Abstract
1,2-Propanediol (1,2-PD) is a major commodity chemical that is currently derived from propylene, a nonrenewable resource. A goal of our research is to develop fermentation routes to 1,2-PD from renewable resources. Here we report the production of enantiomerically pure R-1,2-PD from glucose in Escherichia coli expressing NADH-linked glycerol dehydrogenase genes (E. coli gldA or Klebsiella pneumoniae dhaD). We also show that E. coli overexpressing the E. coli methylglyoxal synthase gene (mgs) produced 1,2-PD. The expression of either glycerol dehydrogenase or methylglyoxal synthase resulted in the anaerobic production of approximately 0.25 g of 1,2-PD per liter. R-1,2-PD production was further improved to 0.7 g of 1,2-PD per liter when methylglyoxal synthase and glycerol dehydrogenase (gldA) were coexpressed. In vitro studies indicated that the route to R-1,2-PD involved the reduction of methylglyoxal to R-lactaldehyde by the recombinant glycerol dehydrogenase and the reduction of R-lactaldehyde to R-1,2-PD by a native E. coli activity. We expect that R-1,2-PD production can be significantly improved through further metabolic and bioprocess engineering.
1,2-Propanediol (1,2-PD; propylene glycol) is a three-carbon diol with a stereogenic center at the central carbon atom. Racemic 1,2-PD is a commodity chemical with an annual production of over 1 billion pounds in the United States (1). The commercial route to racemic 1,2-PD is by the hydration of propylene oxide, which is derived from propylene. There are several routes to 1,2-PD from renewable feedstocks. Hydrogenolysis of sugars at high temperature and under pressure in the presence of a metal catalyst results in the production of a mixture of 1,2-PD and other polyols (21). The resulting 1,2-PD is most likely a racemic mixture. Enantiomerically pure 1,2-PD can be produced by the catalytic hydrogenation of d- or l-lactic acid esters (11), the bioreduction of acetol (22), or the resolution of racemic 1,2-PD (17). A direct fermentation route to S-1,2-PD from 6-deoxyhexose sugars in Escherichia coli is well known (4, 13); however, this is not a feasible commercial route to 1,2-PD since even the least expensive such sugar, l-rhamnose, sells for over $300 per kilogram (Pfanstiehl Laboratories, Waukegan, Ill.). Several organisms are reported to ferment common sugars, such as glucose, to R-1,2-PD (6, 7, 30). For example, the thermophilic bacterium Thermoanaerobacterium thermosaccharolyticum produces R-1,2-PD from glucose, xylose, and several other common sugars (6, 7). However, the titers are fairly low, and the organism is currently not characterized well enough to enable the improvement of the 1,2-PD production by metabolic engineering. A review that focuses on 1,2-PD and its production by microbes was recently published (6).
Wild-type E. coli is not known to produce 1,2-PD from common sugars. Recently, work in our laboratory demonstrated 1,2-PD production in E. coli strains expressing rat lens aldose reductase (7, 25). The recombinant E. coli produces R-1,2-PD in 80% enantiomeric excess (25). The maximum reported titer of 1,2-PD produced by E. coli AG1 expressing aldose reductase alone is less than 0.15 g per liter (25). In addition to R-1,2-PD, acetol is also produced in this fermentation. Aldose reductase is an NADPH-linked reductase with broad substrate specificity and is reported to reduce both methylglyoxal (MG) and acetol (32). However, the turnover efficiency of aldose reductase is 3 orders of magnitude greater for MG than for acetol (32). The main route to 1,2-PD in this organism involves the reduction of MG to acetol by the aldose reductase and further reduction of the acetol to 1,2-PD by a native E. coli activity (25).
In this paper, we describe the production of R-1,2-PD in E. coli strains expressing glycerol dehydrogenase genes. Glycerol dehydrogenase (EC 1.1.1.6), an NADH-dependent enzyme, is known to reduce α-hydroxy ketones to chiral R-1,2-diols (33). We also describe the production of R-1,2-PD in E. coli strains overexpressing MG synthase (EC 4.2.99.11) and both MG synthase and glycerol dehydrogenase. Finally, we explore the use of different fermentation operating conditions, such as the time of induction, to further improve R-1,2-PD production. Some of the early results of this work were briefly summarized in a review of metabolic engineering of propanediol pathways (6).
MATERIALS AND METHODS
Bacterial strains, media, and growth conditions.
E. coli AG1 (F− endA1 hsdR17 [rK−, mK+] supE44 thi-1 recA1 gyrA96 relA1 λ−) (Stratagene, La Jolla, Calif.) was used as the host strain for 1,2-PD production. Strain MG1655 (F− LAM rph-1) was a generous gift of the E. coli Genetic Stock Center (New Haven, Conn.). The standard production medium consisted of the following (per liter): 10 g of glucose, 5 g of yeast extract (Difco Laboratories, Detroit, Mich.), 6 g of Na2HPO4, 3 g of KH2PO4, 1 g of NH4Cl, 0.5 g of NaCl, 2 mmol of MgSO4, 100 mg of ampicillin, and 13.3 g of NaHCO3. Isopropyl-β-d thiogalactopyranoside (IPTG) was added in various amounts to induce gene expression. All fermentations were run at 37°C. Anaerobic fermentations were run in 15-ml Hungate tubes and in 300-ml anaerobic flasks (19) with 10- and 150-ml working volumes, respectively. Inocula for fermentations were grown overnight in Luria-Bertani medium supplemented with 100 μg of ampicillin per ml. The 10-ml fermentation mixtures were inoculated with 100 μl of the overnight culture, and 150-ml fermentation mixtures were inoculated with 1 ml of the overnight culture.
