Abstract
Strain TBP-1, an anaerobic bacterium capable of reductively dehalogenating 2,4,6-tribromophenol to phenol, was isolated from estuarine sediments of the Arthur Kill in the New York/New Jersey harbor. It is a gram-negative, motile, vibrio-shaped, obligate anaerobe which grows on lactate, pyruvate, hydrogen, and fumarate when provided sulfate as an electron acceptor. The organism accumulates acetate when grown on lactate and sulfate, contains desulfoviridin, and will not grow in the absence of NaCl. It will not utilize acetate, succinate, propionate, or butyrate for growth via sulfate reduction. When supplied with lactate as an electron donor, strain TBP-1 will utilize sulfate, sulfite, sulfur, and thiosulfate for growth but not nitrate, fumarate, or acrylate. This organism debrominates 2-, 4-, 2,4-, 2,6-, and 2,4,6-bromophenol but not 3- or 2,3-bromophenol or monobrominated benzoates. It will not dehalogenate monochlorinated, fluorinated, or iodinated phenols or chlorinated benzoates. Together with its physiological characteristics, its 16S rRNA gene sequence places it in the genus Desulfovibrio. The average growth yield of strain TBP-1 grown on a defined medium supplemented with lactate and 2,4,6-bromophenol is 3.71 mg of protein/mmol of phenol produced, and the yield was 1.42 mg of protein/mmol of phenol produced when 4-bromophenol was the electron acceptor. Average growth yields (milligrams of protein per millimole of electrons utilized) for Desulfovibrio sp. strain TBP-1 grown with 2,4,6-bromophenol, 4-bromophenol, or sulfate are 0.62, 0.71, and 1.07, respectively. Growth did not occur when either lactate or 2,4,6-bromophenol was omitted from the growth medium. These results indicate that Desulfovibrio sp. strain TBP-1 is capable of growth via halorespiration.
The field of microbially mediated reductive dehalogenation has made tremendous strides in recent years, largely due to the isolation of a number of anaerobic bacteria that possess the ability to couple the reductive dehalogenation of aromatic and aliphatic compounds to growth. The isolation of the first halorespiring bacterium, Desulfomonile tiedjei, resulted in a new genus (7), and since then a number of other halorespiring bacteria have been isolated (2, 5, 14, 17, 21, 24, 27, 27a, 36, 37, 43). A number of these new isolates have brought about the definition of new genera, including Dehalococcoides (27), Dehalospirillum (37), Desulfitobacterium (2, 4, 14, 27a, 36, 43), and Dehalobacter (17), while some others have added to pre-existing genera, such as Pelobacter (21).
The genus Desulfitobacterium contains six species of halorespiring bacteria: Desulfitobacterium dehalogenans (43), Desulfitobacterium frappieri (2), Desulfitobacterium chlororespirans (36), Desulfitobacterium hafniense (4), Desulfitobacterium sp. strain PCE-S (27a), and Desulfitobacterium sp. strain PCE1 (14). This genus is a member of the Clostridium subphylum; hence, all members are gram positive and three of five (Desulfitobacterium frappieri, Desulfitobacterium dehalogenans, and Desulfitobacterium chlororespirans) are reported to form spores. All except strain PCE-S exhibit the ability to dehalogenate ortho-substituted chlorines from phenols.
The other isolated halorespiring anaerobes fall into different genera, most representing the delta and epsilon subdivisions of the Proteobacteria (5, 7, 21, 37). Two interesting strains are Dehalobacter restrictus and Dehalococcoides ethenogenes. Based upon 16S rRNA gene sequence analysis, Dehalobacter restrictus falls into the fourth subdivision of the gram positive bacteria, yet physiological and biochemical analyses do not support its placement in this subdivision (17). This organism will use only hydrogen or formate as an electron donor and perchloroethene (PCE) or trichloroethene as an electron acceptor and requires fermented yeast extract for growth. Dehalococcoides ethenogenes is similarly metabolically limited. It will utilize only hydrogen as an electron donor and PCE as an electron acceptor and requires extracts from mixed microbial cultures. Analysis of this organism’s 16S rRNA gene sequence indicates that it is a eubacterium, yet is not closely related to any known group (27).
While halorespiring bacteria are phylogenetically diverse based upon their 16S rRNA gene sequences, there is a lack of diversity with respect to the types of compounds which can be dehalogenated. Typically these organisms are capable of using only one of two classes of halogenated compounds as electron acceptors for halorespiration: substituted, single-ring aromatic compounds (e.g., phenols and benzoates) or halogenated alkenes (e.g., PCE). Six of the twelve halorespiring organisms isolated, Desulfitobacterium sp. strain PCE1, Desulfitobacterium frappieri, Desulfitobacterium dehalogenans, Desulfitobacterium chlororespirans, Desulfitobacterium hafniense, Desulfomonile tiedjei, and strain 2CP-1, are capable of dehalogenating substituted aromatic compounds. Desulfomonile tiedjei is also capable of dehalogenating PCE (11). In addition, Desulfitobacterium sp. strain PCE1 can dehalogenate PCE as well as ortho-chlorinated phenols (14).
Although the substrates for these isolates can be divided into two groups based upon their structure (aromatic or aliphatic), much diversity exists with respect to the specific halogen group and the position of halogenation of the target compound. For example, Desulfomonile tiedjei can utilize a variety of halogenated compounds. It can dehalogenate brominated and iodinated benzoates and benzamides regardless of the position of substitution; however, it will remove only meta-substituted chlorines. Also, cell suspensions of Desulfomonile tiedjei grown on 3-chlorobenzoate remove only meta-substituted chlorines from polychlorinated phenols (28). Desulfitobacterium chlororespirans seems more selective than Desulfomonile tiedjei. It will debrominate 2,4,6-tribromophenol (2,4,6-TBP) to 4-bromophenol (4-BP) but will not debrominate 2-BP. Likewise, it will dechlorinate 2,4,6-trichlorophenol to 4-chlorophenol but will not dechlorinate any monochlorinated phenol, nor will it dehalogenate 2-fluoro- or iodophenol. Desulfitobacterium chlororespirans will also dechlorinate 2,3- (to 3-) and 2,6- (to 2-) dichlorophenol as well as 3-chloro-4-hydroxy-benzoate and 3-chloro-4-hydroxy-phenylacetate (36). Strain 2CP-1 appears to have the most limited substrate range of the three. It will dehalogenate 2,6-di- and 2-chlorophenol but is incapable of dehalogenating monosubstituted bromo-, fluoro-, or iodophenols in any position (5). The differences in substrate specificity between these organisms implies that differences exist with respect to their dehalogenating enzymes.
