Summary
Requirements for vesicle fusion within the heart remain poorly understood, despite the multitude of processes that necessitate proper intracellular trafficking within cardiomyocytes. Here, we show that Syntaxin 4 (STX4), a target-Soluble N-ethylmaleimide sensitive factor attachment receptor (t-SNARE) protein, is required for normal vertebrate cardiac conduction and vesicular transport. Two patients were identified with damaging variants in STX4. A patient with a homozygous R240W missense variant displayed biventricular dilated cardiomyopathy, ectopy, and runs of non-sustained ventricular tachycardia, sensorineural hearing loss, global developmental delay, and hypotonia, while a second patient displayed severe pleiotropic abnormalities and perinatal lethality. CRISPR/Cas9-generated stx4 mutant zebrafish exhibited defects reminiscent of these patients’ clinical presentations, including linearized hearts, bradycardia, otic vesicle dysgenesis, neuronal atrophy, and touch insensitivity by 3 days post fertilization. Imaging of Vamp2+ vesicles within stx4 mutant zebrafish hearts showed reduced docking to the cardiomyocyte sarcolemma. Optical mapping of the embryonic hearts coupled with pharmacological modulation of Ca2+ handling together support that zebrafish stx4 mutants have a reduction in L-type Ca2+ channel modulation. Transgenic overexpression of zebrafish Stx4R241W, analogous to the first patient’s STX4R240W variant, indicated that the variant is hypomorphic. Thus, these data show an in vivo requirement for SNAREs in regulating normal embryonic cardiac function and that variants in STX4 are associated with pleiotropic human disease, including cardiomyopathy.
Key words: Syntaxin 4, SNARE, vesicular transport, congenital heart disease, dilated cardiomyopathy, conduction defects, zebrafish, calcium handling
Introduction
Soluble N-ethylmaleimide sensitive factor attachment receptor (SNARE) proteins bridge the vesicle and plasma membrane and are crucial for processes involving intracellular membrane fusion, including neurotransmitter release.1 In humans, variants in the loci of the SNARE proteins SNAP25 (MIM: 600322), VAMP1 (MIM: 185880), VAMP2 (MIM: 185881), and several of their cognates have been reported to result in synaptopathies, a spectrum of diseases that typically manifest in an encephalopathy phenotype composed of severe neural defects, including hypokinesia and epilepsy.2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13 While these syndromes have recently been collectively termed “SNAREopathies,”3 SNAREs ubiquitously regulate vesicle fusion in virtually all cell types by mediating the fusion and subcellar localization of various transmembrane and exocytic components to their membrane targets.14 Therefore, many other extra-neuronal disease pathologies may manifest as “SNAREopathies.” Cardiomyocytes (CMs) are among the most specialized cell types owing to a high degree of organization and the dynamic requirements for their diverse function. Remarkably, while it has long been known that CMs heavily rely on intracellular membrane trafficking, few bona fide trafficking proteins have been identified as having specific functions in the heart.15,16 SNAREs are a promising candidate for the elucidation of how cardiac intracellular trafficking is regulated. Notably, previous in vitro experiments suggest that SNAREs may regulate cardiac ion channel vesicle fusion, sensitivity, or gating behavior.17, 18, 19, 20, 21, 22, 23 Furthermore, all voltage gated Ca2+ channels (VGCCs) are reported to physically interact with the vesicle fusion machinery via common, conserved SNARE-interacting “synprint” domains,17,19 although to date an interaction with cardiac VGCCs has not been examined.19
Here, we report an association between SNARE function within the heart and cardiovascular disease. We identified two patients with unique biallelic pathogenic variants in Syntaxin 4 (STX4 [MIM: 186591]) that showed pleiotropic defects, including one patient with early-onset biventricular dilated cardiomyopathy (DCM) that ultimately necessitated heart transplant. Engineered zebrafish stx4 mutant embryos also have pleiotropic defects, including profound myocardial dysfunction and bradycardia. Within CMs, we found that zebrafish stx4 mutants exhibit a significant reduction in their number of docked vesicles. Mechanistically, optical mapping demonstrated that zebrafish stx4 mutants have aberrant Ca2+ handling. Concordantly, stx4 mutant hearts are highly sensitized to pharmacological inhibition of L-type Ca2+ channels (LTCCs), while agonism of LTCCs can rescue the bradycardia. Transgenic lines expressing the zebrafish Stx4R421W variant (analogous to patient 1’s variant) were unable to fully rescue stx4 mutants, suggesting that the patient’s Stx4R240W allele is hypomorphic. Collectively, these data provide evidence for a conserved requirement for Stx4 in vertebrate cardiac conduction, which may provide mechanistic insights into the etiology of the cardiomyopathies found in humans with damaging STX4 variants.
Material and methods
Ethics statement
Approval of research on human subjects was obtained from the institutional review boards at Cincinnati Children’s Hospital Medical Center and the Ethics Committee of the University Children’s and Women’s Center/University Hospital Carl Gustav Carus at the Technische Universität Dresden performed in compliance with the Declaration of Helsinki, and conformed to the laws and regulations of the respective countries and institutions. Written consent was obtained for all patient images. All zebrafish husbandry and experimental procedures were performed in accordance with approved Institutional Animal Care and Use Committee protocols at the Cincinnati Children’s Hospital Medical Center and the Harvard Medical Area Standing Committee on Animals.
Genetic investigations
For the index patient (patient 1), a clinical SNP microarray (Infinium CytoSNP-850K v1.1 BeadChip [Illumina]) was performed following local protocols by the Cincinnati Children’s Hospital Medical Center cytogenetics lab. Mitochondrial DNA sequencing and clinical whole-exome sequencing (WES) was subsequently performed using a trio-based strategy by MNG Laboratories (Atlanta, GA). WES data processing, sequence alignment to GRCh37, variant filtering, prioritization by allele frequency, prediction of functional impact, and inheritance models were performed using previously reported analysis pipelines.24
Targeted variant testing was performed on patient 1’s unaffected sibling. Testing was approved by the Cincinnati Children’s Hospital Medical Center internal Institutional Review Board (protocol #2020-0390). DNA was extracted from the sibling’s saliva using a DNeasy Blood & Tissue Kit (Qiagen, 69,504). Extracted DNA from patient 1 and DNA from an unaffected individual served as positive and negative controls, respectively. Forward and reverse primers were designed to flank patient 1’s variant (Table S3). PCR products were concentrated using a DNA Clean & Concentrator 5 (Zymo Research). Amplicons were confirmed by Sanger sequencing. Chromatograms were manually reviewed.
For patient 2, prenatal trio-WES was performed clinically on the fetus and on the parents. Trio analysis: Variants found in the patient and in the patient’s parents were compared and filtered for three cases: de novo in the patient, patient is compound heterozygous, patient is homozygous, and the parents are heterozygous. The coding and flanking intronic regions were enriched using in solution hybridization technology and were sequenced using the HiSeq/NovaSeq system (Illumina). Copy number variations (CNV) were computed on uniquely mapping, non-duplicate, high-quality reads using an internally developed method based on sequencing coverage depth. Briefly, reference samples were used to create a model of the expected coverage that represents wet-lab biases as well as inter-sample variation. CNV calling was performed by computing the sample’s normalized coverage profile and its deviation from the expected coverage. Genomic regions are called as variant if they deviate significantly from the expected coverage. Illumina bcl2fastq2 was used to demultiplex sequencing reads. Adapter removal was performed with Skewer.25 The trimmed reads were mapped to the human reference genome (hg19) using the Burrows Wheeler Aligner.26 Reads mapping to more than one location with identical mapping scores were discarded. Read duplicates that likely resulted from PCR amplification were removed. The remaining high-quality sequences were used to determine sequence variants (single nucleotide changes and small insertions/deletions). Only variants (single nucleotide variants [SNVs]/small indels) in the coding region and the flanking intronic regions (±8 base pairs [bp]) with a minor allele frequency (MAF) < 1% were evaluated. Known disease-causing variants (according to the Human Gene Mutation Database) were evaluated in up to ±30 bp of flanking regions and up to 5% MAF. All variants with an MAF <1% (in genes with autosomal recessive heredity) or <0.1% (in genes with dominant heredity) were evaluated, not including variants classified as benign or likely benign according to current literature. In silico prediction of variants was calculated on the basis of the output of the program’s Mutation Taster,27 fathmm/fathmm-MKL coding,28 Mutation Assessor,29 SIFT,30 LRT,31 and PROVEAN32 according to the following criteria: 100% consensus = pathogenic/benign, ≥75% consensus = mostly pathogenic/benign, consensus <75% or no prediction possible = inconsistent. SpliceAI was used to evaluate the consequence of variants on splicing (default [high precision] thresholds: 0.8-1 "splice effect," 0.6–0.8 "possible splice effect," <0.6 "no splice effect).33 For variants within a 0.5 delta score range cutoff with functional analysis unavailable to confirm a predicted splice effect, additional in silico predictions were employed, including MaxEntScan,34 Combined Annotation Dependent Depletion (CADD),35 and MutationTaster.27 All variants were classified and reported based on American College of Medical Genetics and Genomics (ACMG)/Association for Clinical Genomic Sciences-2020v4.01 guidelines.36
Zebrafish husbandry/mutant and transgenic lines used
Adult zebrafish (Danio rerio) were maintained under standard laboratory conditions.37 The zebrafish stx4ci1016 mutant allele was used. The following zebrafish transgenic lines were used: Tg(-5.1myl7:dsRed2-NLS)f2,38 TgBAC(neurod:EGFP)nl1,39 Tg(gata1:dsRed)sd2,40 Tg(kdrl:EGFP)s843,41 Tg(actb2:stx4-IRES-EGFP)ci1018, Tg(actb2:stx4R241W-IRES-EGFP)ci1019. Wild-type (WT) lines used were mixed AB/TU strain.
Generation of transgenic lines
The Tg(actb2:stx4-IRES-EGFP) transgenic line was created using standard Gateway cloning methods and Tol2 mediated transgenesis.42,43 A pME-stx4 middle-entry clone was generated with the coding region of stx4 (ZDB-GENE-030131-2455). Gateway cloning was used to place the p5E-actb2 5′-entry clone,42 pME-stx4 middle-entry clone, and p3E-IRES-EGFPpA 3′-entry clone42 into the pDEST-Tol2 P2a;α-cry:dsRed plasmid.44 To generate the Tg(actb2:stx4R241W-IRES-EGFP) line, a QuikChange II Site-Directed Mutagenesis Kit (Agilent, 200524) was used to introduce the stx4R241W variant into the actb2:stx4-IRES-EGFP plasmid, as the template. Sanger sequencing was used to confirm the sequences of generated plasmids. To generate transgenic embryos, 50 pg of the actb2:stx4-IRES-EGFP or actb2:stx4R241W-IRES-EGFP plasmids and 25 pg of Tol2 mRNA were co-injected into one cell stage stx4+/− heterozygous embryos.45 F1 lines bearing dsRed in the lens and robust ubiquitous EGFP expression in ∼50% of their progeny were selected, indicative of a single insertion, and maintained in hemizygosity in the stx4+/− background. Primers used are listed in Table S3.
Generation of stx4 mutant line
The stx4 mutant allele was created with CRISPR-Cas9, using standard methods.46, 47, 48 Guide RNAs (gRNAs) were designed using ChopChop, templated via PCR as previously described,46 and synthesized using a MEGAshortscript T7 Transcription Kit (Life Technologies, AM1354); 150 pg of each stx4 gRNA and 6 μM EnGen Spy Cas9 NLS (New England Biolabs, M0646M) was co-injected into one cell stage WT embryos. Efficacy of the gRNAs in generating deletions was determined in pooled F0 embryos by PCR. Sequencing the deletion found in F1 progeny identified a 38-bp deletion. Genotyping of the stx4 mutant allele was performed by PCR using stx4-t2-F1 and stx4-t2-R1 primers (Table S3), which produce 250-bp WT and 212-bp mutant products.
In situ hybridization
Whole-mount in situ hybridization (ISH) was performed using NBT/BCIP (Roche, 11383213001 and 11383221001) according to previously established protocols.49 Probes for myl7 (ZDB-GENE-991019) and tbx2b (ZDB-GENE-990726-27) were previously reported. Larvae were imaged using a Zeiss M2BioV12 stereomicroscope equipped with an AxioCam MRc digital camera.
