Abstract
An essential step in SARS-CoV-2 infection is binding the receptor-binding domain (RBD) of the SARS-CoV-2 spike protein to the ACE2 receptor on the surface of host cells. Therefore, variation in this region can have crucial effects on clinical outcomes and the emergence of variants of concern (VOCs) and variants of interest (VOIs). In this cross-sectional descriptive study, 54 patients with SARS-COV-2 infection were enrolled. After collecting samples and identifying the virus using the One-Step Real-Time qRT-PCR technique and confirming the viral infection, the region containing the RBD region for detection of any mutations was amplified using the Nested-PCR method. Finally, to identify probable mutations, the Nested-PCR product was sequenced.
Our data show that the most mutant strains in circulation in our population are the delta variant (90.74%), alpha variant (5.56%), and omicron variant (3.70%), respectively. Pangolin Lineages strains were B.1.1.7(Alpha variant), B.1.617.2(Delta variant) and B.1.1.529(Omicron variant). Also, the mutation profile of variants suggests that N501Y, T478K, and D614G amino acid substitutions, are the significant mutations in the alpha and delta variants that are common with the Omicron variant. The highest frequency of clinical signs in the patients were: lung involvement (42.59%); fever, chills (40.74%); body pain (15%), and other signs (1.67%). Our data revealed that SARS-COV-2 RBD region variation results in substituting essential amino acids and the emergence of the new variant. We can consider it as a predictor for monitoring the emergence of variants of concerns and viral outcomes.
Keywords: SARS-CoV2, Mutations, RBD, SPIKE, Variant
Abbreviations: SARS-COV-2, Severe Acute Respiratory Syndrome Coronavirus 2; RBD, Receptor-Binding-Domain; ACE2, Angiotensin-Converting Enzyme 2
1. Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is the coronavirus that causes coronavirus disease 2019 (COVID-19) and is known to be the agent of the ongoing COVID-19 pandemic. As of September 20, 2020, it has affected more than 21 million people in 221 countries, with approximately 228 million infected, and 4.5 million deaths have been confirmed by official statistics (WHO). This virus belongs to an emerging strain of the coronavirus family and the beta-coronavirus surbecogenus from the order Nidovirales. It is similar to the agent associated with the acute respiratory syndrome (SARS-COV-1). The first new epidemic, identified in 2003, and the second emergence of this viral family in 2009, is known as Middle East Respiratory Coronavirus Syndrome (MERS-COV), was reported [1,2]. The coronavirus encodes four major structural proteins, including spike(S), membrane(M), envelope(E), and nucleocapsid(N). SARS-COV-2 S protein is a large protein ranging from 1160 amino acids to 1400 amino acids [2,3]. This protein is a coronavirus glycoprotein that comprises subunits S1 and S2 and acts as a trimmer on the surface of the virion [4]. Spike protein plays a crucial role in ACE2 receptor binding to host cells (heart, lung, etc.) and virus entry into them. Thus, variations in the genomic encoding region of this protein will have clinical sequels on epidemiological prevalence, virus transmissibility, clinical manifestations, and virus mortality rate [5]. The RBD has a receiver (RBM) and forms the initial contact with the second ACE2 peptidase, which is a distinct furine (arginine-alanine-arginine-arginine) cleavage at amino acid position 682–685 of SARS-CoV-2, between the S1 and S2 domains [6]. A novel evolutionary path by which SARS-CoVS can interface with hACE2 allows for the efficient infection of human cells [7]. Thus, SARS-COV-2 may consider RBD mutations as an interspecific adaptation during transmission. Mutations in RBD increase the stability of the spike virus structure and decrease the binding of vaccine-induced antibodies [8,9]. The virus genome has 29,700 nucleotide sequences, and S protein contains 1273 amino acids with a molecular weight of 140(kDa). By August 2020, approximately 9,654 mutations had occurred in the spike region of the virus, with about 400 mutations and 33 mutations in the RBD region. They relate the highest frequencies to amino acids S477 N, V483A, A344S, and N501Y [10,11]. On September 30, 2020, during a joint study in the Netherlands and the United States by Real Butowt et al., the D614G mutation of SARS-CoV-2 virus spike protein was predominant in the COVID-19 epidemic, and the exact effect of the mutation on the disease phenotype was prominent [12]. Globally, combined genetic, structural, and epidemiological data indicate that the first significant mutation is substituting the amino acid aspartic acid with glycine D614G, leading to clinical complications such as olfactory and taste impairment and increased transmissibility [12]. Another mutation has been identified at the R407I position in RBD, also in India, where there was detected a different mutation at position 930 of the spike protein by substituting A (alanine) for V (valine) A930V [5]. Replacement with valine in the same position, with phylogenetic and functional alteration of the virus, might alter the molecule's dependence on its receptor [5]. Thus, co-mutations in protein S from Indian strains could potentially change fitness and virus entry [5]. Another significant mutation in the category of disturbing variants in the RBD position of amino acid 501 (N501Y) has been identified by substituting N (asparagine) for Y (tyrosine) [13]. There are currently disturbing variants of alpha B.1.1.7 from the United Kingdom, beta B.1.35 from South Africa, gamma P.1 from Japanese travelers who traveled to Brazil, and Delta B.1.617, which were recently identified from India. Only a few Mutations, N479L and T487S in protein S from RBD, are sufficient to increase the human ACE2 affinity field dramatically [14]. For the first time in Botswana and a traveler arriving in Hong Kong from South Africa, a new variant of concern as Omicron strain has been identified [14]. Researchers in South Africa are tracking down a new strain of the SARS-CoV-2 coronavirus; Report the new variant, along with many mutations found in other species, including the delta that it contains, and it appears to be spreading rapidly throughout South Africa. However, mutations in the viral genome that lead to new species are a real challenge in tackling this pandemic worldwide [14].
2. Materials and methods
2.1. Sampling and subjects
This descriptive cross-sectional study collected nasopharyngeal and oropharyngeal specimens from patients suspected of SARS-COV-2 infection from July 2021 to December 2021 (Table 2). The collected samples were then screened using the One-Step Real-Time qRT-PCR technique. After molecular analysis, 54 positive examples were selected from the patients. Before samples, informed consent forms and the medical ethics code approved by the Medical Ethics Committee of Lorestan University of Medical Sciences were obtained (ethics code IR.LUMS.REC.1400.160).
Table 2.
