Abstract
Introduction
Efficient induction of the otic placode, the developmental origin of the inner ear from human pluripotent stem cells (hPSCs), provides a robust platform for otic development and sensorineural hearing loss modelling. Nevertheless, there remains a limited capacity of otic lineage specification from hPSCs by stepwise differentiation methods, since the critical factors for successful otic cell differentiation have not been thoroughly investigated. In this study, we developed a novel differentiation system involving the use of a three-dimensional (3D) floating culture with signalling factors for generating otic cell lineages via stepwise differentiation of hPSCs.
Methods
We differentiated hPSCs into preplacodal cells under a two-dimensional (2D) monolayer culture. Then, we transferred the induced preplacodal cells into a 3D floating culture under the control of the fibroblast growth factor (FGF), bone morphogenetic protein (BMP), retinoic acid (RA) and WNT signalling pathways. We evaluated the characteristics of the induced cells using immunocytochemistry, quantitative PCR (qPCR), population averaging, and single-cell RNA-seq (RNA-seq) analysis. We further investigated the methods for differentiating otic progenitors towards hair cells by overexpression of defined transcription factors.
Results
We demonstrated that hPSC-derived preplacodal cells acquired the potential to differentiate into posterior placodal cells in 3D floating culture with FGF2 and RA. Subsequent activation of WNT signalling induced otic placodal cell formation. By single-cell RNA-seq (scRNA-seq) analysis, we identified multiple clusters of otic placode- and otocyst marker-positive cells in the induced spheres. Moreover, the induced otic cells showed the potential to generate hair cell-like cells by overexpression of the transcription factors ATOH1, POU4F3 and GFI1.
Conclusions
We demonstrated the critical role of FGF2, RA and WNT signalling in a 3D environment for the in vitro differentiation of otic lineage cells from hPSCs. The induced otic cells had the capacity to differentiate into inner ear hair cells with stereociliary bundles and tip link-like structures. The protocol will be useful for in vitro disease modelling of sensorineural hearing loss and human inner ear development and thus contribute to drug screening and stem cell-based regenerative medicine.
Keywords: Human pluripotent stem cell, Floating culture, Otic placode, Single cell RNA-seq, Hair cell
Abbreviations: BMP, bone morphogenetic protein; CHIR, CHIR-99021; EGF, epidermal growth factor; FGF, fibroblast growth factor; hESCs, human embryonic stem cells; hPSCs, human pluripotent stem cells; KSR, KnockOut serum replacement; LDN, LDN-193189; OCT, optimal cutting temperature; PBS, phosphate-buffered saline; qPCR, quantitative PCR; RA, retinoic acid; SB, SB-431542; scRNA-seq, single-cell RNA-sequencing; SEM, scanning electron microscope; SF, serum free; TGFβ, transforming growth factor-β; 2D, two-dimensional; 3D, three-dimensional
Highlights
-
•
A 3D floating culture condition is critical for inducing otic placodal cells from hPSCs-derived preplacodal cells.
-
•
Activation of FGF, RA, WNT signalling pathways is indispensable for differentiating otic lineage under the 3D condition.
-
•
Overexpression of defined transcription factors facilitated the generation of hair cells from hPSCs-derived otic cells.
1. Introduction
Sensorineural hearing loss is caused by cochlear cell defects. Physical or pharmacological damage to mammalian cochlear sensory cells, such as spiral ganglion and hair cells, is irreversible and results in permanent hearing loss [1,2]. Pluripotent stem cells (PSCs) offer a potential resource for regenerative therapies of sensorineural hearing loss, as they can undergo unlimited proliferation and have the potential to differentiate into all three germ cell layers. Induction of cochlear cells from human PSCs (hPSCs) paves the way for inner ear disease modelling and drug development. To facilitate the exploitation of hPSCs for inner ear disease research, rapid, efficient and highly reproducible methods for differentiating hPSCs into otic cell lineages are indispensable.
The development of the early otic lineage has been well studied in various model organisms [3,4] (Fig. 1A). During gastrulation, the ectoderm divides into neural and nonneural segments, and the latter give rise to Six1-and Eya1-positive preplacodal ectoderm, which is the progenitor of all the cranial placodes. Subsequently, the preplacodal region is subdivided into anterior and posterior territories. The anterior placodes develop into the adenohypophyseal, olfactory, lens, and trigeminal placodes, while the posterior placodes become the otic and epibranchial placodes. The otic placode is a thickened epithelium marked by the expression of transcription factors such as Pax8, Pax2, and Dlx5 [5]. Various inner ear cell types, including hair cells, supporting cells, cochlear and vestibular ganglion neurons, and nonsensory cells, are derived from the otic placode.
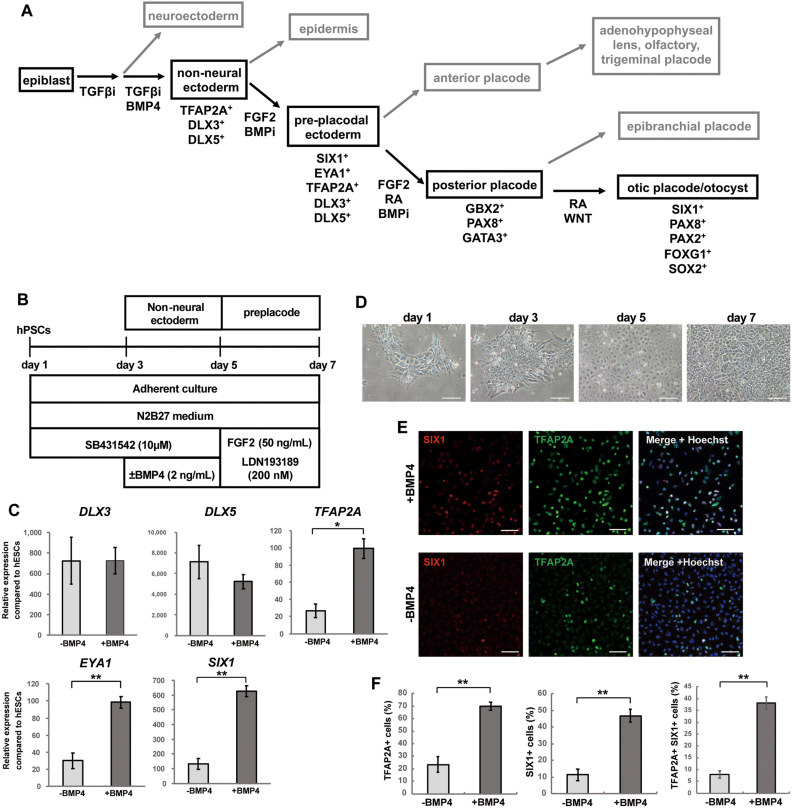
Fig. 1.
Induction of preplacodal ectoderm from hPSCs. (A) Schematic diagram of otic placode development based on studies on nonprimate animal models. Differentiation of the nonneural ectoderm from pluripotent stem cells is mediated by TGFβ signal inhibition with BMP signal activation. Subsequent activation of FGF signalling and inhibition of BMP signalling are necessary for preplacode development. The preplacodal ectoderm is subdivided into anterior and posterior portions. The anterior placode gives rise to the adenohypophyseal, lens, olfactory and trigeminal placodes, while the posterior placode develops into the epibranchial and otic placodes. TGFβi; TGFβ signalling inhibitor, BMPi; BMP signalling inhibitor, FGFi; FGF signalling inhibitor. (B) Schematic figure of preplacodal ectoderm induction via manipulation of TGFβ, BMP, and FGF signalling. Two different culture conditions were compared (with or without BMP4 from Day 3 to Day 5) by analyses of the expression of nonneural ectoderm and preplacode marker genes by qPCR and immunocytochemistry. (C) qRT–PCR of DLX3, DLX5, EYA1, SIX1, and TFAP2A in the human ESC line H9 on Day 7 with or without BMP4 supplementation. The relative gene expression was normalized against that in undifferentiated hESCs. The bars and error bars represent the mean ± SEM (three independent experiments). Asterisks indicate significant differences (∗p < 0.05, ∗∗p < 0.01, two-tailed unpaired Student's t tests). (D) Bright field images of preplacode differentiation on Days 1, 3, 5, and 7 of culture. The cells were treated with SB from Day 1 to Day 2, with SB and BMP4 from Day 3 to Day 4, and with FGF2 and LDN from Day 5 to Day 7 of differentiation. Scale bars, 100 μm. (E) Representative immunocytochemical images for the nonneural ectoderm marker TFAP2A and the preplacode marker SIX1 in H9 hESC-derived preplacodal cells on Day 7 with or without BMP4 supplementation. The nuclei were stained with Hoechst 33,342. Scale bars, 50 μm. (F) Quantitative analysis of TFAP2A+ cells, SIX1+ cells and TFAP2A+/SIX1+ cells among H9 hESC-derived preplacodal cells on Day 7 with or without BMP4 supplementation. The bars and error bars represent the mean ± SEM (three independent experiments). Asterisks indicate significant differences (∗∗p < 0.01, two-tailed unpaired Student's t test).
Generation of the otic cell lineage from human PSCs has been achieved by using two-dimensional (2D) monolayer and 3D culture methods [[6], [7], [8], [9], [10], [11], [12], [13], [14], [15], [16], [17]]. Of all the published protocols, one approach involves stepwise differentiation to recapitulate intravital otic placode development. Ealy et al. used a 2D monolayer culture protocol in combination with cytokine/inhibitor-based control of transforming growth factor-β (TGFβ)/WNT and fibroblast growth factor (FGF)/WNT/bone morphogenetic protein (BMP)/retinoic acid (RA) signalling pathways [11]. However, this protocol resulted in a mixed phenotype of both anterior and posterior placodal cells even after treatment with posteriorizing factors (FGF/WNT/RA). This outcome emphasizes the importance of precise temporal control of critical signals. Moreover, several studies have demonstrated efficient differentiation of hPSCs into SIX1+ preplacodal ectoderm under 2D conditions followed by further differentiation of anterior placode-derived cells such as lens, trigeminal, and anterior pituitary cells [[18], [19], [20]]. However, the ability of preplacodal ectoderm cells to differentiate into posterior placodes, including otic placodes, has not been fully investigated.
In contrast to 2D culture methods, 3D culture systems using mouse and human PSCs can generate PAX8+ preotic posterior placodal cells without treatment with CHIR-99021 (CHIR: a classical WNT signal agonist) and RA, which are important for otic placode development [14,21]. These results imply that the 3D environment itself, or contamination by other lineage cells, such as cranial neural crest cells and/or neuroectoderm-derived cells may contribute to the lineage commitment of posterior placodal cells. Although 3D culture systems offer a promising strategy for otic cell differentiation, the reported induction efficiencies have been variable owing to the spontaneous formation of anterior-posterior, dorsal-ventral, and medial–lateral axes in the floating cell aggregates. The cellular heterogeneity frequently observed in 3D culture imposes limitations on the directed differentiation of otic cells.
To overcome the limitations of both 2D and 3D culture systems, we sought to devise a culture protocol harbouring the advantages of both 2D and 3D environments for otic cell differentiation. Here, we describe the differentiation of hPSCs into a preplacodal ectoderm via stepwise 2D monolayer culture followed by 3D culture of the preplacodal cells to differentiate into the otic cell lineage. We confirmed the characteristics of the induced otic cells by multimodal analyses, including qPCR, immunocytochemistry and single-cell RNA-seq (scRNA-seq). Our data demonstrated the importance of three signalling pathways (FGF, RA and WNT) in human inner ear development. Furthermore, the induced otic cells showed the potential to differentiate into MYO7A+ hair cells with F-actin-rich protrusions by overexpression of the transcription factors ATOH1, POU4F3 and GFI1. Our protocol holds promise for further investigation of the pathophysiology of sensorineural hearing loss and drug screening for related diseases.
2. Materials and methods
2.1. Human PSCs culture
We used the human embryonic stem cell (hESC) line H9. These PSCs were maintained on mitomycin-C-treated SNL feeder cells in DMEM/F12 (FUJIFILM Wako Pure Chemical) hESC medium containing 20% KnockOut serum replacement (Life Technologies), 1% nonessential amino acids (Sigma), 2 mM L-glutamine (Nacalai Tesque), 0.1 mM 2-mercaptoethanol (Sigma), 4 ng/mL FGF2 (PeproTech), 100 U/mL penicillin and 100 μg/mL streptomycin. Cells were passaged onto feeder cells or into feeder-free culture using a dissociation solution containing 0.25% trypsin, 100 mg/mL collagenase type IV (Invitrogen), 1 mM CaCl2 and 20% KnockOut serum replacement (KSR). For feeder-free culture, cells were passaged on Matrigel (Corning, #354277)-coated dishes in SNL feeder-conditioned hESC medium containing 5 ng/mL FGF2.
2.2. Differentiation of otic placodal cells
PSCs in feeder-free culture were dissociated into single cells using Accutase (Nacalai Tesque), resuspended in SNL cell-conditioned hESC medium with 5 ng/mL FGF2 and 10 μM Y-27632 (ROCK inhibitor; FUJIFILM Wako Pure Chemical), and passaged on Matrigel-coated dishes at 15,000–18,000 cells/cm2. After 24 h, the medium was changed to SNL cell-conditioned hESC medium containing 5 ng/mL FGF2 (Day 0). Differentiation was initiated on Day 1 by exchanging the total medium for serum-free (SF) DMEM/F12 medium containing 1x N2 supplement (Invitrogen), 1x B27 supplement (Invitrogen), 1% nonessential amino acids, 1% GlutaMAX (Life Technologies), 0.1 mM 2-mercaptoethanol and 100 μg/mL ampicillin and supplemented with 10 μM SB-431542 (Sigma). From Day 3, the cells were cultured in SF medium supplemented with 10 μM SB-431542 and 2 ng/mL BMP4 (PeproTech). From Day 5, the cells were cultured in SF medium supplemented with 50 ng/mL human FGF2 and 200 nM LDN-193189 (StemRD). Until Day 7, the total medium was changed every day. On Day 7, the cells were dissociated into single cells by Accutase and cultured in an ultra-low-adhesion-6-well plate (Corning) at a density of 150,000 cells/well in DMEM/F12 medium supplemented with N2, B27-vitamin A (Thermo Fisher Scientific), 1% Matrigel, 10 μM Y-27632, 50 ng/mL FGF2, 50 ng/mL FGF3 (R&D Systems), 50 ng/mL FGF10 (PeproTech), 20 ng/mL EGF (PeproTech), 200 nM LDN193189 (StemRD), 50 ng/mL heparan sulfate (Nacalai Tesque) and 200 nM RA (Sigma). On day 10, all spheres in the medium were collected in a 15 mL tube and centrifuged at 120×g for 3 min. After removal of the supernatant, fresh medium was added to the 15 mL tube with gentle pipetting, and the spheres were transferred to a 96-well plate. The spheres in 2 wells of an ultra-low-adhesion 6-well plate were transferred to 96 wells of an ultra-low-adhesion 96-well plate (Corning), and the total medium was changed to DMEM/F12 medium supplemented with 1x N2, 1x B27-vitamin A (Invitrogen), 1% Matrigel, 50 ng/mL FGF3, 50 ng/mL FGF10, 20 ng/mL EGF, 200 nM LDN, 50 ng/mL heparan sulfate, and 500 nM RA. On Day 14, the total medium was replaced with DMEM/F12 medium supplemented with 1x N2, 1x B27-vitamin A, 1% Matrigel, 50 ng/mL FGF3, 50 ng/mL FGF10, 20 ng/mL EGF, 200 nM LDN, 50 ng/mL heparan sulfate, 1 μM RA, and 3 μM CHIR (Focus Biomolecules). On Day 18, the total medium was replaced with DMEM/F12 medium supplemented with 1x N2, 1x B27-vitamin A, 1% Matrigel, 50 ng/mL FGF3, 50 ng/mL FGF10, 20 ng/mL EGF, 200 nM LDN, 50 ng/mL heparan sulfate, 1 μM RA, and 3 μM CHIR. On Days 14 and 18, the spheres were transferred to 96-well plates at an equal ratio. All spheres in medium were first collected in a 15 mL tube. Then, the 96-well plate was rinsed once with PBS, and the rinses were also collected into the same tube and centrifuged at 120×g for 3 min. After removal of the supernatant, fresh medium was added to the 15 mL tube with gentle pipetting, and the spheres were transferred to a 96-well plate.
