Abstract
In the past decade, there has been a shift in research, clinical development, and commercial activity to exploit the many physiological roles of RNA for use in medicine. With the rapid success in the development of lipid–RNA nanoparticles for mRNA vaccines against COVID-19 and with several approved RNA-based drugs, RNA has catapulted to the forefront of drug research. With diverse functions beyond the role of mRNA in producing antigens or therapeutic proteins, many classes of RNA serve regulatory roles in cells and tissues. These RNAs have potential as new therapeutics, with RNA itself serving as either a drug or a target. Here, based on the CAS Content Collection, we provide a landscape view of the current state and outline trends in RNA research in medicine across time, geography, therapeutic pipelines, chemical modifications, and delivery mechanisms.
Introduction
Recent advances in RNA design and delivery have enabled the development of RNA-based medicine for a broad range of applications, including therapeutics, vaccines, and diagnostics. While human RNA medicine has faced many challenges in terms of efficacy and immunogenicity, the recent success of mRNA vaccines against COVID-19 and the approval of new RNA-based drugs provide new momentum to the field. Many classes of RNA play important regulatory roles in cells and tissues, beyond the obvious role of mRNA in protein synthesis. Scientific research, clinical development, and commercial production now focus on exploiting the many roles of RNA for use in biotechnology and medicine. Advances in understanding RNA structure and function are combined with a robust production pipeline to develop clinically effective RNA-related applications.1−12
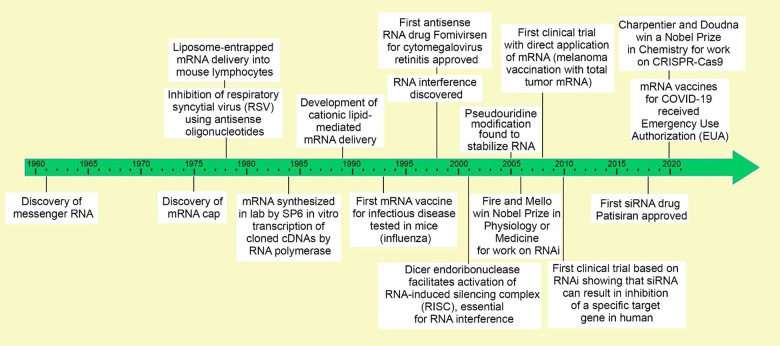
Many key discoveries have contributed to the advancing of RNA medicines we have today. Early research in the 1960s on nucleic acids led to the discovery of mRNA.13 In the next decade, the 5′-cap on mRNA was discovered,14,15 the first liposome-entrapped RNA was delivered into cells,16 and antisense oligomers (ASOs) were used to inhibit Rous sarcoma virus (RSV).17 In the 1980s, in vitro transcription from engineered DNA templates using a bacteriophage SP6 promoter and RNA polymerase18 allowed the manufacture of mRNA and expression of other types of RNA in cell-free systems. Later in the 1980s, the first cationic-lipid-mediated mRNA delivery was achieved.19,20 The discovery of RNA interference (RNAi)21 and the approval of the first antisense RNA drug in the late 1990s22 were key to the development of RNA therapeutics. For their pioneering work on RNAi and the RNA-induced silencing complex (RISC),23 Fire and Mello21 were awarded the Nobel Prize in Physiology and Medicine in 2006. During the 2000s, the discovery of the importance of pseudouridine modification24 and further research on mRNA led to the first human trial of an mRNA vaccine against melanoma in 2008.25 In 2010, a pivotal human clinical trial showed that siRNA could target specific human genes,10 and subsequent pre-clinical research and development led to the approval of the first siRNA drug in 2018.26 Most recently, Doudna and Charpentier were awarded the Nobel Prize in 2020 for CRISPR-Cas9 gene editing. In a CRISPR-Cas9 system, a small piece of RNA with a short “guide” sequence attaches to a specific target sequence of DNA in a genome; the RNA also binds to the Cas9 enzyme—thus, the modified RNA is used to recognize the DNA sequence, and the Cas9 enzyme cuts the DNA at the targeted location.27 Two human mRNA vaccines against COVID-19 received Emergency Use Authorization in 2020, and one of them was finally approved in 2021.28−30 These key milestones and achievements are captured in Figure 1.
Figure 1.
Timeline of major RNA research and development milestones. A more detailed timeline table complete with references is provided as Table S1.
RNA technology provides an innovative approach for developing new drugs for rare or difficult-to-treat diseases. Since 2014, several drugs have been approved to treat macular degeneration, Duchenne muscular dystrophy (DMD), polyneuropathy, and amyotrophic lateral sclerosis.31 Drugs to treat many other diseases, including cancer, hepatic and renal diseases, cardiac diseases, metabolic diseases, blood disorders, respiratory diseases, and autoimmune diseases are currently in various stages of clinical studies, some showing promising results.
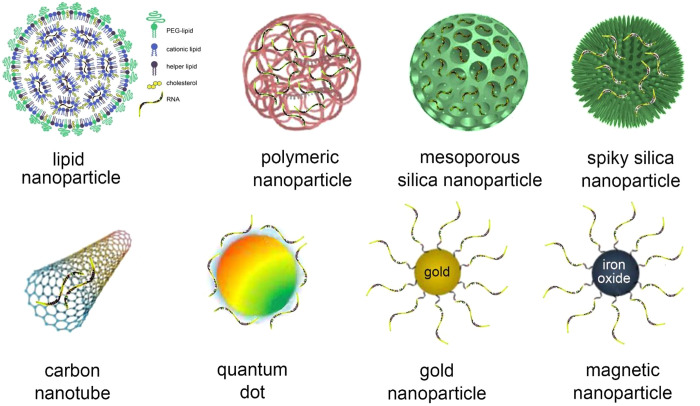
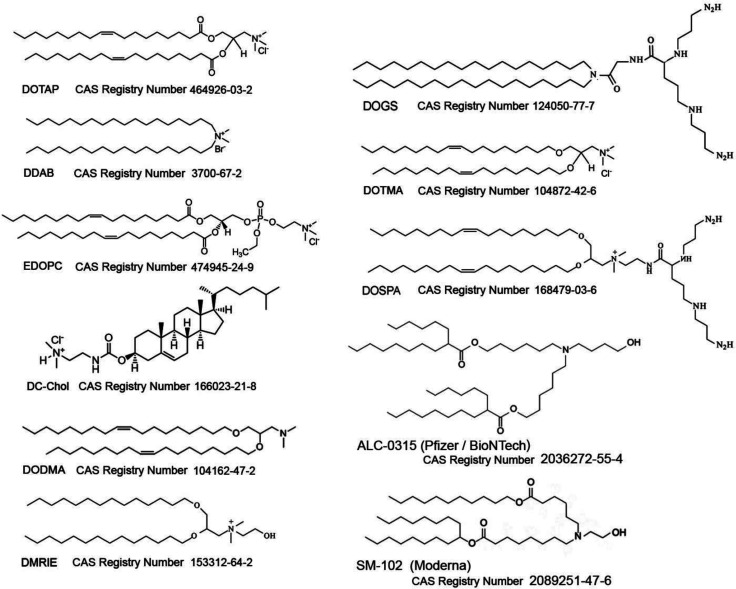
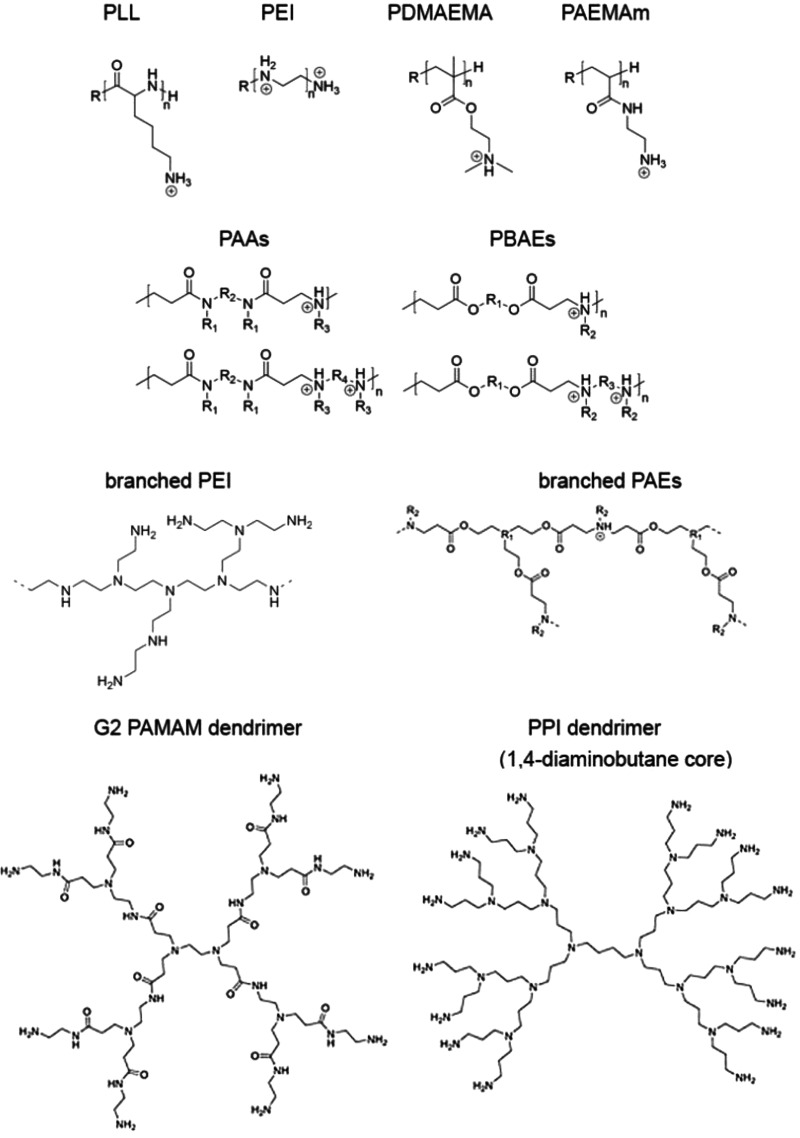
Compared to other biomolecules, RNA molecules are unstable and transient. Foreign RNA molecules, when introduced to the human body, have limited protein expression levels in cells and often trigger immunogenicity in the body. However, these practical problems can often be mitigated by various optimizations on RNA molecules, including chemical modifications, mRNA cap/codon/tail optimization, etc. Leveraging the unique aspects of both the chemical and biological information of the CAS Content Collection,32 this paper focuses on chemical modifications to the base, backbone, sugar, and 5′ or 3′ ligations to other molecules. While chemical modifications increase RNA stability, complexes of RNA within nanoparticles provide further protection. Except for aptamers that bind to a cell surface target, RNA must be delivered into the cell by a carrier. After the RNA is internalized, it must be released from the membrane-bound vesicle or endosome to the cytosol. Without a doubt, the delivery system has been an important research topic for RNA medicine.
In this paper, we reviewed naturally occurring RNAs with their cellular functions and the associated research trends based on the analysis of CAS Content Collection.32 We then appraised different types of RNA in medical applications: their advantages, challenges, and research trends. Subsequently, we assessed the development pipelines of RNA therapeutics and vaccines with company research focuses, disease categories, development stages and publication trends. Finally, we discussed RNA chemical modifications and delivery systems in detail, as they are critical to the success of RNA medicine. We thus revealed the sustained global effort that propelled this field to the cusp of realization for novel medical applications of RNA in many diseases. We hope this review can serve as an easy-to-understand overview so that scientists from many different disciplines can appreciate the current state of the field of RNA medicine and join in solving the remaining challenges for fulfilling its potential.
Types of Naturally Occurring RNA and Their Functions in Biological Systems
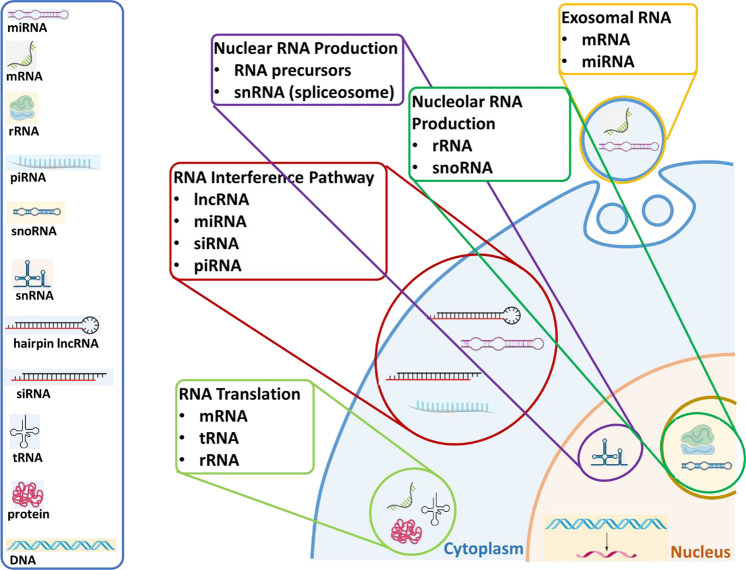
RNA, a versatile macromolecule that is specialized for many functions, can be broadly defined as coding or messenger RNA (mRNA) and non-coding RNA (ncRNA). There are several different types of ncRNA including ribosomal RNA (rRNA),33,34 transfer RNA (tRNA),33,34 small nuclear RNA (snRNA),35−37 small nucleolar RNA (snoRNA),36,38−45 long non-coding RNA (lncRNA),7,46−52 short hairpin RNA (shRNA), micro-RNA (miRNA),53−59 transfer messenger RNA (tmRNA), small interfering RNA (siRNA),60−71 small activating RNA (saRNA), piwi-interacting RNA (piRNA),3,4,72−78 circular RNA (circRNA), ribozymes, and exosomal RNA.79−83 The cellular localizations of different types of RNA are illustrated in Figure 2, and an overview of their functions is provided in the following section.
Figure 2.
Types of naturally occurring RNA and their cellular functions and localizations
Functions of the Naturally Occurring RNA Types
mRNA, which was the first RNA to be characterized, is the initial transcription product of a protein-coding gene and includes both protein-coding exons and non-coding introns. The mature, translatable mRNA must be spliced to remove the introns and, in transcripts that can go through alternative splicing, one or more potential exons. The mature RNA has a 5′-7-methylguanosine cap, a 5′-untranslated region, a start codon (unique sequence of 3 bases) for the translated region of the gene, a stop codon that ends the translated region of the gene and starts the 3′-untranslated region, and a 3′-polyadenosine tail. Gene expression, which is often regulated by the amount of mRNA for the gene, is controlled by the balance between synthesis and degradation of mRNA. Although mRNA is a critically important RNA, it makes up only 1–5% of the RNA in a cell.33,34
ncRNAs, in contrast to mRNA, are the final functional products of the DNA. Although it was thought initially that ncRNAs were non-functional junk RNA, in the 1950s, in the same paper that introduced the phrase “Central Dogma”, Francis Crick correctly hypothesized that the ncRNA might function in the translation of mRNA into protein.84 In 1955, George Palade identified ribosomes as a small particulate component of the cytoplasm that contains RNA, and in 1965, Robert Holley purified a tRNA from yeast and determined the structure.85,86 In the past half-century, many types of ncRNA with various functions have been identified; many are involved in regulating transcription and protein expression in the cell.87−94
rRNA, which constitutes up to 80% of the RNA in an active cell, comprises three rRNAs (the 5S, 5.8S, and 28S) complexed with many proteins to form the large subunit of the ribosome and one rRNA (the 18S) complexed with proteins to form the small subunit of the ribosome. There are also two mitochondrial rRNA genes (the 12S and 16S) which, along with many proteins, form the mitochondrial ribosome. rRNA in the ribosome, acting as a ribozyme, catalyzes peptide bond formation between two amino acids. Synthesis of the large amount of rRNA occurs in the nucleolus, a heterochromatic region found in most nuclei.33,34
tRNA, which makes up 10–15% of the RNA in the cell, translates the mRNA codon sequence for each amino acid. The many different tRNAs, which are usually 75–95 nucleotides, all fold into very similar three-dimensional structures. The 3D structure exposes three unpaired nucleotides that serve as the anti-codon to base pair with the mRNA. Specific amino acids are covalently bound to a tRNA by aminoacyl tRNA synthetases. The specificity between the anti-codon and the bound amino acid is the basis of translation. Each tRNA anticodon can bind to several different mRNA codons. This pairing is based on the wobble rules for the third nucleotide position of the anti-codon, which is often post-transcriptionally modified to allow for wobble-pairing. Common modifications at the third position include 5-methyl-2-thiouridine, 5-methyl-2′-O-methyluridine, 2′-O-methyluridine, 5-methyluridine, 5-hydroxyuridine, hypoxanthine, and lysidine.33,34
snRNAs (∼150 nucleotides) are components of the small nuclear ribonucleoproteins (snRNPs) of the spliceosome. They act as catalysts that splice mRNA into its mature form and they are important in the selection of alternative splicing sequences.35−37
snoRNAs (60–300 nucleotides) are bound to four core proteins and act as guides to correctly target modifications for the maturation of rRNA. They comprise two classes of RNA. C/D snoRNAs participate in the 2-methylation of targeted nucleotides, while H/ACA snoRNAs participate in the modification of uridine to pseudouridine. They help guide the protein to the specific target, rather than catalyzing the reaction directly. As participants in rRNA maturation, snoRNAs are found in the nucleolus.36,38−45
siRNAs are products of double-stranded lncRNAs (e.g., hairpin lncRNA) and are central to RNA interference, which negatively regulates gene expression. Double-stranded RNA (dsRNA), either from genomic lncRNA or dsRNA viruses, is recognized and cleaved by the endonuclease Dicer into 20–24 base-pair sections with short overhangs on both ends. These siRNAs bind the Argonaute protein to form the pre-RISC (RNA-induced silencing complex). Argonaute selects the less thermodynamically stable strand of the siRNA and releases the other strand to form the mature RISC. RISC recognizes mRNA complementary to the single-stranded siRNA, and the Argonaute endonuclease cuts this targeted mRNA, thereby downregulating the gene product. The binding of a RISC to a target mRNA also prevents efficient ribosome binding and translation, further downregulating the gene product. Active RISCs may also affect the transcription of target genes by inducing chromatin reorganization through epigenetic modifications. This can be a defense mechanism against dsRNA viruses or an endogenous gene-regulatory mechanism.60−71
miRNAs are closely related to siRNAs but are formed from pri-miRNAs (primary microRNAs), which are long, imperfectly paired hairpin RNA transcripts. The pri-miRNA is processed first by Drosha nuclease into a ∼70-nucleotide imperfectly paired hairpin pre-miRNA (precursor microRNA) that is then, like siRNA, processed by Dicer to produce the 21–23-bp, mature, double-stranded miRNA that binds to Argonaute to form the RISC. Alternatively, some miRNAs are made from introns in mRNAs. After splicing, that intron is a pre-miRNA that is processed by Dicer to form a RISC. miRNAs form negative gene regulatory networks and intronic miRNAs may regulate and balance potentially competing pathways.53−59
piRNAs, like siRNA and miRNA, negatively regulate gene expression, but they interact with the Piwi class of Argonaute proteins. Unlike siRNA and miRNA, piRNAs (24–31 nucleotides) are produced from long, single-stranded RNA transcripts through an uncharacterized Dicer-independent mechanism. Mature piRNAs bind to Piwi proteins to form RISCs that act primarily as epigenetic regulators of transposons (genetic elements that move around the genome) but may also regulate transposons post-transcriptionally through the ping-pong pathway.3,4,72−78
saRNA, like siRNA, is a ∼21-bp dsRNA long that interacts with Argonaute proteins to form a RISC. Unlike siRNA, saRNA upregulates target gene expression by an unknown mechanism, perhaps activating transcription by targeting the promoter region of the gene. saRNA may be produced endogenously or artificially to strongly activate the target gene.5,95−101
lncRNAs comprise a mixed group of RNAs >200 bp, which differentiates them from short ncRNAs such as snoRNA, siRNA, miRNA, piRNA, etc. lncRNAs have a wide variety of functions including regulation of chromosome architecture and interactions, chromatin remodeling, and positive or negative regulation of transcription, nuclear body architecture, and mRNA stability and turnover.7,46−52
circRNAs are lncRNAs with 5′ and 3′ ends linked covalently to form a continuous circle. circRNAs are broadly expressed in mammalian cells and have shown cell-type and tissue-specific expression patterns.102 Neither the mechanisms leading to circularization of the RNA nor the function of circRNA is known, but the leading hypothesis is that they may serve as miRNA sponges. Many circRNAs contain large numbers of miRNA target sites that may competitively antagonize the ability of miRNA to silence its target genes.
Exosomal RNAs are mRNAs, miRNAs, siRNAs, and lncRNAs that are packaged and exported from the cell through the exosomal pathway. Although they are poorly understood, exosomal RNAs may serve as signaling molecules to regulate gene expression in target cells. These circulating RNAs, especially miRNAs, may serve as diagnostic and/or prognostic targets for various diseases such as cancers.79−83
Antisense RNA (asRNA), also referred to as antisense oligonucleotide (ASO), is a single-stranded RNA that is complementary to a protein-coding messenger RNA (mRNA) with which it hybridizes, and thereby blocks its translation into protein.103
Research Trends on Different Types of RNA As Reflected by Number of Publications
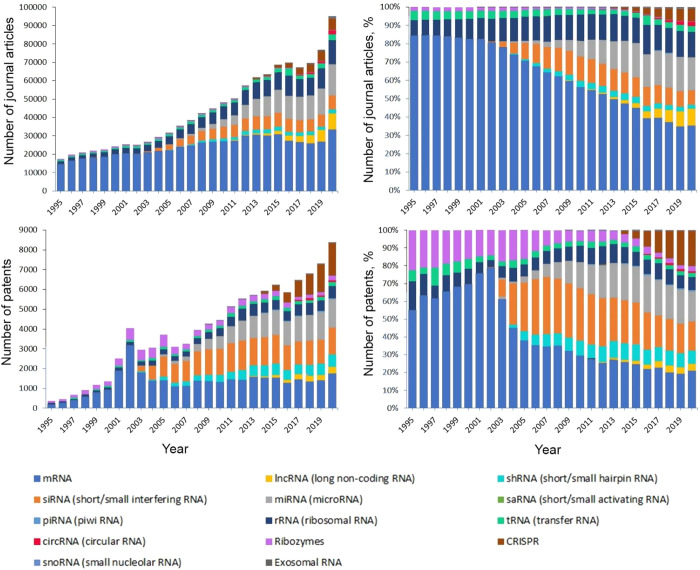
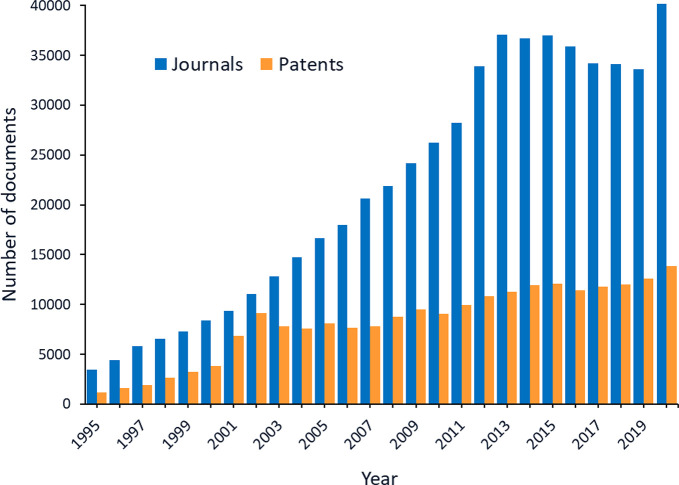
The CAS Content Collection32 is the largest human-curated collection of published scientific knowledge, used for quantitative analysis of global scientific publications against variables such as time, research area, formulation, application, disease association, and chemical composition. We searched the title, abstract, or CAS-indexed terms using RNA-related keywords and their synonyms to identify relevant published documents. Figure 3 shows trends in the number of publications for specific types of RNA. In the past 25 years, the research areas became more diverse as new types of RNA were discovered, and this is reflected in both journal and patent publications, particularly in the areas of siRNA, miRNA, lncRNA, and CRISPR-related research. CRISPR technology has recently increased rapidly in volume of patent publications, and it accounted for 20% of the RNA-related patent publications in 2020.
Figure 3.
Document publication trends for different types of RNA from 1995 to 2020 as found from the CAS Content Collection.32 Top two panels: journal publications in absolute numbers and given year percentages. Bottom two panels: patent publications (counted once per patent family) in absolute numbers and given year percentages.
To better reveal the rising trends of those recently emerged types of RNA, the percentage of document publications of a specific year was calculated within the given type of RNA over the time (Figure 4). Although the cumulative publication numbers for circRNA, exosomal RNA, lncRNA, and CRISPR, are relatively small compared with others (Figure 3), their rates of increase are much faster.
Figure 4.
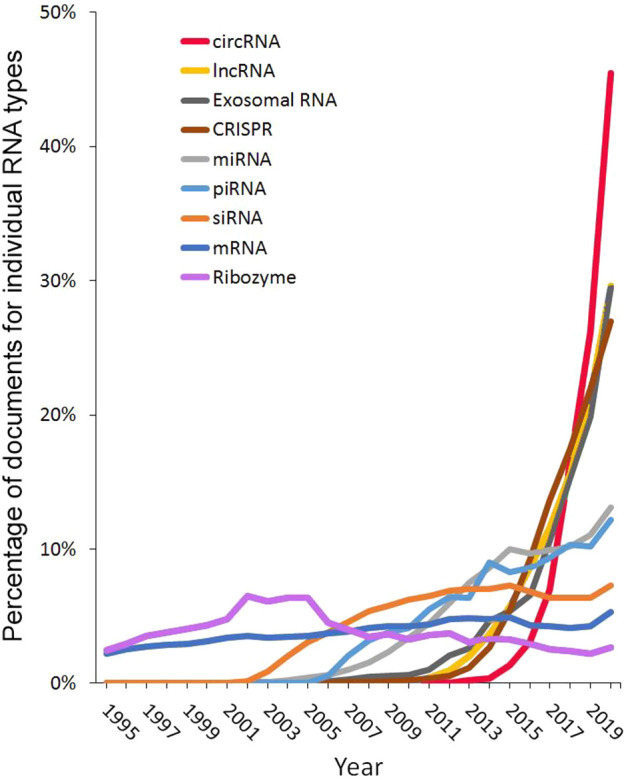
Trends in publication volume for different RNA types in the years 1995–2020. Percentages are calculated with yearly publication numbers for each individual RNA type, normalized by total publications in the years 1995–2020 for the same RNA type. Example: Percentage of circRNA documents in 2020 = (number of circRNA documents in 2020)/(total number of circRNA documents from 1995 to 2020).
Types of RNA Used in Medical Applications, Their Advantages, and Challenges
Types of RNAs and Their Applications in Medicine
mRNA transcripts can act as therapeutic RNAs, diagnostic biomarkers, or therapeutic targets. Translation of an mRNA in the cell can produce a therapeutic protein to replace a defective or missing protein. In the case of vaccines, mRNA translation can generate antigenic targets for the immune system, such as the spike glycoprotein of SARS-CoV-2 in the COVID-19 mRNA vaccines. mRNA may also serve as a therapeutic target for ASOs, siRNA, miRNA, aptamers, and suppressor tRNAs.
In the cell, miRNAs bind to the 3′-untranslated region of mRNAs and target them for degradation by the RISC.104 Because a single miRNA binds to multiple mRNAs, miRNAs serve as regulatory check points. The cellular processes regulated by miRNAs include those involved in many diseases, such as cardiovascular disease, cancer, and disease-related metabolic pathways.105,106 Thus, they can serve as biomarkers for disease diagnosis, as potential drugs, or as attractive targets for other regulatory RNAs.
Although siRNAs, like miRNAs, use the RISC to degrade their target mRNAs, siRNAs bind to specific areas in the mRNA coding region. This target specificity makes them attractive as potential drugs, but off-target effects can negate this advantage. In order to minimize off-target effects, siRNAs are modified to decrease their thermal stability, increase their target specificity, and decrease the stability of their binding of the siRNA to mRNAs that are not an exact match to the intended target mRNA. These usually include 2′-O-methyl and 2′-MOE ribose modifications that introduce steric hindrance to decrease binding affinity (more discussion in the Chemical Modifications section below).104
One of the earliest therapeutic RNAs, ASOs, recognize and bind to complementary DNA or RNA sequences, including mutated sequences that may lead to disease. Upon binding to the mutated sequences, ASOs may facilitate proper mRNA splicing, prevent translation of a defective protein, or target RNAs for degradation.104
In therapeutic antibody–oligonucleotide conjugates (AOCs), the antibody targets the site of interest while carrying an ASO or a siRNA that acts on the targeted region.107 Conjugation of an ASO to an antibody to create an AOC improves the pharmacokinetics of the ASO in vivo by increasing tissue distribution and prolonging gene silencing in multiple tissues.108
The CRISPR-Cas system uses a guide RNA that is either a combination of a trans-activating CRISPR RNA (tracrRNA) and a CRISPR RNA (crRNA) or a joined single guide RNA (sgRNA). The guide RNA directs the CRISPR complex containing a Cas endonuclease to a specific site in the genome for cleavage.6 The ability of the CRISPR-Cas system to create directed double-stranded breaks in DNA allows the repair of genetic mutations. Changing the endonuclease activity of Cas can convert CRISPR-Cas to a system that nicks a single strand of the DNA or that deaminates a specific nucleotide.109 If the Cas endonuclease is inactivated, the system can simply bind to DNA to regulate transcription.104
Aptamers are structure-based rather than sequence-based ligands that neither hybridize with other nucleic acids nor produce proteins. They can be RNA, DNA, RNA/DNA combinations, or even proteins. In vitro systematic evolution of ligands by exponential enrichment (SELEX) is used to identify single-stranded RNA or DNA oligonucleotides with a high affinity for a target. Because their binding depends on their 3D structure, aptamers can bind a wide range of targets, including proteins, cells, microorganisms, chemical compounds, and other nucleic acids.110 Aptamers may also serve as delivery agents for siRNA in nanoparticles for cancer therapy.111
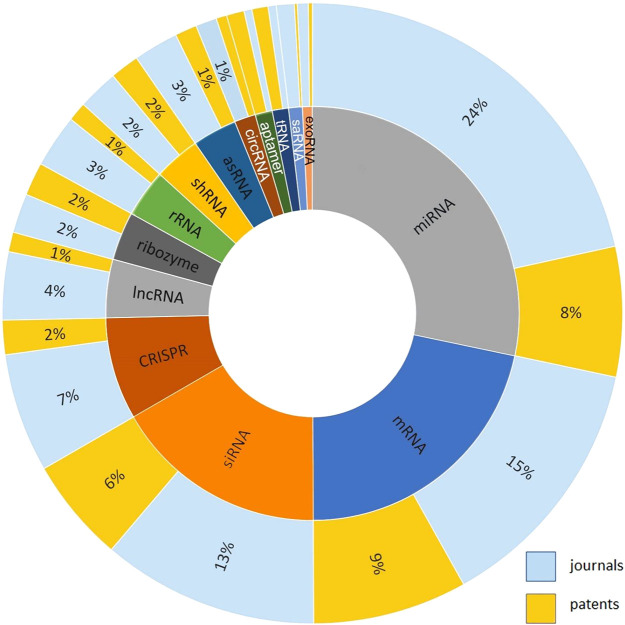
To measure the distribution of research effort using different types of RNA as therapeutics, vaccines, or diagnostics, related documents were extracted accordingly from the CAS Content Collection, and journal and patent publications percentages for each type of RNA were determined as shown in Figure 6.32 miRNA and mRNA, the two most popular therapeutic RNAs in the journal and patent literature, can serve as drugs, disease biomarkers, and drug targets. Together with siRNA, they represent most of the therapeutic RNA patent activity. Approved RNA drugs include mRNAs, siRNAs, ASOs, and aptamers; these RNAs along with CRISPR RNAs and AOCs comprise most of the clinical candidates.
Figure 6.
