Abstract
A single microorganism able to mineralize chloronitrobenzenes (CNBs) has not been reported, and degradation of CNBs by coculture of two microbial strains was attempted. Pseudomonas putida HS12 was first isolated by analogue enrichment culture using nitrobenzene (NB) as the substrate, and this strain was observed to possess a partial reductive pathway for the degradation of NB. From high-performance liquid chromatography-mass spectrometry and 1H nuclear magnetic resonance analyses, NB-grown cells of P. putida HS12 were found to convert 3- and 4-CNBs to the corresponding 5- and 4-chloro-2-hydroxyacetanilides, respectively, by partial reduction and subsequent acetylation. For the degradation of CNBs, Rhodococcus sp. strain HS51, which degrades 4- and 5-chloro-2-hydroxyacetanilides, was isolated and combined with P. putida HS12 to give a coculture. This coculture was confirmed to mineralize 3- and 4-CNBs in the presence of an additional carbon source. A degradation pathway for 3- and 4-CNBs by the two isolated strains was also proposed.
Chloronitrobenzenes (CNBs) are widely used as intermediates in the manufacture of many chlorinated aromatic compounds and are known to be very toxic and resistant to microbial degradation due to the electron-withdrawing properties of nitro and chlorine groups. These compounds have been listed as priority pollutants by the European Economic Community. Much effort has been dedicated to the microbial degradation of nitro- and chloroaromatic compounds, and their degradative pathways have been revealed in detail (5, 11, 14, 22, 23, 27, 30, 32). Even though biodegradation of 3-CNB by sludge samples was attempted (18), studies on the microbial degradation of CNBs have been rare.
Metabolism of chlorinated benzenes has been focused mainly on the oxidative pathway (5, 27, 30, 32). On the other hand, in the case of nitrobenzene (NB), both oxidative and reductive pathways have been reported. Haigler and Spain reported that toluene monooxygenases in Pseudomonas mendocina and Ralstonia pickettii catalyzed the conversion of NB to 3- or 4-nitrophenol, and then these intermediates were further transformed to the corresponding 3- and 4-nitrocatechols by the addition of a hydroxyl group (11). Meanwhile, the dioxygenase-mediated transformation of NB in Pseudomonas putida F1 resulted in cis-1,2-dihydroxynitrocyclohexa-3,5-diene, and then this compound was further converted to 3-nitrocatechol, a dead-end product, by Pseudomonas sp. strain JS150 (11). In our previous work, a hybrid pathway for the degradation of NB was designed in P. putida TB103 (14). In this pathway, NB was initially oxidized to NB-cis-dihydrodiol by toluene dioxygenase, and then this intermediate was converted to catechol by the action of toluate cis-dihydrodiol dehydrogenase. Recently, Nishino and Spain isolated two NB-degrading strains, Pseudomonas pseudoalcaligenes JS45, carrying a partial reductive pathway (22), and Comamonas sp. strain JS765, showing dioxygenase-catalyzed denitration of NB (23).
In this paper, we report the degradation of 3- and 4-CNBs by a coculture of the two isolated strains, P. putida HS12 and Rhodococcus sp. strain HS51. The former was found to possess a partial reductive pathway and to catalyze the conversion of CNBs to chlorohydroxyacetanilides by cometabolism, and these compounds were further degraded by Rhodococcus sp. strain HS51. A degradation pathway for 3- and 4-CNBs by a coculture of the two isolated strains is also proposed based on the identification of metabolic intermediates.
MATERIALS AND METHODS
Chemicals.
NB, 2-, 3-, and 4-CNBs, 2-aminophenol, catechol, and 4-chlorocatechol were purchased from Aldrich (Milwaukee, Wis.), and 4-chloro-2-aminophenol was from Sigma Chemical Co. (St. Louis, Mo.). 5-Chloro-2-hydroxyacetanilide was synthesized from 4-chloro-2-aminophenol by acetylation of 4-chloro-2-aminophenol by the method of Katz and Cohen with a slight modification (15). All other chemicals were of analytical grade.
Isolation and culture conditions.
To screen the CNB-degrading microorganisms, CNBs (20 ppm) were incubated with sludge and soil samples. However, no microbe enrichment was found, even in the presence of yeast extract and other carbon sources (data not shown), which implies that CNBs are not suitable as a sole source of carbon and energy for the isolation of CNB-degrading microorganisms due to their high toxicity. Based on this result, the analogue enrichment technique was employed using NB as the sole carbon, energy, and nitrogen source for the isolation of CNB-degrading microorganisms. For the enrichment, soil or water samples, contaminated with nitroaromatics, was added to 250-ml shake flasks containing 50 ml of nitrogen and carbon-free mineral salts medium, and NB was used to supplement the medium at a final concentration of 1 mM. The mineral salts medium consisted of (per liter of distilled water) 1.0 g of K2HPO4, 0.6 g of NaH2PO4, 0.2 g of MgSO4 · 7H2O, 0.2 g of KCl, 2 mg of yeast extract, and 1 ml of a trace element solution containing 0.05 mg of H3BO3, 0.2 mg of CaSO4, 0.1 mg of CoSO4, 0.2 mg of CuSO4, 3 mg of FeSO4, 0.02 mg of MnCl2, 0.1 mg of NaMoO4 · 2H2O, 0.02 mg of NiCl2, and 0.03 mg of ZnSO4 · 7H2O. The flasks were incubated at 30°C on a rotary shaker at 150 rpm. On NB depletion, subcultures were serially diluted and plated on minimal salts agar containing NB.
Chlorohydroxyacetanilide-degrading microorganisms were also isolated by a procedure similar to that described above, except that chloride-free minimal salts medium supplemented with various chloroaromatics, including chlorohydroxyacetanilides, was used (32).
NB-degrading microorganisms were grown in 500-ml flasks containing 100 ml of minimal salts medium and 1 mM NB. NB was added intermittently after depletion to obtain biomass due to its toxic effect on microbial growth. When necessary, NB-degrading microorganisms were cultured in the minimal salts medium supplemented with 10 mM succinate and 4 mM NH4Cl. Chlorohydroxyacetanilide-degrading microorganisms were cultured in chloride-free minimal salts medium containing 1 mM chlorobenzene and 0.2 mM 5-chloro-2-hydroxyacetanilide.
Identification of isolates.