The optical density (OD) was measured at 660 nm with a Sequoia-Turner (Mountain View, Calif.) model 340 spectrophotometer. One unit of OD at 660 nm corresponds to approximately 0.52 g of dry cell weight.
Description of plasmids.
All cloning vectors and plasmids used in this work are summarized in Table 1. Primers used for the amplification of genes by PCR are listed in Table 2. Standard techniques of recombinant DNA technology, as described by Ausubel et al. (2), were used for all DNA manipulations. Restriction and DNA-modifying enzymes were obtained from Promega (Madison, Wis.) or New England Biolabs (Beverly, Mass.). Taq and Pfu DNA polymerases were obtained from Promega and Stratagene, respectively. All enzymes were used according to the instructions of the manufacturer.
TABLE 1.
Cloning vectors and plasmids used in this work
| Plasmid | Properties | Source or reference |
|---|---|---|
| pSE380 | E. coli cloning vector; trc promoter upstream of MCSa and constitutive lac repressor gene downstream of MCS | Invitrogen |
| pBlueScriptII | E. coli cloning vector; lac promoter upstream of MCS | Stratagene |
| pTC6 | pBR322-based plasmid containing a fragment of K. pneumoniae genomic DNA including dhaD and dhaK | 29 |
| pNEA6 | 4.3-kb SalI-SacI fragment of pTC6 inserted into pSE380; contains dhaD | This work |
| pNEA10 | PCR-derived gldA of E. coli inserted into SalI-BamHI sites of pSE380; contains gldA | This work |
| pNEA11 | Frameshift mutation in gldA of pNEA10 | This work |
| pNEA14 | PCR-derived dhaD of K. pneumoniae inserted into EcoRI-XhoI sites of pSE380; contains dhaD | This work |
| pNEA16 | PCR-derived mgs of E. coli inserted into KpnI-SacI sites of pSE380; contains mgs | This work |
| pNEA30 | PCR-derived mgs of E. coli inserted into KpnI-SacI sites of pNEA10; contains mgs downstream of gldA | This work |
MCS, multiple cloning site.
TABLE 2.
Primers used for PCR amplification
| Primer | Sequence | Restric-tion sitea |
|---|---|---|
| glda_5p | 5′-AGCTCGAGGAGGACTGGAATGCCGCATTTG-3′ | XhoI |
| glda_3p | 5′-CGGGATCCTGGAGTAGGTTATTCCCACTCTTG-3′ | BamHI |
| dhad_5p | 5′-GTCGAATTCCCGCACTTATTTGAGG-3′ | EcoRI |
| dhad_3p | 5′-GGTTTCTCGAGCGCGAATTAACG-3′ | XhoI |
| mgs_5p | 5′-AGCGGTACCTGCTTACAGTAATCTGTAGG-3′ | KpnI |
| mgs_3p | 5′-AACGAGCTCAAACAGGTGGCGGTTTG-3′ | SacI |
Underlined nucleotides represent the restriction sites.
The primers glda_5p and glda_3p were used to amplify the 1,179-bp gldA fragment from genomic E. coli DNA (strain MG1655). The gel-purified PCR fragment was inserted into the SalI-BamHI site of the pSE380 vector (Invitrogen, San Diego, Calif.) to give pNEA10. The resulting plasmid contained gldA under the control of the trc promoter, allowing for high-level expression inducible by IPTG. The fidelity of the PCR was checked by sequencing the entire insert (University of Wisconsin Biotechnology Center—Sequencing Services). A control plasmid (pNEA11) containing a frameshift mutation in glycerol dehydrogenase was constructed by digesting pNEA10 with SalI and then filling in the overhang by using T4 DNA polymerase. The vector was then blunt-end ligated to itself.
The plasmid pTC6 contains a segment of Klebsiella pneumoniae (ATCC 25955) genomic DNA, including dhaD and dhaK (29). A segment of 4.3 kb containing the dhaD gene was obtained by digesting pTC6 with SacI and SalI, and the segment was inserted into the multiple cloning site of pSE380 to give plasmid pNEA6. The complete sequence of the dha regulon is reported in a recent patent on the production of 1,3-PD in E. coli (18). The primers dhad_5p and dhad_3p were used to amplify the 1,148-bp dhaD fragment from genomic K. pneumoniae (ATCC 25955). The gel-purified PCR fragment was inserted at the EcoRI and XhoI sites of pSE380 to obtain pNEA14. The primers mgs_5p and mgs_3p were used to amplify the 549-bp mgs fragment from genomic E. coli DNA. The gel-purified PCR fragment was inserted at the KpnI and SacI restriction sites of pSE380 to obtain pNEA16. The fidelity of the PCR was checked by sequencing the mgs insert. The primers were designed based on the mgs sequence that we obtained from David Harrison at the Medical College of Wisconsin (Milwaukee, Wis.). The sequence later became available from GenBank (accession no. AE000198). Plasmid pNEA30 was constructed by inserting the mgs gene into the KpnI and SacI sites of pNEA10 to obtain a plasmid that cotranscribes both mgs and gldA from the trc promoter.
Source of chemicals.