Immunohistochemistry and CM quantification
Immunohistochemistry (IHC) and CM quantification were performed as previously described.50 Antibodies used included anti-acetylated tubulin, anti-Alcama, anti-cleaved Caspase3, anti-DsRed2, anti-Isl1, anti-sarcomeric myosin heavy chain (Mhc), anti-myosin heavy chain 6 (Myh6, previously called Atrial myosin heavy chain [Amhc]), an anti-zebrafish myosin heavy chain 7 (Myh7, previously called Ventricular myosin heavy chain [Vmhc]),51 anti-Syntaxin 4, and anti-Vamp2. Specifications for all primary and secondary antibodies used are listed in Table S4. DAPI (Life Technologies, 1306) was used at a dilution factor of 1:10,000 to label nuclei, where indicated. Alexa Fluor-555-conjugated α-Bungarotoxin (α-Btx; Invitrogen, B35451) at a concentration of 10 μg/mL was also used. For all IHC, larvae were fixed for 1 h at room temperature in 1% formaldehyde in PBS (v/v %), washed with 0.2% saponin ([w/v %] in PBS), blocked with saponin block (0.2% saponin supplemented with 2 mg/mL bovine serum albumin and 10% [v/v %] heat-inactivated goat serum), post-fixed with 2% paraformaldehyde (v/v %), and stored in PBS, except for larvae labeled with α-Btx or cleaved Caspase-3. For α-Btx labeling, larvae were fixed overnight at 4°C in 4% paraformaldehyde (v/v %) supplemented with 1% DMSO (v/v %) in PBS, washed with PBS, and blocked with incubation buffer (2 mg/mL BSA and 0.5% Triton X-[v/v %] in 0.1M pH 7.4 phosphate buffer), as previously described.52 For cleaved Caspase-3 labeling, larvae were fixed overnight at 4°C in 4% paraformaldehyde, dehydrated in a methanol series at −20°C, washed with PBS supplemented with 0.1% Tween 20 (PBST; v/v %), permeabilized with PBST supplemented with 1% DMSO (v/v %) and 0.3% Triton X-(v/v %), and blocked in PBST supplemented with 10% sheep serum (v/v %), as previously described.53
For CM quantification, embryos were mounted between two coverslips in the same manner as previously described.50 Otherwise, they were immobilized in 15-well angiogenesis μ-Slides (ibidi, 81501) using 1% AquaPor low melt (LM) GTAC agarose (National Diagnostics, EC-204; w/v %) in PBS, once solidified. For CM quantification and Mhy6/Myh7-labeling to assess chamber morphology, micrographs were acquired using a Zeiss M2BioV12 stereomicroscope; otherwise, micrographs were acquired using an inverted, motorized Nikon Eclipse Ti on an A1R confocal microscope equipped with a GaAsP PMT, an HD dual resonant/galvanometric scanner, and an excitation range from 405 to 640 nm, using a Plan Apo 10x/0.45 DIC L; Apo LWD 20x/0.95 WI λS; or Plan Apo VC 60xA/1.2 WI DIC N2 objective. Micrographs were pseudocolored using ImageJ (for images acquired with the Zeiss stereomicroscope) or NIS Elements (for images acquired with the Nikon confocal microscope), if containing red and green channels simultaneously. For Z-stacks acquired with the Nikon confocal microscope, shot noise was removed post-imaging using the Denoise.ai function in NIS Elements.
Quantification of Vamp+ vesicles
To quantify Vamp2+ positive vesicles, embryos were labeled with anti-Vamp2, anti-Alcama, and DAPI and mounted between two coverslips, before being imaged using an inverted, motorized Nikon Eclipse Ti on an A1R confocal microscope equipped with a Plan Apo100x/1.45 λ oil immersion objective. All Z-stacks were subsequently analyzed using Bitplane Imaris 9.3.1 (Oxford Instruments), as detailed below: Total vesicles were modeled using the spot creation tool, while nuclei number and membrane volumes were obtained from surface renderings of DAPI and Alcama labeling, respectively, to ensure a similar proportion of CMs and comparable field of view was quantified between each heart. Total vesicles were normalized to nuclei or membrane volume. To calculate the percent of vesicles close to the CM membrane, the “Spots Close to Surface” XTension was performed with a threshold set to 1.0 μm. The result of this XTension permits for the identification of clusters of objects based on the distance to the selected surface (Alcama-labeled membrane). The resulting spots located inside the threshold-defined region were calculated as a percentage of total vesicle spots, as a surrogate for vesicle clustering. Total vesicles were modeled as surface renderings and a “Surface-Surface coloc” Xtension was performed to permit the masking of two surfaces to find the voxels inside each surface that overlap (Vamp2-labeled vesicles and Alcama-labeled membrane). The resulting new surface generated from these overlapping regions was generated from this Xtension and tabulated, as a surrogate for vesicle docking.
O-dianisidine heme staining
Larval zebrafish were developed in heme staining solution (2.5 mM o-dianisidine, 10 mM sodium acetate pH 5, 0.65% hydrogen peroxide [v/v %], 40% ethanol [v/v %] in water) for 15 min, washed with PBST, subsequently fixed overnight at 4°C in 4% paraformaldehyde, and washed and stored in PBST, as previously described,54 before being imaged by brightfield using a Zeiss M2BioV12 stereomicroscope.
Heart rate measurements
For heart rate measurements, 72 h postfertilization (hpf) larvae were anesthetized with 0.16 mg/mL tricaine (MS-222) and immobilized in 15-well angiogenesis μ-Slides using 1% low melt agarose in embryo water (Milli-Q [Millipore] water supplemented with 0.0001% methylene blue [w/v %] and 0.03% Instant-ocean [w/v %]). Larvae were assayed in embryo water or with drugs at concentrations described below. Hearts were imaged by phase contrast using a Plan Flour 20x/0.5 Ph1 DLL objective on an inverted, motorized Nikon Ti-2 SpectraX Widefield microscope equipped with an Andor Xyla 4.2 megapixel, 16-bit sCMOS monochromatic camera. Larvae were equilibrated for 5 min at thermoneutral conditions (28.5⁰C) in a temperature-controlled chamber prior to high-speed time-lapse imaging. For each acquisition, 500 frames were captured at 2×2 binning with a 17- to 20-ms exposure, using a 12-bit rolling shutter and a 540 Mhz readout. Heart rates were quantified as beats per minute from kymographs obtained from ventricular regions of interest of time measurements that were recorded using NIS Elements.
Drug treatments
At 72 hpf, larvae were treated with the following: 500 μM (±)-isoproterenol hydrochloride (Sigma Aldrich, I5627); 100 nM, 1 μM, 5 μM, or 10 μM of the L-type Ca2+ channel (LTCC) blocker nifedipine (Sigma Aldrich, N7634); 20 μM of LTCC agonist (±)-Bay K-8644 (AG Scientific, B-1019); 10 μM thapsigargin (Sigma Aldrich, T9033) and 10 mM caffeine (Sigma Aldrich, C0750), indicated as TgC; or 10 μM or 20 μM T-type Ca2+ channel antagonist NNC 55-0396 dihydrochloride (Tocris, 2268). All stocks were made with DMSO prior to diluting at the indicated concentrations in embryo water. Control larvae were treated with 1% DMSO in embryo water. Except for isoproterenol treatments, all larvae were treated prior to assaying heart rates for 45 min at 28.5⁰C, similar to what has been reported.55 For isoproterenol treatments, heart rates were assayed both at baseline and following 30 min of treatment at 28.5°C, as previously reported.56
Calcium imaging
Embryonic hearts were explanted with calcium imaging using ratiometric dyes and analysis performed as previously described.55 Briefly, hearts were stained for 20 min with 50 μM Fura-2,AM (Invitrogen) and washed with Normal Tyrode’s solution (NTS; 136 mM Na+, 5.4 mM K+, 1.0 mM Mg2+, 0.3 mM PO43−, 1.8 mM Ca2+, 5.0 mM glucose, and 10.0 mM HEPES, pH 7.4) at room temperature for 45 min. Individual hearts were transferred into perfusion chambers (Warner Instruments, RC-49MFS), containing NTS supplemented with 1 mM Cytochalasin D to inhibit contraction. Perfusion chambers were mounted on a Nikon TE-2000 inverted microscope equipped with a 120 W metal halide lamp (X-Cite 120, Exfo). A high-speed monochromator (Optoscan, Cairn Research Ltd, UK) was used to rapidly switch the excitation wavelength between 340 nm and 380 nm, with a bandwidth of 20 nm and at a frequency of 500 Hz. The excitation light was reflected by a 400-nm cutoff dichroic mirror with the fluorescence emission collected through a 510/580-nm emission filter using a high-speed 80×80 pixel charge-coupled device (CCD) camera (CardioCCD-SMQ, RedShirtImaging) at 14-bit resolution to obtain four frames per acquisition, resulting in a final ratio rate of 125 Hz. For quantification, images were analyzed with MATLAB 2018b (MathWorks) using customized software, as previously described.55
Conventional histopathology and transmission electron microscopy
All biopsy tissue was processed and imaged in a CAP/CLIA-certified laboratory in the Division of Pathology at Cincinnati Children’s, or in the Institute of Pathology at the University Hospital Carl Gustav Carus. For the succinic dehydrogenase staining (SDH), fresh striated muscle biopsy tissue was snap-frozen in 2-methylbutane, sectioned at a thickness of 10 μM, and stained according to standard methodologies.57 For H&E staining, tissues were formalin-fixed, imbedded in paraffin, sectioned to a thickness of 4 to 5 μM, and stained according to standard methodologies.57 Brightfield images were acquired using an ScanScope XT Slide Scanner (Aperio, Leica Biosystems). For ultrastructural examination, muscle tissues were fixed in 3% glutaraldehyde in cacodylate buffer, post-fixed in 1% osmium tetroxide, dehydrated in graded ethanol, epon LX112 resin (Ladd Research Industries Inc) embedded, semithin and ultrathin cut, stained with uranyl acetate and lead citrate, and examined on a Hitachi transmission electron microscope (H-7650; Hitachi High Technologies) equipped with a TEM CCD camera (Advanced Microscopy Techniques), in accordance with standard protocols.58
RT-qPCR
RNA was extracted from whole WT (non-transgenic) and actb2:stx4R241W-IRES-EGFP embryos at 72 hpf using TRIzol (Invitrogen, 15596026), as previously described.59 cDNA was prepared using a SuperScript IV kit (Invitrogen, 18091200) and amplified with oligo d(T)20 primers. RT-qPCR using SYBR green PCR master mix (Applied Biosystems, 4368706) was performed under standard PCR conditions using a Bio-Rad CFX PCR system. Relative expression levels of endogenous stx4 and transgenic stx4R241W-IRES-EGFP were run in technical triplicates, standardized to actb2 (β-actin), and quantified using the 2−ΔΔCT Livak Method.60 Primers used for stx4 (to endogenous stx4) and GFP (to stx4R241W-IRES-EGFP) are listed in Table S3.
Statistical analysis
All statistical analyses were performed using GraphPad Prism (version 9.2.0). Data are represented as mean ± SEM, unless otherwise reported in the figure legend. A minimum of four biological replicates were performed for each experiment. For comparisons between two groups, a 2-tailed Student’s t test was performed with the exception of the survival curve, which employed a Log rank (Mantel-Cox) test. For comparisons involving more than two groups, a one-way ANOVA was performed with Tukey’s multiple comparison test for post hoc analysis to estimate significance of differences from multiple comparisons. For heart rate assays, each arm represents 72 hpf larvae pooled from two to four technical replicates. For all data, p ≤ 0.05 was considered significant. Within graphs p values are indicated as follows: ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001.
