Abstract
X-ray structural determination of segments of the myosin rod has proved difficult because of the strong salt-dependent aggregation properties and repeating pattern of charges on the surface of the coiled-coil that lead to the formation of paracrystals. This problem has been resolved in part through the use of globular assembly domains that improve protein folding and prevent aggregation. The primary consideration now in designing coiled-coil fusion constructs for myosin is deciding where to truncate the coiled-coil and which amino acid residues to include from the folding domain. This is especially important for myosin that contains numerous regions of low predicted coiled-coil propensity. Here we describe the strategy adopted to determine the structure of the region that extends from Arg1677 – Leu1797 that included two areas that do not show a strong sequence signature of a conventional left-handed coiled coil or canonical heptad repeat. This demonstrates again that, with careful choice of fusion constructs, overlapping structures exhibit very similar conformations for the myosin rod fragments in the canonical regions. However, conformational variability is seen around Leu1706 which is a hot spot for cardiomyopathy mutations suggesting that this might be important for function.
Keywords: Coiled-coils, cardiac myosin, fusion proteins, Light Meromyosin, X-ray structural determination, protein structure
Introduction
Muscle myosins are classic examples of proteins that contain an extended coiled-coil. They were among the first proteins to be identified to contain this motif, which holds a special place in the historical development of structural biology (Astbury, 1947; Crick, 1953). The coiled-coil was instrumental in first demonstrating how the packing of amino acid residue side chains influences the quaternary structure in proteins and is found in approximately 3% of all proteins (Crick, 1953; Lupas, 1996b; Moutevelis and Woolfson, 2009; Rackham et al., 2010). It was initially observed in filamentous, cytoskeletal, and structural proteins such as α-keratin, myosin and tropomyosin where the motif can extend over a thousand or more amino acid residues. However, shorter domains are also abundant and commonly direct the oligomerization state of a wide range of proteins including those involved in regulatory systems (Landschulz et al., 1988).
This motif is characterized by an alternating pattern of hydrophobic and hydrophilic residues embedded within a seven-amino acid (heptad) repeat. The position of residues in the motif are designated are designated a, b, c, d, e, f, or g where the a and d positions are normally occupied by branched or unbranched hydrophobic residues respectively in a canonical coiled-coil. This, coupled with a left-handed helical arrangement, creates an extended buried hydrophobic surface area that stabilizes the quaternary arrangement. As such the canonical repeating alternating pattern of hydrophobic and hydrophilic residues is readily detected through sequence analysis (Delorenzi and Speed, 2002; Lupas et al., 1991; Parry et al., 2008). The apparent simplicity of this motif belies the rich diversity of coiled-coils observed in nature (Parry, 2014; Parry et al., 2008; Rose et al., 2005). Even apparently simple coiled-coils, such as that predicted to occur in myosin, show considerable deviation from the canonical heptad repeat (McLachlan and Karn, 1982).
Unlike the fibrous proteins such as tropomyosin and α-keratin, muscle myosin heavy chains also contain a globular motor domain at their N-terminus that binds two light chains and then extends into a long α-helical section of over 1000 amino acid residues that dimerizes to form the coiled-coil or myosin rod. The initial observations that lead to the discovery of the coiled-coil in myosin were based on, hydrodynamic measurements, rotatory dispersion, X-ray diffraction, and electron microscopy (Cohen and Holmes, 1963; Lowey and Cohen, 1962; Slayter and Lowey, 1967). At first sight the myosin rod appeared to be a simple coiled-coil based on the biochemical and physical observations; however once the sequence was known it became clear that it exhibits considerable complexity (McLachlan and Karn, 1982).
As depicted in Fig. 1, the coiled-coil prediction for the cardiac myosin rod shows numerous locations where the sequence deviates from a canonical coiled-coil as indicated by a low coiled-coil propensity due to alterations in the distribution of hydrophobic and hydrophilic residues or where there is a break in the seven-residue heptad repeat (Delorenzi and Speed, 2002; Lupas et al., 1991). There are four locations where an extra or skip residue disrupts the pattern. Structural studies of the regions surrounding these residues shows the first three exhibit very similar X-ray structures though this does not appear to occur in the structure of the thick filament itself determined by cryo-electron microscopy (Hu et al., 2016). In the X-ray structures, the distortion introduced by the insertion of an extra residue results in a stable unwinding of the coiled-coil that extends over ~28 amino acid residues (Taylor et al., 2015). The fourth skip residue introduces a local flexible hinge into the myosin rod. For the other areas that are predicted to exhibit a low coiled-propensity it is unclear whether they represent areas of flexibility or stretches where the structure is distorted or combinations of both scenarios. One solution to this question is to determine the structure of these regions. In this paper, we focus on the structural determination of cardiac myosin between 1690 and 1800 where this lies just prior to Skip 4 (G1807) and includes a region that encompasses a large number of cardiomyopathy and muscle myopathy mutations (Colegrave and Peckham, 2014; Lamont et al., 2014). (Fig. 1)
Figure 1. Predicted coiled-coil propensity for the cardiac myosin rod.
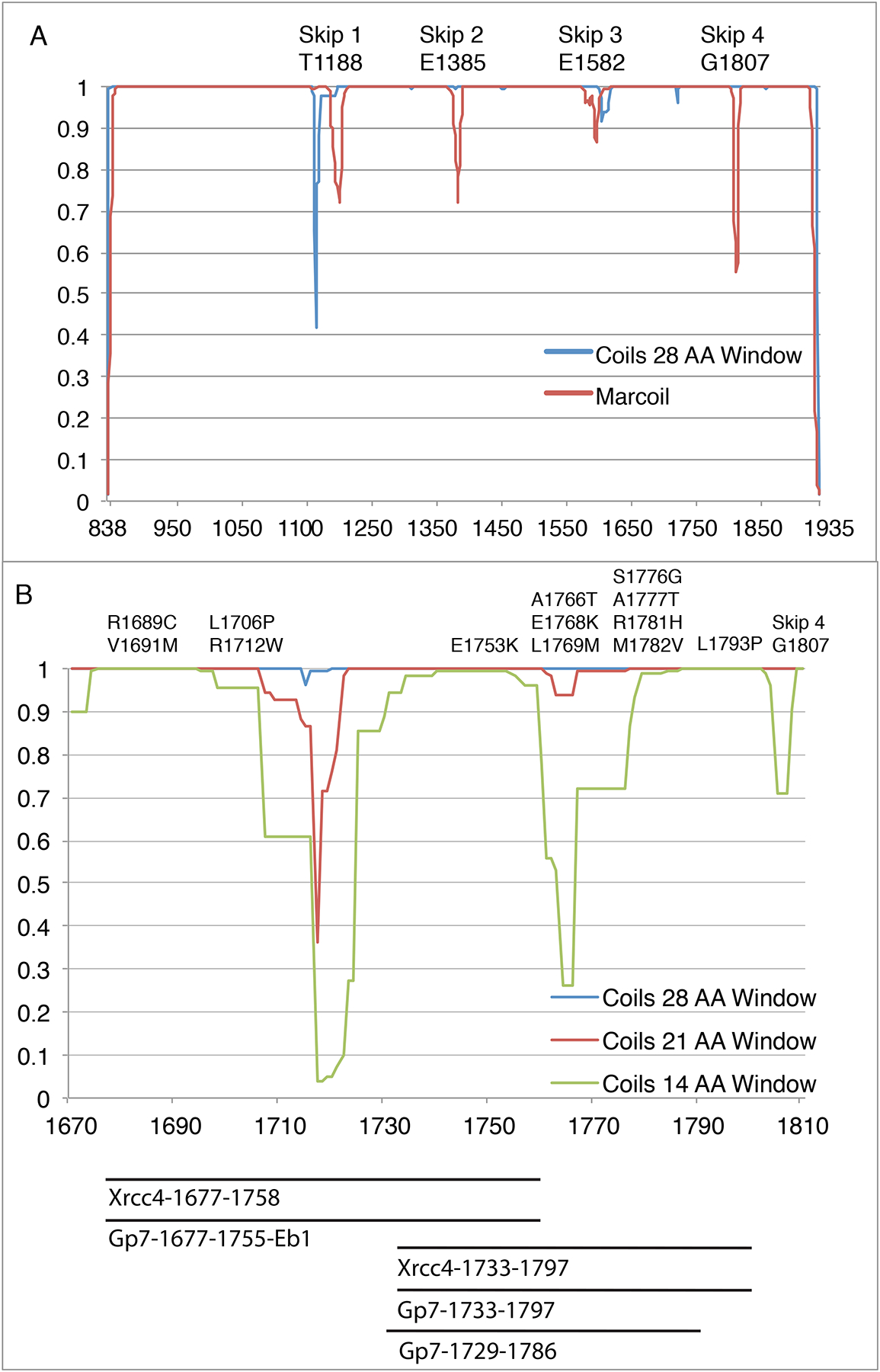
(A) Myosin rod coiled-coil prediction using a 28-amino acid window in COILS (blue) and Marcoil (Red). The COILS algorithm compares a sequence to a database of known parallel two-stranded coiled-coils and derives a similarity score (Lupas, 1996a; Lupas et al., 1991). The 28-amino acid window for coils smooths out most of the local variations in coiled coil propensity except for around the Skip residues which introduce a discontinuity. The Marcoil algorithm is based on a window-less Hidden Markov Model (Delorenzi and Speed, 2002). The propensities relate to the probability that a group of amino acids will adopt the structure of a coiled-coil, but do not indicate stability or structure of the resultant oligomeric state. (B) COILS prediction from myosin rod amino acids 1680–1810. This region contains 13 pathogenic mutation sites, as well as regions of poorly predicted coiled-coil.
