Abstract
Against the backdrop of the second wave of COVID‐19 pandemic in India that started in March 2021, we have monitored the spike (S) protein mutations in all the reported (GISAID portal) whole‐genome sequences of SARS‐CoV‐2 circulating in India from 1 January 2021 to 31 August 2021. In the 43,102 SARS‐CoV‐2 genomic sequences analysed, we have identified 24,260 amino acid mutations in the S protein, based on which 265 Pango lineages could be categorized. The dominant lineage in most of the 28 states of India and its 8 union territories was B.1.617.2 (the delta variant). However, the states Madhya Pradesh, Jammu & Kashmir, and Punjab had B.1.1.7 (alpha variant) as the major lineage, while the Himachal Pradesh state reported B.1.36 as the dominating lineage. A detailed analysis of various domains of S protein was carried out for detecting mutations having a prevalence of >1%; 70, 18, 7, 3, 9, 4, and 1 (N = 112) such mutations were observed in the N‐terminal domain, receptor binding domain, C ‐terminal domain, fusion peptide region, heptapeptide repeat (HR)‐1 domains, signal peptide domain, and transmembrane region, respectively. However, no mutations were recorded in the HR‐2 and cytoplasmic domains of the S protein. Interestingly, 13.39% (N = 15) of these mutations were reported to increase the infectivity and pathogenicity of the virus; 2% (N = 3) were known to be vaccine breakthrough mutations, and 0.89% (N = 1) were known to escape neutralizing antibodies. The biological significance of 82% (N = 92) of the reported mutations is yet unknown. As SARS‐CoV‐2 variants are emerging rapidly, it is critical to continuously monitor local viral mutations to understand national trends of virus circulation. This can tremendously help in designing better preventive regimens in the country, and avoid vaccine breakthrough infections.
Keywords: B.1.617.2, COVID‐19 second wave, India, SARS‐CoV‐2, spike protein variants, whole‐genome
1. INTRODUCTION
As on November 2021, severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) accounted for about 260 million COVID‐19 infections and about 5.2 million deaths worldwide. In India, more than 34 million cases with around half a million deaths were reported in this period (Coronavirus Disease [COVID‐19], n.d.). Viruses (especially RNA viruses) constantly change through mutations (Bessière & Volmer, 2021); a variant has one or more mutations that differentiate it from others in circulation. Owing to the great efforts of many researchers worldwide who are generating and sharing virus whole‐genome sequences publicly, multiple variants of SARS‐CoV‐2 have been documented globally throughout this pandemic. Currently, more than 4 million SARS‐CoV‐2 sequences are available via the Global Initiative on Sharing All Influenza Data (GISAID) (GISAID ‐ Initiative, n.d.). Sequence data greatly enables the identification of mutations that potentially change viral properties and detect emerging SARS‐CoV‐2 variants. The entry of SARS‐CoV‐2 into host cells is mediated by its spike (S) protein binding to the host cell‐surface receptor, angiotensin‐converting enzyme 2 (ACE2) (Zhao et al., 2020). The S protein is heavily glycosylated and induces a protective immune response (F. Li, 2016; Walls et al., 2020). The S protein is divided into two subunits S1 and S2. These two domains are biologically significant. S1 exhibits the function of receptor‐binding and S2 region is associated with the membrane fusion to facilitate cell entry (Xia, 2021). S1 region has four domains, signal peptide (1–13 aa), N‐terminal (14–305 aa), receptor binding (319–541 aa) and C‐ terminal‐ SD1 and SD2 (542–685 aa). The S2 region has five domains, fusion peptide (688–811 aa), heptapeptide repeats 1 and 2 (812–1142 aa), transmembrane (1213–1236 aa), cytoplasmic domain, and extra amino acids (1274–1943 aa) (Winger & Caspari, 2021).
Mutations in S protein were always of major concern as they play a major role in the emergence of new SARS‐CoV‐2 variants (Harvey et al., 2021). The World Health Organization (WHO) classification of variant viruses as variants of concern (VOC) and variants of interest (VOI) is also based on mutations in S protein (Tracking SARS‐CoV‐2 Variants, n.d.) (Table 1).
TABLE 1.
SARS‐CoV‐2 variant classifications as per WHO
| S. No. | Pango lineage | WHO variant classification | Spike mutations |
|---|---|---|---|
| 1 | B.1.525 | VOI | A67V, 69del, 70del, 144del, E484K, D614G, Q677H, F888L |
| 2 | B.1.526 | VOI | L5F, T95I, D253G, E484K, D614G, A701V |
| 3 | B.1.617.1 | VOI | T95I, G142D, E154K, L452R, E484Q, D614G, P681R, Q1071H |
| 4 | B.1.617.3 | VOI | T19R, G142D, L452R, E484Q, D614G, P681R, D950N |
| 5 | B.1.1.7 | VOC | 69del, 70del, 144del, N501Y, A570D, D614G, P681H, T716I, S982A, D1118H |
| 6 | B.1.617.2 | VOC | T19R, T95I, G142D, E156‐, F157‐, R158G, L452R, T478K, D614G, P681R, D950N |
| 7 | B.1.351, B.1.351.2, B.1.351.3 | VOC | D80A, D215G, 241del, 242del, 243del, K417N, E484K, N501Y, D614G, A701V |
| 8 | P.1, P.1.1, P.1.2 | VOC | L18F, T20N, P26S, D138Y, R190S, K417T, E484K, N501Y, D614G, H655Y, T1027I |
In India, the second wave of COVID‐19 emerged in March 2021 (and started declining towards August 2021), with a new set of symptoms affecting the gastrointestinal system along with the previous (first wave) symptoms that primarily included respiratory problems (Kamble et al., 2021). Oxygen requirement, mortality rate and the disease‐spreading rate increased during the second wave even though vaccines like COVAXIN (inactivated whole virus vaccine) and COVISHIELD (adenovirus vectored vaccine) were available and in use since January 2021 (albeit in initial stages by the time second wave emerged in India). In India, an estimate of 62,76,35,244 people completed 2 doses of vaccination; however, vaccine breakthrough infections are not uncommon (Dash et al., 2021). With this perspective, we initiated this study wherein we analysed the S protein sequences of SARS‐CoV‐2 variants reported in India and submitted in the GISAID portal from 1 January 2021 to 31 August 2021. Mutations throughout the S protein domains were studied in detail and analysed (at a translational level) to understand whether the mutations reported affect the virus properties and antigenic consequences such as infectivity, transmissibility, and resistance to the available vaccines, based on the literature available on these mutations.
2. MATERIALS AND METHODS
Whole‐genome sequences (N = 43,102) of SARS‐CoV‐2 variants circulating in India were downloaded from the GISAID website. The GISAID website has an inbuilt algorithm that categorises the Pango lineages and designates them. The same was used in this study. All the available data from 1 January 2021 to 31 August 2021 were analysed in the study. State‐wise statistics and sequence analysis of all the variants were also performed. Each genomic sequence was scrutinised for the acquirement of the data with regard to its: date of collection, location, accession ID, Clade, Pango lineage, and S mutations (Table S1). Each genomic sequence was thoroughly analysed for the Pango lineages and the spike mutations, as they were the main objectives. To study the variants circulating in India, we have segregated the data in a state‐wise pattern. First, every variant observed was counted, and the sequences were noted down. Next, all mutations in the spike region were listed out and calculated, then the percentiles for the enumerated count of all the mutations against more than 40,000 were taken. Briefly, using the excel tool, duplicates were separated. Transpose formula was used to make the total in every single column. All the mutations were arranged in ascending order and segregated into their respective domains in the S protein. All the clinically significant and major mutations were listed out for further analysis. Only mutations with frequencies >1% are analysed in detail. These mutations were studied in detail in S protein region‐1 and S protein region‐2. Based on existing literature, the S protein region's mutations were analysed for increased infectivity, decreased infectivity, and decreased neutralising sensitivity. S protein structure PDBID‐6VXX was downloaded from PDB, and increasing infectivity mutations were mapped. The final statistical data and mutation mapping was formulated into the graphical models using BioRender (https://biorender.com).
3. RESULTS AND DISCUSSION
In 2021, there has been a massive increase in the COVID‐19 community transmission in several countries worldwide (Coronavirus Disease (COVID‐19), n.d.). India also faced a gigantic surge in infectivity and transmissibility of SARS‐CoV‐2. In this study, we have analysed in detail 43,102 genomic sequences of SARS‐CoV‐2 strains circulating in India reported to the GISAID portal from 1 January 2021 to 31 August 2021. We identified the percentage prevalence of Pango lineages, studied the S protein mutations and analysed the structural impact on S protein and clinical significance of the observed S mutations based on existing literature.