Results
Clinical phenotypes of individuals with STX4 variants
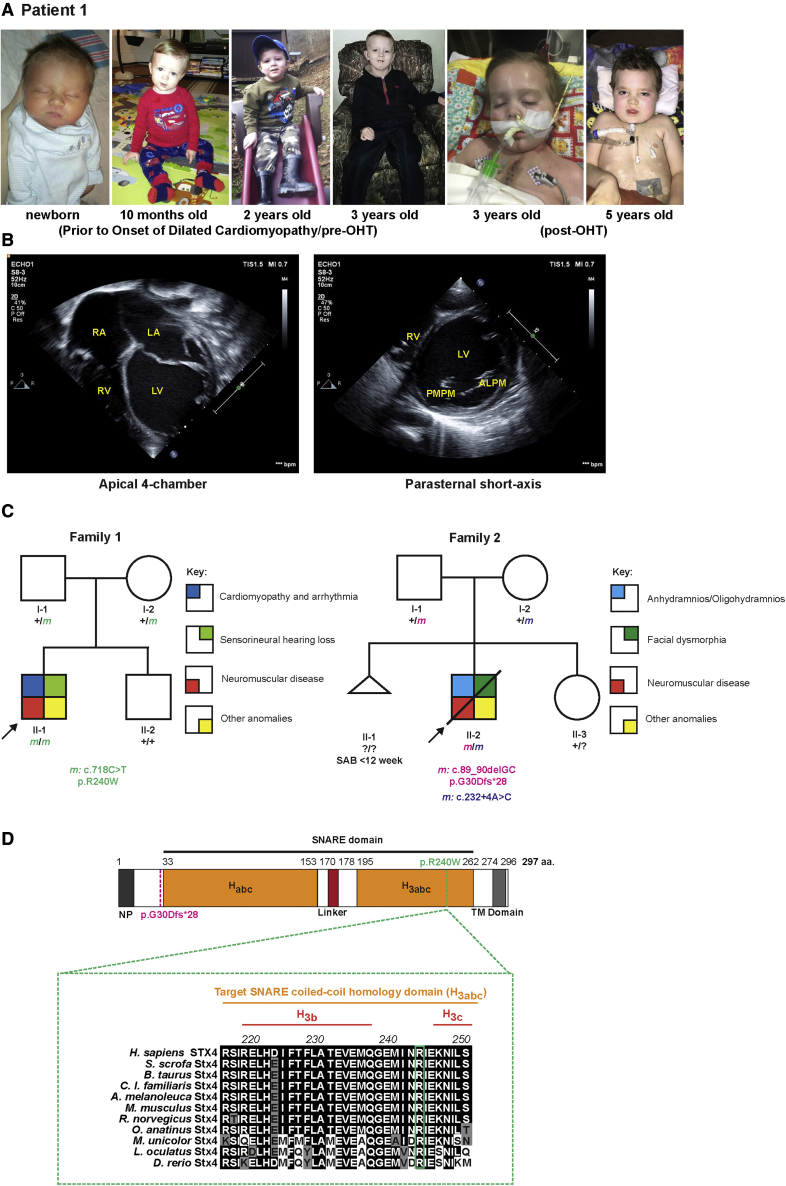
Patient 1 with a prior medical history significant for congenital sensorineural hearing loss, hypotonia, and global developmental delays initially presented with progressive fatigue, dyspnea, hypotension, and severe acidosis at 3 years of age (Figure 1A). Echocardiography showed DCM and severely depressed biventricular systolic function (Figure 1B); patient 1 developed frequent ectopy and ventricular tachycardia. Ultimately, his heart failure and arrhythmia were refractory to medical therapy, and he received an orthotopic heart transplant (OHT) within 2 months from the onset at his initial admission. Consistent with a diagnosis of DCM, post-OHT the explanted heart showed interstitial fibrosis, diminished myocardial cross-striations, loss of myofibrillar volume hypertrophic fibers, and marked variation in fiber size (Figures S1A and S1B). Patient 1’s muscular weakness was progressive after OHT, although no other significant post-operative complications were observed. Skeletal muscle biopsy demonstrated myopathy with fibrosis (Figures S1C and S1D). A cochlear implant installed at age 6 has helped with communication and resulted in marked improvement over patient 1’s previous hearing aids. Additional details of patient 1’s clinical description are provided in “Supplemental note: Case Report for Patient 1.”
Figure 1.
STX4 germline variants in patients
(A) Left to right: Photographs of patient 1 taken before and after onset of DCM, and pre- and post-OHT.
(B) Echocardiograms at onset of patient’s admission due to heart failure. Left: Apical 4-chamber view at end-diastole showed severely dilated, thin-walled left ventricle and severely dilated left and right atria. Right: parasternal short-axis view at the level of the mitral valve at end-diastole showed severely dilated, thin-walled left ventricle. The patient had severe biventricular systolic dysfunction, and evidence for diastolic dysfunction (biatrial dilation). ALPM, anterolateral papillary muscle; LA, left atrium; LV, left ventricle; PMPM, posteromedial papillary muscle; RA, right atrium; RV, right ventricle.
(C) Pedigree segregating the homozygous STX4 c.718C > T; p.R240W variant in family 1 and the c.89_90delGC; p.G30Dfs∗28 and c.232+4A > C alleles in family 2. m, variant allele; ?, unknown carrier; SAB, spontaneous abortion without further data.
(D) Alignment of portion of the STX4 sequences containing the human variant. There is a high degree of conservation overall for STX4 homologues. For instance, mouse Stx4 and zebrafish stx4 respectively share 95.3% and 75.3% amino acid conservation with human STX4. Dashed-magenta line indicates position of p.G30Dfs∗28 allele. Dashed-green line indicates the position of the p.R240W variant and expanded residue sequence indicates the conservation of this residue among the selected vertebrates. The residue positions indicate domain boundaries of the 297 amino acid human STX4 protein.61 Black highlights indicate amino acids that are conserved, gray highlights residues that are chemically similar (conservative), and white highlights display non-conservative variants. Habc, antiparallel three-helix bundle stabilization domain, H3abc, coiled-coil SNARE homology domain; NP, N-terminal peptide, TM, transmembrane.
Due to patient 1’s clinical course, to rule out a known genetic etiology, normal chromosomal microarray, karyotype, Duchenne Muscular Dystrophy (DMD [MIM: 310200]) genetic sequencing, mitochondrial genome sequencing, and metabolic investigations, including amino acids, urine organic acids, and plasma acylcarnitine profiles were performed. As all these assays were negative, an SNP microarray was performed; this identified a 24.8-Mb region of homozygosity, DGV(GRCh 37:Feb.2009) (hg19): arr[hg19] 16p12.1q12.1(24,848,663-49,683,420)x2 Hmz. To refine whether a novel genetic lesion in this region was associated with patient 1’s phenotype, WES was performed. Exome trio analysis detected several variants, including a homozygous missense variant (c.718C > T; p.R240W) in exon 9 of the target-SNARE (t-SNARE) STX4 locus (Table S1), which was confirmed with Sanger sequencing (Figure S2); the trio was of American-European (non-Finnish) descent. Among the detected variants, the STX4 variant was the only candidate locus detected in the region of homozygosity found on the SNP array (Figure 1C). In addition, all other variants were either not concordant with patient 1’s phenotype or zygosity, were predicted to be benign, or failed single-gene deletion/duplication testing by comparative genomic hybridization (Table S1). Notably, this variant is located within the coiled-coil SNARE homology domain of STX4 that is highly conserved among vertebrates (Figure 1D) and was predicted to be damaging by in silico analyses (ACMG Criteria: PM2 PP3).36
A second individual (patient 2) was also identified via GeneMatcher.62 Upon fetal ultrasound at 25+0 weeks of gestation, multiple anomalies were detected, including frontal edema, dilated ductus arteriosus, oligohydramnios, hypoplastic kidneys, dilated echogenic small bowel loops, duodenal atresia, and overlapping fingers. Prenatal trio-WES analysis of the fetus showed compound heterozygous STX4 variants, consisting of two variants (Table S2): c.89_90delGC; p.G30Dfs∗28 and c.232+4A > C. While the former variant is predicted to cause an early truncation, the latter variant occurred at a highly conserved position and is predicted to affect splicing using several in silico analyses, including SpliceAI (donor gain with score of 0.5),33 MaxEntScan (likely disrupting: decreases splicing efficiency as predicted by a MaxEntScan score decrease of 47% [from 7.36 to 3.85]; likely effect: alternative splicing/insertion leading to a frameshift/3′ exon extension),34 CADD (deleterious with a score of 23.7),35 and MutationTaster (disease causing)27 (Figure 1C). This second trio was of European (non-Finnish) descent. No other pathogenic or likely pathogenic variants fitting patient 2’s phenotype were detected (Table S2). Patient 2 was subsequently delivered at 30+4 weeks of pregnancy by secondary cesarean delivery due to complications of anhydramnios and subsequently died 5 days after birth due to multi-organ failure. Additional details of patient 2’s clinical description are provided in Figure S3 and “Supplemental note: Case Report for Patient 2.” Collectively, these clinical data suggest a requirement for STX4 during normal human development and imply a role in cardiac physiology and neuromuscular function.
Zebrafish stx4 mutants have pleiotropic defects analogous to the clinical presentation observed in human patients
Global Stx4 knockout (KO) mice are early embryonic lethal, precluding analysis without conditional alleles.63 In combination with the perinatal lethality of the biallelic variants of patient 2, this suggests that homozygous loss of STX4 function is incompatible with survival in mammals; therefore, we examined the requirements for stx4 in zebrafish, which often can overcome loss-of-function of genes essential for early development due to maternal deposition of RNA and protein, and can survive without a functional heart up to 5 days post fertilization (dpf).64 IHC for Stx4 showed restricted localization in the lateral mesoderm at the 8 somite stage (ss) (Figures S4A and S4A′), and the ventral vasculature, blood progenitors, and endoderm by the 18 s (Figures S4B and S4B′). From 24 to 48 hpf, Stx4 progressively became enriched in the spinal cord, the axonal tracts, and heart (Figures S4C and S4F′). By 72 hpf, Stx4 was expressed in neurons throughout the nervous system (Figures S4H and S4H′).
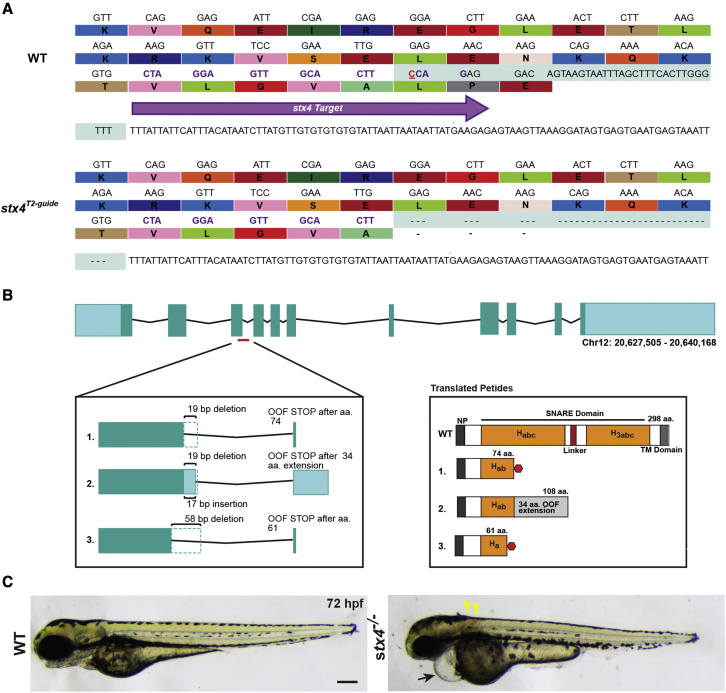
To determine the requirements for stx4 in zebrafish, we generated a stx4 mutant zebrafish allele using CRISPR-Cas9. The allele identified and analyzed has a 38-bp deletion that eliminates the splice donor site of exon 3 (Figure 2A). Sequencing of RNA from the stx4 mutants showed that the resulting allele produces three alternatively spliced transcripts due to the introduction of cryptic splice variants, which are all predicted to generate severely truncated proteins in the Habc stabilization domain (Figure 2B) similar to the earlier truncation observed due to allele 1 of patient 2 (Figure 1D).61 Although overt defects like bradycardia and pericardial edema are already evident in a small proportion of stx4 mutants at 48 hpf, by 72 hpf all stx4 mutants exhibit a complex of defects, including myocardial dysfunction with linearized hearts, pericardial edema, bradycardia, microcephaly, loss of the midbrain/hindbrain boundary, otic vesicle dysgenesis, neuronal atrophy and cell death, and touch insensitivity (Figures 2C and S5A–S5H, Videos S1 and S2). Interestingly, a small proportion of the stx4 mutants (estimated at <10%) have hearts that cease to beat entirely by 72 hpf. In addition, ∼20% of stx4 mutants exhibit hemorrhaging of the intersegmental and cranial vasculature (Figures S5I–S5M). IHC showed diminished expression of Stx4 in the stx4 mutants (Figures 3A–3D′) and stx4 mutants do not survive past 5 dpf (Figure S5N), presumably due to defects in multiple organs, further supporting that this allele results in loss of Stx4. Heterozygous carriers of the zebrafish stx4 allele were equivalent to WT siblings, indicating the allele is recessive. Collectively, the pleiotropic defects found in stx4 mutant zebrafish larvae appear to be reminiscent of the syndromic features observed in patient 1, particularly the cardiac dysfunction, sensorineural hearing loss, global developmental delay, and hypotonia, but also are also consistent with the perinatal lethality observed in patient 2.
Figure 2.
Generation of stx4 mutant zebrafish
(A) CRISPR-Cas9 was used to generate a zebrafish stx4 loss-of-function allele that creates a 38-bp deletion (red underlined cytosine marks the endonuclease cut site), eliminating the splice donor site of exon 3 (protospacer denoted by purple arrow and codons; the last three codons at the splice junction are eliminated in stx4 mutants).
(B) Schematic of transcripts showing the three alternative splice variants generated from this allele. All three transcripts are predicted to be out-of-frame (OOF; left) and to produce truncation products (translated peptides; right). Habc, antiparallel three-helix bundle stabilization domain; H3abc: coiled-coil SNARE homology domain; NP, N-terminal peptide, TM, transmembrane. Red polygon represents a stop codon in the 298 amino acid sized zebrafish Stx4 protein, resulting in early truncation products.
