were developed by adding DAB chromogen-substrates.
Prognosis Value of Target Genes
We used GEPIA (http://gepia.cancer-pku.cn/) to analyze relevance between the expression level of each potential target gene and the prognosis of HNSCC.
Statistical Analysis
Meta-analysis was analyzed using the STATA12.0 software. The I2 index evaluated the underlying heterogeneity of the data. A random-effects model was constructed for data with a P < .05 or I2 > 50%, which indicate high heterogeneity, otherwise, a fixed-effects model was adopted. The standard mean difference (SMD) with 95% CI was used to compare miR-21-5p expression levels in HNSCC and healthy tissues, then was presented as a scatter plot generated using GraphPad Prism 8 software. The remaining statistical analyses were carried out by SPSS 25.0 software. Numerical data were expressed as the mean ± SD (X ± s) or mean ± SEM, the difference between 2 groups was analyzed using the Students T test, while 1-way analysis of variance (ANOVA) test was used to analysis the difference among more than 3 groups. Categorical data were shown as percentages (%) and compared by the Chi-square test. The diagnostic value of miR-21-5p was evaluated using the true negative (TN), false negative (FN), true positive (TP), false positive (FP) results. The sensitivity, specificity, the summary receiver operating characteristic (sROC) curve, positive likelihood ratio (PLR), negative likelihood ratio (NLR), and the diagnostic odds ratio (DOR) were then calculated using the meta-Disc software. MiR-21-5p expression data extracted from GEO and TCGA were normalized to log2. Statistically significant was accepted at P < .05.
Results
MiR-21-5p is Identified as an Upregulated miRNA in HNSCC
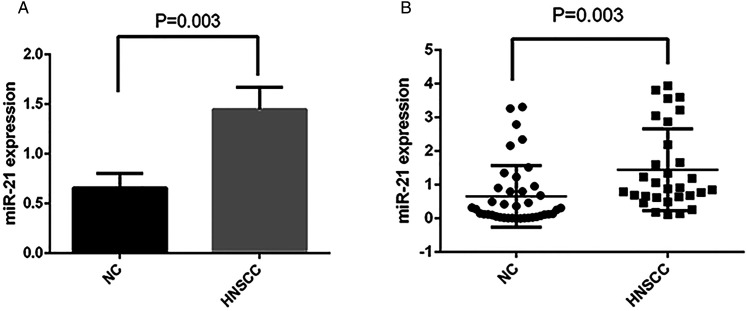
Our RT-qPCR data showed that miR-21-5p was substantially upregulated in tumor tissues during in the HNSCC cohort (n = 30) as compared to non-tumor tissues (n = 40) (P < .01, Figure 1A and B).
Figure 1.
MiR-21-5p levels in HNSCC and NC tissue specimens. RT-qPCR was used to detect miR-21-5p was significantly up-regulated in the tumor tissues of an HNSCC cohort (n = 30) relative to control tissues (n = 40) (P = .003).
Selection of Qualified Literature and Microarray Data Extraction
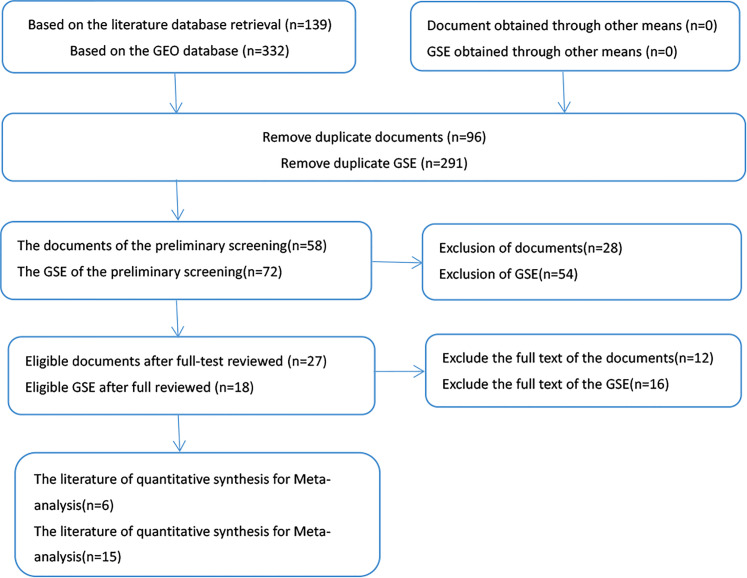
We retrieved 139 in our primary literature search, and we selected 6 papers after careful full-text review according to the inclusion criteria (Figure 2): Brito JA 21 (PMID:24020903), Chang SS 22 (PMID:18798260), Yap T 23 (PMID:29764807), Erkul E 24 (PMID:27545281), Gee HE 25 (PMID:20187102), and Hu A 26 (PMID:23259291). For the GEO microarray datasets, 15 out of 332 met our inclusion criteria: GSE118613, GSE34496, GSE11163, GSE98463, GSE45238, GSE37472, GSE31277-GPL9770, GSE28100, GSE113956, GSE69002, GSE103931, GSE73171, GSE124678, GSE82064, GSE75630. The information is presented in Supplemental Table S1.
Figure 2.
The flow chart of literature and GEO database search as well as selection of relevant studies. Fifteen out of 332 GEO microarray datasets met our inclusion criteria. Meanwhile, 139 articles were initially retrieved, and 6 papers meeting the criteria were included.
Expression Levels and Meta-analysis of miR-21-5p in HNSCC Based on Microarray Data and the Literature
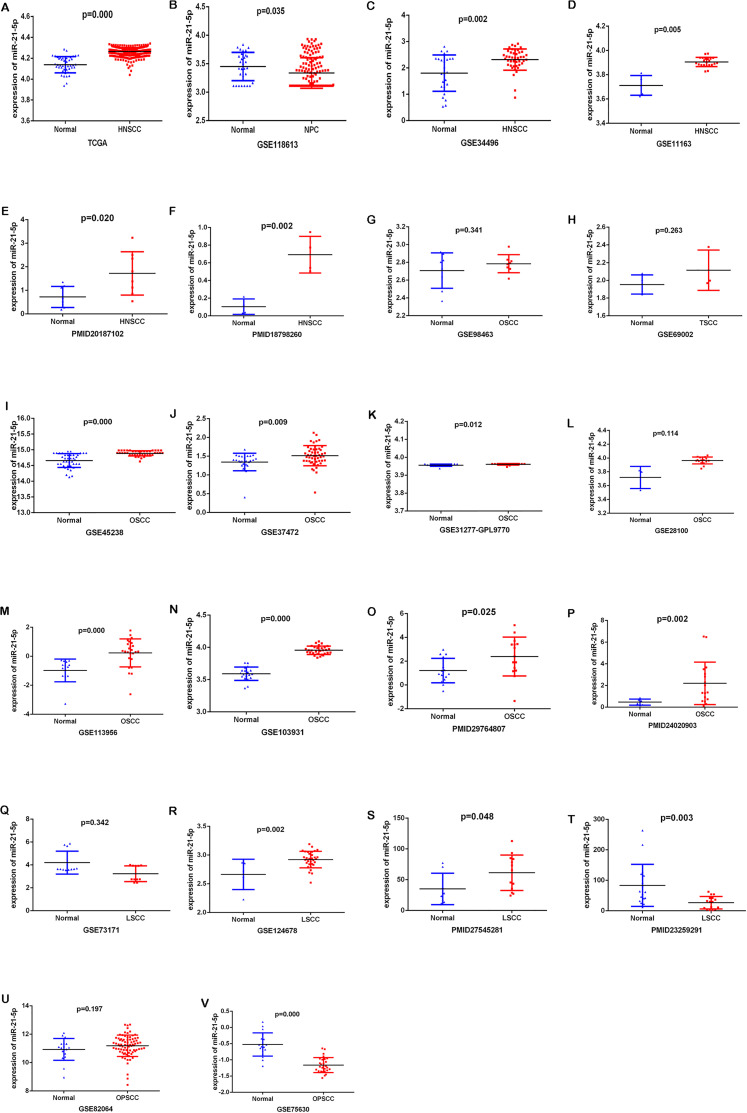
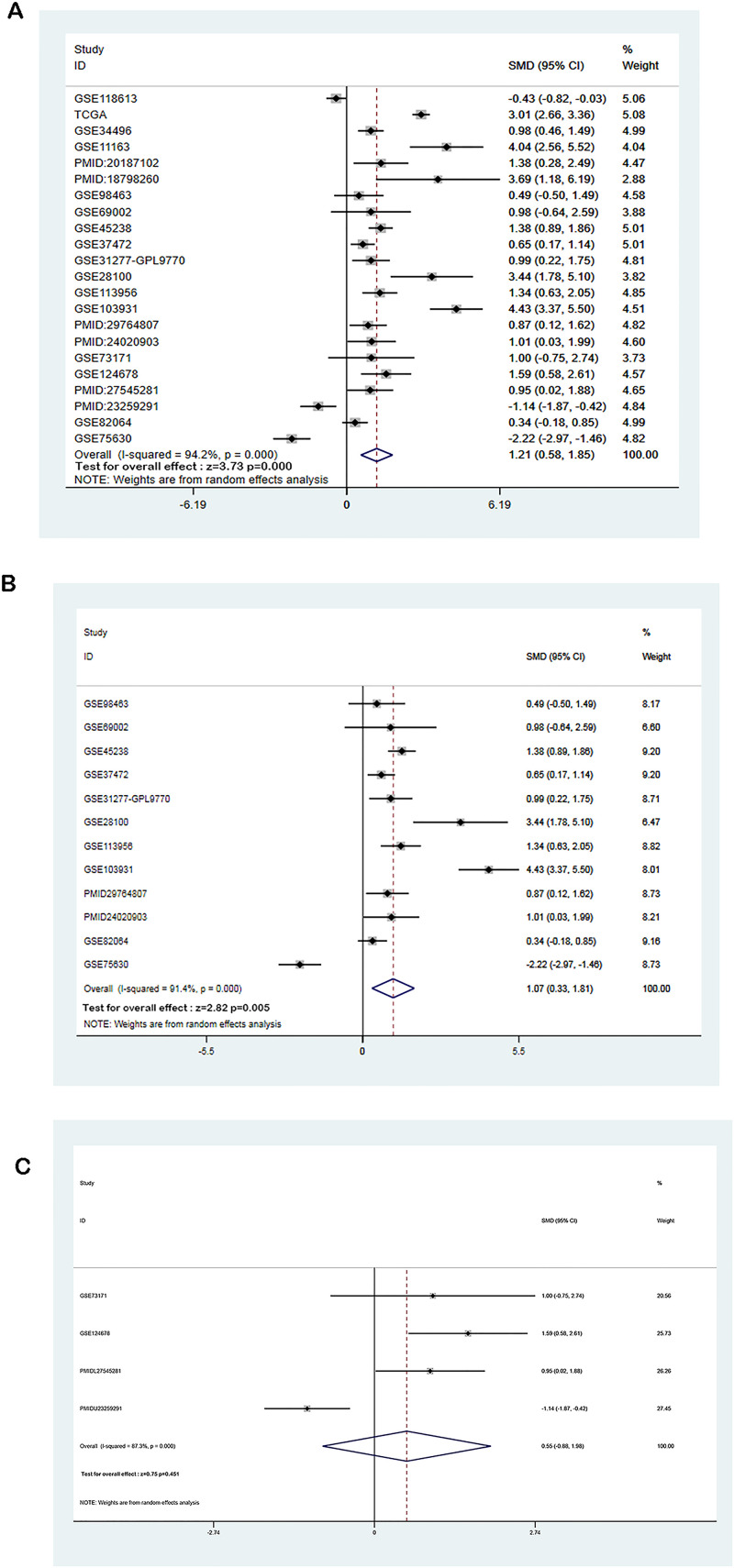
