LETTER
To bolster coronavirus disease 2019 (COVID-19) pandemic mitigation efforts, the U.S. Food and Drug Administration (FDA) issued an emergency use authorization (EUA) for easy-to-use rapid antigen (Ag) tests for the diagnosis and surveillance of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection (1, 2). Unlike sensitive molecular tests that detect multiple SARS-CoV-2 genes, antigen tests target a singular yet genetically conserved nucleocapsid viral protein (3–6). As the pandemic continues, some hypothesized that the performance of available antigen tests may differ between SARS-CoV-2 variants. As of February 2022, the most recent SARS-CoV-2 strains declared variants of concern (VoC) by the U.S. Centers for Disease Control and Prevention are Omicron (strain B.1.1.529) and Delta (strain B.1.617.2) (7). Beyond striking amino acid mutations in the spike protein, Omicron also harbors P13L, Δ31–33, R203K, and G204R nucleocapsid mutations, while Delta strains carry D63G, R203M, and D377Y nucleocapsid mutations (8, 9). However, the limits of detection (LoDs) of many FDA EUA antigen tests were established with gamma-irradiated or heat-inactivated preparations of the USA WA1/2020 (WA1) strain (10) lacking these nucleocapsid mutations. This includes at-home lateral flow tests like the BinaxNOW COVID-19 Ag card (Abbott Diagnostics Scarborough, Inc., Scarborough, ME), the CareStart COVID-19 antigen home test (Access Bio, Inc., Somerset, NJ), and the GenBody Covid-19 Ag test (GenBody, Inc., Chungcheongnam-do, Republic of Korea) and also the LumiraDx SARS-CoV-2 Ag test (LumiraDx UK Ltd., Alloa, Great Britain), a microfluidic immunofluorescence assay for clinical laboratory testing (11–14). In the present study, we used cultured plaque-titered live Omicron, Delta, and WA1 viruses to assess differences in the LoDs with the BinaxNOW, CareStart, GenBody, and LumiraDx tests.
The titers of the Omicron lh01 (NCBI accession number OL719310), Delta (BEI Resources catalog number NR-55671, isolate hCoV-19/USA/MD-HP05285/2021; Johns Hopkins University), and WA1 (10) viruses were determined by a plaque assay (10) and further calibrated with the Abbott RealTime SARS-CoV-2 assay (Abbott Molecular, Inc., Des Plaines, IL) (15). The genomes of the Omicron, Delta, and WA1 viral stocks used in our analysis were also sequenced using the NEBNext ARTIC SARS-CoV-2 companion kit (New England BioLabs, Ipswich, MA) and MinION (Oxford Nanopore Technologies, Oxford, UK) technology (16–20), confirming the lack of mutation acquisition during propagation.
For LoD evaluation, 10-fold serial dilutions in phosphate-buffered saline (PBS) ranging from 2.5 × 104 to 2.5 PFU/mL were applied to swabs in 50-μL volumes and tested in triplicate according to the manufacturers’ instructions (11–14). SteriPack sterile polyester spun nasal swabs (catalog number 60566REVA; LumiraDx UK Ltd., Alloa, Great Britain) and iClean foam swabs (catalog number CY-FS742; Supera, Houston, TX) were used with the LumiraDx test. After identifying the lowest 10-fold dilution with three replicate positive tests, we iteratively tested 3-fold dilutions around this concentration until identifying the lowest dilution (the LoD) in which at least 19 of 20 replicates (≥95%) were positive.
