Abstract
We present a genome assembly from an individual male Arvicola amphibius (the European water vole; Chordata; Mammalia; Rodentia; Cricetidae). The genome sequence is 2.30 gigabases in span. The majority of the assembly is scaffolded into 18 chromosomal pseudomolecules, including the X sex chromosome. Gene annotation of this assembly on Ensembl has identified 21,394 protein coding genes.
Keywords: Arvicola amphibius, European water vole, genome sequence, chromosomal
Species taxonomy
Eukaryota; Metazoa; Chordata; Craniata; Vertebrata; Euteleostomi; Mammalia; Eutheria; Euarchontoglires; Glires; Rodentia; Myomorpha; Muroidea; Cricetidae; Arvicolinae; Arvicola; Arvicola amphibius Linnaeus 1758 (NCBI:txid1047088).
Introduction
The European water vole, Arvicola amphibius Linnaeus 1758, is a small semi-aquatic mammal that lives on the banks of freshwater water courses and in wetlands. A. amphibius is native to Europe, west Asia, Russia and Kazakhstan. While the IUCN Red List of Threatened Species reports that A. amphibius is of “least concern” worldwide, populations in the United Kingdom have declined to such an extent that the species is considered nationally endangered ( Mathews & Harrower, 2020) owing to habitat loss and predation by the American mink, Neovison vison, an invasive alien species. An estimate by Natural England put the 2018 UK population of A. amphibius at 132,000, down from 7.3 million in 1990 ( Strachan, 2004). Water voles are absent from Ireland. There have been a number of conservation projects in the UK aimed at supporting populations of A. amphibius, including efforts at habitat restoration and to control the population of American mink ( Bryce et al., 2011). There are also efforts to reintroduce the water vole in a number of restored urban and wild habitats. This genome sequence will be of use as a reference for researchers that wish to assess the population genomics of A. amphibius and manage reintroductions.
Genome sequence report
The genome was sequenced from a single male A. amphibius collected from the Wildwood Trust, Herne Common, Kent, UK. A total of 45-fold coverage in Pacific Biosciences single-molecule long reads (N50 20 kb) and 52-fold coverage in 10X Genomics read clouds (from molecules with an estimated N50 of 155 kb) were generated. Primary assembly contigs were scaffolded with chromosome conformation Hi-C data. The final assembly has a total length of 2.298 Gb in 216 sequence scaffolds with a scaffold N50 of 138.7 Mb ( Table 1). The majority, 99.4%, of the assembly sequence was assigned to 19 chromosomal-level scaffolds, representing 17 autosomes (numbered by sequence length apart from chromosome 12, which is larger because the previous version of the assembly, mArvAmp1.1, mistakenly labelled this as two separate chromosomes), and the X sex chromosome ( Figure 1– Figure 4; Table 2). The assembly has a BUSCO ( Simao et al., 2015) v5.0.0 completeness of 96.1% using the mammalia_odb10 reference set. While not fully phased, the assembly deposited is of one haplotype. Contigs corresponding to the second haplotype have also been deposited.
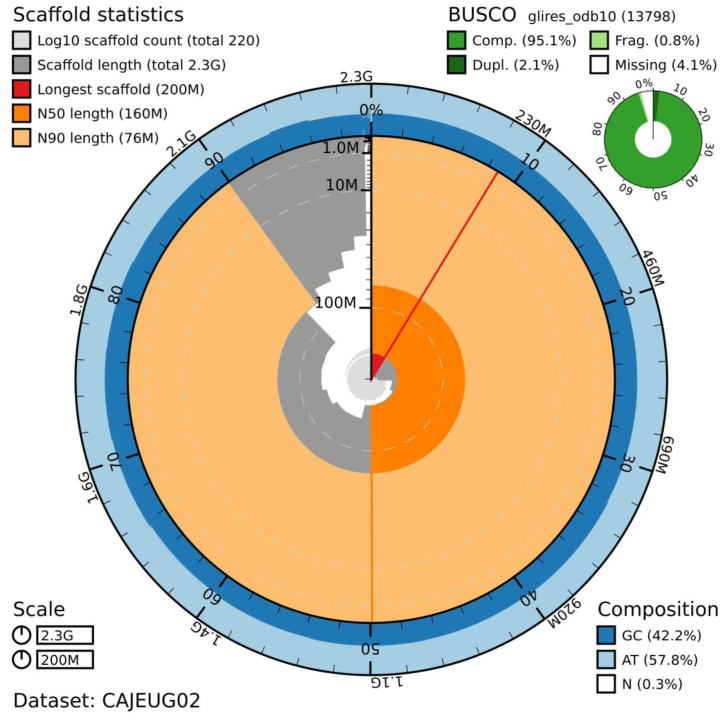
Figure 1. Genome assembly of Arvicola amphibius, mArvAmp1.2: metrics.