Determination of the structure of fragments of the myosin rod has proved difficult for two reasons. First, the myosin rod exhibits a strong salt-dependent aggregation that is in part due to periodic clusters of positively and negatively charged residues that repeat every 28 amino acid residues for the entire length of the myosin (McLachlan and Karn, 1982). As a consequence, fragments of myosin readily form paracrystals that have not proved suitable for structural analysis (Atkinson and Stewart, 1991; Sohn et al., 1997). Second, and perhaps more fundamentally, fragments of extended coiled-coil proteins can be difficult to express in vitro because they lack a trigger sequence that directs assembly (Ciani et al., 2010; Dill et al., 1993; Kammerer et al., 1998; Lupas and Gruber, 2005; Steinmetz et al., 1998; Wu et al., 2000). This problem has been solved for many proteins by including a folding domain, where the most commonly used assembly domain is the ~30 amino acid residue long leucine zipper from the GCN4 transcription factor. The latter yielded the first high resolution structure for a coiled-coil (O’Shea et al., 1991).
One benefit of the GCN4 leucine zipper is that it can be fused to the chosen segment of coiled-coil at either its N-terminal end or its C-terminal end in those cases where a trigger sequence is not needed (Greenfield et al., 1998; Li et al., 2003). The downside of GCN4 is that it necessarily increases the length of the resultant fusion by ~45 Å. This problem can be ameliorated by the alternative use of small globular fusion domains. This approach allowed the structural determinations of the smooth muscle tropomyosin overlap complex, a component of the yeast spindle pole body, and the fragments of cardiac myosin that carry the skip residues (Frye et al., 2010; Klenchin et al., 2011; Korkmaz et al., 2016; Taylor et al., 2015). These domains not only improve the expression levels of soluble proteins in E. coli, but also in many instances increase the likelihood of crystallization (Korkmaz et al., 2016). In all cases, the primary consideration in designing coiled-coil fusion constructs is deciding where to truncate the target protein and what amino acid residues to include from the folding domain. This is especially important for proteins such as tropomyosin and myosin and that contain highly extended coiled-coils. Here, it is necessary to divide the protein into smaller but more manageable fragments (Brown et al., 2005; Brown et al., 2001; Strelkov et al., 2001). Although this is straightforward for segments that show a high coiled-coil propensity, it is problematic for the non-canonical regions. Here we demonstrate a design strategy for fusion constructs that abut such a region in human cardiac β-myosin that exhibits low coiled-coil propensity and the consequences of ill-chosen designs. This region extends from Arg1677 – Leu1797.
Material and methods
Construction and design of coiled-coil fusion protein expression plasmids
All cloning was performed using a modified QuikChange cloning protocol described previously (Klenchin et al., 2011). Briefly, the QuikChange method avoids the introduction of cloning artifacts and allows genes to be inserted directly into vectors via linear amplification resulting in faster preparation of constructs (Chen et al., 2000; van den Ent and Lowe, 2006). This is particularly important for creating fusion proteins that encompass coiled-coils. DNA for the human β-cardiac myosin rod was purchased as an image clone from Open Biosystems. Sequences of clones were verified throughout the open reading frame of each plasmid.
All fusion proteins were cloned into pKLD37, a modified pET31b plasmid (EMD) by QuikChange cloning (Klenchin et al., 2011). All constructs were designed with an N-terminal His-tag followed by an rTEV cleavage site that lead into either GP7 or Xrcc4 fusion proteins (Korkmaz et al., 2016; Morais et al., 2003; Sibanda et al., 2001). The COILS algorithm was used to predict coiled-coil propensity and registration (Lupas et al., 1991). Sequences of cloned constructs are shown in Table 1. The region of myosin spanned by each construct is also shown in Fig. 1C.
Table 1.

|
Myosin segment is underlined.
Residues in bold were not observed in the crystal lattice.
Italic text represents coiled-coil registration as predicted by COILS for both the assembly domain and myosin fragment
Break in the coiled-coil register through the incorrect choice of fusion segments.
Protein expression and purification
Fusion proteins were expressed in an E. coli BL21-CodonPlus (DE3)-RIL cell line (Stratagene). Cells were grown in lysogeny broth (LB) medium in the presence of 100 μg/mL ampicillin and 30 μg/mL chloramphenicol until reaching an A600 between 0.7–1.0, then cooled on ice for 15 minutes and thereafter protein expression was induced by the addition of 1 mM isopropyl β-D-1-thiogalactopyranoside. Cells were incubated with shaking for 16 hours at 16 °C and harvested by centrifugation, washed with 50 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) pH 7.6, 50 mM NaCl, 1 mM ethylenediaminetetraacetic acid (EDTA) at a ratio of 1L of buffer per 6L of culture, and flash-frozen in liquid nitrogen and stored at −80 °C.
All protein purifications were carried out at 4 °C. 10 grams of cells were lysed in 50 mL of lysis buffer consisting of 50 mM HEPES pH 7.6, 20 mM imidazole pH 8.0, 50 mM NaCl, 0.1 mM EDTA, 0.5 mg/mL lysozyme, 1 mM phenylmethylsulfonyl fluoride, 50 nM Leupeptin (Peptide International), 70 nM E-65 (Peptide International), 2 nM Aprotinin (ProSpec), and 2 mM 4-(2-aminoethyl)benzenesulfonyl fluoride (Gold BioTechnology) by sonication. Lysate was clarified by centrifugation at 125,000 g for 30 minutes at 4 °C using a Ti-45 rotor that was pre-cooled 4 °C prior to spinning. Supernatant was loaded over a 5 mL Ni-NTA (nickel-nitrilotriacetic acid) column (QIAGEN) by gravity. The column was washed with 10 column volumes of 300 mM NaCl, 25 mM HEPES pH 7.6, 20 mM imidazole, 1 mM β-mercaptoethanol (BME). The column was washed with 5 column volumes of 300 mM NaCl, 25 mM HEPES pH 7.6, 40 mM imidazole, 1 mM BME. Protein was eluted in five to six column volumes with 300 mM NaCl, 25 mM HEPES pH 7.6, 250 mM imidazole, 1 mM BME. 25 mL of the eluted protein was mixed with recombinant Tobacco Etch Virus protease (rTEV) at a molar ratio of 40:1 and dialyzed for 4 hours at room temperature followed by overnight dialysis at 4 °C in 25 mM HEPES pH 7.6, 100 mM NaCl, 0.1 mM EDTA, 0.5 mM tris(2-carboxyethyl)phosphine (TCEP) (Blommel and Fox, 2007). 5 mL of the original elution was dialyzed against the same buffer, but with no rTEV present to retain the His-tagged protein. After dialysis, NaCl concentration was brought up to 300 mM and rTEV-cleaved protein was loaded over 2 mL Ni-NTA column equilibrated in 300 mM NaCl, 25 mM HEPES pH 7.6, 0.5 mM TCEP. rTEV-cleaved myosin fusion proteins were eluted in 5 column volumes of 300 mM NaCl, 25 mM HEPES pH 7.6, 10 mM imidazole, 0.5 mM TCEP. Residual His-tagged myosin fusion proteins and rTEV were eluted in 5 column volumes of 300 mM NaCl, 25 mM HEPES pH 7.6, 250 mM imidazole, 0.5 mM TCEP. rTEV-cleaved myosin fusion proteins were concentrated to between 10 and 20 mg/mL using an Amicon Ultra-15 30 kDa cutoff (Millipore). Concentrated protein was dialyzed overnight against 100 mM NaCl, 10 mM HEPES pH 7.6, 0.1 mM TCEP at 4°C. The Xrcc4-1677-1758 protein was dialyzed against 300 mM NaCl, 10 mM HEPES pH 7.6, 0.1 mM TCEP at 4°C to prevent aggregation that was observed at lower ionic strengths. Protein was flash-frozen as 30 μL droplets in liquid nitrogen and stored at −80 °C.
Crystallization
The initial crystallization conditions for all constructs were identified with a 144-condition sparse-matrix screen developed in the Rayment laboratory that was applied both at room temperature and 4°C. The initial conditions were refined by varying pH, precipitant concentration, and salt composition. Crystals of GP7-1677-1755-Eb1 were grown at room temperature by vapor diffusion from a 1:1 mixture of 17 mg/ml protein solution and a well solution consisting of 1.6 M ammonium sulfate, 500 mM trimethyl ammonium N-oxide, 100 mM bis-tris propane pH 9.0. Hexagonal rods grew overnight to dimensions of approximately 200 μm × 200 μm × 400 μm. Crystals were frozen by stepwise transfer to a synthetic mother liquor consisting of 1.6 M ammonium sulfate, 1 M trimethyl ammonium N-oxide, 50 mM bis-tris propane, 20% glycerol and soaked for 3 hours. Crystals were then frozen by rapidly plunging them into liquid nitrogen.