As shown in Figure 3A and Table 1, the expression levels of miR-21-5p were significantly higher in HNSCC than in non-tumor controls in the TGCA datasets (P < .001). For GEO datasets and literature, 15 datasets and 6 papers were selected in the present study. The miR-21-5p expression levels in HNSCC in these studies are shown in Figure 3B to V, indicating the overexpression of miR-21-5p in HNSCC while compared with non-tumor controls (P < .05). This result is consistent with those obtained by RT-qPCR. The random-effects model was chosen for high heterogeneity among the studies (I2 = 94.2%, P < .001) (Figure 4A). The combined SMD of miR-21-5p expression in HNSCC was 1.21 (95% CI: 0.58, 1.85) (Figure 4A). Subsequently, we analyzed data obtained from the sample source of all selected literature and microarray datasets and selected 2 subtypes of HNSCC for further analysis (Supplemental Table S2), which had a combined SMD of 1.07 (95% CI: 0.33, 1.81) and 0.55 (95% CI: −0.88, 1.98) (Figure 4B and C). The meta-analysis results indicated that miR-21-5p was substantially upregulated in HNSCC, consistent results were observed in the oral squamous cell carcinoma (OSCC) subtype, but not in laryngeal squamous cell cancer (LSCC).
Figure 3.
The expression data of miR-21-5p in HNSCC from TCGA, GEO, and each included study. (A)TCGA; (B) GSE118613; (C) GSE34496; (D) GSE11163; (E) PMID20187102; (F) PMID18798260; (G) GSE98463; (H) GSE69002; (I) GSE45238; (J) GSE37427; (K) GSE31277-GPL9770; (L) GSE28100; (M) GSE113956; (N) GSE103931; (O) PMID29764807; (P) PMID24020903; (Q) GSE73171; (R) GSE124678; (S) PMID27545281; (T) PMID23259291; (U) GES82064; (V) GSE75630.
Table 1.
The Correlation Between miR-21-5p Expression Levels and Clinicopathological Characteristics Based on TCGA Database.
| Clinicopathological features | Terms | n | Mean ± SD | P-value |
|---|---|---|---|---|
| Unpaired tissue | Normal | 44 | 4.05 ± 0.08 | <.001 |
| HNSCC | 520 | 4.18 ± 0.40 | ||
| Gender | Male | 379 | 4.19 ± 0.04 | .123 |
| Female | 141 | 4.18 ± 0.04 | ||
| Age | <60 | 230 | 4.19 ± 0.04 | .323 |
| ≥60 | 289 | 4.19 ± 0.04 | ||
| Lymphovascular invasion | No | 232 | 4.19 ± 0.04 | .252 |
| Yes | 123 | 4.19 ± 0.04 | ||
| Clinical stage | Ι | 21 | 4.18 ± 0.05 | .001 |
| II | 96 | 4.19 ± 0.04 | ||
| III | 105 | 4.17 ± 0.05 | ||
| IV | 284 | 4.19 ± 0.04 | ||
| Neoplasm histologic grade | G1 | 64 | 4.18 ± 0.04 | .080 |
| G2 | 304 | 4.19 ±0.04 | ||
| G3 | 124 | 4.19 ± 0.05 | ||
| G4 | 7 | 4.15 ± 0.03 | ||
| T stage | T1 | 37 | 4.18 ± 0.05 | .003 |
| T2 | 149 | 4.19 ± 0.04 | ||
| T3 | 138 | 4.18 ± 0.05 | ||
| T4 | 180 | 4.19 ± 0.04 | ||
| N stage | N0 | 242 | 4.19 ± 0.04 | .417 |
| N1 | 84 | 4.18 ± 0.05 | ||
| N2 | 165 | 4.19 ± 0.04 | ||
| N3 | 7 | 5.20 ± 0.02 | ||
| M stage | M0 | 489 | 4.19 ± 0.04 | .761 |
| M1 | 6 | 4.19 ± 0.04 | ||
| Alcohol | NO | 164 | 4.19 ± 0.04 | .918 |
| YES | 347 | 4.19 ± 0.04 | ||
| Perineural invasion | NO | 199 | 4.18 ± 0.04 | .053 |
| YES | 170 | 4.19 ± 0.04 | ||
| Smoking | <3 | 295 | 4.18 ± 0.04 | .015 |
| ≥3 | 212 | 4.19 ± 0.04 | ||
| Status | ALIVE | 301 | 4.19 ± 0.04 | .509 |
| DEAD | 219 | 4.19 ± 0.04 |
Abbreviations: HNSCC, head and neck squamous cell carcinoma; T stage, size or direct extent of the primary tumor; N stage, degree of spread to regional lymph nodes; M stage, presence of distant metastasis; SD, standard deviation.
Figure 4.
Forest plots. The combined SMD of miR-21-5p expression levels in HNSCC from the different sources was 1.21 (95% CI: 0.58, 1.85).
