Abstract
Background
Although coronavirus disease 2019 (COVID‐19) is primarily a respiratory infection, mounting evidence suggests that the gastrointestinal tract is involved in the disease, with gut barrier dysfunction and gut microbiota alterations being related to disease severity. Whether these alterations persist and are related to long‐term respiratory dysfunction remains unknown.
Methods
Plasma was collected during hospital admission and after 3 months from the NOR‐Solidarity trial (n = 181) and analyzed for markers of gut barrier dysfunction and inflammation. At the 3‐month follow‐up, pulmonary function was assessed by measuring the diffusing capacity of the lungs for carbon monoxide (DLCO). Rectal swabs for gut microbiota analyses were collected (n = 97) and analyzed by sequencing the 16S rRNA gene.
Results
Gut microbiota diversity was reduced in COVID‐19 patients with respiratory dysfunction, defined as DLCO below the lower limit of normal 3 months after hospitalization. These patients also had an altered global gut microbiota composition, with reduced relative abundance of 20 bacterial taxa and increased abundance of five taxa, including Veillonella, potentially linked to fibrosis. During hospitalization, increased plasma levels of lipopolysaccharide‐binding protein (LBP) were strongly associated with respiratory failure, defined as pO2/fiO2 (P/F ratio) <26.6 kPa. LBP levels remained elevated during and after hospitalization and were associated with low‐grade inflammation and respiratory dysfunction after 3 months.
Conclusion
Respiratory dysfunction after COVID‐19 is associated with altered gut microbiota and persistently elevated LBP levels. Our results should be regarded as hypothesis generating, pointing to a potential gut–lung axis that should be further investigated in relation to long‐term pulmonary dysfunction and long COVID.
Keywords: microbiome, pulmonary function, SARS‐CoV‐2
Introduction
Although coronavirus disease 2019 (COVID‐19) is primarily a viral respiratory disease caused by severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2), aberrant immune responses to the causative virus, resulting in systemic inflammation and multi‐organ involvement, are central features in severe and critical disease. Mounting evidence suggests that the gastrointestinal (GI) tract is involved in the pathogenesis of COVID‐19 [1, 2].
The GI tract is the largest immunological organ in the body, and its resident microbiota modulates regional as well as systemic host immune responses. It has been hypothesized that the gut microbiota could be a mediator of host inflammatory immune responses during COVID‐19, thereby contributing to the pronounced systemic inflammation observed in patients requiring hospitalization [3, 4].
Recent studies suggest that severe SARS‐CoV‐2 infection could compromise the integrity of the gut–blood barrier, leading to enhanced leakage of microbial products, such as lipopolysaccharides (LPS), possibly affecting the host's response to COVID‐19 through activation of the innate immune system [5, 6]. In a small observational study of hospitalized patients with COVID‐19, we recently reported that elevated plasma levels of LPS‐binding protein (LBP) were associated with elevated cardiac markers [7]. However, knowledge on the relationship between markers of gut barrier dysfunction and the degree of respiratory failure, particularly persistently impaired pulmonary dysfunction following COVID‐19 hospitalization, is scarce.
Patients with COVID‐19 exhibit an altered gut microbiota composition [5, 8], and gut microbiome alterations have recently been found to be related to disease severity [9]. Although these alterations persisted when investigated at a median of 6 days after a negative SARS‐CoV‐2 polymerase chain reaction (PCR) test, it is currently not known whether gut microbiota alterations or gut barrier dysfunction persists long term after hospitalization. Moreover, whether microbiota‐related mechanisms could be involved in the persistent inflammation and pulmonary dysfunction observed in some patients during follow‐up after hospitalization for COVID‐19 is unknown.
The NOR‐Solidarity trial is an independent add‐on study to the WHO Solidarity trial [10], which has recently reported no effects of hydroxychloroquine (HCQ) or remdesivir compared to standard of care (SoC) on clinical outcome, viral clearance, or systemic inflammation in hospitalized patients with COVID‐19 [11]. In the present substudy, we investigated whether gut microbiota collected 3 months after hospital admission, and levels of the gut barrier dysfunction marker LBP measured during and after hospitalization, were related to pulmonary dysfunction after COVID‐19.
Materials and methods
Study design and participants
NOR‐Solidarity is a multicentre, open‐label, adaptive randomized clinical trial evaluating the effect of antiviral drugs on hospitalized patients with COVID‐19 admitted to 23 Norwegian hospitals [11]. In addition, NOR‐Solidarity included collections from a blood biobank and outpatient visits at a 3‐month follow‐up after hospital admission. The study was approved by the Committee for Medical Research Ethics, Region Southeast Norway (118684) and the Norwegian Medicines Agency (20/04950‐23) and registered at ClinicalTrials.gov (NCT04321616). Participants were included from 28 March until 5 October 2020, and all participants >18 years of age admitted to the hospital with PCR‐confirmed SARS‐2‐CoV‐2 infection were eligible for inclusion. Exclusion criteria as described in the original study protocol included severe comorbidity (life expectancy <3 months), high levels of liver transaminases (aspartate aminotransferase [AST]/alanine aminotransferase [ALT] >5 times the upper limit of normal), corrected QT interval time as assessed by electrocardiography >470 ms, pregnancy, breast feeding, acute comorbidity occurrence in a 7‐day period before inclusion, known intolerance to study drugs, concomitant medications interfering with the study drugs, and participation in a confounding trial [11]. All participants provided informed consent prior to inclusion in the study.