Microorganisms isolated from the enrichment culture were identified by using the standard procedures described in Bergey’s Manual of Systematic Bacteriology (25) and tests in Methods for General and Molecular Bacteriology (31). Fatty acid analysis was also performed.
Isolation of metabolites.
Culture media were acidified to pH 2 with HCl and then extracted with an equal volume of ethyl acetate. The solvent phase was dried over anhydrous sodium sulfate and evaporated by using a rotary evaporator. The resulting powder was subjected to high-performance liquid chromatography (HPLC)-mass spectrometry (MS) and 1H nuclear magnetic resonance (1H-NMR) analyses.
Transformation of CNBs by an NB-degrading isolate.
NB-grown cells were washed with 0.02 M phosphate buffer (pH 7.2) and resuspended in 50 ml of nitrogen- and chloride-free minimal salts medium (pH 7.2) supplemented with 10 mM succinate. CNB was added to the cell suspension at a concentration of 0.3 mM. The reaction medium was analyzed by HPLC.
Preparation of cell extracts and enzyme assays.
Cells were harvested by centrifugation (6,000 × g) for 20 min at 4°C, washed with 0.02 M phosphate buffer (pH 7.2), and then resuspended in the same buffer. For preparation of crude extracts, cells were disrupted by a cell disruptor 350 (VWR Scientific Inc.) and centrifuged (13,000 × g) for 30 min at 4°C. Catechol-1,2-dioxygenase and catechol-2,3-dioxygenase were assayed by the method of Schraa et al. (30).
Coculture of the two isolates.
NB- and chlorobenzene-grown cells were combined at a predetermined ratio in 20 ml of nitrogen- and chloride-free minimal salts medium containing 10 mM succinate. CNB was added to the culture medium, and it was incubated at 30°C. The population size of each isolate and concentrations of CNBs, metabolites, and chloride ion were determined with culture time.
Analytical methods.
NB, 2-, 3-, and 4-CNBs, metabolites, and aminophenol were analyzed by HPLC (model LC9A; Shimadzu, Kyoto, Japan) with a UV/VIS detector (model SPD6AV, Shimadzu). An octyldecyl silane column [CLC-ODS(M), 25 cm long; YMC, Wilmington, N.C.] was used with an acetonitrile-water mixture (60:40 [vol/vol]) as the mobile phase. The flow rate of the eluent was 1 ml/min. The eluent was detected at 210 nm. For analysis of 4-chloro-2-aminophenol and 5-chloro-2-hydroxyacetanilide, an acetonitrile-phosphate buffer (50 mM, pH 7.0) mixture (30:70 [vol/vol]) was used as the mobile phase. HPLC-MS analysis was carried out by using an HP1050 series chromatograph (Hewlett-Packard, Avondale, Pa.) coupled with a VG Quattrotriple quadruple mass spectrometer equipped with an atmospheric pressure chemical ionization interface (Fisons Instrument/VG Biotech, Altrincham, United Kingdom). The atmospheric pressure chemical ionization interface was operated in the positive-ion mode. This HPLC-MS system generates a positively charged ion with a mass 1 more than the molecular mass by hydrogen addition. NMR spectra were recorded on a Bruker AMX FT500MHz NMR spectrometer (Bruker GmbH, Karlsruhe, Federal Republic of Germany). Chloride ion concentration was determined by using an ion-selective combination chloride electrode (model 96/17; Orion Research, Inc., Cambridge, Mass.). Ammonia was quantified by the Nessler reaction (8).
RESULTS
Isolation and identification of the NB-degrading microorganism.
As a result of analogue enrichment, an NB-degrading microorganism was isolated from wastewater and soil contaminated with nitroaromatics. The isolated strain was a gram-negative, motile rod, and showed both catalase- and oxidase-positive reactions. In addition, this microorganism utilized glucose, mannitol, N-acetylglucosamine, gluconate, caprate, malate, citrate, and phenylacetate as carbon sources. Fatty acid analysis gave a similarity index of 0.823 to P. putida. From the above results, the NB-degrading isolate was identified as P. putida HS12.
Growth and degradative pathway of P. putida HS12.
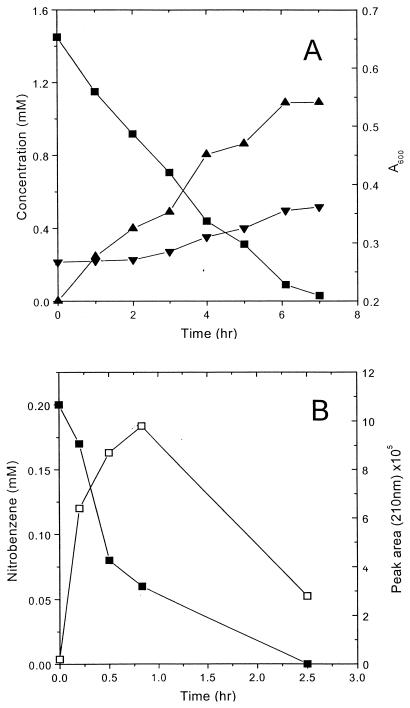
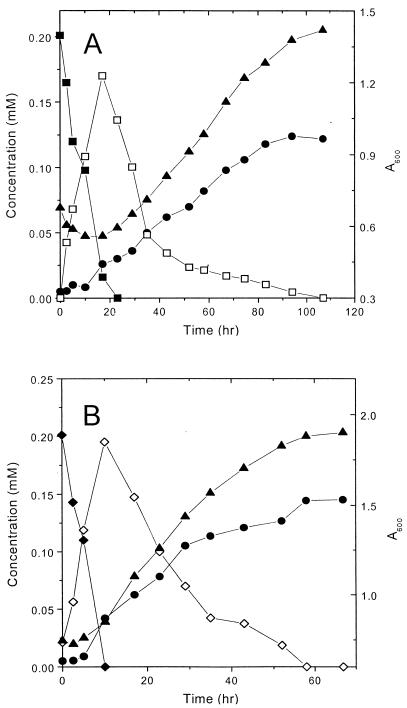
In order to confirm whether the isolated strain can utilize NB as a sole source of carbon, energy, and nitrogen, NB-grown cells of P. putida HS12 were washed and cultured in 0.05 mM phosphate buffer (pH 7.2) containing 1.4 mM NB. As shown in Fig. 1A, NB rapidly decreased with culture time and cell density gradually increased after 2 h of incubation, reaching a maximum in 7 h. Ammonia was detected in the culture medium, but the amount of released ammonia was not stoichiometric to that of NB consumed. This seems to be due to the fact that a significant amount of released ammonia was taken up by the microorganism as a nitrogen source for cell growth. No metabolic intermediate was detected in the culture medium.
