Abstract
OBJECTIVE
To examine the effects of sleep traits on glycated hemoglobin (HbA1c).
RESEARCH DESIGN AND METHODS
This study triangulated evidence across multivariable regression (MVR) and one- (1SMR) and two-sample Mendelian randomization (2SMR) including sensitivity analyses on the effects of five self-reported sleep traits (i.e., insomnia symptoms [difficulty initiating or maintaining sleep], sleep duration, daytime sleepiness, napping, and chronotype) on HbA1c (in SD units) in adults of European ancestry from the UK Biobank (for MVR and 1SMR analyses) (n = 336,999; mean [SD] age 57 [8] years; 54% female) and in the genome-wide association studies from the Meta-Analyses of Glucose and Insulin-Related Traits Consortium (MAGIC) (for 2SMR analysis) (n = 46,368; 53 [11] years; 52% female).
RESULTS
Across MVR, 1SMR, 2SMR, and their sensitivity analyses, we found a higher frequency of insomnia symptoms (usually vs. sometimes or rarely/never) was associated with higher HbA1c (MVR 0.05 SD units [95% CI 0.04–0.06]; 1SMR 0.52 [0.42–0.63]; 2SMR 0.24 [0.11–0.36]). Associations remained, but point estimates were somewhat attenuated after excluding participants with diabetes. For other sleep traits, there was less consistency across methods, with some but not all providing evidence of an effect.
CONCLUSIONS
Our results suggest that frequent insomnia symptoms cause higher HbA1c levels and, by implication, that insomnia has a causal role in type 2 diabetes. These findings could have important implications for developing and evaluating strategies that improve sleep habits to reduce hyperglycemia and prevent diabetes.
Introduction
Experimental studies have shown that reducing sleep duration or interrupting sleep results in increased insulin resistance and higher plasma glucose levels (1). Systematic reviews and meta-analyses of prospective studies (2) have consistently found that both shorter and longer sleep durations are associated with higher risk of type 2 diabetes (T2D) (2). Observational studies have also shown that insomnia (3), daytime napping (4), and chronotype (evening preference) (5) are associated with higher T2D risk. However, causal relations are unclear from these data because of the potential biases from residual confounding (e.g., from physical activity and diet) and reverse causality (e.g., from nocturia and neuropathic pain).
Mendelian randomization (MR) analysis, which uses genetic variants as instrumental variables to appraise causal effects of exposures on outcomes, is less prone to confounding or reverse causality than conventional observational multivariable regression (MVR) (6). MR has different sources of bias relative to MVR; therefore, consistency in the results from these two methods increases confidence when assessing causality (7). Recently published two-sample MR (2SMR) studies found little consistent evidence of causal effects of sleep duration on T2D and/or related glycemic traits (8,9). Prior MR studies have suggested that insomnia might causally increase T2D risk (10,11), but they did not assess whether insomnia influences glycemic levels in the general population. These prior studies had limited statistical power and potential for weak instrument bias (12). Understanding the impact of sleep traits on glycemic levels in the general population could have profound public health implications for the prevention of diabetes.
Our aim was to explore effects of sleep traits (e.g., insomnia (10), sleep duration (13), daytime sleepiness (14), daytime napping (15), and chronotype (16)) on average glycemic levels assessed by glycated hemoglobin (HbA1c) (main outcome) and on glucose (secondary outcome) in the general population.
Research Design and Methods
Study Overview
We assessed relations between sleep traits and measures of glycemia using traditional observational epidemiology: cross-sectional observed confounder-adjusted MVR and 1SMR within the UK Biobank (UKB) (n = 336,999), and 2SMR using genome-wide association studies from the Meta-Analyses of Glucose and Insulin-Related Traits Consortium (MAGIC) (n = ∼46,000) (17,18). Importantly, we undertook extensive sensitivity analyses, including novel methods assessing the robustness of 1SMR to bias by unbalanced horizontal pleiotropy, weak instruments, and winner’s curse (19).
UKB
For our confounder-adjusted MVR and 1SMR analyses, we used data on UKB participants. Between 2006 and 2010, the UKB recruited 503,317 participants age 40–69 years (5.5% response rate from 9.2 million adults), providing sociodemographic and lifestyle data, including sleep traits, as well as nonfasting blood samples for HbA1c, nonfasting glucose, and genotyping. We included 336,999 successfully genotyped White British participants in the final analysis (Supplementary Fig. 1 and Supplementary Material). The UKB has received ethical approval from the U.K. National Health Service National Research Ethics Service (London, U.K.) (ref. 11/NW/0382).
We assessed associations between HbA1c/glucose (outcomes) and seven self-reported sleep traits (exposures): insomnia symptoms frequency (participants were asked, “Do you have trouble falling asleep at night or do you wake up in the middle of the night?”; usually vs. sometimes or rarely/never); total 24-h sleep duration (reported in whole hours), short sleep duration (≤6 vs. 7 or 8 h), and long sleep duration (≥9 vs. 7 or 8 h); daytime sleepiness (“Never/rarely,” “Sometimes,” “Usually,” and “All of the time”) and daytime napping (never/rarely, sometimes, or usually, where category increase reflects more frequent sleepiness/napping); and chronotype (definitely a morning person, more a morning than evening person, do not know, more an evening than morning person, and definitely an evening person, where category increase reflects a tendency toward greater evening preference). Those who responded “do not know” or “prefer not to say” were treated as missing data for all traits, except for chronotype, where “do not know” was treated as an intermediate category (Supplementary Material).
MAGIC
For 2SMR analyses, we obtained sex-combined meta-analysis summary statistics of genetic variants related to HbA1c (n = 46,368) (mean [SD] age 53 [11] years; 52% female) (17) and fasting glucose (n = 46,186) (mean [SD] age 52 [13] years; 56% female) (18) from participants of European descent without diabetes.
Genetic Variants
In 1SMR and 2SMR analyses, we used genetic variants identified from genome-wide association studies (GWAS) of seven self-reported sleep traits passing the GWAS multiple testing P value threshold (<5 ∗ 10−8) (10,13–16) (Supplementary Table 1).