(C) Bright field images of 72 hpf WT/stx4+/− sibling and stx4 mutant larvae. Images of WT sibling embryos in all figures are stx4+/− sibling embryos. Black arrow indicates pericardial edema and linear heart. Yellow arrowheads indicate hemorrhages. stx4 heterozygotes are overtly indistinguishable from WT. Scale bar, 200 μm.
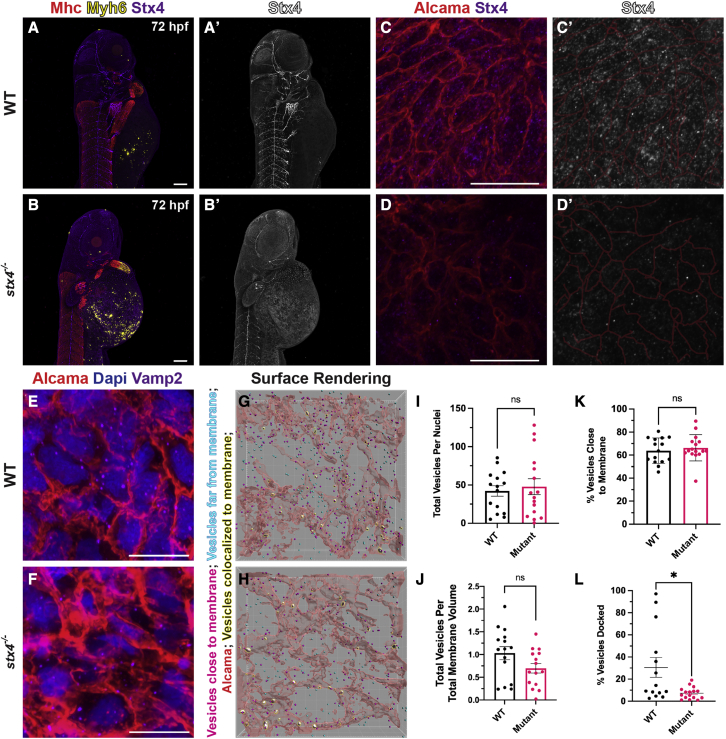
Figure 3.
stx4 mutants exhibit loss of Stx4 expression and function
(A and B) Confocal images of 72 hpf WT or stx4 mutant larvae labeled using IHC markers for Mhc (striated muscle - red), Myh6 (atrial CMs - yellow) and Stx4 (purple). (A′and B′) Single channel confocal images of Stx4. Scale bars, 100 μM.
(C and D) Confocal images of 72 hpf WT and stx4 mutant CM membranes labeled with Alcama (red), and Stx4 (purple). Scale bars, 10 μM (C′and D′) Single channel confocal images of Stx4. Membranes (red lines) from (C and D) are indicated.
(E and F) Confocal images of 72 hpf WT and stx4 mutant CM membranes labeled with DAPI (nuclei; blue), Alcama (red), and Vamp2 (purple). Scale bars, 10 μM.
(G and H) Imaris surface rendering of max intensity projections of Vamp2 from (E and F). Red surfaces indicate Alcama-labeled sarcolemma; yellow-highlighted white surfaces indicate overlapping vesicle surfaces; magenta spots indicate vesicles identified within the threshold as close to the membrane surface; cyan spots indicate vesicles outside this threshold.
(I and J) Total Vamp2+ vesicles from spot rendered puncta normalized to nuclei or cell membrane volume. Vesicles were quantified via spot rendering of puncta labeled with Vamp2. Membrane volumes were obtained from surface renderings of Alcama labeling. Data are represented as the mean ± SEM, n = 15 WT/stx4+/− and n = 16 stx4−/− larvae, Student’s t test.
(K) Vamp2+ vesicles quantified near Alcama-labeled membrane surface threshold of 72 hpf WT or stx4 mutant CMs. Data are represented as the mean ± SEM, n = 15 WT/stx4+/− and n = 16 stx4−/− larvae, Student’s t test.
(L) Quantification of the percent of colocalized Vamp2+ vesicles over total number of vesicles of 72 hpf WT or stx4 mutant CMs. Vesicle surfaces were colocalized to the CM membrane by masking overlapping surfaces. Data are represented as the mean ± SEM, n = 14 WT/stx4+/− and n = 16 stx4−/− larvae, Student’s t test ∗p < 0.05.
stx4 mutant CMs have abrogated vesicle fusion
Given the cardiac abnormalities shared by the index patients and zebrafish stx4 mutants, we next examined Stx4 in the hearts of 72 hpf WT and stx4 mutant larvae in greater detail. As a target-SNARE (t-SNARE), Stx4 mediates vesicular fusion at the cell membrane. As might be expected for a t-SNARE,65,66 we found that in CMs, Stx4 predominantly localized in a punctate pattern in WTs, while its expression was significantly diminished in stx4 mutants (Figures 3C and 3D′). We next assayed vesicle localization by IHC for Vamp2, the cognate vesicle-SNARE (v-SNARE) partner of Stx4 (Figures 3E–3H). Whereas stx4 mutant CMs have a similar number of vesicles as WT (Figures 3I and 3J), fewer vesicles appear “docked” (colocalized) to the CM sarcolemma, despite vesicle “clustering” similar to WT levels as measured by Vamp2+ vesicle proximity to the sarcolemma (Figures 3G, 3H, 3K, and 3L). Collectively, these data support a requirement for Stx4 in promoting vesicle fusion in CMs.
stx4 mutant hearts have marked bradycardia despite normal adrenergic function
As the hearts of 72 hpf zebrafish stx4 mutant larvae are morphologically more linear than WT (Figures S6A–S6D), we assessed if this aberrant morphology reflects a difference in CM number. To quantify CMs, IHC for chamber-specific myosin heavy chains Myh6 and Myh7, which respectively label atrial and ventricular CMs, was used with the Tg(-5.1myl7:dsRed2-NLS) transgene, which is expressed in all CM nuclei. However, no difference in overall or chamber-specific CM number was detected in the hearts of stx4 mutant or WT zebrafish at 72 hpf (Figures S6C–S6G). Similarly, despite the marked bradycardia observed in the mutants (Figures 4A–4C, Videos S1 and S2), the amount of Isl1+ pacemaker CMs67 at the venous pole of the stx4 mutant hearts appeared similar to WT (Figures S6H and S6I). A previous report suggested that zebrafish hearts are not yet innervated by 72 hpf.56 Consistent with this, we failed to detect innervation to the hearts of 72 hpf WT (or stx4 mutant) zebrafish using α-Bungarotoxin (Figures S7A and S7B), acetylated α-Tubulin (Figures S7C and S7D), or the vagus nerve reporter transgene TgBAC(neurod:EGFP) (Figure S7E). However, challenging stx4 mutants with isoproterenol at 72 hpf indicated that they were just as competent to respond to adrenergic stimulation as WT clutch-mates (Figures S7F and S7G). Collectively, these data imply that adrenergic dysfunction does not account for the bradycardia observed in stx4 mutants.
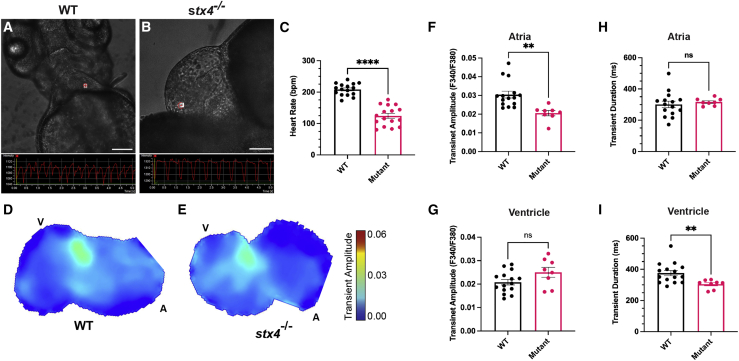
Figure 4.
Stx4 regulates Ca2+ handling in CMs
(A and B) Screenshots from high-speed images of 72 hpf WT and stx4 mutant larvae showing a ventricular region of interest (ROI) and the resulting kymograph used to quantify heart rates. Scale bars, 100 μM.
(C) Heart rates (HR) determined from ventricular ROIs of 72 hpf WT and stx4 mutant larvae captured by high-speed imaging and quantified as beats per minute (bpm). Mutants exhibit an ∼40% reduction in HR versus WT (WT mean HR: 208.4 bpm, Mutant mean HR: 124.3 bpm). Data are represented as the mean ± SEM, n = 16 larvae/group, Student’s t test, ∗∗∗∗p < 0.0001.
(D and E) Maps of Ca2+ transient amplitudes from paced hearts of 72 hpf WT or stx4 mutant larvae. A, atrium; V, ventricle. The color scale depicts Ca2+ transient amplitudes in fluorescence ratio units (F340/F380).
(F and G) Ca2+ transient amplitudes from atrial or ventricular ROIs, respectively.
(H and I) Ca2+ transient durations from atrial and ventricular ROIs. Data are represented as the mean ± SEM, n = 14 WT/stx4+/− and n = 8 stx4−/−, Student’s t test, ∗∗p < 0.01.
stx4 mutants exhibit Ca2+ handling defects and increased sensitization to Ca2+ modulation
As adrenergic function appeared to be preserved by 72 hpf, we explored Ca2+ handling as a potential mechanism for the bradycardia using Fura-2,AM Ca2+ imaging.55 Imaging of 48 hpf explanted hearts extrinsically paced for comparison between stx4 mutant hearts and WT clutch-mates did not reveal any significant differences, consistent with the limited penetrance of pericardial edema and bradycardia at this stage. However, Ca2+ imaging at 72 hpf indicated that the Ca2+-transient amplitude in stx4 mutant atria was significantly diminished compared with WTs (Figures 4D–4G). In addition, the Ca2+-transient duration in paced 72 hpf stx4 mutant ventricles was significantly shorter than in WTs (Figures 4H and 4I). Thus, the cardiac dysfunction observed in stx4 mutants is at least partially due to defects in Ca2+ handling in CMs.
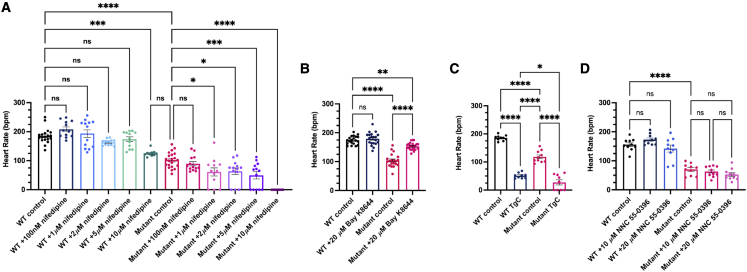
To further characterize the Ca2+ handling abnormalities in stx4 mutants, we exploited pharmacological manipulation of both sarcolemmal Ca2+ uptake and Ca2+-induced-Ca2+ release from the sarcoplasmic reticulum of CMs. We challenged 72 hpf larvae with pharmacological modulators of Ca2+ handling and assayed heart rates only of stx4 mutants with observable heart rates for this analysis. Consistent with the ability of Syntaxins to regulate LTCCs in other tissues,68 stx4 mutants treated with nifedipine, a selective LTCC blocker, were sensitized in a dose-dependent manner and exhibited asystole at concentrations below the threshold for bradycardia in WT clutch-mates (Figure 5A).55 Conversely, Bay K-8644, an LTCC agonist,55 was able to partially rescue the bradycardia phenotype, as the stx4 mutant treated group mean heart rate is restored to ∼90% of untreated WTs, though still statistically different from the untreated WT embryos (Figure 5B). We also challenged larvae with a cocktail of thapsigargin and caffeine (TgC), which blocks the sarco/ER Ca2⁺-ATPase (SERCA) or stimulates Ryanodine receptors, respectively, thereby depleting sarcoplasmic Ca2+ stores.55 TgC produced marked bradycardia in both WT larvae and the majority of the stx4 mutant larvae (a 72.4% reduction in heart rate in WTs versus a 76.8% reduction in stx4 mutants versus controls; Figure 5C), suggesting that baseline SERCA activity is a less likely contributor to the basal bradycardia. We also challenged stx4 mutants with NNC 55-0396 (NNC), a selective T-type Ca2+ channel (TTCC) antagonist.69 However, two different concentrations of NNC, including one previously reported to abolish the cardiac action potential in larval zebrafish,69 were not sufficient to significantly affect the heart rate of WT or stx4 mutant larvae at 72 hpf (Figure 5D), consistent with previous reports regarding the contribution of LTCCs (and to a lesser extent SERCA) to intrinsic cardiac pacing in zebrafish and other vertebrates.70 Together, these data imply that stx4 mutants have reduced sarcolemmal LTCC function, and strongly support this mechanism as a substantive contributor to the cardiac dysfunction observed in the setting of Stx4 loss-of-function.