FIG. 1.
Degradation of NB by NB-grown (A) and succinate-grown (B) cells of P. putida HS12 in phosphate buffer (pH 7.2). The optical density at 600 nm of the biomass in the reaction mixture was 0.12 (B). Symbols: ■, nitrobenzene; ▾, ammonia; ▴, optical density; □, intermediate.
In order to elucidate the degradative pathway of NB in P. putida HS12, succinate-grown cells of P. putida HS12 were incubated in the presence of NB. NB-grown cells completed the degradation of NB in 30 min (data not shown). However, degradation of NB by succinate-grown cells was slow (it took 2.5 h), and in addition, an unidentified metabolite accumulated transiently (Fig. 1B). Prolonged incubation of succinate-grown cells of P. putida HS12 with NB resulted in the complete degradation of NB without any intermediate, which implies that the catabolic pathway of NB in P. putida HS12 is inducible.
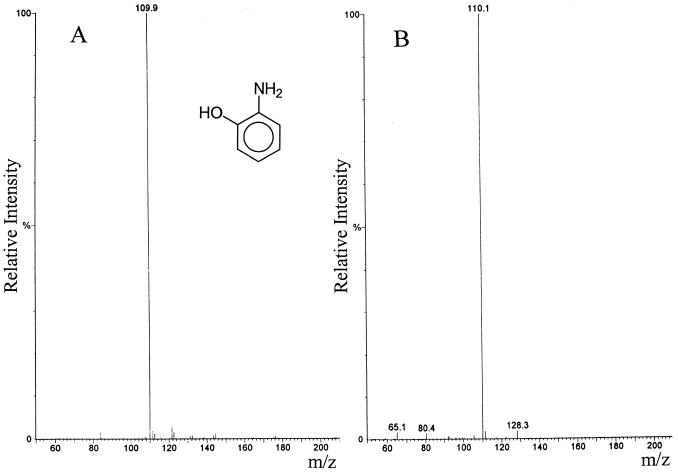
To identify the metabolite formed by uninduced P. putida HS12, the metabolite was separated from the culture medium and analyzed by using HPLC-MS. As shown in Fig. 2, the mass spectrum of the metabolite was consistent with that of authentic 2-aminophenol. Molecular ion peaks were observed at 110 for both authentic 2-aminophenol and the metabolite. The HPLC-MS system used in this work generates a positively charged ion with a mass 1 more than the molecular mass by hydrogen addition. Therefore, a molecular ion peak of 110 corresponds to that of 2-aminophenol. The solvent phase might contain a small amount of impurities, as well as the targeted metabolite, and the peaks above the m/z at 110 seems to be due to the impurities. From the observations that 2-aminophenol accumulated from NB as a metabolite by succinate-grown cells of P. putida HS12 and that ammonia is liberated with the degradation of NB, it seems that P. putida HS12 possesses a partial reductive pathway, as reported by Nishino and Spain (22).
FIG. 2.
Mass spectra of authentic 2-aminophenol (A) and the intermediate (B).
Conversion of CNBs by NB-grown P. putida HS12.
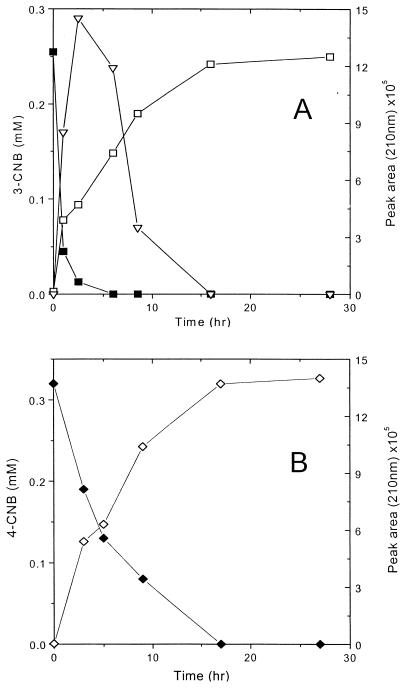
In order to investigate whether P. putida HS12 could attack CNBs, NB-grown cells of P. putida HS12 were incubated with 2-, 3-, or 4-CNB in nitrogen- and chloride-free minimal salts medium supplemented with 10 mM succinate as a maintenance energy source. When P. putida HS12 was incubated with CNBs in the absence of an additional carbon source, severe lysis of cells was observed, and in addition, CNBs were not transformed at all, which seems to be due to the toxic effect of CNBs. In other words, P. putida HS12 requires an additional carbon source for transformation of CNBs, and this indicates that CNBs are partially degraded by P. putida HS12 via cometabolism. For this reason, succinate was added as a carbon source. As shown in Fig. 3, different kinds of metabolites accumulated, depending on the CNB used. In the case of 3-CNB, unknown metabolite I appeared with a rapid decrease of 3-CNB and reached a maximum at 3 h. As intermediate I decreased, another metabolite, II, gradually accumulated in the reaction mixture. The level of intermediate I dropped to 0 in 15 h, but metabolite II remained stable and was not metabolized further (Fig. 3A). For 4-CNB, only one metabolite, metabolite III, was detected as 4-CNB decreased (Fig. 3B). Metabolite III also remained stable in the reaction mixture. On the other hand, when 2-CNB was incubated with NB-grown cells of P. putida H12, conversion of 2-CNB was not completed and severe lysis of cells was observed from the beginning of the reaction. Succinate-grown cells of P. putida HS12 were found to be unable to transform any form of CNB, which implies that CNBs were metabolized through the NB-degrading pathway in induced P. putida HS12. While NB was utilized as a carbon, energy, and nitrogen source, as well as an inducer of the NB catabolic pathway, CNBs did not induce the catabolic pathway and were only transformed into the corresponding metabolites via cometabolism.
FIG. 3.
Transformation of 3-CNB (A) and 4-CNB (B) by NB-grown cells of P. putida HS12. Symbols: ■, 3-CNB; ⧫, 4-CNB; ▿, metabolite I; □, metabolite II; ◊, metabolite III.