Percentage of journal documents and patents for various types of RNA used in medical studies including therapeutics, vaccines, and diagnostics.
Publication Trends for RNAs Used in Medical Applications
The CAS Content Collection32 shows a steady increase in the number of journal articles and patents related to RNA applications in medicine (Figure 5). The peak in patents in 2001–2002 may correlate with the first clinical trials using dendritic cells transfected with mRNA encoding tumor antigens (a therapeutic mRNA cancer vaccine) in 2001.112,113 The spike in journal article numbers in 2020 likely resulted from interest in the COVID-19 mRNA vaccines. The increase in journal articles and patents on therapeutic RNA from 2011 to 2016 can be attributed to initial interest in siRNA and miRNA, which decreased temporarily with the discovery of their off-target effects. Interest in mRNA also increased from 2011 to 2016, then decreased and only recovered once mRNA vaccines took center stage in the fight against COVID-19.
Figure 5.
Numbers of journal documents and patents related to RNAs for medical use by year.
However, other types of RNA are potential therapeutics or targets. These RNAs include (a) shRNA; (b) lncRNA, an RNA whose role in gene regulation is generating increasing interest; (c) circRNA, once thought to be a byproduct of RNA splicing, that may have a role in regulation as a miRNA sponge; (d) saRNA, which regulates gene transcription; and (e) exosomal RNA, which is contained in naturally occurring lipid vesicles called exosomes, which can cross the blood-brain barrier and appear to be vital for cell-to-cell communication, and which transport mainly miRNA.114
Based on the CAS Content Collection data for CRISPR RNA, miRNA, and mRNA used in vaccines, diagnostics, and therapeutics,32 only mRNA has substantial applications in vaccines (Figure 7). However, both miRNA and mRNA have demonstrated their potential as therapeutics and diagnostics.
Figure 7.
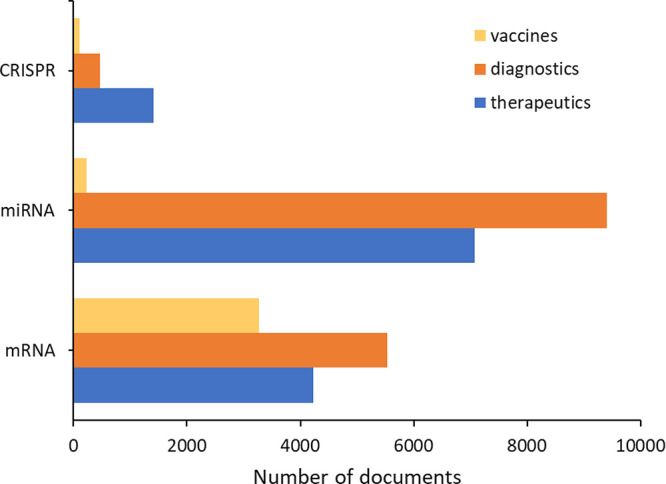
Number of journal publications and patents for mRNA, miRNA, and CRISPR with applications in therapeutics, vaccines, and diagnostics.
The large body of work on miRNA and mRNA in diagnostics may be surprising since these molecules are much more susceptible to nuclease degradation than DNA. However, the essential roles of mRNA and miRNA in cellular metabolism make them excellent biomarkers for the study of normal cellular processes and the diagnosis of disease. Metabolic diseases such as cancer as well as infectious diseases can be diagnosed via miRNA biomarkers by reverse transcriptase-quantitative polymerase chain reaction (RT-qPCR) and lateral flow immunoassays.115,116 High-throughput sequencing of total cellular mRNA pinpoints changes in gene expression that can be used in diagnosis.117 The specificity of the CRISPR system for its target DNA makes it a potential tool with both diagnostic and therapeutic purposes.118 The number of journal publications and patents using RNA for diagnosis demonstrates its power (Figure 7); however, a comprehensive review of RNA as a diagnostic biomarker or, in the case of CRISPR RNA, as a diagnostic agent, is beyond the scope of this review. Here we focus on RNAs as therapeutic agents.
Advantages and Challenges of RNAs as Therapeutics
There are several important advantages to RNA therapeutics. They are: (a) target specific, (b) modular with easy-to-switch sequences, (c) predictable in terms of pharmacokinetics and pharmacodynamics, (d) economical in comparison to antibodies or protein drugs since they are synthesized from widely available synthons on an automated synthesizer, and (e) relatively safe, as most of them do not alter the genome.
Proteins must be designed for synthesis from genes in plasmids, then optimized for expression and purification, and these processes may be different for each protein and challenging to optimize. In contrast, RNA can be easily synthesized and purified by established methods using commercially available reagents and equipment. Small RNAs, such as aptamers, siRNAs, miRNAs, and ASOs can be synthesized using solid-support chemistry in commercial oligonucleotide synthesizers.119,120In vitro transcription using commercial kits121 produces longer RNAs, i.e., mRNAs and lncRNAs. The sequence of the RNA can be changed easily, providing custom molecules targeting different proteins or genes. Thus, the development of RNA therapeutics and vaccines that target disease-specific genes or proteins is relatively fast and straightforward. This was demonstrated by the development, testing, and administration of the COVID-19 mRNA vaccines within a year of the isolation and sequencing of the SARS-CoV-2 viral genome.108
Most RNAs regulate transcription, post-transcriptional processing, and translation, but they do not alter the genome. The exception is the CRISPR-Cas system in which the guide RNA and Cas endonuclease can edit the genome. RNA aptamers, which mimic ligands, regulate post-translational protein activity. The rapid degradation and predictable pharmacokinetics of RNAs give them a safety advantage over gene therapies.104
Despite the attractiveness of the plug-and-play concept of RNA therapeutic drug design, they require testing to determine their efficacy and safety, and cell delivery is difficult because RNA is easily degraded. The therapeutic RNA must penetrate the cell membrane and escape endosomal entrapment.104 Although designed for specific targets, therapeutic RNAs can have off-target effects, limiting their usefulness as drugs. Several of these limitations can be mitigated by chemically modifying the RNA to increase target specificity, lower nuclease susceptibility, and improve cellular uptake.104,108
Types of RNA in Medicine in the Development Pipeline and Their Targeted Diseases
Distribution of Diseases Associated with RNA Medicine in Publications and Patents
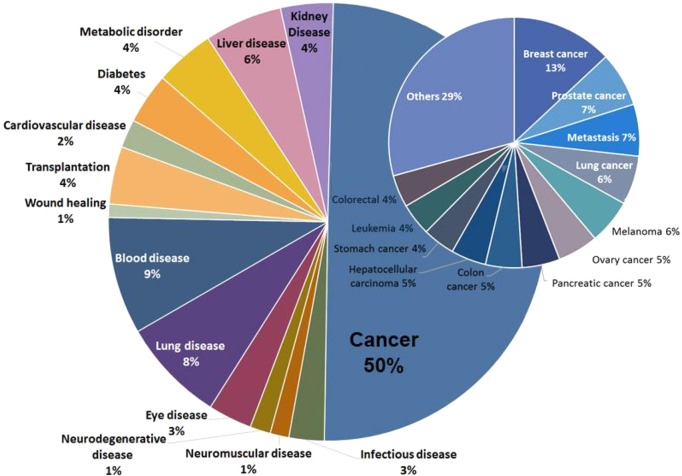
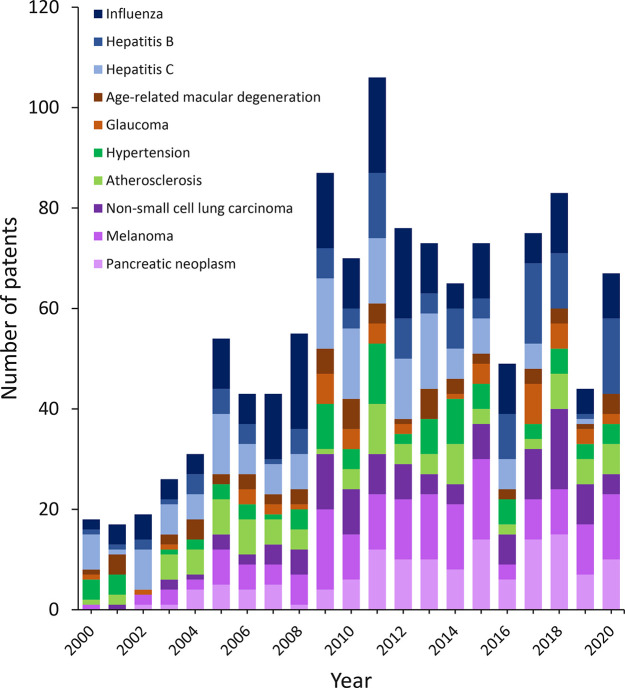
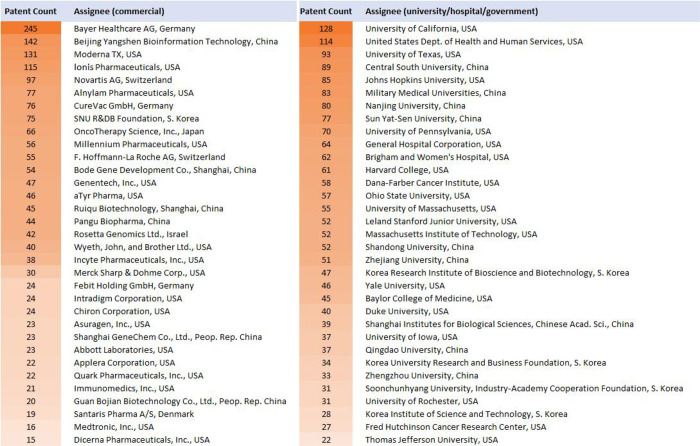
Since the first approved ASO RNA therapeutic in 1998, the research and development of RNA in medical applications has increased. We analyzed data from the CAS Content Collection32 for journal publications and patents on RNAs as therapeutics, vaccines, or diagnostic agents for diseases and found that 50% of the publications are associated with cancer diagnosis or treatment, although lung, liver, and metabolic diseases are also highly represented (Figure 8). There was little correlation between the type of RNA and a targeted disease, indicating that different types of RNA have been explored for many kinds of diseases in the research phase (Figure S1). Infectious diseases and cancer have shown the greatest growth and are the most frequent diseases treated by RNA, followed by eye and cardiovascular diseases, which grew in the first decade of this century and remained relatively stable in the second decade (Figure 9). Specifically, the association of RNA medicine with pancreatic neoplasm, melanoma, non-small-cell lung cancer, hepatitis B, and influenza has increased quickly in the past 20 years. Patent publications for RNA therapeutics for hepatitis C have decreased in recent years, most likely due to the approval of several effective small molecule drugs for hepatitis C. RNA therapeutics for atherosclerosis, hypertension, glaucoma, and age-related macular degeneration research have remained relatively stable. The top 20 patent assignees for patent publications on RNA therapeutics, vaccines, or diagnostics are mostly in the U.S. or China, although a few are in Germany, Korea, Japan, Switzerland, or Israel (Figure 10).
Figure 8.
Percentage of publications associated with RNAs in medical applications
Figure 9.
Yearly number of patent publications on specific diseases targeted by RNA therapeutics, vaccines, and diagnostics.
Figure 10.
Top patent assignees for RNA therapeutics, vaccines, and diagnostics.
Pipeline Dynamics of RNAs for Therapeutics and Vaccines
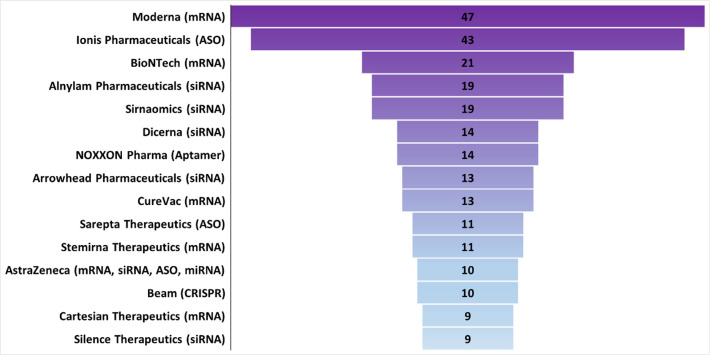
After decades of extensive research, the therapeutic potential of RNAs has led to the development of over 250 therapeutics that are approved or in development (Table S2). Among the top 15 RNA therapeutics companies (Figure 11), which are located worldwide (Figure S2), each mostly focuses on one type of RNA to develop novel RNA therapeutics for treating diseases that range from very rare to common. mRNA and siRNA are the most common RNAs used by the top 15 companies, followed by ASO, CRISPR, and aptamers. Many of these companies are among the top patent assignees for RNA therapeutics, vaccines, and diagnostics in the commercial sector (Figure 10).
Figure 11.
Top pharmaceutical companies ranked by the number of RNA therapeutic and vaccine agents in the development pipeline. Counts include RNA agents in company-announced pre-clinical development, in clinical trials, or approved. A single RNA agent can be counted multiple times when applied to multiple diseases.
Moderna, BioNTech, CureVac, Stemirna Therapeutics, and Cartesian Therapeutics specialize in mRNA related therapeutics or vaccines. Moderna, headquartered in Massachusetts, USA,122 leads this group with over 45 therapeutics in its pipeline (Figure 11) and 131 RNA therapeutic/diagnostic patents (Figure 10). BioNTech and CureVac are both headquartered in Germany.123,124 BioNTech has in its pipeline 21 therapeutics that utilize the immune system to treat cancer and infectious diseases (Figure 11).123 CureVac has 13 therapeutics (Figure 11) along with 76 RNA therapeutic/diagnostic patents (Figure 10) for mRNA medicines.124 Stemirna Therapeutics headquartered in China,125 has a pipeline of 11 mRNA-based therapeutics (Figure 11). Cartesian Therapeutics, headquartered in Maryland, USA,126 has 9 therapeutics and is a pioneer in using mRNA for cell therapies within and beyond oncology, with products in development for autoimmune and respiratory disorders (Figure 11).
Alnylam, Sirnaomics, Arrowhead Pharmaceuticals, Silence Therapeutics, and Dicerna develop siRNAs for their RNA therapeutics. (Effective December 28, 2021, Dicerna is a wholly owned subsidiary of Novo Nordisk. In this paper Dicerna is considered separately.) Alnylam Pharmaceuticals and Dicerna are headquartered in Massachusetts, USA.127,128 Alnylam is the leader in siRNA therapy with over 19 therapeutics in their pipeline and 77 therapeutic/diagnostic patents (Figure 11). Dicerna treats both rare and common diseases with its 14 therapeutics (Figure 11). Sirnaomics, headquartered in Maryland, USA, has developed over 19 therapies for human diseases that have no treatments.129 Arrowhead Pharmaceuticals, headquartered in California, USA,130 treats previously intractable diseases with their 13 therapeutics (Figure 11). Silence Therapeutics, headquartered in London, UK,131 has 9 therapeutics (Figure 11).
Ionis Pharmaceuticals and Sarepta Therapeutics use ASOs for their RNA therapeutics. Ionis Pharmaceuticals is headquartered in California, USA,132 and Sarepta Therapeutics in Massachusetts, USA.133 Ionis has developed over 40 therapeutics (Figure 11) and 115 therapeutic/diagnostic patents (Figure 10). Sarepta uses antisense technology to target neurological diseases, including neuromuscular and neurodegenerative disease, with their 11 therapeutics (Figure 11).
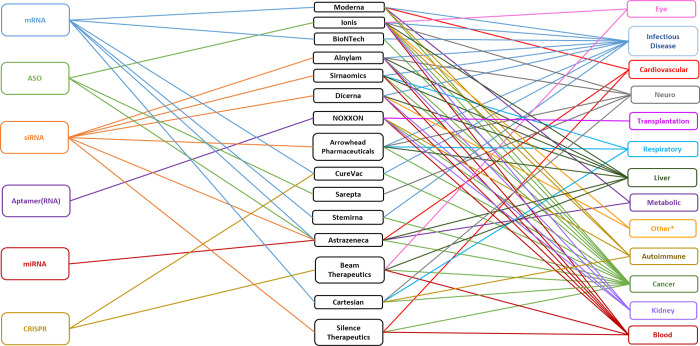
NOXXON Pharma, headquartered in Germany, uses RNA aptamers for their 14 therapeutics (Figure 11).134 Their current pipeline is focused only on oncology, but previously they researched treatments for transplantation, metabolic, blood, autoimmune, and kidney diseases (Figure 12). Beam Therapeutics, headquartered in Massachusetts, USA,135 is pioneering the use of base editing with CRISPR-Cas. Beam has 10 therapeutics in development (Figure 11). AstraZeneca supports multiple RNA platforms through partnerships with many of the top RNA companies.136
Figure 12.
Types of RNA by company and targeted diseases. *Other: acromegaly, hereditary angioedema, and alcohol use disorder.
All but two of the top 15 RNA medicine companies specialize in one type of RNA (Figure 12). AstraZeneca supports multiple RNA platforms and CureVac is partnering with CRISPR Therapeutics for their current pre-clinical CRISPR RNA therapy137 along with their mRNA therapy. Companies typically specialize in one type of RNA but treat multiple diseases. All but two of the top 15 RNA medicine companies cover multiple diseases. Sarepta specializes in neurological and neuromuscular diseases and NOXXON has supported multiple diseases in the past (Figure 12) but is now dedicated exclusively to the treatment of cancer.
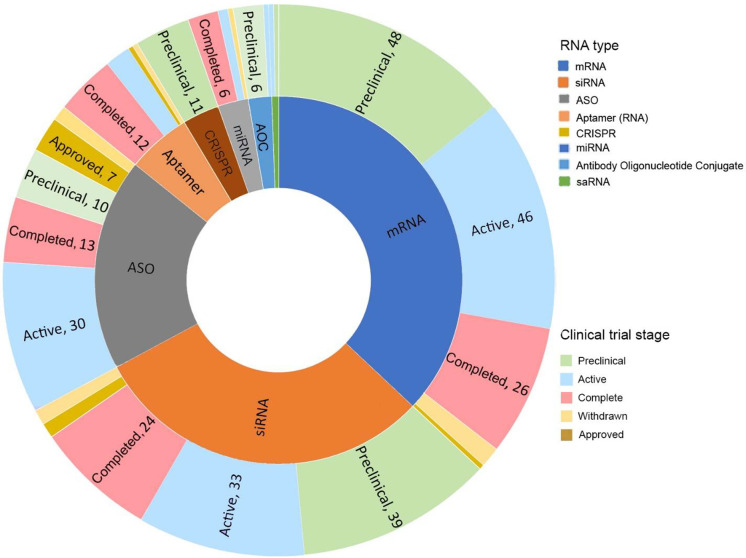
To investigate the research trends among different types of RNA therapeutics, the collected RNA therapeutics were further grouped by their types and development stages (Figures 13 and 14). The newer RNA therapeutics, such as AOCs and CRISPR, often have higher numbers of pre-clinical trials, indicating a great potential for future drug approval. In contrast, the more established types of RNA therapeutics, such as ASO and siRNA, often have a higher percentage of therapeutics on the market and a higher percentage of active and completed clinical trials, suggesting a shifting of research focus on the early development pipeline.
Figure 13.
Counts of potential therapeutics and vaccines in different stages of development (pre-clinical, clinical, completed, withdrawn, and approved) for the various types of RNA. A full list of collected clinical trials is provided in Table S2.
Figure 14.
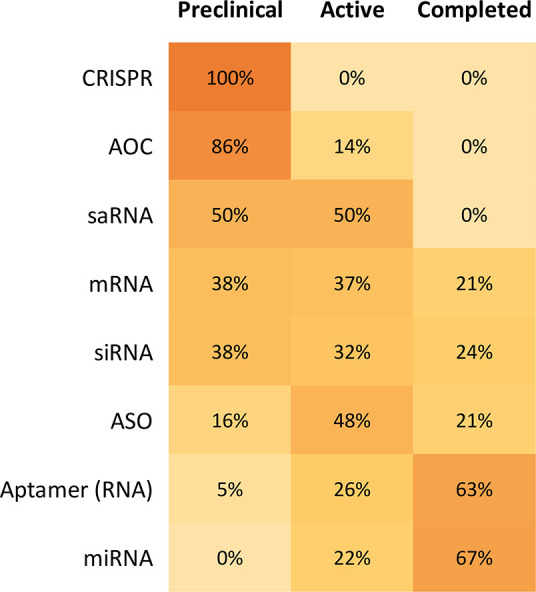
Percentage of pre-clinical, active, and completed clinical trials by RNA type. Figure rows may not sum to 100% because approved and withdrawn clinical trials are not included in this figure.
Disease-Specific RNA Therapeutics and Vaccines
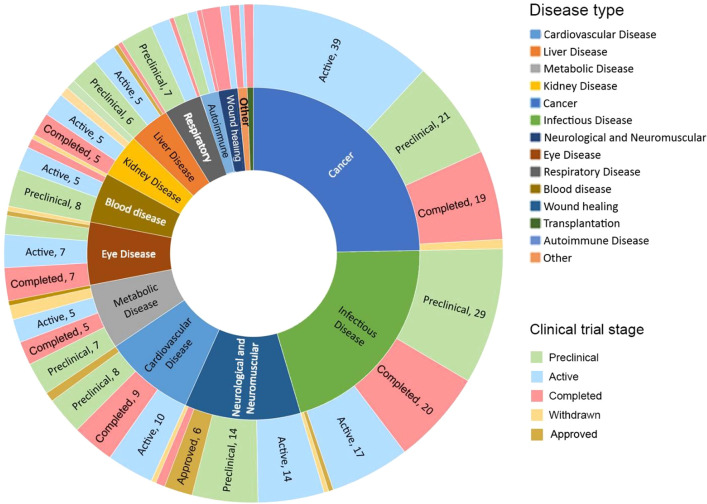
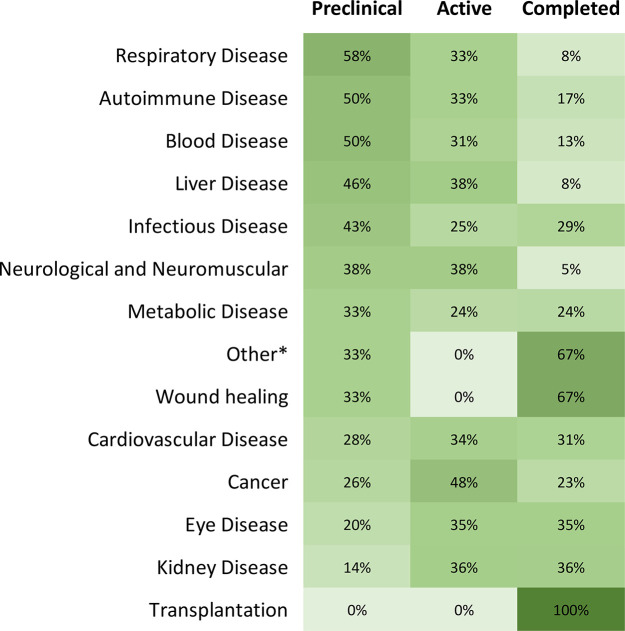
To further assess the pipeline dynamics, the above collected RNA therapeutics (Table S2) were then categorized based on their targeting diseases and development status (Figure 15). Cancer has attracted the highest number of therapeutics and vaccines in the research phase, with infectious diseases in second place. Neurological and neuromuscular diseases have the most approved treatments on the market, followed by cardiovascular and infectious diseases. The COVID-19 pandemic quickly catapulted RNA therapeutics for infectious diseases in both the research phase and approved vaccines to the forefront. While diseases such as familial hypercholesterolemia and DMD have approved RNA therapeutics, blood diseases, cancers, and respiratory diseases currently do not have any approved RNA treatments. Respiratory disease, autoimmune disease, and blood diseases have the highest percentage of therapeutics in pre-clinical trials but so far have no approved treatments. Figure 16 shows the percentages of RNA therapeutics in various development stages based on disease type, revealing places with high activities, as well as places needing more attention.
Figure 15.
Counts of potential therapeutics and vaccines in different development stages (pre-clinical, clinical, completed, withdrawn, and approved) for various disease types. A full list of collected clinical trials is provided in Table S2.
Figure 16.
Percentage of pre-clinical, active, and completed clinical trials by disease type. Figure rows may not sum to 100% because approved and withdrawn clinical trials are not included in this figure. *Other: acromegaly, alcohol use disorder, and hereditary angioedema.
Disease types along with their RNA therapeutics are examined and summarized below (Tables 1–11). These tables and corresponding text are not exhaustive and highlight promising therapeutics in the development stages from the collected RNA therapeutics (Table S2).
Table 1. RNA Therapies for Cardiovascular Diseases.
| cardiovascular disease | drug name/lab code | type of RNA | target | company | development stage | clinical trial number |
|---|---|---|---|---|---|---|
| familial hypercholesterolemia | Leqvio | siRNA | proprotein convertase subtilisin/kexin type 9 | Alnylam/Novartis | FDA approval in 2021139 | |
| cardiovascular disease with high LpA | SLN360 | siRNA | lipoprotein A | Silence Therapeutics | phase I | NCT04606602141 |
| hypertension | Zilebesiran | siRNA | angiotensinogen | Alnylam | phase I | NCT03934307147 |
| ischemic heart disease | AZD8601 | mRNA | vascular endothelial growth factor-A | Moderna/AstraZeneca | phase II | NCT03370887148 |
Table 11. RNA Therapies for Other Diseases.
| other disease | drug name/lab code | type of RNA | target | company | development stage | clinical trial number |
|---|---|---|---|---|---|---|
| keloid scarring | STP705 | siRNA | transforming growth factor beta 1/cyclooxygenase-2 | Sirnaomics | phase II | NCT04844840244 |
| autoimmune diseases | mRNA-6231 | mRNA | interleukin-2 | Moderna | phase I | NCT04916431251 |
| alcohol use disorder | DCR-AUD | siRNA | aldehyde dehydrogenase 2 | Dicerna Pharmaceuticals | phase I | NCT05021640252 |
Cardiovascular diseases, which account for 32% of all deaths, are the leading cause of death worldwide, taking an estimated 17.9 million lives each year.138 Cardiovascular diseases include disorders of the heart and blood vessels. Over 80% of cardiovascular disease deaths are due to heart attacks and strokes, and one-third of these deaths occur prematurely in people under 70 years of age.138
Leqvio is an approved therapeutic for the treatment of the cardiovascular disease, familial hypercholesterolemia (Table 1). Leqvio requires only two doses per year, and it received US FDA approval very recently in December 2021.139 It is a trivalent N-acetylgalactosamine (GalNAc)-conjugated siRNA with three GalNAc molecules clustered and conjugated to one siRNA molecule.140
In addition to this approved one, there are several more promising RNA therapeutics for cardiovascular diseases currently in the clinical trial stages (Table 1). Silence Therapeutics is evaluating the siRNA product SLN360 in a phase I clinical trial for its safety, tolerance, and pharmacodynamic and pharmacokinetic response in individuals with high lipoprotein A (LpA) cardiovascular disease.141 By targeting the LPA gene, SLN360 lowers the levels of LpA, decreasing the risk of heart disease, heart attacks, and strokes.131 Alnylam is also evaluating the siRNA product Zilebesiran that targets angiotensinogen for sustained reduction of hypertension.142 Interim phase I results show >90% reduction in serum angiotensinogen (AGT) for 12 weeks at a dosage of 100 mg or greater given quarterly or biannually.143 This dosing regimen and the continued efficacy and safety of the drug are being evaluated in a phase II study initiated in June 2021 (NCT04936035).143 Zilebesiran also uses Alnylam’s trivalent GalNAc-conjugated siRNA delivery platform.140 AstraZeneca and Moderna are collaborating on the mRNA drug AZD8601 to treat ischemic heart disease.144 AZD8601 targets vascular endothelial growth factor-A (VEGF-A).145 When AZD8601 is injected into the epicardium, VEGF-A is produced close to the damaged heart muscle, allowing cardiac regeneration.146
Metabolic diseases affect over a billion people worldwide by causing too much or too little of essential substances in the body.149 Diabetes alone affects 422 million people worldwide, causing 1.5 million deaths annually.150 IONIS-GCGRRx is an ASO product developed by Ionis Pharmaceuticals that treats diabetes by reducing the production of the glucagon receptor (GCGR) (Table 2).151 Glucagon is a hormone that opposes the action of insulin and stimulates the liver to produce glucose, particularly in patients with type-2 diabetes.152 Waylivra, another ASO product by Ionis Pharmaceuticals targeting apolipoprotein C-III (apoC-III), received European Union (EU) approval in 2019 as a treatment for familial chylomicronemia syndrome (FCS) (Table 2).153 FCS is a disease that prevents the body from breaking down consumed triglycerides. ApoC-III protein, which is produced in the liver, regulates plasma triglyceride levels in FCS patients, and Waylivra (volanesorsen) reduces its mean plasma levels.154
Table 2. RNA Therapies for Metabolic Diseases.
| metabolic disease | drug name/lab code | type of RNA | target | company | development stage | clinical trial number |
|---|---|---|---|---|---|---|
| diabetes | IONIS-GCGRRx | ASO | glucagon receptor GCGR | Ionis Pharmaceuticals | phase II | NCT01885260161 |
| familial chylomicronemia syndrome | Waylivra | ASO | apolipoprotein C-III | Ionis Pharmaceuticals | EU approval in 2019153 | |
| alpha-1 antitrypsin deficiency | ARO-AAT | siRNA | mutant of α1-antitrypsin | Arrowhead Pharmaceuticals | phase II | NCT03946449162 |
| alpha-1 antitrypsin deficiency | unnamed | CRISPR | precise correction of E342K mutation | Beam Therapeutics | pre-clinical157 | |
| methylmalonic acidemia | mRNA-3705 | mRNA | mitochondrial enzyme methylmalonic-CoA mutase | Moderna | phase I/II | NCT04899310163 |
Alpha-1 antitrypsin deficiency (AATD) is a hereditary metabolic disease. Alpha-1 antitrypsin (AAT) is a glycoprotein produced in the liver that travels through the bloodstream to protect the lungs from inflammation.155 Mutations in the SERPINA1 gene cause a deficiency of AAT in the blood, leading to toxic effects in the lungs and the accumulation of high levels of AAT in the liver that cause liver damage.155 ARO-ATT, a siRNA product developed by Arrowhead Pharmaceutical, targets the SERPINA1 mutation, and in pre-clinical studies has shown promise in reducing AAT liver disease (Table 2).156 Beam Therapeutics is developing a CRISPR base editing drug to treat AATD by correcting the E342 K mutation in the SERPINA1 gene.157 Beam’s base editor has two main components, a CRISPR protein bound to a guide RNA and a base editing enzyme, which are fused to form a single protein.158 This fusion allows the precise targeting and editing of a single base pair of DNA, which has not been previously achieved.158 Repairing the mutation would restore normal gene function and eliminate abnormal AAT production. This editing system uses a non-viral lipid nanoparticle delivery system.157
Methylmalonic acidemia (MMA) is a hereditary metabolic disease in which the body is unable to metabolize certain proteins and lipids correctly.159 MMA is caused by mutations in the MMUT, MMAA, MMAB, MMADHC, and MCEE genes. The mutation in the MMUT gene accounts for about 60% of MMA cases. Moderna has developed an mRNA therapeutic mRNA-3705 that targets the MMUT mutation (Table 2).144 mRNA-3705 instructs the cell to restore the missing or dysfunctional proteins that cause MMA. mRNA-3705 entered clinical trials with the first patient treated in August 2021.160
Liver diseases affect more than 1.5 billion people worldwide164 and account for over 2 million deaths per year.165 The siRNA therapeutic GIVLAARI, developed by Alnylam, received FDA approval in 2019 for the treatment of acute hepatic porphyria (Table 3).142 Acute hepatic porphyria is a genetic disease characterized by life-threatening acute attacks and chronic pain.166 GIVLAARI is the first approved GalNAc-conjugated RNA therapeutic.167
Table 3. RNA Therapies for Liver Diseases.
| liver disease | drug name/lab code | type of RNA | target | company | development stage | clinical trial number |
|---|---|---|---|---|---|---|
| acute hepatic porphyria | GIVLAARI | siRNA | 5-aminolevulinic acid synthase 1 mRNA | Alnylam | FDA approval in 2019170 | |
| non-alcoholic fatty liver disease | ION839/AZD2693 | ASO | patatin-like phospholipase domain-containing protein 3 | Ionis/AstraZeneca | phase I | NCT04483947168 |
Non-alcoholic steatohepatitis (NASH) is an accumulation of fat in the liver that causes liver damage. The antisense therapeutic ION839/AZD2693 from Ionis/AstraZeneca entered phase I trials (Table 3) in 2020 in patients with NASH and fibrosis.168 ION839/AZD2693 targets patatin-like phospholipase domain-containing 3 (PNPLA3), reducing its expression.151 Mutation of PNPLA3, which produces a protein that accumulates on the surface of intracellular lipid droplets, is strongly associated with an increased risk for NASH.169
Cancer includes a large group of diseases that are characterized by abnormal cell growth in various parts of the body. It is the second leading cause of death globally, accounting for an estimated 10 million deaths per year with over 19 million new cases diagnosed in 2020.171 Currently there are no approved RNA therapeutics for cancer treatment. NOXXON Pharma’s lead aptamer candidate NOX-A12 is in development as a combination therapy for multiple types of cancers (Table 4).172 It is intended to enhance other anti-cancer treatments without side effects. A phase I/II trial of NOX-A12 in combination with radiotherapy in newly diagnosed brain cancer patients who would not benefit from standard chemotherapy is ongoing.172 Interim data from the study in June 2021 showed tumor reduction in five of six patients, consistent with an anti-cancer immune response, and there were no unexpected adverse events.172 NOXXON is collaborating with Merck in their pancreatic cancer program.172 NOXXON, in a phase I/II combination trial with Merck’s Keytruda, reported success in treating metastatic pancreatic and colorectal cancer and entered a second collaboration with Merck to conduct a phase II study in pancreatic cancer patients.173 NOX-A12 targets C-X-C motif chemokine ligand 12 (CXCL12),172 which, with its receptors, acts as a link between tumor cells and their environment, promotes tumor proliferation, new blood vessel formation, and metastases, and inhibits cell death.174
Table 4. RNA Therapies for Cancers.
| cancer | drug name/lab code | type of RNA | target | company | development stage | clinical trial number |
|---|---|---|---|---|---|---|
| brain cancer/glioblastoma | NOX-A12 | aptamer (RNA) | C-X-C motif chemokine ligand 12 | NOXXON Pharma | phase I/II | NCT04121455179 |
| pancreatic cancer | NOX-A12 | aptamer (RNA) | C-X-C motif chemokine ligand 12 | NOXXON Pharma | phase II | NCT04901741180 |
| solid tumor | STP707 | siRNA | transforming growth factor beta, cyclooxygenase-2 | Sirnaomics | phase I | NCT05037149181 |
| multiple myeloma | Descartes-11 | mRNA | B-cell maturation antigen | Cartesian Therapeutics | phase I/II | NCT03994705182 |
The anti-cancer siRNA product, STP707 by Sirnaomics,175 targets TGF-β1 and COX-2 mRNAs (Table 4). A pre-clinical study demonstrated that knocking down TGF-β1 and COX-2 gene expression simultaneously in the tumor microenvironment increases active T cell infiltration and combining the two siRNAs produces a synergistic effect that diminishes pro-inflammatory factors.176 Descartes-11 developed by Cartesian is a CAR T-cell therapy for treating multiple myeloma, a white blood cell cancer that affects plasma cells (Table 4).177 It recently completed (March 2022) phase II studies with newly diagnosed patients.178 Descartes-11 contains autologous CD8+ T cells engineered with RNA chimeric antigen receptors (CARs) that bind to B-cell maturation antigen (BCMA).177 BCMA is highly expressed in all myeloma cells, and Descartes-11 binds and destroys BCMA-positive myeloma cells.177
Infectious diseases are caused by bacteria, viruses, fungi, or parasites.
