Abstract
In our previous studies, we found that the rs231775 polymorphism of cytotoxic T-lymphocyte antigen 4 (CTLA-4) is associated with risks of different cancer types; however, the association remains controversial and ambiguous, so we conducted an in-depth meta-analysis to verify the association. A complete search of the PubMed, Google Scholar, Embase, Chinese databases, and Web of Science was conducted without regard to language limitations, covering all publications since November 20, 2021. The search criteria for cancer susceptibility associated with the polymorphism in the CTLA-4 gene rs231775 resulted in 87 case-control studies with 29,464 cases and 35,858 controls. The association strength was analyzed using odds ratios and 95% confidence intervals. Overall, we found that the CTLA-4 rs231775 polymorphism may reduce cancer risk. A stratified cancer type analysis showed that CTLA-4 rs231775 polymorphism was a risk factor for colorectal cancer and thyroid cancer; on the other hand, it was a protective factor for breast cancer, liver cancer, cervical cancer, bone cancer, head and neck, and pancreatic cancer. We also classified cancer into five systems and observed an increased association with digestive tract cancer, decreased associations with orthopedic tumors, tumors of the urinary system, and gynecological tumors. In the subgroup based on race, decreased relationships were observed in both Asians and Caucasians. The same decreased association was also shown in the analysis of the source of control analysis. Our present study indicates that the CTLA-4 rs231775 polymorphism contributes to cancer development and aggression.
Keywords: cancer, cytotoxic T-lymphocyte antigen 4, polymorphism, tumor marker, meta-analysis
Introduction
A major obstacle to increasing life expectancy is cancer, which is the primary cause of death worldwide. Cancer, in 112 of 183 countries, is also estimated to be the first or second leading cause of death before the age of 70 and third or fourth in 23 other countries (1), according to the World Health Organization analyses in 2019 (2). Across the globe, the incidence and mortality of cancer are rising rapidly; this is a result of both increasing longevity and population growth as well as changing patterns in the prevalence and distribution of cancer-causing factors, some of which are associated with social and economic development (1). The development of cancer involves multiple factors, including environmental and genetic factors (3).
One of the most common types of germline variants, the SNPs (single nucleotide polymorphisms), play a key role in human diseases, including cancer (2). Many SNPs associated with human cancer were identified through GWAS (genome-wide association studies) in the past decade (4, 5). Recent studies have noted that the expression levels of nearby genes may be influenced by these cancer risk-associated SNPs (4). Cancer treatment includes traditional surgery, chemotherapy, radiotherapy, and so on. In recent years, immunotherapy has gained more attention (6). The CTLA-4 (cytotoxic T-lymphocyte antigen 4) gene is located on chromosome 2q33 and has four exons (7). Cancer cells can acquire immune regulatory surface proteins like CTLA-4, which suppress the activation of immune cells, such as T cells (3, 8). In the early stages of tumorigenesis, it is possible that CTLA-4 may elevate the threshold of activation of T-cells as it inhibits T cell activation and proliferation. Furthermore, the CTLA-4 competitive binding to B7.1 inhibits IL-2 production and proliferation, both of which are essential in down-regulating T cell activity; in turn, this reduces anti-tumor responses and increases cancer susceptibility (5). Several SNPs in the CTLA-4 gene have been widely reported in tumors and non-tumors, such as rs4553808A/G, rs3087243G/A, rs5742909C/T, rs231726A/a, rs17268364, and rs231775A/G (9–13). The Rs231775 (+49) A/G polymorphism is one of the common SNPs in the CTLA-4 gene (4) and has been extensively reported in many types of cancers. Pavkovic et al. first reported a functional SNP in the CTLA-4 gene (rs231775), indicating that the G-allele frequency was highest among chronic lymphocytic leukemia patients who had developed autoimmune hemolytic anemia (14). Since then, the associations among rs231775 polymorphism and other types of cancer have been reported. In addition, Gouda et al. reported that the genotype (GG) was associated with relatively lower CTLA-4 expression levels than the other genotypes (like GC or CC) (11). To evaluate the effects of the functional SNP and cancer susceptibility, we carried out genotyping analyses among rs231775 A/G in 29,464 cases and 35,858 controls. Here, it would be helpful to explain the role of CTLA-4 in immune response control subsequent to completing its function. This is followed by how the polymorphism affects the function as to whether it increases or decreases the affinity of CTLA-4 to its ligand. The variability in the effect of the polymorphism on susceptibility to cancer warrants more in-depth discussions. Finally, we try to find a few potential explanations, which would add value in this regard.
Materials and Methods
Identifying and Evaluating Appropriate Studies
Searches were performed on the Embase, PubMed, Chinese database, Google Scholar, and Web of Science last updated November 20, 2021, using a keyword search that included ‘polymorphism’ or ‘carcinoma’ or ‘CTLA-4’ or ‘cytotoxic T-lymphocyte antigen 4’, or ‘variant’ and ‘cancer’ or ‘tumor’, regardless of language or publication year. These terms led to the retrieval of 592 articles, of which 87 matched the criteria for inclusion. Additionally, we manually searched references of the retrieved or review articles.
Criteria for Inclusion and Exclusion
The following criteria were required to be included in the review: (a) measured cancer risk in relation to CTLA-4 rs231775 polymorphism; (b) case-control studies; and (c) cases and controls have sufficient genotype numbers. Therefore, we also used the following exclusion criteria: (a) no population was used as control, (b) genotype frequency was not available, and (c) previous publications were duplicated.
Extraction of Data
Using the selection criteria, the data were extracted independently by two authors. The following data were collected: last name of the first author, publication year, ethnicity, country of origin, cancer type, the total number of cases and controls, source of controls, Hardy-Weinberg equilibrium (HWE) of controls, and genotyping methods.
Statistical Analysis
The first step was to stratify the subgroups based on cancer type. When a cancer type was reported in only one study, it is classified under the ‘others’ subgroup. In addition, we classified cancer into five systems: digestive tract cancer, orthopedic tumor, tumor of the urinary system, gynecological tumor, and hematological tumor. The ethnicity of the participants was categorized as Asian, Caucasian, and African using two different modes of classification, wherein the source of the control subgroup was analyzed: hospital-based (HB) and population-based (PB). On the basis of genotype frequencies in cases and controls, we calculated OR (odds ratios) with 95% CI (confidence intervals) of the association between CTLA-4 rs231775 polymorphism and the risk for cancer. The overall OR was analyzed using the Z-test (15). Heterogeneity was assessed using chi-square-based Q-tests. The Q-test showed no evidence of heterogeneity among the studies with a P-value greater than 0.05. We used the random-effects model when significant heterogeneity was detected (16); otherwise, the fixed-effects model was applied (16, 17). Using allelic contrast (G-allele vs. A-allele), homozygote comparison (GG vs. AA), dominant genetic model (GG+GA vs. AA), heterozygote comparison (GA vs. AA), and recessive genetic model (GG vs. GA+AA), we investigated the relationship between CTLA-4 rs231775 genetic variants and cancer risk. The Pearson chi-square test was used to calculate HWE in controls at P< 0.05. To estimate the likelihood of publication bias, Egger’s regression test and Begg’s funnel plots were used (18). All statistical assessments for this meta-analysis were conducted using Stata software V 11.0 (StataCorp LP, College Station, TX). We calculated the power and sample size of our meta-analysis using PS: Power and Sample Size Calculation (http://www.powerandsamplesize.com/) (19).
Meta-Regression
The source of publication bias was defined based on a random-effect meta-regression analysis using the publication bias, with publication year as subgroups, ethnicity, source of control, and methods of genotype set as independent variables and the log values regarded as dependent variables (20).
Bioinformatics Analysis
The expression of CTLA-4 between most types of tumors and para-cancerous tissue is shown from the GEPIA website (http://gepia.cancer-pku.cn/). On the same above-mentioned website, you can also find data about CTLA-4 expression levels in each tumor, which includes overall survival and disease-free survival.
Results
Meta-Analysis Study Selection and Characteristics
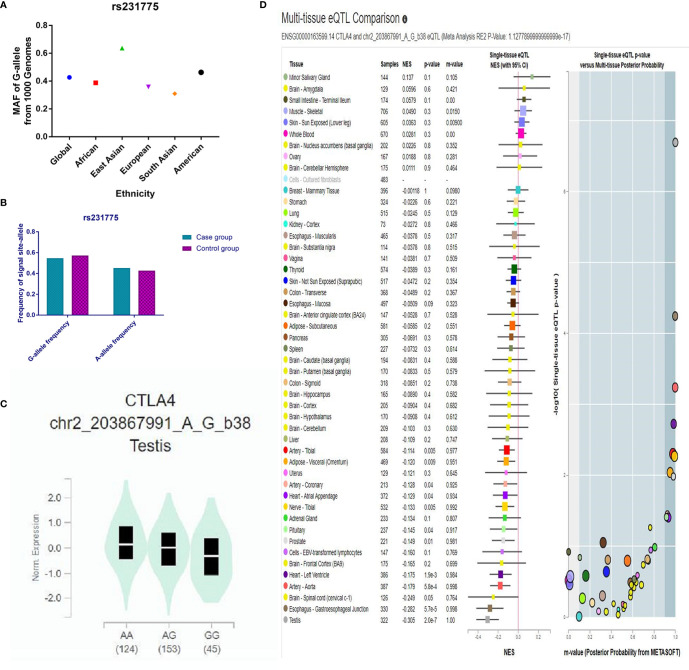
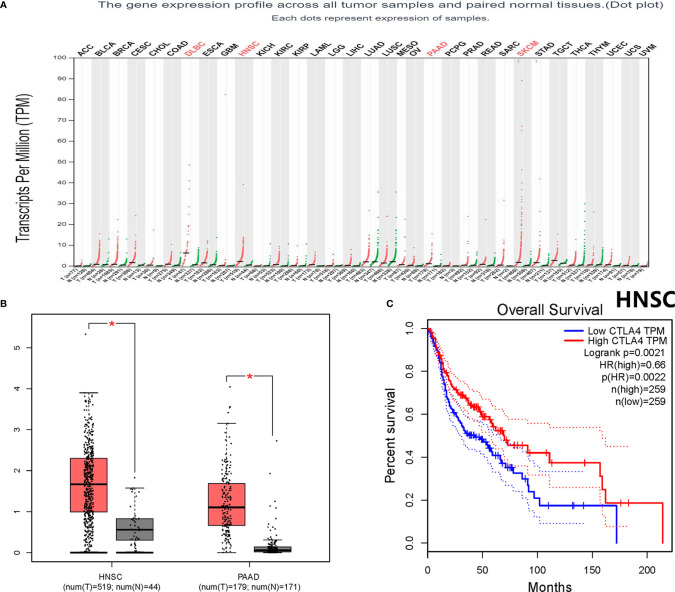
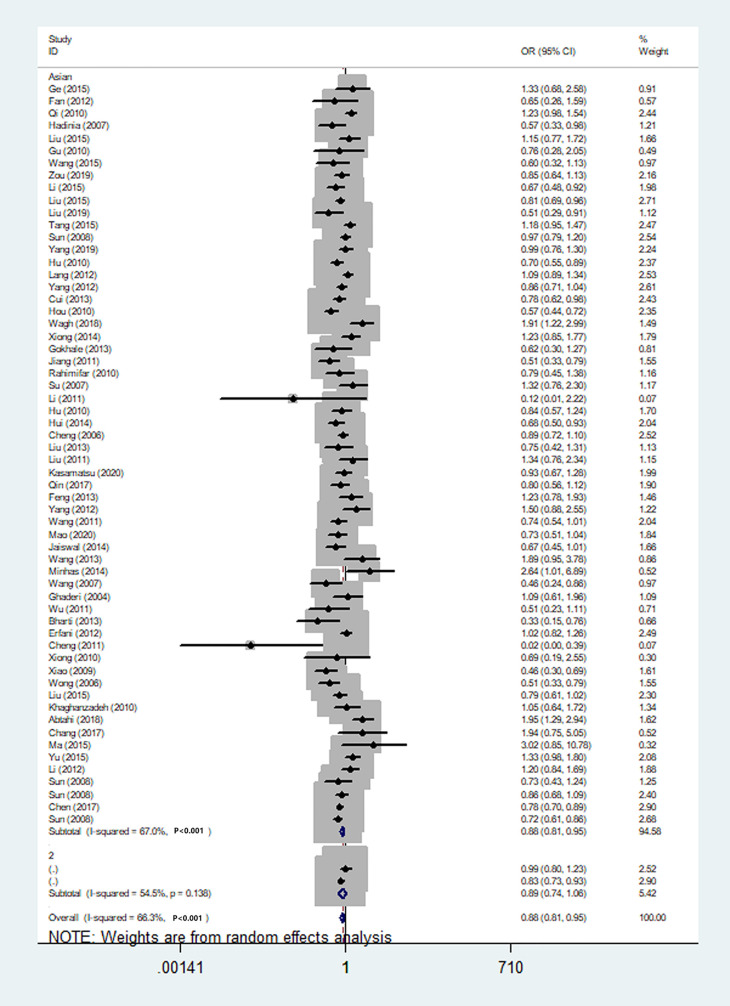
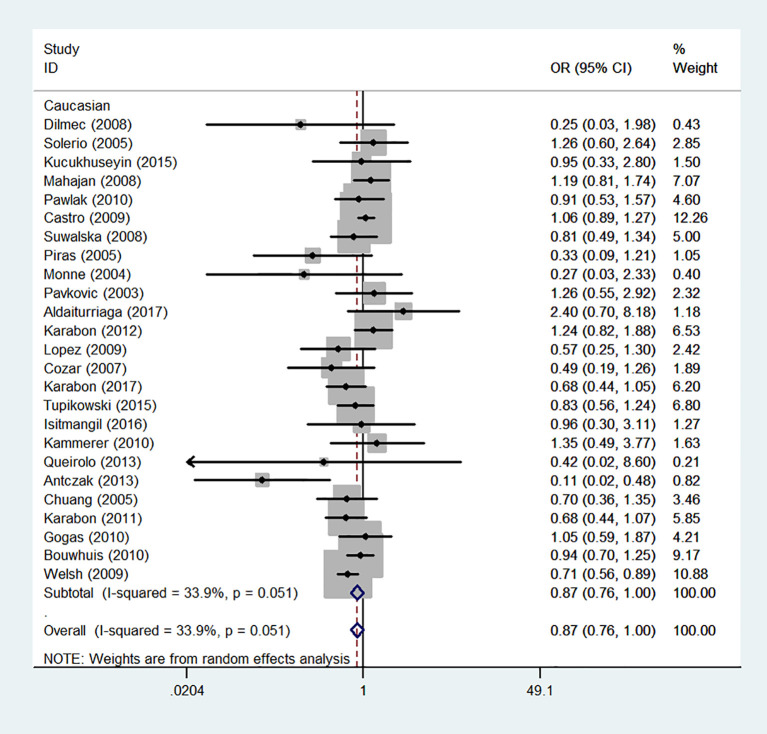
Throughout different databases, 592 articles were identified, and after a meticulous review, we included 87 varying case-control studies for this study (Figure 1). All essential information about included studies is shown in Table 1. Table 1 provides information on the first author, ethnicity, year of publication, cancer type, the numbers of controls and cases, genotyping methods and HWE, and control sources. According to the whole cancer susceptibility search criteria associated with the CTLA-4 rs231775 polymorphism, 87 case-control studies with 35,858 controls and 29,464 cases were retrieved. The controls mainly consisted of healthy populations. Therefore, we have compiled 25 Caucasian, 60 Asian, and 2 African case-control studies for our analyses. The controls in 53 studies came from the source of HB and 34 of PB. We examined the MAF (minor allele frequency) reported for the six major populations globally in the 1000 Genomes Browser (https://www.ncbi.nlm.nih.gov/snp/rs231775) (Figure 2A). Moreover, Asians exhibited significantly higher G-allele frequencies than Caucasian individuals both in cases (59.63% vs. 38.19%, P < 0.001) and controls (62.18% vs. 40.36%, P < 0.001) (Figure 2B). Third, we used the TCGA (The Cancer Genome Atlas) database to search for trends in the frequency of rs231775 polymorphism; our results indicated that the frequency of AA was relatively high compared to other genotypes, as shown in Figure 2C. The polymorphism is associated with prostate, artery, adipose-visceral, heart, nerve, pituitary, testis, and esophagus cancer (https://www.gtexportal.org/home/) (Figure 2D). All the controls except for eight studies were genotyped according to HWE. There is significantly more expression of CTLA-4 in tumor tissues than in normal tissue from four kinds of tumors (melanoma of the skin, head and neck squamous cell carcinoma, lymphoid neoplasm diffuse large B-cell lymphoma, pancreatic adenocarcinoma, P< 0.05, Figures 3A, B). Furthermore, CTLA-4 high expression contributes to a poor overall survival rate in patients with head and neck squamous cell carcinoma (P< 0.01) (Figure 3C).