Statistical Analyses
UKB HbA1c data were right skewed, and units (mmol/mol) differed compared with MAGIC (%). Therefore, we natural log transformed the HbA1c levels in the UKB and then converted them into SD units (HbA1c: 1 SD = 0.15 log mmol/mol). For 2SMR, we also presented results in SD units of the summary data from MAGIC (1 SD = 0.53%; ∼6 mmol/mol). Thus, for all analyses (MVR, 1SMR, and 2SMR), we estimated the mean difference in HbA1c SD between groups for binary exposures and per category increase for categorical exposures. For binary exposures, in MVR and 1SMR, we estimated the average difference in outcome when everyone in the population of interest experienced the exposure (assuming 100% exposure prevalence) compared with when no one experienced the exposure (assuming 0% exposure prevalence) (20).
MVR Model
We considered the following potential confounders in the MVR model: baseline age, sex, smoking, alcohol intake, Townsend residential area deprivation score, education (International Standard Classification of Education code), vigorous physical activity level, diagnosed sleep apnea (ICD-10 code obtained from the Hospital Episode Statistics data), and BMI (Supplementary Material provides references supporting our assumption that these are plausible causes of both sleep traits and glycemic variation). We also adjusted for population stratification (21) in the MVR analyses by including the top 40 genetic principal components of ancestry and UKB assessment center. We ran two MVR models. In model 1, the main model, we adjusted for covariates (above), top 40 genetic principal components, and assessment center (referred to as MVR in Figs. 1–4). In model 2, we additionally adjusted for BMI because of uncertainty as to whether BMI is a confounder or a mediator on the causal pathway between sleep traits and glycemia.
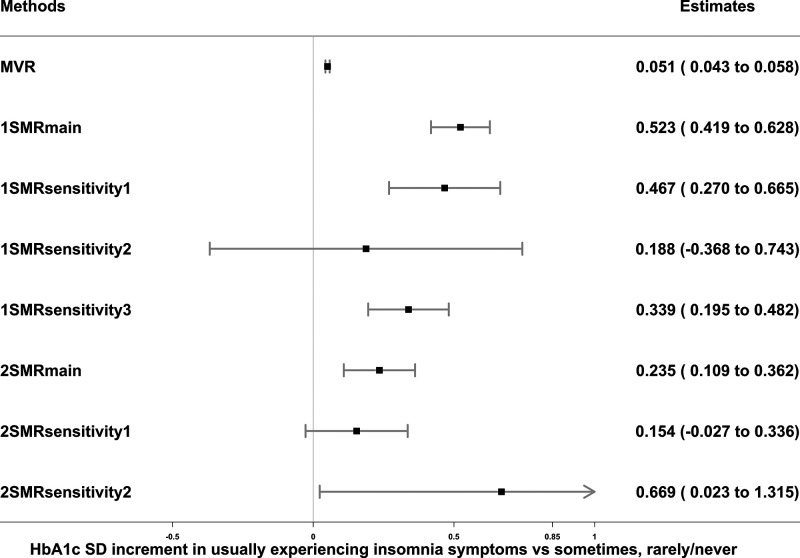
Figure 1.
Associations of insomnia symptoms with HbA1c in observational MVR analysis, 1SMR in the UKB, and 2SMR in MAGIC. Data are SD increment (95% CI) in HbA1c comparing those usually experiencing insomnia vs. sometimes or rarely/never. MVR was adjusted for sex, age, assessment center, 40 genetic principal components, smoking, alcohol intake, Townsend deprivation, education, sleep apnea, and physical activity. 1SMRmain, 1SMRsensitivity1, 1SMRsensitivity2, and 1SMRsensitivity3 were equivalent to two-stage least square, inverse variance–weighted (IVW), MR-Egger, and least absolute deviations regression in 1SMR, respectively. 2SMRmain, 2SMRsensitivity1, and 2SMRsensitivity2 were equivalent to IVW, weighted median, and MR-Egger regression in 2SMR, respectively. 1-SD HbA1c in the UKB is 0.15 log mmol/mol; 1-SD HbA1c in MAGIC is 0.53% (∼6 mmol/mol).
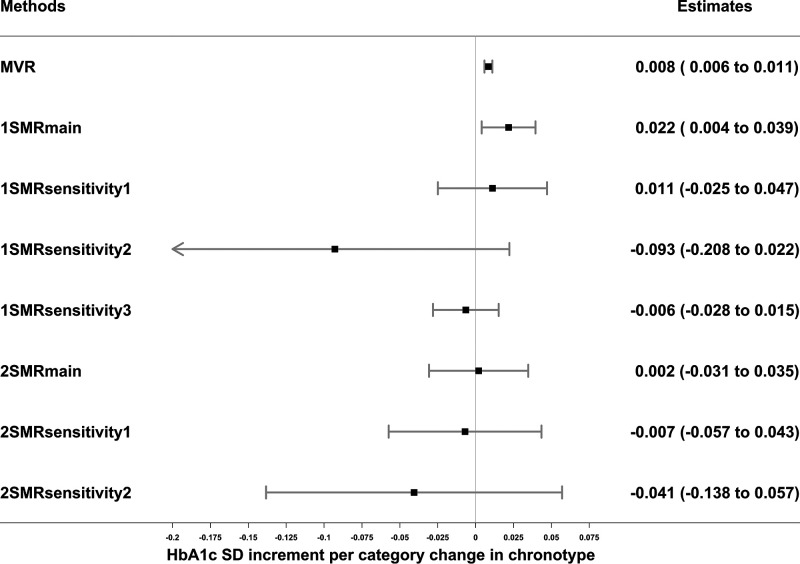
Figure 4.
Associations of chronotype (evening preference) with HbA1c in observational MVR analysis, 1SMR in the UKB, and 2SMR in MAGIC. Data are SD increment (95% CI) in HbA1c in relation to having a higher category of evening preference. MVR was adjusted for sex, age, assessment center, 40 genetic principal components, smoking, alcohol intake, Townsend deprivation, education, sleep apnea, and physical activity. 1SMRmain, 1SMRsensitivity1, 1SMRsensitivity2, and 1SMRsensitivity3 were equivalent to two-stage least square, inverse variance–weighted (IVW), MR-Egger, and least absolute deviations regression in 1SMR, respectively. 2SMRmain, 2SMRsensitivity1, and 2SMRsensitivity2 were equivalent to IVW, weighted median, and MR-Egger regression in 2SMR, respectively. 1-SD HbA1c in the UKB is 0.15 log mmol/mol; 1-SD HbA1c in MAGIC is 0.53% (∼6 mmol/mol).
