Abstract
Inflammatory response following SARS-CoV-2 infection results in substantial increase of amounts of intravascular pro-coagulant extracellular vesicles (EVs) expressing tissue factor (CD142) on their surface. CD142-EV turned out to be useful as diagnostic biomarker in COVID-19 patients. Here we aimed at studying the prognostic capacity of CD142-EV in SARS-CoV-2 infection.
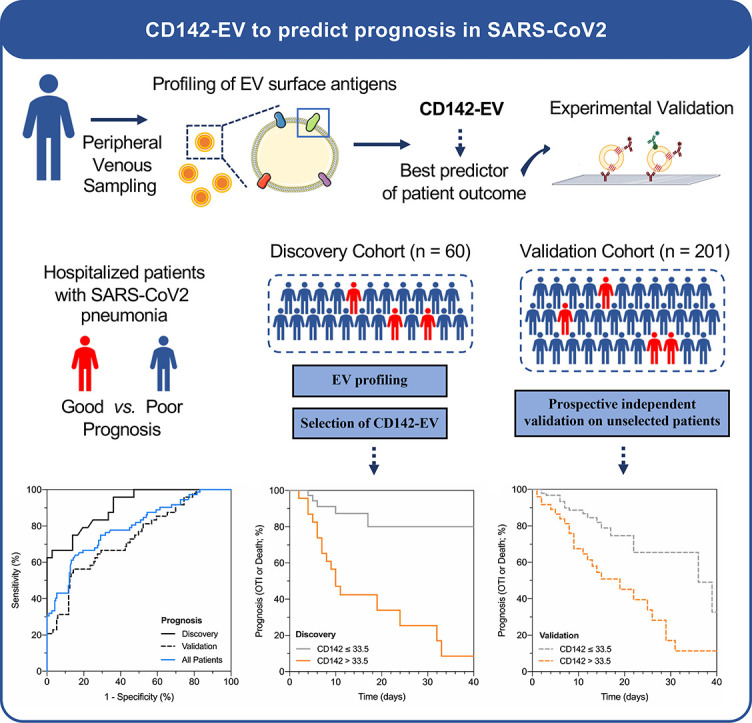
Expression of CD142-EV was evaluated in 261 subjects admitted to hospital for pneumonia and with a positive molecular test for SARS-CoV-2. The study population consisted of a discovery cohort of selected patients (n = 60) and an independent validation cohort including unselected consecutive enrolled patients (n = 201). CD142-EV levels were correlated with post-hospitalization course of the disease and compared to the clinically available 4C Mortality Score as referral.
CD142-EV showed a reliable performance to predict patient prognosis in the discovery cohort (AUC = 0.906) with an accuracy of 81.7%, that was confirmed in the validation cohort (AUC = 0.736). Kaplan-Meier curves highlighted a high discrimination power in unselected subjects with CD142-EV being able to stratify the majority of patients according to their prognosis. We obtained a comparable accuracy, being not inferior in terms of prediction of patients' prognosis and risk of mortality, with 4C Mortality Score. The expression of surface vesicular CD142 and its reliability as prognostic marker was technically validated using different immunocapture strategies and assays.
The detection of CD142 on EV surface gains considerable interest as risk stratification tool to support clinical decision making in COVID-19.
Keywords: CD142, Tissue factor, SARS-CoV2, COVID-19, Extracellular vesicles
Abbreviations: EVs, Extracellular Vesicles; IC, Immuno-Capture; OTI, OroTracheal Intubation; MFI, Median Fluorescence Intensity; nMFI, normalized MFI; SARS-CoV-2, Severe Acute Respiratory Syndrome CoronaVirus 2; TF, Tissue Factor; EV-TF or CD142-EV, Tissue Factor-positive EVs; UC, Ultra Centrifugation; WB, Western Blot
Graphical abstract
1. Introduction
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has infected more than 455 million subjects as of 12 March 2022 (https://coronavirus.jhu.edu). The resulting disease (COVID-19) is associated with high hospitalization rates and an increased risk of respiratory failure, thus determining tremendous burden on the healthcare system of several countries and affecting the best possible care for patients [1,2]. A pragmatic risk score that uses analytic assay to estimate poor outcome from infection may assist medical staff in tailoring management strategies for patients and allocating limited healthcare resources. [3] Several prognostic models have been approached in the past two years to meet the urgent need of an efficient and early prognosis in patients with a confirmed diagnosis of COVID-19 for mortality risk, progression to severe disease and intensive care unit admission. [3] The most frequently used prognostic factors including age, image features, lymphocyte count and C-reactive protein, showed moderate performance in terms of clinical decision making. [3,4] A clinically applicable prediction model with very good discrimination and performance characteristic has recently been validated in large cohort of patients. [5] The 4C Mortality Score including eight variables at hospital admission, outperformed other risk stratification tools and showed clinical decision making utility. [5]
Over the years, several studies have described the potential value of circulating extracellular vesicles (EVs) as prognostic biomarkers. [[6], [7], [8], [9], [10]] Molecular profiles of circulating EVs turned out to be useful as early prediction tool of COVID-19 severity. [11] Very recently, the total number of tissue factor-positive EVs (EV-TF) as well as their enzymatic activity were significantly associated with an increased severity risk in COVID-19 patients. [[12], [13], [14]]
In line with these studies we have lately showed that the expression of TF onto surface of EVs isolated from COVID-19 patients serum was significantly higher than EVs isolated from healthy subjects as well as from those isolated from serum of subjects with pneumonia but different etiology from SARS-CoV-2. [15] Furthermore, the levels of expression of TF-bearing EV (CD142-EV) was significantly correlated with the capacity of EVs to generate factor Xa. [15] Contextually, we produced very preliminary evidence showing that TF was significantly more expressed in severe COVID-19 patients undergoing orotracheal intubation (OTI) and/or death. However, we could not assess the performance of such EV marker as prognostic indicator due to the limited number of included patients. This paved the way for exploring the potential of this specific surface antigen as indicator of disease prognosis in a larger cohort of patients. The scientific endeavor of the present paper relies on the inclusion of 261 laboratory-confirmed COVID-19 patients hospitalized for pneumonia, who underwent blood sampling at time of molecular swab test and were longitudinally monitored to assess the clinical progression of disease. The expression of EV-associated TF was then retrospectively correlated with the course of the disease and its clinical performance was evaluated with the incidence of OTI and/or death as indicator of poor prognosis. The 4C Mortality Score was used as gold standard referral trying to put our experimental tool into scale with a widely validated in-use model. [5] We took advantage from reproducible flow cytometer assay that has been previously standardized and validated for the detection and characterization of EV surface signatures. [[16], [17], [18], [19]]
2. Methods
Supporting data for the present study are available within the article and the supplementary material. Because of their sensitive nature, additional information and single patient data are available from the corresponding author upon reasonable request.
2.1. Patient recruitment
The study was approved by the local ethical. Subjects gave informed consent according to the Declaration of Helsinki. The study population consists of 261 Caucasian white subjects hospitalized for pneumonia and SARS-CoV-2 infection at Internal Medicine Department and Cardiocentro Ticino Institute, Ente Ospedaliero Cantonale, Lugano, Switzerland. All patients were positive for SARS-CoV-2 as for molecular tests (polymerase chain reaction). Serum samples were collected at the time of nasopharyngeal swab sampling. The study population consisted in a discovery cohort (n = 60) composed by selected patients admitted to hospital in March 2020, and in a validation cohort (n = 201) composed by unselected consecutive patients admitted to hospital between April 2020 and May 2020. Patients were included in the study if they met the following criteria: infection by SARS-CoV-2, diagnosis of pneumonia and admission to hospital. Exclusion criteria were: (1) Age lower than 18 years; (2) Pregnancy; (3) Concomitant acute non-respiratory infection; (4) Cancer (active or recent history); (5) Inappropriateness to invasive emergency treatment (i.e., orotracheal intubation, advanced life support). Patients were classified in terms of outcome in good vs. poor prognosis, the latter was defined as need of orotracheal intubation, OTI, and/or death.