We have identified a total of 24,260 mutations in the S protein (Table S2). All of these mutations were further categorised based on Pango nomenclature. A total of 265 Pango lineages could be categorised based on the 24,260 mutations identified (Table 2). The dominant lineage in most of the 28 states of India and its 8 union territories was B.1.617.2 (the delta variant). However, the states Madhya Pradesh, Jammu & Kashmir, and Punjab had B.1.1.7 (alpha variant) as the major lineage, while the state Himachal Pradesh had B.1.36 as the dominating lineage (Figure 1 and Table 2). Furthermore, Pango lineages identified in the country whose prevalence was >1% were also recorded (Figure 2). B.1.617.2‐Delta variant was the major dominating variant (45%), followed by B.1.617‐kappa (9.8 %), B.1.1.7 alpha (8.6 %), B.1 (7%), B.1.36 (4%), B.1.36.29 (2.8%), B.1.1 (2. 7%), B.1.1.216 (1.8%), B.1.1.306 (1.8%), AY12 (1.8%), and AY4 (1.2%). About 3.6% of the variants could not be assigned to any known Pango lineages (Figure 2). The total prevalence of Pango lineages among the different states and union territories of India is detailed in Table S3. It can be seen that two of the variants identified in the study period (B.1.617.2 [45%] and B.1.17 [8.6%]) are classified as VOC (Tracking SARS‐CoV‐2 Variants, n.d.).
TABLE 2.
Pango lineages of SARS CoV‐2 variants in India
| Pango lineage | Total no. obtained | % Prevalence |
|---|---|---|
| B.1.617.2 | 19,613 | 45.5036889239 |
| B.1.617.1 | 4258 | 9.8788919308 |
| B.1.1.7 | 3720 | 8.6306899912 |
| B.1 | 3069 | 7.1203192427 |
| B.1.36 | 1727 | 4.0067746276 |
| None | 1576 | 3.6564428565 |
| B.1.36.29 | 1220 | 2.8304951046 |
| B.1.1 | 1184 | 2.7469722983 |
| AY.12 | 781 | 1.8119808826 |
| B.1.1.216 | 657 | 1.5242912162 |
| B.1.1.306 | 633 | 1.4686093453 |
| AY.4 | 540 | 1.2528420955 |
| B.1.540 | 247 | 0.5730592548 |
| B.1.525 | 241 | 0.5591387871 |
| B.1.1.326 | 240 | 0.5568187091 |
| B.1.617.3 | 236 | 0.5475383973 |
| B.1.36.8 | 228 | 0.5289777737 |
| B.1.351 | 226 | 0.5243376177 |
| B.1.618 | 179 | 0.4152939539 |
| AY.3 | 142 | 0.3294510696 |
| B.1.560 | 138 | 0.3201707577 |
| B.1.604 | 138 | 0.3201707577 |
| B.1.36.10 | 137 | 0.3178506798 |
| B.1.1.220 | 91 | 0.2111270939 |
| B.1.1.46 | 90 | 0.2088070159 |
| A | 75 | 0.1740058466 |
| B.6 | 75 | 0.1740058466 |
| B.1.1.354 | 73 | 0.1693656907 |
| B.1.538 | 68 | 0.1577653009 |
| B.1.153 | 67 | 0.155445223 |
| B.1.533 | 57 | 0.1322444434 |
| AY.6 | 53 | 0.1229641316 |
| B | 50 | 0.1160038977 |
| B.1.210 | 50 | 0.1160038977 |
| B.1.36.22 | 50 | 0.1160038977 |
| B.1.36.18 | 49 | 0.1136838198 |
| B.1.575 | 46 | 0.1067235859 |
| B.1.36.17 | 43 | 0.099763352 |
| B.6.6 | 40 | 0.0928031182 |
| AY.1 | 37 | 0.0858428843 |
| B.1.537 | 32 | 0.0742424945 |
| B.4.7 | 31 | 0.0719224166 |
| B.1.1.25 | 29 | 0.0672822607 |
| B.1.351.3 | 29 | 0.0672822607 |
| B.1.36.38 | 28 | 0.0649621827 |
| B.1.535 | 26 | 0.0603220268 |
| B.1.311 | 25 | 0.0580019489 |
| B.1.468 | 25 | 0.0580019489 |
| B.1.548 | 23 | 0.053361793 |
| B.1.1.44 | 22 | 0.051041715 |
| AY.11 | 20 | 0.0464015591 |
| B.1.1.101 | 19 | 0.0440814811 |
| B.1.195 | 18 | 0.0417614032 |
| A.23.1 | 17 | 0.0394413252 |
| B.1.177 | 17 | 0.0394413252 |
| B.1.36.16 | 17 | 0.0394413252 |
| B.1.2 | 15 | 0.0348011693 |
| B.1.369 | 15 | 0.0348011693 |
| B.1.469 | 14 | 0.0324810914 |
| B.1.1.318 | 13 | 0.0301610134 |
| B.4 | 12 | 0.0278409355 |
| B.1.1.201 | 11 | 0.0255208575 |
| B.1.333 | 10 | 0.0232007795 |
| B.1.459 | 10 | 0.0232007795 |
| B.1.462 | 10 | 0.0232007795 |
| P.2 | 10 | 0.0232007795 |
| B.1.1.174 | 9 | 0.0208807016 |
| B.1.36.33 | 9 | 0.0208807016 |
| B.1.36.35 | 9 | 0.0208807016 |
| B.1.428 | 9 | 0.0208807016 |
| B.1.160 | 8 | 0.0185606236 |
| B.1.222 | 8 | 0.0185606236 |
| B.1.465 | 8 | 0.0185606236 |
| A.29 | 7 | 0.0162405457 |
| B.1.1.274 | 7 | 0.0162405457 |
| B.1.1.419 | 7 | 0.0162405457 |
| B.1.1.431 | 7 | 0.0162405457 |
| B.1.36.19 | 7 | 0.0162405457 |
| B.1.523 | 7 | 0.0162405457 |
| B.1.551 | 7 | 0.0162405457 |
| B.1.145 | 6 | 0.0139204677 |
| B.1.524 | 6 | 0.0139204677 |
| B.1.633 | 6 | 0.0139204677 |
| AY.5 | 5 | 0.0116003898 |
| B.1.1.194 | 5 | 0.0116003898 |
| B.1.1.282 | 5 | 0.0116003898 |
| B.1.1.5 | 5 | 0.0116003898 |
| B.1.111 | 5 | 0.0116003898 |
| B.1.36.24 | 5 | 0.0116003898 |
| B.1.441 | 5 | 0.0116003898 |
| B.1.456 | 5 | 0.0116003898 |
| B.1.569 | 5 | 0.0116003898 |
| B.1.582 | 5 | 0.0116003898 |
| C.36 | 5 | 0.0116003898 |
| A.21 | 4 | 0.0092803118 |
| A.9 | 4 | 0.0092803118 |
| AY.2 | 4 | 0.0092803118 |
| B.1.1.10 | 4 | 0.0092803118 |
| B.1.1.291 | 4 | 0.0092803118 |
| B.1.203 | 4 | 0.0092803118 |
| B.1.221 | 4 | 0.0092803118 |
| B.1.367 | 4 | 0.0092803118 |
| B.1.398 | 4 | 0.0092803118 |
| B.1.470 | 4 | 0.0092803118 |
| B.1.564 | 4 | 0.0092803118 |
| A.27 | 3 | 0.0069602339 |
| AM.4 | 3 | 0.0069602339 |
| B.1.1.307 | 3 | 0.0069602339 |
| B.1.1.398 | 3 | 0.0069602339 |
| B.1.113 | 3 | 0.0069602339 |
| B.1.143 | 3 | 0.0069602339 |
| B.1.177.16 | 3 | 0.0069602339 |
| B.1.214.2 | 3 | 0.0069602339 |
