Highlights
-
•
Deep-learning contouring for radiotherapy is evaluated in the thorax region.
-
•
Specific regions-of-adjustment and variability in editing per organ were found.
-
•
The consistency and accuracy of training data remains crucial to model performance.
-
•
Separate models for specific indications or acquisition protocols may be necessary.
-
•
Subsampling and post-processing make the clinical workflow more efficient.
Keywords: Radiotherapy, Automatic segmentation, Auto-contouring, Deep-learning, Contour adjustment, Thorax organs at risk
Abstract
Background and purpose
User-adjustments after deep-learning (DL) contouring in radiotherapy were evaluated to get insight in real-world editing during clinical practice. This study assessed the amount, type and spatial regions of editing of auto-contouring for organs-at-risk (OARs) in routine clinical workflow for patients in the thorax region.
Materials and methods
A total of 350 lung cancer and 362 breast cancer patients, contoured between March 2020 and March 2021 using a commercial DL-contouring method followed by manual adjustments were retrospectively analyzed. Subsampling was performed for some OARs, using an inter-slice gap of 1–3 slices. Commonly-used whole-organ contouring assessment measures were calculated, and all cases were registered to a common reference shape per OAR to identify regions of manual adjustment. Results were expressed as the median, 10th-90th percentile of adjustment and visualized using 3D renderings.
Results
Per OAR, the median amount of editing was below 1 mm. However, large adjustments were found in some locations for most OARs. In general, enlarging of the auto-contours was needed. Subsampling DL-contours showed less adjustments were made in the interpolated slices compared to simulated no-subsampling for these OARs.
Conclusion
The real-world performance of automatic DL-contouring software was evaluated and proven useful in clinical practice. Specific regions-of-adjustment were identified per OAR in the thorax region, and separate models were found to be necessary for specific clinical indications different from training data. This analysis showed the need to perform routine clinical analysis especially when procedures or acquisition protocols change to have the best configuration of the workflow.
1. Introduction
Deep learning-based (DL) automatic contouring for radiotherapy has evolved over the past years leading to clinical implementation at many institutes and for many treatment sites. Significant time saving compared to both manual contouring and atlas-based contouring methods is achieved [1], [2], [3], [4], [5], [6], [7], [8], [9], [10]. Even though the majority of current clinical tools still use atlas-based auto-contouring, DL-contouring is being offered more often recently [5], [9]. However, clinical implementation of such an automatic contouring software requires thorough validation and quality assurance (QA) [9], [11], [12], [13], [14]. User-adjustments evaluated in routine clinical practice give insight into the real-world model performance. The amount and type of user-adjustments could give more information into acceptance of certain structures by clinical experts, and whether there are regions of structures that need more editing than other regions.
The quality of automatic contouring has been widely studied on the geometrical level by comparing automatic delineations and user-adjustments, but typically using whole-organ measures and performed mostly on a limited patient cohort. A more extended analysis was previously published for head & neck cancer patients by Brouwer et al. [15]. This current study aimed to evaluate the extent of manual adjustments following auto-contouring of organs-at-risk (OARs) in the thorax region in clinical practice for a large patient group. User-adjustments following DLC using a commercial DLC system were assessed retrospectively following 12 months of clinical use. Manual adjustments made during clinical review of the automatically generated contours were assessed and quantified, and regions that required substantial editing per OARs were identified.
2. Materials and methods
2.1. Patient cohort
Since March 2020 a Lung DL-model [4], [16] and since June 2020 a Breast DL-model have been used in our clinical practice. Both models are based on convolutional neural networks with an architecture as described in [6]. CT-scans were acquired using Siemens SOMATOM Drive or Confidence CT-scanners (Siemens Healthineers, Forchheim, Germany) with varying slice-spacing depending on clinical protocol (range:1–3 mm). A total of 415 lung cancer (LC) and 364 breast cancer (BC) cases were included in this study. In our clinical workflow, CT-scans of these patients were exported to Mirada WorkflowBox 2.0–2.4 (Mirada Medical Ltd., Oxford, United Kingdom). The appropriate DL-model (DLCExpert, Mirada Medical Ltd., Oxford, United Kingdom) was automatically selected based on CT-protocol name, and a corresponding DICOM RT Structure Set (RTSS) was returned to the Treatment Planning System (TPS, Eclipse, version 14.0–16.0, Varian, Palo Alto, USA). A radiation therapy technologist (RTT) checked the OARs included in the DL RTSS and manually adjusted where necessary. In this study, all RTTs in our clinic trained to delineate lung and breast patients in clinical practice were included (approximately 40). Both the automatically-generated and the user-adjusted contours were collected for this study. The Institutional Review Board of Maastro approved this retrospective study with project number P0288. A representative example of the DL- and edited contours for a LC and BC patient is shown in Fig. S1 in the Supplementary Material.
