Abstract
Four expression cassettes containing strong fungal promoters, a signal sequence for protein translocation, a KEX protease cleavage site, and a synthetic gene (tha) encoding the sweet protein thaumatin II were used to overexpress this protein in Aspergillus awamori lpr66, a PepA protease-deficient strain. The best expression results were obtained with the gdhA promoter of A. awamori or with the gpdA promoter of Aspergillus nidulans. There was good correlation of tha gene dosage, transcript levels, and thaumatin secretion. The thaumatin gene was expressed as a transcript of the expected size in each construction (1.9 or 1.4 kb), and the transcript levels and thaumatin production rate decayed at the end of the growth phase, except in the double transformant TB2b1-44-GD5, in which secretion of thaumatin continued until 96 h. The recombinant thaumatin secreted by a high-production transformant was purified to homogeneity, giving one major component and two minor components. In all cases, cleavage of the fused protein occurred at the KEX recognition sequence. This work provides new expression systems in A. awamori that result in very high levels of thaumatin production.
Thaumatin is a very sweet protein, originally extracted from the arils of the fruit of Thaumatococcus daniellii Benth (31). It is used as a food sweetener and to increase the palatability of animal feed. The availability of thaumatin of plant origin is very limited (33), and it is notoriously difficult to produce by recombinant DNA methods. Production has been attempted with Escherichia coli (10), Bacillus subtilis (19), Streptomyces lividans (20), Saccharomyces cerevisiae (8), and Aspergillus oryzae (16). A synthetic gene for thaumatin II with fungal codon usage has been synthesized (10), but expression in Aspergillus niger gave poor yields (9).
Expression could be limited by (i) a weak promoter, (ii) copy number, (iii) insertion location, (iv) inefficient processing of the pre-propeptides (26), or (v) bottlenecks in protein traffic and translocation through the membrane systems of the protein secretory pathway (18, 29). Filamentous fungi, particularly Aspergillus awamori, are widely used for expression of heterologous proteins (15, 27, 30). Homologous proteins (e.g., glucoamylase) are usually well expressed in this fungus (up to 20 to 30 mg/ml) (12), but many heterologous proteins are expressed very poorly (29).
Several strong fungal promoters involved in primary and secondary metabolism are now available. These include, among others, the promoters of the glyceraldehyde-3-phosphate dehydrogenase gene (gpdA) of Aspergillus nidulans (22, 23), the glutamate dehydrogenase gene (gdhA) of A. awamori (5), the isopenicillin N-synthase (pcbC) gene of Penicillium chrysogenum (14), and the B2 wide-spectrum esterase gene (cesB) of Acremonium chrysogenum (28).
Efficient production of an heterologous protein may be achieved by increasing gene dosage, although overloading of the secretory pathway may result in abnormal folding and protein degradation (17). Little is known about specific proteolytic cleavage of preproteins during secretion in fungi. One of the best known systems is the KEX protease, which cleaves at Arg-Lys sequences in the polypeptides (4).
Our objectives were (i) to find efficient fungal promoters for overexpression of the plant thaumatin gene, (ii) to determine if the KEX protease processes the fused proteins at the proper sequence to release thaumatin, and (iii) to optimize thaumatin production for commercial applications. We report in this article significant increases in tha gene expression and production of thaumatin by using expression cassettes with different fungal promoters.
MATERIALS AND METHODS
Microbial strains.
An A. awamori lpr66 mutant (deficient in aspergillopepsin A [21]) was used as the host strain for expression studies. Escherichia coli DH5α was used for plasmid amplification and purification.
Media and growth conditions.
A. awamori strains were maintained on solid Power sporulation medium (11) at 30°C for 3 days. Seed cultures of A. awamori lpr66 in CM medium (5 g of malt extract, 5 g of yeast extract, and 5 g of glucose per liter) were inoculated with 106 spores/ml and grown at 28°C in a rotary G10 incubator (New Brunswick Scientific, New Brunswick, N.J.) for 48 h. For thaumatin production studies and for transcription analysis, A. awamori was grown in MDFA defined medium (25), inoculated with a 15% seed culture, and grown at 30°C for 96 to 120 h in a rotary shaker.
Construction of the different cassettes for overexpression of the thaumatin gene.
Four expression cassettes (pBKThb, pCKThb, pGDTh, and pGPThb) were prepared. Each encoded a fusion protein formed by a signal sequence from the A. chrysogenum extracellular esterase B2 and most of the B2 gene as cDNA (665 bp) (except for sequences in the carboxyl-terminal end), a spacer sequence containing a Lys-Arg-Lys-Arg KEX2 processing sequence (18 bp), and the synthetic gene of thaumatin II (627 bp) (10). These constructs were coupled to four different promoter regions. All constructs included a transcription termination signal from the CYC1 gene (285 bp) of Saccharomyces cerevisiae that works well in A. awamori (5) and were introduced in a plasmid that contains a phleomycin resistance gene (ble) or a hygromycin resistance gene (hyg) as a selectable marker.
Southern blotting and hybridization.
Total DNA from A. awamori strains was digested with restriction endonucleases, electrophoresed in 0.7% agarose, and blotted by standard techniques (24). Probes internal to the gdhA and tha genes were mixed and labeled together by nick translation with [32P]dCTP to the same specific activity and hybridized by standard methods (24).