The BlobToolKit Snailplot shows N50 metrics and BUSCO gene completeness. An interactive version of this figure is available at https://blobtoolkit.genomehubs.org/view/Arvicola%20amphibius/dataset/CAJEUG02/snail.
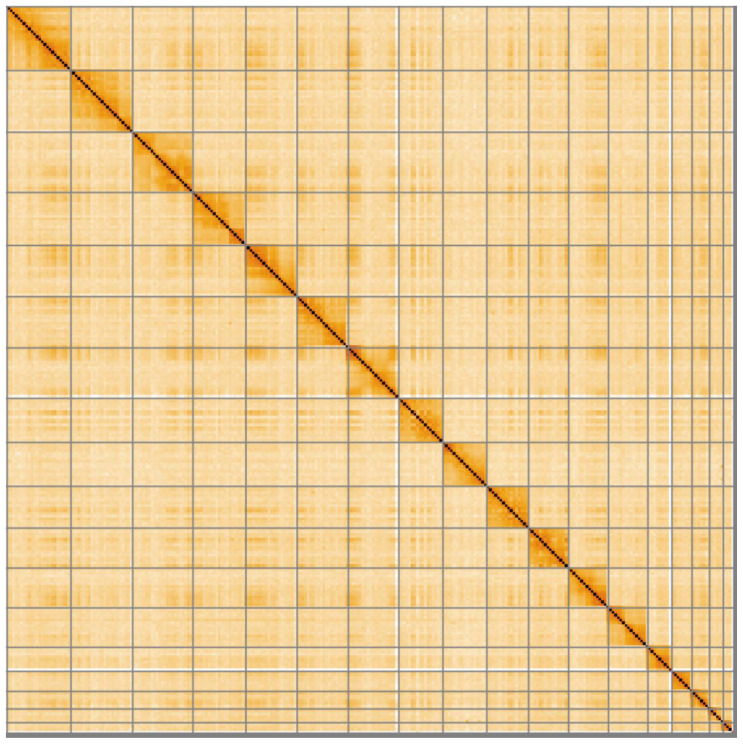
Figure 4. Genome assembly of Arvicola amphibius, mArvAmp1.2: Hi-C contact map.
Hi-C contact map of the mArvAmp1 assembly, visualised in HiGlass.
Table 1. Genome data for Arvicola amphibius, mArvAmp1.2.