Diagnostic Value of miR-21-5p for HNSCC
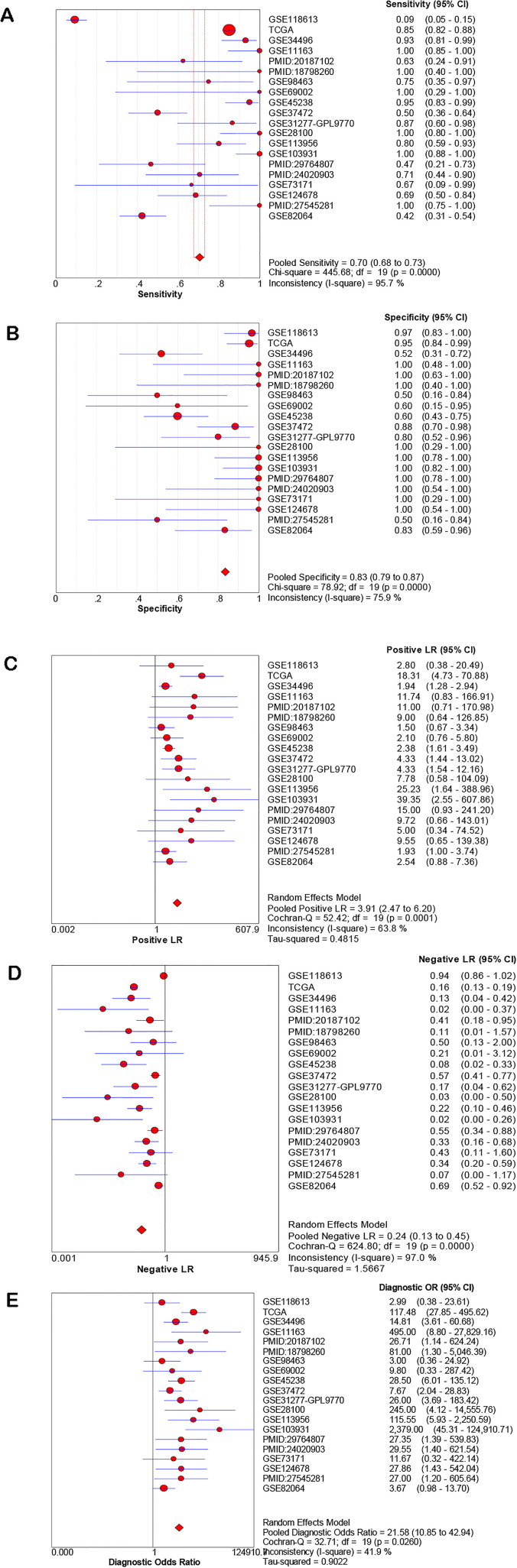
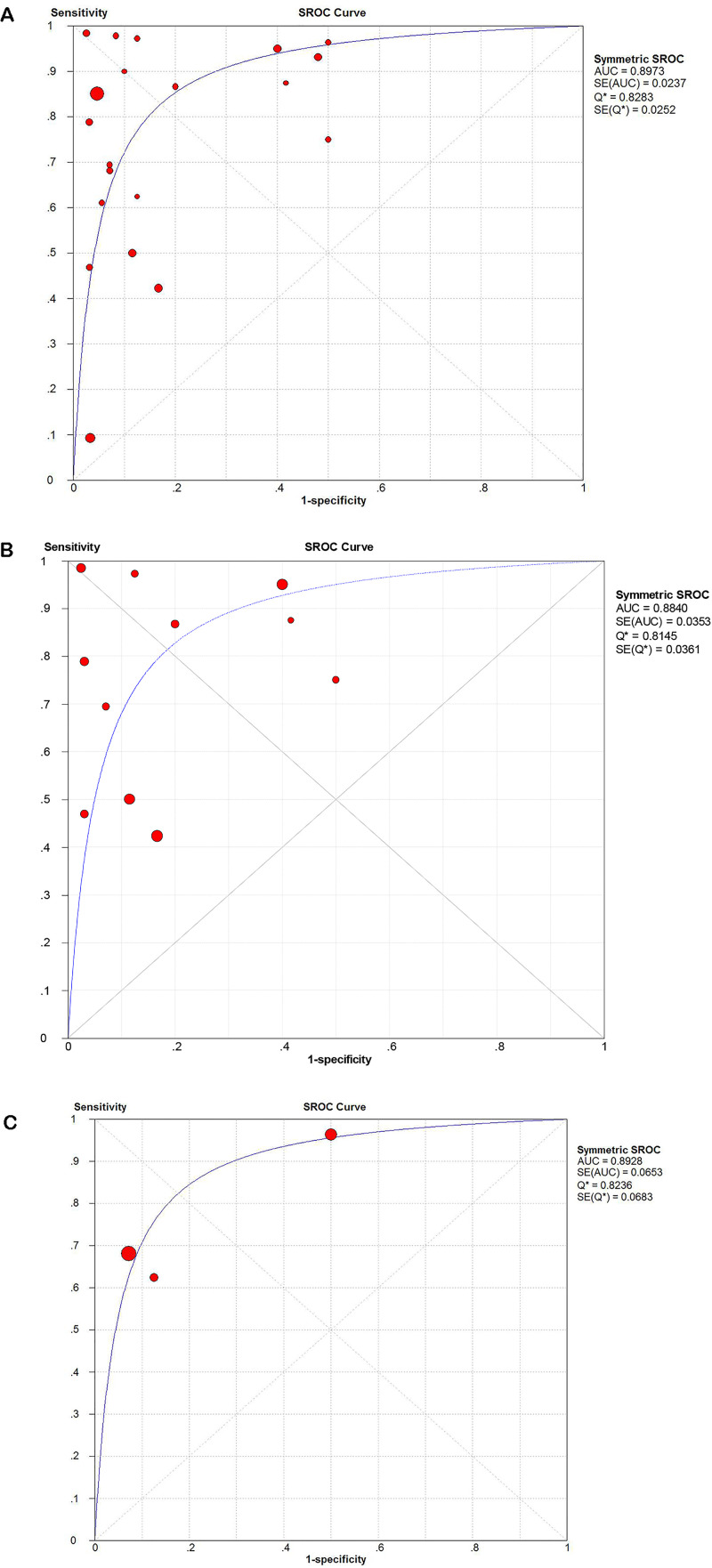
The diagnostic value of miR-21-5p for HNSCC was then analyzed by determining the sensitivity, specificity, PLR, NLR, and DOR, which were 0.83 (95% CI: 0.79-0.87), 0.70 (95% CI: 0.68-0.73), 3.19 (95% CI: 2.47-6.20), 0.24 (95% CI: 0.13-0.45), and 21.58 (95% CI: 10.85-42.94), respectively (Figure 5A to E). The results from each study included in the meta-analysis are displayed in the ROC curve in Supplemental Figure S1. The ROC curve implies that miR-21-5p may act as a potential biomarker to distinguish HNSCC tissues. The sROC curve AUC of HNSCC, as well as its subtypes OSCC and LSCC, were 0.90, 0.88, 0.89, respectively (Figure 6A to C), revealing a high predictive power.
Figure 5.
The diagnostic value of miR-21-5p in HNSCC. (A) Sensitivity, (B) specificity, (C) PLR, (D) NLR, and (E) diagnostic odds ratio.
Figure 6.
Receiver operating characteristic (ROC) curves showing the diagnostic value of miR-21-5p in HNSCC.
Clinical Features of HNSCC Based on the TCGA
As presented in Table 1, the analysis of 520 HNSCC samples, extracted from the TCGA database, revealed a significant association between the expression levels of miR-21-5p and Clinical staging, T staging, as well as smoking history (P < .05), but not with other clinicopathological factors like gender, age, lymphovascular invasion, histologic grade, alcohol use, and peri-neural invasion.