1SMR
We applied unweighted allele scores, generated as the total number of adverse sleep trait–increasing alleles present, for each participant (evening preference alleles for chronotype) (Supplementary Table 1). Two-stage least squares instrumental variable analyses were performed with adjustment for assessment center and 40 genetic principal components to minimize confounding by population stratification (21), as well as baseline age, sex, and genotyping chip to account for known confounders and reduce random variation (referred to as 1SMRmain in Figs. 1–4) (Supplementary Material).
2SMR
We conducted 2SMR analyses of sleep traits with glycemic measures using the summary associations between the genetic instruments and sleep traits identified in the respective GWAS (10,13–15) (sample 1) (Supplementary Table 1) and estimates of the associations between the genetic instruments and glycemic measures (HbA1c and fasting glucose) (17,18) from MAGIC (sample 2) using the TwoSampleMR package in R (version MRCIEU/TwoSampleMR@0.4.26) (22). We used inverse variance–weighted regression under a multiplicative random-effects model (weights are equal to the inverse of variance of associations between single nucleotide polymorphisms [SNP] and outcome) (referred to as 2SMRmain in Figs. 1–4) to obtain causal effects of sleep traits on HbA1c and fasting glucose (Supplementary Material).
Sensitivity Analyses
To account for the potential impact of diabetes treatment and the effect of diabetes on HbA1c or glucose levels, we repeated MVR and 1SMRmain analyses in UKB participants after excluding those with diabetes defined by the Eastwood algorithm (23) and/or those with a baseline HbA1c ≥48 mmol/mol (≥6.5%).
Assessing MR Assumptions and Evaluating Bias
MR analysis requires various assumptions to be satisfied to estimate consistent causal effects: that the genetic instrument is robustly associated with the exposure (instrument strength), that there is no confounding between the genetic instrument and outcome (independence), and that the genetic instrument influences the outcome exclusively through its effect on the exposure (no unbalanced horizontal pleiotropy). Both 1SMR and 2SMR analyses are vulnerable to bias if these assumptions are violated, and they can also be biased by winner’s curse (24). Various steps were taken to assess the MR assumptions and evaluate bias, as outlined below.
Instrument strength was investigated using the first-stage F statistic and R2 values in both 1SMR and 2SMR. For the independence assumption, we restricted analyses to self-reported European ancestry and adjusted for principal components to limit potential confounding of the genetic instrument-outcome association by population stratification. In 1SMR, we examined associations between the allele scores and variables potentially related to HbA1c (smoking, alcohol, deprivation, education, vigorous physical activity, sleep apnea, and BMI) in the UKB to explore violation of the no horizontal pleiotropy assumptions. To assess potential bias resulting from unbalanced horizontal pleiotropy, we explored between-SNP heterogeneity using the Sargan test (25). To further explore this potential bias in 1SMR, we applied a novel method that allows bias introduced by weak instruments (which would bias our results toward the confounded results) (12) and unbalanced horizontal pleiotropy to be addressed in a one-sample setting using 2SMR approaches (19). This method allowed us to report inverse variance–weighted, MR-Egger, and least absolute deviation regression (similar to the weighted median) in our 1SMR analysis. In Figs. 1–4, we refer to these as 1SMRsensitivity1, 1SMRsensitivity2, and 1SMRsensitivity3, respectively (Supplementary Material).
Bias resulting from winner’s curse can occur when the study in which the genetic variants were identified is also used to perform MR analysis (24), as in our 1SMR analysis, in which the results would be expected approach the null. To minimize risk of weak instrument bias, we used unweighted rather than weighted allele scores (26). Furthermore, we identified subsets of genome-wide significant SNP of some sleep traits in other independent GWAS not including UKB participants (Supplementary Material).
In 2SMR, to explore directional horizontal pleiotropy, we applied two pleiotropy-robust MR methods: MR-Egger and weighted median (in Figs. 1–4, we refer to these as 2SMRsensitivity1 and 2SMRsensitivity2).
Additional Analyses
To rule out the potential reverse causality of any sleep traits with regard to HbA1c, we conducted bidirectional MR analyses (1SMR and 2SMR) (Supplementary Material).
As noted above, it is unclear whether BMI is a confounder or mediator in associations between sleep traits and HbA1c. Given that BMI is strongly associated with HbA1c, we agreed that in the context of any MR data supporting causal effects of sleep traits on HbA1c, if the gene allele score were related to BMI, we would further explore this in multivariable MR (MVMR) (27) (Supplementary Material).
Data and Resource Availability
R scripts for the main and sensitivity analyses are available on GitHub at https://github.com/jamesliu0501/sleeptraits_glyacemic_MRproject.git. For statistical code relating to the individual-level data analysis in the UKB, please contact the corresponding author at ieu_james.liu@bristol.ac.uk. Summary data on glycemic traits were downloaded from MAGIC at www.magicinvestigators.org.
Two patients with T2D, under the care of M.K.R., helped develop the original idea for this research, and these discussions highlighted the importance of potential reverse causality and confounding in MVR analyses and hence the need to explore the research questions with additional methods as mentioned above. Participants in the UKB are regularly updated about research undertaken in the study through newsletters and invitations to meetings where scientists using UKB data present their results. The results of this research will be similarly disseminated to study participants, relevant stakeholders, and the broader public as appropriate.
Results
Baseline Characteristics
Among the 336,999 participants, mean (SD) age was 56.9 (8.0) years, and 54% were female (Table 1); 4.6% of participants had diabetes, 3% had an HbA1c value ≥48 mmol/mol, and 0.5% had diagnosed sleep apnea; 28% reported experiencing insomnia symptoms usually; mean sleep duration was 7.2 (1.1) h; 3% reported daytime sleepiness usually; 5% reported daytime napping usually; 24% reported having a definite morning chronotype; and 8% reported having a definite evening chronotype. Median (interquartile) HbA1c was 35 (33, 37) mmol/mol in individuals without diabetes and 50 (43, 59) mmol/mol in those with diabetes; corresponding nonfasting glucose levels were 4.9 (4.6, 5.3) mmol/L and 6.6 (5.3, 9.0) mmol/L.
Table 1.