For the LC group, left lung, right lung, heart, esophagus, spinal cord, and mediastinum envelop were contoured using the Lung DL-model. Training data as well as clinical data included the 50%-expiration phase of a 4DCT-scan of LC patients [4]. Additionally, a 3DCT-scan with intravenous (IV)-contrast was available for fusion with the 4DCT-scan for delineation of the target volume. Heart, esophagus and mediastinum were returned using an inter-slice gap of three slices (i.e. subsampling), editing was allowed and followed by inter-slice contour interpolation of the edited contour. Fewer edits were expected using subsampling followed by interpolation. Automatic post-processing of the DL-contours included removing slices for the heart (superior: 24 mm, inferior:9mm), mediastinum (superior: 21 mm, inferior: 21 mm), and esophagus (superior: 24 mm). This post-processing step was implemented in July 2020 based on interim feedback evaluation from the RTTs.
For the BC group, left lung, right lung, heart, breast clinical target volume (CTV) and contralateral breast were delineated. A dedicated Breast DL-model was used to contour left and right lungs, the breast CTV and the contralateral breast. Training data included a voluntary moderately deep-inspiration breath-hold (vmDIBH) CT-scan for left-sided BC cases and a free-breathing (FB) CT-scan for right-sided BC cases. For the heart and esophagus, the Lung DL-model was used. The thyroid was taken from a head-and-neck DL-model [15]. The esophagus and thyroid were included when periclavicular or parasternal lymph node regions were involved. Heart, CTV, contralateral breast and esophagus were contoured using an inter-slice gap of three slices, the thyroid using an inter-slice gap of one slice. Automatic post-processing of the DL-contours involved removing slices from the CTV and contralateral Breast_L/_R (superior: 12 mm), and from the heart (superior:24 mm). Both the CTV and contralateral Breast_L/_R were expanded in the direction of the body surface (anterior: 10 mm, left/right: 10 mm, respectively).
Following automatic contouring, manual user-adjustments were made where necessary to make the contours clinically acceptable. In clinical practice, the CTV and the contralateral breast structures are cropped with 5 mm to the body contour. DL-contours were similarly cropped.
2.2. Whole-organ evaluation of contour editing
Volumetric Dice Similarity Coefficient (vDSC), mean slice-wise Hausdorff distance (MSHD), surface DSC (sDSC) [17] and added path length (APL) [16] were calculated between the automatic and user-adjusted contours. For subsampled structures, only slices with a DL-contour present were included in the calculation.
2.3. Local evaluation of contour editing
To identify anatomical regions of adjustments in the entire patient population, contours were aligned to common OAR reference shape as proposed by Brouwer et al. [15]. Clinical contours were converted to 3D discrete mesh representations. 3D meshes of an OAR were registered to the reference, and the amount of editing was quantified. Finally, editing statistics were reported as the median and 10-90th percentile range of edits per reference vertex over the patient population. 3D renderings were visualized using 3D slicer (https://www.slicer.org) [18].
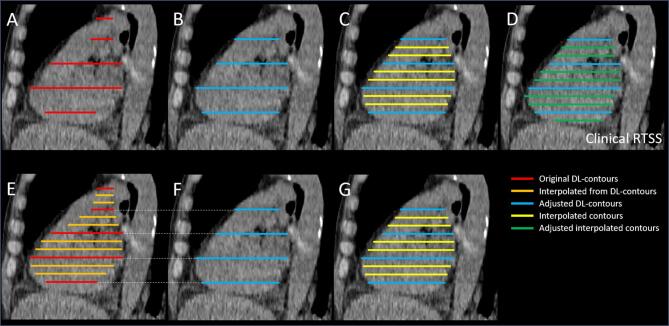
In contrast to previous work by Brouwer et al. (2020), DL-contours were subsampled for some OARs prior to editing. In Fig. 1A-D the clinical workflow of editing these subsampled OARs is presented. To analyze total adjustments for the subsampled OARs, pre-processing was necessary, as illustrated in Fig. 1E-G.
Fig. 1.
Schematic overview of the contour sets considered in this study. A: Sub-sampled DL-contours; B: Manually adjusted DL-contours; C: Manually adjusted DL-contours followed by interpolation; D: Final clinical contour following adjustment of C. In this schematic figure, we assumed no edits were made to the blue contours from C, but in clinical practice it is possible some minor edits are made on these slices after interpolation; E: Interpolated DL-contours for evaluation purposes, for subsampled OARs only to simulate a full DL-contour; F: Subsampling of the clinical contour set (D) at the location of the original DL-contours for analysis, to catch also edits made in the second round of adjusting. This means F is not necessarily the same as B, however only minor differences are expected; G: Interpolation of F to infer edits only on the interpolated contours. Note: amount of edits are computed as signed Euclidean distances between the contour surfaces (in 3D). Abbreviations: DL = Deep-learning, RTSS = RT Structure Set. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
To analyze the edits made to interpolated slices, the contour set from Fig. 1G is compared to Fig. 1D. To analyze the edits made on the original DL-contours, the contour set from Fig. 1E is compared to Fig. 1D. The total editing to the full contour is obtained by combining these two sets of edits on the appropriate slices, done at the vertices level based on the nearest neighbor vertex on either a DL- or interpolated contour.
