Abstract
Background
Non-coding RNAs (ncRNAs), including small nucleolar RNAs (snoRNAs), are widely involved in the physiological and pathological processes of human beings. While up to date, although considerable progress has been achieved in ncRNA-related pathogenesis of non-small cell lung cancer (NSCLC), the underlying mechanisms and biological significance of snoRNAs in NSCLC still need to be further clarified.
Methods
Quantitative real-time polymerase chain reaction or RNAscope was performed to verify the expression of Small Nucleolar RNA, H/ACA Box 38B (SNORA38B) in NSCLC cell lines or clinical samples. BALB/c nude mice xenograft model or C57BL/6J mice syngeneic tumor model were estimated to detect the effects of SNORA38B in tumor growth or tumor immune microenvironment in vivo. Cytometry by time of flight, enzyme-linked immunosorbent assay and flow cytometry assay were conducted to clarify the effects and mechanisms of SNORA38B-mediated tumor immunosuppressive microenvironment. The binding activity between SNORA38B and E2F transcription factor 1(E2F1) was detected by RNA immunoprecipitation and RNA pull-down assays. Then, bioinformatics analysis and chromatin immunoprecipitation were utilized to demonstrate the regulation of GRB2-associated-binding protein 2 (GAB2) by E2F1. Moreover, the combinatorial treatment of SNORA38B locked nucleic acid (LNA) and immune checkpoint blockade (ICB) was used to treat murine Lewis lung carcinoma-derived tumor burden C57BL/6J mice to clarify the effectiveness of targeting SNORA38B in NSCLC immunotherapy.
Results
SNORA38B was found highly expressed in NSCLC tissues and cell lines, and associated with worse prognosis. Further results showed that SNORA38B functioned as an oncogene via facilitating cell proliferation, migration, invasion, and inhibiting cell apoptosis in vitro and promoting tumorigenesis of NSCLC cells in vivo. SNORA38B could also recruit the CD4+FOXP3+ regulatory T cells by triggering tumor cells to secrete interleukin 10, which in turn reduced the infiltration of CD3+CD8+ T cells in NSCLC tumor microenvironment (TME), favoring tumor progression and poorer immune efficacy. Mechanistically, SNORA38B mainly distributed in the nucleus, and promoted NSCLC progression by regulating GAB2 transcription to activate protein kinase B (AKT)/mammalian target of rapamycin (mTOR) pathway through directly binding with E2F1. Moreover, we found that SNORA38B LNAs were able to ameliorate CD3+CD8+ T cell infiltration in TME, which sensitized NSCLC to the treatment of ICB.
Conclusions
In conclusion, our data demonstrated that SNORA38B functioned as an oncogene in NSCLC both in vitro and in vivo at least in part by regulating the GAB2/AKT/mTOR pathway via directly binding to E2F1. SNORA38B could also sensitize NSCLC to immunotherapy, which may be a critical therapeutic target for NSCLC.
Keywords: immunotherapy, lung neoplasms, tumor microenvironment
Key messages.
Small nucleolar RNAs (snoRNAs) have been reported to play critical role on the tumorigenesis in several cancer types, while the detailed functions of snoRNAs in oncogenesis and immunotherapy of non-small cell lung cancer (NSCLC) are remaining to be elucidated.
Small nucleolar RNAs 38B (SNORA38B) is a newly clarified snoRNA that specifically highly expressed and positively corrected with worse prognosis in NSCLC, suggesting SNORA38B could be a potential prognostic biomarker for NSCLC.
SNORA38B facilitated NSCLC progression via directly binding with E2F transcription factor 1 (E2F1) and regulating the GRB2-associated-binding protein 2 (GAB2)/protein kinase B (AKT)/mammalian target of rapamycin (mTOR) signaling, in turn contributing to an immunosuppressive tumor microenvironment in NSCLC.
Targeting SNORA38B by locked nucleic acids (LNAs) attenuated NSCLC tumorigenesis and sensitized NSCLC to immune checkpoint blockade (ICB) treatment, suggesting SNORA38B could be a candidate therapeutic target for treating NSCLC.
Introduction
As one of the dominating cancers, lung cancer has always been a public health issue urgently needing to be solved worldwide.1 The majority (about 85%) of lung cancer is non-small cell lung cancer (NSCLC).2 Although recent researches have made considerable headway in the pathogenesis and treatment of NSCLC,3–6 its detailed mechanisms are still not fully understood. Thus, elucidating the pathogenesis of NSCLC in multiple ways is of great significance to clarify a more sensitive diagnostic biomarker and effective innovative clinical therapy.
Small nucleolar RNAs (snoRNAs) are a class of non-protein coding RNAs widely found in nucleoli.7 Generally, they are divided into two families, C/D-box and H/ACA-box snoRNAs, which play crucial roles in the splicing processing and post-transcriptional modification of ribosomal.8 In decades, increasing evidence has proved that snoRNAs are involved in the initiation and development of cancer. For instance, Siprashvili et al 9 revealed that SNORD50A/B sites are commonly deleted in a variety of human cancer types, and promote tumorigenesis cooperated with Kirsten rat sarcoma viral oncogene mutations. However, Su et al 10 reported that SNORD50A/B could also significantly accelerate p53 wild-type (WT) breast cancer progression via directly interacting with tripartite motif-containing 21 and GMP synthase (GMPS). In addition, SNORD89 was deemed as a biomarker for ovarian cancer, and modulated cancer phenotype through targeting Notch1-c-Myc pathway.11 SNORD42A was reported to guide 18s-U116 2’-O-methylation in leukemia, and affected the synthesis of the translational machinery as well as promoting cell proliferation.12 Furthermore, small nucleolar RNAs 50 (SNORA50), SNORA71A, etc, have also been demonstrated to act as an oncogene or a tumor-suppressive gene in prostate cancer and colorectal cancer, respectively.13 14 In NSCLC, SNORA42 was reported to be activated and regulated cancer development through both p53-dependent and p53-independent pathways via functioning as a p53 regulating factor.15 Recent researches also showed that H/ACA-box snoRNAs, such as SNORA65, SNORA7A and SNORA7B were closely related to NSCLC growth, but those snoRNAs did not participate in regulating cell cycle. Moreover, snoRNA-bound ribonucleoproteins, including NOP10 ribonucleoprotein, were associated with poor prognosis as new potential biomarkers.16 However, the underlying molecular mechanisms of snoRNAs in NSCLC still remain largely unclear, and the exploration for tumor immune environment and convincing innovative treatment-options are still warranted.
Small Nucleolar RNA, H/ACA Box 38B (SNORA38B, NR_003706.2), a member of H/ACA-box family, is located at 17q24.2 with length of 132bp and possessing one exon. It is a highly conserved snoRNA between human and mouse (per cent identity=83.08%). So far, there is no research involved in the regulation of SNORA38B on NSCLC. Thus, elucidating the potential molecular mechanism of SNORA38B and exploring innovative therapies are of great significance for the clinical diagnosis and treatment of NSCLC. In this study, we found that SNORA38B was upregulated in NSCLC and indicated to a poorer prognosis, so we considered whether SNORA38B could act as a biomarker in NSCLC. Functionally, we examined the regulatory effect of SNORA38B on biological behaviors (including cell proliferation, migration, invasion, etc) in NSCLC, and explored its ability to recruit regulatory T cells (Tregs) in tumor microenvironment (TME). Mechanistically, we investigated the details regulatory role of SNORA38B on E2F transcription factor 1 (E2F1)/GRB2-associated-binding protein 2 (GAB2)/protein kinase B (AKT)/mammalian target of rapamycin (mTOR) pathway. Given the essential roles of SNORA38B for contributing to a suppressive immune microenvironment in NSCLC tumors, we tested the sensitization effects of SNORA38B locked nucleic acids (LNAs) on immunotherapy in murine Lewis lung carcinoma (LLC)-derived tumor burden mice, which might provide new insights into SNORA38B as a potential therapeutic target for NSCLC.
Materials and methods
Tissue samples
Normal lung tissues and lung adenocarcinoma tissues arrays (2015 WHO classification), including information such as pathology grade, tumor-node-metastasis and clinical stage, 30 cases/40 cores, replacing BC04119a, were purchased from Biomax.us (cat no. BC04119b). The study protocol was approved by the School of Public Health, Wuhan University. All clinical information is provided in online supplemental table 1.
jitc-2021-004113supp001.xlsx (12.8KB, xlsx)
Bioinformatics analyses
The TCGA-gdc database (https://portal.gdc.cancer.gov/repository) was used to analyze the differentially expressed snoRNA in NSCLC. First, the RNA-sequencing (RNA-seq) data of LUNG-lung adenocarcinoma (LUAD) were selected. R software (V.4.0.2 for Windows) was used to screen out differentially expressed snoRNAs in cancer tissues and adjacent tissues based on RNA-seq data. Then clinical data of NSCLC patients were downloaded to screen out representative and research-significant snoRNAs of NSCLC combined with the patient’s prognosis, and cancer stage, etc. The volcano map, heat map, violin maps and Venn map were drawn using the R package of ggplot2, pheatmap, VennDiagram, etc. Survival analyses of SNORA38B and patients with NSCLC were performed using the package of survival and survminer. We converted the continuous expression of SNORA38B from high to low into categorical expression of ‘high’ and ‘low’ using the medium, then drew a survival curve using ggsurvplot package.
Cell culture and transfection
Ten human NSCLC cell lines (A549, 95D, SK-MES-1, H460, H520, H1299, H157, SPC-A-1, SK-LU-1 and H1975) and three normal lung cell lines (16HBE, Beas-2B and WI-38), were bought from the Institute of Biochemistry and Cell Biology of the Chinese Academy of Sciences (Shanghai, China). Mouse normal lung epithelial cell line MLE-12, and three mouse NSCLC cell lines, including LA-4, LLC and KLN 205, were purchased from American Type Culture Collection. Human NSCLC cell lines and MLE-12 cells were all cultured in RPMI 1640 (C11875500BT, Gibco) medium supplemented with 10% fetal bovine serum (FBS) (10 270–106, Gibco) and 1% penicillin/streptomycin (15 140–122, Gibco). 16HBE, Beas-2B, LA-4 and LLC cells were cultured in DMEM-H (C11995500BT, Gibco) medium with 10% FBS and 1% penicillin/streptomycin. WI-38 and KLN 205 cells were cultured in MEM (41500034, Gibco) medium with 10% FBS and 1% penicillin/streptomycin, ditto. Cells were cultured in humidified air filled with 5% carbon dioxide (CO2) at 37°C. Plasmids used in this research, including pLenti-CMV-human SNORA38B, pLenti-CMV-mouse SNORA38B, pLenti-CMV-E2F1, pLenti-CMV-GAB2, pLenti-CMV-LUC-human SNORA38B, pLenti-CMV-LUC-mouse SNORA38B, plentiCRISPRV2-human SNORA38B-Knockout (KO) #1, plentiCRISPRV2-human SNORA38B-KO #2, plentiCRISPRV2-human SNORA38B-KO #3, plentiCRISPRV2-mouse SNORA38B-KO #1, plentiCRISPRV2-mouse SNORA38B-KO #2, plentiCRISPRV2-mouse SNORA38B-KO #3, plentiCRISPRV2-E2F1-KO #1, plentiCRISPRV2-E2F1-KO #2, plentiCRISPRV2-E2F1-KO #3, plentiCRISPRV2-GAB2-KO #1, plentiCRISPRV2-GAB2-KO #2 and plentiCRISPRV2-GAB2-KO #3, were constructed by ourselves. LNA of human/mouse SNORA38B were purchased from Qiagen (Qiagen). For the construction of stably transfected cell lines, corresponding medium without antibiotics was used to culture the HEK-293FT cells at least 24 hours before transfection. When the cell density reached 70%–80%, HEK-293FT cells were washed twice with cold phosphate-buffered saline (PBS) (pH 7.4) and then co-transfected with lentiviral packaging plasmids (psPAX2 and pMD2.G) and the above plasmids using polyethyleneimine (764965, Millipore) cellular transfection reagent. Replaced with complete medium after 6–8 hours, and continue culturing for 48 hours. Collected the supernatant for lentivirus concentration using polyethylene glycol 8000 (PEG-8000) (1546605, Millipore). A549 and H1975 cells were infected with the concentrated lentivirus with 10 µg/mL polybrene (TR1003G, Millipore) for 24 hours, then 2 ug/mL puromycin (P9620, Millipore) was added to select cells for 3–5 days. The surviving cells are KO/overexpress (OE) cells.
ELISA analysis
For human and mice cells, interleukin 10 (IL-10) ELISA analysis was conducted followed the manuals of human IL-10 Quantikine ELISA Kit (D1000B, R&D Systems) and mouse IL-10 Quantikine ELISA Kit (M1000B, R&D Systems). In brief, cells were cultured in 6-well plate, 50 µl of the culture supernatant was taken from each group for ELISA test according to the steps. Three parallels wells were set up for each group, and finally used 450 nm wavelength to detect the absorbance.