Baseline characteristics of the participants included in final analyses of the UKB (n = 336,999)
| Variable | Data |
|---|---|
| Age, years | 56.9 (8.0) |
| Female sex | 54 |
| Townsend deprivation index | −1.58 (2.93) |
| Diabetes defined by Eastwood algorithm | 4.6 |
| HbA1c ≥48 mmol/mol | 3 |
| Diagnosed sleep apnea | 0.47 |
| BMI, kg/m2 | 27.4 (4.8) |
| Smoking | |
| Current | 10 |
| Former | 35 |
| Alcohol intake* | |
| Daily | 21 |
| One to four times a week | 51 |
| One to three times a month | 11 |
| Never/occasionally | 17 |
| Education (ISCED code) | |
| College or university degree/NVQ, HND, HNC, or equivalent | 48 |
| Other professional equivalent (e.g., nursing, teaching) | 12 |
| A levels/AS levels or equivalent | 5 |
| O levels/GCSE or equivalent/CSE or equivalent | 17 |
| None of the above | 17 |
| Days of vigorous physical activity per week† | |
| 0–1 | 52 |
| 2–3 | 30 |
| 4–7 | 18 |
| Insomnia symptoms | |
| Never/rarely or sometimes | 72 |
| Usually | 28 |
| 24-h sleep duration, h | 7.2 (1.1) |
| Short sleep duration (≤6 h) | 26 |
| Long sleep duration (≥9 h) | 10 |
| Daytime sleepiness | |
| Never/rarely | 77 |
| Sometimes | 20 |
| Usually | 3 |
| All of the time | 0.01 |
| Daytime napping | |
| Never/rarely | 57 |
| Sometimes | 38 |
| Usually | 5 |
| Chronotype | |
| Morning | 24 |
| More morning than evening | 33 |
| Do not know | 10 |
| More evening than morning | 25 |
| Evening | 8 |
| HbA1c, mmol/mol/log HbA1c | |
| All (n = 321,078) | 35 (33, 38)/3.57 (0.15) |
| With diabetes (n = 14,980) | 50 (43, 59)/3.94 (0.24) |
| Without diabetes (n = 306,098) | 35 (33, 37)/3.55 (0.12) |
| Glucose, mmol/L/log glucose | |
| All (n = 293,838) | 4.9 (4.6, 5.3)/1.61 (0.17) |
| With diabetes (n = 13,601) | 6.6 (5.3, 9.0)/1.95 (0.39) |
| Without diabetes (n = 280,237) | 4.9 (4.6, 5.3)/1.60 (0.14) |
Data are presented as %, mean (SD), or median (IQR).
AS, first year of full A level; CSE, Certificate of Secondary Education; GCSE, General Certificate of Secondary Education; ISCED, International Standard Classification of Education; HNC, Higher National Certificate; HND, Higher National Diploma; NVQ, National Vocational Qualification.
Alcohol intake was categorized and adjusted as follows: daily, one or four times a week, once or twice a week, one to three times a month, occasionally, and ever (details in Supplementary Material).
Physical activity was categorized and adjusted in days from 0 to 7 (details in Supplementary Material).
Participants in this study were older and less likely to be female when compared with those who were excluded (n = 165,631) in the whole UKB cohort (age 56.9 vs. 55.8 years; 54% vs. 56% female). Included participants were less likely to be diagnosed with sleep apnea, less deprived, and less likely to smoke (Supplementary Table 2).
Associations of Sleep Traits With HbA1c and Glucose Levels
There was consistent evidence across all analyses that more frequent insomnia symptoms (usually vs. sometimes or rarely/never) resulted in higher HbA1c levels (MVR 0.05 [95% CI 0.04–0.06] SD units; 1SMRmain 0.52 [0.42–0.63]; 2SMRmain 0.24 [0.11–0.36]); results of sensitivity analyses accounting for weak instrument bias, unbalanced horizontal pleiotropy, and winner’s curse were consistent with the 1SMRmain and 2SMRmain results (Fig. 1). With exclusion of participants with diabetes and those with HbA1c ≥48 mmol/mol, results of MVR and 1SMR were attenuated but remained consistent with those of the main analyses (Supplementary Table 3). After accounting for BMI in MVMR, the results were consistent with those of the main analyses (Supplementary Table 4). More frequent insomnia symptoms were associated with higher nonfasting glucose levels in the MVR and 1SMRmain analyses, whereas the positive associations were weaker and consistent with the null in 1SMR pleiotropy-robust sensitivity and in all 2SMR analyses (Supplementary Table 5).
Longer 24-h sleep duration, treated as a continuous variable, was associated with lower HbA1c levels in the MVR (−0.006 [95% CI −0.010 to −0.003] SD units) and 1SMRmain analyses (−0.11 [−0.15 to −0.07]), but were close to the null in the 1SMR pleiotropy-robust sensitivity analyses; 2SMR analyses did not support a strong causal effect (Fig. 2). The inverse associations in MVR and 1SMRmain persisted after excluding participants with diabetes and those with HbA1c ≥48 mmol/mol (Supplementary Table 3).
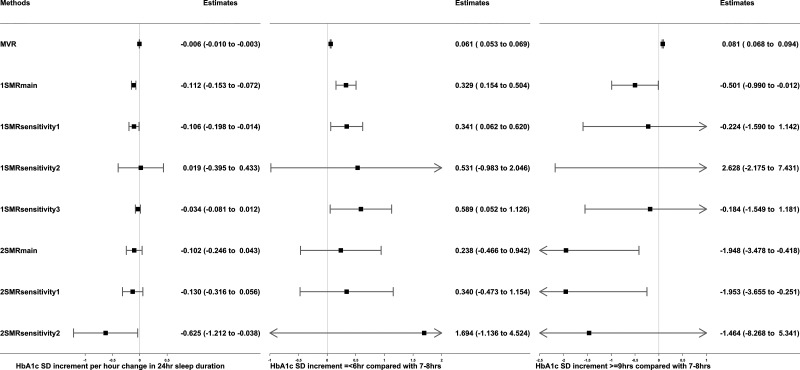
Figure 2.
