Summary
Cancer cell state transitions emerged as powerful mechanisms responsible for drug tolerance and overall poor prognosis; however, evidences were largely missing in oral cancer. Here, by multiplexing phenotypic markers of stem-like cancer cells (SLCCs); CD44, CD24 and aldehyde dehydrogenase (ALDH), we characterized diversity among multiple oral tumor tissues and cell lines. Two distinct patterns of spontaneous transitions with stochastic bidirectional interconversions on ‘ALDH-axis’, and unidirectional non-interconvertible transitions on ‘CD24-axis’ were observed. Interestingly, plastic ‘ALDH-axis’ was harnessed by cells to adapt to a Cisplatin tolerant state. Furthermore, phenotype-specific RNA sequencing suggested the possible maintenance of intermediate hybrid cell states maintaining stemness within the differentiating subpopulations. Importantly, survival analysis with subpopulation-specific gene sets strongly suggested that cell-state transitions may drive non-genetic heterogeneity, resulting in poor prognosis. Therefore, we have described the phenotypic-composition of heterogeneous subpopulations critical for global tumor behavior in oral cancer; which may provide prerequisite knowledge for treatment strategies.
Subject areas: Immunology, Biophysics, Cancer systems biology, Cancer
Graphical abstract
Highlights
-
•
Demonstrated population trajectory driven non-genetic heterogeneity in oral cancer
-
•
Created transition maps for subpopulations using discrete time Markov chain model
-
•
Demonstrated maintenance of stemness in cells undergoing differentiation
-
•
Uniquely expressed genes of these subpopulations associated with disease prognosis
Immunology; Biophysics; Cancer systems biology; Cancer, Stemness
Introduction
Although genetic heterogeneity undeniably confers cell growth and survival advantages, it alone is not sufficient to explain non-genetic heterogeneity aspects including phenotypic and functional heterogeneity, emergence of drug resistance, and metastasis phenotypes within genetically identical cancer sub-clones (Marine et al., 2020; Inde and Dixon, 2018; Jolly et al., 2018). RNA sequencing studies from multiple tumor-types showed that individual tumors with a similar genetic profile may exhibit transcriptomic heterogeneity (Rambow et al., 2019) that may arise because of multiple phenotypic cell states and their diverse interactions with tumor microenvironment (Tirosh et al., 2016; Dalerba et al., 2011; Puram et al., 2017).
The ability of genetically identical cells to exist and switch to multiple phenotypic states is a longstanding notion of developmental biology (Huang, 2009). However, cell state dynamics in context of generating phenotypic heterogeneity in cancer cell populations, independent of their genetic background has started to garner attention. Studies showed that cell state transitions can either occur spontaneously or because of perturbations driven by external triggers (Shlyakhtina et al., 2021; Nguyen et al., 2012; Beck and Blanpain, 2013; Marusyk et al., 2012). Cells existing in each phenotypic state may exert distinct functions, such as therapy resistance, that overall benefits the tumor (Pisco and Huang, 2015). Commonly studied cellular processes associated with phenotypic state transitions are epithelial to mesenchymal transitions (EMT) and cancer stemness, which may be inter-related (Jia et al., 2019). Stemness or stem-like cancer cells (SLCCs) properties in cancer is explained by cancer stem cell model where tumors are organized as hierarchical structures in which only SLCCs are capable of initiating tumors and therefore based on tumorigenic potential they occupy the top of the hierarchy. SLCCs divide asymmetrically to generate themselves and progenitor-like transient amplifying cells with high proliferative capacity as intermediate population. The bulk of the tumor population is generated by the differentiated cells with non-tumorigenic property and are therefore placed in bottom positions (Cole et al., 2020; Bonnet and Dick, 1997). Therefore, SLCCs give rise to the rest of the cellular phenotypes in the hierarchy through self-renewal and differentiation and contribute to intratumoral heterogeneity (Prasetyanti and Medema, 2017). Furthermore, recent evidences suggest that stemness in cancer cells can be achieved over time, even by differentiated cells, suggesting cellular plasticity (Gupta et al., 2011; Dirkse et al., 2019). Hence, the unidirectional rigid hierarchies of SLCC-models are widely debated (Meacham and Morrison, 2013; Barkley and Yanai, 2019).
Multiple studies proved the existence of SLCCs in several solid tumors including head and neck squamous cell carcinoma (HNSCC) using markers; CD44, CD24 and Aldehyde dehydrogenase enzyme activity (ALDH) (Keysar and Jimeno, 2010; Moreb, 2008; Singh et al., 2021); however, phenotypic diversity among SLCCs has been a major challenge in accurately defining these subpopulations (Tang, 2012; Medema, 2013; Gupta et al., 2011; Dirkse et al., 2019; Meacham and Morrison, 2013; Barkley and Yanai, 2019). Tumor cells with high-CD44 expression were the first identified SLCC phenotype in HNSCC (Prince et al., 2007). Later, cells with combined expression for high-CD44 and ALDH enzyme, i.e., CD44+Ve/ALDHHigh cells, were shown to enrich for HNSCC-SLCCs with EMT characteristics (Clay et al., 2010). This subpopulation was shown to have the ability to switch into the more differentiated non-EMT HNSCC cells (Clay et al., 2010). Simultaneously, cells with CD24Low/CD44High cell surface marker profile were reported as representative of SLCCs with EMT phenotypes in oral cancers having the ability to generate intermediate subpopulations with CD24high/CD44Hi phenotype and differentiated populations with CD44Low/CD24Low phenotype (Ghuwalewala et al., 2016; Han et al., 2014). Although studies in breast cancer showed that ALDHHigh and CD24Low/CD44High subpopulations represent distinct non-overlapping subpopulation (Liu et al., 2014); to the best of our knowledge, their combined comprehensive characterization in oral tumors has remained incomplete. Although, co-existence of heterogeneous oral-SLCCs with respect to EMT and their spontaneous interconversions has been reported (Biddle et al., 2011); despite these qualitative demonstrations, studies on population dynamics in a quantitative and unbiased manner are largely missing in oral cancer. Questions such as at what reproducibility these subpopulations arise and the relation between diverse subpopulations with respect to molecular states have remained unanswered in oral cancer.
Landmark studies demonstrated the emergence of therapy resistance among genetically identical clones (Kreso et al., 2013). In addition, under therapy induced stress, cancer cells are reported to harness phenotypic heterogeneity to transit from differentiated drug-sensitive states to stem-like drug-tolerant states as survival strategies (Sharma et al., 2010; Pisco and Huang, 2015; Goldman et al., 2015). Similar studies addressing phenotypic state transitions of oral cancer subpopulations in response to external stress factors such as chemotherapeutic drugs are beginning to be emphasized (Sharma et al., 2018); underscoring the importance of adaptive therapy response among heterogeneous oral cancer cells populations. Thus, SLCC-model provides a conceptual framework for interrogating complex cellular hierarchies, phenotypic heterogeneity and plasticity in cancer.
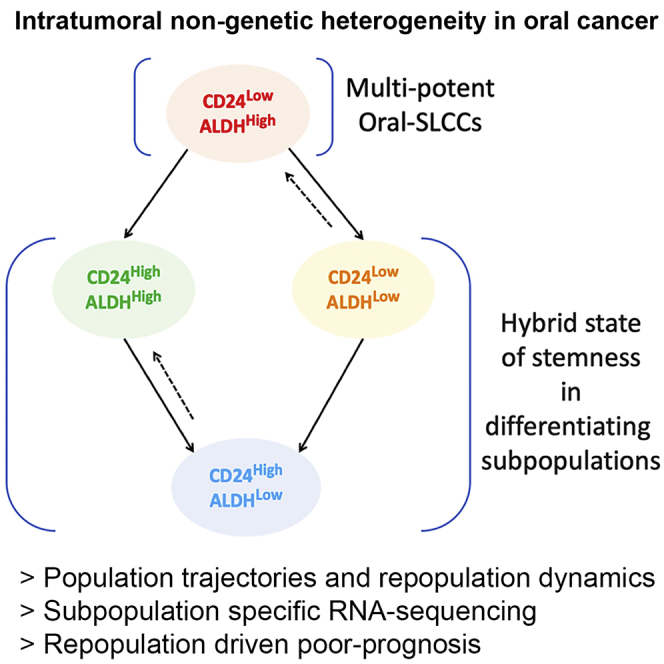
Here, we investigated the cellular states and transitions among oral cancer subpopulations by multiplexing phenotypic markers of SLCCs, CD44, CD24 and ALDH-activity and found the co-existence of ALDHHigh cells overlapping with both CD24High and CD24Low phenotypes in CD44-positive oral cancer cells. RNAseq data of these subpopulations demonstrated intermediate hybrid states of stemness, maintained within differentiating subpopulations, irrespective of their genetic diversity. Intriguingly, using the TCGA-HNSCC patient cohort, we have found poorer prognosis for those patients who had lower expression of transcriptomic signature specific to the reported oral-SLCCs with maximum potency to repopulate other phenotypes. Thus, our work showcased specific cellular transitions leading to transcriptomic heterogeneity and emergence of stemness during differentiation, influencing the tumor behavior in oral cancer patients.
Results
ALDHHigh cells exhibited both CD24Low and CD24High phenotypes
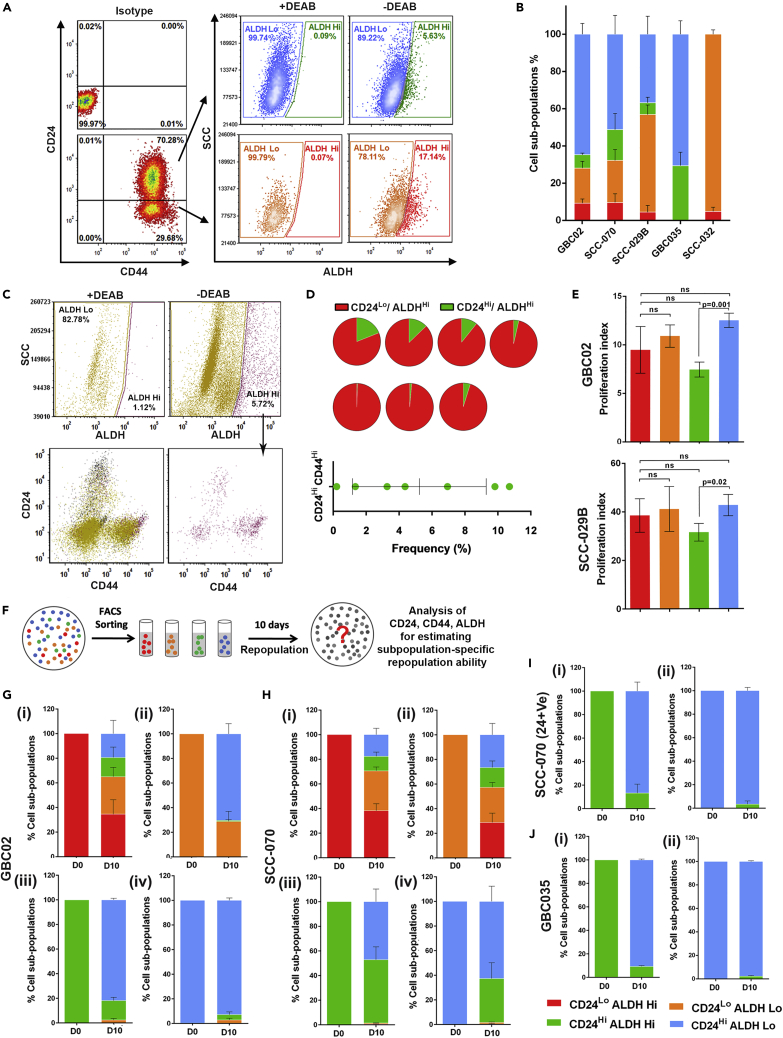
We first examined the co-expression of SLCC markers; CD44, CD24 and ALDH in five different oral cancer cell lines by flow cytometry. All tested cell lines were positive for CD44 marker expression which is in concordance with a previous report where CD44 was constitutively expressed on the surface of HNSCC cell lines (Pries et al., 2008). Conversely, for CD24, three of five cell lines; GBC02, SCC-070 and SCC-029 showed co-existence of both CD24High and CD24Low subpopulations (Figures 1A, 1B, S1A and S1B), whereas, GBC035 and SCC-032 showed predominantly CD24High and CD24Low phenotypes, respectively (Figures 1B, S1C and S1D). Analysis for ALDH-activity by ALDEFLUOR-assay showed that ALDHHigh cells were present in previously reported SLCC-phenotypes, i.e., CD24Low/CD44-positive cells (Ghuwalewala et al., 2016), and also in CD24High/CD44-positive phenotype of oral cancer cells (Han et al., 2014) (Figures 1A, 1B and S1). Hence, based on the differential status of these markers, we identified four subpopulations of CD44-positive cells with marker profiles, CD24Low/ALDHHigh, CD24Low/ALDHLow, CD24High/ALDHHigh, CD24High/ALDHLowwhich were termed ‘Red’, ‘Orange’, ‘Green’ and ‘Blue’ subpopulations, respectively. Frequencies of each of these subpopulations were analyzed in all tested cell lines. Importantly, ALDHHigh phenotypes (‘Red’ and ‘Green’) were maintained at lower frequencies, while ALDHLow subpopulations (‘Orange’ and ‘Blue’) were dominant across different cell lines (Figure 1B). Overall, these results emphasized the co-existence of ALDHHigh cells overlapping with both CD24High and CD24Low phenotypes in CD44-positive oral cancer cells. Furthermore, phenotypes observed in cell lines were also present in primary oral tumors (gating strategy, Figure S2). We found ALDHHigh cells to be primarily enriched in CD24Low phenotype (Figures 1C and1D, pie chart, Figure S3). However, ALDHHigh cells were also enriched in CD24High subpopulations in primary oral tumors at frequencies ranging from 0.3 to 11% (Figures 1C, 1D and S3).
Figure 1.