Intervention and outcomes
In the NOR‐Solidarity study (n = 181), participants were randomized and allocated to one of three treatment arms: (1) local SoC; (2) SoC plus 800 mg of oral HCQ twice daily on day one, followed by 400 mg twice daily for up to 9 days; or (3) SoC plus 200 mg of intravenous remdesivir on day one, followed by 100 mg daily for up to 9 days. All study treatments were stopped at hospital discharge. Since the interventions had no effect on clinical outcome, viral clearance, or systemic inflammation [11], data from the different intervention arms were pooled for analyses in this substudy to examine whether gut microbiota composition after 3 months, and marker of gut barrier dysfunction during hospitalization and after 3 months, had any effect on (i) acute respiratory failure defined as pO2/fiO2 (P/F ratio) <26.6 kPa (<200 mmHg) during hospitalization, and (ii) respiratory dysfunction after 3 months defined as diffusing capacity of the lungs for carbon monoxide (DLCO) below the lower limit of normal (LLN). To ensure the lack of influence of the different treatments, data were also adjusted for the treatment group.
Soluble LPS‐binding protein measurements
Plasma levels of soluble LBP were measured using an enzyme immunoassay, as described in the Supplementary Methods.
Three‐month follow‐up
Three months after hospital admission, 149 participants attended a follow‐up visit that included blood sampling for routine clinical biochemistry and biobanking [11]. Rectal swabs were collected from a subgroup of participants (n = 97). Lung function tests (n = 108), consisting of spirometry and DLCO, were performed at each participating centre according to the guidelines of the European Respiratory Society and the American Thoracic Society [12, 13], as previously described [14]. Studies have shown that DLCO is the pulmonary measure most frequently affected after hospitalization for COVID‐19 [14, 15]. For the purpose of this study, we chose DLCO as a measure of pulmonary function, as previously reported from the NOR‐Solidarity cohort [16]. The DLCO, in per cent of predicted and the LLN, was calculated according to the Global Lung Function Initiative Network (GLI) [17], as previously described [14]. Of the 108 participants who underwent lung function tests, 83 also underwent rectal swab collection.
Gut microbiota analyses
Rectal swabs were stored in a stabilizing transportation medium (soluble Amies, Thermo Scientific™) and frozen at −80°C until analysis. Fecal DNA was extracted using the QIAamp PowerFecal®Pro DNA Kit (Qiagen, Germany), with slight modifications. Briefly, 700 μL of fecal material was pelleted and homogenized in 800 μL of kit solution CD1 using a bead beater (2 × 60 s at 5.5 ms, 20°C) and further processed according to the manufacturer's protocol. Libraries for 16S rRNA amplicon sequencing were generated as previously described [18]. Briefly, the hypervariable regions V3 and V4 of the 16S rRNA gene were amplified using dual‐indexed universal primers 319F (forward) and 806R (reverse), and Phusion High‐Fidelity PCR Master Mix with HF buffer (Thermo Fisher Scientific, USA). The PCR products were cleaned and normalized using a SequalPrep Normalization Plate Kit (Thermo Fisher Scientific, USA). Quality control and quantification of the pooled libraries were performed using an Agilent Bioanalyzer (Agilent Technologies, USA) and Kapa Library Quantification Kit (Kapa Biosystems, London, UK). Sequencing was performed at the Norwegian Sequencing Centre (Oslo, Norway) using the Illumina MiSeq platform and v3 kit (Illumina, San Diego, CA, USA), allowing for 300 bp paired‐end reads.
Sequence processing and bioinformatics
Paired‐end reads were filtered for Illumina Universal Adapters and PhiX, demultiplexed, quality trimmed, and merged using bbduk 38.90, je 1.2, cutadapt 3.3, and bbmerge 38.90, respectively. Denoising to amplicon sequence variants (ASVs) and taxonomic classification were performed using QIIME2 version 2021.2. Contaminants were filtered using the R package microDecon. Diversity analyses were performed on a rarefied (subsampled) dataset with an ASV count of 7123 per sample. Further details are provided in the Supplementary Methods.
Statistical analyses
Baseline characteristics are described as median (25th and 75th percentile [interquartile range]) for continuous variables and percentages for categorical variables. Group‐wise comparisons were performed using a two‐tailed t‐test or Mann–Whitney U test for continuous data and chi‐square or Fisher's exact test (two sided) for categorical data. Predefined outcomes (acute respiratory failure defined as P/F ratio <26.6 kPa during hospitalization and respiratory dysfunction defined as DLCO < LLN after 3 months of follow‐up) were used as dichotomous variables. LBP levels were non‐normally distributed and transformed using log10 for comparisons between groups with linear mixed model analysis, with subjects as random effects and time and respiratory failure as fixed effects (also as interaction). Treatment was included as a covariate. Multivariate analysis of LBP (defined as Z‐score [standardized] log10 LBP) in relation to respiratory failure was performed using binary logistic regression, adjusting for the covariates age, sex, treatment, and known comorbidities in the base model. Correlation analyses were performed using Spearman's rho (ρ) owing to skewed distribution of the data. P‐values were two sided and considered significant at p < 0.05. Group comparisons of LBP levels in relation to respiratory dysfunction were performed using a parametric t‐test of log10‐transformed data. SPSS release 26.0.0.1 and 27.0.0.0 (IBM SPSS Statistics, IBM Corporation, Armonk, NY, USA) were used for the statistical analysis.