2.2. Sample handling
The present study is based on the “re-use” of serum samples from a biological bank (EOLAB - Ente Ospedaliero Cantonale; see Supplementary Methods). Peripheral venous blood samples were collected in serum separator tubes and maintained 30 min at room temperature before centrifugation. After clot formation, blood underwent serial low speed centrifugations at 4 °C (1′600 x g for 10 min; 3′000 x g for 20 min; 10′000 x g for 15 min) to separate serum and to remove cellular debris and larger vesicles. Cleared serum was then aliquoted, stored at −80 °C and never thawed prior to analysis.
2.3. EVs characterization
2.3.1. Bead-based EV surface profiling
Serum samples underwent bead-based EV-capture and flow cytometric analysis by MACSPlex human Exosome Kit (Miltenyi) without further pre-isolation step, as previously described [15,20]. EVs were isolated using capture-beads coated with antibodies coated with 37 different surface antigens and then analyzed after incubation with a detection reagent (labelled antibodies against CD9-CD63-CD81). Median fluorescence intensity (MFI; expressed as arbitrary unit, a.u.) was measured by MACSQuant Analyzer 10 flow cytometer (Miltenyi). For a subset of samples, the analysis of EV surface antigen profile was repeated using CytoFLEX flow cytometer (Beckman Coulter). Expression levels for each EV surface antigen were reported after subtraction for the respective fluorescence values of blank control and normalization for mean MFI for CD9/CD63/CD81 (normalized MFI, nMFI; expressed as percentage, %) [16,17]. A reverse flow cytometric assay was also performed by isolating EVs by capture beads coated with antibodies against CD9-CD63-CD81 (EpCam; JSR Micro) and then incubated with fluorochrome-conjugated antibodies against CD142, and CD63 (as normalizator). MFI was measured CytoFLEX (Beckman Coulter).
2.3.2. Western blot
WB was performed on protein lysate after EV bead-based immuno-capture. Serum aliquots were incubated overnight with MACSPlex capture beads and saline solution. Unbounded fraction was discarded, and samples were lysed in RIPA buffer; total proteins were separated on SDS Page 4–12% gel (BioRad) and signals were detected by Odyssey CLx Detection System (LI-COR Biosciences). Blots for 3 representative samples were incubated with the following primary antibodies: rabbit polyclonal anti-ApoB48, mouse monoclonal anti-GRP94, rabbit monoclonal anti-Alix, rabbit monoclonal anti-CD142, rabbit monoclonal anti-TSG101, rabbit polyclonal anti-Syntenin-1, rabbit monoclonal anti-CD81 (all from Abcam), and rabbit monoclonal anti-Mitofillin (Invitrogen).
2.3.3. Activity assay
The activity assay for CD142 on EVs was performed with Human Tissue Factor Activity Assay (Abcam), according to manufacturer instructions. The protocol assesses amidolytic activity of TF/FVIIa complex to activate factor X (FX) to factor Xa.
2.3.4. Co-localization assay (ExoView)
Co-localization was assessed by on-chip EV analysis using ExoView® R100 Analyzer, as previously described [21,22]. The instrument is able, in a label-free mode, to count and size EVs as small as 50 nm while, upon fluorescence immune-staining, to phenotype EV surface markers. In the present study, a custom assay was developed: chips functionalized by spots containing a 1:1:1 mixture of CD9/CD63/CD81 antibodies (Ancell) were used to capture EVs from serum, in order to minimize any possible bias due to differential tetraspanin expression in EV subpopulations and provide a representative sampling of vesicles from serum. Serum samples were incubated on chips for 2 h at room temperature. After immunocapturing, chips were stained with anti-TF (CD142; Ancell) labelled with CF555 and with a mixture of anti CD9/CD63/CD81 labelled with CF647 (Invitrogen). Simultaneous imaging of spots on the two fluorescence channels allowed us to detect EVs, where surface antigens CD9-CD63-CD81 and CD142 are co-localized.
2.4. Statistical analysis
We expressed variables with a normal distribution as mean ± standard deviation and their analysis was performed by T-student test. We expressed variables with a non-normal distribution as median [interquartile range] and their analysis was performed by Mann-Whitney test. Categorical variables were expressed as absolute number (percentage) and analyzed by Chi square test (or Fisher test, when appropriated). P-value of less than 0.05 were considered significant. Logistic regression analysis was performed to assess the association of EV surface antigens with the outcome of patients. Hazard ratios (HRs) were evaluated together with their 95% confidence intervals. Receiver Characteristics Operating (ROC) curves were drawn to estimate the area under the curve (AUC) for EV surface antigens, to estimate their prediction performance (patient outcome). Statistics was performed by IBM SPSS Statistics 26 (IBM, New York, USA) and GraphPad PRISM 8.0 (La Jolla, California). For Estimation of study power see detailed methods in supplementary file.
3. Results
3.1. Characteristics of the study cohorts
We enrolled a total of 261 subjects with SARS-CoV-2 infection confirmed by PCR molecular test and admitted to hospital with a diagnosis of pneumonia (Table 1 ). Mean age was 68 years, 65.5% were males, 62.1% displayed bilateral pneumonia, and 1.1% suffered from pulmonary embolism at hospitalization. Patients were stratified according to their outcome: 36% needed to be treated with high flow O2, 13.8% underwent OTI, while the overall mortality was 19.2%. A poor prognosis, defined as needed of OTI or death, was reported in 72 patients (27.6%); the median time from hospitalization to OTI/death was 7 days. As expected, the median duration of hospitalization was longer for patients with a poor prognosis compared to those with a good one (14 vs. 8 days).
Table 1.
Patient characteristics.
| Variable | All Patients [n = 261] |
Good Prognosis [n = 189] |
Poor Prognosis [n = 72] |
P-value |
|---|---|---|---|---|
| Age (years) | 68 ± 13.4 | 68 ± 13.6 | 71 ± 12.8 | 0.100 |
| Sex (Male; %) | 171 (65.5) | 119 (63.0) | 52 (72.2) | 0.160 |
| BMI (Kg/sqm) | 27.4 ± 5.67 | 27.0 ± 5.60 | 28.2 ± 5.78 | 0.204 |
| Bilateral Pneumonia (%) | 162 (62.1) | 104 (55.0) | 58 (80.6) | <0.001 |
| Pulmonary Embolism (%) | 3 (1.1) | 2 (1.1) | 1 (1.4) | 1.000 |
| Respiratory rate (a.p.m.) | 22 ± 5.0 | 20 ± 4.0 | 25 ± 5.6 | <0.001 |
| Peripheral O2 saturation (%) | 92 ± 4.1 | 93 ± 3.5 | 90 ± 4.9 | <0.001 |
| GCS (<15; n) | 28 (10.7) | 16 (8.5) | 12 (16.7) | 0.056 |
| Anamnesis | ||||
| CKD (%) | 44 (16.9) | 16 (8.5) | 28 (38.9) | <0.001 |
| Hypertension (%) | 144 (55.2) | 101 (53.4) | 43 (59.7) |
0.362 |
| Chronic Pulmonary Disease (%) | 46 (17.6) | 30 (15.9) | 16 (22.2) | 0.229 |