Demographic information and mutations profiles in Lorestan patients.
| Patient | C Collection date | Gender | Hospital | Outpatient | Vaccinated | Lineage | Distinct Nucleotides Mutations | Distinct Amino Acid Mutations | |
|---|---|---|---|---|---|---|---|---|---|
| 1 | July 2021 | Male | Yes | – | No | – | B.1.1.7 | c.23063 A > T | S: p. Asn501Tyr |
| c.23271C > A | S: p. Ala570Asp | ||||||||
| c.23403 A > G | S: p. Asp614Gly | ||||||||
| 2 | August 2021 | Female | – | Yes | Yes | Two dosages | B.1.617.2 | c.22917 T > G | S: p. Leu452Arg |
| c.22995C > A | S: p. Thr478Lys | ||||||||
| c.23403 A > G | S: p. Asp614Gly | ||||||||
| c.23032 T > C | S: p. Phe490Phe | ||||||||
| 3 | August 2021 | Male | – | Yes | No | – | B.1.617.2 | c.22917 T > G | S: p. Leu452Arg |
| c.22995C > A | S: p. Thr478Lys | ||||||||
| c.23403 A > G | S: p. Asp614Gly | ||||||||
| 4 | August 2021 | Female | – | Yes | No | – | B.1.617.2 | c.22995C > A | S: p. Thr478Lys |
| c.23340 G > C | S: p. Gly > Ala | ||||||||
| 5 | August 2021 | Female | Yes | – | No | – | B.1.617.2 | c.22917 T > G | S: p. Leu452Arg |
| c.22995C > A | S: p. Thr478Lys | ||||||||
| c.22839incC | Coding Region | ||||||||
| 6 | August 2021 | Male | – | Yes | No | – | B.1.617.2 | c.22917 T > G | S: p. Leu452Arg |
| c.22995C > A | S: p. Thr478Lys | ||||||||
| c.23403 A > G | S: p. Asp614Gly | ||||||||
| c.23491incT | Coding Region | ||||||||
| 7 | August 2021 | Male | – | Yes | No | – | B.1.617.2 | c.22917 T > G | S: p. Leu452Arg |
| c.22995C > A | S: p. Thr478Lys | ||||||||
| c.23403 A > G | S: p. Asp614Gly | ||||||||
| 8 | September 2021 | Female | – | Yes | No | – | B.1.617.2 | c.22917 T > G | S: S: p. Leu452Arg S S: p. Thr478Lys |
| c.22995C > A | |||||||||
| 9 | September 2021 | Male | – | Yes | No | – | B.1.617.2 | c.22995C > A | S: p. Thr478Lys |
| c.23403 A > G | S: p. Asp614Gly | ||||||||
| c.23420 G > A | S: p. Val620Ile | ||||||||
| c.23422C > A | S: p. Val620Val | ||||||||
| 10 | September 2021 | Female | – | Yes | No | – | B.1.617.2 | c.22917 T > G | S: p. Leu452Arg |
| c.22995C > A | S: p. Thr478Lys | ||||||||
| c.23403 A > G | S: p. Asp614Gly | ||||||||
| c.23401 G > T | S: p. Gln613His | ||||||||
| 11 | September 2021 | Male | – | Yes | Yes | One dosage | B.1.617.2 | c.22917 T > G | S: p. Leu452Arg |
| c.22995C > A | S: p. Thr478Lys | ||||||||
| c.23403 A > G | S: p. Asp614Gly | ||||||||
| 12 | September 2021 | Male | Yes | Yes | One dosage | B.1.617.2 | c.22917 T > G | S: p. Leu452Arg | |
| c.22995C > A | S: p. Thr478Lys | ||||||||
| 13 | December 2021 | Female | – | Yes | No | – | B.1.617.2 | c.23403 A > G | S: p. Asp614Gly Coding Region |
| c.23446incA | |||||||||
| c.23468incT | |||||||||
| c.23546incT | |||||||||
| c.23561incG | |||||||||
| c.23568incG | |||||||||
| 14 | September 2021 | Male | – | Yes | No | – | B.1.617.2 | c.22917 T > G | S: p. Leu452Arg |
| c.22995C > A | S: p. Thr478Lys | ||||||||
| c.23032 T > C | S: p. Phe490Phe | ||||||||
| 15 | September 2021 | Male | – | Yes | No | – | B.1.617.2 | c.22917 T > G | S: p. Leu452Arg |
| c.22995C > A | S: p. Thr478Lys | ||||||||
| c.22895 G > A | S: p. Val445Ile | ||||||||
| 16 | September 2021 | Female | – | Yes | No | – | B.1.617.2 | c.22917 T > G | S: p. Leu452Arg |
| c.22995C > A | S: p. Thr478Lys | ||||||||
| c.23403 A > G | S: p. Asp614Gly | ||||||||
| 17 | September 2021 | Male | – | Yes | Yes | Two dosages | B.1.617.2 | c.22995C > A | S: p. Thr478Lys |
| c.23403 A > G | S: p. Asp614Gly | ||||||||
| 18 | September 2021 | Male | – | Yes | No | – | B.1.617.2 | c.22917 T > G | S: p. Leu452Arg |
| c.22995C > A | S: p. Thr478Lys | ||||||||
| 19 | September 2021 | Male | – | Yes | No | – | B.1.617.2 | c.22995C > A | S: p. Thr478Lys |
| 20 | November 2021 | Male | – | Yes | No | – | B.1.617.2 | c.22995C > A | S: p. Thr478Lys |
| 21 | November 2021 | Male | – | Yes | No | – | B.1.617.2 | c.22917 T > G | S: p. Leu452Arg |
| c.22995C > A | S: p. Thr478Lys | ||||||||
| 22 | November 2021 | Female | – | Yes | Yes | One dosage | B.1.617.2 | c.22995C > A | S: p. Thr478Lys |
| 23 | November 2021 | Male | Yes | - - | No | --- -- | B.1.617.2 | c.22917 T > G | S: p. Leu452Arg |
| c.22995C > A | S: p. Thr478Lys | ||||||||
| c.23086C > T | S: p. Tyr508Tyr | ||||||||
| 24 | November 2021 | Male | Yes | Yes | One dosage | B.1.617.2 | c.22917 T > G | S: p. Leu452Arg S: p. Thr478Lys S: p. Asp614Gly |
|
| c.22995C > A | |||||||||
| c.23403 A > G | |||||||||
| 25 | September 2021 | Male | Yes | No | – | B.1.617.2 | c.22995C > A | S: p. Thr478Lys | |
| c.23403 A > G | S: p. Asp614Gly | ||||||||
| 26 | October 2021 | Male | Yes | No | – | B.1.617.2 | c.22917 T > G | S: p. Leu452Arg | |