R-, S-, and RS-lactaldehyde were synthesized by the reaction of ninhydrin with l-, d-, and dl-threonine, respectively, by slight modification of the method of Zagalak et al. (34). A 100 mM sodium phosphate buffer at pH 5.8 replaced the citrate buffer in order to avoid the presence of any organic compound other than lactaldehyde. The lactaldehyde concentration was estimated by the reduction to 1,2-PD with KBH4 and then measuring the 1,2-PD concentration by high-performance liquid chromatography (HPLC) as described below. R-1,2-PD, produced and purified from a T. thermosaccharolyticum fermentation (7, 26), was from laboratory stock. Racemic 1,2-PD, S-1,2-PD, and MG were purchased from Sigma Chemicals (St. Louis, Mo.). The MG was purified by collecting fractions from the HPLC column as described below. Acetol was purchased from TCI America (Portland, Oreg.).
Preparation of cell extracts.
Cells were grown anaerobically in 150-ml standard production medium to late exponential phase and were collected by centrifugation at 3,000 × g for 10 min at 4°C with a Beckman (Fullerton, Calif.) model J2-21 centrifuge. The cell pastes were washed twice in 100 mM potassium phosphate buffer at pH 7.2. The washed cell pastes were resuspended in 1 to 2 ml of the appropriate assay resuspension buffer. The cells were then disrupted by sonication on ice for 5 min at a duty cycle of 70% with 1-s cycles. The cell debris was removed by centrifugation in a microcentrifuge at 16,000 × g for 15 min.
HPLC analysis.
The fermentation products were quantified with a Waters Alliance Integrity system (Milford, Mass.) equipped with a refractive index detector, a photodiode array detector, and an Aminex HPX-87H (Bio-Rad, Hercules, Calif.) organic acids column. The mobile phase was a 0.01 N sulfuric acid solution. The flow rate was 0.5 ml per min, and the column temperature was 40°C. Compounds were identified by coelution with authentic standards. A secondary identification of compounds, as well as the purification of MG, was done with a Waters Sugar Pak II column at 90°C with water as the mobile phase at a flow rate of 0.5 ml per min and using the refractive index detector. Prior to analysis, all samples were filtered through 0.45-μm-pore-size membranes (Gelman Sciences, Ann Arbor, Mich.).
Purification of 1,2-PD from fermentation broth.
1,2-PD was purified by rotary evaporation as described previously (25). An initial volume of 150 ml of fermentation broth was reduced to approximately 5 ml.
Determination of the enantiomeric purity of 1,2-PD.
The enantiomeric purity of the 1,2-PD was determined by gas chromatography with a chiral capillary column (Chirasil-DEX CB capillary column; Chrompack Inc., Raritan, N.J.). Prior to injection on the column, the fermentation broth was purified and then diluted to contain approximately 1 g of 1,2-PD per liter of methanol. A 1-μl sample was injected into the column at a temperature of 75°C. The carrier was helium at 60 lb/in2 and the injector split was 100:1 with an injector temperature of 250°C. The products were detected with a flame ionization detector at 275°C.
Assays.
Glycerol dehydrogenase activity was assayed by measuring either the initial rate of reduction of NAD+ or the initial rate of oxidation of NADH with a spectrophotometer (Varian Carry-1 Bio, Sugar Land, Tex.) at a wavelength of 340 nm and a constant temperature of 37°C. A total of 3.0 ml of 100 mM potassium phosphate buffer at pH 7.2 for the reduction assay or 3.0 ml of 100 mM sodium carbonate-bicarbonate buffer at pH 9.0 for the oxidation assay was added to quartz cuvette cells. The buffer was allowed to equilibrate to the assay temperature. A total of 50 μl of 10 mg of NADH per ml of potassium phosphate buffer was added to the assay mixture for the substrate reduction assay. A total of 100 μl of 30 mg of NAD+ per ml of sodium carbonate-bicarbonate buffer was added to the assay mixture for the substrate oxidation assay. The background activity was recorded after the addition of 1 to 10 μl of cell extracts, prepared as described above, to the assay solution. Assays were initiated by the addition of one of the following substrates: glycerol, 1,2-PD, or acetol. The assay mixtures were stirred with microstirrers during the assays.
For the enzyme assays, 1 U was defined as the number of micromoles of NAD+ reduced or NADH oxidized per minute at 37°C. Total protein concentrations in cell extracts were determined by using the Bradford assay (Bio-Rad) with bovine serum albumin as the standard.
Purified glycerol dehydrogenase from K. pneumoniae was obtained from Boehringer Mannheim (Indianapolis, Ind.). A mixture containing approximately 1 g of substrates per liter of 100 mM potassium phosphate buffer (pH 7.2), with 1.5 U of enzyme and 42 mM of NADH, was incubated at 37°C for 6 h. Control assays contained no NADH or no enzyme. Products were identified by HPLC as described above. This assay was also done with 100 μl of E. coli AG1::pSE380 and AG1::pNEA10 cell extracts instead of the purified glycerol dehydrogenase. Cells were grown in 150 ml of standard production medium to late exponential phase, and cell extracts were prepared as described above.
MG synthase activity was assayed as described by Hopper and Cooper (15).
RESULTS
Overexpression of glycerol dehydrogenase in E. coli.
E. coli strains overexpressing the glycerol dehydrogenase gene from E. coli (gldA) or from K. pneumoniae (dhaD) produced 1,2-PD as a product of glucose fermentation. Properties of E. coli AG1 expressing gldA grown in 150-ml anaerobic flasks are shown in Table 3. The results indicate that the expression of glycerol dehydrogenase leads to the production of 1,2-PD, which increased with the addition of the inducer IPTG. The three different oxidoreductase activities were measured in crude extracts of the cells from these fermentations (Table 3). Three activities were measured because glycerol dehydrogenase is known to have a broad substrate range and various assays of its activity have been reported (24, 28). The activities were higher in the strains transformed with pNEA10 than with the control plasmid and further increased in the induced case. The leaky expression of genes was expected and observed with pSE380-based plasmids, since the trc promoter is very strong and not tightly repressed.