Subpopulations exhibited distinct repopulation ability
(A) Representative FACS dot plots of GBC02 cell line’s CD24/CD44 staining (Left bottom) with respect to Isotype control (Left top). ALDEFLUOR (ALDH) phenotype of CD24High/CD44+Ve subpopulation (Right top) and CD24Low/CD44+Ve subpopulation (Right bottom) in presence or absence of DEAB, an inhibitor of ALDH enzyme used as a negative experimental control.
(B) Frequency distribution of the four subpopulations CD24Low/ALDHHigh(Red), CD24Low/ALDHLow (Orange), CD24High/ALDHHigh (Green) and CD24High/ALDHLow (Blue) in five genetically distinct oral cancer cell lines.
(C) Representative FACS dot plots of a freshly resected and digested patient tumor. Hematopoietic lineage negative cells segregated into ALDHHigh and ALDHLow cells in presence or absence of DEAB (top). CD24/CD44 phenotype of all lineage negative cells (bottom left). CD24/CD44 phenotype of ALDHHigh cells only (bottom right).(D) Pie charts showing frequencies of CD24High/ALDHHigh (Green) and CD24Low/ALDHHigh (Red) subpopulations from seven freshly resected human oral tumor samples and frequencies of CD24High/ALDHHigh cells from all seven oral tumor patient samples.
(E) Graphs showing proliferation index of GBC02 and SCC029 cell lines based on CellTrace-Violet dye dilution assay.
(F) Schematic of repopulation experiments.
(G–J) Repopulation frequencies of each subpopulation in (I) Red (ii) Orange (iii) Green and (iv) Blue sorted cells on Day-0 and Day-10 of sorting in GBC02 (F), SCC-070 (G), SCC-070 (CD24+) sub-line (H) and GBC035 (I). Error bars represent mean ± SEM from three biological repeats.
Collectively, results provided evidence for existence of heterogeneous ALDHHigh cells with diverse CD24 phenotypes in primary oral tumors similar to in vitro cell lines.
Subpopulations exhibited distinct repopulation abilities
Maintenance of subpopulations at different frequencies in a mixed population could be because of differences in their proliferative potential. However, in ‘Cell Trace-Violet’ dye dilution assay, we did not observe significant differences in proliferation indices among the four subpopulations in both tested cell-lines, GBC02 and SCC-029B (Figure 1E). This observation prompted us to probe the repopulation ability of these four subpopulations. The term ‘repopulation’ used here is depicting the ability of each of the subpopulation to possibly generate any of the four tested phenotypes during independent regrowth when plated individually after cell sorting (Figure 1F). As explained in the methods section, the repopulation-ability was tested by sorting each of these subpopulations after plating them separately at low density and allowed to proliferate and form colonies. Cells were collected and tested for the combined CD24, CD44 expression and ALDH-activity. Repopulation results from GBC02, SCC-029B and SCC-070 cell lines showed that the ‘Red’ subpopulation was most ‘potent’ which efficiently reproduced all four subpopulations. Of interest, ‘Green’ subpopulation could only give rise to itself and its ALDHLow counterpart, showing ‘commitment’ to generate only CD24High phenotype in all tested cell lines (Figures 1G, 1H, S4A, S4B and S4C). Although the ALDHLow-‘Orange’ subpopulations efficiently repopulated the ALDHLow subpopulations of ‘Orange’ and ‘Blue’ phenotypes; the ‘Blue’ subpopulation showed the phenomena of ‘CD24High specification’ by repopulating itself with higher efficiency (Figures 1G, 1H, S4A, S4B and S4C). Of note, the ALDHLow (‘Orange’ or ‘Blue’) subpopulations showed higher plasticity and generated ALDHHigh phenotypes at higher frequency in SCC-070 cell line as compared to other cell lines (Figures 1H and S4B). Distinct from other cell lines, SCC-032 could not generate the CD24High (‘Green’ or ‘Blue’) from CD24Low(‘Red’ or ‘Orange’) subpopulations. Also, similar to the ‘Orange’ subpopulation of SCC-070, the ‘Orange’ subpopulations of SCC-032 demonstrated the capacity to recapitulate both ‘Red’ and ‘Orange’ subpopulations (Figure S4D).
So far, our repopulation results with CD44High/CD24Low were in concordance with previous reports in oral cancer cell lines, where this subpopulation represented putative Oral-SLCC phenotype whereas, CD44High/CD24High was also suggested to be an intermediate cell state (Ghuwalewala et al., 2016) with tumorigenic ability (Han et al., 2014). To confirm the property of CD24High cells to be more committed subpopulation, we developed a separate sub-line termed‘SCC-070 (CD24+)’, by sorting and propagating CD24High cells repopulated from ‘Red’ subpopulation of SCC-070 cells. Interestingly, though CD24High cells were originated from CD24Low subpopulation, these cells did not regenerate CD24Low cells or parent phenotype (Figures 1I and S5A(1)), confirming the committed property of CD24High subpopulation. Furthermore, the specification of ‘Blue’ subpopulation could also be confirmed (Figures 1I and S5A(2)). These results were further supported by an independent patient derived cell line, GBC035, which predominantly has phenotypically homogeneous CD24High phenotype. Similar to SCC-070 (CD24+) sub-line, sorted ‘Green’ and ‘Blue’ subpopulations of GBC035 also showed ‘Green’ subpopulation to be committed to generate only CD24High subpopulations; whereas ‘Blue’ subpopulation to have specification for generating itself (Figures 1J and S5B).
Overall, two distinct patterns of repopulations in oral cancer cells were observed; 1) ‘Fixed’ CD24-Axis, where transitions from CD24Low to CD24High were largely unidirectional and non-interconvertible, and 2) ‘Plastic’ ALDH-axis, where transitions from ALDHHigh to ALDHLow and vice-versa were frequent, although with certain cell-line specific trends. Therefore, in contrast to breast cancer studies where cells with ALDHHigh and CD24Low/CD44High phenotype represent distinct subpopulations (Liu et al., 2014), we are reporting the overlapping subpopulations on ALDH-axis in oral cancer. Furthermore, subpopulation-dichotomy for ALDHHigh subpopulation having more potent CD24Low subpopulations with more committed CD24High counterpart in oral cancer cells was observed.
All subpopulations exhibited enrichment of spheroid forming cells and maintained repopulation patterns
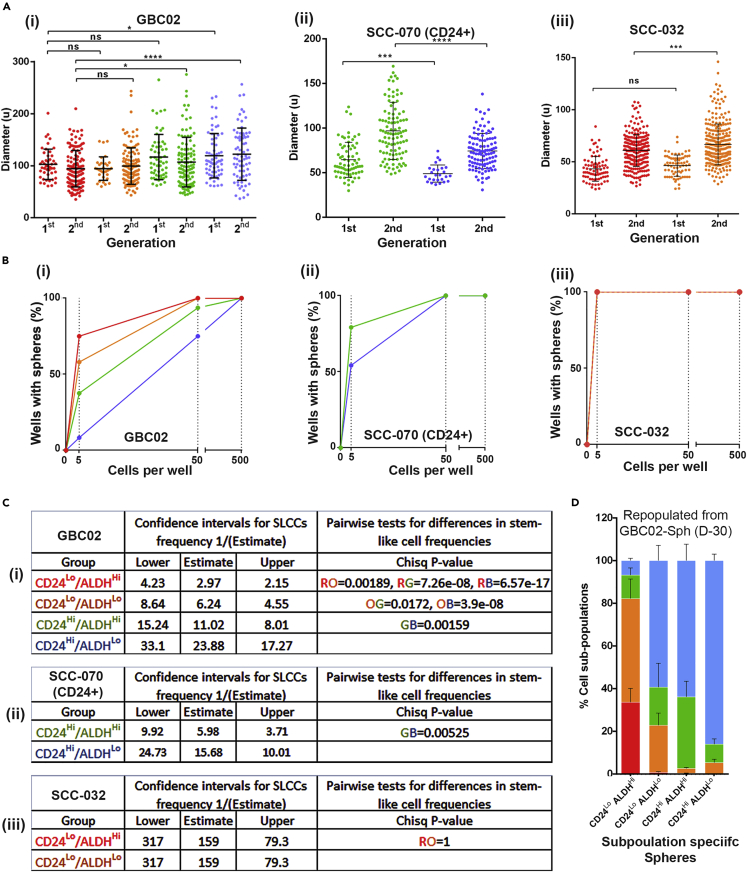
We next tested all four subpopulations for in vitro spheroid formation assays in three cell lines to demonstrate the stem/progenitor-like or differentiation properties among these subpopulations. The spheroids are reported to be generated by only stem/progenitor-like cells, capable of long-term regenerating ability in adherence independent, serum free conditions and successfully utilized to identify and enumerate SLCCs in a variety of cancers including oral cancer (Pastrana et al., 2011; Ghuwalewala et al., 2016; Biddle et al., 2011). Surprisingly all four subpopulations from GBC02 cell lines (Figure 2A-i), ‘Green’ and ‘Blue’ subpopulations of SCC-070 (CD24 + Ve) sub-line (Figure 2A-ii) and ‘Red’ and ‘Orange’ subpopulations of SCC-032 (Figure 2A-iii) demonstrated spheroid-forming potential in tested two generations of spheroid propagation. Next, more critical evaluation of their spheroid-forming efficiencies was performed by limiting dilution spheroid formation assays. This revealed the highest enrichment of spheroid-forming cells (1 in 3 cells) in the ‘Red’ subpopulation. However, interestingly, the ‘Orange’ (1 in 6 cells), ‘Green’ (1 in 11 cells) and ‘Blue’ (1 in 24 cells) subpopulations also demonstrated the enrichment of spheroid-forming cells (Figures 2B-i and 2C-i). These differences in efficiency were significant as tested by pairwise comparisons (Figure 2C-i, last column). Similar to these observations; SCC-070 (CD24 + Ve) sub-line also demonstrated spheroid-forming efficiency of ‘Green’ (1 in six cells) to be higher than ‘Blue’ subpopulation (1 in 16 cells) (Figures 2B-ii and 2C-ii). Interconverting ‘Red’ and ‘Orange’ subpopulations of SCC032 showed similar (1 in 159 cells) spheroid-forming efficiency (Figures 2B-iii and 2C-iii).
Figure 2.
Enrichment of cells with spheroid forming ability by all four subpopulations
(A) Size of the four subpopulations spheroids from GBC02, SCC-070 CD24+ and SCC-032 cell lines.
(B) Second generation spheroid formation frequencies in limiting dilution assay from GBC02, SCC-070 CD24+ and SCC-032 cell lines.
(C) Estimation of SLCC frequencies in each subpopulation of (1) GBC02, (2) SCC-070 CD24+ and (3) SCC-032 cell lines using ELDA software.
(D) Frequencies of each subpopulation in colonies generated from Red, Orange, Green and Blue subpopulation specific single spheroids after 30 days (D-30) of sorting. Error bars represent mean ± SEM from four biological repeats and p values are denoted as: ∗∗∗∗p < 0.0001; ∗∗∗p < 0.001; ∗∗p < 0.01; ∗p < 0.05.
We next tested if clonal spheroid cultures of these four subpopulations exhibit similar patterns of repopulation as previously observed for 2D cultures (Figure 1G). Intriguingly, with close resemblance to the repopulation ability in 2D colonies, each clonal spheroid cultures of these subpopulations exhibited a similar pattern of repopulation (Figures 2D, S6A, and S6B). Spheroids generated from ‘Red’ subpopulation remained the most potent to generate all four subpopulations, whereas the ‘Green’ subpopulation spheroids remained committed to generate CD24Highsubpopulations and ‘Blue’ demonstrated its specificity to mainly maintain itself, efficiently (Figures 2D and S6B). These results suggested that, 'Red' subpopulation exerts its supremacy with highest spheroid forming efficiency and repopulation potency, compared to other subpopulations. However, importantly other subpopulations too showed spheroid formation potential, albeit at a lower frequency than the ‘Red’. Overall, data from all different cell lines emphasized that CD24High cells possessed the ability to initiate spheroid formation despite being committed to repopulate only CD24High cells.
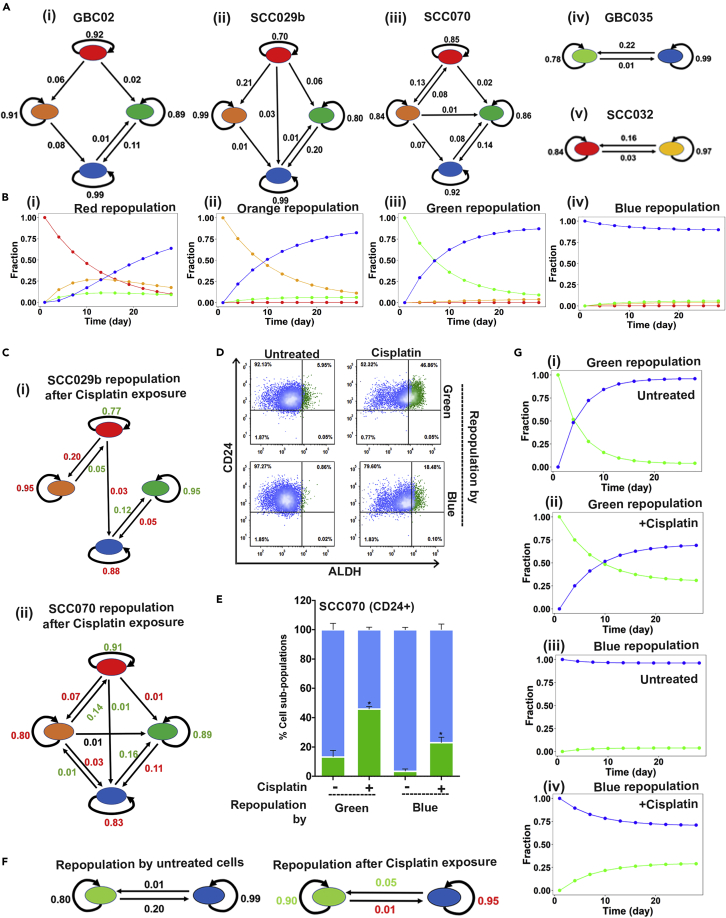
Quantitative model of phenotype-transition dynamics
Next, to demonstrate all possible transition paths which these subpopulations may take in terms of self-maintenance or conversions resulting in the observed repopulation dynamics, we applied a Discrete Time Markov Chain (DTMC) model for a simple linear description of the evolution of phenotypic heterogeneity based on their mutual transitions. Assuming that the transition rates are constant over time, we used CellTrans (Buder et al., 2017) package to infer daily transition probabilities of all four subpopulation among each other in GBC02, SCC- 029B, SCC-070, GBC035 and SCC032 cell lines. Results were represented as transition graphs (Figures 3A- i, 3ii, 3iii and3iv). All subpopulations showed high sustenance probabilities (probability with which the cells maintain their identity), with ‘Blue’ subpopulations having the maximum. Also, the majority of transition paths led to the ‘Blue’ subpopulation with very low transition probability from ‘Blue’ subpopulation to other subpopulations (efflux probability). Thus, quantitative modeling combined with the experimental observation explained the low efficiency of transition to other subpopulations from sorted ‘Blue’ subpopulation.