| Project accession data | |
|---|---|
| Assembly identifier | mArvAmp1.2 |
| Species | Arvicola amphibius |
| Specimen | mArvAmp1 |
| NCBI taxonomy ID | txid1047088 |
| BioProject | PRJEB39550 |
| BioSample ID | SAMEA994740 |
| Isolate information | Male; blood sample |
| Raw data accessions | |
| PacificBiosciences SEQUEL I | ERX3146757-ERX3146763 |
| 10X Genomics Illumina | ERX3163119-ERX3163121, ERX3341539-ERX3341546 |
| Hi-C Illumina | ERX3338011, ERX3338012 |
| BioNano | ERZ1392829 |
| Genome assembly | |
| Assembly accession | GCA_903992535.2 |
| Accession of alternate haplotype | GCA_903992525.1 |
| Span (Mb) | 2,298 |
| Number of contigs | 1,085 |
| Contig N50 length (Mb) | 5.4 |
| Number of scaffolds | 216 |
| Scaffold N50 length (Mb) | 138.7 |
| Longest scaffold (Mb) | 199.8 |
| BUSCO * genome score | C:96.1%[S:94.1%,D:2.0%],F:0.8%,M:3.1%,n:9226 |
| Genome annotation | |
| Number of protein-coding genes | 21,394 |
| Average length of protein-coding gene (bp) | 1,700 |
| Average number of exons per gene | 11 |
| Average exon size (bp) | 208 |
| Average intron size (bp) | 4,995 |
* BUSCO scores based on the mammalia_odb10 BUSCO set using v5.0.0. C= complete [S= single copy, D=duplicated], F=fragmented, M=missing, n=number of orthologues in comparison. A full set of BUSCO scores is available at https://blobtoolkit.genomehubs.org/view/Arvicola%20amphibius/dataset/CAJEUG02/busco.
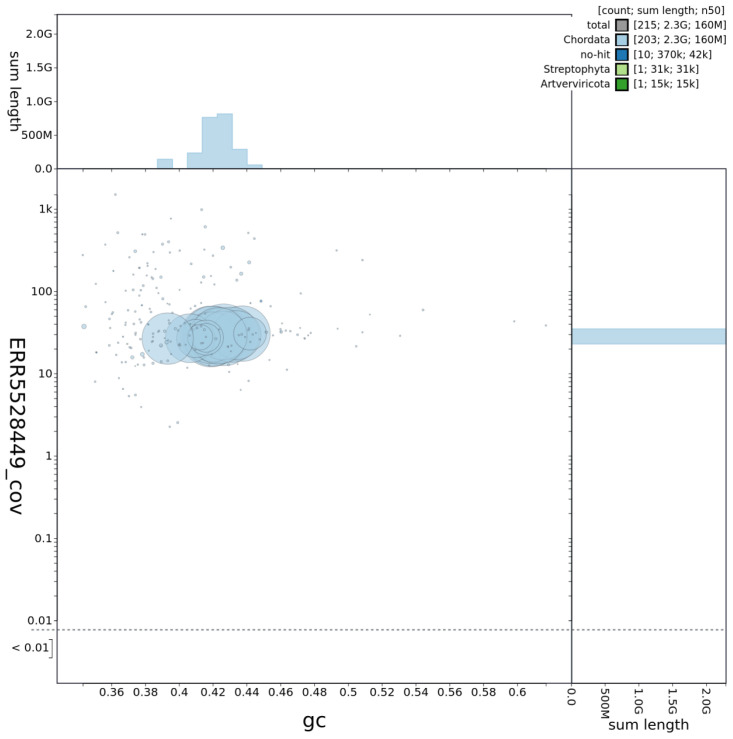
Figure 2. Genome assembly of Arvicola amphibius, mArvAmp1.2: GC coverage.
BlobToolKit GC-coverage plot. An interactive version of this figure is available at https://blobtoolkit.genomehubs.org/view/Arvicola%20amphibius/dataset/CAJEUG02/blob.
Table 2. Chromosomal pseudomolecules in the genome assembly of Arvicola amphibius, mArvAmp1.2.