Figure 5.
Stx4 regulates L-type voltage gated Ca2+ channel activity
(A) Heart rates (HRs) determined from ventricular regions of interest of 72 hpf WT and stx4 mutant larvae following a dose-response treatment with 1% DMSO/embryo water (control), 100 nM nifedipine, 1 μM nifedipine, 2 μM nifedipine, 5 μM nifedipine, or 10 μM nifedipine for 45 min. Data are represented as the mean ± SEM, n = 10–21 larvae/group, one-way ANOVA, ∗p < 0.05, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
(B) HRs determined from 72 hpf WT and stx4 mutant larvae treated with either 1% DMSO/embryo water (control) or 20 μM Bay K-8644. Control stx4 mutants exhibit an ∼40% reduction in mean HR versus WT (WT control mean HR: 174.2 bpm, stx4 mutant control mean HR: 104.1 bpm). By contrast, stx4 mutants treated with 20 μM Bay K-8644 (mean HR: 153.1 bpm) are rescued to ∼90% of the WT control HR and ∼150% of the untreated stx4 mutant HR, while HR of WTs treated with 20 μM Bay K-8644 are similar to untreated WT controls (mean HR: 179.3 bpm). Data are represented as the mean ± SEM, n = 20–34 larvae/group, one-way ANOVA, ∗∗p < 0.01, ∗∗∗∗p < 0.0001.
(C) HRs determined from WT and stx4 mutant larvae treated with either 1% DMSO/embryo water (control) or a mixture of 10 μM thapsigargin and 10 mM caffeine (TgC). Data are represented as the mean ± SEM, n = 9–10 larvae/group, one-way ANOVA, ∗p < 0.05, ∗∗∗∗p < 0.0001.
(D) HRs determined from a dose-response of WT and stx4 mutant larvae treated with either 1% DMSO/embryo water (control), 10 μM NNC 55-0396, or 20 μM NNC 55-0396. Data are represented as the mean ± SEM, n = 10 larvae/group, one-way ANOVA, ∗∗∗∗p < 0.0001.
The analogous zebrafish Stx4R241W allele is functionally hypomorphic
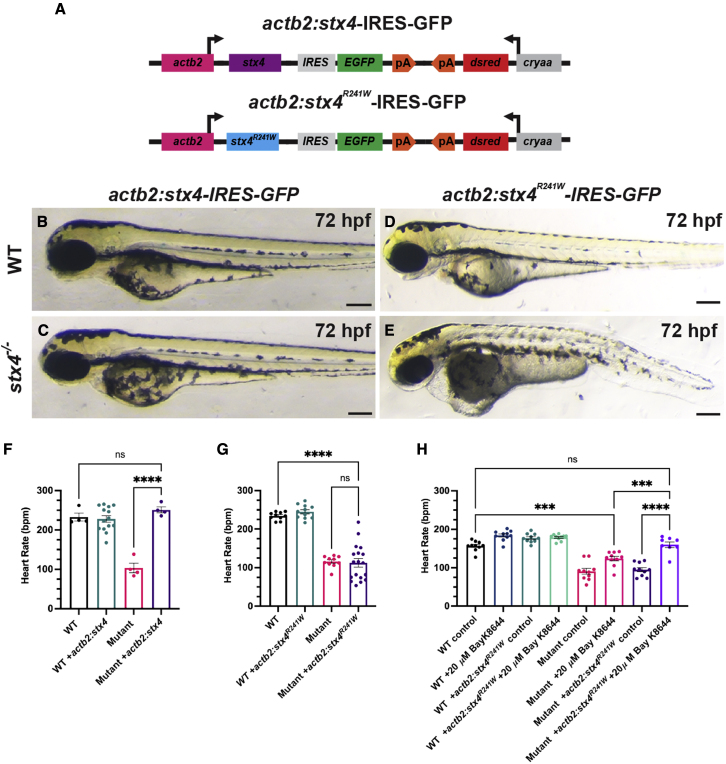
Given the discordance between the predominantly cardiac manifestations emerging in the third year of life observed with the homozygous STX4R240W variant of patient 1 and the perinatal lethality of patient 2 (with the premature truncation variant in trans with a splice variant resulting in a donor gain in intron 3), we next investigated whether features of the pleotropic stx4 mutant syndrome might be rescued and whether patient 1’s STX4R240W variant functions as a hypomorphic allele. Thus, we generated stable transgenic lines ubiquitously expressing WT zebrafish Stx4, or Stx4 with a R241W variant, which is the analogous point-mutation in zebrafish to that observed in patient 1 (Figure 6A). Importantly, hemizygous expression of either transgene in WT or stx4 heterozygous larvae does not cause gain-of-function or dominant negative phenotypes (Figures 6B and 6D). Hemizygous expression of the actb2:stx4-IRES-EGFP transgene was sufficient to rescue the pleiotropic stx4 mutant defects (Figures 6B and 6C), including the bradycardia (Figure 6F). However, hemizygous expression of the actb2:stx4R241W-IRES-EGFP transgene was not able to rescue the mutants (Figures 6D and 6E), despite being expressed approximately at the levels of one WT stx4 allele (Figure S8). In contrast to heterozygous stx4 larvae and stx4 mutant larvae hemizygous for the actb2:stx4-IRES-EGFP transgene, stx4 mutant larvae hemizygous for the actb2:stx4R241W-IRES-EGFP transgene showed a high degree of variability in their bradycardia (Figure 6G), which suggested that the variant likely produces a hypomorphic protein, consistent with patient 1’s survival into childhood and the progressive onset of their syndrome. Finally, we treated stx4 mutants bearing hemizygous expression of actb2:stx4R241W-IRES-EGFP with Bay K-8644 to ascertain whether this allele might be responsive to modulation of LTCC function. Remarkably, treatment with Bay K-8644 was able to fully rescue the bradycardia of stx4 mutant larvae with hemizygous expression of the actb2:stx4R241W-IRES-EGFP, as their heart rates were indistinguishable from WT (Figure 6H). Therefore, together these data support that the variant is hypomorphic and present the possibility that an LTCC agonist may therapeutically rescue some aspects of the dysfunction due to the R240W allele.
Figure 6.
The zebrafish Stx4R241W variant is hypomorphic
(A) Schematic of constructs used to generate transgenic zebrafish expressing WT Stx4 (actb2:stx4-IRES-GFP) and the zebrafish Stx4R241W variant (actb2:stx4R241W-IRES-GFP), which is equivalent to the patient 1 STX4R240W variant.
(B–E) Images of 72 hpf WT and stx4 mutants hemizygous for the actb2:stx4-IRES-GFP and actb2:stx4R241W-IRES-GFP transgenes. Scale bar, 200 μM.
(F and G) Heart rates determined from ventricular regions of interest of 72 hpf WT and stx4 mutant larvae lacking and hemizygous for actb2:stx4-IRES-GFP or actb2:stx4R241W-IRES-GFP. Data are represented as the mean ± SEM, n = (F) 4–14 and (G) 10–18 larvae/group, respectively, one-way ANOVA, ∗∗∗∗p < 0.0001.
(H) Heart rates from ventricular ROIs determined from 72 hpf WT and stx4 mutant larvae lacking and hemizygous for actb2:stx4R241W-IRES-GFP treated with either 1% DMSO/embryo water (control) or 20 μM Bay K-8644. Data are represented as the mean ± SEM, n = 8–10 larvae/group, one-way ANOVA, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
Discussion
In the present study, we report an index patient (patient 1) with a complex syndrome consisting of global developmental delays, sensorineural hearing loss, hypotonia, frequent ectopy, and biventricular DCM that were refractory to treatment and necessitated heart transplant. WES revealed a homozygous, non-conservative missense variant in STX4 that was predicted by Polyphen-2 and SIFT to be probably damaging and deleterious, respectively.30,71 Although three heterozygous alleles of the same residue detected in patient 1 are readily identifiable in the gnomAD database (single nucleotide variants: 16-31050877-C-T[GRCh37] and 16-31039556-C-T[GRCh38], gnomAD v.2.1.1 and v.3.1.2, respectively),72 no additional homozygous variants are available, which is unsurprising, given that this allele is rare with a global MAF of 0.00000795. Recently, an association between Silver-Russell syndrome (SRS [MIM: 180860]) and other SNVs in STX4 has also been suggested73; however, we did not detect SRS features in patient 1, such as characteristic facies, nor analogous features in stx4 mutant zebrafish, aside from a general failure to thrive. Thus, to the best of our knowledge, we are reporting the first direct association of STX4 in human disease and an SNARE protein affecting cardiac function in vivo.
To date, there has been limited understanding of the mechanism by which STX4 proteins might affect cardiac function. Although Stx4 has been proposed to affect release of atrial natriuretic factor in cultured CMs,74,75 this role is unlikely to account for patient 1’s cardiac defects, given their complex physiology.76 In addition to the aforementioned early embryonic lethality of Stx4 global-KO mice, which is reminiscent of the compound heterozygous truncating allele that we identified in patient 2 (perinatally lethal, with even more severe, pleotropic abnormalities), heterozygous Stx4 mice merely exhibit impaired glucose tolerance due to reduced Glut4 translocation in skeletal muscle.63 While this is consistent with STX4’s role in GLUT4 trafficking in adipocytes,77, 78, 79 similar to heterozygous stx4 mutant zebrafish, these mice display no other overt defects.63 Additionally, a recent study that aimed to uncover the role of SNAREs in cardiac insulin resistance and Glut4 trafficking failed to detect differential expression of Stx4 between control, C57BL/KsJ-leprdb/leprdb, or high-fat diet fed mice.80 Conditional Stx4 KO mice have also been used to investigate Stx4’s requirement in bone matrix deposition.81,82 Despite these proposed roles for STX4, patient 1 did not exhibit metabolic or bone mineralization defects, the former of which is another common feature of SRS.
Because cardiac dysfunction was the major feature of patient 1’s acute presentation and because reduced-LV function was also identified perinatally in patient 2, indicating that progressive loss of STX4 function may present along a spectrum manifesting in cardiac dysfunction, we aimed to understand the requirement for Stx4 in the heart. While decreased cardiac function has been associated as a potential morbidity in synaptopathies, including Otahara and West Syndrome,83,84 the regulation of proper cardiac function by SNARE proteins had not been previously established in vivo. Importantly, in both of these reports, one a clinical case report83 and the other a zebrafish model study,84 the effects on cardiac function were secondary due to neuronal input. In vitro, studies conducted in HEK 293 cells and Xenopus laevis oocytes have suggested that SNARE proteins play a role in regulating ion channel vesicle fusion as well as can exhibit independent allosteric interactions with ion channels to modulate sensitivity or gating behavior.18, 19, 20, 21, 22, 23 For instance, Syntaxin 1A is reported to modulate different steps of the exocytosis and gating of several cardiac ion channels, including Cav (L-, N-, and T-type), Kv (Kv2.1, Kv4.2, Kv4.3, Kv7.1, and Kv11.1), and KATP (Kir6.2), by direct interaction.17, 18, 19, 20, 21, 22, 23 Notably, although some of these proteins are implicated as causes of cardiac channelopathies,19,85,86 many have extracardiac functions. While these previous studies suggested that SNARE proteins may play a role in normal cardiac function, we investigated potential mechanistic links between the variants and the relevant cardiac outcomes. In contrast to previous Stx4 loss-of-function models, stx4 mutant zebrafish develop overt pleiotropic abnormalities that are analogous to both of the patients (a cardiac syndrome and early developmental lethality by 5 dpf, respectively), suggesting a highly conserved requirement for STX4 during vertebrate development. Notably, the syndromic phenotype becomes fully penetrant by 72 hpf in zebrafish stx4 mutant larvae, indicating that maternal stx4 mRNA likely accounts for a delay in the onset of the syndrome.87 The expression of Stx4 in neural tissues and the heart is consistent with the conservation of the predominantly neuromuscular/neurosensory and cardiac syndrome of the zebrafish stx4 mutants and both index patients.