Results
Baseline characteristics
The NOR‐Solidarity trial design and main results have recently been published [11]. A total of 181 randomized patients were included in the present study, and 149 completed the 3‐month follow‐up, of which 108 underwent pulmonary function testing and 97 underwent rectal swab collection for gut microbiota analyses (Fig. 1). Baseline characteristics for the main study group and the subgroups of patients with and without respiratory failure, as well as those with microbiota samples available, are given in Table 1. In the main study group, participants were 59 years old on average and mostly male (66%), and the median body mass index (BMI) was 27.4 kg/m2 (27% obese, BMI >30 kg/m2). The use of antibiotics during hospitalization was reported in 48% of the participants, and 68% reported any known comorbidities, with hypertension (31%), obesity (27%), and diabetes mellitus (17%) as the most common, whereas only 6% reported the presence of chronic pulmonary disease.
Fig. 1.
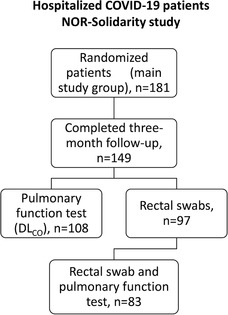
Flow chart of patients included in the present add‐on study based on the original NOR‐Solidarity study protocol.
Table 1.
Results are shown as median values (25th to 75th percentile), unless otherwise specified. Respiratory failure during period of hospitalization is defined as P/F ratio < 26.6 kPa. P‐values refer to chi‐square or Fisher's exact test (two sided) for categorical data, or two‐tailed t‐test or Mann–Whitney U test for continuous data, comparing patients with and without respiratory failure during hospitalization
| Main study group | Microbiota subgroup | Respiratory failure | No respiratory failure | ||
|---|---|---|---|---|---|
| (n = 181) | (n = 97) | (n = 60) | (n = 121) | p | |
| Age, years | 59 (50–71) | 57 (48–65) | 62 (56–75) | 55 (47–65) | <0.001 |
| Male gender (%) | 119 (66) | 60 (62) | 41 (68) | 78 (65) | 0.623 |
| Body mass index (BMI), kg/m2 | 27.4 (24.7–30.9) | 27.7 (25.0–31.6) | 28.1 (25.2–31.9) | 27.1 (24.5–29.8) | 0.512 |
| Antibiotics use (%) | 87 (48) | 41 (42) | 42 (70) | 44 (36) | <0.001 |
| Treatment group | 0.444 | ||||
| Standard of care, SoC (%) | 87 (48) | 47 (49) | 25 (42) | 62 (51) | |
| SoC + hydroxychloroquine (%) | 52 (29) | 33 (34) | 20 (33) | 32 (26) | |
| SoC + remdesivir (%) | 42 (23) | 17 (18) | 15 (25) | 27 (22) | |
| Comorbidities | |||||
| Any known comorbidities (%) | 122 (68) | 66 (68) | 42 (70) | 80 (66) | 0.611 |
| Chronic pulmonary disease (%) | 10 (6) | 5 (5) | 4 (7) | 6 (5) | 0.731 |
| Hypertension (%) | 55 (31) | 28 (29) | 21 (35) | 34 (28) | 0.308 |
| Chronic cardiac disease (%) | 28 (16) | 12 (12) | 13 (22) | 15 (12) | 0.124 |
| Diabetes mellitus (%) | 31 (17) | 19 (20) | 15 (25) | 16 (13) | 0.057 |
| Obesity, BMI >30 kg/m2 (%) | 44 (27) | 28 (30) | 18 (30) | 26 (22) | 0.264 |
| Symptom duration prior to admission, days | 7 (5–10) | 7 (5–10) | 7 (5–10) | 7 (5–10) | 0.814 |
| Length of hospitalization, days | 6 (4–11) | 6 (3–11) | 13 (10–25) | 5 (3–8) | <0.001 |
| Admission to ICU (%) | 35 (19) | 18 (19) | 34 (19) | 1 (1) | <0.001 |
| Invasive mechanical ventilation (%) | 19 (10) | 11 (10) | 19 (32) | 0 | <0.001 |
| P/F‐ratio at admission, kPa | 42.1 (32.0–48.1) | 42.4 (33.4–49.6) | 30.1 (23.4–38.6) | 45.2 (40.1–51.9) | <0.001 |
| C‐reactive protein, mg/L | 70.0 (36.3–70.0) | 66.0 (33.5–122.5) | 113 (65.5–163.5) | 57 (28.3–110.8) | <0.001 |
| White blood cell count, ×109/L | 6.2 (4.7–8.6) | 5.8 (4.3–7.6) | 8.1 (5.5–9.8) | 5.7 (4.5–7.2) | <0.001 |
| Neutrophil count, ×109/L | 4.3 (3.0–6.6) | 3.7 (2.4–6.2) | 6.4 (4.0–8.0) | 3.7 (2.7–5.6) | <0.001 |
| Ferritin, μg/L | 626 (325–1209) | 608 (298–1017) | 1015 (534–1481) | 516 (257–1037) | <0.001 |
| D‐dimer, mg/L | 0.7 (0.5–1.2) | 0.7 (0.4–1.1) | 1.1 (0.5–1.7) | 0.6 (0.4–0.9) | <0.001 |
| Procalcitonin, μg/L | 0.13 (0.10–0.21) | 0.10 (0.10–0.20) | 0.20 (0.12–0.51) | 0.10 (0.10–0.17) | <0.001 |
| Viral load (log10/1000) | 1.9 (0.7–3.1) | 1.7 (0.0–2.9) | 2.5 (1.0–3.2) | 1.7 (0.6–3.1) | 0.111 |
| Lipopolysaccharide‐binding protein, μg/ml | 16.3 (8.5–26.1) | 17.1 (7.3–26.3) | 23.5 (12.1–32.3) | 12.9 (7.2–21.6) | <0.001 |
Abbreviations: P/F‐ratio, pO2/fiO2‐ratio; ICU, intensive care unit.