TABLE 3.
1,2-PD titers and oxidoreductase and MG synthase activities for glucose fermentations with transformed E. coli AG1 strainsa
| Plasmid | IPTG added (mM) | 1,2-PD (g/liter) | Sp act (U/mg of protein) ofb:
|
|||
|---|---|---|---|---|---|---|
| Acetol | Glycerol | 1,2-PD | DHAPc | |||
| pSE380 | 0 | 0.01 | 0.02 | 0.10 | 0.08 | 0.2 |
| pNEA10 | 0 | 0.10 | 0.16 | 0.48 | 0.32 | – |
| 0.10 | 0.22 | 0.88 | 2.69 | 2.35 | – | |
| pNEA30 | 0 | 0.36 | 0.13 | – | – | – |
| 0.10 | 0.70 | 0.92 | – | – | 5.7 | |
Plasmid pSE380 is the control; pNEA10 encodes gldA; pNEA30 encodes gldA and mgs. All cells were grown in 300-ml anaerobic flasks, and IPTG was added after 12 h of fermentation. Specific oxidoreductase activities for the reduction of acetol and the oxidation of glycerol and racemic 1,2-PD were determined.
–, not measured.
MG synthase activities were determined in a separate experiment with DHAP as the substrate.
E. coli AG1 transformed with plasmids containing the glycerol dehydrogenase gene from K. pneumoniae (dhaD) (either pNEA6 or pNEA14) also produced 1,2-PD (Table 4). As with gldA, some 1,2-PD was produced in the absence of the inducer. The titers of 1,2-PD increased when the inducer was added and were comparable to the titers obtained with gldA (Table 3). We do not know why pNEA6 and pNEA14 gave somewhat different results (Table 4).
TABLE 4.
1,2-PD titers for glucose fermentations of E. coli AG1 transformed with various plasmidsa
| Plasmid | Expressed gene | IPTGb | 1,2-PD (g/liter) |
|---|---|---|---|
| pNEA6 | dhaD | − | 0.24 ± 0.01 |
| + | 0.34 ± 0.02 | ||
| pNEA14 | dhaD | − | 0.13 ± 0.01 |
| + | 0.25 ± 0.02 | ||
| pNEA16 | mgs | − | 0.19 ± 0.01 |
| + | 0.26 ± 0.01 |
Fermentations were carried out in the standard production medium in 10-ml Hungate tubes for 24 h at 37°C. Results are given as averages ± standard deviation of three replicates.
IPTG (0.1 mM) was added to the medium at 12 h (+) or was withheld (−).
Determination of the enantiomeric purity of the 1,2-PD.
The enantiomeric purity of the 1,2-PD produced by the engineered E. coli strains was determined by gas chromatography with a chiral capillary column. Samples of 1,2-PD purified from fermentation broth gave a single peak. By spiking the samples with R- or S-1,2-PD, we determined that the fermentation product was R-1,2-PD. Since no S-1,2-PD was detected, the enantiomeric purity of the R-1,2-PD must be nearly 100%. We detected only R-1,2-PD in fermentations of all our engineered E. coli strains.
Determination of the metabolic pathway to R-1,2-PD in the transformed E. coli.
To elucidate the pathway for 1,2-PD production in E. coli expressing glycerol dehydrogenase genes, we carried out in vitro studies of glycerol dehydrogenase and E. coli cell extracts with several potential intermediates of the 1,2-PD pathway. A commercial preparation of K. pneumoniae glycerol dehydrogenase (Boehringer Mannheim) or E. coli AG1::pSE380 cell extracts were incubated with MG, R-lactaldehyde, S-lactaldehyde, or acetol, and the resulting products were determined by HPLC. The results are summarized in Table 5. For example, when incubated with glycerol dehydrogenase MG was converted almost completely to lactaldehyde, with only a small amount further reduced to 1,2-PD. When more NADH and enzyme were added, no additional lactaldehyde was converted and no additional 1,2-PD was produced. The results in Table 5 indicate that the route to R-1,2-PD in our recombinant E. coli probably involves the reduction of MG to R-lactaldehyde by the overexpressed glycerol dehydrogenase followed by the reduction of the R-lactaldehyde to R-1,2-PD by a native E. coli activity.
TABLE 5.
In vitro studies to elucidate the pathway to R-1,2-PD in engineered E. colia
| Substrate | Incubation of substrate with glycerol dehydrogenase
|
Incubation of substrate with cell extracts
|
||||
|---|---|---|---|---|---|---|
| Cofactor added | Product | % Conversion | Cofactor added | Product | % Conversion | |
| MG | NADH | Lactaldehyde | 95 | NADH | Lactaldehyde | 35 |
| 1,2-PD | <5 | 1,2-PD | 40 | |||
| MG | None | None detected | No conversion | |||
| R-lactaldehyde | NADH | None detected | No conversion | NADH | 1,2-PD | 75 |
| S-lactaldehyde | NADH | 1,2-PD | 100 | NADH | 1,2-PD | 90 |
| S-lactaldehyde | None | None detected | No conversion | |||
| Acetol | NADH | 1,2-PD | 100 | NADH | 1,2-PD | 100 |
| Acetol | None | None detected | No conversion | |||
Various substrates at approximately 1-g-per-liter concentrations were incubated with purified glycerol dehydrogenase or E. coli AG1::pSE380 cell extracts. The percentages of conversion were measured by HPLC as described in Materials and Methods. For controls, substrates were incubated without the cofactor or without the enzyme, and no substrate degradation was measured. For cell extracts, substrates were incubated with the cofactor.