| Diabetes (%) | 66 (25.3) | 45 (23.8) | 21 (29.2) | 0.373 |
| Smoking habit (%) | 36 (13.8) | 20 (10.6) | 16 (22.2) | 0.015 |
| CHF (%) | 20 (7.7) | 10 (5.3) | 10 (13.9) | 0.020 |
| CAD (%) | 47 (18.0) | 25 (13.2) | 22 (30.6) | 0.001 |
| Liver Disease (%) | 51 (19.5) | 29 (15.3) | 22 (30.6) | 0.006 |
| Chronic Neurological Disease (%) | 58 (22.2) | 36 (19.0) | 22 (30.6) | 0.046 |
| Dementia (%) | 39 (14.9) | 22 (11.6) | 17 (23.6) | 0.015 |
| Autoimmune Disease (%) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1.000 |
| HIV/AIDS (%) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1.000 |
| Cancer (%) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1.000 |
| Obesity (%) | 52 (19.9) | 34 (18.0) | 18 (25.0) | 0.205 |
| Number of Comorbidities (n) | 1 [0; 2] | 1 [0; 2] | 2 [1; 4] | <0.001 |
| Arterial blood gas assay | ||||
| pCO2 (KPa) | 4.5 [4.1; 5.0] | 4.5 [4.1; 4.9] | 4.7 [4.0; 5.2] | 0.171 |
| pO2 (KPa) | 9.1 [8.1; 10.5] 23.6 ± 3.10 |
9.3 [8.5; 10.8] 23.8 ± 2.74 |
8.6 [7.7; 10.1] 23.0 ± 3.85 |
0.003 |
| Bicarbonate (mmol/L) | 0.163 | |||
| Lactic acid (mmol/L) | 1.3 ± 0.80 | 1.2 ± 0.69 | 1.5 ± 1.02 | 0.049 |
| Biochemical parameters | ||||
| Haemoglobin (g/L) | 139 ± 18.6 | 141 ± 17.4 | 135 ± 21.1 | 0.052 |
| PLTS (*10E9/L) | 190 ± 73.3 | 187 ± 71.4 | 199 ± 77.7 | 0.238 |
| `WBC (*10E9/L) | 6.6 ± 3.30 | 6.4 ± 3.08 | 6.9 ± 3.80 | 0.242 |
| Neutrophils (*10E9/L) | 5.3 ± 4.22 | 5.3 ± 4.74 | 5.5 ± 3.61 | 0.806 |
| Lymphocytes (*10E9/L) | 1.0 ± 0.92 | 1.1 ± 0.84 | 0.9 ± 0.81 | 0.533 |
| Monocytes (*10E9/L) | 0.4 ± 0.23 | 0.4 ± 0.19 | 0.4 ± 0.30 | 0.868 |
| Eosinophils (*10E9/L) | 0.06 ± 0.043 | 0.06 ± 0.034 | 0.07 ± 0.029 | 0.780 |
| Basophils (*10E9/L) | 0.06 ± 0.051 | 0.05 ± 0.032 | 0.06 ± 0.028 | 0.905 |
| C-reactive protein (mg/L) | 47 [21; 98] | 40 [18; 86] | 69 [35; 137] | 0.003 |
| D-dimer (mg/L) | 0.76 [0.50; 1.41] | 0.67 [0.48; 1.15] | 1.08 [0.66; 2.35] | <0.001 |
| PT-INR (a.u.) |
1.2 ± 0.62 |
1.2 ± 0.53 |
1.4 ± 0.81 |
0.097 |
| aPTT (sec) | 32 ± 7.1 | 31 ± 6.1 | 34 ± 8.8 | 0.031 |
| LDH (U/L) | 467 [381; 633] | 462 [375; 601] | 484 [392; 776] | 0.079 |
| Urea (mmol/L) | 7.2 ± 5.06 | 6.4 ± 3.50 | 9.4 ± 7.38 | 0.001 |
| Troponin I (ng/L) | 14 [8; 29] | 12 [6; 19] | 25 [13; 66] | <0.001 |
| Outcome | ||||
| 4C Mortality Score (n) | 9 ± 4.0 | 8 ± 3.7 | 11 ± 3.8 | <0.001 |
| Hospitalization (days) | 9 [2; 16] | 8 [2; 14] | 14 [7; 26] |
<0.001 |
| Time to OTI / Death (days) | N.A. | N.A. | 7 [4; 12] | N.A. |
| Low-flow O2 Treatment (%) | 222 (85.1) | 158 (83.6) | 64 (88.9) | 0.284 |
| High-flow O2 Treatment (%) | 94 (36.0) | 41 (21.7) | 53 (73.6) | <0.001 |
| Orotracheal Intubation (%) | 36 (13.8) | N.A. | 36 (50.0) |
N.A. |
| Death (%) | 50 (19.2) | N.A. | 50 (69.4) | N.A. |
Clinical and biochemical characteristics of patients admitted to hospital for SARS-CoV2 infection and pneumonia (n = 261) stratified according to prognosis; a poor prognosis is defined as need of orotracheal intubation (OTI) or death. GCS, Glasgow Coma Scale; CKD, Chronic Kidney Disease (defined as eGFR <60 mL/min); CHF, Chronic Heart Failure (defined as ejection fraction <35%), CAD, Coronary Artery Disease; Liver disease, defined as chronic hepatitis or cirrhosis with or without portal hypertension; Chronic neurological disease, defined as presence of Parkinson disease, Alzheimer's disease, history of major cerebrovascular accident; HIV/AIDS, infection by Human Immunodeficiency Virus, Acquired Immunodeficiency Syndrome; WBC, White Blood Cells; PT-INR, Thrombin Time - International Normalized Ratio; aPTT, activated Partial Thromboplastin Time; LDH, Lactate Dehydrogenase; N.A., Not Applicable. 4C Mortality Score was calculated as detailed in Knight SR et al. 2020. [5] Comorbidities were defined using the Charlson comorbidities index. [36] A p < 0.05 was considered significant and shown in bold.
Patients with a poor prognosis showed a higher incidence of bilateral pneumonia, a higher respiratory rate, and a lower peripheral O2 saturation at admission. Moreover, they suffered from a higher number of comorbidities, and in particular chronic kidney disease, chronic heart failure, coronary artery disease, liver disease, chronic neurological conditions, and dementia. Concerning biochemical parameters, patients with a poor prognosis displayed higher values of lactic acid, C-reactive protein, D-dimer, aPTT, urea, and troponin I, and lower levels of pO2 at arterial blood gas analysis (p < 0.05 for all comparisons; Table 1); no difference was found evaluating levels of haemoglobin, white blood cells and platelets count.
The study population consisted in a first cohort of selected patients (discovery cohort; n = 60), which was used to identify the detection threshold of CD142 expressed as nMFI (see methods) and corresponding to the expression level of such antigen onto surface of EV, that could be used as cut-off value to predict SARS-CoV-2 prognosis. Following the same criteria of inclusion as for the discovery cohort, a second prospective group composed by unselected consecutive patients was included as validation cohort (n = 201). An overview of study design is depicted in Online Fig. 1. Characteristics of discovery and validation cohorts are reported in Online Tables 1–3.
3.2. EV profiling and selection of CD142-EV as biomarker to predict patient outcome
The bead-based immunocapture flow cytometric assay used for EV profiling was first validated for its specificity to bind EVs by western blot for specific markers and potential contaminants and flow cytometry for tetraspanins expression on EV surface (Supplementary Results; Online Fig. 2). We then evaluated the expression of 37 EV surface antigens in all recruited patients (discovery and validation cohort; n = 261) and compared their fluorescence levels in patients with SARS-CoV2 infection after stratification for prognosis (Online Tables 4–5) and mortality (Online Tables 6–7). The EV surface signature in patients stratified according to their prognosis is shown in Online Figs. 3 and 4.
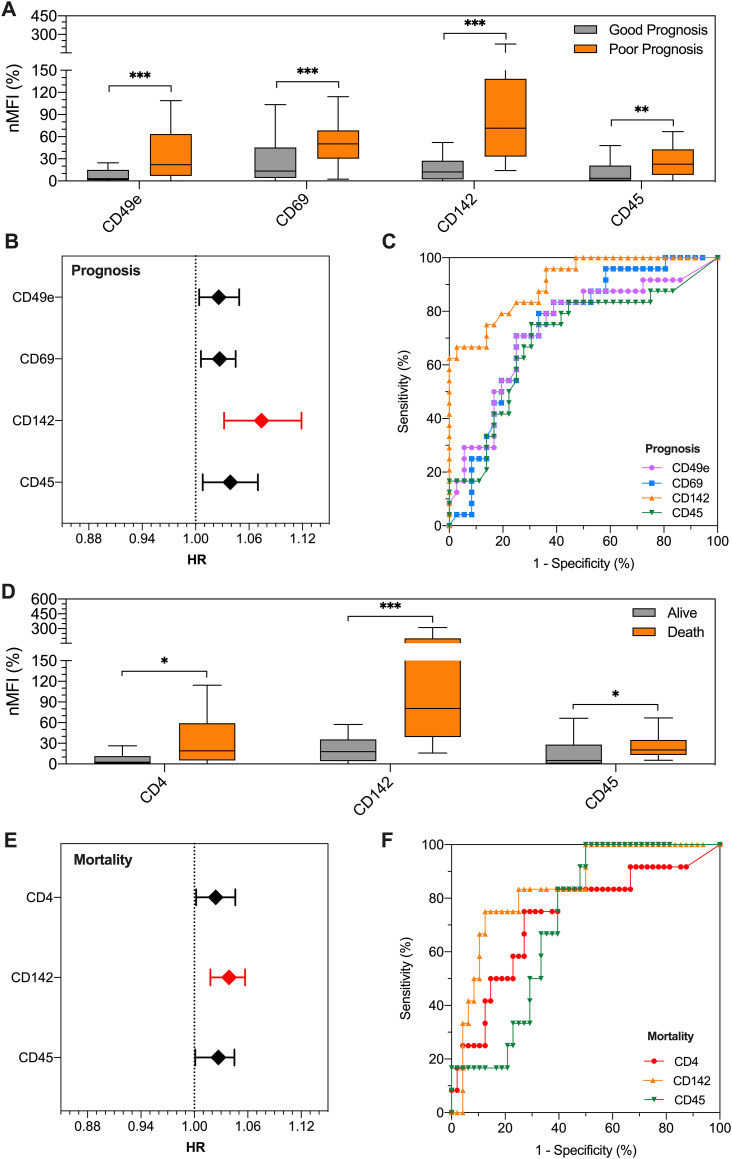
In the discovery cohort (n = 60), among the differentially expressed surface epitopes in patients with good vs. poor prognosis (CD49e, CD69, CD142, and CD45; see Supplementary Results, Fig. 1A and Online Table 4), CD142-EV displayed the strongest association with prognosis with a hazard ratio (HR) of 1.074 (95% CI 1.032–1.119) at regression models, thus meaning that for each single unit of increase in nMFI for this EV marker, the risk of a poor prognosis increased of 7.4% (Fig. 1B and Online Table 8). At ROC curve analysis, CD142-EV had an AUC of 0.906 (95% CI 0.833–0.979) with an accuracy of 81.7% (Fig. 1C and Online Table 9), using a cut-off value of 33.5 (nMFI, %).