Patients with respiratory failure were older and hospitalized for longer than patients without, with a higher need for intensive care unit (ICU) admission and invasive mechanical ventilation, more frequent use of antibiotics, and higher levels of inflammatory markers. The clinical characteristics at the 3‐month follow‐up are presented in Table S1. Patients with respiratory dysfunction after 3 months were older and had been hospitalized for longer than those without, with a higher frequency of pre‐existing pulmonary disease and a greater need for ICU admission and invasive mechanical ventilation during hospitalization. However, the levels of inflammatory markers after 3 months were similar in both the groups.
Respiratory dysfunction after 3 months is associated with altered gut microbiota composition and reduced bacterial diversity in previously hospitalized patients with COVID‐19
At the 3‐month follow‐up, 25 out of the 83 (30%) previously hospitalized patients with COVID‐19, with both rectal swabs and DLCO measurements available, had respiratory dysfunction defined as DLCO < LLN. Notably, these patients had altered global gut microbiota composition as assessed by beta diversity (Bray–Curtis [Fig. 2a]). It is important to note that patients who received antibiotics during hospitalization had overlapping global microbiota compositions with those not receiving antibiotics (Fig. 2b). Moreover, there was no statistical interaction between antibiotic use during hospitalization and the association between global microbiota composition and respiratory dysfunction at the 3‐month follow‐up, although such an interaction could not be ruled out due to the limited sample size.
Fig. 2.
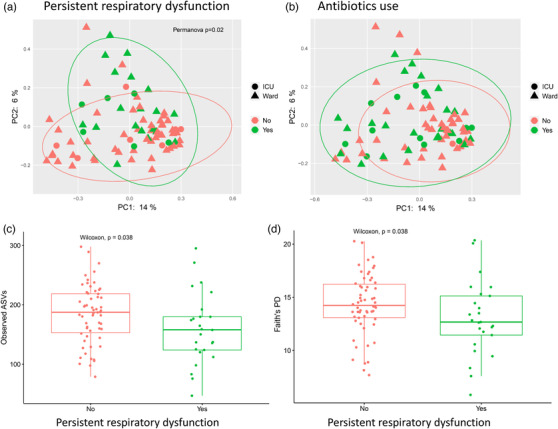
Gut microbiota diversity in patients with or without respiratory dysfunction at the 3‐month follow‐up. Beta diversity by principal coordinate analysis showing Bray–Curtis distances separating patients with or without respiratory dysfunction (diffusing capacity of the lungs for carbon monoxide < lower limit of normal, n = 83) (a), with overlapping microbiota composition in relation to antibiotic use (b). Alpha diversity measured with observed ASVs (c) and Faith`s PD (d) in patients with or without respiratory dysfunction. Abbreviations: ASVs, amplicon sequence variants; PD, phylogenetic diversity.
Patients with respiratory dysfunction 3 months after hospital admission also had reduced bacterial alpha diversity (Faith's phylogenetic diversity [PD] and observed ASVs) compared to patients with respiratory function in the normal range (Fig. 2c,d). Moreover, alpha diversity measures were inversely associated with the need for invasive mechanical ventilation (Faith's PD and observed ASVs), length of hospitalization (Shannon index), and diagnosis of diabetes mellitus, but were not associated with other comorbidities, age, sex, BMI, treatment groups, inflammatory markers, or viral load (Table S2). Although not statistically significant, there was a tendency toward reduced bacterial alpha diversity in patients treated with antibiotics during hospitalization.
At the taxonomic level, screening analysis using linear discriminant analysis effect size (LEfSe) suggested that respiratory dysfunction after 3 months was associated with increased relative abundance of 5 taxa and reduced relative abundance of 20 taxa (Fig. 3a,b), including Erysipelotrichaceae UCG‐003 and several members of the Lachnospiraceae family (Coprococcus, Eubacterium_ventriosum_group, Lachnospiraceae_FCS020_group, Lachnospiraceae_ND3007_group, Fusicatenibacter) and Ruminococcaceae family (Subdoligranulum, Ruminococcus), which are potential producers of butyrate, the main energy source for enterocytes. We subsequently performed the same analysis using the ALDEx2 algorithm (filtered to p < 0.05), reducing the number of genera identified by both LEfSe and ALDEx2 as associated with respiratory dysfunction to three: reduced abundance of Erysipelotrichaceae UCG‐003 and increased abundance of Veillonella and Flavonifractor (Fig. 3c). The group‐wise abundance distributions of the three genera are shown in Fig. S3. The largest effect size and lowest p‐value from ALDEx2 was found for Veillonella, an anerobic opportunistic pathogen previously reported to be increased in patients with COVID‐19 [8] and associated with tissue fibrosis [19].
Fig. 3.
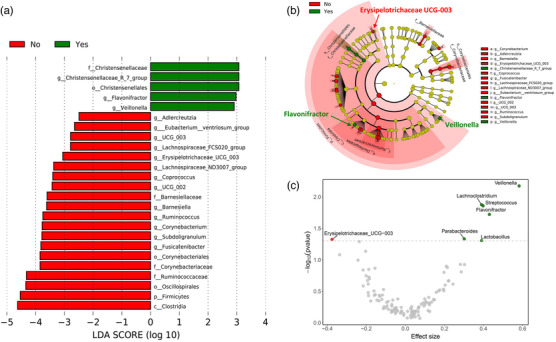
Gut microbial composition in patients with respiratory dysfunction at the 3‐month follow‐up (diffusing capacity of the lungs for carbon monoxide < lower limit of normal, n = 83). (a) LDA score of taxa abundance differences using LefSe analysis. (b) Taxonomic cladogram highlighting differentially abundant taxa (p < 0.05) by LEfSe. (c) Volcano plot from ALDEx2 analysis showing effect size representing the median of “difference in clr (centered log‐ratio) values between groups divided by largest difference in clr values within group" on log2‐scale by p‐value of differentially abundant genera. Abbreviations: LDA, linear discriminant analysis; LEfSe, linear discriminant analysis effect size.