Overexpression of E. coli MG synthase in E. coli.
MG is an important intermediate in the 1,2-PD pathway. Therefore, we investigated the influence of overexpression of the E. coli MG synthase gene (mgs) on 1,2-PD production. Our primary goal was to further enhance the production of 1,2-PD in strains already overexpressing a glycerol dehydrogenase gene. However, we found that E. coli AG1 transformed with pNEA16, a plasmid containing the E. coli mgs gene under the control of the trc promoter, produced levels of 1,2-PD similar to those produced by strains expressing gldA or dhaD alone (Table 4).
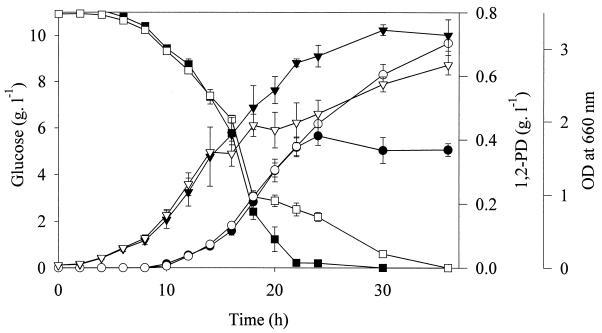
We also constructed a plasmid in which mgs and gldA were coexpressed (pNEA30). The coexpression of both genes resulted in much improvement of the 1,2-PD yield and titers (Table 3). Time courses for two sets of batch fermentations with E. coli AG1 containing pNEA30 are shown in Fig. 1. When no IPTG was added to the fermentation medium, approximately 0.4 g of 1,2-PD per liter was produced, and the glucose was depleted by 30 h. When 0.1 mM of IPTG was added 12 h into the fermentation, the glucose consumption was slowed, and 0.7 g of 1,2-PD per liter was produced by 36 h. The addition of IPTG slowed cell growth, but the final OD at 660 nm in both cases was approximately 3.
FIG. 1.
Time course of two sets of 150-ml anaerobic batch fermentations of E. coli carrying pNEA30 expressing the genes for E. coli glycerol dehydrogenase (gldA) and E. coli MG synthase (mgs). Concentrations in the flasks are shown for glucose (■), 1,2-PD (●), and cells as optical density (▾) with no IPTG addition and for glucose (□), 1,2-PD (○), and cells as optical density (▿) with 0.1 mM IPTG added at 12 h after inoculation. Error bars are standard deviations of three replicates.
To confirm the functional expression of MG synthase, its activity was measured in strains transformed with pNEA16 and pNEA30. The MG synthase activity was found to be over 25-fold greater than the 0.2 U of total protein per mg measured with strain AG1 transformed with a control plasmid, pSE380.
Influence of the amount of IPTG added and the timing of its addition on 1,2-PD production.
In initial screening of fermentations with E. coli AG1::pNEA10 expressing gldA, we found that when 0.05 mM or more of IPTG was added at the time of inoculation, very little cell growth or product formation occurred (the OD at 660 nm was less than 0.3 and the total fermentation products amounted to less than 0.2 g of products per liter). We also observed that increased IPTG levels increased enzyme activity over fivefold but increased 1,2-PD titer and yields only slightly more than twofold (Table 3). To further investigate the effect of IPTG on 1,2-PD production, fermentations were run with E. coli AG1::pNEA30 expressing both gldA and mgs in which either the IPTG level or the time of addition were varied (Table 6). Cell growth and product formation were very sensitive to IPTG added at the time of inoculation. Growth and production were good for 0.001 mM of IPTG but dropped significantly for 0.05 and 0.1 mM. In a separate experiment, also shown in Table 6, a fixed amount of IPTG (0.1 mM) was added at different times after inoculation. The later the induction, the better the growth and product formation. The inducer was not added after 12 h because by then most of the glucose had been consumed and the fermentation was nearly complete.
TABLE 6.
Effect of amount of IPTG and time of addition on 1,2-PD production and OD for E. coli AG1::pNEA30a
| IPTG (mM) | Time addedb (h) | Residual glucose (g/liter) | 1,2-PD (g/liter) | OD660 |
|---|---|---|---|---|
| 0.0 | 1.39 ± 0.55 | 0.44 ± 0.02 | 1.78 ± 0.12 | |
| 0.001 | 0 | 1.39 ± 0.77 | 0.56 ± 0.04 | 1.64 ± 0.06 |
| 0.05 | 0 | 9.07 ± 0.11 | 0.03 ± 0.01 | 0.42 ± 0.01 |
| 0.1c | 0 | 9.32 ± 0.18 | 0.03 ± 0.01 | 0.32 ± 0.03 |
| 0.1c | 0 | 8.82 ± 0.02 | 0.10 ± 0.01 | 0.36 ± 0.02 |
| 4 | 7.65 ± 0.27 | 0.16 ± 0.04 | 0.68 ± 0.01 | |
| 6 | 6.31 ± 0.26 | 0.24 ± 0.02 | 0.77 ± 0.11 | |
| 8 | 2.21 ± 0.05 | 0.25 ± 0.02 | 0.98 ± 0.20 | |
| 12 | 1.55 ± 0.22 | 0.49 ± 0.03 | 1.41 ± 0.25 |
Fermentations were carried out in the standard medium in 10-ml working Hungate tubes for 24 h at 37°C. Results are given as averages ± standard deviation of three replicates.