DNA sequencing.
DNA fragments that contain the connections between the different promoters and the B2 gene and between the B2 gene and the thaumatin gene were subcloned into pBluescript KS+ and sequenced by using the GeneAmp PCR 2400 system coupled to the ABI-PRISM 310 automatic sequencer (Perkin-Elmer, Norwalk, Conn.). Computer analyses of nucleotide and amino acid sequences were made with the DNASTAR Programs (DNASTAR, Inc., London, United Kingdom).
Isolation of RNA, Northern hybridization, and slot blotting.
Total RNA of A. awamori strains was obtained from mycelia grown for 24, 48, or 72 h in MDFA medium (25) by the phenol-sodium dodecyl sulfate (SDS) method (1). For Northern analysis, total RNA (5 μg) was run on a 1.2% agarose–formaldehyde gel. The gel was blotted onto a nylon filter (Nytran 0.45; Schleicher and Schuell) by standard methods (24). The RNA was fixed by UV irradiation with a UV-Stratalinker 2400 lamp (Stratagene, La Jolla, Calif.). For slot blotting, the RNA (5 μg) was loaded on a filter (Nytran 0.45) by vacuum in a Bio-Dot SF microfiltration apparatus (Bio-Rad) and fixed by UV irradiation as described above. The filters were prehybridized for 3 h at 42°C in a mixture of 50% formamide, 5× Denhardt’s solution, 5× SSPE (1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA [pH 7.7]), 0.1% SDS, and 500 μg of denatured salmon sperm DNA per ml and hybridized in the same buffer containing 100 μg of denatured salmon sperm DNA per ml at 42°C for 18 h, with an internal fragment (0.33-kb NcoI) of the synthetic thaumatin gene used as a probe. The filter was washed once in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–0.1% SDS at 42°C for 15 min, once in 0.1× SSC–0.1% SDS at 42°C for 15 min, and once more in 0.1× SSC–0.1% SDS at 65°C for 15 min and then autoradiographed with Amersham X-ray film.
Transformation of A. awamori lpr66.
A. awamori lpr66 was transformed by the method of Yelton et al. (32). Transformants were selected in tryptic soy agar (Difco, Detroit, Mich.) with 10% sucrose supplemented with 25 μg of phleomycin or 100 μg of hygromycin per ml for double transformation experiments with pGD71.
Antithaumatin polyclonal antibodies.
Thaumatin (from Thaumatococcus daniellii [Sigma, Saint Louis, Mo.]) was purified by electroelution of the 22-kDa thaumatin band from a SDS–15% polyacrylamide gel. Antithaumatin antibodies were obtained by immunizing New Zealand rabbits by standard procedures (7), as described in detail elsewhere (14).
Immunoblotting.
Immunoblot analysis of A. awamori cell extracts and culture supernatants was made after protein separation by electrophoresis on an SDS–15% polyacrylamide gel. The supernatants were concentrated 10-fold with trichloroacetic acid at 10%. In culture broths in which the production of thaumatin was high, 3 μl of supernatant was loaded directly (without concentration) on a 15% polyacrylamide gel. After SDS polyacrylamide gel electrophoresis (PAGE), the proteins were transferred to a polyvinylidene difluoride membrane (PVDF) (Immobilon-P; Millipore) by using a Minitransblot electroblotting system (Bio-Rad). The membranes were treated with antithaumatin antibodies (serum dilution, 1:15,000) in 50 mM Tris-HCl (pH 8.0) with 150 mM sodium chloride (TBS), containing Tween 20 at 0.2% for 1 h, and for 30 min with the antirabbit commercial alkaline phosphatase conjugate (1:5,000) in the same buffer. The membranes were treated with a BCIP-NBT (5-bromo-4-chloro-3-indolylphosphate toluidinium–nitroblue tetrazolium) solution for alkaline phosphatase (Sigma) until the color was developed.
Quantification of thaumatin by ELISA.
Thaumatin in the culture medium of A. awamori lpr66 transformants was quantified by an enzyme-linked immunoassay (ELISA). The wells of plates (Nunc Immunoplates) were coated with dilutions of the supernatant samples (1:10 to 1:1,280) overnight at 4°C. The plates were washed three times with phosphate-buffered saline (PBS) plus 0.1% Tween 20 (PBS-T), blocked with PBS-T containing 5% dry milk for 1 h, and washed three times with PBS-T. The thaumatin was measured by addition of a 1:10,000 dilution of the rabbit antithaumatin antiserum for 1 h followed by a 1:5,000 dilution of a goat anti-rabbit commercial alkaline phosphatase conjugate (Sigma) for 30 min. The antigen-antibody complexes were quantified by using a stabilized substrate solution for alkaline phosphatase (Sigma) at 405 and 620 nm in a Scanning Autoreader and Microplate Workstation (CERES 900C; Bio-Tek Instruments). Decreasing concentrations of thaumatin II from Sigma (1 μg/ml to 1.6 ng/ml) were used as a standard.
Purification of the recombinant thaumatin.