| B.1.258 | 3 | 0.0069602339 |
| B.1.260 | 3 | 0.0069602339 |
| B.1.351.2 | 3 | 0.0069602339 |
| B.1.393 | 3 | 0.0069602339 |
| B.1.432 | 3 | 0.0069602339 |
| B.1.466.1 | 3 | 0.0069602339 |
| B.1.466.2 | 3 | 0.0069602339 |
| B.1.526 | 3 | 0.0069602339 |
| B.1.94 | 3 | 0.0069602339 |
| A.2 | 2 | 0.0046401559 |
| AE.2 | 2 | 0.0046401559 |
| AY.10 | 2 | 0.0046401559 |
| AY.3.1 | 2 | 0.0046401559 |
| B.1.1.132 | 2 | 0.0046401559 |
| B.1.1.192 | 2 | 0.0046401559 |
| B.1.1.231 | 2 | 0.0046401559 |
| B.1.1.254 | 2 | 0.0046401559 |
| B.1.1.315 | 2 | 0.0046401559 |
| B.1.1.327 | 2 | 0.0046401559 |
| B.1.1.350 | 2 | 0.0046401559 |
| B.1.1.353 | 2 | 0.0046401559 |
| B.1.1.369 | 2 | 0.0046401559 |
| B.1.1.401 | 2 | 0.0046401559 |
| B.1.1.402 | 2 | 0.0046401559 |
| B.1.1.411 | 2 | 0.0046401559 |
| B.1.1.462 | 2 | 0.0046401559 |
| B.1.1.523 | 2 | 0.0046401559 |
| B.1.1.8 | 2 | 0.0046401559 |
| B.1.1.83 | 2 | 0.0046401559 |
| B.1.170 | 2 | 0.0046401559 |
| B.1.177.7 | 2 | 0.0046401559 |
| B.1.201 | 2 | 0.0046401559 |
| B.1.243 | 2 | 0.0046401559 |
| B.1.258.20 | 2 | 0.0046401559 |
| B.1.302 | 2 | 0.0046401559 |
| B.1.346 | 2 | 0.0046401559 |
| B.1.36.26 | 2 | 0.0046401559 |
| B.1.36.31 | 2 | 0.0046401559 |
| B.1.36.7 | 2 | 0.0046401559 |
| B.1.395 | 2 | 0.0046401559 |
| B.1.453 | 2 | 0.0046401559 |
| B.1.476 | 2 | 0.0046401559 |
| B.1.557 | 2 | 0.0046401559 |
| B.1.596 | 2 | 0.0046401559 |
| B.1.609 | 2 | 0.0046401559 |
| B.1.619 | 2 | 0.0046401559 |
| B.1.629 | 2 | 0.0046401559 |
| B.29 | 2 | 0.0046401559 |
| B.40 | 2 | 0.0046401559 |
| B.49 | 1 | 0.002320078 |
| C.11 | 1 | 0.002320078 |
| P.1.1 | 1 | 0.002320078 |
| A.2.5.1 | 1 | 0.002320078 |
| A.22 | 1 | 0.002320078 |
| A.23 | 1 | 0.002320078 |
| AE.4 | 1 | 0.002320078 |
| AM.2 | 1 | 0.002320078 |
| AY.7 | 1 | 0.002320078 |
| B.1.1.1 | 1 | 0.002320078 |
| B.1.1.116 | 1 | 0.002320078 |
| B.1.1.117 | 1 | 0.002320078 |
| B.1.1.121 | 1 | 0.002320078 |
| B.1.1.135 | 1 | 0.002320078 |
| B.1.1.141 | 1 | 0.002320078 |
| B.1.1.148 | 1 | 0.002320078 |
| B.1.1.157 | 1 | 0.002320078 |
| B.1.1.161 | 1 | 0.002320078 |
| B.1.1.164 | 1 | 0.002320078 |
| B.1.1.200 | 1 | 0.002320078 |
| B.1.1.213 | 1 | 0.002320078 |
| B.1.1.214 | 1 | 0.002320078 |
| B.1.1.228 | 1 | 0.002320078 |
| B.1.1.262 | 1 | 0.002320078 |
| B.1.1.263 | 1 | 0.002320078 |
| B.1.1.27 | 1 | 0.002320078 |
| B.1.1.28 | 1 | 0.002320078 |
| B.1.1.294 | 1 | 0.002320078 |
| B.1.1.308 | 1 | 0.002320078 |
| B.1.1.310 | 1 | 0.002320078 |
| B.1.1.312 | 1 | 0.002320078 |
| B.1.1.317 | 1 | 0.002320078 |
| B.1.1.33 | 1 | 0.002320078 |
| B.1.1.364 | 1 | 0.002320078 |
| B.1.1.365 | 1 | 0.002320078 |
| B.1.1.366 | 1 | 0.002320078 |
| B.1.1.372 | 1 | 0.002320078 |
| B.1.1.374 | 1 | 0.002320078 |
| B.1.1.378 | 1 | 0.002320078 |
| B.1.1.406 | 1 | 0.002320078 |
| B.1.1.413 | 1 | 0.002320078 |
| B.1.1.416 | 1 | 0.002320078 |
| B.1.1.433 | 1 | 0.002320078 |
| B.1.1.452 | 1 | 0.002320078 |
| B.1.1.487 | 1 | 0.002320078 |
| B.1.1.500 | 1 | 0.002320078 |
| B.1.1.526 | 1 | 0.002320078 |
| B.1.1.57 | 1 | 0.002320078 |
| B.1.1.712 | 1 | 0.002320078 |
| B.1.1.74 | 1 | 0.002320078 |
| B.1.1.75 | 1 | 0.002320078 |
| B.1.1.97 | 1 | 0.002320078 |
| B.1.119 | 1 | 0.002320078 |
| B.1.149 | 1 | 0.002320078 |
| B.1.151 | 1 | 0.002320078 |
| B.1.164 | 1 | 0.002320078 |
| B.1.177.18 | 1 | 0.002320078 |
| B.1.177.19 | 1 | 0.002320078 |
| B.1.177.4 | 1 | 0.002320078 |
| B.1.177.87 | 1 | 0.002320078 |
| B.1.179 | 1 | 0.002320078 |
| B.1.184 | 1 | 0.002320078 |
| B.1.225 | 1 | 0.002320078 |
| B.1.231 | 1 | 0.002320078 |
| B.1.232 | 1 | 0.002320078 |
| B.1.239 | 1 | 0.002320078 |
| B.1.289 | 1 | 0.002320078 |
| B.1.349 | 1 | 0.002320078 |
| B.1.36.2 | 1 | 0.002320078 |
| B.1.36.28 | 1 | 0.002320078 |
| B.1.36.36 | 1 | 0.002320078 |
| B.1.36.9 | 1 | 0.002320078 |
| B.1.362 | 1 | 0.002320078 |
| B.1.371 | 1 | 0.002320078 |
| B.1.378 | 1 | 0.002320078 |
| B.1.380 | 1 | 0.002320078 |
| B.1.382 | 1 | 0.002320078 |
| B.1.397 | 1 | 0.002320078 |
| B.1.409 | 1 | 0.002320078 |
| B.1.411 | 1 | 0.002320078 |
| B.1.413 | 1 | 0.002320078 |
| B.1.427 | 1 | 0.002320078 |
| B.1.429 | 1 | 0.002320078 |
| B.1.438 | 1 | 0.002320078 |
| B.1.450 | 1 | 0.002320078 |
| B.1.460 | 1 | 0.002320078 |
| B.1.496 | 1 | 0.002320078 |
| B.1.527 | 1 | 0.002320078 |
| B.1.545 | 1 | 0.002320078 |
| B.1.562 | 1 | 0.002320078 |
| B.1.594 | 1 | 0.002320078 |
| B.1.600 | 1 | 0.002320078 |
| B.1.617 | 1 | 0.002320078 |
| B.1.620 | 1 | 0.002320078 |
| B.1.93 | 1 | 0.002320078 |
| B.1.96 | 1 | 0.002320078 |
| B.3 | 1 | 0.002320078 |
| B.6.3 | 1 | 0.002320078 |
| C.1 | 1 | 0.002320078 |
| C.36.3 | 1 | 0.002320078 |
| L.3 | 1 | 0.002320078 |
| Q.1 | 1 | 0.002320078 |
| R.1 | 1 | 0.002320078 |
| TOTAL = 265 | 43102 | 100% |
FIGURE 1.
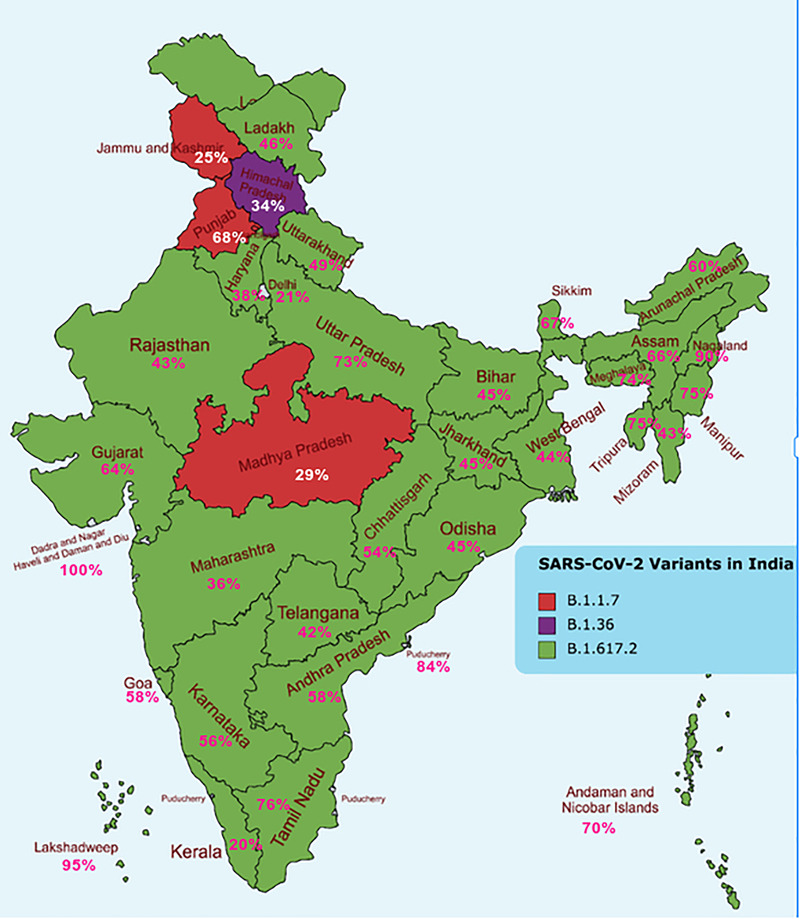
Dominant lineages in each of the states of India during January–August 2021. In the Indian map, the most prevalent lineages were shown. Green indicates B.1.617.2 (the delta variant), red indicates B.1.1.7 (alpha variant), and purple indicates B.1.36 lineage
FIGURE 2.