In addition to modeling features of both identified patients, loss of Stx4 leads to severe bradycardia in zebrafish larvae, at least partially due to aberrations in Ca2+ handling that are directly attributable to reduced LTCC activity. These results indicate at least a functional interaction between Stx4 and LTCCs within CMs. Evidence for this is bolstered by our finding that vesicle docking is significantly reduced in CMs of stx4 mutants, concomitant with Ca2+ handling defects. Notably, it has been reported that approximately 80% of early CM Ca2+ in zebrafish is mediated by Ca2+ influx via LTCCs,70 which are known to play a crucial role in zebrafish heart development and human disease.88 We also observed a significant decrease in the ventricular Ca2+ transient duration of stx4 mutants. The changes in Ca2+ transient amplitude and duration that we observed imply substantial reduction in total Ca2+ flux in each cardiac cycle, which would be predicted to affect not only pacemaker CM rates but also atrial and ventricular CM contractility, as observed in the clinic. These data also support the possibility that STX4 functionally interacts and affects LTCCs in a conserved fashion among vertebrates. Interestingly, a recent report has implicated an association between CACNA1C and neurodevelopmental abnormalities and epilepsy (including West Syndrome), suggesting a possible common axis of interaction between SNAREs and LTCCs, given the parity of the syndromic features observed from both protein classes.89
Despite the effects on Ca2+ handling in the stx4 zebrafish mutants, we presently cannot rule out that aspects of the cardiac phenotype and dysfunction are non-cell autonomous. For example, pericardial edema, a common defect observed in zebrafish mutants with cardiovascular dysfunction, may contribute to the morphologically linearized hearts in the stx4 mutants. In addition, a cardiac-specific myl7:stx4-IRES-EGFP transgenic line that we created failed to rescue the overt cardiac defects in the stx4 mutants (data not shown). Although one interpretation of this result is that it supports there may be non-autonomous effects contributing to the cardiac defects observed in the stx4 mutants, there are caveats to using this transgenic CM-specific rescue approach and line, such as whether the timing and levels of transgenic stx4 expression reflect endogenous stx4 within CMs. Unfortunately, we could not determine if there was rescue of Ca2+ transients, despite the lack of morphological rescue, as the spectral emission of EGFP and Fura-2,AM overlap. Thus, the variables involved with this experiment prevent us from definitively concluding there are non-autonomous cardiac defects from stx4 loss. Nevertheless, the stx4 zebrafish mutants also have defects in endothelial integrity, as indicated by hemorrhages, suggesting vascular defects as a possibility that could non-autonomously contribute to the cardiac dysfunction. However, while negative modulation of LTCC activity may reflexively increase heart rate due to vasodilation of the coronary arteries and peripheral vasculature in humans,90 zebrafish lack coronary vasculature until ∼7 weeks post fertilization.91 In addition, stx4 mutants exhibit asystole upon treatment with nifedipine, suggesting that the effect of LTCC modulation is predominantly chronotropic and acts specifically in the heart. Therefore, our data are consistent with at least the cardiac Ca2+ handling defects observed in stx4 mutants being cell autonomous within CMs; however, further analysis is needed to discern if there are additional cell non-autonomous influences of Stx4 on the heart.
Our data also support that patient 1’s STX4R240W variant functions as a hypomorph, given that ubiquitous expression of zebrafish Stx4R241W is not sufficient to rescue the syndromic phenotype of stx4 mutant zebrafish. However, treatment with Bay K-8644 is sufficient to rescue the bradycardia of stx4 mutants hemizygous for the actb2:stx4R241W-IRES-EGFP transgene to rates indistinguishable from WTs, while stx4 homozygous mutants treated with Bay K-8644 is only sufficient to restore their heart rate to ∼90% of WTs, supporting some functionality of the variant. Collectively these data suggest that arrythmias due to loss or partial abrogation of STX4 function may be ameliorated by modulation of Ca2+ levels in CMs. While the heterozygous stx4 larvae do not have overt defects, a caveat of the current analysis using the hemizygous actb2:stx4R241W-IRES-EGFP transgenic line is that it may be expressed at slightly lower levels than predicted for one endogenous WT allele of stx4 (Figure S8), suggesting that higher levels of this variant may confer greater compensatory functionality in the absence of WT stx4 alleles. It should also be noted that variants in other genes that stabilize or traffic ion channels, such as Ankrin-B (ANK2 [MIM: 106410]) and Caveolin-3 (CAV3 [MIM: 601253]) have been implicated in channelopathies,92, 93, 94, 95, 96 indicating that monogenic lesions, such as the one observed in patient 1, might plausibly be linked to arrhythmogenic disorders. This is notable as congenital heart diseases (CHDs), which are the most common birth defects and account for nearly one-third of all major congenital anomalies,97,98 often lead to arrhythmia by adulthood.99, 100, 101, 102 As such disorders often present as sudden cardiac arrest and are typically lethal, the collective impact of congenital arrhythmogenic disorders (CADs) is significant.103,104 While further study of the role of Stx4 in the heart may provide additional mechanistic information, our data modeling previously unreported human disease variants using zebrafish demonstrate a conserved requirement for SNARE proteins in vertebrate heart development, and highlight new potential avenues targeting SNARE proteins in the treatment of CHD and CADs.
Acknowledgments
We thank Terri L. VanDyke for technical assistance. We thank the Cincinnati Children’s Confocal Imaging Core for their assistance with imaging and data interpretation. We thank the Cincinnati Children’s Division of Pathology for their assistance in the processing and scanning of human tissue samples. This work was supported by funding from the National Institutes of Health (R01 HL137766, R01 HL141186, and R01 HL154522 to J.S.W.; T32 HL125204 and T32 GM063483 to E.P.; R24 OD017870 to C.A.M.; K08 HL143177-01A1 to K.N.W.), the Leducq Foundation (to C.A.M.), the American Heart Association (predoctoral fellowship 829174 to E.P.), and the Albert J. Ryan Foundation (to E.P.).
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xhgg.2022.100115.
Web resources
The following web-based resources were used:
Burrows-Wheeler Aligner. http://bio-bwa.sourceforge.net.
CADD. https://cadd.gs.washington.edu/
ChopChop. http://chopchop.cbu.uib.no.
Developmental Studies Hybridoma Bank. https://dshb.biology.uiowa.edu.
Ensembl. http://useast.ensembl.org/index.html.
Fathmm. http://fathmm.biocompute.org.uk.
GeneMatcher. https://genematcher.org
gnomAD/ExAC. https://gnomad.broadinstitute.org.
ImageJ. https://imagej.nih.gov/ij/download.html.
Imaris Open. https://imaris.oxinst.com/open.
MaxEntScan. http://hollywood.mit.edu/burgelab/maxent/Xmaxentscan_scoreseq.html.
MutationAssessor. http://mutationassessor.org/r3.
MutationTaster. www.mutationtaster.org.
Online Mendelian Inheritance in Man. http://www.omim.org.
PolyPhen-2. http://genetics.bwh.harvard.edu/pph2.
PROVEAN. http://provean.jcvi.org/index.php.
SIFT. https://sift.bii.a-star.edu.sg.
Skewer. https://sourceforge.net/projects/skewer.
Varsome. https://varsome.com.
The Zebrafish Information Network. https://zfin.org.
Supplemental information
Data and code availability
This study did not generate/analyze [datasets/code].
References
- 1.Rizo J., Xu J. The synaptic vesicle release machinery. Annu. Rev. Biophys. 2015;44:339–367. doi: 10.1146/annurev-biophys-060414-034057. [DOI] [PubMed] [Google Scholar]
- 2.Salpietro V., Lin W., Delle Vedove A., Storbeck M., Liu Y., Efthymiou S., Manole A., Wiethoff S., Ye Q., Saggar A., et al. Homozygous mutations in VAMP 1 cause a presynaptic congenital myasthenic syndrome. Ann. Neurol. 2017;81:597–603. doi: 10.1002/ana.24905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Verhage M., Sørensen J.B. SNAREopathies: diversity in mechanisms and symptoms. Neuron. 2020;107:22–37. doi: 10.1016/j.neuron.2020.05.036. [DOI] [PubMed] [Google Scholar]
- 4.Salpietro V., Malintan N.T., Llano-Rivas I., Spaeth C.G., Efthymiou S., Striano P., Vandrovcova J., Cutrupi M.C., Chimenz R., David E., et al. Mutations in the neuronal vesicular SNARE VAMP2 affect synaptic membrane fusion and impair human neurodevelopment. Am. J. Hum. Genet. 2019;104:721–730. doi: 10.1016/j.ajhg.2019.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fukuda H., Imagawa E., Hamanaka K., Fujita A., Mitsuhashi S., Miyatake S., Mizuguchi T., Takata A., Miyake N., Kramer U., et al. A novel missense SNAP25b mutation in two affected siblings from an Israeli family showing seizures and cerebellar ataxia. J. Hum. Genet. 2018;63:673–676. doi: 10.1038/s10038-018-0421-3. [DOI] [PubMed] [Google Scholar]
- 6.Torres V.I., Vallejo D., Inestrosa N.C. Emerging synaptic molecules as candidates in the etiology of neurological disorders. Neural Plast. 2017;2017:1–25. doi: 10.1155/2017/8081758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Devaux J., Dhifallah S., De Maria M., Stuart-Lopez G., Becq H., Milh M., Molinari F., Aniksztejn L. A possible link between KCNQ2- and STXBP1-related encephalopathies: STXBP1 reduces the inhibitory impact of syntaxin-1A on M current. Epilepsia. 2017;58:2073–2084. doi: 10.1111/epi.13927. [DOI] [PubMed] [Google Scholar]
- 8.Saitsu H., Kato M., Mizuguchi T., Hamada K., Osaka H., Tohyama J., Uruno K., Kumada S., Nishiyama K., Nishimura A., et al. De novo mutations in the gene encoding STXBP1 (MUNC18-1) cause early infantile epileptic encephalopathy. Nat. Genet. 2008;40:782–788. doi: 10.1038/ng.150. [DOI] [PubMed] [Google Scholar]
- 9.Hamada N., Iwamoto I., Tabata H., Nagata K.-I. MUNC18-1 gene abnormalities are involved in neurodevelopmental disorders through defective cortical architecture during brain development. Acta Neuropathol. Commun. 2017;5:92. doi: 10.1186/s40478-017-0498-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ma H., Feng S., Deng X., Wang L., Zeng S., Wang C., Ma X., Sun H., Chen R., Du S., et al. A PRRT2 variant in a Chinese family with paroxysmal kinesigenic dyskinesia and benign familial infantile seizures results in loss of interaction with STX1B. Epilepsia. 2018;59:1621–1630. doi: 10.1111/epi.14511. [DOI] [PubMed] [Google Scholar]
- 11.Klöckner C., Sticht H., Zacher P., Popp B., Babcock H.E., Bakker D.P., Barwick K., Bonfert M.V., Bönnemann C.G., Brilstra E.H., et al. De novo variants in SNAP25 cause an early-onset developmental and epileptic encephalopathy. Genet. Med. 2021;23:653–660. doi: 10.1038/s41436-020-01020-w. [DOI] [PubMed] [Google Scholar]
- 12.Rohena L., Neidich J., Truitt Cho M., Gonzalez K.D., Tang S., Devinsky O., Chung W.K. Mutation in SNAP25 as a novel genetic cause of epilepsy and intellectual disability. Rare Dis. 2013;1:e26314. doi: 10.4161/rdis.26314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heyne H.O., Singh T., Stamberger H., Abou Jamra R., Caglayan H., Craiu D., De Jonghe P., Guerrini R., Helbig K.L., Koeleman B.P.C., et al. De novo variants in neurodevelopmental disorders with epilepsy. Nat. Genet. 2018;50:1048–1053. doi: 10.1038/s41588-018-0143-7. [DOI] [PubMed] [Google Scholar]
- 14.Südhof T.C., Rothman J.E. Membrane fusion: grappling with SNARE and SM proteins. Science. 2009;323:474–477. doi: 10.1126/science.1161748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saffitz J.E. Protein trafficking in cardiovascular disease: how the science has evolved and where it must go. Trends Cardiovasc. Med. 2015;25:390–391. doi: 10.1016/j.tcm.2015.01.002. [DOI] [PubMed] [Google Scholar]