All of the previously reported halorespiring anaerobic bacteria have been isolated from soils, compost, sewage sludge, or sediments from freshwater environments. Marine and estuarine sediments would seem to be good sources of halorespiring bacteria given their exposure to numerous brominated, iodinated, and chlorinated organic compounds (12, 18, 35). In fact, reductive dehalogenation of bromophenols (19, 30), bromobenzoates (30), chlorophenols (26), and polychlorinated biphenyls (1) has been demonstrated in marine and estuarine sediments. A bacterium, strain DSL-1, that debrominates 2,4,6-TBP to 4-BP has been isolated from sediments inhabited with bromophenol-producing marine worms (39). In contrast to the halorespirers, this organism does not couple the dehalogenation of 2,4,6-TBP to growth.
In this report we describe the enrichment and isolation of strain TBP-1, a bacterium capable of reductive dehalogenation, from estuarine sediments. This obligately anaerobic bacterium removes bromines substituted in the ortho- and para-positions of brominated phenols. It does not dehalogenate monochlorinated, fluorinated, or iodinated phenols. Physiological and molecular analyses indicate that strain TBP-1 is a member of the genus Desulfovibrio. Strain TBP-1 possesses the ability to grow by coupling the oxidation of lactate to the reductive dehalogenation of 2,4,6-TBP.
MATERIALS AND METHODS
Enrichment preparation.
Anaerobic sediments from the Arthur Kill in the New York/New Jersey harbor estuary (16) were used as an inoculum. One hundred grams of previously collected sediment (45) was diluted, under a mixture of nitrogen and carbon dioxide (70:30), in 1.0 liter of anaerobic media composed of (in grams per liter) KCl (1.3), KH2PO4 (0.2), NaCl (21.0), NaHCO3 (7.5), NH4Cl (0.3), CaCl2 · 2H2O (0.15), MgCl2 · 6H2O (3.0), resazurin (0.001), and Na2S · 9H2O (0.5); a 0.1-ml/liter concentration of vitamin stock, containing (in milligrams per liter) folic acid (20), pyridoxine HCl (100), riboflavin (50), biotin (20), thiamine (50), nicotinic acid (50), pantothenic acid (50), vitamin B12 (1.0), p-aminobenzoic acid (50), and thiotic acid (50); and a 0.1-ml/liter concentration of metals stock (prepared in dilute HCl), containing (in grams per liter) H3BO4 (0.062), MnCl2 · 4H2O (0.098), FeCl2 · 4H2O (1.49), CoCl2 · 6H2O (0.119), NiCl2 · 6H2O (0.237), CuCl2 (0.134), and ZnCl2 (0.068). Thirty-milliliter aliquots of this slurry were distributed in 60-ml serum bottles that had been purged with N2-CO2. Bottles were crimp-sealed with butyl rubber septa. Succinate was added from a sterile stock solution to a final concentration of approximately 5 mM. Initial 2,4,6-TBP (Aldrich, Milwaukee, Wis.) addition and periodic resupplementation to active enrichments were performed with a concentrated stock solution prepared in 0.1 N NaOH. Each set of conditions and controls were prepared in triplicate. All enrichments and subsequent isolation and characterization experiments were incubated statically in the dark at 30°C.
Isolation of strain TBP-1.
The medium used for all isolation experiments was the same as that used for the initial enrichments. Media prepared for isolation experiments were supplemented with acetate (25 to 35 mM) and hydrogen. Hydrogen was supplied by evacuating cultures under vacuum and repressurizing to 14 lb/in2 with H2-CO2 (80:20). Soft agar shake tubes (28 ml, total volume; 10 ml of medium) were prepared with Noble agar (Difco, Detroit, Mich.) at a final concentration of 0.8%. Culture purity was checked by phase-contrast light microscopy and confirmed by growth on medium D plus 2.5% NaCl (33), modified Baar’s medium with 2.5% NaCl (15), and anaerobically prepared tryptic soy broth (Difco).
Strain characterization.
Electron donors and acceptors used for isolate characterization were added from concentrated aqueous stocks to a final concentration of 10 mM unless otherwise indicated. Elemental sulfur was added (0.1 ml per 10 ml) as an aqueous slurry. The inocula for donor and acceptor experiments were grown with limiting concentrations of lactate or sulfate, respectively, or centrifuged and resuspended in anaerobic medium unamended with any donor or acceptor. Strain TBP-1 was determined to be capable of utilizing various electron donors and acceptors based upon their ability to support growth (increase in total protein) and sulfate consumption (for donors) or lactate consumption (for acceptors) relative to controls lacking either donor or acceptor, depending upon the experiment. Each donor and acceptor was tested in at least three separate experiments.
The various haloaromatics tested (see Table 1) were added from stocks prepared in 0.1 N NaOH to a final concentration of 100 to 200 μM. Cultures were supplied hydrogen and acetate or lactate as electron donors and incubated for 7 to 10 days. Dehalogenated products (phenol or benzoate) were assayed by high-pressure liquid chromatography (HPLC). Compounds that were dehalogenated were added to cultures a second time to ensure that activity could be maintained. The presence of desulfoviridin was tested for by the technique of Postgate (32). Gram staining was done by the modified Hucker method (8).
TABLE 1.
Dehalogenation of halogenated phenols and benzoates by strain TBP-1a
| Compound | Dehalogenation observed |
|---|---|
| 2-BP | + |
| 3-BP | − |
| 4-BP | + |
| 2,3-DBP | − |
| 2,4-DBP | + |
| 2,6-DBP | + |
| 2,4,6-TBP | + |
Strain TBP-1 was supplied with H2 and acetate or lactate and assayed after 7 to 10 days of incubation at 30°C. Positive cultures were refed haloaromatic compounds after initial consumption to confirm activity. The following compounds were not dehalogenated by strain TBP-1: 2-, 3-, 4-, 2,4-, and 2,4,6-chlorophenol; 2-, 3-, and 4-fluorophenol; 2-, 3-, and 4-iodophenol; 2-, 3-, and 4-bromobenzoate; and 2-, 4-, and 2,4-chlorobenzoate.
Halorespiration experiments.
Strain TBP-1 was inoculated into media supplemented with lactate (30 to 40 mM) and either 2,4,6-TBP, 4-BP, or sulfate. In addition, it was tested with lactate alone and with 2,4,6-TBP alone. Cultures with lactate and bromophenols were resupplemented with the appropriate bromophenol once or twice daily for approximately 4 days after the onset of dehalogenating activity. After the consumption of approximately 4 mM bromophenol or sulfate, 20 ml of all cultures were centrifuged at 10,000 × g for 25 min, decanted, and resuspended in 1.0 ml of sterile, unamended medium for protein analysis.