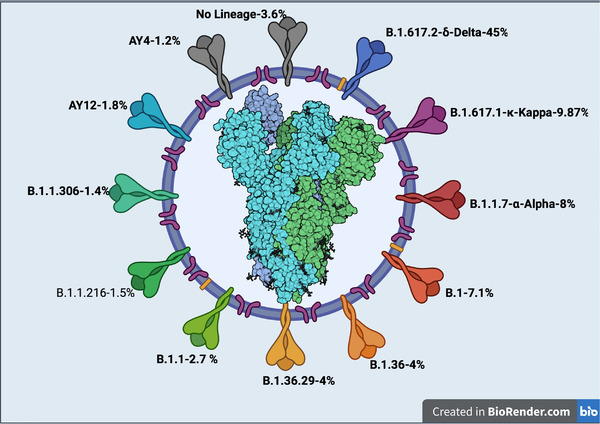
Dominant Pango lineages in India during January–August 2021. Pango lineages whose prevalence was >1% were shown. The numbers in the outer circle refers to percentages of Pango lineages
3.1. Clinical significance of the reported S mutations
Of the 24,260 mutations recorded in the S protein, mutations that possibly are clinically significant by playing a crucial role in altering the biological properties of the virus were analysed based on existing literature. We observed 70 mutations with >1% prevalence in the N‐terminal domain, 18 in the receptor binding domain (RBD), 8 in the C‐terminal domain (CTD), 4 in the signal peptide domain, 9 in HR‐1, 3 in fusion peptide region, and 1 in the transmembrane region of the S protein (Table S4). Mutations with a prevalence of >1% were not observed in the HR‐2 and cytoplasmic domains of S protein. All the clinically significant mutations (as per published literature) were recorded, and their percentiles were calculated to understand the significance of such mutations in the Indian context during the second wave of COVID‐19 in India. Of the 112 mutations recorded throughout the S protein with >1% prevalence in Indian populations, 15 (T19R, D63G, V70del, G142D, D377Y, L452R, T478K, E484Q, E484K, N501Y, A570D, D614G, Q677H, P681H, P681R) were known to be involved in increasing viral pathogenicity, 3 (E156G, F157del, R158del) were known to be vaccine breakthrough mutations, 1 mutation (Y144del) was known to be involved in the escape of the virus to neutralizing antibodies, and 1 (R385K) mutation was known to be involved in decreased virulence (Figure 3). The biological significance of 92 of the recorded mutations is not yet known. In India, 13 S protein mutations were correlated with the massive surge during the second wave of infections in April 2021; the most prevalent mutations included D614G, L452R, P681R, G142D, and Q1071H (Venkatakrishnan et al., 2021). It has been reported that B.1.617.1 and B.1.617.2 were the predominant circulating strains in the western part of India since January 2021, and that E484Q, D614G, and P681R were the common mutations of these strains (Cherian et al., 2021).
FIGURE 3.
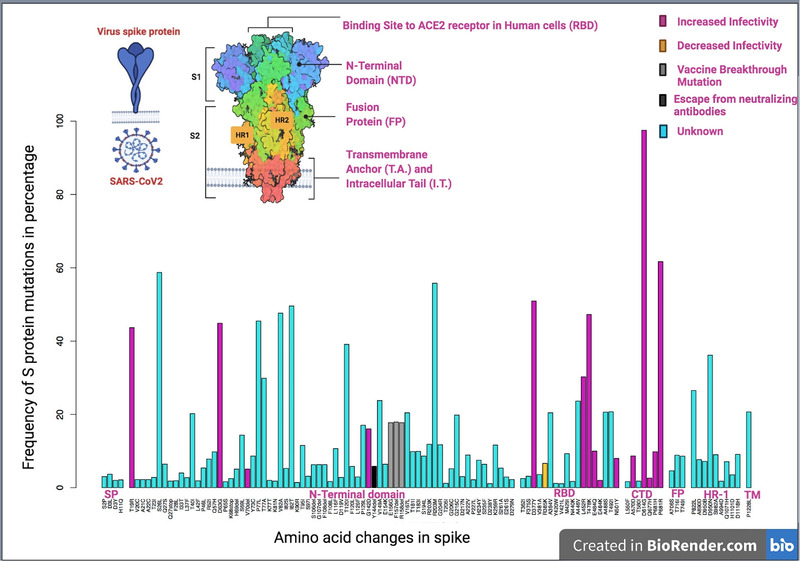
Spike mutations of >1% prevalence in India, January–August 2021. Percentage prevalence of mutations across the domains of S protein whose prevalence was >1% were shown. A specific colour was assigned for each respective category of amino acid mutations, increase of infectivity (pink), decrease the infectivity (yellow), vaccine breakthrough mutations(grey), escaping neutralizing antibodies (black), and those whose role is yet unknown are shown in turquoise. X‐axis refers to amino acid changes in S protein, Y‐axis refers to frequency of S protein mutations in percentages. SP: signal peptide sequence domain, RBD: receptor binding domain, CTD: C‐terminal domain, FP: fusion peptide domain, HR‐1: heptapeptide repeat domain, TM: transmembrane domain
3.2. N‐terminal domain
N‐terminal domain of S protein in the Indian variants recorded 70 mutations with >1% prevalence, and most of these mutations were shown to be clinically significant. P77L mutation is dominant in the delta variants (Patchett et al., 2021), but the clinical significance is unknown. Y144del mutation was known to be involved in escaping neutralizing antibodies (R. Wang et al., 2021). E156G, F157del, G142D mutations were known to be involved in vaccine breakthrough infections (Mishra et al., 2021; Shen et al., 2021). In our study, these mutations are found at 17% prevalence in Indian population and are likely to be clinically significant as these mutations are involved in reduced sensitivity to existing vaccines (Liu et al., 2021). T19R mutation (found at 43.66% prevalence in the current study) is found in Delta plus variant, which is classified as a VOC (Jordan et al., 2021). D63G and V70del were reported to increase viral pathogenicity (X.‐J. Yang, [Link], [Link]). S26L, T77A, K81N, and V82A mutations observed in the Indian variants in our study are at significant prevalence percentages, but their role in the infectivity of SARS‐CoV‐2 has not been reported yet. Furthermore, L42F, P46L, S97I, F108L, L116F, D119V, F120L, P65S, and T183I are not reported yet as significant in terms of viral infectivity.
3.3. Receptor binding domain
Eighteen mutations were recorded in the receptor‐binding domain with >1% prevalence. The high percentile mutations in the receptor‐binding domain reported in our study are P323L, D377Y, T478K, L452R, A446V, N501Y, and E484K. L452R and T478K are mutations already reported in the Indian variants and are known to increase the infectivity of the virus with decreased binding to monoclonal antibodies (mAbs), which may affect their neutralization potential (Cherian et al., 2021). Mutations at L452R, D377Y reported in our study were known to increase viral infectivity in a worldwide scenario (Motozono et al., 2021). E484Q mutation was reported as a neutralization‐resistant mutation that can increase the viral infectivity (Verghese et al., 2021). The E484K substitution alone conferred resistance to several monoclonal antibodies (Ku et al., 2021) while N501Y mutation was reported to increase the interacting force between S protein's RBD and ACE2 receptor and was reported to increase viral infectivity (Santos & Passos, 2021). One mutation in the receptor‐binding domain R385K was reported to decrease the viral infectivity (Miljanovic et al., 2021).