For the subsampled structures, further analysis was carried out to estimate the number of required edits when no sub-sampling would have been performed. A comparison of the interpolated DL-contour (Fig. 1E) to the clinical contour (Fig. 1D) provides a good estimate of the amount of editing when sub-sampling is not performed.
2.4. Excluded datasets
In total, 65 and 2 cases were excluded from the LC and BC group respectively, because of an incorrect identification of the location of one or both lungs in the first stage of the region detection algorithm, negatively influencing the accuracy of the other OARs. In our clinic, an extended scan range in longitudinal direction for breast patients eligible for proton therapy (to avoid collision of the treatment machine with the body) was performed. In some of these elongated scans, one or both automatically generated breasts were predicted in the pelvic region for 14 cases. These breast CTVs and/or contralateral breasts were excluded. Because the esophagus and the heart in the BC group were contoured using the Lung DL-model, and following the same exclusion criteria as for the LC group, 10 cases for these OARs were excluded. In all cases where automatic contouring was failing, the RTT manually recontoured the OAR; these cases were excluded for further evaluation in this study.
2.5. Statistical analysis
All results were expressed as mean ± standard deviation or median (range). Grouped data were tested for statistical significance using two-sided Wilcoxon-Mann-Whitney tests. Analyses were performed in Matlab 2020a (The Mathworks Inc., Natick, MA, USA) and SciPy version 1.5.2.
3. Results
Comparable or better results were found for whole-organ results of user-adjustments in the LC group compared to the BC group (Table 1). Statistically significant differences were found between LC and BC group for the same OARs for the left lung, heart, and esophagus (Table 1).
Table 1.
Whole-organ results of both the LC and BC group. Count is the number of patients included in the analysis per structure. Number of structures where the DL-contour can be considered as completely wrong (defined as vDSC < 0.2) were also added. Number of slices added and deleted represent slices outside of the range of the automatically generated contour. Median (10-90th percentile). *Statistically significant difference (p < 0.05) between lung cancer (LC) and breast cancer (BC) group. Abbreviations: vDSC = Volumetric Dice Similarity Coefficient, sDSC = surface DSC, MSHD = mean slice-wise Hausdorff distance, APL = added path length.
| LC group | Count | # patients (%) vDSC < 0.2 | vDSC [-] | sDSC [-] | MSHD [cm] | APL [cm] | # Slices Added [-] |
# Slices Deleted [-] |
|---|---|---|---|---|---|---|---|---|
| Left Lung | 350 | 0 (0%) | 1.00 (0.99–1.00) | 0.99 (0.94–1.00) | 0.09 (0.00–0.34)* | 30.60 (0.00–329.70) | 0 (0–0) | 0 (0–0) |
| Right Lung | 350 | 0 (0%) | 1.00 (0.99–1.00) | 0.99 (0.94–1.00) | 0.11 (0.00–0.50) | 40.25 (0.00–430.20) | 0 (0–0) | 0 (0–1) |