Analytical methods.
Sediment slurry (from enrichments) or liquid-only (all other experiments) samples (0.5 to 1.0 ml) were drawn through the stoppers by using degassed 1.0-ml sterile syringes with 16-gauge needles and centrifuged for 2 min in a benchtop microcentrifuge. The supernatant was filtered with a 0.45-μm-pore-size Millex syringe filter (Millipore) prior to analysis. All haloaromatics were measured with an HPLC (Beckman, Fullerton, Calif.) equipped with a 4.6-mm by 25-cm Ultrasphere C18 column (Beckman) with UV detection. The initial mobile-phase composition was 40% mobile phase A (40:57:3, methanol-water-acetic acid) and 60% mobile phase B (80:18:2, methanol-water-acetic acid) at a flow rate of 1.0 ml/min. This was changed to 35% A–65% B over 5 min with a linear gradient. The mobile phase was held at this composition for 5 min and then changed to 20% A and 80% B over 2 min with a linear gradient. This condition was maintained for another 11.5 min, at which point the analysis was stopped. At 7.8 min in the run, the UV detector was switched from 280 to 295 nm to increase the sensitivity for the detection of the di- and tribrominated phenols. The retention times (in minutes) under these conditions were as follows: phenol, 4.5; 2-BP, 7.2; 4-BP, 9.1; 2.4-dibromophenol (2,4-DBP), 15.6; 2,6-DBP, 11.9; and 2,4,6-TBP, 21.5. Twenty-microliter injections were performed with an autosampler (Gilson Medical Electronics, Inc., Middleton, Wis.).
Lactate and acetate were analyzed for with an HPLC (Beckman) equipped with a 7.8-mm by 30-cm Rezex organic acids column (Phenomenex, Torrance, Calif.) with UV detection at 210 nm. The mobile phase consisted of 0.005 N H2SO4 at a flow rate of 0.5 ml/min. The column was maintained at room temperature. Twenty-microliter injections were performed with an autosampler (Gilson).
Sulfate analyses were performed on a Dionex (Sunnyvale, Ca.) model 100 ion chromatograph with an IonPac AS9 column by using conductivity detection. The eluant was 2 mM Na2CO3–0.75 mM NaHCO3 at 2.0 ml/min. The regenerant consisted of 25 mM H2SO4 under He at 8 lb/in2.
Protein concentrations in unfiltered samples were determined by the Bradford method (3). Briefly, samples were diluted 1:1 in 2 N NaOH and incubated at 70°C for 45 min. Samples were then centrifuged in a benchtop microcentrifuge (3 min) and 100 μl was reacted with 1.0 ml of dilute protein dye reagent (Bio-Rad, Hercules, Calif.). Absorbance at 595 nm was read after 5 min.
16S rRNA sequencing and analysis.
DNA was extracted from cells (31) and further purified with a Gene-Clean kit (Bio 101) before amplification. 16S rDNA was amplified with the universal eubacterial primers (27 Forward [5′ AGA GTT TGA TCC TGG CTC AG 3′] and 1525 Reverse [5′ AAG GAG GTG WTC CAR CC 3′]) (22). The following amplification parameters were used: initial denaturation at 95°C for 5 min, 20 to 30 cycles of 94°C for 0.5 min, 55°C for 0.5 min, and 72°C for 1.5 min, and a final extension at 72°C for 10 min, all amplified in a Gene Amp PCR system 2400 thermal cycler (Perkin-Elmer, Foster City, Calif.).
The PCR product was purified with a QIAquick PCR purification kit (Qiagen Inc., Chatsworth, Calif.), and the sequence was determined by automated techniques (Perkin-Elmer-ABI, Foster City, CA) with the following primers: 27 Forward (5′ AGA GTT TGA TCC TGG CTC AG 3′), 519 Reverse (5′ GWA TTA CCG CGG CKG CTG 3′), 530 Forward (5′ GTG CCA GCM GCC GC GG 3′), 907 Reverse (5′ CCG TCA ATT CMT TTR AGT TT 3′), 926 Forward (5′ AAA CTY AAA KGA ATT GAC GG 3′), and 1492 Reverse (5′ GGT TAC CTT GTT ACG ACT T 3′) (22).
The complete sequence was aligned to related organisms with the Ribosomal Database Project (25) alignment function and Genetic Data Environment software, version 2.2 (38). Phylogenetic analysis was performed by the neighbor-joining method with programs in the PHYLIP package, version 3.5c (13). Bootstrap values were calculated with SEQBOOT, and a distance matrix was constructed with DNADIST employing a Kimura 2-parameter model and a transition/transversion ratio of 2.0. The tree was drawn with NEIGHBOR and CONSENSE (13).
Nucleotide sequence accession number.
The GenBank accession number for the 16S rRNA gene sequence of strain TBP-1 is AF090830.
RESULTS
Enrichment and isolation of strain TBP-1.
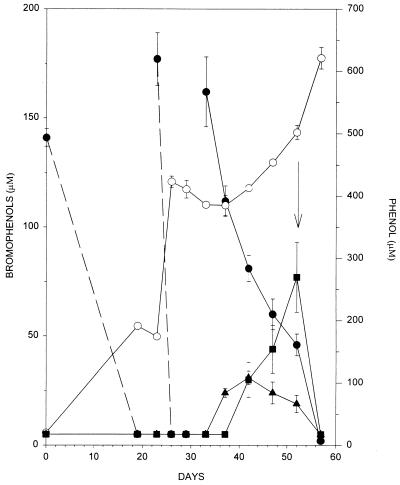
Initial enrichment cultures were inoculated with sediments from the Arthur Kill and supplemented with succinate and 2,4,6-TBP. As seen in Fig. 1, complete debromination of 2,4,6-TBP to phenol occurred prior to our first sampling point, which was at 19 days. No brominated intermediates were initially observed. Autoclaved controls exhibited no loss of 2,4,6-TBP and no production of phenol (data not shown). Dehalogenation to phenol continued after subsequent refeeding of 2,4,6-TBP. After the second resupplementation of 2,4,6-TBP, the rate of 2,4,6-TBP loss (debromination) slowed and partially debrominated intermediates (2,4-DBP and 4-BP) were observed to accumulate. The addition of succinate at 52 days resulted in the rapid dehalogenation of these intermediates and the residual 2,4,6-TBP. The transient accumulation of 2,4-DBP and 4-BP suggests that removal of ortho-substituted bromines precedes para-debromination. The rapid dehalogenation of intermediates after succinate resupplementation suggests that the debrominating system was carbon and/or electron limited.
FIG. 1.