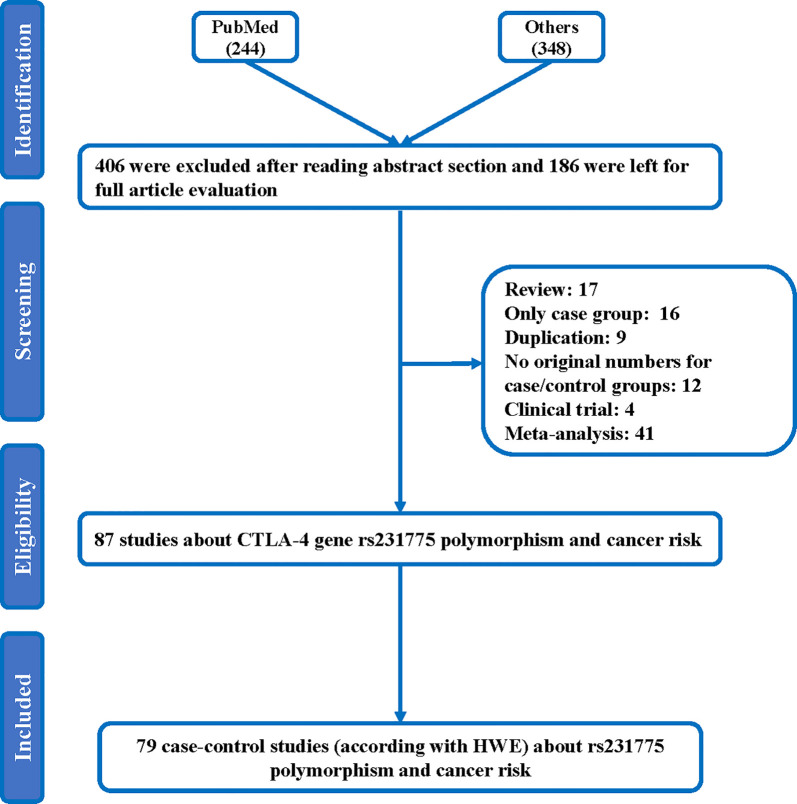
Figure 1.
Flowchart illustrating the search strategy used to identify association studies for CTLA-4 rs231775 polymorphism and the total cancer risk.
Table 1.
Characteristics of studies of the CTLA-4 gene rs231775 A/G polymorphism and cancer risk included in our meta-analysis.
| First author | Year | Origin | Cancer type (1) | Cancer type (2) | Ethnicity | Source | Case | Control | HWE | Method |
|---|---|---|---|---|---|---|---|---|---|---|
| Ge et al. (21) | 2015 | China | Colorectal | Digestive tract cancer | Asian | HB | 572 | 626 | 0.095 | PCR-RFLP |
| Fan et al. (22) | 2012 | China | Colorectal | Digestive tract cancer | Asian | HB | 291 | 352 | 0.059 | PCR-RFLP |
| Qi et al. (23) | 2010 | China | Colorectal | Digestive tract cancer | Asian | HB | 124 | 407 | 0.902 | PCR-LDR |
| Hadinia et al. (24) | 2007 | Iran | Colorectal | Digestive tract cancer | Asian | HB | 105 | 190 | 0.097 | PCR-RFLP |
| Liu et al. (25) | 2015 | China | Liver | Digestive tract cancer | Asian | HB | 80 | 78 | 0.966 | PCR-RFLP |
| Gu et al. (26) | 2010 | China | Liver | Digestive tract cancer | Asian | HB | 367 | 407 | 0.902 | PCR-LDR |
| Wang et al. (27) | 2015 | China | Colorectal | Digestive tract cancer | Asian | HB | 311 | 289 | 0.001 | TaqMan |
| Dilmec et al. (28) | 2008 | Turkey | Colorectal | Digestive tract cancer | Caucasian | HB | 56 | 162 | 0.058 | PCR-RFLP |
| Solerio et al. (29) | 2005 | Italy | Colorectal | Digestive tract cancer | Caucasian | HB | 132 | 238 | 0.618 | PCR-RFLP |
| Zou et al. (30) | 2019 | China | Colorectal | Digestive tract cancer | Asian | PB | 979 | 1299 | 0.430 | SNPscan Kit |
| Li et al. (31) | 2015 | China | Colorectal | Digestive tract cancer | Asian | PB | 231 | 325 | 0.057 | PCR-RFLP |
| Liu et al. (32) | 2015 | China | Esophageal | Digestive tract cancer | Asian | PB | 604 | 664 | 0.283 | PCR-LDR |
| Liu et al. (33) | 2019 | China | Gastric | Digestive tract cancer | Asian | PB | 487 | 1470 | 0.926 | SNPscan Kit |
| Tang et al. (34) | 2015 | China | Gastric | Digestive tract cancer | Asian | PB | 330 | 590 | 0.179 | PCR-LDR |
| Sun et al. (35) | 2008 | China | Gastric | Digestive tract cancer | Asian | PB | 530 | 530 | 0.974 | PCR-RFLP |
| Yang et al. (36) | 2019 | China | Liver | Digestive tract cancer | Asian | PB | 575 | 920 | 0.893 | SNPscan Kit |
| Hu et al. (37) | 2010 | China | Liver | Digestive tract cancer | Asian | PB | 853 | 854 | 0.476 | TaqMan |
| Lang et al. (38) | 2012 | China | Pancreatic | Digestive tract cancer | Asian | PB | 602 | 651 | 0.056 | PCR-RFLP |
| Yang et al. (39) | 2012 | China | Pancreatic | Digestive tract cancer | Asian | PB | 368 | 926 | 0.828 | PCR-RFLP |
| Cui et al. (40) | 2013 | China | Colorectal | Digestive tract cancer | Asian | PB | 128 | 205 | <0.001 | PCR-RFLP |
| Hou et al. (41) | 2010 | China | Gastric | Digestive tract cancer | Asian | PB | 205 | 262 | 0.001 | PCR-RFLP |
| Kucukhuseyin et al. (42) | 2015 | Turkey | Colorectal | Digestive tract cancer | Caucasian | PB | 80 | 115 | 0.467 | PCR-RFLP |
| Mahajan et al. (43) | 2008 | Poland | Gastric | Digestive tract cancer | Caucasian | PB | 301 | 411 | 0.393 | TaqMan |
| Wagh et al. (44) | 2018 | Indian | Cervical | Gynecological tumor | Asian | HB | 92 | 57 | 0.405 | PCR-RFLP |
| Xiong et al. (45) | 2014 | China | Cervical | Gynecological tumor | Asian | HB | 365 | 421 | 0.056 | TaqMan |
| Gokhale et al. (46) | 2013 | Indian | Cervical | Gynecological tumor | Asian | HB | 104 | 162 | 0.239 | PCR-RFLP |
| Jiang et al. (47) | 2011 | China | Cervical | Gynecological tumor | Asian | HB | 100 | 110 | 0.473 | PCR-RFLP |
| Rahimifar et al. (48) | 2010 | Iran | Cervical | Gynecological tumor | Asian | HB | 55 | 110 | 0.658 | PCR-RFLP |
| Su et al. (49) | 2007 | China | Cervical | Gynecological tumor | Asian | HB | 139 | 375 | 0.351 | PCR-RFLP |
| Pawlak et al. (50) | 2010 | Poland | Cervical | Gynecological tumor | Caucasian | HB | 141 | 217 | 0.610 | PCR-RFLP |
| Li et al. (51) | 2011 | China | Cervical | Gynecological tumor | Asian | PB | 314 | 320 | 0.339 | PCR-RFLP |
| Hu et al. (37) | 2010 | China | Cervical | Gynecological tumor | Asian | PB | 696 | 709 | 0.483 | TaqMan |
| Castro et al. (52) | 2009 | Sweden | Cervical | Gynecological tumor | Caucasian | PB | 953 | 1715 | 0.118 | Multiplex PCR |
| Khorshied et al. (53) | 2013 | Egypt | Lymphoma | Hematological tumors | African | HB | 181 | 200 | 0.416 | PCR-RFLP |
| Hui et al. (54) | 2014 | China | Leukemia | Hematological tumors | Asian | HB | 86 | 112 | 0.137 | PCR-RFLP |
| Cheng et al. (55) | 2006 | China | Lymphoma | Hematological tumors | Asian | HB | 62 | 250 | 0.323 | PCR-RFLP |
| Suwalska et al. (56) | 2008 | Poland | Leukemia | Hematological tumors | Caucasian | HB | 170 | 224 | 0.524 | SNaPshot |
| Piras et al. (57) | 2005 | Italy | Lymphoma | Hematological tumors | Caucasian | HB | 100 | 128 | 0.199 | PCR-RFLP |
| Monne et al. (58) | 2004 | Italy | Lymphoma | Hematological tumors | Caucasian | HB | 44 | 76 | 0.837 | PCR-RFLP |
| Pavkovic et al. (59) | 2003 | Macedonia | Lymphoma | Hematological tumors | Caucasian | HB | 130 | 100 | 0.533 | PCR-RFLP |
| Liu et al. (60) | 2013 | China | Lymphoma | Hematological tumors | Asian | PB | 291 | 300 | 0.163 | PCR–LDR |
| Liu et al. (61) | 2011 | China | Bone | Orthopedic tumor | Asian | HB | 267 | 282 | 0.053 | PCR-RFLP |
| Kasamatsu et al. (62) | 2020 | Japan | Myeloma | Orthopedic tumor | Asian | HB | 124 | 211 | 0.556 | PCR-RFLP |
| Qin et al. (63) | 2017 | China | Myeloma | Orthopedic tumor | Asian | HB | 86 | 154 | 0.201 | TaqMan |
| Aldaiturriaga et al. (64) | 2017 | Spain | Bone | Orthopedic tumor | Caucasian | HB | 66 | 125 | 0.101 | PCR-RFLP |
| Feng et al. (65) | 2013 | China | Bone | Orthopedic tumor | Asian | PB | 308 | 362 | 0.055 | PCR-RFLP |
| Yang et al. (66) | 2012 | China | Bone | Orthopedic tumor | Asian | PB | 223 | 302 | 0.054 | PCR-RFLP |
| Wang et al. (67) | 2011 | China | Bone | Orthopedic tumor | Asian | PB | 205 | 216 | 0.130 | PCR-RFLP |
| Karabon et al. (68) | 2012 | Poland | Bone | Orthopedic tumor | Caucasian | PB | 199 | 368 | 0.213 | PCR-RFLP |
| Mao et al. (69) | 2020 | China | Bladder | Tumor of urinary tract | Asian | HB | 354 | 434 | 0.812 | PCR-RFLP |
| Jaiswal et al. (70) | 2014 | Indian | Bladder | Tumor of urinary tract | Asian | HB | 212 | 200 | 0.981 | PCR-RFLP |
| Wang et al. (71) | 2013 | China | Bladder | Tumor of urinary tract | Asian | HB | 300 | 300 | 0.005 | PCR-RFLP |
| Lopez et al. (72) | 2009 | Spain | Renal | Tumor of urinary tract | Caucasian | HB | 125 | 176 | 0.766 | TaqMan |
| Cozar et al. (73) | 2007 | Spain | Renal | Tumor of urinary tract | Caucasian | HB | 96 | 176 | 0.766 | PCR-RFLP |
| Karabon et al. (74) | 2017 | Poland | Prostate | Tumor of urinary tract | Caucasian | PB | 301 | 301 | 0.503 | PCR-RFLP |
| Tupikowski et al. (75) | 2015 | Poland | Renal | Tumor of urinary tract | Caucasian | PB | 236 | 505 | 0.607 | TaqMan |
| Babteen et al. (76) | 2020 | Egypt | Breast | African | HB | 93 | 179 | 0.164 | TaqMan | |
| Minhas et al. (77) | 2014 | Indian | Breast | Asian | HB | 250 | 250 | 0.197 | PCR-RFLP | |
| Wang et al. (78) | 2007 | China | Breast | Asian | HB | 117 | 148 | 0.926 | PCR-RFLP | |