Potetial Target Genes of miR-21-5p in HNSCC
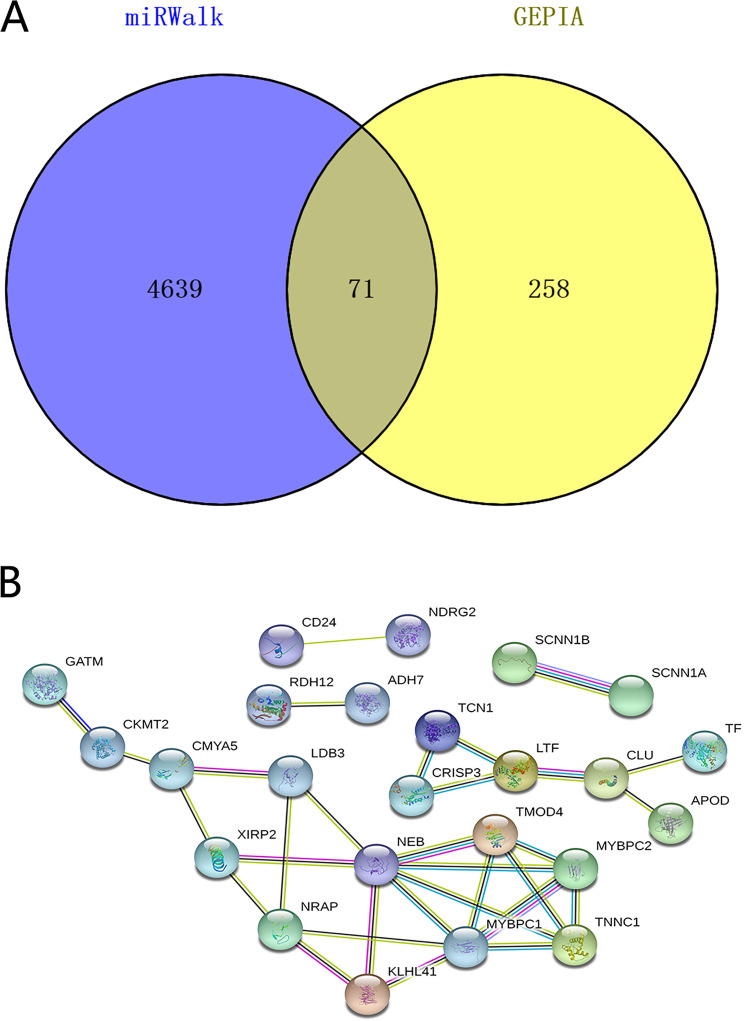
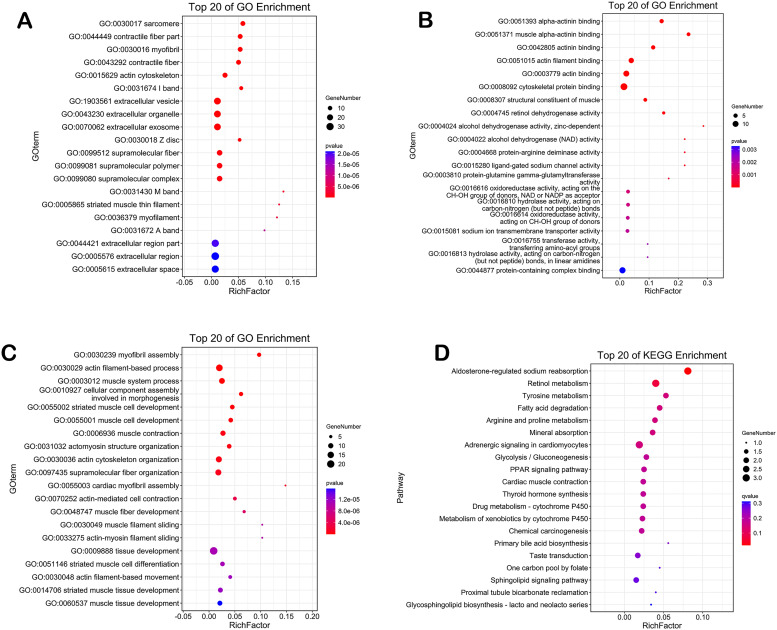
Potential target genes of miR-21-5p in HNSCC were predicted by differential expression analysis based on TCGA database and 12 target gene prediction platforms. GEPIA and MiRWalk identified 329 and 4710 DEGs in HNSCC tissues, respectively. Of these, 71 overlapped genes were putative as possible target genes of miR-21-5p (Figure 7A, Supplemental Table S3). GO analysis of these target genes is shown in Figure 8A to C; Supplemental Table S4. The most significant BP GO was myofibril assembly, actin filament-based process, muscle system process, cellular component assembly involved in morphogenesis. For CC, most of the target genes were enriched in the contractile fiber, myofibril, contractile fiber part, sarcomere. For MF, the most enriched terms were alpha-actinin binding, muscle alpha-actinin binding, actinin binding, actin filament binding. On the other hand, the KEGG pathway enrichment analysis indicated that miR-21-5p destabilization could affect aldosterone-regulated sodium reabsorption and retinol metabolism in HNSCC (Figure 8D, Supplemental Table S5). Those significant KEGG pathways and GO terms may play an vital role in the molecular mechanism of miR-21-5p in HNSCC patients, which might provide novel insights into therapy.
Figure 7.
(A) Venn diagram showing overlapping miR-21-5p target genes. (B) PPI network showing the connections among the 24 miR-21-5p targets. The nodes represent target genes and the lines are associations between genes.
Figure 8.
Significantly enriched annotation of (Gene Ontology) GO categories and (Kyoto Encyclopedia of Genes and Genomes) KEGG pathways of potential targets of miR-21-5p in HNSCC. (A) Biological processes (BP), (B) cellular components (CC), (C) molecular factors (MF), (D) KEGG pathway.
PPI Network Analysis
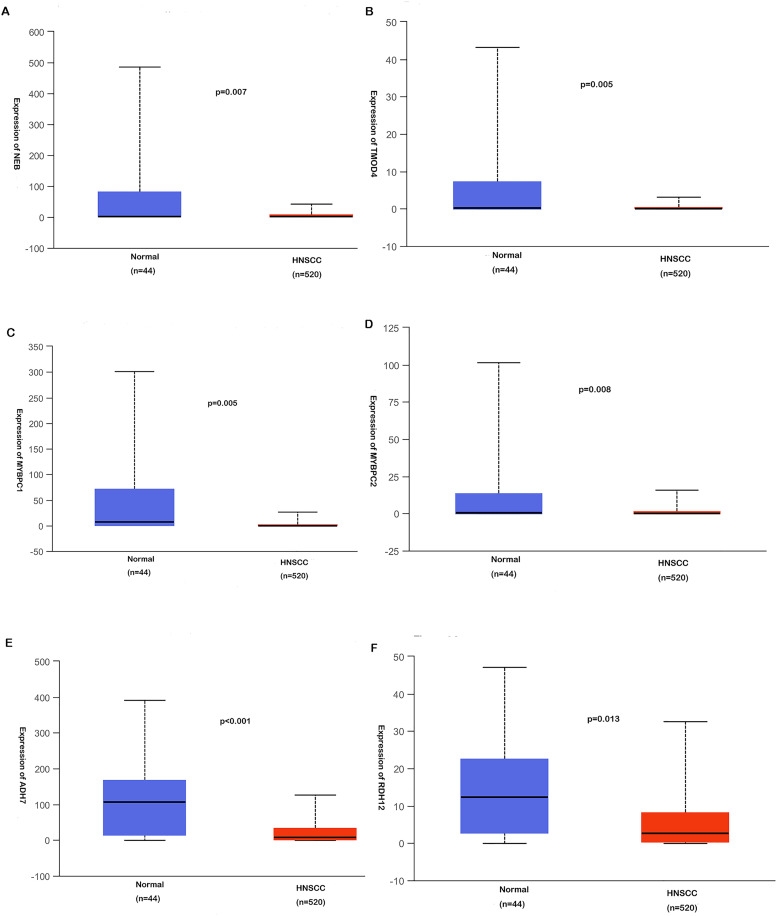
PPI network was performed by STRING based on the overlapped DEGs (Figure 7B). Based on the PPI network analysis and the KEGG pathway enrichment analysis, 4 genes with a high degree, as well as 2 genes involved in retinol metabolism, were selected as target genes, including NEB, TMOD4, MYBPC1, MYBPC2, ADH7, and RDH12. Those 6 target genes may be the potential target genes of miR-21-5p and may be involved in the regulatory mechanisms in HNSCC.