3.4. C‐terminal domain
Seven mutations were recorded in the CTD (with >1% prevalence) and five of these mutations that is, A570D, D614G, Q677H, P681H, and P681R are known to increase virus infectivity. A570D increases virus infectivity by enhancing host receptor binding, and can also act as a deleterious immune escape mutation (X. Yang et al., 2020). D614G mutation was shown to increase virion S protein infectivity and density (Zhang et al., 2020), Q677H was shown to enhance viral infectivity and confer neutralizing antibody resistance (Zeng et al., 2021). P681H mutation was reported as a worldwide emerging mutation that increases the infectivity of SARS‐CoV‐2 (Maison et al., 2021). P681R was shown to have played a crucial role in the alpha to delta replacement and increase in infectivity and pathogenicity of the virus (Liu et al., 2021).
3.5. Fusion peptide domain
Three mutations with >1% prevalence, that is, T716I, T749I, A706S were recorded in the fusion peptide region; the role of these mutations in the pathogenicity of SARS CoV‐2 is yet unknown.
3.6. HR‐1 domain
Nine mutations were recorded in the HR‐1 region of the spike protein (with >1% prevalence). S982A mutation was reported in the antigenic sites of alpha, which might be involved in increasing viral infectivity (Y. Wang et al., 2021).
3.7. Transmembrane domain
Only one mutation, that is, P1228L was recorded in the transmembrane domain, and the clinical significance of this mutation is not reported yet.
3.8. Signal peptide domain
Four mutations were recorded in the signal peptide domain: S2P, D3L, D3Y, H11Q. The biological significance of all these mutations is not reported yet.
3.9. Structural significance of the reported mutations on the S protein
Among the 15 mutations that were reported to increase the infectivity, 4 mutations (T19R, D63G, V70 del, and G142D) were located on the N‐terminal domain of the spike protein, which is involved in the host cell attachment through diverse polysaccharide moieties. Most of the VOC have mutations in the N‐terminal domain to escape the neutralization (McCallum et al., 2021). T19R mutation is found on the surface patch targeted by most NTD‐neutralizing antibodies (Planas et al., 2021). D63G mutation is important as it is directly involved in the RNA binding (Dinesh et al., 2020). V70del is present in the prominent exterior loop of the spike and is required for the efficient cell entry and increases cleavage of S2 thus enhancing spike infectivity (Meng et al., 2021). SARS‐CoV‐2 variants with a mutation at G142D showed resistance to the mAbs (Suryadevara et al., 2021). Six mutations that increase infectivity were found in the RBD, which interacts with host receptor ACE2. L452R mutation increases spike stability and viral infectivity, thereby increasing viral infection (Motozono et al., 2021). Mutation T478K is located on the interface with ACE2. Amino acid change from threonine to lysine is known to be involved in enhancement of electrostatic potential of S protein and hence was predicted to affect the spike–ACE2 interaction (Giacomo et al., 2021). Mutation at E484 (either with Q or K) has shown higher binding affinity for the ACE2 cell receptor resulting in enhanced transmissibility (Augusto et al., 2022). Variant with N501Y substitution was found to increase the viral transmissibility; substitution with tyrosine allowing more interaction with ACE2 receptor may lead to higher binding affinity to the host cell receptor (Zhou et al., 2021).
The CTD harbours A570D mutation, which is found to be more infectious in the pseudoviral assay. Structural analysis on A570D revealed that this mutation introduces the molecular switch for the opening and closing of RBD (X.‐J. Yang, [Link], [Link]). D614G mutation is more infectious and several studies indicated that this mutation is associated with increased viral load in the patients infected with COVID‐19 as D614G shifts S protein conformation towards an ACE2‐binding fusion state (Yurkovetskiy et al., 2020). It was demonstrated that Q677H mutation increases viral infectivity and syncytium formation (Zeng et al., 2021). Mutations P681R and P681H are located in the polybasic S1/S2 furin cleavage site and were reported to enhance the fusogenic activity of the spike protein (Tao et al., 2021).
Three mutations in the N‐terminal domain, E156G, F157del, and R158del revealed less flexibility compared to the Wuhan strain. Docking studies of these mutants with monoclonal antibodies reported low binding affinity compared to the Wuhan strain; this may be a possible case of immune escape (Chaudhari et al., 2021). Y144 del has been found to abrogate binding to neutralizing antibodies (McCarthy et al., 2021).
The initial contact between RBD of the S‐protein and ACE2 is through the peptidase domain (PD) (Adhikari et al., 2020). The possible binding mechanism is of pivotal importance and has vital implications for vaccine design. The presence of an unexpected furin cleavage site at the S1/S2 boundary of SARS‐CoV‐2 S, which is cleaved during biosynthesis, is a novel feature setting this virus apart from SARS‐CoV and SARSr‐CoVs (Walls et al., 2020). The boundary of the (uncoupled) N‐terminal region lies in physical proximity to the furin‐targeted motif RRAR, which is essential for pre‐activation of SARS‐CoV‐2 spike protein through proteolysis (Serapian et al., 2020). A second proteolytic cleavage at site S2′ releases the fusion peptide, which penetrates the host cell membrane, preparing it for fusion (Apellániz et al., 2014).
S glycoproteins are densely decorated by heterogeneous N‐linked glycans protruding from the trimer surface. These oligosaccharides participate in S folding, affect priming by host proteases, and might modulate the antibody recognition (Walls et al., 2020). Glycan acts as a shield that protects epitope from antibody binding. Site‐specific glycan analysis of SARS‐CoV‐2 S protein was performed in which glycopeptides are generated using proteases and analysed by liquid chromatography–mass spectrometry; as a result, 22 N‐linked glycosylation sequons per protomer were determined (Watanabe et al., 2020).
One study demonstrated that the simulated models of N‐glycan at position 165 and 234 act as RBD modulators on its binding with ACE2. On addition of N‐glycans, RBD is stabilized and the removal of N‐glycans showed a reduced ACE2 binding, which revealed conformational shift of RBD (Casalino et al., 2020).
E484 is an immunodominant spike protein residue in RBD region with various substitutions (E484A, E484D, E484G, and E484K) (Harvey et al., 2021). E484K introduces a residue with a charge opposite to the wild‐type, which would significantly alter the electrostatic complementarity of antibody binding to this region (Andreano et al., 2021). Also, the same group in another study reported that mutation E484K can lead to a fourfold decrease in neutralization activity of convalescent plasma to SARS‐CoV‐2 (Andreano et al., 2021). ΔY144 deletion was found to alter the N3 NTD loop (140–156 residues) and can abolish the neutralizing effect of various antibodies (Harvey et al., 2021).
An in‐silico study predicted that replacement of aspartic acid to glycine (D614G) leads to the loss of hydrogen bond interactions formed with valine at position 859, thereby eliminating the hydrogen bonding between S1 and S2 domain, leading to increased main‐chain flexibility enabling a more favourable orientation of Q613, possibly facilitating cleavage by TMPRSS2 by perturbing its affinity with the S1‐furin cleavage site (Raghav et al., 2020).
One previous study performed an unbiased multi‐microsecond molecular dynamics of 7 glycosylated S‐protein variants derived from cryo‐EM structure 6VSB and applied a matrix of low coupling energy (MLCE)‐based approach to predict potential changes in immunogenic regions on each variant (Triveri et al., 2021). In this study, reference S structure used was the dominant D614G variant. According to the MLCE study, contiguous residues uncoupled from the S protein core shrink in number compared to D614G S; their experimental data also confirms that variant evades Abs binding to the shrunk or lost epitopes. The number of residues defining the epitope located in the long RBD loop (residues 417−503) is much lower in mutants 501Y.V2 (South African), B1.1.28 (gamma), and N439K. Also, they predict that N439K RBD forms a new interaction with the human ACE2 receptor (hACE2) and has an enhanced affinity for hACE2 (Triveri et al., 2021).
Two different kinds of vaccines, COVAXIN ((a whole‐virion inactivated SARS‐CoV‐2 antigen [Strain: NIV‐2020‐7700]) and COVISHIELD (a recombinant, replication‐deficient chimpanzee adenovirus vector encoding the SARS‐CoV‐2 Spike (S) glycoprotein) have been in use in India since January 2021. Both these vaccines are based on the original variant of SARS‐CoV‐2. The second wave originated during March 2021 and lasted till August 2021. In this context, it is crucial to identify the various variants that were circulating and were responsible for the increased surge in the pandemic during 2021. Such knowledge can also help to understand vaccine breakthrough infections. For example, in our study, B.1.617.1 variant showed co‐occurrence of three critical mutations, L452R, E484Q, and P681R in the S protein. E484K, T95I, and Y144del were identified as vaccine breakthrough mutations (Hacisuleyman et al., 2021; Wu et al., 2021). Variants including A475V, L452R, V483A, and F490L were known to be involved in the putative antibody resistance (Q. Li et al., 2020). It is alarming that these mutations were also identified in the present study with significant prevalence rates: E484K (2.03%), T95I (11.5%), Y144del (5.86%), L452R (30.25%).