Western blotting assay
Western blotting assay was performed according to the previously described protocol.17 18 In detail, cells were cultured in 6-well plate to 70%–90% confluent. Radio immunoprecipitation assay buffer (50 mM Tris (pH 7.4), 150 mM NaCl, 1 mM EDTA, 1% Nonidet P-40, 1 mM NaF, and 0.5% sodium deoxycholate) mixed with protease inhibitor cocktail (P1005, Beyotime Biotechnology) was used to lyse the cells. After detecting the concentration of lysate from different cells using enhanced BCA protein assay kit (P0010, Beyotime Biotechnology) and denaturing them at 95°C for 5 min, the proteins were then loaded on 4%–12% SDS-polyacrylamide gel (P0065A, Beyotime Biotechnology) and separated in 80V for 20 min, and in 120V for 60 min. Then, trans-blot turbo (Bio-Rad) was used to transfer the protein onto nitrocellulose filter membranes (HATF00010, Millipore). Afterwards, the membranes were placed into a petri dish containing blocking solution, and sealed with shaking on a decolorizing shaker at room temperature for 1 hour. The following primary antibodies were applied: anti-p-AKT (T308), anti-p-AKT (S473), anti-AKT, anti-p-mTOR (S2448), anti-mTOR, anti-p-ULK1 (S757), anti-ULK1, anti-p-70S6K (T421/424), anti-p-70S6K (T389), anti-70S6K, anti-p-4E-BP1 (T34/46), anti-4E-BP1, anti-p-elF4E (S209), anti-elF4E, anti-p-S6 (S240/244), anti-S6, anti-p53, anti-E2F1, anti-GAB2, anti-Actin, detailed information is listed in online supplemental table 2. Actin was used as a reference protein. After incubating overnight at 4°C, the membranes were washed three times with Tris-buffered saline with 0.1% Tween 20 detergent (TBST) and incubated with the corresponding secondary antibody for 1 hour at room temperature. Detail information of secondary antibody is listed in online supplemental table 2. After washing with TBST for three times again, Beyo ECL plus (P0018, Beyotime Biotechnology) was used to enhance the chemiluminescence and the membranes were then imaged with chemiluminescence imaging system (G: BOX Chemi XRQ, Syngene).
jitc-2021-004113supp002.xlsx (14.3KB, xlsx)
RNA isolation and quantitative real-time polymerase chain reaction
RNA was isolated from A549 and H1975 cells using TRIzol Reagent (15596026, Invitrogen) and then reverse transcribed into complementary DNA using PrimeScript RT reagent Kit (RR037B, TAKARA) in 37°C for 15 min and 85°C for 5 s. The quantitative real-time PCR (qRT-PCR) was performed using 2×SYBR Green qPCR Master Mix (K1070, APExBIO). Related primer sequences are listed in the online supplemental table 3. The formula of 2−ΔΔCt was utilized to calculate the relative expression levels of genes. Statistical analysis was conducted using the fold change.
jitc-2021-004113supp003.xlsx (14.9KB, xlsx)
In vivo murine models and treatment procedures
For tumor growth assay, 4−6 weeks old male BALB/c nude mice (weight ~20 g) were purchased from Hubei Research Center of Laboratory Animal (Wuhan, China). About 2×106 A549 cells stably expressed luciferase-labeled SNORA38B KO/SNORA38B OE or their control vectors were suspended in 100 µl PBS and injected into nude mice subcutaneously (n=6). Bioluminescence (BLI) assay was performed every 4 days, and caliper was used to measure the length (L) and width (W) of tumor side. The mice were anesthetized and sacrificed on the day 24, and tumors were separated for section staining, TUNEL assay, etc.
For lung metastasis assay, 4–6 weeks old male BALB/c nude mice (weight ~20 g) were purchased from Hubei Research Center of Laboratory Animal. About 1×106 A549 cells stably transfected with SNORA38B KO/SNORA38B OE or their control vectors were suspended in 100 µl PBS and injected lateral tail-vein into nude mice (n=6). Signs of metastasis formation in mice including labored breathing, weight loss and hunched posture. Mice were monitored two to three times a week, and were anesthetized and sacrificed after 24 days post injection, then subjected to lung tissue fixation. Metastatic lung tumor burdens were analyzed using H&E staining together with Visiopharm Image Analysis (Visiopharm, Denmark).
For tumor microenvironmental assay, 4–6 weeks old male C57BL/6J mice (weight ~20 g) were purchased from Hubei Research Center of Laboratory Animal. About 1×106 LLC cells stably transfected with luciferase-labeled SNORA38B KO/SNORA38B OE or their control vectors were suspended in 100 µl PBS and injected subcutaneously into nude mice (n=5). BLI assay was performed every 4 days, and caliper was used to measure the length (L) and width (W) of tumor size. The mice were anesthetized and sacrificed on the day 20, and tumors were separated for section staining, flow cytometry, etc.
For in vivo combinatorial treatment of ICB plus LNA assay, 4–6 weeks old male C57BL/6J mice (weight ~20 g) were purchased from Hubei Research Center of Laboratory Animal and divided randomly into four groups: PBS (Scr LNA+αIgG (Clone: TNP6A7, BE0089, BioXcell; 10 mg/kg.bw)), LNA (LNA+αIgG), immune-checkpoint blockade (ICB) (Scr LNA+ICB (anti-programmed cell death protein-1 (PD-1) (Clone: J43, BE0146, BioXcell; 10 mg/kg.bw)+anti-cytotoxic T-lymphocytes-associated protein 4 (CTLA-4) (Clone: 9D9, BE0164, BioXcell; 10 mg/kg.bw)), LNA+ICB (LNA (5 mg/kg.bw)+ICB (n=5 for each group)). LNAs of SNORA38B were synthesized by Exiqon, and were dissolved in RNase/DNase free water with indicated concentration. About 1×106 luciferase-labeled LLC cells were suspended in 100 µl PBS and injected subcutaneously into mice. The treatment of PBS, LNA or ICB was conducted at the day 4 post LLC inoculation by i.p. every 4 days. BLI assay was performed every 4 days, and caliper was used to measure the length (L) and width (W) of tumor size. The mice were anesthetized and sacrificed on the day 24, and tumors were separated for section staining, flow cytometry, etc.
To test whether SNORA38B increased Tregs via secreting IL-10 in NSCLC, 4–6 weeks old male C57BL/6J mice (weight ~20 g) were divided randomly into three groups: CTL (Blank OE+Vehicle), SNORA38B OE (SNORA38B OE+Vehicle), SNORA38B OE +anti-IL-10R ((SNORA38B OE+anti-IL-10R (Clone: 1B1.3A, BE0050, BioXcell; 10 mg/kg.bw)), n=5 for each group. The treatment of vehicle or anti-IL-10R was conducted at the day 4 post LLC inoculation by i.p. every 4 days.
RNA pull-down assays
Restriction endonucleases EcoRI (R6011, Promega) and SalI (R6051, Promega) were used to linearized the pGEM-3Z vector (P2151, Promega), then pGEM-3Z-SNORA38B plasmid was constructed. Biotinylated sense and anti-sense SNORA38B were transcribed respectively in vitro using Megascript T7 transcription kit (AM1334, Invitrogen) and the Megascript SP6 transcription kit (AM1330, Invitrogen) together with the usage of 10×Biotin RNA Labeling Mix (11685597910, Sigma). After purification by NucleoSpin RNA Plus (740984, TaKaRa), the biotinylated sense SNORA38B RNA and its biotinylated antisense control RNA were incubated with A549 and H1975 cells lysate in NT2 buffer (50 mM Tris-HCl (pH 7.4), 150 mM NaCl, 1 mM MgCl 2, 0.05% NP-40, with 1×protease inhibitor cocktail) at 4°C over night. Then, 40 µl Streptavidin Beads T3 (K1301, APExBIO) were added into each binding reaction, and incubated at room temperature with rotation for 2 hours. Finally, the magnetic beads were washed three times with NT2 buffer and digested by proteinase K. Then, western blotting was conducted to identify the separated protein.
RNA fluorescence in situ hybridization
LNAs of SNORA38B and U6 coupled with fluorescein were synthesized by Exiqon and then used in RNA fluorescence in situ hybridization (FISH) assay.19 In brief, A549 cells were fixed with 4% formaldehyde and handled by 1% pepsin working solution, then were denatured using ethanol with 70%, 90% and 100%. Rinse once with saline-sodium citrate (SSC) formamide buffer then rehydrate with SSC formamide solution for 15 min at room temperature. Next, 40 nM LNA FISH probes were used to incubate the cells with the hybridized mixture at 37°C for 2 hours, followed by washing twice with SSC formamide buffer at 37°C, and covering with Antifade Mounting Medium with DAPI (P0131, Beyotime Biotechnology) for collaborative detection under the confocal laser scanning microscope (Leica-LCS-SP8-STED).
Nuclear and cytoplasmic RNA fractionation analysis
PARIS kit (AM1921, Thermo Fisher Scientific) was used to separate RNA from nucleus and cytoplasm according to the instruction. Then TRIzol was used to extract RNA from cytoplasm and nucleus, respectively. 18S, U6 and SNORA38B level in the nucleus and cytoplasm of A549 and H1975 cells were detected using qRT-PCR. Related primer sequences are listed in the online supplemental table 3.
Transwell migration/invasion assay
Transwell migration/invasion assays were conducted according to the previously described protocols.20 In detail, 8 µm pore size culture inserts (3422; Costar) were placed into the wells of 24-well culture plates to separate the upper and the lower chambers. Complete medium 600 µl was added into the lower chamber. To measure cell migration, 3×105 A549 and H1975 cells transfected with SNORA38B KO, SNORA38B OE or their corresponding vector plasmid were seeded into the upper chambers of transwell plates for 24 hours incubation. For cell invasion assay, Matrigel (356234, BD Biosciences) was used to coat the pore polyester membrane insert uniformly, and then 6×105 cells were incubated into the upper chambers for 24 hours. Then 0.1% crystal violet was used to stain the migratory and invasive cells, and inverted microscope was used to calculate the migrated and invaded cells.
Terminal deoxyribonucleotide transferase-mediated dUTP nick end labeling
One-step Terminal deoxyribonucleotide transferase-mediated dUTP nick end labeling (TUNEL) apoptosis assay kit (C1090, Beyotime Biotechnology) was utilized to examine the cell apoptosis in SNORA38B KO/OE tumor tissue isolated from tumor-burden nude mice. In detail, tumor tissue sections were dewaxed and rehydrated using xylene, 90% ethanol, 70% ethanol and distilled water. After that, 20 µg/mL DNase-free proteinase K (ST532, Beyotime Biotechnology) was used for 15 min at 37℃ and then washed twice with PBS to remove proteinase K. Mixed the TUNEL working solution according to the instructions, and then the working mixtures were added to tissue sections followed by an incubation for 1 hour in 37°C at dark. After washing with PBS for three times, Antifade Mounting Medium with DAPI (P0131, Beyotime Biotechnology) was added onto the sections and then detected the fluorescence signal.
5-ethynyl-2’-deoxyuridine assay
A549 and H1975 SNORA38B-OE/KO cells were seeded on sterile cover glasses placed in the 24-well plates overnight, and incubated with 50 µM 5-ethynyl-2’-deoxyuridine (EdU) (C10310-3, Cell-Light EdU Apollo488 In Vitro Kit, RiBoBio) in the culture medium for 2 hours. After washing with 1×PBS, cells were then fixed for 20 min in 4% paraformaldehyde and permeabilized with 0.5% Triton X-100 in 1×PBS for 10 min. Then, cells were incubated with 1×Apollo reaction buffer for 30 min at dark, followed by three times of washing using 0.5% Triton X-100 in 1×PBS, and another three times of washing by 1×PBS. Next, mounted the coverslip onto the slides with Antifade Mounting Medium with DAPI (P0131, Beyotime Biotechnology) and visualized using fluorescence microscope (Olympus 600 auto-biochemical analyzer). Ten microscopic fields were taken for calculating EdU, and images were analyzed using with Image-Pro Plus software.
Chromatin immunoprecipitation (ChIP) assay
EZ-ChIP kit (17–371, Millipore) was utilized to perform ChIP assay. First, SNORA38B OE and vector expressed A549 and H1975 cells were cultured. The cross-linked chromatin-protein complexes were prepared using 1% formaldehyde, then were sonicated into ~100 to ~500 bp fragments. After purification with 10 000 g centrifugation at 4°C for 10 min, anti-E2F1 (3742, Cell Signaling Technology) was added to incubate overnight at 4°C, while normal rabbit IgG (2729S, Cell Signaling Technology) was used as control. The next day, after a 2-hour rotation incubation with magnetic protein A/G beads at 4°C, 5M NaCl was used to delink the cross-linking overnight again. Finally, DNA was recovered by Gel Extraction kit (D2500, Omega Bio-Tek), and the binding activity between E2F1 protein and GAB2 promoter was detected by qPCR.
Flow cytometry
Cell apoptosis and Ki67 staining
Cell apoptosis was detected using FITC Annexin V Apoptosis Detection Kit I (556547, BD Biosciences). In detail, SNORA38B KO/OE A549 and H1975 cells were digested with trypsin, washed twice by ice-cold PBS, and then re-suspended into cell staining buffer (420201, BioLegend). For cell apoptosis staining, 5 µl of membrane protein V-FITC and 5 µl of propidium iodide were added into 100 µl of cell suspension in dark and incubated at room temperature for 15 min ready for flow cytometry analysis. For Ki67 staining, 5 µl of Ki-67-PE was added into 100 µl of cell suspension in dark and incubated at room temperature for 15 min, washed twice by ice-cold PBS and then ready for flow cytometry analysis.
Immune cells
First, tumors were isolated from tumor burden C57BL/6J mice. After washing with PBS and removing the surrounding connective tissue, tumors were minced in experimental medium with scalpel to obtain ~1–3 mm3 pieces, and then the minced tumor tissues were disassociated by 1 mg/mL collagenase II (C6885, Sigma-Aldrich) and 100 Kunitz/mL DNase I (DN25, Sigma-Aldrich) on a nutating platform (18 rpm) in an incubator (37°C, 5% CO2) for 60 min. Then, the mixture was filtered using 70 µm cell strainer (352350, BD Falcon) and re-suspended in warm fresh complete medium, so that the tumor single cell suspension was completed. For flow cytometry staining, RBC Lysis Buffer (420301, BioLegend) was first used to lyse red blood cells, then, 1×106 cells per condition were subjected to blockade with TruStain FcX (anti-mouse CD16/32) antibody (101319, BioLegend) for 10 min on ice. Cell staining buffer (420201, BioLegend) with proper antibodies was added for cell surface staining at room temperature for 1 hour. Mouse antibodies anti-CD3+, CD8+ and CD4+ were used, see online supplemental table 2 in detail, and the fluorescence minus 1 staining corresponding to each condition served as a control. After standing of the surface markers, 0.5 mL/tube Fixation Buffer (420801, BioLegend) was used to fix cells in the dark for 20 min at room temperature. Then fixed cells were permeabilized and washed in Intracellular Staining Perm Wash Buffer (421002, BioLegend) twice, and resuspended with the FOXP3 antibody diluted in Intracellular Staining Perm Wash Buffer, see online supplemental table 2 in detail. Zombie Violet fixable viability dyes (423113, BioLegend) were used to remove dead cells. Flow cytometry was conducted on an LSRII (BD Biosciences) to count the target cells, and FlowJo (TreeStar) was used to analyze the data.