Dehalogenation of 2,4,6-TBP in enrichments inoculated with Arthur Kill sediments and supplemented with succinate. Closed circles, 2,4,6-TBP; open circles, phenol; squares, 4-BP; triangles, 2,4-DBP. Dotted lines are used to indicate that these lines do not represent actual rates of activity since all of the 2,4,6-TBP was consumed prior to our first sampling point after feeding. Data are the average of triplicates, and error bars represent 1 standard deviation. Arrow indicates time of succinate resupplementation. Sterile controls did not exhibit 2,4,6-TBP loss or phenol production (data not shown).
At 114 days one of the replicate enrichment cultures was serially diluted to 10−12 in anaerobic media supplemented with 2,4,6-TBP and hydrogen and acetate. Succinate was replaced as the carbon and electron donor in all further isolation activities in an attempt to minimize the growth of nonhalorespiring bacteria. The highest dilution to express dehalogenating activity after 51 days of incubation (10−5) was further diluted 1,000-fold. This culture was incubated for 9 days and then diluted yet again by a factor of 10. After growing for 29 days with periodic refeeding of 2,4,6-TBP this culture was serially diluted into anaerobic shake tubes containing 0.8% Noble agar. The shake tubes were incubated for 18 days, after which point nine colonies were picked, two from the 10−3 dilution and seven from the 10−2 dilution. All had the same off-white, diffuse, round morphology. Eight of the nine colonies expressed dehalogenating activity. One isolate was chosen for further study and designated TBP-1. Purity was checked by culturing the isolate on a variety of complex anaerobic media (see Materials and Methods). Under all growth conditions only one cellular morphology was observed, as determined by phase-contrast light microscopy.
Initial characterization of strain TBP-1.
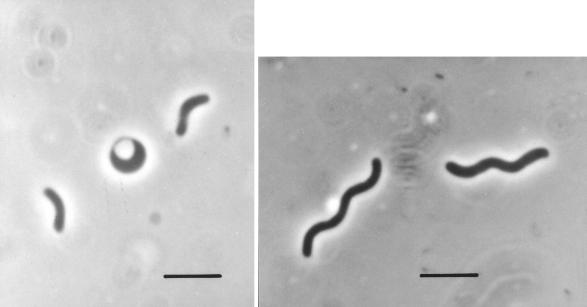
Strain TBP-1 is a gram-negative, vibrio-shaped bacterium approximately 1 by 4 μm in size (Fig. 2). It is motile and tested positive for the presence of desulfoviridin, suggesting that it is a member of the genus Desulfovibrio. Older cultures of TBP-1 do not appear to form spores; however, they undergo distinct morphological changes. As the culture ages, the cells change from a vibrio shape to a spherical form which appears to be devoid of a cell wall. These observations are consistent with morphological variations observed with a number of Desulfovibrio species (33). Growth did not occur on modified Baar’s medium with 2.5% NaCl when the culture was incubated statically with a headspace of air rather than argon, indicating that strain TBP-1 cannot grow aerobically.
FIG. 2.
Phase-contrast light micrographs of strain TBP-1 grown on lactate and sulfate. Spherical body illustrates morphological transformation that occurs in older cultures. Bar, 5 μm.
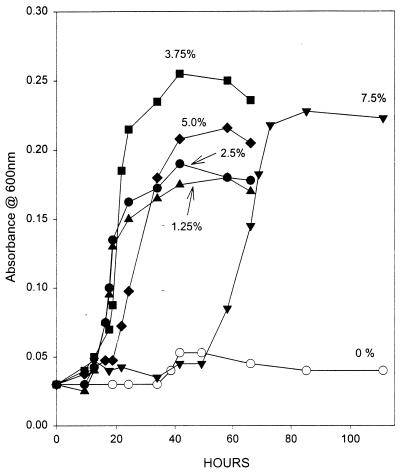
The effects of various NaCl concentrations on the growth of strain TBP-1 on modified Baar’s medium at 30°C are illustrated in Fig. 3. The highest growth rate (0.20 h−1) and yield were achieved with 3.75% NaCl. The specific growth rates with 1.25, 2.5, 5.0, and 7.5% NaCl were 0.16, 0.17, 0.09, and 0.07 h−1, respectively. Growth occurred after 10 h of incubation in cultures with 1.25, 2.5, and 3.75% NaCl, while lag periods of 19 and 50 h were observed prior to the onset of growth for the 5.0 and 7.5% NaCl cultures, respectively. No growth occurred in the absence of NaCl.
FIG. 3.
Growth of strain TBP-1 on modified Baar’s medium at 30°C with increasing concentrations of NaCl (indicated for each curve). Data are the averages of duplicates.
Various electron donors and acceptors were tested for their ability to support the growth of strain TBP-1. Lactate, pyruvate, fumarate, formate, and hydrogen supported sulfate consumption and growth. All of these donors except fumarate (not tested) support dehalogenation of 2,4,6-TBP. Acetate accumulates in the culture medium when strain TBP-1 is provided lactate and sulfate, indicating that it is an incomplete oxidizer. Acetate, propionate, butyrate, and succinate do not support growth, sulfate reduction, or debromination of 2,4,6-TBP. Sulfate, sulfite (5 mM), thiosulfate, and sulfur support lactate consumption and growth, while nitrate, acrylate, and fumarate do not.
The ability of strain TBP-1 to dehalogenate a variety of substituted phenols and benzoates was examined and summarized in Table 1. Only brominated phenols with bromines in the ortho and/or para position were dehalogenated. The bromine in the ortho position of 2,3-bromophenol was not removed. Furthermore, none of the other halogenated aromatics could serve as substrates. These results suggest that only bromines in the ortho or para positions will be removed and that meta-substituted bromines prevent the dehalogenation of adjacent ortho-substituted bromines.
Growth via halorespiration.
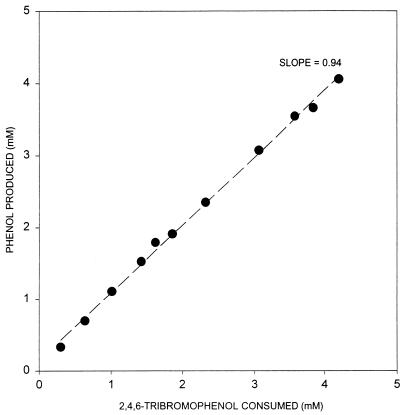
Figure 4 shows that strain TBP-1 consumes 2,4,6-TBP with the stoichiometric accumulation of phenol. Duplicate cultures were refed 2,4,6-TBP a total of 10 times, each refeeding occurring after the complete consumption of the previous addition. The cumulative amount of 2,4,6-TBP consumed is plotted against the cumulative amount of phenol produced. The slope of the regression line is 0.94, indicating stoichiometric conversion of 2,4,6-TBP to phenol. Stoichiometric production of phenol also demonstrates that no abiotic loss of phenol occurred. Similarly, phenol loss has never been observed in cultures of TBP-1. The medium contained only bicarbonate and bromophenol as potential electron acceptors. Since no methane was produced we postulated that dehalogenation was occurring by one of two mechanisms: either tribromophenol was serving as an electron acceptor in halorespiration and was required for growth on lactate, or the organism was growing fermentatively on lactate and dehalogenating the bromophenol by a mechanism that was not linked to growth.