Circulating levels of LBP in relation to acute respiratory failure during hospitalization
As microbiota traits associated with respiratory dysfunction were potentially related to a reduced capacity for butyrate production, which is vital for a functional gut barrier, we hypothesized that gut barrier dysfunction could also be related to respiratory failure in acute COVID‐19. As microbiota samples from the acute phase of disease were not available, we explored whether levels of circulating LBP were associated with acute respiratory failure, defined as a P/F ratio <26.6 kPa, which occurred in 60 patients (33%) during hospitalization. LBP levels remained consistently elevated during hospitalization in patients with acute respiratory failure compared to patients without, although with a declining trend over time (Fig. 4a). Moreover, LBP levels were consistently inversely associated with P/F ratio during the hospitalization period (p < 0.001, linear mixed model, intercept of all three time points; Fig. 4b). In multivariate logistic regression adjusted for age, sex, comorbidity, and treatment, LBP was associated with acute respiratory failure during hospitalization (adjusted odds ratio [aOR] 1.98 [1.15–3.40]) per standard deviation of log10 LBP. Further adjustment for smoking and antibiotic use had little impact on this association (aOR 1.77 [1.01–3.10]).
Fig. 4.
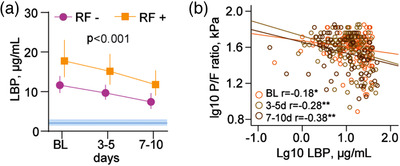
Temporal profile of soluble lipopolysaccharide‐binding protein (LBP) levels (n = 144) in relation to acute respiratory failure (n = 44) during hospitalization (pO2/fiO2 [P/F] ratio < 26.6 kPa) (a). The p‐value reflects the overall effect of respiratory failure (RF) from the repeated measures regression analysis. Blue areas reflect levels in age‐ and sex‐matched healthy controls (n = 24). (b) Spearman correlations between LBP and P/F ratio at different time points (baseline orange; days 3–5, yellow green; days 7–10, brown) during hospitalization. *p < 0.05, **p < 0.005. Observations per time point: Baseline (BL), n = 144; 3–5 days, n = 134; 7–10 days, n = 84.
The association of LBP with respiratory dysfunction and inflammation after 3 months
Notably, LBP levels after 3 months remained significantly higher in patients than in age‐ and sex‐matched healthy controls, where patients with respiratory dysfunction (DLCO < LLN) had numerically higher levels than patients with normal respiratory function (Fig. S1). Moreover, LBP levels correlated negatively with the percent predicted values of DLCO (ρ = −0.27, p = 0.031). Notably, we found no correlation between LBP and measures of microbiota diversity in the subgroup with both microbiota and lung function tests available. Plasma LBP levels at admission and after 3 months were significantly associated with several clinical markers of systemic inflammation, including C‐reactive protein (CRP) levels and white blood cell and neutrophil counts (Fig. S2), although these inflammatory markers were not related to respiratory dysfunction after 3 months (Table S1).
Discussion
In this substudy of the NOR‐Solidarity trial, we investigated the potential gut–lung axis during and after hospitalization for COVID‐19. Our results can be summarized as follows: (i) 3 months after hospitalization for COVID‐19, patients with respiratory dysfunction (DLCO < LLN) showed a lower microbial diversity and an altered global gut microbiota composition than patients with normal respiratory function; (ii) these microbiota alterations included reduced abundance of Erysipelotrichaceae UCG‐003 and increased abundance of Veillonella and Flavonifractor; (iii) during hospitalization, increased plasma levels of LBP were significantly associated with acute respiratory failure; and (iv) LBP levels remained elevated during and after hospitalization, and were significantly associated with persistent low‐grade inflammation and respiratory dysfunction at the 3‐month follow‐up.
In a recent report, Yeoh et al. reported the downregulation of several gut commensals with known immunomodulatory potential in patients with COVID‐19, including Faecalibacterium prausnitzii and Eubacterium rectale, which remained low in samples collected up to 30 days after disease resolution [9]. Here, we extend these findings by showing that altered gut microbiota, including decreased alpha diversity 3 months after hospital admission, was associated with impaired pulmonary function at this time point. We also found that several members of the Lachnospiraceae and Ruminococcaceae families, such as Coprococcus and Ruminococcus, which are known producers of butyrate, were reduced in patients with COVID‐19 with pulmonary impairment after 3 months. Butyrate has local immunomodulatory effects in the gut mucosa and, as the main energy substrate for enterocytes, is vital for gut barrier maintenance [20]. Interestingly, LBP levels as a potential indirect marker of gut leakage were associated with impaired pulmonary function, not only during hospitalization (P/F ratio < 26.6 kPa, respiratory failure), but also after 3 months (inverse correlation with DLCO).
COVID‐related dysbiosis reported by Yeoh et al. correlated with several inflammatory markers, in line with our finding that LBP is associated with acute inflammation during hospitalization. Notably, we now show that LBP levels are associated with persistent low‐grade inflammation even after 3 months. A potential role of gut barrier dysfunction as a driver of the multisystem inflammatory syndrome occurring in pediatric COVID‐19 has been recently reported [21], and our findings suggest that similar mechanisms could be relevant in adult patients. In some chronic inflammatory conditions related to immunodeficiencies, we had also seen a direct correlation between microbiota composition and markers of gut barrier function [22, 23], but this was not seen in patients with COVID‐19 at 3 months.