| Ghaderi et al. (79) | 2004 | Iran | Breast | Asian | HB | 197 | 151 | 0.716 | PCR-RFLP | |
| Wu et al. (80) | 2011 | China | Glioma | Asian | HB | 653 | 665 | 0.841 | PCR-LDR | |
| Bharti et al. (81) | 2013 | Indian | Head and neck | Asian | HB | 130 | 180 | 0.622 | PCR-RFLP | |
| Erfani et al. (82) | 2012 | Iran | Head and neck | Asian | HB | 80 | 85 | 0.531 | PCR-RFLP | |
| Cheng et al. (83) | 2011 | China | Head and neck | Asian | HB | 205 | 205 | 0.054 | PCR-RFLP | |
| Xiong et al. 45) | 2010 | China | Head and neck | Asian | HB | 365 | 421 | 0.056 | PCR-RFLP | |
| Xiao et al. (84) | 2009 | China | Head and neck | Asian | HB | 457 | 485 | 0.730 | PCR-RFLP | |
| Wong et al. (85) | 2006 | China | Head and neck | Asian | HB | 118 | 147 | 0.314 | PCR-RFLP | |
| Liu et al. (86) | 2015 | China | Lung | Asian | HB | 231 | 250 | 0.059 | PCR-RFLP | |
| Khaghanzadeh et al. (87) | 2010 | Iran | Lung | Asian | HB | 123 | 122 | 0.763 | PCR-RFLP | |
| Abtahi et al. (88) | 2018 | Iran | Thyroid | Asian | HB | 164 | 100 | 0.965 | PCR-RFLP | |
| Chang et al. (89) | 2017 | China | Thyroid | Asian | HB | 324 | 350 | 0.062 | PCR-RFLP | |
| Ma et al. (90) | 2015 | China | Lung | Asian | HB | 528 | 600 | 0.031 | PCR-RFLP | |
| Isitmangil et al. (91) | 2016 | Turkey | Breast | Caucasian | HB | 79 | 76 | 0.402 | PCR-RFLP | |
| Kammerer et al. (92) | 2010 | Germany | Head and neck | Caucasian | HB | 83 | 40 | 0.287 | RT-PCR | |
| Queirolo et al. (93) | 2013 | Italy | Melanoma | Caucasian | HB | 14 | 45 | 0.802 | PCR-RFLP | |
| Antczak et al. (94) | 2013 | Poland | Lung | Caucasian | HB | 71 | 104 | 0.001 | TaqMan | |
| Chuang et al. (95) | 2005 | Germany | Thymoma | Caucasian | HB | 125 | 173 | 0.015 | PCR-RFLP | |
| Yu et al. (96) | 2015 | China | Breast | Asian | PB | 376 | 366 | 0.962 | PCR-RFLP | |
| Li et al. (97) | 2012 | China | Breast | Asian | PB | 576 | 553 | 0.739 | PCR-RFLP | |
| Sun et al. (35) | 2008 | China | Breast | Asian | PB | 2097 | 2140 | 0.053 | PCR-RFLP | |
| Sun et al. (35) | 2008 | China | Head and neck | Asian | PB | 1010 | 1008 | 0.684 | PCR-RFLP | |
| Chen et al. (98) | 2017 | China | Lung | Asian | PB | 520 | 1028 | 0.950 | SNPscan Kit | |
| Sun et al. (35) | 2008 | China | Lung | Asian | PB | 2205 | 2153 | 0.103 | PCR-RFLP | |
| Karabon et al. (99) | 2011 | Poland | Lung | Caucasian | PB | 208 | 324 | 0.089 | PCR-RFLP | |
| Gogas et al. (100) | 2010 | Greece | Melanoma | Caucasian | PB | 286 | 288 | 0.465 | Multiplex PCR | |
| Bouwhuis et al. (101) | 2010 | Germany | Melanoma | Caucasian | PB | 762 | 734 | 0.956 | TaqMan | |
| Welsh et al. (102) | 2009 | USA | Skin | Caucasian | PB | 1581 | 819 | 0.004 | TaqMan | |
HB, hospital-based; PB, population-based; SOC; source of control; PCR-RFLP, polymerase chain reaction followed by restriction fragment length polymorphism; PCR-LDR, polymerase chain reaction by ligase detection reaction; HWE, Hardy-Weinberg equilibrium of the control group.
Figure 2.
(A) The MAF of minor-allele (mutant-allele) for CTLA-4 rs231775 polymorphism from the 1000 Genomes online database. (B) The frequency about G-allele or A-allele both in case and control groups. (C) The distribution of each genotype from online GTEx Portal (https://www.gtexportal.org/home/). (D) The risk frequency of rs231775 polymorphism in several diseases from the TCGA database.
Figure 3.
Bioinformatics analysis about the CTLA-4 gene. (A) The CTLA-4 gene expression profile across all tumor samples and paired normal tissues. (B) CTLA-4 gene expression both in HNSC and PAAD. *P < 0.05. (C) Overall survival analysis for HNSC. HR, hazard ratio; ACC, adrenocortical carcinoma; BLCA, bladder urothelial carcinoma; BRCA, breast invasive carcinoma; CESC, cervical squamous cell carcinoma and endocervical adenocarcinoma; CHOL, cholangiocarcinoma; COAD, colon adenocarcinoma; DLBC, lymphoid neoplasm diffuse large B-cell lymphoma; ESCA, esophageal carcinoma; GBM, glioblastoma multiforme; HNSC, head and neck squamous cell carcinoma; KICH, kidney chromophobe; KIRC, kidney renal clear cell carcinoma; KIRP, kidney renal papillary cell carcinoma; LAML, acute myeloid leukemia; LGG, brain lower grade glioma; LIHC, liver hepatocellular carcinoma; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma; MESO, mesothelioma; OV, ovarian serous cystadenocarcinoma; PAAD, pancreatic adenocarcinoma; PCPG, pheochromocytoma and paraganglioma; PRAD, prostate adenocarcinoma; READ, rectum adenocarcinoma; SARC, sarcoma; SKCM, skin cutaneous melanoma; STAD, stomach adenocarcinoma; TGCT, testicular germ cell tumors; THCA, thyroid carcinoma; THYM, thymoma; UCEC, uterine corpus endometrial carcinoma; UCS, uterine carcinosarcoma; UVM, uveal melanoma.
Meta-Analysis
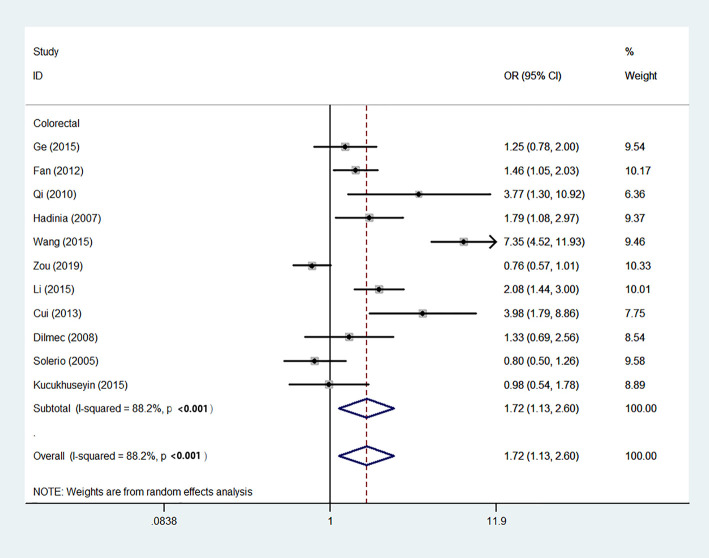
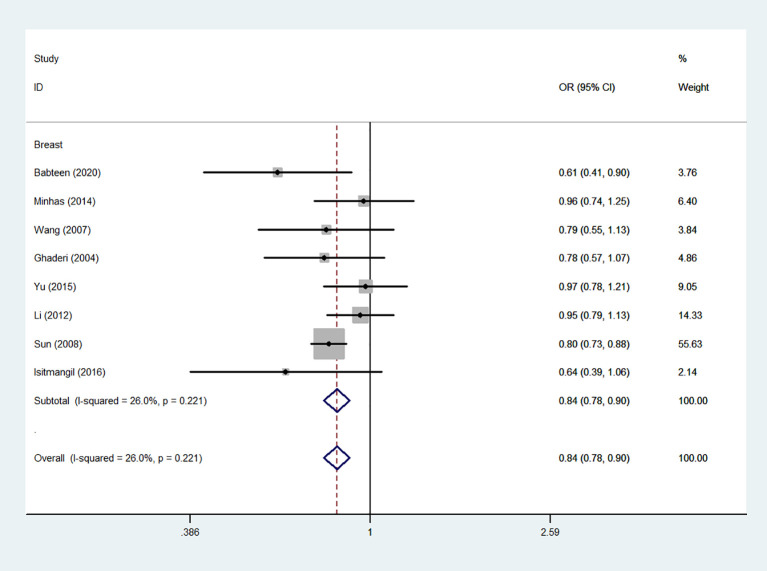
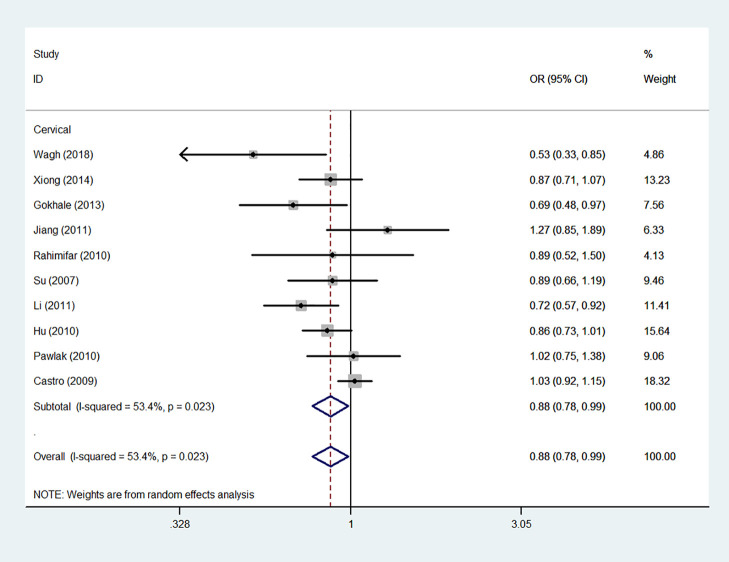
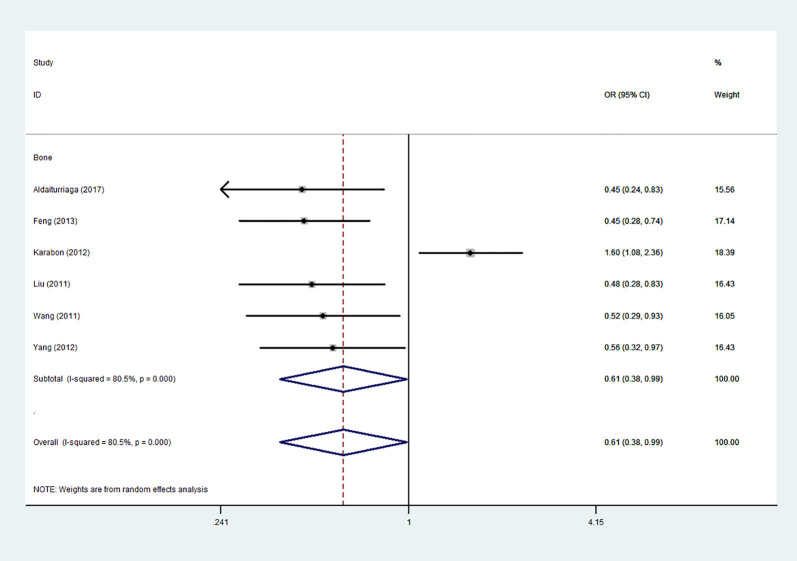
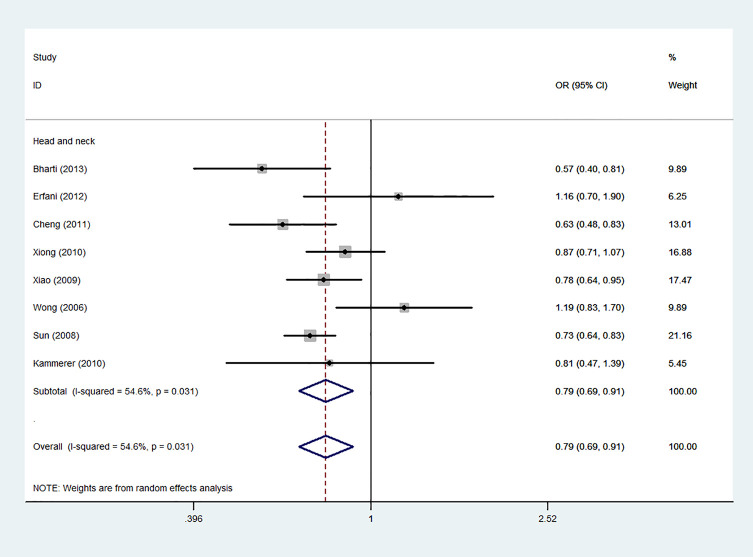
Using 29,464 cases and 35,858 controls, the overall risk of CTLA-4 rs231775 is summarized in Table 2. CTLA-4 rs231775 polymorphism appears to decrease cancer risk in overall genetic models (G-allele vs. A-allele, OR = 0.94, 95%CI = 0.90-1.00, Pheterogeneity< 0.001, P = 0.037; GG vs. AA, OR = 0.86, 95%CI = 0.76-0.96, Pheterogeneity< 0.001, P = 0.010; GG vs. GA+AA, OR = 0.88, 95%CI = 0.82-0.94, Pheterogeneity< 0.001, P < 0.001). There were significant associations between CTLA-4 polymorphisms and two types of cancers (colorectal cancer: GA vs. AA, OR = 1.72, 95%CI = 1.13-2.60, Pheterogeneity< 0.001, P = 0.011; GG+GA vs. AA, OR = 1.52, 95%CI = 1.08-2.15, Pheterogeneity< 0.001, P = 0.017, Figure 4; thyroid cancer: G-allele vs. A-allele, OR = 1.50, 95%CI = 1.22-1.85, Pheterogeneity= 0.134, P< 0.001). On the other hand, significantly decreased associations were detected in six kinds of cancer (breast cancer: G-allele vs. A-allele, OR = 0.84, 95%CI = 0.78-0.90, Pheterogeneity= 0.221, P< 0.001, Figure 5; liver cancer: G-allele vs. A-allele, OR = 0.89, 95%CI = 0.82-0.98, Pheterogeneity= 0.151, P = 0.018; cervical cancer: G-allele vs. A-allele, OR = 0.88, 95%CI = 0.78-0.99, Pheterogeneity= 0.023, P = 0.028, Figure 6; bone cancer: GG+GA vs. AA, OR = 0.61, 95%CI = 0.38-0.99, Pheterogeneity< 0.001, P = 0.044, Figure 7; head and neck: G-allele vs. A-allele, OR = 0.79, 95%CI = 0.69-0.91, Pheterogeneity= 0.031, P =0.001, Figure 8; pancreatic cancer: G-allele vs. A-allele, OR = 0.72, 95%CI = 0.57-0.91, Pheterogeneity= 0.049, P = 0.006).
Table 2.
Stratified analysis of CTLA-4rs231775 A/G variation on cancer susceptibility.
| Variables | N | Case/ | G-allele vs. A-allele | GA vs. AA | GG vs. AA | GG+GA vs. AA | GG vs. GA+AA | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs231775 A/G | Control | OR (95%CI) | Ph | P | OR (95%CI) | Ph | P | OR (95%CI) | Ph | P | OR (95%CI) | Ph | P | OR (95%CI) | Ph | P | ||
| Total | 87 | 29464/35858 | 0.94 (0.90-1.00) | <0.001 | 0.037 | 1.01 (0.92-1.12) | <0.001 | 0.773 | 0.86 (0.76-0.96) | <0.001 | 0.010 | 0.96 (0.87-1.05) | <0.001 | 0.353 | 0.88 (0.82-0.94) | <0.001 | <0.001 | |
| HWE | 79 | 26215/33106 | 0.93 (0.89-0.98) | ≤0.001 | 0.011 | 0.97 (0.88-1.06) | ≤0.001 | 0.480 | 0.83 (0.74-0.93) | ≤0.001 | 0.001 | 0.92 (0.84-1.01) | ≤0.001 | 0.091 | 0.88 (0.82-0.94) | ≤0.001 | ≤0.001 | |
| Cancer Type (1) | ||||||||||||||||||