To conclude, we have identified the total number of circulating SARS‐CoV‐2 lineages reported during the second wave of the COVID‐19 pandemic in India. Further, the S mutations associated with the rise in COVID‐19 cases were investigated in detail. Importantly, we have identified 15 mutations that were known to be involved in increasing viral pathogenicity (T19R, D63G, V70del, G142D, D377Y, L452R, T478K, E484Q, E484K, N501Y, A570D, D614G, Q677H, P681H, P681R); 3 that were known to be vaccine breakthrough mutations (E156G, F157del, R158del); and 1 mutation that was known to escape neutralizing antibodies (Y144del). Since new variants will continue to emerge, it is crucial to identify and monitor mutations in the S protein, and any functionally significant co‐occurring modifications. Going forward, experimental assessment of SARS‐CoV‐2 S protein mutations can be beneficial from a public health point of view to understand vaccine breakthrough infections, and to devise better preventive strategies by individual nations.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
ETHICS STATEMENT
The authors confirm that the ethical policies of the journal, as noted on the journal's author guidelines page, have been adhered to. No ethical approval was required as this article only uses whole genome sequences deposited on GISAID portal that are publicly available with no original research data.
AUTHOR CONTRIBUTIONS
Radhakrishna Muttineni conceptualized the study. Binitha R.N., Jaslin Panyam, Aravind Vemula, Shashi Mohan Singh, Subin Balachandran, and Sandra K.P. collected data and did the preliminary analysis. Radhakrishna Muttineni, Binitha R.N., Kalyani Putty, Kavitha Marapakala, Viroji Rao S.T., and Anand Kumar Kondapi analysed the data and wrote the manuscript.
Supporting information
Supplementary table 1: Details of 43102 SARS CoV‐2 whole genome sequences used in the study.
Supplementary table 2: Details of 24260 S protein mutations identified in the study
Supplementary table 3: Individual state‐wide distribution of SARS CoV‐2 pango lineages in India
Supplementary table 4: Spike mutations with >1% prevalence across the domains
ACKNOWLEDGEMENT
The authors would like to acknowledge GISAID and researchers worldwide depositing their sequences in GISAID.
Muttineni, R. , R.N., B. , Putty, K. , Marapakala, K. , K.P., S. , Panyam, J. , Vemula, A. , Singh, S. M. , Balachandran, S. , S.T., V. R. , & Kondapi, A. K. (2022). SARS‐CoV‐2 variants and spike mutations involved in second wave of COVID‐19 pandemic in India. Transboundary and Emerging Diseases, 1–16. 10.1111/tbed.14508
Radhakrishna Muttineni, Binitha R.N, Kalyani Putty, Kavitha Marapakala, and Sandra K.P contributed equally to this work.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are openly available in GISAID at https://www.gisaid.org.
REFERENCES
- Adhikari, P. , Li, N. , Shin, M. , Steinmetz, N. F. , Twarock, R. , Podgornik, R. , & Ching, W. Y. (2020). Intra‐ and intermolecular atomic‐scale interactions in the receptor binding domain of SARS‐CoV‐2 spike protein: Implication for ACE2 receptor binding. Physical Chemistry Chemical Physics, 22(33), 18272–18283. 10.1039/D0CP03145C [DOI] [PubMed] [Google Scholar]
- Andreano, E. , Piccini, G. , Licastro, D. , Casalino, L. , Johnson, N. V. , Paciello, I. , Monego, S. D. , Pantano, E. , Manganaro, N. , Manenti, A. , Manna, R. , Casa, E. , Hyseni, I. , Benincasa, L. , Montomoli, E. , Amaro, R. E. , McLellan, J. S. , & Rappuoli, R. (2021). SARS‐CoV‐2 escape from a highly neutralizing COVID‐19 convalescent plasma. Proceedings of the National Academy of Sciences of the United States of America, 118(36). 10.1073/PNAS.2103154118/-/DCSUPPLEMENTAL [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apellániz, B. , Huarte, N. , Largo, E. , & Nieva, J. L. (2014). The three lives of viral fusion peptides. Chemistry and Physics of Lipids, 181, 40–55. 10.1016/J.CHEMPHYSLIP.2014.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augusto, G. , Mohsen, M. O. , Zinkhan, S. , Liu, X. , Vogel, M. , & Bachmann, M. F. (2022). In vitro data suggest that Indian delta variant B.1.617 of SARS‐CoV‐2 escapes neutralization by both receptor affinity and immune evasion. Allergy, 77(1), 111–117. 10.1111/ALL.15065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessière, P. , & Volmer, R. (2021). From one to many: The within‐host rise of viral variants. PLoS Pathogens, 17(9). 10.1371/JOURNAL.PPAT.1009811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casalino, L. , Gaieb, Z. , Goldsmith, J. A. , Hjorth, C. K. , Dommer, A. C. , Harbison, A. M. , Fogarty, C. A. , Barros, E. P. , Taylor, B. C. , Mclellan, J. S. , Fadda, E. , & Amaro, R. E. (2020). Beyond shielding: The roles of glycans in the SARS‐CoV‐2 spike protein. ACS Central Science, 6(10), 1722–1734. 10.1021/ACSCENTSCI.0C01056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhari, A. M. , Kumar, D. , Joshi, M. , Patel, A. , & Joshi, C. (2021). E156/G and Arg158, Phe‐157/del mutation in NTD of spike protein in B.1.167.2 lineage of SARS‐CoV‐2 leads to immune evasion through antibody escape. BioRxiv. 10.1101/2021.06.07.447321 [DOI] [Google Scholar]
- Cherian, S. , Potdar, V. , Jadhav, S. , Yadav, P. , Gupta, N. , Das, M. , Rakshit, P. , Singh, S. , Abraham, P. , Panda, S. , & Team, N. (2021). SARS‐CoV‐2 spike mutations, L452R, T478K, E484Q and P681R, in the second wave of COVID‐19 in Maharashtra, India. Microorganisms 2021, 9(7), 1542. 10.3390/MICROORGANISMS9071542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coronavirus disease (COVID‐19) . (n.d.). https://www.who.int/emergencies/diseases/novel‐coronavirus‐2019
- Dash, G. C. , Subhadra, S. , Turuk, J. , Parai, D. , Rath, S. , Sabat, J. , Rout, U. K. , Kanungo, S. , Choudhary, H. R. , Nanda, R. R. , Pattnaik, M. , Pati, S. , & Bhattacharya, D. (2021). Breakthrough SARS‐CoV‐2 infections among BBV‐152 (COVAXIN®) and AZD1222 (COVISHIELD TM) recipients: Report from the eastern state of India. Journal of Medical Virology, 94(3), 1201–1205. 10.1002/JMV.27382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinesh, D. C. , Chalupska, D. , Silhan, J. , Koutna, E. , Nencka, R. , Veverka, V. , & Boura, E. (2020). Structural basis of RNA recognition by the SARS‐CoV‐2 nucleocapsid phosphoprotein. PLoS Pathogens, 16(12), e1009100. 10.1371/JOURNAL.PPAT.1009100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giacomo, S. D.i , Mercatelli, D. , Rakhimov, A. , & Giorgi, F. M. (2021). Preliminary report on severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) spike mutation T478K. Journal of Medical Virology, 93(9), 5638–5643. 10.1002/JMV.27062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- GISAID ‐ Initiative . (n.d.). https://www.gisaid.org/