The above results indicate that NB-grown cells of P. putida HS12 transform 2-, 3-, and 4-CNBs to different metabolites by cometabolism when supplemented with an energy source. Neither ammonia nor chloride ion was detected during the conversion of CNBs by P. putida HS12, which suggests that these metabolites still retain aromatic ring structures with ammonia and chlorine.
Identification of metabolites produced from CNBs.
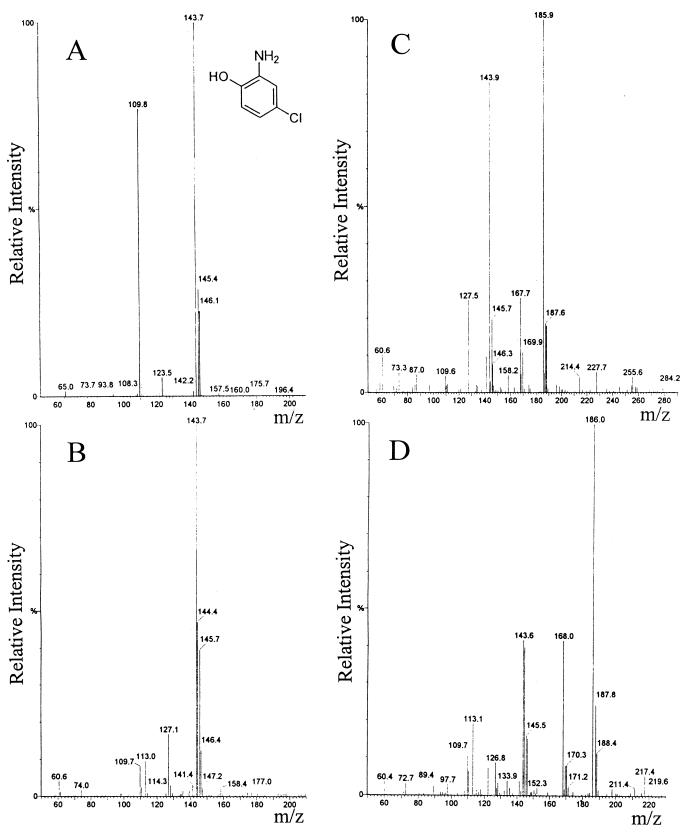
To identify the metabolites formed from CNBs by NB-grown P. putida HS12, the metabolites were separated from the reaction mixture and subjected to HPLC-MS and 1H-NMR analyses. Since NB was found to be degraded via 2-aminophenol through a partial reductive pathway in P. putida HS12, these metabolites were first compared with authentic 4-chloro-2-aminophenol. As shown in Fig. 4A and B, metabolite I showed a mass spectrum almost identical to that of 4-chloro-2-aminophenol. The molecular ion peak of authentic 4-chloro-2-aminophenol was observed at m/z 144, and the peak at m/z 146 resulted from the characteristic M/(M+2) ratio of 3:1 due to the presence of 35Cl and 37Cl. On the other hand, the molecular ion peaks of metabolites II and III appeared at m/z 186 (Fig. 4C and D). The peak at m/z 168 seems to result from the liberation of water from the metabolites, and the base peak at m/z 144 is likely to be due to the loss of an acetyl group. Because chlorine was eliminated from the aromatic ring, no isotopic pattern of chlorine was observed for the fragments lower than m/z 110. From the above results, metabolite I is identified as 4-chloro-2-aminophenol, and metabolites II and III also seem to have structures similar to that of 4-chloro-2-aminophenol, except for an additional acetyl group.
FIG. 4.
Mass spectra of authentic 4-chloro-2-aminophenol (A), metabolite I (B), metabolite II (C), and metabolite III (D).
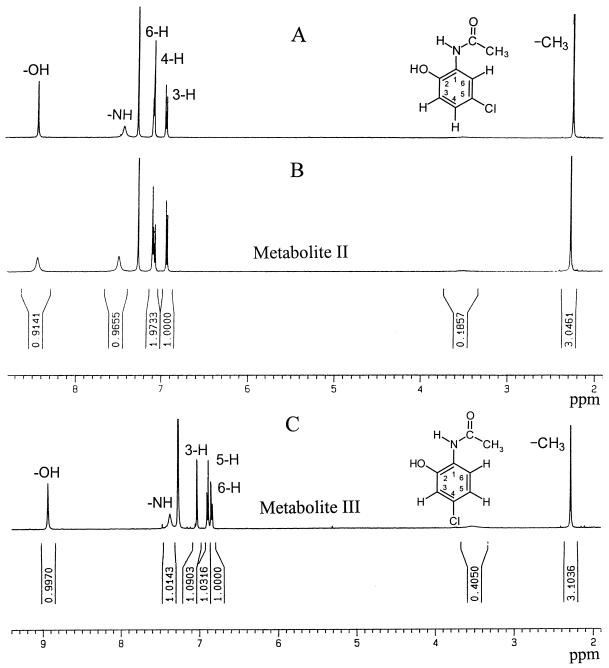
The proton NMR spectrum of metabolite II was found to be identical to that of the predicted compound, 5-chloro-2-hydroxyacetanilide (Fig. 5A and B). In the proton NMR spectra of metabolite II and 5-chloro-2-hydroxyacetanilide, the peak for 3H at 3.27 ppm indicates the presence of a methyl group of acetamide. The doublet at 6.94 ppm shows the splitting of the aromatic proton of H-3 by the neighboring proton of H-4 that also has a doublet at 7.07 ppm. The aromatic protons of H-3 and H-4 have the same coupling constant, and this indicates that these protons are adjacent to each other. The proton of H-6 is not split and appears as a singlet at 7.09 ppm. The downfield absorptions at 7.42 and 8.42 ppm are due to the protons of amino and hydroxyl groups, respectively, showing singlet signals. In the proton NMR spectrum of metabolite III, the protons of the methyl group were also shown at 2.27 ppm (Fig. 5C). The two doublet signals at 6.84 and 6.89 ppm possessing the same coupling constant represent the adjacent aromatic protons of H-5 and H-6. The singlet signals at 7.02, 7.37, and 8.93 ppm are due to the proton of H-3, the proton of the amino group, and the proton of the hydroxyl group, respectively.
FIG. 5.
1H-NMR spectra of chemically synthesized 5-chloro-2-hydroxyacetanilide (A), metabolite II (B), and metabolite III (C).
Isolation of a chlorohydroxyacetanilide-degrading microorganism.