- 16.Xiao S., Shaw R.M. Cardiomyocyte protein trafficking: relevance to heart disease and opportunities for therapeutic intervention. Trends Cardiovasc. Med. 2015;25:379–389. doi: 10.1016/j.tcm.2014.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Atlas D. Voltage-gated calcium channels function as Ca2+-activated signaling receptors. Trends Biochem. Sci. 2014;39:45–52. doi: 10.1016/j.tibs.2013.12.005. [DOI] [PubMed] [Google Scholar]
- 18.He Y., Kang Y., Leung Y.-M., Xia F., Gao X., Xie H., Gaisano H.Y., Tsushima R.G. Modulation of Kv2.1 channel gating and TEA sensitivity by distinct domains of SNAP-25. Biochem. J. 2006;396:363–369. doi: 10.1042/BJ20051478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chao C.C.T., Mihic A., Tsushima R.G., Gaisano H.Y. SNARE protein regulation of cardiac potassium channels and atrial natriuretic factor secretion. J. Mol. Cell. Cardiol. 2011;50:401–407. doi: 10.1016/j.yjmcc.2010.11.018. [DOI] [PubMed] [Google Scholar]
- 20.Ng B., Kang Y., Xie H., Sun H., Gaisano H.Y. Syntaxin-1A inhibition of P-1075, cromakalim, and diazoxide actions on mouse cardiac ATP-sensitive potassium channel. Cardiovasc. Res. 2008;80:365–374. doi: 10.1093/cvr/cvn210. [DOI] [PubMed] [Google Scholar]
- 21.Pasyk E.A., Kang Y., Huang X., Cui N., Sheu L., Gaisano H.Y. Syntaxin-1A binds the nucleotide-binding folds of sulphonylurea receptor 1 to regulate the KATPChannel. J. Biol. Chem. 2004;279:4234–4240. doi: 10.1074/jbc.M309667200. [DOI] [PubMed] [Google Scholar]
- 22.Wu J., Cui N., Piao H., Wang Y., Xu H., Mao J., Jiang C. Allosteric modulation of the mouse Kir6.2 channel by intracellular H+ and ATP. J. Physiol. 2002;543:495–504. doi: 10.1113/jphysiol.2002.025247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yamakawa T., Saith S., Li Y., Gao X., Gaisano H.Y., Tsushima R.G. Interaction of syntaxin 1A with the N-terminus of Kv4.2 modulates channel surface expression and gating. Biochemistry. 2007;46:10942–10949. doi: 10.1021/bi7006806. [DOI] [PubMed] [Google Scholar]
- 24.Retterer K., Juusola J., Cho M.T., Vitazka P., Millan F., Gibellini F., Vertino-Bell A., Smaoui N., Neidich J., Monaghan K.G., et al. Clinical application of whole-exome sequencing across clinical indications. Genet. Med. 2016;18:696–704. doi: 10.1038/gim.2015.148. [DOI] [PubMed] [Google Scholar]
- 25.Jiang H., Lei R., Ding S.W., Zhu S. Skewer: a fast and accurate adapter trimmer for next-generation sequencing paired-end reads. BMC Bioinformatics. 2014;15:182. doi: 10.1186/1471-2105-15-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schwarz J.M., Cooper D.N., Schuelke M., Seelow D. MutationTaster2: mutation prediction for the deep-sequencing age. Nat. Methods. 2014;11:361–362. doi: 10.1038/nmeth.2890. [DOI] [PubMed] [Google Scholar]
- 28.Shihab H.A., Rogers M.F., Gough J., Mort M., Cooper D.N., Day I.N.M., Gaunt T.R., Campbell C. An integrative approach to predicting the functional effects of non-coding and coding sequence variation. Bioinformatics. 2015;31:1536–1543. doi: 10.1093/bioinformatics/btv009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reva B., Antipin Y., Sander C. Determinants of protein function revealed by combinatorial entropy optimization. Genome Biol. 2007;8 doi: 10.1186/gb-2007-8-11-r232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vaser R., Adusumalli S., Leng S.N., Sikic M., Ng P.C. SIFT missense predictions for genomes. Nat. Protoc. 2016;11:1–9. doi: 10.1038/nprot.2015.123. [DOI] [PubMed] [Google Scholar]
- 31.Chun S., Fay J.C. Identification of deleterious mutations within three human genomes. Genome Res. 2009;19:1553–1561. doi: 10.1101/gr.092619.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Choi Y., Chan A.P. PROVEAN web server: a tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics. 2015;31:2745–2747. doi: 10.1093/bioinformatics/btv195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jaganathan K., Kyriazopoulou Panagiotopoulou S., McRae J.F., Darbandi S.F., Knowles D., Li Y.I., Kosmicki J.A., Arbelaez J., Cui W., Schwartz G.B., et al. Predicting splicing from primary sequence with deep learning. Cell. 2019;176:535–548.e24. doi: 10.1016/j.cell.2018.12.015. [DOI] [PubMed] [Google Scholar]
- 34.Yeo G., Burge C.B. Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. Proc. Annu. Int. Conf. Comput. Mol. Biol. RECOMB. 2003;11:322–331. doi: 10.1089/1066527041410418. [DOI] [PubMed] [Google Scholar]
- 35.Rentzsch P., Schubach M., Shendure J., Kircher M. CADD-Splice-improving genome-wide variant effect prediction using deep learning-derived splice scores. Genome Med. 2021;13 doi: 10.1186/s13073-021-00835-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Richards S., Aziz N., Bale S., Bick D., Das S., Gastier-Foster J., Grody W.W., Hegde M., Lyon E., Spector E., et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of medical genetics and genomics and the association for molecular Pathology. Genet. Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Westerfield M. University of Oregon Press; Eugene: 2007. The Zebrafish Book: A Guide for the Laboratory Use of Zebrafish (Danio rerio) [Google Scholar]
- 38.Mably J.D., Burns C.G., Chen J.-N., Fishman M.C., Mohideen M.-A.P. Heart of glass regulates the concentric growth of the heart in zebrafish. Curr. Biol. 2003;13:2138–2147. doi: 10.1016/j.cub.2003.11.055. [DOI] [PubMed] [Google Scholar]
- 39.Obholzer N., Wolfson S., Trapani J.G., Mo W., Nechiporuk A., Busch-Nentwich E., Seiler C., Sidi S., Söllner C., Duncan R.N., et al. Vesicular glutamate transporter 3 is required for synaptic transmission in zebrafish hair cells. J. Neurosci. 2008;28:2110–2118. doi: 10.1523/JNEUROSCI.5230-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Traver D., Paw B.H., Poss K.D., Penberthy W.T., Lin S., Zon L.I. Transplantation and in vivo imaging of multilineage engraftment in zebrafish bloodless mutants. Nat. Immunol. 2003;4:1238–1246. doi: 10.1038/ni1007. [DOI] [PubMed] [Google Scholar]
- 41.Beis D., Bartman T., Jin S.W., Scott I.C., D’Amico L.A., Ober E.A., Verkade H., Frantsve J., Field H.A., Wehman A., et al. Genetic and cellular analyses of zebrafish atrioventricular cushion and valve development. Development. 2005;132:4193–4204. doi: 10.1242/dev.01970. [DOI] [PubMed] [Google Scholar]
- 42.Kwan K.M., Fujimoto E., Grabher C., Mangum B.D., Hardy M.E., Campbell D.S., Parant J.M., Yost H.J., Kanki J.P., Chien C.-B. The Tol2kit: a multisite gateway-based construction kit forTol2 transposon transgenesis constructs. Dev. Dyn. 2007;236:3088–3099. doi: 10.1002/dvdy.21343. [DOI] [PubMed] [Google Scholar]
- 43.Villefranc J.A., Amigo J., Lawson N.D. Gateway compatible vectors for analysis of gene function in the zebrafish. Dev. Dyn. 2007;236:3077–3087. doi: 10.1002/dvdy.21354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mandal A., Rydeen A., Anderson J., Sorrell M.R.J., Zygmunt T., Torres-Vázquez J., Waxman J.S. Transgenic retinoic acid sensor lines in zebrafish indicate regions of available embryonic retinoic acid. Dev. Dyn. 2013;242:989–1000. doi: 10.1002/dvdy.23987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Higashijima S., Okamoto H., Ueno N., Hotta Y., Eguchi G. High-frequency generation of transgenic zebrafish which reliably express GFP in whole muscles or the whole body by using promoters of zebrafish origin. Dev. Biol. 1997;192:289–299. doi: 10.1006/dbio.1997.8779. [DOI] [PubMed] [Google Scholar]
- 46.Talbot J.C., Amacher S.L. A streamlined CRISPR pipeline to reliably generate zebrafish frameshifting alleles. Zebrafish. 2014;11:583–585. doi: 10.1089/zeb.2014.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hruscha A., Krawitz P., Rechenberg A., Heinrich V., Hecht J., Haass C., Schmid B. Efficient CRISPR/Cas9 genome editing with low off-target effects in zebrafish. Development. 2013;140:4982–4987. doi: 10.1242/dev.099085. [DOI] [PubMed] [Google Scholar]
- 48.Gagnon J.A., Valen E., Thyme S.B., Huang P., Ahkmetova L., Pauli A., Montague T.G., Zimmerman S., Richter C., Schier A.F. Efficient Mutagenesis by Cas9 protein-mediated oligonucleotide insertion and large-scale Assessment of single-guide RNAs. PLoS One. 2014;9:e98186. doi: 10.1371/journal.pone.0098186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Oxtoby E., Jowett T. Cloning of the zebrafish krox-20 gene ( krx-20 ) and its expression during hindbrain development. Nucleic Acids Res. 1993;21:1087–1095. doi: 10.1093/nar/21.5.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Waxman J.S., Keegan B.R., Roberts R.W., Poss K.D., Yelon D. Hoxb5b acts downstream of retinoic acid signaling in the forelimb field to restrict heart field potential in zebrafish. Dev. Cell. 2008;15:923–934. doi: 10.1016/j.devcel.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Song Y.C., Dohn T.E., Rydeen A.B., Nechiporuk A.V., Waxman J.S. HDAC1-mediated repression of the retinoic acid-responsive gene ripply3 promotes second heart field development. PLOS Genet. 2019;15:e1008165. doi: 10.1371/journal.pgen.1008165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jing L. Flourescent immunostaining protocol for a-bungorotoxin (AChRs) in zebrafish. BIO-PROTOCOL. 2012;2:1–5. [Google Scholar]
- 53.Duong T.B., Ravisankar P., Song Y.C., Gafranek J.T., Rydeen A.B., Dohn T.E., Barske L.A., Crump J.G., Waxman J.S. Nr2f1a balances atrial chamber and atrioventricular canal size via BMP signaling-independent and -dependent mechanisms. Dev. Biol. 2018;434:7–14. doi: 10.1016/j.ydbio.2017.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Detrich H.W., Kieran M.W., Chan F.Y., Barone L.M., Yee K., Rundstadler J.A., Pratt S., Ransom D., Zon L.I. Intraembryonic hematopoietic cell migration during vertebrate development. Proc. Natl. Acad. Sci. U. S. A. 1995;92:10713–10717. doi: 10.1073/pnas.92.23.10713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Panáková D., Werdich A.A., MacRae C.A. Wnt11 patterns a myocardial electrical gradient through regulation of the L-type Ca2+ channel. Nature. 2010;466:874–878. doi: 10.1038/nature09249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Burkhard S.B., Bakkers J. Spatially resolved RNA-sequencing of the embryonic heart identifies a role for Wnt/β-catenin signaling in autonomic control of heart rate. Elife. 2018;7:1–19. doi: 10.7554/eLife.31515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sheehan D.C., Hrapchak B.B., Enterline H.T. Battelle Press; 1987. Theory and Practice of Histotechnology. [Google Scholar]
- 58.Woods A.E., Stirling J.W. In: Bancroft’s Theory and Practice of Histological Techniques. Suvarna S.K., Layton C., Bancroft J.D., editors. Elsevier; 2013. Transmission electron microscopy; pp. 493–538. [Google Scholar]
- 59.D’Aniello E., Rydeen A.B., Anderson J.L., Mandal A., Waxman J.S. Depletion of retinoic acid receptors initiates a novel positive feedback mechanism that promotes teratogenic increases in retinoic acid. Plos Genet. 2013;9:e1003689. doi: 10.1371/journal.pgen.1003689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 61.Misura K.M.S., Scheller R.H., Weis W.I. Three-dimensional structure of the neuronal-Sec1-syntaxin 1a complex. Nature. 2000;404:355–362. doi: 10.1038/35006120. [DOI] [PubMed] [Google Scholar]