| Myeloma | 2 | 210/365 | 1.17 (0.91-1.51) | 0.896 | 0.209 | 0.91 (0.53-1.56) | 0.138 | 0.737 | 1.22 (0.71-2.11) | 0.420 | 0.478 | 1.05 (0.63-1.75) | 0.232 | 0.858 | 1.33 (0.94-1.89) | 0.578 | 0.104 | |
| Bladder cancer | 3 | 866/934 | 1.19 (0.73-1.95) | <0.001 | 0.481 | 1.24 (1.01-1.51) | 0.086 | 0.040 | 1.38 (0.41-4.64) | <0.001 | 0.603 | 1.24 (0.79-1.97) | 0.004 | 0.353 | 1.27 (0.42-3.820 | 0.002 | 0.668 | |
| Breast cancer | 8 | 3785/3863 | 0.84 (0.78-0.90) | 0.221 | <0.001 | 0.86 (0.69-1.07) | 0.021 | 0.169 | 0.67 (0.57-0.80) | 0.134 | <0.001 | 0.81 (0.58-1.37) | 0.022 | 0.042 | 0.79 (0.71-0.87) | 0.370 | <0.001 | |
| Colorectal cancer | 11 | 3009/4208 | 1.15 (0.98-1.35) | <0.001 | 0.094 | 1.72 (1.13-2.61) | <0.001 | 0.011 | 1.24 (0.81-1.90) | <0.001 | 0.319 | 1.52 (1.08-2.15) | <0.001 | 0.017 | 0.91 (0.71-1.16) | <0.001 | 0.440 | |
| Liver cancer | 4 | 1875/2259 | 0.89 (0.82-0.98) | 0.151 | 0.018 | 0.76 (0.62-0.94) | 0.870 | 0.010 | 0.74 (0.60-0.90) | 0.360 | 0.003 | 0.75 (0.61-0.91) | 0.618 | 0.004 | 0.92 (0.81-1.04) | 0.164 | 0.187 | |
| Gastric cancer | 5 | 1853/3263 | 1.07 (0.85-1.35) | <0.001 | 0.552 | 1.33 (0.87-2.01) | 0.001 | 0.186 | 1.15 (0.75-1.80) | 0.001 | 0.513 | 1.23 (0.81-1.87) | <0.001 | 0.094 | 0.94 (0.83-1.06) | 0.052 | 0.325 | |
| Cervical cancer | 10 | 2959/4196 | 0.88 (0.78-0.99) | 0.023 | 0.028 | 0.88 (0.70-1.10) | 0.013 | 0.257 | 0.70 (0.52-0.94) | 0.006 | 0.017 | 0.83 (0.66-1.03) | 0.008 | 0.094 | 0.83 (0.70-0.99) | 0.039 | 0.043 | |
| Thyroid cancer | 2 | 488/450 | 1.50 (1.22-1.85) | 0.134 | <0.001 | 1.96 (1.34-2.87) | 0.812 | 0.001 | 2.42 (1.48-3.95) | 0.400 | <0.001 | 2.13 (1.48-3.07) | 0.805 | <0.001 | 1.40 (1.05-1.88) | 0.217 | 0.024 | |
| Other cancers | 5 | 3264/2622 | 0.94 (0.87-1.01) | 0.065 | 0.094 | 1.00 (0.78-1.29) | 0.030 | 0.991 | 0.79 (0.7-0.93) | 0.109 | 0.005 | 0.92 (0.81-1.04) | 0.063 | 0.179 | 0.88 (0.69-1.11) | 0.011 | 0.279 | |
| Lung cancer | 7 | 3886/4581 | 0.95 (0.73-1.24) | <0.001 | 0.724 | 0.98 (0.69-1.40) | <0.001 | 0.927 | 0.97 (0.57-1.65) | <0.001 | 0.901 | 0.94 (0.62-1.43) | <0.001 | 0.774 | 1.01 (0.75-1.35) | <0.001 | 0.968 | |
| Bone cancer | 6 | 1268/1655 | 0.82 (0.63-1.05) | 0.004 | 0.051 | 0.63 (0.40-1.00) | 0.001 | 0.051 | 0.64 (0.38-1.09) | 0.001 | 0.102 | 0.61 (0.38-0.99) | <0.001 | 0.044 | 0.81 (0.69-0.95) | 0.125 | 0.011 | |
| Renal cancer | 3 | 457/857 | 0.85 (0.72-1.00) | 0.143 | 0.056 | 0.92 (0.71-1.17) | 0.125 | 0.485 | 0.71 (0.49-1.03) | 0.272 | 0.069 | 0.85 (0.67-1.08) | 0.109 | 0.185 | 0.73 (0.52-1.02) | 0.485 | 0.062 | |
| Leukemia | 2 | 256/336 | 0.91 (0.72-1.15) | 0.987 | 0.432 | 1.10 (0.74-1.66) | 0.362 | 0.634 | 0.88 (0.54-1.43) | 0.592 | 0.607 | 1.01 (0.69-1.48) | 0.499 | 0.966 | 0.78 (0.53-1.14) | 0.84 | 0.197 | |
| Head and neck | 8 | 2448/2571 | 0.79 (0.69-0.91) | 0.031 | 0.001 | 0.92 (0.68-1.24) | 0.004 | 0.577 | 0.60 (0.43-0.84) | 0.034 | 0.003 | 0.80 (0.60-1.06) | 0.004 | 0.123 | 0.69 (0.53-0.88) | 0.017 | 0.003 | |
| Lymphoma | 6 | 808/1054 | 0.91 (0.63-1.33) | <0.001 | 0.625 | 0.99 (0.55-1.77) | <0.001 | 0.974 | 1.12 (0.60-2.08) | 0.040 | 0.726 | 0.96 (0.53-1.76) | <0.001 | 0.899 | 1.00 (0.79-1.27) | 0.264 | 0.985 | |
| Melanoma | 3 | 1062/1067 | 1.04 (0.92-1.19) | 0.486 | 0.504 | 1.14 (0.95-1.37) | 0.306 | 0.165 | 1.00 (0.76-1.33) | 0.767 | 0.983 | 1.11 (0.93-1.32) | 0.349 | 0.233 | 0.95 (0.73-1.23) | 0.814 | 0.706 | |
| Pancreatic cancer | 2 | 970/1577 | 0.72 (0.57-0.91) | 0.049 | 0.006 | 0.70 (0.53-0.92) | 0.766 | 0.009 | 0.51 (0.38-0.67) | 0.173 | <0.001 | 0.60 (0.46-1.00) | 0.347 | <0.001 | 0.67 (0.57-0.79) | 0.063 | <0.001 | |
| Cancer Type (2) | ||||||||||||||||||
| Orthopedic tumor | 8 | 1478/2020 | 0.88 (0.73-1.06) | 0.001 | 0.192 | 0.68 (0.46-0.99) | 0.001 | 0.048 | 0.74 (0.47-1.16) | ≤0.001 | 0.192 | 0.87 (0.62-1.21) | 0.006 | 0.408 | 0.94 (0.75-1.17) | 0.032 | 0.562 | |
| Tumor of urinary tract | 7 | 1624/2002 | 0.96 (0.76-1.22) | ≤0.001 | 0.755 | 1.06 (0.86-1.32) | ≤0.001 | 0.553 | 0.86 (0.53-1.39) | 0.002 | 0.540 | 0.55 (0.42-0.71) | ≤0.001 | ≤0.001 | 0.84 (0.56-1.26) | 0.009 | 0.398 | |
| Digestive tract cancer | 23 | 8311//11971 | 1.02 (0.92-1.13) | ≤0.001 | 0.692 | 1.25 (0.99-1.59) | ≤0.001 | 0.061 | 0.99 (0.79-1.25) | ≤0.001 | 0.952 | 1.32 (1.04-1.67) | ≤0.001 | 0.022 | 0.91 (0.80-1.02) | ≤0.001 | 0.098 | |
| Gynecological tumor | 10 | 2959/4196 | 0.87 (0.78-0.99) | 0.023 | 0.028 | 0.87 (0.69-1.10) | 0.013 | 0.257 | 0.70 (0.52-0.94) | 0.006 | 0.017 | 0.92 (0.74-1.14) | 0.014 | 0.427 | 0.83 (0.69-0.99) | 0.039 | 0.043 | |
| Hematological tumors | 8 | 1064/1390 | 0.93 (0.71-1.21)≤ | 0.001 | 0.577 | 1.04 (0.68-1.59) | 0.001 | 0.839 | 1.07 (0.69-1.65) | 0.069 | 0.755 | 0.82 (0.43-1.57) | ≤0.001 | 0.556 | 0.93 (0.76-1.14) | 0.349 | 0.480 | |
| Ethnicity | ||||||||||||||||||
| Asian | 60 | 22851/27839 | 0.96 (0.90-1.02) | <0.001 | 0.187 | 1.06 (0.93-1.20) | <0.001 | 0.368 | 0.86 (0.75-1.00) | <0.001 | 0.053 | 0.99 (0.88-1.12) | <0.001 | 0.903 | 0.87 (0.81-0.95) | <0.001 | 0.001 | |
| African | 2 | 274/379 | 0.95 (0.41-2.19) | 0.001 | 0.900 | 0.93 (0.28-3.10) | <0.001 | 0.910 | 0.92 (0.25-3.36) | 0.027 | 0.904 | 0.93 (0.27-3.13) | <0.001 | 0.902 | 1.02 (0.60-1.72) | 0.208 | 0.949 | |
| Caucasian | 25 | 6339/7640 | 0.90 (0.81-0.99) | <0.001 | 0.037 | 0.95 (0.83-1.09) | <0.001 | 0.447 | 0.88 (0.74-1.04) | 0.013 | 0.128 | 0.90 (0.78-1.04) | <0.001 | 0.143 | 0.89 (0.81-0.97) | 0.051 | 0.010 | |
| Source of control | ||||||||||||||||||
| HB | 53 | 9844/12125 | 0.94 (0.86-1.03) | <0.001 | 0.196 | 1.03 (0.89-1.19) | <0.001 | 0.684 | 0.88 (0.73-1.06) | <0.001 | 0.185 | 0.97 (0.84-1.12) | <0.001 | 0.705 | 0.88 (0.77-1.00) | <0.001 | 0.046 | |
| PB | 34 | 19620/23733 | 0.93 (0.87-1.00) | <0.001 | 0.036 | 0.98 (0.87-1.11) | <0.001 | 0.761 | 0.82 (0.71-0.95) | <0.001 | 0.007 | 0.93 (0.82-1.05) | <0.001 | 0.241 | 0.86 (0.81-0.93) | <0.001 | <0.001 | |
Ph: the value of Q-test for the heterogeneity test; P: Z-test for the statistical significance of the OR.
Figure 4.
Forest plot of the association between the CTLA-4 gene rs231775 polymorphism and colorectal cancer risk (G-allele vs. A-allele).
Figure 5.
Forest plot of the association between the CTLA-4 gene rs231775 polymorphism and breast cancer risk (G-allele vs. A-allele).
Figure 6.
Forest plot of the association between the CTLA-4 gene rs231775 polymorphism and cervical cancer risk (G-allele vs. A-allele).
Figure 7.
Forest plot of the association between the CTLA-4 gene rs231775 polymorphism and bone cancer risk (G-allele vs. A-allele).
Figure 8.
Forest plot of the association between the CTLA-4 gene rs231775 polymorphism and head and neck cancer risk (G-allele vs. A-allele).
We also classified tumors into five systems and observed a significant association between the polymorphism and digestive tract cancer (GG+GA vs. AA, OR = 1.32, 95%CI = 1.04-1.67, Pheterogeneity< 0.001, P = 0.022), however, decreased associations were observed in three kinds of systems (orthopedic tumor: GG vs. AA: OR = 0.68, 95%CI = 0.46-0.99, Pheterogeneity= 0.001, P = 0.048; urinary tract tumor: GG+GA vs. AA, OR = 0.55, 95%CI = 0.42-0.71, Pheterogeneity< 0.001, P < 0.001; gynecological tumor: G-allele vs. A-allele, OR = 0.87, 95%CI = 0.78-0.99, Pheterogeneity= 0.023, P = 0.028). In spite of variations in the frequency of occurrence of this sequence variant among ethnic groups, decreased cancer risk in both Asian (GG vs. GA+ AA, OR = 0.87, 95%CI = 0.81-0.95, Pheterogeneity< 0.001, P=0.001, Figure 9) and Caucasian (GG vs. GA+ AA, OR = 0.89, 95%CI = 0.81-0.97, Pheterogeneity= 0.051, P=0.010, Figure 10) populations was observed. On the basis of stratification by source of control, we evaluated an OR for the rs231775 polymorphism of CTLA-4, and found a decreased association in a recessive genetic model (HB: OR = 0.88, 95%CI = 0.77-1.00, Pheterogeneity< 0.001, P = 0.046; PB: OR = 0.86, 95%CI = 0.81-0.93, Pheterogeneity< 0.001, P <0.001) (Table 2).