SARS-CoV-2 has infected over 549 million people, causing over 6.1 million deaths worldwide.183 The COVID-19 pandemic brought the first approved mRNA vaccine to market. BioNTech/Pfizer’s COVID-19 BNT162/Comirnaty vaccine was given U.S. FDA emergency use authorization in 2020 and approved by the FDA in 2021 (Table 5).184 Moderna’s COVID-19 mRNA-1273/Spikevax vaccine was also given US FDA emergency use authorization in 2020 and approved by the FDA in 2022 (Table 5).185 Globally over 11.39 billion COVID-19 vaccine doses have been administered.186
Table 5. RNA Therapies and Vaccines for Infectious Diseases.
| infectious disease | drug name/lab code | type of RNA | target | company | development stage | year (first posted) | clinical trial number |
|---|---|---|---|---|---|---|---|
| COVID-19 | BNT162/Comirnaty | mRNA | SARS-CoV-2 spike protein | BioNTech/Pfizer | FDA approval in 2021184 | ||
| COVID-19 | mRNA-1273/Spikevax | mRNA | SARS-CoV-2 spike protein | Moderna | FDA approval in 2022185 | ||
| hepatitis B viral | AB-729 | siRNA | hepatitis B viral surface antigen | Arbutus Biopharma Corporation | phase I | 2021 | NCT04775797192 |
| influenza | mRNA-1010 | mRNA | Moderna | phase I/II | 2021 | NCT04956575193 | |
| respiratory syncytial virus | unnamed | mRNA | CureVac | pre-clinical137 | |||
| respiratory syncytial virus | mRNA-1345 | mRNA | Moderna | phase I | 2021 | NCT04528719194 |
Arbutus Biopharma Corporation developed AB-729, an RNA interference (RNAi) therapeutic specifically designed to reduce all hepatitis B virus (HBV) antigens, including hepatitis B surface antigen (HBsAg) (Table 5).187 HBsAg interferes with host immune response,188 and preliminary data indicate that long-term suppression of HBsAg with AB-729 results in an increased HBV-specific immune response.187
Moderna’s first quadrivalent seasonal influenza mRNA vaccine candidate mRNA-1010 is in phase I/II trials (Table 5).144 mRNA-1010 targets influenza lineages recommended by the World Health Organization (WHO) for the prevention of influenza, including seasonal influenza A H1N1 and H3N2 and influenza B Yamagata and Victoria.189 CureVac has a mRNA prophylactic vaccine for respiratory syncytial virus (RSV), a common respiratory virus that can cause serious illness in infants and older adults, in their pipeline under pre-clinical development (Table 5).137 Moderna has also developed a vaccine for RSV.144 Moderna’s mRNA-1345 (Table 5) is currently in a phase I trial to evaluate its tolerance and reactogenicity in children, younger adults, and older adults.190,191
Neuromuscular diseases affect the function of muscles and nerves that communicate sensory information to the brain.195 They affect the brain as well as the nerves found throughout the body and the spinal cord.196 These types of diseases have the greatest number of approved RNA therapeutics. Approved treatments for Duchenne muscular dystrophy include Sarepta’s ASO therapeutics Exondys 51, which targets exon 51 of the dystrophin gene, receiving FDA approval in 2016,197 Vyondys 53, which targets exon 53 of the dystrophin gene, receiving FDA approval in 2019,198 and NS Pharma’s Viltepso, also targeting exon 53 of the dystrophin gene, receiving FDA approval in 2020 (Table 6).199 Ionis Pharmaceuticals received FDA approval in 2016 for ASO therapeutic Spinraza targeting survival of motor neuron 2 (SMN2) for the treatment of spinal muscular atrophy (Table 6).200 Their second therapeutic approved by the FDA in 2018 was the ASO neurological therapeutic Tegsedi, which targets hepatic production of transthyretin (TTR) and is used for the treatment of hATTR amyloidosis-polyneuropathy, a disease due to mutations in the gene encoding TTR that leads to abnormal amyloid deposits on nerves.201 Onpattro by Alnylam is another approved RNA drug for the treatment of hATTR amyloidosis-polyneuropathy by targeting hepatic production of transthyretin (Table 6).202 Onpattro, which was FDA approved in 2018 (Table 6), was the first siRNA drug.26 Sarepta Therapeutics has two other ASO therapeutics for the treatment of DMD, SRP-5044 and SRP-5050 (targeting exon 44 and exon 50 of the dystrophin gene, respectively), that are in pre-clinical studies (Table 6).203
Table 6. RNA Therapies for Neuromuscular and Neurological Diseases.
| neurological and neuromuscular disease | drug name/lab code | type of RNA | target | company | development stage | clinical trial number |
|---|---|---|---|---|---|---|
| Duchenne muscular dystrophy | Exondys 51 | ASO | exon 51 dystrophin | Sarepta Therapeutics | FDA approval in 2016197 | |
| Duchenne muscular dystrophy | Vyondys 53 | ASO | exon 53 dystrophin | Sarepta Therapeutics | FDA approval in 2019198 | |
| Duchenne muscular dystrophy | Viltepso | ASO | exon 53 dystrophin | NS Pharma | FDA approval in 2020199 | |
| spinal muscular atrophy | Spinraza | ASO | survival of motor neuron 2 mRNA | Ionis Pharmaceuticals | FDA approval in 2016200 | |
| hATTR amyloidosis-polyneuropathy | Tegsedi | ASO | transthyretin mRNA | Ionis Pharmaceuticals | FDA approval in 2018201 | |
| hATTR amyloidosis-polyneuropathy | Onpattro | siRNA | transthyretin mRNA | Alnylam | FDA approval in 2018207 | |
| amyotrophic lateral sclerosis | Tofersen | ASO | superoxide dismutase 1 | Ionis Pharmaceuticals/Biogen | phase III | NCT04856982208 |
| Duchenne muscular dystrophy | SRP-5044 | ASO | exon 44 dystrophin | Sarepta Therapeutics | pre-clinical203 | |
| Duchenne muscular dystrophy | SRP-5050 | ASO | exon 50 dystrophin | Sarepta Therapeutics | pre-clinical203 | |
| myotonic dystrophy type 1 | AOC 1001 | antibody–oligonucleotide conjugate | myotonic dystrophy protein kinase | Avidity Biosciences | phase I/II | NCT05027269209 |
| Duchenne muscular dystrophy | AOC 1044 | antibody–oligonucleotide conjugate | exon 44 dystrophin | Avidity Biosciences | pre-clinical205 |
Amyotrophic lateral sclerosis (ALS) is a progressive neurodegenerative disease that affects nerve cells in the brain and spinal cord.204 Ionis Pharmaceuticals in partnership with Biogen has an ASO investigational drug, Tofersen, in phase III clinical trials (Table 6).151 Tofersen targets superoxide dismutase 1 (SOD1), the second most common and best understood genetic cause of ALS.
Avidity Biosciences has a pipeline of antibody oligo-nucleotide conjugate (AOC) therapeutics focused on neuromuscular diseases. Their leading candidate, AOC 1001, is a siRNA conjugated with a monoclonal antibody (mAb) (Table 6).205 AOC 1001 has an ongoing phase I/II trial in adults with myotonic dystrophy type 1 (DM1). DM1 is a progressive neuromuscular disease that impacts skeletal and cardiac muscle. DM1 is caused by an abnormal number of CUG triplet repeats in the myotonic dystrophy protein kinase gene (DMPK), reducing muscleblind-like protein (MBNL) activity and disrupting muscle development.206 AOC 1001 is designed to reduce DMPK levels and CUG triplet repeats so that MBNL can perform normally.205 Avidity Biosciences has also developed an AOC to treat DMD.205 The oligonucleotides are designed to promote skipping of specific exons to produce dystrophin in patients with DMD. Their leading DMD drug candidate, AOC 1044, can induce exon skipping specifically for exon 44 (Table 6), and clinical trials are planned for 2022.205
Eye diseases including visual impairment affect 2.2 billion people globally.210 Macular degeneration, which causes loss in the center of the field of vision and is irreversible, affects 196 million people.211 Macugen, developed by Gilead Sciences (Table 7), was the first FDA-approved (2014) RNA aptamer treatment for neovascular age-related macular degeneration.212 Macugen targets the Vascular endothelial growth factor (VEGF) protein in the eye, reducing the growth of blood vessels to control the leakage and swelling that cause vision loss.213 As age-related macular degeneration (AMD) progresses, geographic atrophy (GA), a chronic progressive degeneration of the macula, can develop.214 Zimura by Iver Bio is another RNA aptamer therapeutic and is currently in a phase III clinical trial (Table 7). Zimura inhibits complement component 5,215 which is involved in the development and progression of AMD. In clinical trials, Zimura slowed the progression of GA over 12 months in individuals with age-related macular degeneration.214 QR-504a from ProQR Therapeutics is an investigational RNA ASO therapeutic that is in ongoing phase I/II clinical trials.216 QR-504a is designed to slow vision loss in individuals with Fuchs endothelial corneal dystrophy (FECD) which results from trinucleotide repeat expansion mutations in the Transcription factor 4 (TCF4) gene.217
Table 7. RNA Therapies for Eye Diseases.
| eye disease | drug name/lab code | type of RNA | target | company | development stage | clinical trial number |
|---|---|---|---|---|---|---|
| macular degeneration | Macugen | aptamer (RNA) | vascular endothelial growth factor protein | Gilead Sciences | FDA approval in 2014212 | |
| geographic atrophy related to age-related macular degeneration | Zimura | aptamer (RNA) | complement component 5 | IVERIC Bio | phase III | NCT04435366215 |
| Fuchs endothelial corneal dystrophy | QR-504a | ASO | transcription factor 4 | ProQR Therapeutics | phase I/II | NCT05052554216 |
Kidney disease affects over 850 million people worldwide.218 Primary hyperoxaluria type 1 (PH1) is a genetic kidney disease characterized by the overproduction of oxalate, which leads to kidney stones, kidney failure, and systemic oxalosis.218 The siRNA therapeutic Oxlumo was developed by Alynlam and approved by the FDA in 2020 (Table 8).219 Oxlumo targets the hydroxy acid oxidase 1 (HAO1) gene, which encodes glycolate oxidase.219 Oxlumo reduced urinary oxalate excretion in patients with progressive kidney failure in PH1.219,220 The majority of patients had normal or near-normal levels of oxalate after 6 months of treatment. Autosomal dominant polycystic kidney disease (ADPKD) is caused by mutations in the PKD1 or PKD2 gene.221 It is characterized by cysts within the kidneys, often leading to kidney failure. To treat ADPKD, Regulus designed RGLS4326, a second-generation oligonucleotide that inhibits miR-17 (Table 8), which produces kidney cysts. RGLS4326 binds to miR-17 microRNAs, inhibits miR-17 activity, and reduces disease progression.222 Immunoglobulin A (IgA) nephropathy is a chronic kidney disease that is caused by deposits of protein IgA inside the glomeruli of the kidneys.223 Overproduction of complement factor B (FB) is associated with increased IgA nephropathy. Ionis has partnered with Roche to develop a ligand-conjugated ASO therapeutic, IONIS-FB-LRx, which targets FB to reduce the production of IgA and alleviate the symptoms of IgA nephropathy (Table 8).151
Table 8. RNA Therapies for Kidney Diseases.
| kidney disease | drug name/lab code | type of RNA | target | company | development stage | clinical trial number |
|---|---|---|---|---|---|---|
| primary hyperoxaluria type 1 | Oxlumo | siRNA | hydroxy acid oxidase 1 | Alnylam | FDA approval in 2020 | NCT04152200224 |
| autosomal dominant polycystic kidney disease | RGLS4326 | oligonucleotide | miR-17 | Regulus | phase I | NCT04536688225 |
| IGA nephropathy | IONIS-FB-LRx | ASO | complement factor B | Ionis/Roche | phase II | NCT04014335226 |
Respiratory diseases affect more than 1 billion people worldwide.227 COVID-19 brought acute respiratory distress syndrome (ARDS) to the forefront of medical news during the pandemic. ARDS is a severe inflammatory lung disease with a mortality rate of over 40%.228 Inflammation leads to lung tissue injury and leakage of blood and plasma into air spaces, resulting in low oxygen levels and often requiring mechanical ventilation.228 Descartes-30 is an engineered mRNA cell therapy product by Cartesian Therapeutics (Table 9). It comprises human mesenchymal stem cells that secret two human DNases for degrading ARDS-causing neutrophil extracellular traps (NETS).177 Descartes-30 is currently in phase I/II clinical trials.229
Table 9. RNA Therapies for Respiratory Diseases.
| respiratory disease | drug name/lab code | type of RNA | target | company | development stage | clinical trial number |
|---|---|---|---|---|---|---|
| acute respiratory distress syndrome | Descartes-30 | mRNA | neutrophil extracellular traps | Cartesian Therapeutics | phase I/II | NCT04524962229 |
| cystic fibrosis | MRT5005 | mRNA | cystic fibrosis transmembrane conductance regulator | Translate Bio | phase I/II | NCT03375047236 |
| cystic fibrosis | ARO-ENaC | siRNA | epithelial sodium channel α subunit | Arrowhead Pharmaceuticals | phase I/IIa | NCT04375514237 |
Cystic fibrosis is caused by a dysfunctional cystic fibrosis transmembrane conductance regulator (CFTR) protein, resulting from mutations in the CFTR gene.230 Without CFTR, mucus in various organs including the lungs is extremely thick and sticky. MRT5005, developed by Translate Bio, is an mRNA therapeutic (Table 9) that bypasses this mutation by delivering mRNA encoding a fully functional CFTR protein to the cells in the lungs through nebulization.231 Although initial interim results from clinical studies showed promise,232 results from the second interim phase I/II clinical trial showed no increase in the lung function of individuals receiving MRT5005.233 The siRNA therapeutic ARO-ENaC is designed by Arrowhead Pharmaceuticals to reduce epithelial sodium channel alpha subunit (αENaC) in the lungs and airways (Table 9). Increased ENaC contributes to airway dehydration and increased mucus.234 However, Arrowhead paused a phase I/II study of ARO-ENaC in July 2021 after safety studies showed local lung inflammation in rats.235
Blood diseases affect one or more components of blood. Sickle cell disease is a type of blood disease that causes red blood cells to become misshapen and break down.238 Beta-thalassemia syndromes are a group of blood diseases that result in reduced levels of hemoglobin in red blood cells.239 BEAM-101 (Table 10), Beam Therapeutics leading ex vivo base editor, is a patient-specific, autologous hematopoietic investigational cell therapy. BEAM-101 introduces base edits that mimic the single nucleotide polymorphisms found in individuals with hereditary persistence of fetal hemoglobin (HPFH), which could alleviate the effects of mutations causing sickle cell disease or beta-thalassemia since the fetal hemoglobin does not become misshapen.240 Beam plans to initiate a phase I/II clinical trial to assess the safety and efficacy of BEAM-101 for the treatment of sickle cell disease.240 BEAM-101 uses an electroporation delivery system.157 Silence Therapeutics has RNA therapeutics for blood diseases such as thalassemia, myelodysplastic syndrome, and rare iron-loading anemias.241 Their siRNA therapeutic SLN124 targets transmembrane serine protease 6 (TMPRSS6) (Table 10), a gene that prevents the liver from producing hepcidin.241 Phase I clinical trial data showed that SLN124 improved red blood cell production and reduced anemia by increasing the levels of hepcidin.241,242
Table 10. RNA Therapies for Blood Disease.
| blood disease | drug name/lab code | type of RNA | target | company | development stage | clinical trial number |
|---|---|---|---|---|---|---|
| sickle cell disease/beta-thalassemia | BEAM-101 | CRISPR | fetal hemoglobin activation | Beam Therapeutics | pre-clinical157 | |
| thalassemia/low-risk myelodysplastic syndrome | SLN124 | siRNA | transmembrane serine protease 6 | Silence Therapeutics | phase I | NCT04718844243 |
Keloid scarring is a type of pathological condition that forms abnormally thick scarring after a skin injury. STP705, from Sirnaomics, is a siRNA therapeutic in phase II clinical trial for the treatment of keloid scars (Table 11).244 STP705 targets both TGF-β1 and COX-2 gene expression with polypeptide nanoparticle-enhanced delivery.245 The synergistic effect of simultaneous silencing TGF-β1 and COX-2 may reverse skin fibrotic scarring by decreasing inflammation and activating fibroblast apoptosis. This mechanism of action of STP705 can be widely applied for the treatment of other fibrotic conditions.245
Autoimmune diseases are characterized by immune activation in response to normal antigens.246 mRNA-6231 from Moderna is an mRNA encoding IL-2 mutein (Table 11) that activates and expands the regulatory T cell population and dampens self-reactive lymphocytes to help restore normal immune function.247
Alcohol use disorder (AUD) is the inability to control alcohol use despite adverse social, occupational, or health consequences. According to the World Health Organization, AUD affects over 283 million people globally.248 Alcohol is metabolized in the liver to acetaldehyde via alcohol dehydrogenase and then to acetic acid by aldehyde dehydrogenase 2 (ALDH2).249 Inhibiting ALDH2 results in unpleasant symptoms due to the increased acetaldehyde when alcohol is not fully metabolized.250 The siRNA therapeutic DCR-AUD by Dicerna knocks down ALDH2 protein expression (Table 11) in the liver, which results in increased acetaldehyde levels that discourage continued alcohol use in AUD patients.250
Chemical Modifications for Improving RNA Stability and Target Specificity
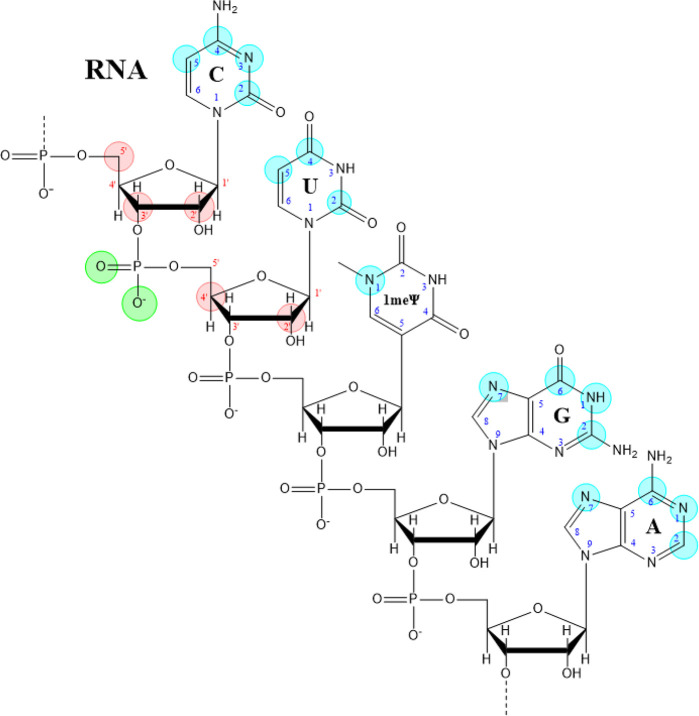
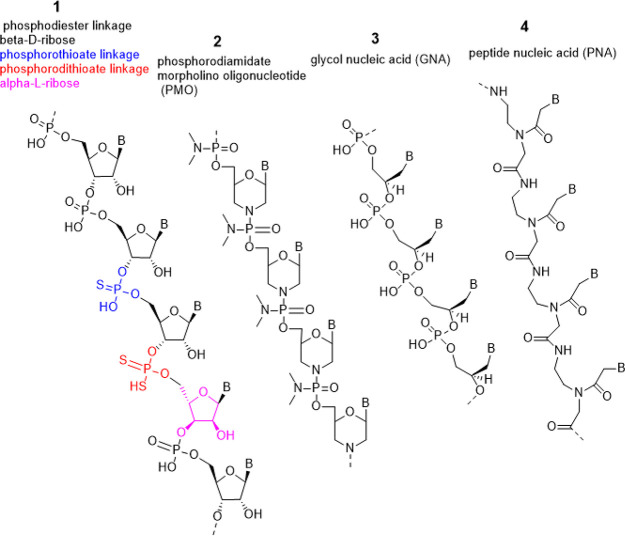
RNA is composed of nucleosides that consist of a nucleic acid base attached to a d-ribose through a β-N-glycosyl bond between the ribose and a pyrimidine base (uracil and cytosine) or a purine base (adenosine and guanosine). The nucleosides are connected by a phosphodiester bond using the phosphate between the 3′ and 5′ carbons on adjacent ribose molecules (Figure 17).253 RNAs can be modified on the nucleic acid base and on the phosphate and the ribose of the sugar–phosphate backbone, as illustrated in Figure 17 with color circles.
Figure 17.
An example of RNA structure and modification sites. Green circles, modification sites on the phosphate; red circles, attachment sites for modifications on the ribose; blue circles, attachment sites for modifications on the nucleic acid base: 1-methylpseudouridine (1meΨ), cytosine (C), uridine (U), guanosine (G), and adenosine (A).
The nucleic acid side chains extending from the sugar–phosphate backbone form hydrogen bonds between the nucleic acid bases of complementary RNA chains; U bonds with A, and G bonds with C. Double-stranded RNA structures form as a result of intramolecular and intermolecular base pairing. Base pairing between loops of double-stranded stem-loops can yield 3D structures, and triple helixes can form within single strands or between multiple strands.
Chemical modifications on RNA can improve the stability and reduce the immunogenicity of therapeutic RNAs. RNA is very susceptible to nucleases and hydrolysis by basic compounds. Chemical modification of RNA protects the vulnerable sugar–phosphate backbone from nuclease degradation and lowers the risk of off-target effects. For RNAs that form a duplex with a target sequence, mutations that lower the melting temperature of the duplex destabilize the complex and improve target specificity by decreasing base pairing with non-target RNA. RNA modifications can also improve the delivery of the RNA into the cell through the plasma membrane and enhance the activity of the RNA.104
Nucleic acid base modifications
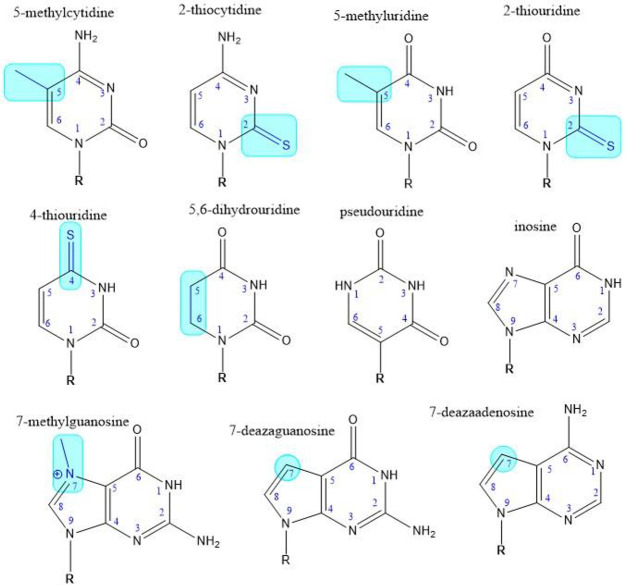
Figure 18 shows chemical structures of commonly seen base modifications and rare bases. Modifications include methylation, replacement of oxygen with sulfur, and replacement of a nitrogen within the ring system with carbon. Cytidines or uridines can be methylated at the N-5 position to be 5-methylcytidines or 5-methyluridines. Cytidines can have the oxygen replaced with a sulfur at the N-2 position to be 2-thiocytidines. When uridines have the oxygen replaced with a sulfur at either the N-2 or the N-4 position, they become 2-thiouridines or 4-thiouridines respectively. Uridines can also be reduced on the base ring and become 5,6-dihydrouridines. Another commonly seen base modification, methylation at the N-7 position of the guanosine, occurs in mRNA cap structure. The guanosines can also be modified at the N-7 position by converting the nitrogen to a carbon, and become 7-deazaguanosine. Similar modifications can happen to adenosines, resulting in 7-methyladenosines and 7-deazaadenosines. The rare bases, pseudouridine and inosine, are also seen in modified RNA sequences. Additional modified or rare bases are shown in Figure S7. According to the data in the CAS Content Collection,32 the use of RNA modifications has been increasing since 1995 (Figure S6).
Figure 18.
Examples of modified and rare bases. R = d-ribose; locations of modifications are shown in blue.
Base modifications that interfere with the formation of hydrogen bonds can thermally destabilize duplex formation with the target, and thus improve target specificity by limiting off-target binding.254 In addition, modifications improve the performance of therapeutic RNA. Replacing uridine with the modified base 1-methylpseudouridine (Figure 17) in therapeutic mRNAs, such as the COVID-19 vaccines (Pfizer’s Comirnaty and Moderna’s Spikevac), improves translation and lowers cytotoxic side effects and immune responses to the mRNA.255 Both Pfizer’s Comirnaty and Moderna’s Spikevac mRNA vaccines also use a 7-methylguanosine cap linked by a 5′-triphosphate to the 5′ end of the mRNA, replicating the naturally occurring mRNA caps that prevent degradation of the 5′ end of the mRNA.256
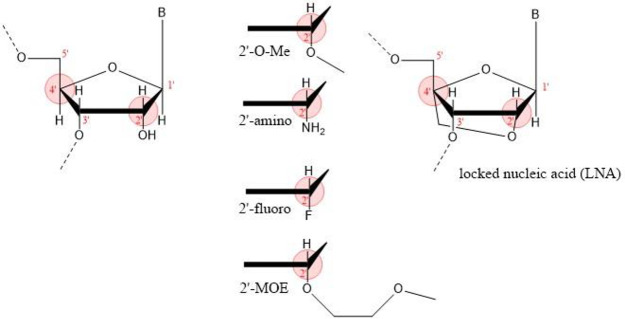
Modifications on Ribose
The hydroxyl group on the C-2′ position of the ribose destabilizes RNA compared to DNA. Modification of this hydroxyl protects against nuclease digestion and can lower the thermal stability of duplexes formed with a target RNA. Figure 19 shows the chemical structures of these modifications on ribose. The most common modifications at the C-2′ position include 2′-O-methyl, 2′-amine, 2′-fluoro, and 2′-O-methoxyethyl (2′-MOE). Another modification at the C-2′ position is the LNA in which a 2′-O,4′-C-methylene bridge connects the 2′ position to the 4′ position on the ribose.257
Figure 19.
Common modifications on the 2′-hydroxyl group of d-ribose. B = nucleic acid base.
In addition, the 5′ and 3′ ends of RNA can be modified using cleavable linkers attached to GalNAc groups or lipophilic moieties to target the therapeutic RNA to the desired tissue. The linkers are cleaved by acid pH, redox potential, or degradative enzymes in cells but not in serum or blood. Cleavable linkers include acid-cleavable groups, ester-based cleavable groups, and peptide-based cleavable groups.254
Backbone Modifications
Modifications to the phosphate group in the sugar–phosphate backbone can improve the resistance of therapeutic RNAs to extracellular and intracellular nucleases. In addition, the negative charge on the phosphate group interferes with the delivery of the RNA into the cell through the lipid bilayer membrane, which is impermeable to polar molecules. Thus, replacing the oxygens on the phosphates with neutral groups or complexing the phosphate groups to cations like sodium can improve RNA delivery.258
A widely used backbone modification, phosphorothioate, as shown in Figure 20, which replaces an oxygen in the phosphate group with sulfur, reduces the activity of extracellular and intracellular nucleases.259 RNA molecules with phosphorothioate linkages at the ends resist exonucleases, whereas RNA molecules with phosphorothioate linkages within the RNA resist endonucleases. However, the sulfur on the phosphate group creates stereogenic α-phosphorus atoms resulting in diastereomers with different functional properties that can affect duplex formation. Careful spacing of the phosphorothioate linkages within the RNA can ameliorate this problem.257,258
Figure 20.
Examples of modified RNA backbones. Backbone 1 shows phosphate–ribose backbone linkages, which include the classic phosphodiester (black), phosphorothioate (blue), and phosphorodithioate (red). The purple ribose, α-l-ribose, has an alternative stereochemistry compared to the normal β-d-ribose moieties shown in black. Backbone 2 is a phosphorodiamidate morpholino (PMO) backbone, backbone 3 is (R)-glycol nucleic acid ((R)-GNA), and backbone 4 is peptide nucleic acid (PNA). B = nucleic acid base.