- Hacisuleyman, E. , Hale, C. , Saito, Y. , Blachere, N. E. , Bergh, M. , Conlon, E. G. , Schaefer‐Babajew, D. J. , DaSilva, J. , Muecksch, F. , Gaebler, C. , Lifton, R. , Nussenzweig, M. C. , Hatziioannou, T. , Bieniasz, P. D. , & Darnell, R. B. (2021). Vaccine breakthrough infections with SARS‐CoV‐2 variants. National Library of Medicine, 384(23), 2212–2218. 10.1056/NEJMOA2105000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey, W. T. , Carabelli, A. M. , Jackson, B. , Gupta, R. K. , Thomson, E. C. , Harrison, E. M. , Ludden, C. , Reeve, R. , Rambaut, A. , Peacock, S. J. , & Robertson, D. L. (2021). SARS‐CoV‐2 variants, spike mutations and immune escape. Nature Reviews Microbiology, 19(7), 409–424. 10.1038/s41579-021-00573-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan, S. C. , Shin, B.‐H. , Gadsden, T.‐A. M. , Chu, M. , Petrosyan, A. , Le, C. N. , Zabner, R. , Oft, J. , Pedraza, I. , Cheng, S. , Vo, A. , Ammerman, N. , Plummer, J. , Ge, S. , Froch, M. , Berg, A. , Toyoda, M. , & Zhang, R. (2021). T cell immune responses to SARS‐CoV‐2 and variants of concern (Alpha and Delta) in infected and vaccinated individuals. Cellular & Molecular Immunology, 18(11), 2554–2556. 10.1038/s41423-021-00767-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamble, P. , Daulatabad, V. , John, N. , & John, J. (2021). Synopsis of symptoms of COVID‐19 during second wave of the pandemic in India. Hormone Molecular Biology and Clinical Investigation. 10.1515/HMBCI-2021-0043 [DOI] [PubMed] [Google Scholar]
- Meng, B. , Kemp, S. A. , Papa, G. , Datir, R. , Ferreira, I. A.T.M. , Marelli, S. , Harvey, W. T. , Lytras, S. , Mohamed, A. , Gallo, G. , Thakur, N. , Collier, D. A. , Mlcochova, P. , Duncan, L. M. , Carabelli, A. M. , Kenyon, J. C. , Lever, A. M. , De Marco, A. , Saliba, C. , … Wright, Sean (2021). Recurrent emergence of SARS‐CoV‐2 spike deletion H69/V70 and its role in the Alpha variant B.1.1.7. Cell Reports, 35, (13), 109292. 10.1016/j.celrep.2021.109292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku, Z. , Xie, X. , Davidson, E. , Ye, X. , Su, H. , Menachery, V. D. , Li, Y. , Yuan, Z. , Zhang, X. , Muruato, A. E. , i Escuer, A. G. , Tyrell, B. , Doolan, K. , Doranz, B. J. , Wrapp, D. , Bates, P. F. , McLellan, J. S. , Weiss, S. R. , Zhang, N. , … An, Z. (2021). Molecular determinants and mechanism for antibody cocktail preventing SARS‐CoV‐2 escape. Nature Communications, 12(1), 1–13. 10.1038/s41467-020-20789-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, F. (2016). Structure, function, and evolution of coronavirus spike proteins. Annual Review of Virology, 3, 237–261. 10.1146/ANNUREV-VIROLOGY-110615-042301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Q. , Wu, J. , Nie, J. , Zhang, L. , Hao, H. , Liu, S. , Zhao, C. , Zhang, Q. , Liu, H. , Nie, L. , Qin, H. , Wang, M. , Lu, Q. , Li, X. , Sun, Q. , Liu, J. , Zhang, L. , Li, X. , Huang, W. , & Wang, Y. (2020). The impact of mutations in SARS‐CoV‐2 spike on viral infectivity and antigenicity. Cell, 182(5), 1284–1294.e9. 10.1016/J.CELL.2020.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y. , Liu, J. , Johnson, B. A. , Xia, H. , Ku, Z. , Schindewolf, C. , Widen, S. G. , An, Z. , Weaver, S. C. , Menachery, V. D. , Xie, X. , & Shi, P.‐Y. (2021). Delta spike P681R mutation enhances SARS‐CoV‐2 fitness over Alpha variant. BioRxiv. 10.1101/2021.08.12.456173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maison, D. P. , Ching, L. L. , Shikuma, C. M. , & Nerurkar, V. R. (2021). Genetic characteristics and phylogeny of 969‐bp S gene sequence of SARS‐CoV‐2 from Hawai‘i reveals the worldwide emerging P681H mutation. Hawai'i Journal of Health & Social Welfare, 80(3), 52. [PMC free article] [PubMed] [Google Scholar]
- McCallum, M. , De Marco, A. , Lempp, F. A. , Tortorici, M. A. , Pinto, D. , Walls, A. C. , Beltramello, M. , Chen, A. , Liu, Z. , Zatta, F. , Zepeda, S. , di Iulio, J. , Bowen, J. E. , Montiel‐Ruiz, M. , Zhou, J. , Rosen, L. E. , Bianchi, S. , Guarino, B. , Fregni, C. S. , … Veesler, D. (2021). N‐terminal domain antigenic mapping reveals a site of vulnerability for SARS‐CoV‐2. Cell, 184(9), 2332–2347.e16. 10.1016/J.CELL.2021.03.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy, K. R. , Rennick, L. J. , Nambulli, S. , Robinson‐McCarthy, L. R. , Bain, W. G. , Haidar, G. , & Paul Duprex, W. (2021). Recurrent deletions in the SARS‐CoV‐2 spike glycoprotein drive antibody escape. Science (New York, N.Y.), 371(6534), 1139–1142. 10.1126/SCIENCE.ABF6950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miljanovic, D. , Milicevic, O. , Loncar, A. , Abazovic, D. , Despot, D. , & Banko, A. (2021). The first molecular characterization of Serbian SARS‐CoV‐2 isolates from a unique early second wave in Europe. Frontiers in Microbiology, 0, 1526. 10.3389/FMICB.2021.691154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra, T. , Joshi, G. , Kumar, A. , Dalavi, R. , Pandey, P. , Shukla, S. , Mishra, R. K. , & Chande, A. (2021). B.1.617.3 SARS CoV‐2 spike E156G/Δ157‐158 mutations contribute to reduced neutralization sensitivity and increased infectivity. BioRxiv. 10.1101/2021.10.04.463028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motozono, C. , Toyoda, M. , Zahradnik, J. , Saito, A. , Nasser, H. , Tan, T. S. , Ngare, I. , Kimura, I. , Uriu, K. , Kosugi, Y. , Yue, Y. , Shimizu, R. , Ito, J. , Torii, S. , Yonekawa, A. , Shimono, N. , Nagasaki, Y. , Minami, R. , Toya, T. , … Sato, K. (2021). SARS‐CoV‐2 spike L452R variant evades cellular immunity and increases infectivity. Cell Host & Microbe, 29(7), 1124–1136.e11. 10.1016/J.CHOM.2021.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patchett, S. , Lv, Z. , Rut, W. , Békés, M. , Drag, M. , Olsen, S. K. , & Huang, T. T. (2021). A molecular sensor determines the ubiquitin substrate specificity of SARS‐CoV‐2 papain‐like protease. Cell Reports, 36(13), 109754. 10.1016/J.CELREP.2021.109754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planas, D. , Veyer, D. , Baidaliuk, A. , Staropoli, I. , Guivel‐Benhassine, F. , Rajah, M. M. , Planchais, C. , Porrot, F. , Robillard, N. , Puech, J. , Prot, M. , Gallais, F. , Gantner, P. , Velay, A. , Le Guen, J. , Kassis‐Chikhani, N. , Edriss, D. , Belec, L. , Seve, A. , … Schwartz, O. (2021). Reduced sensitivity of SARS‐CoV‐2 variant Delta to antibody neutralization. Nature, 596(7871), 276–280. 10.1038/S41586-021-03777-9 [DOI] [PubMed] [Google Scholar]
- Raghav, S. , Ghosh, A. , Turuk, J. , Kumar, S. , Jha, A. , Madhulika, S. , Priyadarshini, M. , Biswas, V. K. , Shyamli, P. S. , Singh, B. , Singh, N. , Singh, D. , Datey, A. , Avula, K. , Smita, S. , Sabat, J. , Bhattacharya, D. , Kshatri, J. S. , Vasudevan, D. , … Parida, A. (2020). Analysis of Indian SARS‐CoV‐2 genomes reveals prevalence of D614G mutation in spike protein predicting an increase in interaction with TMPRSS2 and virus infectivity. Frontiers in Microbiology, 11, 2847. 10.3389/FMICB.2020.594928/BIBTEX [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos, J. C. , & Passos, G. A. (2021). The high infectivity of SARS‐CoV‐2 B.1.1.7 is associated with increased interaction force between Spike‐ACE2 caused by the viral N501Y mutation. BioRxiv. 10.1101/2020.12.29.424708 [DOI] [Google Scholar]
- Serapian, S. A. , Marchetti, F. , Triveri, A. , Morra, G. , Meli, M. , Moroni, E. , Sautto, G. A. , Rasola, A. , & Colombo, G. (2020). The answer lies in the energy: How simple atomistic molecular dynamics simulations may hold the key to epitope prediction on the fully glycosylated SARS‐CoV‐2 spike protein. The Journal of Physical Chemistry Letters, 11, 8093. 10.1021/acs.jpclett.0c02341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, L. , Triche, T. J. , Bard, J. D. , Biegel, J. A. , Judkins, A. R. , & Gai, X. (2021). Spike protein NTD mutation G142D in SARS‐CoV‐2 Delta VOC lineages is associated with frequent back mutations, increased viral loads, and immune evasion. MedRxiv. 10.1101/2021.09.12.21263475 [DOI] [Google Scholar]