Time of IPTG addition after inoculation with an overnight culture.
Two separate experiments with 0.1 mM IPTG were performed.
DISCUSSION
We have shown that E. coli strains in which the genes for glycerol dehydrogenase, MG synthase, or both are overexpressed produce 1,2-PD as a fermentation product of glucose. In all cases we tested, the 1,2-PD was the R enantiomer. Coexpression of the glycerol dehydrogenase and MG synthase genes gave better results than when they were expressed individually.
We used two different glycerol dehydrogenase genes. The first gene, gldA from E. coli, is repressed in native E. coli during anaerobic glucose fermentation (16, 31). The second, dhaD, is involved in anaerobic glycerol metabolism in K. pneumoniae (10). E. coli strains overexpressing either gene produced 1,2-PD with similar titers. The fact that both genes gave similar results was expected, since the genes have a high level of homology (64% DNA sequence identity as determined by the Wisconsin Package [Genetics Computer Group, Madison, Wis.]) and both enzymes have similar catalytic properties (20, 27, 28).
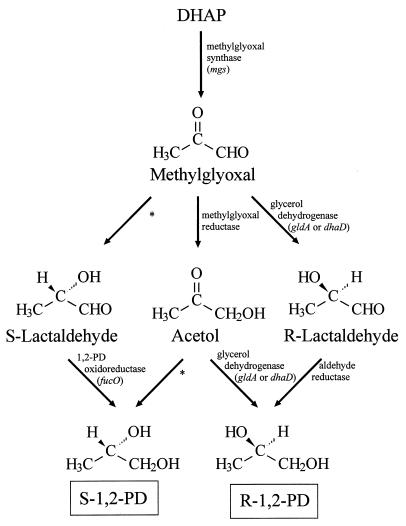
Experiments to elucidate the biochemical pathways to 1,2-PD in the recombinant strains were carried out. Several possible routes are shown in Fig. 2. Since in all cases only R-1,2-PD was detected, only routes to this enantiomer are discussed here. E. coli is well known to convert dihydroxyacetone phosphate (DHAP) to MG via the action of MG synthase (8, 15). Normally, MG is converted to d-lactate via the glyoxalase system (8). We propose that in our recombinant strains some of the MG is converted to R-1,2-PD. One route for this conversion is the reduction of the aldehyde group of MG to give acetol, followed by the stereospecific reduction of the ketone group of acetol to give R-1,2-PD. This route has been proposed for T. thermosaccharolyticum (7). A second route is the stereospecific reduction of the ketone group of MG to give R-lactaldehyde, followed by the reduction of the aldehyde group of R-lactaldehyde to give R-1,2-PD. This route has been shown in Clostridium sphenoides (30).
FIG. 2.
Metabolic pathways to the enantiomers of 1,2-PD from the common precursor MG. In our engineered pathway, MG synthase converts DHAP to MG, which is then reduced to R-lactaldehyde by glycerol dehydrogenase. A native E. coli enzyme is responsible for the reduction of R-lactaldehyde to R-1,2-PD. ∗, possible activity, but not yet reported in literature.
Based on the literature and our experimental results, the most likely pathway to R-1,2-PD in our E. coli strains expressing the glycerol dehydrogenase genes involves R-lactaldehyde as the intermediate. We propose that the recombinant glycerol dehydrogenase reduces MG to R-lactaldehyde, which is reduced by a native E. coli activity to R-1,2-PD. This route is consistent with our finding that glycerol dehydrogenase reduces MG to lactaldehyde rather than acetol. We deduce that the lactaldehyde is the R-enantiomer because R-1,2-PD is the ultimate fermentation product and because the purified glycerol dehydrogenase reduces S-lactaldehyde but not the lactaldehyde formed when the enzyme reduces MG. Since glycerol dehydrogenase does not reduce R-lactaldehyde, a native E. coli activity must carry out this reduction. We do not know the identity of this enzyme, but one possibility is alcohol dehydrogenase. Alcohol dehydrogenase reduces the aldehyde group of acetaldehyde to give ethanol (12), and catalyzes the oxidation of 1,2-PD with NAD+ (23). A well-studied enzyme in E. coli, 1,2-PD oxidoreductase, reduces S-lactaldehyde to S-1,2-PD; this is probably not the activity in question, however, as it does not reduce R-lactaldehyde (4).
MG synthase converts DHAP, a glycolytic intermediate, to MG. Cells normally utilize the glyoxalase system to convert the small amounts of MG formed under physiological conditions to d-lactate. When we overexpressed MG synthase in E. coli, 1,2-PD was produced and the titers were as high as those obtained with glycerol dehydrogenase expression. This indicates that in addition to R-lactaldehyde reductase activities, E. coli has a native MG reductase activity.
In order to improve the 1,2-PD titers obtained with the expression of either glycerol dehydrogenase or MG synthase, we constructed pNEA30 to express both genes. This construct improved 1,2-PD titers to approximately 0.7 g per liter from the approximately 0.2 g per liter obtained with the expression of either enzyme alone. We also tested the coexpression of dhaD and mgs in a different construct, but since the 1,2-PD yield was lower than with pNEA30 (data not shown), we used pNEA30 for further studies.
