Electronic health records have the potential to improve patient care while reducing the burden of documentation on clinicians. Smartforms are tools for standardized data collection within the Epic electronic health record. We developed an inflammatory bowel disease (IBD) Smartform and tested the impact of direct patient entry of symptom data via the internet or in-office tablet computers on visit length, patient and provider satisfaction, and changes in the tablet-associated microbiome.
METHODS
This research was determined to be exempt by the University of Pennsylvania Institutional Review Board. The IBD Smartform includes current patient symptom and historical data (eg, disease distribution). Patients can self-report current IBD symptoms before their scheduled clinic visits utilizing the Symptom Questionnaire, either via an electronic patient portal or at the office using a computer. Patients’ responses feed automatically into the IBD Smartform, allowing for immediate use in clinical notes by providers.
In February 2015, the IBD Smartform was launched and the Symptom Questionnaire was added in February 2017. Initially, the Symptom Questionnaire could only be completed via the patient portal. Since July 2018, patients were provided the opportunity to complete the Symptom Questionnaire on tablet computers located in the waiting room of two IBD clinics: the Perelman Center for Advanced Medicine (PCAM) and the Penn-Presbyterian Medical Center (PPMC).
Data for individual outpatient visits (between July 2017 and October 2019; Fig. 1A) were queried to compute mean visit lengths for new patient visits (NPV) and return patient visits (RPV), both related and unrelated to IBD. Patients’ visit satisfaction before the implementation of the tablets (February-May 2018) and during the postimplementation period (October-December 2018) was assessed. Using a Likert scale, IBD providers from the PCAM and PPMC clinics were surveyed on their satisfaction with patient use of the Symptom Questionnaire.
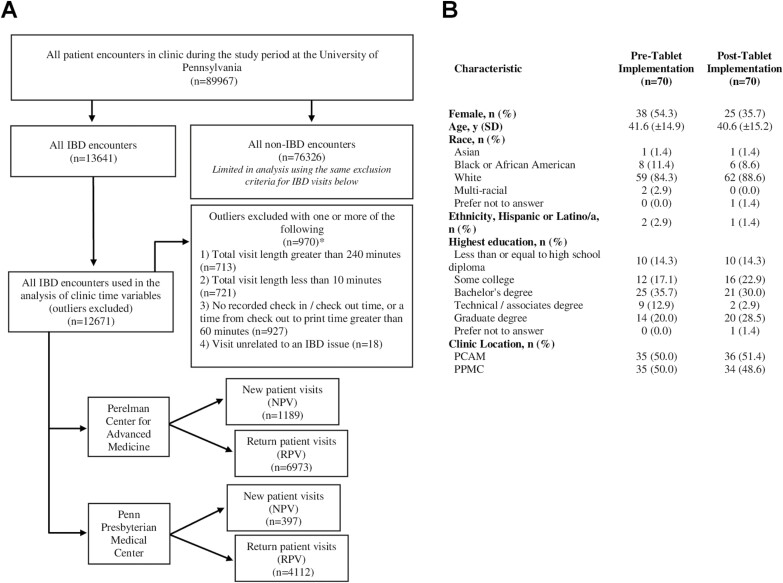
FIGURE 1.
Patient populations included in the study. A, Flow diagram of clinic population used for visit length calculations and analysis across tablet implementation. B, Characteristics of survey participants pre- and posttablet implementation (n = 140). *Groups are not mutually exclusive.
Microbiome samples were collected using sterile cotton swabs premoistened with distilled water that were then immediately stored in cryo-vials and frozen at –80°C pending sequencing. Before use in the clinic, the tablets were swabbed across the screen, sides, and back surface. As comparators, swabs from examination rooms, intake rooms, and provider stethoscopes were also collected. Air swabs were collected as controls. Repeat microbiome samples were collected after 6 months of tablet use. At PCAM, clinic staff cleaned tablets daily using PDI Easy Screen cleaning wipes. At PPMC, clinic staff cleaned tablets 2 to 3 times per week using germicidal disposable cloths (Sani-Cloth Plus).