Of note, LBP is an acute‐phase protein produced in the liver [24], and as such, could be more related to acute inflammation than gut barrier alterations. Whether gut microbiota alterations were related to gut barrier dysfunction could not be determined in the present study. However, whereas pulmonary dysfunction was related to gut microbiota alterations and inversely correlated with LBP levels, no associations with inflammatory markers, including CRP levels, were detected at the 3‐month follow‐up.
Other microbiota‐related traits might also be relevant. The largest effect size observed in patients with pulmonary dysfunction was the increased relative abundance of Veillonella, which has previously been linked to several disease states of the lung and liver, where fibrosis is central to pathogenesis [19, 25, 26]. Interestingly, an increased relative abundance of Veillonella has recently been reported in patients with COVID‐19 [27, 28]. However, whether this bacterial genus is relevant for fibrosis development after COVID‐19, either as a contributing factor or as a consequence of a fibrotic process, cannot be answered in an observational study. A future study would benefit from parallel evaluation of the respiratory tract microbiome, as Veillonella has been found to be enriched in airway samples of patients with idiopathic pulmonary fibrosis [29].
The present study has several limitations. Due to the logistic constraints of launching a randomized trial during the first wave of the pandemic, the gut microbiota samples and pulmonary function tests were only performed at the 3‐month follow‐up. Therefore, we cannot relate long‐term microbiota alterations to potential microbiota alterations or the potential gut–lung axis during hospitalization. In addition, as the analysis of microbiota composition was performed in only one cohort, and without a validation panel, these data must be considered explorative.
Furthermore, whether the observed alterations in microbiota composition in this trial are related to COVID‐19 per se or mirror other factors, such as pre‐existing comorbidities, length of hospital stay, need for ICU, organ support, or other treatment, is not clear. However, our findings suggest a possible impact of invasive mechanical ventilation and length of hospitalization on gut microbial diversity after 3 months, which could reflect disease severity, treatment during hospitalization, or a combination. Notably, the use of antibiotics was significantly more frequent in patients with respiratory failure during hospitalization. The use of antibiotics was also potentially related to lower alpha diversity measures in the gut microbiota samples, but not significantly, possibly due to the limited sample size. Although power calculations should preferably have been performed, this was not possible as the NOR‐Solidarity trial was executed as an add‐on trial of the WHO Solidarity trial, and the decision to stop further inclusion was made after advice from an independent data and safety monitoring board.
Finally, the compositional alterations found in the rectal specimens could have been supported by measuring butyrate levels in plasma or fecal samples. However, this was not performed because of the poor detection rate in previously analyzed plasma samples of other patient cohorts [30], as well as the presence of preservation liquid in the rectal swab samples, which precludes measurements of short‐chain fatty acids.
The study also has obvious strengths, including standardized data capture in a randomized trial with longitudinal biobanking as well as comprehensive long‐term follow‐up with blood tests, microbiota sampling, and assessment of pulmonary function by DLCO. To our knowledge, our study is the first to potentially link pulmonary function to gut microbiota alterations in COVID‐19.
In conclusion, the decreased microbial diversity and compositional gut microbiota alterations in patients with respiratory dysfunction after 3 months, as well as the association of persistently raised LBP levels with these clinical features, point to a potential gut–lung axis in COVID‐19. These observations could be related not only to acute respiratory failure during hospitalization but also to long‐term COVID‐19 morbidity. However, owing to power limitations, the cross‐sectional nature of the microbiota and lung function assessments, and the interpretation of LBP as a gut leakage marker beyond an inflammatory marker, the present study should be considered exploratory. Nevertheless, our findings warrant further research on the potential role of gut microbiota composition and gut barrier dysfunction in relation to long‐term pulmonary dysfunction and long COVID.
Author contributions
Beate Vestad, Thor Ueland, Pål Aukrust, Johannes Roksund Hov, and Marius Trøseid were responsible for study conception and execution of the present substudy. Andreas Barratt‐Due, Trine Kåsine, Inge Christoffer Olsen, Anne Ma Dyrhol‐Riise, Katerina Nezvalova Henriksen, Pål Aukrust, and Marius Trøseid were responsible for the management, coordination, research activity planning, and execution of the NOR‐Solidarity trial. Tøri Vigeland Lerum and Ole Henning Skjønsberg were responsible for the 3‐month follow‐up protocol for pulmonary function. Tuva Børresdatter Dahl, Andreas Barratt‐Due, Bente Halvorsen, Trine Ranheim, and Pål Aukrust coordinated collection and storage of the biobank material. Anne Ma Dyrhol‐Riise, Birgitte Stiksrud, Kristian Tonby, Hedda Hoel, Anders Tveita, Ravinea Manotheepan, Mette Haugli, Ragnhild Eiken, Åse Berg, Anders Benjamin Kildal, Asgeir Johannessen, Lars Thoresen, Hilde Skudal, Bård Reiakvam Kittang, Roy Bjørkholt Olsen, Carl Magnus Ystrøm, Nina Vibeche Skei, Raisa Hannula, Saad Aballi, and Reidar Kvåle were locally responsible for conducting the trial at the various included hospitals providing rectal swab material. Beate Vestad performed DNA extraction and library preparation of microbiota samples. Thor Ueland, Tove Lekva, and Annika Elisabeth Michelsen performed the gut leakage marker analyses. Beate Vestad and Thor Ueland performed the statistical analyses. Kristian Holm performed the bioinformatics analyses of the microbiota data. Beate Vestad and Marius Trøseid drafted the manuscript. Thor Ueland, Andreas Barratt‐Due, Pål Aukrust, and Johannes Roksund Hov critically revised the manuscript. All authors revised and approved the final version of the manuscript.