Other backbone modifications replace the d-ribose with either an l-ribose or a non-ribose moiety (Figure 20). Phosphorodiamidate morpholino oligonucleotides (PMOs) contain morpholino groups linked by phosphorodiamidate groups rather than ribose linked by phosphates. Glycol nucleic acids (GNAs) have a backbone of repeating glycerol units linked by phosphodiester bonds.260 In peptide nucleic acids (PNAs) the sugar–phosphate backbone is replaced with a flexible N-(2-aminoethyl)glycine polymer with the nucleobases attached via a methylene carbonyl linkage.261 PMOs, GNAs, and PNAs all resist nuclease degradation. PNA forms duplexes with complementary DNA or RNA with higher affinity and specificity than unmodified DNA–DNA or DNA–RNA duplexes. However, duplexes containing PNAs, PMOs, and LNAs resist RNase H degradation, inhibiting gene knockdown through targeted mRNA degradation.257
Trends of RNA Chemical Modifications
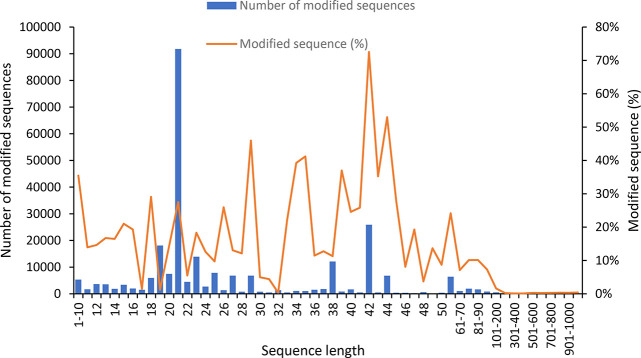
Sequence data for RNAs and RNA modifications are annotated and collected with CAS Registry Sequence Guidelines262 from published documents and stored in the CAS Content Collection.32 To better understand the chemical modification trends on RNA molecules, we extracted ∼170,000 modified RNA sequences from the CAS Content Collection. Figure 21 shows the number of the modified RNA sequences and distribution of these modified sequences along the sequence length. The predominance of modified nucleotide RNAs for lengths of 18–27 bases reflects the fact that this sequence length is commonly used in siRNAs and ASOs; processed, naturally occurring double-stranded siRNAs are typically 21 or 23 bp long. The double-stranded nature of siRNAs accounts for the large number of modifications for nucleotides with a length of 42 and 44; two 21-nucleotide RNAs produce 42 nucleotides of RNA, while a 21-nucleotide and a 23-nucleotide RNA produce 44 nucleotides. The percentage of modified RNA sequences in the total RNA sequences was also shown along the sequence length, suggesting that sequences less than 100 base pairs are more frequently being modified than the longer sequences. This drop-off observed in RNA modification for RNAs over 100 nt is likely an artifact of the methods used to synthesize artificial RNA and the methods used to sequence naturally occurring RNAs. Small artificial RNAs are produced by solid-support synthesis or other chemical methods; larger RNAs are produced by in vitro transcription. Chemical synthesis has more capacity to add modified nucleotides to an RNA; in vitro transcription faces the limitations posed by the use of enzymes. Furthermore, naturally occurring RNA sequences are generally characterized by reverse transcription to cDNA, amplification, and then sequencing. This process would obscure modification present on the initial RNA.
Figure 21.
RNA sequences containing modifications and their distribution with respect to sequence lengths (from the CAS Content Collection). Blue bars: absolute number of modified RNA sequences; orange line: percentage of modified RNA sequences in the total RNA sequences with same sequence length.
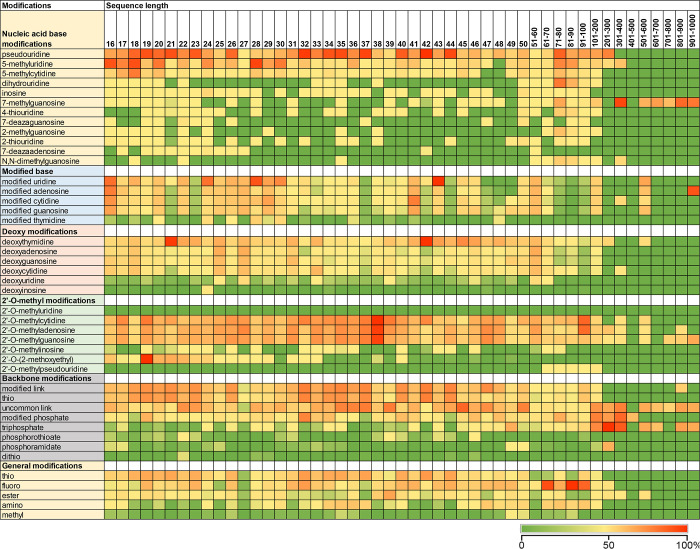
Chemical modifications were further analyzed using this data set with specific types of modifications and sequence lengths. Out of 145 curated sequence modifications, 117 sequence modifications were identified in the modified RNA data set. A heatmap (Figure 22) covering the most popular modifications was constructed based on the relative frequencies of specific types of modification in the total modification events in that sequence length. The heat map of modification type versus sequence length shows a sharp change in the types of modifications in sequences <200 nucleotides vs >200 nucleotides. Sequences >200 nucleotides are modified much less than shorter sequences. This set of longer RNAs, which includes lncRNAs and mRNAs, is either transcribed in vivo or produced using in vitro transcription. The most common modifications contained in the longer sequences are triphosphates and 7-methylguanosines, suggesting that they are mRNAs with 5′ end caps consisting of 7-methylguanosine linked to the 5′ end of the mRNA with a triphosphate group. In addition, modified adenosine is observed in longer sequences, particularly in sequences from 901 to 1000 nucleotides; this would include the mRNA modification, 6-methyladenosine.263,264 Since therapeutic mRNAs are translated by the ribosomes to produce an active protein, excessive modification might provide steric hindrance that inhibits translation.
Figure 22.
Frequencies of modifications of RNA and their distributions based on sequence lengths (CAS Content Collection).
Common nucleobase modifications for small RNAs (those <200 nucleotides in length) are 5-methyluridine (m5U) and 5-methylcytosine (m5C). The rare base pseudouridine appeared frequently in the database, followed by the rare bases dihydrouracil and inosine. Bases in which the N-7 position is converted to a carbon (7-deazaguanine and 7-deazaadenine) appeared with some frequency, as did the replacement by sulfur of the oxygen double-bonded to the N-4 or N-2 position of uracil or the N-2 position of cytosine. Less common modifications included benzoyl groups attached to N-3 in cytosine and the nitrogens in adenosine.
Modifications at the C-2′ ribose position are also common, with 2′-O-methyl and 2′-deoxy modifications as the most highly represented and 2′-fluoro and 2′-MOE somewhat less. The prevalence of thymidine (deoxythymidine) at sequence lengths of 21 and 42 is because thymidine at the 3′ end of artificial siRNAs protects them from exonucleases. Artificial siRNAs are predominantly 21-mers, and since they are double-stranded, they are highly represented at both 21 and 42 nucleotides.
Phosphorothioate is the most common phosphate modification for the sugar–phosphate backbone, although phosphorodithioate also occurs. Altered ribose stereochemistry, such as l-ribose in place of d-ribose, is a relatively rare modification, as are replacement backbones, such as PMO, GNA, and PNA.
Heat maps for modifications versus disease targets show little correlation between the disease and the modifications on the therapeutic RNA (Figures S3 and S4). Overall, there is little correlation between the type of RNA and the targeted diseases (Figure S1), suggesting any RNA can be explored to target any disease.
Approved RNA medicines show a correlation between the type of RNA and specific modifications (Table 12). Of the approved RNA therapeutics, only ASOs have m5C and m5U. The ASOs have either a PMO backbone, a phosphorothioate backbone where all the nucleotides contain 2′-MOE groups, or a phosphorothioate backbone with blocks of nucleotides at the 5′ and 3′ ends containing 2′-MOE groups and internal 2′-deoxyribonucleotide residues. While the 2′-MOE groups protect the ASO from degradation, they also prevent RNase H digestion of the target mRNA. Thus, an ASO with an internal gap between the protected ends permits RNase H recognition of the target, which is the case for three of the four ASOs with 2′-MOE groups (Table 12).257 ASOs with PMO backbones (see Figure 21) regulate their targets through steric hindrance, not nuclease digestion.265 For the five ASOs in Table 12, three have phosphorothioate backbones and two, Exondys 51 and Vyondys 53, have PMO backbones together with a triethylene glycol extension at their 5′ end (Figure 23).
Table 12. Number of RNA Modifications in Approved RNA Therapeuticsa.
The color intensity reflects the number of RNA modifications.
Figure 23.
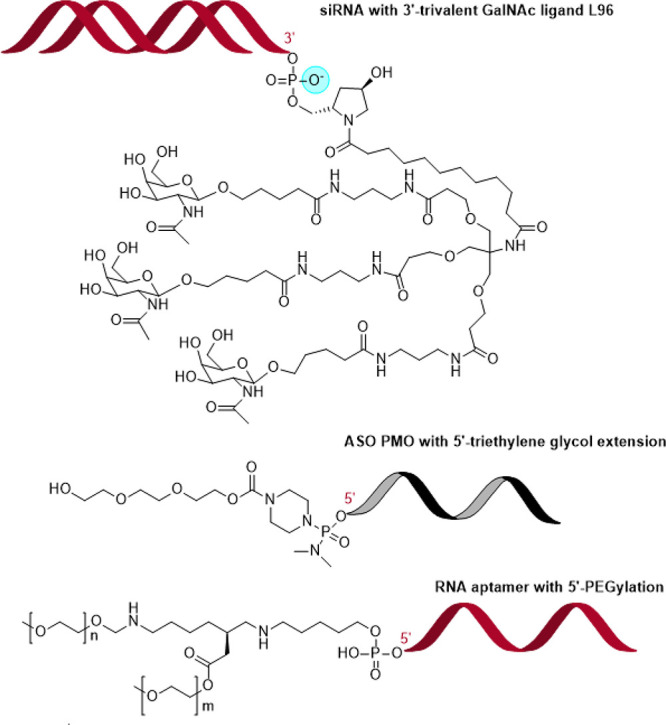
Examples of terminal modifications for therapeutic RNAs: siRNA with 3′ tripartite GalNAc ligand L96 (O in the blue circle can be replaced with S); ASO with a PMO backbone (black) and triethylene glycol; RNA aptamer with double 5′-PEGylation.268−270
siRNAs have a moderate number of phosphorothioate linkages, with 2′-fluoro and 2′-O-methyl modification of the ribose, and are often modified on their 3′ ends. Three of the approved therapeutic siRNAs, Leqvio, Givlaari/Girosiran, and Oxlumo, are 3′-glycosylated with the trivalent GalNAc conjugate266 shown in Figure 23. This trivalent branched linker containing GalNAc residues targets the siRNAs to hepatocytes to treat liver diseases.267 The other approved siRNA, Onpattro, has two 3′-terminal thymidine (dT) residues on both RNA strands to protect the siRNA from exonucleases.
Aptamers have ester linkages to protective groups and can have extensive sugar–phosphate backbone modifications, such as PEGylation (the covalent attachment of polyethylene glycol) on their 5′ ends. Because aptamers bind to their targets as a result of their tertiary structure and do not rely on nucleic acid hybridization for function, they have fewer constraints on modifications compared to other therapeutic RNAs.104 The single FDA-approved aptamer, Macugen, has extensive 2′-O-methyl and 2′-fluoro modifications, a 3′-to-3′ link to thymidine at its 3′ end, as well as double PEGylation at its 5′ end (Figure 23).
The mRNA vaccines BNT162/Comirnaty and mRNA-1273/Spikevax have 7-methylguanosine caps connected to the 5′ end of the mRNA by a triphosphate group and have all their uridines replaced with 1-methylpseudouridine, which improves mRNA stability and translation. In addition, both contain a 2′-O-methyl modification to the nucleotide immediately following the 7-methylguanosine cap to improve stability to base and nuclease digestion.
In summary, chemical modifications protect therapeutic RNAs from exonucleases, endonucleases, and the cellular environment, and they enhance pharmaceutical activity. The choice of the backbone determines whether an ASO blocks cellular processes such as translation, transcription, or splicing, or targets an RNA for nuclease digestion. The 2′-ribose modifications on siRNAs mitigate off-target effects by lowering the thermal stability, thereby enhancing target-specific binding. 1-methylpseudouridine improves stability and translation of therapeutic mRNAs. Since therapeutic RNAs are extensively modified, they frequently are not designated as RNA vs DNA but have names such as ASO (antisense oligonucleotide) and siNA (short interfering nucleic acid) for siRNAs.271
RNA Delivery Systems
RNA therapeutics, which are hydrophilic and negatively charged, cannot diffuse across cell membranes; thus, they require delivery vectors and/or chemical modification to reach their targets. When administered systemically, RNA delivery systems need to protect the RNA against serum nucleases, bypass the immune system, avoid non-specific interactions with serum proteins, and block renal clearance.11 While biological barriers such as immunogenicity and nucleases are usually addressed by modifying the RNA chemically, encapsulation of RNA into nanocarriers can both protect and deliver RNA to cells.272 Nanomaterials with biodegradability, biocompatibility, and low toxicity are used as RNA carriers. These include lipids, chitosan, cyclodextrin, polyethylenimine (PEI), poly(lactic-co-glycolic acid), dendrimers, magnetic nanoparticles, carbon nanotubes, gold nanoparticles, silica nanoparticles, and others (Figure 24).
Figure 24.
Examples of RNA nanocarriers.
Research publications on RNA delivery systems
Nearly 7000 scientific publications on RNA delivery systems, including patents and non-patents (journal articles, books, dissertations, meeting abstracts, etc.), are in the CAS Content Collection.32,273 Publications of studies involving RNA carriers are dominated by lipid nanoparticles, followed closely by polymeric nanocarriers (Figure 25).
Figure 25.
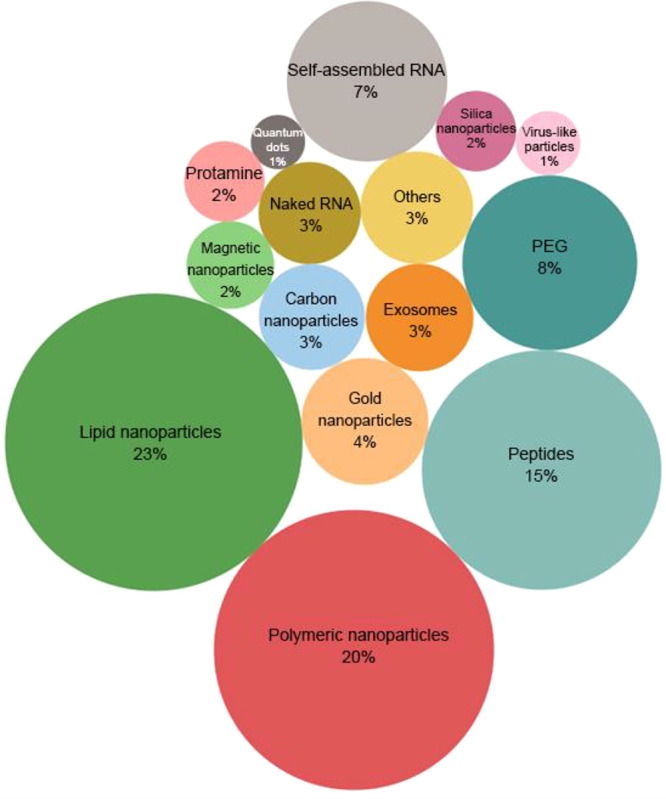
Percentage distribution of RNA nanocarrier-related documents in the CAS Content Collection.32
Lipid Nanoparticles
Lipid nanoparticles, comprising stable complexes between synthetic cationic lipids and anionic nucleic acids, are currently the most widely used non-viral delivery system for nucleic acid drugs and vaccines.19,274−276 The advantages of lipid systems include ease of production, biodegradability, protection of entrapped nucleic acids from nuclease degradation and renal clearance, promotion of cellular uptake, and endosomal escape.277 Liposomes have been recognized for a long time for their role as immunological adjuvants in vaccines. As early as 1974 it was reported that they enhance the immune response to liposome-entrapped tetanus toxoid.278 Subsequently, this was shown to apply to many other antigens including mRNA vaccine products, such as the spike protein of the coronavirus.279−281
Since the first cationic lipids successfully delivered plasmids into cells in 1987, many more have been synthesized and tested as nucleic acid carriers.19 Cationic lipids differ from natural lipids by having an ionizable (cationic) headgroup in place of the zwitterionic or anionic headgroup of the natural lipids. They include two hydrophobic alkyl chains or a cholesterol moiety, a positively charged polar headgroup, and a linker joining them. Ionizable lipids are positively charged inside the cell but are neutral in the bloodstream due to pH differences, and they are less toxic than non-ionizable cationic lipids.282 Examples of cationic lipid structures used as vectors for gene delivery are provided in Figure 26, and a comprehensive list, including the chemical structures of the ∼50 most frequently used cationic lipids in drug delivery,32 is available.276
Figure 26.
Examples of cationic lipids used as nucleic acid carriers. (An extensive list with the structures of the most frequently used cationic lipids in lipid nanoparticle pharmaceutical formulations according to the CAS Content Collection is available276).
The head groups of cationic lipids are typically amine derivatives such as primary, secondary, and tertiary amines (e.g., DOGS, DC-Chol), quaternary ammonium (e.g., DOTMA, DOTAP, DORIE, DMRIE), combinations of amines (e.g., DOSPA, GAP-DLRIE), and amidinium salts (e.g., diC14-amidine) (Figure 26). Guanidine and imidazole groups283 as well as pyridinium, piperazine, and amino acid headgroups such as lysine, arginine, ornithine, and tryptophan284,285 have also been used. Headgroups with multiple cationic charges such as DOSPA, DOGS286 may be more efficient than single-charged lipids287 since the multiple charges of the headgroups condense nucleic acids. However, multivalent cationic lipids bind strongly to the nucleic acid, preventing intracellular release, and they tend to form toxic micelles.288 Combinations of quaternary amines and polyamines enhance transfection efficiency. The first cationic lipid to combine these two functionalities, Lipofectamine, is the long-standing gold standard in nucleic acid delivery efficiency. It comprises DOSPA formulated with the helper lipid dioleoylphosphatidylethanolamine (DOPE).289
Recently, LNPs have been in the global spotlight as a vital component of COVID-mRNA vaccines, playing a key role as an immunological adjuvant, in effectively protecting and transporting mRNA to cells.276,279,290 These vaccines deliver mRNA encoding the SARS-CoV-2 spike protein into the cytoplasm of host cells, which is then translated and acts as an antigen for the development of an immune response. Both vaccines contain ionizable cationic lipids enabling RNA complexation. The hydrocarbon chains of the proprietary cationic lipids of both these vaccines are optimized to enable safe clearance and mRNA delivery efficiency. They also contain a PEGylated lipid to confer “invisibility” from antibody opsonization and phagocytosis. Packing of RNA into lipid nanoparticles is aided by distearoylphosphatidylcholine and cholesterol.
Complexation with cationic lipids stabilizes nucleic acids, protects them from nuclease degradation, and facilitates delivery to the target cells. Lipid nanoparticles carrying nucleic acids adsorb to the cell membrane, are endocytosed, and release the nucleic acids into the cell. Cell membranes have negative charges that attract the cationic lipid nanoparticles and drive the adsorption and fusion of the lipid nanoparticle with the cell membrane. The anionic lipids of the cells are thought to neutralize the charge of the cationic lipid carriers, thereby eliminating the electrostatic interactions between the lipid carriers and the nucleic acids, and facilitating the release of the nucleic acids. Neutralization of the cationic lipids also destroys the nanoparticles by promoting the formation of non-lamellar structures,291 which are associated with the efficacy of cationic lipid carriers in delivering nucleic acids into cells.292 Despite their benefits, lipid nanoparticles show cell toxicity and stimulate the release of systemic inflammatory cytokines, and lipid aggregates may accumulate in the liver and spleen causing hepatotoxicity.276,293
Nearly 20 years after scientists first prepared liposomes (the first-generation lipid nanoparticles), they found similar ∼100 nm extracellular lipid vesicles in most eukaryotic cells. They are exosomes that form naturally in the endosomes and are released from cells normally or as a result of some pathologies. They are formed by invaginations of the endosomal membrane and are subsequently released in the extracellular space via fusion with the plasma membrane.294,295 Although their functions are largely unknown, they are believed to be associated with important physiological processes such as the regulation of intercellular communications and signaling, and of transmission of macromolecules between cells.296 Exosomes, which are secreted by most cells, may be important in intercellular communication and signaling, and in the transport of proteins, lipids, and nucleic acids between cells. Since they are rich in mRNAs and small RNAs and can transport their contents to recipient cells, they are considered good candidates for RNA delivery. Exosome-mediated delivery of β-secretase-1 siRNA (targeting the enzyme β-secretase, which is important in Alzheimer’s disease) to the brain resulted in highly efficient β-secretase-1 gene knockdown in the mouse brain cortex.297 Electroporation has been applied to load exosomes with exogenous siRNA.298,299
Exosomes exhibit certain advantages over conventional delivery systems. They are natural transporters300 and hence less likely to be toxic or to cause immune responses. Moreover, exosomes can cross biological barriers such as the blood-brain barrier. The unique membrane composition of exosomes is key to their ability to enter target cells. The membranes of exosomes are enriched in cholesterol and phosphatidylserine.301 However, vesicles comprising only the lipids from the exosomal membrane are unable to fuse with cells, indicating that exosomal membrane proteins are also important for their activity.301 A notable advantage of the exosomes as compared to other nanoparticle delivery vehicles is that they do not lead to a harmful accumulation of therapeutic RNAs in the liver.302,303 In addition to exosomes, other biological carriers obtained from living organisms may include red blood cells, extracellular vesicles, platelet membrane-coating vehicles, red blood cell membrane coating nanoparticles, cancer cell membrane-coated nanoparticles, and macrophage-based vehicles.304,305 Red blood cells are appropriate as carriers because they lack both mitochondrial and nuclear DNA, so the risk of horizontal gene transfer is avoided.306 Macrophages are attractive carriers for solid tumor targeting RNA delivery due to their natural capacities to reside into tumors throughout tumor progression.307
Polymeric Nanoparticles
Polymers comprise the second largest group of nucleic acid delivery vehicles after lipids. Cationic polymers form stable complexes (polyplexes) with anionic nucleic acids and offer a versatile, scalable, and easily adjustable platform for efficient nucleic acid delivery, while minimizing immune responses and cellular toxicity.308
Linear cationic polymers are the most widely studied polymeric nucleic acid carriers.309 PEI, poly(l-lysine) (PLL), poly(2-(dimethylamino)ethyl methacrylate) (PDMAEMA), poly(2-aminoethyl methacrylamide) (PAEMA), poly(amidoamines) (PAAs), and poly(β-amino esters) (PBAEs) have been studied for drug delivery (Figure 27). These linear polymers differ in their cation types (primary amines such as PLL, secondary amines such as PEI, or tertiary amines such as PDMAEMA), and their cationic charge density.
Figure 27.
Examples of structures of polymeric nucleic acid carriers.
Block linear copolymers comprising polycationic homopolymers and non-ionic hydrophilic blocks compact nucleic acids into polyplex micelles. PEG, hydrophilic acrylamide, acrylate, and methacrylate polymers can arrange the nucleic acid in a micelle core and create a hydrophilic protective shell. For example, the PEG-b-PLL diblock copolymer assembles into monodisperse micelles with ASOs.310
Branched polycations include a type that has randomly distributed secondary chains branching from the primary polymer backbone. There are also dendrimers—polymers with fractal branching from a core. Nucleic acid carriers include branched PEI, branched PBAEs, as well as PLL, PAMAM, and poly(propyleneimine) (PPI) dendrimers (Figure 27). Branched PEI, one of the most widely studied polycationic nucleic acid carriers, comprises primary, secondary, and tertiary amines, with different pKa values. Branched PEI provides efficient nucleic acid binding and extensive buffering capacity, which likely contribute to its outstanding performance.311
PAMAM dendrimers, the most widely used dendrimer for nucleic acid delivery, comprise a hydrogen-bonding amide and tertiary amine groups in their cores and primary amine end groups in their coronas. Structural modifications have been attempted to lessen potential toxicity, enhance circulation time, and/or enhance targeting.312−315
Natural polymers including proteins and polysaccharides, particularly cationic collagen derivatives and chitosan, have been studied as nucleic acid delivery vehicles.316 Chitosan, a linear cationic polysaccharide, has been formulated into polyplexes to deliver miRNA to osteopathic tumors, for multiple myeloma, and for bone regeneration.317−319 An advantage of chitosan is that nucleic acids easily dissociate from the polymer upon cellular internalization.320 Cationic collagen derivatives have been used for nucleic acid delivery to articular cartilage, for bone regeneration, and for treating tumor metastases.321−323
Cyclodextrins (CDs) are natural carbohydrate polymers for the delivery of therapeutic nucleic acids324 that have shown clinical success.325 CDs are biocompatible cyclic oligosaccharides with a hydrophilic exterior surface and a hydrophobic interior cavity. Cationic CD derivatives assemble with siRNA via electrostatic interactions into stable gene delivery nanostructures of 50–200 nm.326
Similar to cationic lipid nanoparticles, polymeric nucleic acid carriers bind non-specifically to the negatively charged cell membrane via electrostatic interactions327 and are internalized by endocytosis.328 Following endocytosis into early endosomes, polyplexes move through the progressively more acidic endosomes (pH 6.0–4.8). Escaping endosomal confinement is critical for nucleic acid delivery; therefore, scientists are developing and modifying polymer-based systems to promote endosomal escape.327,329−331
Peptides
Peptides are structurally and functionally versatile, biocompatible, and can target cells; thus, they are attractive RNA carriers. Peptides that penetrate the cellular membrane and transfer to the cytoplasm, CPPs, are used most frequently for RNA delivery (Table 13). CPPs have variable sequences, lengths, and polarities, and they can enter cells via multiple pathways, such as by forming a hole in the membrane or by endocytosis. CPPs deliver nucleic acids into cells via chemical conjugation or non-covalent complex formation with nucleic acids. Electrostatic and hydrophobic interactions between CPPs and nucleic acids result in self-assembly into peptide-based nanoparticles.332,333 Amphipathic CPPs have been developed using hydrophilic and hydrophobic domains to provide both nucleic acid complexation and membrane interactions.334 CPPs are promising, non-viral alternatives to lipid- or polymer-based carriers of therapeutic nucleic acids.12
Table 13. Examples of CPPs Used in RNA Delivery335.
| CPP | amino acid sequence |
|---|---|
| Natural CPPs | |
| HIV Tat | YGRKKRRQRRR |
| HIV Rev | TRQARRNRRRRWRERQR |
| FHV coat | RRRRNRTRRNRRRVR |
| Penetratin | RQIKIWFQNRRMKWKK |
| MPG | GALFLGFLGAAGSTMGAWSQPKKKRKV |
| Polyarginines | |
| PR9 | FFLIPKGRRRRRRRRR |
| SR9 | RRRRRRRRR |
| IR9 | GLFEAIEGFIENGWEGMIDGWYGRRRRRRRRR |
| HR9 | CHHHHHRRRRRRRRRHHHHHC |
| Artificial/Engineered CPPs | |
| Transportan | CLIKKALAALAKLNIKLLYGASNLTWG |
| CADY | GLWRALWRLLRSLWRLLWRA |
| C6 | RLLRLLLRLWRRLLRLLR |
| PF20 | LLKLLKKLLKLLKKLLKLL |
| NAP | KALKLKLALALLAKLKLA |
| POD | GGG[ARKKAAKA]4 |
The Tat peptide originated from the Tat protein of the human immunodeficiency virus (HIV).336−338 The positively charged oligoarginine peptides promote internalization by forming hydrogen bonds with the membrane sulfates and the nucleic acid phosphate groups.339,340 The histidine-rich peptide is efficient in siRNA delivery.341 The skin permeating and cell entering (SPACE) peptides facilitate the penetration of conjugated cargoes into the epidermis and dermis.339,340,342 Recently, fusion peptides were developed comprising SPACE and cationic oligoarginine linked by a GCG sequence.343 Nanocomplexes of fusion peptides and siRNA exhibited enhanced cellular uptake, gene silencing, and retention, possibly due to the synergistic effect of the oligoarginine and SPACE peptides. Oligoarginine electrostatically attracts siRNAs to form nanocomplexes, and the SPACE peptide interacts with the cellular membrane via hydrogen bonding.
Non-covalent complexation of cationic peptides that comprise hydrophilic and hydrophobic domains with RNA provide efficient gene silencing.344,345 The hydrophilic domain of positively charged amino acids, such as arginine, lysine, and histidine, provides a net positive charge of at least +8 that condenses the RNA and promotes hydrogen bonding with the anionic cellular membrane to stimulate cellular uptake.346 The hydrophobic peptide domain of tryptophan and phenylalanine enhances interactions with the lipid bilayer. Terminal modification of the cationic peptides with hydrophobic molecules such as cholesterol, stearic acid, or cholic acid increases the hydrophobicity.347,348 The other terminus of the peptide can be modified with hydrophilic molecules such as PEG. These peptides form a micelle-like structure with siRNA and deliver them efficiently to target cells.347,348
Protamines are naturally occurring cationic peptides involved in the condensation of chromatin during spermatogenesis. Comprising >65% positively charged arginines, protamines form non-covalent electrostatic complexes with nucleic acids and protect them from enzymatic degradation. Although protamines are intrinsically disordered peptides, in the presence of nucleic acids they switch from a random coil to a structure with one or more α-helices.349 Protamine–RNA complexes show limited efficacy, possibly due to the tight interaction between the protamine and RNA.350
Other Carriers
Gold nanoparticles (AuNP) carriers are biocompatible, protect RNA from degradation, and have tunable shape, size, and optical properties.351,352 Two approaches for the design of AuNP nucleic acid carriers are covalent attachment and supramolecular assembly. To covalently conjugate RNA with the AuNP core, a thiol group is introduced onto the RNA353 to create RNA monolayers by linking RNAs directly to the AuNP surface via Au–thiol bonds. The dense shell of oligonucleotides on the surface of the AuNPs inhibits nuclease degradation. Since AuNP–RNA conjugates can be delivered transdermally, they may be useful for gene therapy to treat cutaneous tumors, skin inflammation, and other skin disorders. To electrostatically attract the negatively charged nucleic acids, AuNPs have been functionalized by structurally modifying them, adding positively charged molecules such as polymers and amino acids.354
Spherical nucleic acids are 3D nucleic acid nanostructures that are functionalized, packed densely, and oriented spherically around a nanoparticle core.355 Since they are resistant to nuclease degradation and are taken up efficiently by cells they show promise as therapeutic agents.356 Despite their high negative charge (zeta potential less than −30 mV), they rapidly and efficiently enter cells by caveolin-mediated endocytosis and regulate gene expression. Immunomodulatory spherical nucleic acids that stimulate or regulate immunity by engaging toll-like receptors are a promising immunotherapy for cancers including triple-negative breast cancer, prostate cancer, and melanoma.357
Magnetic nanoparticles deliver nucleic acid by magnetofection, which is the application of a magnetic field.358,359 The nanoparticles need functionalization (i.e., surface modifications) to enhance their performance as a magnetic gene delivery vector. The earliest magnetic core–shell nanoparticles for gene delivery were iron oxide nanoparticles stabilized with high molecular weight PEI, with a hydrodynamic diameter of ∼200 nm and positive zeta potential.360 Nucleic acids with magnetic nanoparticles increased transfection efficiency by about 1000-fold compared to non-magnetic vectors. A variety of formulations of PEI-coated superparamagnetic iron oxide nanoparticles complexed with nucleic acids have been reported.361 Other polymers and lipids have also been used to functionalize iron oxide nanoparticles for nucleic acid delivery.362−366 These include biodegradable polylactide magnetic nanoparticles containing oleate-coated magnetite and surface-modified with PEI-oleate for nucleic acid binding,367 and mesoporous silica nanoparticles decorated with magnetite nanocrystals to yield highly efficient transfection agents.368
Mesoporous silica nanoparticles (MSNs) are hollow particles that can encapsulate a wide range of drugs after surface modification. Silica nanoparticles allow slow release of the cargo as they degrade slowly in the body to non-toxic byproducts. The advantages of MSNs as drug delivery vehicles include a large surface area, tunable pore sizes, simple surface modifications, and efficient encapsulation of cargo molecules.369 Mesoporous and solid silica nanoparticles are synthesized using the sol–gel method in which a silica precursor is hydrolyzed and then condensed to generate spherical particles. Tetraethyl orthosilicate, the usual silica precursor, provides good control by modification of the synthesis conditions. Silica nanoparticles with a spiky nanotopography can enlarge the surface area for binding RNA molecules. To bind the negatively charged nucleic acids and enhance cellular uptake, silica particles are modified with positively charged compounds such as PLL or PEI, or with branched PEI to enhance the binding of the nucleic acids.370 After cell entry by endocytosis, the acidic pH in the endosomes changes the charge of PEI resulting in the dissociation and release of the RNA.