Figure 3.
Spontaneous and Cisplatin induced population dynamics
(A) The phenotypic transition graphs of (1) GBC02, (2) SCC029, (3) SCC070, (4) GBC035 and (5) SCC032. Arrows represent the direction of transition and the corresponding number show the calculated transition probabilities. This number in green and red colors represents the gain and loss in transition probabilities in response to Cisplatin as compared to untreated condition. Lack of arrows between two subpopulations indicates that the transition probability in that direction is less than 0.01.
(B) The phenotypic transition trajectories predicted based on the transition graph for GBC02 cell line’s (1) Red (2) Orange (3) Green and (4) Blue subpopulations repopulations.
(C) The phenotypic transition graphs of SCC029 and SCC070 subpopulations treated with 2μM Cisplatin showing increased influx and sustenance probability of the green state and its reduced outflux, overall increasing the fraction of green subpopulation underCisplatin treatment.
(D) Representative dot plots of SCC070 CD24+ sub-line’s repopulation showing increase in Green cells in sorted Green (top) and Blue (bottom) subpopulations in untreated and 2μM Cisplatin treated conditions.
(E) Repopulation frequencies of SCC070 CD24+ sub-line’s Green and Blue subpopulations in untreated versus Cisplatin (2μM) treated conditions. Statistical comparisons were made between each subpopulation in untreated versus Cisplatin treatment conditions (e.g. Green untreated versus Green Cisplatin treated). Error bars represent mean ± SEM from four biological repeats and ∗ indicates p value ≤0.05.
(F) The phenotypic transition graph of Green and Blue subpopulations of SCC070 (CD24+) sub-line in untreated and Cisplatin treated conditions.
(G) The phenotypic transition trajectories of (1, 3) Untreated Green and Blue cells and (2, 4) Cisplatin treated Green and Blue cells repopulations of SCC070 CD24+ sub-line showing increased fraction of Green cells in the population upon Cisplatin treatment.
Using these calculated transition probabilities, we next generated trajectories of evolution of population heterogeneity starting from sorted homogeneous subpopulations using the calculated transition probabilities. As shown in Figure 3B for GBC02 cell line, when starting from a ‘Red’ sorted subpopulation, reduction in the fraction of ‘Red’ subpopulation was associated with an initial rise and fall in the ‘Orange’ subpopulation and rise in ‘Blue’ subpopulation, indicating a transition from CD24Low to CD24High phenotype (Figure 3B- i). Furthermore, initial population of homogeneous CD24High phenotypes (‘Blue’ or ‘Green’ subpopulations) failed to give rise to CD24Low phenotypes, whereas the homogeneous ‘Green’ subpopulations CD44+/CD24High/ALDHHigh could give rise to CD44+/CD24High/ALDHLow ‘Blue’ subpopulation and vice-versa (Figures 3B-iii and3iv). These trajectories revealed the direction(s) of repopulation ability along the ‘CD24 and ALDH-axes’. Together, these results, obtained across different cell lines (Figures S7A-S7D) were in consistence with our experimental observations of unidirectional transition on ‘CD24-axis’ and bidirectional plasticity on ‘ALDH-axis’.
Next, we performed sensitivity analysis to characterize the effects of cell-to-cell variability in transition rates on emergence of observed phenotypic compositions. To do so, we shuffled the values in the transition matrices (that contain calculated transition rates), i.e. these in silico experiments created ‘hypothetical’ scenarios where, for instance, the influx and efflux rates were swapped and the effect on phenotypic distributions was calculated (see STAR Methods section for more details) (Figures S7E–S7N). We found that as long as high influx and low efflux of the ‘Blue’ subpopulation rates were maintained, blue population retained its dominance. These observations identified the necessary and sufficient conditions enabling the ‘Blue’ subpopulation to be more dominant than the red, orange, and green.
Cisplatin alters phenotype-interconversion on ALDH-Axis
Our spontaneous repopulation results showed that ALDHHigh and ALDHLow cells repopulate each other with inter-convertible transitions (‘Red’ to ‘Orange’ or ‘Green’ to ‘Blue’ subpopulations); whereas, CD24Low cells repopulates CD24High cells, (Red to Green or Orange to Blue) without interconvertibility. Previous studies showed that external cues such as chemotherapy-induced stress propels cells to transit into a more drug tolerant state (Pisco and Huang, 2015; Sharma et al., 2010) often associated with stem-like phenotypes. Therefore, we explored the influence of chemotherapy on state transitions of the four subpopulations from SCC-029B and SCC-070 cell lines, without killing the drug sensitive cells or selecting drug resistant clones. We administered sub-lethal, low dose of Cisplatin (2μM) as a pulse, to model the effect of external environmental drug stress. Exposure to Cisplatin during repopulation resulted in ‘Red’ subpopulation retaining higher frequency of itself and generating significantly higher frequency of ‘Green’ subpopulations as compared to the ‘Red’ subpopulation unexposed to Cisplatin, for both cell lines (Figures S8 and S9). Similarly, isolated ‘Green’ subpopulation exposed to Cisplatin during repopulation retained itself at a significantly higher frequency as compared to untreated in both cell lines (Figures S8 and S9). Moreover, the ALDHLow (‘Orange’ and ‘Blue’) subpopulations showed significant increase in ALDHHigh subpopulations (‘Red’ and ‘Green’ respectively) when exposed to Cisplatin compared to untreated cells (Figures S8 and S9). Therefore, our data suggested that the Cisplatin treatment enriches ALDHHigh subpopulations which may be a result of conversion from ALDHLow subpopulations, possibly because of their intrinsic plasticity. Furthermore, it is important to note that even under drug induced stress, CD24High subpopulation retained its commitment and did not generate CD24Low subpopulations.
Using the quantitative approach described earlier, we compared the transition rates in response to Cisplatin treatment with those in repopulation of treatment naive subpopulations. Cisplatin treatment increased the sustenance rates for both ALDHHigh subpopulations (‘Red’ and ‘Green’) and reduced the efflux probability from ‘Red’ or ‘Green’ subpopulation to any other subpopulations in both SCC-029B and SCC-070 cell lines (Figures 3C-i and3ii). Further supporting the transition of ALDHLow subpopulations to ALDHHigh subpopulations upon Cisplatin treatment, repopulation from the ‘Orange’ and ‘Blue’ subpopulations in both SCC-029B and SCC-070 cell lines were in stark contrast to observations in untreated conditions. The quantitative model demonstrated a higher probability of transition from ‘Orange’ to ‘Red’ and ‘Blue’ to ‘Green’ subpopulations which was either absent or at a lower probability in untreated cells (Figure S10).
To confirm that the bidirectional plasticity of ALDH-Axis is harnessed by ALDHLow cells to accumulate ALDHHigh subpopulation but the CD24-axis remains unidirectional even in response to Cisplatin, we next treated Green and Blue subpopulations of SCC-070 (CD24 + Ve) sub-line. As anticipated, the ‘Green’ subpopulation maintained itself at higher frequency and ‘Blue’ subpopulation repopulated higher frequency of ‘Green’ cells with Cisplatin. Importantly, both ‘Blue’ and ‘Green’ subpopulations failed to give rise to CD24Low phenotypes even in response to Cisplatin treatment (Figures 3D and 3E). Assessment of this population dynamics revealed reduced sustenance and increased efflux probabilities of ‘Blue’ subpopulation, but increased sustenance probability of the ‘Green’ subpopulations in Cisplatin treated conditions (Figure 3F). Dynamic trajectories plotted to predict the evolution of subpopulations under untreated and Cisplatin treated conditions predicted that, Cisplatin treated ‘Green’ subpopulation maintained itself at higher fraction, whereas ‘Blue’ subpopulation generated ‘Green’ subpopulation at higher probabilities (Figures 3G-ii and3G-iv) as compared to their untreated counterparts (Figures 3G-i and 3G-iii). These results strongly demonstrated the plastic nature of ALDH-Axis to be responsible for accumulation of ALDHHigh subpopulations under low dose, Cisplatin treatment condition; however unidirectional transitions along the CD24-Axis (CD24Low to CD24High) were strictly maintained.
Transcriptome profiling revealed hybrid cell states among subpopulations
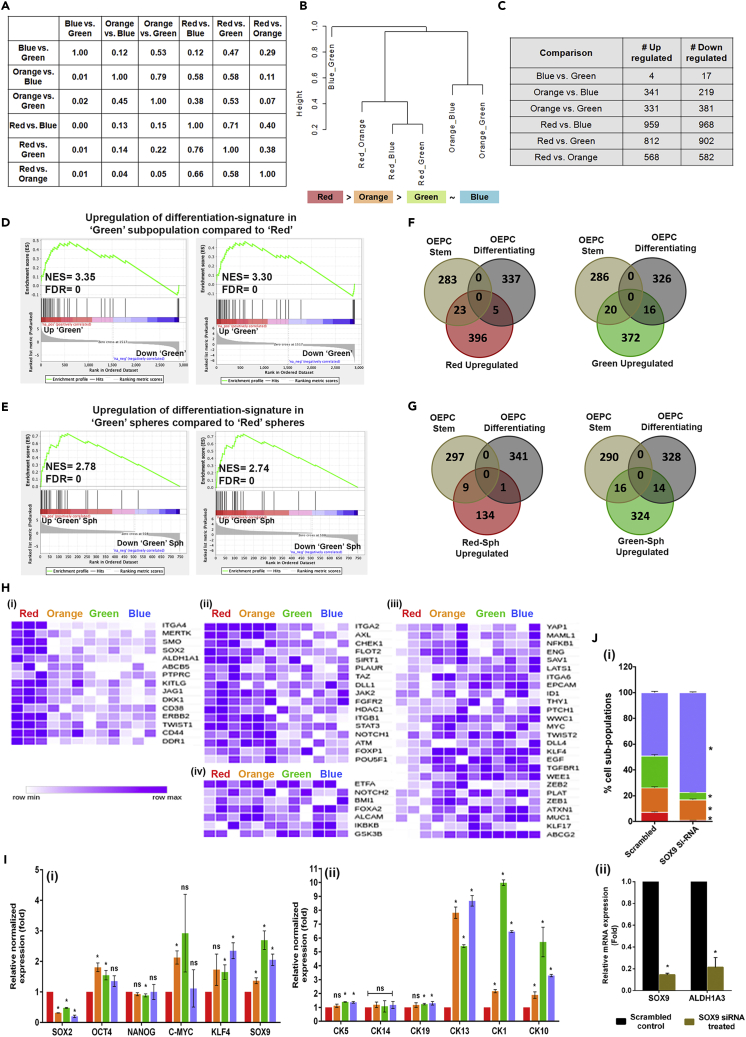
The observed dynamic relationship among subpopulations prompted us to determine the molecular association between them. Toward this, we performed RNA sequencing of these four subpopulations isolated from 2D cultures in three biological repeats and 3D-spheriods generated from ‘Red’ and ‘green’ subpopulations of GBC02. Based on the overlap between sets of differentially expressed genes (DEGs) obtained from pair-wise comparisons we found varying degrees of overlap in DEGs between different comparisons (Figure 4A). For instance, 79% of overlap was seen for DEGs between ‘Orange’ versus ‘Blue’ subpopulations and ‘Orange’ versus ‘Green’ subpopulations. Similarly, 76% of DEGs for ‘Red’ versus ‘Green’ subpopulations were found in ‘Red’ versus ‘Blue’ DEG analysis. Conversely, 71% of DEGs noted in ‘Red’ and ‘Blue’ were witnessed in ‘Red’ versus ‘Green’ DEGs-list (Figure 4A). This pattern of overlap pointed out that ‘Red’ subpopulation may be approximately equidistant from ‘Blue’ and ‘Green’ subpopulations. Similarly, ‘Orange’ may also be approximately equidistant from ‘Green’ and ‘Blue’ subpopulations. Furthermore, the DEGs between ‘Blue’ and ‘Green’ subpopulations had very few genes in common when overlapped with any of the five other DEG lists (see column 1 in Figure 4A), indicating that these subpopulations may be transcriptionally most similar to one another. These overlaps in DEGs, coupled with quantifying distances among these comparisons enabled us in arranging these subpopulations in a hierarchical structure with ‘Red’ subpopulations being the most upstream, followed by ‘Orange’, ‘Green’ and ‘Blue’ (Figure 4B).
Figure 4.
Subpopulation specific transcriptome analysis
(A and B) Overlap in DE-Gs of different pairwise comparisons. Proportion of overlap between each pairwise comparison (A).Hierarchical structure of different cell subpopulations (B).
(C) Pairwise comparisons of differentially expressed genes of four subpopulations of GBC02 cell line.
(D) Gene set enrichment analysis (GSEA) with genes up-regulated in ‘Green’ subpopulation as compared to ‘Red’ subpopulation from GBC02 monolayer cultures showing enrichment of genes involved in Keratinization and Cornified envelope formation.
(E) Gene set enrichment analysis (GSEA) with genes up-regulated in Green subpopulation as compared to Red subpopulation from GBC02 3D spheroids showing enrichment of genes involved in keratinization and cornified envelope formation.