Validation of the miR-21-5p Target Genes
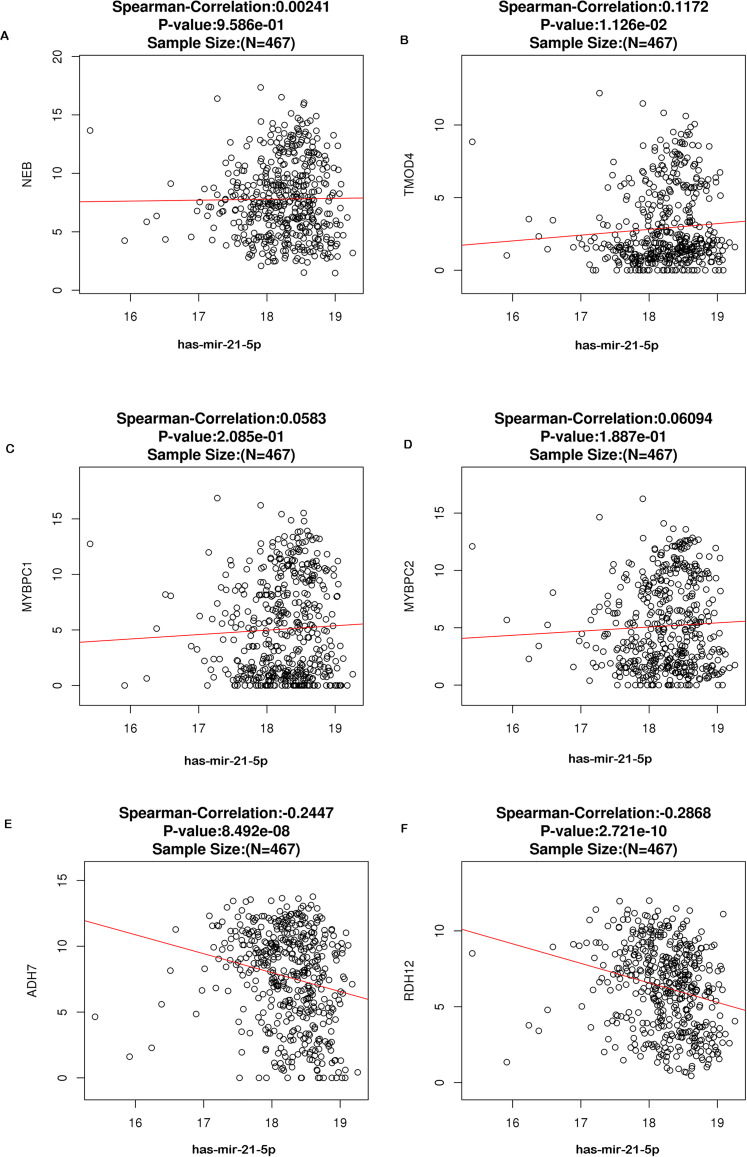
Six potential target genes of miR-21-5p associated with HNSCC were identified to be downregulated in HNSCC tissues as compared to control. The correlation between 6 potential target genes and miR-21-5p expression levels in HNSCC is shown in Figure 9A to F. Due to mir-21-5p is upregulated in HNSCC, the genes with decreased expression in HNSCC are most likely to act as target genes of miR-21-5p. Then Spearman's correlation analysis revealed that both ADH7 and RDH12 were negatively correlated with miR-21-5p expression in HNSCC significantly, with R = −.245, P < .001, and R = −.087, P < .001, respectively (Figure 10E and F). However, the negative correlation of miR-21-5p with NEB, TMOD4, MYBPC1, or MYBPC2 did not exhibit statistical significance (Figure 10A to D).
Figure 9.
The expression levels of miR-21-5p potential target genes in HNSCC.
Figure 10.
Spearman analysis shows the association between miR-21-5p expression levels and the 6 identified target genes.
Validation of ADH7 and RDH12 Expression in HNSCC
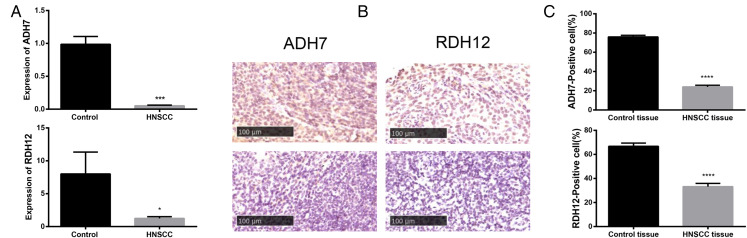
Based on the validation of the miR-21-5p target genes, 2 potential target genes (ADH7 and RDH12) involved in retinol metabolism aroused our interest. RT-qPCR and IHC were performed to further verify the expression level of target genes in HNSCC. As shown in Figure 11A, expression of ADH7 mRNA and RDH12 mRNA in HNSCC was significantly lower than non-tumor control (P < .05). Meanwhile, IHC results show that the expression of ADH7 protein and RDH12 protein in HNSCC tissue was lower than that in non-tumor controls (Figure 11B and C).
Figure 11.
(A) Expression of ADH7 mRNA and RDH12 mRNA in HNSCC determined by qRT-PCR, (B) immunohistochemistry of ADH7 and RDH12. Scale bar = 100 μm, (C) and its quantification.
Evaluation of Prognostic Value of Target Genes
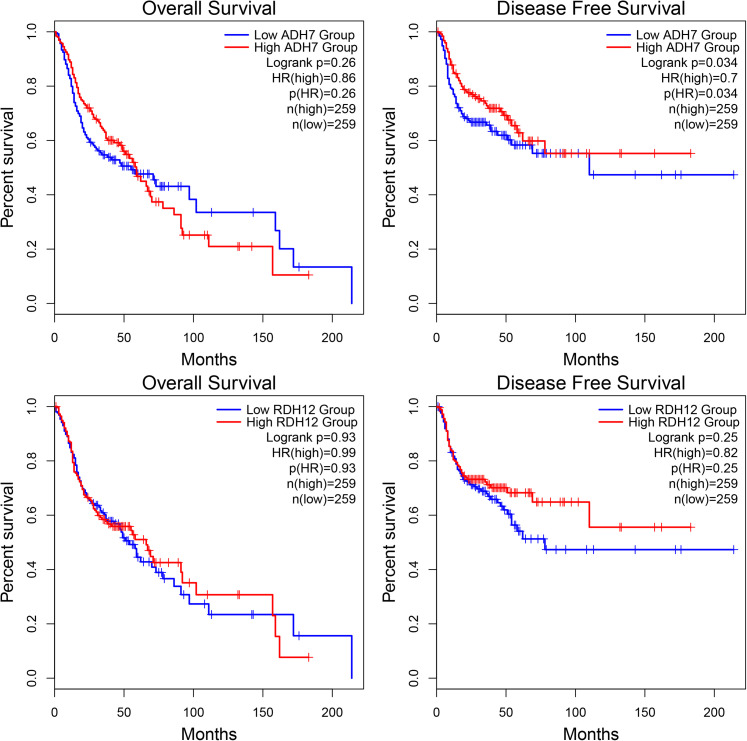
In order to further explore the prognostic value of the 2 potential target genes in HNSCC, Kaplan–Meier curve analysis was conducted with 518 HNSCC cases. High ADH7 expression was found to be significantly correlated with better DFS (HR = 0.7, P = .034), but no statistical significance was observed for OS (Figure 12A and B). And no statistically significant was found in the survival analysis for RDH12 (Figure 12C and D).
Figure 12.
The survival curves of 2 validated target genes of miR-21-5p associated with retinol metabolism.
Discussion
HNSCC is a worldwide public health problem, with an increasing incidence worldwide. In order to improve the diagnosis and treatment of HNSCC, it is necessary to find reliable predictive biomarkers and to understand the molecular mechanisms of the interaction of HNSCC.
It has been reported that miR-21-5p overexpression in multiple head and neck malignancies, such as OSCC, LSCC, and tonsil SCC,10,27–29 where it is correlated with poor prognosis.30–32 In addition, miR-21-5p may also play an oncogenic role in HNSCC, wherein it facilitates cancerous cell proliferation, invasion, and migration, and inhibits apoptosis.30,33 However, the molecular mechanisms related to Mir-21-5p in HNSCC have not been fully elucidated. To this end, we performed a meta-analysis to determine the prognostic value of miR-21-5p in HNSCC and investigated potential molecular mechanisms of action using bioinformatics.
Based on the miRNA expression data of HNSCC and control tissues from GEO datasets, TCGA database, and 6 published studies, we concluded that miR-21-5p levels were significantly elevated in HNSCC and is, therefore, likely oncogenic. In addition, data from TCGA indicated that miR-21-5p overexpression was significantly relevant with clinical staging, T staging, as well as smoking history in patients with HNSCC. Wang et al. 33 demonstrated that miR-21-5p was positively correlated with the apoptotic index and is useful in assessing the proliferative inhibition in tongue squamous cell carcinoma. Sondermann et al. 34 reported a prognostic role of miR-21-5p in predicting the recurrence of papillary thyroid carcinoma, while Zhou et al. 35 reported miR-21-5p as a diagnostic biomarker for laryngeal cancer.