CyTOF run and sample normalization
Cytometry by time of flight (CyTOF) was conducted according to the previously described protocol.3 In brief, cells were stained with antibodies conjugated to metal isotopes, and then subjected to quadrupole (TOF) mass spectrometry analysis. In the very beginning, LLC cells were stably transfected with SNORA38B KO/OE or their corresponding vectors and then injected into C57BL/6J mice. Before reaching the endpoint, tumors were digested into single cell using 0.25% trypsin and stained with cisplatin, blocked with TruStain FcX antibody and incubated with surface antibodies listed in online supplemental table 4 for 30 min in dark. After that, cells were fixed in fixation/permeabilization buffer and intracellular antigens were stained using intracellular antibodies listed in online supplemental table 4 for 2 hours at 4°C. Afterwards, cells were washed twice and incubated in Intercalator (Cell-ID Intercalator-Ir, 201 192A, Fluidigm). Finally, cells were washed and collected to run on a Helios mass cytometer (Fluidigm), with re-suspended and spiked the equilibration beads into each sample for signal normalization. TH1 (type 1 helper T cells, marked as CD45+CD4+T-bet+), TH2 (type 2 helper T cells, marked as CD45+CD4+GATA3+), Treg (regulatory T cells, marked as CD45+CD4+CD25+FOXP3+), TH17 (type 2 helper T cells, marked as CD45+CD4+RORγt+), activated CD8+ T cell (marked as CD45+CD3+CD8+Granzyme B+), B cell (marked as CD45+CD19+), DC (dendritic cells) cell (marked as CD45+CD11c+), NK (natural killer, marked as CD45+NK1.1+) cell, MDSC (myeloid-derived suppressor cells, marked as CD45+CD11b+Gr1+), M1 (classically-activated macrophages, marked as CD45+F4/80+CD86+INOS+), M2 (alternatively-activated macrophages, marked as CD45+ F4/80+CX3CR1+Arg1+) cell subtypes were analyzed. The annotations for specific cell subtypes are as below: Active CD8+ T cells: clusters 34, 2, 36; Treg cells: cluster 9, 13; TH1: cluster; TH1: cluster 18; TH2: cluster 27; TH17: cluster 7; B cells: cluster 32; DC cells: cluster 21; NK cells: clusters 6, 22; MDSC cells: clusters 23, 25.
jitc-2021-004113supp004.xlsx (14.2KB, xlsx)
RNAScope
SNORA38B expression in human lung cancer tissues and mouse lung tumor tissues (derived from the KP mice (Krastm4Tyj Trp53tm1Brn/J, Stock No: 032435) were detected using RNAScope. In detail, 6–8 week-old mice were infected with about 2.5×107 adeno 5 (Ad5)-CMV-Cre virus per mouse by the intratracheal intubation method to get the mouse lung tumor. RNAScope probes (designed by Advanced Cell Diagnostics) were used to detect SNORA38B following RNAScope 2.5 High-Definition Assay kit (Advanced Cell Diagnostics) based on the manufacturer’s procedures. Image quantifications were conducted using ImageJ software.
Prediction of E2F1-SNORA38B interactions
To elucidate the effect of SNORA38B binding on the E2F1, three-dimensional (3D) models were constructed based on the crystal structure of full-length E2F1 (https://www.alphafold.ebi.ac.uk/entry/Q01094), and the structure was optimized using MODELLER software (V.9.24).21 All default parameters were used unless otherwise stated. The derived structure was further prepared and refined using Molecular Operating Environment (MOE) (V.2020.09) software. To model E2F1-SNORA38B interactions, the 3D structures of SNORA38B were built using the MC-fold/MC-Sym (https://www.major.iric.ca/MC-Fold/) pipeline.22 These 3D RNA structures and the 3D E2F1 protein model described above were used as the input ligand and receptor, respectively, for docking study. Next, protein-RNA docking was performed using multiple web servers, including 3dRPC,23 HDOCK,24 25 and NPDock.26 Energy minimizations of the docked complex structures were then conducted using MOE (V.2020.09). For each docking experiment, the top 10 poses with the best scores were selected and examined for interactions using PyMOL (Schrödinger; V.2.4), along with the final image/figure generation.
Multiplex immunohistochemistry and immunofluorescence staining
Multiplex immunohistochemistry staining (mIHC) assay was performed according to the previously described protocols.27–29 In detail, human NSCLC TMA sections were divided into SNORA38B high/low and were repaired with 0.01M Sodium Citrate buffer (pH=6.0). Then PBS with 0.2% triton X-100 was used to permeate cell/nuclear membrane for FOXP3 staining. Tumor sections were incubated with anti-EpCAM, anti-CD8, and anti-FOXP3 overnight at 4°C according to vendor’s instruction of Opal Polaris 7 Color Automation IHC Detection Kit (NEL871001KT, PerkinElmer) and imaged with PE Vectra Polaris Automated Quantitative Pathology Imaging System (PerkinElmer). Immunofluorescence was conducted as follws: cells were seeded on sterile glass coverslips (poly-L-lysine coated), and then were fixed with freshly prepared 4% paraformaldehyde in neutral PBS at room temperature for 10 minutes. Next, the coverslips were incubated in 0.5% Triton X-100 in PBS at room temperature for five minutes, followed by blocking buffer (5% normal serum prepared in PBS) for one hour at room temperature. Then, incubating cells with indicated primary and secondary antibodies, after which the slides were stained with DAPI and then subjected to image capture using the confocal laser scanning microscope (Leica-LCS-SP8-STED).Statistical analysis was performed as previously described. Details of the antibodies used are listed in online supplemental table 2.
Statistical analysis
For normally distributed data, two-tailed Student’s t-test was used for pairwise comparison, one-way analysis of variance was used for the comparison of the data with three groups and above. Kaplan-Meier estimate was used for the survival analysis of patients with NSCLC. Assays were performed triple and statistical analysis was performed using SPSS V.23.0 software. Differences were considered significant when p<0.05.
Results
SNORA38B was highly expressed in NSCLC and associated with poor prognosis
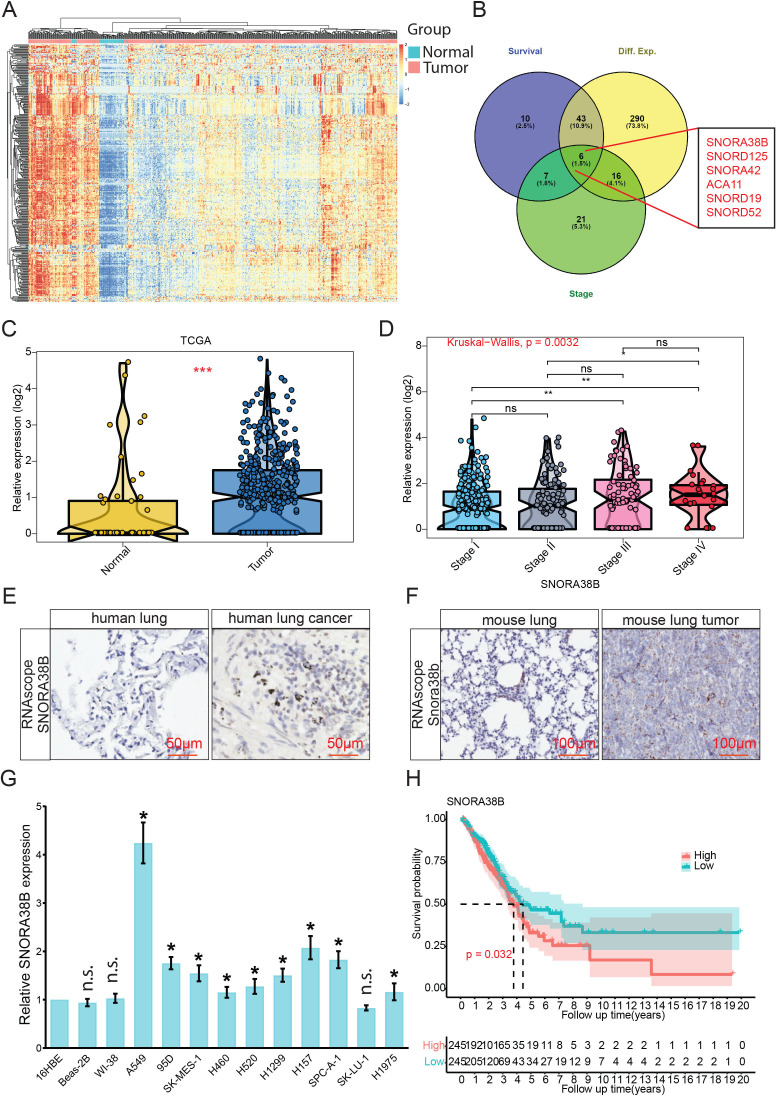
We first explored the differential expression of snoRNA in NSCLC samples and normal samples using The Cancer Genome Atlas (TCGA) (https://portal.gdc.cancer.gov/ repository) (figure 1A, online supplemental figure S1A). Six differentially expressed snoRNAs including SNORA38B, SNORA125, SNORA42, ACA11, SNORA19 and SNORA52, were screened out according to the interactive analysis for selecting snoRNAs using gene expression, cancer stages and survival rates (figure 1B). Among them, SNORA38B was the most differentially expressed one, hence it was selected for the further study. Our data revealed that SNORA38B level in NSCLC was significantly higher than that in adjacent normal counterparts (figure 1C, online supplemental figure S1B) and exhibited a positive correlation with clinical stages. Especially, there were significant differences between stage I and stage III, stage I and stage IV, stage II and stage IV (figure 1D, and online supplemental figure S1C). We further verified SNORA38B level in human lung cancer and mouse lung tumor tissues using RNAscope. Results demonstrated that higher signals of SNORA38B were detected in human lung cancer tissues (n=20) and mouse lung tumor tissues (n=10) than those in normal human tissues (n=10) or mouse (n=10) lung tissues (figure 1E,F, online supplemental figure S1D,E). In addition, qRT-PCR also demonstrated that SNORA38B was significantly higher expressed in NSCLC cell lines (A549, 95D, SK-MES-1, H460, H520, H1299, H157, SPC-A-1 and H1975) than those in normal lung cell lines (16HBE, Beas-2B and WI-38) (*p<0.05, **p<0.01, ***p<0.001, ****p<0.0001, figure 1G). To further evaluate the implications of SNORA38B in clinical diagnosis and treatment, patients with lung cancer were divided into high-SNORA38B and low-SNORA38B expression groups, then the correlation between SNORA38B level and overall survival ratio was evaluated using Kaplan-Meier survival assay by R software. Results indicated that patients with high-SNORA38B expressed group showed shorter median survival time than those with low-SNORA38B expressed group (figure 1H). Therefore, our findings demonstrated that SNORA38B was highly expressed in NSCLC and associated with poor prognosis.
Figure 1.
SNORA38B was highly expressed in NSCLC and associated with poor prognosis. (A) Heatmap of differential gene expression in lung cancer tissues compared with adjacent normal tissues using TCGA database. Mapping by R software (V.4.0.2). (B) Six differentially expressed small nucleolar RNAs that associated with lung cancer stage and survival were screened out using Venn diagram. Mapping by R software. (C) Violin Plot of SNORA38B expression in tumor samples compared with adjacent normal samples using TCGA database. Mapping by R software. (D) Violin Plot of SNORA38B expression in different stages of NSCLC. Mapping by R software. (E–F) Representative images of SNORA38B expression in human lung/mouse tumor tissues and human/mouse lung tissues using RNAscope. Bar=50/100 µm. (G) Expression levels of SNORA38B in normal lung cell lines (16HBE, BEAS-2B, WI-38) and NSCLC cell lines (A549, 95D, SK-MES-1, H460, H520, H1299, H157, SPC-A-1, SK-LU-1 and H1975) were examined using qRT-PCR. (H) Kaplan-Meier survival analysis showed the correlation between SNORA38B expression and patient prognosis. Mapping by R software. Assays were conducted in triple. *p<0.05, **p<0.01, ***p<0.001, means±SD was shown. Statistical analysis was performed by Student’s t-test, Kruskal-Wallis, one-way analysis of variance and the log-rank test. NSCLC, non-small cell lung cancer; SNORNA, small nucleolar RNA; TCGA, The Cancer Genome Atlas.
jitc-2021-004113supp005.pdf (6.3MB, pdf)
SNORA38B accelerated cell proliferation, migration, invasion and inhibited cell apoptosis in vitro
To explore the roles of SNORA38B in NSCLC cell growth, A549 and H1975 cells were transfected with Scr-KO or three independent SNORA38B-KO vectors, that is, KO-1, KO-2, and KO-3. Results of qRT-PCR indicated that KO-1 exhibited the most knockout efficiency, so it was selected to represent SNORA38B-KO (online supplemental figure S2A). Besides, transfection of pLenti-CMV-SNORA38B (SNORA38B-OE) effectively improved the expression of SNORA38B in A549 and H1975 cells (online supplemental figure S2B). Subsequently, EdU staining assay was used to identify whether SNORA38B facilitated cell growth in A549 and H1975 cells. Results demonstrated that SNORA38B-KO significantly inhibited cells growth while SNORA38B-OE significantly improved the cell proliferation of A549 and H1975 cells compared with the control groups (online supplemental figure S2C–F). Ki-67 is a cell proliferation-related antigen expressed in active phase of cell cycle.30 We used Ki-67 antibody to detect the growth of A549 cells transfected with SNORA38B-KO, SNORA38B-OE or their counterparts and results indicated that SNORA38B significantly increased the expression of Ki-67 of A549 cells (online supplemental figure S2G,H). These results confirmed that SNORA38B facilitated NSCLC cell proliferation. Then, the roles of SNORA38B on the migration and invasion of A549 and H1975 cells were explored using transwell assay. Our data showed that elevated expression of SNORA38B promoted the migration and invasion of NSCLC cells, whereas knockout of SNORA38B inhibited the cell migration and invasion, compared with their control groups (online supplemental figure S2I–M). Furthermore, we investigated the effect of SNORA38B on NSCLC cell apoptosis. Flow cytometry analysis demonstrated that the apoptostic cells in A549 and H1975 cells were with about 2.1-fold of increase in SNORA38B-KO versus Scr-KO, while SNORA38B-OE inhibited cell apoptosis (online supplemental figure S3A–C). In general, our findings confirmed that SNORA38B could significantly promote cell proliferation, migration and invasion, and repress cell apoptosis in vitro.