Calcium phosphate nanoparticles are promising non-viral vectors for gene therapy, an emerging strategy for effective bone repair and regeneration. The calcium phosphate naturally present in bones makes calcium phosphate nanoparticles a good choice for RNA delivery for bone repair.371 There is particular interest in the use of calcium phosphate nanoparticles with RNAi to regulate gene expression in bone tissue engineering because of their osteoinductivity, osteoconductivity, and affinity for nucleic acids.372 Calcium phosphate nanoparticles are endocytosed by cells and dissolve in the acidic endosomes and lysosomes, resulting in the release of the nucleic acid cargo.373 Calcium phosphate nanocarriers are biocompatible, biodegradable, non-toxic and non-immunogenic, inexpensive, and easily synthesized. However, they become unstable over time because of the growth of crystals,371 and they provide only limited nuclease protection. Surface functionalization of calcium phosphate particles can facilitate their endocytosis and subsequent endosomal escape.371,374
Although many vehicles or ligands have been used for the delivery of nucleic acids, they may exhibit low immunocompatibility, toxicity, poor stability, inefficient drug release, or restricted tissue accessibility.375 However, RNA itself can self-assemble into complex programmable structures.376,377 Ligand-conjugated RNA nanoparticles based on bacteriophage phi29 packaging RNA provide a targeted ribozyme delivery system.378 RNA nanoparticles can also carry CpG DNA to macrophages.377 Origami-like RNA nanostructures are stable and efficient as drug carriers for controlled drug release.379,380
Virus-like particles (VLPs) are organized protein complexes resembling a native virus capsid. VLPs are either naturally occurring empty virus shells or are synthesized by the expression of viral structural proteins that self-assemble into a virus-like structure. VLPs are attractive drug delivery platforms due to their biocompatibility, biodegradability, and targeting ability.381 VLPs produced in vivo are used for various medical applications such as drug and vaccine delivery,382 including the COVID-19 vaccine Novavax (NVX-CoV2373) currently in phase III trials.383In vivo and in vitro cargo loading has been demonstrated for VLPs.384
Quantum dots (QDs) are semiconductor crystalline nanoparticles with unique tunable optical properties including nearly 100-fold greater brightness and 1000-fold better stability against photobleaching versus organic dyes and luminescent proteins. Their fluorescence emission can be adjusted by the particle size, known as the quantum size effect.385 QDs emit narrow wavelength bands under a wide excitation range, can be appropriately functionalized, and are desirable vectors for imaging-guided therapy. Since they can efficiently deliver RNAs into target cells and can be used to track the RNA distribution in cells,386 they may serve as an effective theranostic RNA delivery agent.387,388 The application of theranostic agents with appropriate targeted, controlled delivery and imaging capabilities has the potential to significantly advance gene therapy.
Carbon nanotubes (CNTs)—tubes made of carbon with nanosized diameters—have attracted increasing attention in biomedicine because of their unique structures and properties, being the strongest and stiffest materials yet found in terms of tensile strength and elastic modulus. With functionalization, CNTs have been used as nanocarriers for nucleic acids including siRNA, oligonucleotides, and RNA/DNA aptamers.389 Amino-functionalized multiwalled CNTs deliver proprietary toxic siRNA to human lung carcinoma cells.390 Appropriately designed amino-functional segments on the CNTs promote internalization in the cell and gene silencing.391 Cationic single-walled CNTs deliver siRNA to Lewis lung carcinoma cells.392
Despite the widespread use of carriers for RNA delivery, naked RNA has also been delivered in vivo,393 including delivery of mRNA by intramuscular, subcutaneous, or intradermal injections; the latter can promote wound healing by in situ expression of specific proteins in the skin.394−398 Local injections circumvent complications associated with systemic administration such as clearance from the bloodstream.398 Naked mRNA administered subcutaneously produces a more efficient translation of the protein than mRNA-loaded nanoparticles.399,400
PEGylation allows protein drugs to avoid the immune response,401 and it also improves the surface properties of biomolecules and drug delivery systems. It blocks surface access by steric hindrance, increasing the circulation time in blood.402 Thus, it may improve pharmacokinetic properties and enhance the efficacy of drugs, including RNA therapeutics.403 PEGylation of the RNA nanocarriers reduces non-specific interactions with serum proteins and prevents recognition by the immune system, increasing the blood circulation time. PEGylation stabilizes RNAs, with longer PEGs stabilizing oligonucleotides better than shorter ones.404 The aptamer drug Macugen comprises an oligonucleotide modified with branched 40 kDa PEG at the 5′ terminus, thus enhancing the nuclease resistance of the PEG-aptamer conjugate. PEGylation markedly improves the pharmacokinetics and boosts the neutralizing activity of anti-IL-17A aptamers, an important advance in the development of therapeutic aptamers.405
The advantages of the various RNA carriers are summarized in Table 14.
Table 14. Advantages of the RNA Carriers.
| RNA delivery system | advantages |
|---|---|
| lipid nanoparticles | • ease of production |
| • biodegradability | |
| • enhanced RNA stability | |
| • improved cellular uptake and intracellular release | |
| polymeric nanoparticles | • versatile, adaptable, and scalable |
| • low immunogenicity and cellular toxicity | |
| peptides | • versatile |
| • biocompatible | |
| • high target specificity | |
| gold nanoparticles | • adaptable |
| • biocompatible | |
| • protects RNA from degradation | |
| exosomes | • low toxicity and immunogenicity |
| • unique membrane composition enhances entrance into target cells | |
| • no accumulation of therapeutic RNAs in liver | |
| magnetic nanoparticles | • highly efficient transfection efficiency |
| carbon nanotubes | • strong, yet highly flexible with high tensile strength |
| virus-like particles | • biocompatibility |
| • biodegradability | |
| • high target specificity | |
| silica nanoparticles | • large surface area and tunable pore size |
| • simple surface modifications | |
| • efficient encapsulation of cargo compounds | |
| quantum dots | • stability |
| • adaptability | |
| • most desirable for imaging-guided therapies | |
| PEGylation | • reduced immune response |
| • improves pharmacokinetic properties | |
| • enhanced efficacy | |
| • enhanced RNA stability | |
| self-assembled RNA/naked RNA | • highly customizable |
| • can provide a more efficient translation than mRNA-loaded nanoparticles for certain routes of drug administration | |
Conclusions
Our understanding of the types and functions of RNA has increased dramatically over the past 50 years. Originally identified simply as coding or non-coding, RNA was recognized early as central to the process of transcribing and translating a gene into a protein. Over the last few decades, many new types of RNA have been discovered with specific functions, including snRNA, snoRNA, shRNA, miRNA, tmRNA, siRNA, saRNA, piRNA, and circRNA. These RNA molecules participate in a complex network that regulates how RNA is used in the cell and modifies gene expression. The multiple functions of RNA provide multiple pathways for exploiting RNA as a therapeutic molecule.
RNA can be engineered for its intended therapeutic functions by in vitro transcription or solid-support synthesis. Therapeutic mRNAs, which are translated in vivo to deliver a therapeutic or vaccine protein, require extensive replacement of uridine with 1-methylpseudouridine to decrease the immune response to the mRNA and improve the translation. ASOs and therapeutic siRNAs have phosphorothioate and 2′-ribose modifications to protect the RNA from degradation and to improve target specificity. Aptamers, whose 3D structure rather than primary sequence determines their function, are heavily modified. For all RNAs, the degree of modification can be adjusted to enhance the effectiveness of the therapeutic RNA.
There are now approved RNA medicines for cardiovascular, metabolic, liver, infectious, neurological, neuromuscular, kidney, and eye diseases, with many more in the research phase. Although there are many types of therapeutic RNAs, ASOs dominate the approved drugs, followed by siRNAs. CRISPR and AOC therapeutics currently have high research interests, still in the pre-clinical stage. The top RNA medicine companies such as Moderna, Ionis, BioNTech, Alnylam, and Sirnaomics all specialize in one type of RNA, but research and treat as many as nine different diseases. RNA therapeutics have the potential to treat a wide range of diseases, from the most common to the extremely rare.
RNA carriers are important in overcoming the physiological barriers of systemic administration of RNA-based drugs. However, they provide a challenge in the translation of RNA medicines to the clinical setting. After the recent success of the lipid nanoparticle-based mRNA vaccines against COVID-19, lipid carriers have become the primary RNA delivery vehicle. However, since lipid nanoparticles can be toxic to cells and stimulate the release of systemic inflammatory cytokines, interest in natural transporters such as exosomes is gaining momentum. Other non-toxic and non-immunogenic carriers, such as structurally and functionally versatile peptides, are also attracting attention. Novel technologies, including carbon nanotubes and other inorganic biocompatible polymers, carbon nanodots, and functionalized or hybrid systems, are also exciting avenues of exploration and evaluation.
Our growing understanding of the many types and functions of RNA has been combined with the ability to synthesize modified RNAs with improved stability and pharmaceutical activity. Nanotechnology-based systems to deliver those RNAs to the cell have resulted in an explosion of new therapeutic options for diseases ranging from viral infections to cancer. This arsenal of multiple specifically targeted RNA therapies has the potential to revolutionize the treatment of human disease.
Acknowledgments
We sincerely appreciate Zach Baum for help with RNA modification data handling, Laura Czuba for project coordination, and Peter Jap and Cristina Tomeo for insightful discussion. We are also grateful to Manuel Guzman, Gilles Georges, Michael Dennis, Carmin Gade, Dawn George, Cynthia Casebolt, and Hong Xie for executive sponsorship.
Glossary
Abbreviations Used
- AAT
alpha-1 antitrypsin
- AATD
alpha-1 antitrypsin deficiency
- AOC
antibody–oligonucleotide conjugate
- APOCIII
apolipoprotein C-III
- ASO
antisense oligomer
- asRNA
antisense RNA
- AUD
alcohol use disorder
- circRNA
circular RNA
- CMV
cytomegalovirus
- CNT
carbon nanotube
- CRISPR
clustered regularly interspaced short palindromic repeats
- crRNA
CRISPR RNA
- DMD
Duchenne muscular dystrophy
- FDA
U.S. Food and Drug Administration
- GNA
glycol nucleic acid
- LNA
locked nucleic acid
- lncRNA
long non-coding RNA
- miRNA
micro-RNA
- MOE
2′-O-(2-methoxyethyl)
- mRNA
messenger RNA
- ncRNA
non-coding RNA
- PAA
poly(amidoamine)
- PAEMA
poly(2-aminoethyl methacrylamide)
- PBAE
poly(β-amino ester)
- PDMAEMA
poly(2-(dimethylamino)ethyl methacrylate)
- PEI
polyethylene imine
- piRNA
piwi-interacting RNA
- PLL
poly(l-lysine)
- PMO
phosphorodiamidate morpholino oligonucleotide
- PNA
peptide nucleic acid
- RISC
RNA-induced silencing complex
- RNAi
RNA interference
- rRNA
ribosomal RNA
- RSV
respiratory syncytial virus
- RT-qPCR
reverse transcriptase-quantitative polymerase chain reaction
- saRNA
small activating RNA
- SELEX
systematic evolution of ligands by exponential enrichment
- sgRNA
single guide RNA
- shRNA
short hairpin RNA
- siRNA
short/small interfering RNA
- snoRNA
small nucleolar RNA
- snRNA
small nuclear RNA
- tmRNA
transfer messenger RNA
- tracrRNA
trans-activating CRISPR RNA
- tRNA
transfer RNA
Biographies
Janet M. Sasso received her B.S. in biology and Master of Public Health from The Ohio State University. Janet has been employed with CAS since 2006 and works within the Life Sciences Department. She is currently an Information Scientist, curating information in the areas of pharmacological, toxicological, and biological sciences for the CAS Content Collection. Her research interests include pharmacology, global public health equity, and evidence-based public health education.
Barbara J. B. Ambrose joined CAS in 1986 while earning her Ph.D. from The Ohio State University for work on DNA sequencing. She curates journal documents and patents in molecular genetics for the Life Sciences Department, with an emphasis on those documents containing DNA and RNA sequences. In March 2020, she joined the Coronavirus Task Force established at CAS to expedite processing of COVID-19-related journal articles and information. In addition, she currently serves as a data steward for the Life Sciences Department.
Rumiana Tenchov (Koynova) holds Ph.D. and D.Sc. degrees in biophysics from the Bulgarian Academy of Sciences. She has been a research faculty member at Northwestern University and The Ohio State University and is currently an Information Scientist at CAS. Her research interests include lipid membrane structure and assembly, lipid nanoparticles, cationic lipids/DNA assemblies and DNA delivery for transfection, thermodynamics and stability of proteins, and scientific information analysis in a broad range of life sciences. Current H-index 37 according to WoS.
Ruchira S. Datta received a B.S. in mathematics from Caltech and an M.S. in computer science and Ph.D. in mathematics from UC Berkeley, specializing in game theory. Dr. Datta worked as a software engineer at Google before doing 9 years of postdoctoral research in computational biology in the Berkeley Phylogenomics Group at UC Berkeley, the Center for Evolution and Cancer at UCSF, and the Mathematical Biosciences Institute at Ohio State. Dr. Datta works as principal at Datta Enterprises LLC (consulting and intellectual property) and joined CAS as a Senior Data Scientist in 2019. Currently Dr. Datta applies machine learning to Search & Relevance for the CAS SciFindern scientific discovery platform. Dr. Datta’s research interests include predictive immunology and mathematical ecosociology: the dynamics of interacting populations of agents.
Matthew T. Basel is an assistant professor at Kansas State University, where he runs a lab focused on cancer modeling and nano-targeted cancer therapies and also teaches physiology and biomedicine to graduate, veterinary, and physician assistant students. He received his Ph.D. in biological chemistry at Kansas State University.
Robert K. DeLong studied nucleic acid biophysics and biochemistry for his Ph.D. at Johns Hopkins University, taking antisense oligomers in vivo pharmacokinetics/pharmacodynamics for his post-doc. He then led groups in biopharma supporting nucleic acid vaccine nanoparticles’ pre-clinical to clinical translation. Transitioning to academia, he was tenured in biomedical sciences in 2013, then moved to Kansas State University, building his own lab focused on RNA interaction, stabilization, and delivery by nanoparticles. Research support has been received from NSF, NIH, CF Foundation, HHMI, APS, Johnson Cancer Research Center, corporate sponsorships, and state and local agencies. He has six patents, three pending, 57 peer-reviewed publications, one textbook, three book chapters, and four review articles and has given 84 proceedings, including several Cold Spring Harbor Laboratory, Gordon Research conferences, and NIH presentations.
Qiongqiong Angela Zhou is the scientific content creation and development supervisor at CAS. She earned her Ph.D. in molecular pharmacology and toxicology from the University of Southern California. She was further trained as a cell biologist at Johns Hopkins School of Medicine and worked in the area of cancer biology at Harvard Medical School. She was a faculty member at Missouri State University and Denison University prior to joining CAS. With a mission to inform and inspire the global community through the power of science, she and her CAS colleagues have published multiple scientific trend reports on emerging topics based on the CAS Content Collection.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.jmedchem.2c00024.
Figure S1, number of documents for various RNA types applied in medical studies in the years of 1995–2020, categorized by the type of diseases; Figure S2, top companies with RNA therapeutics and vaccines in clinical trials; Figure S3, frequencies of various types of modifications on RNA sequences and their distributions with respect to disease types obtained from the CAS Content Collection; Figure S4, frequencies of various types of modifications on RNA sequences and their distributions with respect to specific diseases acquired from the CAS Content Collection; Figure S5, the co-occurrence of RNA modifications on the same sequences; Figure S6, number of documents per year related to modified RNAs (data were obtained from a SciFindern search); Figure S7, modified and rare nucleic acid bases; Table S1, timeline and milestones of RNA research and development (PDF)
Table S2, listing RNA therapies and vaccines for various diseases in the development stages (XLSX)
Author Contributions
† J.M.S., B.J.B.A., and R.T. contributed equally to this paper.
The authors declare no competing financial interest.
Supplementary Material
References
- Titze-de-Almeida S. S.; Brandão P. R. d. P.; Faber I.; Titze-de-Almeida R. Leading RNA Interference Therapeutics Part 1: Silencing Hereditary Transthyretin Amyloidosis, with a Focus on Patisiran. Molecular Diagnosis Therapy 2020, 24, 49–59. 10.1007/s40291-019-00434-w. [DOI] [PubMed] [Google Scholar]
- Dolgin E. Injection of Hope. Nature 2019, 574, S10–S12. 10.1038/d41586-019-03072-8. [DOI] [PubMed] [Google Scholar]
- Ozata D. M.; Gainetdinov I.; Zoch A.; O’Carroll D.; Zamore P. D. Piwi-Interacting RNAs: Small RNAs with Big Functions. Nat. Rev. Genet. 2019, 20, 89–108. 10.1038/s41576-018-0073-3. [DOI] [PubMed] [Google Scholar]
- Ramat A.; Simonelig M. Functions of Piwi Proteins in Gene Regulation: New Arrows Added to the piRNA Quiver. Trends Genet. 2021, 37, 188–200. 10.1016/j.tig.2020.08.011. [DOI] [PubMed] [Google Scholar]
- Yoon S.; Rossi J. J. Therapeutic Potential of Small Activating RNAs (saRNAs in Human Cancers. Curr. Pharm. Biotechnol. 2018, 19, 604–610. 10.2174/1389201019666180528084059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang F.; Doudna J. A. CRISPR–Cas9 Structures and Mechanisms. Annu. Rev. Biophys. 2017, 46, 505–529. 10.1146/annurev-biophys-062215-010822. [DOI] [PubMed] [Google Scholar]
- Yao R. W.; Wang Y.; Chen L. L. Cellular Functions of Long Noncoding RNAs. Nat. Cell Biol. 2019, 21, 542–551. 10.1038/s41556-019-0311-8. [DOI] [PubMed] [Google Scholar]
- Li X.; Yang L.; Chen L. L. The Biogenesis, Functions, and Challenges of Circular RNAs. Mol. Cell 2018, 71, 428–442. 10.1016/j.molcel.2018.06.034. [DOI] [PubMed] [Google Scholar]
- Wong E.; Goldberg T. Mipomersen (Kynamro): A Novel Antisense Oligonucleotide Inhibitor for the Management of Homozygous Familial Hypercholesterolemia. P & T: a peer-reviewed journal for formulary management 2014, 39, 119–122. [PMC free article] [PubMed] [Google Scholar]
- Davis M. E.; Zuckerman J. E.; Choi C. H. J.; Seligson D.; Tolcher A.; Alabi C. A.; Yen Y.; Heidel J. D.; Ribas A. Evidence of RNAi in Humans from Systemically Administered siRNA Via Targeted Nanoparticles. Nature 2010, 464, 1067–1070. 10.1038/nature08956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehead K. A.; Langer R.; Anderson D. G. Knocking Down Barriers: Advances in siRNA Delivery. Nat. Rev. Drug Discov. 2009, 8, 129–138. 10.1038/nrd2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts T. C.; Langer R.; Wood M. J. A. Advances in Oligonucleotide Drug Delivery. Nat. Rev. Drug Discov. 2020, 19, 673–694. 10.1038/s41573-020-0075-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S.; Meselson M.; Jacob F. An Unstable Intermediate Carrying Information from Genes to Ribosomes for Protein Synthesis. Nature 1961, 190, 576–581. 10.1038/190576a0. [DOI] [PubMed] [Google Scholar]
- Muthukrishnan S.; Both G. W.; Furuichi Y.; Shatkin A. J. 5′-Terminal 7-Methylguanosine in Eukaryotic Messenger-RNA Is Required for Translation. Nature 1975, 255, 33–37. 10.1038/255033a0. [DOI] [PubMed] [Google Scholar]
- Furuichi Y.; Miura K. I. Blocked Structure at 5′ Terminus of Messenger-RNA from Cytoplasmic Polyhedrosis-Virus. Nature 1975, 253, 374–375. 10.1038/253374a0. [DOI] [PubMed] [Google Scholar]
- Dimitriadis G. J. Translation of Rabbit Globin mRNA Introduced by Liposomes into Mouse Lymphocytes. Nature 1978, 274, 923–924. 10.1038/274923a0. [DOI] [PubMed] [Google Scholar]
- Zamecnik P. C.; Stephenson M. L. Inhibition of Rous Sarcoma Virus Replication and Cell Transformation by a Specific Oligodeoxynucleotide. Proc. Natl. Acad. Sci. U. S. A. 1978, 75, 280–284. 10.1073/pnas.75.1.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieg P. A.; Melton D. A. Functional Messenger-RNAs Are Produced by SP6 Invitro Transcription of Cloned cDNAs. Nucleic Acids Res. 1984, 12, 7057–7070. 10.1093/nar/12.18.7057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felgner P. L.; Gadek T. R.; Holm M.; Roman R.; Chan H. W.; Wenz M.; Northrop J. P.; Ringold G. M.; Danielsen M. Lipofection - a Highly Efficient, Lipid-Mediated DNA-Transfection Procedure. Proc. Natl. Acad. Sci. U. S. A. 1987, 84, 7413–7417. 10.1073/pnas.84.21.7413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone R. W.; Felgner P. L.; Verma I. M. Cationic Liposome-Mediated RNA Transfection. Proc. Natl. Acad. Sci. U. S. A. 1989, 86, 6077–6081. 10.1073/pnas.86.16.6077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire A.; Xu S. Q.; Montgomery M. K.; Kostas S. A.; Driver S. E.; Mello C. C. Potent and Specific Genetic Interference by Double-Stranded RNA in Caenorhabditis Elegans. Nature 1998, 391, 806–811. 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Piascik P. Fomiversen Sodium Approved to Treat CMV Retinitis. J. Am. Pharm. Assoc. (Wash) 1999, 39, 84–85. 10.1016/S1086-5802(16)30428-4. [DOI] [PubMed] [Google Scholar]
- Bernstein E.; Caudy A. A.; Hammond S. M.; Hannon G. J. Role for a Bidentate Ribonuclease in the Initiation Step of RNA Interference. Nature 2001, 409, 363–366. 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- Kariko K.; Buckstein M.; Ni H. P.; Weissman D. Suppression of RNA Recognition by Toll-Like Receptors: The Impact of Nucleoside Modification and the Evolutionary Origin of RNA. Immunity 2005, 23, 165–175. 10.1016/j.immuni.2005.06.008. [DOI] [PubMed] [Google Scholar]
- Weide B.; Pascolo S.; Scheel B.; Derhovanessian E.; Pflugfelder A.; Eigentler T. K.; Pawelec G.; Hoerr I.; Rammensee H.-G.; Garbe C. Direct Injection of Protamine-Protected mRNA: Results of a Phase 1/2 Vaccination Trial in Metastatic Melanoma Patients. J. Immunotherapy 2009, 32, 498–507. 10.1097/CJI.0b013e3181a00068. [DOI] [PubMed] [Google Scholar]
- Adams D.; Gonzalez-Duarte A.; O’Riordan W. D.; Yang C. C.; Ueda M.; Kristen A. V.; Tournev I.; Schmidt H. H.; Coelho T.; Berk J. L.; Lin K. P.; Vita G.; Attarian S.; Planté-Bordeneuve V.; Mezei M. M.; Campistol J. M.; Buades J.; Brannagan T. H. III; Kim B. J.; Oh J.; Parman Y.; Sekijima Y.; Hawkins P. N.; Solomon S. D.; Polydefkis M.; Dyck P. J.; Gandhi P. J.; Goyal S.; Chen J.; Strahs A. L.; Nochur S. V.; Sweetser M. T.; Garg P. P.; Vaishnaw A. K.; Gollob J. A.; Suhr O. B. Patisiran, an RNAi Therapeutic, for Hereditary Transthyretin Amyloidosis. N. Engl. J. Med. 2018, 379, 11–21. 10.1056/NEJMoa1716153. [DOI] [PubMed] [Google Scholar]
- Allen D.; Rosenberg M.; Hendel A. Using Synthetically Engineered Guide RNAs to Enhance CRISPR Genome Editing Systems in Mammalian Cells. Front. Genome Editing 2021, 2, 617910. 10.3389/fgeed.2020.617910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaccines and Related Biological Products Advisory Committee Meeting. Moderna COVID-19 Vaccine. Fda Briefing Document. https://www.fda.gov/media/144434/download (accessed December 22, 2020).
- Emergency Use Authorization (Eua) of the Pfizer-Biontech COVID-19 Vaccine to Prevent Coronavirus Disease 2019 (COVID-19) in Individuals 16 Years of Age and Older. https://www.fda.gov/media/144414/download (accessed December 22, 2020).
- Fda Approves First COVID-19 Vaccine. https://www.fda.gov/news-events/press-announcements/fda-approves-first-covid-19-vaccine (accessed Dec 27, 2021).
- Castanotto D.; Zhang X.; Rüger J.; Alluin J.; Sharma R.; Pirrotte P.; Joenson L.; Ioannou S.; Nelson M. S.; Vikeså J.; Hansen B. R.; Koch T.; Jensen M. A.; Rossi J. J.; Stein C. A. A Multifunctional Lna Oligonucleotide-Based Strategy Blocks Ar Expression and Transactivation Activity in PCa Cells. Molecular Therapy - Nucleic Acids 2021, 23, 63–75. 10.1016/j.omtn.2020.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CAS Content Collection. https://www.cas.org/about/cas-content (accessed Jan 5, 2022).