FIG. 4.
Stoichiometric dehalogenation of 2,4,6-TBP to phenol by strain TBP-1. Dotted line represents the regression line of the average cumulative 2,4,6-TBP consumed by duplicate cultures plotted against the average cumulative phenol produced. Stoichiometric phenol production demonstrates that dehalogenation of 2,4,6-TBP was complete and that no phenol was consumed by or lost from these cultures.
In order to test these hypotheses, strain TBP-1 was grown in the presence of excess lactate (approximately 38 mM) and one of the following: sulfate, 2,4,6-TBP, or 4-BP. The bromophenols were added once or twice daily (0.4 to 0.5 mM) to avoid high levels (>1.5 mM), which we have observed to be toxic to TBP-1. The initial sulfate concentration was 4.5 mM. The results from this experiment are shown in Table 2. Cultures incubated with only lactate or 2,4,6-TBP failed to grow, indicating that TBP-1 can neither grow fermentatively on lactate or 2,4,6-TBP nor dehalogenate 2,4,6-TBP in the absence of an electron donor.
TABLE 2.
Protein yield for strain TBP-1 grown on lactate with various electron acceptorsa
| Electron donor | Electron acceptor | Amt (mmol) of:
|
Amt (mg) of protein | Yield (mg of protein/mmolb)
|
||
|---|---|---|---|---|---|---|
| Phenol produced | Electrons utilized for reductionc | e− | Phenol | |||
| Lactate | 4-BP | 4.67 ± 0.08 | 9.33 ± 0.15 | 6.61 ± 0.36 | 0.71 ± 0.05 | 1.42 ± 0.10 |
| Lactate | 2,4,6-TBP | 3.67 | 22.0 | 13.6 | 0.62 | 3.71 |
| Lactate | Sulfate | NA | 35.1 ± 7.0 | 37.0 ± 4.98 | 1.07 ± 0.10 | NA |
| None | 2,4,6-TBP | 0.01 ± 0.006 | 0.30 ± 0.01 | <0.5 | NA | NA |
| Lactate | None | NA | NA | 0.70 ± 0.20 | NA | NA |
4-BP and 2,4,6-TBP cultures were refed to 400 to 500 μM nine and ten times, respectively. The initial sulfate concentration was 4.5 mM. Values are the average of triplicates ± 1 standard deviation, except data for 2,4,6-TBP, which are averages of duplicates. NA, not applicable.
Millimole of electrons utilized to reduce electron acceptors (e−) or millimole of phenol produced (p).
Calculated from concentrations of dehalogenation products present or total amount of sulfate consumed at conclusion of experiment, based upon two electrons per bromine removed and eight electrons per sulfate consumed. Calculations were as follows: for lactate–4-BP cultures, 2 × millimoles of phenol; for lactate–2,4,6-TBP cultures, 6 × millimoles of phenol; for lactate-sulfate cultures, 8 × millimoles of sulfate consumed; and for 2,4,6-TBP-only cultures, (6 × millimoles of phenol) + (2 × millimoles of 4-BP) + (4 × millimoles of 2,4-DBP).
When TBP-1 was grown on 2,4,6-TBP, the average growth yield, 3.71 mg of protein per mmol of phenol produced, was 2.6 times that obtained when it was grown on 4-BP (1.42). Since this is reasonably close to the increase in number of bromines removed per molecule, it indicates a proportional increase in the electrons transferred and hence the energy available. This is further underscored by the similarity in average growth yields, 0.71 and 0.62, when the growth yields are normalized to millimoles of electrons transferred for growth on 4-BP and 2,4,6-TBP, respectively. Therefore, growth of strain TBP-1 in the presence of brominated phenols and lactate is dependent upon dehalogenation, and growth increases proportionally with the number of bromines removed. In contrast, the average growth yield per millimole of electrons with sulfate (1.07) is higher than with brominated phenols (0.67, average of 2,4,6- and 4-BP results).
Phylogeny.
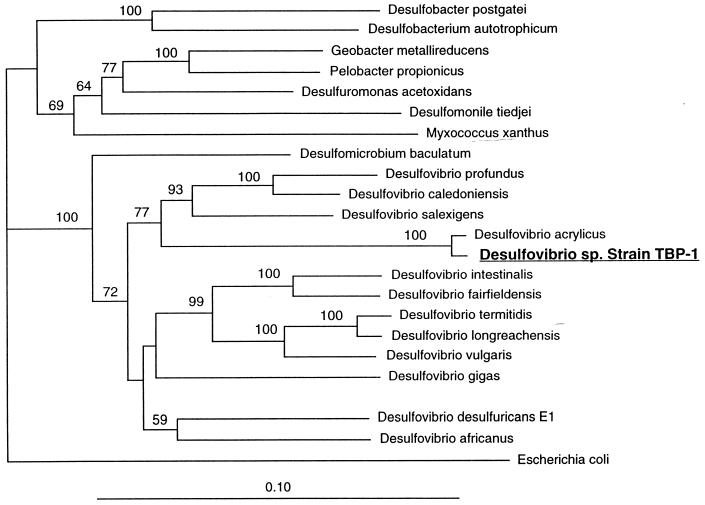
The morphological and physiological characteristics of strain TBP-1 suggest that it is a member of the genus Desulfovibrio. This is further supported by the organisms 16S rRNA gene sequence. Sequence analysis of 1,435 bp indicates that strain TBP-1 is a member of the delta subdivision of the Proteobacteria within the family Desulfovibrionaceae (Fig. 5) (6). Strain TBP-1 clustered with members of the genus Desulfovibrio, with its closest relative being Desulfovibrio acrylicus (98.9%).
FIG. 5.
Phylogenetic tree of strain TBP-1 and other related species based on 16S rRNA gene sequences. Bootstrap values at nodes are the percentages of 100 iterations. Values less than 50% are not included. The scale bar indicates the estimated number of substitutions per 100 positions.