| c.22995C > A | S: p. Thr478Lys | ||||||||
| 27 | October 2021 | Male | Yes | – | No | – | B.1.617.2 | c.22917 T > G | S: p. Leu452Arg |
| c.22995C > A | S: p. Thr478Lys | ||||||||
| c.23403 A > G | S: p. Asp614Gly | ||||||||
| 28 | September 2021 | Male | – | Yes | No | – | B.1.617.2 | c.22917 T > G | S: p. Leu452Arg |
| c.22995C > A | S: p. Thr478Lys | ||||||||
| c.23403 A > G | S: p. Asp614Gly | ||||||||
| 29 | December 2021 | Female | – | Yes | No | – | B.1.617.2 | c.22917 T > G | S: p. Leu452Arg |
| c.22995C > A | S: p. Thr478Lys | ||||||||
| 30 | December 2021 | Male | – | Yes | No | – | B.1.617.2 | c.22995C > A | S: p. Thr478Lys |
| c.23403 A > G | S: p. Asp614Gly | ||||||||
| 31 | November 2021 | Female | – | Yes | No | – | B.1.617.2 | c.22917 T > G | S: p. Leu452Arg |
| c.22995C > A | S: p. Thr478Lys | ||||||||
| 32 | December 2021 | Female | – | Yes | No | – | B.1.617.2 | c.22917 T > G | S: p. Leu452Arg |
| c.22995C > A | S: p. Thr478Lys | ||||||||
| 33 | October 2021 | Female | – | Yes | No | – | B.1.617.2 | c.22995C > A | S: p. Thr478Lys |
| 34 | July 2021 | Male | Yes | – – |
No | – | B.1.1.7 | c.23271C > A | S: p. Ala570Asp |
| c.23403 A > G | S: p. Asp614Gly | ||||||||
| c.23063 A > T | S: p. Asn501Tyr | ||||||||
| c.23477 G > A | S: p. Gly639Ser | ||||||||
| c.22912 T > A | S: p. Asn > Lys | ||||||||
| c.22834incA | Coding Region | ||||||||
| 35 | December 2021 | Female | – | Yes | No | – | B.1.617.2 | c.22917 T > G | S: p. Leu452Arg |
| c.22995C > A | S: p. Thr478Lys | ||||||||
| c.23403 A > G | S: p. Asp614Gly | ||||||||
| 36 | December 2021 | Female | – | Yes | No | – | B.1.617.2 | c.22917 T > G | S: p. Leu452Arg |
| c.22995C > A | S: p. Thr478Lys | ||||||||
| c.23403 A > G | S: p. Asp614Gly | ||||||||
| 37 | July 2021 | Male | Yes | – | No | – | B.1.1.7 | c.23063 A > T | S: p. Asn501Tyr |
| c.23271C > A | S: p. Ala570Asp | ||||||||
| c.23403 A > G | S: p. Asp614Gly | ||||||||
| c.23491incT | Coding Region | ||||||||
| c.23557incC | Coding Region | ||||||||
| 38 | December 2021 | Male | – | Yes | No | – | B.1.617.2 | c.22917 T > G | S: p. Leu452Arg |
| c.22995C > A | S: p. Thr478Lys | ||||||||
| c.23403 A > G | S: p. Asp614Gly | ||||||||
| c.23409incA | Coding Region | ||||||||
| c.23454incC | Coding Region | ||||||||
| 39 | August 2021 | Male | Yes | – | No | – | B.1.617.2 | c.22917 T > G | S: p. Leu452Arg |
| c.22995C > A | S: p. Thr478Lys | ||||||||
| c.23403 A > G | S: p. Asp614Gly | ||||||||
| c.23459incT | Coding Region | ||||||||
| 40 | November 2021 | Female | – | Yes | No | – | B.1.617.2 | c.22917 T > G | S: p. Leu452Arg |
| c.22995C > A | S: p. Thr478Lys | ||||||||
| c.23403 A > G | S: p. Asp614Gly | ||||||||
| 41 | October 2021 | Male | – | Yes | No | – | B.1.617.2 | c.22917 T > G | S: p. Leu452Arg |
| c.22995C > A | S: p. Thr478Lys | ||||||||
| c.23403 A > G | S: p. Asp614Gly | ||||||||
| c.23472incC | Coding Region | ||||||||
| c.23491incT | Coding Region | ||||||||
| c.23495incA | Coding Region | ||||||||
| 42 | December 2021 | Female | Yes | Yes | Two dosages | B.1.617.2 | c.22917 T > G | S: p. Leu452Arg | |
| c.22995C > A | S: p. Thr478Lys | ||||||||
| c.23403 A > G | S: p. Asp614Gly | ||||||||
| 43 | September 2021 | Male | – | Yes | No | – | B.1.617.2 | c.22995C > A | S: p. Thr478Lys |
| c.23403 A > G | S: p. Asp614Gly | ||||||||
| 44 | December 2021 | Female | – | Yes | No | – | B.1.617.2 | c.22917 T > G | S: p. Leu452Arg |
| c.22995C > A | S: p. Thr478Lys | ||||||||
| c.23403 A > G | S: p. Asp614Gly | ||||||||
| 45 | October 2021 | Female | – | Yes | No | – | B.1.617.2 | c.22917 T > G | S: p. Leu452Arg |
| c.22995C > A | S: p. Thr478Lys | ||||||||
| c.23403 A > G | S: p. Asp614Gly | ||||||||
| 46 | November 2021 | Female | Yes | Yes | Two dosages | B.1.617.2 | c.22917 T > G | S: p. Leu452Arg | |
| c.22995C > A | S: p. Thr478Lys | ||||||||
| c.23511incT | Coding Region | ||||||||
| c.23521incT | Coding Region | ||||||||
| c.23553incT | Coding Region | ||||||||
| 47 | December 2021 | Male | – | Yes | No | – | B.1.617.2 | c.22917 T > G | S: p. Leu452Arg |
| c.22995C > A | S: p. Thr478Lys | ||||||||
| c.23403 A > G | S: p. Asp614Gly | ||||||||
| c.23568incG | Coding Region | ||||||||
| 48 | December 2021 | Female | – | Yes | No | – | B.1.617.2 | c.22917 T > G | S: p. Leu452Arg |
| c.22995C > A | S: p. Thr478Lys | ||||||||
| c.23403 A > G | S: p. Asp614Gly | ||||||||
| 49 | December 2021 | Female | – | Yes | No | – | B.1.617.2 | c.22917 T > G | S: p. Leu452Arg |
| c.22995C > A | S: p. Thr478Lys | ||||||||
| c.23403 A > G | S: p. Asp614Gly | ||||||||
| c.23568incG | Coding Region | ||||||||
| 50 | December 2021 | Female | Yes | – | Yes | One dosage | B.1.617.2 | c.22917 T > G | S: p. Leu452Arg |