- Suryadevara, N. , Shrihari, S. , Gilchuk, P. , VanBlargan, L. A. , Binshtein, E. , Zost, S. J. , Nargi, R. S. , Sutton, R. E. , Winkler, E. S. , Chen, E. C. , Fouch, M. E. , Davidson, E. , Doranz, B. J. , Chen, R. E. , Shi, P. Y. , Carnahan, R. H. , Thackray, L. B. , Diamond, M. S. , & Crowe, J. E. (2021). Neutralizing and protective human monoclonal antibodies recognizing the N‐terminal domain of the SARS‐CoV‐2 spike protein. Cell, 184(9), 2316. 10.1016/J.CELL.2021.03.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao, K. , Tzou, P. L. , Nouhin, J. , Gupta, R. K. , de Oliveira, T. , Kosakovsky Pond, S. L. , Fera, D. , & Shafer, R. W. (2021). The biological and clinical significance of emerging SARS‐CoV‐2 variants. Nature Reviews. Genetics, 22(12), 757–773. 10.1038/S41576-021-00408-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tracking SARS‐CoV‐2 variants . (n.d.). https://www.who.int/en/activities/tracking‐SARS‐CoV‐2‐variants/
- Triveri, A. , Serapian, S. A. , Marchetti, F. , Doria, F. , Pavoni, S. , Cinquini, F. , Moroni, E. , Rasola, A. , Frigerio, F. , & Colombo, G. (2021). SARS‐CoV‐2 spike protein mutations and escape from antibodies: A computational model of epitope loss in variants of concern. Journal of Chemical Information and Modeling, 61, 4700. 10.1021/ACS.JCIM.1C00857/SUPPL_FILE/CI1C00857_SI_001.PDF [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatakrishnan, A. J. , Anand, P. , Lenehan, P. , Ghosh, P. , Suratekar, R. , Siroha, A. , Chowdhury, D. R. , O'Horo, J. C. , Yao, J. D. , Pritt, B. S. , Norgan, A. , Hurt, R. T. , Badley, A. D. , Halamka, J. D. , & Soundararajan, V. (2021). Antigenic minimalism of SARS‐CoV‐2 is linked to surges in COVID‐19 community transmission and vaccine breakthrough infections. MedRxiv. 10.1101/2021.05.23.21257668 [DOI] [Google Scholar]
- Verghese, M. , Jiang, B. , Iwai, N. , Mar, M. , Sahoo, M. K. , Yamamoto, F. , Mfuh, K. O. , Miller, J. , Wang, H. , Zehnder, J. , & Pinsky, B. A. (2021). A SARS‐CoV‐2 variant with L452R and E484Q neutralization resistance mutations. Journal of Clinical Microbiology, 59(7). 10.1128/JCM.00741-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls, A. C. , Park, Y. J. , Tortorici, M. A. , Wall, A. , McGuire, A. T. , & Veesler, D. (2020). Structure, function, and antigenicity of the SARS‐CoV‐2 spike glycoprotein. Cell, 181(2), 281–292.e6. 10.1016/J.CELL.2020.02.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, R. , Zhang, Q. , Ge, J. , Ren, W. , Zhang, R. , Lan, J. , Ju, B. , Su, B. , Yu, F. , Chen, P. , Liao, H. , Feng, Y. , Li, X. , Shi, X. , Zhang, Z. , Zhang, F. , Ding, Q. , Zhang, T. , Wang, X. , & Zhang, L. (2021). Analysis of SARS‐CoV‐2 variant mutations reveals neutralization escape mechanisms and the ability to use ACE2 receptors from additional species. Immunity, 54(7), 1611–1621.e5. 10.1016/J.IMMUNI.2021.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y. , Wu, J. , Zhang, L. , Zhang, Y. , Wang, H. , Ding, R. , Nie, J. , Li, Q. , Liu, S. , Yu, Y. , Yang, X.‐M. , Qu, X. , Duan, K. & Huang, W. (2021). The infectivity and antigenicity of epidemic SARS‐CoV‐2 variants in the United Kingdom. 10.21203/RS.3.RS-153108/V1 [DOI] [PMC free article] [PubMed]
- Watanabe, Y. , Allen, J. D. , Wrapp, D. , McLellan, J. S. , & Crispin, M. (2020). Site‐specific glycan analysis of the SARS‐CoV‐2 spike. Science (New York, N.Y.), 369(6501), 330–333. 10.1126/SCIENCE.ABB9983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winger, A. , & Caspari, T. (2021). The spike of concern—The novel variants of SARS‐CoV‐2. Viruses 2021, 13(6), 1002. 10.3390/V13061002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, K. , Werner, A. P. , Moliva, J. I. , Koch, M. , Choi, A. , Stewart‐Jones, G. B. E. , Bennett, H. , Boyoglu‐Barnum, S. , Shi, W. , Graham, B. S. , Carfi, A. , Corbett, K. S. , Seder, R. A. , & Edwards, D. K. (2021). mRNA‐1273 vaccine induces neutralizing antibodies against spike mutants from global SARS‐CoV‐2 variants. BioRxiv. 10.1101/2021.01.25.427948 [DOI] [Google Scholar]
- Xia, X. (2021). Domains and functions of spike protein in Sars‐Cov‐2 in the context of vaccine design. Viruses, 13(1). 10.3390/V13010109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, X.‐J. (2021a). SARS‐COV‐2 δ variant drives the pandemic in the USA through two subvariants. 10.21203/RS.3.RS-986605/V1 [DOI]
- Yang, X.‐J. (2021b). Delta‐1 variant of SARS‐COV‐2 acquires spike V1264L and drives the pandemic in Indonesia, Singapore and Malaysia. 10.21203/RS.3.RS-999390/V1 [DOI]
- Yang, X. , Yu, Y. , Xu, J. , Shu, H. , Xia, J. , Liu, H. , Wu, Y. , Zhang, L. , Yu, Z. , Fang, M. , Yu, T. , Wang, Y. , Pan, S. , Zou, X. , Yuan, S. , & Shang, Y. (2020). Clinical course and outcomes of critically ill patients with SARS‐CoV‐2 pneumonia in Wuhan, China: A single‐centered, retrospective, observational study. The Lancet Respiratory Medicine, 8(5), 475–481. 10.1016/S2213-2600(20)30079-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yurkovetskiy, L. , Wang, X. , Pascal, K. E. , Tomkins‐Tinch, C. , Nyalile, T. P. , Wang, Y. , Baum, A. , Diehl, W. E. , Dauphin, A. , Carbone, C. , Veinotte, K. , Egri, S. B. , Schaffner, S. F. , Lemieux, J. E. , Munro, J. B. , Rafique, A. , Barve, A. , Sabeti, P. C. , Kyratsous, C. A. , … Luban, J. (2020). Structural and functional analysis of the D614G SARS‐CoV‐2 spike protein variant. Cell, 183(3), 739–751.e8. 10.1016/J.CELL.2020.09.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng, C. , Evans, J. P. , Faraone, J. N. , Qu, P. , Zheng, Y.‐M. , Saif, L. , Oltz, E. M. , Lozanski, G. , Gumina, R. J. , & Liu, S.‐L. (2021). Neutralization of SARS‐CoV‐2 variants of concern harboring Q677H. MBio, 12(5). 10.1128/MBIO.02510-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, L. , Jackson, C. B. , Mou, H. , Ojha, A. , Peng, H. , Quinlan, B. D. , Rangarajan, E. S. , Pan, A. , Vanderheiden, A. , Suthar, M. S. , Li, W. , Izard, T. , Rader, C. , Farzan, M. , & Choe, H. (2020). SARS‐CoV‐2 spike‐protein D614G mutation increases virion spike density and infectivity. Nature Communications, 11(1), 1–9. 10.1038/s41467-020-19808-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, X. , Chen, D. , Szabla, R. , Zheng, M. , Li, G. , Du, P. , Zheng, S. , Li, X. , Song, C. , Li, R. , Guo, J.‐T. , Junop, M. , Zeng, H. , & Lin, H. (2020). Broad and differential animal angiotensin‐converting enzyme 2 receptor usage by SARS‐CoV‐2. Journal of Virology, 94(18). 10.1128/JVI.00940-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, H. Y. , Ji, C. Y. , Fan, H. , Han, N. , Li, X. F. , Wu, A. , & Qin, C. F. (2021). Convergent evolution of SARS‐CoV‐2 in human and animals. Protein & Cell, 12(11), 832–835. 10.1007/S13238-021-00847-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary table 1: Details of 43102 SARS CoV‐2 whole genome sequences used in the study.
Supplementary table 2: Details of 24260 S protein mutations identified in the study
Supplementary table 3: Individual state‐wide distribution of SARS CoV‐2 pango lineages in India
Supplementary table 4: Spike mutations with >1% prevalence across the domains
Data Availability Statement
The data that support the findings of this study are openly available in GISAID at https://www.gisaid.org.