DISCUSSION
Strain TBP-1, isolated from sediments of the New York/New Jersey harbor estuary, is an obligately anaerobic bacterium capable of reductively dehalogenating ortho- and para-substituted bromophenols. Growth of strain TBP-1 on lactate depends on the presence of NaCl and a suitable electron acceptor. When 2,4,6-TBP and 4-BP are used as electron acceptors, growth occurs and stoichiometric levels of phenol are produced. Growth does not occur in the absence of a utilizable electron acceptor and neither dehalogenation nor growth occurs when strain TBP-1 is incubated with only 2,4,6-TBP. These results therefore demonstrate that strain TBP-1 can grow via halorespiration.
In comparison to other halorespiring organisms, TBP-1 has a low growth yield when it uses bromophenols (0.67 mg of protein per mmol of electrons utilized for dehalogenation). For example, growth yields for Desulfitobacterium sp. strain PCE1 (on 3-chloro-4-hydroxybenzoate), Desulfomonile tiedjei (on 3-chlorobenzoate), and Dehalobacter restrictus (on PCE) were reported to be 0.8 (14), 1.05 (17), and 1.4 (29) mg of protein/mmol of electrons utilized, respectively. Likewise, yields ranging from 1.6 to 5.65 mg of cells (dry weight) per mmol of electrons utilized (originally reported as grams per mole of electron pairs utilized) were obtained for Desulfitobacterium dehalogenans (23). Mackiewicz and Weigel indicated that, assuming that the protein yield is 50% of the total dry-weight biomass, this range of values (0.8 to 2.83 g of protein per mol of electrons utilized) agrees with those obtained for Desulfomonile tiedjei and Dehalobacter restrictus.
There are a number of possible explanations for the observed differences in growth yield. On a thermodynamic basis, the structure of the halogenated compound dictates the amount of energy available from the cleavage of a carbon-halide bond. For instance, the dechlorination of monosubstituted anilines (124.6 kJ/reaction when hydrogen is used as the electron donor) and 2-hydroxy-5-chlorobenzoate (144.4 kJ/reaction) yields approximately 22 and 10% less energy, respectively, than the dechlorination of monosubstituted phenols (160.7 kJ/reaction) (10, 41). This suggests that growth yield differences between halorespirers may be the result of structural differences between, and thus the amount of energy available from, the halogenated electron acceptors.
In addition, different experimental techniques may also contribute to the observed differences in growth yield. Mohn and Tiedje (29) discussed the possibility that the differences in growth rate and yield they observed and the values obtained by Dolfing (9) for Desulfomonile tiedjei were a result of differences in medium composition. Indeed, the cell yield data we report may be a result of suboptimal salinity. Further characterization of TBP-1 on modified Baar’s medium indicated that optimal growth took place at 3.75% salinity, not 2%, which was used in the yield experiments.
Variations in the reported growth yields may also be a result of the age at which the cultures were harvested for biomass determinations. Mackiewicz and Wiegel reported a nonlinear relationship between measured growth and amount of substrate utilized for Desulfitobacterium dehalogenans. The authors speculated that the decrease in growth yield with increasing culture age was a result of higher maintenance energy demands during late exponential growth than in early exponential growth (23). In comparison, the values for Desulfomonile tiedjei, Dehalobacter restrictus, and Desulfitobacterium sp. strain PCE1 were made as endpoint measurements with all cultures apparently in some stage of stationary growth.
Finally, it is possible that periodic limitation of substrate (2,4,6-TBP) caused discontinuous, and thus less efficient, growth of TBP-1, resulting in the relatively lower growth yield. Despite resupplementation of the cultures twice daily, 4-BP and 2,4,6-TBP were at times limiting during the course of the experiment. The observation that higher growth yields for TBP-1 were obtained on sulfate, an energetically less favorable acceptor (10, 42) that was in excess, than on 2,4,6-TBP and 4-BP, is consistent with this hypothesis. In addition, yields for Desulfitobacterium dehalogenans with fumarate, nitrate, sulfite, and 3-chloro-4-hydroxyphenylacetate as electron acceptors, provided in excess, were similar (25.9, 23.8, 23.3, and 24.2 g of biomass/mol of ATP formed) (23), suggesting that for strain TBP-1, 2,4,6-TBP limitation may indeed have contributed to the lower growth yields. While numerous factors may be involved in determining the cellular yields for these different bacteria growing with halogenated substrates as terminal electron acceptors, it is clear that all of these microorganisms are capable of halorespiration.
Morphological, physiological, and phylogenetic characteristics support the placement of strain TBP-1 in the genus Desulfovibrio. It also has high (98.9%) 16S rRNA gene sequence homology to Desulfovibrio acrylicus. The latter organism was isolated from marine sediments based upon its ability to cleave dimethylsulfoniopropionate and couple the reduction of acrylate to growth (44). Desulfovibrio sp. strain TBP-1 does not reduce acrylate. Furthermore, Desulfovibrio sp. strain TBP-1 grows on fumarate and sulfate while Desulfovibrio acrylicus does not. Also, while TBP-1 can grow on modified Baar’s medium with 7.5% NaCl, Desulfovibrio acrylicus cannot grow on lactate and acrylate with more than 4% NaCl in the growth medium. These two strains therefore, have several key differences. Whether they should be considered separate species would depend on further physiological and molecular comparisons.
The types of halogenated electron acceptors used by Desulfovibrio sp. strain TBP-1 is limited. It will use only bromophenols with bromines in the ortho and/or para position without adjacent bromines. It sequentially dehalogenates 2,4,6-TBP to phenol. On the other hand, Desulfitobacterium chlororespirans (36) and strain DSL-1 (39) both only partially dehalogenate 2,4,6-TBP to 4-BP. Furthermore, Desulfitobacterium chlororespirans, a halorespiring bacterium, cannot dehalogenate 2-BP even though it removes the ortho-substituted bromines from 2,4,6-BP (36). In contrast, strain DSL-1 is reported to be a fermentative bacterium which grows only on yeast extract and constitutively expresses debrominating activity (39). The different degrees of substrate specificity exhibited by the various debrominating strains suggests broad diversity with respect to their dehalogenating enzymes.