Culture broths of strain TB2b1-44 grown in 5-liter fermentors (BiofloII, New Brunswick, N.J.) were used for purification of the recombinant protein. The broth (500 ml) was filtered through Nytal cloth (pore diameter, 30 μm), and the proteins of the culture were precipitated with solid ammonium sulfate in the range of 20 to 50% of saturation. The resulting precipitate was resuspended in 25 mM sodium phosphate buffer (pH 7.0). The sample was desalted by filtration through a Sephadex G-25 column (Pharmacia) and eluted with the same buffer. The protein solution was loaded onto a carboxymethyl-Sepharose column (23.5 by 1.6 cm) and washed with the same buffer until the A280 was less than 0.1. The thaumatin was eluted with a linear gradient of 0 to 400 mM sodium chloride (flow rate, 0.5 ml/min). Fractions of 5 ml were collected, and the fractions containing thaumatin were used for subsequent analysis.
N-terminal amino acid sequence.
Purified recombinant thaumatin (12, 16, and 6 μg, of each thaumatin component, respectively) was loaded onto an SDS-PAGE (15% polyacrylamide) gel. After electrophoresis, the proteins were transferred to a PVDF membrane and sequenced on an automated protein sequencer 473A (Applied Biosystems, Inc., Foster City, Calif.).
RESULTS
Construction of expression plasmids and transformation of A. awamori.
Four expression plasmids, pBKThb, pCKThb, pGDTh, and pGPThb, were constructed for secretion of thaumatin protein to culture medium (Fig. 1). All plasmid constructions contain a cDNA sequence encoding the signal sequence and the first 311 amino acids (except pCKThb, which contains 141 amino acids) of the amino-terminal end of the B2 esterase of A. chrysogenum, a spacer sequence containing two cleavage sites for the KEX2-like protease (Lys-Arg) in tandem, the synthetic gene of thaumatin II with optimized codon usage for fungi (9), and the CYC1 transcriptional terminator of S. cerevisiae.
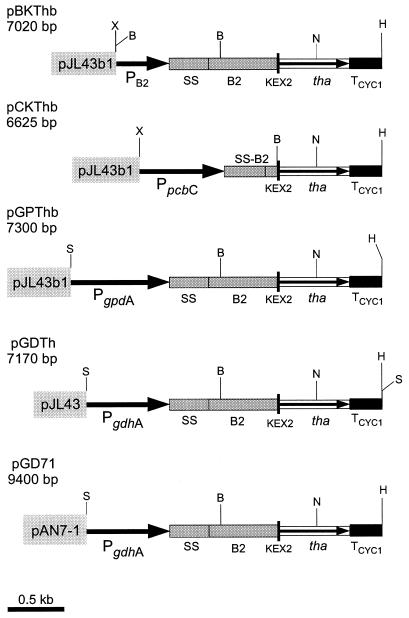
FIG. 1.
Physical map of plasmids pBKThb, pCKThb, pGPThb, pGDTh, and pGD71 constructed to express the thaumatin gene from efficient transcription initiation regions (thick arrows): PB2, 477 bp; PpcbC, 761 bp; PgpdA, 880 bp; PgdhA, 750 bp. The signal sequence and amino-terminal region of the B2 protein of A. chrysogenum (SS-B2) are indicated by shaded boxes. The KEX2 sequence is indicated as a black vertical thick line, and the synthetic gene of thaumatin II (tha) is shown by a thin arrow inside the open box. TCYC1, transcriptional terminator of S. cerevisiae. Vector pAN7-1 contains a hygromycin resistance marker, whereas pJL43 and pJL43b1 contain the phleomycin resistance marker. The following restriction sites were used: HindIII (H), BamHI (B), SalI (S), XbaI (X), and NcoI (N).
This cassette was fused to four fungal promoters, namely the B2 promoter of A. chrysogenum (pBKThb), the pcbC promoter of P. chrysogenum (pCKThb), the gdhA promoter of A. awamori (pGDTh), and the gpdA promoter of A. nidulans (pGPTh).
All constructs were introduced into the A. awamori lpr66 strain, containing an inactive aspergillopepsin A (PepA) (21) by using ble as the selectable marker.
Production of thaumatin by transformants with different expression cassettes.
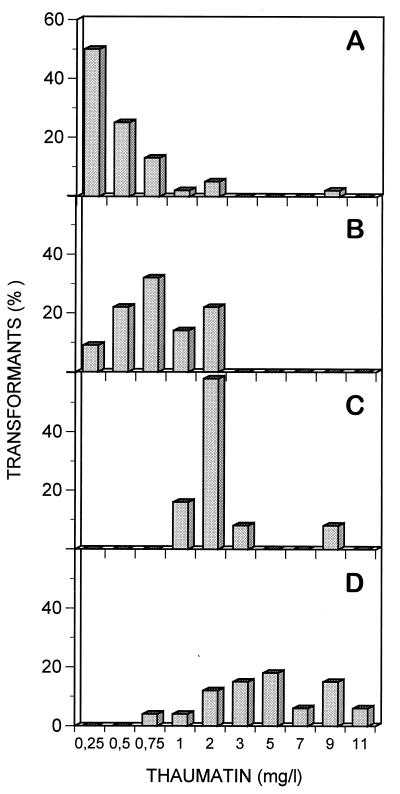
The frequency distribution of thaumatin secreted by the lpr66 transformants in flask cultures in MDFA medium is summarized in Fig. 2. Thaumatin production was observed with all four promoters (Fig. 2), but the best results were obtained with the gdhA promoter of A. awamori (Fig. 2C) and the gpdA promoter of A. nidulans (Fig. 2D). With these two promoters, a significant fraction of transformants yielded more than 2 mg of thaumatin per liter, with some yields as high as 9 to 11 mg/liter.