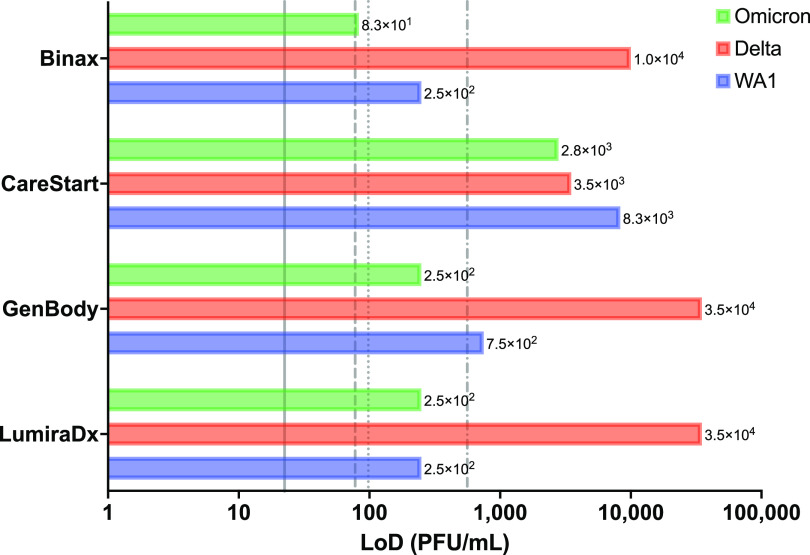
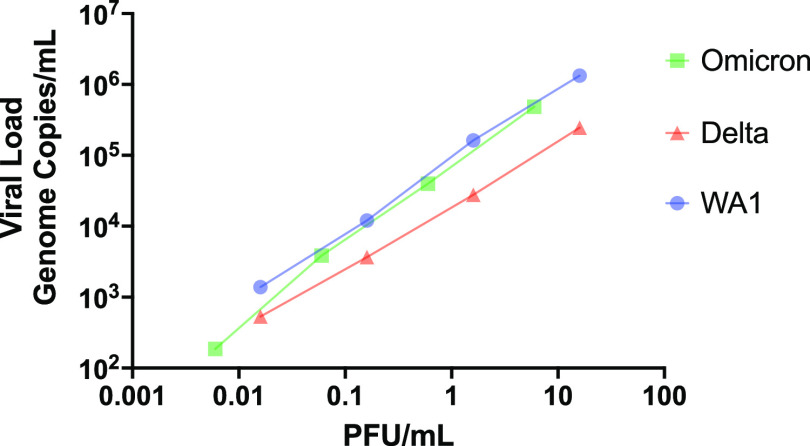
We found that Omicron had a 95% LoD threshold similar to or lower than that of WA1 for all four tests (Fig. 1). In contrast, the LoDs were 40- to 140-fold higher for Delta than for WA1 for every test examined except for CareStart (Fig. 1). The equal detection of all three variants by CareStart and the only relatively modest increase in the ratio of PFU to genome copies per milliliter for Delta (Fig. 2) suggest that this represents a true reduction in analytical sensitivity for Delta rather than an artifact of enhanced plaquing efficiencies for Delta relative to antigen levels and associated levels of genome copies. We previously found that the CareStart and LumiraDx antigen tests were excellent in the detection of presumptively WA1-infected individuals (15). We expect, however, that the observed magnitude of the loss in Delta sensitivity could result in a >20% loss in the detection of potentially infectious individuals based on our previous examination of the effect of LoD on clinical sensitivity (21). Nevertheless, the most infectious individuals should still be detected.
FIG 1.
Limits of detection of antigen tests. Shown are the limits of detection (LoDs) in PFU per milliliter determined in our analysis (bars). Vertical lines reference the manufacturer-reported LoDs in the respective instructions for use (IFU) documents (11–14), converted from 50% tissue culture infective doses (TCID50) per milliliter to PFU per milliliter by multiplying the TCID50 per milliliter by 0.7, a standard conversion based on the Poisson distribution, for BinaxNOW (dotted line) (1.4 × 102 TCID50/mL; 9.8 × 101 PFU/mL), CareStart (dashed and dotted line) (8.0 × 102 TCID50/mL; 5.6 × 102 PFU/mL), GenBody (dashed line) (1.1 × 102 TCID50/mL; 7.8 × 101 PFU/mL), and LumiraDx (solid line) (3.2 × 101 TCID50/mL; 2.2 × 101 PFU/mL).
FIG 2.
Correlation of PFU per milliliter and viral load in genome copies per milliliter. Stocks of each strain were serially diluted 10-fold in PBS and analyzed by PFU (10) and calibrated reverse transcription-quantitative PCR (RT-qPCR) assays (15). Both axes are on a log10 scale.
Of note, our use of live virus, analyte volume, and swab type may explain the slight discrepancy with the manufacturers’ determined LoDs. Our results for variant detection were also not completely consistent with similar reports, but these studies either fell short of the FDA’s EUA requirement of 20 LoD replicates, examined tests unavailable in the United States, and/or tested gamma-irradiated or heat-killed virus, inactivation processes which may artifactually affect test performance (22–24). In summary, we demonstrate that the rapid antigen tests evaluated detect Omicron effectively. However, our unexpected findings of decreased detection of Delta virus suggest that antigen test performance needs to be reevaluated for emerging variants to ensure that they still meet the intended public health testing goals of the pandemic.