| Heart | 348 | 0 (0%) | 0.89 (0.77–0.94) | 0.43 (0.15–0.71)* | 1.13 (0.58–2.18)* | 234.15 (102.28–416.67)* | 2 (1–6.7) | 2 (0–5) |
| Spinal Cord | 333 | 1 (0.3%) | 1.00 (0.93–1.00) | 0.99 (0.71–1.00) | 0.00 (0.00–0.13) | 8.50 (0.00–306.50) | 0 (0–7) | 0 (0–4) |
| Esophagus | 348 | 11 (3.2%) | 0.73 (0.32–0.89) | 0.41 (0.17–0.78) | 0.32 (0.15–0.57) | 78.15 (27.92–238.80) | 2 (0–30) | 2 (0–5) |
| Mediastinum | 349 | 0 (0%) | 0.93 (0.85–0.96) | 0.39 (0.15–0.59) | 0.99 (0.64–1.62) | 468.70 (293.76–819.02) | 2 (0–9.6) | 4 (0–9) |
| BC group | ||||||||
| Left Lung | 362 | 0 (0%) | 1.00 (0.99–1.00) | 0.99 (0.71–1.00) | 0.13 (0.00–0.68)* | 51.30 (0.00–1027.85) | 0 (0–0) | 0 (0–0) |
| Right Lung | 362 | 0 (0%) | 1.00 (0.99–1.00) | 0.99 (0.71–1.00) | 0.14 (0.00–0.77) | 60.15 (0.00–1166.73) | 0 (0–0) | 0 (0–0) |
| Heart | 352 | 0 (0%) | 0.91 (0.75–0.95) | 0.61 (0.27–0.80)* | 0.75 (0.37–1.28)* | 149.75 (63.88–299.30)* | 2 (0–7.3) | 1 (0–3) |
| Left CTV | 164 | 7 (4.3%) | 0.88 (0.40–0.95) | 0.58 (0.15–0.77) | 1.32 (0.67–2.97) | 250.65 (123.51–670.28) | 2 (0–8.2) | 2 (0–7) |
| Right CTV | 135 | 4 (3.0%) | 0.88 (0.62–0.95) | 0.55 (0.18–0.75) | 1.15 (0.63–2.31) | 282.50 (139.10–552.60) | 2 (0–8) | 1 (0–3) |
| Left Contralateral Breast | 156 | 1 (0.6%) | 0.90 (0.79–0.96) | 0.63 (0.38–0.81) | 1.08 (0.54–1.74) | 223.70 (100.96–398.01) | 1.5 (0–5) | 1 (0–4) |
| Right Contralateral Breast | 188 | 0 (0%) | 0.90 (0.77–0.97) | 0.60 (0.32–0.83) | 0.91 (0.41–2.00) | 240.65 (99.41–470.15) | 1 (0–5) | 1 (0–4) |
| Esophagus | 93 | 13 (14.0%) | 0.60 (0.15–0.90) | 0.33 (0.07–0.80) | 0.33 (0.15–0.68) | 75.60 (20.40–262.44) | 2 (0–48.2) | 2 (0–5) |
| Thyroid | 94 | 5 (5.3%) | 0.66 (0.25–0.91) | 0.34 (0.07–0.73) | 0.85 (0.28–1.80) | 47.95 (21.88–98.30) | 1 (0–4) | 0 (0–2) |
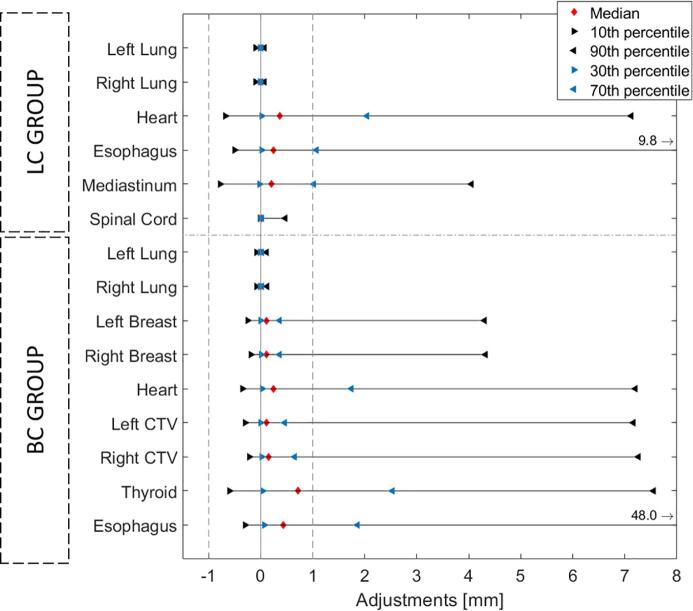
Per OAR, the median local amount of editing was low, i.e. < 1 mm (Fig. 2). For most structures, an asymmetric distribution of editing was observed, and for most vertex points per structure, the median of adjustments was within 1 mm as well (Fig. 3, Fig. 4). Almost all median adjustments were positive, showing that overall editing of the DL-contours was to enlarge the automatic contour. Histograms of the adjustments per OAR can be found in Figs. S2-S3 in the Supplementary Material.
Fig. 2.
Spatial adjustments showing median, 10, 30, 70 and 90 percentiles of the adjustments over all cases per organ at risk (OAR), for the lung cancer (LC) and breast cancer (BC) group. For the esophagus in both LC and BC group, the 90th percentile was cut from the axis at respectively 9.8 and 48.0 mm.
Fig. 3.
Spatial adjustments showing median and 10–90th percentile range projected on the reference shape of each organ at risk (OAR) in the lung cancer (LC) group. Positive adjustments reflect an enlargement outward of the DL-contour. The 10–90th percentile range represents the variability in adjustment between cases.