FIG. 2.
Frequency distribution of thaumatin production by transformants with different promoter regions in the thaumatin expression cassettes. (A) Constructs with the B2 promoter of A. chrysogenum. (B) clones with the pcbC promoter of P. chrysogenum. (C) clones with the gdhA promoter of A. awamori. (D) clones with the gpdA promoter of A. nidulans. The transformants were grown in MDFA medium at 30°C for 5 days, and thaumatin was quantified by the ELISA procedure.
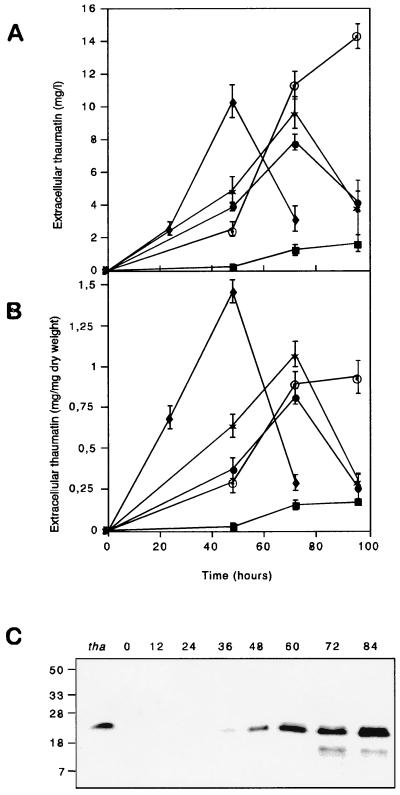
One transformant, TB2b1-44, which contains the expression cassette controlled by the B2 promoter, was retransformed with plasmid pGD71, which contains the expression cassette with the gdhA promoter and the hygromycin resistance gene as a selectable marker. Double transformants with higher levels of thaumatin production were selected and cultured in shake flasks with MDFA medium modified to permit maximum transcription of the promoter in the expression cassette (Table 1). The highest levels of thaumatin in most cases were detected at 72 h of incubation (Fig. 3). After this time, thaumatin levels dropped markedly, except in the double transformant TB2b1-44-GD5, in which production continued to increase. TGDTh-4, which contained the gdhA promoter-thaumatin fusion, had the highest level of secreted thaumatin at 48 h, with ammonium sulfate as a nitrogen source, in agreement with the known early expression of this gene in A. awamori (5).
TABLE 1.
Thaumatin production by selected transformants with different thaumatin expression cassettesa
| Transformant | Promoter of expression cassette | N source (80 mM) | C source | Thaumatin production (mg/liter) |
|---|---|---|---|---|
| TB2b1-44 | B2 | l-Asn | 2.7% Glc–3.6% sucrose | 7.7 ± 0.5 |
| TCTh-21 | pcbC | l-Asn | 2.7% Glc–3.6% sucroseb | 1.6 ± 0.2 |
| TGP-3 | gpdA | l-Asn | 1% Glc–3.6% sucrose | 9.6 ± 0.2 |
| TGDTh-4 | gdhA | (NH4)2SO4 | 1% Glc–3.6% sucrosec | 10 ± 0.7 |
| TB2b1-44-GD4 | B2, gdhA | l-Asn | 2.7% Glc–3.6% sucrose | 10 ± 0.4 |
| TB2b1-44-GD5 | B2, gdhA | l-Asn | 2.7% Glc–3.6% sucrose | 14 ± 1.1 |
| TB2b1-44-GD6 | B2, gdhA | l-Asn | 2.7% Glc–3.6% sucrose | 11 ± 0.8 |
| TB2b1-44-GD7 | B2, gdhA | l-Asn | 2.7% Glc–3.6% sucrose | 14 ± 1.3 |
The transformants were grown in MDFA medium for 96 h in an orbital incubator at 30°C and 250 rpm. Thaumatin was quantified by ELISA as described in Materials and Methods.
The pcbC promoter is induced by lactose, but this transformant showed a poor growth with lactose as the carbon source, and thaumatin production was very low.
The MDFA was buffered with 2% MOPS (morpholine propanesulfonic acid) to prevent acidification of medium.
FIG. 3.
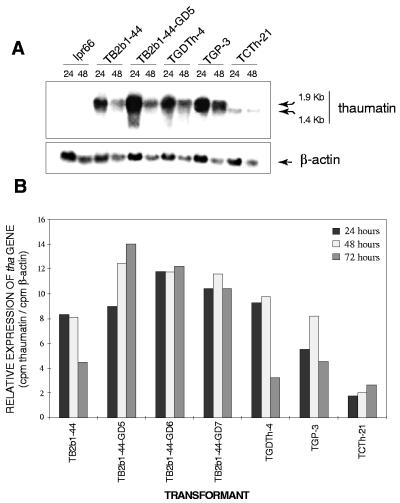
Time course of thaumatin production by selected transformants with the different promoters in flask cultures with the nitrogen and carbon sources shown in Table 1. (A) Total production of thaumatin. (B) Specific production of thaumatin. ⧫, TGDTh-4; ●, TB2b1-44; ✶, TGP-3; ■, TCTh-21; ⊙, TB2b1-44-GD5. The vertical thin lines indicate the standard deviation of the four determinations at each sample point. (C) Immunoblot detection of thaumatin produced at different times during fermentation by strain TB2b1-44-GD5 (A). The first lane contains a control of purified 22-kDa thaumatin from T. daniellii (100 ng).