SNORA38B accelerated tumor growth and metastasis in vivo
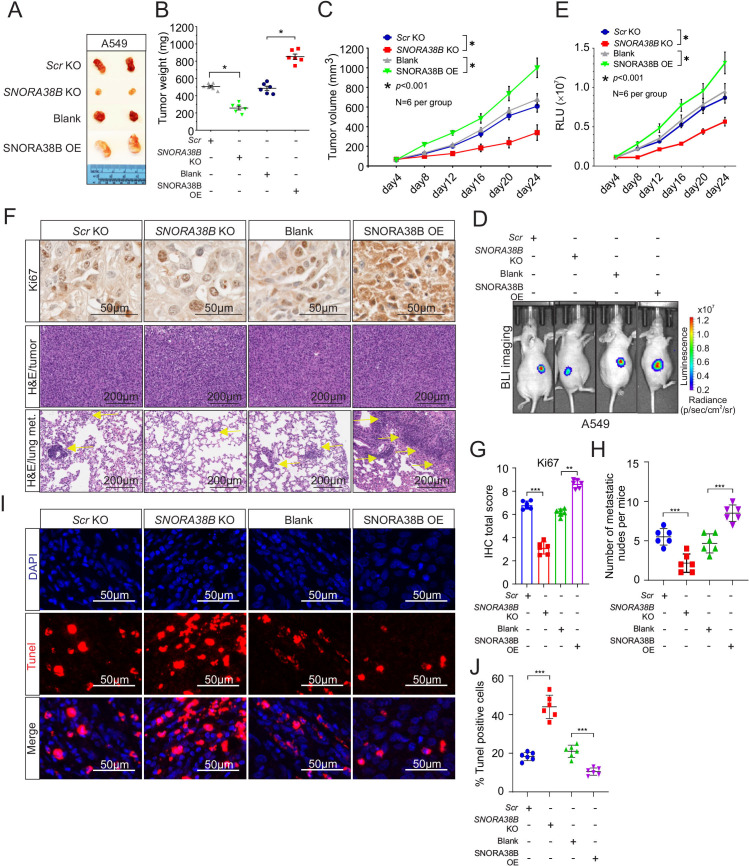
In order to explore the carcinogenicity of SNORA38B in vivo, BALB/c nude mice xenograft model was established using A549 cells. In comparison to the control group, tumor volume and tumor weight of SNORA38B-KO group were significantly decreased, while on the contrary, SNORA38B-OE increased tumor volume and tumor weight (figure 2A–C). To further clarify the oncogenic effect of SNORA38B in vivo, SNORA38B-KO/OE plasmid labeled with luciferase reporter gene were transfected into A549 cells and injected into nude mice. Before reaching the end point, bioluminescence imaging was used to detect the tumor region for nude mice every 4 days. Our data indicated that SNORA38B-KO suppressed tumor growth, while SNORA38B-OE significantly accelerated tumor development in vivo (figure 2D,E), suggesting that SNORA38B could facilitate the tumorigenicity of A549 cells in vivo. Besides, Ki-67 staining and H&E staining were performed using tumor tissues from tumor-burdened nude mice. As expected, Ki-67-positive cells in SNORA38B-OE treated group were increased significantly (figure 2F,G), together with the increased tumor density and the number of lung metastatic nodules (figure 2F,H), while the diametrically opposite results were obtained in SNORA38B-KO group. In addition, results of TUNEL assay also proved that SNORA38B could negatively regulate the cell apoptosis of tumor cells in vivo (figure 2I,J). Collectively, our results indicated that SNORA38B accelerated tumor growth and metastasis of human NSCLC cells in mouse model.
Figure 2.
SNORA38B accelerated tumor growth and metastasis in vivo. (A) Representative image of SNORA38B KO/ SNORA38B OE A549 xenograft tumors separated from nude mice. (B) Tumor weight of separated tumors, n=6 per group. (C) Tumor volume measured every 4 days using caliper (for L and W) and calculated according to formula 0.5×L×W2. n=6 per group. (D–E) Representative images and quantification of BLI in the luciferase reporter gene labeled A549 tumor regions. BLI was examined every 4 days before reaching the observation endpoint. n=6 per group. (F) Representative images of immunohistochemistry staining for Ki67, and H&E staining in tumor region and metastases. The arrows point to the lung metastatic nodule. Bar=50/200 µm. (G) Quantitative results of Ki-67 detection assay. (H) Numbers of lung metastasis per mice. (I–J) Representative results of Terminal deoxyribonucleotide transferase-mediated dUTP nick end labeling assay using tumor tissue sections separated from nude mice. Bar=50 µm. Assays were conducted in triplicate. *p<0.05, **p<0.01, ***p<0.001. Means±SD was shown. Statistical analysis was subjected to one-way analysis of variance. Comparison between-group (SNORA38B KO and Scr KO, SNORA38B OE and vector OE) using LSD method. BLI, bioluminescence imaging; KO, knockout; OE, overexpress; SNORNA, small nucleolar RNA; RLU, relative light units; LSD, the Fisher Least Significant Difference.
SNORA38B accumulated Treg infiltration by promoting the secretion of IL-10 from lung tumor
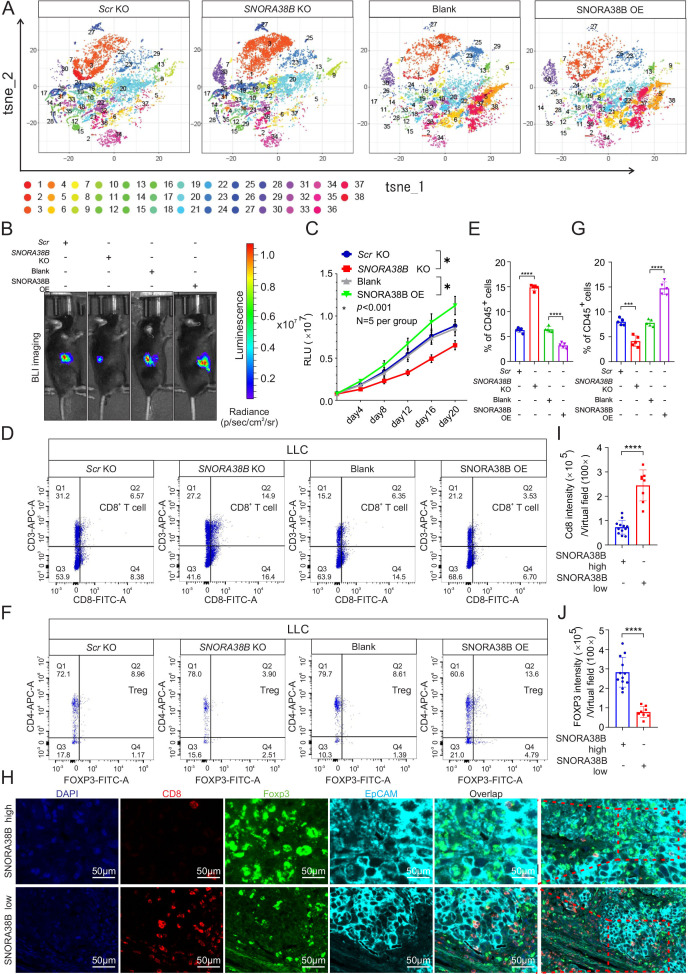
It is well-known that different classes of immune cells tend to accumulate in and around the tumors, forming an important immune part of the TME. In the past few years, immune checkpoint blockade (ICB), especially anti-PD-1/CTLA-4, have been widely used clinically to treat patients with advanced lung tumor and are promising for several solid tumors.31 Our above results demonstrated that SNORA38B was highly expressed in NSCLC and promoted tumor progression both in vitro and in vivo. Therefore, we further explored the effect of SNORA38B in the immune environment of NSCLC. We first established the syngeneic tumor mouse model via injecting luciferase-labeled SNORA38B KO/OE LLC cells into C57BL/6J mice. First, the images and quantification of BLI showed that SNORA38B accelerated the tumor growth in vivo, which were consistent with our results above (figure 3B,C). Then, the infiltration of immune cell populations in tumors was analyzed. After digesting tumors as single cell suspension, CyTOF analysis was performed using 33 types of antibodies. Results indicated that SNORA38B-KO group showed higher CD3+CD8+ T cells infiltration and lower CD4+FOXP3+ Treg cells infiltration than in the Scr-KO group, while SNORA38B-OE showed the exact opposite results (figure 3A, and online supplemental figure S4A,B,E and F). These findings seem to explain that SNORA38B could act as a biomarker for predicting the TME of NSCLC. In addition, we also indicated that SNORA38B could reduce NK cells infiltration, while with no significant effect on MDSCs, DC cells, B cells and subsets of CD4+ T cells including TH1, TH2 and TH17 (online supplemental figure S4C,D,G–K). Then, flow cytometry analysis was performed using isolated single cells to further verify the infiltration of CD4+FOXP3+ Tregs and CD3+CD8+ T cells in tumors. Results demonstrated that SNORA38B OE accelerated CD4+FOXP3+ Tregs infiltration in the LLC-derived tumor burden C57BL/6J mice, simultaneously inhibited the aggregation of CD3+CD8+ T cells, while SNORA38B KO accumulated CD3+CD8+ T cells but suppressed CD4+FOXP3+ Tregs (figure 3D–G, online supplemental figure S5A,B). Furthermore, the results of mIHC staining using the isolated burden tumors showed the same trend (online supplemental figure S5C). In order to further clarify the immunosuppressive effects of SNORA38B, 20 samples of human NSCLC were divided into SNORA38B high (n=12) and SNORA38B low (n=8) base on SNORA38B level. As expected, the results of mIHC staining showed that CD8 was suppressed but FOXP3 was accumulated in the high SNORA38B expression group, while the low SNORA38B expression group indicated completely opposite results (figure 3H–J). These data suggested that SNORA38B may play critical roles in contributing to immunosuppressive TME on NSCLC via recruiting Tregs and reducing the infiltration of tumor killer cells including CD3+CD8+ T cells.
Figure 3.
SNORA38B promoted tumor progression in C57BL/6J mice and regulated Treg infiltration. (A) Representative images of cytometry by time of flight analysis with 33 types of antibodies using PhenoGraph. Thirty-eight cells clusters are represented by different colors, t-distributed stochastic neighbor embedding (t-SNE)-1 and t-SNE-2 are shown. (B–C) Representative images and quantification of BLI in the luciferase reporter gene labeled SNORA38B KO/ OE LLC tumor regions in C57BL/6J mice. BLI was exmained every 4 days before reaching the observation endpoint. n=5 per group. (D–G) Flow cytometry detection of CD3+CD8+ T cells, CD4+FOXP3+ Treg and their proportion in leukocyte in tumors isolated from C57BL/6J mice. (H) Representative images of mIHC staining using human lung cancer samples with high or low SNORA38B expression. CD8, FOXP3, and EpCAM were shown in different colors. Bar=50 µm. n=20. (I–J) Quantification of CD8 and FOXP3 in mIHC staining using human lung cancer samples with high or low SNORA38B expression. n=20. *p<0.05,***p<0.001,****p<0.0001, means±SD was shown. Statistical analysis was performed by Student’s t-test and one-way analysis of variance, comparison between-group using LSD method. BLI, bioluminescence imaging; KO, knockout; mIHC, multiplex immunohistochemistry; NSCLC, non-small cell lung cancer; OE, overexpress; SNORNA, small nucleolar RNA; TCGA, The Cancer Genome Atlas; Treg, regulatory T cells; t-SNE, t-distributed stochastic neighbor embedding; LSD, the Fisher Least Significant Difference; LLC, murine Lewis lung carcinoma.