The 16S rRNA marker gene sequencing of microbiome samples was completed using the 16S rRNA gene V1-V2 variable region as previously described.1 Purified 16S amplicon products were pooled and sequenced on the Illumina MiSeq instrument. Sequence data were processed using QIIME,2 read pairs were processed with DADA2,3 and taxonomy was assigned using the Greengenes reference database.4 A phylogenetic tree was inferred from the sequence data using MAFFT.5
Pre- and postimplementation visit lengths were compared using the unpaired t test. In addition, we compared the change in visit length between the pre- and postperiods using segmented regression.
Alpha diversity was assessed by the expected number of operational taxonomic units, the Shannon index, and Faith’s phylogenetic diversity. Beta diversity was assessed by unweighted UniFrac distances.6,7 Principal coordinate analysis was used to display the UniFrac distances. Community-level differences between sample groups were assessed using the permutational multivariate analysis of variance test, which allows sample-sample distances to be applied to an analysis of variance–like framework.8 The differential abundance of specific taxa was assessed using linear mixed-effects models.
RESULTS
We compared the postimplementation mean visit length to the preimplementation mean visit length. At PCAM, the postimplementation mean visit length decreased for NPVs and RPVs (3.3 minutes, P < 0.001 and 6.3 minutes, P < 0.001, respectively). At PPMC, the mean visit length for NPVs increased by 3.1 minutes (P = 0.24) and for RPVs it decreased by 2.4 minutes (P < 0.001). However, using segmented regression, we found that these reductions in IBD visit length postimplementation (vs preimplementation) were not statistically significant (P > 0.05 for all comparisons), suggesting that visit length was already decreasing and continued to do so.
Visit satisfaction surveys were completed by 140 patients (70 pre- and 70 posttablet implementation; Fig. 1B). Of the patients who completed the postimplementation survey, 25.7% used the patient portal and 38.6% used a tablet in the clinic. There was no statistically significant difference in patient satisfaction postimplementation for clinic wait time (P = 0.42), provider courtesy (P = 1.00), confidence in the provider’s medical expertise (P = 1.00), belief that concerns were heard by the provider (P = 1.00), questions that were answered in an understandable way (P = 1.00), sufficient time spent with the provider (P = 0.22), understanding of treatment plans (P = 1.00), and quality of care (P = 1.00). Repeating the analysis and limiting it to those who completed the IBD Smartform Symptom Questionnaire in the posttablet implementation period resulted in similar findings (data not shown).
Nine IBD providers were surveyed about their satisfaction with the Symptom Questionnaire. Eight of the 9 providers (89%) agreed that completion of the questionnaire had improved the quality of in-office discussions and documentation along with patient recall, organization, and articulation of current symptoms. Of the 9 providers, 78% agreed that the questionnaire had improved overall in-office efficiency, 78% agreed that tablet implementation had been effectively integrated into the clinic workflow, and 67% agreed that clinic staff had effectively distributed tablets in the waiting areas.
We carried out 16S rRNA marker gene sequencing and found that prominent taxa pre- and postimplementation included Propionibacterium, Corynebacterium, Streptococcus, Staphylococcus, and Pseudomonas. Propionibacterium was the most abundant taxon and accounted for >20% of reads in 71% of samples from the clinic.