In current study, 2 novel candidate target genes (ADH7 and RDH12), related to the retinol metabolism pathway, were identified to be the potential target genes of miR-21-5P in HNSCC. It is reported that retinoids, a hormone important for cellular differentiation, has the ability to regulate cell proliferation, differentiation, and epithelial-mesenchymal transformation through specific nuclear retinoic acid receptors.36,37 Retinol metabolism, through its main active metabolite retinoids, plays critical physiological roles in many biological processes, including vision, immunity, reproduction, neural function, gene transcription, cell proliferation, differentiation, and apoptosis.38,39 The metabolism and distribution of retinol are crucial for tumor formation and development, as it is involved in the inhibition of tumor growth and proliferation, and inhibits chemical-induced cancer.40,41 After uptake by target cells, retinol is first oxidized to retinaldehyde and then to its active derivative retinoids through several enzymatic steps.42,43
ADH7 (alcohol dehydrogenase 7), a maximal activity retinol dehydrogenase, participates in the synthesis of retinoids. Previous studies have reported that ADH7 can be used as a prognostic biomarker for non-small cell lung cancer 44 and lung squamous cell cancer, 45 and high expression of ADH7 may be associated with better prognosis of LUSC patients. Wei et al. reported that the ADH7 variant was associated with HNSCC risk. 46 Another study discovered that ADH7 might act as an essential role in the differentiation of melanoma. 47 RDH12 is an all-trans-retinol dehydrogenase that metabolizes retinoids. 48 Low expression of RDH12 can lead to dysregulation of cell proliferation and differentiation and the occurrence and development of gastric cancer. 49 Peng found that decreased expression of RDH12 is negatively correlated with tumor size and depth of cervical invasion and plays a critical role in the prognosis of cervical SCC. 50
So far, the correlation between miR-21-5p and ADH7 or RDH12 in HNSCC has not been mentioned. In current study, we found that ADH7 and RDH12 related to retinol metabolism were down-regulated in HNSCC. What's more, we further verified the down-regulated expression of ADH7 and RDH12 in HNSCC by RT-qPCR and IHC. In the survival analysis, we found that ADH7 was significantly correlated with the DFS of HNSCC patients. And high expression of ADH7 predicted better DFS in HNSCC patients. Therefore, we supposed that miR-21-5p may target at ADH7, RDH12 and participate in regulation of retinol metabolism, which might affect the prognosis of HNSCC.
In summary, due to the present study based on online, comprehensive, large databases, our findings are useful at the bioinformatics. There is no doubt that more evidence is needed for future research. However, the limitations of our study should be addressed. On the one hand, the number of sample size for validation of target genes was relatively small. On the other hand, there might have been hidden biases regarding grouping and blinding of data among the studies. In addition, we excluded studies in languages other than English and Chinese, which might contribute to an additional bias. Therefore, experimental studies on the molecular biological function of the target genes in miR-21-5p are needed to confirm our data. In addition, the diagnostic and prognostic value of miR-21-5P should be validated further in prospective studies in large patient cohorts.
Conclusions
In current study, we found that miR-21-5p was up-regulated in HNSCC compared to control. ADH7 and RDH12 are potentially important target genes of miR-21-5p. We assumed that miR-21-5p may target at ADH7, RDH12 and participate in regulation of retinol metabolism, which might affect the prognosis of HNSCC. And high expression of ADH7 may be associated with prognosis in patients with HNSCC. These findings might pave the way for molecular mechanism exploration and provide new targets for the clinical treatment of HNSCC. Further studies in vitro and in vivo are needed to elucidate the potential molecular mechanism underlying the involvement of miR-21-5p in HNSCC. And larger-scale studies are needed to verify its diagnostic value.
Supplemental Material
Supplemental material, sj-png-1-tct-10.1177_15330338221081245 for Investigation of miR-21-5p Key Target Genes and Pathways in Head and Neck Squamous Cell Carcinoma Based on TCGA Database and Bioinformatics Analysis by Mingjun Shen, Ziyan Zhou, Bai bei Li, Meixin Lv, Chunling Feng, Sixia Chen, Shuo Shi, Min Kang and Tingting Zhao in Technology in Cancer Research & Treatment
Supplemental material, sj-docx-2-tct-10.1177_15330338221081245 for Investigation of miR-21-5p Key Target Genes and Pathways in Head and Neck Squamous Cell Carcinoma Based on TCGA Database and Bioinformatics Analysis by Mingjun Shen, Ziyan Zhou, Bai bei Li, Meixin Lv, Chunling Feng, Sixia Chen, Shuo Shi, Min Kang and Tingting Zhao in Technology in Cancer Research & Treatment
Supplemental material, sj-docx-3-tct-10.1177_15330338221081245 for Investigation of miR-21-5p Key Target Genes and Pathways in Head and Neck Squamous Cell Carcinoma Based on TCGA Database and Bioinformatics Analysis by Mingjun Shen, Ziyan Zhou, Bai bei Li, Meixin Lv, Chunling Feng, Sixia Chen, Shuo Shi, Min Kang and Tingting Zhao in Technology in Cancer Research & Treatment
Supplemental material, sj-docx-4-tct-10.1177_15330338221081245 for Investigation of miR-21-5p Key Target Genes and Pathways in Head and Neck Squamous Cell Carcinoma Based on TCGA Database and Bioinformatics Analysis by Mingjun Shen, Ziyan Zhou, Bai bei Li, Meixin Lv, Chunling Feng, Sixia Chen, Shuo Shi, Min Kang and Tingting Zhao in Technology in Cancer Research & Treatment
Supplemental material, sj-docx-5-tct-10.1177_15330338221081245 for Investigation of miR-21-5p Key Target Genes and Pathways in Head and Neck Squamous Cell Carcinoma Based on TCGA Database and Bioinformatics Analysis by Mingjun Shen, Ziyan Zhou, Bai bei Li, Meixin Lv, Chunling Feng, Sixia Chen, Shuo Shi, Min Kang and Tingting Zhao in Technology in Cancer Research & Treatment
Supplemental material, sj-docx-6-tct-10.1177_15330338221081245 for Investigation of miR-21-5p Key Target Genes and Pathways in Head and Neck Squamous Cell Carcinoma Based on TCGA Database and Bioinformatics Analysis by Mingjun Shen, Ziyan Zhou, Bai bei Li, Meixin Lv, Chunling Feng, Sixia Chen, Shuo Shi, Min Kang and Tingting Zhao in Technology in Cancer Research & Treatment
Abbreviations
- HNSCC
head and neck squamous cell carcinoma
- OSCC
oral squamous cell carcinoma
- LSCC
laryngeal squamous cell carcinoma
- miR
microRNA
- RT-qPCR
real-time RT-polymerase chain reaction
- PPI
protein–protein interaction
- GO
gene ontology
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- GEO
gene expression omnibus
- TCGA
The Cancer Genome Atlas
- DAVID
Database for Annotation, Visualization and Integrated Discovery
- BP
biological process
- CC
cellular component
- MF
molecular function
Footnotes
Authors’ Contributions: TTZ and MK designed the study and accessed the relevant information. MJS and ZYZ designed the study, analyzed data, and wrote the manuscript. BBL and MXL collected and analyzed the data. CLF, SXC, and SS critically revised the manuscript. All authors read and approved the final manuscript.
Authors’ Note: The study was approved by the Human Research Ethics Committee of the First Affiliated Hospital of Guangxi Medical University, and the approval number was 2020(KY-E-181) and 2021(K-Y-E153). Written informed consent was obtained from all patients or their families prior to enrolment in the present study.
Declaration of Conflicting Interests: The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: The publication cost of this article was funded by grants from the National Natural Science Foundation of China (No. 81460460,81760542), and The Research Foundation of the Science and Technology Department of Guangxi Province, China (grant Nos. 2016GXNSFAA380252, 2018AB61001, and 2014GXNSFBA118114); the design of the study and collection, analysis, and interpretation of data was supported by the Research Foundation of the Health Department of Guangxi Province, China (No. S2018087), Guangxi Medical University Training Program for Distinguished Young Scholars (2017), and Medical Excellence Award Funded by the Creative Research Development Grant from the First Affiliated Hospital of Guangxi Medical University (2016); the writing of the manuscript was supported by Guangxi Medical High-level Talents Training Program, The central government guide local science and technology development projects (ZY18057006).