| c.22995C > A | S: p. Thr478Lys | ||||||||
| c.23403 A > G | S: p. Asp614Gly | ||||||||
| c.23568incG | Coding Region | ||||||||
| 51 | October 2021 | Male | – | Yes | Yes | One dosage | B.1.617.2 | c.22917 T > G | S: p. Leu452Arg |
| c.22995C > A | S: p. Thr478Lys | ||||||||
| c.23403 A > G | S: p. Asp614Gly | ||||||||
| 52 | December 2021 | Male | – | Yes | Yes | Two dosages | B.1.617.2 | c.22917 T > G | S: p. Leu452Arg |
| c.22995C > A | S: p. Thr478Lys | ||||||||
| c.23403 A > G | S: p. Asp614Gly | ||||||||
| 53 | December 2021 | Male | – | Yes | Yes | Two dosage | B.1.1.529 | c.22995C > A | S: p. Thr478Lys |
| c.23403 A > G | S: p. Asp614Gly | ||||||||
| c.23063 A > T | S: p. Asn501Tyr | ||||||||
| c.23202C > A | S: p. Thr547Lys | ||||||||
| 54 | December 2021 | Male | – | Yes | Yes | Two dosages | B.1.1.529 | c.22995C > A | S: p. Thr478Lys |
| c.23403 A > G | S: p. Asp614Gly | ||||||||
| c.23063 A > T | S: p. Asn501Tyr | ||||||||
| c.23048 G > A | S: p. Gly496Ser | ||||||||
2.2. RNA extraction, cDNA synthesis
ACCORDING TO THE INSTRUMENT, THE total RNA of collected samples of subjects was extracted by (QIAamp Viral RNA Kits). Extracted RNA was then kept at −80 c for further assays. Subsequently, cDNA synthesis of extracted RNA was performed using SuperScriptTM III Reverse Transcriptase first strand kit as described in the manufacturing steps.
2.3. Molecular assay and sequencing
The RBD domain sequences of SARS-CoV-2 spike protein were selected in the first step. It is published in the gene bank with the latest and most complete version of the virus genome with access number NC_045512.2. Furthermore, the S protein genome with access number NC_045512.2, which is in the full version of the viral genome with nucleotide sequence (21563 … 25384) in the NCBI database, was targeted. The first nucleotide locus of the primer was designed at position 22771, and the first nucleotide locus of the reverse primer was located at position 23576 of the SARS-COV-2 virus S1 protein. Using the designed external primer, an internal primer of 712 bp sequence was separated from the target of the S1 subunit-containing RBM, RBD, to the end of S1. Next, the primer sequences were designed using Gene Runner, Primer3, Oligo Analyzer, Reverse Complement software, and NCBI BLAST primer sequences to amplify the RBD gene in the desired region (Table 1 ). In the forward positions 22771 and reverse 23576 nucleotides primers of the RBD region in spike protein, we designed the SARS-COV-2 virus. An internal primer (Table 1) was also designed with the 712 bp sequence at position 22864 of the first nucleotide from the primer and the same reverse primer position.
Table 1.
Pairs of primers used for SARS-CoV2 RBD (spike) gene NESTED-PCR amplification.
| SARS-COV-2-F1 | 5-AGGTGATGAAGTCAGACAAATCG-3 |
| SARS-COV-2-R | 3-5-CGCATATACCTGCACCAATGG |
| INNNER-COV-2-F2 | 5-AGCTTGGAATTCTAACAATCTTGAT-3 |
| SARS-COV-2-R | 5-CGCATATACCTGCACCAATGG-3 |
Using the Nested-PCR amplification method in the first step (PCR1), Master Mix 2x 12/5(μl), DEPCE water free from RNase 7.1(μl), forward and reverse primers of each 0/7 For each PCR reaction, 21(μl) was mixed with 4(μl) of cDNA for a final of 25(μl) volume. The negative control sample of water and the positive control sample included the SARS-COV-2 genome. Overall, the genome segment was isolated with 805bp length (product PCR) on the electrophoresis agarose gel at 1.5% in the first round of PCR. (Step 2) amplification Master Mix 2x 12.5(μl), DEPCE water free from RNase 8.3(μl), forward and reverse primers of each 0/7. For each PCR reaction, 21.5(μl was mixed with 3.5(μl) of cDNA for a final of 25(μl) volume. The negative control sample of water and the positive control sample included the SARS-COV-2 genome. Overall, the genome segment was isolated with 712bp length (product PCR) on the electrophoresis agarose gel at 1.5% in the first round of PCR. Set up program (PCR1), first stage Initial Denaturation, temperature 94 0C for 5 min with one cycle. The second stage of several parts of Denaturation, 94 0C temperature for 30Sec, annealing temperature 57 0Cfor 30Sec, extension temperature 72 0C for 1Min, all three parts of the second stage had 35 cycles of repetition. The (PCR2) setup program is similar to all (PCR1) steps, except; with this difference, an annealing temperature of 56 0C was performed. Patient genome products were then sent for sequencing and accuracy. Finally, using SnapGene software, the total alignment of the RBD sequence of S protein and the final sequences of Nested-PCR products, the results of the observed mutations with a quality of 20 and higher were reported.