- Cooper G. M., RNA Synthesis and Processing. In The Cell: A Molecular Approach; Sinauer Associates: Sunderland, MA, 2000. [Google Scholar]
- Alberts B.; Johnson A.; Lewis J.; et al. How Cells Read the Genome: From DNA to Protein. In Molecular Biology of the Cell, 4th ed.; New York: Garland Science,New York: 2002. [Google Scholar]
- Valadkhan S.; Gunawardane L. S. Role of Small Nuclear RNAs in Eukaryotic Gene Expression. Essays Biochem. 2013, 54, 79–90. 10.1042/bse0540079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matera A. G.; Terns R. M.; Terns M. P. Non-Coding RNAs: Lessons from the Small Nuclear and Small Nucleolar RNAs. Nat. Rev. Mol. Cell Biol. 2007, 8, 209–220. 10.1038/nrm2124. [DOI] [PubMed] [Google Scholar]
- Morais P.; Adachi H.; Yu Y. T. Spliceosomal snRNA Epitranscriptomics. Front. Genet. 2021, 12, 652129. 10.3389/fgene.2021.652129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wajahat M.; Bracken C. P.; Orang A. Emerging Functions for snoRNAs and snoRNA-Derived Fragments. Int. J. Mol. Sci. 2021, 22, 10193. 10.3390/ijms221910193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Werf J.; Chin C. V.; Fleming N. I. snoRNA in Cancer Progression, Metastasis and Immunotherapy Response. Biology (Basel) 2021, 10, 809. 10.3390/biology10080809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang J.; Wen J.; Huang Z.; Chen X. P.; Zhang B. X.; Chu L. Small Nucleolar RNAs: Insight into Their Function in Cancer. Front. Oncol. 2019, 9, 587 10.3389/fonc.2019.00587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy K.; Chanfreau G. F. The Diverse Functions of Fungal RNAse Iii Enzymes in RNA Metabolism. Enzymes 2012, 31, 213–235. 10.1016/B978-0-12-404740-2.00010-0. [DOI] [PubMed] [Google Scholar]
- Williams G. T.; Farzaneh F. Are snoRNAs and snoRNA Host Genes New Players in Cancer?. Nat. Rev. Cancer 2012, 12, 84–88. 10.1038/nrc3195. [DOI] [PubMed] [Google Scholar]
- Holley C. L.; Topkara V. K. An Introduction to Small Non-Coding RNAs: miRNA and snoRNA. Cardiovasc. Drugs Ther. 2011, 25, 151–159. 10.1007/s10557-011-6290-z. [DOI] [PubMed] [Google Scholar]
- Bachellerie J. P.; Cavaillé J.; Hüttenhofer A. The Expanding snoRNA World. Biochimie 2002, 84, 775–790. 10.1016/S0300-9084(02)01402-5. [DOI] [PubMed] [Google Scholar]
- Ro-Choi T. S. Nucleolar snoRNA and Ribosome Production. Mol. Cells 1997, 7, 451–467. [PubMed] [Google Scholar]
- Bridges M. C.; Daulagala A. C.; Kourtidis A. Lnccation: lncRNA Localization and Function. J. Cell Biol. 2021, 220, e202009045. 10.1083/jcb.202009045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrè F.; Colantoni A.; Helmer-Citterich M. Revealing Protein-lncRNA Interaction. Brief Bioinform. 2016, 17, 106–116. 10.1093/bib/bbv031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jathar S.; Kumar V.; Srivastava J.; Tripathi V. Technological Developments in lncRNA Biology. Adv. Exp. Med. Biol. 2017, 1008, 283–323. 10.1007/978-981-10-5203-3_10. [DOI] [PubMed] [Google Scholar]
- Li J.; Meng H.; Bai Y.; Wang K. Regulation of lncRNA and Its Role in Cancer Metastasis. Oncol. Res. 2016, 23, 205–217. 10.3727/096504016X14549667334007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng W. X.; Koirala P.; Mo Y. Y. lncRNA-Mediated Regulation of Cell Signaling in Cancer. Oncogene 2017, 36, 5661–5667. 10.1038/onc.2017.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Statello L.; Guo C. J.; Chen L. L.; Huarte M. Gene Regulation by Long Non-Coding RNAs and Its Biological Functions. Nat. Rev. Mol. Cell Biol. 2021, 22, 96–118. 10.1038/s41580-020-00315-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z.; Jiang S.; Shang J.; Jiang Y.; Dai Y.; Xu B.; Yu Y.; Liang Z.; Yang Y. lncRNA: Shedding Light on Mechanisms and Opportunities in Fibrosis and Aging. Ageing Res. Rev. 2019, 52, 17–31. 10.1016/j.arr.2019.04.001. [DOI] [PubMed] [Google Scholar]
- Backes C.; Meese E.; Keller A. Specific miRNA Disease Biomarkers in Blood, Serum and Plasma: Challenges and Prospects. Mol. Diagn. Ther. 2016, 20, 509–518. 10.1007/s40291-016-0221-4. [DOI] [PubMed] [Google Scholar]
- Bernardo B. C.; Ooi J. Y.; Lin R. C.; McMullen J. R. miRNA Therapeutics: A New Class of Drugs with Potential Therapeutic Applications in the Heart. Future Med. Chem. 2015, 7, 1771–1792. 10.4155/fmc.15.107. [DOI] [PubMed] [Google Scholar]
- Cai Y.; Yu X.; Hu S.; Yu J. A Brief Review on the Mechanisms of miRNA Regulation. Genomics Proteomics Bioinformatics 2009, 7, 147–154. 10.1016/S1672-0229(08)60044-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao T.; Zhen X. C. Dysregulation of miRNA and Its Potential Therapeutic Application in Schizophrenia. CNS Neurosci. Ther. 2018, 24, 586–597. 10.1111/cns.12840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y. S.; Dutta A. Micrornas in Cancer. Annu. Rev. Pathol. 2009, 4, 199–227. 10.1146/annurev.pathol.4.110807.092222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu T. X.; Rothenberg M. E. Microrna. J. Allergy Clin. Immunol. 2018, 141, 1202–1207. 10.1016/j.jaci.2017.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari A.; Mukherjee B.; Dixit M. Microrna Key to Angiogenesis Regulation: miRNA Biology and Therapy. Curr. Cancer Drug Targets 2018, 18, 266–277. 10.2174/1568009617666170630142725. [DOI] [PubMed] [Google Scholar]
- Piatek M. J.; Werner A. Endogenous siRNAs: Regulators of Internal Affairs. Biochem. Soc. Trans. 2014, 42, 1174–1179. 10.1042/BST20140068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooke S. T.; Witztum J. L.; Bennett C. F.; Baker B. F. RNA-Targeted Therapeutics. Cell Metab. 2018, 27, 714–739. 10.1016/j.cmet.2018.03.004. [DOI] [PubMed] [Google Scholar]
- Schütze N. siRNA Technology. Mol. Cell. Endocrinol. 2004, 213, 115–119. 10.1016/j.mce.2003.10.078. [DOI] [PubMed] [Google Scholar]
- Rossi J. J.; Rossi D. J. siRNA Drugs: Here to Stay. Mol. Ther. 2021, 29, 431–432. 10.1016/j.ymthe.2021.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pushparaj P. N.; Aarthi J. J.; Manikandan J.; Kumar S. D. siRNA, miRNA, and shRNA: In Vivo Applications. J. Dent. Res. 2008, 87, 992–1003. 10.1177/154405910808701109. [DOI] [PubMed] [Google Scholar]
- Qureshi A.; Tantray V. G.; Kirmani A. R.; Ahangar A. G. A Review on Current Status of Antiviral siRNA. Rev. Med. Virol 2018, 28, e1976. 10.1002/rmv.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B.; Weng Y.; Xia X. H.; Liang X. J.; Huang Y. Clinical Advances of siRNA Therapeutics. J. Gene Med. 2019, 21, e3097. 10.1002/jgm.3097. [DOI] [PubMed] [Google Scholar]
- Gavrilov K.; Saltzman W. M. Therapeutic siRNA: Principles, Challenges, and Strategies. Yale J. Biol. Med. 2012, 85, 187–200. [PMC free article] [PubMed] [Google Scholar]
- Hu B.; Zhong L.; Weng Y.; Peng L.; Huang Y.; Zhao Y.; Liang X. J. Therapeutic siRNA: State of the Art. Signal Transduct. Target. Ther. 2020, 5, 101 10.1038/s41392-020-0207-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A.; Trivedi P.; Jain N. K. Advances in siRNA Delivery in Cancer Therapy. Artif Cells Nanomed. Biotechnol. 2018, 46, 274–283. 10.1080/21691401.2017.1307210. [DOI] [PubMed] [Google Scholar]
- Nikam R. R.; Gore K. R. Journey of siRNA: Clinical Developments and Targeted Delivery. Nucleic Acid Ther. 2018, 28, 209–224. 10.1089/nat.2017.0715. [DOI] [PubMed] [Google Scholar]
- Saw P. E.; Song E. W. siRNA Therapeutics: A Clinical Reality. Sci. China Life Sci. 2020, 63, 485–500. 10.1007/s11427-018-9438-y. [DOI] [PubMed] [Google Scholar]
- Czech B.; Munafò M.; Ciabrelli F.; Eastwood E. L.; Fabry M. H.; Kneuss E.; Hannon G. J. piRNA-Guided Genome Defense: From Biogenesis to Silencing. Annu. Rev. Genet. 2018, 52, 131–157. 10.1146/annurev-genet-120417-031441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizu H.; Nagao A.; Siomi H. Gatekeepers for Piwi-piRNA Complexes to Enter the Nucleus. Curr. Opin. Genet. Dev. 2011, 21, 484–490. 10.1016/j.gde.2011.05.001. [DOI] [PubMed] [Google Scholar]
- Iwasaki Y. W.; Siomi M. C.; Siomi H. Piwi-Interacting RNA: Its Biogenesis and Functions. Annu. Rev. Biochem. 2015, 84, 405–433. 10.1146/annurev-biochem-060614-034258. [DOI] [PubMed] [Google Scholar]
- Lenart P.; Novak J.; Bienertova-Vasku J. Piwi-piRNA Pathway: Setting the Pace of Aging by Reducing DNA Damage. Mech. Ageing Dev. 2018, 173, 29–38. 10.1016/j.mad.2018.03.009. [DOI] [PubMed] [Google Scholar]
- Liu Y.; Dou M.; Song X.; Dong Y.; Liu S.; Liu H.; Tao J.; Li W.; Yin X.; Xu W. The Emerging Role of the piRNA/piwi Complex in Cancer. Mol. Cancer 2019, 18, 123 10.1186/s12943-019-1052-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mani S. R.; Juliano C. E. Untangling the Web: The Diverse Functions of the piwi/piRNA Pathway. Mol. Reprod. Dev. 2013, 80, 632–664. 10.1002/mrd.22195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Z. H.; Zhao Y. M. The Regulatory Functions of piRNA/piwi in Spermatogenesis. Yi Chuan 2017, 39, 683–691. 10.16288/j.yczz.17-245. [DOI] [PubMed] [Google Scholar]
- Chang W.; Wang J. Exosomes and Their Noncoding RNA Cargo Are Emerging as New Modulators for Diabetes Mellitus. Cells 2019, 8, 853. 10.3390/cells8080853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu Y. L.; Li H.; Ng P. L. A.; Kong S. T.; Zhang H.; Lin Y.; Tai W. C. S.; Yu A. C. S.; Yim A. K. Y.; Tsang H. F.; Cho W. C. S.; Wong S. C. C. The Potential of Circulating Exosomal RNA Biomarkers in Cancer. Expert Rev. Mol. Diagn. 2020, 20, 665–678. 10.1080/14737159.2020.1745064. [DOI] [PubMed] [Google Scholar]
- Mathivanan S.; Ji H.; Simpson R. J. Exosomes: Extracellular Organelles Important in Intercellular Communication. J. Proteomics 2010, 73, 1907–1920. 10.1016/j.jprot.2010.06.006. [DOI] [PubMed] [Google Scholar]
- Xie Y.; Dang W.; Zhang S.; Yue W.; Yang L.; Zhai X.; Yan Q.; Lu J. The Role of Exosomal Noncoding RNAs in Cancer. Mol. Cancer 2019, 18, 37. 10.1186/s12943-019-0984-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J.; Li S.; Li L.; Li M.; Guo C.; Yao J.; Mi S. Exosome and Exosomal Microrna: Trafficking, Sorting, and Function. Genomics Proteomics Bioinformatics 2015, 13, 17–24. 10.1016/j.gpb.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crick F. H. On Protein Synthesis. Symp. Soc. Exp Biol. 1958, 12, 138–163. [PubMed] [Google Scholar]
- Palade G. E. A Small Particulate Component of the Cytoplasm. Journal of Biophysical and Biochem. Cytol. 1955, 1, 59–68. 10.1083/jcb.1.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holley R. W.; Apgar J.; Everett G. A.; Madison J. T.; Marquisee M.; Merrill S. H.; Penswick J. R.; Zamir A. Structure of a Ribonucleic Acid. Science 1965, 147, 1462–1465. 10.1126/science.147.3664.1462. [DOI] [PubMed] [Google Scholar]
- Esteller M. Non-Coding RNAs in Human Disease. Nat. Rev. Genet. 2011, 12, 861–874. 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- Hombach S.; Kretz M. Non-Coding RNAs: Classification, Biology and Functioning. Adv. Exp. Med. Biol. 2016, 937, 3–17. 10.1007/978-3-319-42059-2_1. [DOI] [PubMed] [Google Scholar]
- Yang J. X.; Rastetter R. H.; Wilhelm D. Non-Coding RNAs: An Introduction. Adv. Exp. Med. Biol. 2016, 886, 13–32. 10.1007/978-94-017-7417-8_2. [DOI] [PubMed] [Google Scholar]
- Xu J.; Bai J.; Xiao J. Computationally Modeling ncRNA-ncRNA Crosstalk. Adv. Exp. Med. Biol. 2018, 1094, 77–86. 10.1007/978-981-13-0719-5_8. [DOI] [PubMed] [Google Scholar]
- Liu N.; Wang Z. Z.; Zhao M.; Zhang Y.; Chen N. H. Role of Non-Coding RNA in the Pathogenesis of Depression. Gene 2020, 735, 144276. 10.1016/j.gene.2019.144276. [DOI] [PubMed] [Google Scholar]
- Anastasiadou E.; Jacob L. S.; Slack F. J. Non-Coding RNA Networks in Cancer. Nat. Rev. Cancer 2018, 18, 5–18. 10.1038/nrc.2017.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panni S.; Lovering R. C.; Porras P.; Orchard S. Non-Coding RNA Regulatory Networks. Biochim. Biophys. Acta Gene Regul. Mech. 2020, 1863, 194417. 10.1016/j.bbagrm.2019.194417. [DOI] [PubMed] [Google Scholar]
- Mattick J. S.; Makunin I. V. Non-Coding RNA. Hum. Mol. Genet. 2006, 15, R17–R29. 10.1093/hmg/ddl046. [DOI] [PubMed] [Google Scholar]
- Jiao A. L.; Slack F. J. RNA-Mediated Gene Activation. Epigenetics 2014, 9, 27–36. 10.4161/epi.26942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwok A.; Raulf N.; Habib N. Developing Small Activating RNA as a Therapeutic: Current Challenges and Promises. Ther. Deliv. 2019, 10, 151–164. 10.4155/tde-2018-0061. [DOI] [PubMed] [Google Scholar]
- Laham-Karam N.; Laitinen P.; Turunen T. A.; Ylä-Herttuala S. Activating the Chromatin by Noncoding RNAs. Antioxid. Redox Signal. 2018, 29, 813–831. 10.1089/ars.2017.7248. [DOI] [PubMed] [Google Scholar]
- Li L. C. Small RNA-Guided Transcriptional Gene Activation (RNAa) in Mammalian Cells. Adv. Exp. Med. Biol. 2017, 983, 1–20. 10.1007/978-981-10-4310-9_1. [DOI] [PubMed] [Google Scholar]
- Li S. M.; Hu J. Small Activating RNAs: Towards Development of New Therapeutic Agents and Clinical Treatments. Curr. Pharm. Biotechnol. 2018, 19, 622–630. 10.2174/1389201019666180802145134. [DOI] [PubMed] [Google Scholar]
- Zheng L.; Wang L.; Gan J.; Zhang H. RNA Activation: Promise as a New Weapon against Cancer. Cancer Lett. 2014, 355, 18–24. 10.1016/j.canlet.2014.09.004. [DOI] [PubMed] [Google Scholar]
- Zhao X.; Voutila J.; Habib N.; et al. RNA Activation. In Innovative Medicine: Basic Research and Development; Nakao K., Minato N., Uemoto S., Eds.; Springer: Tokyo, 2015. [PubMed] [Google Scholar]
- Yu C. Y.; Kuo H. C. The Emerging Roles and Functions of Circular RNAs and Their Generation. J. Biomed Sci. 2019, 26, 29. 10.1186/s12929-019-0523-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eguchi Y.; Itoh T.; Tomizawa J.-i. Antisense RNA. Annu. Rev. Biochem. 1991, 60, 631–652. 10.1146/annurev.bi.60.070191.003215. [DOI] [PubMed] [Google Scholar]
- Damase T. R.; Sukhovershin R.; Boada C.; Taraballi F.; Pettigrew R. I.; Cooke J. P. The Limitless Future of RNA Therapeutics. Front. Bioeng. Biotechnol. 2021, 9, 628137. 10.3389/fbioe.2021.628137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna J.; Hossain G. S.; Kocerha J. The Potential for Microrna Therapeutics and Clinical Research. Front. Genet. 2019, 10, 00478. 10.3389/fgene.2019.00478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobba C. M.; Fei Q.; Shukla V.; Lee H.; Patel P.; Putman R. K.; Spitzer C.; Tsai M.; Wewers M. D.; Lee R. J.; Christman J. W.; Ballinger M. N.; Ghadiali S. N.; Englert J. A. Nanoparticle Delivery of Microrna-146a Regulates Mechanotransduction in Lung Macrophages and Mitigates Injury During Mechanical Ventilation. Nat. Commun. 2021, 12, 289. 10.1038/s41467-020-20449-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dovgan I.; Koniev O.; Kolodych S.; Wagner A. Antibody–Oligonucleotide Conjugates as Therapeutic, Imaging, and Detection Agents. Bioconjugate Chem. 2019, 30, 2483–2501. 10.1021/acs.bioconjchem.9b00306. [DOI] [PubMed] [Google Scholar]
- DeLong R. Ushering in a New Era of RNA-Based Therapies. Commun. Biol. 2021, 4, 577. 10.1038/s42003-021-02150-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D. R.; Levy J.; Ma Y.; Wei H.; Comander J.; Tachida Y.; Pierce E. A.; Liu Q.; Pendse N.. Targeted Base Editing of the Ush2a Gene. Patent WO2021222318, 2021.
- Darmostuk M.; Rimpelova S.; Gbelcova H.; Ruml T. Current Approaches in SELEX: An Update to Aptamer Selection Technology. Biotechnol. Adv. 2015, 33, 1141–1161. 10.1016/j.biotechadv.2015.02.008. [DOI] [PubMed] [Google Scholar]
- Lamberti G.; Barba A. A. Drug Delivery of siRNA Therapeutics. Pharmaceutics 2020, 12, 178. 10.3390/pharmaceutics12020178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiser A.; Coleman D.; Dannull J.; Yancey D.; Maurice M. A.; Lallas C. D.; Dahm P.; Niedzwiecki D.; Gilboa E.; Vieweg J. Autologous Dendritic Cells Transfected with Prostate-Specific Antigen RNA Stimulate CTL Responses against Metastatic Prostate Tumors. J. Clin. Invest. 2002, 109, 409–417. 10.1172/JCI0214364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahin U.; Karikó K.; Türeci Ö. mRNA-Based Therapeutics — Developing a New Class of Drugs. Nat. Rev. Drug Discovery 2014, 13, 759–780. 10.1038/nrd4278. [DOI] [PubMed] [Google Scholar]
- Saeedi S.; Israel S.; Nagy C.; Turecki G. The Emerging Role of Exosomes in Mental Disorders. Transl. Psychiatry 2019, 9, 122. 10.1038/s41398-019-0459-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condrat C. E.; Thompson D. C.; Barbu M. G.; Bugnar O. L.; Boboc A.; Cretoiu D.; Suciu N.; Cretoiu S. M.; Voinea S. C. miRNAs as Biomarkers in Disease: Latest Findings Regarding Their Role in Diagnosis and Prognosis. Cells 2020, 9, 276. 10.3390/cells9020276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tribolet L.; Kerr E.; Cowled C.; Bean A. G. D.; Stewart C. R.; Dearnley M.; Farr R. J. MicroRNA Biomarkers for Infectious Diseases: From Basic Research to Biosensing. Front. Microbiol. 2020, 11, 01197. 10.3389/fmicb.2020.01197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravkin H. D.; Givton O.; Geffen D. B.; Rubin E. Direct Comparison Shows That mRNA-Based Diagnostics Incorporate Information Which Cannot Be Learned Directly from Genomic Mutations. BMC Bioinformatics 2020, 21, 196. 10.1186/s12859-020-3512-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminski M. M.; Abudayyeh O. O.; Gootenberg J. S.; Zhang F.; Collins J. J. CRISPR-Based Diagnostics. Nat. Biomed. Eng. 2021, 5, 643–656. 10.1038/s41551-021-00760-7. [DOI] [PubMed] [Google Scholar]
- Mermade 192x Synthesizer. https://shop.biosearchtech.com/nucleic-acid-chemistry-reagents-and-instruments/dna-and-rna-synthesis-instruments-and-accessories/dna-and-rna-oligonucleotide-synthesizers/mermade-192x/mermade-192x-synthesizer/p/NACINS-007 (accessed Nov 20, 2021).
- Tremblay F.; Bondurant L. D.; McIninch J. D.; Castoreno A.; Schlegel M. K.; Kaittanis C.. New Double Stranded RNA Agent Capable of Inhibiting Expression of Apolipoprotein C3 in Cell, Used for Treating Subject Having Disorder E.G. Hypertriglyceridemia, Non-Alcoholic Fatty Liver Disease, and Obesity That Would Benefit from Reduction in Apolipoprotein C3 Expression. Patents WO2021167841-A1, US2021292756-A1, US11162103-B2, 2021
- The Basics: In Vitro Transcription. https://www.thermofisher.com/us/en/home/references/ambion-tech-support/probe-labeling-systems/general-articles/the-basics-in-vitro-transcription.html (accessed 25 Nov 2021).
- Moderna . https://www.modernatx.com/about-us (accessed Nov 20, 2021).
- Biontech . https://biontech.de/ (accessed Nov 20, 2021).
- Curevac . https://www.curevac.com/en/ (accessed Nov 20, 2021).
- Stemirna Therapeutics . https://www.stemirna.com/en/about/index.aspx (accessed Nov 20, 2021).
- Cartesian Therapeutics . https://www.cartesiantherapeutics.com/rna-cell-therapy/ (accessed Nov 20, 2021).
- Alnylam https://www.alnylam.com/about-alnylam/ (accessed Nov 20, 2021).
- Dicerna https://dicerna.com/ (accessed Nov 20, 2021).
- siRNAomics https://sirnaomics.com/overview/ (accessed Nov 20, 2021).
- Arrowhead Pharmaceuticals . https://arrowheadpharma.com/about/ (accessed Nov 20, 2021).
- Silence Therapeutics . https://silence-therapeutics.com/about-us/default.aspx (accessed Nov 20, 2021).
- Ionis: A Force for Life. https://www.ionispharma.com/about/ (accessed 20 Niv 2021).
- Sarepta Therapeutics . https://www.sarepta.com/about-us (accessed Nov 20, 2021).
- Noxxon Pharma . https://www.noxxon.com/ (accessed Nov 20, 2021).
- Beam Therapeutics . https://beamtx.com/ (accessed Nov 20, 2021).
- Astrazeneca . https://www.astrazeneca.com/our-company.html (accessed Nov 20, 2021).
- Curevac Pipeline. https://www.curevac.com/en/pipeline/ (accessed Nov 20, 2021).
- Cardiovascular Diseases. https://www.who.int/health-topics/cardiovascular-diseases#tab=tab_1 (accessed Nov 20, 2021).
- Fda Approves Novartis Leqvio (Inclisiran), First-in-Class siRNA to Lower Cholesterol and Keep It Low with Two Doses a Year. https://www.novartis.com/news/media-releases/fda-approves-novartis-leqvio-inclisiran-first-class-sirna-lower-cholesterol-and-keep-it-low-two-doses-year (accessed Dec 27, 2021).
- Delivery Platforms. https://www.alnylam.com/our-science/sirna-delivery-platforms/ (accessed Nov 20, 2021).
- Study to Investigate Safety, Tolerability, Pk and Pd Response of Sln360 in Subjects with Elevated Lipoprotein(a). https://clinicaltrials.gov/ct2/show/NCT04606602?term=NCT04606602&draw=2&rank=1 (accessed Nov 20, 2021).
- Alnylam Pipeline. https://www.alnylam.com/alnylam-rnai-pipeline/ (accessed Nov 20, 2021).
- Zilebesiran (Aln-Agt), in Development for the Treatment of Hypertension. https://www.alnylam.com/wp-content/uploads/2021/06/2021-RNAi-Roundtables-Zilebesiran_FINAL.pdf (accessed Nov 20, 2021).
- Moderna’s Pipeline. https://www.modernatx.com/pipeline (accessed Nov 20, 2021).
- Anttila V.; Saraste A.; Knuuti J.; Jaakkola P.; Hedman M.; Svedlund S.; Lagerström-Fermér M.; Kjaer M.; Jeppsson A.; Gan L.-M. Synthetic mRNA Encoding Vegf-a in Patients Undergoing Coronary Artery Bypass Grafting: Design of a Phase 2a Clinical Trial. Mol. Ther. - Methods Clin. Dev. 2020, 18, 464–472. 10.1016/j.omtm.2020.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taimeh Z.; Loughran J.; Birks E. J.; Bolli R. Vascular Endothelial Growth Factor in Heart Failure. Nat. Rev. Cardiol. 2013, 10, 519–530. 10.1038/nrcardio.2013.94. [DOI] [PubMed] [Google Scholar]
- A Study to Evaluate ALN-AGT01 in Patients with Hypertension. https://clinicaltrials.gov/ct2/show/NCT03934307?term=NCT03934307&draw=2&rank=1 (accessed Nov 20, 2021).
- Azd8601 Study in Cabg Patients. https://clinicaltrials.gov/ct2/show/NCT03370887?term=NCT03370887&draw=2&rank=1 (accessed Nov 20, 2021).
- Saklayen M. G. The Global Epidemic of the Metabolic Syndrome. Curr. Hypertens. Rep. 2018, 20, 12. 10.1007/s11906-018-0812-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diabetes. https://www.who.int/health-topics/diabetes#tab=tab_1 (accessed Nov 20, 2021).
- The Ionis Antisense Pipeline. https://www.ionispharma.com/ionis-innovation/pipeline/ (accessed Nov 20, 2021).
- Morgan E. S.; Tai L. J.; Pham N. C.; Overman J. K.; Watts L. M.; Smith A.; Jung S. W.; Gajdošík M.; Krššák M.; Krebs M.; Geary R. S.; Baker B. F.; Bhanot S. Antisense Inhibition of Glucagon Receptor by Ionis-Gcgr(Rx) Improves Type 2 Diabetes without Increase in Hepatic Glycogen Content in Patients with Type 2 Diabetes on Stable Metformin Therapy. Diabetes Care 2019, 42, 585–593. 10.2337/dc18-1343. [DOI] [PubMed] [Google Scholar]
- Paik J.; Duggan S. Volanesorsen: First Global Approval. Drugs 2019, 79, 1349–1354. 10.1007/s40265-019-01168-z. [DOI] [PubMed] [Google Scholar]
- Witztum J. L.; Gaudet D.; Freedman S. D.; Alexander V. J.; Digenio A.; Williams K. R.; Yang Q.; Hughes S. G.; Geary R. S.; Arca M.; Stroes E. S. G.; Bergeron J.; Soran H.; Civeira F.; Hemphill L.; Tsimikas S.; Blom D. J.; O’Dea L.; Bruckert E. Volanesorsen and Triglyceride Levels in Familial Chylomicronemia Syndrome. N. Engl. J. Med. 2019, 381, 531–542. 10.1056/NEJMoa1715944. [DOI] [PubMed] [Google Scholar]
- Strnad P.; McElvaney N. G.; Lomas D. A. Alpha(1)-Antitrypsin Deficiency. N Engl J. Med. 2020, 382, 1443–1455. 10.1056/NEJMra1910234. [DOI] [PubMed] [Google Scholar]
- Wooddell C. I.; Blomenkamp K.; Peterson R. M.; Subbotin V. M.; Schwabe C.; Hamilton J.; Chu Q.; Christianson D. R.; Hegge J. O.; Kolbe J.; Hamilton H. L.; Branca-Afrazi M. F.; Given B. D.; Lewis D. L.; Gane E.; Kanner S. B.; Teckman J. H. Development of an RNAi Therapeutic for Alpha-1-Antitrypsin Liver Disease. JCI Insight 2020, 5, 135348. 10.1172/jci.insight.135348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Precise, Versatile Editing Platform. https://beamtx.com/our-portfolio/ (accessed Nov 20, 2021).
- Understanding the Human Genome. https://beamtx.com/our-science/ (accessed Nov 20, 2021).
- Methylmalonic Acidemia. https://medlineplus.gov/genetics/condition/methylmalonic-acidemia/ (accessed Nov 20, 2021).
- Moderna Announces First Patient Dosed in Phase 1/2 Study of mRNA-3705 for Methylmalonic Acidemia. https://investors.modernatx.com/news-releases/news-release-details/moderna-announces-first-patient-dosed-phase-12-study-mrna-3705 (accessed Nov 20, 2021).
- Safety, Tolerability and Efficacy of Isis-Gcgrrx in Type 2 Diabetes. https://clinicaltrials.gov/ct2/show/NCT01885260?term=NCT01885260&draw=2&rank=1 (accessed Nov 20, 2021).
- Study of Aro-Aat in Patients with Alpha-1 Antitrypsin Deficiency Associated Liver Disease (Aatd). https://clinicaltrials.gov/ct2/show/NCT03946449?term=NCT03946449&draw=2&rank=1 (accessed Nov 20, 2021).
- A Study to Assess Safety, Pharmacokinetics, and Pharmacodynamics of mRNA-3705 in Participants with Isolated Methylmalonic Acidemia. https://clinicaltrials.gov/ct2/show/NCT04899310?term=NCT04899310&draw=2&rank=1 (accessed Nov 20, 2021).
- Moon A. M.; Singal A. G.; Tapper E. B. Contemporary Epidemiology of Chronic Liver Disease and Cirrhosis. Clin Gastroenterol Hepatol 2020, 18, 2650–2666. 10.1016/j.cgh.2019.07.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asrani S. K.; Devarbhavi H.; Eaton J.; Kamath P. S. Burden of Liver Diseases in the World. J. Hepatol 2019, 70, 151–171. 10.1016/j.jhep.2018.09.014. [DOI] [PubMed] [Google Scholar]
- Acute Hepatic Porphyria (Ahp). https://liverfoundation.org/acute-hepatic-porphyria-ahp/#what-are-some-symptoms-of-ahp (accessed Nov 20, 2021).
- Alnylam Announces Approval of Givlaari (Givosiran) by the U.S. Food and Drug Administration (FDA). https://investors.alnylam.com/press-release?id=24281 (accessed Nov 20, 2021).
- A Study to Assess Safety, Tolerability, Pk and Pd of Azd2693 in Non-Alcoholic Steatohepatitis Patients. https://clinicaltrials.gov/ct2/show/NCT04483947?term=NCT04483947&draw=2&rank=1 (accessed Nov 20, 2021).
- BasuRay S.; Wang Y.; Smagris E.; Cohen J. C.; Hobbs H. H. Accumulation of Pnpla3 on Lipid Droplets Is the Basis of Associated Hepatic Steatosis. Proc. Natl. Acad. Sci. U. S. A. 2019, 116, 9521–9526. 10.1073/pnas.1901974116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FDA Approves Givosiran for Acute Hepatic Porphyria. https://www.fda.gov/drugs/resources-information-approved-drugs/fda-approves-givosiran-acute-hepatic-porphyria (accessed Nov 20, 2021).
- Sung H.; Ferlay J.; Siegel R. L.; Laversanne M.; Soerjomataram I.; Jemal A.; Bray F. Global Cancer Statistics 2020: Globocan Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA: A Cancer Journal for Clinicians 2021, 71, 209–249. 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- Noxxon ’S Pipeline. https://www.noxxon.com/index.php?option=com_content&view=article&id=15&Itemid=503 (accessed Nov 20, 2021).
- Nox-A12 https://www.noxxon.com/index.php?option=com_content&view=article&id=21&Itemid=478 (accessed Nov 20, 2021).
- Guo F.; Wang Y.; Liu J.; Mok S. C.; Xue F.; Zhang W. Cxcl12/Cxcr4: A Symbiotic Bridge Linking Cancer Cells and Their Stromal Neighbors in Oncogenic Communication Networks. Oncogene 2016, 35, 816–826. 10.1038/onc.2015.139. [DOI] [PubMed] [Google Scholar]
- siRNAomics Pipeline. https://sirnaomics.com/pipeline/ (accessed Nov 20, 2021).
- siRNAomics Receives Fda Approval of Ind for Phase 1 Clinical Trial of Systemic RNAi Therapeutic Stp707 for Solid Tumor Treatment. https://sirnaomics.com/news/sirnaomics-receives-fda-approval-of-ind-for-phase-1-clinical-trial-of-systemic-rnai-therapeutic-stp707-for-solid-tumor-treatment/ (accessed Nov 20, 2021).
- Cartesian’s Pipeline. https://www.cartesiantherapeutics.com/clinical-trials/ (accessed Nov 20, 2021).
- What Is Multiple Myeloma? https://www.cancer.org/cancer/multiple-myeloma/about/what-is-multiple-myeloma.html (accessed Nov 20, 2021).
- Glioblastoma Treatment with Irradiation and Olaptesed Pegol (Nox-A12) in Mgmt Unmethylated Patients (Gloria). https://clinicaltrials.gov/ct2/show/NCT04121455?term=NCT04121455&draw=2&rank=1 (accessed Nov 20, 2021).
- Olaptesed with Pembrolizumab and Nanoliposomal Irinotecan or Gemcitabine/Nab-Paclitaxel in Mss Pancreatic Cancer (Optimus). https://clinicaltrials.gov/ct2/show/NCT04901741?term=NCT04901741&draw=2&rank=1 (accessed Nov 20, 2021).
- Ph. 1, Evaluation of Safety, Tolerability, PK, Anti-Tumor Activity of STP707 IV in Subjects with Solid Tumors. https://clinicaltrials.gov/ct2/show/NCT05037149?term=NCT05037149&draw=2&rank=1 (accessed Nov 20, 2021).
- Descartes-11 in Multiple Myeloma. https://clinicaltrials.gov/ct2/show/NCT03994705?term=NCT03994705&draw=2&rank=1 (accessed Nov 20, 2021).
- Who Coronavirus (COVID-19) Dashboard. https://covid19.who.int/ (accessed Feb 23, 2022).
- Pfizer-Biontech COVID-19 Vaccine Comirnaty® Receives Full U.S. FDA Approval for Individuals 16 Years and Older. https://www.pfizer.com/news/press-release/press-release-detail/pfizer-biontech-covid-19-vaccine-comirnatyr-receives-full (accessed Nov 20, 2021).
- Moderna COVID-19 Vaccine. https://www.fda.gov/emergency-preparedness-and-response/coronavirus-disease-2019-covid-19/moderna-covid-19-vaccine (accessed Nov 20, 2021).
- Coronavirus (COVID-19) Vaccinations. https://ourworldindata.org/covid-vaccinations?country=OWID_WRL (accessed Feb 23, 2022).
- Ab-729 (GalNAc-RNAi). http://www.arbutusbio.com/portfolio/ab-729-galnac-rnai.php (accessed Nov 20, 2021).
- Kapoor R.; Kottilil S. Strategies to Eliminate Hbv Infection. Future Virol 2014, 9, 565–585. 10.2217/fvl.14.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moderna Announces First Participant Dosed in Phase 1/2 Study of Its Quadrivalent Seasonal Flu mRNA Vaccine. https://investors.modernatx.com/news-releases/news-release-details/moderna-announces-first-participant-dosed-phase-12-study-its (accessed Nov 20, 2021).
- Moderna Receives Fda Fast Track Designation for Respiratory Syncytial Virus (RSV) Vaccine (mRNA-1345). https://investors.modernatx.com/news-releases/news-release-details/moderna-receives-fda-fast-track-designation-respiratory (accessed Nov 20, 2021).
- Xu L.; Ma Z.; Li Y.; Pang Z.; Xiao S. Antibody Dependent Enhancement: Unavoidable Problems in Vaccine Development. Adv. Immunol. 2021, 151, 99–133. 10.1016/bs.ai.2021.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safety, Tolerability, and Pharmacokinetics of Ab-836 in Healthy Subjects and Subjects with Chronic Hbv Infection. https://clinicaltrials.gov/ct2/show/NCT04775797?term=NCT04775797&draw=2&rank=1 (accessed Nov 20, 2021).
- A Study of mRNA-1010 Seasonal Influenza Vaccine in Healthy Adults. https://clinicaltrials.gov/ct2/show/NCT04956575?term=NCT04956575&draw=2&rank=1 (accessed Nov 20, 2021).
- A Dose Escalation Study to Evaluate Safety, Reactogenicity, and Immunogenicity of mRNA-1345 in Healthy Adults and in Children Who Are Respiratory Syncytial Virus (Rsv)-Seropositive. https://clinicaltrials.gov/ct2/show/NCT04528719?term=NCT04528719&draw=2&rank=1 (accessed Nov 20, 2021).