We observed that the amount of IPTG and the timing of its addition were important factors for achieving elevated 1,2-PD titers. For the previously reported aldose reductase system, IPTG could be added at the time of inoculation, and the addition of IPTG at later times did not improve 1,2-PD production (25). We found that the early addition of IPTG, and consequently the early induction of glycerol dehydrogenase from pNEA10, inhibited cell growth. Further induction studies performed with pNEA30, a plasmid containing both glycerol dehydrogenase and MG synthase, showed that only very small amounts of IPTG could be added at the time of inoculation without inhibiting cell growth. The inhibition of cell growth may be caused by the toxicity of expressed genes; moreover, the early expression of MG synthase may lead to accumulation of toxic levels of MG. We found that the best 1,2-PD productions were obtained when IPTG was added during the late growth phase.
The effect of IPTG addition at 12 h on cell growth and 1,2-PD production is shown in Fig. 1. The addition of IPTG resulted in slower cell growth. Although glucose consumption was also slowed down after the addition of IPTG, the rate of 1,2-PD production remained the same as in the case of no IPTG addition. The addition of the inducer increased the yield of 1,2-PD on consumed glucose, resulting in higher final 1,2-PD titers.
We used the pSE380 expression system for our constructions. Vector pSE380 contains the strong regulated trp-lac fusion promoter (trc promoter). The promoter is very strong and it is expected that even uninduced cells may have a low level of expression. This may be the reason for the low level of expression of gldA we observe with uninduced cultures (Table 3). This low level of expression is sufficient for 1,2-PD production. We have also used a maximum of 0.1 mM IPTG for inducing higher expression levels, which is at least 10-fold lower than the 1 to 5 mM IPTG used to fully induce pSE380-based plasmids. A high level of IPTG is used only when overexpression of proteins is desired for purification or other purposes. The gldA expression level we observed with low levels of IPTG-induced cultures is sufficient to obtain a compromise between sustained cell growth and production of 1,2-PD.
The results in this article demonstrate a significant improvement in the production of 1,2-PD compared to a previously reported engineered E. coli system expressing aldose reductase and MG synthase (25). Our titers are also higher than those for the first reports of the production of a related compound, 1,3-PD, in a recombinant E. coli (around 0.1 g of 1,3-PD per liter) (18). In metabolic engineering, the initial step is to construct a functional pathway to the desired product. Further work is then required to improve the production of the product. The prospect for improving the production of 1,2-PD is good. E. coli is capable of growth with up to 120 g of exogenously added 1,2-PD per liter (6). The maximum theoretical yield of 1,2-PD for anaerobic fermentation is 0.51 g of 1,2-PD per g of glucose consumed (6). There are several obvious strategies for improving 1,2-PD production. For example, it should be possible to enhance the native E. coli activity for the reduction of R-lactaldehyde with an overexpressed heterologous activity and thus overexpress all the activities in the pathway from DHAP. It should also be possible to modify E. coli to be a better host strain for 1,2-PD production, such as by deleting genes for competing pathways. We expect that with further metabolic and bioprocess engineering, fermentation will provide an important route to 1,2-PD from renewable resources.
ACKNOWLEDGMENTS
This work was partially supported by the Wisconsin Center for Dairy Research and the Environmental Protection Agency grant R824726.
REFERENCES
- 1.Anonymous. Propylene glycol: chemical profile. Chem Market Rep. 1998;254:33. [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1994. [Google Scholar]
- 3.Boronat A, Aguilar J. Rhamnose-induced propanediol oxidoreductase in Escherichia coli: purification, properties, and comparison with the fucose-induced enzyme. J Bacteriol. 1979;140:320–326. doi: 10.1128/jb.140.2.320-326.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boronat A, Aguilar J. Metabolism of l-fucose and l-rhamnose in Escherichia coli: differences in induction of propanediol oxidoreductase. J Bacteriol. 1981;147:181–185. doi: 10.1128/jb.147.1.181-185.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bullock W O, Fernandez J M, Short J M. XL1-Blue: a high efficiency plasmid transformation recA Escherichia coli strain with beta-galactosidase selection. BioTechniques. 1987;5:376–378. [Google Scholar]
- 6.Cameron D C, Altaras N E, Hoffman M L, Shaw A J. Metabolic engineering of propanediol pathways. Biotechnol Prog. 1998;14:116–125. doi: 10.1021/bp9701325. [DOI] [PubMed] [Google Scholar]
- 7.Cameron D C, Cooney C L. A novel fermentation: the production of (R)-1,2-propanediol and acetol by Clostridium thermosaccharolyticum. Bio/Technology. 1986;4:651–654. [Google Scholar]
- 8.Cooper R A. Metabolism of methylglyoxal in microorganisms. Annu Rev Microbiol. 1984;38:49–68. doi: 10.1146/annurev.mi.38.100184.000405. [DOI] [PubMed] [Google Scholar]
- 9.Ferguson G P, Chacko A D, Lee C, Booth I R. The activity of the high-affinity K+ uptake system Kdp sensitizes cells of Escherichia coli to methylglyoxal. J Bacteriol. 1996;178:3957–3961. doi: 10.1128/jb.178.13.3957-3961.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Forage R G, Lin E C C. dha system mediating aerobic and anaerobic dissimilation of glycerol in Klebsiella pneumoniae NCIB 418. J Bacteriol. 1982;151:591–599. doi: 10.1128/jb.151.2.591-599.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fryzuk M D, Bosnich B. Asymmetric synthesis. An asymmetric homogeneous hydrogenation catalyst which breeds its own chirality. J Am Chem Soc. 1978;100:5491–5494. [Google Scholar]
- 12.Goodlove P E, Cunningham P R, Parker J, Clark D P. Cloning and sequence analysis of the fermentative alcohol-dehydrogenase-encoding gene of Escherichia coli. Gene. 1989;85:209–214. doi: 10.1016/0378-1119(89)90483-6. [DOI] [PubMed] [Google Scholar]
- 13.Hacking A J, Lin E C C. Disruption of the fucose pathway as a consequence of genetic adaptation to propanediol as a carbon source in Escherichia coli. J Bacteriol. 1976;126:1166–1172. doi: 10.1128/jb.126.3.1166-1172.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hanahan D. Studies on the transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 15.Hopper D J, Cooper R A. The regulation of Escherichia coli methylglyoxal synthase: a new control site in glycolysis? FEBS Lett. 1971;13:213–216. doi: 10.1016/0014-5793(71)80538-0. [DOI] [PubMed] [Google Scholar]
- 16.Jin R Z, Tang J C T, Lin E C C. Experimental evolution of a novel pathway for glycerol dissimilation in Escherichia coli. J Mol Evol. 1983;19:429–436. doi: 10.1007/BF02102318. [DOI] [PubMed] [Google Scholar]
- 17.Kometani T, Morita Y, Furui H, Yoshii H, Matsuno R. Preparation of chiral 1,2-alkanediols with baker’s yeast-mediated oxidation. Chem Lett. 1993;12:2123–2124. [Google Scholar]
- 18.Laffend, L. A., V. Nagarajan, and C. E. Nakamura. November 1996. Bioconversion of a fermentable carbon source to 1,3-propanediol by a single microorganism. Patent Cooperation Treaty (PCT) Int. Appl. WO9635796.