ACKNOWLEDGMENTS
We thank Ginkgo Bioworks for providing CareStart and GenBody SARS-CoV-2 antigen test kits and also Abbott and LumiraDx for providing their respective SARS-CoV-2 antigen test kits used in this study.
Contributor Information
James E. Kirby, Email: jekirby@bidmc.harvard.edu.
Phyllis J. Kanki, Email: pkanki@hsph.harvard.edu.
Melissa B. Miller, UNC School of Medicine
REFERENCES
- 1.US Food and Drug Administration. 2022. In vitro diagnostics EUAs—antigen diagnostic tests for SARS-CoV-2. US Food and Drug Administration, Silver Spring, MD. https://www.fda.gov/medical-devices/coronavirus-disease-2019-covid-19-emergency-use-authorizations-medical-devices/in-vitro-diagnostics-euas-antigen-diagnostic-tests-sars-cov-2. Accessed 24 January 2022. [Google Scholar]
- 2.Centers for Disease Control and Prevention. 2022. Overview of testing for SARS-CoV-2, the virus that causes COVID-19. Centers for Disease Control and Prevention, Atlanta, GA. https://www.cdc.gov/coronavirus/2019-ncov/hcp/testing-overview.html. Accessed 24 January 2022. [Google Scholar]
- 3.Jegerlehner S, Suter-Riniker F, Jent P, Bittel P, Nagler M. 2021. Diagnostic accuracy of a SARS-CoV-2 rapid antigen test in real-life clinical settings. Int J Infect Dis 109:118–122. 10.1016/j.ijid.2021.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Allan-Blitz LT, Klausner JD. 2021. A real-world comparison of SARS-CoV-2 rapid antigen testing versus PCR testing in Florida. J Clin Microbiol 59:e01107-21. 10.1128/JCM.01107-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pray IW, Ford L, Cole D, Lee C, Bigouette JP, Abedi GR, Bushman D, Delahoy MJ, Currie D, Cherney B, Kirby M, Fajardo G, Caudill M, Langolf K, Kahrs J, Kelly P, Pitts C, Lim A, Aulik N, Tamin A, Harcourt JL, Queen K, Zhang J, Whitaker B, Browne H, Medrzycki M, Shewmaker P, Folster J, Bankamp B, Bowen MD, Thornburg NJ, Goffard K, Limbago B, Bateman A, Tate JE, Gieryn D, Kirking HL, Westergaard R, Killerby M, CDC COVID-19 Surge Laboratory Group. 2021. Performance of an antigen-based test for asymptomatic and symptomatic SARS-CoV-2 testing at two university campuses—Wisconsin, September-October 2020. MMWR Morb Mortal Wkly Rep 69:1642–1647. 10.15585/mmwr.mm695152a3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grifoni A, Sidney J, Zhang Y, Scheuermann RH, Peters B, Sette A. 2020. A sequence homology and bioinformatic approach can predict candidate targets for immune responses to SARS-CoV-2. Cell Host Microbe 27:671–680.e2. 10.1016/j.chom.2020.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Centers for Disease Control and Prevention. 2021. SARS-CoV-2 variant classifications and definitions. Centers for Disease Control and Prevention, Atlanta, GA. https://www.cdc.gov/coronavirus/2019-ncov/variants/variant-classifications.html#anchor_1632154493691. Accessed 14 March 2022. [Google Scholar]
- 8.Nextstrain. 2022. Genomic epidemiology of novel coronavirus—global subsampling. https://nextstrain.org/ncov/gisaid/global. Accessed 21 January 2022.