Figure 9.
Forest plot of cancer risk associated with the CTLA-4 gene rs231775 polymorphism in Asians (G-allele vs. A-allele model).
Figure 10.
Forest plot of cancer risk associated with the CTLA-4 gene rs231775 polymorphism in Caucasians (G-allele vs. A-allele model).
Meta-Regression
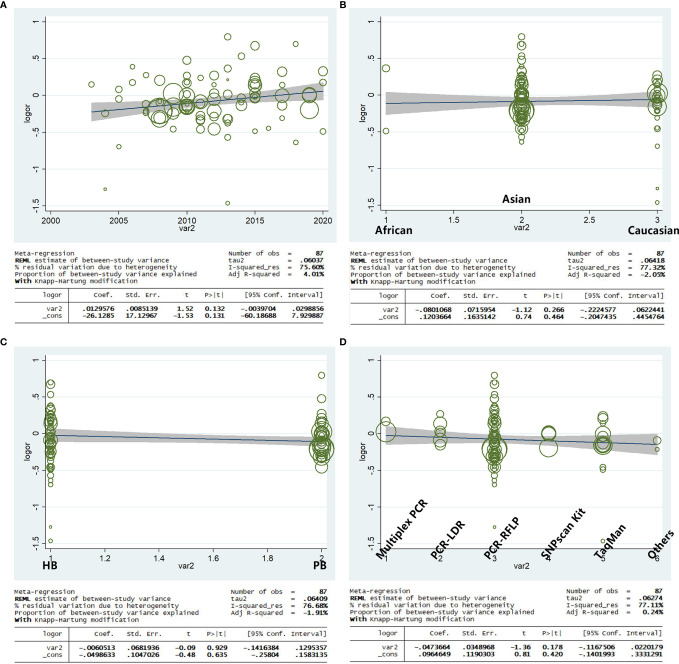
Based on the year of publication, ethnicity, genotype methods, and source of control, a meta-regression analysis indicated that there was a significant association for the allele model (A-allele vs. G-allele) with a regression coefficient of 0.131, 0.464, 0.635, and 0.420, respectively, this suggests that the heterogeneity from the rs231775 polymorphism in cancer could not result from the year of publication, ethnicity, source of control, or genotype methods subgroups (Figures 11A–D) if the heterogeneity was found in the current study.
Figure 11.
Random-effect meta-regression of log odds ratio vs. publication year (A), regular ethnicity (B), source of control (C), and genotype methods (D), respectively.
Discussion
Nearly 9 million people die of cancer each year worldwide (103). In the challenge of cancer treatment, immunotherapy has attracted remarkable interest among scientists because of its ability to kill tumor cells directly (14, 104). The Treg cell population expresses a number of immune-modulatory receptors, including CTLA-4, programmed cell death protein 1, and the vascular endothelial growth factor receptor (105). Activated T and Treg cells (106) express CTLA-4. Atkins et al. demonstrated improvement in the rate of survival of non-small cell lung cancer, renal cell carcinoma, melanoma, and head and neck squamous cell cancer by blocking the CTLA-4 immune checkpoint, which showed that the CTLA-4 gene is a promising target gene in the future treatment for cancer (107).
Previously, several meta-analyses were focused on the CTLA-4 polymorphisms, which showed the vital role of CTLA-4 in the susceptibility to many diseases, such as cancer. It was documented that the immune related gene CTLA-4 rs5742909 polymorphism had a significantly increased association with cervical carcinogenesis. Dai et al. found the CTLA-4 rs3087243 polymorphism may reduce breast cancer risk, however, rs4553808 may increase breast cancer risk in different ethnicity or genetic models (108, 109). Another polymorphism rs231775 is the most common SNP that has been reported in many tumors, however, a clear conclusion has not been gained yet despite few meta-analyses (110, 111).
Based on 87 case-control studies, we carried out a meta-analysis, which showed CTLA-4 rs231775 polymorphism plays an important role in cancer risks. According to the results, CTLA-4 rs231775 is strongly associated with the maximum cancer risk. Second, both Asian and Caucasian populations were significantly less likely to develop cancer when individuals carry the rs231775 G-allele. Last, individuals with the rs231775G allele may be at a lower risk for cancer in both HB and PB studies. The results of these studies recommend that the rs231775 polymorphism may contribute to cancer development. Next, based on the stratified cancer type analysis, CTLA-4 rs231775 polymorphism was found to be a risk factor for thyroid cancer and colorectal cancer; that is, in individuals carrying the G-allele, the risk of being diagnosed with cancer is increased; on the other hand, it proved to be a protective factor for liver cancer, breast cancer, cervical cancer, head and neck cancer, bone cancer, and pancreatic cancer, in other words, individuals carrying G-allele may have a lower risk of being diagnosed with cancer. However, no association was detected between this SNP and myeloma, bladder cancer, gastric cancer, lung cancer, renal cancer, leukemia, lymphoma, or melanoma. Some of the reasons why the same gene polymorphism plays different roles in different cancer types may be the difference in the pathogenesis of each kind of cancer, and the same gene and its polymorphism may have different functions and susceptibility.
Gene polymorphisms have the important property of their incidence varying widely across different ethnic populations or races. Based on the subgroup analysis by ethnicity, CTLA-4 rs231775 polymorphism was observed to be significantly associated with lower cancer risks in Asians and Caucasians, but not Africans, suggesting genetic diversity across ethnic groups. This difference can be explained by two factors: genetic and environmental differences among different ethnic groups, and linkage disequilibrium patterns between different populations. Polymorphisms may be related to the presence of closer causal variants in varying populations.
The meta-analysis we performed has certain limitations. To begin with, interactions between gene-environment, gene-gene, or different polymorphic loci of the same gene can modulate the risk for cancer, so researchers should investigate these factors in the future. Moreover, other covariates such as age, sex, family history, environmental factors, cancer stage, and lifestyle should be considered. Furthermore, the control group did not comprise strictly healthy controls. Even so, the meta-analysis we conducted has two advantages. First, data from numerous studies were pooled, significantly increasing the power of the analysis. Second, our selection criteria led to a satisfactory quality of case-control studies that are included in the current meta-analysis. Finally, the strength of the current study as per the software is ‘1’, which indicates the conclusions from our study are convincing and clear.
Conclusion
The meta-analysis in the current study suggests a significant association between CTLA-4 rs231775 polymorphism and some types of cancer and overall risk for cancer. Consequently, more large-scale studies, which are well-designed, are needed, with a focus on gene-environment and gene-gene interactions. Future research should provide a more comprehensive clarity of the association between the CTLA-4 rs231775 polymorphism and the risk of developing cancer.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding authors.
Ethics Statement
Ethical review and approval was not required for this animal study, in accordance with the local legislation and institutional requirements.
Author Contributions
HW, YF, and HZ were major contributors in writing the manuscript. HW and YF created all the figures. HZ performed the literature search. LZ, YC and YM made substantial contributions to the design of the manuscript and revised it critically for important intellectual content. All authors have read and approved the final version of this manuscript.