Western blot analysis of the supernatants showed that recombinant thaumatin migrated at the position of pure plant thaumatin (22 kDa), indicating that the KEX2 cleavage site is adequately recognized for removal of the B2 carrier protein even in transformants with high thaumatin yields (Fig. 3C). Smaller immunoreactive proteins may be observed migrating ahead of the thaumatin, suggesting that some proteolytic degradation by A. awamori proteases other than PepA still occurs.
Intracellular levels of thaumatin in the best producer strains were determined. Proteins in the soluble fraction of cell extracts from samples taken at different culture times were resolved by SDS-PAGE and had immunoreactive bands usually smaller than that of thaumatin in cultures with efficient expression (data not shown), indicating that intracellular thaumatin degradation is occurring. The intracellular thaumatin was about 3% of the total intracellular protein (200 ng in a 6-μg protein sample); i.e., a significant amount of thaumatin remained unsecreted.
Copy number of the thaumatin cassette correlates with thaumatin production.
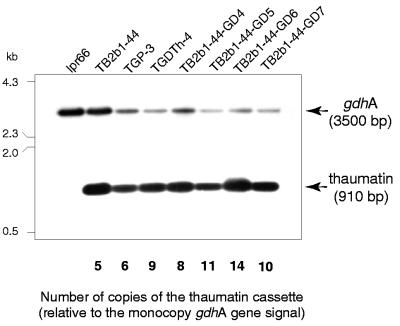
The copy number of the integrated thaumatin cassette was analyzed by Southern hybridization with a probe (332-bp NcoI fragment internal to the thaumatin gene) for the transformants with higher thaumatin production from each expression cassette and for three strains derived from transformant TB2b1-44 by retransformation with plasmid pGD71. Hybridization with a 694-bp PvuII fragment of the gdhA gene, which is a single copy in the A. awamori genome (5), was used as a control to estimate the number of thaumatin cassettes in each transformant. Copy number was calculated as the ratio in the phosphorimager between the radioactive signal of the band corresponding to the thaumatin gene and that of the band corresponding to the gdhA gene.
DNA of the lpr66 control strain had a 3.5-kb band corresponding to the gdhA gene but did not hybridize with a 910-bp BamHI-NcoI DNA fragment from the thaumatin gene expression cassette (Fig. 4). All transformants hybridized strongly with this probe, indicating the presence of multiple copies of the thaumatin gene, ranging from 5 copies in strain TB2b1-44 to 14 copies in the retransformed strain TB2b1-44-GD6. There was a good correlation between the copy number of the expression cassette and extracellular production of thaumatin. No rearrangements of the expression cassette (as indicated by the lack of hybridizing bands of sizes different from that of the cassette) were observed in any of the transformants.
FIG. 4.
Southern blot hybridizations of A. awamori lpr66 transformants and an untransformed control strain (lpr66) with a 32P-labeled 332-bp NcoI fragment internal to tha and a 694-bp PvuII probe from the gdhA gene. Genomic DNA was digested with the restriction enzymes NcoI and BamHI, and the fragments were separated on a 0.8% agarose gel and transferred to a nylon filter. Numbers on the left indicate the molecular size (in kilobases) of λ-HindIII standards. The copy number of the thaumatin gene in each transformant (relative to one copy per genome of gdhA) is indicated on the bottom of the figure.
The thaumatin gene is expressed as a monocistronic transcript in multicopy transformants.
Expression of the thaumatin gene in the different thaumatin-producing strains was analyzed by Northern hybridization of RNA samples taken at several times during fermentation with the same probe used for Southern blot analysis. A probe containing the A. nidulans β-actin gene was used to control the amount of RNA.
The amount of tha gene transcript present in the transformants (Fig. 5A) is higher than the level of β-actin mRNA. The highest level of the thaumatin transcript was observed in the double transformant TB2b1-44-GD5. The fused B2-thaumatin mRNA was transcribed correctly, having the expected transcript size of 1.9 kb. A smaller thaumatin mRNA (1.4 kb) was found in the TCTh-21 transformant, with the pcbC promoter, because the DNA fragment corresponding to the B2 gene in this cassette has only 105 bp of the 5′ end of the gene instead of the 615 bp in the other expression cassettes.
FIG. 5.
(A) Northern blot analysis of the transcripts of the thaumatin and β-actin genes in transformants with the different promoters. Total RNA was extracted from A. awamori transformants grown in MDFA medium and harvested at 24 and 48 h. The probes used were a 332-bp NcoI fragment internal to tha and an 834-bp NcoI-KpnI fragment of the β-actin gene from A. nidulans. The size of the thaumatin transcript (in kilobases) is indicated by arrows on the right. (B) Relative levels of thaumatin gene expression of different transformants at 24, 48, and 72 h in MDFA medium normalized with respect to the β-actin signal as a control.
Time course of expression of the thaumatin cassette.