(F and G) Venn diagram showing overlap of basal OEPC and basal differentiating gene sets with Red versus Green comparison upregulated genes in Red and Green sorted subpopulations monolayer cultures (F) and Red and Green 3D spheroids from GBC02 cell line (G).
(H) Heatmaps of mean normalized expression values from GBC02 monolayer cultures for genes expressed distinctly in (1) ‘Red’ (2) both ‘Red’ and ‘Orange’ subpopulations (3) commonly in ‘Orange’, ‘Green’ and ‘Blue’ subpopulations and (4) among all the four subpopulations.
(I) qRT-PCR of the four sorted subpopulations from GBC02 monolayer cultures for various (1) stemness genes and (2) differentiating cytokeratins.
(J) (1) Graph showing decreased frequency in ALDHHigh subpopulations in SCC-070 cell line upon siRNA mediated silencing of SOX9. Statistical comparisons were made between each subpopulation in scrambled versus SOX9 siRNA treatment conditions (ex; Green scrambled versus Green SOX9 siRNA). Error bars represent mean ± SEM from three biological repeats and ∗ indicates p value ≤0.05. (2) qRT-PCR after siRNA knockdown of SOX9 in SCC-070 cell line. Error bars represent mean ± SD from three biological repeats and ∗ indicates p value ≤0.05 for qRT-PCR experiments.
Because the ‘Red’ subpopulation was the most distinct from other subpopulations; the pairwise comparisons were made to tabulate DE-Gs between ‘Orange’, ‘Green’, ‘Blue’ with ‘Red’ subpopulations. This showed differentially expression of 568 upregulated and 582 downregulated genes between ‘Red’ and ‘Orange’ subpopulations; 812 upregulated and 908 downregulated genes between ‘Red’ and ‘Green’ subpopulations and 959 upregulated and 968 downregulated genes between ‘Red’ and ‘Blue’ subpopulations with log fold change of more than two and adjusted p value of less than 0.05 (Figure 4C and Table S1). Differentially expressed genes from pairwise comparisons are also depicted as volcano plots (Figure S11). Interestingly, ALDHHigh (‘Red’ and ‘Green’) subpopulations showed significantly higher expression of ALDH1A3 as compared with their respective ALDHLow subpopulations, ‘Orange’ and ‘Blue’ (Figure S11). Gene set enrichment analysis (GSEA) was performed from larger list of DEGs (q < 0.1, log2FC > 0.5) between Red and Orange, Red and Green, Red and Blue subpopulations to derive biological inferences of differentially expressed genes among subpopulations. We observed depletion for GPCR-related gene set in all three subpopulations compared to the ‘Red’ subpopulation, indicating loss of GPCR-related signaling may change transcription state from ‘Red’ to other subpopulations (Figure S12A). Intriguingly, we observed enrichment of signatures related to keratinization and cornified envelope with the DEGs in ‘Green’ compared to the ‘Red’ subpopulations (Figure 4D). Similarly, 3D-spheroid cultures of the ‘Green’ subpopulation showed an enrichment of these signatures compared to 3D-spheroid cultures generated from ‘Red’ subpopulation (Figure 4E). This suggested the onset of differentiation in ‘Green’ subpopulation, even when maintained in 3D spheroid cultures.
A recent study by Jones et al. (2019) reported diversity within basal layer oral epithelial progenitor cells (OEPCs) in normal oral mucosa of mouse, using single cell RNA sequencing (Jones et al., 2019). This study has demonstrated the maintenance of stemness with onset of differentiation in subsets of basal layer keratinocytes in mouse oral mucosa (Jones et al., 2019). Therefore; to explore similarities, we overlapped our DEGs with the gene sets of stem/progenitors and differentiating cells of basal layer keratinocytes. Interestingly, from the DEGs between ‘Red’ and ‘Green’ subpopulations; the upregulated genes in ‘Red’ subpopulation had significantly higher overlap with OEPC-stem cell cluster (Figure 4F); whereas, the upregulated genes in ‘Green’ subpopulation had almost equal number of genes overlapped to both OEPC-stem as well as differentiating cell clusters (Figure 4F). Very similar patterns were obtained with DEGs between 3D-spheroids generated from ‘Red’ and ‘Green’ subpopulations (Figure 4G). Similar to the ‘Green’ subpopulation, upregulated genes in ‘Orange’ and ‘Blue’ subpopulations as compared to the ‘Red’ also had equal overlaps with the OEPC-stem and differentiating cell clusters (Figure S12B). Furthermore, we explored the enrichment of transcription factors (TFs) specific to stem/progenitors and differentiating clusters from this study. Interestingly, all four subpopulations showed mosaic expression pattern of these TFs (Figure S12C). This prompted us to explore if our four subpopulations exhibit the signature of stem cells. Hence, we performed single sample gene set enrichment analysis (ssGSEA) utilizing the previously reported gene-sets for Adult Tissue Stem Cells (ATSC) [30]. All four subpopulations as well as 3D-spheroids of ‘Red’ and ‘Green’ subpopulations showed positive enrichment with no significant differences among them for the stemness signature (Figure S12D). Encouraged by these results, we further evaluated expression pattern of 65 different genes which are studied for their roles in stemness maintenance in various cancer types. Interestingly, we found specific subsets of genes expressed distinctly in ‘Red’ (Figure 4H(i)); or both ‘Red’ and ‘Orange’ (Figure 4H(ii)); or commonly in ‘Orange’, ‘Green’ and ‘Blue’ subpopulations (Figure 4H(iii), or among all four subpopulations (Figure 4H(iv)). This suggested that stemness may be maintained in these subpopulations and regulated by distinct gene expression networks.
To define the cell state specific gene expression signatures of these four subpopulations, we generated a list of unique-DEGs specific to each subpopulation (Table S2). Since, the ‘Green’ and ‘Blue’ subpopulations showed similar gene expression, the ‘Blue’ subpopulation was excluded from this analysis. Gene Ontology (GO) analysis for subpopulation specific unique-DEGs using the cytoscape tool-BiNGO resulted in overrepresentation of specific pathways for these subpopulations (Tables S3, S4, and S5). Interestingly, while the ‘Red’ subpopulation with CD24Low/ALDHHigh phenotype overrepresented the process of organ development and lipid metabolism, the CD24High/ALDHHigh, ‘Green’ subpopulation had overrepresentation of organ development with differentiation processes.
Based on these results, we inferred that the spontaneous emergence of CD24High/ALDHHigh subpopulation from the CD24Low/ALDHHigh is a process of cellular differentiation, retaining mixed transcriptional signatures of Intermediate states of stemness and differentiation. We validated these results in spheroids generated from the four subpopulations of GBC02 by qRT-PCR for TFs responsible for maintenance of stemness (Figure 4I-i) as well as cytokeratin markers associated with basal and differentiated suprabasal layer of keratinocytes (Figure 4I-ii). Although all four subpopulations-derived spheroids maintained basal cytokeratin markers CK14, CK5, CK19 at similar levels; ‘Green’, ‘Orange’ and ‘Blue’ subpopulations expressed differentiation markers CK13, CK1 and CK10 at significantly higher levels compared to the ‘Red’ spheroids. Interestingly, while KLF4, cMYC, OCT4 and NANOG expression was variable among all subpopulations; SOX2 was significantly downregulated while SOX9 was expressed at higher level in ‘Green’, ‘Blue’ and ‘Orange’ spheroids compared to the ‘Red’ spheroids. siRNA mediated knockdown of SOX9 expression in SCC-070 cells indeed displayed alteration in phenotype distribution (Figure 4J-i) and lower expression of ALDH1A3 (Figure 4J-ii), suggesting the crucial role of SOX9 in maintaining ALDHHigh phenotypes.
Evaluation of prognostic value of population-specific gene expression pattern
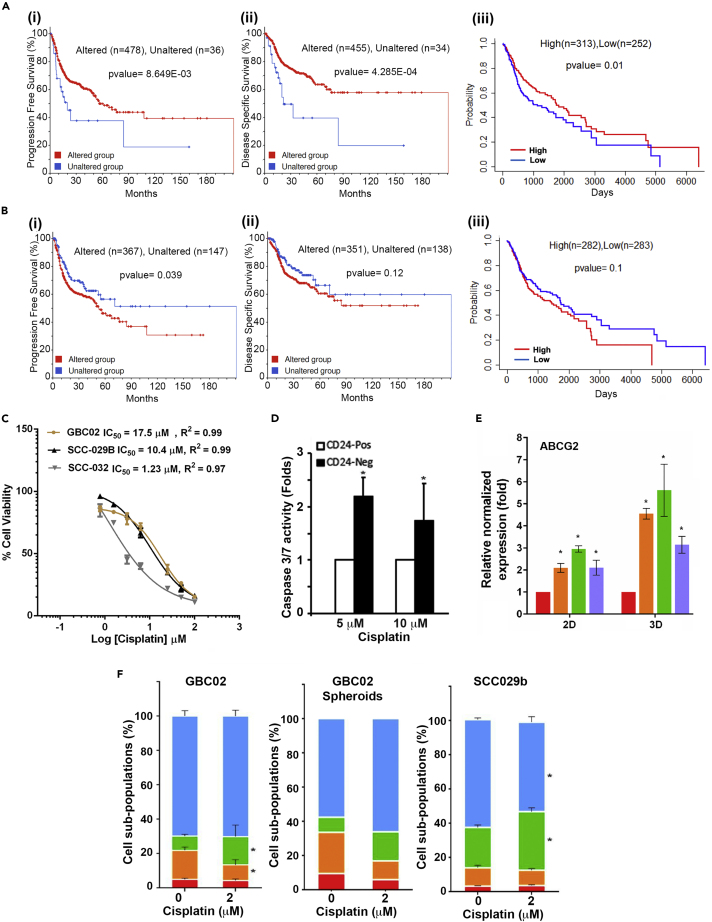
To explore the functional relevance of cell state transitions, we utilized unique-DEGs to test the correlation with clinical outcome of HNSCC patient cohort in TCGA-HNSCC, PanCancer Atlas study. This preliminary analysis is performed using cBioPortal web tool (Gao et al., 2013; https://www.cbioportal.org) to explore the correlation of specific DEGs with prognosis of the patient cohort. To compare patient outcome, the gene list with ‘n’ number of subpopulation-specific upregulated or downregulated gene-list was put in the query. After running the query, the survival data was checked. The ‘cases with alteration’ were patients with high or low expression (±2.0 Z score) for quarried genes in the gene-list and the ‘cases without alteration’ are those with no change in expression from the mean expression in the cohort. Patients with alterations in upregulated unique-DEGs for 'Red’ subpopulation correlated significantly with better prognosis (Figures 5A-i and5ii); whereas, alterations in downregulated unique-DE-Gs for Red’ subpopulation, significantly correlated with poorer progression and disease-free survival (Figures 5B-i and5ii). We verified this result with more systematic approach where survival analysis was performed using the most upregulated or downregulated unique-DE-Gs with ‘ssGSEA score’. First, top ‘n’ number of up or down regulated genes from unique-DE-Gs list for each subpopulation were sorted based on specific fold change (sFC) values. Next, the ssGSEA score was calculated for the respective gene-list with ‘n’ number of genes for individual patient and mean ssGSEA score was calculated across all patients in the cohort. Patients with their individual ssGSEA scores more than the mean were classified as ‘high’, and the others as ‘low’. The survival of these groups was estimated using Kaplan-Meier (KM) curves and Cox-regression analyses. Importantly, among all the comparisons as given in Figure S13-A; statistically significant difference in survival was obtained from the gene list with the top 50 or 80 upregulated unique DEGs from ‘Red’ subpopulation (Figures 5A-iii andS13A). Conversely, patients with higher ssGSEA-score calculated using the gene list of 70 most downregulated unique DEGs from ‘Red’ subpopulation showed a trend of poorer prognosis (Figures 5B-iii and S13B).
Figure 5.
Prognostic significance of emergence of CD24High Cisplatin tolerant subpopulations
(A) (1)Kaplan Meier curves for progression free survival and (2) Disease specific survival for HNSCC patients having altered (Red line) or unchanged (Blue line) expression for top 60 upregulated genes in ‘Red’ subpopulation. (3) Kaplan Meier curves of HNSCC patients segregated into High expression (Red line) and Low expression (Blue line) groups based on top 50 uniquely up-regulated genes of ‘Red’ subpopulation.
(B) (1) Kaplan Meier curves for progression free survival and (2) Disease specific survival for HNSCC patients having altered (Red-line) or unchanged (Blue-line) expression for top 30 downregulated genes in ‘Red’ subpopulation. (3) Kaplan Meier curves of HNSCC patients segregated into High expression (‘Red’) and Low expression (‘Blue’) groups based on top 70 uniquely down-regulated genes of ‘Red’ subpopulation.
(C) Cell survival percentages and the IC50 values of GBC02, SCC-029 and SCC-032 cell lines 3D-spheroid cultures with Cisplatin treatment.
(D) Graph showing higher Caspase 3/7 activity in GBC02 cell line’s CD24Low (CD24-Neg) cells compared to CD24High (CD24-Pos) cells upon Cisplatin treatment for 48 h.
(E) qRT-PCR result of ABCG2 expression in GBC02 2D sorted and 3D spheroid subpopulations.
(F) (1) Graph showing significant increase in ‘Green’ subpopulation upon Cisplatin treatment (2 μM) for 48 h in GBC02 cell line. Error bars represent Mean ± SEM from three biological repeats. Similar increase in ‘Green’ cells with 48 h Cisplatin treatment (2 μM) in (2) GBC02 3D spheroids (3) SCC-029 parent cell line.
Error bars represent mean ± SD from two to three independent experiments performed in triplicates and ∗ indicates p value ≤0.05.