Funding
This work was supported by National Natural Science Foundation (No. 81802576), Wuxi Commission of Health and Family Planning (No. T202024, J202012, Z202011), the Science and Technology Development Fund of Wuxi (No. WX18IIAN024, N20202021), and Jiangnan University Wuxi School of Medicine (No. 1286010242190070) and Wuxi “Taihu Talent Program”-High-end Talent in Medical and Healthentalent plan of Taihu Lake in Wuxi (Double Hundred Medical Youth Professionals Program) from Health Committee of Wuxi (No. BJ2020061).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors are thankful for the guidance of Professor YC.
References
- 1. Bray F, Laversame M, Weiderpass E. The Ever-Increasing Importance of Cancer as a Leading Cause of Premature Death Worldwide. Cancer (2021) 127(16):3029–30. doi: 10.1002/cncr.33587 [DOI] [PubMed] [Google Scholar]
- 2. (WHO)., W.H.O . Global Health Estimates 2020: Deaths by Cause, Age, Sex, by Country and by Region 2000-2019 (2020). WHO; (Accessed December 11, 2020). [Google Scholar]
- 3. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin (2021) 71(3):209–49. doi: 10.3322/caac.21660 [DOI] [PubMed] [Google Scholar]
- 4. Sud A, Kinnersley B, Houlston RS. Genome-Wide Association Studies of Cancer: Current Insights and Future Perspectives. Nat Rev Cancer (2017) 17(11):692–704. doi: 10.1038/nrc.2017.82 [DOI] [PubMed] [Google Scholar]
- 5. Visscher PM, Wray NR, Zhang Q, Sklar P, McCarthy MI, Brown MA, et al. 10 Years of GWAS Discovery: Biology, Function, and Translation. Am J Hum Genet (2017) 101(1):5–22. doi: 10.1016/j.ajhg.2017.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gersten O, Wilmoth JR. The Cancer Transition in Japan Since 1951. Demogr Res (2002) 7:271–306. doi: 10.4054/DemRes.2002.7.5 [DOI] [Google Scholar]
- 7. Omran AR. The Epidemiologic Transition. A Theory of the Epidemiology of Population Change. 1971. Bull World Health Organ (2001) 79(2):161–70. [PMC free article] [PubMed] [Google Scholar]
- 8. Shin JH, Jeong J, Maher SE, Lee HW, Lim J, Bothwell ALM. Colon Cancer Cells Acquire Immune Regulatory Molecules From Tumor-Infiltrating Lymphocytes by Trogocytosis. Proc Natl Acad Sci USA (2021) 118(48):e2110241118. doi: 10.1073/pnas.2110241118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Li J, Wang W, Sun Y, Zhu Y. CTLA-4 Polymorphisms and Predisposition to Digestive System Malignancies: A Meta-Analysis of 31 Published Studies. World J Surg Oncol (2020) 18(1):55. doi: 10.1186/s12957-020-1806-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhuo C, Yi T, Wei C, Wu X, Cen X, Feng S, et al. Association of Cytotoxic T Lymphocyte-Associated Protein 4 Gene -1772T/C Polymorphism With Gastric Cancer Risk: A Prisma-Compliant Meta-Analysis. Medicine (Baltimore) (2020) 99(50):e23542. doi: 10.1097/md.0000000000023542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gouda NS, Fawzy MS, Toraih EA. Impact of Cytotoxic T-Lymphocyte-Associated Protein 4 Codon 17 Variant and Expression on Vitiligo Risk. J Clin Lab Anal (2021) 35(6):e23777. doi: 10.1002/jcla.23777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Qi YY, Zhao XY, Liu XR, Wang YN, Zhai YL, Zhang XX, et al. Lupus Susceptibility Region Containing CTLA4 Rs17268364 Functionally Reduces CTLA4 Expression by Binding EWSR1 and Correlates IFN-α Signature. Arthritis Res Ther (2021) 23(1):279. doi: 10.1186/s13075-021-02664-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhou B, Chen M, Shang S, Zhao J. Association of CTLA-4 Gene Polymorphisms and Alopecia Areata: A Systematic Review and Meta-Analysis. Biomarkers (2022) 1–11. doi: 10.1080/1354750x.2022.2046855 [DOI] [PubMed] [Google Scholar]
- 14. Sobhani N, Tardiel-Cyril DR, Davtyan A, Generali D, Roudi R, Li Y. CTLA-4 in Regulatory T Cells for Cancer Immunotherapy. Cancers (Basel) (2021) 13(6):1440. doi: 10.3390/cancers13061440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Higgins JP, Thompson SG. Quantifying Heterogeneity in a Meta-Analysis. Stat Med (2002) 21(11):1539–58. doi: 10.1002/sim.1186 [DOI] [PubMed] [Google Scholar]
- 16. Mantel N, Haenszel W. Statistical Aspects of the Analysis of Data From Retrospective Studies of Disease. J Natl Cancer Inst (1959) 22(4):719–48. [PubMed] [Google Scholar]
- 17. DerSimonian R, Laird N. Meta-Analysis in Clinical Trials. Control Clin Trials (1986) 7(3):177–88. doi: 10.1016/0197-2456(86)90046-2 [DOI] [PubMed] [Google Scholar]
- 18. Hayashino Y, Noguchi Y, Fukui T. Systematic Evaluation and Comparison of Statistical Tests for Publication Bias. J Epidemiol (2005) 15(6):235–43. doi: 10.2188/jea.15.235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Shieh G. Power and Sample Size Calculations for Comparison of Two Regression Lines With Heterogeneous Variances. PloS One (2018) 13(12):e0207745. doi: 10.1371/journal.pone.0207745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Musso G, Sircana A, Saba F, Cassader M, Gambino R. Assessing the Risk of Ketoacidosis Due to Sodium-Glucose Cotransporter (SGLT)-2 Inhibitors in Patients With Type 1 Diabetes: A Meta-Analysis and Meta-Regression. PloS Med (2020) 17(12):e1003461. doi: 10.1371/journal.pmed.1003461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ge J, Zhu L, Zhou J, Li G, Li Y, Li S, et al. Association Between Co-Inhibitory Molecule Gene Tagging Single Nucleotide Polymorphisms and the Risk of Colorectal Cancer in Chinese. J Cancer Res Clin Oncol (2015) 141(9):1533–44. doi: 10.1007/s00432-015-1915-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fan Z, Fu J, Chen T, Wang G. A Study of Relationship Between Cytotoxic T−lymphocyte Antigen−4+49A>G Gene Polymorphism and Colorectal Cancer. Heilongjiang Med J (2012) 36:810–1. doi: 10.3969/j.issn.1004-5775.2012.11.003 [DOI] [Google Scholar]
- 23. Qi P, Ruan CP, Wang H, Zhou FG, Xu XY, Gu X, et al. CTLA-4 +49a>G Polymorphism is Associated With the Risk But Not With the Progression of Colorectal Cancer in Chinese. Int J Colorectal Dis (2010) 25(1):39–45. doi: 10.1007/s00384-009-0806-z [DOI] [PubMed] [Google Scholar]
- 24. Hadinia A, Hossieni SV, Erfani N, Saberi-Firozi M, Fattahi MJ, Ghaderi A. CTLA-4 Gene Promoter and Exon 1 Polymorphisms in Iranian Patients With Gastric and Colorectal Cancers. J Gastroenterol Hepatol (2007) 22(12):2283–7. doi: 10.1111/j.1440-1746.2007.04862.x [DOI] [PubMed] [Google Scholar]
- 25. Liu Z, Song Z, Sun J, Sun F, Li C, Sun J, et al. Association Between CTLA-4 Rs231775 Polymorphism and Hepatocellular Carcinoma Susceptibility. Int J Clin Exp Pathol (2015) 8(11):15118–22. [PMC free article] [PubMed] [Google Scholar]
- 26. Gu X, Qi P, Zhou F, Ji Q, Wang H, Dou T, et al. +49g > A Polymorphism in the Cytotoxic T-Lymphocyte Antigen-4 Gene Increases Susceptibility to Hepatitis B-Related Hepatocellular Carcinoma in a Male Chinese Population. Hum Immunol (2010) 71(1):83–7. doi: 10.1016/j.humimm.2009.09.353 [DOI] [PubMed] [Google Scholar]
- 27. Wang L, Jing F, Su D, Zhang T, Yang B, Jiao S, et al. Association Between CTLA-4 Rs231775 Polymorphism and Risk of Colorectal Cancer: A Meta Analysis. Int J Clin Exp Med (2015) 8(1):650–7. [PMC free article] [PubMed] [Google Scholar]
- 28. Dilmec F, Ozgonul A, Uzunkoy A, Akkafa F. Investigation of CTLA-4 and CD28 Gene Polymorphisms in a Group of Turkish Patients With Colorectal Cancer. Int J Immunogenet (2008) 35(4-5):317–21. doi: 10.1111/j.1744-313X.2008.00782.x [DOI] [PubMed] [Google Scholar]
- 29. Solerio E, Tappero G, Iannace L, Matullo G, Ayoubi M, Parziale A, et al. CTLA4 Gene Polymorphism in Italian Patients With Colorectal Adenoma and Cancer. Dig Liver Dis (2005) 37(3):170–5. doi: 10.1016/j.dld.2004.10.009 [DOI] [PubMed] [Google Scholar]
- 30. Zou C, Qiu H, Tang W, Wang Y, Lan B, Chen Y. CTLA4 Tagging Polymorphisms and Risk of Colorectal Cancer: A Case-Control Study Involving 2,306 Subjects. Onco Targets Ther (2018) 11:4609–19. doi: 10.2147/OTT.S173421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Li X, Zhao L, Zhong H, Fang T, Liu W. Correlative Study on CTLA−4+49a>G Polymorphism Gene Polymorphism and Colorectal Cancer. Prog Mod BioMed (2015) 15:2−824. doi: 10.13241/j.cnki.pmb.2015.05.007 [DOI] [Google Scholar]
- 32. Liu C, Wang Y, Jiang H, Tang W, Chen S, Kang M, et al. Association Between Cytotoxic T-Lymphocyte Antigen 4 (CTLA-4) +49 G>A (Rs231775) Polymorphism and Esophageal Cancer: From a Case-Control Study to a Meta-Analysis. Int J Clin Exp Med (2015) 8(10):17664–73. [PMC free article] [PubMed] [Google Scholar]
- 33. Liu J, Tang W, Lin W, Wang Y, Chen Y, Wang J, et al. Lack of Association Between CTLA-4 Genetic Polymorphisms and Noncardiac Gastric Cancer in a Chinese Population. DNA Cell Biol (2019) 38(5):443–8. doi: 10.1089/dna.2018.4555 [DOI] [PubMed] [Google Scholar]
- 34. Tang W, Wang Y, Chen S, Lin J, Chen B, Yu S, et al. Investigation of Cytotoxic T-Lymphocyte Antigen 4 Polymorphisms in Gastric Cardia Adenocarcinoma. Scand J Immunol (2016) 83:212–8. doi: 10.1111/sji.12409 [DOI] [PubMed] [Google Scholar]
- 35. Sun T, Zhou Y, Yang M, Hu Z, Tan W, Han X, et al. Functional Genetic Variations in Cytotoxic T-Lymphocyte Antigen 4 and Susceptibility to Multiple Types of Cancer. Cancer Res (2008) 68(17):7025–34. doi: 10.1158/0008-5472.CAN-08-0806 [DOI] [PubMed] [Google Scholar]
- 36. Yang J, Liu J, Chen Y, Tang W, Liu C, Sun Y, et al. Association of CTLA-4 Tagging Polymorphisms and Haplotypes With Hepatocellular Carcinoma Risk: A Case-Control Study. Medicine (Baltimore) (2019) 98(29):e16266. doi: 10.1097/MD.0000000000016266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hu L, Liu J, Chen X, Zhang Y, Liu L, Zhu J, et al. CTLA-4 Gene Polymorphism +49 a/G Contributes to Genetic Susceptibility to Two Infection-Related Cancers-Hepatocellular Carcinoma and Cervical Cancer. Hum Immunol (2010) 71(9):888–91. doi: 10.1016/j.humimm.2010.05.023 [DOI] [PubMed] [Google Scholar]