We used slot blots to assess the temporal expression of the thaumatin gene (Fig. 5B). The level of thaumatin mRNA was highest at 24 to 48 h in transformants TGDTh-4, TB2b1-44, and TGP-3. Transformant TCTh-21 (with the pcbC promoter) had poor expression throughout the fermentation. Transformants TB2b1-44-GD5, -GD6, and -GD7, which contained two distinct thaumatin expression cassettes, did not have a decrease in thaumatin expression at 72 h.
The three forms of recombinant thaumatin differ in one amino acid at the amino-terminal end.
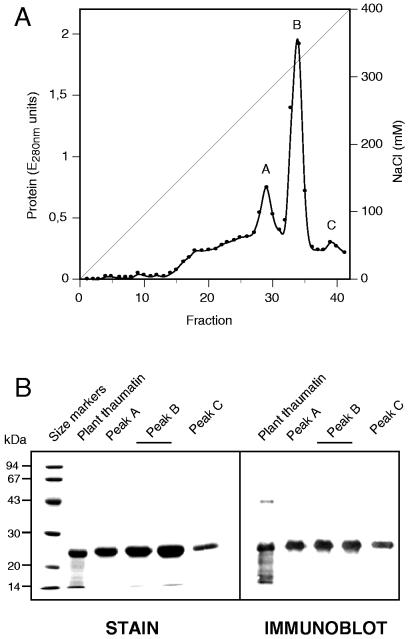
During elution of the recombinant thaumatin from a carboxymethyl-Sepharose column, three peaks (named A, B, and C) were detected (Fig. 6A); all of them correspond to thaumatin according to SDS-PAGE and Western blot analysis (Fig. 6B). The efficiency of recovery was about 70%.
FIG. 6.
(A) Elution profile of recombinant thaumatin from a carboxymethyl-Sepharose column with an NaCl gradient (0 to 400 mM). Peaks A, B, and C represent the three purified forms of thaumatin. (B) SDS-PAGE (left) and immunoblotting (right) analysis of purified recombinant thaumatin. Lanes: 1, molecular mass markers (Pharmacia low-molecular-weight calibration kit); 2, commercial thaumatin from Thaumatococcus (Sigma); 3, fraction 29 from carboxymethyl-Sepharose elution (peak A); 4 and 5, fractions 33 and 34, respectively (peak B); 6, fraction 39 (peak C).
The amino-terminal end of each form was sequenced (Fig. 7). Peak A (elution at 280 mM NaCl; 18 to 20% of total purified thaumatin) corresponded to thaumatin II with an additional Arg residue (originating from the KEX2 processing linker); peak B (elution at 330 mM NaCl; 75 to 78% of the total) had an additional Lys-Arg fused to thaumatin II, and peak C (elution at 380 mM NaCl and only 3 to 5% of total purified thaumatin) had an amino-terminal Arg-Lys-Arg addition (Fig. 7). Identical results were obtained with thaumatin purified from supernatants from strain TGDTh-4. All three forms of purified thaumatin had a sweet taste.
FIG. 7.
Amino acid sequence of the B2-KEX2-thaumatin fusions showing the cleavage sites. Major and minor vertical arrows correspond to the cleavage sites forming thaumatins A, B, and C as shown by Edman determination of the amino-terminal sequences.
DISCUSSION
The bottlenecks that limit gene expression and heterologous protein secretion in A. awamori are a subject of great interest. An efficient system for secretion of thaumatin has been developed by combining (i) strong fungal promoters, (ii) double transformants with different expression cassettes, and (iii) a cleavage sequence for the KEX proteolytic system. The high production of thaumatin (up to 14 μg/ml in shake flasks) has been confirmed in fermentors (21) (nearly 100 μg/ml) and has a potential application at the commercial scale.
Effective expression of the thaumatin gene was observed at the mRNA level in A. awamori, particularly in expression cassettes containing the gdhA promoter of A. awamori and the gpdA promoter of A. nidulans. In contrast, the pcbC promoter of P. chrysogenum (involved in penicillin biosynthesis) and the B2 esterase promoter of A. chrysogenum were not as efficient. These results may reflect the requirement of species-specific transcriptional factors that are required to activate expression of the penicillin genes (2) or the B2 esterase gene, respectively (14). The homologous Aspergillus promoters appear to be the best choice, since adequate transcriptional factors do occur in the host A. awamori strain.
It is interesting to note that strain TGDTh-4 containing the expression cassette with the gdhA promoter gives a higher secretion at 48 h with ammonium sulfate as the nitrogen source. This A. awamori promoter, which belongs to the NADP-dependent glutamate dehydrogenase gene, is expressed early during the growth phase of this fungus (5).
There is a clear decay in gene expression and thaumatin secretion after 72 h in many of the clones, except in the double-transformant strain TB2b1-44-GD5, in which constructs with two different promoters were used. Expression of the gdhA promoter is growth phase associated, whereas the esterase B2 promoter is largely expressed when the growth phase has been completed (28). The combination of early and late promoters may provide a valuable tool with which to prolong gene expression during fermentation. However, late promoters (e.g., those of the secondary metabolism) must be recognized by the A. awamori transcriptional machinery.
In vivo cleavage is required to release thaumatin from the fused carrier protein. In fungi, this is achieved by the endogenous KEX proteolytic system that recognizes the lysine-arginine dipeptide (4, 6). Although the A. awamori KEX proteolytic system has not been characterized, it seems to be related to the S. cerevisiae KEX2 protease, a Ca2+-dependent membrane-bound serine endoprotease that is present in a late Golgi compartment (3, 13, 26). We used two tandemly repeated Lys-Arg pairs to facilitate recognition and cleavage. Three recombinant forms of secreted thaumatin were purified from the A. awamori culture broths. All of them were cleaved at the expected KRKR sequence and differed by one amino acid.