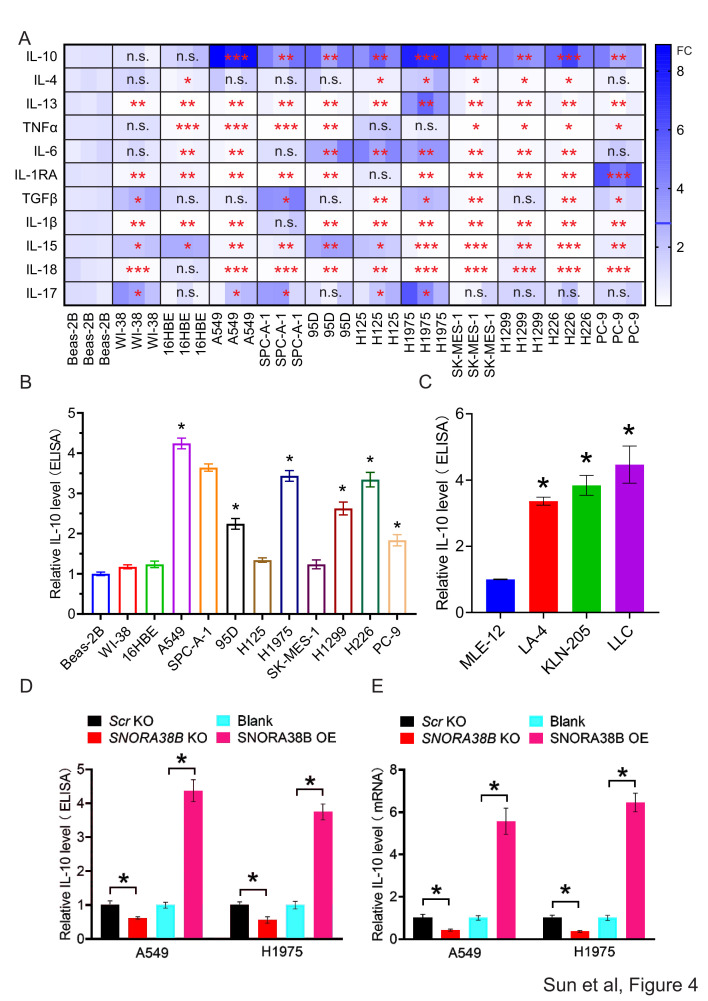
Cytokines are a kind of small molecular proteins with biological activity produced by varieties of cells, which play important parts in mediating inflammation and regulating the proliferation and differentiation of immune cells. Previous studies have shown that immunosuppressive cytokines such as IL-10, IL-6, transforming growth factor-beta (TGF-beta), etc, have a regulatory effect on the function of Tregs and CD8+ T cells in tumor immunity.32–35 In order to confirm whether SNORA38B could recruit CD4+FOXP3+ Tregs by regulating inhibitory cytokines, we first tested the messenger RNA (mRNA) level of 11 cytokines in human normal lung cells and NSCLC cells using qRT-PCR. Our data showed that IL-10 mRNA expression exhibited the most significant difference between normal lung cells and NSCLC cells (figure 4A). To further verify this result, we performed ELISA to examine IL-10 level in the supernatant of normal lung cells and NSCLC cells from both human and mice. Results indicated that NSCLC cells generally showed a significantly high level of IL-10 than normal lung cells, expcept H125 and SK-MES-1 cells (figure 4B,C). In addition, SNORA38B-KO group showed lower IL-10 secretion than control group, while SNORA38B-OE treated group accumulated higher IL-10 level in both A549 and H1975 cells (figure 4D). The same results were showed in mRNA level of IL-10 (figure 4E). Additionally, an in vivo validation was performed using SNORA38B-OE LLC cells and IL-10 blockade in C57BL/6J mice. Our data revealed that SNORA38B OE significantly increased the Tregs infiltration, but could not increase the Tregs infiltration under the IL-10 blockade treatment situation (online supplemental figure S6A–C). Therefore, our findings indicated that SNORA38B could accumulate Tregs recruitment to participate in the formation of immunosuppressive TME in NSCLC at least in part by promoting the secretion of IL-10 from lung tumors.
Figure 4.
SNORA38B promoted IL-10 secretion in NSCLC cells. (A) The mRNA level of cytokines in NSCLC cells and normal lung cells using qRT-PCR. The expression differences are shown by fold change (FC) in different colors. (B–C) Relative IL-10 level in the supernatant of human or mouse NSCLC cells and normal lung cells using ELISA. Human/mouse normal lung cell Beas-2B and MLE-12 were used as control. (D) Relative IL-10 level in the supernatant of A549 and H1975 cells transfected with SNORA38B KO, SNORA38B OE or its counterparts using ELISA. (E) Relative IL-10 mRNA level in A549 and H1975 cells transfected with SNORA38B KO, SNORA38B OE or its counterparts using qRT-PCR. Assays were conducted in triple. *p<0.05, **p<0.01., ***p<0.001., ****p<0.001, means±SD was shown. Statistical analysis was subjected to Student’s t-test and one-way analysis of variance. IL, interleukin; KO, knockout; mRNA, messenger RNA; NSCLC, non-small cell lung cancer; OE, overexpress; qRT-PCR, quantitative real-time PCR; SNORNA, small nucleolar RNA; TGF, transforming growth factor; TNF, tumor necrosis factor; FC, fold change; ELISA, enzyme-linked immunosorbent assay.
SNORA38B activated the AKT/mTOR and its downstream pathways
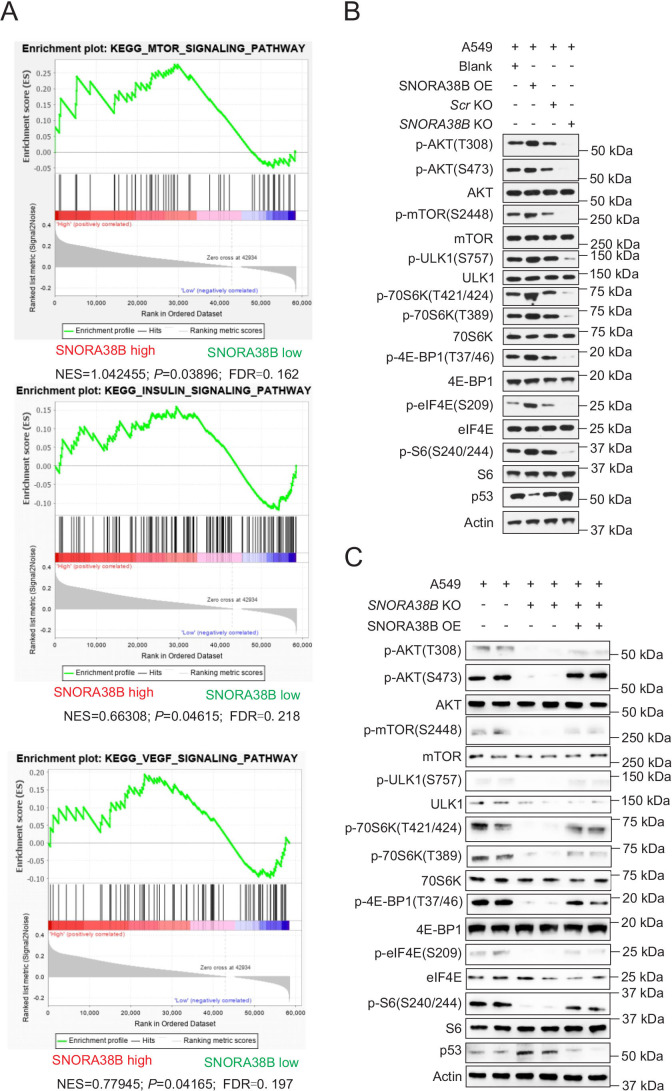
In order to explore the possible molecular mechanisms involved in SNORA38B on NSCLC, gene set enrichment analysis was performed. Our data indicated that gene sets related to lung tumors of SNORA38B higher group were generally enriched in mTOR signaling pathway, insulin and vascular endothelial growth factor signaling pathway (figure 5A). mTOR is a type of Serine/Threonine protein kinases downstream of phosphoinositide 3-kinase (PI3K)/AKT pathway, which plays essential roles on facilitating cell proliferation, invasion, metastasis, and inhibiting autophagy of tumor cells.36 In order to figure out whether SNORA38B is involved in the regulation of the mTOR pathway, western blotting assay was conducted to examine the activity of downstream effectors of AKT/mTOR pathway in A549 and H1975 cells treated with SNORA38B-KO or SNORA38B-OE, respectively. Our data showed that lower expression of p-AKT(S473), p-AKT(T308), p-mTOR (S2448), p-ULK1(S757), p-70S6K (T389), p-70S6K (T421/424), p-eIF4E (S209), p-S6 (S240/244), p-4E-BP1 (T37/46) while higher level of p53 were detected in SNORA38B-KO treated group than in its counterpart, while the opposite results were obtained in SNORA38B-OE group (figure 5B, and online supplemental figure S7A). In addition, overexpressed SNORA38B could rescue the low expression of signal molecules above caused by SNORA38B-KO, and correspondingly reduced the activity of downstream p53 activity in A549 cells (figure 5C). Hence, our findings demonstrated that SNORA38B promoted NSCLC progression through activating the AKT/mTOR and its downstream pathways.
Figure 5.
SNORA38B activated the AKT/mTOR and its downstream effectors. (A) Gene set enrichment analysis of gene sets including SNORA38B high and SNORA38B low in NSCLC. (B) Western blotting analysis of AKT/mTOR and its downstream effectors in A549 cells transfected with SNORA38B KO, SNORA38B OE or their counterparts. (C) Rescue experiment using SNORA38B KO and SNORA38B OE in A549 cell was conducted to detect the regulation of AKT/mTOR and its downstream effectors by SNORA38B. Assays were conducted in triple. AKT, protein kinase B; FDR, false discovery rate; KO, knockout; mTOR, mammalian target of rapamycin; NES, normalized enrichment score; OE, overexpress; SNORNA, small nucleolar RNA.
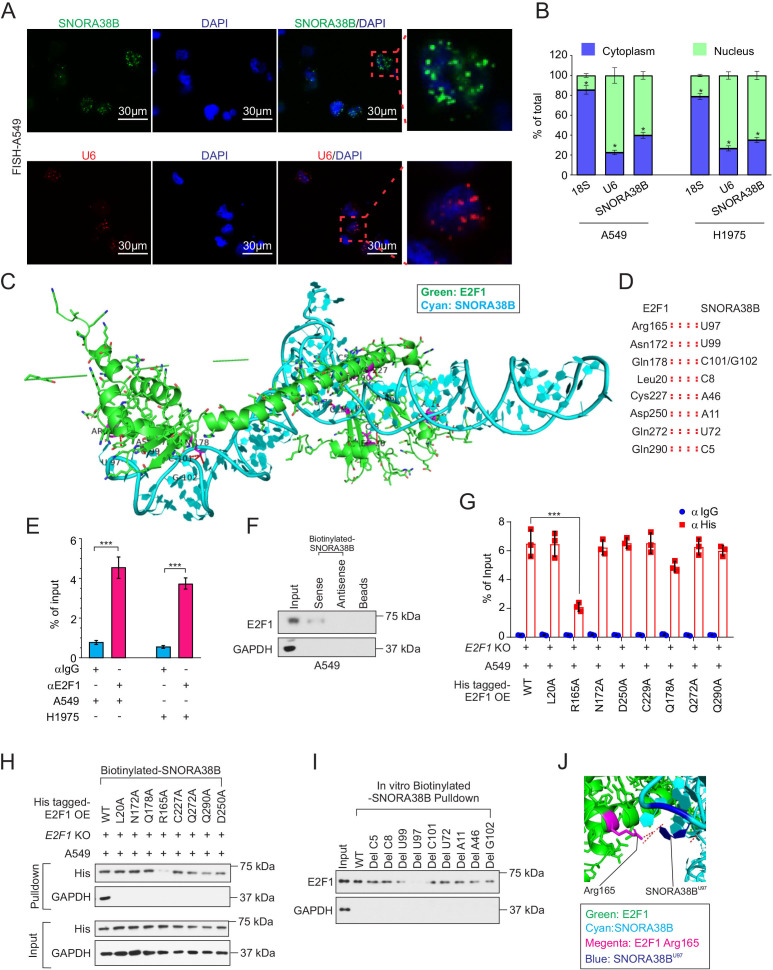
SNORA38B mainly located in nucleus and directly bound with E2F1
To further clarify the oncogenic molecular mechanisms of SNORA38B in NSCLC, secondary structure of human and mouse SNORA38B were first predicted using the RNA Central website (https://rnacentral.org/rna/URS00006C5BAC/9606; https://rnacentral.org/rna/URS0001BBFB4B/10090) (online supplemental figure S8A). Then, we assessed the subcellular localization of SNORA38B in A549 and H1975 cells using RNA FISH and nuclear and cytoplasmic RNA fractionation analysis. Our data indicated that SNORA38B mainly located in the nucleus, suggesting it may play a cancer-promoting effect by regulation of gene transcription (figure 6A,B). Our above results have proven that SNORA38B could promote NSCLC development through activating AKT/mTOR pathway. According to the previous studies, E2F upregulated the activity of AKT by targeting transcriptional regulation of the adaptor protein GRB2-associated-binding protein 2 (GAB2).37 E2F family of transcription factors participated in handles of essential biological processes of the human beings, such as cell proliferation, differentiation, apoptosis, etc. Researches have shown that abnormally upregulated E2F1 was generally associated with the malignant progression and poor prognosis of cancer.38 Using the crystal structure information of E2F1, protein structural bioinformatics analysis suggested that SNORA38B nucleotides C5, C8, A11, A46, U72, U97, U99, C101/G102 associated with E2F1 amino acids Gln290, Leu20, Asp250, Cys227, Gln272, Arg165, Asn172, Gln178 (figure 6C,D). To identify the binding activity between E2F1 and SNORA38B, RIP assay was conducted and found that SNORA38B was enriched in anti-E2F1 antibody group, in both A549 and H1975 cells (figure 6E). Moreover, RNA-pulldown assay was performed and further confirmed that SNORA38B could directly bind with E2F1 (figure 6F, and online supplemental figure S7B). To further clarify the detailed binding sites between SNORA38B and E2F1, we first constructed his-tagged E2F1-WT or E2F1-mutants (L20A, R165A, N172A, D250A, C227A, Q178A, Q272A, and Q290A) stable expressed while E2F1-KO A549 and H1975 cells, and then RIP assay using anti-His or anti-IgG was conducted to identify the binding activities between SNORA38B with his-tagged E2F1-WT or E2F1-mutants. Results demonstrated that R165A mutation significantly decreased the enrichment of SNORA38B in anti-His group (figure 6G, online supplemental figure S7C). In addition, biotinylated-SNORA38B based RNA pulldown using the cell lysis of his-tagged E2F1-WT or E2F1-mutants (L20A, R165A, N172A, D250A, C227A, Q178A, Q272A, and Q290A) stable expressed while E2F1-KO A549 and H1975 cells, and results demonstrated that E2F1 R165A mutation attenuated the binding activity between E2F1 and SNORA38B groups (figure 6H, online supplemental figure S7D). Furthermore, in vitro RNA pulldown using biotinylated-SNORA38B-WT or biotinylated-SNORA38B-mutants and bacterial-derived E2F1 protein revealed that depletion of U97, U99 and C101 could significantly suppress the binding activity between E2F1 and SNORA38B (figure 6I). The most effective binding pair between E2F1 and SNORA38B, namely E2F1 Arg165 and SNORA38B U97, was shown in figure 6J. Taking all into account, SNORA38B was able to directly bind with EF21.