- Neuromuscular Disorders. https://www.cedars-sinai.org/health-library/diseases-and-conditions/n/neuromuscular-disorders.html (accessed Nov 20, 2021).
- Neurological Disorders. https://dphhs.mt.gov/schoolhealth/chronichealth/neurologicaldisorders (accessed Nov 20, 2021).
- Fda Grants Accelerated Approval to First Drug for Duchenne Muscular Dystrophy. https://www.fda.gov/news-events/press-announcements/fda-grants-accelerated-approval-first-drug-duchenne-muscular-dystrophy (accessed Nov 20, 2021).
- Fda Grants Accelerated Approval to First Targeted Treatment for Rare Duchenne Muscular Dystrophy Mutation. https://www.fda.gov/news-events/press-announcements/fda-grants-accelerated-approval-first-targeted-treatment-rare-duchenne-muscular-dystrophy-mutation (accessed Nov 20, 2021).
- Fda Approves Targeted Treatment for Rare Duchenne Muscular Dystrophy Mutation. https://www.fda.gov/news-events/press-announcements/fda-approves-targeted-treatment-rare-duchenne-muscular-dystrophy-mutation (accessed Feb 23, 2022).
- Ottesen E. W. Iss-N1Makes the First Fda-Approved Drug for Spinal Muscular Atrophy. Transl. Neurosci. 2017, 8, 1–6. 10.1515/tnsci-2017-0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gales L. Tegsedi (Inotersen): An Antisense Oligonucleotide Approved for the Treatment of Adult Patients with Hereditary Transthyretin Amyloidosis. Pharmaceuticals 2019, 12, 78. 10.3390/ph12020078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Aguado I.; Rodriguez-Castejon J.; Vicente-Pascual M.; Rodriguez-Gascon A.; Solinis M. A.; del Pozo-Rodriguez A. Nanomedicines to Deliver mRNA: State of the Art and Future Perspectives. Nanomaterials 2020, 10, 364. 10.3390/nano10020364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Building an Industry-Leading Genetic Medicine Pipeline. https://www.sarepta.com/products-pipeline/pipeline (accessed Nov 20, 2021).
- What Is Als? https://www.als.org/understanding-als/what-is-als (accessed Nov 20, 2021).
- Delivering on the RNA Revolution. https://www.aviditybiosciences.com/pipeline/pipeline-overview/ (accessed Nov 20, 2021).
- Holt I.; Jacquemin V.; Fardaei M.; Sewry C. A.; Butler-Browne G. S.; Furling D.; Brook J. D.; Morris G. E. Muscleblind-Like Proteins: Similarities and Differences in Normal and Myotonic Dystrophy Muscle. Am. J. Pathol. 2009, 174, 216–227. 10.2353/ajpath.2009.080520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akinc A.; Maier M. A.; Manoharan M.; Fitzgerald K.; Jayaraman M.; Barros S.; Ansell S.; Du X. Y.; Hope M. J.; Madden T. D.; Mui B. L.; Semple S. C.; Tam Y. K.; Ciufolini M.; Witzigmann D.; Kulkarni J. A.; van der Meel R.; Cullis P. R. The Onpattro Story and the Clinical Translation of Nanomedicines Containing Nucleic Acid-Based Drugs. Nat. Nanotechnol. 2019, 14, 1084–1087. 10.1038/s41565-019-0591-y. [DOI] [PubMed] [Google Scholar]
- A Study of Biib067 When Initiated in Clinically Presymptomatic Adults with a Confirmed Superoxide Dismutase 1 Mutation (Atlas). https://clinicaltrials.gov/ct2/show/NCT04856982?term=NCT04856982&draw (accessed Nov 20, 2021).
- Study of Aoc 1001 in Adult Myotonic Dystrophy Type 1 (Dm1) Patients (Marina). https://clinicaltrials.gov/ct2/show/NCT05027269?term=NCT05027269&draw=2&rank=1 (accessed Nov 20, 2021).
- Blindness and Vision Impairment. https://www.who.int/news-room/fact-sheets/detail/blindness-and-visual-impairment (accessed Nov 20, 2021).
- Wong W. L.; Su X.; Li X.; Cheung C. M.; Klein R.; Cheng C. Y.; Wong T. Y. Global Prevalence of Age-Related Macular Degeneration and Disease Burden Projection for 2020 and 2040: A Systematic Review and Meta-Analysis. Lancet Glob. Health 2014, 2, e106–16. 10.1016/S2214-109X(13)70145-1. [DOI] [PubMed] [Google Scholar]
- Nagpal M.; Nagpal K.; Nagpal P. N. A Comparative Debate on the Various Anti-Vascular Endothelial Growth Factor Drugs: Pegaptanib Sodium (Macugen), Ranibizumab (Lucentis) and Bevacizumab (Avastin). Indian J. Ophthalmol. 2007, 55, 437–439. 10.4103/0301-4738.36478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamis A. P.; Altaweel M.; Bressler N. M.; Cunningham E. T. Jr.; Davis M. D.; Goldbaum M.; Gonzales C.; Guyer D. R.; Barrett K.; Patel M. Changes in Retinal Neovascularization after Pegaptanib (Macugen) Therapy in Diabetic Individuals. Ophthalmology 2006, 113, 23–28. 10.1016/j.ophtha.2005.10.012. [DOI] [PubMed] [Google Scholar]
- Jaffe G. J.; Westby K.; Csaky K. G.; Monés J.; Pearlman J. A.; Patel S. S.; Joondeph B. C.; Randolph J.; Masonson H.; Rezaei K. A. C5 Inhibitor Avacincaptad Pegol for Geographic Atrophy Due to Age-Related Macular Degeneration: A Randomized Pivotal Phase 2/3 Trial. Ophthalmology 2021, 128, 576–586. 10.1016/j.ophtha.2020.08.027. [DOI] [PubMed] [Google Scholar]
- A Phase 3 Safety and Efficacy Study of Intravitreal Administration of Zimura (Complement C5 Inhibitor). https://clinicaltrials.gov/ct2/show/NCT04435366?term=NCT04435366&draw=2&rank=1 (accessed Nov 20, 2021).
- Study with Qr-504a to Evaluate Safety, Tolerability & Corneal Endothelium Molecular Biomarker(S) in Subjects with Fecd3 (Fuchs Focus). https://clinicaltrials.gov/ct2/show/NCT05052554 (accessed Nov 20, 2021).
- Qr-504a for Fuchs Endothelial Corneal Dystrophy. https://www.proqr.com/qr-504a-for-fuchs-endothelial-corneal-dystrophy (accessed Nov 20, 2021).
- 850 Million People Worldwide Have Kidney Disease. https://www.webmd.com/kidney-stones/news/20180705/850-million-people-worldwide-have-kidney-disease (accessed Nov 20, 2021).
- Scott L. J.; Keam S. J. Lumasiran: First Approval. Drugs 2021, 81, 277–282. 10.1007/s40265-020-01463-0. [DOI] [PubMed] [Google Scholar]
- Garrelfs S. F.; Frishberg Y.; Hulton S. A.; Koren M. J.; O’Riordan W. D.; Cochat P.; Deschênes G.; Shasha-Lavsky H.; Saland J. M.; Van’t Hoff W. G.; Fuster D. G.; Magen D.; Moochhala S. H.; Schalk G.; Simkova E.; Groothoff J. W.; Sas D. J.; Meliambro K. A.; Lu J.; Sweetser M. T.; Garg P. P.; Vaishnaw A. K.; Gansner J. M.; McGregor T. L.; Lieske J. C. Lumasiran, an RNAi Therapeutic for Primary Hyperoxaluria Type 1. N. Engl. J. Med. 2021, 384, 1216–1226. 10.1056/NEJMoa2021712. [DOI] [PubMed] [Google Scholar]
- Lee E. C.; Valencia T.; Allerson C.; Schairer A.; Flaten A.; Yheskel M.; Kersjes K.; Li J.; Gatto S.; Takhar M.; Lockton S.; Pavlicek A.; Kim M.; Chu T.; Soriano R.; Davis S.; Androsavich J. R.; Sarwary S.; Owen T.; Kaplan J.; Liu K.; Jang G.; Neben S.; Bentley P.; Wright T.; Patel V. Discovery and Preclinical Evaluation of Anti-Mir-17 Oligonucleotide Rgls4326 for the Treatment of Polycystic Kidney Disease. Nat. Commun. 2019, 10, 4148. 10.1038/s41467-019-11918-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schairer A.; Valencia T.; Lockton S.; Gatto S.; Kim M.; Wallace D.; Lee E.. Efficacy of RGLS4326 in Human Primary 3D-Cyst Cultures Derived from Autosomal Dominant Polycystic Kidney Disease (ADPKD) Donors http://regulusrx.com/wp-content/uploads/2018/11/ASN-2018-RGLS4326-Poster-3021589-FINAL.pdf (accessed Nov 20, 2021).
- Ji F. P.; Wen L.; Zhang Y. P.; Liu E. P.; Wen J. G., Serum Complement Factor B Is Associated with Disease Activity and Progression of Idiopathic Membranous Nephropathy Concomitant with Iga Nephropathy. Int. Urol. Nephrol. 2021. 10.1007/s11255-021-02997-2 [DOI] [PubMed] [Google Scholar]
- A Study to Evaluate Lumasiran in Patients with Advanced Primary Hyperoxaluria Type 1 (Illuminate-C). https://clinicaltrials.gov/ct2/show/NCT04152200?term=NCT04152200&draw=2&rank=1 (accessed Nov 20, 2021).
- A Study of Rgls4326 in Patients with Autosomal Dominant Polycystic Kidney Disease. https://clinicaltrials.gov/ct2/show/NCT04536688?term=NCT04536688&draw=2&rank=1 (accessed Nov 20, 2021).
- A Study to Evaluate the Effectiveness and Safety of IONIS-FB-LRx, an Antisense Inhibitor of Complement Factor B, in Adult Participants with Primary IgA Nephropathy. https://clinicaltrials.gov/ct2/show/NCT04014335?term=NCT04014335&draw=2&rank=1 (accessed Nov 20, 2021).
- The Global Impact of Respiratory Disease. https://www.who.int/gard/publications/The_Global_Impact_of_Respiratory_Disease.pdf (accessed Nov 20, 2021).
- Cartesian Therapeutics Initiates Clinical Trial of First RNA-Engineered Cell Therapy for Acute Respiratory Distress Syndrome and COVID-19 https://www.prnewswire.com/news-releases/cartesian-therapeutics-initiates-clinical-trial-of-first-rna-engineered-cell-therapy-for-acute-respiratory-distress-syndrome-and-covid-19-301121921.html (accessed Nov 20, 2021).
- Study of Descartes-30 in Acute Respiratory Distress Syndrome. https://clinicaltrials.gov/ct2/show/NCT04524962?term=NCT04524962&draw=2&rank=1 (accessed Nov 20, 2021).
- Ratjen F.; Bell S. C.; Rowe S. M.; Goss C. H.; Quittner A. L.; Bush A. Cystic Fibrosis. Nat. Rev. Dis. Primers 2015, 1, 15010–15010. 10.1038/nrdp.2015.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Translate Bio Pipeline - Present and Future Focus. https://translate.bio/pipeline/ (accessed Nov 20, 2021).
- Chow M. Y. T.; Qiu Y.; Lam J. K. W. Inhaled RNA Therapy: From Promise to Reality. Trends Pharmacol. Sci. 2020, 41, 715–729. 10.1016/j.tips.2020.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Translate Bio Announces Results from Second Interim Data Analysis from Ongoing Phase 1/2 Clinical Trial of Mrt5005 in Patients with Cystic Fibrosis (CF). https://investors.translate.bio/news-releases/news-release-details/translate-bio-announces-results-second-interim-data-analysis (accessed Nov 20, 2021).
- Arrowhead’s Pipeline - Novel Drugs Totreat Intractable Diseases. https://arrowheadpharma.com/pipeline/ (accessed Nov 20, 2021).
- Arrowhead Pauses Aro-Enac Phase 1/2 Clinical Study. https://ir.arrowheadpharma.com/news-releases/news-release-details/arrowhead-pauses-aro-enac-phase-12-clinical-study (accessed Nov 20, 2021).
- Study to Evaluate the Safety & Tolerability of Mrt5005 Administered by Nebulization in Adults with Cystic Fibrosis (Restore-Cf). https://clinicaltrials.gov/ct2/show/NCT03375047?term=NCT03375047&draw=2&rank=1 (accessed Nov 20, 2021).
- Study of Aro-Enac in Healthy Volunteers and in Patients with Cystic Fibrosis. https://clinicaltrials.gov/ct2/show/NCT04375514?term=NCT04375514&draw=2&rank=1 (accessed Nov 20, 2021).
- Salinas Cisneros G.; Thein S. L. Recent Advances in the Treatment of Sickle Cell Disease. Front. Physiol. 2020, 11, 435. 10.3389/fphys.2020.00435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galanello R.; Origa R. Beta-Thalassemia. Orphanet J. Rare Dis. 2010, 5, 11. 10.1186/1750-1172-5-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beam Therapeutics Provides Business and Pipeline Updates and Reports Third Quarter 2021 Financial Results. https://investors.beamtx.com/node/7596 (accessed Nov 20, 2021).
- Silence Therapeutics Pipeline - Our mRNAi Gold Pipeline Targets. https://silence-therapeutics.com/our-pipeline/default.aspx (accessed Nov 20, 2021).
- SLN-124. https://thalassaemia.org.cy/clinical-trial-updates/sln-124/ (accessed Nov 20, 2021).
- A Study Investigate the Safety, Tolerability, Pharmacokinetic, and Pharmacodynamic Response of Sln124 in Adults with Alpha/Beta-Thalassaemia and Very Low- and Low-Risk Myelodysplastic Syndrome. https://clinicaltrials.gov/ct2/show/NCT04718844?term=NCT04718844&draw=2&rank=1 (accessed Nov 20, 2021).
- A Study for Safety and Efficacy Evaluation of Various Doses of Stp705 in Reducing Keloid Recurrence. https://clinicaltrials.gov/ct2/show/NCT04844840?term=NCT04844840&draw=2&rank=1 (accessed Nov 20, 2021).
- siRNAomics Doses First Patient in Phase 2 Study of Stp705 for Keloid Scar Prevention. https://sirnaomics.com/news/sirnaomics-doses-first-patient-in-phase-2-study-of-stp705-for-keloid-scar-prevention/ (accessed Nov 20, 2021).
- Janeway C. A. Jr.; Travers P; Walport M.; et al. Autoimmune Responses Are Directed against Self Antigens. Immunobiology: The Immune System in Health and Disease, 5th ed.; Garland Science, New York, 2001. [Google Scholar]
- Moderna Builds on Clinical Validation of Systemic Delivery with Two Additional Development Candidates in New Autoimmune Therapeutic Area. https://investors.modernatx.com/news-releases/news-release-details/moderna-builds-clinical-validation-systemic-delivery-two (accessed Nov 20, 2021).
- Global Alcohol Action Plan 2022–2030 to Strengthen Implementation of the Global Strategy to Reduce the Harmful Use of Alcohol. https://cdn.who.int/media/docs/default-source/alcohol/action-plan-on-alcohol_first-draft-final_formatted.pdf?sfvrsn=b690edb0_1&download=true (accessed Nov 20, 2021).
- Alcohol Use Disorder (AUD). https://dicerna.com/patients/alcohol-use-disorder-aud/ (accessed Nov 20, 2021).
- A New and Unique Approach to Potentially Improving Treatment Outcomes for People with Alcohol Use Disorder (Aud). https://dicerna.com/pipeline/dcr-aud/ (accessed Nov 20, 2021).
- A Study to Evaluate the Safety, Tolerability, Pharmacokinetics, and Pharmacodynamics of mRNA-6231 in Healthy Adults. https://clinicaltrials.gov/ct2/show/NCT04916431?term=NCT04916431&draw=2&rank=1 (accessed Nov 20, 2021).
- Study of DCR-AUD in Healthy Volunteers. https://clinicaltrials.gov/ct2/show/NCT05021640?term=NCT05021640&draw=2&rank=1 (accessed Nov 20, 2021).
- Lehninger A. L.Biochemistry: The Molecular Basis of Cell Structure and Function, 2nd ed.; Worth Publishers: New York, NY, 1975. [Google Scholar]
- McIninch J. D.; Keating M.; Schlegel M. K.; Castoreno A.; Jadhav V. R.; Kaittanis C.; Castellanos-Rizaldos E.; Pandya B. A.. Compositions and Methods for Silencing VEGF-A Expression. Patent WO/2021/163066, 2021.
- Parr C. J. C.; Wada S.; Kotake K.; Kameda S.; Matsuura S.; Sakashita S.; Park S.; Sugiyama H.; Kuang Y.; Saito H. N1-Methylpseudouridine Substitution Enhances the Performance of Synthetic mRNA Switches in Cells. Nucleic Acids Res. 2020, 48, e35–e35. 10.1093/nar/gkaa070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shatkin A. J. Capping of Eucaryotic mRNAs. Cell 1976, 9, 645–653. 10.1016/0092-8674(76)90128-8. [DOI] [PubMed] [Google Scholar]
- Chan J. H.; Lim S.; Wong W. F. Antisense Oligonucleotides: From Design to Therapeutic Application. Clin. Exp. Pharmacol. Physiol. 2006, 33, 533–540. 10.1111/j.1440-1681.2006.04403.x. [DOI] [PubMed] [Google Scholar]
- Phosphorothioate Oligonucleotides. https://www.sigmaaldrich.com/US/en/technical-documents/technical-article/genomics/gene-expression-and-silencing/phosphorothioates (accessed Nov 20, 2021).
- Putney S. D.; Benkovic S. J.; Schimmel P. R. A DNA Fragment with an Alpha-Phosphorothioate Nucleotide at One End Is Asymmetrically Blocked from Digestion by Exonuclease Iii and Can Be Replicated in Vivo. Proc. Natl. Acad. Sci. U. S. A. 1981, 78, 7350–7354. 10.1073/pnas.78.12.7350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantley W.; McIninch J. D.; Castoreno A.; Kaittanis C.; Schlegel M. K.. Compositions and Methods for Silencing Scn9a Expression. Patent WO/2021/207189, 2021.
- Nielsen P. E.; Egholm M. An Introduction to Peptide Nucleic Acid. Curr. Issues Mol. Biol. 1999, 1, 89–104. [PubMed] [Google Scholar]
- Blocking Groups and Annotations for Registry Sequences. https://www.cas.org/sites/default/files/documents/blocking.pdf (accessed Jan 1, 2022).
- Chao Y.; Li H.-B.; Zhou J. Multiple Functions of RNA Methylation in T Cells: A Review. Front. Immunol. 2021, 12, 627455. 10.3389/fimmu.2021.627455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry R. P.; Kelley D. E. Existence of Methylated Messenger RNA in Mouse L Cells. Cell 1974, 1, 37–42. 10.1016/0092-8674(74)90153-6. [DOI] [Google Scholar]
- Summerton J. Morpholino Antisense Oligomers: The Case for an RNAse H-Independent Structural Type. Biochim. Biophys. Acta 1999, 1489, 141–158. 10.1016/S0167-4781(99)00150-5. [DOI] [PubMed] [Google Scholar]
- Fitzgerald K.; Tremblay F.; McIninch J. D.. G Protein-Coupled Receptor 146 (Gpr146) iRNA Compositions and Methods of Use Thereof. Patent WO/2021/174056, 2021.
- Springer A. D. GalNAc-siRNA Conjugates: Leading the Way for Delivery of RNAi Therapeutics. Nucleic Acid Ther. 2018, 28, 109–118. 10.1089/nat.2018.0736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald K.; Tremblay F.; McIninch J. D.. G Protein-Coupled Receptor 146 (Gpr146) iRNA Compositions and Methods of Use Thereof. Patent WO/2021/174056, 2021.
- Pubchem Substance Record for Sid 135267507, Aiw6036fas. https://pubchem.ncbi.nlm.nih.gov/substance/135267507 (accessed Dec 9, 2021).
- Pegaptanib . https://en.wikipedia.org/wiki/Pegaptanib#/media/File:Pegaptanib_sodium_skeletal.svg (accessed Dec 8, 2021).
- Vieira Araujo Soares Da Silva P. M.siRNA Molecules, Methods of Production and Uses Thereof. U.S. Patent US20210332364, 2021.
- Kanasty R.; Dorkin J. R.; Vegas A.; Anderson D. Delivery Materials for siRNA Therapeutics. Nat. Mater. 2013, 12, 967–977. 10.1038/nmat3765. [DOI] [PubMed] [Google Scholar]
- CASData. https://www.cas.org/cas-data (accessed Nov 25, 2021).
- Yang N. Nonviral Gene Delivery System. Int. J. Pharmaceutical Investigation 2012, 2, 97–98. 10.4103/2230-973X.104388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koynova R.; Tenchov B.. Cationic Lipids: Molecular Structure/Transfection Activity Relationships and Interactions with Biomembranes. In Nucleic Acid Transfection; Bielke W., Erbacher C., Eds.; Springer-Verlag: Berlin, Heidelberg, 2010; Vol. 296, pp 51–93. [DOI] [PubMed] [Google Scholar]
- Tenchov R.; Bird R.; Curtze A. E.; Zhou Q. Lipid Nanoparticles—from Liposomes to mRNA Vaccine Delivery, a Landscape of Research Diversity and Advancement. ACS Nano 2021, 15, 16982–17015. 10.1021/acsnano.1c04996. [DOI] [PubMed] [Google Scholar]
- Mitchell M. J.; Billingsley M. M.; Haley R. M.; Wechsler M. E.; Peppas N. A.; Langer R. Engineering Precision Nanoparticles for Drug Delivery. Nat. Rev. Drug Disc. 2021, 20, 101–124. 10.1038/s41573-020-0090-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allison A. C.; Gregoriadis G. Liposomes as Immunological Adjuvants. Nature 1974, 252, 252–252. 10.1038/252252a0. [DOI] [PubMed] [Google Scholar]
- Gregoriadis G. Liposomes and mRNA: Two Technologies Together Create a COVID-19 Vaccine. Med. Drug Disc. 2021, 12, 100104. 10.1016/j.medidd.2021.100104. [DOI] [Google Scholar]
- Attia M. A.; Essa E. A.; Elebyary T. T.; Faheem A. M.; Elkordy A. A. Brief on Recent Application of Liposomal Vaccines for Lower Respiratory Tract Viral Infections: From Influenza to COVID-19 Vaccines. Pharmaceuticals (Basel) 2021, 14, 1173. 10.3390/ph14111173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosales-Mendoza S.; Wong-Arce A.; de Lourdes; Betancourt-Mendiola M.. RNA-Based Vaccines against Sars-Cov-2. In Biomedical Innovations to Combat COVID-19; Rosales-Mendoza S., Comas-Garcia M., Gonzalez-Ortega O., Eds.; Academic Press, 2022; Chap. 8, pp 129–152. [Google Scholar]
- Hajj K. A.; Ball R. L.; Deluty S. B.; Singh S. R.; Strelkova D.; Knapp C. M.; Whitehead K. A. Branched-Tail Lipid Nanoparticles Potently Deliver mRNA in Vivo Due to Enhanced Ionization at Endosomal pH. Small 2019, 15, e1805097. 10.1002/smll.201805097. [DOI] [PubMed] [Google Scholar]
- Heyes J. A.; Niculescu-Duvaz D.; Cooper R. G.; Springer C. J. Synthesis of Novel Cationic Lipids: Effect of Structural Modification on the Efficiency of Gene Transfer. J. Med. Chem. 2002, 45, 99–114. 10.1021/jm010918g. [DOI] [PubMed] [Google Scholar]
- Karmali P. P.; Chaudhuri A. Cationic Liposomes as Non-Viral Carriers of Gene Medicines: Resolved Issues, Open Questions, and Future Promises. Med. Res. Rev. 2007, 27, 696–722. 10.1002/med.20090. [DOI] [PubMed] [Google Scholar]
- Karmali P. P.; Kumar V. V.; Chaudhuri A. Design, Syntheses and in Vitro Gene Delivery Efficacies of Novel Mono-, Di- and Trilysinated Cationic Lipids: A Structure-Activity Investigation. J. Med. Chem. 2004, 47, 2123–2132. 10.1021/jm030541+. [DOI] [PubMed] [Google Scholar]
- Behr J P; Demeneix B; Loeffler J P; Perez-Mutul J Efficient Gene-Transfer into Mammalian Primary Endocrine-Cells with Lipopolyamine-Coated DNA. Proc. Natl. Acad. Sci. U. S. A. 1989, 86, 6982–6986. 10.1073/pnas.86.18.6982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari M. E.; Nguyen C. M.; Zelphati O.; Tsai Y. L.; Felgner P. L. Analytical Methods for the Characterization of Cationic Lipid Nucleic Acid Complexes. Hum. Gene Ther. 1998, 9, 341–351. 10.1089/hum.1998.9.3-341. [DOI] [PubMed] [Google Scholar]
- de Lima M. C. P.; Neves S.; Filipe A.; Duzgunes N.; Simoes S. Cationic Liposomes for Gene Delivery: From Biophysics to Biological Applications. Curr. Med. Chem. 2003, 10, 1221–1231. 10.2174/0929867033457430. [DOI] [PubMed] [Google Scholar]
- Wheeler C. J.; Felgner P. L.; Tsai Y. J.; Marshall J.; Sukhu L.; Doh S. G.; Hartikka J.; Nietupski J.; Manthorpe M.; Nichols M.; Plewe M.; Liang X. W.; Norman J.; Smith A.; Cheng S. H. A Novel Cationic Lipid Greatly Enhances Plasmid DNA Delivery and Expression in Mouse Lung. Proc. Natl. Acad. Sci. U. S. A. 1996, 93, 11454–11459. 10.1073/pnas.93.21.11454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y.; Tenchov R.; Smoot J.; Liu C.; Watkins S.; Zhou Q. A Comprehensive Review of the Global Efforts on COVID-19 Vaccine Development. ACS Central Sci. 2021, 7, 512–533. 10.1021/acscentsci.1c00120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarahovsky Y. S.; Arsenault A. L.; MacDonald R. C.; McIntosh T. J.; Epand R. M. Electrostatic Control of Phospholipid Polymorphism. Biophys. J. 2000, 79, 3193–3200. 10.1016/S0006-3495(00)76552-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koynova R.; Wang L.; MacDonald R. C. An Intracellular Lamellar - Nonlamellar Phase Transition Rationalizes the Superior Performance of Some Cationic Lipid Transfection Agents. Proc. Natl. Acad. Sci. U. S. A. 2006, 103, 14373–14378. 10.1073/pnas.0603085103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedmi R.; Ben-Arie N.; Peer D. The Systemic Toxicity of Positively Charged Lipid Nanoparticles and the Role of Toll-Like Receptor 4 in Immune Activation. Biomaterials 2010, 31, 6867–6875. 10.1016/j.biomaterials.2010.05.027. [DOI] [PubMed] [Google Scholar]
- Kalluri R.; LeBleu V. S. The Biology, Function, and Biomedical Applications of Exosomes. Science 2020, 367, aau6977. 10.1126/science.aau6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar J. R. Q&A: What Are Exosomes, Exactly?. BMC Biol. 2016, 14, 46. 10.1186/s12915-016-0268-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y.; Liu Y.; Liu H.; Tang W. H. Exosomes: Biogenesis, Biologic Function and Clinical Potential. Cell Biosci. 2019, 9, 19. 10.1186/s13578-019-0282-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Erviti L.; Seow Y.; Yin H.; Betts C.; Lakhal S.; Wood M. J. A. Delivery of siRNA to the Mouse Brain by Systemic Injection of Targeted Exosomes. Nat. Biotechnol. 2011, 29, 341–345. 10.1038/nbt.1807. [DOI] [PubMed] [Google Scholar]
- Shtam T. A.; Kovalev R. A.; Varfolomeeva E. Y.; Makarov E. M.; Kil Y. V.; Filatov M. V. Exosomes Are Natural Carriers of Exogenous siRNA to Human Cells in Vitro. Cell Commun. Signal. 2013, 11, 88. 10.1186/1478-811X-11-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamichhane T. N.; Raiker R. S.; Jay S. M. Exogenous DNA Loading into Extracellular Vesicles Via Electroporation Is Size-Dependent and Enables Limited Gene Delivery. Mol. Pharmaceutics 2015, 12, 3650–3657. 10.1021/acs.molpharmaceut.5b00364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X.; Brown B. A.; Siegel A. P.; El Masry M. S.; Zeng X.; Song W.; Das A.; Khandelwal P.; Clark A.; Singh K.; Guda P. R.; Gorain M.; Timsina L.; Xuan Y.; Jacobson S. C.; Novotny M. V.; Roy S.; Agarwal M.; Lee R. J.; Sen C. K.; Clemmer D. E.; Ghatak S. Exosome-Mediated Crosstalk between Keratinocytes and Macrophages in Cutaneous Wound Healing. ACS Nano 2020, 14, 12732–12748. 10.1021/acsnano.0c03064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subra C.; Laulagnier K.; Perret B.; Record M. Exosome Lipidomics Unravels Lipid Sorting at the Level of Multivesicular Bodies. Biochimie 2007, 89, 205–212. 10.1016/j.biochi.2006.10.014. [DOI] [PubMed] [Google Scholar]
- Ha D.; Yang N.; Nadithe V. Exosomes as Therapeutic Drug Carriers and Delivery Vehicles across Biological Membranes: Current Perspectives and Future Challenges. Acta Pharm. Sin B 2016, 6, 287–296. 10.1016/j.apsb.2016.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butreddy A.; Kommineni N.; Dudhipala N. Exosomes as Naturally Occurring Vehicles for Delivery of Biopharmaceuticals: Insights from Drug Delivery to Clinical Perspectives. Nanomaterials (Basel) 2021, 11, 1481. 10.3390/nano11061481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y.; Sun C.; Wang C.; Jankovic K. E.; Dong Y. Lipids and Lipid Derivatives for RNA Delivery. Chem. Rev. 2021, 121, 12181–12277. 10.1021/acs.chemrev.1c00244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C.; Zhang Y.; Dong Y. Lipid Nanoparticle-mRNA Formulations for Therapeutic Applications. Acc. Chem. Res. 2021, 54, 4283–4293. 10.1021/acs.accounts.1c00550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J.; Kundrat L.; Pishesha N.; Bilate A.; Theile C.; Maruyama T.; Dougan S. K.; Ploegh H. L.; Lodish H. F. Engineered Red Blood Cells as Carriers for Systemic Delivery of a Wide Array of Functional Probes. Proc. Natl. Acad. Sci. U. S. A. 2014, 111, 10131–10136. 10.1073/pnas.1409861111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S.; Feng S.; Ding L.; Liu Y.; Zhu Q.; Qian Z.; Gu Y. Nanomedicine Engulfed by Macrophages for Targeted Tumor Therapy. Int. J. Nanomed. 2016, 11, 4107–4124. 10.2147/IJN.S110146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar R.; Santa Chalarca C. F.; Bockman M. R.; Bruggen C. V.; Grimme C. J.; Dalal R. J.; Hanson M. G.; Hexum J. K.; Reineke T. M. Polymeric Delivery of Therapeutic Nucleic Acids. Chem. Rev. 2021, 121, 11527–11652. 10.1021/acs.chemrev.0c00997. [DOI] [PubMed] [Google Scholar]