2.4. Phylogenetic analysis
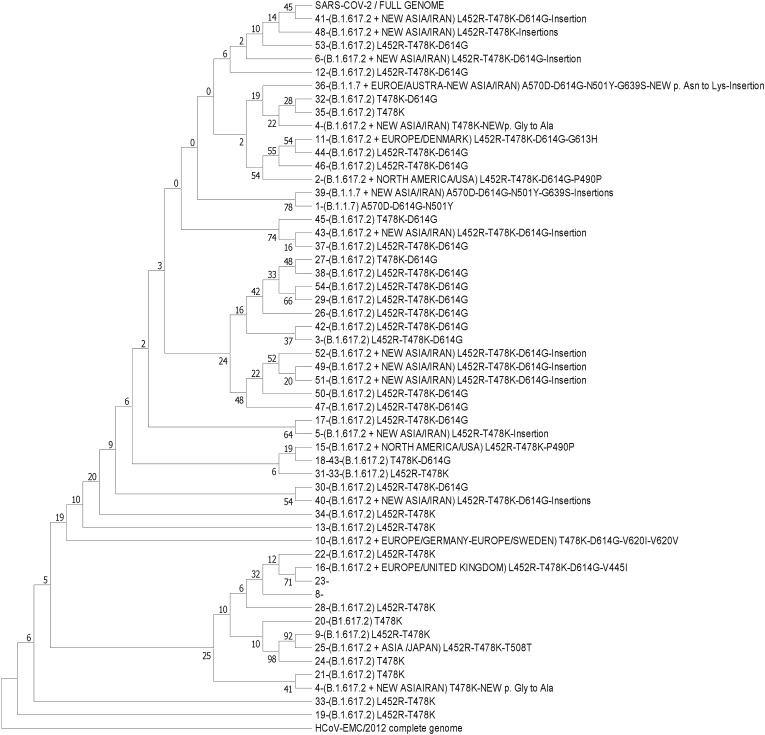
Phylogenetic and sequence analysis based on entire genomes and RBDs from the different isolates were included in this study. The phylogeny tree (Fig. 4) shows the clade discrepancies, and discontinuous lines show the equivalent taxon between each tree [15]. A total of 54 sequences resulted from the sequences of the current study. Sequences were clustered using Mega11 software. Sequence sequencing and alignment were performed by the ClustalW method with the Wuhan reference genome. Finally, evolutionary history was inferred using the Maximum Likelihood method (∼1000 bootstrap) and Tamura-Nei model.
Fig. 4.
Phylogenetic tree of variants.
2.5. Statistical and data analysis
The non-parametric ×2 test of significance was used to determine the differences in variables, such as the effect of gender, age, vaccination, and mutations on mortality in our population. The variants were called, and consensus sequences of all genomes were generated as per Centers for Diseases Control and Prevention (CDC, USA) guidelines [16]. The assembled genomes were classified into PANGO lineages using the Pangolin v3.1.5 and pangoLEARN model dated 12.20.2021 [17].
3. Results
3.1. Demographics summary
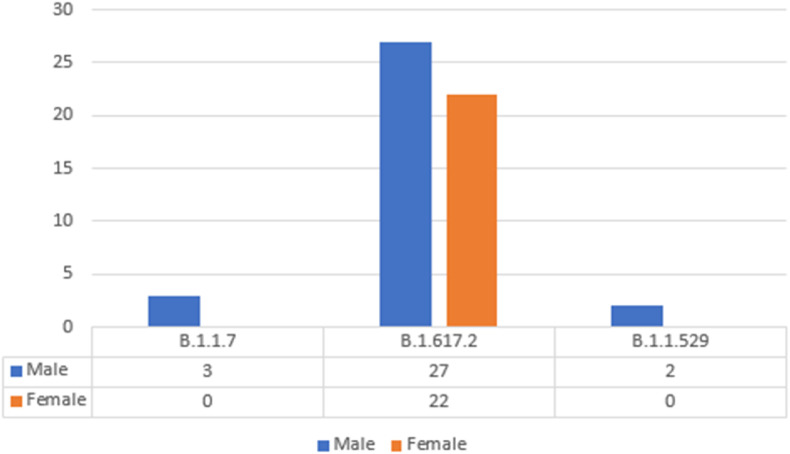
According to the patient's metadata report (Table 2 ), 54 nasopharyngeal and oropharyngeal samples were taken between July 2021 and December 2021. The highest prevalence of the disease (Fig. 1 ) was in men (59.26%) compared to women (40.74%), with a mean age of 41 years for men and 34 years for women. The samples covered five regions out of 11 districts in all parts of Lorestan province. A summary of patient demographics is shown in Table 2; the minimum age of patients was one year, and the maximum period was 84 years.
Fig. 1.
Differentiation of Lineage Pangolin with the gender of patients.
3.1.1. Phylogenetic classification and diversity
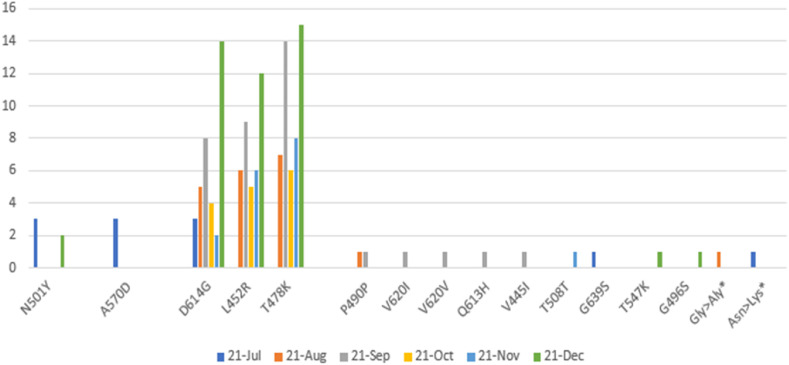
This study examined the phylodynamic distribution (Table 2), relative distribution, Pangolin lineages, or prevalence of SARS-COV-2, and the virus mutation distributed in our population from July 4, 2021, to December 31, 2021. Sequences confirmed the introduction of both VOC species by overcoming delta species over alpha. In several significant samples, in addition to the delta variant, since the beginning of January, several mutations of the omicron variant omicron N501Y, T478K, G496S, T547k have been added to this type in patients in this area. Two patients, examining more mutations in these two patients, the N501Y mutation (asparagine to tyrosine) present in the Omicron variant was also detected earlier in the Alpha variant. The D614G mutation (substitution of aspartic acid to glycine) located in the S1 subunit of the Omicron variant is shared with Alpha and Delta variants. Among all the mutations, the crucial mutations in the RBD of the Omicron variant are T478K and N501Y. We found two types of mutations in the RBD: G496S region, S1: T547K region associated with BA.1 species from Lineage B.1.1.529 with several standards and virulent mutations B.1.617. 2 is included. Mutations in these two diseases have shown evolution and a new variant species that will soon dominate the previous species. This finding indicates that the transfer of society still exists in the country. The prevalence of VOCs with Pangolin lineages (Fig. 3 ) B.1.1.7/Alpha (5.56%), B.1.617.2/Delta (90.74%), B.1.1.529/Omicron (3.70%) and switching at four-month intervals was the main prominent feature. However, by the end of December, only B.1.617.2/Delta was found in most sequences, indicating a significant VOC for monitoring in this region for the coming months.