The identification of dehalogenating activity in marine environments (1, 19, 30) suggests that marine and estuarine systems are potentially sources of halorespiring bacteria. The isolation of Desulfovibrio sp. strain TBP-1, a salinity-dependent halorespiring bacterium, from estuarine sediments supports this likelihood. It is interesting that based on recent phylogenetic evidence, other dehalogenating bacteria related to TBP-1 may be present in marine and estuarine environments. For example, an isolate from San Francisco Harbor dechlorinates 2,6-di- and 2-chlorophenol and has been tentatively identified as a Desulfovibrio (based upon 16S rRNA sequence); however, it utilizes acetate for growth (40). It also cannot dehalogenate bromophenols. In addition, four distinct clones from a highly enriched Arthur Kill sediment culture which dehalogenates 2-BP were obtained from a library generated by amplification of the 16S rRNA genes. One of the four clones has high sequence homology to members of the genus Desulfovibrio while a second has homology to members of the family Desulfobacteriaceae (20). Similarly, molecular characterization of a highly enriched 2,3,5,6-tetrachlorobiphenyl-dechlorinating community inoculated with Baltimore Harbor sediments indicates the presence of members of the delta subdivision of the Proteobacteria (34). The organisms responsible for the dehalogenation reactions in the enrichments of Knight et al. (20) and Pulliam Holoman et al. (34), however, have not been determined. The reports of various dehalogenating cultures from marine and estuarine sediments suggest that these environments are an underexplored source of anaerobic bacteria with diverse dehalogenating capabilities.
ACKNOWLEDGMENTS
We thank Norberto Palleroni for photographing strain TBP-1; Lee Kerkhof and Victoria Knight for assisting in phylogenetic analyses; Imelda Harjono, Vivian Chu, and Roza Wojcik for technical assistance; and Beau Ranheim, the crew of Osprey (New York City Department of Environmental Protection), and Monica Togna for sediment collection.
This research was funded, in part, by grants R-823575 and R-819679 from the United States Environmental Protection Agency.
REFERENCES
- 1.Berkaw M, Sowers K R, May H D. Anaerobic ortho dechlorination of polychlorinated biphenyls by estuarine sediments from Baltimore Harbor. Appl Environ Microbiol. 1996;62:2534–2539. doi: 10.1128/aem.62.7.2534-2539.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bouchard B, Beaudet R, Villemur R, McSween G, Lepine F, Bisaillon J G. Isolation and characterization of Desulfitobacterium frappieri sp. nov., an anaerobic bacterium which reductively dechlorinates pentachlorophenol to 3-chlorophenol. Int J Syst Bacteriol. 1996;46:1010–1015. doi: 10.1099/00207713-46-4-1010. [DOI] [PubMed] [Google Scholar]
- 3.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 4.Christiansen N, Ahring B K. Desulfitobacterium hafniense sp. nov., an anaerobic, reductively dechlorinating bacterium. Int J Syst Bacteriol. 1996;46:442–448. [Google Scholar]
- 5.Cole J R, Cascarelli A L, Mohn W W, Tiedje J M. Isolation and characterization of a novel bacterium growing via reductive dehalogenation of 2-chlorophenol. Appl Environ Microbiol. 1994;60:3536–3542. doi: 10.1128/aem.60.10.3536-3542.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Devereux R, He S-H, Doyle C L, Orkland S, Stahl D A, LeGall J, Whitman W B. Diversity and origin of Desulfovibrio species: phylogenetic definition of a family. J Bacteriol. 1990;172:3609–3619. doi: 10.1128/jb.172.7.3609-3619.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DeWeerd K A, Mandelco L, Tanner R S, Woese C R, Suflita J M. Desulfomonile tiedjei gen. nov. and sp. nov., a novel anaerobic, dehalogenating, sulfate-reducing bacterium. Arch Microbiol. 1990;154:23–30. [Google Scholar]
- 8.Doetsch R N. Determinative methods of light microscopy. In: Gerhardt P, Murray R G E, Costilow R N, Nester E W, Wood W A, Krieg N R, Phillips G B, editors. Manual of methods for general microbiology. Washington, D.C: American Society for Microbiology; 1981. pp. 21–33. [Google Scholar]
- 9.Dolfing J. Reductive dechlorination of 3-chlorobenzoate is coupled to ATP production and growth in an anaerobic bacterium, strain DCB-1. Arch Microbiol. 1990;153:264–266. doi: 10.1007/BF00249079. [DOI] [PubMed] [Google Scholar]
- 10.Dolfing J, Harrison B K. Gibbs free energy of formation of halogenated aromatic compounds and their potential role as electron acceptors in anaerobic environments. Environ Sci Technol. 1992;26:2213–2218. [Google Scholar]
- 11.Fathepure B Z, Nengu J P, Boyd S A. Anaerobic bacteria that dechlorinate perchloroethene. Appl Environ Microbiol. 1987;53:2671–2674. doi: 10.1128/aem.53.11.2671-2674.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Faulkner D J. Interesting aspects of marine natural products chemistry. Tetrahedron. 1977;33:1421–1443. [Google Scholar]
- 13.Felsenstein J. PHYLIP (phylogeny inference package), version 3.5c. Seattle: Department of Genetics, University of Washington; 1993. [Google Scholar]
- 14.Gerritse J, Renard V, Gomes T M P, Lawson P A, Collins M D, Gottschal J C. Desulfitobacterium sp. strain PCE1, an anaerobic bacterium that can grow by reductive dechlorination of tetrachloroethene or ortho-chlorinated phenols. Arch Microbiol. 1996;165:132–140. doi: 10.1007/s002030050308. [DOI] [PubMed] [Google Scholar]
- 15.Gherna R, Pienta P, Cote R, editors. American Type Culture Collection catalogue of bacteria and phages. 18th ed. Rockville, Md: American Type Culture Collection; 1992. [Google Scholar]
- 16.Gunster D G, Bonnevie N L, Gillis C A, Wenning R J. Assessment of chemical loadings to Newark Bay, New Jersey from petroleum and hazardous accidents occurring from 1986 to 1991. Ecotoxicol Environ Safety. 1993;25:202–213. doi: 10.1006/eesa.1993.1019. [DOI] [PubMed] [Google Scholar]
- 17.Holliger C. Reductive dehalogenation by anaerobic bacteria. Ph.D. thesis. Wageningen, The Netherlands: Wageningen Agricultural University; 1992. [Google Scholar]
- 18.King G M. Inhibition of microbial activity in marine sediments by a bromophenol from a hemichordate. Nature. 1986;323:257–259. [Google Scholar]
- 19.King G M. Dehalogenation in marine sediments containing natural sources of halophenols. Appl Environ Microbiol. 1988;54:3079–3085. doi: 10.1128/aem.54.12.3079-3085.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Knight, V. K., L. J. Kerkhof, and M. M. Häggblom. 1998. Personal communication.
