Abstract
Vibrio vulnificus is an estuarine bacterium that is capable of causing a rapidly fatal infection in humans. A randomly amplified polymorphic DNA (RAPD) PCR protocol was developed for use in detecting V. vulnificus, as well as other members of the genus Vibrio. The resulting RAPD profiles were analyzed by using RFLPScan software. This RAPD method clearly differentiated between members of the genus Vibrio and between isolates of V. vulnificus. Each V. vulnificus strain produced a unique band pattern, indicating that the members of this species are genetically quite heterogeneous. All of the vibrios were found to have amplification products whose sizes were within four common molecular weight ranges, while the V. vulnificus strains had an additional two molecular weight range bands in common. All of the V. vulnificus strains isolated from clinical specimens produced an additional band that was only occasionally found in environmental strains; this suggests that, as is the case with the Kanagawa hemolysin of Vibrio parahaemolyticus, the presence of this band may be correlated with the ability of a strain to produce an infection in humans. In addition, band pattern differences were observed between encapsulated and nonencapsulated isogenic morphotypes of the same strain of V. vulnificus.
Many members of the genus Vibrio have been implicated in both human diseases and marine animal diseases. One member of this genus, Vibrio vulnificus, is ubiquitous in coastal waters and is responsible for more than 95% of all seafood-related deaths (11, 13). V. vulnificus infection can occur either after ingestion of raw or undercooked shellfish, particularly oysters, or through entry via a flesh wound (11). The people vulnerable to infection include those with chronic diseases involving elevated serum iron levels, immune function abnormalities, and other chronic disorders (11).
The same isolate of V. vulnificus has been observed to have two different colony morphologies when it is grown on solid nutrient media (14, 24). The first morphotype is termed opaque, and the surfaces of the cells are covered with a polysaccharide capsule. The translucent morphotype lacks a polysaccharide capsule. All virulent strains of V. vulnificus are opaque morphotype strains, which indicates that the capsule plays a role in the virulence of the organism (1, 6, 9, 14). Opaque strains of V. vulnificus have been observed to lose their capsule; they become translucent and lose their virulence (14). However, the reverse situation (translucent cells gaining a capsule) generally has not been observed. It has also been reported that more than 90% of environmental V. vulnificus strains are opaque morphotype strains (16), yet these strains have been found to be highly variable in terms of virulence (8, 15), suggesting that factors other than the presence of a capsule also contribute to the virulence of the organism. Because of the severity of V. vulnificus infections, a reliable method for rapid identification of virulent strains of this organism is needed.
Randomly amplified polymorphic DNA (RAPD) PCR (5, 19, 20, 21, 22) is a technique that is known to be a sensitive method for detecting slight genetic differences between samples. We optimized a RAPD method suitable for distinguishing various Vibrio species from one another, as well as for differentiating between V. vulnificus strains. Furthermore, we investigated the ability of the RAPD method to detect genetic differences between opaque and translucent morphotypes of the same isolate of V. vulnificus. Finally, we investigated the possibility that clinical isolates of V. vulnificus may produce a unique RAPD band pattern that could be used to differentiate virulent strains from avirulent strains.
MATERIALS AND METHODS
Bacterial stains and culture preparation.
A total of 16 Vibrio species (Table 1), as well as 39 clinical isolates and 30 environmental isolates of V. vulnificus, were used in the present study. Translucent morphotypes were isolated as spontaneous mutants of the opaque morphotypes in the laboratory of Ronald Siebling at Louisiana State University. All cells were grown to the stationary phase at 22°C with aeration in heart infusion (Difco Laboratories, Detroit, Mich.) broth. These cultures were used as the sources of template DNA for RAPD analysis.
TABLE 1.
Vibrio species used for RAPD analysis
| Strains used |
|---|
| V. aestuarianus OSU-65 |
| V. alginolyticus ATCC 17749 |
| V. anguillarum ATCC 19264 |
| V. cholerae 2741–80 |
| V. fluvialis 5440 |
| V. fischeri B64 |
| V. furnisii 8760 |
| V. harveyi ATCC 25919 |
| V. leiognathi B474 |
| V. metschnikovii ATCC 7708 |
| V. mimicus ATCC 33653 |
| V. natriegens ATCC 14048 |
| V. nigripulchritudo ATCC 27043 |
| V. parahaemolyticus ATCC 17802 |
| V. pelagius 25916 |
| V. proteolyticus ATCC 15538 |
| V. vulnificusa |
A total of 70 V. vulnificus strains from clinical and environmental sources were used.