| INSDC accession | Chromosome | Size (Mb) | GC% |
|---|---|---|---|
| LR862380.1 | 1 | 200.53 | 42.7 |
| LR862381.2 | 2 | 193.96 | 41.9 |
| LR862382.1 | 3 | 189.60 | 42.0 |
| LR862383.1 | 4 | 161.33 | 43.8 |
| LR862384.1 | 5 | 160.72 | 42.6 |
| LR862385.2 | 6 | 158.92 | 43.1 |
| LR862386.1 | 7 | 138.66 | 41.9 |
| LR862388.1 | 8 | 131.41 | 42.0 |
| LR862389.2 | 9 | 125.83 | 43.4 |
| LR862390.1 | 10 | 125.09 | 42.6 |
| LR862391.2 | 11 | 123.99 | 40.7 |
| LR862392.2 | 12 | 166.75 | 42.3 |
| LR862393.1 | 13 | 75.71 | 41.0 |
| LR862394.1 | 14 | 63.16 | 41.6 |
| LR862395.1 | 15 | 55.45 | 44.2 |
| LR862397.2 | 17 | 42.65 | 41.6 |
| LR862398.1 | 18 | 33.21 | 41.2 |
| LR862387.1 | X | 137.70 | 39.3 |
Gene annotation
The Ensembl gene annotation system ( Aken et al., 2016) was used to generate annotation for an earlier version of the Arvicola amphibius assembly ( GCA_903992535.1). Annotation was created primarily through alignment of transcriptomic data to the genome, with gap filling via protein to-genome alignments of a select set of vertebrate proteins from UniProt ( UniProt Consortium, 2019) and coordinate mapping of GENCODE ( Frankish et al., 2019) mouse reference annotations via a pairwise whole genome alignment. The resulting Ensembl annotation includes 34,750 transcripts assigned to 21,394 coding and 2,252 non-coding genes ( Arvicola amphibius - Ensembl Rapid Release).
Methods
A blood sample was taken from a live male A. amphibius specimen that was part of the captive breeding population of Wildwood Trust, Herne Common, Kent, UK (latitude 51.33181, longitude 1.11443). DNA was extracted using an agarose plug extraction from a blood sample following the Bionano Prep Animal Tissue DNA Isolation Soft Tissue Protocol. Pacific Biosciences CLR long read and 10X Genomics read cloud sequencing libraries were constructed according to the manufacturers’ instructions. Sequencing was performed by the Scientific Operations DNA Pipelines at the Wellcome Sanger Institute on Pacific Biosciences SEQUEL I and Illumina HiSeq X instruments. Hi-C data were generated using the Dovetail v1.0 kit and sequenced on HiSeq X. Ultra-high molecular weight DNA was extracted using the Bionano Prep Animal Tissue DNA Isolation Soft Tissue Protocol and assessed by pulsed field gel and Qubit 2 fluorimetry. DNA was labeled for Bionano Genomics optical mapping following the Bionano Prep Direct Label and Stain (DLS) Protocol and run on one Saphyr instrument chip flowcell.
Assembly was carried out following the Vertebrate Genome Project pipeline v1.6 ( Rhie et al., 2020) with Falcon-unzip ( Chin et al., 2016), haplotypic duplication was identified and removed with purge_dups ( Guan et al., 2020) and a first round of scaffolding carried out with 10X Genomics read clouds using scaff10x. Hybrid scaffolding was performed using the BioNano DLE-1 data and BioNano Solve. Scaffolding with Hi-C data ( Rao et al., 2014) was carried out with SALSA2 ( Ghurye et al., 2019). The Hi-C scaffolded assembly was polished with arrow using the PacBio data, then polished with the 10X Genomics Illumina data by aligning to the assembly with longranger align, calling variants with freebayes ( Garrison & Marth, 2012) and applying homozygous non-reference edits using bcftools consensus. Two rounds of the Illumina polishing were applied. The assembly was checked for contamination and corrected using the gEVAL system ( Chow et al., 2016) as described previously ( Howe et al., 2021). Manual curation was performed using evidence from Bionano (using the Bionano Access viewer), using HiGlass and Pretext, and by taking marker data and inspecting 10X barcode overlap using longranger. Figure 1– Figure 3 were generated using BlobToolKit ( Challis et al., 2020). Table 3 contains a list of all software tool versions used, where appropriate.