Crystals of Xrcc4-1677-1758 were grown at room temperature by vapor diffusion from a 1:1 mixture of 15 mg/mL protein solution and a well solution consisting of 8–10% methyl ether polyethylene glycol (MEPEG) 5K, 300 mM glycine, bis-tris propane pH 7.0, 1.5–3.0% (w/v) jeffamine M-600. After streaking with a cat whisker, box-shaped crystals grew overnight, followed by thin rods over the course of approximately two weeks. The rods diffracted significantly better than the box-shaped crystals, and were cryoprotected by stepwise transfer to a synthetic mother liquor consisting of 18% MEPEG 5K, 100 mM sodium chloride, 300 mM glycine, 100 mM bis-tris propane pH 7.0, 15% ethylene glycol. Crystals were frozen by rapidly plunging them into liquid nitrogen.
Crystals of GP7-1729-1787 were grown at room temperature by vapor diffusion from a 1:1 mixture of 15 mg/mL protein solution and a well solution consisting of 18% pentaerythritol ethoxylate 797, 150 mM ammonium thiocyanate, 100 mM sodium acetate pH 5.0. Plate-shaped crystals grew rapidly overnight to dimensions of 400 μm × 200 μm × 100 μm. Crystals were very sensitive to manipulation, would begin to crack 4–5 days after the initiation of crystallization, and dissolved upon transfer to various cryoprotectant solutions. Crystals were cryoprotected by addition of 10 μL 14% pentaerythritol ethoxylate 797, 110 mM ammonium thiocyanate, 75 mM sodium acetate pH 5.0, 25% ethylene glycol to the drop containing the crystals, followed by rapidly plunging them into liquid nitrogen.
Crystals of His-tagged-GP7-1733-1797 were grown at room temperature by vapor diffusion from at 1:1 mixture of 15 mg/mL protein solution and a polyethylene glycol solution consisting of 14% (w/v) MEPEG 2K, 1.5% (w/v) myo-inositol, 100 mM HEPES pH 7.5, 50 mM MgCl2. Plate-shaped crystals grew over the course of 2–3 days and reached average dimensions of 800 μm × 200 μm × 50 μm. Crystals were cryoprotected by transferring to a synthetic mother liquor solution of 20% MEPEG 2K, 1.5% (w/v) myo-inositol, 100 mM HEPES pH 7.5, 50 mM MgCl2, 15% ethylene glycol followed by rapidly plunging them into liquid nitrogen.
Crystals of Xrcc4-1733-1797 were grown at room temperature by vapor diffusion from a 1:1 mixture of 16% (w/v) MEPEG 5K, 300 mM glycine, 100 mM triethanolamine pH 7.5.Over the course of 2 days, spade-shaped crystals grew to dimensions of approximately 400 μm × 200 μm × 100 μm. Crystals were cryoprotected by transferring to a synthetic mother liquor of 20% (w/v) MEPEG 5K, 250 mM glycine, 100 mM NaCl, 60 mM triethanolamine pH 7.5, 12% ethylene glycol (w/v) followed by rapidly plunging them into liquid nitrogen.
Data collection and structure determination
X-ray diffraction data were collected at beamline SBC 19-ID (Advanced Photon Source, Argonne National Laboratory, Argonne, IL). Integration and scaling were performed using HKL3000 and HKL2000 (Minor et al., 2006; Otwinowski and Minor, 1997). X-ray data set statistics are shown in Table 3. The structures of Xrcc4-1677-1758, Gp7-1677-1758-EB1, GP7-1729-1787, His-tagged-GP7-1733-1797, and Xrcc4-1733-1797 were solved using molecular replacement using residues 2–49 of GP7 (PDB ID: 1NO4 (Morais et al., 2003)) or residues 1–131 of Xrcc4 (PDB ID: 1IK9 (Sibanda et al., 2001)) using Phaser MR software (McCoy et al., 2007). Density modification was performed by Parrot (Zhang et al., 1997). The coiled-coil regions of myosin were built through cycles of model building in Coot, followed by restrained refinement in RefMac 5.6 (Emsley et al., 2010; Vagin et al., 2004). Final refinements were performed in Phenix using PhenixRefine (Adams et al., 2010).
Table 3.
X-ray data collection and refinement statistics
| Crystallographic Data | Xrcc4-1677-1758 | GP7-1677-1755-EB1 | GP7-1729-1786 | Xrcc4-1733-1797 | GP7-1733-1797 |
| Space group | P 212121 | P 6122 | P 1 21 1 | P 212121 | P 212121 |
| Cell dimensions (Å) | 85.1, 102.6, 135.0 | 141.8, 141.8, 87.4 | 24.2, 73.0, 114.5 | 44.0, 71.5, 173.1 | 26.5, 93.7, 234.1 |
| Resolution range (Å) | 50–3.5 | 50–3.1 | 50.00–2.3 | 100–2.43 | 100–2.9 |
| Completeness (%) (Last shell) | 100.0 (100.0) | 99.7 (96.2) | 97.8 (97.0) | 99.6 (97.2) | 99.8 (99.9) |
| Rsym (%) (Last shell) | 0.139 (0.807) | 0.117 (0.667) | 0.071 (0.47) | 0.046 (0.20) | 0.108 (0.401) |
| Mean I/σ (last shell) | 17.1 (3.2) | 24.2 (4.5) | 18.1 (4.7) | 53.6 (7.9) | 19.3 (4.1) |
| No. of unique reflections | 16299 | 9815 | 17830 | 21429 | 13797 |
| Chains per asymmetric unit | 4 | 1 | 4 | 2 | 4 |
| Matthews coefficient (Å3 Da−1) | 3.1 | 6.3 | 2.0 | 3.0 | 2.4 |
| Solvent content (%) | 60.1 | 80.4 | 39.5 | 59.6 | 48.1 |
| Multiplicity (Last shell) | 9.5 (9.8) | 19.6 (19.5) | 4.1 (3.2) | 12.0 (5.9) | 5.7 (5.5) |
| Refinement | |||||
| No. of residues | 794 | 104 | 366 | 391 | 467 |
| Water molecules | 0 | 8 | 43 | 155 | 13 |
| Rwork (%) | 0.22 (0.26) | 0.22 (0.25) | 0.22 (0.25) | 0.19 (0.25) | 0.25 (0.31) |
| Rfree (%) | 0.26 (0.32) | 0.24 (0.27) | 0.26 (0.30) | 0.24 (0.32) | 0.30 (0.33) |
| Average B-factor (Å2) | 52.6 | 61.4 | 49.3 | 48.0 | 60.3 |
| r.m.s.d on bond lengths (Å) | 0.002 | 0.003 | 0.002 | 0.002 | 0.003 |
| r.m.s.d on bond angles (Å) | 0.42 | 0.45 | 0.42 | 0.44 | 0.58 |
| Ramachandran plot (%) | |||||
| Preferred | 95.0 | 99.0 | 98.9 | 96.9 | 99.8 |
| Allowed | 4.6 | 1 | 1.1 | 2.8 | 0.2 |
| Outliers | 0.4 | 0 | 0 | 0.3 | 0 |
Values in parentheses are for the outermost resolution shell.
Results and discussion
The region of human cardiac β-myosin targeted in this investigation extends from Arg1677 – Leu1797 where this overlaps with the previously determined structure for the segment that extends from Ala1777 – Thr1854 and includes Skip 4 (Gly1807) (Taylor et al., 2015). As seen in Fig. 1B, this segment includes two regions where the coiled-coil propensity drops considerably from that seen for most of the myosin rod. Construction of fusion proteins that cover this region therefore creates an opportunity to see how the deviations from a canonical heptad motif are accommodated within a larger structure. It also provides a test case on which to develop a robust strategy for structural determinations type of coiled-coil. Finally, it allows an examination of the effects of terminating coiled-coils in regions of low predicted propensity.
Realistically, two constructs are needed to span this region of myosin since previous experience suggests that fusion proteins that contain 55–75 amino acid residues of myosin yield the best crystals (Korkmaz et al., 2016; Taylor et al., 2015). Longer constructs do not crystallize well, whereas shorter constructs yield less information given that the first heptad after the fusion junction is influenced by the assembly domain itself (Korkmaz et al., 2016). A second strategical restraint is the requirement that each segment of myosin included in a fusion protein overlap with the preceding and succeeding constructs to provide overlapping structural coverage of the myosin rod and allow assembly of the final model. This strategy was adopted to provide insight into any fusion artifacts and end effects (Korkmaz et al., 2016). Finally, from first principles it would appear wiser to create constructs in which the fusion point is located in a region of high coiled-coil propensity since these regions are expected to exhibit the conformation of a normal coiled-coil, such as that seen in the fusion proteins. It is likely that the geometry of the coiled-coil would be similar for both the assembly domain and the target and that the resultant structure would be representative of the real structure of the myosin rod. Conversely, choosing to fuse a myosin rod segment that has lower coiled-coil propensity to an assembly protein that has a high coiled-coil propensity might eliminate any natural distortion present in the real structure of the myosin rod, where the latter would be of interest. Examination of the coiled-coil propensity (Fig. 1B) reveals a limited range of fusion points that can accommodate all of the issues raised above.