2.3. Differentiation of inner ear hair cells
Based on the results of scRNA-seq, for hair cell differentiation, the concentrations of FGF3 and FGF10 were reduced from 50 ng/mL to 25 ng/mL (from Day 7 to Day 22), and the concentrations of RA were optimized to 200 nM (from Day 7 to Day 14) and 20 nM (from Day 14 to Day 22). From Day 22, the total medium was replaced with DMEM/F12 medium supplemented with 1x N2, 1x B27-vitamin A, 1% Matrigel, 20 ng/mL EGF, 200 nM LDN, and 3 μM CHIR. From Day 26, the total medium was replaced with DMEM/F12 medium supplemented with 1x N2, 1x B27-vitamin A, 1% Matrigel, 20 ng/mL EGF, 200 nM LDN, and 1.5 μM CHIR. On Days 22 and 26, the spheres were transferred to 96-well plates at an equal ratio. All spheres in medium were first collected in a 15 mL tube. Then, the 96-well plate was rinsed once with PBS, and the rinses were also collected into the same tube and centrifuged at 120×g for 3 min. After the removal of supernatant, fresh medium was added to the 15 mL tube with gentle pipetting, and the spheres were passed to a 96-well plate. On Day 28 or 30, the spheres were dissected with a tungsten needle, and the epithelial parts of the spheres were collected. The isolated epithelial aggregates were embedded in 25 μl of 100% Matrigel by the hanging drop method on prewarmed 24-well plates. After 30 min, DMEM/F12 medium supplemented with 1x N2, 1x B27-vitamin A, 1% Matrigel, 20 ng/mL EGF, and 1.5 μM CHIR was added to each well, and the spheres were cultured under hyperoxia. The total medium was changed every other day. After 10 days of Matrigel embedding culture, the spheres were dissected to remove Matrigel and the overproliferated nonepithelial cells using a tungsten needle. Then, the dissected spheres were dissociated into single cells using TrypLE Express at 37 °C for 40 min. The dissociated cells were resuspended in DMEM/F12 medium supplemented with 1x N2, 1x B27-vitamin A, 20 ng/mL EGF, 1.5 μM CHIR and 10 μM Y-27632 and seeded onto a polyornithine-fibronectin-coated 8-well chamber (IWAKI) at 7.2 × 104–9.6 × 104 cells/well. The cells were infected with lentivirus (MOI = 5). After 2 days, the total medium was changed to DMEM/F12 medium supplemented with 1x N2, 1x B27-vitamin A, 20 ng/mL EGF, and 1.5 μM CHIR. Half of the medium was changed every other day.
2.4. Immunocytochemistry
Monolayer-cultured cells and floating spheres were fixed in 4% paraformaldehyde at 4 °C overnight. The fixed spheres were cryoprotected in a solution of 30% sucrose and embedded in optimal cutting temperature (OCT) compound (Sakura Finetek). The frozen samples were sectioned into 20 μm sections using a cryostat. For immunostaining, the cells were subjected to antigen retrieval in 10 mM citrate buffer (pH 6.0) for 20 min at 80 °C and permeabilized with 0.3% Triton-X in phosphate-buffered saline (PBS) for 5 min. The cells were blocked with 10% normal donkey serum (Nacalai Tesque) in PBS for 1 h and incubated overnight at 4 °C with the following primary antibodies: mouse anti-SIX1 (Atlas Antibodies; AMAb90544; 1:200), mouse anti-E-cadherin (BD; 610,182; 1:500), rabbit anti-PAX8 (Abcam; ab191870; 1:500), rabbit anti-TFAP2A (Abcam; ab108311; 1:200), rabbit anti-PAX2 (Abcam; ab79389; 1:500), rabbit anti-PAX6 (MBL; PD022; 1:200), goat anti-SOX2 (Santa Cruz; sc-17320; 1:100), goat anti-SOX2 (R&D; AF2018; 1:500), rabbit anti-FOXG1 (Abcam; ab196868; 1:200), mouse anti-EPCAM (Cell Signaling; 5198S; 1:500), rabbit anti-JAG1 (Abcam; ab109536; 1:200), and mouse anti-MYO7A (DSHB; 138–1; 1:50). On the following day, the cells were washed with PBS and incubated with secondary antibodies conjugated with Alexa Fluor 488, 555, or 647 (Thermo Fisher Scientific; 1:1,000) for 1 h at room temperature.
For F-actin labelling, the cells were incubated with Alexa Fluor 555 phalloidin (Invitrogen; A34055; 1:400) with other secondary antibodies for 1 h at room temperature. For coimmunostaining with PAX2 and PAX8 antibodies, after labelling with the PAX2 antibody and secondary antibody, the cells were incubated with an Alexa Fluor 555-conjugated rabbit anti-PAX8 antibody (Abcam; ab217733; 1:500) overnight at 4 °C. The nuclei were stained with Hoechst 33,342 (Thermo Fisher Scientific; 1:1,000). The samples were analysed using an LSM700 confocal laser scanning microscope (Carl Zeiss).
2.5. Quantitative RT-PCR (qPCR)
RNA was isolated using an RNeasy Mini Kit (QIAGEN) according to the manufacturer's instructions. PolyA + RNA was reverse-transcribed by Prime Script II (Toyobo). cDNA was synthesized from 500 ng of RNA. qRT–PCR was performed using TB Green Premix Ex Taq II (TaKaRa Bio) on the ViiA 7 Real-Time PCR System (Thermo Fisher Scientific) according to the manufacturer's instructions. The sequences of the primers used in this study are listed in Table 1. ACTB (Figs. 1B, 2B-D, 3B, 5B, Fig. S1B-D) and HPRT1 (Fig. 5B) expression was used for normalization.
Table 1.
The primer sequences for qPCR analysis
| Transcript | Forward | Reverse |
|---|---|---|
| PAX2 | TCAAGTCGAGTCTATCTGCATCC | CATGTCACGACCAGTCACAAC |
| PAX8 | GCCCAGTGTCAGCTCCATTA | GCTGTCCATAGGGAGGTTGAA |
| GATA3 | CCTAAGGTGGTTGTGCTCGG | CACAGGCTGCAGGAATAGGG |
| SIX1 | ACAAGAACGAGAGCGTACTCA | CTCCACGTAATGCGCCTTCA |
| DLX3 | TACCCTGCCCGAGTCTTCTG | TGGTGGTAGGTGTAGGGGTTC |
| DLX5 | GTCTTCAGCTACCGATTCTGAC | CTTTGCCATAGGAAGCCGAG |
| FBXO2 | GTGTCGCAAAGCACAGGTC | CGGACAGTAGCTTAACGGTGAG |
| EYA1 | GGACTATCCGTCTTATCCCAGT | GCTGCTGGTCATATAATGTGCTG |
| ACTB | TGAAGTGTGACGTGGACATC | GGAGGAGCAATGATCTTGAT |
| GBX2 | GACGAGTCAAAGGTGGAAGAC | GATTGTCATCCGAGCTGTAGTC |
| JAG1 | CGGGAACATACTGCCATGAAAATA | ATGCACTTGTAGGAGTTGACACCA |
| OTX2 | CAAAGTGAGACCTGCCAAAAAGA | TGGACAAGGGATCTGACAGTG |
| TFAP2A | CTCCGCCATCCCTATTAACAAG | GACCCGGAACTGAACAGAAGA |
| PAX6 | TGGGCAGGTATTACGAGACTG | ACTCCCGCTTATACTGGGCTA |
| SOX2 | GCTGGCAGACCTACATGAAC | GGGAGGAAGAGGTAACCACA |
| HPRT1 | TGACACTGGCAAAACAATGCA | GGTCCTTTTCACCAGCAAGCT |
Fig. 5.
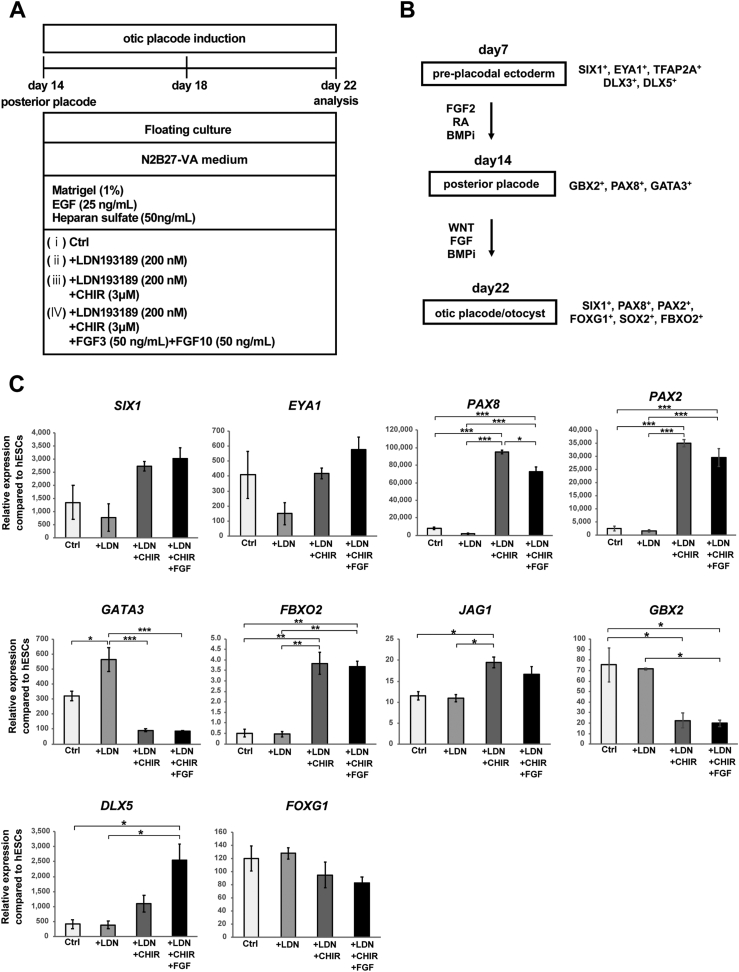
Differentiation of otic placodes from hPSCs via manipulation of BMP, WNT, and FGF signalling. (A) Schematic figure of the culture conditions used to investigate the roles of BMP, WNT, and FGF signalling in the differentiation of the otic placode. Four different culture conditions (Ctrl, +LDN, +LDN + CHIR, +LDN + CHIR + FGF3/FGF10) were compared to analyse the effects of BMP, WNT, and FGF signal manipulation on otic placode marker expression. (B) Signalling pathways important for deriving otic cell lineages from preplacodal cells and representative marker genes of each cell lineage. (C) qPCR analysis of the otic plate markers SIX1, EYA1, PAX8, PAX2, GATA3, FBXO2, JAG1, GBX2, DLX5, and FOXG1 on Day 22 of culture with or without LDN, CHIR, or FGF3/FGF10 from Day 14 to Day 22. The relative gene expression levels were normalized against those of undifferentiated hESCs. The bars and error bars represent the mean ± SEM (three independent experiments). Asterisks indicate significant differences (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, one-way ANOVA with Tukey's post hoc test).
2.6. Single cell dissociation and single-cell RNA sequencing
On Day 22, spheres with or without RA treatment were washed once with PBS, incubated with Accutase for 35 min at 37 °C and dissociated into single cells by tapping and pipetting. The cells were spun down at 120×g for 5 min, resuspended in PBS with 1% BSA, and checked for cell viability. For scRNA-seq, dissociated cells were processed with a 10x Genomics Chromium platform following the manufacturer's instructions. Briefly, ∼13,200 cells were loaded to yield an estimated recovery of 8,000 cells per sample onto a Chromium Single Cell G Chip (10x Genomics), and GEMs (Gel Beads in Emulsion) were generated using the Chromium Controller. Libraries were constructed with a Chromium Next GEM Single Cell 3′ Kit ver. 3.1 (10x Genomics) and sequenced on a HiSeq X instrument (Illumina).
2.7. scRNA-seq data analysis
The Cell Ranger pipeline (v6.0.2) was used to map reads to the GRCh38 human reference genome and generate cell-by-gene unique molecular identifier (UMI) count matrices. The count matrices were analysed using the Seurat (v4.0.5) R (v4.1.2) package. Cells with a total UMI count >64,819 (RA-), 62,106 (RA+), or cells with a mitochondrial read percentage >18% (RA-), 12% (RA+) were excluded. Doublets were detected by the scDblFinder (v1.8.0) package and subsequently removed. The gene UMI counts were normalized by the total UMI counts per cell, multiplied by a scaling factor of 10,000, and log-transformed. The top 3,000 variable genes were used for principal component analysis. Leiden clustering with the default cut-offs and uniform manifold approximation and projection (UMAP) visualization were performed on the first 30 principal components. Genes specifically expressed in each cell cluster were identified using the FindAllMarkers function. Then, the cluster cell types were manually annotated based on the cluster-specific gene lists.
2.8. Cloning of plasmids and lentivirus production
To construct the CAG-APG (ATOH1, POU4F3 and GFI1) lentiviral vector, the hATOH1-P2A-hPOU4F3-T2A-hGFI1 sequence from pPB-TRE3G-APG (customized by VectorBuilder) was cloned into CS-CA-MCS (kindly provided by Dr. Hiroyuki Miyoshi, Keio University) by a restriction enzyme-based method. The CAG-APG lentivirus was produced as described previously. Briefly, packaging vectors such as pCAG-HIVgp, pCMV-VSV-G-RSV-Rev (both kindly provided by Dr. Hiroyuki Miyoshi, Keio University) and CAG-APG lentivirus vector were transfected into HEK293T cells using polyethyleneimine at subconfluency in DMEM with high glucose (Thermo Fisher Scientific) supplemented with 10% FBS (Sigma Aldrich) and 1% penicillin/streptomycin (Nacalai Tesque). After 24 h, the medium was replaced with 10% FBS DMEM with high glucose containing 10 μM forskolin (Sigma Aldrich). Two days after the medium was changed, the supernatant was collected, ultracentrifuged at 25,000 rpm for 2 h at 4 °C, and concentrated by ultrafiltration using Amicon Ultra 0.5 mL filters (Millipore). The titres were determined with a qPCR Lentivirus Titration Kit (Applied Biological Materials).