Because bulk RNAseq data of the tumor represents the transcriptome make up of the milieu of diverse cells; its match with any specific and unique gene signature of a subpopulation may be possible if the sample is represented by that specific subpopulation at a higher proportion. Similarly, samples with a mix of more diverse subpopulations will show poorer match with the subpopulation specific signature. Therefore, the observation where patients with higher ‘Red’-subpopulation specific DEGs showing good prognosis; one of the possible interpretations may be that the ‘Red’ subpopulation in these tumors lacked the ability to generate other subpopulations and thus remained more homogeneous (enriched with ‘Red’ subpopulation) as compared to those where ‘Red’ subpopulation was able to generate heterogeneous subpopulations, resulting in loss of ‘Red’-subpopulation specific gene signatures and in turn achieving higher intratumoral heterogeneity (ITH). Hence, poor prognosis could be correlated with higher ITH. To explore this postulation, we calculated IC50 value of Cisplatin among three different oral cancer cell lines with distinct ‘Red’ subpopulation driven heterogeneity pattern (Figure 5C). We found that 3D-spheroid cultures of heterogeneous cell lines GBC02 and SCC-029B (where the four subpopulations originates from ‘Red’ subpopulation) exhibited higher IC50 of Cisplatin compared to SCC-032 cell line (where ‘Red’ subpopulation fails to generate ‘Green’ subpopulation (Figure S14A), highlighting the importance of spontaneous generation of CD44+Ve/CD24High/ALDHHigh ‘Green’ subpopulation from CD44+Ve/CD24Low/ALDHHigh ‘Red’ subpopulation (Figure 5C). Next, we tested the relative Cisplatin sensitivity of CD24High and CD24Low subtypes of cells isolated from GBC02 cell line. Interestingly, upon Cisplatin treatment CD24Low subtypes showed significantly higher Caspase 3/7 activity, indicative of higher induction of apoptosis compared to CD24High subtypes of cells (Figure 5D). RT-PCR analysis revealed highest expression of drug efflux gene ABCG2 in the ‘Green’ subpopulations among all four in both 2D and 3D-spheroid cultures of GBC02 in treatment naive condition (Figure 5E), which may be a possible explanation for Cisplatin refraction by CD24Highsubpopulations. Furthermore, the heterogeneous parent cultures in 2D as well as 3D-spheroid conditions, significantly enriched for CD44+Ve/CD24High/ALDHHigh ‘Green’ subpopulation after exposure to sub-lethal dose of Cisplatin (2 μM) (Figures 5F and S14B). Therefore, spontaneous transition of CD24Low to CD24High subtype and Cisplatin-induced plasticity of ALDHLow to ALDHHigh state collectively result in accumulation of CD44+Ve/CD24High/ALDHHigh‘Green’ subpopulation as Cisplatin tolerant state of cells, resulting in better survival of cells.
Discussion
Phenotypic markers have been successfully employed to study ITH, biology of phenotype switching, and cancer cells survival strategies in response to therapies (Gupta et al., 2011; Goldman et al., 2015; Chaffer et al., 2011). High ALDH activity (ALDHHigh) or CD24LowCD44High surface marker profiles are individually used to describe SLCCs phenotypes in multiple tumors (Clay et al., 2010; Ghuwalewala et al., 2016; Liu et al., 2014; Han et al., 2014). Here, using CD44, CD24, and ALDH markers, we have characterized diversity among CD44-positive oral cancer cells for their distinct expression of CD24 and ALDH-activity. Our results showed enrichment of ALDHHigh cells in both well regarded CD24Low/CD44High SLCC phenotype and in relatively underexplored CD24High/CD44High phenotype, suggesting stable co-existence of phenotypically heterogeneous ALDHHigh subpopulations in oral tumors. This is contrary to the report where ALDHHigh and CD24LowCD44High phenotypes represent distinct non-overlapping subpopulations in breast cancer (Liu et al., 2014). Furthermore, in accordance with the model of stochastic interconversions of cell states recapitulating the heterogeneous subpopulations reported in the breast cancer (Gupta et al., 2011), we observed bidirectional phenotype switching on ALDH axis. However, importantly, the population trajectory results also highlighted the self-sufficiency of CD44+ve/CD24High subpopulations to maintain itself in long-term 2D and 3D cultures without being dependent on or switching to the CD44+ve/CD24Low subpopulations under both regular growth conditions as well as drug induced stress conditions; suggesting a strict unidirectional switching on CD24-axis. Opposed to the ALDH-axis interconversions, our observations that CD24High cells were committed to repopulate only CD24High compartment gives key insights about oral subpopulations hierarchies and emphasizes that the transitions and emergence of cell-states may not be stochastically equal on all different cellular axes.
Previous studies have reported that CD24High cells represent terminally differentiated luminal phenotypes in both normal and cancerous breast tissues (Shipitsin et al., 2007; Sleeman et al., 2005). However, there is increasing appreciation that CD24High cells are stem-like cells in many cancers including melanoma, breast, oral, pancreatic, ovarian and colorectal cancer (Rostoker et al., 2015; Tang et al., 2018; Han et al., 2014; Li et al., 2007; Yeung et al., 2010). In oral cancer, CD24High cells are reported to be highly angiogenic under in vivo conditions (Zimmerer et al., 2017). Furthermore, CD24High cells exhibited resistance to Cisplatin and hence were proposed to be used as a predictive marker of Cisplatin treatment response in HNSCC (Modur et al., 2016). Moreover, CD24+ve cells are recently found as novel multipotent dental pulp regenerating stem cells in mouse and human (Chen et al., 2020; Liang et al., 2022). Therefore, the CD24High subpopulations may carry out important tumor promoting functions in oral cancer. Therefore, in addition to the widely reported subpopulation with CD44+ve/CD24Low phenotype as putative SLCCs in a variety of solid tumors including oral cancer, our results provide evidence for CD44+ve/CD24High phenotype to be a hybrid cell state. Although similar observations were made in a previous study (Han et al., 2014) here we have provided a deeper understanding of these cell states with experimental evidence of population trajectories, drug tolerance and transcriptomic makeup as discussed below.
Recently, heterogeneity within mice oral mucosal basal layer has been reported (Jones et al., 2019), where in addition to harboring oral epithelial progenitor cells (OEPCs), the basal layer also accommodates maturing keratinocytes; thus, revealing a continuum of cell differentiation states within it. Interestingly, some of these differentiating keratinocytes continued to co-express high levels of cytokeratin 14 (CK14) and genes associated with both OEPCs and differentiation processes, representing transitional intermediated cell states. Furthermore, most of the basal layer cells were found to be cycling. We could draw similarities between this tissue hierarchy in normal mice oral mucosa and population trajectories to oral cancer cells. As anticipated, the ‘Red’ subpopulation with CD44+Ve/CD24Low/ALDHHigh phenotype showed greater similarity with the transcriptome of OEPCs, supporting the reports of enrichment of putative Oral-SLCCs in CD44+Ve/ALDHHigh cell population. Strikingly, other three subpopulations emerging from the ‘Red’ subpopulation had transcriptome overlaps with both OEPCs and differentiating cells of oral mucosal basal layer cells, representing the transitional intermediated state of oral cancer cells, maintaining stemness. The GSEA result with the differentially expressed genes between ‘Red’ and ‘Green’ subpopulation clearly showed the onset of differentiation in ‘Green’ subpopulation. Thus, the nonreversible transition of CD24Low to CD24High phenotype cells is possibly because of the onset of differentiation to CD24High cells. Also, the DEGs between ALDHHigh (‘Red’) and ALDHLow (‘Orange’ and ‘Blue’) subpopulations did not result in any specific gene sets enrichment (data not shown). This could be because of the plastic nature of ALDHLow subtype of cells which may have cells with mixed transcriptome states masked in the bulk RNA-sequence data and needs exploration in future.
Furthermore, while SOX2 was highly expressed in ‘Red’ subpopulation, other subpopulations showed downregulated SOX2 and upregulated SOX9 expression. In concordance with a recent report where depletion of SOX9 had resulted in downregulation of ALDH1 in both SOX2 and SOX9 expressing oral cancer cells (Sharma et al., 2018), we too observed downregulation of ALDH1A1 expression and loss of ALDHHigh ‘Red’ and ‘Green’ subpopulations upon SOX9 silencing. Both SOX2 and SOX9 expression are known to play important roles in maintaining stemness in multiple cancers (Zhang et al., 2020; Aguilar-Medina et al., 2019). Upregulated SOX9 expression is linked with lineage infidelity driving wound repair and cancer of squamous cells (Ge et al., 2017). Therefore, we may suggest that the ‘Green’,‘Orange’ and ‘Blue’ subpopulations may represent intermediate transitional cell states, enriched with SOX9-positive alternate state of stemness. In vitro spheroid formation results from multiple independent oral cancer cells lines strongly supported this possibility and must be explored in future. Our study provided evidence of distinct phenotypic states of cells mapped to the transcriptome states demonstrating the cellular hierarchy in oral cancer. Furthermore, contrary to the popular notion where stem-like cancer cells are limited to specific phenotypic compartments (e.g. ‘Red’ subpopulation), our study provides strong support to the possibility of the existence of hybrid states of stemness in alternate phenotypes of differentiating oral cancer cells, irrespective of their genetic diversity.
Because the impact of ITH on clinical outcomes is a major focus of cancer research (Morris et al., 2016; Stanta and Bonin, 2018); tumor samples are usually employed to estimate the extent of ITH using genomics approaches (Sharma et al., 2019; Karaayvaz et al., 2018). However, efforts on developing genomics-based data for investigating ITH and its clinical significance using isogenic subpopulations from patient derived cell cultures, have been limiting. Patient-derived cell cultures not only serve as essential tools for modeling disease heterogeneity but also are essential for functional validation of molecular mechanisms associated with observed complexities. Evident from our study, patients with high-abundance of ‘Red’ subpopulation specific top-unique DEGs were identified to have better prognosis in HNSCC-TCGA cohort. As, SOX2High/SOX9Low gene expression pattern was higher in ‘Red’ subpopulation and loss of SOX2 and gain of SOX9 was the pattern in ‘Green’,‘orange’ and ‘Blue’ subpopulation in our study, this had a strong concordance with a recent study which reported better prognosis in SOX2High/SOX9Low expressing group compared to SOX2Low/SOX9High expressing group in HNSCC-TCGA patient cohorts (Sharma et al., 2018). Therefore, our in vitro cell culture model-based study has revealed the dynamic property of subpopulations of oral cancer cells to generate ITH; as possible non-genetic mechanism, responsible for worse prognosis in oral cancer patient. Gene signatures or markers associated with stem-like state are contrastingly reported for its prognostic value in oral cancer. Although higher expression of CD44, OCT4 or Integrin-β1 was correlated with poor clinical outcome in HNSCC; however, in the same study contrastingly, patients with high expression of ALDH1 showed better treatment outcome and favorable prognosis (Koukourakis, 2012). Therefore, these observations also indicate that oral cancer stem cells are more diverse. Thus, collectively with the evidences from us and others we support the notion of emergence of hybrid cell state as poor prognostic factor and urge for more systematic studies in the oral cancer stem cells research, in future.
Furthermore, to develop drug tolerance, one of the emerging possibilities is the transition of cancer cells across a continuum of states in response to chemotherapy (Pisco and Huang, 2015). Indeed; in earlier studies, cells with CD24High-phenotype marked a transient chemo-resistant cell state in lung cancer (Sharma et al., 2010) and breast cancer (Goldman et al., 2015). Also, increased CD24 expression with OCT4 and NANOG is recently correlated with poor chemo-radiotherapy response and unfavorable prognosis in oral cancer (Mishra et al., 2020). Therefore, phenotype switching may act as a robust mechanism for cancer cells to rapidly acquire a drug tolerant state in changing environment, from where these cells may eventually emerge as a drug-resistant population in relapsed tumor. In our study, we have demonstrated that transient exposure to low doses of Cisplatin was sufficient to induce transition of ALDHLow subpopulations (‘Orange’ and ‘Blue’) into the ALDHHigh subpopulations (‘Red’ and ‘Green’). Using mathematical modeling and further validation with experiments using CD24Low-depleted cells, we demonstrated that the accumulation of drug tolerant ALDHHigh subpopulations was a result of the induction of phenotype switching rather than selection under low dose of Cisplatin treatment. Moreover, a higher dose had resulted in increased induction of apoptosis in cells with CD24Low phenotype. Better drug efflux because of higher expression of ABCG2 could be one of the possible mechanisms for higher drug tolerance in these hybrid cell states, although emerged from putative oral-SLCCs (‘Red’ subpopulations). The emergence of drug tolerant state in breast cancer is also found to be the transition of a non-stem cell population to CD44High/CD24High cell phenotype (Goldman et al., 2015).Supporting our observation; the Cisplatin selected oral cancer cells were found to express higher levels of SOX9 in earlier study (Sharma et al., 2018); which is similar to our results of high SOX9 expression in CD24High subpopulations. Furthermore, the balance between SOX2 and SOX9 levels is found to play key role in lung tissue development and morphogenesis (Rockich et al., 2013). Also, SOX2 is shown to epigenetically suppress the levels of SOX9 in breast cancer cells and determines the cancer cell plasticity and metastatic spread to the lungs (Lin et al., 2016). Collectively, we suggest that the higher expression of ABCG2 and SOX9, transition of phenotypes to ALDHHigh state as well as selection of CD24High cells by Cisplatin at higher dose, collectively might have resulted in emergence of CD44+ve/CD24High/ALDHHigh cell phenotype to be the drug tolerant cell state in our experiments. However, we are currently studying the possible mechanisms by which this hybrid cell state had emerged as drug tolerant population in oral cancer cells.
In summary, our study has identified the maintenance of stem-like-state throughout the process of cellular differentiation in oral cancer cells. Furthermore, irrespective of the genetic diversity, we unmasked the ability of oral cancer cells to harness its property of cell-state transition and gain adaptive Cisplatin tolerance. We have also suggested the clinical implications of ITH achieved because of the cell-state transitions using theoretical and experimental studies. We propose that the characteristics of these phenotypic subgroups may be optimum for estimating the intratumoral heterogeneity in oral cancer patients for its clinical significance, which needs further investigations.