A large number of constructs were created and tested for expression, solubility, ability to crystallize, and their diffraction properties (Table 2). The variations in constructs included the beginning and ending point of the segment, the nature of the N-terminal assembly domain, and the absence or presence of an additional C-terminal blocking domain (Eb1) (Frye et al., 2010). As can be seen, most constructs yield soluble protein that crystallizes, but only a limited number gave crystals that diffract to high or even moderate resolution. As indicated earlier, structural determination of two segments is required to cover this gap. The first of these covers the gap between Arg1677 and Ala1758
Table 2.
Table of constructs prepared during the course of this study and their properties.
| Name | Fusion (Amino Acids) | Myosin (Amino Acids) | Soluble? | Crystallized? | Diffraction? | Myosin Initial Amino Acid Coiled-coil Propensity * |
Myosin Final Amino Acid Coiled-coil Propensity* |
|---|---|---|---|---|---|---|---|
| Gp7-MyH7-1660-1758-Eb1 | Gp7 (2–49) Eb1 (214–256) |
1660–1758 | Yes | Yes | No visible diffraction | 0.865 | 0.962 |
| Gp7-MyH7-1677-1755-Eb1 | Gp7 (2–49) Eb1 (207–256) |
1677–1755 | Yes | Yes | 3.0 Å | 1.000 | 0.986 |
| Gp7-MyH7-1729-1786 | Gp7 (2–49) | 1729–1786 | Yes | Yes | 2.3 Å | 0.854 | 0.996 |
| Gp7-MyH7-1733-1797 | Gp7 (2–49) | 1733–1797 | Yes | Yes | 3.0 Å | 0.943 | 0.998 |
| Xrcc4-MyH7-1677-1754-Eb1 | Xrcc4 (2–132) Eb1 (214–256) |
1677–1754 | Yes | Yes | <6 Å | 1.000 | 0.990 |
| Xrcc4-MyH7-1677-1758 | Xrcc4 (2–132) | 1677–1758 | Yes | Yes | No visible diffraction | 1.000 | 0.962 |
| Xrcc4-MyH7-1677-1758 | Xrcc4 (2–132) | 1677–1758 | Yes | Yes | 3.5 Å | 1.000 | 0.962 |
| Xrcc4-MyH7-1678-1737 | Xrcc4 (2–140) | 1678–1737 | Yes | No | n/a | 1.000 | 0.982 |
| Xrcc4-MyH7-1678-1751 | Xrcc4 (2–140) | 1678–1751 | Yes | Yes | 6 Å | 1.000 | 0.997 |
| Xrcc4-MyH7-1732-1790 | Xrcc4 (2–137) | 1732–1790 | Yes | No | n/a | 0.943 | 0.998 |
| Xrcc4-MyH7-1733-1797 | Xrcc4 (2–132) | 1733–1797 | Yes | Yes | 2.5 Å | 0.943 | 0.998 |
Names in Bold indicate structures determined during study.
Coiled-coil propensity was determined for cardiac myosin using a 14 amino acid window in the COILS server.
Conformational flexibility accompanies coil prediction
Two structures were obtained for the human cardiac β-myosin rod fragment that covers the region between Arg1677 and Ala1758 utilizing different blocking domains. A total of six constructs were made before successful fusion domains that yielded a structure for this region were found. Multiple structures are beneficial for non-canonical regions since they allow an assessment of whether the observed structure of the myosin fragment is reflective of its conformation in the complete molecule or is influenced by the assembly domains. In this instance, alternative domains were included at the N-terminus (Gp7 or Xrcc4) and the C-terminus was either blocked with Eb1 or free (Fig. 2A and 2B). These constructs are defined as Gp7-Myh7-1677-1755-Eb1 and Xrcc4-Myh7-1677-1758.
Figure 2. Structures and comparison of the fusion proteins covering Arg1677 - Ala1758.
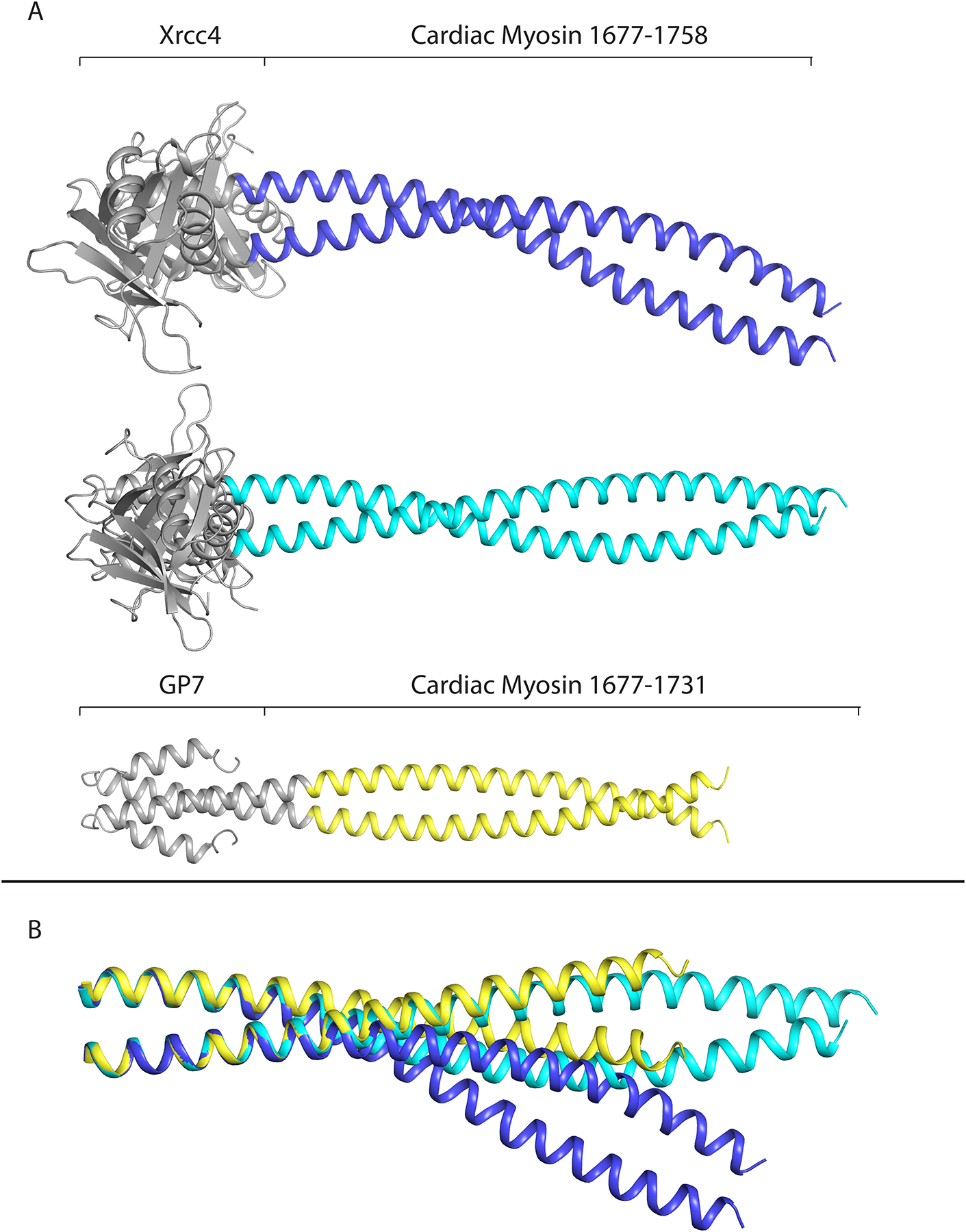
(A) Structures of cardiac myosin containing residues 1677–1758 using both Xrcc4 and Gp7/Eb1 fusion domains. The Eb1 domain in the Gp7-1677-1758-Eb1 structure was not resolved. (B) The three structural conformations overlapped from residues 1677–1691. When aligned with this segment, the three conformations diverge most strongly around residue L1706, highlighting the flexible properties of this region of myosin. Figures 2–5 were prepared with Pymol (DeLano, 2002).
The Gp7-Myh7-1677-1755-Eb1 protein contained Myh7 amino acids 1677–1755 with an N-terminal fusion of residues 2–47 of Gp7 and a C-terminal fusion to residues 207–256 of Eb1. The structure was determined to a resolution of 3.1 Å and had clear coiled-coil electron density to residue 1731, with poorly resolved electron density from residues 1732–1755 and the Eb1 fusion domain. Eb1 was attached at the C-terminus of the construct with the aim to avoid any unwinding of helices due to a drop in coiled coil-propensity from residues 1707–1731 as was predicted by the COILS server (Lupas et al. 1991). This structure displayed nearly ideal canonical coiled-coil behaviour with a superhelical frequency ω0 of −3.4°/residue, helical frequency ω1 of 102.9°/residue, Zoff of 0°/residue, superhelical pitch of 156 Å, and 0.51 Å root mean square deviation (RMSD) from an ideal coiled-coil. (coiled-coil Crick Parameterization (CCCP) Server) (Grigoryan and Degrado, 2011). The dimeric coiled-coiled nature of this structure was also enforced by the P6122 hexagonal space group, which contained only one α-helix per asymmetric unit, and hence requires a two-fold related protomer to form the coiled-coil. Because of the unique packing of the crystal lattice, the C-terminal remainder of the protein after residue 1731 was positioned in the core of a helical lattice of myosin dimers. The resulting drop in electron density after residue 1731 was most likely influenced by a break in the canonical coiled-coil influenced by the hexagonal restrictions of the P6122 space group.