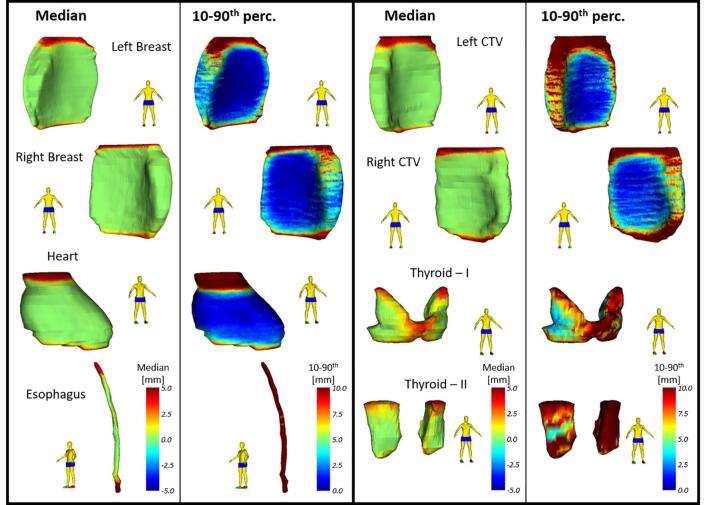
Fig. 4.
Spatial adjustments showing median and 10–90th percentile range projected on the reference shape of each organ at risk (OAR) in the breast cancer (BC) group. Positive adjustments reflect an enlargement outward of the deep learning (DL)-contour. The 10–90th percentile range represents the variability in adjustment between cases. Both lungs were not shown because they showed similar results as for the lung cancer (LC) group. Left and Right Breast represent the contralateral breast, Left and Right CTV represent the CTV breast. Note: for the thyroid there are two topologies because of anatomical variations in contouring.
For both lungs (LC and BC) and spinal cord (LC), median and 10-90th percentile range were close to zero, showing almost no editing was performed. For the heart (LC and BC) and mediastinum (LC), most adjustments were located in the superior and inferior region of the structure; the middle part of the structures were mostly accepted with only a few edits. For the left and right contralateral breasts most editing was also performed in the superior and inferior regions; the middle part was almost perfect. In the left and right CTV, a similar pattern was found, and additional adjustments were more carried out in the medial and lateral side of the CTV.
Largest 10-90th percentile range was found for the esophagus, with adjustments up to several centimeters, indicating the DL-contour was either missing or mostly wrong. Higher variability was found in the BC group compared to the LC group. The different thyroid topologies did not result in different findings, adjustments were highest in the superior region. For the topology with the connecting region present, a high amount of adjustment was found there.
For the sub-sampled structures, more adjustments would have been made if editing was performed on the full DL-contours. The amount of adjustment estimates without DL-contour sub-sampling are shown in Figs S4 and S5 in the Supplementary Material. This resembles the alternative situation of editing the full DL-structure. As can be seen, more adjustments would have been made compared to when editing is performed on a sub-sampled structure (see Fig. 3, Fig. 4), showing that using sub-sampling improves the clinical workflow in terms of adjustments needed and consequently efficiency.
Some specific observations were made showing where the DL-model could not contour an OAR of clinical quality. From the total number of BC cases, 65 were scanned and treated after a mastectomy. Upon feedback from the RTTs, it was seen that the Breast DL-model underperformed for both breasts because the shape of the target volume was different compared to the training set of the model, and they might not use the structure in clinical practice. For the esophagus, it was seen that for a subgroup of cases (LC:71 cases (20.4%), BC:18 cases (19.4%)), the inferior part of the esophagus was missing; the DL-contour only included the superior part.
4. Discussion
A small systematic overall editing (median < 1 mm) of manual adjustments was found following DL-contouring of OARs in LC and BC cases. The variability in editing (10-90th percentile range) was low for the lungs and spinal cord, indicating these contours are generally accepted and do not need much editing. There was an asymmetric distribution of editing at the superior and inferior region of the structures (Figs. 2 and S2-3). From post-processing at the top and bottom of OARs, this was expected. Besides this, there is a known systematic trend of under-segmentation of DL-contours for the currently-used implementation [15].
For the contralateral breasts and breast CTVs, most edits were found on the superior and inferior side. To a smaller extent edits were made on the medial and lateral sides. Delineation guidelines denote both breasts should be delineated medially lateral to the medial perforating mammary vessels; maximally to the edge of the sternal bone, and laterally up to the lateral thoracic artery [19]. These regions are not always anatomically clearly defined, but rather visual markers, and consequently inter-observer variability is higher [20]. For the contralateral breast, the medial side is most relevant since it is closest to the CTV. No edits were found on the anterior side of the contralateral breasts since we cropped the DL-contours to the body outline in a pre-processing step. On the posterior side only a few edits were performed, showing the ability of the DL-model to distinguish the transition between the breast and the pectoralis muscle accurately. We instructed the RTTs afterwards to not use the breast CTV structure for these patients having a mastectomy, and to delineate from scratch. A solution to restore performance in this category of patients could be a separate post-mastectomy DL-model.