Limitations of the study
This study provides the prerequisite knowledge about the phenotype composition of subpopulations, critical for global tumor behavior in oral cancer; however, our results are predominantly based on the in vitro cell culture models. Therefore, our assumption of relating ITH with the observed poor prognosis still needs validation in patient tissue samples. Furthermore, our observations of Cisplatin induced plasticity to accumulate drug-resistant state of stem-like cells are still limited to the in vitro studies. Experiments are required to validate our findings in animal models.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Alexa Fluor®647 Mouse Anti-Human CD24 (Clone: ML5) | BD Biosciences | Cat # 561644 RRID: AB_10894010 |
| BV786 Mouse Anti-Human CD44 (Clone: G44-26) | BD Biosciences | Cat # 564942 RRID: AB_2739019 |
| Human Hematopoietic Lineage Antibody Cocktail, APC | eBioscience | Cat # 22-7776-72 RRID: AB_2043865 |
| BV421 Mouse Anti-Human CD24 (Clone: ML5) | BD Biosciences | Cat # 562789 RRID: AB_2737796 |
| FITC Mouse Anti-Human CD44 (Clone: G44-26) | BD Biosciences | Cat # 555478 RRID: AB_395870 |
| Biological samples | ||
| Surgically resected human oral tumor tissues | Tata Medical Centre, Kolkata, India | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Propidium iodide | Sigma | Cat # P4170 |
| Collagenase/Hyaluronidase Mix | Stem Cell Technologies | Cat # 07912 |
| DNase-1 | Stem Cell Technologies | Cat # 07900 |
| ACK Lysing Buffer | Thermo Fisher Scientific | Cat # A10492-01 |
| Human Fc Block | BD Biosciences | Cat # 564220 |
| HBSS Buffer | Thermo Fisher Scientific | Cat # 14175 |
| HEPES | Thermo Fisher Scientific | Cat # 15630 |
| Minimum Essential Media | Thermo Fisher Scientific | Cat #11095-080 |
| Non-essential amino acids | Thermo Fisher Scientific | Cat # 11140-050 |
| L-glutamine | Thermo Fisher Scientific | Cat # 25030-081 |
| Accutase dissociation reagent | Thermo Fisher Scientific | Cat # A11105-01 |
| EGF | Thermo Fisher Scientific | Cat # PHG0311 |
| bFGF | Thermo Fisher Scientific | Cat # PHG0263 |
| B-27 | Thermo Fisher Scientific | Cat # 12587-010 |
| Hydrocortisone | Sigma | Cat # H0888 |
| Geltrex | Thermo Fisher Scientific | Cat # A14132-02 |
| EpiLife Media | Thermo Fisher Scientific | Cat # MEP1500CA |
| DMEM/F12K Media | Thermo Fisher Scientific | Cat # 11330 |
| Antibiotic-Antimycotic Mix | Thermo Fisher Scientific | Cat # 15240-062 Cat # 50-0640 |
| Dispase | Stem cell technologies | Cat # 07923 |
| Collagenase | Stem cell technologies | Cat # 07909 |
| Trypsin-EDTA | Thermo Fisher Scientific | Cat # 25300 |
| Trypsin Neutralizer Solution | Thermo Fisher Scientific | Cat # R002100 |
| Cisplatin | Sigma | Cat # P4394 |
| Critical commercial assays | ||
| ALDEFLUOR assay Kit | Stem Cell Technologies | Cat. # 01700 |
| CellTrace™ Violet Cell Proliferation Kit | ThermoFisher Scientific | Cat # C34557 |
| Apo-ONE® Homogeneous Caspase-3/7 Assay |
Promega | Cat # G7792 |
| RNeasy Mini/MicroKits | Qiagen | Cat # 74134 Cat # 74034 |
| SsoEva Green SYBR Mix | BioRad | Cat # 172-5203 |
| Deposited data | ||
| Dataset of The Cancer Genome Atlas (TCGA) Head and Neck Cancer patient cohort | https://www.cbioportal.org | N/A |
| All original RNAseq data | ENA | ENA: PRJEB52288 |
| Experimental models: Cell lines | ||
| Human oral cancer: GBC02 cells | NIBMG, India | Vipparthi et al. (2021) |
| Human oral cancer: GBC035 cells | NIBMG, India | Vipparthi et al. (2021) |
| Human oral cancer: SCC-029b cells | Susanne M. Gollin Lab, University of Pittsburgh, USA. | White et al. (2007) |
| Human oral cancer: SCC-070 cells | Susanne M. Gollin Lab, University of Pittsburgh, USA. | White et al. (2007) |
| Human oral cancer: SCC-084 cells | Susanne M. Gollin Lab, University of Pittsburgh, USA. | White et al. (2007) |
| Human oral cancer: SCC-032 cells | Susanne M. Gollin Lab, University of Pittsburgh, USA. | White et al. (2007) |
| Oligonucleotides | ||
| Primer Table | This paper | Table S6 |
| Software and algorithms | ||
| FCS Express 5 software | Denovo software | https://denovosoftware.com/ |
| Extreme Limiting Dilution Analysis (ELDA) software | https://bioinf.wehi.edu.au/software/elda/ | N/A |
| Image J (1.52p) software | https://imagej.nih.gov/ij/ | N/A |
| Venn diagrams online tool | http://bioinformatics.psb.ugent.be/webtools/Venn/ | N/A |
| Gene Set Enrichment Analysis software GSEA version 4.0.3 |
http://www.broad.mit.edu/gsea/ | N/A |
| CellTrans R package | Buder et al. (2017) | N/A |
Resource availability
Lead contact
-
•
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Sandeep Singh (ss5@nibmg.ac.in)
Materials availability
-
•
This study did not generate new unique reagents
Experimental model and subject details
Oral cancer cell lines, GBC02 and GBC035 used in this study were recently established by us (Vipparthi et al., 2021). GBC02 cell line were maintained in EpiLife media (Cat # MEP1500CA, Thermo) supplemented with 1× antibiotics mix (15240-062, Thermo) and 2% fetal bovine serum (FBS) (Cat # 16140071, Thermo), 0.4 μg/mL Hydrocortisone (Cat # H0888, Sigma), 1X B-27 (Cat # 12587-010, Thermo), 20 ng/mL EGF (Cat # PHG0311, Thermo), 20 ng/mL bFGF (Cat # PHG0263, Thermo). GBC035 cell line was maintained in DMEM/F12K media (Cat # 11330, Thermo) supplemented with 10% FBS and 1X antibiotics mix. Other oral cancer cell lines SCC-029, SCC-070, SCC-084 and SCC-032 were kindly provided by Dr. Susanne M. Gollin, University of Pittsburgh, USA (White et al., 2007). These cell lines were maintained in MEM (Minimum Essential Media, Cat #11095-080, Thermo) with non-essential amino acids (Cat # 11140-050, Thermo), L-glutamine (Cat # 25030-081, Thermo) and 10% FBS and 1X antibiotics mix. Cells were maintained in the incubator with 5% CO2 at 37°C. Human subjects were included in this study (EC/GOVT/01/12) after approval obtained from institutional ethics committee of National Institute of Biomedical Genomics (NIBMG) and the institutional review board of Tata Medical Center (TMC), Kolkata, India. All recruited subjects were above the age of 18 years. Recruitment of these subjectswere not influenced by their age, sex, or gender. Out of these eight recruited subjects, seven were males and one female.
Method details
Dissociation of primary oral tumor cells
Primary tumor tissue samples were collected after taking informed consent from patients (Vipparthi et al., 2021; Patel et al., 2018). Following enzymatic digestion of tumor tissues using 1× Collagenase/Hyaluronidase and DNase-1 (Cat # 07912 and 07900; Stem cell technologies), dissociated tumor cells were collected in a different tube and filtered through 40 μm strainer. Ice-cold serum-free growth media was used to rinse the strainer’s membrane to avoid cell loss. Next, cell suspension was centrifuged at 500g for 5 min and supernatant was discarded. Pellet was resuspended in ACK lysis buffer (Cat # A10492-01; Gibco) for 1 min at room temperature to lyse RBCs. It is followed by a quick centrifugation for 2 min, discarding of the lysis buffer and two washes with ice-cold HBSS buffer (Cat # 14175; Thermo) containing 0.5% FBS and 10mM HEPES (Cat # 15630, Thermo) (HBSS+). Cells were finally resuspended in ice-cold HBSS + buffer and counted using trypan blue on a hemocytometer. For downstream flow cytometry analysis, dissociated tumor cells were Fc receptor blocked by incubating 1∗106 cells/50-100 μL of staining buffer with 2.5 μg of Human Fc Block (Cat # 564220; BD) for 10 min.
Flow cytometry for combined ALDEFLUOR assay and antibody immunophenotyping
Single cell suspensions were prepared by resuspending 1∗106 cells/mL (for cell lines) or 1∗106 cells/100 μL (for dissociated primary tumor cells) of Aldefluor assay buffer. Aldefluor assay was performed as per the manufacturer’s protocol (Cat. # 01700, Stem Cell Technologies). For negative control, Diethyl aminobenzaldehyde (DEAB), an inhibitor of Aldehyde dehydrogenase enzymes, was used. Few cells (for instance; 2∗105 cells in 200 μL assay buffer) were separated in a tube labeled ‘with DEAB’ and 1 μL of DEAB reagent per 100 μL of sample was added and incubated for 5 min at 37°C. Next, Aldefluor reagent, BODIPY-aminoacetaldehyde (BAAA) was added in both ‘with DEAB’ and ‘without DEAB’ tubes at 5 μL/mL of assay buffer. They were incubated for 1 h at 37°C, by intermittent mixing and inverting of the tubes every 15 min. After 1 h, cells were washed with ice-cold Aldefluor assay buffer for 5 min at 400 rcf and supernatant is discarded. Furthermore, ‘with DEAB’ control tube is sub-divided into two tubes labeled ‘with DEAB only’ and ‘with DEAB + Antibody’. CD24-Alexa647 (Cat # 561644; Clone: ML5; BD) and CD44-BV786 (Cat # 564942; Clone: G44-26; BD) antibodies were added in ‘without DEAB’ and ‘with DEAB + Antibody’ tubes and incubated for 30 min at 4°C. In case of primary tumor cells, hematopoietic lineage cocktail antibodies (Cat # 22-7776-72; eBioscience) were added along with CD24-BV421 (Cat #562789; BD) and CD44-BV786 (Cat #564942; BD) antibodies. After incubation, cells are washed twice with ice-cold Aldefluor buffer, filtered and resuspended at 3∗106 cells/mL. Propidium iodide (Cat # P4170; Sigma) at final concentration of 2 μg/mL was added to exclude dead cells from the analysis. Flow cytometry data was acquired using BD FACS Aria Fusion cytometer and analysis was performed using FCS Express 5 (DeNovo Software).
Cell sorting
Sorting buffer was prepared by adding 5% FBS and 1X antibiotic-antimycotic mix (Cat # 15240-062, 50-0640; Thermo) to HBSS + buffer. FACS tubes with sorting buffer were kept at 4°C, 2 h prior to cell sorting. Gating was set as per the experimental choice and sort layouts were prepared. For isolating the four subpopulations, cells were sorted four ways into the collection tubes containing sorting buffer, using BD FACS Aria Fusion flow cytometer.
Analysis of frequencies of four subpopulations
Percentages of four phenotypic subpopulations were obtained from the gating statistics of flow cytometry dot plots, analyzed by FCS Express 5 Software. Percentages were summed up to 100% and plotted as stacked bar graphs, using graph pad prism software.
Cell trace (CT)-Violet dye dilution assay combined with marker phenotyping
Equal numbers of parental cells were seeded in triplicates (1X105 cells per 60mm dish). 48 h later CT-Violet dye staining (Cat #C34557, Invitrogen) was performed, followed by Aldefluor and CD24 immuno-staining on one of the stained plates, for recording baseline fluorescence signal at Day 0. Other plates were allowed to proliferate for 4 days and subsequently subjected to Aldefluor and CD24 immuno-staining. Proliferation index was calculated using FCS express software based on fluorescence intensities at Day-0 and Day-4. Readings from three independent plates were taken to determine average proliferation index and S.D.
Repopulation assay
To study the ability of each of the subpopulation to regenerate all or any of these subpopulations ‘repopulation assay’ was conducted. Sorted cells were centrifuged at 400 rcf for 5 min, supernatant was discarded and cells were resuspended in their respective growth media. Sorted subpopulation at more than 99% purity were plated in wells of a 24-well culture plate at low density (5000 cells/well) in triplicates to allow them to proliferate and form colonies without the need of trypsinization (as trypsinization and re-plating might have introduced disturbance, stress and experimental variations). Cells were allowed to grow un-interrupted for 10 days to allow cells to undergo doubling cycles and form colonies with media change once in every three days. On 10th day post-sorting and plating, cells were harvested using Accutase dissociation reagent and re-analyzed for Aldefluor assay, CD24 and CD44 immuno-phenotyping as described above.
Cisplatin treatment and assessment of its impact during repopulation
After plating cells for repopulation, on 6th day post-sort, growth media containing 2μM Cisplatin was added to cells. Following 48 h of treatment (i.e., 8th day post-sort), Cisplatin was removed and cells were recovered by adding fresh growth media devoid of Cisplatin, for next 48 h. On 10th day post-sort, cells were harvested and re-analyzed for Aldefluor assay, CD24 and CD44 immuno-phenotyping as described above.
Spheroid formation assay
5-10 cells/μL were resuspended in growth media without serum and with 20ng/mL EGF, bFGF, 1× B-27 and 0.4μg/mL hydrocortisone (Merck, Sigma) along with 1.25% geltrex (Cat # A14132-02, Thermo) and plated in multiple wells of ultra-low attachment plate (Corning). Growth factors were supplemented every alternate day. Spheroids formation was continued for 7-10 days. Spheroids with size ≥60 μm were counted for calculating efficiency. Size of spheroids was counted using ImageJ software. Limiting dilution assays were conducted in same manner with varying cell numbers ranging from 5-500/well, as depicted in results section. For estimation of SLCCs frequencies in each subpopulation, ELDA (Extreme Limiting Dilution Analysis) was performed by entering the limiting dilution spheroid data in the website http://bioinf.wehi.edu.au/software/elda/.