The Xrcc4-Myh7-1677-1758 protein was designed with an N-terminal fusion of amino acids 2–132 of Xrcc4 to Myh7 amino acids 1677–1758. After trying similar Xrcc4 fusions (Table 2), the following surface mutations were introduced to the Xrcc4 domain of the Xrcc4-Myh7-1677-1758 protein: E29K, E51K, D57A, D58T, E62N, C93R, E98K. These residues were chosen with the goal of changing the predicted isoelectric point of the fusion domain from ~ pI ~5.0 to ~9.0. The residues were chosen by examining the structure of Xrcc4 (RCSB accession number 1IK9 (Sibanda et al., 2001)) seeking surface residues that were not involved in intra- or inter-molecular interactions such as salt-bridges or hydrogen bonds. Cys94 was replaced with an arginine in order to create a cysteine-free construct. It was hypothesized that these changes would force the protein to crystallize under different conditions. Indeed, with these surface mutations, the structure of the Xrcc4-1677-1758 protein was determined to a resolution of 3.5 Å and contained two coiled-coils with different conformations in the asymmetric unit. The coiled-coil of chains AB had a superhelical frequency ω0 of −3.6°/residue, helical frequency ω1 of 102.9°/residue, Zoff of −1.6 Å/residue, superhelical pitch of 144 Å, and 2.5 Å RMSD from ideal coiled-coil. (CCCP Server). This differed slightly from chains CD, which had a superhelical frequency ω0 of −3.7 °/residue, helical frequency ω1 of 102.9°/residue, Zoff of 0.6 Å/residue, superhelical pitch of 141 Å and 1.9 Å RMSD from ideal coiled-coil. (CCCP Server).
When aligned using residues 1677–1691 the three conformations deviate most strongly around residue L1706, a site of a known L1706P mutation responsible for Laing distal myopathy that results in poorly organized bare-zones when incorporated in thick filaments (Buvoli et al., 2012; Meredith et al., 2004) (Fig. 2C). Together these three conformations highlight the flexible properties of the myosin rod around residue L1706 which may have implications on why a proline can be inserted into the rod at this position without complete loss of function or assembly of the thick filament. It is possible that inherent flexibility of this region can accommodate the mutation where insertion of a proline in more canonical regions might have a far larger effect on stability. This also highlights the importance of determining multiple structures for problematic regions of coiled-coils.
Design mismatch in coiled-coil registry can result in tetrameric conformations
During the design of the GP7-1729-1787 protein, the myosin fragment was inadvertently placed out of heptad coiled-coil register with the GP7 fusion domain. This resulted in the placement of residue (Lys1729) from myosin, that is predicted to occupy a c position in the coiled-coil, adjacent to Lys48 of Gp7 where this is predicted to occupy an f position. The net result of this mistake is equivalent to an insertion of four residues or deletion of three which amounts to the introduction of a “stutter” in the coiled-coil heptad repeat (Lupas and Gruber, 2005). Surprisingly, the resultant protein was stable in solution and overexpressed to a great degree in E. coli cells, despite the mismatch in coiled-coil registration. However, the crystals of this material proved to be very sensitive to even minor physical disturbances.
The structural determination revealed that this construct formed an anti-parallel four-helix bundle in the crystal lattice as opposed to a dimeric coiled-coil (Fig. 3A). This overall unwinding of the coiled-coil is consistent with that expected from the introduction of a stammer in the heptad repeat (Lupas and Gruber, 2005). Interestingly, the disruption in coiled-coil registry did not appear to affect the GP7 α-helix leading into the myosin domain, but instead propagated into the dimeric interaction between myosin chains, resulting in an almost parallel orientation of the dimeric coiled-coil from residues Lys1729 - Glu1743.
Figure 3. Mismatch of the coiled-coil registration in the fusion protein design yields a tetrameric assembly.
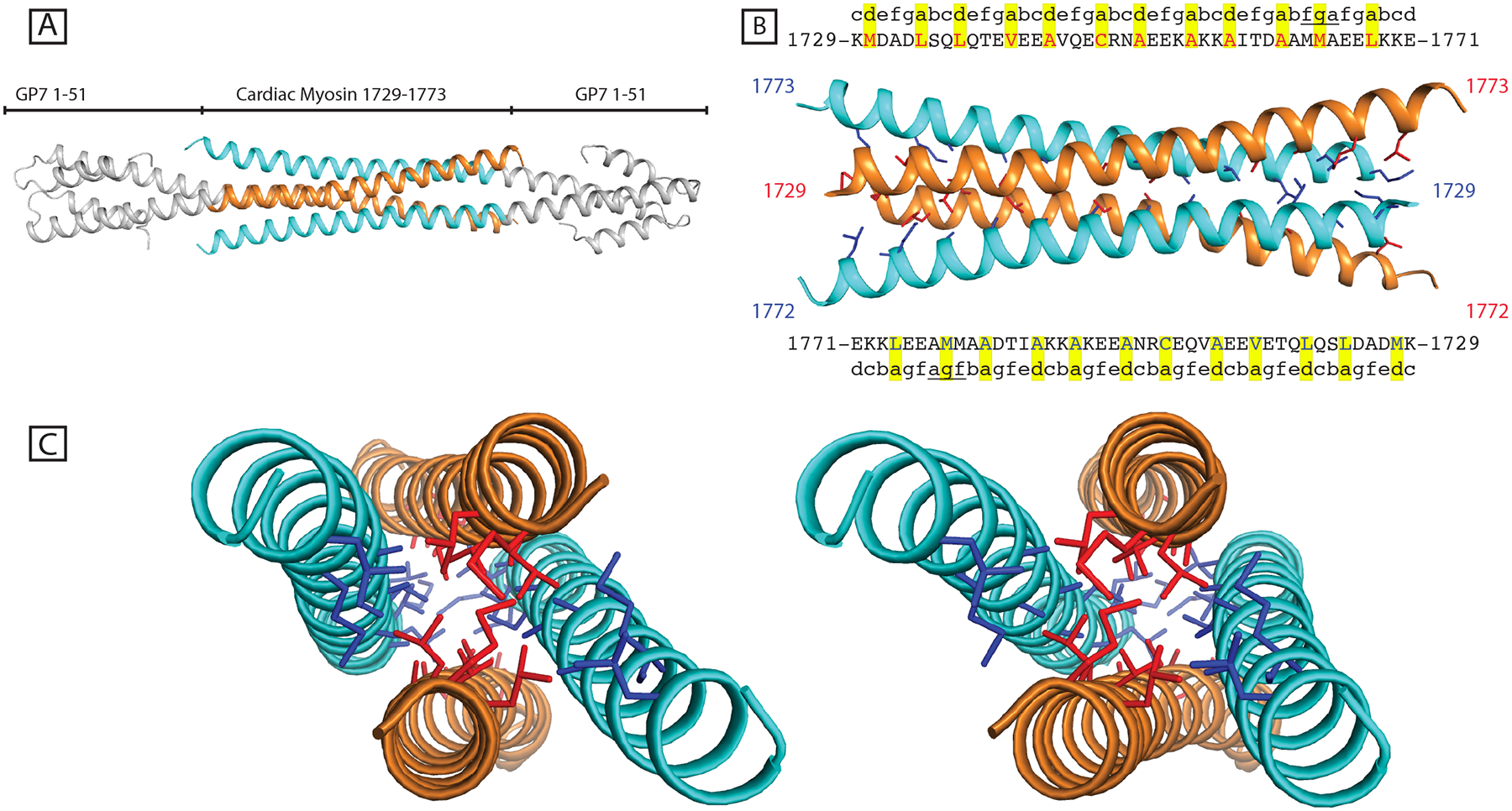
(A) A coiled-coil mismatch in the fusion between Gp7 and myosin rod resulted in a tetrameric artifact of anti-parallel coiled-coil. (B) The core residues are highlighted and shown in red and blue for the respective dimers. The predicted coiled-coil register for the intact myosin rod is shown along with the amino acid labels. A predicted deviation from a canonical coiled-coil is underlined between 1662–1673. However, in this structure the M1664 is in the d position, not g as predicted by COILS. (C) Stereo view of the four-helix bundle along the hydrophobic core.
The deviation from dimeric conformation starts most clearly at Met1730 which in this construct is forced to occupy an a position instead of the d position inherent to the myosin coiled-coil. The bulky methionine side chains are seen to be buried between the α-helices and force them apart. The adopted a position is consistent with the heptad register of Gp7, but means that the next hydrophobic residue along the chain in myosin is not consistent with a canonical coiled-coil. This residue, Leu1734, now lies in an e position and hence is surface exposed. This break in heptad registration, that leaves hydrophobic residues surface-exposed propagates, along the α-helix and allows the formation of tetramer with symmetry related chains (Fig. 3B and 3C). Additionally, COILS predicted lower coiled-coil propensity and a disruption of the heptad registration between residues Thr1760 - Ala1777, which likely further contributed to the conformational flexibility of this protein and its ability to form a tetramer. Interpretable electron density was only visible for residues Ala1729 - Gln1773 of myosin even though it included the region Ala1729 - Glu1787. The electron density for Gp7 was well-resolved.