RAPD analysis.
Ten 10-bp oligonucleotide primers (Genosys Biotechnologies, Inc., The Woodlands, Tex.) with G+C contents of 50% were screened for the ability to provide a suitable band pattern with various V. vulnificus strains. The primer selected had the following sequence: 5′GGATCTGAAC3′. Each 25.0-μl RAPD reaction mixture contained the following reagents: 2.5 μl of 10× reaction buffer (100 mM Tris-HCl [pH 8.3], 500 mM KCl, 15 mM MgCl2, 0.01% gelatin) (Promega, Madison, Wis.), 2.0 μl of sterile H2O, 3.5 μl of 25 mM MgCl2, 8.0 μl of a solution containing each of the deoxynucleoside triphosphates (Promega) at a concentration of 5 mM, 3.0 μl of primer (Biosynthesis, Lewisville, Tex.), 5.0 U of Taq DNA polymerase (Promega), and 5.0 μl of cell culture. The reaction mixtures were overlaid with 20.0 μl of sterile mineral oil (Sigma Chemical Co., St. Louis, Mo.) to seal them and to prevent evaporation in the thermal cycler. Thermal cycling was performed with a model PHC-3 thermal cycler (Techne, Princeton, N.J.). The cycling profile was as follows: one cycle consisting of 94°C for 5 min, 45 cycles consisting of 94°C for 1 min, 36°C for 1 min, and 72°C for 2 min, and a final cycle consisting of 72°C for 5 min. The RAPD products were electrophoresed by using a Fisher Biotech Small Horizontal Gel System (Fisher Scientific, Pittsburgh, Pa.) at 60 V for approximately 3 h on a 2.0% agarose gel containing ethidium bromide (2.5 μl of a 10-mg/ml solution) and were photographed with a Polaroid model ASP Quick Shooter camera (International Biotechnologies, Inc., New Haven, Conn.) under UV light. A 123-bp ladder (Sigma) was used as a molecular weight marker. The RAPD method was used with all strains at least three times.
Computer analysis of RAPD profiles.
All of the gels were scanned with an ImageMaster DTS scanner (Pharmacia, Uppsala, Sweden). A 123-bp ladder was included every three or four lanes on all gels as a standard molecular weight marker. Images were calibrated and data analysis was performed by using RFLPScan software (Scanalytics, Billerica, Mass.). A match tolerance equivalent to 1.0% of the molecular weight of each band was used.
RESULTS
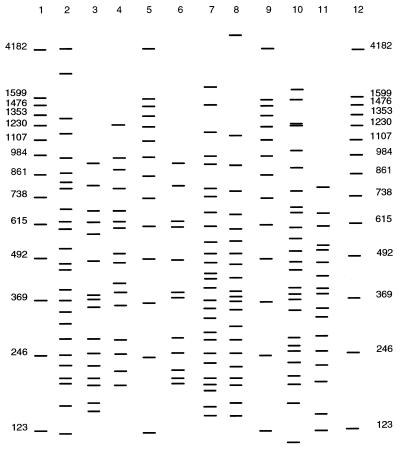
A RAPD method was developed and optimized for use with Vibrio species, including V. vulnificus. When this method was applied to various gram-positive and gram-negative organisms (Staphylococcus aureus, Bacillus subtilis, Escherichia coli, and Pseudomonas aeruginosa), no RAPD profiles were observed. All of the Vibrio species tested produced unique RAPD profiles. Representative profiles of various vibrios are shown in Fig. 1. Four bands in specific molecular weight ranges were identified as bands that were common to all species of this genus (Table 2). Furthermore, when 70 V. vulnificus isolates were analyzed (Fig. 2), bands in an additional two molecular weight ranges (492 to 464 and 264 to 244 bp) were produced by all of the isolates of this species. While all of the V. vulnificus strains examined produced bands in these six ranges, all of the V. vulnificus isolates could be clearly distinguished from one another based on their RAPD profiles. In addition to the two unique V. vulnificus bands, one additional molecular weight range band was present in all 31 clinical isolates tested. This band, at 200 to 178 bp, was found in only 3 of the 39 environmental isolates tested.