Associations of sleep duration traits (24-h sleep duration, short sleep duration, and long sleep duration) with HbA1c in observational MVR analysis, 1SMR in the UKB, and 2SMR in MAGIC. Data are SD increment (95% CI) in HbA1c in relation to differences in sleep duration traits (per hour increase in sleep duration, sleep duration ≤6 vs. 7–8 h, sleep duration ≥9 vs. 7–8 h). MVR was adjusted for sex, age, assessment center, 40 genetic principal components, smoking, alcohol intake, Townsend deprivation, education, sleep apnea, and physical activity. 1SMRmain, 1SMRsensitivity1, 1SMRsensitivity2, and 1SMRsensitivity3 were equivalent to two-stage least square, inverse variance–weighted (IVW), MR-Egger, and least absolute deviations regression in 1SMR, respectively. 2SMRmain, 2SMRsensitivity1, and 2SMRsensitivity2 were equivalent to IVW, weighted median, and MR-Egger regression in 2SMR, respectively. 1-SD HbA1c in the UKB is 0.15 log mmol/mol; 1-SD HbA1c in MAGIC is 0.53% (∼6 mmol/mol).
In the MVR analysis, both short (≤6 h) and long sleep duration (≥9 h), treated as binary variables, were associated with higher HbA1c when compared with having a normal 7–8 h per day sleep duration (short sleep 0.06 [95% CI 0.05–0.07] SD units]; long sleep 0.08 [0.07–0.09]). However, in the 1SMRmain analysis, there was evidence that short sleep duration increased HbA1c (0.33 [0.15–0.50]) and that long sleep duration reduced HbA1c (−0.50 [−0.99 to −0.01]). The corresponding 1SMR pleiotropy-robust sensitivity analyses showed a similar direction but with wide CIs. The 2SMR estimates did not support any associations of short sleep duration with HbA1c but indicated that longer sleep was associated with lower HbA1c (Fig. 2). After excluding participants with diabetes and those with HbA1c ≥48 mmol/mol, the positive associations of short sleep with HbA1c remained, but the associations of long sleep with HbA1c were attenuated toward the null (Supplementary Table 3). For nonfasting glucose, longer 24-h sleep duration and short and long sleep were all positively associated with glucose levels in MVR, whereas 1SMR, 2SMR, and all their sensitivity analyses did not consistently support causal effects (Supplementary Table 5).
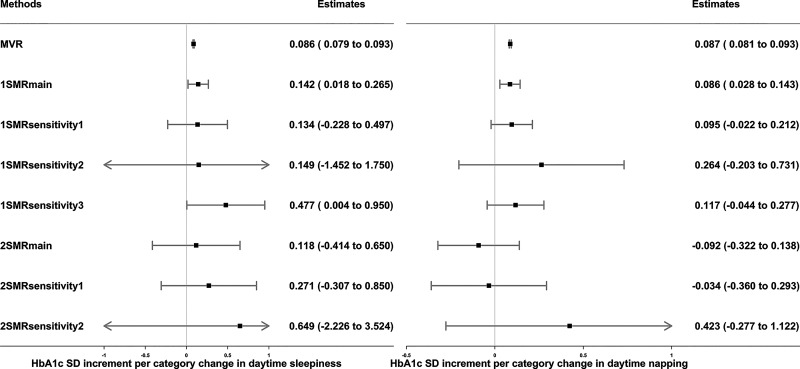
MVR and 1SMRmain showed that more frequent daytime sleepiness (MVR 0.086 [95% CI 0.079–0.093] SD units; 1SMRmain 0.14 [0.02–0.27]) and daytime napping (MVR 0.087 [0.081–0.093]; 1SMRmain 0.09 [0.03–0.14]) were associated with higher HbA1c levels, although 1SMR sensitivity analyses, including those exploring winner’s curse and weak instrument bias, and all 2SMR analyses were not supportive of causal effects (Fig. 3). Excluding participants with diabetes and those with HbA1c ≥48 mmol/mol, the MVR estimates remained, but the main 1SMR estimates were attenuated toward the null (Supplementary Table 3). There was limited evidence of a causal effect of daytime sleepiness or daytime napping on glucose (Supplementary Table 5).
Figure 3.
Associations of daytime sleepiness and daytime napping with HbA1c in observational MVR analysis, 1SMR in the UKB, and 2SMR in MAGIC. Data are SD increment (95% CI) in HbA1c in relation to higher frequencies of daytime sleepiness or daytime napping. MVR was adjusted for sex, age, assessment center, 40 genetic principal components, smoking, alcohol intake, Townsend deprivation, education, sleep apnea, and physical activity. 1SMRmain, 1SMRsensitivity1, 1SMRsensitivity2, and 1SMRsensitivity3 were equivalent to two-stage least square, inverse variance–weighted (IVW), MR-Egger, and least absolute deviations regression in 1SMR, respectively. 2SMRmain, 2SMRsensitivity1, and 2SMRsensitivity2 were equivalent to IVW, weighted median, and MR-Egger regression in 2SMR, respectively. 1-SD HbA1c in the UKB is 0.15 log mmol/mol; 1-SD HbA1c in MAGIC is 0.53% (∼6 mmol/mol).
Evening preference was associated with higher HbA1c levels in MVR (0.008 [95% CI 0.006–0.011] SD units) and 1SMRmain (0.022 [0.004–0.039]). However, these results were attenuated and included the null in all 1SMR pleiotropy-robust sensitivity analyses (including accounting for winner’s curse) and in 2SMR analyses (Fig. 4). MVR and 1SMRmain estimates included the null when excluding participants with diabetes and those with HbA1c ≥48 mmol/mol (Supplementary Table 3). MVR and 1SMRmain estimates indicated evening preference was associated with higher glucose levels, whereas the 1SMR sensitivity analyses and 2SMR estimates did not support an association (Supplementary Table 5). Most of the MVR estimates of sleep traits with HbA1c and glucose were attenuated to some extent after adjusting for BMI (Supplementary Tables 3 and 5).
A detailed description of approaches used to test MR assumptions can be found in the Supplementary Material.
Conclusions
Across different study designs and their sensitivity analyses with different assumptions, we found consistent evidence that frequent insomnia symptoms increase HbA1c levels. Evidence supporting causal effects of other sleep traits on HbA1c and glucose levels was weaker and less consistent.