The determination of this structure suggests that long coiled-coil fusion proteins may be capable of forming higher symmetry complexes when there is a mismatch in the coiled-coil registry. This observation is supported by numerous published mutations in GCN4, a short but strong coiled-coil, that result in higher order assemblies (Ciani et al., 2010; Woolfson, 2005). Furthermore, it suggests the presence of a dimeric fusion domain does not guarantee a dimeric assembly in the crystal lattice and emphasizes the importance of choosing the correct fusion points when designing expression constructs.
Changing globular domains does not significantly alter the target coiled-coil
To avoid the tetrameric crystallographic assembly, new constructs were designed using both the GP7 and Xrcc4 as assembly domains at the N-terminus. In addition to changing the fusion domains, the myosin target region was shifted downstream to cover Asp1733 - Leu1797 in both recombinant proteins. The latter was done to move the C-terminal end of the fragment ten residues further away from the area of low coiled-coil propensity centered at ~Asp1774. Amino acids 2–131 of Xrcc4 were fused to amino acids of 1733–1797 of cardiac myosin to make the Xrcc4-1733-1797 construct. Amino acids 2–47 of Gp7 were fused to amino acids 1733–1797 of cardiac myosin to make the Gp7-1733-1797 construct. Coiled-coil register was confirmed using COILS (Lupas et al., 1991). Both proteins crystallized readily, and structures of both the Xrcc4 fusion and the Gp7 fusion were solved to 2.5 Å and 2.9 Å resolution respectively (Fig. 4A and 4B). Many crystal forms were screened for the Gp7-1733-1797 construct, and while many diffracted past 2.9 Å resolution, most suffered from high mosaicity and anisotropic diffraction that made structure determination exceedingly difficult. The Gp7-1733-1797 structure exhibited interpretable electron density for the entire length of the coil, whereas the Xrcc4-1733-1797 construct showed interpretable electron density to residue 1795. The Xrcc4-1733-1797 structure contained one dimer in the asymmetric unit, while the Gp7-1733-1797 structure contained two non-crystallographically related dimers in the asymmetric unit.
Figure 4. Structures and comparison of the dimeric fusion proteins covering.
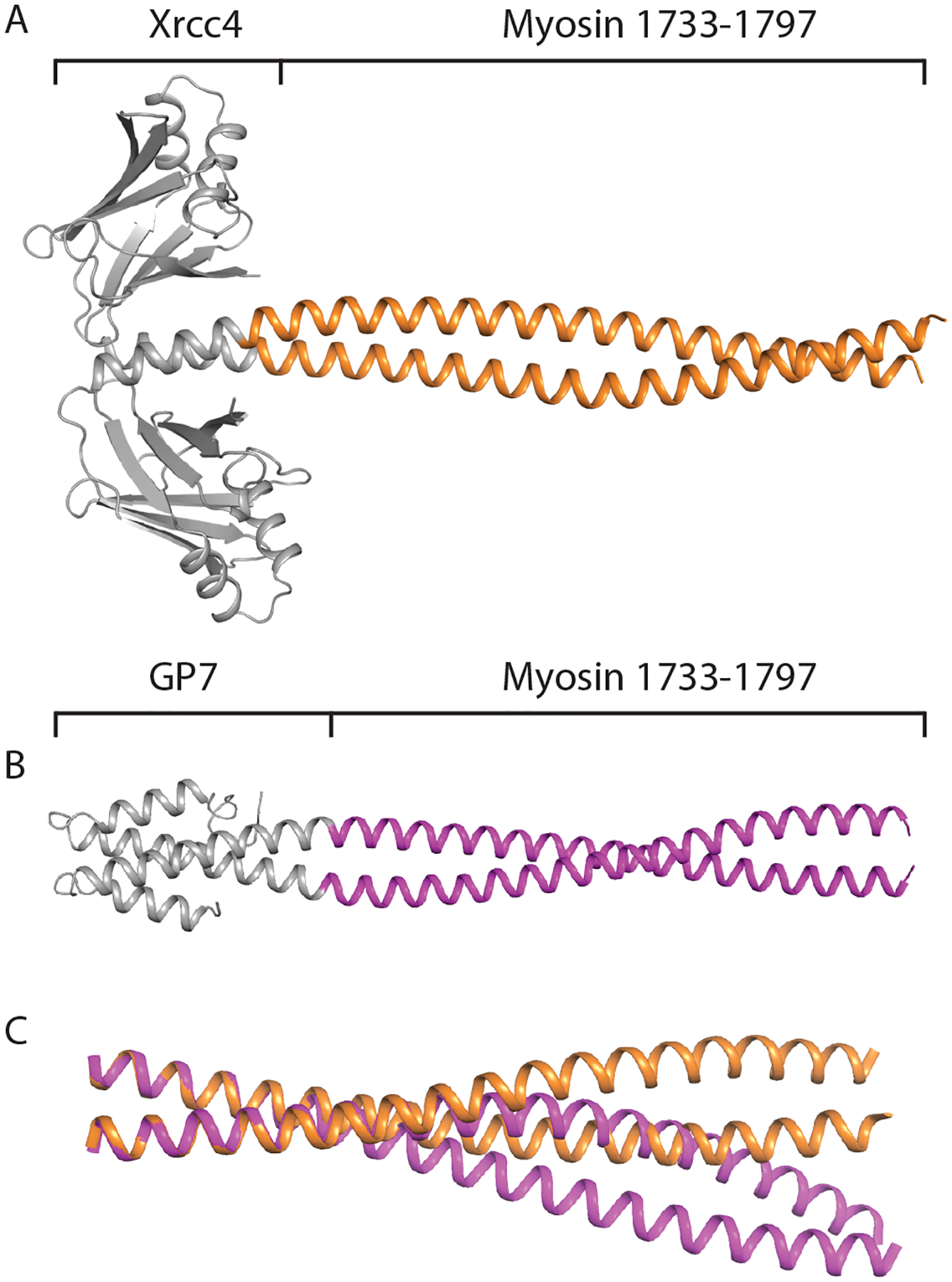
Asp1733 - Leu1797. (A) Structure of the coiled-coil domain of myosin rod from amino acids 1733–1797 fused to Xrcc4 fusion domain. (B) Structure of myosin rod from amino acids 1733–1797 fused to Gp7 fusion domain. (C) Aligned structure of the coiled-coil domains of the Xrcc4-1733-1797 structure and Gp7-1733-1797 structure. The two conformations of myosin were aligned from amino acids 1733–1746, and the deviation in coiled-coil pitch is clearly visible between the two conformations.
Using the Superpose alignment program, the myosin coiled-coil component of the two NCS related dimers in the asymmetric unit of the Gp7-1733-1797 overlapped with an RMSD of 0.25 Å as would be expected (Krissinel and Henrick, 2004). When the Gp7-1733-1797 myosin-coiled coil from amino acids 1733–1797 was aligned to the Xrcc4-1733-1797 coiled-coil using Superpose, the two structures overlapped with an RMSD of 1.73 Å, indicating conformational differences. Furthermore, the Xrcc4-1733-1797 coiled-coil domain had a superhelical pitch of 163 Å, while the Gp7-1733-1797 coiled-coil had a superhelical pitch of 146 Å, suggesting the Xrcc4-1733-1797 coiled-coil conformation was wound less tightly than the Gp7-1733-1797 coiled-coil. When the structures are overlapped from residues Asp1733 - Gln1746 with an RMSD of 0.23 Å, the deviation in pitch is clearly visible. Together, these conformations support structural flexibility throughout the region from Ala1690 - Ala1800 of cardiac myosin.
As noted earlier, the coiled-coil propensity drops considerably between Thr1760 and Ala1777 accompanied by a predicted disruption in the heptad repeat. Examination of the structures reveals that the distribution of side chains follows that of a canonical coiled-coil, to a first approximation which was unexpected based on the coiled-coil predictions. Many of the core residues in this segment are alanine (1751,1755, 1758, 1762, and 1766) which is also consistent with conformational variation (Brown, 2010). Alanine clustering has been invoked in the bending of tropomyosin around actin and may play a role in thick filament assembly (Brown et al., 2001). There are also four places following the alanine cluster where hydrophobic residues are surface exposed.
Assembly of an overall model for residues Arg1677 – Gly1807 of human β-cardiac myosin
The four overlapping fragments of human β-cardiac myosin reported here were assembled by concatenation with the previously reported fragment that includes Skip 4 (Fig. 5) (Taylor et al., 2015). Given the apparent conformational variation observed within the body of the fragments it is not possible or appropriate to generate a single model to represent this region since this would not necessarily be representative of the structure in solution or in the filament. However, the current models represent a good starting point for the molecular dynamics approach to building a complete model for the myosin rod as described earlier (Korkmaz et al., 2016).
Figure 5. Composite preliminary models for the myosin rod extending from Arg1677 – Gly1807.