2.9. Scanning electron microscopy (SEM)
Cells were fixed in 2.5% glutaraldehyde for 12 h at 4 °C, washed in 0.1 M PB, and postfixed in 1% OsO4 for 2 h at 4 °C. The cells were dehydrated with a series of increasing concentrations of ethanol (50%, 70%, 80%, 90%, 95% and 100%), freeze-dried with 100% t-butyric alcohol (VFD-21S, vacuum device), and coated with platinum-palladium by a heavy metal coater (SC-701, Sanyu Electron). The samples were analysed using field emission scanning electron microscopy (SU6600 Hitachi High-Technologies) with 7.0 keV acceleration.
2.10. Statistical analysis
All values are expressed as the mean ± SEM. The relative gene expression levels and the number of placode marker-positive cells in each group were compared statistically with two-tailed unpaired Student's t tests or one-way ANOVA followed by a post hoc Tukey's test. A p value of <0.05 was considered to indicate statistical significance.
3. Results
3.1. Differentiation of preplacodal ectoderm from human PSCs
Initially, we sought to differentiate hPSCs (H9 ESCs) into preplacodal ectoderm in a 2D monolayer culture by manipulating TGFβ, BMP, and FGF signalling. Previous studies have demonstrated that the TGF-β inhibitor SB-431542 (SB) supplemented with BMP4 can promote nonneural ectoderm differentiation from PSCs [18,20,22,23]. After the specification of nonneural ectoderm, FGF activation and BMP inhibition promote the induction of preplacodal cells [13,14,20,21] (Fig. 1A). To differentiate hPSCs into preplacodal ectoderm, we initially treated the cells with SB for 2 days and then treated them with SB and BMP4 for 2 days to induce nonneural ectoderm. From Day 5, we treated the cells with FGF2 and the BMP inhibitor LDN-193189 (LDN) to derive preplacodal ectoderm (Fig. 1B). qPCR analysis showed that supplementation with BMP4 from Day 3 to Day 5 triggered significant upregulation of the nonneural ectoderm marker TFAP2A and the preplacodal markers SIX1 and EYA1 [24,25] on Day 7 (Fig. 1C). The expression levels of the nonneural ectoderm markers DLX5 and DLX3 [24,25] were dramatically upregulated in both BMP4-treated and untreated conditions (Fig. 1C). In the process of differentiation, we observed dynamic morphological changes in the cultured cells from Day 3 to Day 7 (Fig. 1D). In particular, the cells changed from having a high nucleus/cytoplasm ratio with prominent nucleoli to having a polygonal shape. Moreover, immunocytochemical analysis showed that supplementation with BMP4 during preplacodal induction (Day 3 to Day 5) significantly increased the number of SIX1+ cells (–BMP4, 11.3 ± 3.41%; +BMP4, 46.8 ± 3.76%; p = 0.005), TFAP2A+ cells (–BMP4, 23.3 ± 6.15%; +BMP4, 69.8 ± 3.49%; p = 0.006) and SIX1+/TFAP2A+ cells (–BMP4, 7.83 ± 1.56%; +BMP4, 38.2 ± 2.55%; p = 0.001) (Fig. 1E and F). These results demonstrated that efficient preplacode induction from hPSCs was successfully achieved by transient activation of BMP signalling.
3.2. FGF2 and RA treatments are critical for deriving posterior placodal cells from induced preplacodal cells in a 3D culture
Previous studies have shown that the expression of anterior placodal markers does not decrease after preplacode induction under 2D conditions using posteriorizing cues such as FGF, WNT, and RA [11]. 3D culture systems permit induction of PAX8+ posterior placodal cells after supplementation with FGF2 and LDN [14], suggesting that the 2D monolayer culture system tends to promote anterior placode differentiation from preplacodal cells. We therefore postulated that the 3D culture condition might facilitate the posteriorization of preplacodal cells derived from 2D culture. Various studies have shown that posterior placodal fate, including the otic placode fate, is determined by FGF and RA signalling [[26], [27], [28], [29], [30], [31], [32], [33]]. Therefore, to enhance posterior placode differentiation, we used a 3D floating culture system with FGF2 and RA treatments. During the 3D floating culture, we added Matrigel and epidermal growth factor (EGF) to support epithelial cell survival and proliferation. On Day 7, we transferred the induced preplacodal cells to a single-cell suspension culture with differentiation medium containing Matrigel, EGF, and LDN (Fig. 2A).
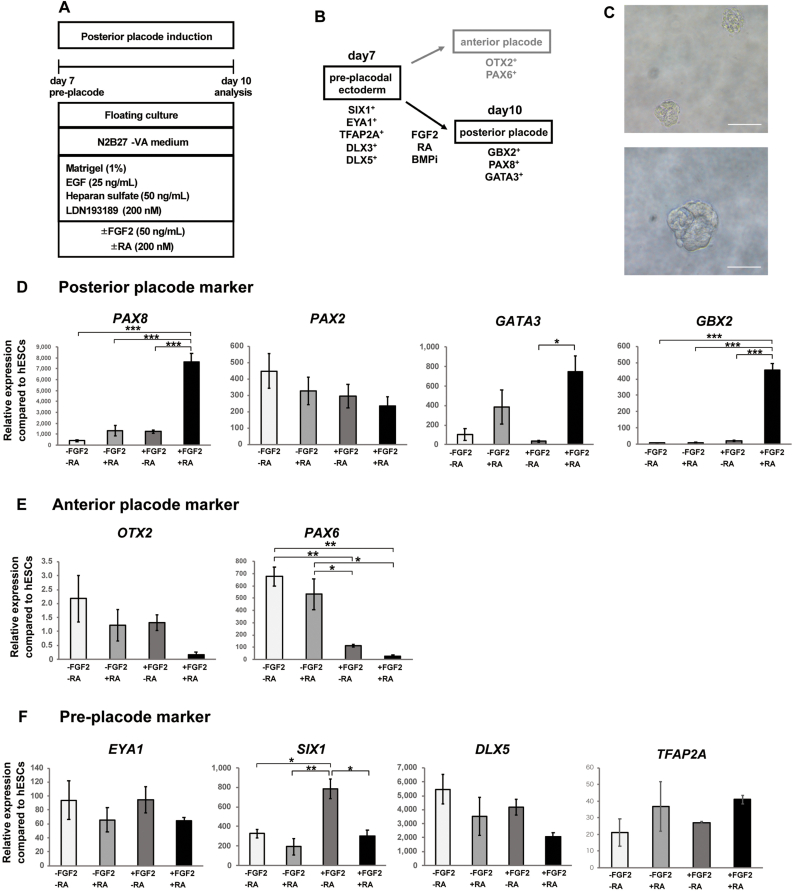
Fig. 2.
Generation of posterior placodal cells from preplacodal ectoderm using a 3D floating culture system. (A) Schematic figure of the differentiation of posterior placodes from hPSC-derived preplacodal cells. (B) Specification of anterior and posterior placodal cells from preplacodal cells. Representative pre-, anterior, and posterior placodal marker genes are shown. (C) Representative bright field images of floating spheres on Day 10. Scale bars, upper 100 μm, lower 50 μm. (D–F) qPCR analysis of posterior placode markers (PAX8, PAX2, GATA3, GBX2) (D), anterior placode markers (OTX2, PAX6) (E), and preplacode markers (EYA1, SIX1, DLX5, TFAP2A) (F) on Day 10 of culture. The relative gene expression levels were normalized against those of undifferentiated hESCs. The bars and error bars represent the mean ± SEM (three independent experiments). Asterisks indicate significant differences (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, one-way ANOVA with Tukey's post hoc test).
To assess the effects of the initial FGF2 and RA treatment, the cells were cultured in the presence or absence of 50 ng/mL FGF2 and 200 nM RA. Then, we compared the expression levels of the preplacode markers SIX1, EYA1, and DLX5; the posterior placode markers GBX2, GATA3, PAX2, and PAX8 [3,34]; and the anterior placode markers PAX6 and OTX2 [3] on Day 10 (Fig. 2B). We generated approximately 7,480 ± 1,121 spheres (n = 5 independent experiments, ± S.D.) per well in a 6-well ultra-low-attachment plate from the start of culture (Fig. 2C). We found that FGF2- and RA-treated cells showed significant increases in the expression of the posterior placode markers GBX2, GATA3, and PAX8 (Fig. 2D). Treatment with either FGF2 or RA alone did not significantly increase the expression of these markers (Fig. 2D). One of the otic placode markers, PAX2, did not show a significant change in expression after FGF2 and RA treatment (Fig. 2D). We found that the expression of the anterior placode markers PAX6 and OTX2 was decreased in FGF2- and RA-treated cells compared to untreated cells (Fig. 2E).
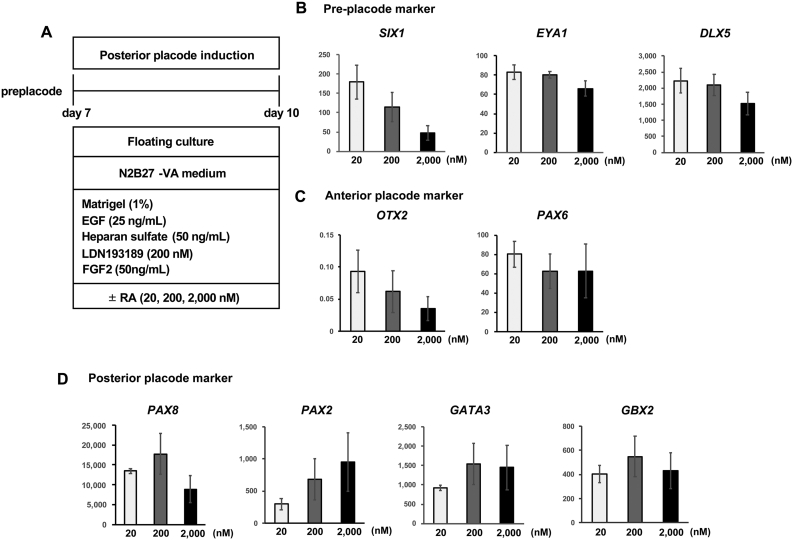
We also found that the expression of the preplacode markers SIX1, EYA1, and DLX5 was downregulated upon RA treatment (Fig. 2F). Next, to confirm the sensitivity of preplacode cells to RA, we compared marker gene expression after treating the cells with different RA concentrations (20 nM, 200 nM, 2,000 nM; Fig. 3A). qPCR analysis showed that the highest concentration of RA (2,000 nM) inhibited the expression of the pretreatment markers SIX1 and DLX5 (Fig. 3B), while PAX2 expression was upregulated by RA treatment in a concentration-dependent manner (Fig. 3D). While the expression of posterior placode markers did not show further upregulation at the highest RA concentration (2,000 nM) compared to the 200 nM RA-treated condition (Fig. 3D), the expression of the anterior placode marker OTX2 was downregulated at the highest RA concentration. In contrast, the expression of another anterior placode marker, PAX6, did not decrease upon treatment with 2,000 nM RA compared to 200 nM RA (Fig. 3C). These results suggest that floating culture with FGF2 and RA supplementation promotes the posteriorization of PSC-derived preplacodal cells. Although the highest RA concentration tested (2,000 nM) negatively affected the expression of several otic placodal marker genes, treatment with a high concentration of RA enhanced the gene expression of PAX2 (Fig. 3). Therefore, we gradually increased the RA concentration or withdrew RA at the time of medium change (on Day 10 and/or Day 14) for the subsequent experiments.
Fig. 3.
Effect of RA on otic placode marker expression. (A) Schematic figure of differentiation of the posterior placode from hPSC-derived preplacodal cells. Three different culture conditions (20 nM RA, 200 nM RA, and 2,000 nM RA) were compared to investigate the effect of RA on posterior placode marker expression. (B–D) qPCR analysis of preplacode markers (EYA1, SIX1, and DLX5) (B), anterior placode markers (OTX2 and PAX6) (C), and posterior placode markers (PAX8, PAX2, GATA3, and GBX2) (D) on Day 10 of culture. The relative gene expression levels were normalized against those of undifferentiated hESCs. The bars and error bars represent the mean ± SEM (three independent experiments).
3.3. 3D floating culture with BMP inhibition is important for maintaining several otic placodal marker expression
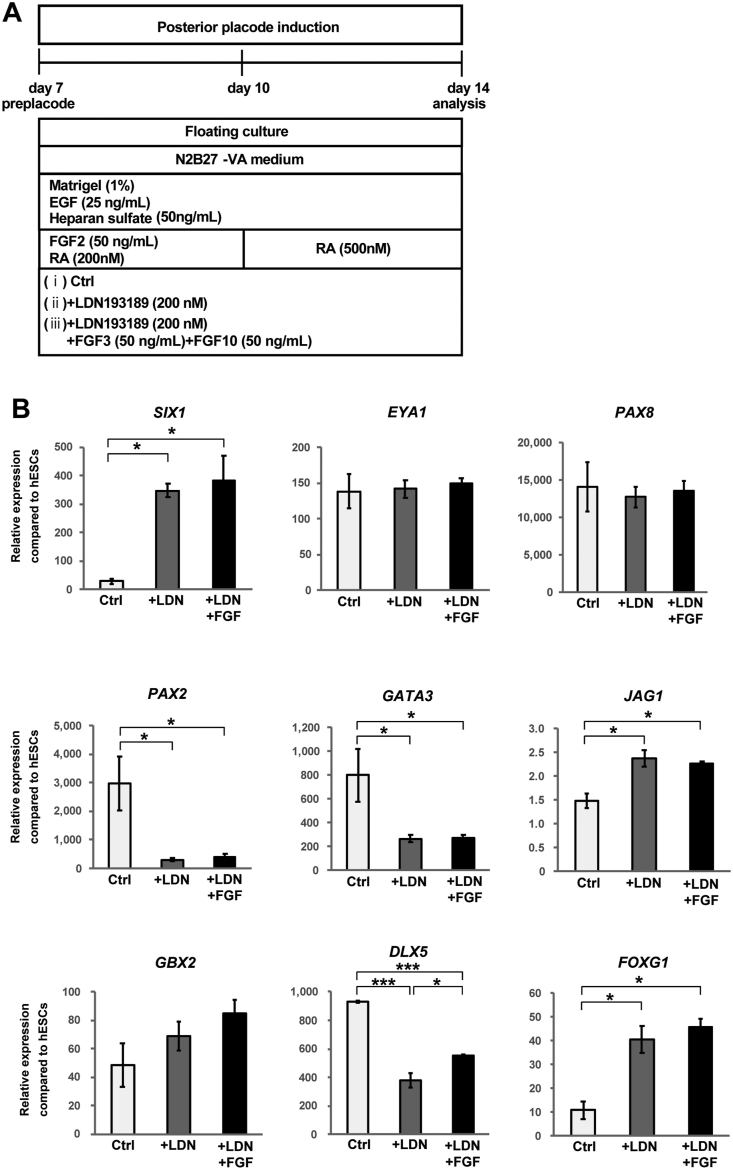
During otic development, the activation of FGF signalling and the inhibition of BMP signalling play significant roles in otic lineage specification (Fig. 1A). The BMP signal is important for anterior placode and epibranchial placode differentiation [[35], [36], [37], [38]], and specific FGF ligands such as FGF3 and FGF10 are important for otic placode development in mice [5,39]. Therefore, in addition to supplementation with FGF2 and RA, we investigated the effects of a BMP inhibitor and FGF3/FGF10 from Day 7 to Day 14 of the 3D floating culture. The expression of posterior placode marker genes was compared on Day 14 among 3 different culture conditions, as follows: control conditions (without LDN and FGF3/FGF10 supplementation), LDN treatment, and LDN + FGF3/FGF10 treatment from Day 7 to Day 14 (Fig. 4A). Inhibition of BMP signalling from Day 7 allowed the cells to maintain the expression of several otic placode markers, SIX1, FOXG1, and JAG1 [40,41], on Day 14 (Fig. 4B). LDN supplementation did not significantly affect the expression of EYA1, PAX8, or GBX2 but did reduce the expression of PAX2, GATA3, and DLX5. Supplementation of both LDN and FGF3/FGF10 from Day 7 to Day 14 did not induce any detectable changes in the expression of posterior placode marker genes. These results indicate that BMP inhibition is critical for the expression of several otic placode marker genes. However, FGF3 and FGF10 did not further increase otic placode marker expression.