For the heart and mediastinum, most edits and highest variability were seen in the superior and inferior parts. The superior extension of these OARs is prone to high inter-observer variability. For the heart, a mean dose constraint is often used in clinical practice, and it has been shown that the impact of contour variations on the mean dose is only relevant when the heart overlaps with the PTV [21]. For the mediastinum a maximum dose constraint is considered in clinical practice, indicating only the part of the contour close to the PTV is relevant and should be adjusted. Different topologies were seen in the delineations of the mediastinum, only one would be expected based on the delineation guidelines. We chose to use the topology that corresponds best to the guidelines. It is not always straightforward in the individual patient anatomy to correctly follow the guidelines, by doing these types of studies, we were able to bring this back to the attention of the RTTs.
For the esophagus, most editing was seen in the inferior region. Here the esophagus crosses the diaphragm and enters a region in the abdomen where it is surrounded by soft tissue. An accurate delineation of the esophagus is difficult there, which was reflected in an increase of the amount of adjustment. Especially the transition from esophagus to stomach can be difficult to contour. Manual adjustments to the DL-contour are also prone to inter-observer variations introducing uncertainty to esophagus contours in the inferior region.
We performed a more thorough post-hoc analysis, looking at whether the quality of the CT-scan had an influence on the prediction of the esophagus. The training data of the Lung DL-model was based on the 50%-expiration phase of a 4DCT-scan. A vmDIBH CT-scan is used for left-sided BC cases and a FB CT-scan is used for right-sided BC cases. Consequently, the esophagus adjustments can be split according to the side of breast cancer. It was found that the esophagus adjustments were somewhat lower for vmDIBH-scans compared to FB-scans (1 ± 17 mm vs. 14 ± 27 mm, p = 0.095, see Fig. 5), showing that the quality of the CT-scan impacted the prediction of the esophagus. Thus, even though the anatomical region is similar, taking organs from a model trained with a different type of data (FB/vmDIBH vs 4DCT), does not result in contours of similar quality. This factor needs to be considered regarding the transferability of models based on differences in acquisition protocols. A possible solution could be the training of more robust models using different types of CT-scans and acquisition protocols which could lead to increased DL-contour quality over the range of CT acquisition parameters. Alternatively, acquisition specific models could be developed.
Fig. 5.
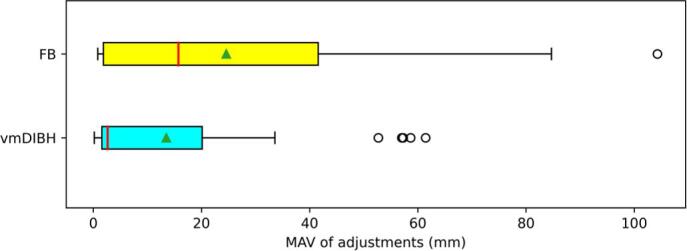
Mean Absolute Value (MAV) of the adjustments for the esophagus in the breast cancer group, separated for left/right CTV breast showing difference in CT scan that is used. For left-sided breast cancer patients, a voluntary moderately deep-inspiration breath-hold (vmDIBH) CT-scan is used, for right-sided breast cancer patients, a free-breathing (FB) CT-scan is used. The vertical red line in the boxplot represent the median value, the green triangles represent the mean value. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
For the thyroid, substantial variability in adjustments was found (median:0.72 mm, 10-90th percentile range:8.15 mm). This OAR is mostly used during plan optimization as an anatomical region to avoid, the exact location of the contour is of less relevance. In future it could be investigated if the non-adjusted DL-contour of such regions of avoidance could be already sufficient in the treatment planning stage.
When procedures or acquisition protocols change, or when different post-processing is implemented, it is needed to (periodically) investigate the results using a continuous monitoring system or logbook, and ask for feedback from the users [12]. Specifically, for post-processing, some more iterations and careful analysis could find the ideal cut-off point in the post-processing to have as few edits as possible. The same holds for the impact of subsampling, which has been shown in Fig. S4-5 in the Supplementary Material. This showed estimates of expected edits when no subsampling was performed. The benefit of subsampling depends highly on the size of the OAR and on the DL-model performance for that OAR. For large OARs that need much editing, subsampling is more beneficial (e.g. the heart) than for smaller OARs or large OARs where DL-model performance is almost perfect (e.g. lungs or spinal cord).