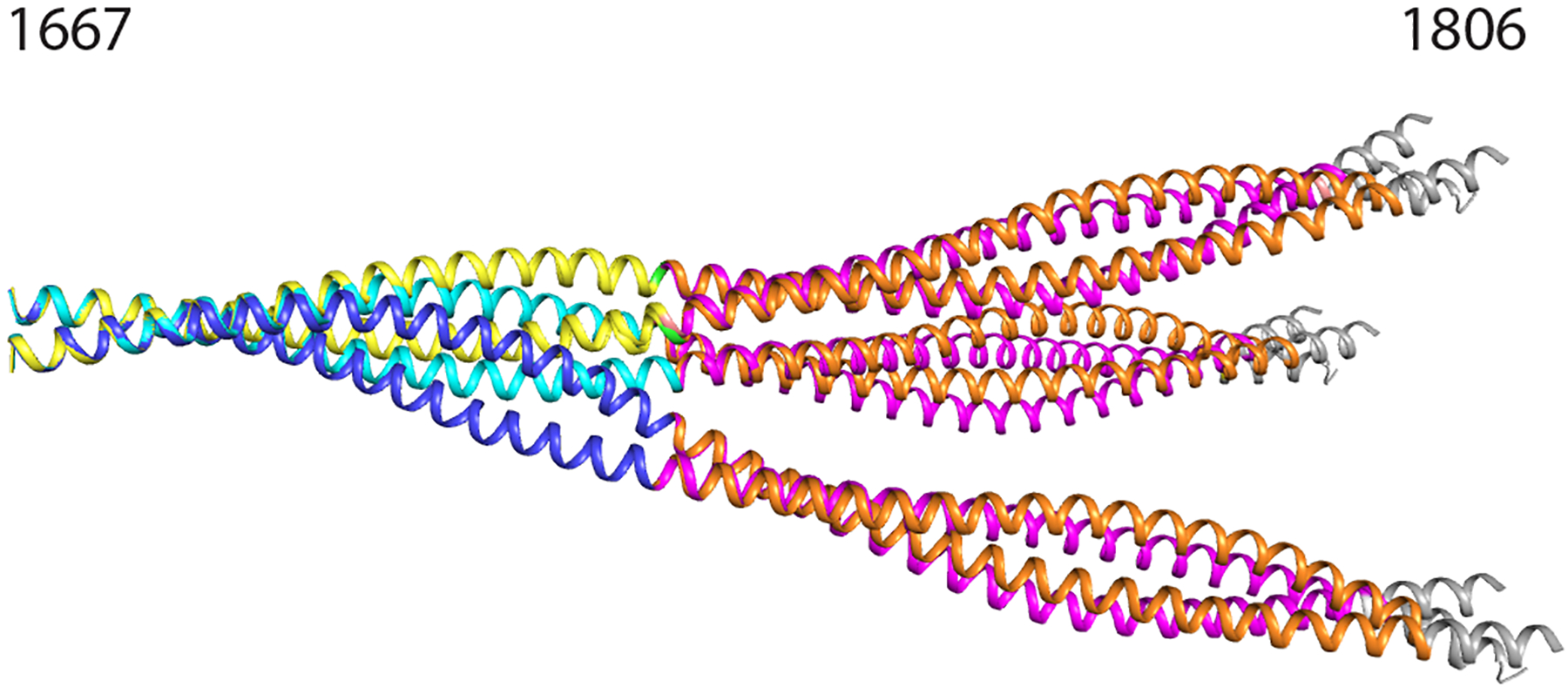
Composite model showing six conformations of myosin rod between amino acids 1677–1807. Models were overlapped from residues 1677–1691. Blue chains represent chains A and B of Xrcc4-1677-1758. Cyan chains represent chains C and D of Xrcc4-1677-1758. Yellow chains represent Gp7-1677-1755-Eb1. Orange chains represent Xrcc4-1733-1797. Magenta chains represent Gp7-1733-1797. Grey chains represent residues 1798–1807 of the published structure up to the skip 4 residue (PDB ID: 4XA6).
Conclusions
Utilizing globular coiled-coil fusion proteins to obtain crystal structures is an effective method to obtain structural data on coiled-coil domains. Using this method, we have structural evidence suggesting the myosin rod is highly flexible in the region between residues 1680–1800, Three conformations were obtained between residues 1677–1758, which deviated most strongly around residue L1706, the site of a pathogenic proline mutation. Finally, the use of coiled-coil fusion domains while effective is not without issue. We have shown that when regions of poor coiled-coil are placed out of coiled-coil register with fusion domains, they have the ability to organize into higher symmetry conformations. To avoid tetrameric artifacts as shown in the Gp7-1729-1787 structure, care must be taken to ensure that the designed fusion protein is indeed in the appropriate coiled-coil register. Finally, the presence of a dimeric fusion domain does not guarantee a dimeric assembly in the crystal lattice and emphasizes the importance of choosing the correct fusion points when designing expression constructs.
Acknowledgements
We thank Drs. Keenan Taylor and Mark Seeger for helpful discussions. We also thank Dr. James Thoden for maintenance of our X-ray diffraction facility.
Funding Sources
This work was supported in part by NIH grant R21 HL111237 to IR. Use of the Structural Biology ID19 and BM19 beamlines, Argonne National Laboratory Advanced Photon Source was supported by the U.S. Department of Energy, Office of Energy Research, under Contract No. W-31-109-ENG-38.
Abbreviations
- HEPES
4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid
- EDTA
ethylenediaminetetraacetic acid
- BME
β-mercaptoethanol
- rTEV
recombinant Tobacco Etch Virus protease
- TCEP
tris(2-carboxyethyl)phosphine
- MEPEG
methyl ether polyethylene glycol
- RMSD
root mean square deviation
- rTEV
recombinant Tobacco Etch Virus protease
- TCEP
tris(2-carboxyethyl)phosphine
- MEPEG
methyl ether polyethylene glycol
Footnotes
X-ray coordinate depositions
X-ray coordinates for Xrcc4-1677-1758, GP7-1677-1755-EB1, GP7-1729-1786, Xrcc4-1733-1797, and GP7-1733-1797 have been deposited with the Research Collaboratory for Structural Bioinformatics as Protein Data Bank entries XXXX, YYYY, YYY1, YYY2, and ZZZZ.
References
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung L-W, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH, 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallographica Section D 66, 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astbury WT, 1947. On the structure of biological fibres and the problem of muscle. Proc R Soc Lond B Biol Sci 134, 303–328. [DOI] [PubMed] [Google Scholar]
- Atkinson SJ, Stewart M, 1991. Expression in Escherichia coli of fragments of the coiled-coil rod domain of rabbit myosin: influence of different regions of the molecule on aggregation and paracrystal formation. J Cell Sci 99 (Pt 4), 823–836. [DOI] [PubMed] [Google Scholar]
- Blommel PG, Fox BG, 2007. A combined approach to improving large-scale production of tobacco etch virus protease. Protein Expr Purif 55, 53–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JH, 2010. How sequence directs bending in tropomyosin and other two-stranded alpha-helical coiled coils. Protein Sci 19, 1366–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JH, Zhou Z, Reshetnikova L, Robinson H, Yammani RD, Tobacman LS, Cohen C, 2005. Structure of the mid-region of tropomyosin: bending and binding sites for actin. Proc Natl Acad Sci U S A 102, 18878–18883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JH, Kim KH, Jun G, Greenfield NJ, Dominguez R, Volkmann N, Hitchcock-DeGregori SE, Cohen C, 2001. Deciphering the design of the tropomyosin molecule. Proc Natl Acad Sci U S A 98, 8496–8501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buvoli M, Buvoli A, Leinwand LA, 2012. Effects of pathogenic proline mutations on myosin assembly. J Mol Biol 415, 807–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen GJ, Qiu N, Karrer C, Caspers P, Page MG, 2000. Restriction site-free insertion of PCR products directionally into vectors. Biotechniques 28, 498–505. [DOI] [PubMed] [Google Scholar]
- Ciani B, Bjelic S, Honnappa S, Jawhari H, Jaussi R, Payapilly A, Jowitt T, Steinmetz MO, Kammerer RA, 2010. Molecular basis of coiled-coil oligomerization-state specificity. Proc Natl Acad Sci U S A 107, 19850–19855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen C, Holmes KC, 1963. X-ray diffraction evidence for alpha-helical coiled-coils in native muscle. J Mol Biol 6, 423–432. [DOI] [PubMed] [Google Scholar]
- Colegrave M, Peckham M, 2014. Structural implications of beta-cardiac myosin heavy chain mutations in human disease. Anat Rec (Hoboken) 297, 1670–1680. [DOI] [PubMed] [Google Scholar]
- Crick FHC, 1953. The Packing of a-Helices: simple coiled coils: Simple Coiled-Coils. Acta Cryst, 689–697. [Google Scholar]
- DeLano WL 2002. The PyMOL Molecular Graphics System.