Figure 6.
SNORA38B directly bound with E2F1. (A) Position of SNORA38B/U6 in A549 cells shown by RNA fluorescence in situ hybridization analysis (FISH). Bar=30 µm. (B) Position of SNORA38B in A549 and H1975 cells shown by nuclear and cytoplasmic RNA fractionation analysis. 18S and U6 were used as cytoplasmic and nuclear internal controls, respectively. (C–D) The whole view of SNORA38B’s interaction with the regulatory domain of E2F1. Green: E2F1; Cyan: SNORA38B. (E) RIP detecting the binding enrichment of E2F1 with SNORA38B in A549 and H1975 cells; anti-IgG as controls. (F) RNA pulldown assay detected the interaction of SNORA38B and E2F1 in A549 cells using antisense small nucleolar RNA and beads as controls. (G) RIP assay detecting the binding activities between his-tagged E2F1-WT or E2F1-mutants with SNORA38B in E2F1-WT or E2F1-mutants treated E2F1-KO A549 cells. (H) RNA pulldown assay detected the interaction of SNORA38B and E2F1-WT or E2F1-mutants in E2F1 WT or mutants treated E2F1-KO A549 cells. (I) In vitro RNA pulldown assay detected the interaction of SNORA38B-WT or SNORA38B-mutants; Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) as a negative control. (J) Magnified view of SNORA38B’s interaction with the regulatory domain of E2F1. SNORA38B U97 forms hydrogen bonds and polar interaction with Arg165. Color representation: green indicates E2F1; cyan indicates SNORA38B; magenta indicates Arg165; blue indicates SNORA38B U97. *p<0.05, ***p<0.001, means±SD was shown. Statistical analysis was subjected to Student’s t-test and one-way analysis of variance . Assays were conducted in triple. GAPDH, glyceraldehyde 3-phosphate dehydrogenase; KO, knockout; OE, overexpress; RIP, RNA immunoprecipitation; SNORNA, small nucleolar RNA; WT, wild-type.
SNORA38B activated GAB2/AKT/mTOR pathway through interacting with E2F1
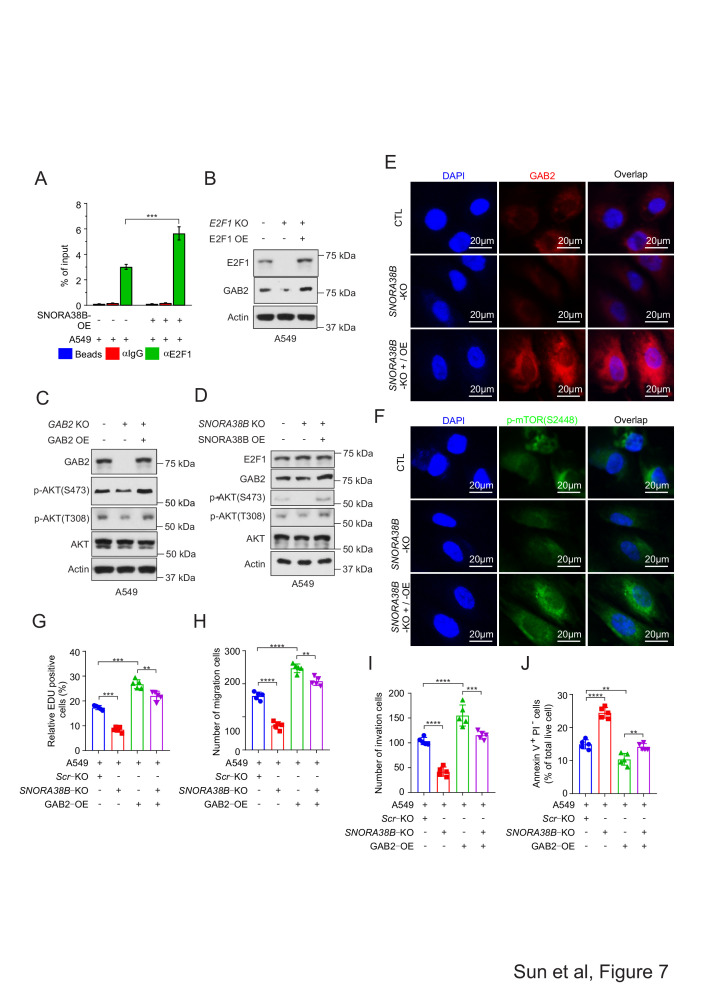
Then, we suspected that the interaction between SNORA38B and E2F1 could facilitate the binding activity between E2F1 and the promoter regions of GAB2 gene. To valid this hypothesis, ChIP assay was conducted using anti-E2F1 and anti-IgG to incubate with SNORA38B-OE or vector expressed A549 and H1975 cell lysis, and results indicated that SNORA38B treatment significantly increased the binding activity between E2F1 and the promoter regions of GAB2 gene (figure 7A, and online supplemental figure S7E). To validate the effectiveness of the E2F1/GAB2/AKT axis, E2F1 KO, E2F1 OE, GAB2 KO, and GAB2 OE were transfected into A549 and H1975 cells, respectively, then western blotting assay was conducted to analyze the expression of downstream proteins. In consistency with our hypothesis, E2F1 KO decreased the protein level of GAB2 while overexpressed E2F1 restored it (figure 7B, online supplemental figures S7F and S8B,C). Similarly, GAB2 KO inhibited the AKT phosphorylation at S473 and T308 sites, while GAB2 OE increased the expression of those proteins (figure 7C, and online supplemental figures S7G and S8D,E). Besides, expression of GAB2, p-AKT (both S473 and T308) were decreased in SNORA38B-KO group, but increased in SNORA38B OE treated group in A549 and H1975 cells (figure 7D, online supplemental figures S7H). Overall, our above results suggested that SNORA38B had a favorable effect on GAB2/AKT axis.
Figure 7.
SNORA38B positively regulated GAB2/AKT pathway and facilitated NSCLC cells proliferation, migration, and invasion. (A.) Binding activity between GAB2 promoters and E2F1 was shown using ChIP assay in A549 cells. Beads and anti-IgG were utilized as controls. (B.) Western blotting assay of E2F1 and GAB2 in A549 cells transfected with E2F1 KO/OE. Actin was used as loading control. (C). Western blotting assay of GAB2, AKT and p-AKT (S473 and T308) in A549 cells transfected with GAB2 KO/OE. Actin was used as loading control. (D.) Western blotting assay of E2F1, GAB2, AKT and p-AKT (S473 and T308) in A549 cells transfected with SNORA38B KO/OE. Actin was used as loading control. (E–F.) Immunofluorescence analysis of GAB2 and p-mTOR in A549 cells transfected with SNORA38B KO and SNORA38B KO +SNORA38B OE. (G). Proliferation assay of A549 cells transfected with Scr KO, SNORA38B KO, GAB2 OE and SNORA38B KO +GAB2 OE. EdU staining assay was used and the positive cells were quantified. (H–I.) Migration and invasion of A549 cells transfected with Scr KO, SNORA38B KO, GAB2 OE and SNORA38B KO +GAB2 OE. Transwell migration and invasion assay were performed. The migrated and invaded cells were quantified. (J.) Cell apoptotic analysis of A549 cells transfected with Scr KO, SNORA38B KO, GAB2 OE and SNORA38B KO +GAB2 OE. Annexin V-FITC/PI staining was performed, apoptostic cells were quantified. Assays were conducted in triple. **p<0.01, ***p<0.001, ****p<0.0001, means±SD was shown. Statistical analysis was subjected to Student’s t-test and one-way analysis of variance (comparison between-group using LSD method). AKT, protein kinase B; ChIP, chromatin immunoprecipitation; CTL, Scr-KO + Blank vector; EdU, 5-ethynyl-2’-deoxyuridine; E2F, E2F transcription factor; GAB2, GRB2-associated-binding protein 2; KO, knockout; LSD, the Fisher's least significant difference; NSCLC, non-small cell lung cancer; OE, overexpress; p-mTOR, phosphorylation of mammalian target of rapamycin; Scr, Scramble; SNORA, small nucleolar RNA.
To further clarify the regulatory role of GAB2 and mTOR by SNORA38B in NSCLC cells, immunofluorescence (IF) staining was used to detect GAB2 and p-mTOR(S2448) expression in A549 cells transfected with SNORA38B KO and SNORA38B KO +SNORA38B OE. Results showed that SNORA38B KO could significantly reduce the signal of GAB2 and p-mTOR, while co-transfected with SNORA38B OE could rescue those inhibitory effects (figure 7E,F). Interestingly, in A549 and H1975 cells treated with SNORA38B KO +GAB2 OE, the growth-promoting and anti-apoptotic functions of GAB2 could be effectively reversed by the anti-growth and pro-apoptotic effects of SNORA38B KO treatment (figure 7G, H, I and J, online supplemental figure S8F–I). Together, our data demonstrated that SNORA38B played oncogenic effects through directly binding with E2F1 and thereby activating the GAB2/AKT/mTOR pathway.
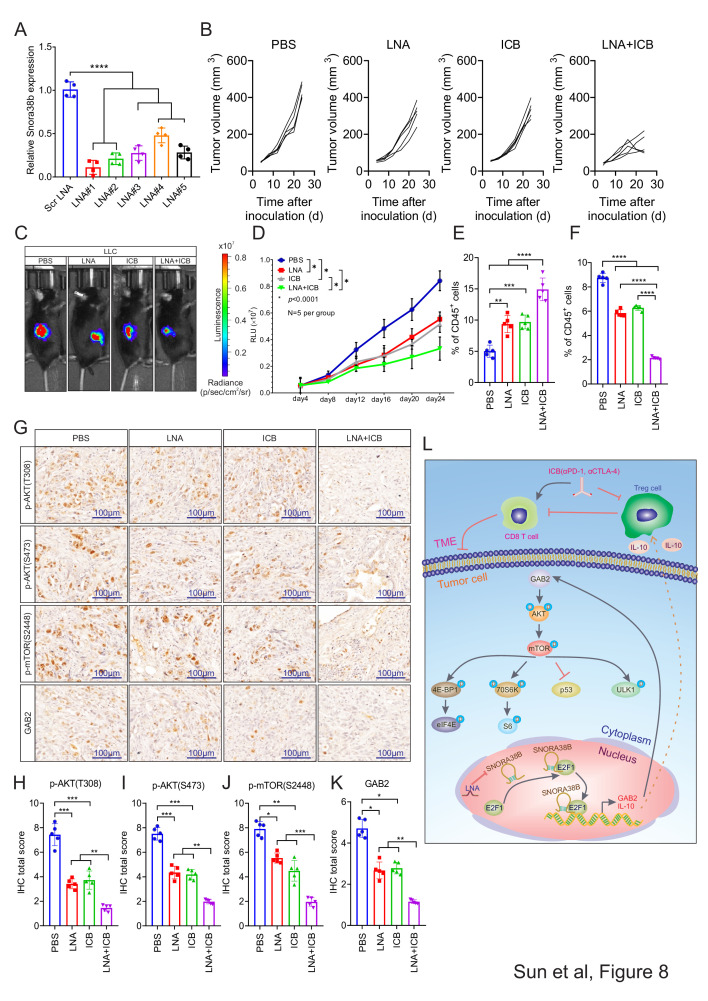
SNORA38B LNAs sensitized NSCLC to ICBs treatment
LNA is a modified RNA nucleoside analog with excellent recognition and strong affinity for DNA and RNA, which could be expected to be used in oligonucleotide therapy for cancer.39 Current researches have shown that LNA is able to knock down the target gene stably as well as effectively inhibiting tumor growth.27 40 Although ICB therapy is only suitable for a small number of patients with NSCLC currently, we speculated that combinatorial treatment of SNORA38B LNAs and ICB (αPD-1, αCTLA-4) may significantly reverse this status given the critical roles of SNORA38B in TME. To confirm our hypothesis, we first verified the knockdown effects of LNAs on SNORA38B RNA levels. Our results indicated that SNORA38B LNAs significantly reduced the expression of SNORA38B, especially, LNA#1 showed the most significant effect (figure 8A). Therefore, LNA#1 was used as the representative SNORA38B LNA. Then, syngeneic tumor mouse models were established using luciferase-labeled LLC cell lines with C57BL/6J mice. Four days after tumor inoculation, tumor-bearing C57BL/6J mice were treated by PBS, SNORA38B LNA, ICB or SNORA38B LNA+ICB via i.p. every 4 days for six doses, respectively. Results showed that SNORA38B LNA+ICB treatment significantly suppressed the tumor growth, and its inhibitory effect was stronger than those in SNORA38B LNA or ICB treatment alone (figure 8C,D). The isolated tumors from LNA+ICB group also showed significantly smaller sizes than from other three groups (figure 8B). The combinatorial treatment using LNA and ICB exhibited minimal effects on histological analysis of major organs, including heart, kidney, liver, spleen, lung and pancreas (online supplemental figure S9). We then digested tumors into single cells for flow cytometry analysis, and performed IHC staining with the isolated formalin-fixed paraffin-embedded tumor sections from C57BL/6J mice. Our results demonstrated that the combinatorial treatment of SNORA38B LNA and ICB significantly promoted the infiltration of CD3+CD8+ T cells and remarkably suppressed the recruitment of CD4+FOXP3+ Tregs (figure 8E,F, online supplemental figure S10A–D). In addition, IHC staining showed a substantially lower signal of p-AKT (T308, S473), p-mTOR (S2448), GAB2 in SNORA38B LNA+ICB treatment group than those in PBS, LNA or ICB treatment groups (figure 8G, H, I, J and K). Taken together, our research demonstrated that SNORA38B contributed to NSCLC initiation and progression via directly interacting with E2F1 and then activating GAB2/AKT/mTOR pathway and its downstream effectors, as well as accumulating Tregs in TME of NSCLC by promoting IL-10 secretion (figure 8L). Hence, targeting SNORA38B by LNAs further sensitized lung tumors to the ICB treatments.