- Zhu L.; Mahato R. I. Lipid and Polymeric Carrier-Mediated Nucleic Acid Delivery. Expert Opin. Drug Deliv. 2010, 7, 1209–1226. 10.1517/17425247.2010.513969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kataoka K.; Togawa H.; Harada A.; Yasugi K.; Matsumoto T.; Katayose S. Spontaneous Formation of Polyion Complex Micelles with Narrow Distribution from Antisense Oligonucleotide and Cationic Block Copolymer in Physiological Saline. Macromolecules 1996, 29, 8556–8557. 10.1021/ma961217+. [DOI] [Google Scholar]
- Neu M.; Fischer D.; Kissel T. Recent Advances in Rational Gene Transfer Vector Design Based on Poly(Ethylene Imine) and Its Derivatives. J. Gene Med. 2005, 7, 992–1009. 10.1002/jgm.773. [DOI] [PubMed] [Google Scholar]
- Vandamme T. F.; Brobeck L. Poly(Amidoamine) Dendrimers as Ophthalmic Vehicles for Ocular Delivery of Pilocarpine Nitrate and Tropicamide. J. Controlled Release 2005, 102, 23–38. 10.1016/j.jconrel.2004.09.015. [DOI] [PubMed] [Google Scholar]
- Urbiola K.; Blanco-Fernández L.; Ogris M.; Rödl W.; Wagner E.; Tros de Ilarduya C. Novel PAMAM-PEG-Peptide Conjugates for siRNA Delivery Targeted to the Transferrin and Epidermal Growth Factor Receptors. J. Personalized Med. 2018, 8, 4. 10.3390/jpm8010004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh P.; Gupta U.; Asthana A.; Jain N. K. Folate and Folate–PEG–PAMAM Dendrimers: Synthesis, Characterization, and Targeted Anticancer Drug Delivery Potential in Tumor Bearing Mice. Bioconjugate Chem. 2008, 19, 2239–2252. 10.1021/bc800125u. [DOI] [PubMed] [Google Scholar]
- Tang Y.; Li Y.-B.; Wang B.; Lin R.-Y.; van Dongen M.; Zurcher D. M.; Gu X.-Y.; Banaszak Holl M. M.; Liu G.; Qi R. Efficient in Vitro siRNA Delivery and Intramuscular Gene Silencing Using PEG-Modified PAMAM Dendrimers. Mol. Pharmaceutics 2012, 9, 1812–1821. 10.1021/mp3001364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piotrowski-Daspit A. S.; Kauffman A. C.; Bracaglia L. G.; Saltzman W. M. Polymeric Vehicles for Nucleic Acid Delivery. Adv. Drug Deliv. Rev. 2020, 156, 119–132. 10.1016/j.addr.2020.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaur S.; Wen Y.; Song J. H.; Parikh N. U.; Mangala L. S.; Blessing A. M.; Ivan C.; Wu S. Y.; Varkaris A.; Shi Y.; Lopez-Berestein G.; Frigo D. E.; Sood A. K.; Gallick G. E. Chitosan Nanoparticle-Mediated Delivery of miRNA-34a Decreases Prostate Tumor Growth in the Bone and Its Expression Induces Non-Canonical Autophagy. Oncotarget 2015, 6, 29161. 10.18632/oncotarget.4971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosco D.; Cilurzo F.; Maiuolo J.; Federico C.; Di Martino M. T.; Cristiano M. C.; Tassone P.; Fresta M.; Paolino D. Delivery of Mir-34a by Chitosan/Plga Nanoplexes for the Anticancer Treatment of Multiple Myeloma. Sci. Rep. 2015, 5, 17579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X.; Gu S.; Chen B.-F.; Shen W.-L.; Yin Z.; Xu G.-W.; Hu J.-J.; Zhu T.; Li G.; Wan C.; Ouyang H.-W.; Lee T.-L.; Chan W.-Y. Nanoparticle Delivery of Stable Mir-199a-5p Agomir Improves the Osteogenesis of Human Mesenchymal Stem Cells Via the Hif1a Pathway. Biomaterials 2015, 53, 239–250. 10.1016/j.biomaterials.2015.02.071. [DOI] [PubMed] [Google Scholar]
- Köping-Höggård M.; Tubulekas I.; Guan H.; Edwards K.; Nilsson M.; Vårum K. M.; Artursson P. Chitosan as a Nonviral Gene Delivery System. Structure–Property Relationships and Characteristics Compared with Polyethylenimine in Vitro and after Lung Administration in Vivo. Gene Ther. 2001, 8, 1108–1121. 10.1038/sj.gt.3301492. [DOI] [PubMed] [Google Scholar]
- Curtin C. M.; Tierney E. G.; McSorley K.; Cryan S.-A.; Duffy G. P.; O’Brien F. J. Combinatorial Gene Therapy Accelerates Bone Regeneration: Non-Viral Dual Delivery of Vegf and Bmp2 in a Collagen-Nanohydroxyapatite Scaffold. Adv. Healthcare Mater. 2015, 4, 223–227. 10.1002/adhm.201400397. [DOI] [PubMed] [Google Scholar]
- Capito R. M.; Spector M. Collagen Scaffolds for Nonviral IGF-1 Gene Delivery in Articular Cartilage Tissue Engineering. Gene Ther. 2007, 14, 721–732. 10.1038/sj.gt.3302918. [DOI] [PubMed] [Google Scholar]
- Cohen-Sacks H.; Elazar V.; Gao J.; Golomb A.; Adwan H.; Korchov N.; Levy R. J.; Berger M. R.; Golomb G. Delivery and Expression of pDNA Embedded in Collagen Matrices. J. Controlled Release 2004, 95, 309–320. 10.1016/j.jconrel.2003.11.001. [DOI] [PubMed] [Google Scholar]
- Aoife M. O.; Mahony; Martin J. O.; Neill; Bruno M. D. C. G.; Raphael D.; John F. C.; Caitriona M. O.; Driscoll Cyclodextrins for Non-Viral Gene and siRNA Delivery. Pharmaceutical Nanotechnol. 2013, 1, 6–14. 10.2174/2211738511301010006. [DOI] [Google Scholar]
- Chaturvedi K.; Ganguly K.; Kulkarni A. R.; Kulkarni V. H.; Nadagouda M. N.; Rudzinski W. E.; Aminabhavi T. M. Cyclodextrin-Based siRNA Delivery Nanocarriers: A State-of-the-Art Review. Expert Opin. Drug Deliv. 2011, 8, 1455–1468. 10.1517/17425247.2011.610790. [DOI] [PubMed] [Google Scholar]
- Singh R. P.; Hidalgo T.; Cazade P.-A.; Darcy R.; Cronin M. F.; Dorin I.; O’Driscoll C. M.; Thompson D. Self-Assembled Cationic B-Cyclodextrin Nanostructures for siRNA Delivery. Mol. Pharmaceutics 2019, 16, 1358–1366. 10.1021/acs.molpharmaceut.8b01307. [DOI] [PubMed] [Google Scholar]
- Mintzer M. A.; Simanek E. E. Nonviral Vectors for Gene Delivery. Chem. Rev. 2009, 109, 259–302. 10.1021/cr800409e. [DOI] [PubMed] [Google Scholar]
- El-Sayed A.; Harashima H. Endocytosis of Gene Delivery Vectors: From Clathrin-Dependent to Lipid Raft-Mediated Endocytosis. Mol. Ther. 2013, 21, 1118–1130. 10.1038/mt.2013.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bus T.; Traeger A.; Schubert U. S. The Great Escape: How Cationic Polyplexes Overcome the Endosomal Barrier. J. Mater. Chem. B 2018, 6, 6904–6918. 10.1039/C8TB00967H. [DOI] [PubMed] [Google Scholar]
- Degors I. M. S.; Wang C.; Rehman Z. U.; Zuhorn I. S. Carriers Break Barriers in Drug Delivery: Endocytosis and Endosomal Escape of Gene Delivery Vectors. Acc. Chem. Res. 2019, 52, 1750–1760. 10.1021/acs.accounts.9b00177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang K.; Such G. K.; Zhu Z.; Dodds S. J.; Johnston A. P. R.; Cui J.; Ejima H.; Caruso F. Engineering Cellular Degradation of Multilayered Capsules through Controlled Cross-Linking. ACS Nano 2012, 6, 10186–10194. 10.1021/nn3039353. [DOI] [PubMed] [Google Scholar]
- Boisguérin P.; Deshayes S.; Gait M. J.; O’Donovan L.; Godfrey C.; Betts C. A.; Wood M. J. A.; Lebleu B. Delivery of Therapeutic Oligonucleotides with Cell Penetrating Peptides. Adv. Drug Deliv. Rev. 2015, 87, 52–67. 10.1016/j.addr.2015.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurrikoff K.; Langel Ü. Recent Cpp-Based Applications in Medicine. Expert Opin. Drug Deliv. 2019, 16, 1183–1191. 10.1080/17425247.2019.1665021. [DOI] [PubMed] [Google Scholar]
- Boisguérin P.; Konate K.; Josse E.; Vivès E.; Deshayes S. Peptide-Based Nanoparticles for Therapeutic Nucleic Acid Delivery. Biomedicines 2021, 9, 583. 10.3390/biomedicines9050583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y.-W.; Lee H.-J.; Tolliver L. M.; Aronstam R. S. Delivery of Nucleic Acids and Nanomaterials by Cell-Penetrating Peptides: Opportunities and Challenges. BioMed. Res. Int. 2015, 2015, 834079. 10.1155/2015/834079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivès E.; Brodin P.; Lebleu B. A Truncated Hiv-1 Tat Protein Basic Domain Rapidly Translocates through the Plasma Membrane and Accumulates in the Cell Nucleus. J. Biol. Chem. 1997, 272, 16010–16017. 10.1074/jbc.272.25.16010. [DOI] [PubMed] [Google Scholar]
- Yi A.; Sim D.; Lee Y.-J.; Sarangthem V.; Park R.-W. Development of Elastin-Like Polypeptide for Targeted Specific Gene Delivery in Vivo. J. Nanobiotechnol. 2020, 18, 15. 10.1186/s12951-020-0574-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasai H.; Inoue K.; Imamura K.; Yuvienco C.; Montclare J. K.; Yamano S. Efficient siRNA Delivery and Gene Silencing Using a Lipopolypeptide Hybrid Vector Mediated by a Caveolae-Mediated and Temperature-Dependent Endocytic Pathway. J. Nanobiotechnol. 2019, 17, 11. 10.1186/s12951-019-0444-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo F.; Fu Q.; Zhou K.; Jin C.; Wu W.; Ji X.; Yan Q.; Yang Q.; Wu D.; Li A.; Yang G. Matrix Metalloprotein-Triggered, Cell Penetrating Peptide-Modified Star-Shaped Nanoparticles for Tumor Targeting and Cancer Therapy. J. Nanobiotechnol. 2020, 18, 48. 10.1186/s12951-020-00595-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren E.; Torchilin V. P. Cell-Penetrating Peptides: Breaking through to the Other Side. Trends Mol. Med. 2012, 18, 385–393. 10.1016/j.molmed.2012.04.012. [DOI] [PubMed] [Google Scholar]
- Langlet-Bertin B.; Leborgne C.; Scherman D.; Bechinger B.; Mason A. J.; Kichler A. Design and Evaluation of Histidine-Rich Amphipathic Peptides for siRNA Delivery. Pharm. Res. 2010, 27, 1426–1436. 10.1007/s11095-010-0138-2. [DOI] [PubMed] [Google Scholar]
- Hsu T.; Mitragotri S. Delivery of siRNA and Other Macromolecules into Skin and Cells Using a Peptide Enhancer. Proc. Natl. Acad. Sci. U. S. A. 2011, 108, 15816–15821. 10.1073/pnas.1016152108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu Y. C.; Kim K. A.; Kim B. C.; Wang H.-M. D.; Hwang B. H. Novel Fusion Peptide-Mediated siRNA Delivery Using Self-Assembled Nanocomplex. J. Nanobiotechnol. 2021, 19, 44. 10.1186/s12951-021-00791-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla R. S.; Qin B.; Cheng K. Peptides Used in the Delivery of Small Noncoding RNA. Mol. Pharmaceutics 2014, 11, 3395–3408. 10.1021/mp500426r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simeoni F.; Morris M. C.; Heitz F.; Divita G. Insight into the Mechanism of the Peptide-Based Gene Delivery System Mpg: Implications for Delivery of siRNA into Mammalian Cells. Nucleic Acids Res. 2003, 31, 2717–2724. 10.1093/nar/gkg385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang B.; Guo H. Y.; Zhang M.; Jiang L.; Ren F. Z. The Six Amino Acid Antimicrobial Peptide bLFcin6 Penetrates Cells and Delivers siRNA. Febs J. 2013, 280, 1007–1017. 10.1111/febs.12093. [DOI] [PubMed] [Google Scholar]
- Qin B.; Chen Z.; Jin W.; Cheng K. Development of Cholesteryl Peptide Micelles for siRNA Delivery. J. Controlled Release 2013, 172, 159–168. 10.1016/j.jconrel.2013.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J.; Cheng W. P.; Gu J.; Ding C.; Qu X.; Yang Z.; O’Driscoll C. Systemic Delivery of Therapeutic Small Interfering RNA Using a pH-Triggered Amphiphilic Poly-L-Lysine Nanocarrier to Suppress Prostate Cancer Growth in Mice. Eur. J. Pharm. Sci. 2012, 45, 521–532. 10.1016/j.ejps.2011.11.024. [DOI] [PubMed] [Google Scholar]
- Warrant R. W.; Kim S.-H. A-Helix–Double Helix Interaction Shown in the Structure of a Protamine-Transfer RNA Complex and a Nucleoprotamine Model. Nature 1978, 271, 130–135. 10.1038/271130a0. [DOI] [PubMed] [Google Scholar]
- Schlake T.; Thess A.; Fotin-Mleczek M.; Kallen K. J. Developing mRNA-Vaccine Technologies. RNA Biol. 2012, 9, 1319–1330. 10.4161/rna.22269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosi N. L.; Giljohann D. A.; Thaxton C. S.; Lytton-Jean A. K.; Han M. S.; Mirkin C. A. Oligonucleotide-Modified Gold Nanoparticles for Intracellular Gene Regulation. Science 2006, 312, 1027–30. 10.1126/science.1125559. [DOI] [PubMed] [Google Scholar]
- Artiga A.; Serrano-Sevilla I.; De Matteis L.; Mitchell S. G.; de la Fuente J. M. Current Status and Future Perspectives of Gold Nanoparticle Vectors for siRNA Delivery. J. Mater. Chem. B 2019, 7, 876–896. 10.1039/C8TB02484G. [DOI] [PubMed] [Google Scholar]
- Giljohann D. A.; Seferos D. S.; Prigodich A. E.; Patel P. C.; Mirkin C. A. Gene Regulation with Polyvalent siRNA–Nanoparticle Conjugates. J. Am. Chem. Soc. 2009, 131, 2072–2073. 10.1021/ja808719p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y.; Jiang Z.; Saha K.; Kim C. S.; Kim S. T.; Landis R. F.; Rotello V. M. Gold Nanoparticles for Nucleic Acid Delivery. Mol. Ther.: J. Am. Soc. Gene Ther. 2014, 22, 1075–1083. 10.1038/mt.2014.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler J. I.; Auyeung E.; Mirkin C. A. Spherical Nucleic Acids. J. Am. Chem. Soc. 2012, 134, 1376–1391. 10.1021/ja209351u. [DOI] [PubMed] [Google Scholar]
- Seferos D. S.; Giljohann D. A.; Hill H. D.; Prigodich A. E.; Mirkin C. A. Nano-Flares: Probes for Transfection and mRNA Detection in Living Cells. J. Am. Chem. Soc. 2007, 129, 15477–15479. 10.1021/ja0776529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S.; Qin L.; Yamankurt G.; Skakuj K.; Huang Z.; Chen P.-C.; Dominguez D.; Lee A.; Zhang B.; Mirkin C. A. Rational Vaccinology with Spherical Nucleic Acids. Proc. Natl. Acad. Sci. U. S. A. 2019, 116, 10473–10481. 10.1073/pnas.1902805116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plank C.; Zelphati O.; Mykhaylyk O. Magnetically Enhanced Nucleic Acid Delivery. Ten Years of Magnetofection—Progress and Prospects. Adv. Drug Deliv. Rev. 2011, 63, 1300–1331. 10.1016/j.addr.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mykhaylyk O.; Sanchez-Antequera Y.; Vlaskou D.; Cerda M. B.; Bokharaei M.; Hammerschmid E.; Anton M.; Plank C. Magnetic Nanoparticle and Magnetic Field Assisted siRNA Delivery in Vitro. Methods Mol. Biol. 2015, 1218, 53–106. 10.1007/978-1-4939-1538-5_5. [DOI] [PubMed] [Google Scholar]
- Scherer F.; Anton M.; Schillinger U.; Henke J.; Bergemann C.; Krüger A.; Gänsbacher B.; Plank C. Magnetofection: Enhancing and Targeting Gene Delivery by Magnetic Force in Vitro and in Vivo. Gene Ther. 2002, 9, 102–109. 10.1038/sj.gt.3301624. [DOI] [PubMed] [Google Scholar]
- Steitz B.; Hofmann H.; Kamau S. W.; Hassa P. O.; Hottiger M. O.; von Rechenberg B.; Hofmann-Amtenbrink M.; Petri-Fink A. Characterization of Pei-Coated Superparamagnetic Iron Oxide Nanoparticles for Transfection: Size Distribution, Colloidal Properties and DNA Interaction. J. Magn. Magn. Mater. 2007, 311, 300–305. 10.1016/j.jmmm.2006.10.1194. [DOI] [Google Scholar]
- Tai M. F.; Chi K. M.; Lau K. H. W.; Baylink D. J.; Chen S. T. Generation of Magnetic Retroviral Vectors with Magnetic Nanoparticles. Rev. Adv. Mater. Sci. 2003, 5, 319–323. [Google Scholar]
- Ito A.; Takahashi T.; Kameyama Y.; Kawabe Y.; Kamihira M. Magnetic Concentration of a Retroviral Vector Using Magnetite Cationic Liposomes. Tissue Eng. C: Methods 2009, 15, 57–64. 10.1089/ten.tec.2008.0275. [DOI] [PubMed] [Google Scholar]
- Namiki Y.; Namiki T.; Yoshida H.; Ishii Y.; Tsubota A.; Koido S.; Nariai K.; Mitsunaga M.; Yanagisawa S.; Kashiwagi H.; Mabashi Y.; Yumoto Y.; Hoshina S.; Fujise K.; Tada N. A Novel Magnetic Crystal–Lipid Nanostructure for Magnetically Guided in Vivo Gene Delivery. Nat. Nanotechnol. 2009, 4, 598–606. 10.1038/nnano.2009.202. [DOI] [PubMed] [Google Scholar]
- Pan B.; Cui D.; Sheng Y.; Ozkan C.; Gao F.; He R.; Li Q.; Xu P.; Huang T. Dendrimer-Modified Magnetic Nanoparticles Enhance Efficiency of Gene Delivery System. Cancer Res. 2007, 67, 8156–8163. 10.1158/0008-5472.CAN-06-4762. [DOI] [PubMed] [Google Scholar]
- Zheng X.; Lu J.; Deng L.; Xiong Y.; Chen J. Preparation and Characterization of Magnetic Cationic Liposome in Gene Delivery. Int. J. Pharm. 2009, 366, 211–217. 10.1016/j.ijpharm.2008.09.019. [DOI] [PubMed] [Google Scholar]
- Chorny M.; Fishbein I.; Alferiev I.; Levy R. J. Magnetically Responsive Biodegradable Nanoparticles Enhance Adenoviral Gene Transfer in Cultured Smooth Muscle and Endothelial Cells. Mol. Pharmaceutics 2009, 6, 1380–1387. 10.1021/mp900017m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yiu H. H.; McBain S. C.; Lethbridge Z. A.; Lees M. R.; Dobson J. Preparation and Characterization of Polyethylenimine-Coated Fe3o4-Mcm-48 Nanocomposite Particles as a Novel Agent for Magnet-Assisted Transfection. J. Biomed Mater. Res. A 2010, 92, 386–392. 10.1002/jbm.a.32363. [DOI] [PubMed] [Google Scholar]
- Hom C.; Lu J.; Liong M.; Luo H.; Li Z.; Zink J. I.; Tamanoi F. Mesoporous Silica Nanoparticles Facilitate Delivery of siRNA to Shutdown Signaling Pathways in Mammalian Cells. Small 2010, 6, 1185–1190. 10.1002/smll.200901966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H.; Yu M.; Lu Y.; Gu Z.; Yang Y.; Zhang M.; Fu J.; Yu C. Plasmid DNA Delivery: Nanotopography Matters. J. Am. Chem. Soc. 2017, 139, 18247–18254. 10.1021/jacs.7b08974. [DOI] [PubMed] [Google Scholar]
- Bose S.; Tarafder S. Calcium Phosphate Ceramic Systems in Growth Factor and Drug Delivery for Bone Tissue Engineering: A Review. Acta Biomater. 2012, 8, 1401–1421. 10.1016/j.actbio.2011.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levingstone T. J.; Herbaj S.; Redmond J.; McCarthy H. O.; Dunne N. J. Calcium Phosphate Nanoparticles-Based Systems for RNAi Delivery: Applications in Bone Tissue Regeneration. Nanomaterials (Basel) 2020, 10, 146. 10.3390/nano10010146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feray B.Gene Delivery by Hydroxyapatite and Calcium Phosphate Nanoparticles: A Review of Novel and Recent Applications. 10.5772/intechopen.71062 (accessed Aug 6, 2021). [DOI]
- Wagner D. E.; Bhaduri S. B. Progress and Outlook of Inorganic Nanoparticles for Delivery of Nucleic Acid Sequences Related to Orthopedic Pathologies: A Review. Tissue Eng. B: Reviews 2012, 18, 1–14. 10.1089/ten.teb.2011.0081. [DOI] [PubMed] [Google Scholar]
- van den Boorn J. G.; Schlee M.; Coch C.; Hartmann G. siRNA Delivery with Exosome Nanoparticles. Nat. Biotechnol. 2011, 29, 325–326. 10.1038/nbt.1830. [DOI] [PubMed] [Google Scholar]
- Lu H.; Wang J.; Wang T.; Zhong J.; Bao Y.; Hao H. Recent Progress on Nanostructures for Drug Delivery Applications. J. Nanomater. 2016, 2016, 5762431. 10.1155/2016/5762431. [DOI] [Google Scholar]
- Khisamutdinov E. F.; Li H.; Jasinski D. L.; Chen J.; Fu J.; Guo P. Enhancing Immunomodulation on Innate Immunity by Shape Transition among RNA Triangle, Square and Pentagon Nanovehicles. Nucleic Acids Res. 2014, 42, 9996–10004. 10.1093/nar/gku516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeprich S.; Zhou Q.; Guo S.; Shu D.; Qi G.; Wang Y.; Guo P. Bacterial Virus Phi29 pRNA as a Hammerhead Ribozyme Escort to Destroy Hepatitis B Virus. Gene Ther. 2003, 10, 1258–1267. 10.1038/sj.gt.3302002. [DOI] [PubMed] [Google Scholar]
- Geary C.; Rothemund P. W. K.; Andersen E. S. A Single-Stranded Architecture for Cotranscriptional Folding of RNA Nanostructures. Science 2014, 345, 799–804. 10.1126/science.1253920. [DOI] [PubMed] [Google Scholar]
- Li H.; Lee T.; Dziubla T.; Pi F.; Guo S.; Xu J.; Li C.; Haque F.; Liang X.-J.; Guo P. RNA as a Stable Polymer to Build Controllable and Defined Nanostructures for Material and Biomedical Applications. Nano Today 2015, 10, 631–655. 10.1016/j.nantod.2015.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohovie M. J.; Nagasawa M.; Swartz J. R. Virus-Like Particles: Next-Generation Nanoparticles for Targeted Therapeutic Delivery. Bioeng. Transl. Med. 2017, 2, 43–57. 10.1002/btm2.10049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian C.; Liu X.; Xu Q.; Wang Z.; Chen J.; Li T.; Zheng Q.; Yu H.; Gu Y.; Li S.; Xia N. Recent Progress on the Versatility of Virus-Like Particles. Vaccines 2020, 8, 139. 10.3390/vaccines8010139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keech C.; Albert G.; Cho I.; Robertson A.; Reed P.; Neal S.; Plested J. S.; Zhu M.; Cloney-Clark S.; Zhou H.; Smith G.; Patel N.; Frieman M. B.; Haupt R. E.; Logue J.; McGrath M.; Weston S.; Piedra P. A.; Desai C.; Callahan K.; Lewis M.; Price-Abbott P.; Formica N.; Shinde V.; Fries L.; Lickliter J. D.; Griffin P.; Wilkinson B.; Glenn G. M. Phase 1–2 Trial of a Sars-Cov-2 Recombinant Spike Protein Nanoparticle Vaccine. N. Engl. J. Med. 2020, 383, 2320–2332. 10.1056/NEJMoa2026920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajaj S.; Banerjee M. Engineering Virus Capsids into Biomedical Delivery Vehicles: Structural Engineering Problems in Nanoscale. J. Biomed. Nanotechnol. 2015, 11, 53–69. 10.1166/jbn.2015.1959. [DOI] [PubMed] [Google Scholar]
- Resch-Genger U.; Grabolle M.; Cavaliere-Jaricot S.; Nitschke R.; Nann T. Quantum Dots Versus Organic Dyes as Fluorescent Labels. Nat. Methods 2008, 5, 763–775. 10.1038/nmeth.1248. [DOI] [PubMed] [Google Scholar]
- Wegner K. D.; Hildebrandt N. Quantum Dots: Bright and Versatile in Vitro and in Vivo Fluorescence Imaging Biosensors. Chem. Soc. Rev. 2015, 44, 4792–4834. 10.1039/C4CS00532E. [DOI] [PubMed] [Google Scholar]
- Knipe J. M.; Peters J. T.; Peppas N. A. Theranostic Agents for Intracellular Gene Delivery with Spatiotemporal Imaging. Nano Today 2013, 8, 21–38. 10.1016/j.nantod.2012.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z.; Liu G.; Zheng H.; Chen X. Rigid Nanoparticle-Based Delivery of Anti-Cancer siRNA: Challenges and Opportunities. Biotechnology Advances 2014, 32, 831–843. 10.1016/j.biotechadv.2013.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y. D.; Park T. E.; Singh B.; Maharjan S.; Choi Y. J.; Choung P. H.; Arote R. B.; Cho C. S. Nanoparticle-Mediated Delivery of siRNA for Effective Lung Cancer Therapy. Nanomedicine (Lond) 2015, 10, 1165–88. 10.2217/nnm.14.214. [DOI] [PubMed] [Google Scholar]
- Podesta J. E.; Al-Jamal K. T.; Herrero M. A.; Tian B.; Ali-Boucetta H.; Hegde V.; Bianco A.; Prato M.; Kostarelos K. Antitumor Activity and Prolonged Survival by Carbon-Nanotube-Mediated Therapeutic siRNA Silencing in a Human Lung Xenograft Model. Small 2009, 5, 1176–1185. 10.1002/smll.200990047. [DOI] [PubMed] [Google Scholar]
- Al-Jamal K. T.; Toma F. M.; Yilmazer A.; Ali-Boucetta H.; Nunes A.; Herrero M.-A.; Tian B.; Eddaoudi A.; Al-Jamal W.T.; Bianco A.; Prato M.; Kostarelos K. Enhanced Cellular Internalization and Gene Silencing with a Series of Cationic Dendron-Multiwalled Carbon Nanotube:siRNA Complexes. FASEB J. 2010, 24, 4354–4365. 10.1096/fj.09-141036. [DOI] [PubMed] [Google Scholar]
- Zhang Z.; Yang X.; Zhang Y.; Zeng B.; Wang S.; Zhu T.; Roden R. B.; Chen Y.; Yang R. Delivery of Telomerase Reverse Transcriptase Small Interfering RNA in Complex with Positively Charged Single-Walled Carbon Nanotubes Suppresses Tumor Growth. Clin. Cancer Res. 2006, 12, 4933–4939. 10.1158/1078-0432.CCR-05-2831. [DOI] [PubMed] [Google Scholar]
- Wadhwa A.; Aljabbari A.; Lokras A.; Foged C.; Thakur A. Opportunities and Challenges in the Delivery of mRNA-Based Vaccines. Pharmaceutics 2020, 12, 102. 10.3390/pharmaceutics12020102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardi N.; Tuyishime S.; Muramatsu H.; Kariko K.; Mui B. L.; Tam Y. K.; Madden T. D.; Hope M. J.; Weissman D. Expression Kinetics of Nucleoside-Modified mRNA Delivered in Lipid Nanoparticles to Mice by Various Routes. J. Controlled Release 2015, 217, 345–351. 10.1016/j.jconrel.2015.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C.; Maruggi G.; Shan H.; Li J. Advances in mRNA Vaccines for Infectious Diseases. Front. Immunol. 2019, 10, 594. 10.3389/fimmu.2019.00594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardi N.; Hogan M. J.; Pelc R. S.; Muramatsu H.; Andersen H.; DeMaso C. R.; Dowd K. A.; Sutherland L. L.; Scearce R. M.; Parks R.; Wagner W.; Granados A.; Greenhouse J.; Walker M.; Willis E.; Yu J.-S.; McGee C. E.; Sempowski G. D.; Mui B. L.; Tam Y. K.; Huang Y.-J.; Vanlandingham D.; Holmes V. M.; Balachandran H.; Sahu S.; Lifton M.; Higgs S.; Hensley S. E.; Madden T. D.; Hope M. J.; Karikó K.; Santra S.; Graham B. S.; Lewis M. G.; Pierson T. C.; Haynes B. F.; Weissman D. Zika Virus Protection by a Single Low-Dose Nucleoside-Modified mRNA Vaccination. Nature 2017, 543, 248–251. 10.1038/nature21428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakrewsky M.; Kumar S.; Mitragotri S. Nucleic Acid Delivery into Skin for the Treatment of Skin Disease: Proofs-of-Concept, Potential Impact, and Remaining Challenges. J. Controlled Release 2015, 219, 445–456. 10.1016/j.jconrel.2015.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golombek S.; Pilz M.; Steinle H.; Kochba E.; Levin Y.; Lunter D.; Schlensak C.; Wendel H. P.; Avci-Adali M. Intradermal Delivery of Synthetic mRNA Using Hollow Microneedles for Efficient and Rapid Production of Exogenous Proteins in Skin. Mol. Ther. - Nucleic Acids 2018, 11, 382–392. 10.1016/j.omtn.2018.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Lint S.; Goyvaerts C.; Maenhout S.; Goethals L.; Disy A.; Benteyn D.; Pen J.; Bonehill A.; Heirman C.; Breckpot K.; Thielemans K. Preclinical Evaluation of Trimix and Antigen mRNA-Based Antitumor Therapy. Cancer Res. 2012, 72, 1661–1671. 10.1158/0008-5472.CAN-11-2957. [DOI] [PubMed] [Google Scholar]
- Sultana N.; Magadum A.; Hadas Y.; Kondrat J.; Singh N.; Youssef E.; Calderon D.; Chepurko E.; Dubois N.; Hajjar R. J.; Zangi L. Optimizing Cardiac Delivery of Modified mRNA. Mol. Ther. 2017, 25, 1306–1315. 10.1016/j.ymthe.2017.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis F. F. The Origin of Pegnology. Adv. Drug Deliv Rev. 2002, 54, 457–458. 10.1016/S0169-409X(02)00021-2. [DOI] [PubMed] [Google Scholar]
- Zalipsky S. Chemistry of Polyethylene-Glycol Conjugates with Biologically-Active Molecules. Adv. Drug Deliv. Rev. 1995, 16, 157–182. 10.1016/0169-409X(95)00023-Z. [DOI] [Google Scholar]
- Ikeda Y.; Nagasaki Y. Impacts of PEGylation on the Gene and Oligonucleotide Delivery System. J. Appl. Polym. Sci. 2014, 131, 40293. 10.1002/app.40293. [DOI] [Google Scholar]
- Bouchard P. R.; Hutabarat R. M.; Thompson K. M. Discovery and Development of Therapeutic Aptamers. Annu. Rev. Pharmacol. Toxicol. 2010, 50, 237–257. 10.1146/annurev.pharmtox.010909.105547. [DOI] [PubMed] [Google Scholar]
- Haruta K.; Otaki N.; Nagamine M.; Kayo T.; Sasaki A.; Hiramoto S.; Takahashi M.; Hota K.; Sato H.; Yamazaki H. A Novel PEGylation Method for Improving the Pharmacokinetic Properties of Anti-Interleukin-17a RNA Aptamers. Nucleic Acid Ther. 2017, 27, 36–44. 10.1089/nat.2016.0627. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.