- 21.Krumholz L R, Sharp R, Fishbain S S. A freshwater anaerobe coupling acetate oxidation to tetrachloroethylene dehalogenation. Appl Environ Microbiol. 1996;62:4108–4113. doi: 10.1128/aem.62.11.4108-4113.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lane D J. 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M, editors. Nucleic acid techniques in bacterial systematics. New York, N.Y: John Wiley and Sons; 1991. pp. 115–175. [Google Scholar]
- 23.Mackiewicz M, Wiegel J. Comparison of energy and growth yields for Desulfitobacterium dehalogenans during utilization of chlorophenol and various traditional electron acceptors. Appl Environ Microbiol. 1998;64:352–355. doi: 10.1128/aem.64.1.352-355.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Madsen T, Licht D. Isolation and characterization of an anaerobic chlorophenol-transforming bacterium. Appl Environ Microbiol. 1992;58:2874–2878. doi: 10.1128/aem.58.9.2874-2878.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maidak B L, Olsen G J, Larsen N, Overbeek R, McCaughey M J, Woese C R. The ribosomal database project (RDP) Nucleic Acids Res. 1997;25:109–111. doi: 10.1093/nar/25.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Masunaga S, Susarla S, Gundersen J L, Yonezawa Y. Pathway and rate of chlorophenol transformation in anaerobic estuarine sediments. Environ Sci Technol. 1996;30:1253–1260. [Google Scholar]
- 27.Maymo-Gatell X, Tandoi V, Gossett J M, Zinder S H. Isolation of a bacterium that reductively dechlorinates tetrachloroethene to ethene. Science. 1997;276:1568–1571. doi: 10.1126/science.276.5318.1568. [DOI] [PubMed] [Google Scholar]
- 27a.Miller E, Wohlfarth G, Diekert G. Comparative studies on tetrachloroethene reductive dechlorination mediated by Desulfitobacterium sp. strain PCE-S. Arch Microbiol. 1997;168:513–519. doi: 10.1007/s002030050529. [DOI] [PubMed] [Google Scholar]
- 28.Mohn W W, Kennedy K J. Reductive dehalogenation of chlorophenols by Desulfomonile tiedjei DCB-1. Appl Environ Microbiol. 1992;58:1367–1370. doi: 10.1128/aem.58.4.1367-1370.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mohn W W, Tiedje J M. Strain DCB-1 conserves energy for growth from reductive dechlorination coupled to formate oxidation. Arch Microbiol. 1990;153:267–271. doi: 10.1007/BF00249080. [DOI] [PubMed] [Google Scholar]
- 30.Monserrate E, Häggblom M M. Dehalogenation and biodegradation of brominated phenols and benzoic acids under iron-reducing, sulfidogenic and methanogenic conditions. Appl Environ Microbiol. 1997;63:3911–3915. doi: 10.1128/aem.63.10.3911-3915.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Phelps C D, Kerkhof L J, Young L Y. Molecular characterization of a sulfate-reducing consortium which mineralizes benzene. FEMS Microbiol Ecol. 1998;27:269–279. [Google Scholar]
- 32.Postgate J R. A diagnostic reaction for Desulfovibrio desulphuricans. Nature. 1959;183:481–482. doi: 10.1038/183481b0. [DOI] [PubMed] [Google Scholar]
- 33.Postgate J R. Genus Desulfovibrio. In: Kreig N R, Holt J G, editors. Bergey’s manual of systematic bacteriology. Vol. 1. Baltimore, Md: Williams & Wilkins; 1984. pp. 666–672. [Google Scholar]
- 34.Pulliam Holoman T R, Elberson M A, Cutter L A, May H D, Sowers K R. Characterization of a defined 2,3,5,6-tetrachlorobiphenyl-ortho-dechlorinating microbial community by comparative sequence analysis of genes coding for 16S rRNA. Appl Environ Microbiol. 1998;64:3359–3367. doi: 10.1128/aem.64.9.3359-3367.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rivera P L, Astudillo L, Rovirosa J, San-Martin A. Halogenated monoterpenes of the red alga Shottera nicaensis. Biochem Syst Ecol. 1987;15:3–4. [Google Scholar]
- 36.Sanford R A, Cole J R, Löffler F E, Tiedje J M. Characterization of Desulfitobacterium chlororespirans sp. nov., which grows by coupling the oxidation of lactate to the reductive dechlorination of 3-chloro-4-hydroxybenzoate. Appl Environ Microbiol. 1996;62:3800–3808. doi: 10.1128/aem.62.10.3800-3808.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Scholz-Muramatsu H, Neumann A, Meßmer M, Moore E, Diekert G. Isolation and characterization of Dehalospirillum multivorans gen. nov., sp. nov., a tetrachloroethene-utilizing, strictly anaerobic bacterium. Arch Microbiol. 1995;163:48–56. [Google Scholar]
- 38.Smith S W, Overbeek R, Woese C R, Gilbert W, Gillevet P M. The Genetic Data Environment: an expandable GUI for multiple sequence analysis. CABIOS. 1994;10:671–675. doi: 10.1093/bioinformatics/10.6.671. [DOI] [PubMed] [Google Scholar]
- 39.Steward C C, Dixon T C, Chen Y P, Lovell C R. Enrichment and isolation of a reductively debrominating bacterium from the burrow of a bromometabolite-producing marine hemichordate. Can J Microbiol. 1992;41:637–642. [Google Scholar]
- 40.Sun B, Cole J R. Enrichment and isolation of anaerobic dechlorinating microorganisms from marine sediments. Poster Q-193. Abstracts of the 98th General Meeting of the American Society for Microbiology. Washington, D.C: American Society for Microbiology; 1998. [Google Scholar]
- 41.Susarla S, Masunaga S, Yonezawa Y. Redox potential as a parameter to predict the reductive dechlorination pathway of chloroanilines in anaerobic environments. Microb Ecol. 1997;33:252–256. doi: 10.1007/s002489900028. [DOI] [PubMed] [Google Scholar]
- 42.Thauer R K, Jungermann K, Decker K. Energy conservation in chemotrophic anaerobic bacteria. Microbiol Rev. 1977;41:100–180. doi: 10.1128/br.41.1.100-180.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Utkin I, Woese C, Wiegel J. Isolation and characterization of Desulfitobacterium dehalogenans gen. nov., sp. nov., an anaerobic bacterium which reductively dechlorinates chlorophenolic compounds. Int J Syst Bacteriol. 1994;44:612–619. doi: 10.1099/00207713-44-4-612. [DOI] [PubMed] [Google Scholar]
- 44.van der Maarel M J E C, van Bergeijk S, van Werkhoven A F, Laverman A M, Meojer W G, Stam W T, Hansen T A. Cleavage of dimethylsulfoniopropionate and reduction of acrylate by Desulfovibrio acrylicus sp. nov. Arch Microbiol. 1996;166:109–115. [Google Scholar]
- 45.Zhang X, Young L Y. Carboxylation as an initial reaction in the anaerobic metabolism of naphthalene and phenanthrene by sulfidogenic consortia. Appl Environ Microbiol. 1997;63:4759–4764. doi: 10.1128/aem.63.12.4759-4764.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]