Conflict of interests
The authors declare no competing interests.
Funding
This work was supported by the National Clinical Therapy Research in the Specialist Health Services (KLINBEFORSK), Norway, and the South‐Eastern Norway Regional Health Authority (grant number 2021071). The microbiota analyses were funded by the strategic research area “Personalized microbiota therapy in clinical medicine” at Oslo University Hospital. The funders had no role in the study design, data collection, data analysis, data interpretation, or writing of the manuscript. The corresponding author had full access to all data in the study and had final responsibility for the decision to submit for publication.
Supporting information
Supplementary Figure 1. Levels of LBP measured at the three‐month follow‐up in patients with or without persistent respiratory dysfunction. LBP, lipopolysaccharide‐binding protein.
Supplementary Figure 2. Correlation heatmap of LBP levels with inflammation markers at corresponding time points. LBP, lipopolysaccharide‐binding protein.
Supplementary Figure 3. Abundance distribution histograms of altered genera associated with respiratory dysfunction after three months.
Supplementary Table 1. Clinical characteristics at the three‐month follow‐up.
Supplementary Table 2. Associations of bacterial alpha diversity measures with clinical baseline characteristics.
Supplementary methods. Soluble LPS‐binding protein (LBP) measurements, sequence processing, and bioinformatics (gut microbiota analyses).
Acknowledgments
We thank the WHO Solidarity and NOR‐Solidarity study groups for the opportunity to perform this add‐on study. We would also like to thank Karoline Hansen Skåra and Azita Rashidi from the Institute of Internal Medicine, Oslo University Hospital, Rikshospitalet, for their contributions to the biobank collection, and Mona Skjelland for access to the control plasma biobank. Moreover, we thank Fredrik Müller and Cahtrine Fladeby from the Department of Microbiology, Oslo University Hospital, for access to the viral analysis data.
Vestad B, Ueland T, Lerum TV, Dahl TB, Holm K, Barratt‐Due A, et al. Respiratory dysfunction three months after severe COVID‐19 is associated with gut microbiota alterations. J Intern Med. 2022;291:801–812.
Contributor Information
Beate Vestad, Email: beate.vestad@medisin.uio.no.
Marius Trøseid, Email: marius.troseid@medisin.uio.no.
References
- 1. Galanopoulos M, Gkeros F, Doukatas A, Karianakis G, Pontas C, Tsoukalas N, et al. Covid‐19 pandemic: pathophysiology and manifestations from the gastrointestinal tract. World J Gastroenterol. 2020;26(31):4579–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Tabary M, Khanmohammadi S, Araghi F, Dadkhahfar S, Tavangar SM. Pathologic features of COVID‐19: a concise review. Pathol Res Pract. 2020;216(9):153097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ferreira C, Viana SD, Reis F. Gut microbiota dysbiosis‐immune hyperresponse‐inflammation triad in coronavirus disease 2019 (COVID‐19): impact of pharmacological and nutraceutical approaches. Microorganisms. 2020;8(10):1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Vignesh R, Swathirajan CR, Tun ZH, Rameshkumar MR, Solomon SS, Balakrishnan P. Could perturbation of gut microbiota possibly exacerbate the severity of COVID‐19 via cytokine storm? Front Immunol. 2020;11:607734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zuo T, Zhang F, Lui GCY, Yeoh YK, Li AYL, Zhan H, et al. Alterations in gut microbiota of patients with COVID‐19 during time of hospitalization. Gastroenterology. 2020;159(3):944–55.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Giron LB, Dweep H, Yin X, Wang H, Damra M, Goldman AR, et al. Plasma markers of disrupted gut permeability in severe COVID‐19 patients. Front Immunol. 2021;12:686240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hoel H, Heggelund L, Reikvam DH, Stiksrud B, Ueland T, Michelsen AE, et al. Elevated markers of gut leakage and inflammasome activation in COVID‐19 patients with cardiac involvement. J Intern Med. 2021;289(4):523–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gu S, Chen Y, Wu Z, Chen Y, Gao H, Lv L, et al. Alterations of the gut microbiota in patients with coronavirus disease 2019 or H1N1 influenza. Clin Infect Dis. 2020;71(10):2669–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yeoh YK, Zuo T, Lui GC, Zhang F, Liu Q, Li AY, et al. Gut microbiota composition reflects disease severity and dysfunctional immune responses in patients with COVID‐19. Gut. 2021;70(4):698–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pan H, Peto R, Henao‐Restrepo AM, Preziosi MP, Sathiyamoorthy V, Karim QA, et al. Repurposed antiviral drugs for COVID‐19—interim WHO solidarity trial results. N Engl J Med. 2021;384(6):497–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Barratt‐Due A, Olsen IC, Nezvalova‐Henriksen K, Kåsine T, Lund‐Johansen F, Hoel H, et al. Evaluation of the effects of remdesivir and hydroxychloroquine on viral clearance in COVID‐19: a randomized trial. Ann Intern Med. 2021;174:1261–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Graham BL, Brusasco V, Burgos F, Cooper BG, Jensen R, Kendrick A, et al. 2017 ERS/ATS standards for single‐breath carbon monoxide uptake in the lung. Eur Respir J. 2017;49(1):1600016. [DOI] [PubMed] [Google Scholar]