Fig. 4.
Differentiation of posterior placodal cells via manipulation of BMP and FGF signalling. (A) Schematic figure of the culture conditions used to investigate the roles of BMP and FGF signalling in the differentiation of the posterior placode. Three different culture conditions (Ctrl, +LDN, +LDN + FGF3/FGF10) were compared to explore the effects of manipulating BMP and FGF signalling on posterior placode marker expression. (B) qPCR analysis of the preplacodal ectoderm and otic placode genes SIX1, EYA1, PAX8, PAX2, GATA3, JAG1, GBX2, DLX5, and FOXG1 on Day 14 of culture with or without LDN or FGF3/FGF10 from Day 7 to Day 14. The relative gene expression levels were normalized against those of undifferentiated hESCs. The bars and error bars represent the mean ± SEM (three independent experiments). Asterisks indicate significant differences (∗p < 0.05, ∗∗∗p < 0.001, one-way ANOVA with Tukey's post hoc test).
3.4. 3D floating culture with CHIR treatment is important for otic placodal marker upregulation
Although the expression of several otic placodal genes was upregulated by Day 10 to Day 14, one of the otic placodal markers, PAX2, was not significantly upregulated after treatment with FGF3 and FGF10, which induce otic differentiation. WNT signalling is required to differentiate the posterior placode into the PAX2+ otic placode, while inhibition of the WNT signal results in epibranchial placode development [26,42]. Therefore, to differentiate PAX2+ otic placodal cells from hPSC-derived posterior placodes, we treated the cells with the WNT agonist CHIR from Day 14. Since BMP inhibition from Day 7 to Day 14 enhanced the expression of several otic placodal markers at Day 14 (Fig. 4B), we exposed the cells continuously to the BMP inhibitor LDN. Another candidate factor for the upregulation of PAX2 is FGF signalling. FGF10 is expressed in the ventromedial part of the otocyst, which is a PAX2-positive area, and hindbrain-derived FGF3 is important for the development of the cochlea [43]. Therefore, to investigate the possible roles of FGF3 and FGF10 in otic placode induction, we started to culture Day 14 cells in the presence of FGF3/FGF10 as well as LDN and CHIR. To evaluate the necessity of these factors in otic placode induction, we compared four different culture conditions to evaluate otic gene expression at Day 22, as follows: control conditions, LDN treatment, LDN + CHIR treatment, and LDN + CHIR + FGF3/FGF10 treatment from Day 14 to Day 22 (Fig. 5A and B). The expression of the otic lineage markers SIX1, EYA1, PAX8, PAX2, GATA3, FBXO2, JAG1, GBX2, DLX5, and FOXG1 [5,40,41,[44], [45], [46], [47]] was analysed on Day 22 by qPCR (Fig. 5C). With the exception of GATA3, we did not detect upregulation of the otic marker genes in LDN-treated conditions compared to the control condition. CHIR treatment resulted in upregulation of SIX1, PAX8, PAX2, FBXO2, and DLX5, although GATA3 and GBX2 were downregulated. LDN + CHIR + FGF3/FGF10-treated spheres showed significantly greater DLX5 expression than cultures without FGF3/FGF10. These results show that CHIR enhances the differentiation of posterior placodal cells into otic placodal cells in 3D floating culture. However, LDN and FGF3/FGF10 supplementation did not further enhance the expression of most otic marker genes.
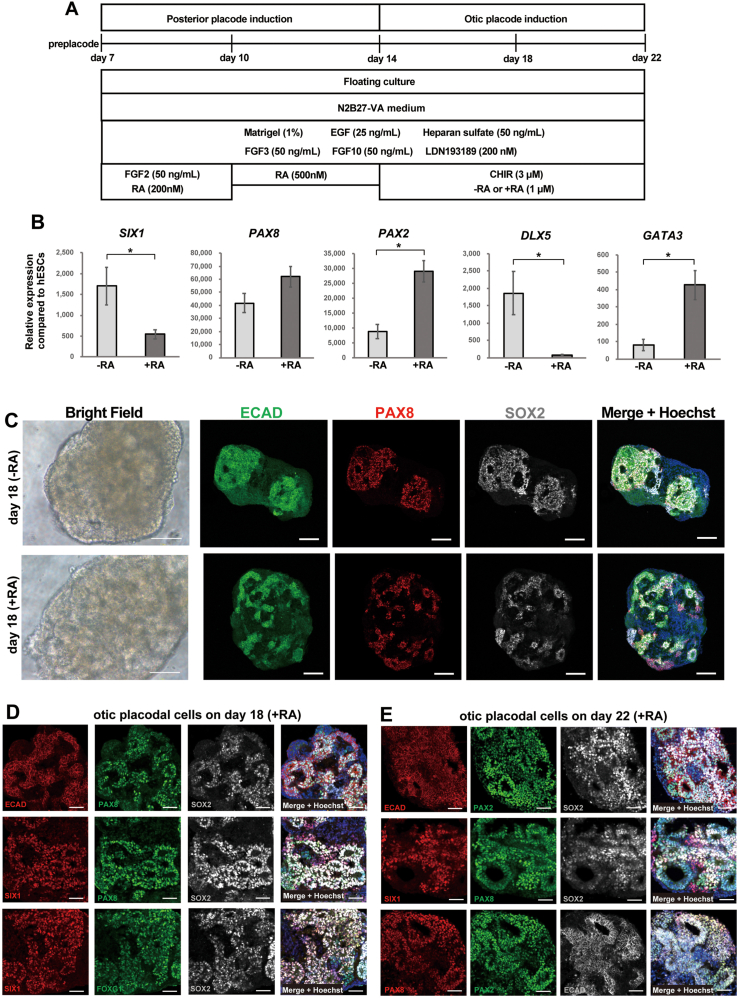
3.5. Effect of RA treatment in the otic placode differentiation from posterior placodal cells
We found that RA is necessary to induce the differentiation of posterior placodal cells from hPSC-derived preplacodal cells. Previous studies have demonstrated that RA also has a significant role in otocyst development. In zebrafish, ectopic RA generates otocyst-like structures in the preplacodal domain. In contrast, inhibition of RA signalling results in failure to maintain otic placodal fate [32]. These observations suggest that RA might further increase otic placode gene expression beyond the increase achieved with CHIR treatment.
To examine the effect of RA on otic placode differentiation from posterior placodal cells, we cultured the cells in the presence of RA with the addition of LDN, CHIR, and FGF3/FGF10 from Day 14 to Day 22 (Fig. 6A). We found that RA-treated spheres showed significant increases in the expression of PAX2 and GATA3. In contrast, RA treatment reduced the expression of SIX1 and DLX5 (Fig. 6B). These results confirm that the expression of the otocyst genes SIX1, DLX5, GATA3, and PAX2 can be controlled by RA signalling.
Fig. 6.
Otic placode marker expression of the induced cells in the presence or absence of RA from Day 14. (A) Schematic figure of the culture conditions used to investigate the role of RA in the differentiation of the otic placode. The cells were treated with LDN, CHIR, and FGF3/FGF10 from Day 14 to Day 22 of differentiation. Culture conditions with or without RA (1 μM) treatment were compared. (B) qPCR analysis of the otic placode markers SIX1, PAX8, PAX2, DLX5, and GATA3 on Day 22 of cultures with or without RA treatment from Day 14 to Day 22. The relative gene expression levels were normalized against those of undifferentiated hESCs. The bars and error bars represent the mean ± SEM (three independent experiments). Asterisks indicate significant differences (∗p < 0.05, two-tailed unpaired Student's t test). (C) Representative bright field images and low-magnification immunocytochemical images on Day 18 of spheres with or without RA treatment. The nuclei were stained with Hoechst 33,342. Scale bars, bright field images, 100 μm; immunostaining images, 100 μm. (D) Representative immunocytochemical images of induced cells on Day 18 in cultures treated with RA. The cells were coimmunostained for multiple otic placodal markers (ECAD/PAX8/SOX2, SIX1/PAX8/SOX2, or SIX1/FOXG1/SOX2). The nuclei were stained with Hoechst 33,342. Scale bars, 50 μm. (E) Representative immunocytochemical images of induced cells on Day 22 of culture treated with RA. The cells were coimmunostained for multiple otic placodal markers (ECAD/PAX2/SOX2, SIX1/PAX8/SOX2, or PAX8/PAX2/SOX2). The nuclei were stained with Hoechst 33,342. Scale bars, 50 μm.
3.6. Immunocytochemical analysis of otic placode markers in the induced cells
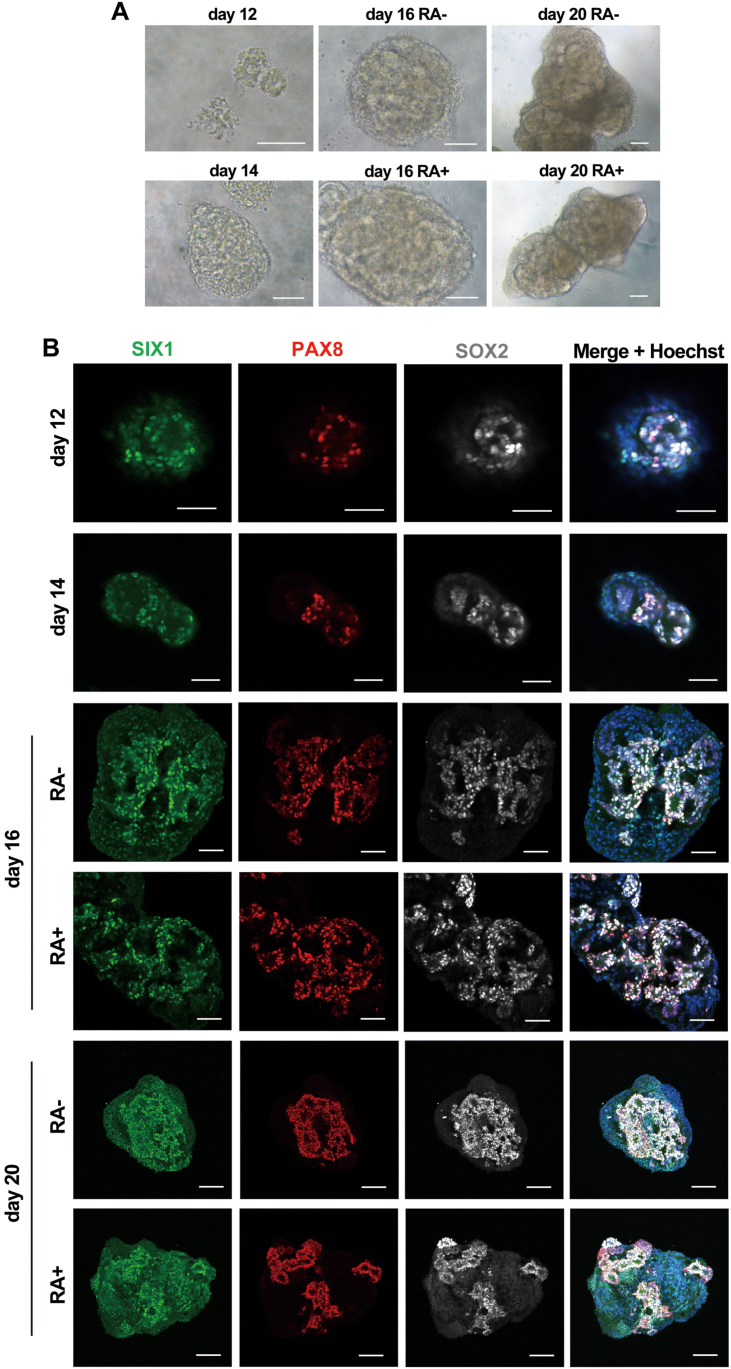
Next, we performed immunocytochemical analysis of the induced cells at various timepoints to assess the presence of otic marker proteins (Fig. 6A). From Day 14, the cells were cultured under treatment with LDN, CHIR, and FGF3/FGF10 and with or without RA. The presence of otocyst marker proteins was evaluated by coimmunostaining for SIX1, PAX8, PAX2, ECAD, FOXG1, and SOX2. On Day 12, tiny epithelial cell spheres were positive for SIX1, PAX8 and SOX2 (Fig. 7A and B). We detected a gradual increase in the size of SIX1-, PAX8-and SOX2-positive cell aggregates during a floating culture from Day 14 to Day 20 (Fig. 7). On Days 16, 18 and 20 of sphere formation with or without RA treatment, we identified epithelial cell structures under bright field microscopy (Fig. 6C, Fig. 7A–B). We found that PAX8+/SOX2+ cells were also positive for the epithelial cell marker ECAD (Fig. 6C). Immunostaining for various otocyst markers on Days 18 and 22 with RA treatment revealed that ECAD+ epithelial cells were also positive for the otic placode markers PAX8, PAX2, and SOX2 (Fig. 6D and E). SIX1+ placodal cells also showed positive staining for the otic placode markers PAX8, FOXG1, and SOX2 (Fig. 6D and E).
Fig. 7.
Immunocytochemical analysis of the posterior placodal and otic placodal cells induced from H9 hESCs. (A) Representative bright field images of floating spheres on Days 12, 14, 16 and 20. Scale bars, 100 μm. (B) Representative immunocytochemical images of induced cells on Days 12, 14, 16 and 20 of culture. The cells were coimmunostained for the otic placodal markers SIX1, PAX8 and SOX2. The nuclei were stained with Hoechst 33,342. Scale bars, Days 12, 14 and 16, 50 μm; Day 20, 100 μm.