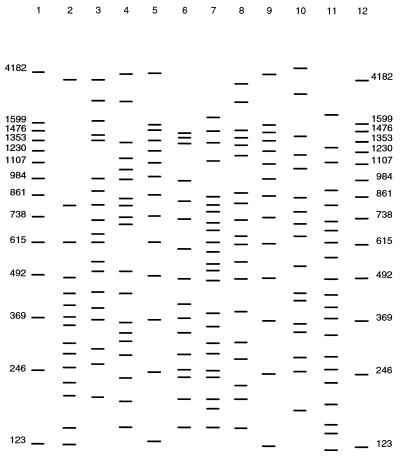
FIG. 1.
Representative RAPD profiles obtained for various Vibrio species. Lanes 1, 5, 9, and 12, 123-bp ladder; lane 2, V. parahaemolyticus; lane 3, V. nigripulchritudo; lane 4, V. anguillarum; lane 6, V. fluvialis; lane 7, V. alginolyticus; lane 8, V. natriegens; lane 10, V. mimicus; lane 11, V. harveyi.
TABLE 2.
Common molecular weight range bands in Vibrio species as determined by computer analysis
| Mol wt range (bp) | Organisms found in |
|---|---|
| 853–714 | All Vibrio species |
| 706–509 | All Vibrio species |
| 426–328 | All Vibrio species |
| 319–273 | All Vibrio species |
| 492–273 | V. vulnificus (all isolates) |
| 264–244 | V. vulnificus (all isolates) |
| 200–178 | V. vulnificus (clinical isolates only) |
FIG. 2.
Representative RAPD profiles of clinical and environmental V. vulnificus isolates. Lanes 1, 5, 9, and 12, 123-bp ladder; lane 2, strain EDL174 (clinical); lane 3, strain E4125 (clinical); lane 4, strain 371 (environmental); lane 6, strain M06 (clinical); lane 7, strain CVD713 (clinical); lane 8, strain H3308 (clinical); lane 10, strain B9629 (clinical); lane 11, strain 890 (environmental).
RAPD analysis of opaque and translucent isogenic morphotypes of 12 V. vulnificus strains revealed differences in the RAPD profiles obtained for each pair of morphotypes (Fig. 3). Occasionally, the two morphotypes produced nearly identical profiles, but in most cases there were detectable differences between the two morphotypes. The opaque morphotypes always produced complete band profiles, while the translucent morphotypes often produced band profiles with fewer bands than the opaque morphotype profiles, as well as unique bands not found in the opaque profiles. As was the case with all of the Vibrio species and V. vulnificus isolates examined, the RAPD profiles of the opaque and translucent morphotypes were consistently reproducible.
FIG. 3.
Representative RAPD profiles of opaque and translucent morphotypes of V. vulnificus isolates. Lanes 1, 6, and 11, 123-bp ladder; lane 2, strain 549 (opaque); lane 3, strain 549 (translucent); lane 4, strain 1007 (opaque); lane 5, strain 1007 (translucent); lane 7, strain 707 (opaque); lane 8, strain 707 (translucent); lane 9, strain 1002 (opaque); lane 10, strain 1002 (translucent).
DISCUSSION
When the RAPD method was used, all of the Vibrio species could be clearly differentiated from one another. Furthermore, all isolates could be identified as members of the genus Vibrio based on the presence of amplification fragments whose molecular weights were within four molecular weight ranges. V. vulnificus isolates produced a unique band profile with two additional molecular weight range bands, suggesting that the RAPD method may be an important tool for differentiating V. vulnificus from other members of the genus Vibrio. The finding that each V. vulnificus isolate produced a unique RAPD profile also suggests that V. vulnificus isolates are genetically very heterogeneous, which is consistent with the results of previous studies of V. vulnificus in which researchers used arbitrarily primed PCR (3), ribotyping (7), clamped homogeneous electric field gel electrophoresis (4), pulsed-field gel electrophoresis (17), and amplified fragment length polymorphisms (2).
Previous studies have revealed no significant differences between clinical and environmental isolates of V. vulnificus based on biochemical test results, antimicrobial susceptibility patterns, or virulence characteristics (15, 18). Other studies have shown that a capsule is required for V. vulnificus virulence (1, 6, 9, 14) and that 90% of environmental isolates have capsules (16). Nevertheless, the virulence of these isolates is variable (8, 15), suggesting that the presence of a capsule is not the sole requirement for virulence. Attempts in our lab to correlate particular capsular types with virulence have been unsuccessful (10). The fact that the 200- to 178-bp segment was always present in the RAPD profiles of clinical V. vulnificus isolates and only occasionally present in the RAPD profiles of environmental isolates suggests that this segment of DNA may in some way influence the virulence of an isolate and thus the ability of the organism to infect human hosts. A recent study performed with V. vulnificus resulted in a similar conclusion; most clinical isolates contained a certain iron acquisition gene (viuB), while most environmental strains did not contain this gene (3). This gene may also serve as a target for identification of virulent strains of V. vulnificus.