Primary spheroids generated were collected and centrifuged down at 800 rcf for 5 min. Supernatant was discarded and to the pellet, Dispase (Cat # 07923; Stem cell technologies), Collagenase (Cat # 07909; Stem cell technologies) mix in 1:1 ratio was added to digest geltrex by incubating for 10-15 min at 37°C. Trypsin neutralizing solution (TNS) was added to stop over-digestion and centrifuged for 5 min at 800 rcf. Spheroids were further suspended in Trypsin-EDTA for 2-5 min to obtain single cells. Reaction was stopped by another round of TNS addition and removal of supernatant. Finally, single cells were resuspended in fresh, serum free, spheroid formation media described above and gently pipetted for homogeneity. They were plated for secondary spheroid formation at the concentrations of 5-500 cells/well.
3D repopulation assay
Sorted cells were plated for spheroid formation (5 cells/μL) as described above and their growth was followed for 7-10 days. Single spheroids >60 μm size from each subpopulation were carefully picked by observing under the microscope and plated in wells of a 96 well plate with one spheroid per well. They were cross checked microscopically and wells with only single spheroids were followed up. Once colonies grew, they were transferred onto dishes with a bigger surface and the repopulation analysis was performed from 4 independent single spheroid colonies from each subpopulation.
RNA isolation, qPCR
RNA isolation and qPCR were performed, using RNeasy mini/micro kit and SsoEva Green SYBR mix on CFX96 Real-Time PCR system (BioRad) respectively. Relative gene expression fold changes were calculated by 2−ΔΔCT method. Sequences of the primers used are provided as Table S6.
siRNA mediated gene expression knockdown
Cells were transfected with 10nM of siRNA designed against SOX9 or non-targeting siRNA negative control/scrambled (Eurogentec) using INTERFERin kit (Polypus) as per manufacturer’s instructions. Downstream assays were performed 48 h post transfection.
Discrete time Markov chain model construction
A discrete time Markov chain model was constructed to explain the repopulation patterns of different phenotypes. Assumptions of the model are as follows:
-
1.
Repopulation is because of transition of different phenotypes between each other. Other processes like cell division do not affect the phenotypic heterogeneity.
-
2.
Transition probabilities among the phenotypes are independent of time and of each other. To infer the transition probabilities, the package CellTrans (Buder et al., 2017)(R 3.6.4) was applied on repopulation data. Briefly, the package uses repopulation data at time ti obtained starting from a sorted population to construct a phenotypic proportion matrix (). Using the theory of Markov chains, it calculates the transition matrix () corresponding to the unit time by left-multiplying the proportion matrix with the identity matrix and taking a tithroot of the resultant matrix.
The repopulation trajectories in Figure 4 were constructed using the transition matrix using the following formula:
where is the population composition (a row vector) at time and is the population composition at time .
For sensitivity analysis of the estimated transition matrices, we generated random transition matrices by randomly shuffling the rows of the estimated TM and then randomly shuffling the columns.
Transition matrix randomization
From the Markovian model, we were able to obtain transition matrices and predicted possible trajectories leading to steady states. To understand our predictions further and to verify whether the predicted trajectories are unique to the transition matrix and hence to the experimental data, we randomized transition matrices and compared the matrices and the steady states with those of “WT” matrix. To maintain transition matrix property, i.e., all row-sums must be equal to one, the randomization was performed by shuffling rates in a single row and shuffling entire rows (Figure S7E).
We measured the distance between steady states and the element-wise distance between the transition matrices using a standard distance metric: Euclidean distance. Based on these distances, we clustered the transition matrix-steady state pairs using k-means function of R, as demonstrated for the case of GBC02 in Figure S7E. Further analysis of these clusters in terms of mean and standard deviation values of steady states (Figures S7G and S7H) and transition rates (Figures S7I and S7J) were performed.
RNA sequence data generation, analysis, and identification of differentially expressed genes
For RNA sequencing studies, RNA is extracted from FACS isolated four subpopulations of the GBC02 cell line grown in 2D monolayer cultures, in triplicates, and also from the ‘Red’ and ‘Green’ spheroids generated from sorted cells of GBC02 cell line, in duplicates.
Whole RNA was extracted from three replicates for each of the cell-lines followed by a quality assessment using Agilent Bioanalyser-2100 with Agilent nano Kit (Agilent Technologies). For each sample, library was prepared by taking 400ng of RNA and sequencing was performed using Illumina Hiseq-2500 to obtain 50 million paired end reads. Raw data QC was performed through FastQC, and the RNA sequence data was aligned to hg19 primary assembly using STAR aligner (version 2.6.0) with GRCh37.87 (Ensembl) transcript model. An average of 87% uniquely mapped reads were obtained from each sample. HTSeq (version 0.11.0) was used to generate count data. Protein coding genes that expressed in at least two of three biological replicates in case or control group were taken for differential gene expression (DGE) analysis. DGE analysis was performed through DESeq2 method with the raw count data. A gene was considered to be differentially expressed if |log2 fold change| > 1 with FDR corrected p value < 0.1 is obtained. All downstream analysis was performed in R-3.5.1 and ggplot2 package was used for generation of plots.
Extraction of cell population-specific unique DEGs
Unique DEGs were obtained for each of the cell subpopulations based on all pairwise comparisons. Genes were selected as unique DEGs for a cell population i as following - (1) by taking common set of n different set of DEGs (all pairwise comparison of cell population i with other cell population). (2) by excluding genes from common sets that are differentially expressed for any other pairwise comparison.
| (Equation 1) |
| (Equation 2) |
| (Equation 3) |
Here, i1 to in are set of DEGs of cell population i as compared to other n numbers of different cell populations and j1 to jn are set of DEGs for all other cell population pairwise comparisons.
Calculation of population-specific fold change
Cell population-specific fold change for each unique DEG for a given cell population was calculated as follows:
| (Equation 4) |
sFCa is specific fold change of gene “a ” in X cell population.Where,
Xavg– Mean expression of gene “a” in the X cell population
Ymin– Minimum of average expression of gene “a” in other cell populations
Ymax– Maximum of average expression of gene “a” in other cell populations
Single sample gene set enrichment analysis (ssGSEA) scores and survival analysis utilizing population-specific signature
The scores were calculated using GSVA R Bioconductor package and “ssGSEA” method was employed in the estimation of gene-set enrichment scores for each patient. To identify the role of different subpopulation-specific gene signatures on patient survival, dataset of The Cancer Genome Atlas (TCGA) Head and Neck Cancer patient cohort was used. A particular patient was segregated in ‘High’ if this individual showed ssGSEA score higher than the mean ssGSEA scores value calculated across all patients for a gene-set of ‘n’number of genes; otherwise it was segregated in ‘Low’. To perform survival analysis R tool, survival was used.
This analysis was performed as follows:
-
(1)
Select top n number of up and down regulated genes from unique DEG list sorted based on specific Fold Change (sFC) values.
-
(2)
For each patient, calculate ssGSEA score for the given gene-set with ‘n’number of genes.
-
(3)
Across all patients in the cohort, calculate the mean ssGSEA score.
-
(4)
Patients with their individual ssGSEA scores more than this mean are classified as 'high', and the other set as 'low'.
-
(5)
Perform survival analysis between the two patient groups.
Quantification and statistical analysis
All quantitative data were presented as means ± S.E.M or ± S.D, from experiments performed in two or three biological repeats in triplicates, as given. Statistical analysis was performed using two-sided paired or unpaired t-tests. p values ≤0.05 were considered statistically significant. All the statistical details of experiments can be found in the respective methodology or figure legends.
Acknowledgment
This work is supported by the grant (IA/I/13/1/500908) received from Wellcome trust-DBT India Alliance. KV acknowledges DST-INSPIRE, SG acknowledges DBT and AKP acknowledges ICMR for fellowship support. We thank Professors Saumitra Das, Partha P Majumder and Dr. Kartiki Desai for scientific discussions and Dr. Arindam Maitra for assistance with transcriptomics platform and Ms. Bhaswati Tarafdar for her assistance with flow cytometry.
Author contributions
Conceptualization, S.S., K.V.; Methodology, K.V., A.K.P., K.H., and P.C.; Investigation, K.V., A.K.P., and S.G; Formal Analysis, K.V., K.H., P.C., A.G., N.K.B, M.K.J., and S.S.; Writing – Original Draft, K.V. and S.S.; Writing – Review and Editing, K.V., S.G., K.H., P.C., A.G., N.K.B, M.K.J, and S.S.; Resources, S.S., N.K.B., M.K.J., R.S., P.A.; Supervision, S.S., M.K.J; Funding Acquisition, S.S.
Declaration of interests
The authors declare no potential conflicts of interest.
Published: May 20, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.104317.
Supplemental information
Data and code availability
-
•
RNAseq data have been deposited at ENA database and are publicly available as of the date of publication. Accession numbers are listed in the key resources table.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Aguilar-Medina M., Avendano-Felix M., Lizarraga-Verdugo E., Bermudez M., Romero-Quintana J.G., Ramos-Payan R., Ruiz-Garcia E., Lopez-Camarillo C. SOX9 stem-cell factor: clinical and functional relevance in cancer. J. Oncol. 2019;2019:6754040. doi: 10.1155/2019/6754040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkley D., Yanai I. Plasticity and clonality of cancer cell states. Trends Cancer. 2019;5:655–656. doi: 10.1016/j.trecan.2019.09.002. [DOI] [PubMed] [Google Scholar]
- Beck B., Blanpain C. Unravelling cancer stem cell potential. Nat. Rev. Cancer. 2013;13:727–738. doi: 10.1038/nrc3597. [DOI] [PubMed] [Google Scholar]
- Biddle A., Liang X., Gammon L., Fazil B., Harper L.J., Emich H., Costea D.E., Mackenzie I.C. Cancer stem cells in squamous cell carcinoma switch between two distinct phenotypes that are preferentially migratory or proliferative. Cancer Res. 2011;71:5317–5326. doi: 10.1158/0008-5472.can-11-1059. [DOI] [PubMed] [Google Scholar]
- Bonnet D., Dick J.E. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat. Med. 1997;3:730–737. doi: 10.1038/nm0797-730. [DOI] [PubMed] [Google Scholar]
- Buder T., Deutsch A., Seifert M., Voss-Bohme A. CellTrans: an R package to quantify stochastic cell state transitions. Bioinform. Biol. Insights. 2017;11 doi: 10.1177/1177932217712241. [DOI] [Google Scholar]
- Chaffer C.L., Brueckmann I., Scheel C., Kaestli A.J., Wiggins P.A., Rodrigues L.O., Brooks M., Reinhardt F., Su Y., Polyak K., et al. Normal and neoplastic nonstem cells can spontaneously convert to a stem-like state. Proc. Natl. Acad. Sci. 2011;108:7950–7955. doi: 10.1073/pnas.1102454108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Fu H., Wu X., Duan Y., Zhang S., Hu H., Liao Y., Wang T., Yang Y., Chen G., et al. Regeneration of pulpo-dentinal-like complex by a group of unique multipotent CD24a+ stem cells. Sci. Adv. 2020;6:eaay1514. doi: 10.1126/sciadv.aay1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clay M.R., Tabor M., Owen J.H., Carey T.E., Bradford C.R., Wolf G.T., Wicha M.S., Prince M.E. Single marker identification of head and neck squamous cell carcinoma cancer stem cells with aldehyde dehydrogenase. Head Neck. 2010;32:1195–1201. doi: 10.1002/hed.21315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole A.J., Fayomi A.P., Anyaeche V.I., Bai S., Buckanovich R.J. An evolving paradigm of cancer stem cell hierarchies: therapeutic implications. Theranostics. 2020;10:3083–3098. doi: 10.7150/thno.41647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalerba P., Kalisky T., Sahoo D., Rajendran P.S., Rothenberg M.E., Leyrat A.A., Sim S., Okamoto J., Johnston D.M., Qian D., et al. Single-cell dissection of transcriptional heterogeneity in human colon tumors. Nat. Biotechnol. 2011;29:1120–1127. doi: 10.1038/nbt.2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dirkse A., Golebiewska A., Buder T., Nazarov P.V., Muller A., Poovathingal S., Brons N.H.C., Leite S., Sauvageot N., Sarkisjan D., et al. Stem cell-associated heterogeneity in Glioblastoma results from intrinsic tumor plasticity shaped by the microenvironment. Nat. Commun. 2019;10:1787–1816. doi: 10.1038/s41467-019-09853-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J., Aksoy B.A., Dogrusoz U., Dresdner G., Gross B., Sumer S.O., Sun Y., Jacobsen A., Sinha R., Larsson E., et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge Y., Gomez N.C., Adam R.C., Nikolova M., Yang H., Verma A., Lu C.P.-J., Polak L., Yuan S., Elemento O., Fuchs E. Stem cell lineage infidelity drives wound repair and cancer. Cell. 2017;169:636–650.e14. doi: 10.1016/j.cell.2017.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghuwalewala S., Ghatak D., Das P., Dey S., Sarkar S., Alam N., Panda C.K., Roychoudhury S. CD44highCD24low molecular signature determines the cancer stem cell and EMT phenotype in oral squamous cell carcinoma. Stem Cel. Res. 2016;16:405–417. doi: 10.1016/j.scr.2016.02.028. [DOI] [Google Scholar]