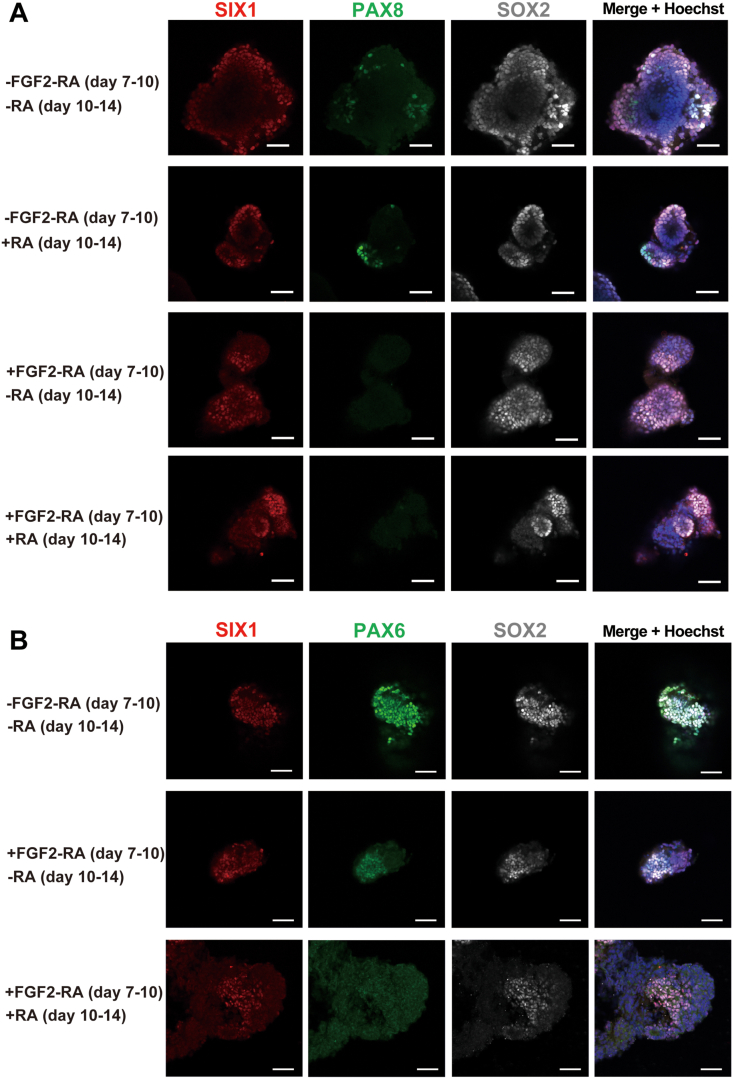
Next, to determine whether the differentiation of SIX1+/PAX8+ cells resulted from the posteriorization of preplacodal cells, we performed an immunocytochemical analysis of otic marker proteins in spheres that had not been cultured with FGF2 and RA during the initial phase of 3D floating culture. From Day 14, the cells were cultured as described in Fig. 6A. In the absence of FGF2 and RA, several SIX1+ cells were identified, but few cells were colocalized with PAX8+ cells at Day 18 (Fig. 8A). In contrast, most SIX1+ cells were coimmunostained for the anterior placode marker PAX6 (Fig. 8B). In cultures with FGF2 but without RA, several SIX1+ cells were present; however, PAX8+ cells were rarely observed, while most PAX6+ cells were colocalized with SIX1+ cells. As expected from the qPCR analysis of Day 10 spheres (Fig. 2C), we did not detect PAX6+ cells in spheres at Day 18 after treatment with either FGF2 or RA (Fig. 8B). These results demonstrate that activation of FGF and WNT signalling is not sufficient to induce SIX1+/PAX8+ otic placodal cells. Supplementation of RA at early phases of the 3D floating culture is necessary for the induction of SIX1+/PAX8+ posterior placodal cells.
Fig. 8.
Requirement for FGF2 and RA during the initial phases of floating culture for the induction of otic placodal cells. (A) Representative immunocytochemical images of SIX1, PAX8, and SOX2 in spheres at Day 18. Four different culture conditions (with or without FGF2 and RA on Day 7 to Day 10, with or without RA on Day 10 to Day 14) were compared to analyse the effects of FGF2 and RA signal manipulation on otic placode marker expression. The nuclei were stained with Hoechst 33,342. Scale bars, 50 μm. (B) Representative immunocytochemical images of SIX1, PAX6, and SOX2 in spheres at Day 18. Three different culture conditions (with or without FGF2 and RA on Day 7 to Day 10, with or without RA on Day 10 to Day 14) were compared to analyse the effects of FGF2 and RA signal manipulation on anterior placode marker expression. The nuclei were stained with Hoechst 33,342. Scale bars, 50 μm.
3.7. scRNA-seq analysis of the induced otic placodal cells
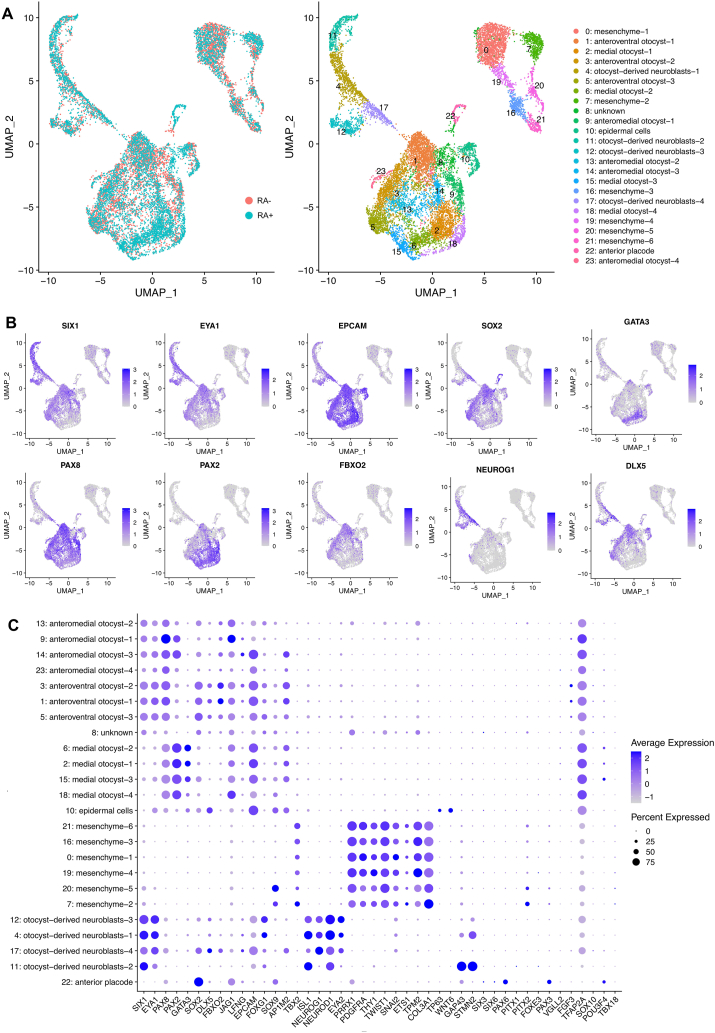
Furthermore, to gain insights into the cellular heterogeneity in the induced spheres, we carried out scRNA-seq on Day 22 of spheres with or without RA treatments from Day 14 to Day 22. After excluding low-quality cells, we obtained the gene expression profiles of 7,155 and 6,277 cells with and without RA treatment, respectively. We then integrated the two datasets, clustered the cells (Methods; Fig. 9A) and annotated the cell type of each cluster based on manual curation of cluster-specific genes against the known marker genes of otic and other cell lineages.
Fig. 9.
Identification of otocyst cell populations in the induced spheres by scRNA-seq. (A) UMAP plot of integrated datasets from RA-treated and untreated RA cells. Overlaid UMAP plots of RA-treated and untreated sphere data are shown in the left panel. The cell cluster identities are coloured and annotated based on the expression of markers for otic placodes/otocysts, otocyst-derived neuroblasts, mesenchymal cells, epidermal progenitors and anterior placodes (right panel). (B) UMAP plots for the otic placode/otocyst or otocyst-derived neuroblast markers SIX1, EYA1, EPCAM, SOX2, GATA3, PAX8, PAX2, FBXO2, NEUROG1 and DLX5 showing enriched expression patterns. Each dot is coloured according to the selected gene expression level. (C) The dot plot shows the expression of selected otic placode/otocyst, otocyst-derived neuroblast and off-target lineage (mesenchyme, epidermal cells, neurons, anterior placode, epibranchial placode, cranial neural crest and peri-otic mesenchyme) marker genes. The spot size represents the percentage of cells expressing a selected gene, and the colour intensity indicates the z score-scaled gene expression level.
One of the major cell types represented otic placode/otocyst-like cells and was composed of 11 clusters (anteromedial otocyst-1–4, anteroventral otocyst-1–3, medial otocyst-1–4 in Fig. 9A). These cells were positive for the markers SIX1, EYA1, PAX2, PAX8, FBXO2, SOX2, DLX5, FOXG1, JAG1, EPCAM, NOTCH1, LFNG, SOX9, AP1M2 and TBX2 [41,44,45,[48], [49], [50], [51], [52]]. We observed heterogeneous expression of various otocyst marker genes among these clusters (Fig. 9B and C). For example, we found that otocyst marker genes such as SIX1, EYA1, PAX8, SOX2, DLX5, FBXO2, JAG1 and EPCAM were highly expressed in anteroventral otocyst-1–3. In medial otocyst-1–4, PAX8, PAX2, GATA3 and JAG1 were highly expressed, while the gene expression of SIX1, EYA1, SOX2, DLX5, and FBXO2 was downregulated compared with that in the anteromedial and anteroventral otocyst clusters. In otocyst-derived neuroblasts-1–4, NEUROG1, NEUROD1, SIX1, EYA2, FOXG1 and ISL1 [50] were highly expressed. Several cells of these neuroblast clusters were also positive for otocyst markers, such as EPCAM, FBXO2 and DLX5. Therefore, these neuroblast clusters (otocyst-derived neuroblasts-1–4) were seemingly the delaminated cell populations from the otic epithelial cells. Of note, cells in anteromedial otocyst-1–4, anteroventral otocyst-1–3 and otocyst-derived neuroblasts-1, 3, and 4 expressed FBXO2, a specific marker of otocysts in E10.5 mice [46,50].
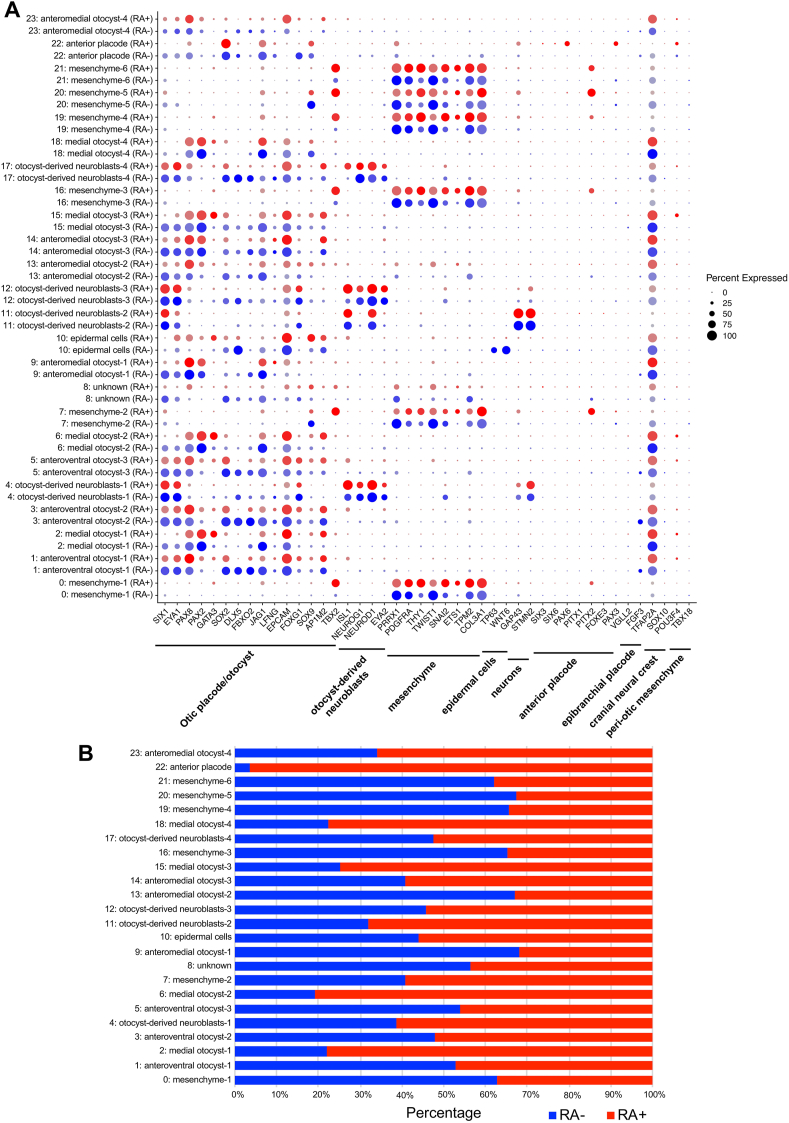
Off-target cell populations were detected in spheres with or without RA treatment (Fig. 9C), including a PRRX1+/PDGFRA+ mesenchymal cell cluster (mesenchyme-1–6), a TP63+/WNT6+ epidermal cell cluster, a PAX3+/PAX6+ anterior placode-like cell cluster and one unknown cell cluster. The mesenchymal cell clusters were also positive for THY1, TWIST1, SNAI2, ETS1, TPM2, and COL3A1, but the cranial neural crest cell populations positive for TFAP2A and SOX10 or the peri-otic mesenchymal cell clusters expressing POU3F4 and TBX18 were not identified in either culture condition.
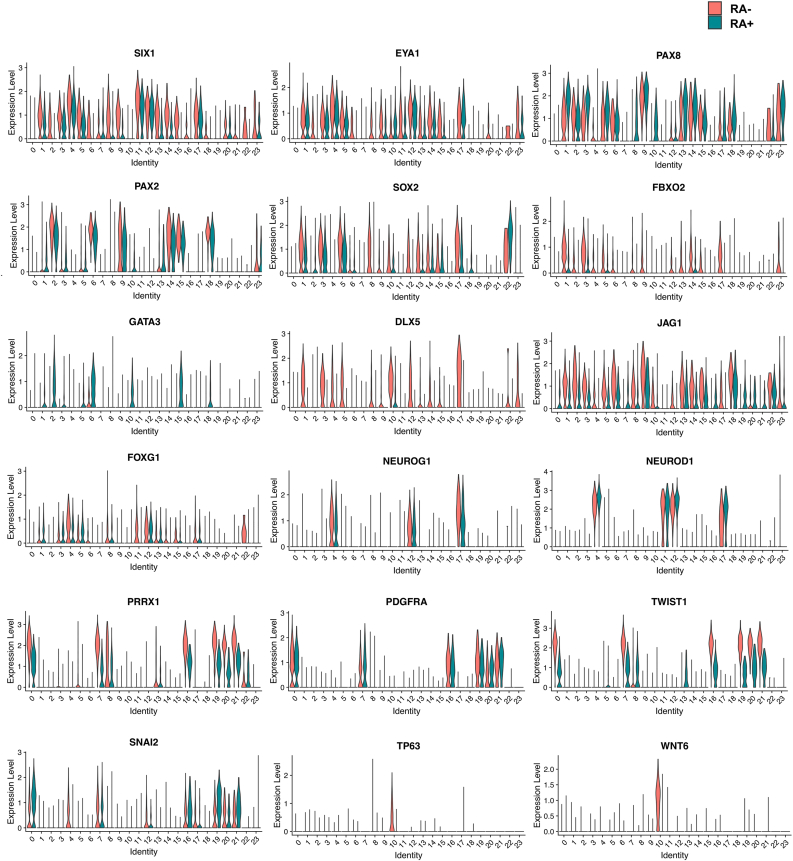
We revealed the differences in expression patterns of several otic marker genes in the otocyst cell clusters between RA-treated and untreated cells, such as SIX1, EYA1, PAX2, GATA3, DLX5, SOX2, FBXO2, JAG1 and AP1M2 (Fig. 10, Fig. 11). In particular, cells without RA treatment showed increased expression of the anterior and ventral otocyst markers SIX1, SOX2, FBXO2 and JAG1, while the number of PAX2-expressing medial otocyst cells was increased among RA-treated cells. These data suggested that a large number of cells were categorized in otic cell lineages but that the otic gene expression profiles were variable among clusters. Additionally, treatment with RA affected the cell populations positive for various otocyst genes such as SIX1, DLX5, GATA3, FBXO2, SOX2 and PAX2.