- 19.Lamed R J, Zeikus J G. Glucose fermentation pathway of Thermoanaerobium brockii. J Bacteriol. 1980;141:1251–1257. doi: 10.1128/jb.141.3.1251-1257.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee L G, Whitesides G M. Preparation of optically active 1,2-diols and α-hydroxy ketones using glycerol dehydrogenase as catalyst: limits to enzyme catalyzed synthesis due to noncompetitive and mixed inhibition by product. J Org Chem. 1986;51:25–36. [Google Scholar]
- 21.Lenth C W, Du Puis R N. Polyhydric alcohol production by hydrogenolysis of sugars in the presence of copper-aluminum oxide. Indust Eng Chem. 1945;37:152–157. [Google Scholar]
- 22.Levene P A, Walti A. l-Propylene glycol. In: Blatt A H, editor. Organic syntheses collective. 2. J. New York, N.Y: Wiley & Sons, Inc.; 1943. pp. 545–547. [Google Scholar]
- 23.Machate T, Kettrup A. Spectrophotometric method for the determination of 1,2-propylene glycol. Fresenius Z Anal Chem. 1998;360:137–138. [Google Scholar]
- 24.Marshall J H, May J W, Sloan J. Purification and properties of glycerol:NAD+ 2-oxidoreductase (glycerol dehydrogenase) from Schizosaccharomyces pombe. J Gen Microbiol. 1985;131:1581–1588. doi: 10.1099/00221287-135-3-697. [DOI] [PubMed] [Google Scholar]
- 25.Shaw A J. Metabolic engineering of methylglyoxal metabolism in Escherichia coli. Ph.D. thesis. University of Wisconsin, Madison; 1997. [Google Scholar]
- 26.Simon E S, Whitesides G M, Cameron D C, Weitz D J, Cooney C L. A combined microbial/chemical synthesis of (+)-(R)-methyloxirane having high enantiomeric excess. J Org Chem. 1987;52:4042–4044. [Google Scholar]
- 27.Tang C-T, Ruch F E, Jr, Lin E C C. Purification and properties of a nicotinamide adenine dinucleotide-linked dehydrogenase that serves an Escherichia coli mutant for glycerol catabolism. J Bacteriol. 1979;140:182–187. doi: 10.1128/jb.140.1.182-187.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tang J C T, Forage R G, Lin E C C. Immunochemical properties of NAD+-linked glycerol dehydrogenases from Escherichia coli and Klebsiella pneumoniae. J Bacteriol. 1982;152:1169–1174. doi: 10.1128/jb.152.3.1169-1174.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tong I-T. Microbial production of 1,3-propanediol in Escherichia coli: a model system for metabolic engineering. Ph.D. thesis. University of Wisconsin, Madison; 1992. [Google Scholar]
- 30.Tran-Din K, Gottschalk G. Formation of d(−)-1,2-propanediol and d(−)-lactate from glucose by Clostridium sphenoides under phosphate limitation. Arch Microbiol. 1985;142:87–92. [Google Scholar]
- 31.Truniger V, Boos W. Mapping and cloning of gldA, the structural gene of the Escherichia coli glycerol dehydrogenase. J Bacteriol. 1994;176:1796–1800. doi: 10.1128/jb.176.6.1796-1800.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vander Jagt D L, Robinson B, Taylor K K, Hunsaker L. Reduction of trioses by NADPH-dependent aldo-keto reductases. Aldose reductase, methylglyoxal and diabetic complications. J Biol Chem. 1992;267:4364–4369. [PubMed] [Google Scholar]
- 33.Wong C-H. Enzymes in synthetic organic chemistry. London, United Kingdom: Elsevier Science Ltd.; 1994. [Google Scholar]
- 34.Zagalak B, Frey P A, Karabatsos G L, Abeles R H. The stereochemistry of the conversion of d and l 1,2-propanediols to propionaldehyde. J Biol Chem. 1966;241:3028–3035. [PubMed] [Google Scholar]