In order to degrade CNBs by a coculture, we attempted to isolate a microorganism which is able to degrade the 5- and 4-chloro-2-hydroxyacetanilides produced from 3- and 4-CNBs by P. putida HS12. A microorganism that degrades 4- and 5-chloro-2-hydroxyacetanilides and releases chloride ion was isolated from soils contaminated with chloroaromatics. The isolated strain was a gram-positive, acid fastness-negative organism and showed an elementary branching-rod-coccus growth cycle. From the analysis of its membrane composition, this strain was found to have meso-diaminopimelic acid, tuberculostearic acid, and glycolate. Arabinose and galactose were mainly observed as sugar components of the membrane. Fatty acid analysis gave a similarity index of 0.648 to Rhodococcus rhodochrous. From the above results, the isolate was identified as Rhodococcus sp. strain HS51.
Degradation of CNBs by a coculture.
For the degradation of CNBs, a coculture of the two isolated strains was carried out. NB-grown cells of P. putida HS12 and chlorobenzene-grown cells of Rhodococcus sp. strain HS51 were combined at a ratio of 1:2 and incubated with 3- or 4-CNB in nitrogen- and chloride-free minimal salts medium in the presence of 10 mM succinate. Since P. putida HS12 was observed to convert CNBs by cometabolism, an additional carbon source was required, and succinate was used to supplement the culture medium. Figure 6 shows the changes in the optical density of total cells and concentrations of CNBs, intermediates, and chloride ion in the coculture medium. The concentration of 3-CNB decreased to 0 in 22 h, and that of metabolite II (5-chloro-2-hydroxyacetanilide) increased and reached a maximum (Fig. 6A). The level of metabolite II gradually lowered due to its uptake by Rhodococcus sp. strain HS51, and at the same time, chloride ion and ammonia accumulated in the culture medium, which indicates mineralization of 3-CNB by the coculture of the two isolated microorganisms. At the beginning of the culture, a decrease in the total biomass was observed, and this was found to be due mainly to lysis of P. putida HS12 by 3-CNB from the viable cell count. After 20 h, 5-chloro-2-hydroxyacetanilide was degraded and the total biomass gradually increased. The ratio of P. putida HS12 to Rhodococcus sp. strain HS51 was estimated to be about 4:3 by viable cell count. Cells of P. putida HS12 showed fast growth on succinate and ammonia liberated from 5-chloro-2-hydroxyacetanilide, but the growth of the Rhodococcus sp. was slow due to the low rate of 5-chloro-2-hydroxyacetanilide degradation. The difference between the populations of the two microorganisms seems to be due mainly to the different growth rates of the microorganisms on succinate. It was revealed that the specific growth rate of P. putida HS12 on succinate was five times that of Rhodococcus sp. strain HS51, and this indicates that most of the succinate was used by P. putida HS12 (data not shown). A similar profile was observed for 4-CNB, except that no significant decrease in total biomass was observed at the beginning of the culture (Fig. 6B).
FIG. 6.
Degradation of 3-CNB (A) and 4-CNB (B) by a coculture of P. putida HS12 and Rhodococcus sp. strain HS51. Symbols: ■, 3-CNB; ⧫, 4-CNB; □, 5-chloro-2-hydroxyacetanilide; ⧫, 4-chloro-2-hydroxyacetanilide; ●, chloride ions; ▴, optical density.
To get some insight into the ring cleavage pathways of P. putida HS12 and Rhodococcus sp. strain HS51 for chlorohydroxyacetanilides, we assayed the activities of enzymes involved in the meta, ortho, and modified ortho cleavage pathways. As shown in Table 1, no catechol-2,3-dioxygenase activity was observed in crude extracts of Rhodococcus sp. strain HS51, but catechol-1,2-dioxygenase activity toward both catechol and 4-chlorocatechol was detected. Meanwhile, P. putida HS12 exhibited only low specific activity of catechol-2,3-dioxygenase toward catechol. Catechol-1,2-dioxygenase in Rhodococcus sp. strain HS51 was induced by chlorobenzene, but no induction of catechol-2,3-dioxygenase by NB in P. putida HS12 occurred. This is probably because catechol-2,3-dioxygenase is not involved in the catabolic pathway of NB. These results strongly imply that Rhodococcus sp. strain HS51 possesses the modified ortho cleavage pathway, whereas P. putida HS12 has only the meta cleavage pathway. Formation of chlorohydroxyacetanilides from CNBs by P. putida HS12 and subsequent degradation of chlorohydroxyacetanilides by Rhodococcus sp. strain HS51 in a coculture also support the above presumption.
TABLE 1.
Enzyme activities in cell extracts
| Enzyme and test substrate | Sp act (μmol/min/mg of protein)
|
|
|---|---|---|
| P. putida HS12a | Rhodococcus sp. strain HS51b | |
| Catechol-1,2-dioxygenase | ||
| Catechol | 0.002 | 0.167 |
| 4-Chlorocatechol | <0.001 | 0.047 |
| Catechol-2,3-dioxygenase, catechol | 0.010 | <0.001 |
Pseudomonas putida HS12 grown on NB was used as the enzyme source.
Rhodococcus sp. strain HS51 grown on chlorobenzene was used as the enzyme source.
DISCUSSION
In this study, we demonstrated that degradation of CNBs is achieved by a coculture of the two isolated strains P. putida HS12 and Rhodococcus sp. strain HS51. NB-grown cells of P. putida HS12 were found to be able to transform 3- and 4-CNBs to 5- and 4-chloro-2-hydroxyacetanilide via 4- and 5-chloro-2-aminophenol, respectively, by cometabolism. From the release of ammonia and the transient accumulation of 2-aminophenol, this microorganism was assumed to possess a partial reductive pathway for NB, similar to that of P. pseudoalcaligenes JS45 reported by Nishino and Spain (22). Nitrobenzene nitroreductase is known to have a rather relaxed substrate specificity, and this enzyme seems to attack 3- and 4-CNBs, resulting in the formation of 3- and 4-chlorohydroxylaminobenzenes. Nitrobenzene nitroreductase in P. pseudoalcaligenes JS52 was reported to transform nitro groups of 2,4,6-trinitrotoluene to the corresponding hydroxylamino groups (7). In the case of P. putida HS12, 4-chloro-2-aminophenol was detected instead of chloroaniline intermediates, which indicates that 3- and 4-chlorohydroxylaminobenzenes undergo an enzyme-catalyzed rearrangement to 4- and 5-chloro-2-aminophenol rather than reduction to the corresponding 3- and 4-chloroanilines. Thus, NB-grown cells of P. putida HS12 seem to transform 3- and 4-CNBs to 4- and 5-chloro-2-aminophenols by using the catalytic enzymes involved in the degradation of NB.