Because most user-adjustments were found within 1 mm, one can ask whether these adjustments were clinically meaningful. Multiple studies have already shown that small variability in contouring does not always result in significant dosimetric differences for the majority of organs [3], [22], [23], whereas errors close to high dose regions or very large errors should always be investigated [24], [25], [26]. We previously published recommendations for user-adjustments following auto-contouring for OARs in non-small cell lung cancer cases including the distance to the PTV, using the same Mirada DL-contouring solution as in this study [21]. Considering these guidelines, most of the adjustments performed do not have any clinical impact. The current clinical workflow needs to be revised in terms of the order of delineation of OARs and target volume. Trained RTTs can then accept contours within these thresholds to make the clinical workflow more efficient and to decrease inter-observer variability. For this current study, a dosimetric analysis is out-of-scope.
To conclude, training data for any DL-model remains crucial to the quality of the contours, as shown here. User-adjustments remain necessary to adhere to clinical guidelines and ensure quality of clinical contours. Designing separate models for specific clinical indications, changes in contouring guidelines or for different CT-scan acquisition protocols could be necessary to generate contours of higher clinical quality. Subsampling and post-processing of automatically generated contours can reduce editing needed and make the clinical workflow more efficient.
Declaration of Competing Interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: Maastro has licensing and research agreements with Mirada Medical Ltd.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.phro.2022.04.008.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Cardenas C.E., Yang J., Anderson B.M., Court L.E., Brock K.B. Advances in auto-segmentation. Semin Radiat Oncol. 2019;29:185–197. doi: 10.1016/j.semradonc.2019.02.001. [DOI] [PubMed] [Google Scholar]
- 2.Van der Veen J., Willems S., Robben D., Crijns W., Maes F., Nuyts S. Benefits of deep learning for delineation of organs at risk in head and neck cancer. Radiother Oncol. 2019;138:68–74. doi: 10.1016/j.radonc.2019.05.010. [DOI] [PubMed] [Google Scholar]
- 3.van Dijk L.V., Van den Bosch L., Aljabar P., Peressutti D., Both S., Steenbakkers Roel J.H.M., et al. Improving automatic delineation for head and neck organs at risk by Deep Learning Contouring. Radiother Oncol. 2020;142:115–123. doi: 10.1016/j.radonc.2019.09.022. [DOI] [PubMed] [Google Scholar]
- 4.Lustberg T., van Soest J., Gooding M., Peressutti D., Aljabar P., van der Stoep J., et al. Clinical evaluation of atlas and deep learning based automatic contouring for lung cancer. Radiother Oncol. 2018;126:312–317. doi: 10.1016/j.radonc.2017.11.012. [DOI] [PubMed] [Google Scholar]
- 5.Sharp G., Fritscher K.D., Pekar V., Peroni M., Shusharina N., Veeraraghavan H., et al. Vision 20/20: perspectives on automated image segmentation for radiotherapy. Med Phys. 2014;41:1–13. doi: 10.1118/1.4871620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang J., Veeraraghavan H., Armato S.G., Farahani K., Kirby J.S., Kalpathy-Kramer J., et al. Autosegmentation for thoracic radiation treatment planning: a grand challenge at AAPM 2017. Med Phys. 2018;45:4568–4581. doi: 10.1002/mp.13141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cha E., Elguindi S., Onochie I., Gorovets D., Deasy J.O., Zelefsky M., et al. Clinical implementation of deep learning contour autosegmentation for prostate radiotherapy. Radiother Oncol. 2021;159:1–7. doi: 10.1016/j.radonc.2021.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen X., Sun S., Bai N., Han K., Liu Q., Yao S., et al. A deep learning-based auto-segmentation system for organs-at-risk on whole-body computed tomography images for radiation therapy. Radiother Oncol. 2021;160:175–184. doi: 10.1016/j.radonc.2021.04.019. [DOI] [PubMed] [Google Scholar]
- 9.Robert C., Munoz A., Moreau D., Mazurier J., Sidorski G., Gasnier A., et al. Clinical implementation of deep-learning based auto-contouring tools–Experience of three French radiotherapy centers. Cancer/Radiothérapie. 2021;1:1–6. doi: 10.1016/j.canrad.2021.06.023. [DOI] [PubMed] [Google Scholar]
- 10.Fu Y., Lei Y., Wang T., Curran W.J., Liu T., Yang X. A review of deep learning based methods for medical image multi-organ segmentation. Phys Med. 2021;85:107–122. doi: 10.1016/J.EJMP.2021.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Valentini V., Boldrini L., Damiani A., Muren L.P. Recommendations on how to establish evidence from auto-segmentation software in radiotherapy. Radiother Oncol. 2014;112:317–320. doi: 10.1016/j.radonc.2014.09.014. [DOI] [PubMed] [Google Scholar]