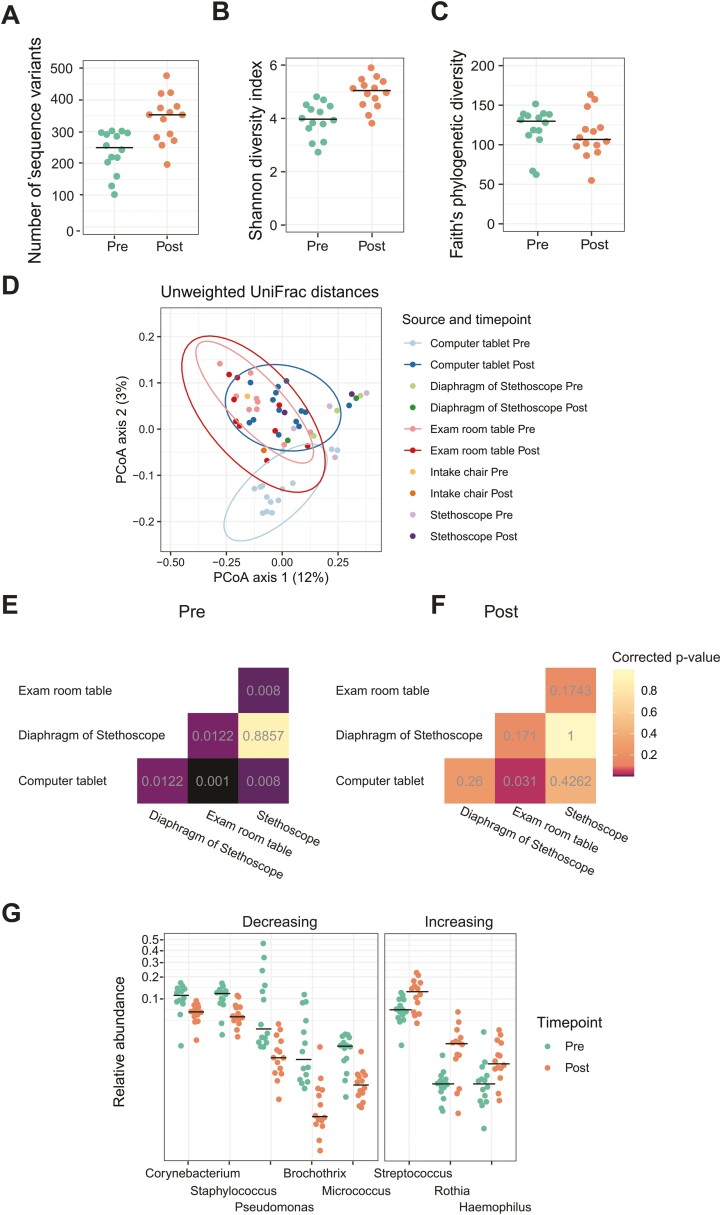
We observed an increase in diversity from pre- to posttablet implementation as quantified by the number of unique 16S sequence variants (P < 0.001; Fig. 2A) and the Shannon index (P < 0.001; Fig. 2B). The Shannon index measures both the number of taxa and the evenness of their abundance distribution, whereas the number of unique sequence variants approximates the number of bacterial species present. We did not observe a difference in Faith’s phylogenetic diversity measure (P = 0.46; Fig. 2C), indicating that increased diversity was not specific to 1 lineage of bacteria.
FIGURE 2.
Microbiome changes after implementation of the tablet computers. A, Number of sequence variants. B, Shannon diversity index. C, Faith’s phylogenetic diversity. D, Beta diversity assessed with unweighted UniFrac distances for the tablet computers and other clinic surfaces. E, PERMANOVA P values for comparisons of communities preimplementation of the tablet computers. F, PERMANOVA P values for comparisons of communities postimplementation of the tablet computers. G, Differential abundance of specific taxa on the tablet computers pre- and postimplementation. PERMANOVA indicates permutational multivariate analysis of variance.
We compared the similarity of bacterial communities among clinical samples using the unweighted UniFrac distance (Fig. 2D). Microbiome samples collected from tablets preimplementation had a microbial community composition different from that of the clinic environment (Fig. 2E). The microbial community composition posttablet implementation did not differ from that of other clinical surfaces except for the examination room table (P = 0.03; Fig. 2F).
Unlike with other clinic surfaces, the community composition of tablet samples changed from pre- to postimplementation (P = 0.002). The relative abundance of Streptococcus, Rothia, and Haemophilus increased, and the relative abundance of Corynebacterium, Staphylococcus, Pseudomonas, Brochothrix, and Micrococcus decreased (P < 0.05; Fig. 2G).