- 62.Sobreira N., Schiettecatte F., Valle D., Hamosh A. GeneMatcher: a matching tool for connecting investigators with an interest in the same gene. Hum. Mutat. 2015;36:928–930. doi: 10.1002/humu.22844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yang C., Coker K.J., Kim J.K., Mora S., Thurmond D.C., Davis A.C., Yang B., Williamson R.A., Shulman G.I., Pessin J.E. Syntaxin 4 heterozygous knockout mice develop muscle insulin resistance. J. Clin. Invest. 2001;107:1311–1318. doi: 10.1172/JCI12274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stainier D.Y., Fouquet B., Chen J.N., Warren K.S., Weinstein B.M., Meiler S.E., Mohideen M.A., Neuhauss S.C., Solnica-Krezel L., Schier A.F., et al. Mutations affecting the formation and function of the cardiovascular system in the zebrafish embryo. Development. 1996;123:285–292. doi: 10.1242/dev.123.1.285. [DOI] [PubMed] [Google Scholar]
- 65.Bertuccio C.A., Wang T.T., Hamilton K.L., Rodriguez-Gil D.J., Condliffe S.B., Devor D.C. Plasma membrane insertion of KCa2.3 (SK3) is dependent upon the SNARE proteins, syntaxin-4 and SNAP23. PLoS One. 2018;13:1–20. doi: 10.1371/journal.pone.0196717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Predescu S.A., Predescu D.N., Shimizu K., Klein I.K., Malik A.B. Cholesterol-dependent syntaxin-4 and SNAP-23 clustering regulates caveolar fusion with the endothelial plasma membrane. J. Biol. Chem. 2005;280:37130–37138. doi: 10.1074/jbc.M505659200. [DOI] [PubMed] [Google Scholar]
- 67.Tessadori F., van Weerd J.H., Burkhard S.B., Verkerk A.O., de Pater E., Boukens B.J., Vink A., Christoffels V.M., Bakkers J. Identification and functional characterization of cardiac pacemaker cells in zebrafish. PLoS One. 2012;7:1–9. doi: 10.1371/journal.pone.0047644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yang S.N., Larsson O., Bränström R., Bertorello A.M., Leibiger B., Leibiger I.B., Moede T., Köhler M., Meister B., Berggren P.O. Syntaxin 1 interacts with the L(D) subtype of voltage-gated Ca(2+) channels in pancreatic beta cells. Proc. Natl. Acad. Sci. U. S. A. 1999;96:10164–10169. doi: 10.1073/pnas.96.18.10164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Alday A., Alonso H., Gallego M., Urrutia J., Letamendia A., Callol C., Casis O. Ionic channels underlying the ventricular action potential in zebrafish embryo. Pharmacol. Res. 2014;84:26–31. doi: 10.1016/j.phrs.2014.03.011. [DOI] [PubMed] [Google Scholar]
- 70.Bovo E., Dvornikov A.V., Mazurek S.R., de Tombe P.P., Zima A.V. Mechanisms of Ca2+ handling in zebrafish ventricular myocytes. Pflügers Arch. - Eur. J. Physiol. 2013;465:1775–1784. doi: 10.1007/s00424-013-1312-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Adzhubei I.A., Schmidt S., Peshkin L., Ramensky V.E., Gerasimova A., Bork P., Kondrashov A.S., Sunyaev S.R. A method and server for predicting damaging missense mutations. Nat. Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Karczewski K.J., Francioli L.C., Tiao G., Cummings B.B., Alföldi J., Wang Q., Collins R.L., Laricchia K.M., Ganna A., Birnbaum D.P., et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581:434–443. doi: 10.1038/s41586-020-2308-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Alhendi A.S.N., Lim D., McKee S., McEntagart M., Tatton-Brown K., Temple I.K., Davies J.H., Mackay D.J.G. Whole-genome analysis as a diagnostic tool for patients referred for diagnosis of Silver-Russell syndrome: a real-world study. J. Med. Genet. 2021 doi: 10.1136/jmedgenet-2021-107699. jmedgenet-2021-107699. [DOI] [PubMed] [Google Scholar]
- 74.Peters C.G., Miller D.F., Giovannucci D.R. Identification, localization and interaction of SNARE proteins in atrial cardiac myocytes. J. Mol. Cell. Cardiol. 2006;40:361–374. doi: 10.1016/j.yjmcc.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 75.Ferlito M., Fulton W.B., Zauher M.A., Marbán E., Steenbergen C., Lowenstein C.J. VAMP-1, VAMP-2, and syntaxin-4 regulate ANP release from cardiac myocytes. J. Mol. Cell. Cardiol. 2010;49:791–800. doi: 10.1016/j.yjmcc.2010.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gottlieb S.S., Kukin M.L., Ahern D., Packer M. Prognostic importance of atrial natriuretic peptide in patients with chronic heart failure. J. Am. Coll. Cardiol. 1989;13:1534–1539. doi: 10.1016/0735-1097(89)90344-6. [DOI] [PubMed] [Google Scholar]
- 77.Grusovin J., Macaulay S.L. Snares for GLUT4--mechanisms directing vesicular trafficking of GLUT4. Front Biosci. 2003;8:d620–d641. doi: 10.2741/1052. [DOI] [PubMed] [Google Scholar]
- 78.Tellam J.T., Macaulay S.L., McIntosh S., Hewish D.R., Ward C.W., James D.E. Characterization of Munc-18c and syntaxin-4 in 3T3-L1 adipocytes. Putative role in insulin-dependent movement of GLUT-4. J. Biol. Chem. 1997;272:6179–6186. doi: 10.1074/jbc.272.10.6179. [DOI] [PubMed] [Google Scholar]
- 79.Volchuk A., Wang Q., Ewart H.S., Liu Z., He L., Bennett M.K., Klip A. Syntaxin 4 in 3T3-L1 adipocytes: regulation by insulin and participation in insulin-dependent glucose transport. Mol. Biol. Cell. 1996;7:1075–1082. doi: 10.1091/mbc.7.7.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bowman P.R.T., Smith G.L., Gould G.W. Cardiac SNARE expression in Health and disease. Front. Endocrinol. (Lausanne). 2019;10:1–10. doi: 10.3389/fendo.2019.00881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kawai S., Michikami I., Kitagaki J., Hata K., Kiyonari H., Abe T., Amano A., Wakisaka S. Syntaxin 4a regulates matrix vesicle-mediated bone matrix production by osteoblasts. J. Bone Miner. Res. 2017;32:440–448. doi: 10.1002/jbmr.3056. [DOI] [PubMed] [Google Scholar]
- 82.Sanchez E., Gonzalez E.A., Moreno D.S., Cardenas R.A., Ramos M.A., Davalos A.J., Manllo J., Rodarte A.I., Petrova Y., Moreira D.C., et al. Syntaxin 3, but not syntaxin 4, is required for mast cell–regulated exocytosis, where it plays a primary role mediating compound exocytosis. J. Biol. Chem. 2019;294:3012–3023. doi: 10.1074/jbc.RA118.005532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Jansen K., Vandeput S., Van Huffel S., Lagae L. Cardiac autonomic dysfunction in West syndrome. Epilepsy Res. 2012;102:167–172. doi: 10.1016/j.eplepsyres.2012.05.010. [DOI] [PubMed] [Google Scholar]
- 84.Grone B.P., Marchese M., Hamling K.R., Kumar M.G., Krasniak C.S., Sicca F., Santorelli F.M., Patel M., Baraban S.C. Epilepsy, behavioral abnormalities, and physiological comorbidities in syntaxin-binding protein 1 (STXBP1) mutant zebrafish. PLoS One. 2016;11:1–25. doi: 10.1371/journal.pone.0151148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cooper E.C. National Center for Biotechnology Information (US)); 2012. Potassium Channels (Including KCNQ) and Epilepsy. [PubMed] [Google Scholar]
- 86.Ravens U. Ionic basis of cardiac electrophysiology in zebrafish compared to human hearts. Prog. Biophys. Mol. Biol. 2018;138:38–44. doi: 10.1016/j.pbiomolbio.2018.06.008. [DOI] [PubMed] [Google Scholar]
- 87.Ciruna B., Weidinger G., Knaut H., Thisse B., Thisse C., Raz E., Schier A.F. Production of maternal-zygotic mutant zebrafish by germ-line replacement. Proc. Natl. Acad. Sci. U. S. A. 2002;99:14919–14924. doi: 10.1073/pnas.222459999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rottbauer W., Baker K., Wo Z.G., Mohideen M.A., Cantiello H.F., Fishman M.C. Growth and function of the embryonic heart depend upon the cardiac-specific L-type calcium channel alpha1 subunit. Dev. Cell. 2001;1:265–275. doi: 10.1016/s1534-5807(01)00023-5. [DOI] [PubMed] [Google Scholar]
- 89.Rodan L.H., Spillmann R.C., Kurata H.T., Lamothe S.M., Maghera J., Jamra R.A., Alkelai A., Antonarakis S.E., Atallah I., Bar-Yosef O., et al. Phenotypic expansion of CACNA1C-associated disorders to include isolated neurological manifestations. Genet. Med. 2021;23:1922–1932. doi: 10.1038/s41436-021-01232-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Soward A.L., Vanhaleweyk G.L.J., Serruys P.W. The haemodynamic effects of nifedipine, verapamil and diltiazem in patients with coronary artery disease. A. Review. Drugs. 1986;32:66–101. doi: 10.2165/00003495-198632010-00004. [DOI] [PubMed] [Google Scholar]
- 91.Hata Y., Yoshida K., Kinoshita K., Nishida N. Epilepsy-related sudden unexpected death: targeted molecular analysis of inherited heart disease genes using next-generation DNA sequencing. Brain Pathol. 2017;27:292–304. doi: 10.1111/bpa.12390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Harrison M.R.M., Bussmann J., Huang Y., Zhao L., Osorio A., Burns C.G., Burns C.E., Sucov H.M., Siekmann A.F., Lien C.L. Chemokine-guided angiogenesis directs coronary vasculature formation in zebrafish. Dev. Cell. 2015;33:442–454. doi: 10.1016/j.devcel.2015.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Spears D.A., Gollob M.H. Genetics of inherited primary arrhythmia disorders. Appl. Clin. Genet. 2015;8:215–233. doi: 10.2147/TACG.S55762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kline C.F., Mohler P.J. Defective interactions of protein partner with ion channels and transporters as alternative mechanisms of membrane channelopathies. Biochim. Biophys. Acta - Biomembr. 2014;1838:723–730. doi: 10.1016/j.bbamem.2013.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Mohler P.J., Schott J.J., Gramolini A.O., Dilly K.W., Guatimosim S., DuBell W.H., Song L.S., Haurogné K., Kyndt F., Ali M.E., et al. Ankyrin-B mutation causes type 4 long-QT cardiac arrhythmia and sudden cardiac death. Nature. 2003;421:634–639. doi: 10.1038/nature01335. [DOI] [PubMed] [Google Scholar]
- 96.Vatta M., Ackerman M.J., Ye B., Makielski J.C., Ughanze E.E., Taylor E.W., Tester D.J., Balijepalli R.C., Foell J.D., Li Z., et al. Mutant caveolin-3 induces persistent late sodium current and is associated with long-QT syndrome. Circulation. 2006;114:2104–2112. doi: 10.1161/CIRCULATIONAHA.106.635268. [DOI] [PubMed] [Google Scholar]
- 97.van der Linde D., Konings E.E.M., Slager M.A., Witsenburg M., Helbing W.A., Takkenberg J.J.M., Roos-Hesselink J.W. Birth prevalence of congenital heart disease worldwide: a systematic review and Meta-analysis. J. Am. Coll. Cardiol. 2011;58:2241–2247. doi: 10.1016/j.jacc.2011.08.025. [DOI] [PubMed] [Google Scholar]
- 98.Bernier P.L., Stefanescu A., Samoukovic G., Tchervenkov C.I. The challenge of congenital heart disease worldwide: epidemiologic and demographic facts. Semin. Thorac. Cardiovasc. Surg. Pediatr. Card. Surg. Annu. 2010;13:26–34. doi: 10.1053/j.pcsu.2010.02.005. [DOI] [PubMed] [Google Scholar]
- 99.Khairy P., Balaji S. Cardiac arrhythmias in congenital heart diseases. Indian Pacing Electrophysiol. J. 2009;9:299–317. PMCID: PMC2766579. [PMC free article] [PubMed] [Google Scholar]
- 100.Triedman J.K. Arrhythmias in adults with congenital heart disease. Heart. 2002;87:383–389. doi: 10.1136/heart.87.4.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Tavera M.C., Foresti S., Cappato R. Arrhythmia in congenital heart disease. Eur. Cardiol. Rev. 2007;3:110. [Google Scholar]
- 102.Hernández-Madrid A., Paul T., Abrams D., Aziz P.F., Blom N.A., Chen J., Chessa M., Combes N., Dagres N., Diller G., et al. Arrhythmias in congenital heart disease: a position paper of the European heart rhythm association (EHRA), association for European paediatric and congenital cardiology (AEPC), and the European society of cardiology (ESC) working group on grown-up congenital heart disease, endorsed by HRS, PACES, APHRS, and SOLAECE. EP Eur. 2018;20:1719–1753. doi: 10.1093/europace/eux380. [DOI] [PubMed] [Google Scholar]
- 103.Virani S.S., Alonso A., Aparicio H.J., Benjamin E.J., Bittencourt M.S., Callaway C.W., Carson A.P., Chamberlain A.M., Cheng S., Delling F.N., et al. Heart disease and stroke statistics—2021 update. Circulation. 2021;143:E254–E743. doi: 10.1161/CIR.0000000000000950. [DOI] [PubMed] [Google Scholar]
- 104.Krahn A.D., Healey J.S., Chauhan V., Birnie D.H., Simpson C.S., Champagne J., Gardner M., Sanatani S., Exner D.V., Klein G.J., et al. Systematic assessment of patients with unexplained cardiac arrest: cardiac arrest survivors with preserved ejection fraction registry (CASPER) Circulation. 2009;120:278–285. doi: 10.1161/CIRCULATIONAHA.109.853143. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study did not generate/analyze [datasets/code].