- 38. Lang C, Chen L, Li S. Cytotoxic T-Lymphocyte Antigen-4 +49G/A Polymorphism and Susceptibility to Pancreatic Cancer. DNA Cell Biol (2012) 31(5):683–7. doi: 10.1089/dna.2011.1417 [DOI] [PubMed] [Google Scholar]
- 39. Yang M, Sun T, Zhou Y, Wang L, Liu L, Zhang X, et al. The Functional Cytotoxic T Lymphocyte-Associated Protein 4 49G-to-A Genetic Variant and Risk of Pancreatic Cancer. Cancer (2012) 118(19):4681–6. doi: 10.1002/cncr.27455 [DOI] [PubMed] [Google Scholar]
- 40. Cui J, Ma H, Cui W, Ma W, Qi Y. Association of Cytotoxic T Lymphocyte Antigen-4 Gene Polymorphism With Susceptibility on Colorectal Cancer. Chin J Curr Adv Gen Surg (2013) 16(2):127–30. doi: 10.3969/j.issn.1009-9905.2013.02.013 [DOI] [Google Scholar]
- 41. Hou R, Cao B, Chen Z, Li Y, Ning T, Li C, et al. Association of Cytotoxic T Lymphocyte-Associated Antigen-4 Gene Haplotype With the Susceptibility to Gastric Cancer. Mol Biol Rep (2010) 37(1):515–20. doi: 10.1007/s11033-009-9705-1 [DOI] [PubMed] [Google Scholar]
- 42. Kucukhuseyin O, Turan S, Yanar K, Arikan S, Duzkoylu Y, Aydin S, et al. Individual and Combined Effects of CTLA4-CD28 Variants and Oxidant-Antioxidant Status on the Development of Colorectal Cancer. Anticancer Res (2015) 35(10):5391–400. [PubMed] [Google Scholar]
- 43. Mahajan R, El-Omar EM, Lissowska J, Grillo P, Rabkin CS, Baccarelli A, et al. Genetic Variants in T Helper Cell Type 1, 2 and 3 Pathways and Gastric Cancer Risk in a Polish Population. Jpn J Clin Oncol (2008) 38(9):626–33. doi: 10.1093/jjco/hyn075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wagh P, Kulkarni P, Kerkar S, Warke H, Chaudhari H, Deodhar K, et al. Polymorphisms in Cytotoxic T-Lymphocyte Associated Antigen 4 Gene Does Not Affect Scytotoxic T-Lymphocyte Associated Antigen 4 Levels in Human Papillomavirus-Infected Women With or Without Cervical Cancer. Indian J Med Microbiol (2018) 36(2):207–10. doi: 10.4103/ijmm.IJMM_17_220 [DOI] [PubMed] [Google Scholar]
- 45. Xiong YH, He L, Fei J. Genetic Variations in Cytotoxic T-Lymphocyte Antigen-4 and Susceptibility to Cervical Cancer. Int Immunopharmacol (2014) 18(1):71–6. doi: 10.1016/j.intimp.2013.10.018 [DOI] [PubMed] [Google Scholar]
- 46. Gokhale P, Kerkar S, Tongaonkar H, Salvi V, Mania-Pramanik J. CTLA-4 Gene Polymorphism at Position +49 a>G in Exon 1: A Risk Factor for Cervical Cancer in Indian Women. Cancer Genet (2013) 206(5):154–61. doi: 10.1016/j.cancergen.2013.04.003 [DOI] [PubMed] [Google Scholar]
- 47. Jiang L, Luo RY, Zhang W, Wang LR, Wang F, Cheng YX. [Single Nucleotide Polymorphisms of CTLA4 Gene and Their Association With Human Cervical Cancer]. Zhonghua Yi Xue Yi Chuan Xue Za Zhi (2011) 28(3):313–7. doi: 10.3760/cma.j.issn.1003-9406.2011.03.017 [DOI] [PubMed] [Google Scholar]
- 48. Rahimifar S, Erfani N, Sarraf Z, Ghaderi A. Ctla-4 Gene Variations may Influence Cervical Cancer Susceptibility. Gynecol Oncol (2010) 119(1):136–9. doi: 10.1016/j.ygyno.2010.06.006 [DOI] [PubMed] [Google Scholar]
- 49. Su TH, Chang TY, Lee YJ, Chen CK, Liu HF, Chu CC, et al. CTLA-4 Gene and Susceptibility to Human Papillomavirus-16-Associated Cervical Squamous Cell Carcinoma in Taiwanese Women. Carcinogenesis (2007) 28(6):1237–40. doi: 10.1093/carcin/bgm043 [DOI] [PubMed] [Google Scholar]
- 50. Pawlak E, Karabon L, Wlodarska-Polinska I, Jedynak A, Jonkisz A, Tomkiewicz A, et al. Influence of CTLA-4/CD28/ICOS Gene Polymorphisms on the Susceptibility to Cervical Squamous Cell Carcinoma and Stage of Differentiation in the Polish Population. Hum Immunol (2010) 71(2):195–200. doi: 10.1016/j.humimm.2009.11.006 [DOI] [PubMed] [Google Scholar]
- 51. Li H, Zhou YF, Guo HY, Sun T, Zhang WH, Lin DX. Association Between CTLA-4 Gene Polymorphism and Susceptibility to Cervical Cancer. Zhonghua Zhong Liu Za Zhi (2011) 33(9):681–4. doi: 10.3760/cma.j.issn.0253-3766.2011.09.009 [DOI] [PubMed] [Google Scholar]
- 52. Castro FA, Haimila K, Sareneva I, Schmitt M, Lorenzo J, Kunkel N, et al. Association of HLA-DRB1, Interleukin-6 and Cyclin D1 Polymorphisms With Cervical Cancer in the Swedish Population–a Candidate Gene Approach. Int J Cancer (2009) 125(8):1851–8. doi: 10.1002/ijc.24529 [DOI] [PubMed] [Google Scholar]
- 53. Khorshied MM, Gouda HM, Khorshid OM. Association of Cytotoxic T-Lymphocyte Antigen 4 Genetic Polymorphism, Hepatitis C Viral Infection and B-Cell non-Hodgkin Lymphoma: An Egyptian Study. Leuk Lymphoma (2014) 55(5):1061–6. doi: 10.3109/10428194.2013.820294 [DOI] [PubMed] [Google Scholar]
- 54. Hui L, Lei Z, Peng Z, Ruobing S, Fenghua Z. Polymorphism Analysis of CTLA-4 in Childhood Acute Lymphoblastic Leukemia. Pak J Pharm Sci (2014) 27(4 Suppl):1005–13. doi: 10.4314/tjpr.v13i7.24 [DOI] [PubMed] [Google Scholar]
- 55. Cheng TY, Lin JT, Chen LT, Shun CT, Wang HP, Lin MT, et al. Association of T-Cell Regulatory Gene Polymorphisms With Susceptibility to Gastric Mucosa-Associated Lymphoid Tissue Lymphoma. J Clin Oncol (2006) 24(21):3483–9. doi: 10.1200/JCO.2005.05.5434 [DOI] [PubMed] [Google Scholar]
- 56. Suwalska K, Pawlak E, Karabon L, Tomkiewicz A, Dobosz T, Urbaniak-Kujda D, et al. Association Studies of CTLA-4, CD28, and ICOS Gene Polymorphisms With B-Cell Chronic Lymphocytic Leukemia in the Polish Population. Hum Immunol (2008) 69(3):193–201. doi: 10.1016/j.humimm.2008.01.014 [DOI] [PubMed] [Google Scholar]
- 57. Piras G, Monne M, Uras A, Palmas A, Murineddu M, Arru L, et al. Genetic Analysis of the 2q33 Region Containing CD28-CTLA4-ICOS Genes: Association With non-Hodgkin's Lymphoma. Br J Haematol (2005) 129(6):784–90. doi: 10.1111/j.1365-2141.2005.05525.x [DOI] [PubMed] [Google Scholar]
- 58. Monne M, Piras G, Palmas A, Arru L, Murineddu M, Latte G, et al. Cytotoxic T-Lymphocyte Antigen-4 (CTLA-4) Gene Polymorphism and Susceptibility to non-Hodgkin's Lymphoma. Am J Hematol (2004) 76(1):14–8. doi: 10.1002/ajh.20045 [DOI] [PubMed] [Google Scholar]
- 59. Pavkovic M, Georgievski B, Cevreska L, Spiroski M, Efremov DG. CTLA-4 Exon 1 Polymorphism in Patients With Autoimmune Blood Disorders. Am J Hematol (2003) 72(2):147–9. doi: 10.1002/ajh.10278 [DOI] [PubMed] [Google Scholar]
- 60. Liu J, Liu J, Song B, Wang T, Liu Y, Hao J, et al. Genetic Variations in CTLA-4, TNF-Alpha, and LTA and Susceptibility to T-Cell Lymphoma in a Chinese Population. Cancer Epidemiol (2013) 37(6):930–4. doi: 10.1016/j.canep.2013.08.011 [DOI] [PubMed] [Google Scholar]
- 61. Liu Y, He Z, Feng D, Shi G, Gao R, Wu X, et al. Cytotoxic T-Lymphocyte Antigen-4 Polymorphisms and Susceptibility to Osteosarcoma. DNA Cell Biol (2011) 30(12):1051–5. doi: 10.1089/dna.2011.1269 [DOI] [PubMed] [Google Scholar]
- 62. Kasamatsu T, Awata M, Ishihara R, Murakami Y, Gotoh N, Matsumoto M, et al. PDCD1 and PDCD1LG1 Polymorphisms Affect the Susceptibility to Multiple Myeloma. Clin Exp Med (2020) 20(1):51–62. doi: 10.1007/s10238-019-00585-4 [DOI] [PubMed] [Google Scholar]
- 63. Qin XY, Lu J, Li GX, Wen L, Liu Y, Xu LP, et al. CTLA-4 Polymorphisms Are Associated With Treatment Outcomes of Patients With Multiple Myeloma Receiving Bortezomib-Based Regimens. Ann Hematol (2018) 97(3):485–95. doi: 10.1007/s00277-017-3203-7 [DOI] [PubMed] [Google Scholar]
- 64. Bilbao-Aldaiturriaga N, Patino-Garcia A, Martin-Guerrero I, Garcia-Orad A. Cytotoxic T Lymphocyte-Associated Antigen 4 Rs231775 Polymorphism and Osteosarcoma. Neoplasma (2017) 64(2):299–304. doi: 10.4149/neo_2017_218 [DOI] [PubMed] [Google Scholar]
- 65. Feng D, Yang X, Li S, Liu T, Wu Z, Song Y, et al. Cytotoxic T-Lymphocyte Antigen-4 Genetic Variants and Risk of Ewing's Sarcoma. Genet Test Mol Biomarkers (2013) 17(6):458–63. doi: 10.1089/gtmb.2012.0488 [DOI] [PubMed] [Google Scholar]
- 66. Yang S, Wang C, Zhou Y, Sun G, Zhu D, Gao S. Cytotoxic T-Lymphocyte Antigen-4 Polymorphisms and Susceptibility to Ewing's Sarcoma. Genet Test Mol Biomarkers (2012) 16(10):1236–40. doi: 10.1089/gtmb.2012.0129 [DOI] [PubMed] [Google Scholar]
- 67. Wang W, Wang J, Song H, Liu J, Song B, Cao X. Cytotoxic T-Lymphocyte Antigen-4 +49G/A Polymorphism is Associated With Increased Risk of Osteosarcoma. Genet Test Mol Biomarkers (2011) 15(7-8):503–6. doi: 10.1089/gtmb.2010.0264 [DOI] [PubMed] [Google Scholar]
- 68. Karabon L, Pawlak-Adamska E, Tomkiewicz A, Jedynak A, Kielbinski M, Woszczyk D, et al. Variations in Suppressor Molecule Ctla-4 Gene are Related to Susceptibility to Multiple Myeloma in a Polish Population. Pathol Oncol Res (2012) 18(2):219–26. doi: 10.1007/s12253-011-9431-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Mao F, Niu XB, Gu S, Ji L, Wei BJ, Wang HB. CTLA-4 +49a/G Polymorphism Increases the Susceptibility to Bladder Cancer in Chinese Han Participants: A Case-Control Study. Dis Markers (2020) 2020:8143158. doi: 10.1155/2020/8143158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Jaiswal PK, Singh V, Mittal RD. Cytotoxic T Lymphocyte Antigen 4 (CTLA4) Gene Polymorphism With Bladder Cancer Risk in North Indian Population. Mol Biol Rep (2014) 41(2):799–807. doi: 10.1007/s11033-013-2919-2 [DOI] [PubMed] [Google Scholar]
- 71. Wang L, Su G, Zhao X, Cai Y, Cai X, Zhang J, et al. Association Between the Cytotoxic T-Lymphocyte Antigen 4 +49A/G Polymorphism and Bladder Cancer Risk. Tumour Biol (2014) 35(2):1139–42. doi: 10.1007/s13277-013-1152-x [DOI] [PubMed] [Google Scholar]
- 72. Saenz Lopez P, Vazquez Alonso F, Romero JM, Carretero R, Tallada Bunuel M, Ruiz Cabello F, et al. [Polymorphisms in Inflammatory Response Genes in Metastatic Renal Cancer]. Actas Urol Esp (2009) 33(5):474–81. doi: 10.1016/s0210-4806(09)74180-4 [DOI] [PubMed] [Google Scholar]
- 73. Cozar JM, Romero JM, Aptsiauri N, Vazquez F, Vilchez JR, Tallada M, et al. High Incidence of CTLA-4 AA (CT60) Polymorphism in Renal Cell Cancer. Hum Immunol (2007) 68(8):698–704. doi: 10.1016/j.humimm.2007.05.002 [DOI] [PubMed] [Google Scholar]
- 74. Karabon L, Tupikowski K, Tomkiewicz A, Partyka A, Pawlak-Adamska E, Wojciechowski A, et al. Is the Genetic Background of Co-Stimulatory CD28/CTLA-4 Pathway the Risk Factor for Prostate Cancer? Pathol Oncol Res (2017) 23(4):837–43. doi: 10.1007/s12253-016-0180-4 [DOI] [PubMed] [Google Scholar]