- Goldman A., Majumder B., Dhawan A., Ravi S., Goldman D., Kohandel M., Majumder P.K., Sengupta S. Temporally sequenced anticancer drugs overcome adaptive resistance by targeting a vulnerable chemotherapy-induced phenotypic transition. Nat. Commun. 2015;6:6139. doi: 10.1038/ncomms7139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta P.B., Fillmore C.M., Jiang G., Shapira S.D., Tao K., Kuperwasser C., Lander E.S. Stochastic state transitions give rise to phenotypic equilibrium in populations of cancer cells. Cell. 2011;147:1197–1644. doi: 10.1016/j.cell.2011.11.008. [DOI] [Google Scholar]
- Han J., Fujisawa T., Husain S.R., Puri R.K. Identification and characterization of cancer stem cells in human head and neck squamous cell carcinoma. BMC Cancer. 2014;14:173–211. doi: 10.1186/1471-2407-14-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S. Non-genetic heterogeneity of cells in development: more than just noise. Development. 2009;136:3853–3862. doi: 10.1242/dev.035139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inde Z., Dixon S.J. The impact of non-genetic heterogeneity on cancer cell death. Crit. Rev. Biochem. Mol. Biol. 2018;53:99–114. doi: 10.1080/10409238.2017.1412395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia D., Li X., Bocci F., Tripathi S., Deng Y., Jolly M.K., Onuchic J.N., Levine H. Quantifying cancer epithelial-mesenchymal plasticity and its association with stemness and immune response. J. Clin. Med. 2019;8:725. doi: 10.3390/jcm8050725. [DOI] [Google Scholar]
- Jolly M.K., Kulkarni P., Weninger K., Orban J., Levine H. Phenotypic plasticity, bet-hedging, and androgen independence in prostate cancer: role of non-genetic heterogeneity. Front. Oncol. 2018;8:50. doi: 10.3389/fonc.2018.00050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones K.B., Furukawa S., Marangoni P., Ma H., Pinkard H., D'urso R., Zilionis R., Klein A.M., Klein O.D. Quantitative clonal analysis and single-cell transcriptomics reveal division Kinetics, hierarchy, and fate of oral epithelial progenitor cells. Cell Stem Cell. 2019;24:183–192.e8. doi: 10.1016/j.stem.2018.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karaayvaz M., Cristea S., Gillespie S.M., Patel A.P., Mylvaganam R., Luo C.C., Specht M.C., Bernstein B.E., Michor F., Ellisen L.W. Unravelling subclonal heterogeneity and aggressive disease states in TNBC through single-cell RNA-seq. Nat. Commun. 2018;9:3588. doi: 10.1038/s41467-018-06052-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keysar S.B., Jimeno A. More than markers: biological significance of cancer stem cell-defining molecules. Mol. Cancer Ther. 2010;9:2450–2457. doi: 10.1158/1535-7163.mct-10-0530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koukourakis M.I. Cancer stem cell phenotype relates to radio-chemotherapy outcome in locally advanced squamous cell head–neck cancer. Br. J. Cancer. 2012;106:846–853. doi: 10.1038/bjc.2012.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreso A., O'brien C.A., Van Galen P., Gan O.I., Notta F., Brown A.M.K., Ng K., Ma J., Wienholds E., Dunant C., et al. Variable clonal repopulation dynamics influence chemotherapy response in colorectal cancer. Science. 2013;339:543–548. doi: 10.1126/science.1227670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Heidt D.G., Dalerba P., Burant C.F., Zhang L., Adsay V., Wicha M., Clarke M.F., Simeone D.M. Identification of pancreatic cancer stem cells. Cancer Res. 2007;67:1030–1037. doi: 10.1158/0008-5472.can-06-2030. [DOI] [PubMed] [Google Scholar]
- Liang J., Zhao Y.-J., Li J.-Q., Lan L., Tao W.-J., Wu J.-Y. A pilot study on biological characteristics of human CD24 (+) stem cells from the apical papilla. J. Dental Sci. 2022;17:264–275. doi: 10.1016/j.jds.2021.01.012. [DOI] [Google Scholar]
- Lin S.-C., Chou Y.-T., Jiang S.S., Chang J.-L., Chung C.-H., Kao Y.-R., Chang I.-S., Wu C.-W. Epigenetic switch between SOX2 and SOX9 regulates cancer cell plasticity. Cancer Res. 2016;76:7036–7048. doi: 10.1158/0008-5472.can-15-3178. [DOI] [PubMed] [Google Scholar]
- Liu S., Cong Y., Wang D., Sun Y., Deng L., Liu Y., Martin-Trevino R., Shang L., Mcdermott S.P., Landis M.D., et al. Breast cancer stem cells transition between epithelial and mesenchymal states reflective of their normal counterparts. Stem Cell Rep. 2014;2:78–91. doi: 10.1016/j.stemcr.2013.11.009. [DOI] [Google Scholar]
- Marine J.-C., Dawson S.-J., Dawson M.A. Non-genetic mechanisms of therapeutic resistance in cancer. Nat. Rev. Cancer. 2020;20:743–756. doi: 10.1038/s41568-020-00302-4. [DOI] [PubMed] [Google Scholar]
- Marusyk A., Almendro V., Polyak K. Intra-tumour heterogeneity: a looking glass for cancer? Nat. Rev. Cancer. 2012;12:323–334. doi: 10.1038/nrc3261. [DOI] [PubMed] [Google Scholar]
- Meacham C.E., Morrison S.J. Tumour heterogeneity and cancer cell plasticity. Nature. 2013;501:328–337. doi: 10.1038/nature12624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema J.P. Cancer stem cells: the challenges ahead. Nat. Cel. Biol. 2013;15:338–344. doi: 10.1038/ncb2717. [DOI] [Google Scholar]
- Mishra S., Tiwari V., Arora A., Gupta S., Anand N., Husain N. Increased expression of Oct4, nanog and CD24 predicts poor response to chemo-radiotherapy and unfavourable prognosis in locally advanced oral squamous cell carcinoma. Asian Pac. J. Cancer Prev. 2020;21:2539–2547. doi: 10.31557/apjcp.2020.21.9.2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modur V., Joshi P., Nie D., Robbins K.T., Khan A.U., Rao K. CD24 expression may play a role as a predictive indicator and a modulator of cisplatin treatment response in head and neck squamous cellular carcinoma. PLoS One. 2016;11:e0156651. doi: 10.1371/journal.pone.0156651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreb J.S. Aldehyde dehydrogenase as a marker for stem cells. Curr. Stem Cel. Res. Ther. 2008;3:237–246. doi: 10.2174/157488808786734006. [DOI] [Google Scholar]
- Morris L.G.T., Riaz N., Desrichard A., Senbabaoglu Y., Hakimi A.A., Makarov V., Reis-Filho J.S., Chan T.A. Pan-cancer analysis of intratumor heterogeneity as a prognostic determinant of survival. Oncotarget. 2016;7:10051–10063. doi: 10.18632/oncotarget.7067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen L.V., Vanner R., Dirks P., Eaves C.J. Cancer stem cells: an evolving concept. Nat. Rev. Cancer. 2012;12:133–143. doi: 10.1038/nrc3184. [DOI] [PubMed] [Google Scholar]
- Pastrana E., Silva-Vargas V., Doetsch F. Eyes wide open: a critical review of sphere-formation as an assay for stem cells. Cell Stem Cell. 2011;8:486–498. doi: 10.1016/j.stem.2011.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel A.K., Vipparthi K., Thatikonda V., Arun I., Bhattacharjee S., Sharan R., Arun P., Singh S. A subtype of cancer-associated fibroblasts with lower expression of alpha-smooth muscle actin suppresses stemness through BMP4 in oral carcinoma. Oncogenesis. 2018;7:78. doi: 10.1038/s41389-018-0087-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisco A.O., Huang S. Non-genetic cancer cell plasticity and therapy-induced stemness in tumour relapse: 'What does not kill me strengthens me. Br. J. Cancer. 2015;112:1725–1732. doi: 10.1038/bjc.2015.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasetyanti P.R., Medema J.P. Intra-tumor heterogeneity from a cancer stem cell perspective. Mol. Cancer. 2017;16:41–49. doi: 10.1186/s12943-017-0600-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pries R., Witrkopf N., Trenkle T., Nitsch S.M., Wollenberg B. Potential stem cell marker CD44 is constitutively expressed in permanent cell lines of head and neck cancer. vivo. 2008;22:89–92. [Google Scholar]
- Prince M.E., Sivanandan R., Kaczorowski A., Wolf G.T., Kaplan M.J., Dalerba P., Weissman I.L., Clarke M.F., Ailles L.E. Identification of a subpopulation of cells with cancer stem cell properties in head and neck squamous cell carcinoma. Proc. Natl. Acad. Sci. 2007;104:973–978. doi: 10.1073/pnas.0610117104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puram S.V., Tirosh I., Parikh A.S., Patel A.P., Yizhak K., Gillespie S., Rodman C., Luo C.L., Mroz E.A., Emerick K.S., et al. Single-cell transcriptomic analysis of primary and metastatic tumor ecosystems in head and neck cancer. Cell. 2017;171:1611–1624.e24. doi: 10.1016/j.cell.2017.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambow F., Marine J.-C., Goding C.R. Melanoma plasticity and phenotypic diversity: therapeutic barriers and opportunities. Genes Dev. 2019;33:1295–1318. doi: 10.1101/gad.329771.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockich B.E., Hrycaj S.M., Shih H.P., Nagy M.S., Ferguson M.A.H., Kopp J.L., Sander M., Wellik D.M., Spence J.R. Sox9 plays multiple roles in the lung epithelium during branching morphogenesis. Proc. Natl. Acad. Sci. 2013;110:E4456–E4464. doi: 10.1073/pnas.1311847110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rostoker R., Abelson S., Genkin I., Ben-Shmuel S., Sachidanandam R., Scheinman E.J., Bitton-Worms K., Orr Z.S., Caspi A., Tzukerman M., LeRoith D. CD24+ cells fuel rapid tumor growth and display high metastatic capacity. Breast Cancer Res. 2015;17:78. doi: 10.1186/s13058-015-0589-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A., Cao E.Y., Kumar V., Zhang X., Leong H.S., Wong A.M.L., Ramakrishnan N., Hakimullah M., Teo H.M.V., Chong F.T., et al. Longitudinal single-cell RNA sequencing of patient-derivedprimary cells reveals drug-induced infidelity in stem cell hierarchy. Nat. Commun. 2018;9:4931. doi: 10.1038/s41467-018-07261-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A., Merritt E., Hu X., Cruz A., Jiang C., Sarkodie H., Zhou Z., Malhotra J., Riedlinger G.M., De S. Non-genetic intra-tumor heterogeneity is a major predictor of phenotypic heterogeneity and ongoing evolutionary dynamics in lung tumors. Cell Rep. 2019;29:2164–2174.e5. doi: 10.1016/j.celrep.2019.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S.V., Lee D.Y., Li B., Quinlan M.P., Takahashi F., Maheswaran S., Mcdermott U., Azizian N., Zou L., Fischbach M.A., et al. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell. 2010;141:69–80. doi: 10.1016/j.cell.2010.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shipitsin M., Campbell L.L., Argani P., Weremowicz S., Bloushtain-Qimron N., Yao J., Nikolskaya T., Serebryiskaya T., Beroukhim R., Hu M., et al. Molecular definition of breast tumor heterogeneity. Cancer Cell. 2007;11:259–273. doi: 10.1016/j.ccr.2007.01.013. [DOI] [PubMed] [Google Scholar]
- Shlyakhtina Y., Moran K.L., Portal M.M. Genetic and non-genetic mechanisms underlying cancer evolution. Cancers. 2021;13:1380. doi: 10.3390/cancers13061380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh P., Augustine D., Rao R.S., Patil S., Awan K.H., Sowmya S.V., Haragannavar V.C., Prasad K. Role of cancer stem cells in head-and-neck squamous cell carcinoma–A systematic review. J. Carcinogenesis. 2021;20:12. doi: 10.4103/jcar.jcar_14_20. [DOI] [Google Scholar]
- Sleeman K.E., Kendrick H., Ashworth A., Isacke C.M., Smalley M.J. CD24 staining of mouse mammary gland cells defines luminal epithelial, myoepithelial/basal and non-epithelial cells. Breast Cancer Res. 2005;8:R7. doi: 10.1186/bcr1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanta G., Bonin S. Overview on clinical relevance of intra-tumor heterogeneity. Front. Med. 2018;5:85. doi: 10.3389/fmed.2018.00085. [DOI] [Google Scholar]
- Tang D.G. Understanding cancer stem cell heterogeneity and plasticity. Cell Res. 2012;22:457–472. doi: 10.1038/cr.2012.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang M.-R., Guo J.-Y., Di Wang N.X., Xu N. Identification of CD24 as a marker for tumorigenesis of melanoma. OncoTargets Ther. 2018;11:3401–3406. doi: 10.2147/ott.s157043. [DOI] [Google Scholar]
- Tirosh I., Venteicher A.S., Hebert C., Escalante L.E., Patel A.P., Yizhak K., Fisher J.M., Rodman C., Mount C., Filbin M.G., et al. Single-cell RNA-seq supports a developmental hierarchy in human oligodendroglioma. Nature. 2016;539:309–313. doi: 10.1038/nature20123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vipparthi K., Patel A.K., Ghosh S., Das S., Das C., Das K., Sarkar A., Thatikonda V., Pal B., Remani A.S.K.N., et al. Two novel cell culture models of buccal mucosal oral cancer from patients with no risk-habits of tobacco smoking or chewing. Oral Oncol. 2021;113:105131. doi: 10.1016/j.oraloncology.2020.105131. [DOI] [PubMed] [Google Scholar]
- White J.S., Weissfeld J.L., Ragin C.C.R., Rossie K.M., Martin C.L., Shuster M., Ishwad C.S., Law J.C., Myers E.N., Johnson J.T., Gollin S.M. The influence of clinical and demographic risk factors on the establishment of head and neck squamous cell carcinoma cell lines. Oral Oncol. 2007;43:701–712. doi: 10.1016/j.oraloncology.2006.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung T.M., Gandhi S.C., Wilding J.L., Muschel R., Bodmer W.F. Cancer stem cells from colorectal cancer-derived cell lines. Proc. Natl. Acad. Sci. 2010;107:3722–3727. doi: 10.1073/pnas.0915135107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Xiong X., Sun Y. Functional characterization of SOX2 as an anticancer target. Signal Transduction Targeted Ther. 2020;5:135. doi: 10.1038/s41392-020-00242-3. [DOI] [Google Scholar]
- Zimmerer R.M., Ludwig N., Kampmann A., Bittermann G., Spalthoff S., Jungheim M., Gellrich N.-C., Tavassol F. CD24+ tumor-initiating cells from oral squamous cell carcinoma induce initial angiogenesis in vivo. Microvasc. Res. 2017;112:101–108. doi: 10.1016/j.mvr.2017.03.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
RNAseq data have been deposited at ENA database and are publicly available as of the date of publication. Accession numbers are listed in the key resources table.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.