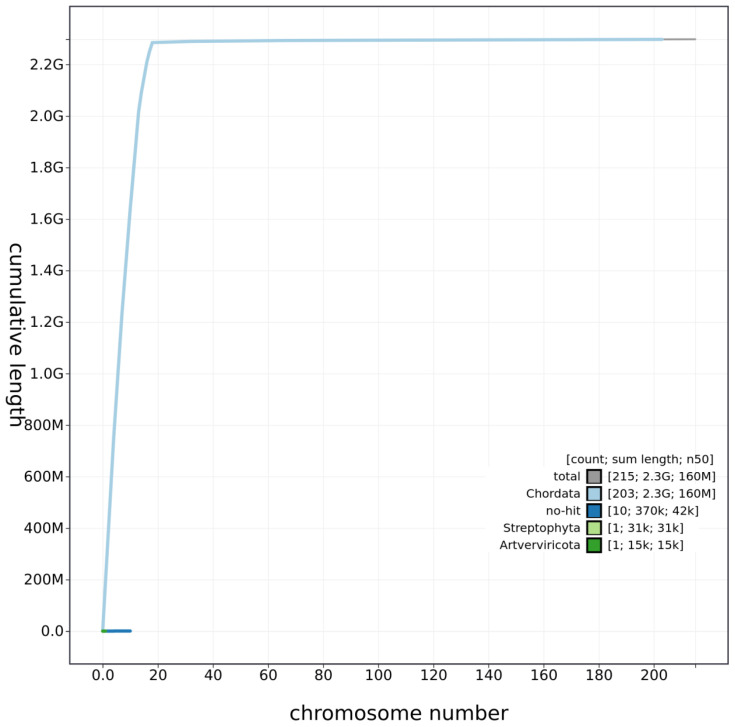
Figure 3. Genome assembly of Arvicola amphibius, mArvAmp1.2: cumulative sequence.
BlobToolKit cumulative sequence plot. An interactive version of this figure is available at https://blobtoolkit.genomehubs.org/view/Arvicola%20amphibius/dataset/CAJEUG02/cumulative.
Table 3. Software tools used.
| Software tool | Version | Source |
|---|---|---|
| Falcon-unzip | falcon-kit 1.8.0 | ( Chin et al., 2016) |
| purge_dups | 1.2.3-b542dbf | ( Guan et al., 2020) |
| SALSA2 | 2.2-14-g974589f | ( Ghurye et al., 2019) |
| scaff10x | 4.2 | https://github.com/wtsi-hpag/Scaff10X |
| Bionano Solve | 3.3_10252018 | N/A |
| arrow | gcpp 1.9.0-SL-release-
8.0.0+1-37-gd7b188d |
https://github.com/PacificBiosciences/GenomicConsensus |
| longranger align | 2.2.2 |
https://support.10xgenomics.com/genome-exome/software/
pipelines/latest/advanced/other-pipelines |
| freebayes | 1.3.1-17-gaa2ace8 | ( Garrison & Marth, 2012) |
| bcftools consensus | 1.9-78-gb7e4ba9 | http://samtools.github.io/bcftools/bcftools.html |
| gEVAL | N/A | ( Chow et al., 2016) |
| HiGlass | 1.11.6 | ( Kerpedjiev et al., 2018) |
| PretextView | 0.0.4 | https://github.com/wtsi-hpag/PretextMap |
| BlobToolKit | 2.5 | ( Challis et al., 2020) |
Data availability
Underlying data
European Nucleotide Archive: Arvicola amphibius (European water vole) genome assembly, mArvAmp1. Accession number PRJEB39550.
The genome sequence is released openly for reuse. The Arvicola amphibius genome sequencing initiative is part of the Wellcome Sanger Institute’s “ 25 genomes for 25 years” project. It is also part of the Vertebrate Genome Project (VGP) ordinal references programme and the Darwin Tree of Life (DToL) project. All raw data and the assembly have been deposited in the ENA. The genome will be annotated and presented through the Ensembl pipeline at the European Bioinformatics Institute. Raw data and assembly accession identifiers are reported in Table 1.
Acknowledgements
We thank Mike Stratton and Julia Wilson for their continuing support for the 25 genomes for 25 years project.
Funding Statement
This work was supported by the Wellcome Trust through core funding to the Wellcome Sanger Institute (206194) and the Darwin Tree of Life Discretionary Award (218328).