Fig. 1.
EV surface antigens associated to patient outcome.
Profiling of EV surface antigens in patients admitted to hospital for SARS-CoV2 infection and pneumonia in the discovery cohort (n = 60). Patients were stratified for outcome (good prognosis, grey, vs. poor prognosis, orange; a poor prognosis is defined as need of orotracheal intubation or death) and mortality. Median fluorescence intensity (MFI) was analyzed after normalization by the average MFI of CD9-CD63-CD81 (normalized MFI; nMFI, %). (A) Expression levels of EV surface antigens differentially expressed in patients with good vs. poor prognosis; (B) Association of EV surface antigens with patient outcome (good vs. poor prognosis; a poor prognosis is defined as need of orotracheal intubation, OTI, or death). Hazard ratios (HRs) are shown together with their 95% confidence intervals. (C) ROC curves for EV surface antigens discriminating patients according to prognosis. (D) Expression levels of EV surface antigens differentially expressed in patients stratified for mortality. (E) Association of EV surface antigens with mortality. Hazard ratios (HRs) are shown together with their 95% confidence intervals. (F) ROC curves for EV surface antigens discriminating patients according to mortality. Statistics is reported in Online Tables 4-6-8-9-10. * p < 0.01; **p < 0.01; ***p < 0.001.
After stratification for mortality, CD4, CD142 and CD45 were highly expressed in deceased patients (see Supplementary Results, Fig. 1D and Online Table 6). CD142-EV was again di best predictor, with an HR of 1.039 (95% CI 1.018–1.057), thus meaning an increase of 3.9% in mortality rate, for each unit of increase in nMFI of the marker (Fig. 1E and Online Table 8). The analysis of ROC curve showed an AUC of 0.842 (95% CI 0.727–0.957) with an accuracy of 85% to predict mortality (Fig. 1F and Online Table 10), using a cut-off value of 52.8 (nMFI, %).
Although a specific tissue-cellular source tracing of circulating EVs is beyond the scope of the present work, we estimated the origin of analyzed EVs by grouping surface antigens according to their putative parental cells (Online Fig. 5). The majority of EVs express endothelial (CD31, CD62P, CD105, CD146) or platelet (CD41b, CD42a, CD62P) surface antigens. Interestingly, the trends of the two subpopulations (endothelial and platelet derived EV) related differently to prognosis with an increased trend toward the poor prognosis for endothelial EVs (24.3 and 29.6% good vs poor prognosis respectively) and decreased one for platelet-derived EV (from 38.9%to 27%). Finally, EVs associated with inflammatory cells mainly T-lymphocytes, B- lymphocytes, NK cells increased in patients with a poor prognosis (27.5 and 33.7% good vs poor prognosis respectively).
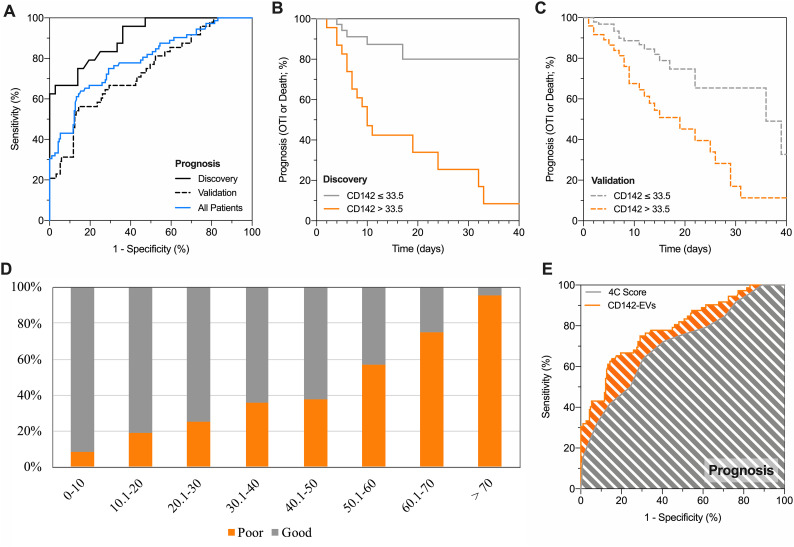
3.3. CD142-EV discriminates SARS-CoV2 patients according to prognosis
In the discovery cohort a cut-off greater than 33.5 (nMFI, %) for CD142-EV correctly identified 18 out of 24 patients with a poor prognosis (sensitivity 75%), while those with an nMFI equal or lower to 33.5 displayed a good prognosis in 31 out of 36 cases (specificity 86.1%; Fig. 2A and Online Tables 11-12). The potential of CD142-EV as discriminant for subjects belonging to the discovery cohort was further assessed by Kaplan-Meier curves showing a log-rank of 4.75 (95% CI 2.09–10.81; Fig. 2B). By applying the same cut-off in the validation cohort (unselected subjects), we were still able to correctly classify 131 out of 153 patients with a good prognosis (specificity 85.6%) and a high negative predictive value (86.2%). The overall accuracy was 78.6%, with a negligible overfitting bias (3.1%) when compared with accuracy in the discovery cohort (Fig. 2A). Kaplan-Meier curves further confirmed a high discrimination power, with CD142-EV able to correctly stratify 158 out of 201 patients according to prognosis (good vs. poor prognosis; log-rank = 2.22–95% CI 1.23–3.99; Fig. 2C).
Fig. 2.
CD142-EV to predict patient prognosis.
Performance of CD142 expressed on EV surface (CD142-EV) to predict outcome (poor prognosis vs. good prognosis) in patients with SARS-CoV2 infection and pneumonia (Discovery cohort, n = 60; Validation cohort, n = 201; All patients, n = 261); a poor prognosis is defined as need of orotracheal intubation or death. Median fluorescence intensity (MFI) was analyzed after normalization by the average MFI of CD9-CD63-CD81 (normalized MFI; nMFI, %) for each EV antigen. (A) ROC curves showing performance of CD142-EV to predict patient prognosis: AUC at discovery = 0.906 (0.833–0.979); AUC at validation = 0.736 (0.654–0.818); AUC in all patients = 0.792 (0.728–0.855). (B) Kaplan-Meier curves for CD142-EV; the cut-off (nMFI = 33.5%) to discriminate patient outcome (good vs. poor prognosis; discovery cohort) was defined by analysis of ROC curves. HR (log-rank) = 4.75 (95% CI 2.09–10.81). (C) Kaplan-Meier curves for CD142-EV; the cut-off (nMFI = 33.5%) to discriminate patient outcome (good vs. poor prognosis; validation cohort) was defined by analysis of ROC curves. HR (log-rank) = 2.22 (95% CI 1.23–3.99). (D) Stratification of patients according to levels of expression of CD142 on EV surface and patient prognosis (good prognosis, grey; poor prognosis, orange) on the combined discovery and validation cohorts. (E) ROC curve analysis: prediction of patient prognosis; CD142-EV vs. 4C Score [5]. Statistics is reported in Online Tables 11-12-13.