Fig. 3.
The trend of major mutations in RBD (spike) AA from July 2021 to December 2021.
3.2. Amino acid substitutions and relation to variants
Overly, the 144 amino acid (AA) substitutions observed in 54 SARS-COV-2 sequences in our region (Fig. 3) were analyzed in this study. While most mutations have accumulated in the respective lines, some have recurred and have been stable. Our study identified all three VOCs; B.1.1.7/Alpha (D614G, A570D, N501Y), B.1.617.2/Delta (D614G, L452R, T478K) and B.1.1.529/Omicron (D614G, T478K, G496S, T547K) with two new substitutions S: p. Asn > Lys with nucleotide position c.22912T > A in RBD region and S: p. Gly > Ala with nucleotide position c.23340G > C after RBD in subunit S1 with several insertions, in S Protein, observed and reported. Several rare mutations reported from other regions (P490P, V620I, V620V, Q613H, V445I, Y508Y, G639S) have been observed and reported as single, double, or triple mutations in the relevant strains.
This figure shows the comparison between the sexes of patients with pangolin lineages. We examined three types of delta strains (B.1.617.2), alpha (B.1.1.7), Omicron (B.1.1.529) distributed from Lorestan patients in Iran from July 4, 2021, to December 31, 2021. Our data showed the most mutations in the delta strain, males (50%) and females (40.74%), and in the alpha strain, males (100%).
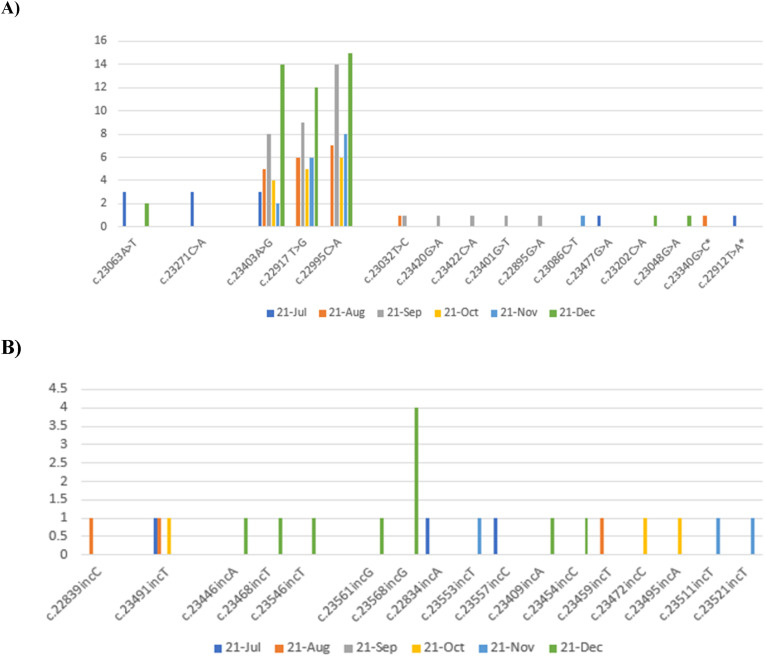
In [Fig. 2], our data are shown as Transition and Transversion. Fig. 2 A (Transition) describes the most common mutations in our region with their nucleotide position for six months, from July 4, 2021, to December 20, 2021. The most frequent RBD nucleotide mutations to the end of subunit S1 of SARS-COV-2 spike protein (respectively): c.22995C > A, c.22917T > G, c.23403A > G related to delta strain during Sep-2021, Dec-2021 and Dec-2021 reached their peak. In late December 2021, the omicron strain appeared. Among our data, we observed and reported two new nucleotide mutation sites, c.22912T > A and c.23340G > C. In Fig. 2B(Transversion), the most frequent mutations were included: c.23568incG and c.23491incT, during Dec-2021 and Aug-2021, respectively.
Fig. 2.
The trend of major mutations (Transition & Transversion) in the RBD region from July 2021 to December 2021.
As shown in Fig. 3, the most frequent amino acid mutations in the RBD region from spike protein to the end of the S1 subunit in 54 patients in Lorestan province for six months is shown. In Jul-2021, the most frequent mutations were A570D and N501Y AA substitution from the alpha lineage. By mid-December 2021, AA was the biggest substitution for the Delta T478K, L452R, and D614G lineages. In late December 2021, the Omicron strain also appeared.
The evolutionary history was inferred using the Maximum Likelihood method (∼1000 bootstrap) and Tamura-Nei model [18]. The tree with the highest log likelihood (−87760.55) is shown [Fig. 4]. The percentage of trees in which the associated taxa clustered together is displayed next to the branches. Initial tree(s) for the heuristic search were obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using the Tamura-Nei model and then selecting the topology with a superior log-likelihood value. This analysis involved 55 nucleotide sequences. There was a total of 30176 positions in the final dataset. Evolutionary studies were conducted in MEGA11 [19].
The evolutionary history was inferred using the Neighbor-Joining method [20]. The optimal tree is shown [Fig. 5 ]. The evolutionary distances were computed using the Maximum Composite Likelihood method [21] and are in the units of the number of base substitutions per site. This analysis involved 55 nucleotide sequences. Codon positions included were 1st+2nd+3rd + Noncoding. All ambiguous positions were removed for each sequence pair (pairwise deletion option). There was a total of 30176 positions in the final dataset. Evolutionary analyses were conducted in MEGA11 [19].
Fig. 5.
Phylogenetic tree with the lineage of patients in our population.
4. Discussion
RBD is responsible for the interaction with the cellular receptor (ACE2) and is considered a key target to induce immune responses and anti-viral therapy via blocking the receptor binding [5]. Reports have revealed that a few mutations (N479L and T487S) in the spike protein within RBD are sufficient to dramatically increase the human ACE2 affinity [4,22,23]. Thus, mutations in spike protein and RBD can result in the emergence of variants that lead to altered cell tropism or virulence [5]. Eventually, mutations helped the virus evolve into a dominant variant in circulation among host populations. Out of 54 patients studied, the most common clinical symptoms were: pulmonary involvement, 42.59%, fever and chills, 40.74%, body pain 15%, and other symptoms such as sweating, cough, and diarrhea 1.67%. Patients with two mutations in L452R, and T478K, had clinical signs such as 84% pulmonary involvement and 80.95% fever and chills. The most many mutations were in the alpha and delta viruses. Most of the lineage was related to B.1.1.617 delta virus type.