- Delorenzi M, Speed T, 2002. An HMM model for coiled-coil domains and a comparison with PSSM-based predictions. Bioinformatics 18, 617–625. [DOI] [PubMed] [Google Scholar]
- Dill KA, Fiebig KM, Chan HS, 1993. Cooperativity in protein-folding kinetics. Proc Natl Acad Sci U S A 90, 1942–1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K, 2010. Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66, 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frye J, Klenchin VA, Rayment I, 2010. Structure of the tropomyosin overlap complex from chicken smooth muscle: insight into the diversity of N-terminal recognition. Biochemistry 49, 4908–4920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenfield NJ, Montelione GT, Farid RS, Hitchcock-DeGregori SE, 1998. The structure of the N-terminus of striated muscle alpha-tropomyosin in a chimeric peptide: nuclear magnetic resonance structure and circular dichroism studies. Biochemistry 37, 7834–7843. [DOI] [PubMed] [Google Scholar]
- Grigoryan G, Degrado WF, 2011. Probing designability via a generalized model of helical bundle geometry. J Mol Biol 405, 1079–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, Taylor DW, Reedy MK, Edwards RJ, Taylor KA, 2016. Structure of myosin filaments from relaxed Lethocerus flight muscle by cryo-EM at 6 A resolution. Sci Adv 2, e1600058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kammerer RA, Schulthess T, Landwehr R, Lustig A, Engel J, Aebi U, Steinmetz MO, 1998. An autonomous folding unit mediates the assembly of two-stranded coiled coils. Proc Natl Acad Sci U S A 95, 13419–13424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klenchin VA, Frye JJ, Jones MH, Winey M, Rayment I, 2011. Structure-function analysis of the C-terminal domain of CNM67, a core component of the Saccharomyces cerevisiae spindle pole body. The Journal of biological chemistry 286, 18240–18250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korkmaz EN, Taylor KC, Andreas MP, Ajay G, Heinze NT, Cui Q, Rayment I, 2016. A composite approach towards a complete model of the myosin rod. Proteins 84, 172–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krissinel E, Henrick K, 2004. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr D Biol Crystallogr 60, 2256–2268. [DOI] [PubMed] [Google Scholar]
- Lamont PJ, Wallefeld W, Hilton-Jones D, Udd B, Argov Z, Barboi AC, Bonneman C, Boycott KM, Bushby K, Connolly AM, Davies N, Beggs AH, Cox GF, Dastgir J, DeChene ET, Gooding R, Jungbluth H, Muelas N, Palmio J, Penttila S, Schmedding E, Suominen T, Straub V, Staples C, Van den Bergh PY, Vilchez JJ, Wagner KR, Wheeler PG, Wraige E, Laing NG, 2014. Novel mutations widen the phenotypic spectrum of slow skeletal/beta-cardiac myosin (MYH7) distal myopathy. Human mutation 35, 868–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landschulz WH, Johnson PF, McKnight SL, 1988. The leucine zipper: a hypothetical structure common to a new class of DNA binding proteins. Science 240, 1759–1764. [DOI] [PubMed] [Google Scholar]
- Li Y, Brown JH, Reshetnikova L, Blazsek A, Farkas L, Nyitray L, Cohen C, 2003. Visualization of an unstable coiled coil from the scallop myosin rod. Nature 424, 341–345. [DOI] [PubMed] [Google Scholar]
- Lowey S, Cohen C, 1962. Studies on the structure of myosin. J Mol Biol 4, 293–308. [DOI] [PubMed] [Google Scholar]
- Lupas A, 1996a. Prediction and analysis of coiled-coil structures. Methods Enzymol 266, 513–525. [DOI] [PubMed] [Google Scholar]
- Lupas A, 1996b. Coiled coils: new structures and new functions. Trends in biochemical sciences 21, 375–382. [PubMed] [Google Scholar]
- Lupas A, Van Dyke M, Stock J, 1991. Predicting coiled coils from protein sequences. Science 252, 1162–1164. [DOI] [PubMed] [Google Scholar]
- Lupas AN, Gruber M, 2005. The structure of alpha-helical coiled coils. Advances in protein chemistry 70, 37–78. [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ, 2007. Phaser crystallographic software. J Appl Crystallogr 40, 658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLachlan AD, Karn J, 1982. Periodic charge distributions in the myosin rod amino acid sequence match cross-bridge spacings in muscle. Nature 299 226–231 [DOI] [PubMed] [Google Scholar]
- Meredith C, Herrmann R, Parry C, Liyanage K, Dye DE, Durling HJ, Duff RM, Beckman K, de Visser M, van der Graaff MM, Hedera P, Fink JK, Petty EM, Lamont P, Fabian V, Bridges L, Voit T, Mastaglia FL, Laing NG, 2004. Mutations in the slow skeletal muscle fiber myosin heavy chain gene (MYH7) cause laing early-onset distal myopathy (MPD1). Am J Hum Genet 75, 703–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minor W, Cymborowski M, Otwinowski Z, Chruszcz M, 2006. HKL-3000: the integration of data reduction and structure solution--from diffraction images to an initial model in minutes. Acta Crystallogr. D Biol. Crystallogr 62, 859–866. [DOI] [PubMed] [Google Scholar]
- Morais MC, Kanamaru S, Badasso MO, Koti JS, Owen BA, McMurray CT, Anderson DL, Rossmann MG, 2003. Bacteriophage phi29 scaffolding protein gp7 before and after prohead assembly. Nat Struct Biol 10, 572–576. [DOI] [PubMed] [Google Scholar]
- Moutevelis E, Woolfson DN, 2009. A periodic table of coiled-coil protein structures. J Mol Biol 385, 726–732. [DOI] [PubMed] [Google Scholar]
- O’Shea EK, Klemm JD, Kim PS, Alber T, 1991. X-ray structure of the GCN4 leucine zipper, a two-stranded, parallel coiled coil. Science 254, 539–544. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W, 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods in Enzymology 276, 307–326. [DOI] [PubMed] [Google Scholar]
- Parry DA, 2014. Fifty years of fibrous protein research: a personal retrospective. J Struct Biol 186, 320–334. [DOI] [PubMed] [Google Scholar]
- Parry DA, Fraser RD, Squire JM, 2008. Fifty years of coiled-coils and alpha-helical bundles: a close relationship between sequence and structure. J Struct Biol 163, 258–269. [DOI] [PubMed] [Google Scholar]
- Rackham OJ, Madera M, Armstrong CT, Vincent TL, Woolfson DN, Gough J, 2010. The evolution and structure prediction of coiled coils across all genomes. J Mol Biol 403, 480–493. [DOI] [PubMed] [Google Scholar]
- Rose A, Schraegle SJ, Stahlberg EA, Meier I, 2005. Coiled-coil protein composition of 22 proteomes--differences and common themes in subcellular infrastructure and traffic control. BMC evolutionary biology 5, 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibanda BL, Critchlow SE, Begun J, Pei XY, Jackson SP, Blundell TL, Pellegrini L, 2001. Crystal structure of an Xrcc4-DNA ligase IV complex. Nat Struct Biol 8, 1015–1019. [DOI] [PubMed] [Google Scholar]
- Slayter HS, Lowey S, 1967. Substructure of the myosin molecule as visualized by electron microscopy. Proc Natl Acad Sci U S A 58, 1611–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohn RL, Vikstrom KL, Strauss M, Cohen C, Szent-Gyorgyi AG, Leinwand LA, 1997. A 29 residue region of the sarcomeric myosin rod is necessary for filament formation. J Mol Biol 266, 317–330. [DOI] [PubMed] [Google Scholar]
- Steinmetz MO, Stock A, Schulthess T, Landwehr R, Lustig A, Faix J, Gerisch G, Aebi U, Kammerer RA, 1998. A distinct 14 residue site triggers coiled-coil formation in cortexillin I. Embo J 17, 1883–1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strelkov SV, Herrmann H, Geisler N, Lustig A, Ivaninskii S, Zimbelmann R, Burkhard P, Aebi U, 2001. Divide-and-conquer crystallographic approach towards an atomic structure of intermediate filaments. J Mol Biol 306, 773–781. [DOI] [PubMed] [Google Scholar]
- Taylor KC, Buvoli M, Korkmaz EN, Buvoli A, Zheng Y, Heinze NT, Cui Q, Leinwand LA, Rayment I, 2015. Skip residues modulate the structural properties of the myosin rod and guide thick filament assembly. Proc Natl Acad Sci U S A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagin AA, Steiner RA, Lebedev AA, Potterton L, McNicholas S, Long F, Murshudov GN, 2004. REFMAC5 dictionary: organization of prior chemical knowledge and guidelines for its use. Acta Crystallogr D Biol Crystallogr 60, 2184–2195. [DOI] [PubMed] [Google Scholar]
- van den Ent F, Lowe J, 2006. RF cloning: a restriction-free method for inserting target genes into plasmids. J Biochem Biophys Methods 67, 67–74. [DOI] [PubMed] [Google Scholar]
- Woolfson DN, 2005. The design of coiled-coil structures and assemblies. Adv Protein Chem 70, 79–112. [DOI] [PubMed] [Google Scholar]
- Wu KC, Bryan JT, Morasso MI, Jang SI, Lee JH, Yang JM, Marekov LN, Parry DA, Steinert PM, 2000. Coiled-coil trigger motifs in the 1B and 2B rod domain segments are required for the stability of keratin intermediate filaments. Mol Biol Cell 11, 3539–3558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang KY, Cowtan K, Main P, 1997. Combining constraints for electron-density modification. Methods Enzymol 277, 53–64. [DOI] [PubMed] [Google Scholar]