Comparison With Other Studies
Our findings support previous observational (3) and MR (10,11) studies showing that insomnia is associated with higher T2D risks. Here, we extended these findings by showing an effect of frequent insomnia symptoms on HbA1c in the wider population and after excluding people with diabetes. The finding that insomnia was more strongly linked to hyperglycemia than other sleep traits is in keeping with previous observational data (28). The mechanisms underlying insomnia symptoms with increasing HbA1c are unclear. They could include mediation by depression, anxiety, bipolar disorders, or alterations of sleep physiology (29), with the genetic correlations between insomnia and depression-related traits being stronger than those seen with other sleep-related traits (10). Several hormones that sleep disturbance may influence (e.g., evening cortisol, nighttime growth hormone, ghrelin), together with possible influences on brain glucose utilization, alterations in the sympatho-vagal balance, and proinflammatory processes, may also be relevant (30). Whether these hypothesized mechanisms are causal requires further exploration.
With regard to 24-h sleep duration and short and long sleep, prior 2SMR studies (8,9) found limited evidence of causal effects of these exposures on HbA1c, glucose, or T2D. Our MVR estimates suggest a U-shaped association between sleep duration and HbA1c/glucose levels, which is consistent with previous observational studies (2). However, our MR estimates strengthen previous null MR findings (8,9). The discrepancies between the MVR and MR estimates are likely to be explained by confounding. For example, health status (e.g., baseline chronic diseases) was not accounted for in our study or previous studies using MVR models. This might influence both sleep duration and glycemic levels.
Our findings suggest that previous observational estimates (31,32) linking evening preference and daytime napping with hyperglycemia are also potentially influenced by confounding, because we found no convincing evidence of a causal effect, and previous MR findings are inconsistent (16,33).
Strengths and Limitations
Key strengths of this study are 1) the comparison of results from different methods with differing key sources of bias (7); 2) the range of MR sensitivity analyses performed, including applying novel methods accounting for biases resulting from unbalanced horizontal pleiotropy, weak instruments, and winner’s curse in a one-sample study (19); and 3) the use of large cohorts.
MVR analyses are susceptible to residual confounding. For example, sleep apnea is a plausible confounder (34), but we were only able to adjust for physician-diagnosed sleep apnea. This means that participants with undiagnosed sleep apnea would be treated as not having this condition, which would not be fully accounted for. The MVR analysis used cross-sectional data. It is possible that HbA1c levels could influence sleep through mechanisms including neuropathic pain and nocturia. However, we explored this potential reverse causality using bidirectional 1SMR and 2SMR and found no evidence of an effect of HbA1c on insomnia symptoms (Supplementary Table 6). Because a diabetes diagnosis and use of diabetes medication could alter glycemic levels, we performed subsequent analyses excluding these individuals, which attenuated the estimates. The attenuation of the association between insomnia and HbA1c might be because this association is somewhat stronger in those with diabetes. However, much larger samples than those currently available would be needed to test a different magnitude of association between those with and without diabetes (i.e., an interaction between diabetes status and insomnia in relation to HbA1c). It should also be noted that while there was some attenuation, statistically the results with and without those with diabetes included were consistent (the 95% CI overlap considerably). In addition, we cannot rule out the possibility that excluding/restricting people with diabetes could introduce collider bias (35), although the effect is uncertain.
All sleep traits used were self-reported, so they might have been subject to measurement error. Because participants would not know their value for the genetic IV, any such error would be expected to be random in relation to our outcomes, which would be expected to bias toward the null. For the majority of our MR analyses, the F statistics and R2 suggested that weak instrument bias (12) was unlikely to be substantial (Supplementary Tables 1 and 7). This is supported by the consistency of results in sensitivity analyses, which explored potential bias resulting from weak instruments. We found that some of the genetic instruments were associated with risk factors for HbA1c (Supplementary Table 8), which could violate the MR assumptions through horizontal pleiotropy. This was particularly the case for the insomnia symptoms genetic instrument, which has been shown to be associated with some HbA1c risk factors (36). These associations might reflect vertical pleiotropy, in which they are part of the causal path from frequent insomnia symptoms to HbA1c (e.g., the allele score might be associated with BMI because frequent insomnia symptoms result in higher BMI). Such vertical pleiotropy would not bias our causal estimates. The associations might also highlight specific horizontal pleiotropic paths. However, for insomnia, sensitivity analyses in both 1SMR and 2SMR, except Sargan tests, suggested no direct (unbalanced) horizontal pleiotropy (Supplementary Tables 3 and 5). Additionally, MVMR did not support BMI resulting in horizontal pleiotropy, because results were consistent with the main analysis results.
While we explored consistency across the three methods by focusing on the point estimates, we acknowledge that we had greater statistical power for the MVR and 1SMR than the 2SMR. It would be valuable to explore 2SMR analyses with larger outcome sample summary data. Furthermore, we cannot be certain whether the effects represent specific relations of any sleep trait. For example, there is modest genetic overlap of insomnia symptoms with sleep duration (e.g., effect estimates of 15 of 78 loci predicting 24-h sleep duration were attenuated by 15–25% upon adjustment for insomnia (13); 14 of 49 sleep duration loci have been found to overlap with insomnia (10). However, there are no identical SNPs predicting both insomnia symptoms and 24-h sleep duration among the SNPs in our analysis (Supplementary Table 10).
The low participation rate in the UKB (37) may cause selection bias (35) in observational and MR analyses. Different population sources were used in the 2SMR design, which may be less susceptible to selection bias, although in general, our 2SMR estimates provided limited supporting evidence for causality. Participants in all the study designs were predominantly of European ancestry. Generalizing our findings to other ethnicities requires further validation. Additionally, we acknowledge it would be interesting to see whether the effects of sleep traits on the underlying continuous traits of dysglycemia also influence the risk of incident T2D.
Implications
Two approaches provide some insights into the potential clinical significance of our MR data, although we believe that the true impact of insomnia interventions on HbA1c will only be fully understood through clinical trials.
First, our MR estimates suggest that treating insomnia symptoms could have a greater effect on reducing HbA1c than a substantial degree of weight loss. For example, a previous MR study suggested that 1-SD reduction in BMI would cause a 0.062% reduction in HbA1c (Diabetes Control and Complications Trial [DCCT]–derived units ∼0.12 SD) (38). In our study, we estimated that reducing insomnia symptoms from usually to sometimes or rarely/never would reduce HbA1c by between 0.24 and 0.52 SD units (∼0.13% and 0.28% DCCT units for 2SMRmain and 1SMRmain analyses, respectively). This represents a greater HbA1c reduction through a hypothetical insomnia treatment than the reduction expected in response to 1-SD reduction in BMI (∼5 kg/m2) (e.g., equivalent to 14-kg weight loss in a person of 1.7-m average height).