According to the phylogenetic tree sequence map, lineages are divided into evolved and immature groups. Most of the sequence mutations in our patients were initially detected by the alpha variant with the emergence of a new nucleotide locus-specific to Iran, which overcame alpha with Lineage B.1.617.2 strain delta. We observed men with a minimum of 5–10% pulmonary involvement on the average at 41 years who had not received any vaccine. Among the patients (Table 2 and Fig. 2A and B), delta strains showed a complete stream with a new nucleotide locus from Iran and the Lineage Pangolin strain called B.1.617.2. Out of 54 patients studied (Table 2), 21 patients with evolved variants B.1.617.2, B.1.1.7 and B.1.1.529 Lineages and 33 patients according to Lineage B.1.617.2. We observed a new nucleotide mutation (Fig. 2A) at position 22912 in RBD of SARS-COV-2 virus with substitution of thymine to adenine c.22912T > A with a significant level (P < 0.001) and insertion in B.1.1.7 lineage, respectively. Also, we observed and reported the location of another 23340 new mutations (Fig. 2A) in the SARS-COV-2 spike protein replacing guanine with cytosine c.23340G > C in Lineage B.1.617.2. We also observed and reported cases of a new Iranian mutation that has not yet been registered. Also, in patients 34 and 37, we developed an evolved form of B.1.1.7 Lineage, one case with a global and known 614G position mutation with several new Iranian insertions, and another case of an undiagnosed new mutation, a rare strain from Europe-Australia. The remaining nine patients observed an evolved form of Lineage B.1.617.2 with insertions of several similar and new cases. Patients 2–14 similar to each other and 9,10, 15, and 23 evolved forms of Lineage B.1.617.2 were associated with the rare strain of North America-USA, Europe-Germany, and Europe-Sweden and Europe-Denmark and the Europe-United Kingdom and Asia-Japan, respectively. In a similar study, Massab Umair et al. identified the dominant variants in Pakistan. From these positive spike samples, 22 samples were processed for whole-genome sequencing. Among VOCs, 45.5% (n = 10) belonged to B.1.351 followed by B.1.617.2, 36% (n = 8). The delta variant cases were reported mostly from Islamabad (n = 5; 63%), followed by Peshawar and Azad Kashmir (n = 1; 13% each). Beta variant cases originated from Islamabad (n = 5; 56%), Peshawar (n = 2; 22%), Azad Kashmir, and Rawalpindi (n = 1; 11% each). The mutation profile of delta variant isolates reported significant mutations, L452R, T478K, P681R, and D950 N [24]. In another similar study, Sarah Cherian et al. examined mutations and variants prevalent in India [25]. Phylogenetic analysis revealed that newly identified lineages B.1.617.1 and B.1.617.2 were predominantly circulating [25]. The signature mutations possessed by these strains were L452R, T478K, E484Q, D614G, and P681R in the spike protein, including within the receptor-binding domain (RBD) [25]. Xiaoying Shen et al. showed in a study that B.1.1.7 is still sensitive to neutralization, albeit at a moderate level (∼SIM; 2 times), by serum samples from people with mRNA vaccine (mRNA-1273), (Moderna) and reduced protein nanoparticle vaccines (NVX -Cov2373, Novavax) [26]. Globally, L452R has emerged independently since November/December 2020, playing an important role in virus escape or stability [27]. Chris Davis et al. Believe that vaccines play an important role in preventing hospitalization and mortality from SARS-COV-2 infection. Still, new mutated antigen profiles pose a serious threat to the emergence of new strains of the virus. Which reduces their effectiveness [28]. Also, it has recently been shown that; L484Q and L452R in the RBD region of the SARS-COV-2 delta variant cause an increase in the viral pathogenesis disease and high re-infections [11]. Fattahi, Zohreh, et al. genomic sequencing and phylogenetic analysis suggested that SARS‐CoV‐2 entered in late 2019/early 2020 in Iran and circulated among vulnerable patients. The increase in the frequency of D614G mutations and B.1* lineages from mid‐May onwards predicted a rapid viral transmission followed by a considerable change in the composition of viral lineages circulating in the country [29]. According to the results of our phylogenetic tree (Fig. 4), 62.50% of 100% of patients cared for Thr478Lys amino acid mutation. All patients had Leu452Arg mutation and Thr478Lys mutation simultaneously. In this study (Table 2), 68.29% of patients had not received any vaccine and had at least 30% lung involvement, and 31.71% of patients who received the vaccine did not have 30% pulmonary involvement. We predict that the delta strain may be one of the agents of death and hospitalization in Iran and our region from September to October. In our January study, we identified several suspected cases of mutations in the Omicron variant in patients in this area. New mutation cases can be considered VOI and will be a global warning to convert VOI to VOC in the future. Given that the majority of patients had not received any vaccine, the development of the delta may lead to increased transmission and an increase in the number of re-infections and even soon, with the outbreak of the new omicron VOC variant, Therefore, considering the new variant of Omicron, the importance of this study implies for a third dose of vaccination in the country due to the development of virus strains emerging variants, long-term effects of the disease in some patients. We predict a high probability of emerging diseases due to SARS-COV-2 variants in the future.
In conclusion, in the present study (Fig. 2), the detected mutations in L452R could cause the virulence of lung involvement and clinical symptoms. Also (Fig. 3), we have shown that significant amino acid substitution in the SARS-COV-2 RBD region leads to the emergence of new strains. Therefore, it can be considered a valid partial genome evaluation to identify a new species and predict clinical outcomes.
Ethics approval and consent to participate
All the procedures performed in the study were following the ethical standards of the local ethics committee of Lorestan University of Medical Sciences (IR.LUMS.REC.1400.160), as well as the 1964 Helsinki declaration. Written informed consent was obtained from all subjects.
CRediT authorship contribution statement
Faezeh Hajizadeh: Methodology, Investigation, Formal analysis. Sayyad Khanizadeh: Validation, Supervision, Project administration, Conceptualization. Hamidreza Khodadadi: Validation, Software, Formal analysis. Yaser Mokhayeri: Formal analysis, Data curation. Mehdi Ajorloo: Writing – original draft, Visualization. Asra Malekshahi: Writing – original draft, Software, Investigation. Ezatoallah Heydari: Methodology, Investigation, Formal analysis.
Declaration of competing interest
The authors declare that they have no any competing interests.