Nitroaromatic compounds are metabolized by two general catabolic pathways, oxidative and reductive pathways. In the oxidative pathway, the nitro group is eliminated as nitrite. Many nitroaromatic compounds, such as 4-chloro-2-nitrophenol (3), NB (11, 14, 23), 2-nitrotoluene (13), 1,3-dinitrobenzene (6), 2,4-dinitrotoluene (33), m-nitrobenzoic acid (21), and o-nitrophenol (36), have been reported to be mineralized via common intermediates, substituted catechols, by an oxygenase-catalyzed oxidative pathway. On the other hand, reduction of nitroaromatic compounds is more complex. Although hydroxylamino aromatic compounds are key intermediates in the reductase-catalyzed reduction of nitroaromatic compounds, three groups of reduction metabolites can be produced from hydroxylamino aromatic compounds. In the first pathway, hydroxylamino aromatics can be transformed to dihydroxyl aromatic compounds and elimination of ammonia occurs at the same time. It has been reported that 4-hydroxylaminobenzoate reduced from 4-nitrobenzoate and 4-nitrotoluene is converted to 3,4-dihydroxybenzoate, which is easily attacked by a dioxygenase-catalyzing ring cleavage reaction (10, 12, 20, 26). In the second pathway, hydroxylamino aromatic compounds can undergo an enzyme-catalyzed rearrangement to aminophenolic compounds. Schenzle et al. reported that while Ralstonia eutropha JMP 134 transformed NB to a mixture of 2-aminophenol and 4-aminophenol, only aminohydroquinone, an ortho-aminophenolic compound, was identified as a metabolite of 3-nitrophenol (29). Similarly, enzyme-catalyzed rearrangement of hydroxylaminobenzene to 2-aminophenol was observed in the partial reductive degradation of NB by P. pseudoalcaligenes JS45 (22). In the third pathway, hydroxylamino aromatic compounds are more reduced to amino aromatic compounds, most of which are not biodegradable and are easily polymerized in the presence of oxygen. The reductive transformation of 2,4-dinitrotoluene and 2,4,6-trinitrotoluene was reported to result in the formation of amino-substituted toluenes (7, 9, 24, 35). It was also reported that Pseudomonas sp. strain CBS3 converts various nitroaromatic compounds to the corresponding amino aromatic compounds under aerobic resting conditions (28). Beunink and Rehm found that 4-chloro-2-nitrophenol is transformed to 4-chloro-2-aminophenol and then completely degraded by a coupled reductive and oxidative pathway (2). On the other hand, a mixture of ortho- and para-aminophenolic compound and aminoaromatic compound were detected as intermediates in the metabolism of 4-CNB by Rhodosporidium sp. (4). In this case, 4-chlorohydroxylaminobenzene partially reduced from 4-CNB is converted to 5-chloro-2-aminophenol, 4-hydroxylaniline, and 4-chloroaniline, which implies that Rhodosporidium sp. performs both complete reduction and enzyme-catalyzed rearrangement of nitroaromatic compounds.
Beunink and Rehm reported that 4-chloro-2-aminophenol is metabolized by Alcaligenes sp. strain TK-2 isolated by the filter-mating technique (2), but the detailed metabolic pathway of 4-chloro-2-aminophenol has not been revealed. Lendenmann and Spain showed that ring cleavage of 4-chloro-2-aminophenol is catalyzed by 2-aminophenol-1,6-dioxygenase and the resulting 2-amino-4-chloromuconic semialdehyde is spontaneously converted to 4-chloropicolinic acid (17). Meanwhile, when NB-grown P. putida HS12 was incubated with 4-chloro-2-aminophenol, stoichiometric formation of 5-chloro-2-hydroxyacetanilide was observed (data not shown). Neither 2-amino-4-chloromuconic semialdehyde nor 4-chloropicolinic acid was detected.
Except for some compounds, such as 4-chloro-2-nitrophenol (3), NB (22), and 3-nitrophenol (29), most of the arylamines and some of the aminophenolic compounds reduced from nitroaromatics have been reported to be acetylated rather than degraded further. Acetylation of the aromatic amine groups has been observed in the metabolism of 4-CNB (4), 4-chloro-2-nitrophenol (2), 2,4-dinitrotoluene (24), 3-nitrophenol (29), and 2,4,6-trinitrotoluene (9). Even though 4-chloro-2-hydroxyacetanilide was reported in the metabolism of 4-chloroaniline by Fusarium oxysporum (16) and that of 4-CNB by Rhodosporidium sp. (4), this compound is not a major metabolite, and other acetylated metabolites, including 4-chloroacetanilide and 4-hydroxyacetanilide, were also detected. On the other hand, metabolism of 3- and 4-CNBs by NB-grown P. putida HS12 is very simple and clear. In other words, 3- and 4-CNBs were converted to the corresponding 5- and 4-chloro-2-hydroxyacetanilides in a stoichiometric manner, and no other metabolite was detected. From this observation, it is likely that NB-grown cells of P. putida HS12 transform 3- and 4-CNBs to 4- and 5-chloro-2-aminophenols through a partial reductive pathway, and these metabolites are further acetylated to produce 5- and 4-chloro-2-hydroxyacetanilides as final metabolites. Acetamide groups are known to be less toxic than amino groups, and acetylation of amino groups has been considered as a detoxification mechanism in microorganisms (34).
The catabolic pathway for most acetylated metabolites, including 4-acetamide-2-aminotoluene, 4-acetamide-2-amino-6-nitrotoluene, 4-chloroacetanilide, 4-chloro-2-hydroxyacetanilide, 4-hydroxyacetanilide, and N-acetylaminohydroquinone, has not been elucidated yet. In this study, we observed degradation of 4- and 5-chloro-2-hydroxyacetanilides by Rhodococcus sp. strain HS51. Although extradiol ring cleavage of chloroaromatics has been reported recently (1, 19), most of the chloroaromatic compounds are known to be degraded through the modified ortho cleavage pathway (5, 27, 30, 32). 4-Chloro- and 5-chloro-2-hydroxyacetanilides, as chloroaromatics, were metabolized not by P. putida HS2 carrying the meta cleavage pathway but by Rhodococcus sp. strain HS51 possessing the modified ortho cleavage pathway. However, slow degradation of chlorohydroxyacetanilides suggests that 4- and 5-chloro-2-hydroxyacetanilides may not be physiological substrates for Rhodococcus sp. strain HS51. When considering the chemical structure of chlorohydroxyacetanilides, ortho ring cleavage of chlorohydroxyacetanilides by catechol-1,2-dioxygenase of Rhodococcus sp. strain HS51 seems to be an unusual case, and more detailed study of the ring cleavage reaction mediated by Rhodococcus sp. strain HS51 is required.