Availability of Data and Materials: All data generated or analyzed during this study are included in this published article.
ORCID iDs: Ziyan Zhou https://orcid.org/0000-0002-2306-6307
Supplemental Material: Supplemental material for this article is available online.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394-424. doi: 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- 2.Vigneswaran N, Williams MD. Epidemiologic trends in head and neck cancer and aids in diagnosis. Oral Maxillofac Surg Clin North Am. 2014;26(2):123-141. doi: 10.1016/j.coms.2014.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61(2):69-90. doi: 10.3322/caac.20107 [DOI] [PubMed] [Google Scholar]
- 4.Leemans CR, Braakhuis BJ, Brakenhoff RH. The molecular biology of head and neck cancer. Nat Rev Cancer. 2011;11(1):9-22. doi: 10.1038/nrc2982 [DOI] [PubMed] [Google Scholar]
- 5.Chen D, Cabay RJ, Jin Y, et al. MicroRNA deregulations in head and neck squamous cell carcinomas. J Oral Maxillofac Res. 2013;4(1):e2. doi: 10.5037/jomr.2013.4102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.A LM, L AC, M ME, et al. MiR-21 as prognostic biomarker in head and neck squamous cell carcinoma patients undergoing an organ preservation protocol. Oncotarget. 2017;8(6):9911-9921. doi: 10.18632/oncotarget.14253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dai X, Fang M, Li S, Yan Y, Zhong Y, Du B. miR-21 is involved in transforming growth factor β1-induced chemoresistance and invasion by targeting PTEN in breast cancer. Oncol Lett. 2017;14(6):6929-6936. doi: 10.3892/ol.2017.7007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wei WF, Han LF, Liu D, et al. Orthotopic xenograft mouse model of cervical cancer for studying the role of MicroRNA-21 in promoting lymph node metastasis. Int J Gynecol Cancer: Official J Int Gynecol Cancer Soc. 2017;27(8):1587-1595. doi: 10.1097/igc.0000000000001059 [DOI] [PubMed] [Google Scholar]
- 9.Yuan Y, Xu XY, Zheng HG, et al. Elevated miR-21 is associated with poor prognosis in non-small cell lung cancer: a systematic review and meta-analysis. Eur Rev Med Pharmacol Sci . 2018;22(13):4166-4180. doi: 10.26355/eurrev_201807_15410 [DOI] [PubMed] [Google Scholar]
- 10.Re M, Magliulo G, Gioacchini FM, et al. Expression levels and clinical significance of miR-21-5p, miR-let-7a, and miR-34c-5p in laryngeal squamous cell carcinoma. BioMed Res Int. 2017;2017:3921258. doi: 10.1155/2017/3921258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yoon JS, Kim G, Lee YR, et al. Clinical significance of microRNA-21 expression in disease progression of patients with hepatocellular carcinoma. Biomark Med. 2018;12(10):1105-1114. doi: 10.2217/bmm-2018-0096 [DOI] [PubMed] [Google Scholar]
- 12.Yu X, Wu Y, Liu Y, et al. miR-21, miR-106b and miR-375 as novel potential biomarkers for laryngeal squamous cell carcinoma. Curr Pharm Biotechnol. 2014;15(5):503-508. doi: 10.2174/1389201015666140519110616 [DOI] [PubMed] [Google Scholar]
- 13.Chen ZJ, Yu TW, Cabay RJ, et al. miR-486-3p, miR-139-5p, and miR-21 as biomarkers for the detection of oral tongue squamous cell carcinoma. Biomark Cancer. 2017 Jan;9:1-8. doi: 10.4137/bic.S40981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gao L, Zhang LJ, Li SH, et al. Role of miR-452-5p in the tumorigenesis of prostate cancer: a study based on the cancer genome Atl(TCGA), gene expression omnibus (GEO), and bioinformatics analysis. Pathol Res Pract. 2018;214(5):732-749. doi: 10.1016/j.prp.2018.03.002 [DOI] [PubMed] [Google Scholar]
- 15.Liu C, Tong Z, Tan J, Xin Z, Wang Z, Tian L. MicroRNA-21-5p targeting PDCD4 suppresses apoptosis via regulating the PI3K/AKT/FOXO1 signaling pathway in tongue squamous cell carcinoma. Exp Ther Med. 2019;18(5):3543-3551. doi: 10.3892/etm.2019.7970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dwee H, Gretz N., miRWalk2.0: a comprehensive atlas of microRNA-target interactions. Nat Methods. 2015;12(8):697. doi: 10.1038/nmeth.3485 [DOI] [PubMed] [Google Scholar]
- 17.Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25(1):25-29. doi: 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kanehisa M , Goto S, Sato Y, Furumichi M, et al. KEGG For integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2012;40(Database issue):D109-D114. doi: 10.1093/nar/gkr988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chandrashekar DS, Bhuwan B, Balasubramanya SAH, et al. UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017;19(8):649-658. doi: 10.1016/j.neo.2017.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vasaikar SV, Straub P, Wang J, et al. Linkedomics: analyzing multi-omics data within and across 32 cancer types. Nucleic Acids Res. 2018 Jan 4 ;46(D1):D956-D963. doi: 10.1093/nar/gkx1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brito JA, Gomes CC, Guimarães AL, et al. Relationship between microRNA expression levels and histopathological features of dysplasia in oral leukoplakia. J Oral Pathol Med. 2014;43(3):211-216. doi: 10.1111/jop.12112 [DOI] [PubMed] [Google Scholar]
- 22.Chang SS, Jiang WW, Smith I, et al. MicroRNA alterations in head and neck squamous cell carcinoma. Int J Cancer. 2008;123(12):2791-2797. doi: 10.1002/ijc.23831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yap T, Koo K, Cheng L, et al. Predicting the presence of oral squamous cell carcinoma using commonly dysregulated MicroRNA in oral swirls. Cancer Prev Res (Phila). 2018;11(8):491-502. doi: 10.1158/1940-6207.Capr-17-0409 [DOI] [PubMed] [Google Scholar]
- 24.Erkul E, Yilmaz I, Gungor A, Kurt O, Laryngoscope BMJT. MicroRNA-21 in laryngeal squamous cell carcinoma: diagnostic and prognostic features. Laryngoscope. 2017;127(2):E62-E66. doi: 10.1002/lary.26226 [DOI] [PubMed] [Google Scholar]
- 25.Gee HE, Camps C, Buffa FM, et al. hsa-mir-210 is a marker of tumor hypoxia and a prognostic factor in head and neck cancer. Cancer. 2010;116(9):2148-2158. doi: 10.1002/cncr.25009 [DOI] [PubMed] [Google Scholar]
- 26.Hu A, Jin X. Expression of mir-21 and mir-375 in laryngeal squamous cell carcinoma. Lin chuang er bi yan hou tou jing wai ke za zhi=J Clin Otorhinolaryngol Head Neck Surg. 2012;26(18):788-792.PMID: 23259291. [PubMed] [Google Scholar]
- 27.He Q, Chen Z, Cabay RJ, et al. microRNA-21 and microRNA-375 from oral cytology as biomarkers for oral tongue cancer detection. Oral Oncol. 2016 Jun;57:15-20. doi: 10.1016/j.oraloncology.2016.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang X, Gee H, Rose B, et al. Regulation of the tumour suppressor PDCD4 by miR-499 and miR-21 in oropharyngeal cancers. BMC Cancer. 2016. Feb 11;16:86. doi: 10.1186/s12885-016-2109-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lamperska KM, Kozlowski P, Kolenda T, et al. Unpredictable changes of selected miRNA in expression profile of HNSCC. Cancer Biomark. 2016;16(1):55-64. doi: 10.3233/cbm-150540 [DOI] [PubMed] [Google Scholar]