Acknowledgements
The authors thank the Hepatitis Research Center the Lorestan University of Medical Sciences, Khorramabad, Iran and Genetic Reference Laboratory, Khorramabad, Iran for the services to this research.
References
- 1.Wong N.A., Saier M.H., Jr. The SARS-coronavirus infection cycle: a survey of viral membrane proteins, their functional interactions and pathogenesis. Int. J. Mol. Sci. 2021;22(3):1308. doi: 10.3390/ijms22031308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mousavizadeh L., Ghasemi S. Genotype and phenotype of COVID-19: their roles in pathogenesis. J. Microbiol. Immunol. Infect. 2021;54(2):159–163. doi: 10.1016/j.jmii.2020.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dhama K., Khan S. Vol. 33. 2020. Coronavirus Disease 2019-COVID-19. (4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Wilde A.H., et al. Host factors in coronavirus replication. Curr. Top. Microbiol. Immunol. 2018;419:1–42. doi: 10.1007/82_2017_25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Saha P., et al. A virus that has gone viral: amino acid mutation in S protein of Indian isolate of Coronavirus COVID-19 might impact receptor binding, and thus, infectivity. Biosci. Rep. 2020;40(5) doi: 10.1042/BSR20201312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pillay T.S. Gene of the month: the 2019-nCoV/SARS-CoV-2 novel coronavirus spike protein. J. Clin. Pathol. 2020;73(7):366–369. doi: 10.1136/jclinpath-2020-206658. [DOI] [PubMed] [Google Scholar]
- 7.Sheahan T., et al. Mechanisms of zoonotic severe acute respiratory syndrome coronavirus host range expansion in human airway epithelium. J. Virol. 2008;82(5):2274–2285. doi: 10.1128/JVI.02041-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sheahan T., et al. Mechanisms of zoonotic severe acute respiratory syndrome coronavirus host range expansion in human airway epithelium. J. Virol. 2008;82(5):2274–2285. doi: 10.1128/JVI.02041-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li F., et al. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science. 2005;309(5742):1864–1868. doi: 10.1126/science.1116480. [DOI] [PubMed] [Google Scholar]
- 10.Guruprasad L. Human SARS CoV-2 spike protein mutations. Proteins. 2021;89(5):569–576. doi: 10.1002/prot.26042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mohammadi M., Shayestehpour M., Mirzaei H. The impact of spike mutated variants of SARS-CoV2 [Alpha, Beta, Gamma, Delta, and Lambda] on the efficacy of subunit recombinant vaccines. Braz. J. Infect. Dis. 2021;25(4):101606. doi: 10.1016/j.bjid.2021.101606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Butowt R., Bilinska K., von Bartheld C.S. Vol. 11. 2020. Chemosensory Dysfunction in COVID-19: Integration of Genetic and Epidemiological Data Points to D614G Spike Protein Variant as a Contributing Factor; pp. 3180–3184. (20) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jangra, S., et al., SARS-CoV-2 spike E484K mutation reduces antibody neutralisation. The Lancet Microbe. [DOI] [PMC free article] [PubMed]
- 14.Callaway E. Heavily mutated Omicron variant puts scientists on alert. Nature. 2021;600(7887):21. doi: 10.1038/d41586-021-03552-w. [DOI] [PubMed] [Google Scholar]
- 15.Huddleston J., et al. Augur: a bioinformatics toolkit for phylogenetic analyses of human pathogens. J. Open-Source Software. 2021;6(57):2906. doi: 10.21105/joss.02906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Paden C., et al. Rapid, sensitive, full-genome sequencing of severe acute respiratory syndrome coronavirus 2. Emerging Infectious Dis. J. 2020;26(10):2401. doi: 10.3201/eid2610.201800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.O'Toole Á., Scher E. Vol. 7. 2021. Assignment of Epidemiological Lineages in an Emerging Pandemic using the Pangolin Tool. (2) veab064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tamura K., Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 1993;10(3):512–526. doi: 10.1093/oxfordjournals.molbev.a040023. [DOI] [PubMed] [Google Scholar]
- 19.Tamura K., Stecher G., Kumar S. MEGA11: molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021;38(7):3022–3027. doi: 10.1093/molbev/msab120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saitou N., Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4(4):406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 21.Tamura K., Nei M., Kumar S. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc. Natl. Acad. Sci. U. S. A. 2004;101(30):11030–11035. doi: 10.1073/pnas.0404206101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Islam M.R., et al. Genome-wide analysis of SARS-CoV-2 virus strains circulating worldwide implicates heterogeneity. Sci. Rep. 2020;10(1):14004. doi: 10.1038/s41598-020-70812-6. 14004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Salehi N., Amiri-Yekta A., Totonchi M. Profiling of initial available SARS-CoV-2 sequences from Iranian related COVID-19 patients. Cell J. 2020;22(Suppl 1):148–150. doi: 10.22074/cellj.2020.7524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Umair M., et al. medRxiv; 2021. Detection and Whole-Genome Sequencing of SARS-CoV-2 B.1.617.2 and B.1.351 Variants of Concern from Pakistan during the COVID-19 Third Wave; p. 2021. 07.14.21259909. [Google Scholar]
- 25.Cherian S., et al. SARS-CoV-2 spike mutations, L452R, T478K, E484Q and P681R, in the second wave of COVID-19 in Maharashtra, India. Microorganisms. 2021;9(7) doi: 10.3390/microorganisms9071542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shen X., et al. SARS-CoV-2 variant B.1.1.7 is susceptible to neutralizing antibodies elicited by ancestral spike vaccines. Cell Host Microbe. 2021;29(4):529–539. doi: 10.1016/j.chom.2021.03.002. e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tchesnokova V., et al. Vol. 59. 2021. Acquisition of the L452R Mutation in the ACE2-Binding Interface of Spike Protein Triggers Recent Massive Expansion of SARS-CoV-2 Variants; p. e0092121. (11) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Davis C., et al. Reduced neutralisation of the Delta (B.1.617.2) SARS-CoV-2 variant of concern following vaccination. PLoS Pathog. 2021;17(12):e1010022. doi: 10.1371/journal.ppat.1010022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fattahi Z., et al. SARS-CoV-2 outbreak in Iran: the dynamics of the epidemic and evidence on two independent introductions. Transboundary Emerging Dis. 2022;69(3):1375–1386. doi: 10.1111/tbed.14104. [DOI] [PMC free article] [PubMed] [Google Scholar]