DISCUSSION
This quality improvement initiative explored the effects of the Symptom Questionnaire on visit length, patient and provider satisfaction, and changes in the tablet-associated microbiome. We found that visit length was declining before the implementation of the tablets and continued to do so after implementation. Patient satisfaction was high during the preimplementation period and remained unchanged postimplementation. Providers generally felt that the Symptom Questionnaire improved the patient encounter. As expected, the microbiota composition of the tablet evolved to become similar to that of other surfaces in the clinic.
A majority of IBD providers agreed that having data from the questionnaire increased the quality and efficiency of visits. Patients had comparable satisfaction pre- and postimplementation of the tablets. The process of completing the questionnaire in the waiting room may have led to the perception of decreased wait times for patients while simultaneously providing providers with valuable information before the visit. However, providers’ responses indicated a need for better logistics of tablet implementation because some felt that tablets had not been effectively distributed by clinic staff.
Mostly skin flora were detected on the tablets after they had been in circulation at the clinics for 6 months, similar to prior research.9 The microbiota community on the tablets increasingly resembled other components of the clinical setting, such as examination tables and stethoscopes. We were unable to assay for viruses using 16S sequencing; however, this type of assay should be examined in future studies.
The COVID-19 pandemic could impact patients’ acceptance of using tablet computers in health care settings. The routine cleaning of tablets that was conducted during the study period will likely need to be enhanced to mitigate COVID-19 risk (ie, disinfecting between each patient use). This process could potentially impact workflow. Fortunately, the Symptom Questionnaire can be completed on one’s home computer or cell phone, thereby reducing the need to rely on in-office computers; this development may also have implications for making symptom information available to providers during telemedicine visits, which have gained prominence during the COVID-19 pandemic.
CONCLUSIONS
We determined from our study that IBD providers found the IBD Symptom Questionnaire tool to be useful and further increased the efficiency of their patient visits while not compromising the quality of their visits or prolonging visit length. Patients’ satisfaction was high. Analysis of potential microbiome changes on the tablets in the IBD clinic mostly revealed the growth of skin flora after 6 months. Although not directly studied here, the collection of patients’ symptoms in routine clinical care using standardized nomenclature should be highly valuable for future research.
ACKNOWLEDGMENTS
The authors acknowledge the following individuals for their assistance in implementing this initiative: Kristi Delp, Richard Duerr, Chenda Neth, Afi Peele, Jason Swoger, Lauren Wilson, and Ariel Myatt. Epic licensees can install the Epic IBD Smartform by accessing SLG 1636438.
Supported by: This work was supported by a grant from the Crohn’s & Colitis Foundation and a Sherman Prize to Dr. Lewis.
REFERENCES
- 1. Giron LB, Tanes CE, Schleimann MH, et al. Sialylation and fucosylation modulate inflammasome-activating eIF2 Signaling and microbial translocation during HIV infection. Mucosal Immunol. 2020;13:753–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bolyen E, Rideout JR, Dillon MR, et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol. 2019;37:852–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Callahan BJ, McMurdie PJ, Rosen MJ, et al. DADA2: High-resolution sample inference from Illumina amplicon data. Nat Methods. 2016;13:581–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. McDonald D, Price MN, Goodrich J, et al. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. Isme J. 2012;6:610–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol. 2005;71:8228–8235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lozupone CA, Hamady M, Kelley ST, et al. Quantitative and qualitative beta diversity lead to different insights into factors that structure microbial communities. Appl Environ Microbiol. 2007;73:1576–1585. [DOI] [PMC free article] [PubMed]
- 8. Anderson MJ. A new method for non-parametric multivariate analysis of variance. Austral Ecol. 2001;26:32–46. [Google Scholar]
- 9. Lax S, Sangwan N, Smith D, et al. Bacterial colonization and succession in a newly opened hospital. Sci Transl Med. 2017;9(391):eaah6500. [DOI] [PMC free article] [PubMed] [Google Scholar]