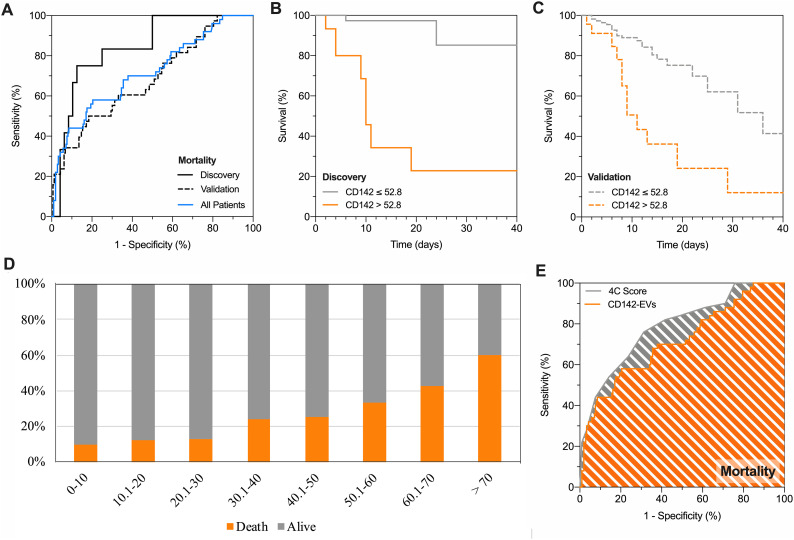
3.4. CD142-EV discriminates SARS-CoV2 patients according to mortality
Considering the outcome of survival as for the ROC analysis, the nMFI value of 52.8% was selected as critical cut-off for CD142-EV (Fig. 3A;). Such value allowed the classification of 51 out of 60 patients in the discovery cohort, resulting in an accuracy of 85%, with a sensitivity and specificity of 75.0% and 87.5%, respectively (Online Tables 11-12). Kaplan-Meier curves showed that CD142-EV was able to stratify patient according to their mortality in discovery cohort with a log-rank = 11.30 (95% CI 2.82–45.34 Fig. 3B). At validation, we correctly predicted the survival of 152 out of 163 patients (specificity 94.3%), once again with a high negative predictive value (85.9%) and an overall accuracy of 82.1% with an overfitting bias of 2.9% (Fig. 3A). The discrimination power according to mortality (Kaplan-Meier curves) was consistent with 165 out of 201 patients correctly predicted (survival; log-rank = 3.37–95% CI 1.27–8.93; Fig. 3C).
Fig. 3.
CD142-EV to predict patient prognosis and mortality.
Performance of CD142 expressed on EV surface (CD142-EV) to predict mortality (death vs. alive) in patients with SARS-CoV2 infection and pneumonia (Discovery cohort, n = 60; Validation cohort, n = 201; All patients, n = 261). Median fluorescence intensity (MFI) was analyzed after normalization by the average MFI of CD9-CD63-CD81 (normalized MFI; nMFI, %) for each EV antigen. (A) ROC curves showing performance of CD142-EV to predict mortality: AUC at discovery = 0.842 (0.727–0.957); AUC at validation = 0.682 (0.585–0.779); AUC in all patients = 0.714 (0.630–0.798). (B) Kaplan-Meier survival curves for CD142-EV; the cut-off (nMFI = 52.8%) to predict patient mortality (discovery cohort) was defined by analysis of ROC curves. HR (log-rank) = 11.30 (95% CI 2.82–45.34). (C) Kaplan-Meier survival curves for CD142-EV; the cut-off (nMFI = 52.8%) to predict patient mortality (validation cohort) was defined by analysis of ROC curves. HR (log-rank) = 3.37 (95% CI 1.27–8.93). (D) Stratification of patients according to mortality (alive, grey; death, orange). (E) ROC curve analysis: prediction of mortality; CD142-EV vs. 4C Score [5]. Statistics is reported in Online Tables 11-12-13.
3.5. CD142-EV predict prognosis and mortality in SARS-CoV2
Having assessed the performance of CD142-EV as prognostic tool, we applied such unconventional biomarker to the entire population of included patients (discovery plus validation cohort), to assess patient distribution according to outcome and expression levels of CD142-EV. The likelihood of a poor prognosis, as well as mortality, gradually increased at the increase of nMFI for CD142-EV (Figs. 2D-3D). Among patients with lowest score (CD142-EV ≤ 10) 107 out of 117 displayed good prognosis (91.5%), and 108 out of 120 (90.0%) were alive at follow-up. Thus, translating in a very high specificity and negative predictive value (Online Table 13). Conversely, among patients with the highest score (CD142-EV >70), 22 out of 23 (95.7%) displayed a poor prognosis and 15 out of 25 (60%) deceased at follow-up, with a very high sensitivity and positive predictive value (Online Table 13). For each patient, we calculated the 4C (Coronavirus Clinical Characterization Consortium) Mortality score as described in Knight et al. [5] 4C Mortality score was then used as referral to estimate the potential application of our experimental model based on CD142-EV in predicting patient prognosis and mortality (Online Fig. 6; Online Table 11). Considering all patients CD142-EV showed a higher accuracy compared to 4C score in predicting patient prognosis (AUC 0.792 vs. 0.705 – p = 0.044; accuracy 79.3% vs. 67.8%; Fig. 2E), whereas the overall accuracy was comparable when predicting mortality (AUC 0.714 vs. 0.786; p = 0.131; accuracy 82.3% vs. 73.9%; Fig. 3E). Diagnostic performance and confusion matrix of CD142-EV and 4C Mortality score to predict either patient prognosis or mortality, are summarized in Online Table 12.
3.6. CD142-EV experimental validation as biomarker in SARS-CoV2
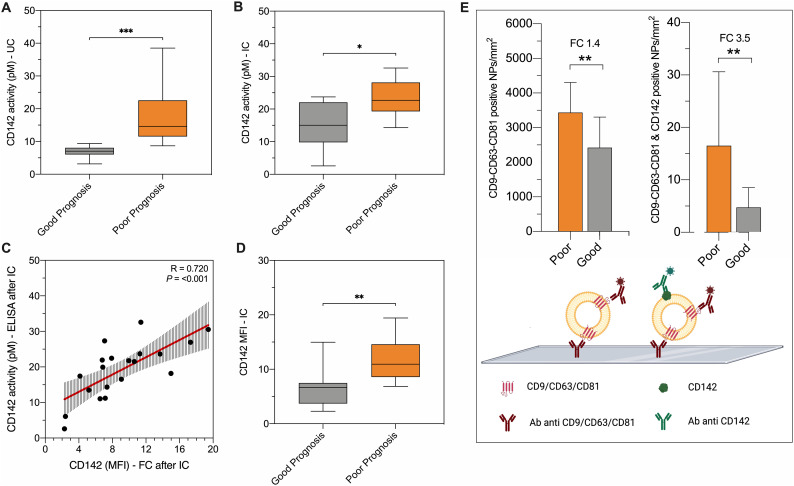
We have previously shown that TF expressed on the surface of EVs possess enzymatic activity which directly correlate with its level of expression. [15] Here, we assessed whether such activity has also potential as prognostic marker, as further confirmation of CD142-EV as predictor of patient outcome in SARS-CoV2. EVs isolated by ultracentrifugation (UC) and by immuno-capture beads (IC) from serum of 20 randomly selected patients from the validation cohort (10 with a good prognosis and 10 with a poor prognosis) were quantitatively measured for CD142 enzymatic activity. Both, EVs enriched by classical UC or using IC showed an augmented CD142 activity when isolated from serum of patients with poor vs. good prognosis (p < 0.05; Fig. 4A-B). Notably, CD142-EV level of expression measured at flow cytometry directly correlated to CD142 activity measured by ELISA (R = 0.720; p < 0.001; Fig. 4C).
Fig. 4.
Experimental validation with different techniques.