An important question is whether, under high-secretion conditions, the A. awamori secretion system is overloaded. A significant proportion of thaumatin remained intracellular in the high-production strains that was not observed in low-production strains. These results suggest that, under efficient expression conditions, protein secretion may be a bottleneck for thaumatin production.
No major unspecific proteolytic degradation was observed by immunoblotting of the secreted thaumatin. As shown in Fig. 3C, one satellite immunoreactive band of 14 kDa was observed in the high-thaumatin-secretion strains, suggesting that, in the PepA-defective lpr66 strain, other proteases (26) have only a limited effect on unspecific thaumatin degradation. The secreted thaumatin was sweet, indicating conservation of the conformational state of the recombinant thaumatin. In conclusion, A. awamori appears to provide an excellent system for thaumatin secretion.
ACKNOWLEDGMENTS
This work was supported by a Concerted Grant (95/075) of CDTI (Madrid) and Urquima, S.A. (Barcelona, Spain). F. J. Moralejo received a fellowship of the Diputación de León.
We thank I. Faus for continuous support; J. Velasco for help with the amino-terminal end sequencing; and B. Martín, J. Merino, and M. Corrales for technical assistance.
REFERENCES
- 1.Ausubel F M, Brent R, Kingston R E, Moore D, Smith J A, Seidman J G, Struhl K. Current protocols in molecular biology. New York, N.Y: Wiley; 1989. [Google Scholar]
- 2.Brakhage A A. Molecular regulation of β-lactam biosynthesis in filamentous fungi. Microbiol Mol Biol Rev. 1998;62:547–585. doi: 10.1128/mmbr.62.3.547-585.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brenner C, Fuller R S. Structural and enzymatic characterization of a purified prohormone-processing enzyme: secreted, soluble Kex2 protease. Proc Natl Acad Sci USA. 1992;89:922–926. doi: 10.1073/pnas.89.3.922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Calmels T P G, Martín F, Durand H, Tiraby G. Proteolytic events in the processing of secreted proteins in fungi. J Biotechnol. 1991;17:51–66. doi: 10.1016/0168-1656(91)90026-r. [DOI] [PubMed] [Google Scholar]
- 5.Cardoza R E, Moralejo F J, Gutiérrez S, Casqueiro J, Fierro F, Martín J F. Characterization and nitrogen-source regulation at the transcriptional level of the gdhA gene of Aspergillus awamori encoding an NADP-dependent glutamate dehydrogenase. Curr Genet. 1998;34:50–59. doi: 10.1007/s002940050365. [DOI] [PubMed] [Google Scholar]
- 6.Contreras R, Carrez D, Kinghorn J R, van den Hondel C A M J J, Fiers W. Efficient Kex2-like processing of a glucoamylase-interleukin-6 fusion protein by Aspergillus nidulans and secretion of mature interleukin-6. Bio/Technology. 1991;9:378–381. doi: 10.1038/nbt0491-378. [DOI] [PubMed] [Google Scholar]
- 7.Dunbar B S, Schwoebel E D. Preparation of polyclonal antibodies. Methods Enzymol. 1990;182:663–670. doi: 10.1016/0076-6879(90)82051-3. [DOI] [PubMed] [Google Scholar]
- 8.Edens L, Born I, Ledeboer A M, Maat J, Toonen M Y, Visser C, Verrips C T. Synthesis and processing of the plant protein thaumatin in yeast. Cell. 1984;37:629–633. doi: 10.1016/0092-8674(84)90394-5. [DOI] [PubMed] [Google Scholar]
- 9.Faus I, del Moral C, Adroer N, del Río J L, Patińo C, Sisniega H, Casas C, Bladé J, Rubio V. Secretion of the sweet-tasting protein thaumatin by recombinant strains of Aspergillus niger var. awamori. Appl Microbiol Biotechnol. 1998;49:393–398. doi: 10.1007/s002530051188. [DOI] [PubMed] [Google Scholar]
- 10.Faus I, Patińo C, del Río J L, del Moral C, Sisniega H, Rubio V. Expression of a synthetic gene encoding the sweet-tasting protein thaumatin in Escherichia coli. Biochem Biophys Res Commun. 1996;229:121–127. doi: 10.1006/bbrc.1996.1767. [DOI] [PubMed] [Google Scholar]
- 11.Fernández F J, Gutierrez S, Velasco J, Montenegro E, Marcos A T, Martín J F. Molecular characterization of three loss-of-function mutations in the isopenicillin N-acyltransferase gene (penDE) of Penicillium chrysogenum. J Bacteriol. 1994;176:4941–4948. doi: 10.1128/jb.176.16.4941-4948.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Finkelstein D B, Rambosek J, Crawford M S, Soliday C L, McAda P C, Leach J. Protein secretion in Aspergillus niger. In: Hershberger C L, Queener S W, Hegeman G, editors. Genetics and molecular biology of industrial microorganisms. Washington, D.C: American Society for Microbiology; 1989. pp. 295–300. [Google Scholar]