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
[version 1; peer review: 3 approved]
References
- Aken BL, Ayling S, Barrell D, et al. : The Ensembl Gene Annotation System. Database (Oxford). 2016;2016:baw093. 10.1093/database/baw093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryce R, Oliver MK, Davies L, et al. : Turning Back the Tide of American Mink Invasion at an Unprecedented Scale through Community Participation and Adaptive Management. Biological Conservation. 2011;144(1):575–83. 10.1016/j.biocon.2010.10.013 [DOI] [Google Scholar]
- Challis R, Richards E, Rajan J, et al. : BlobToolKit - Interactive Quality Assessment of Genome Assemblies. G3 (Bethesda). 2020;10(4):1361–74. 10.1534/g3.119.400908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin CS, Peluso P, Sedlazeck FJ, et al. : Phased Diploid Genome Assembly with Single-Molecule Real-Time Sequencing. Nat Methods. 2016;13(12):1050–54. 10.1038/nmeth.4035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow W, Brugger K, Caccamo M, et al. : gEVAL — a Web-Based Browser for Evaluating Genome Assemblies. Bioinformatics. 2016;32(16):2508–10. 10.1093/bioinformatics/btw159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankish A, Diekhans M, Ferreira AM, et al. : GENCODE Reference Annotation for the Human and Mouse Genomes. Nucleic Acids Res. 2019;47(D1):D766–73. 10.1093/nar/gky955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrison E, Marth G: Haplotype-Based Variant Detection from Short-Read Sequencing. arXiv:1207.3907. 2012. Reference Source [Google Scholar]
- Ghurye J, Rhie A, Walenz BP, et al. : Integrating Hi-C Links with Assembly Graphs for Chromosome-Scale Assembly. PLoS Comput Biol. 2019;15(8):e1007273. 10.1371/journal.pcbi.1007273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan D, McCarthy SA, Wood J, et al. : Identifying and Removing Haplotypic Duplication in Primary Genome Assemblies. Bioinformatics. 2020;36(9):2896–98. 10.1093/bioinformatics/btaa025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe K, Chow W, Collins J, et al. : Significantly Improving the Quality of Genome Assemblies through Curation. Gigascience. 2021;10(1):giaa153. 10.1093/gigascience/giaa153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerpedjiev P, Abdennur N, Lekschas F, et al. : HiGlass: Web-Based Visual Exploration and Analysis of Genome Interaction Maps. Genome Biol. 2018;19(1):125. 10.1186/s13059-018-1486-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews F, Harrower C: IUCN - compliant Red List for Britain’s Terrestrial Mammals.Assessment by the Mammal Society under contract to Natural England, Natural Resources Wales and Scottish Natural Heritage. Natural England, Peterborough,2020. Reference Source [Google Scholar]
- Rao SSP, Huntley MH, Durand NC, et al. : A 3D Map of the Human Genome at Kilobase Resolution Reveals Principles of Chromatin Looping. Cell. 2014;159(7):1665–80. 10.1016/j.cell.2014.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhie A, McCarthy SA, Fedrigo O, et al. : Towards Complete and Error-Free Genome Assemblies of All Vertebrate Species. bioRxiv. 2020; 2020.05.22.110833. 10.1101/2020.05.22.110833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simão FA, Waterhouse RM, Ioannidis P, et al. : BUSCO: Assessing Genome Assembly and Annotation Completeness with Single-Copy Orthologs. Bioinformatics. 2015;31(19):3210–12. 10.1093/bioinformatics/btv351 [DOI] [PubMed] [Google Scholar]
- Strachan R: Conserving Water Voles: Britain’s Fastest Declining Mammal. Water Environ J. 2004;18(1):1–4. 10.1111/j.1747-6593.2004.tb00483.x [DOI] [Google Scholar]
- UniProt Consortium: UniProt: A Worldwide Hub of Protein Knowledge. Nucleic Acids Res. 2019;47(D1):D506–15. 10.1093/nar/gky1049 [DOI] [PMC free article] [PubMed] [Google Scholar]