Second, based on further analysis (Supplementary Material), we estimate that ∼27,300 (95% CI 26,800–27,500) individuals with frequent insomnia symptoms would be free from having diabetes (HbA1c ≥48 mmol/mol) in the U.K. if an effective intervention was delivered to the 25 million adults between 40 and 70 years of age (39). This estimate is based on the 28% of the population who reported usually experiencing insomnia symptoms in the UKB cohort, which is similar to the recorded national surveys of the adult population in the U.K. that range from 5 to 38% (40).
In conclusion, we present robust evidence across MVR, 1SMR, and 2SMR studies that frequent insomnia symptoms cause higher HbA1c. Lifestyle and/or pharmacological interventions that improve insomnia might therefore have benefits in preventing T2D. Understanding the mechanisms underlying the effect of insomnia symptoms on hyperglycemia could help identify therapeutic strategies or new drug targets for preventing T2D.
Article Information
Acknowledgments. The authors thank the participants and researchers from the UKB who contributed or collected data. The authors also thank Dr. Eleanor Sanderson (Medical Research Council Integrative Epidemiology Unit, University of Bristol) for providing statistical support on multivariable MR. Data on glycemic traits were contributed by MAGIC investigators and downloaded from www.magicinvestigators.org. This research was conducted using the UKB Resource under application 6818.
Funding. This work is supported by a Diabetes UK grant (17/0005700), which funds salaries of J.L., A.K.W., and S.E.J. J.L., R.C.R., and D.A.L. work in a unit that is funded by the University of Bristol and the U.K. Medical Research Council (MRC) (MC_UU_00011/1 and MC_UU_00011/6), and contribution of D.A.L. to this paper was support by a grant from the British Heart Foundation (AA/18/7/34219). D.A.L. is an NIHR Senior Investigator (NF-0616-10102) and British Heart Foundation Chair in Cardiovascular Science and Clinical Epidemiology (CH/F/20/90003). R.C.R. is a de Pass Vice Chancellor’s research fellow at the University of Bristol. H.S.D. and R.S. are funded by National Institutes of Health (NIH) grants R01DK107859, and R.S. is funded by NIH grant R01DK105072. R.S. is a recipient of the Phyllis and Jerome Lyle Rappaport Massachusetts General Hospital Research Scholar Award. J.B. is funded by an Establishing Excellence in England (E3) grant awarded to the University of Exeter. D.W.R. is funded by MRC programme grant MR/P023576/1 and is a Wellcome Investigator, Wellcome Trust (107849/Z/15/Z and 107849/A/15/Z). C.B. is supported by the Wellcome Trust via a PhD (218495/Z/19/Z).
Duality of Interest. M.K.R. reports receiving research funding from Novo Nordisk and consultancy fees from Novo Nordisk and Roche Diabetes Care and has a modest shareholding in GlaxoSmithKline, all unrelated to this work. J.B. reports receiving consultancy fees from Novartis, unrelated to this work. D.A.L. has received support from Roche Diagnostics and Medtronic Ltd for research unrelated to this work. No other potential conflicts of interest relevant to this article were reported.
Author Contributions. J.L. affirms that this manuscript is an honest, accurate, and transparent account of the study being reported, that no important aspects of the study have been omitted, and that any discrepancies from the study as planned (and, if relevant, registered) have been explained. J.L. and R.C.R. carried out the data analysis. M.K.R. conceived the idea for the study. M.K.R., R.A.E., D.W.R., S.G.A., M.J.C., M.N.W., T.M.F., A.R.W., J.B., and D.A.L. obtained funds for the project. R.C.R., J.B., D.A.L., and M.K.R. designed the study, including developing the analysis plan. J.B. and C.B. developed the novel one-sample pleiotropy robust analyses. J.L. and R.C.R. carried out the data analysis. J.L., R.C.R., J.B., D.A.L., and M.K.R. wrote the initial draft, with subsequent input from other authors. All authors made critical revisions to the paper. J.L., R.C.R., and D.A.L. are the guarantors of this work and, as such, had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis.
Footnotes
J.L. and R.C.R. contributed equally as first authors.
D.A.L. and M.K.R. contributed equally as senior authors.
This article contains supplementary material online at https://doi.org/10.2337/figshare.17054093.