Additional studies in our laboratory are concentrating on the role that the fragment from clinical isolates may play in virulence. It is possible that this fragment could be used as a probe to determine whether environmental strains are virulent. The fact that only about 8% of the environmental strains contained this DNA fragment suggests the possibility that, like strains of Vibrio parahaemolyticus (12), only certain strains of V. vulnificus may be capable of causing human infection. If this is the case, then the commonly employed mouse models used to predict the virulence of V. vulnificus strains (23) may not be valid. These models often predict that nearly all strains are virulent.
Differences in the RAPD profiles of opaque and translucent isogenic isolates may eventually lead to an understanding of the genetic mechanism responsible for the change from opaque morphotype to translucent morphotype. This mechanism is poorly understood, and various attempts to determine the trigger for this event have been unsuccessful. It has been suggested previously that the trigger for the switching event may be a chromosomal rearrangement or genetic inversion (16). Our results indicate that this phenomenon is influenced by permanent genetic changes which can be detected by the production of unique RAPD profiles by isogenic morphotypes of the same isolate of V. vulnificus. The presence of such dramatic differences between opaque and translucent morphotypes further suggests that global changes may occur instead of a single, simple event.
The findings of this study suggest that the RAPD method is indeed a useful tool for identification and differentiation of various Vibrio species, as well as for differentiation of clinical and environmental V. vulnificus isolates. The genetic differences between opaque and translucent isolates of V. vulnificus will be explored further. The finding that the 200- to 178-bp segment of DNA is found in all clinical isolates and in only a few environmental strains should serve as a focus for further studies involving sequencing and probe production.
ACKNOWLEDGMENTS
This study was supported, in part, by a grant (R-MG-94-21) from the North Carolina Sea Grant Program.
We thank Ronald Siebling at Louisiana State University for providing the opaque and translucent morphotypes of V. vulnificus used in this study.
REFERENCES
- 1.Amako D, Okada K, Miake S. Presence of a capsule in Vibrio vulnificus. J Gen Microbiol. 1984;130:2741–2743. doi: 10.1099/00221287-130-10-2741. [DOI] [PubMed] [Google Scholar]
- 2.Arias C R, Verdonck L, Swings J, Garay E, Aznar R. Intraspecific differentiation of Vibrio vulnificus biotypes by amplified fragment length polymorphism and ribotyping. Appl Environ Microbiol. 1997;63:2600–2606. doi: 10.1128/aem.63.7.2600-2606.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bej A K, Harold N, Vickery M C L, Brasher C, Jeffreys A, Jones D D, DePaola A, Cook D W. Abstracts of the 97th General Meeting of the American Society for Microbiology 1997. Washington, D.C: American Society for Microbiology; 1997. Use of PCR to determine genomic diversity and distribution of siderophore-mediated iron acquisition genes in clinical and environmental isolates of Vibrio vulnificus, abstr. Q-177; p. 485. [Google Scholar]
- 4.Buchreiser C, Murphree V V, Tamplin R L, Kasper C W. Multiple Vibrio vulnificus strains in oysters as demonstrated by clamped homogeneous electric field gel electrophoresis. Appl Environ Microbiol. 1995;61:1163–1168. doi: 10.1128/aem.61.3.1163-1168.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caetano-Anolles G, Bassam B J, Gresshoff P M. DNA amplification fingerprinting using very short oligonucleotide primers. Bio/Technology. 1991;9:553–557. doi: 10.1038/nbt0691-553. [DOI] [PubMed] [Google Scholar]
- 6.Hayat U K, Reddy G P, Bush C A, Johnson J A, Wright A C, Morris J G., Jr Capsular types of Vibrio vulnificus: an analysis of strains from clinical and environmental sources. J Infect Dis. 1993;168:758–762. doi: 10.1093/infdis/168.3.758. [DOI] [PubMed] [Google Scholar]