- 13.Fuller R S, Sterne R E, Thorner J. Enzymes required for yeast prohormone processing. Annu Rev Physiol. 1988;50:345–362. doi: 10.1146/annurev.ph.50.030188.002021. [DOI] [PubMed] [Google Scholar]
- 14.Gutiérrez S, Velasco J, Marcos A T, Fernández F J, Fierro F, Barredo J L, Díez B, Martín J F. Expression of the cefG gene is limiting for cephalosporin biosynthesis in Acremonium chrysogenum. Appl Microbiol Biotechnol. 1997;48:606–614. doi: 10.1007/s002530051103. [DOI] [PubMed] [Google Scholar]
- 15.Gwynne D I, Devchand M. Expression of foreign proteins in the genus Aspergillus. In: Bennet J W, Klich M A, editors. Aspergillus, the biology and industrial applications. London, United Kingdom: Butterworth; 1992. pp. 203–214. [PubMed] [Google Scholar]
- 16.Hahm Y T, Batt C A. Expression and secretion of thaumatin from Aspergillus oryzae. Agric Biol Chem. 1990;54:2513–2520. [Google Scholar]
- 17.Hammond C, Helenius A. Quality control in the secretory pathway. Curr Opin Cell Biol. 1995;7:523–529. doi: 10.1016/0955-0674(95)80009-3. [DOI] [PubMed] [Google Scholar]
- 18.Hijarrubia M J, Casqueiro J, Gutiérrez S, Fernández F J, Martin J F. Characterization of the bip gene of Aspergillus awamori encoding a protein with an HDEL retention signal homologous to the mammalian BiP involved in polypeptide secretion. Curr Genet. 1997;32:139–146. doi: 10.1007/s002940050258. [DOI] [PubMed] [Google Scholar]
- 19.Illingworth C, Larson G, Hellekant G. Secretion of the sweet-tasting plant protein thaumatin by Bacillus subtilis. Biotechnol Lett. 1988;10:587–592. [Google Scholar]
- 20.Illingworth C, Larson G, Hellekant G. Secretion of the sweet-tasting plant protein thaumatin by Streptomyces lividans. J Ind Microbiol. 1989;4:37–42. [Google Scholar]
- 21.Moralejo, F. J., R. E. Cardoza, S. Gutiérrez, and J. F. Martin. Unpublished data.
- 22.Punt P J, Dingemanse M A, Kuyvenhoven J, Soede R D M, Pouwels P H, von den Hondel C A M J J. Functional elements in the promoter region of the Aspergillus nidulans gpdA gene, encoding glyceraldehyde-3-phosphate dehydrogenase. Gene. 1990;93:101–109. doi: 10.1016/0378-1119(90)90142-e. [DOI] [PubMed] [Google Scholar]
- 23.Punt P J, Kramer C, Kuyvenhoven A, Pouwels P H, van den Hondel C A M J J. An upstream activating sequence from the Aspergillus nidulans gpdA gene. Gene. 1992;120:67–73. doi: 10.1016/0378-1119(92)90010-m. [DOI] [PubMed] [Google Scholar]
- 24.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 25.Shen Y-Q, Heim J, Solomon N A, Wolfe S, Demain A L. Repression of β-lactam production in Cephalosporium acremonium by nitrogen sources. J Antibiot. 1984;37:503–511. doi: 10.7164/antibiotics.37.503. [DOI] [PubMed] [Google Scholar]
- 26.Van den Homberg J P T W, van de Vondervoort P J I, Fraissinet-Tachet L, Visser J. Aspergillus as a host for heterologous protein production: the problem of proteases. Trends Biotechnol. 1997;15:256–263. doi: 10.1016/s0167-7799(97)01020-2. [DOI] [PubMed] [Google Scholar]
- 27.van den Hondel C A M J J, Punt P J, van Gorcom R F M. Heterologous gene expression in filamentous fungi. In: Bennett J W, Lasure L L, editors. More gene manipulation in fungi. San Diego, Calif: Academic Press; 1991. pp. 396–428. [Google Scholar]
- 28.Velasco, J., S. Gutiérrez, and J. F. Martín. Unpublished data.
- 29.Verdoes J C, Punt P J, van den Hondel C A M J J. Molecular genetic strain improvement for the over-production of fungal proteins by filamentous fungi. Appl Microbiol Biotechnol. 1995;43:195–205. [Google Scholar]
- 30.Visser J, Bussink H J, Witteveen D. Gene expression in filamentous fungi. In: Smith A, editor. Gene expression in recombinant microorganisms. New York, N.Y: Marcel Dekker Inc.; 1995. pp. 241–308. [PubMed] [Google Scholar]
- 31.Witty M, Higginbotham J D. Thaumatin. Boca Raton, Fla: CRC Press; 1994. [Google Scholar]
- 32.Yelton M M, Hamer J E, Timberlake W E. Transformation of Aspergillus nidulans by using a trpC plasmid. Proc Natl Acad Sci USA. 1984;81:1470–1474. doi: 10.1073/pnas.81.5.1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zemanek E, Wasserman B P. Issues and advances in the use of transgenic organisms for the production of thaumatin, the intensely sweet protein from Thaumatococcus daniellii. Crit Rev Food Sci Nutr. 1995;35:455–466. doi: 10.1080/10408399509527709. [DOI] [PubMed] [Google Scholar]