- 9.Suratekar R, Ghosh P, Niesen MJ, Donadio G, Anand P, Soundararajan V, Venkatakrishnan AJ. 2022. High diversity in Delta variant across countries revealed by genome‐wide analysis of SARS‐CoV‐2 beyond the spike protein. Mol Syst Biol 18:e10673. 10.15252/msb.202110673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harcourt J, Tamin A, Lu X, Kamili S, Sakthivel SK, Murray J, Queen K, Tao Y, Paden CR, Zhang J, Li Y, Uehara A, Wang H, Goldsmith C, Bullock HA, Wang L, Whitaker B, Lynch B, Gautam R, Schindewolf C, Lokugamage KG, Scharton D, Plante JA, Mirchandani D, Widen SG, Narayanan K, Makino S, Ksiazek TG, Plante KS, Weaver SC, Lindstrom S, Tong S, Menachery VD, Thornburg NJ. 2020. Severe acute respiratory syndrome coronavirus 2 from patient with coronavirus disease, United States. Emerg Infect Dis 26:1266–1273. 10.3201/eid2606.200516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abbott Diagnostics Scarborough, Inc. 2022. BinaxNOW COVID-19 Ag card—instructions for use. Abbott Diagnostics Scarborough, Inc, Scarborough, ME. https://www.fda.gov/media/141570/download. Accessed 21 January 2022. [Google Scholar]
- 12.Access Bio, Inc. 2021. CareStart COVID-19 antigen test—instructions for use. Access Bio, Inc, Somerset, NJ. https://www.fda.gov/media/142919/download. Accessed 21 January 2022. [Google Scholar]
- 13.GenBody, Inc. 2021. GenBody COVID-19 AG—instructions for use. GenBody, Inc, Chungcheongnam-do, Republic of Korea. https://www.fda.gov/media/150788/download. Accessed 19 February 2022. [Google Scholar]
- 14.LumiraDx UK Ltd. 2022. SARS-CoV-2 antigen (Ag) product insert. LumiraDx UK Ltd, Alloa, Great Britain. https://www.lumiradx.com/assets/pdfs/covid-19-antigen-test/sars-cov-2-antigen-product-insert/sars-cov-2-ag-test-strip-product-insert-eua.pdf?v=6. Accessed 21 January 2022. [Google Scholar]
- 15.Kirby JE, Riedel S, Dutta S, Arnaout R, Cheng A, Ditelberg S, Hamel DJ, Chang C, Kanki PJ. 2021. SARS-CoV-2 antigen tests predict infectivity based on viral culture: comparison of antigen, PCR viral load, and viral culture testing on a large sample cohort. medRxiv. 10.1101/2021.12.22.21268274. [DOI] [PMC free article] [PubMed]
- 16.ARTIC. 2022. The ARTIC field bioinformatics pipeline. https://github.com/artic-network/fieldbioinformatics. Accessed 23 March 2022.
- 17.Li H. 2018. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 34:3094–3100. 10.1093/bioinformatics/bty191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.ARTIC. 2022. ARTIC nanopore protocol for nCoV2019 novel coronavirus. https://github.com/artic-network/artic-ncov2019. Accessed 23 March 2022.
- 19.Aksamentov I, Roemer C, Hodcroft EB, Neher RA. 2021. Nextclade: clade assignment, mutation calling and quality control for viral genomes. J Open Source Softw 6:3773. https://joss.theoj.org/papers/10.21105/joss.03773. [Google Scholar]
- 20.Rambaut A, Holmes EC, O’Toole Á, Hill V, McCrone JT, Ruis C, du Plessis L, Pybus OG. 2020. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat Microbiol 5:1403–1407. 10.1038/s41564-020-0770-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arnaout R, Lee RA, Lee GR, Callahan C, Cheng A, Yen CF, Smith KP, Arora R, Kirby JE. 2021. The limit of detection matters: the case for benchmarking severe acute respiratory syndrome coronavirus 2 testing. Clin Infect Dis 73:e3042–e3046. 10.1093/cid/ciaa1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Regan J, Flynn JP, Choudhary MC, Uddin R, Lemieux J, Boucau J, Bhattacharyya RP, Barczak AK, Li JZ, Siedner MJ. 2022. Detection of the omicron variant virus with the Abbott BinaxNow SARS-CoV-2 rapid antigen assay. Open Forum Infect Dis 9:ofac022. 10.1093/ofid/ofac022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bekliz M, Perez-Rodriguez F, Puhach O, Adea K, Melancia SM, Baggio S, Corvaglia A-R, Jacquerioz-Bausch F, Alvarez C, Essaidi-Laziosi M, Escadafal C, Kaiser L, Eckerle I. 2022. Sensitivity of SARS-CoV-2 antigen-detecting rapid tests for Omicron variant. medRxiv. 10.1101/2021.12.18.21268018. [DOI] [PMC free article] [PubMed]
- 24.Deerain J, Druce J, Tran T, Batty M, Yoga Y, Fennell M, Dwyer DE, Kok J, Williamson DA. 2022. Assessment of the analytical sensitivity of ten lateral flow devices against the SARS-CoV-2 omicron variant. J Clin Microbiol 60:e02479-21. 10.1128/jcm.02479-21. [DOI] [PMC free article] [PubMed] [Google Scholar]