The discriminant performance of CD142-EV was experimentally validated by different techniques in patients with SARS-CoV2 infection: good prognosis (grey; n = 10) vs. poor prognosis (orange; n = 10). (A-B) CD142 activity per particle measured by ELISA (pM per 109 particles), after EV isolation by ultracentrifugation (UC) or immunocapture (IC using beads covered by antibodies against CD9-CD63-CD81). (C) Correlation between CD142 activity per particle (pM) and CD142 MFI at flow cytometry after IC. (D) CD142-EV MFI after IC (direct staining after immuno-capture, using beads covered by antibodies against CD9-CD63-CD81). (E) Colocalization of tetraspanins (CD9-CD63-CD81) and CD142 was assessed by ExoView® R100 Analyzer. Data are reported for mean number of nanoparticles (NPs) per mm2 for vesicles labelled with fluorochrome-conjugated antibodies against CD9-CD63-CD81 and for the double positive for CD9-CD63-CD81 and CD142.
To overcome possible methodological- or instrumental-related biases, the expression of surface vesicular CD142 was also measured by using a different flow cytometer (CytoFLEX) with a reverse immunocapture strategy. Indeed, EVs were captured by using beads coated with antibodies direct against tetraspanins and immuno-stained for CD142. We confirmed that the level of expression of CD142 was significantly higher in EVs from patients with poor vs. good prognosis regardless EV-binding protocol of (p < 0.01; Fig. 4D). We also compared EV profiling obtained by CytoFLEX and MACSQuant flow cytometers in a subset of samples (n = 5; Online Fig. 7): coefficients of variation for CD9-CD63-CD81 and CD142 nMFI were 7.4% and 9.5%, respectively.
To exclude major impacts on EV profiling related to the choice of serum as starting material, we compared the expression of surface antigens on EVs from serum and patient-matched plasma samples in a small cohort of healthy volunteers (n = 10; Online Fig. 8A). No differences were found for any of the 37 EV antigens. Coefficients of variation for mean nMFI of CD9-CD63-CD81 and CD142 were 6.6% and 12.0%, respectively (Online Fig. 8B-C).
Finally, we further assessed the co-localization of tetraspanins with CD142 by ExoView® Analyzer which allowed the immunocapture of EVs onto silico chip and the simultaneously detection of surface antigens CD9, CD63, CD81 and CD142, (Fig. 4E). The assay confirmed that EV specific tetraspanins are mainly co-expressed with tissue factor. By quantifying the degree of expression of each marker, we could further confirm that the number of total tetraspanin positive EVs as well as the number of CD142-bearing EV, were both increased in patients with more severe disease (FC 1.4 - p = 0.005, and FC 3.5 - p = 0.002, respectively).
4. Discussion
We have addressed the potential value of CD142-EV as prognostic biomarker in a cohort of patients admitted to hospital for pneumonia and SARS-CoV-2 infection. Both the discovery and the validation cohorts were tailored on reliable estimation of minimum number of subjects to be included on the base of our previous pilot study. [15] By using this prospective cohort of unselected patients consecutively recruited, we obtained an overall accuracy of 78.6% and 82.1% in predicting patient prognosis and mortality, respectively. Noteworthy, CD142-EV reached a reliable grade of “generalizability” as prognostic marker since the overfitting bias was negligible when comparing accuracy in the discovery and validation cohorts (ranging between 2.9 and 3.1%).
CD142-EV performed well against the clinically applied 4C Mortality Score, which is currently one of the most robustly validated COVID-19 prognostic model. [5] When considering all patients, we obtained an overall comparable accuracy, being not inferior in terms of prediction of patients' prognosis (overall accuracy 79.3% CD142-EV vs. 67.8% 4C Mortality Score) and risk of mortality (overall accuracy 82.3%% CD142-EV vs. 73.9% 4C Mortality Score). CD142-EV displayed a very high specificity and negative predictive value (ranging between 83.4 and 93.3%). However, as compared to 4C Mortality score it shows lower sensitivity and positive predictive value (ranging between 34.2 and 78.2%), thus making CD142-EV mainly suitable to rule out severe cases. CD142-EV also performed well in stratifying patients according to their risk of a poor prognosis. The likelihood of a poor outcome (OTI and/or death) gradually increases with CD142-EV expression and therefore it was suitable for the quantification of a discrete risk. For instance, patients with a CD142-EV nMFI ranging between 20 and 30 display a likelihood of 25% and 13% in terms of poor prognosis and mortality respectively. On the other hand, patients with CD142-EV ranging between 60 and 70 will have a poor prognosis in 75% of cases, with a mortality of 42.9%.
Although patients for validation have been enrolled by avoiding selection bias, the cohort still suffer of the limited number of enrolled subjects. Such small cohort of validation also represents the main limit for more in-depth comparison with 4C Mortality Score that included more than 22,000 subjects in validation. A further limitation includes the fact that the study was temporally and geographically narrowed. Infection rates and patients' characteristics might change by time and geography during a pandemic. Here we could not show robustness of the CD142-EV over time and geography.
We do not add substantial advancing in the debate concerning whether it is better to measure levels of TF activity or TF protein as marker of thrombotic risk, [23,24] however we clearly show that the TF protein level on the surface of EV consistently predict the severity of COVID-19 disease. We have also shown that CD142-bearing EVs have an augmented enzymatic activity when isolated from serum of patients with severe disease and a poor prognosis, regardless the method of isolation. Finally, we showed that the level of expression of TF strongly correlates with its activity in COVID-19 patients and it is hampered when using specific antibody that causes steric hindrance with the enzymatic site of the TF. [15] It is plausible, and some recently published data come in help supporting this hypothesis, that both parameters are associated with severity of disease in COVID-19 patients. [12,13,25] The discrepancy between protein expression and activity, due to the presence of undefined portion of intravascular TF present as inactive or encrypted state, [26] is negligible when referring to EVs. The cytokines storm [27] as well as the hyper-activation of platelets [12] occurring in these patients may dramatically contribute to increase the release of EVs with pro-coagulant activity, thus expressing TF in a decrypted state. [28] Not only, an increased risk of thrombotic cardiovascular events can be a direct consequence of such mechanism in COVID-19 patients. Circulating EVs may spread systemic inflammation, leading to vascular damage, endothelial dysfunction and in loop to the activation of coagulation cascade. This may ultimately contribute to increase risk of cardiovascular disease and heart-brain disorders in patients with SARS-CoV2 infection. [29] A dysfunction of heart-brain axis may lead to severe multi-organs failure and worsen the outcome of COVID-19 patients. [[29], [30], [31]] In this view, EVs have a potential for accurate stratification of patients according to the respective cardiovascular risk, [32] supporting medical staff in tailoring management strategies for patients.
As respect to Guervilly et al. we found significant increase in the total amount of circulating EVs in patients with poor versus good prognosis. The apparent discrepancy might be explained by the fact that we only addressed concentration (expressed as nMFI) of CD9; CD81 and CD63 positive EVs, while a direct FC assay as in Guervilly et al. can account for enumeration of large vesicles that can be negative for tetraspanins while still expressing TF. [25] A second possible explanation reside in the starting material as EVs source: we used serum in stand of plasma. We are aware that this aspect may represent a weakness of the study, however we have previously shown that the profiling of EVs from serum has good potential as biomarker, showing consistent diagnostic and prognostic performances, in line with gold-standard biomarkers. [33] Both plasma and serum have been used in previous studies. [34]. In the present manuscript we compared the expression of surface antigens on EVs from serum and patient-matched plasma samples in a small cohort of healthy volunteers, without finding differences for any of the 37 evaluated EV antigens. Above all, the prognostic performance of serum CD142-EV is in line with others regardless EV's sources [12,14,25]. Anyway, the methodological assessment of the most appropriate biological fluid is beyond the scope of the present study.
5. Conclusions
The aim of the present study was to give clinical relevance to a biomarker that can be useful to assess the risk of negative outcome and to prompt the adoption of strategies to treat the disease. Indeed, the detection of CD142 on the surface of EV is a cost-effective and rapid test that can be available at time of admission by using conventional flow cytometer. The method used is well standardized from our group [20,35] as well as from other independent groups [16,17] from sample preparation to data analysis, ensuring that results can be reproducible and shared among different laboratories. We believe that such analysis gains considerable interest as risk stratification tool to support frontline clinical decision making.