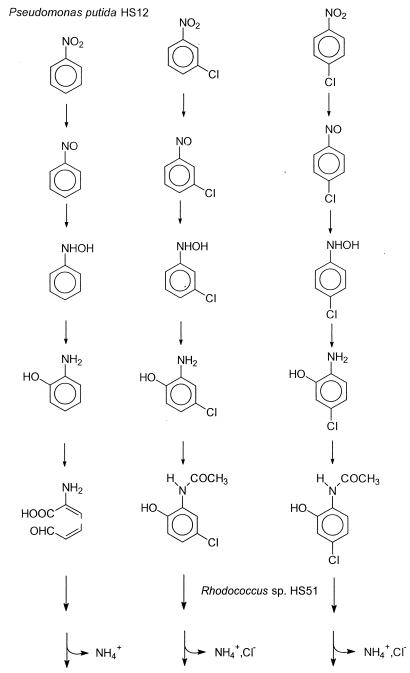
Based on the identification of metabolic intermediates formed by P. putida HS12 and Rhodococcus sp. strain HS51, we propose here the metabolic pathway for 3- and 4-CNBs by a coculture of the two isolated strains (Fig. 7). In other words, 3- and 4-CNBs are mineralized via 5- and 4-chloro-2-hydroxyacetanilides by partial reduction, acetylation, and subsequent ring cleavage in a coculture of P. putida HS12 and Rhodococcus sp. strain HS51. In this metabolism, the degradation rates of 4- and 5-chloro-2-hydroxyacetanilides were found to be quite low compared to the transformation rates of 3- and 4-chloronitrobenzenes, suggesting that the ring cleavage reaction by Rhodococcus sp. strain HS51 is a limiting step in the degradation of CNBs. Either isolation of a microorganism with high ring cleavage activity or incorporation of a gene manipulation technique would improve the overall rate of degradation by a coculture.
FIG. 7.
Proposed pathway for the mineralization of 3- and 4-CNBs by the sequential action of the two isolated strains, P. putida HS12 and Rhodococcus sp. strain HS51.
In this work, degradation of CNBs was achieved by a coculture of Rhodococcus sp. strain HS51 and NB-grown P. putida HS12. Since CNBs do not induce the enzymes involved in their breakdown in P. putida HS12, a coculture cannot be maintained on succinate and CNBs. Therefore, NB-grown P. putida HS12 was initially used for the coculture. However, in reality, CNBs are synthesized from NB by chlorination and usually coexist with NB, which is an inducer of the catabolic pathway of NB in P. putida HS12. In this regard, the coculture system developed here is thought to be useful in removing CNBs from the environment.
REFERENCES
- 1.Arensdorf J J, Focht D D. A meta cleavage pathway for 4-chlorobenzoate, an intermediate in the metabolism of 4-chlorobiphenyl by Pseudomonas cepacia P166. Appl Environ Microbiol. 1995;61:443–447. doi: 10.1128/aem.61.2.443-447.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beunink J, Rehm H-J. Coupled reductive and oxidative degradation of 4-chloro-2-nitrophenol by a co-immobilized mixed culture system. Appl Microbiol Biotechnol. 1990;34:108–115. doi: 10.1007/BF00170933. [DOI] [PubMed] [Google Scholar]
- 3.Bruhn C, Bayly R C, Knackmuss H-J. The in vivo construction of 4-chloro-2-nitrophenol assimilatory bacteria. Arch Microbiol. 1988;150:171–177. [Google Scholar]
- 4.Corbett M D, Corbett B R. Metabolism of 4-chloronitrobenzene by the yeast Rhodosporidium sp. Appl Environ Microbiol. 1981;41:942–949. doi: 10.1128/aem.41.4.942-949.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De Bont J A M, Vorage M J A W, Hartmans S, van den Tweel W J J V. Microbial degradation of 1,3-dichlorobenzene. Appl Environ Microbiol. 1986;52:677–680. doi: 10.1128/aem.52.4.677-680.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dickel O, Knackmuss H-J. Catabolism of 1,3-dinitrobenzene by Rhodococcus sp. QT-1. Arch Microbiol. 1991;157:76–79. doi: 10.1007/BF00245339. [DOI] [PubMed] [Google Scholar]
- 7.Fiorella P D, Spain J C. Transformation of 2,4,6-trinitrotoluene by Pseudomonas putida JS52. Appl Environ Microbiol. 1997;63:2007–2015. doi: 10.1128/aem.63.5.2007-2015.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gerhardt P, Murray R G E, Wood W A, Krieg N R, editors. Methods for general and molecular bacteriology. Washington, D.C: American Society for Microbiology; 1994. pp. 541–542. [Google Scholar]
- 9.Gilcrease P C, Murphy V G. Bioconversion of 2,4-diamino-6-nitrotoluene to a novel metabolite under anoxic and aerobic conditions. Appl Environ Microbiol. 1995;61:4209–4212. doi: 10.1128/aem.61.12.4209-4214.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Groenewegen P E J, Breeuwer P, van Helvoort J M L M, Langenhoff A A M, de Vries F P, de Bont J A M. Novel degradative pathway of 4-nitrobenzoate in Comamonas acidovorans NBA-10. J Gen Microbiol. 1992;138:1599–1605. doi: 10.1099/00221287-138-8-1599. [DOI] [PubMed] [Google Scholar]
- 11.Haigler B E, Spain J C. Biotransformation of nitrobenzene by bacteria containing toluene degradative pathways. Appl Environ Microbiol. 1991;57:3156–3162. doi: 10.1128/aem.57.11.3156-3162.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haigler B E, Spain J C. Biodegradation of 4-nitrotoluene by Pseudomonas sp. strain 4NT. Appl Environ Microbiol. 1993;59:2239–2243. doi: 10.1128/aem.59.7.2239-2243.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haigler B E, Wallace W H, Spain J C. Biodegradation of 2-nitrotoluene by Pseudomonas sp. strain JS42. Appl Environ Microbiol. 1994;60:3466–3469. doi: 10.1128/aem.60.9.3466-3469.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jung K-H, Lee J-Y, Kim H-S. Biodegradation of nitrobenzene through a hybrid pathway in Pseudomonas putida. Biotechnol Bioeng. 1995;48:625–630. doi: 10.1002/bit.260480610. [DOI] [PubMed] [Google Scholar]