- 13. Graham BL, Steenbruggen I, Miller MR, Barjaktarevic IZ, Cooper BG, Hall GL, et al. Standardization of spirometry 2019 update. An official American Thoracic Society and European Respiratory Society technical statement. Am J Respir Crit Care Med. 2019;200(8):e70–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lerum TV, Aalokken TM, Bronstad E, Aarli B, Ikdahl E, Lund KMA, et al. Dyspnoea, lung function and CT findings 3 months after hospital admission for COVID‐19. Eur Respir J. 2021;57(4):2003448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Guler SA, Ebner L, Aubry‐Beigelman C, Bridevaux P‐O, Brutsche M, Clarenbach C, et al. Pulmonary function and radiological features 4 months after COVID‐19: first results from the national prospective observational Swiss COVID‐19 lung study. Eur Respir J. 2021;57(4):2003690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lerum TV, Maltzahn NN, Aukrust P, Trøseid M, Henriksen KN, Kåsine T, et al. Persistent pulmonary pathology after COVID‐19 is associated with high viral load, weak antibody response, and high levels of matrix metalloproteinase‐9. Sci Rep. 2021;11(1):23205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Stanojevic S, Graham BL, Cooper BG, Thompson BR, Carter KW, Francis RW, et al. Official ERS technical standards: Global Lung Function Initiative reference values for the carbon monoxide transfer factor for Caucasians. Eur Respir J. 2017;50(3):1700010. [DOI] [PubMed] [Google Scholar]
- 18. Fadrosh DW, Ma B, Gajer P, Sengamalay N, Ott S, Brotman RM, et al. An improved dual‐indexing approach for multiplexed 16S rRNA gene sequencing on the Illumina MiSeq platform. Microbiome. 2014;2(1):6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kummen M, Holm K, Anmarkrud JA, Nygård S, Vesterhus M, Høivik ML, et al. The gut microbial profile in patients with primary sclerosing cholangitis is distinct from patients with ulcerative colitis without biliary disease and healthy controls. Gut. 2017;66(4):611–9. [DOI] [PubMed] [Google Scholar]
- 20. Gelpi M, Vestad B, Hansen SH, Holm K, Drivsholm N, Goetz A, et al. Impact of human immunodeficiency virus—related gut microbiota alterations on metabolic comorbid conditions. Clin Infect Dis. 2020;71:e359–67. [DOI] [PubMed] [Google Scholar]
- 21. Yonker LM, Gilboa T, Ogata AF, Senussi Y, Lazarovits R, Boribong BP, et al. Multisystem inflammatory syndrome in children is driven by zonulin‐dependent loss of gut mucosal barrier. J Clin Invest. 2021;131:e149633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nowak P, Troseid M, Avershina E, Barqasho B, Neogi U, Holm K, et al. Gut microbiota diversity predicts immune status in HIV‐1 infection. AIDS. 2015;29(18):2409–18. [DOI] [PubMed] [Google Scholar]
- 23. Jorgensen SF, Troseid M, Kummen M, Anmarkrud JA, Michelsen AE, Osnes LT, et al. Altered gut microbiota profile in common variable immunodeficiency associates with levels of lipopolysaccharide and markers of systemic immune activation. Mucosal Immunol. 2016;9(6):1455–65. [DOI] [PubMed] [Google Scholar]
- 24. Zweigner J, Schumann RR, Weber JR. The role of lipopolysaccharide‐binding protein in modulating the innate immune response. Microbes Infect. 2006;8(3):946–52. [DOI] [PubMed] [Google Scholar]
- 25. Thavamani A, Salem I, Sferra TJ, Sankararaman S. Impact of altered gut microbiota and its metabolites in cystic fibrosis. Metabolites. 2021;11(2):123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Enaud R, Hooks KB, Barre A, Barnetche T, Hubert C, Massot M, et al. Intestinal inflammation in children with cystic fibrosis is associated with Crohn's‐like microbiota disturbances. J Clin Med. 2019;8(5):645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tao W, Zhang G, Wang X, Guo M, Zeng W, Xu Z, et al. Analysis of the intestinal microbiota in COVID‐19 patients and its correlation with the inflammatory factor IL‐18. Med Microecol. 2020;5:100023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. de Oliveira GLV, Oliveira CNS, Pinzan CF, de Salis LVV, Cardoso CRB. Microbiota modulation of the gut–lung axis in COVID‐19. Front Immunol. 2021;12:635471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Molyneaux PL, Cox MJ, Willis‐Owen SA, Mallia P, Russell KE, Russell A‐M, et al. The role of bacteria in the pathogenesis and progression of idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. 2014;190(8):906–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mayerhofer CCK, Kummen M, Holm K, Broch K, Awoyemi A, Vestad B, et al. Low fibre intake is associated with gut microbiota alterations in chronic heart failure. ESC Heart Fail. 2020;7:456–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Levels of LBP measured at the three‐month follow‐up in patients with or without persistent respiratory dysfunction. LBP, lipopolysaccharide‐binding protein.
Supplementary Figure 2. Correlation heatmap of LBP levels with inflammation markers at corresponding time points. LBP, lipopolysaccharide‐binding protein.
Supplementary Figure 3. Abundance distribution histograms of altered genera associated with respiratory dysfunction after three months.
Supplementary Table 1. Clinical characteristics at the three‐month follow‐up.
Supplementary Table 2. Associations of bacterial alpha diversity measures with clinical baseline characteristics.
Supplementary methods. Soluble LPS‐binding protein (LBP) measurements, sequence processing, and bioinformatics (gut microbiota analyses).