Figure 8.
SNORA38B LNAs sensitized NSCLC to ICBs treatment. (A) Transfection efficiency of LNAs in LLC cells. (B) Tumor volume examined in LLC derived tumors in tumor burden C57BL/6J mice according to formula 0.5×L×W2. n=5 per group. (C–D) Representative images and quantification of BLI in the luciferase reporter gene labeled LLC tumor regions in C57BL/6J mice treated with PBS, LNA, ICB or LNA+ICB. Signals were recorded every 4 days before reaching the observation endpoint. n=5 per group. (E–F) Flow cytometry detection of CD3+CD8+ T cells, CD4+FOXP3+ Tregs and their percentage of CD45+ cells isolated from LLC cells injected C57BL/6J mice treated with PBS, LNA, ICB or LNA+ICB. (G) IHC staining of AKT/mTOR/GAB2 signing pathway with the isolated burden tumors from C57BL/6J mice treated with PBS, LNA, ICB or LNA+ICB. Bar=100 µm. (H–K) Quantification of IHC staining of AKT/mTOR/GAB2 signing pathway. (L) Basic summary of the carcinogenic mechanism of SNORA38B in NSCLC. SNORA38B directly bound to the transcription factor E2F1, then accelerated the transcription of downstream GAB2 gene, activated AKT/mTOR pathway and its downstream effectors to perform cancer-promoting effects. Besides, SNORA38B recruited Tregs to mediate immunosuppression by promoting the secretion of IL-10. Knockdown of SNORA38B using LNAs could sensitize NSCLC to ICB treatment. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001. Assays were conducted in triple. Means±SD was shown. Statistical analysis was subjected to one-way analysis of variance. Comparison between-group using LSD method. BLI, bioluminescence imaging; CTLA-4, cytotoxic T-lymphocytes-associated protein 4; E2F, E2F transcription factor; GAB2, GRB2-associated-binding protein 2; ICB, immune checkpoint blockade; IHC, immunohistochemistry; IL, interleukin; LLC, murine Lewis lung carcinoma; LNA, locked nucleic acid; LSD, the Fisher's least significant difference; NSCLC, non-small cell lung cancer; p-AKT, phosphorylation of protein kinase B; PBS, phosphate-buffered saline; PD-1, programmed cell death protein-1; p-mTOR, phosphorylation of mammalian target of rapamycin; Scr, scramble; SNORNA, small nucleolar RNA; TME, tumor microenvironment; Treg, regulatory T cells;.
Discussion
SnoRNA is a class of newly discovered small non-coding RNA existing widely in nucleolus with the length in 60–300 nt.8 With the increasing studies of snoRNAs, they have been found involved in many processes during human biological activities, including ribosomal RNA (rRNA) processing, RNA splicing, translation regulation, and so on.41–44 Dysregulated snoRNAs acted as important markers for breast cancer,45 lung cancer,46 lymphoma,47 etc. In this study, we found that SNORA38B was significantly upregulated in NSCLC tissues, facilitated tumor development by promoting cell proliferation, migration, invasion, metastasis and inhibiting cell apoptosis in vivo and in vitro, which suggested that it may be another useful biomarker of NSCLC.
Studies have shown that snoRNAs involved in post-transcriptional epigenetic modification of rRNAs, thereby affecting translation of mRNA. Liu et al revealed that SNORA23 could reduce the 28S rRNA 2’-O-ribose methylation through directly binding with, and then significantly inhibiting the phosphorylation of downstream 4E-BP1, resulting in damage to the initiation of ribosomal translation and inhibition of the occurrence for liver cancer.48 However, SNORA42A could increase the specificity of ribosomal protein translation through 18s-U116 methylation and maintain the cell proliferation of leukemia.12 In addition to the translation-affected functions above, post-transcriptional modifications to mRNAs by SNORA74B or other C/D-box snoRNAs have also been reported in tumor progression, and these dysregulations primarily resulted in the activation of mTOR signaling pathway.49–51 As a member of H/ACA box snoRNAs family, SNORA38B owned conserved double-hairpin structure with H box sequence-containing hinge region in human and mouse.52 In our current study, we found that mTOR pathway was also activated by SNORA38B. Further verification showed that the downstream protein 4E-BP1, 70S6K, ULK1 were phosphorylated and p53 activity was inhibited. 70S6K and 4E-BP1 are two direct effectors downstream of mTOR, which play essential roles in translational initiation, protein synthesis, cell cycle, cell migration, etc.53 Phosphorylation of 4E-BP1 accelerated the translation of mRNA containing 5’TOP sequences, which indirectly promoted the translation of ribosomal proteins containing the similar structure,54 55 while 70S6K directly regulated rRNA synthesis.56 ULK1 is a core protein in autophagy signaling pathway with serine/threonine kinase activity. Studies have shown that mTOR could bind to serine at position 757 of ULK1 to inhibit interaction between ULK1-AMPK, leading to the inactivation of ULK1 and finally turning off autophagy signal and inhibiting cell apoptosis.57 58 As a tumor suppressive gene, the inhibition of p53 activity greatly enhanced the cancer-promoting effect in different cancer types.59 60 Furthermore, p53 could also directly bind to and inhibit rRNA methyl-transferase fibrillarin expression, resulting in dysregulated rRNA methylation and leading to the increased intrinsic ribosome activity and accelerated cellular self-renewal.61 62 In our study, as nuclear localized snoRNA, SNORA38B was found directly binding to transcriptional factor E2F1, and regulated the transcription of the downstream for GAB2 gene. GAB2 has been implicated essential for signaling pathways of ERK and PI3K-AKT in various cancer types, especially in breast cancer.63 GAB2 was also reported to upregulate mmp2/9 proteins in NSCLC, resulting in accelerated migration and invasion.64 We found that, as downstream gene of transcriptional factor E2F1, GAB2 was activated and associated with a range of malignant phenotypes in NSCLC cells. Meanwhile, it activated AKT/mTOR pathway, which further contributed to the malignancy of cancer cells. Therefore, our results demonstrated that SNORA38B played oncogenic roles through the GAB2/AKT/mTOR axis.
One of the core functions of the immune system is to phagocytize and destroy allogeneic cells in human bodies. Changes in the immune microenvironment may be closely related to the development and deterioration of tumors by reducing the sensitivity of T lymphocyte to cancer cells. To date, accumulated studies have shown that non-coding RNAs, such as long non-coding RNAs, microRNAs, etc, play important roles in tumor immune escape by regulating the proportion of different immune cell groups and the activities of cytokines in TME.3 27 29 65 However, the regulatory mechanism of TME related to snoRNA is still under elucidating. Our present study demonstrated that SNORA38B could promote the secretion of IL-10 to recruit CD3+FOXP3+ Tregs and thereby reduce CD3+CD8+ T cells infiltration, leading to a tumor immunosuppressive microenvironment. Besides, as an immunosuppressive cytokine, IL-10 maintained the expression and activity of FOXP3 in Tregs,32 and promoted the expression of TGF-beta by IL-10 positive feedback, so that facilitating tumor progression cooperatively.66 67 Moreover, rapamycin was widely used as an effective immunomodulator in treatment anti-rejection and antitumor. Previous studies have shown that mTORC1 promoted IL-10 production by intestinal DCs in intestinal,68 and this regulatory effect may also exist in immune cells in TME, which further increasing the recruitment of Tregs synergistically with SNORA38B in NSCLC. In this study, SNORA38B was blocked by LNAs in LLC-derived tumor-burden C57BL/6J mice, and we found that SNORA38B-based LNA could effectively improve the immune environment and sensitize NSCLC to ICB treatment. Hence, we believed that SNORA38B might be one of the biomarkers for evaluating the prognosis and therapeutic target for NSCLC. However, there are still some limitations for our study: are there any other potential mechanisms that make SNORA38B’ oncogenic pathway more comprehensive? The clinical evidence that combinatorial treatment of SNORA38B LNAs and ICB in NSCLC are relatively weak. The answers to the above questions and the clinical evidence will provide more useful evidence for SNORA38B as a therapeutic target for NSCLC.
Conclusions
Taken all into account, our results indicated that SNORA38B was highly expressed in NSCLC tissues and cell lines, and higher expressed of SNORA38B was related to the poorer prognosis of patients with NSCLC. SNORA38B could facilitate cell proliferation, migration, invasion, and suppress cell apoptosis both in vivo and in vitro, thereby exerting its oncogenic effects. In addition, we also found that CD4+FOXP3+ Tregs infiltration was increased while CD3+CD8+ T cells infiltration was decreased in SNORA38B-enriched NSCLC TME due to the secretion of IL-10. Besides, our data showed that SNORA38B activated the GAB2/AKT/mTOR axis and its downstream effectors by directly binding to the transcription factor E2F1. Finally, we found that SNORA38B LNA could sensitize NSCLC to ICBs treatment, suggesting that SNORA38B might be a potential therapeutic target of NSCLC.
Footnotes
YZ, SL and WH contributed equally.
Correction notice: This article has been corrected after publication to remove Dejia Liu from the author list as they did not fulfil BMJ’s authorship criteria.
Contributors: CS conceived and designed the research. YZ, SL, YS, JZ, FZ, YZ, JW, ML, JC, and HQ conducted the experiments and analyzed the data. WH conducted the bioinformatics analysis. YZ, SL, YS, JZ, FZ, YZ, JW, ML, JC, and HQ acquired and analyzed the data. CS, DL, YZ, and SL wrote the manuscript. CS and SL obtained the funding to support the project and supervised the development of the work. All authors read and approved the final version of the manuscript. CS and DL are responsible for the overall content as guarantors.
Funding: This study was funded by the National Natural Science Foundation of China (No. 81802285), the Fundamental Research Funds for the Central Universities (No. 2015305020202, 2042018kf0025), China Postdoctoral Science Foundation (No. 2017 M620340), National Postdoctoral Program for Innovative Talents (No. BX201700178), Health Commission of Hubei Province scientific research project (grant No. WJ2019Q039), Natural Science Foundation of Hubei Province (No. 2020CFB564), the Hubei Province Key Laboratory of Occupational Hazard Identification and Control (No. OHIC2017Y02), the ChuTian Scholarship, the Wuhan University Startup Funds, the China Scholarship Council (No. 201806275014), and the Independent Research Funds of School of Public Health at Wuhan University (No. ZZKY0014) to CS. It was also supported by the Health Commission of Wuhan City Scientific Research Project (No. WG18Q01) to SL.
Competing interests: The authors declare that they have no competing interests.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Data availability statement
All data generated or analyzed during this study are included in this published article and its supplementary information files. Raw data are available on reasonable request.
Ethics statements
Patient consent for publication
Not applicable.
Ethics approval
All animal experiments were conducted with the approval of the Institutional Animal Care and Use Committee of Wuhan University. Ethical approval was obtained from the ethical committee of the Wuhan University.