- 7.Høi L A, Dalsgaard A, Larsen J L, Warner J M, Oliver J D. Comparison of ribotyping and randomly amplified polymorphic DNA PCR for characterization of Vibrio vulnificus. Appl Environ Microbiol. 1997;63:1674–1678. doi: 10.1128/aem.63.5.1674-1678.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kaysner C A, Abeyta C, Wekell M M, DePaola A, Scott R F, Leitch J M. Virulent strains of Vibrio vulnificus isolated from estuaries of the United States west coast. Appl Environ Microbiol. 1987;53:1349–1351. doi: 10.1128/aem.53.6.1349-1351.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kreger A, DeChalet L, Shirley P. Interaction of Vibrio vulnificus with human polymorphonuclear leukocytes: association of virulence with resistance to phagocytosis. J Infect Dis. 1981;144:244–248. doi: 10.1093/infdis/144.3.244. [DOI] [PubMed] [Google Scholar]
- 10.Linkous D, Simpson L M, Oliver J D. Abstracts of the 97th General Meeting of the American Society for Microbiology 1997. Washington, D.C: American Society for Microbiology; 1997. Comparison of pathogenicity among Vibrio vulnificus strains based on capsular and LPS serotypes, abstr. B-210; p. 65. [Google Scholar]
- 11.Oliver J D. Vibrio vulnificus. In: Doyle M, editor. Foodborne bacterial pathogens. New York, N.Y: Marcel Dekker, Inc.; 1989. pp. 569–599. [Google Scholar]
- 12.Oliver J D, Kaper J B. Vibrio species. In: Doyle M, Beuchat L R, Montville T J, editors. Food microbiology: fundamentals and frontiers. Washington, D.C: ASM Press; 1997. pp. 228–264. [Google Scholar]
- 13.Oliver J D, Warner R A, Cleland D R. Distribution of Vibrio vulnificus and other lactose-fermenting vibrios in the marine environment. Appl Environ Microbiol. 1983;45:985–998. doi: 10.1128/aem.45.3.985-998.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Simpson L M, White V K, Zane S F, Oliver J D. Correlation between virulence and colony morphology in Vibrio vulnificus. Infect Immun. 1987;55:269–272. doi: 10.1128/iai.55.1.269-272.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stelma G N, Jr, Reyes A L, Peeler J T, Johnson C H, Spaulding P L. Virulence characteristics of clinical and environmental isolates of Vibrio vulnificus. Appl Environ Microbiol. 1992;58:2776–2782. doi: 10.1128/aem.58.9.2776-2782.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tamplin M L. Proceedings of the 1994 Vibrio vulnificus Workshop. U.S. Washington, D.C: Food and Drug Administration; 1995. The ecology of Vibrio vulnificus; pp. 75–86. [Google Scholar]
- 17.Tamplin M L, Jackson J K, Buchreiser C, Murphree R L, Portier K M, Gangar V, Miller L G, Kaspar C W. Pulsed-field gel electrophoresis and ribotype profiles of clinical and environmental Vibrio vulnificus isolates. Appl Environ Microbiol. 1996;62:3572–3580. doi: 10.1128/aem.62.10.3572-3580.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tison D L, Kelly M T. Virulence of Vibrio vulnificus strains from marine environments. Appl Environ Microbiol. 1986;52:1004–1006. doi: 10.1128/aem.51.5.1004-1006.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang G, Whittam T S, Berg C M, Berg D E. RAPD (arbitrary primer) PCR is more sensitive than multilocus enzyme electrophoresis for distinguishing related bacterial strains. Nucleic Acids Res. 1993;21:5930–5933. doi: 10.1093/nar/21.25.5930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Welsh J, McClelland M. Fingerprinting genomes using PCR with arbitrary primers. Nucleic Acids Res. 1990;18:7213–7218. doi: 10.1093/nar/18.24.7213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Welsh J, McClelland M. The characterization of pathogenic microorganisms by genomic fingerprinting using arbitrarily primed polymerase chain reaction (AP-PCR) In: Persing D H, Smith T F, Tenover F C, White T J, editors. Diagnostic molecular microbiology. Washington, D.C: ASM Press; 1993. pp. 595–602. [Google Scholar]
- 22.Williams J G K, Kubelik A R, Livak K J, Rafalski J A, Tingey S V. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res. 1990;18:6531–6535. doi: 10.1093/nar/18.22.6531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wright A C, Simpson L M, Oliver J D. Role of iron in the pathogenesis of Vibrio vulnificus infections. Infect Immun. 1981;34:503–507. doi: 10.1128/iai.34.2.503-507.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yoshida S I, Ogawa M, Mizuguchi Y. Relation of capsular material and colony opacity to the virulence of Vibrio vulnificus. Infect Immun. 1985;47:446–451. doi: 10.1128/iai.47.2.446-451.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]