- 12.Vandewinckele L., Claessens M., Dinkla A., Brouwer C., Crijns W., Verellen D., et al. Overview of artificial intelligence-based applications in radiotherapy: Recommendations for implementation and quality assurance. Radiother Oncol. 2020;153:55–66. doi: 10.1016/j.radonc.2020.09.008. [DOI] [PubMed] [Google Scholar]
- 13.Jia X., Ren L., Cai J. Clinical implementation of AI technologies will require interpretable AI models. Med Phys. 2020;47:1–4. doi: 10.1002/mp.13891. [DOI] [PubMed] [Google Scholar]
- 14.Wong J., Huang V., Wells D., Giambattista J., Giambattista J., Kolbeck C., et al. Implementation of deep learning-based auto-segmentation for radiotherapy planning structures: a workflow study at two cancer centers. Radiat Oncol. 2021;16:1–10. doi: 10.1186/s13014-021-01831-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brouwer C.L., Boukerroui D., Oliveira J., Looney P., Steenbakkers R.J.H.M., Langendijk J.A., et al. Assessment of manual adjustment performed in clinical practice following deep learning contouring for head and neck organs at risk in radiotherapy. Phys Imaging Radiat Oncol. 2020;16:54–60. doi: 10.1016/j.phro.2020.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vaassen F., Hazelaar C., Vaniqui A., Gooding M., van der Heyden B., Canters R., et al. Evaluation of measures for assessing time-saving of automatic organ-at-risk segmentation in radiotherapy. Phys Imaging Radiat Oncol. 2020;13:1–6. doi: 10.1016/j.phro.2019.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nikolov S, Blackwell S, Mendes R, De Fauw J, Meyer C, Hughes C, et al. Deep learning to achieve clinically applicable segmentation of head and neck anatomy for radiotherapy. ArXiv:180904430 2018:1–31.
- 18.Fedorov A., Beichel R., Kalpathy-Cramer J., Finet J., Fillion-Robin J., Pujol S., et al. 3D Slicer as an image computing platform for the Quantitative Imaging Network. Magn Reson Imaging. 2012;30:1323–1341. doi: 10.1016/j.mri.2012.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Offersen B., Boersma L., Kirkove C., Hol S., Aznar M., Sola A., et al. ESTRO consensus guideline on target volume delineation for elective radiation therapy of early stage breast cancer, version 1.1. Radiother Oncol. 2016;118:205–208. doi: 10.1016/j.radonc.2015.12.027. [DOI] [PubMed] [Google Scholar]
- 20.Batumalai V., Koh E.S., Delaney G.P., Holloway L.C., Jameson M.G., Papadatos G., et al. Interobserver variability in clinical target volume delineation in tangential breast irradiation: a comparison between radiation oncologists and radiation therapists. Clin Oncol. 2011;23:108–113. doi: 10.1016/j.clon.2010.10.004. [DOI] [PubMed] [Google Scholar]
- 21.Vaassen F., Hazelaar C., Canters R., Peeters S., Petit S., van Elmpt W. The impact of organ-at-risk contour variations on automatically generated treatment plans for NSCLC. Radiother Oncol. 2021;163:136–142. doi: 10.1016/j.radonc.2021.08.014. [DOI] [PubMed] [Google Scholar]
- 22.van Rooij W., Dahele M., Ribeiro Brandao H., Delaney A.R., Slotman B.J., Verbakel W.F. Deep learning-based delineation of head and neck organs at risk: geometric and dosimetric evaluation. Int J Radiat Oncol Biol Phys. 2019;104:677–684. doi: 10.1016/j.ijrobp.2019.02.040. [DOI] [PubMed] [Google Scholar]
- 23.Simoes R., Wortel G., Wiersma T.G., Janssen T.M., van der Heide U.A., Remeijer P. Geometrical and dosimetric evaluation of breast target volume auto-contouring. Phys Imaging Radiat Oncol. 2019;12:38–43. doi: 10.1016/j.phro.2019.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cui Y., Chen W., Kong F.M., Olsen L.A., Beatty R.E., Maxim P.G., et al. Contouring variations and the role of atlas in non-small cell lung cancer radiation therapy: Analysis of a multi-institutional preclinical trial planning study. Pract Radiat Oncol. 2015;5:e67–e75. doi: 10.1016/j.prro.2014.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim K., Chun M., Jin H., Jung W., Shin K.H., Shin S.S., et al. Inter-institutional variation in intensity-modulated radiotherapy for breast cancer in Korea (KROG 19-01) Anticancer Res. 2021;41:3145–3152. doi: 10.21873/anticanres.15100. [DOI] [PubMed] [Google Scholar]
- 26.Thor M., Apte A., Haq R., Iyer A., LoCastro E., Deasy J.O. Using auto-segmentation to reduce contouring and dose inconsistency in clinical trials: the simulated impact on RTOG 0617. Int J Radiat Oncol Biol Phys. 2021;109:1619–1626. doi: 10.1016/j.ijrobp.2020.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.