References
- 1. Herbst RS, Morgensztern D, Boshoff C. The biology and management of non-small cell lung cancer. Nature 2018;553:446–54. 10.1038/nature25183 [DOI] [PubMed] [Google Scholar]
- 2. Gridelli C, Rossi A, Carbone DP, et al. Non-small-cell lung cancer. Nat Rev Dis Primers 2015;1:15009. 10.1038/nrdp.2015.9 [DOI] [PubMed] [Google Scholar]
- 3. Sun C-C, Zhu W, Li S-J, et al. FOXC1-mediated LINC00301 facilitates tumor progression and triggers an immune-suppressing microenvironment in non-small cell lung cancer by regulating the HIF1α pathway. Genome Med 2020;12:77. 10.1186/s13073-020-00773-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Jonna S, Subramaniam DS. Molecular diagnostics and targeted therapies in non-small cell lung cancer (NSCLC): an update. Discov Med 2019;27:167–70. [PubMed] [Google Scholar]
- 5. Pan J, Fang S, Tian H, et al. lncRNA JPX/miR-33a-5p/Twist1 axis regulates tumorigenesis and metastasis of lung cancer by activating Wnt/β-catenin signaling. Mol Cancer 2020;19:9. 10.1186/s12943-020-1133-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Osmani L, Askin F, Gabrielson E, et al. Current WHO guidelines and the critical role of immunohistochemical markers in the subclassification of non-small cell lung carcinoma (NSCLC): moving from targeted therapy to immunotherapy. Semin Cancer Biol 2018;52:103–9. 10.1016/j.semcancer.2017.11.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Esteller M. Non-coding RNAs in human disease. Nat Rev Genet 2011;12:861–74. 10.1038/nrg3074 [DOI] [PubMed] [Google Scholar]
- 8. Bratkovič T, Rogelj B. Biology and applications of small nucleolar RNAs. Cell Mol Life Sci 2011;68:3843–51. 10.1007/s00018-011-0762-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Siprashvili Z, Webster DE, Johnston D, et al. The noncoding RNAs SNORD50A and SNORD50B bind K-Ras and are recurrently deleted in human cancer. Nat Genet 2016;48:53–8. 10.1038/ng.3452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Su X, Feng C, Wang S, et al. The noncoding RNAs SNORD50A and SNORD50B-mediated TRIM21-GMPS interaction promotes the growth of p53 wild-type breast cancers by degrading p53. Cell Death Differ 2021;28:2450–64. 10.1038/s41418-021-00762-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhu W, Niu J, He M, et al. SNORD89 promotes stemness phenotype of ovarian cancer cells by regulating Notch1-c-Myc pathway. J Transl Med 2019;17:259. 10.1186/s12967-019-2005-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pauli C, Liu Y, Rohde C, et al. Site-specific methylation of 18S ribosomal RNA by SNORD42A is required for acute myeloid leukemia cell proliferation. Blood 2020;135:2059–70. 10.1182/blood.2019004121 [DOI] [PubMed] [Google Scholar]
- 13. Zhang Z, Tao Y, Hua Q, et al. SNORA71A promotes colorectal cancer cell proliferation, migration, and invasion. Biomed Res Int 2020;2020:8284576. 10.1155/2020/8284576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dong X-Y, Rodriguez C, Guo P, et al. SnoRNA U50 is a candidate tumor-suppressor gene at 6q14.3 with a mutation associated with clinically significant prostate cancer. Hum Mol Genet 2008;17:1031–42. 10.1093/hmg/ddm375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mei Y-P, Liao J-P, Shen J, et al. Small nucleolar RNA 42 acts as an oncogene in lung tumorigenesis. Oncogene 2012;31:2794–804. 10.1038/onc.2011.449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cui C, Liu Y, Gerloff D, et al. NOP10 predicts lung cancer prognosis and its associated small nucleolar RNAs drive proliferation and migration. Oncogene 2021;40:909–21. 10.1038/s41388-020-01570-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lin A, Hu Q, Li C, et al. The LINK-A lncRNA interacts with PtdIns(3,4,5)P 3 to hyperactivate AKT and confer resistance to AKT inhibitors. Nat Cell Biol 2017;19:238–51. 10.1038/ncb3473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lin A, Li C, Xing Z, et al. The LINK-A lncRNA activates normoxic HIF1α signalling in triple-negative breast cancer. Nat Cell Biol 2016;18:213–24. 10.1038/ncb3295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sun C-C, Zhang L, Li G, et al. The lncRNA PDIA3P interacts with miR-185-5p to modulate oral squamous cell carcinoma progression by targeting cyclin D2. Mol Ther Nucleic Acids 2017;9:100–10. 10.1016/j.omtn.2017.08.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sun C, Li S, Zhang F, et al. Long non-coding RNA NEAT1 promotes non-small cell lung cancer progression through regulation of miR-377-3p-E2F3 pathway. Oncotarget 2016;7:51784–814. 10.18632/oncotarget.10108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol 1993;234:779–815. 10.1006/jmbi.1993.1626 [DOI] [PubMed] [Google Scholar]
- 22. Parisien M, Major F. The MC-fold and MC-Sym pipeline infers RNA structure from sequence data. Nature 2008;452:51–5. 10.1038/nature06684 [DOI] [PubMed] [Google Scholar]
- 23. Huang Y, Li H, Xiao Y. 3dRPC: a web server for 3D RNA-protein structure prediction. Bioinformatics 2018;34:1238–40. 10.1093/bioinformatics/btx742 [DOI] [PubMed] [Google Scholar]
- 24. Yan Y, Tao H, He J, et al. The HDOCK server for integrated protein-protein docking. Nat Protoc 2020;15:1829–52. 10.1038/s41596-020-0312-x [DOI] [PubMed] [Google Scholar]
- 25. Yan Y, Zhang D, Zhou P, et al. HDOCK: a web server for protein-protein and protein-DNA/RNA docking based on a hybrid strategy. Nucleic Acids Res 2017;45:W365–73. 10.1093/nar/gkx407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tuszynska I, Magnus M, Jonak K, et al. NPDock: a web server for protein-nucleic acid docking. Nucleic Acids Res 2015;43:W425–30. 10.1093/nar/gkv493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hu Q, Ye Y, Chan L-C, et al. Oncogenic lncRNA downregulates cancer cell antigen presentation and intrinsic tumor suppression. Nat Immunol 2019;20:835–51. 10.1038/s41590-019-0400-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Huang D, Chen J, Yang L, et al. NKILA lncRNA promotes tumor immune evasion by sensitizing T cells to activation-induced cell death. Nat Immunol 2018;19:1112–25. 10.1038/s41590-018-0207-y [DOI] [PubMed] [Google Scholar]
- 29. Zheng Y, Tian X, Wang T, et al. Long noncoding RNA PVT1 regulates the immunosuppression activity of granulocytic myeloid-derived suppressor cells in tumor-bearing mice. Mol Cancer 2019;18:61. 10.1186/s12943-019-0978-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sobecki M, Mrouj K, Colinge J, et al. Cell-cycle regulation accounts for variability in Ki-67 expression levels. Cancer Res 2017;77:2722–34. 10.1158/0008-5472.CAN-16-0707 [DOI] [PubMed] [Google Scholar]
- 31. Xia L, Liu Y, Wang Y. PD-1/PD-L1 blockade therapy in advanced non-small-cell lung cancer: current status and future directions. Oncologist 2019;24:S31–41. 10.1634/theoncologist.2019-IO-S1-s05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Murai M, Turovskaya O, Kim G, et al. Interleukin 10 acts on regulatory T cells to maintain expression of the transcription factor FOXP3 and suppressive function in mice with colitis. Nat Immunol 2009;10:1178–84. 10.1038/ni.1791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lippitz BE. Cytokine patterns in patients with cancer: a systematic review. Lancet Oncol 2013;14:e218–28. 10.1016/S1470-2045(12)70582-X [DOI] [PubMed] [Google Scholar]
- 34. Konkel JE, Chen W. Balancing acts: the role of TGF-β in the mucosal immune system. Trends Mol Med 2011;17:668–76. 10.1016/j.molmed.2011.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Vignali DAA, Collison LW, Workman CJ. How regulatory T cells work. Nat Rev Immunol 2008;8:523–32. 10.1038/nri2343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Liu GY, Sabatini DM. mTOR at the nexus of nutrition, growth, ageing and disease. Nat Rev Mol Cell Biol 2020;21:183–203. 10.1038/s41580-019-0199-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chaussepied M, Ginsberg D. Transcriptional regulation of AKT activation by E2F. Mol Cell 2004;16:831–7. 10.1016/j.molcel.2004.11.003 [DOI] [PubMed] [Google Scholar]
- 38. Sun C-C, Zhou Q, Hu W, et al. Transcriptional E2F1/2/5/8 as potential targets and transcriptional E2F3/6/7 as new biomarkers for the prognosis of human lung carcinoma. Aging 2018;10:973–87. 10.18632/aging.101441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Petersen M, Wengel J. LNA: a versatile tool for therapeutics and genomics. Trends Biotechnol 2003;21:74–81. 10.1016/S0167-7799(02)00038-0 [DOI] [PubMed] [Google Scholar]
- 40. Herkt M, Thum T. Pharmacokinetics and proceedings in clinical application of nucleic acid therapeutics. Mol Ther 2021;29:521–39. 10.1016/j.ymthe.2020.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bratkovič T, Božič J, Rogelj B. Functional diversity of small nucleolar RNAs. Nucleic Acids Res 2020;48:1627–51. 10.1093/nar/gkz1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kiss T. Small nucleolar RNAs: an abundant group of noncoding RNAs with diverse cellular functions. Cell 2002;109:145–8. 10.1016/s0092-8674(02)00718-3 [DOI] [PubMed] [Google Scholar]
- 43. McMahon M, Contreras A, Ruggero D. Small RNAs with big implications: new insights into H/ACA snoRNA function and their role in human disease. Wiley Interdiscip Rev RNA 2015;6:173–89. 10.1002/wrna.1266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Watkins NJ, Bohnsack MT. The box C/D and H/ACA snoRNPs: key players in the modification, processing and the dynamic folding of ribosomal RNA. Wiley Interdiscip Rev RNA 2012;3:397–414. 10.1002/wrna.117 [DOI] [PubMed] [Google Scholar]
- 45. Schulten H-J, Bangash M, Karim S, et al. Comprehensive molecular biomarker identification in breast cancer brain metastases. J Transl Med 2017;15:269. 10.1186/s12967-017-1370-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Gao L, Ma J, Mannoor K, et al. Genome-wide small nucleolar RNA expression analysis of lung cancer by next-generation deep sequencing. Int J Cancer 2015;136:E623–9. 10.1002/ijc.29169 [DOI] [PubMed] [Google Scholar]
- 47. Valleron W, Ysebaert L, Berquet L, et al. Small nucleolar RNA expression profiling identifies potential prognostic markers in peripheral T-cell lymphoma. Blood 2012;120:3997–4005. 10.1182/blood-2012-06-438135 [DOI] [PubMed] [Google Scholar]
- 48. Liu Z, Pang Y, Jia Y, et al. SNORA23 inhibits HCC tumorigenesis by impairing the 2'-O-ribose methylation level of 28S rRNA. Cancer Biol. Med. 2021;18. 10.20892/j.issn.2095-3941.2020.0343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Qin Y, Meng L, Fu Y, et al. SNORA74B gene silencing inhibits gallbladder cancer cells by inducing PHLPP and suppressing Akt/mTOR signaling. Oncotarget 2017;8:19980–96. 10.18632/oncotarget.15301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Matsuoka T, Yashiro M. The role of PI3K/Akt/mTOR signaling in gastric carcinoma. Cancers 2014;6:1441–63. 10.3390/cancers6031441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Su H, Xu T, Ganapathy S, et al. Elevated snoRNA biogenesis is essential in breast cancer. Oncogene 2014;33:1348–58. 10.1038/onc.2013.89 [DOI] [PubMed] [Google Scholar]
- 52. Ganot P, Caizergues-Ferrer M, Kiss T. The family of box ACA small nucleolar RNAs is defined by an evolutionarily conserved secondary structure and ubiquitous sequence elements essential for RNA accumulation. Genes Dev 1997;11:941–56. 10.1101/gad.11.7.941 [DOI] [PubMed] [Google Scholar]
- 53. Pópulo H, Lopes JM, Soares P. The mTOR signalling pathway in human cancer. Int J Mol Sci 2012;13:1886–918. 10.3390/ijms13021886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Pause A, Belsham GJ, Gingras AC, et al. Insulin-dependent stimulation of protein synthesis by phosphorylation of a regulator of 5'-cap function. Nature 1994;371:762–7. 10.1038/371762a0 [DOI] [PubMed] [Google Scholar]
- 55. Meyuhas O, Dreazen A. Ribosomal protein S6 kinase from TOP mRNAs to cell size. Prog Mol Biol Transl Sci 2009;90:109–53. 10.1016/S1877-1173(09)90003-5 [DOI] [PubMed] [Google Scholar]
- 56. Gentilella A, Kozma SC, Thomas G. A liaison between mTOR signaling, ribosome biogenesis and cancer. Biochim Biophys Acta 2015;1849:812–20. 10.1016/j.bbagrm.2015.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kim J, Kundu M, Viollet B, et al. AMPK and mTOR regulate autophagy through direct phosphorylation of ULK1. Nat Cell Biol 2011;13:132–41. 10.1038/ncb2152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Wang C, Cigliano A, Jiang L, et al. 4EBP1/eIF4E and p70S6K/RPS6 axes play critical and distinct roles in hepatocarcinogenesis driven by AKT and N-Ras proto-oncogenes in mice. Hepatology 2015;61:200–13. 10.1002/hep.27396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Bykov VJN, Eriksson SE, Bianchi J, et al. Targeting mutant p53 for efficient cancer therapy. Nat Rev Cancer 2018;18:89–102. 10.1038/nrc.2017.109 [DOI] [PubMed] [Google Scholar]
- 60. Mantovani F, Collavin L, Del Sal G. Mutant p53 as a guardian of the cancer cell. Cell Death Differ 2019;26:199–212. 10.1038/s41418-018-0246-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Liu Z, Pang Y, Jia Y, et al. SNORA23 inhibits HCC tumorigenesis by impairing the 2'-O-ribose methylation level of 28S rRNA. Cancer Biol Med 2021;19:104–19. 10.20892/j.issn.2095-3941.2020.0343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Marcel V, Ghayad SE, Belin S, et al. P53 acts as a safeguard of translational control by regulating fibrillarin and rRNA methylation in cancer. Cancer Cell 2013;24:318–30. 10.1016/j.ccr.2013.08.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Adams SJ, Aydin IT, Celebi JT. GAB2-a scaffolding protein in cancer. Mol Cancer Res 2012;10:1265–70. 10.1158/1541-7786.MCR-12-0352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Xu LJ, Wang YC, Lan HW, et al. Grb2-associated binder-2 gene promotes migration of non-small cell lung cancer cells via Akt signaling pathway. Am J Transl Res 2016;8:1208–17. [PMC free article] [PubMed] [Google Scholar]
- 65. Zhou J, Li X, Wu X, et al. Exosomes released from tumor-associated macrophages transfer miRNAs that induce a Treg/Th17 cell imbalance in epithelial ovarian cancer. Cancer Immunol Res 2018;6:1578–92. 10.1158/2326-6066.CIR-17-0479 [DOI] [PubMed] [Google Scholar]
- 66. Mocellin S, Marincola FM, Young HA. Interleukin-10 and the immune response against cancer: a counterpoint. J Leukoc Biol 2005;78:1043–51. 10.1189/jlb.0705358 [DOI] [PubMed] [Google Scholar]
- 67. Batlle E, Massagué J. Transforming growth factor-β signaling in immunity and cancer. Immunity 2019;50:924–40. 10.1016/j.immuni.2019.03.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Ohtani M, Hoshii T, Fujii H, et al. Cutting edge: mTORC1 in intestinal CD11c+ CD11b+ dendritic cells regulates intestinal homeostasis by promoting IL-10 production. J Immunol 2012;188:4736–40. 10.4049/jimmunol.1200069 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
jitc-2021-004113supp001.xlsx (12.8KB, xlsx)
jitc-2021-004113supp002.xlsx (14.3KB, xlsx)
jitc-2021-004113supp003.xlsx (14.9KB, xlsx)
jitc-2021-004113supp004.xlsx (14.2KB, xlsx)
jitc-2021-004113supp005.pdf (6.3MB, pdf)
Data Availability Statement
All data generated or analyzed during this study are included in this published article and its supplementary information files. Raw data are available on reasonable request.