Fig. 10.
Comparison of otic placode/otocyst marker gene expression levels in otocyst cell clusters between RA-treated and untreated RA cells by scRNA-seq. The violin plot shows the expression of selected otic placode/otocyst (SIX1, EYA1, PAX8, PAX2, SOX2, FBXO2, GATA3, DLX5, JAG1 and FOXG1), otocyst-derived neuroblast (NEUROG1 and NEUROD1), mesenchyme (PRRX1, PDGFRA, TWIST1 and SNAI2) and epidermal cell (TP63 and WNT6) marker genes.
Fig. 11.
Comparison of otic placode/otocyst marker gene expression levels in otocyst cell clusters between RA-treated and untreated RA cells by scRNA-seq. (A) The dot plot shows the expression of selected otic placode/otocyst and otocyst-derived neuroblast and off-target lineage (mesenchyme, epidermal cells, neurons, anterior placode, epibranchial placode, cranial neural crest and peri-otic mesenchyme) marker genes. The spot size represents the percentage of cells expressing a selected gene. The blue and red spots indicate non-RA-treated cells (RA-) and RA-treated cells (RA+), respectively. (B) The bar plot shows the relative proportions of cells with and without RA treatment in each cluster. The blue and red bars indicate non-RA-treated cells (RA-) and RA-treated cells (RA+), respectively.
3.8. Differentiation of sensory epithelial cells from the induced otic placodal/otocyst cells
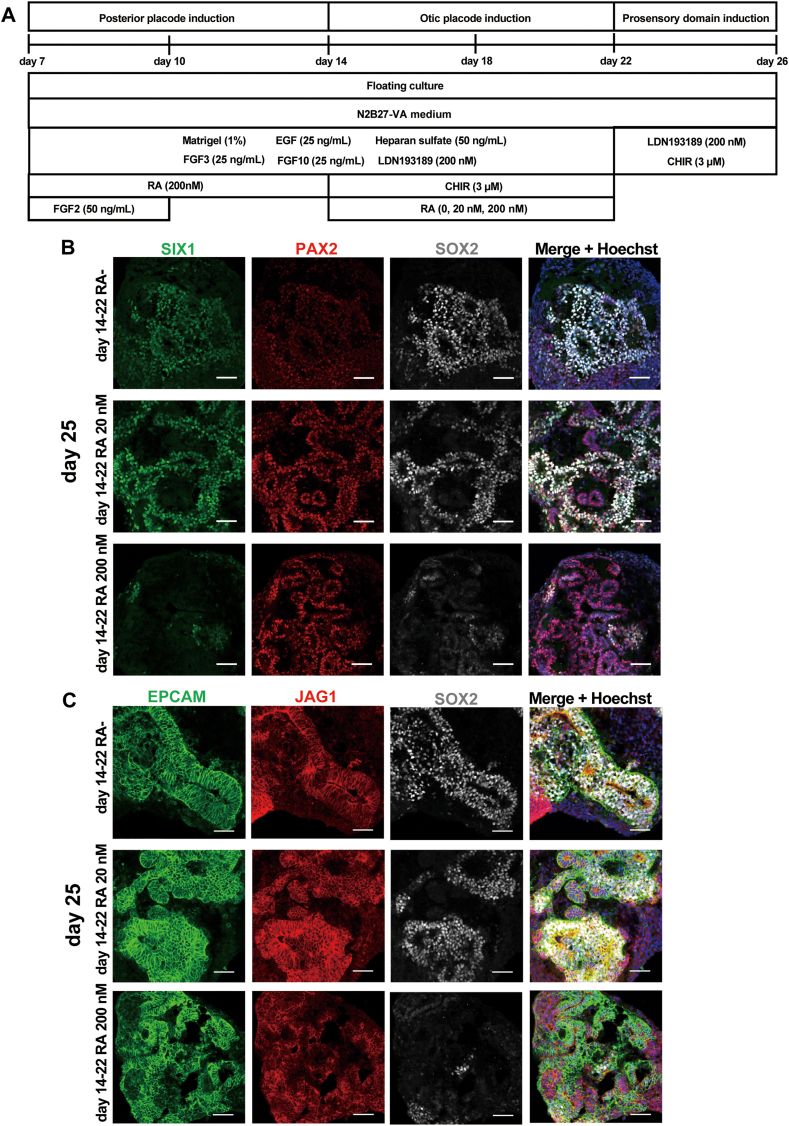
Finally, we attempted to differentiate the induced otic placodal/otocyst cells into prosensory and sensory epithelial cells. We used a cocktail of recombinant proteins and chemicals during a floating culture from Day 7, with partially modified culture conditions based on the gene expression profiles (Fig. 6, Fig. 9, Fig. 10, Fig. 11). First, the qPCR data suggested that treatment with FGF3 and FGF10 only enhanced the expression of the dorsal otocyst marker DLX5. Although FGF3 and FGF10 are important for otic placode development in mice, considering that these factors are potential nonsensory epithelial cell inducers, we slightly modified the concentrations of both recombinant proteins from 50 ng/mL to 25 ng/mL during the floating culture from Day 7 to Day 22. Second, our scRNA-seq data suggested that RA affected the marker gene expression of the population of anterior otocysts on Day 22. We initially aimed to derive PAX2-positive cells by gradually increasing the concentration of RA from Day 7 to Day 22. However, this method downregulated the neurosensory anterior otocyst markers SIX1, SOX2 and FBXO2 (Fig. 10, Fig. 11). According to the qPCR and scRNA-seq data, complete withdrawal of RA from Day 14 resulted in reduced PAX2 gene expression and an increased DLX5+ cell population. Therefore, to achieve prosensory cell differentiation from the induced otic placodal/otocyst cells, we validated the concentration of RA during a floating culture from Day 7 to Day 22 (Fig. 12). First, we fixed the concentration of RA from Day 7 to Day 14 at 200 nM. In development, low-concentration and transient RA exposure support the development of anterior otocysts [53]; thus, we evaluated the conditions of no RA treatment (RA–), low-concentration RA (20 nM) treatment and high-concentration RA (200 nM) treatment during differentiation from Day 14 to Day 22. From Day 22, we continuously treated the cells with LDN and CHIR to guide the cells towards prosensory cell lineages, since BMP signalling is important for the development of dorsal otocysts [54] and WNT activity is critical for the specification of the prosensory domain in the chick otocyst [55]. Due to the overproliferation of nonepithelial cells and increased debris caused by 3 μM CHIR treatment after Day 26, we decreased the concentration of CHIR to 1.5 μM from Day 26 of differentiation. We compared EPCAM-, JAG1-, SIX1-, PAX2-and SOX2-immunopositive cells [44,47,51,56,57] on Day 25 under treatment with various concentrations of RA (Fig. 12). We found that the number of SIX1+/SOX2+ cells was decreased by treatment with a high concentration of RA (200 nM); in contrast, we detected more SIX1+/SOX2+ cells or JAG1+/SOX2+ cells in spheres treated with 20 nM RA or not treated with RA than in spheres treated with 200 nM RA (Fig. 12B and C). We also found that the PAX2 signal intensity was reduced in SIX1+/SOX2+ cells under RA-untreated conditions on Day 25 (Fig. 12B), while under 20 nM RA treatment, most PAX2-positive cells merged with SOX2-positive cells. These data suggest that low-concentration RA (20 nM) treatment from Day 14 to Day 22 can maintain the protein expression of prosensory domain markers such as SIX1, JAG1, PAX2 and SOX2 during the additional period of floating culture from Day 22.
Fig. 12.
Immunocytochemical analysis of the otocyst-like cells induced from H9 hESCs. (A) Schematic figure of the culture conditions used to differentiate prosensory cells from induced otic placodal/otocyst cells. The cells were grown in culture with 25 ng/mL FGF3, 25 ng/mL FGF50 from Day 7 to Day 22, and 200 nM RA from Day 7 to Day 14. Three different culture conditions were compared (20 nM RA, 200 nM RA or no RA treatment from Day 14 to Day 22). (B) Representative immunocytochemical images of induced cells on Day 25 of culture. The cells were coimmunostained for the otic placodal markers SIX1, PAX2 and SOX2. The nuclei were stained with Hoechst 33,342. Scale bars, 50 μm. (C) Representative immunocytochemical images of induced cells on Day 25 of culture. The cells were coimmunostained for the otic placodal markers EPCAM, JAG1 and SOX2. The nuclei were stained with Hoechst 33,342. Scale bars, 50 μm.
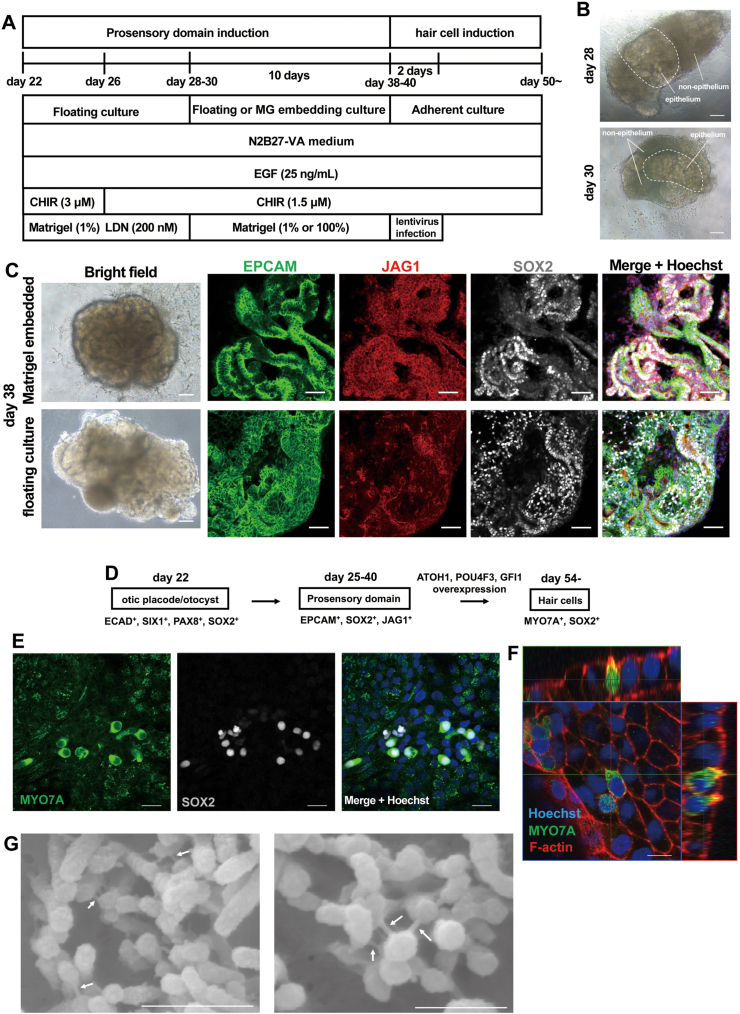
Because continuous degradation of spheres from the surface was frequently observed and since these degraded nonepithelial cells seemed to be off-target mesenchymal cells or neuroblasts, we dissected and isolated the epithelial cell populations using a tungsten needle without enzymatic dissociation (Fig. 13B). To support the growth of isolated epithelial cell aggregates, we embedded the spheres into Matrigel and cultured them under hyperoxic (40% O2) conditions (Fig. 13A). Through this method, we maintained the expression of both SOX2 and JAG1 in EPCAM+ epithelial cells (Fig. 13C). Continuous floating culture without dissection reduced the population of JAG1+/SOX2+ cells (Fig. 13C). We also examined the expression of the inner ear hair cell markers BRN3C and MYO7A. However, cells positive for these hair cell markers were not observed on Day 60 (data not shown). We inferred that there may have been a lack of important secretion factors or environmental factors for the generation of sensory hair cells in our culture system.
Fig. 13.
Inner ear sensory epithelial cell differentiation from induced otic progenitor cells. (A) Schematic figure of the culture conditions used to differentiate inner ear hair cells. From Day 28 or 30, the spheres were dissected, the epithelial cell aggregates were isolated and cultured under 100% Matrigel embedding conditions. After 10 days, the Matrigel was removed using a tungsten needle and dissociated into single cells. The cells were infected with lentivirus (MOI = 5) to overexpress ATOH1, POU4F3 and GFI1. (B) Representative bright field images of induced cells on Day 28 and 30 of culture. The epithelial parts of the spheres were marked by white dotted line. Scale bars, 100 μm. (C) Representative immunocytochemical images of induced cells on Day 38 of culture. The spheres were dissected, and epithelial cells were isolated on Day 28. The continuous floating culture was compared with Matrigel-embedded culture. The cells were coimmunostained for prosensory cell markers (EPCAM/JAG1/SOX2). The nuclei were stained with Hoechst 33,342. Scale bars, 50 μm. (D) Schematic diagram of the differentiation of prosensory and sensory hair cells from hPSC-derived otic placodal/otocyst cells. Representative otocyst and hair cell marker genes are shown. (E) Representative immunocytochemical images of induced cells on Day 52 of culture. The spheres were dissected, and epithelial cells were isolated on Day 28. The cells were coimmunostained for the hair cell marker MYO7A and immature or vestibular hair cells and the supporting cell marker SOX2. The nuclei were stained with Hoechst 33,342. Scale bars, 20 μm. (F) Representative immunocytochemical images of the induced cells on Day 56 of culture. The spheres were dissected, and epithelial cells were isolated on Day 28. The cells were coimmunostained for the hair cell marker MYO7A and F-actin and labelled with Alexa 555-conjugated phalloidin. The nuclei were stained with Hoechst 33,342. Scale bar, 10 μm. (G) Representative scanning electron microscopy (SEM) image of the induced cells on Day 57 of culture. In this experiment, the spheres were dissected, and epithelial cells were isolated on Day 30. After 10 days, the epithelial aggregates were collected and infected with lentivirus to express ATOH1, POU4F3 and GFI1. Tip link or side link-like structures were visible between stereocilia (white arrows). Scale bars, left 500 nm, right 300 nm.