Author contributions
J.B. data generation and interpretation, statistical analysis, manuscript writing. E.C., L.G.G., E.R., T.F.S., and E.F., patient data collection. E.P., and G.Ma. sample collection. A.B. statistical analysis. C.B., S.B., E.L., V.B., R.F., A.G., M.C., G.V., and G.Me. data generation and interpretation, critical revision. S.M., and L.B. study design, data interpretation, manuscript writing. All authors read and approved the final version of the manuscript.
Funding
This study was supported by research funding from Fidinam Foundation (Lugano, Switzerland). This work was partially funded from the European Union's Horizon 2020 research and innovation program under grant agreements No. 951768 (project MARVEL).
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Visual abstract was produced using Servier Medical Art (https://smart.servier.com/).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.vph.2022.106999.
Appendix A. Supplementary data
Supplementary material
References
- 1.Arabi Y.M., Murthy S., Webb S. COVID-19: a novel coronavirus and a novel challenge for critical care. Intensive Care Med. 2020;46(5):833–836. doi: 10.1007/s00134-020-05955-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang X., Yu Y., Xu J., Shu H., Xia J., Liu H., Wu Y., Zhang L., Yu Z., Fang M., Yu T., Wang Y., Pan S., Zou X., Yuan S., Shang Y. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. Lancet Respir Med. 2020;8(5):475–481. doi: 10.1016/S2213-2600(20)30079-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wynants L., Van Calster B., Collins G.S., Riley R.D., Heinze G., Schuit E., Bonten M.M.J., Dahly D.L., Damen J.A.A., Debray T.P.A., de Jong V.M.T., De Vos M., Dhiman P., Haller M.C., Harhay M.O., Henckaerts L., Heus P., Kammer M., Kreuzberger N., Lohmann A., Luijken K., Ma J., Martin G.P., McLernon D.J., Navarro C.L. Andaur, Reitsma J.B., Sergeant J.C., Shi C., Skoetz N., Smits L.J.M., Snell K.I.E., Sperrin M., Spijker R., Steyerberg E.W., Takada T., Tzoulaki I., van Kuijk S.M.J., van Bussel B., van der Horst I.C.C., van Royen F.S., Verbakel J.Y., Wallisch C., Wilkinson J., Wolff R., Hooft L., Moons K.G.M., van Smeden M. Prediction models for diagnosis and prognosis of covid-19: systematic review and critical appraisal. BMJ. 2020;369 doi: 10.1136/bmj.m1328. m1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gupta R.K., Marks M., Samuels T.H.A., Luintel A., Rampling T., Chowdhury H., Quartagno M., Nair A., Lipman M., Abubakar I., van Smeden M., Wong W.K., Williams B., Noursadeghi M., U.C.-R. Group Systematic evaluation and external validation of 22 prognostic models among hospitalised adults with COVID-19: an observational cohort study. Eur Respir J. 2020;56(6) doi: 10.1183/13993003.03498-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Knight S.R., Ho A., Pius R., Buchan I., Carson G., Drake T.M., Dunning J., Fairfield C.J., Gamble C., Green C.A., Gupta R., Halpin S., Hardwick H.E., Holden K.A., Horby P.W., Jackson C., McLean K.A., Merson L., Nguyen-Van-Tam J.S., Norman L., Noursadeghi M., Olliaro P.L., Pritchard M.G., Russell C.D., Shaw C.A., Sheikh A., Solomon T., Sudlow C., Swann O.V., Turtle L.C., Openshaw P.J., Baillie J.K., Semple M.G., Docherty A.B., Harrison E.M., I.C. investigators Risk stratification of patients admitted to hospital with covid-19 using the ISARIC WHO Clinical Characterisation Protocol: development and validation of the 4C Mortality Score. BMJ. 2020;370 doi: 10.1136/bmj.m3339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zacharia E., Zacharias K., Papamikroulis G.A., Bertsias D., Miliou A., Pallantza Z., Papageorgiou N., Tousoulis D. Cell-derived microparticles and acute coronary syndromes: is there a predictive role for microparticles? Curr Med Chem. 2020;27(27):4440–4468. doi: 10.2174/0929867327666191213104841. [DOI] [PubMed] [Google Scholar]
- 7.Chiva-Blanch G., Crespo J., Suades R., Arderiu G., Padro T., Vilahur G., Cubedo J., Corella D., Salas-Salvado J., Aros F., Martinez-Gonzalez M.A., Ros E., Fito M., Estruch R., Badimon L. CD142+/CD61+, CD146+ and CD45+ microparticles predict cardiovascular events in high risk patients following a Mediterranean diet supplemented with nuts. Thromb Haemost. 2016;116(1):103–114. doi: 10.1160/TH16-02-0130. [DOI] [PubMed] [Google Scholar]
- 8.Huo S., Krankel N., Nave A.H., Sperber P.S., Rohmann J.L., Piper S.K., Heuschmann P.U., Landmesser U., Endres M., Siegerink B., Liman T.G. Endothelial and leukocyte-derived microvesicles and cardiovascular risk after stroke: PROSCIS-B. Neurology. 2021;96(6):e937–e946. doi: 10.1212/WNL.0000000000011223. [DOI] [PubMed] [Google Scholar]
- 9.Camera M., Brambilla M., Canzano P., Cavallotti L., Parolari A., Tedesco C.C., Zara C., Rossetti L., Tremoli E. Association of microvesicles with graft patency in patients undergoing CABG surgery. J Am Coll Cardiol. 2020;75(22):2819–2832. doi: 10.1016/j.jacc.2020.03.073. [DOI] [PubMed] [Google Scholar]
- 10.Vacchi E., Burrello J., Di Silvestre D., Burrello A., Bolis S., Mauri P., Vassalli G., Cereda C.W., Farina C., Barile L., Kaelin-Lang A., Melli G. Immune profiling of plasma-derived extracellular vesicles identifies Parkinson disease. Neurol Neuroimmunol Neuroinflamm. 2020;7(6) doi: 10.1212/NXI.0000000000000866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fujita Y., Hoshina T., Matsuzaki J., Yoshioka Y., Kadota T., Hosaka Y., Fujimoto S., Kawamoto H., Watanabe N., Sawaki K., Sakamoto Y., Miyajima M., Lee K., Nakaharai K., Horino T., Nakagawa R., Araya J., Miyato M., Yoshida M., Kuwano K., Ochiya T. Early prediction of COVID-19 severity using extracellular vesicle COPB2. J Extracell Vesicles. 2021;10(8) doi: 10.1002/jev2.12092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Canzano P., Brambilla M., Porro B., Cosentino N., Tortorici E., Vicini S., Poggio P., Cascella A., Pengo M.F., Veglia F., Fiorelli S., Bonomi A., Cavalca V., Trabattoni D., Andreini D., Omodeo Sale E., Parati G., Tremoli E., Camera M. Platelet and endothelial activation as potential mechanisms behind the thrombotic complications of COVID-19 patients. JACC Basic Transl Sci. 2021;6(3):202–218. doi: 10.1016/j.jacbts.2020.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rosell A., Havervall S., von Meijenfeldt F., Hisada Y., Aguilera K., Grover S.P., Lisman T., Mackman N., Thalin C. Patients with COVID-19 have elevated levels of circulating extracellular vesicle tissue factor activity that is associated with severity and mortality-brief report. Arterioscler Thromb Vasc Biol. 2021;41(2):878–882. doi: 10.1161/ATVBAHA.120.315547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krishnamachary B., Cook C., Kumar A., Spikes L., Chalise P., Dhillon N.K. Extracellular vesicle-mediated endothelial apoptosis and EV-associated proteins correlate with COVID-19 disease severity. J Extracell Vesicles. 2021;10(9) doi: 10.1002/jev2.12117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Balbi C., Burrello J., Bolis S., Lazzarini E., Biemmi V., Pianezzi E., Burrello A., Caporali E., Grazioli L.G., Martinetti G., Fusi-Schmidhauser T., Vassalli G., Melli G., Barile L. Circulating extracellular vesicles are endowed with enhanced procoagulant activity in SARS-CoV-2 infection. EBioMedicine. 2021;67 doi: 10.1016/j.ebiom.2021.103369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koliha N., Wiencek Y., Heider U., Jungst C., Kladt N., Krauthauser S., Johnston I.C., Bosio A., Schauss A., Wild S. A novel multiplex bead-based platform highlights the diversity of extracellular vesicles. J Extracell Vesicles. 