- 30.Min A, Zhu C, Pang S, Rajthala S, Costea DE, international SDJBr. MicroRNAs as important players and biomarkers in oral carcinogenesis. Biomed Res Int. 2015;2015:186904. doi: 10.1155/2015/186904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hu A, Huang JJ, Xu WH, et al. miR-21 and miR-375 microRNAs as candidate diagnostic biomarkers in squamous cell carcinoma of the larynx: association with patient survival. Am J Transl Res. 2014;6(5):604-613. PMID:25360224 [PMC free article] [PubMed] [Google Scholar]
- 32.Hedbäck N, Jensen DH, Specht L, et al. MiR-21 expression in the tumor stroma of oral squamous cell carcinoma: an independent biomarker of disease free survival. PLoS One. 2014;9(4):e95193. doi: 10.1371/journal.pone.0095193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Y, Zhu Y, Lv P, et al. The role of miR-21 in proliferation and invasion capacity of human tongue squamous cell carcinoma in vitro. Int J Clin Exp Pathol. 2015;8(5):4555-4563. PMID:26191145 [PMC free article] [PubMed] [Google Scholar]
- 34.Sondermann A, Andreghetto FM, Moulatlet AC, et al. MiR-9 and miR-21 as prognostic biomarkers for recurrence in papillary thyroid cancer. Clin Exp Metastasis. 2015;32(6):521-530. doi: 10.1007/s10585-015-9724-3 [DOI] [PubMed] [Google Scholar]
- 35.Zhou P, Zeng F, Liu J, Lv D, et al. Correlation between miR-21 expression and laryngeal carcinoma risks: a meta-analysis. J Evid Based Med. 2016;9(1):32-37. doi: 10.1111/jebm.12184 [DOI] [PubMed] [Google Scholar]
- 36.Sporn MB , Roberts AB. Role of Retinoids in Differentiation and Carcinogenesis. J Natl Cancer Inst. 1984 Dec;73(6):1381-7.PMID: 6595447. [PubMed]
- 37.Abu J, Batuwangala M, et al. Retinoic acid and retinoid receptors: potential chemopreventive and therapeutic role in cervical cancer. Lancet Oncol. 2005 Sep;6(9):712-20. doi: 10.1016/S1470-2045(05)70319-3 [DOI] [PubMed]
- 38.Noy N. Between death and survival: retinoic acid in regulation of apoptosis. Annu Rev Nutr. 2010 Aug 21;30:201-217. doi: 10.1146/annurev.nutr.28.061807.155509 [DOI] [PubMed] [Google Scholar]
- 39.Tang XH, Gudas LJ. Retinoids, retinoic acid receptors, and cancer. Annu Rev Pathol. 2011;6:345-364. doi: 10.1146/annurev-pathol-011110-130303 [DOI] [PubMed] [Google Scholar]
- 40.Barnard JH, Collings JC, Whiting A, Przyborski SA, Marder TB. Synthetic retinoids: structure-activity relationships. Chemistry (Weinheim an der Bergstrasse, Germany). 2009;15(43):11430-11442. doi: 10.1002/chem.200901952 [DOI] [PubMed] [Google Scholar]
- 41.Das BC, Thapa P, Karki R, et al. Retinoic acid signaling pathways in development and diseases. Bioorg Med Chem. 2014;22(2):673-683. doi: 10.1016/j.bmc.2013.11.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lidén M, Eriksson U. Understanding retinol metabolism: structure and function of retinol dehydrogenases. J Biol Chem. 2006;281(19):13001-4. doi: 10.1074/jbc.R500027200 [DOI] [PubMed] [Google Scholar]
- 43.Marceau G, Gallot D, Lemery D, Metabolism of retinol during mammalian placental and embryonic development. Vitam Horm. 2007;75:97-115. doi: 10.1016/s0083-6729(06)75004-x [DOI] [PubMed] [Google Scholar]
- 44.Wang P, Zhang L, Huang C, et al. Distinct prognostic values of alcohol dehydrogenase family members for non-small cell lung cancer. Med Sci Monit. 2018 May 29;24:3578-3590. doi: 10.12659/msm.910026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xie ZC, Li TT, Gan BL, et al. Investigation of miR-136-5p key target genes and pathways in lung squamous cell cancer based on TCGA database and bioinformatics analysis. Pathol Res Pract. 2018;214(5):644-654. doi: 10.1016/j.prp.2018.03.028 [DOI] [PubMed] [Google Scholar]
- 46.Wei S, Liu Z, Zhao H, et al. A single nucleotide polymorphism in the alcohol dehydrogenase 7 gene (alanine to glycine substitution at amino acid 92) is associated with the risk of squamous cell carcinoma of the head and neck. Cancer. 2010;116(12):2984-2992. doi: 10.1002/cncr.25058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen X, Yang M, Han W, et al. Differentiation-inducing and anti-proliferative activities of isoliquiritigenin and all-trans-retinoic acid on B16F0 melanoma cells: mechanisms profiling by RNA-seq. Gene. 2016;592(1):86-98. doi: 10.1016/j.gene.2016.07.052 [DOI] [PubMed] [Google Scholar]
- 48.Duester G. Biochemistry DGJEjo. Families of retinoid dehydrogenases regulating vitamin A function: production of visual pigment and retinoic acid. 2000;267(14):4315-4324. doi: 10.1046/j.1432-1327.2000.01497.x [DOI] [PubMed] [Google Scholar]
- 49.Kropotova ES, Zinov’eva OL, Zyrianova AF, et al. Expression of genes involved in retinoic acid biosynthesis in human gastric cancer. Mol Biol (Mosk). 2013;47(2):317-330. doi: 10.7868/s0026898413020079 [DOI] [PubMed] [Google Scholar]
- 50.Peng G, Dan W, Jun W, et al. Transcriptome profiling of the cancer and adjacent nontumor tissues from cervical squamous cell carcinoma patients by RNA sequencing. Tumour Biol, J Int Soc Oncodevelopmental Biol Med. 2015;36(5):3309-3317. doi: 10.1007/s13277-014-2963-0 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-png-1-tct-10.1177_15330338221081245 for Investigation of miR-21-5p Key Target Genes and Pathways in Head and Neck Squamous Cell Carcinoma Based on TCGA Database and Bioinformatics Analysis by Mingjun Shen, Ziyan Zhou, Bai bei Li, Meixin Lv, Chunling Feng, Sixia Chen, Shuo Shi, Min Kang and Tingting Zhao in Technology in Cancer Research & Treatment
Supplemental material, sj-docx-2-tct-10.1177_15330338221081245 for Investigation of miR-21-5p Key Target Genes and Pathways in Head and Neck Squamous Cell Carcinoma Based on TCGA Database and Bioinformatics Analysis by Mingjun Shen, Ziyan Zhou, Bai bei Li, Meixin Lv, Chunling Feng, Sixia Chen, Shuo Shi, Min Kang and Tingting Zhao in Technology in Cancer Research & Treatment
Supplemental material, sj-docx-3-tct-10.1177_15330338221081245 for Investigation of miR-21-5p Key Target Genes and Pathways in Head and Neck Squamous Cell Carcinoma Based on TCGA Database and Bioinformatics Analysis by Mingjun Shen, Ziyan Zhou, Bai bei Li, Meixin Lv, Chunling Feng, Sixia Chen, Shuo Shi, Min Kang and Tingting Zhao in Technology in Cancer Research & Treatment
Supplemental material, sj-docx-4-tct-10.1177_15330338221081245 for Investigation of miR-21-5p Key Target Genes and Pathways in Head and Neck Squamous Cell Carcinoma Based on TCGA Database and Bioinformatics Analysis by Mingjun Shen, Ziyan Zhou, Bai bei Li, Meixin Lv, Chunling Feng, Sixia Chen, Shuo Shi, Min Kang and Tingting Zhao in Technology in Cancer Research & Treatment
Supplemental material, sj-docx-5-tct-10.1177_15330338221081245 for Investigation of miR-21-5p Key Target Genes and Pathways in Head and Neck Squamous Cell Carcinoma Based on TCGA Database and Bioinformatics Analysis by Mingjun Shen, Ziyan Zhou, Bai bei Li, Meixin Lv, Chunling Feng, Sixia Chen, Shuo Shi, Min Kang and Tingting Zhao in Technology in Cancer Research & Treatment
Supplemental material, sj-docx-6-tct-10.1177_15330338221081245 for Investigation of miR-21-5p Key Target Genes and Pathways in Head and Neck Squamous Cell Carcinoma Based on TCGA Database and Bioinformatics Analysis by Mingjun Shen, Ziyan Zhou, Bai bei Li, Meixin Lv, Chunling Feng, Sixia Chen, Shuo Shi, Min Kang and Tingting Zhao in Technology in Cancer Research & Treatment