Finally, to explore the potential of hair cell differentiation of the induced JAG1+/SOX2+ prosensory cells, we overexpressed defined transcription factors, ATOH1, POU4F3 and GFI1 [[58], [59], [60]], via lentiviral transduction. After 10 days of Matrigel embedding culture, we recovered the spheres from Matrigel by dissection, dissociated the spheres into single cells and infected the cells with lentivirus under 2D culture conditions (Fig. 13D). After 14 days of lentivirus infection, we detected MYO7A+/SOX2+ cells, indicating the differentiation of immature hair cells or vestibular hair cells (Fig. 13E). We also observed F-actin rich stereociliary bundle-like structures in MYO7A+ cells (Fig. 13F). Scanning electron microscopy analysis showed that stereociliary bundle protrusions had tip link or side link-like structures (Fig. 13G) [75]. These data demonstrate that sensory epithelial cells such as hair cells can be generated by transgene overexpression in induced prosensory cells.
4. Discussion
In this study, we examined the temporal dynamics of pre-, posterior, and otic placodal marker gene expression to explore the capacity of hPSC-derived preplacodal cells to undergo otic lineage differentiation. Following efficient induction of preplacodal cells from hPSCs by 2D culture, 3D suspension culture of the preplacodal cells enabled further otic lineage differentiation. We also demonstrated that activation of FGF and RA signalling in the 3D cultures enhanced the expression levels of posterior placode marker genes. Subsequent activation of WNT signalling increased the expression levels of the otic pathway and otocyst markers. In both cells with and cells without RA treatment from Day 14 of culture, the populations of otic placodal/otocyst cells and otocyst-derived neuroblast marker-expressing cells were identified by scRNA-seq analysis. Although the otic placode/otocyst cells were the major cell populations in the induced spheres, we found heterogeneity in these otic epithelial cells upon comparison of multiple otocyst marker genes. We finally demonstrated the hair cell differentiation capacity of the induced otocyst-like cells by forced expression of defined transcription factors, such as ATOH1, BRN3C and GFI1.
The FGF and WNT signalling pathways play pivotal roles in otic placode development from the preotic posterior placodal region (Fig. 1A). The induction of the preotic region is initiated by FGF signalling; WNT activation mediates otic placode specification and inhibits epibranchial placode differentiation [26,42]. Before the differentiation of the otic placode, WNT signalling suppresses preplacodal development [61]. After the differentiation of the preplacode, WNT signalling mediates trigeminal placode differentiation [62,63]. Therefore, to induce otic differentiation from the preplacode, it is necessary to activate WNT signalling in the critical time window of posterior placode differentiation.
The segregation of the anterior-posterior axis in the preplacodal domain is determined by the homeobox transcription factors Gbx2 and Otx2. While Gbx2 is necessary for otic specification, Otx2 is important for the specification of anterior placodes, including the trigeminal placode [64]. However, the signalling that regulates the expression of Otx2 and Gbx2 in the preplacodal domain is largely unknown.
In the present study, we demonstrated that a 3D suspension culture provides a supportive environment for the efficient differentiation of the posterior placode from preplacodal cells. Our qPCR data showed that a suspension culture supplemented with FGF2 and RA dramatically upregulated GBX2, GATA3, and PAX8 expression and downregulated OTX2 and PAX6 expression. We speculate that previous reports of mixed cellular phenotypes, including both anterior and posterior placode marker gene expression, in induced placodal cells might result from increased expression levels of OTX2 in 2D culture [11]. In contrast, FGF2 may enhance GBX2 expression in hPSC-derived preplacodal cells of cellular aggregations [14]. Although a study using an invertebrate model has proven the necessity of RA for the development of the homologous organ of the vertebrate posterior placode [33], a direct role of RA in the specification of anterior-posterior placodal identity has not been confirmed in vertebrates, especially mammalian species. Our findings suggest that both FGF and RA signalling regulate the positional identity of the preplacode during the differentiation of human PSCs into the cranial placode.
We examined the identity of the induced cells on Day 22 using scRNA-seq analysis. Cell clustering analysis revealed that, with or without RA treatment from Day 14, the majority of the cells were identified as otic placode/otocyst or otocyst-derived neuroblasts, despite their heterogeneous expression patterns of otocyst marker genes. Since the expression of known otocyst marker genes, such as DLX5, GBX2, PAX8, and PAX2, is spatially and temporally restricted in rodents [5], our scRNA-seq data may represent the presence of different regions of otocyst cells in each cluster. Importantly, in most otocyst cell clusters, we detected the expression of FBXO2, which is expressed specifically in the mouse otocyst at E10.5 and is highly expressed in the anteromedial portion of the otocyst [46,50]. Further optimization of the culture method may be required to decrease the heterogeneity of the otocyst lineage cell populations. One difficulty in evaluating the spatiotemporal identity of the induced otic cells is the lack of scRNA-seq data from human in vivo counterparts. In the future, the accurate regional identity of each otocyst cluster derived in vitro should be clarified by comparison with transcriptome data from in vivo human otic placode/otocyst cells.
Although the continuous floating culture failed to maintain prosensory marker gene expression around Day 30, we succeeded in culturing SOX2+/JAG1+ cells containing aggregates until Day 38 with several optimizations of the floating culture from Day 7 to Day 22. However, we did not observe hair cell marker-positive cells in these epithelial cell aggregates in the continuous floating culture. A previous study described the generation of early placodal cell lineages from hPSCs using 3D culture at a single-cell resolution [65]. The scRNA-seq data from that study showed that the organoid contained major off-target ectodermal cell lineages, such as neuroectoderm, cranial neural crest, epidermis and anterior placode lineages. We expect that these off-target cells may be supportive and help to guide the differentiation of otic placode/otocyst cells and further differentiation of sensory epithelial cells via self-organization-based culture [14]. On the other hand, based on our scRNA-seq analysis (Fig. 9, Fig. 11), the induced spheres in this study did not contain high percentages of major ectodermal lineage cells other than posterior placodal cells, and the mesenchymal cell clusters seemed to be off-target cells. Therefore, the indispensable factors that guide the otocyst cells towards a sensory hair cell fate may have been missing in our floating sphere system. Because of the lack of knowledge about the mechanisms of cochlear and vestibular prosensory domain specification and hair cell differentiation from prosensory cells, especially for the vestibular system, the generation of inner ear hair cells from induced otocyst cells without self-organization-based culture is still challenging. The specification mechanisms may involve currently unknown key secretion factors, appropriate mechanical stimuli, and cell-to-cell contacts or conjugations.
For substitution of secretion factor-based differentiation, we switched to using transgene-based differentiation of hair cells from the induced otocyst cells. Three transcription factors, ATOH1, POU4F3 and GFI1, are important for inner ear hair cell differentiation, survival and maturation [[66], [67], [68], [69], [70], [71], [72], [73], [74]]. Several reports have shown the direct induction of inner ear hair cells from PSCs or fibroblasts via overexpression of these transcription factors [[58], [59], [60]]. Although MYO7A+ cells have been observed among cells directly converted from human fibroblasts, stereociliary bundle-like structures have not been detected in inner ear hair cell-like cells via a direct conversion approach [59]. In this study, we confirmed the generation of inner ear sensory epithelial cells with stereocilia-like protrusions from the induced otocyst cells by overexpression of ATOH1, POU4F3 and GFI1. According to previously reported hair cell differentiation protocols, we started to express transcription factors at the expected time (around Day 40) of hair cell marker upregulation [7,14]. Future studies regarding the process of human inner ear hair cell differentiation will help us formulate chemically defined culture conditions or more sophisticated transcription factor-based methods for rapid induction of hair cells.
5. Conclusions
In summary, we have established a novel differentiation method for generating otic placodal cells and sensory epithelial cells from hPSCs. We have demonstrated that the combination of initial 2D culture and subsequent 3D culture with FGF2 and RA supplementation successfully induces an otic placode fate. Further studies are required to improve the protocol to enable morphologically and functionally mature inner ear sensory hair cell differentiation from hPSCs. Nevertheless, we believe that the method described here will enable significant progress in research on human inner ear development and disease modelling of sensorineural hearing loss.
Author's contributions
TS: conception and design, collection and assembly of data, data analysis and interpretation, and manuscript writing. SY: manuscript review and editing. MI: conception and design, and financial support. CCH: data analysis and interpretation, manuscript editing. IK: technical support for scRNA-seq. SS: technical support for electron microscopy. MH, CS, KO and MF: data interpretation, reviewed the manuscript and financial support. JWS: collection and assembly of data, data interpretation. HO: manuscript editing, final approval of manuscript. All authors read and approved the final manuscript.
Declaration of competing interest
H.O. is a founding scientist and scientific advisor of SanBio Co. Ltd. and K Pharma Inc. M.H., K.O. and M.F. are co-founders of Otolink Inc. The other authors indicate no potential conflicts of interest.
Acknowledgements
We greatly thank Dr. Takashi Sasaki and Yoshinari Ando for kind technical support for RNA-seq analysis. We also thank all the laboratory members of H.O. for their encouragement and generous support for the current study. This study was supported by Grant-in-Aid for JSPS Fellows (20J12301 to T.S.) from Japan Society for the Promotion of Science (JSPS), the Doctoral Student Grant-in-Aid Program from Keio University (to T.S.), grant from Keio University Global Research Institute (KGRI) (to H.O.), a grant from the Research Center Network for Realization of Regenerative Medicine by AMED Japan under the grants JP21bm0204001 to H.O. and JP21bm0804023 to M.I and Grant-in-Aid for Scientific Research (A) 18H04065, 19H05473 and 21H04839 (to M.F. and H.O.) and Grant-in-Aid for Challenging Research (Exploratory) 21K19580 (to C.S.) from MEXT KAKENHI.
Footnotes
Peer review under responsibility of the Japanese Society for Regenerative Medicine.
References
- 1.Wong A.C., Ryan A.F. Mechanisms of sensorineural cell damage, death and survival in the cochlea. Front Aging Neurosci. 2015;7:58. doi: 10.3389/fnagi.2015.00058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Müller U., Barr-Gillespie P.G. New treatment options for hearing loss. Nat Rev Drug Discov. 2015;14:346–365. doi: 10.1038/nrd4533. [DOI] [PubMed] [Google Scholar]
- 3.Saint-Jeannet J.P., Moody S.A. Establishing the pre-placodal region and breaking it into placodes with distinct identities. Dev Biol. 2014;389:13–27. doi: 10.1016/j.ydbio.2014.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schlosser G. Induction and specification of cranial placodes. Dev Biol. 2006;294:303–351. doi: 10.1016/j.ydbio.2006.03.009. [DOI] [PubMed] [Google Scholar]
- 5.Wright T.J., Mansour S.L. Fgf3 and Fgf10 are required for mouse otic placode induction. Dev. 2003;130:3379–3390. doi: 10.1242/dev.00555. [DOI] [PubMed] [Google Scholar]
- 6.Chen W., Jongkamonwiwat N., Abbas L., Eshtan S.J., Johnson S.L., Kuhn S., et al. Restoration of auditory evoked responses by human ES-cell-derived otic progenitors. Nature. 2012;490:278–282. doi: 10.1038/nature11415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ronaghi M., Nasr M., Ealy M., Durruthy-Durruthy R., Waldhaus J., Diaz G.H., et al. Inner ear hair cell-like cells from human embryonic stem cells. Stem Cell Dev. 2014;23:1275–1284. doi: 10.1089/scd.2014.0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gunewardene N., Bergen N.V., Crombie D., Needham K., Dottori M., Nayagam B.A. Directing human induced pluripotent stem cells into a neurosensory lineage for auditory neuron replacement. Biores Open Access. 2014;3:162–175. doi: 10.1089/biores.2014.0019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ohnishi H., Skerleva D., Kitajiri S., Sakamoto T., Yamamoto N., Ito J., et al. Limited hair cell induction from human induced pluripotent stem cells using a simple stepwise method. Neurosci Lett. 2015;599:49–54. doi: 10.1016/j.neulet.2015.05.032. [DOI] [PubMed] [Google Scholar]
- 10.Ding J., Tang Z., Chen J., Shi H., Wang C., Zhang C., et al. Induction of differentiation of human embryonic stem cells into functional hair-cell-like cells in the absence of stromal cells. Int J Biochem Cell Biol. 2016;81:208–222. doi: 10.1016/j.biocel.2015.11.012. [DOI] [PubMed] [Google Scholar]
- 11.Ealy M., Ellwanger D.C., Kosaric N., Stapper A.P., Heller S. Single-cell analysis delineates a trajectory toward the human early otic lineage. Proc Natl Acad Sci U S A. 2016;113:8508–8513. doi: 10.1073/pnas.1605537113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hosoya M., Fujioka M., Sone T., Okamoto S., Akamatsu W., Ukai H., et al. Cochlear cell modeling using disease-specific iPSCs unveils a degenerative phenotype and suggests treatments for congenital progressive hearing loss. Cell Rep. 2017;18:68–81. doi: 10.1016/j.celrep.2016.12.020. [DOI] [PubMed] [Google Scholar]