- 15.Katz L, Cohen M S. Benzoxazole derivatives. I. 2-Mercaptobenzoxazoles. J Org Chem. 1954;19:758–766. [Google Scholar]
- 16.Kaufman D D, Plimmer J R, Klingebiel U I. Microbial oxidation of 4-chloroaniline. J Agric Food Chem. 1973;21:127–132. doi: 10.1021/jf60185a028. [DOI] [PubMed] [Google Scholar]
- 17.Lendenmann U, Spain J C. 2-Aminophenol 1,6-dioxygenase: a novel aromatic ring cleavage enzyme purified from Pseudomonas pseudoalcaligenes JS45. J Bacteriol. 1996;178:6227–6232. doi: 10.1128/jb.178.21.6227-6232.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Livingston A G. A novel membrane bioreactor for detoxifying industrial wastewater. II. Biodegradation of 3-chloronitrobenzene in an industrially produced wastewater. Biotechnol Bioeng. 1993;41:927–936. doi: 10.1002/bit.260411003. [DOI] [PubMed] [Google Scholar]
- 19.Mars A E, Kasberg T, Kaschabek S R, van Agteren M H, Janssen D B, Reineke W. Microbial degradation of chloroaromatics: use of the meta-cleavage pathway for mineralization of chlorobenzene. J Bacteriol. 1997;179:4530–4537. doi: 10.1128/jb.179.14.4530-4537.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Michan C, Delgado A, Haidour A, Lucchesi G, Ramos J L. In vivo construction of a hybrid pathway for metabolism of 4-nitrotoluene in Pseudomonas fluorescens. J Bacteriol. 1997;179:3036–3038. doi: 10.1128/jb.179.9.3036-3038.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nadeau L J, Spain J C. Bacterial degradation of m-nitrobenzoic acid. Appl Environ Microbiol. 1995;61:840–843. doi: 10.1128/aem.61.2.840-843.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nishino S F, Spain J C. Degradation of nitrobenzene by a Pseudomonas pseudoalcaligenes. Appl Environ Microbiol. 1993;59:2520–2525. doi: 10.1128/aem.59.8.2520-2525.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nishino S F, Spain J C. Oxidative pathway for the biodegradation of nitrobenzene by Comamonas sp. strain JS765. Appl Environ Microbiol. 1995;61:2308–2313. doi: 10.1128/aem.61.6.2308-2313.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Noguera D R, Freedman D L. Reduction and acetylation of 2,4-dinitrotoluene by a Pseudomonas aeruginosa strain. Appl Environ Microbiol. 1996;62:2257–2263. doi: 10.1128/aem.62.7.2257-2263.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Palleroni N J. Gram-negative aerobic rods and cocci. In: Krieg N R, Holt J G, editors. Bergey’s manual of systematic bacteriology. Vol. 1. Baltimore, Md: The Williams & Wilkins Co.; 1984. pp. 140–199. [Google Scholar]
- 26.Rhys-Williams W, Taylor S C, Williams P A. A novel pathway for the catabolism of 4-nitrotoluene by Pseudomonas. J Gen Microbiol. 1993;139:1967–1972. doi: 10.1099/00221287-139-9-1967. [DOI] [PubMed] [Google Scholar]
- 27.Sander P, Wittich R-M, Fortnagel P, Wilkes H, Francke W. Degradation of 1,2,4-trichloro- and 1,2,4,5-tetrachlorobenzene by Pseudomonas strains. Appl Environ Microbiol. 1991;57:1430–1440. doi: 10.1128/aem.57.5.1430-1440.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schackmann A, Muller R. Reduction of nitroaromatic compounds by different Pseudomonas species under aerobic conditions. Appl Microbiol Biotechnol. 1991;34:809–813. [Google Scholar]
- 29.Schenzle A, Lenke H, Fischer P, Williams P A, Knackmuss H-J. Catabolism of 3-nitrophenol by Ralstonia eutropha JMP 134. Appl Environ Microbiol. 1997;63:1421–1427. doi: 10.1128/aem.63.4.1421-1427.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schraa G, Boone M L, Jetten M S M, van Neerven A R W, Colberg P J, Zehnder A J B. Degradation of 1,4-dichlorobenzene by Alcaligenes sp. strain A175. Appl Environ Microbiol. 1986;52:1374–1381. doi: 10.1128/aem.52.6.1374-1381.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smibert R M, Krieg N R. Phenotypic characterization. In: Gerhardt P, Murray R G E, Wood W A, Krieg N R, editors. Methods for general and molecular bacteriology. Washington, D.C: American Society for Microbiology; 1994. pp. 607–654. [Google Scholar]
- 32.Spain J C, Nishino S F. Degradation of 1,4-dichlorobenzene by a Pseudomonas sp. Appl Environ Microbiol. 1987;53:1010–1019. doi: 10.1128/aem.53.5.1010-1019.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Spanggord R J, Spain J C, Nishino S F, Mortelmans K E. Biodegradation of 2,4-dinitrotoluene by a Pseudomonas sp. Appl Environ Microbiol. 1991;57:3200–3205. doi: 10.1128/aem.57.11.3200-3205.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tweedy B G, Loeppky C, Ross J A. Metobromuron: acetylation of the aniline moiety as a detoxification mechanism. Science. 1970;168:482–483. doi: 10.1126/science.168.3930.482. [DOI] [PubMed] [Google Scholar]
- 35.Vanderberg L A, Perry J J, Unkefer P J. Catabolism of 2,4,6-trinitrotoluene by Mycobacterium vaccae. Appl Microbiol Biotechnol. 1995;43:937–945. doi: 10.1007/BF02431931. [DOI] [PubMed] [Google Scholar]
- 36.Zeyer J, Kearney P C. Degradation of o-nitrophenol and m-nitrophenol by a Pseudomonas putida. J Agric Food Chem. 1984;32:238–242. [Google Scholar]