2016;5:29975. doi: 10.3402/jev.v5.29975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wiklander O.P.B., Bostancioglu R.B., Welsh J.A., Zickler A.M., Murke F., Corso G., Felldin U., Hagey D.W., Evertsson B., Liang X.M., Gustafsson M.O., Mohammad D.K., Wiek C., Hanenberg H., Bremer M., Gupta D., Bjornstedt M., Giebel B., Nordin J.Z., Jones J.C., El Andaloussi S., Gorgens A. Systematic methodological evaluation of a multiplex bead-based flow cytometry assay for detection of extracellular vesicle surface signatures. Front Immunol. 2018;9:1326. doi: 10.3389/fimmu.2018.01326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vacchi E., Burrello J., Burrello A., Bolis S., Monticone S., Barile L., Kaelin-Lang A., Melli G. Profiling inflammatory extracellular vesicles in plasma and cerebrospinal fluid: an optimized diagnostic model for Parkinson’s disease. Biomedicines. 2021;9(3) doi: 10.3390/biomedicines9030230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Castellani C., Burrello J., Fedrigo M., Burrello A., Bolis S., Di Silvestre D., Tona F., Bottio T., Biemmi V., Toscano G., Gerosa G., Thiene G., Basso C., Longnus S.L., Vassalli G., Angelini A., Barile L. Circulating extracellular vesicles as non-invasive biomarker of rejection in heart transplant. J Heart Lung Transplant. 2020;39(10):1136–1148. doi: 10.1016/j.healun.2020.06.011. [DOI] [PubMed] [Google Scholar]
- 20.Burrello J., Bianco G., Burrello A., Manno C., Maulucci F., Pileggi M., Nannoni S., Michel P., Bolis S., Melli G., Vassalli G., Albers G.W., Cianfoni A., Barile L., Cereda C.W. Extracellular vesicle surface markers as a diagnostic tool in transient ischemic attacks. Stroke. 2021;52(10):3335–3347. doi: 10.1161/STROKEAHA.120.033170. STROKEAHA120033170. [DOI] [PubMed] [Google Scholar]
- 21.Gori A., Romanato A., Greta B., Strada A., Gagni P., Frigerio R., Brambilla D., Vago R., Galbiati S., Picciolini S., Bedoni M., Daaboul G.G., Chiari M., Cretich M. Membrane-binding peptides for extracellular vesicles on-chip analysis. J Extracell Vesicles. 2020;9(1):1751428. doi: 10.1080/20013078.2020.1751428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Daaboul G.G., Gagni P., Benussi L., Bettotti P., Ciani M., Cretich M., Freedman D.S., Ghidoni R., Ozkumur A.Y., Piotto C., Prosperi D., Santini B., Unlu M.S., Chiari M. Digital detection of exosomes by interferometric imaging. Sci Rep. 2016;6:37246. doi: 10.1038/srep37246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mackman N., Hisada Y., Grover S.P., Rosell A., Havervall S., von Meijenfeldt F., Aguilera K., Lisman T., Thalin C. Response by Mackman et al to letter regarding article, “patients with COVID-19 have elevated levels of circulating extracellular vesicle tissue factor activity that is associated with severity and mortality-brief report”. Arterioscler Thromb Vasc Biol. 2021;41(6):e381–e382. doi: 10.1161/ATVBAHA.121.316203. [DOI] [PubMed] [Google Scholar]
- 24.Brambilla M., Canzano P., Becchetti A., Tremoli E., Camera M. Letter by Brambilla et al regarding article, “patients with COVID-19 have elevated levels of circulating extracellular vesicle tissue factor activity that is associated with severity and mortality-brief report”. Arterioscler Thromb Vasc Biol. 2021;41(6):e379–e380. doi: 10.1161/ATVBAHA.121.316188. [DOI] [PubMed] [Google Scholar]
- 25.Guervilly C., Bonifay A., Burtey S., Sabatier F., Cauchois R., Abdili E., Arnaud L., Lano G., Pietri L., Robert T., Velier M., Papazian L., Albanese J., Kaplanski G., Dignat-George F., Lacroix R. Dissemination of extreme levels of extracellular vesicles: tissue factor activity in patients with severe COVID-19. Blood Adv. 2021;5(3):628–634. doi: 10.1182/bloodadvances.2020003308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rao L.V., Kothari H., Pendurthi U.R. Tissue factor encryption and decryption: facts and controversies. Thromb Res. 2012;129(Suppl. 2):S13–S17. doi: 10.1016/j.thromres.2012.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang L., Xie X., Tu Z., Fu J., Xu D., Zhou Y. The signal pathways and treatment of cytokine storm in COVID-19. Signal Transduct Target Ther. 2021;6(1):255. doi: 10.1038/s41392-021-00679-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang J., Pendurthi U.R., Yi G., Rao L.V.M. SARS-CoV-2 infection induces the activation of tissue factor-mediated coagulation by activation of acid sphingomyelinase. Blood. 2021;138(4):344–349. doi: 10.1182/blood.2021010685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lionetti V., Bollini S., Coppini R., Gerbino A., Ghigo A., Iaccarino G., Madonna R., Mangiacapra F., Miragoli M., Moccia F., Munaron L., Pagliaro P., Parenti A., Pasqua T., Penna C., Quaini F., Rocca C., Samaja M., Sartiani L., Soda T., Tocchetti C.G., Angelone T. Understanding the heart-brain axis response in COVID-19 patients: a suggestive perspective for therapeutic development. Pharmacol Res. 2021;168 doi: 10.1016/j.phrs.2021.105581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moccia F., Gerbino A., Lionetti V., Miragoli M., Munaron L.M., Pagliaro P., Pasqua T., Penna C., Rocca C., Samaja M., Angelone T. COVID-19-associated cardiovascular morbidity in older adults: a position paper from the Italian Society of Cardiovascular Researches. Geroscience. 2020;42:1021–1049. doi: 10.1007/s11357-020-00198-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Razeghian-Jahromi I., Zibaeenezhad M.J., Lu Z., Zahra E., Mahboobeh R., Lionetti V. Angiotensin-converting enzyme 2: a double-edged sword in COVID-19 patients with an increased risk of heart failure. Heart Fail Rev. 2021;26(2):371–380. doi: 10.1007/s10741-020-10016-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Burrello J., Burrello A., Vacchi E., Bianco G., Caporali E., Amongero M., Airale L., Bolis S., Vassalli G., Cereda C.W., Mulatero P., Bussolati B., Camici G.G., Melli G., Monticone S., Barile L. Supervised and unsupervised learning to define the cardiovascular risk of patients according to an extracellular vesicle molecular signature. Transl Res. 2022;244:114–125. doi: 10.1016/j.trsl.2022.02.005. [DOI] [PubMed] [Google Scholar]
- 33.Burrello J., Bolis S., Balbi C., Burrello A., Provasi E., Caporali E., Gauthier L.G., Peirone A., D’Ascenzo F., Monticone S., Barile L., Vassalli G. An extracellular vesicle epitope profile is associated with acute myocardial infarction. J Cell Mol Med. 2020;24(17):9945–9957. doi: 10.1111/jcmm.15594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Witwer K.W., Buzas E.I., Bemis L.T., Bora A., Lasser C., Lotvall J., Nolte-'t Hoen E.N., Piper M.G., Sivaraman S., Skog J., Thery C., Wauben M.H., Hochberg F. Standardization of sample collection, isolation and analysis methods in extracellular vesicle research. J Extracell Vesicles. 2013;2 doi: 10.3402/jev.v2i0.20360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Burrello J., Tetti M., Forestiero V., Biemmi V., Bolis S., Pomatto M.A.C., Amongero M., Di Silvestre D., Mauri P., Vassalli G., Camussi G., Williams T.A., Mulatero P., Barile L., Monticone S. Characterization of circulating extracellular vesicle surface antigens in patients with primary aldosteronism. Hypertension. 2021;78(3):726–737. doi: 10.1161/HYPERTENSIONAHA.121.17136. HYPERTENSIONAHA12117136. [DOI] [PubMed] [Google Scholar]
- 36.Charlson M.E., Pompei P., Ales K.L., MacKenzie C.R. A new method of classifying prognostic comorbidity in longitudinal studies: development and validation. J Chronic Dis. 1987;40(5):373–383. doi: 10.1016/0021-9681(87)90171-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material