- 13.Matsuoka A.J., Morrissey Z.D., Zhang C., Homma K., Belmadani A., Miller C.A., et al. Directed differentiation of human embryonic stem cells toward placode-derived spiral ganglion-like sensory neurons. Stem Cells Transl Med. 2017;6:923–936. doi: 10.1002/sctm.16-0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koehler K.R., Nie J., Longworth-Mills E., Liu X.P., Lee J., Holt J.R., et al. Generation of inner ear organoids containing functional hair cells from human pluripotent stem cells. Nat Biotechnol. 2017;35:583–589. doi: 10.1038/nbt.3840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lahlou H., Nivet E., Lopez-Juarez A., Fontbonne A., Assou S., Zine A. Enriched differentiation of human otic sensory progenitor cells derived from induced pluripotent stem cells. Front Mol Neurosci. 2018;11:452. doi: 10.3389/fnmol.2018.00452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jeong M., O'Reilly M., Kirkwood N.K., Al-Aama J., Lako M., Kros C.J., et al. Generating inner ear organoids containing putative cochlear hair cells from human pluripotent stem cells. Cell Death Dis. 2018;9:922. doi: 10.1038/s41419-018-0967-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mattei C., Lim R., Drury H., Nasr B., Li Z., Tadros M.A., et al. Generation of vestibular tissue-like organoids from human pluripotent stem cells using the rotary cell culture system. Front Cell Dev Biol. 2019;7:25. doi: 10.3389/fcell.2019.00025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dincer Z., Piao J., Niu L., Ganat Y., Kriks S., Zimmer B., et al. Specification of functional cranial placode derivatives from human pluripotent stem cells. Cell Rep. 2013;5:1387–1402. doi: 10.1016/j.celrep.2013.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leung A.W., Kent Morest D., Li J.Y. Differential BMP signaling controls formation and differentiation of multipotent preplacodal ectoderm progenitors from human embryonic stem cells. Dev Biol. 2013;379:208–220. doi: 10.1016/j.ydbio.2013.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tchieu J., Zimmer B., Fattahi F., Amin S., Zeltner N., Chen S., et al. A modular platform for differentiation of human PSCs into all major ectodermal lineages. Cell Stem Cell. 2017;21:399–410. doi: 10.1016/j.stem.2017.08.015. e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koehler K.R., Mikosz A.M., Molosh A.I., Patel D., Hashino E. Generation of inner ear sensory epithelia from pluripotent stem cells in 3D culture. Nature. 2013;500:217–221. doi: 10.1038/nature12298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smith J.R., Vallier L., Lupo G., Alexander M., Harris W.A., Pedersen R.A. Inhibition of Activin/Nodal signaling promotes specification of human embryonic stem cells into neuroectoderm. Dev Biol. 2008;313:107–117. doi: 10.1016/j.ydbio.2007.10.003. [DOI] [PubMed] [Google Scholar]
- 23.Chambers S.M., Fasano C.A., Papapetrou E.P., Tomishima M., Sadelain M., Studer L. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat Biotechnol. 2009;27:275–280. doi: 10.1038/nbt.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Streit A. The preplacodal region: an ectodermal domain with multipotential progenitors that contribute to sense organs and cranial sensory ganglia. Int J Dev Biol. 2007;51:447–461. doi: 10.1387/ijdb.072327as. [DOI] [PubMed] [Google Scholar]
- 25.Grocott T., Tambalo M., Streit A. The peripheral sensory nervous system in the vertebrate head: a gene regulatory perspective. Dev Biol. 2012;370:3–23. doi: 10.1016/j.ydbio.2012.06.028. [DOI] [PubMed] [Google Scholar]
- 26.Freter S., Muta Y., Mak S.S., Rinkwitz S., Ladher R.K. Progressive restriction of otic fate: the role of FGF and Wnt in resolving inner ear potential. Dev. 2008;135:3415–3424. doi: 10.1242/dev.026674. [DOI] [PubMed] [Google Scholar]
- 27.Ladher R.K., Wright T.J., Moon A.M., Mansour S.L., Schoenwolf G.C. FGF8 initiates inner ear induction in chick and mouse. Genes Dev. 2005;19:603–613. doi: 10.1101/gad.1273605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Phillips B.T., Storch E.M., Lekven A.C., Riley B.B. A direct role for Fgf but not Wnt in otic placode induction. Development. 2004;131:923–931. doi: 10.1242/dev.00978. [DOI] [PubMed] [Google Scholar]
- 29.Pirvola U., Spencer-Dene B., Xing-Qun L., Kettunen P., Thesleff I., Fritzsch B., et al. FGF/FGFR-2(IIIb) signaling is essential for inner ear morphogenesis. J Neurosci. 2000;20:6125–6134. doi: 10.1523/JNEUROSCI.20-16-06125.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Park B.Y., Saint-Jeannet J.P. Hindbrain-derived Wnt and Fgf signals cooperate to specify the otic placode in Xenopus. Dev Biol. 2008;324:108–121. doi: 10.1016/j.ydbio.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martin K., Groves A.K. Competence of cranial ectoderm to respond to Fgf signaling suggests a two-step model of otic placode induction. Dev. 2006;133:877–887. doi: 10.1242/dev.02267. [DOI] [PubMed] [Google Scholar]
- 32.Hans S., Westerfield M. Changes in retinoic acid signaling alter otic patterning. Dev. 2007;134:2449–2458. doi: 10.1242/dev.000448. [DOI] [PubMed] [Google Scholar]
- 33.Sasakura Y., Kanda M., Ikeda T., Horie T., Kawai N., Ogura Y., et al. Retinoic acid-driven Hox1 is required in the epidermis for forming the otic/atrial placodes during ascidian metamorphosis. Dev. 2012;139:2156–2160. doi: 10.1242/dev.080234. [DOI] [PubMed] [Google Scholar]
- 34.Birol O., Ohyama T., Edlund R.K., Drakou K., Georgiades P., Groves A.K. The mouse Foxi3 transcription factor is necessary for the development of posterior placodes. Dev Biol. 2016;409:139–151. doi: 10.1016/j.ydbio.2015.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wawersik S., Purcell P., Rauchman M., Dudley A.T., Robertson E.J., Maas R. BMP7 acts in murine lens placode development. Dev Biol. 1999;207:176–188. doi: 10.1006/dbio.1998.9153. [DOI] [PubMed] [Google Scholar]
- 36.Sjödal M., Edlund T., Gunhaga L. Time of exposure to BMP signals plays a key role in the specification of the olfactory and lens placodes ex vivo. Dev Cell. 2007;13:141–149. doi: 10.1016/j.devcel.2007.04.020. [DOI] [PubMed] [Google Scholar]
- 37.Holzschuh J., Wada N., Wada C., Schaffer A., Javidan Y., Tallafuss A., et al. Requirements for endoderm and BMP signaling in sensory neurogenesis in zebrafish. Dev. 2005;132:3731–3742. doi: 10.1242/dev.01936. [DOI] [PubMed] [Google Scholar]
- 38.Sun S.K., Dee C.T., Tripathi V.B., Rengifo A., Hirst C.S., Scotting P.J. Epibranchial and otic placodes are induced by a common Fgf signal, but their subsequent development is independent. Dev Biol. 2007;303:675–686. doi: 10.1016/j.ydbio.2006.12.008. [DOI] [PubMed] [Google Scholar]
- 39.Alvarez Y., Alonso M.T., Vendrell V., Zelarayan L.C., Chamero P., Theil T., et al. Requirements for FGF3 and FGF10 during inner ear formation. Dev. 2003;130:6329–6338. doi: 10.1242/dev.00881. [DOI] [PubMed] [Google Scholar]
- 40.Hébert J.M., McConnell S.K. Targeting of cre to the Foxg1 (BF-1) locus mediates loxP recombination in the telencephalon and other developing head structures. Dev Biol. 2000;222:296–306. doi: 10.1006/dbio.2000.9732. [DOI] [PubMed] [Google Scholar]
- 41.Jayasena C.S., Ohyama T., Segil N., Groves A.K. Notch signaling augments the canonical Wnt pathway to specify the size of the otic placode. Dev. 2008;135:2251–2261. doi: 10.1242/dev.017905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ohyama T., Mohamed O.A., Taketo M.M., Dufort D., Groves A.K. Wnt signals mediate a fate decision between otic placode and epidermis. Dev. 2006;133:865–875. doi: 10.1242/dev.02271. [DOI] [PubMed] [Google Scholar]
- 43.Urness L.D., Wang X., Doan H., Shumway N., Noyes C.A., Gutierrez-Magana E., et al. Spatial and temporal inhibition of FGFR2b ligands reveals continuous requirements and novel targets in mouse inner ear morphogenesis. Dev. 2018;145 doi: 10.1242/dev.170142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zheng W., Huang L., Wei Z.B., Silvius D., Tang B., Xu P.X. The role of Six1 in mammalian auditory system development. Dev. 2003;130:3989–4000. doi: 10.1242/dev.00628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zou D., Silvius D., Rodrigo-Blomqvist S., Enerbäck S., Xu P.X. Eya1 regulates the growth of otic epithelium and interacts with Pax2 during the development of all sensory areas in the inner ear. Dev Biol. 2006;298:430–441. doi: 10.1016/j.ydbio.2006.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hartman B.H., Durruthy-Durruthy R., Laske R.D., Losorelli S., Heller S. Identification and characterization of mouse otic sensory lineage genes. Front Cell Neurosci. 2015;9:79. doi: 10.3389/fncel.2015.00079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lawoko-Kerali G., Rivolta M.N., Holley M. Expression of the transcription factors GATA3 and Pax2 during development of the mammalian inner ear. J Comp Neurol. 2002;442:378–391. doi: 10.1002/cne.10088. [DOI] [PubMed] [Google Scholar]
- 48.Riccomagno M.M., Martinu L., Mulheisen M., Wu D.K., Epstein D.J. Specification of the mammalian cochlea is dependent on Sonic hedgehog. Genes Dev. 2002;16:2365–2378. doi: 10.1101/gad.1013302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Riccomagno M.M., Takada S., Epstein D.J. Wnt-dependent regulation of inner ear morphogenesis is balanced by the opposing and supporting roles of Shh. Genes Dev. 2005;19:1612–1623. doi: 10.1101/gad.1303905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Durruthy-Durruthy R., Gottlieb A., Hartman B.H., Waldhaus J., Laske R.D., Altman R., et al. Reconstruction of the mouse otocyst and early neuroblast lineage at single-cell resolution. Cell. 2014;157:964–978. doi: 10.1016/j.cell.2014.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nagao K., Zhu J., Heneghan M.B., Hanson J.C., Morasso M.I., Tessarollo L., et al. Abnormal placental development and early embryonic lethality in EpCAM-null mice. PLoS One. 2009;4 doi: 10.1371/journal.pone.0008543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kaiser M., Wojahn I., Rudat C., Lüdtke T.H., Christoffels V.M., Moon A., et al. Regulation of otocyst patterning by Tbx2 and Tbx3 is required for inner ear morphogenesis in the mouse. Dev. 2021:148. doi: 10.1242/dev.195651. [DOI] [PubMed] [Google Scholar]
- 53.Bok J., Raft S., Kong K.A., Koo S.K., Dräger U.C., Wu D.K. Transient retinoic acid signaling confers anterior-posterior polarity to the inner ear. Proc Natl Acad Sci U S A. 2011;108:161–166. doi: 10.1073/pnas.1010547108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ohta S., Wang B., Mansour S.L., Schoenwolf G.C. BMP regulates regional gene expression in the dorsal otocyst through canonical and non-canonical intracellular pathways. Dev. 2016;143:2228–2237. doi: 10.1242/dev.137133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Żak M., Daudet N. A gradient of Wnt activity positions the neurosensory domains of the inner ear. Elife. 2021;10 doi: 10.7554/eLife.59540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hartman B.H., Reh T.A., Bermingham-McDonogh O. Notch signaling specifies prosensory domains via lateral induction in the developing mammalian inner ear. Proc Natl Acad Sci U S A. 2010;107:15792–15797. doi: 10.1073/pnas.1002827107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kiernan A.E., Xu J., Gridley T. The Notch ligand JAG1 is required for sensory progenitor development in the mammalian inner ear. PLoS Genet. 2006;2:e4. doi: 10.1371/journal.pgen.0020004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Costa A., Sanchez-Guardado L., Juniat S., Gale J.E., Daudet N., Henrique D. Generation of sensory hair cells by genetic programming with a combination of transcription factors. Dev. 2015;142:1948–1959. doi: 10.1242/dev.119149. [DOI] [PubMed] [Google Scholar]
- 59.Duran Alonso M.B., Lopez Hernandez I., de la Fuente M.A., Garcia-Sancho J., Giraldez F., Schimmang T. Transcription factor induced conversion of human fibroblasts towards the hair cell lineage. PLoS One. 2018;13 doi: 10.1371/journal.pone.0200210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Menendez L., Trecek T., Gopalakrishnan S., Tao L., Markowitz A.L., Yu H.V., et al. Generation of inner ear hair cells by direct lineage conversion of primary somatic cells. Elife. 2020;9 doi: 10.7554/eLife.55249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Litsiou A., Hanson S., Streit A. A balance of FGF, BMP and WNT signalling positions the future placode territory in the head. Dev. 2005;132:4051–4062. doi: 10.1242/dev.01964. [DOI] [PubMed] [Google Scholar]
- 62.Lassiter R.N., Dude C.M., Reynolds S.B., Winters N.I., Baker C.V., Stark M.R. Canonical Wnt signaling is required for ophthalmic trigeminal placode cell fate determination and maintenance. Dev Biol. 2007;308:392–406. doi: 10.1016/j.ydbio.2007.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Canning C.A., Lee L., Luo S.X., Graham A., Jones C.M. Neural tube derived Wnt signals cooperate with FGF signaling in the formation and differentiation of the trigeminal placodes. Neural Dev. 2008;3:35. doi: 10.1186/1749-8104-3-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Steventon B., Mayor R., Streit A. Mutual repression between Gbx2 and Otx2 in sensory placodes reveals a general mechanism for ectodermal patterning. Dev Biol. 2012;367:55–65. doi: 10.1016/j.ydbio.2012.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lee J., Rabbani C.C., Gao H., Steinhart M.R., Woodruff B.M., Pflum Z.E., et al. Hair-bearing human skin generated entirely from pluripotent stem cells. Nature. 2020;582:399–404. doi: 10.1038/s41586-020-2352-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Xiang M., Gan L., Li D., Chen Z.Y., Zhou L., O'Malley B.W., et al. Essential role of POU-domain factor Brn-3c in auditory and vestibular hair cell development. Proc Natl Acad Sci U S A. 1997;94:9445–9450. doi: 10.1073/pnas.94.17.9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Xiang M., Gao W.Q., Hasson T., Shin J.J. Requirement for Brn-3c in maturation and survival, but not in fate determination of inner ear hair cells. Dev. 1998;125:3935–3946. doi: 10.1242/dev.125.20.3935. [DOI] [PubMed] [Google Scholar]
- 68.Wallis D., Hamblen M., Zhou Y., Venken K.J., Schumacher A., Grimes H.L., et al. The zinc finger transcription factor Gfi1, implicated in lymphomagenesis, is required for inner ear hair cell differentiation and survival. Dev. 2003;130:221–232. doi: 10.1242/dev.00190. [DOI] [PubMed] [Google Scholar]
- 69.Pan N., Jahan I., Kersigo J., Duncan J.S., Kopecky B., Fritzsch B. A novel Atoh1 "self-terminating" mouse model reveals the necessity of proper Atoh1 level and duration for hair cell differentiation and viability. PLoS One. 2012;7 doi: 10.1371/journal.pone.0030358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cai T., Seymour M.L., Zhang H., Pereira F.A., Groves A.K. Conditional deletion of Atoh1 reveals distinct critical periods for survival and function of hair cells in the organ of Corti. J Neurosci. 2013;33:10110–10122. doi: 10.1523/JNEUROSCI.5606-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chonko K.T., Jahan I., Stone J., Wright M.C., Fujiyama T., Hoshino M., et al. Atoh1 directs hair cell differentiation and survival in the late embryonic mouse inner ear. Dev Biol. 2013;381:401–410. doi: 10.1016/j.ydbio.2013.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hertzano R., Montcouquiol M., Rashi-Elkeles S., Elkon R., Yücel R., Frankel W.N., et al. Transcription profiling of inner ears from Pou4f3(ddl/ddl) identifies Gfi1 as a target of the Pou4f3 deafness gene. Hum Mol Genet. 2004;13:2143–2153. doi: 10.1093/hmg/ddh218. [DOI] [PubMed] [Google Scholar]
- 73.Woods C., Montcouquiol M., Kelley M.W. Math1 regulates development of the sensory epithelium in the mammalian cochlea. Nat Neurosci. 2004;7:1310–1318. doi: 10.1038/nn1349. [DOI] [PubMed] [Google Scholar]
- 74.Bermingham N.A., Hassan B.A., Price S.D., Vollrath M.A., Ben-Arie N., Eatock R.A., et al. Math1: an essential gene for the generation of inner ear hair cells. Sci. 1999;284:1837–1841. doi: 10.1126/science.284.5421.1837. [DOI] [PubMed] [Google Scholar]
- 75.Ikäheimo K., Herranen A., Iivanainen V., Lankinen T., Aarnisalo A.A., Sivonen V., et al. MANF supports the inner hair cell synapse and the outer hair cell stereocilia bundle in the cochlea. Life Sci Alliance. 2021;5(2):e202101068. doi: 10.26508/lsa.202101068. [DOI] [PMC free article] [PubMed] [Google Scholar]