- 75. Tupikowski K, Partyka A, Kolodziej A, Dembowski J, Debinski P, Halon A, et al. CTLA-4 and CD28 Genes' Polymorphisms and Renal Cell Carcinoma Susceptibility in the Polish Population–a Prospective Study. Tissue Antigens (2015) 86(5):353–61. doi: 10.1111/tan.12671 [DOI] [PubMed] [Google Scholar]
- 76. Babteen NA, Fawzy MS, Alelwani W, Alharbi RA, Alruwetei AM, Toraih EA, et al. Signal Peptide Missense Variant in Cancer-Brake Gene CTLA4 and Breast Cancer Outcomes. Gene (2020) 737:144435. doi: 10.1016/j.gene.2020.144435 [DOI] [PubMed] [Google Scholar]
- 77. Minhas S, Bhalla S, Shokeen Y, Jauhri M, Saxena R, Verma IC, et al. Lack of Any Association of the CTLA-4 +49 G/A Polymorphism With Breast Cancer Risk in a North Indian Population. Asian Pac J Cancer Prev (2014) 15(5):2035–8. doi: 10.7314/apjcp.2014.15.5.2035 [DOI] [PubMed] [Google Scholar]
- 78. Wang L, Li D, Fu Z, Li H, Jiang W, Li D. Association of CTLA-4 Gene Polymorphisms With Sporadic Breast Cancer in Chinese Han Population. BMC Cancer (2007) 7:173. doi: 10.1186/1471-2407-7-173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Ghaderi A, Yeganeh F, Kalantari T, Talei AR, Pezeshki AM, Doroudchi M, et al. Cytotoxic T Lymphocyte Antigen-4 Gene in Breast Cancer. Breast Cancer Res Treat (2004) 86(1):1–7. doi: 10.1023/B:BREA.0000032918.89120.8e [DOI] [PubMed] [Google Scholar]
- 80. Wu Q, Zhan X, Dou T, Chen H, Fan W, Zhou K, et al. CTLA4 A49G Polymorphism Shows Significant Association With Glioma Risk in a Chinese Population. Biochem Genet (2011) 49(3-4):190–201. doi: 10.1007/s10528-010-9398-0 [DOI] [PubMed] [Google Scholar]
- 81. Bharti V, Mohanti BK, Das SN. Functional Genetic Variants of CTLA-4 and Risk of Tobacco-Related Oral Carcinoma in High-Risk North Indian Population. Hum Immunol (2013) 74(3):348–52. doi: 10.1016/j.humimm.2012.12.008 [DOI] [PubMed] [Google Scholar]
- 82. Erfani N, Haghshenas MR, Hoseini MA, Hashemi SB, Khademi B, Ghaderi A. Strong Association of CTLA-4 Variation (CT60A/G) and CTLA-4 Haplotypes With Predisposition of Iranians to Head and Neck Cancer. Iran J Immunol (2012) 9(3):188–98 [PubMed] [Google Scholar]
- 83. Cheng XL, Ning T, Xu CQ, Li Y, Zhu BS, Hou FT, et al. Haplotype Analysis of CTLA4 Gene and Risk of Esophageal Squamous Cell Carcinoma in Anyang Area of China. Hepatogastroenterology (2011) 58(106):432–7. doi: 10.1016/j.dld.2010.10.013 [DOI] [PubMed] [Google Scholar]
- 84. Xiao M, Qi F, Chen X, Luo Z, Zhang L, Zheng C, et al. Functional Polymorphism of Cytotoxic T-Lymphocyte Antigen 4 and Nasopharyngeal Carcinoma Susceptibility in a Chinese Population. Int J Immunogenet (2010) 37(1):27–32. doi: 10.1111/j.1744-313X.2009.00888.x [DOI] [PubMed] [Google Scholar]
- 85. Wong YK, Chang KW, Cheng CY, Liu CJ. Association of CTLA-4 Gene Polymorphism With Oral Squamous Cell Carcinoma. J Oral Pathol Med (2006) 35(1):51–4. doi: 10.1111/j.1600-0714.2005.00377.x [DOI] [PubMed] [Google Scholar]
- 86. Liu HN, Su JL, Zhou SH, Liu LJ, Qie P. Cytotoxic T Lymphocyte-Associated Antigen-4 +49A>G Polymorphism and the Risk of non-Small Cell Lung Cancer in a Chinese Population. Int J Clin Exp Med (2015) 8(7):11519–23. [PMC free article] [PubMed] [Google Scholar]
- 87. Khaghanzadeh N, Erfani N, Ghayumi MA, Ghaderi A. CTLA4 Gene Variations and Haplotypes in Patients With Lung Cancer. Cancer Genet Cytogenet (2010) 196(2):171–4. doi: 10.1016/j.cancergencyto.2009.09.001 [DOI] [PubMed] [Google Scholar]
- 88. Abtahi S, Izadi Jahromi F, Dabbaghmanesh MH, Malekzadeh M, Ghaderi A. Association Between CTLA-4 + 49a > G and - 318C > T Single-Nucleotide Polymorphisms and Susceptibility to Thyroid Neoplasm. Endocrine (2018) 62(1):159–65. doi: 10.1007/s12020-018-1663-8 [DOI] [PubMed] [Google Scholar]
- 89. Chang DF, Chen XH, Huang J, Sun YM, Zhu DY, Xu ZQ. CTLA-4 Gene Polymorphisms Associate With Efficacy of Postoperative Radioiodine-131 for Differentiated Thyroid Carcinoma. Future Oncol (2017) 13(12):1057–68. doi: 10.2217/fon-2016-0399 [DOI] [PubMed] [Google Scholar]
- 90. Ma Y, Liu X, Zhu J, Li W, Guo L, Han X, et al. Polymorphisms of Co-Inhibitory Molecules (CTLA-4/PD-1/PD-L1) and the Risk of non-Small Cell Lung Cancer in a Chinese Population. Int J Clin Exp Med (2015) 8(9):16585–91. [PMC free article] [PubMed] [Google Scholar]
- 91. Isitmangil G, Gurleyik G, Aker FV, Coskun C, Kucukhuseyin O, Arikan S, et al. Association of CTLA4 and CD28 Gene Variants and Circulating Levels of Their Proteins in Patients With Breast Cancer. In Vivo (2016) 30(4):485–93. [PubMed] [Google Scholar]
- 92. Kammerer PW, Toyoshima T, Schoder F, Kammerer P, Kuhr K, Brieger J, et al. Association of T-Cell Regulatory Gene Polymorphisms With Oral Squamous Cell Carcinoma. Oral Oncol (2010) 46(7):543–8. doi: 10.1016/j.oraloncology.2010.03.025 [DOI] [PubMed] [Google Scholar]
- 93. Queirolo P, Morabito A, Laurent S, Lastraioli S, Piccioli P, Ascierto PA, et al. Association of CTLA-4 Polymorphisms With Improved Overall Survival in Melanoma Patients Treated With CTLA-4 Blockade: A Pilot Study. Cancer Invest (2013) 31(5):336–45. doi: 10.3109/07357907.2013.793699 [DOI] [PubMed] [Google Scholar]
- 94. Antczak A, Pastuszak-Lewandoska D, Gorski P, Domanska D, Migdalska-Sek M, Czarnecka K, et al. Ctla-4 Expression and Polymorphisms in Lung Tissue of Patients With Diagnosed non-Small-Cell Lung Cancer. BioMed Res Int (2013) 2013:576486. doi: 10.1155/2013/576486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Chuang WY, Strobel P, Gold R, Nix W, Schalke B, Kiefer R, et al. A CTLA4high Genotype is Associated With Myasthenia Gravis in Thymoma Patients. Ann Neurol (2005) 58(4):644–8. doi: 10.1002/ana.20577 [DOI] [PubMed] [Google Scholar]
- 96. Yu H, Yang J, Jiao S, Li Y, Zhang W, Wang J. Cytotoxic T Lymphocyte Antigen 4 Expression in Human Breast Cancer: Implications for Prognosis. Cancer Immunol Immunother (2015) 64(7):853–60. doi: 10.1007/s00262-015-1696-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Li D, Zhang Q, Xu F, Fu Z, Yuan W, Li D, et al. Association of CTLA-4 Gene Polymorphisms With Sporadic Breast Cancer Risk and Clinical Features in Han Women of Northeast China. Mol Cell Biochem (2012) 364(1-2):283–90. doi: 10.1007/s11010-012-1228-8 [DOI] [PubMed] [Google Scholar]
- 98. Chen S, Wang Y, Chen Y, Lin J, Liu C, Kang M, et al. Investigation of Cytotoxic T-Lymphocyte Antigen-4 Polymorphisms in non-Small Cell Lung Cancer: A Case-Control Study. Oncotarget (2017) 8(44):76634–43. doi: 10.18632/oncotarget.20638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Karabon L, Pawlak E, Tomkiewicz A, Jedynak A, Passowicz-Muszynska E, Zajda K, et al. CTLA-4, CD28, and ICOS Gene Polymorphism Associations With non-Small-Cell Lung Cancer. Hum Immunol (2011) 72(10):947–54. doi: 10.1016/j.humimm.2011.05.010 [DOI] [PubMed] [Google Scholar]
- 100. Gogas H, Dafni U, Koon H, Spyropoulou-Vlachou M, Metaxas Y, Buchbinder E, et al. Evaluation of Six CTLA-4 Polymorphisms in High-Risk Melanoma Patients Receiving Adjuvant Interferon Therapy in the He13A/98 Multicenter Trial. J Transl Med (2010) 8:108. doi: 10.1186/1479-5876-8-108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Bouwhuis MG, Gast A, Figl A, Eggermont AM, Hemminki K, Schadendorf D, et al. Polymorphisms in the CD28/CTLA4/ICOS Genes: Role in Malignant Melanoma Susceptibility and Prognosis? Cancer Immunol Immunother (2010) 59(2):303–12. doi: 10.1007/s00262-009-0751-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Welsh MM, Applebaum KM, Spencer SK, Perry AE, Karagas MR, Nelson HH. CTLA4 Variants, UV-Induced Tolerance, and Risk of non-Melanoma Skin Cancer. Cancer Res (2009) 69(15):6158–63. doi: 10.1158/0008-5472.CAN-09-0415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Miller KD, Fidler-Benaoudia M, Keegan TH, Hipp HS, Jemal A, Siegel RL. Cancer Statistics for Adolescents and Young Adults 2020. CA Cancer J Clin (2020) 70(6):443–59. doi: 10.3322/caac.21637 [DOI] [PubMed] [Google Scholar]
- 104. Speiser DE, Ho PC, Verdeil G. Regulatory Circuits of T Cell Function in Cancer. Nat Rev Immunol (2016) 16(10):599–611. doi: 10.1038/nri.2016.80 [DOI] [PubMed] [Google Scholar]
- 105. Walunas TL, Lenschow DJ, Bakker CY, Linsley PS, Freeman GJ, Green JM, et al. Pillars Article: CTLA-4 can Function as a Negative Regulator of T Cell Activation. Immunity (1994) 1:405–13. doi: 10.1016/1074-7613(94)90071-X [DOI] [PubMed] [Google Scholar]
- 106. Buchbinder E, Hodi FS. Cytotoxic T Lymphocyte Antigen-4 and Immune Checkpoint Blockade. J Clin Invest (2015) 125(9):3377–83. doi: 10.1172/jci80012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Matheu MP, Othy S, Greenberg ML, Dong TX, Schuijs M, Deswarte K, et al. Imaging Regulatory T Cell Dynamics and CTLA4-Mediated Suppression of T Cell Priming. Nat Commun (2015) 6:6219. doi: 10.1038/ncomms7219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Dai Z, Tian T, Wang M, Liu X, Lin S, Yang P, et al. CTLA-4 Polymorphisms Associate With Breast Cancer Susceptibility in Asians: A Meta-Analysis. PeerJ (2017) 5:e2815. doi: 10.7717/peerj.2815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Das AP, Saini S, Agarwal SM. A Comprehensive Meta-Analysis of non-Coding Polymorphisms Associated With Precancerous Lesions and Cervical Cancer. Genomics (2022) 114(3):110323. doi: 10.1016/j.ygeno.2022.110323 [DOI] [PubMed] [Google Scholar]
- 110. Hu P, Liu Q, Deng G, Zhang J, Liang N, Xie J, et al. The Prognostic Value of Cytotoxic T-Lymphocyte Antigen 4 in Cancers: A Systematic Review and Meta-Analysis. Sci Rep (2017) 7:42913. doi: 10.1038/srep42913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Fang M, Huang W, Mo D, Zhao W, Huang R. Association of Five Snps in Cytotoxic T-Lymphocyte Antigen 4 and Cancer Susceptibility: Evidence From 67 Studies. Cell Physiol Biochem (2018) 47(1):414–27. doi: 10.1159/000489953 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding authors.