References
- 1. Nedeltcheva AV, Kessler L, Imperial J, Penev PD. Exposure to recurrent sleep restriction in the setting of high caloric intake and physical inactivity results in increased insulin resistance and reduced glucose tolerance. J Clin Endocrinol Metab 2009;94:3242–3250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Shan Z, Ma H, Xie M, et al. Sleep duration and risk of type 2 diabetes: a meta-analysis of prospective studies. Diabetes Care 2015;38:529–537 [DOI] [PubMed] [Google Scholar]
- 3. Green MJ, Espie CA, Popham F, Robertson T, Benzeval M. Insomnia symptoms as a cause of type 2 diabetes Incidence: a 20 year cohort study. BMC Psychiatry 2017;17:94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hublin C, Lehtovirta M, Partinen M, Koskenvuo M, Kaprio J. Napping and the risk of type 2 diabetes: a population-based prospective study. Sleep Med 2016;17:144–148 [DOI] [PubMed] [Google Scholar]
- 5. Vetter C, Devore EE, Ramin CA, Speizer FE, Willett WC, Schernhammer ES. Mismatch of sleep and work timing and risk of type 2 diabetes. Diabetes Care 2015;38:1707–1713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lawlor DA, Harbord RM, Sterne JA, Timpson N, Davey Smith G. Mendelian randomization: using genes as instruments for making causal inferences in epidemiology. Stat Med 2008;27:1133–1163 [DOI] [PubMed] [Google Scholar]
- 7. Lawlor DA, Tilling K, Davey Smith G. Triangulation in aetiological epidemiology. Int J Epidemiol 2016;45:1866–1886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wang J, Kwok MK, Au Yeung SL, et al. Sleep duration and risk of diabetes: observational and Mendelian randomization studies. Prev Med 2019;119:24–30 [DOI] [PubMed] [Google Scholar]
- 9. Bos MM, van Heemst D, Donga E, et al. The association between habitual sleep duration and sleep quality with glycemic traits: assessment by cross-sectional and Mendelian randomization analyses. J Clin Med 2019;8:682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jansen PR, Watanabe K, Stringer S, et al.; 23andMe Research Team . Genome-wide analysis of insomnia in 1,331,010 individuals identifies new risk loci and functional pathways. Nat Genet 2019;51:394–403 [DOI] [PubMed] [Google Scholar]
- 11. Yuan S, Larsson SC. An atlas on risk factors for type 2 diabetes: a wide-angled Mendelian randomisation study. Diabetologia 2020;63:2359–2371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Burgess S; CRP CHD Genetics Collaboration . Avoiding bias from weak instruments in Mendelian randomization studies. Int J Epidemiol 2011;40:755–764 [DOI] [PubMed] [Google Scholar]
- 13. Dashti HS, Jones SE, Wood AR, et al. Genome-wide association study identifies genetic loci for self-reported habitual sleep duration supported by accelerometer-derived estimates. Nat Commun 2019;10:1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wang H, Lane JM, Jones SE, et al. Genome-wide association analysis of self-reported daytime sleepiness identifies 42 loci that suggest biological subtypes. Nat Commun 2019;10:3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dashti HS, Daghlas I, Lane J, et al. Genetic determinants of daytime napping and effects on cardiometabolic health. medRxiv 2020:2020.2006.2012.20129858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jones SE, Lane JM, Wood AR, et al. Genome-wide association analyses of chronotype in 697,828 individuals provides insights into circadian rhythms. Nat Commun 2019;10:343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Soranzo N, Sanna S, Wheeler E, et al.; WTCCC . Common variants at 10 genomic loci influence hemoglobin A1(C) levels via glycemic and nonglycemic pathways [published correction appears in Diabetes. 2011;60:1050–1051]. Diabetes 2010;59:3229–3239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dupuis J, Langenberg C, Prokopenko I, et al.; DIAGRAM Consortium; GIANT Consortium; Global BPgen Consortium; Anders Hamsten on behalf of Procardis Consortium; MAGIC investigators . New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat Genet 2010;42:105–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Barry C, Liu J, Richmond R, et al. Exploiting collider bias to apply two-sample summary data Mendelian randomization methods to one-sample individual level data. PLoS Genet 2021;17:e1009703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Burgess S, Labrecque JA. Mendelian randomization with a binary exposure variable: interpretation and presentation of causal estimates. Eur J Epidemiol 2018;33:947–952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bycroft C, Freeman C, Petkova D, et al. The UK Biobank resource with deep phenotyping and genomic data. Nature 2018;562:203–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hemani G, Zheng J, Elsworth B, et al. The MR-Base platform supports systematic causal inference across the human phenome. Elife 2018;7:e34408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Eastwood SV, Mathur R, Atkinson M, et al. Algorithms for the capture and adjudication of prevalent and incident diabetes in UK Biobank. PLoS One 2016;11:e0162388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Taylor AE, Davies NM, Ware JJ, VanderWeele T, Smith GD, Munafò MR. Mendelian randomization in health research: using appropriate genetic variants and avoiding biased estimates. Econ Hum Biol 2014;13:99–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Baum CF, Schaffer ME, Stillman S. Instrumental variables and GMM: estimation and testing. Stata J 2003;3:1–31 [Google Scholar]
- 26. Burgess S, Dudbridge F, Thompson SG. Combining information on multiple instrumental variables in Mendelian randomization: comparison of allele score and summarized data methods. Stat Med 2016;35:1880–1906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sanderson E, Davey Smith G, Windmeijer F, Bowden J. An examination of multivariable Mendelian randomization in the single-sample and two-sample summary data settings. Int J Epidemiol 2019;48:713–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bin YS. Is sleep quality more important than sleep duration for public health? Sleep (Basel) 2016;39:1629–1630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lind MJ, Gehrman PR. Genetic pathways to insomnia. Brain Sci 2016;6:64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Van Cauter E, Spiegel K, Tasali E, Leproult R. Metabolic consequences of sleep and sleep loss. Sleep Med 2008;9(Suppl. 1):S23–S28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Knutson KL, Wu D, Patel SR, et al. Association between sleep timing, obesity, diabetes: the Hispanic Community Health Study/Study of Latinos (HCHS/SOL) cohort study. Sleep (Basel) 2017;40:zsx014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Guo VY, Cao B, Wong CKH, Yu EYT. The association between daytime napping and risk of diabetes: a systematic review and meta-analysis of observational studies. Sleep Med 2017;37:105–112 [DOI] [PubMed] [Google Scholar]
- 33. Adams CD, Neuhausen SL. Evaluating causal associations between chronotype and fatty acids and between fatty acids and type 2 diabetes: a Mendelian randomization study. Nutr Metab Cardiovasc Dis 2019;29:1176–1184 [DOI] [PubMed] [Google Scholar]
- 34. Humer E, Pieh C, Brandmayr G. Metabolomics in sleep, insomnia and sleep apnea. Int J Mol Sci 2020;21:7244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Munafò MR, Tilling K, Taylor AE, Evans DM, Davey Smith G. Collider scope: when selection bias can substantially influence observed associations. Int J Epidemiol 2018;47:226–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lane JM, Jones SE, Dashti HS, et al.; HUNT All In Sleep . Biological and clinical insights from genetics of insomnia symptoms. Nat Genet 2019;51:387–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sudlow C, Gallacher J, Allen N, et al. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med 2015;12:e1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hu X, Zhuang X-D, Mei W-Y, et al. Exploring the causal pathway from body mass index to coronary heart disease: a network Mendelian randomization study. Ther Adv Chronic Dis 2020;11:2040622320909040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Office for National Statistics . Overview of the UK population: August 2019. Accessed 22 December 2020. Available from https://www. ons.gov.uk/peoplepopulationandcommunity/populationandmigration/populationestimates/ articles/overviewoftheukpopulation/august 2019
- 40. Calem M, Bisla J, Begum A, et al. Increased prevalence of insomnia and changes in hypnotics use in England over 15 years: analysis of the 1993, 2000, and 2007 National Psychiatric Morbidity Surveys. Sleep (Basel) 2012;35:377–384 [DOI] [PMC free article] [PubMed] [Google Scholar]