Abstract
Gene Ontology (GO) is widely used in the biological domain. It is the most comprehensive ontology providing formal representation of gene functions (GO concepts) and relations between them. However, unintentional quality defects (e.g. missing or erroneous relations) in GO may exist due to the large size of GO concepts and complexity of GO structures. Such quality defects would impact the results of GO-based analyses and applications. In this work, we introduce a novel evidence-based lexical pattern approach for quality assurance of GO relations. We leverage two layers of evidence to suggest potentially missing relations in GO as follows. We first utilize related concept pairs (i.e. existing relations) in GO to extract relationship-specific lexical patterns, which serve as the first layer evidence to automatically suggest potentially missing relations between unrelated concept pairs. For each suggested missing relation, we further identify two other existing relations as the second layer of evidence that resemble the difference between the missing relation and the existing relation based on which the missing relation is suggested. Applied to the 15 December 2021 release of GO, this approach suggested a total of 866 potentially missing relations. Local domain experts evaluated the entire set of potentially missing relations, and identified 821 as missing relations and 45 indicate erroneous existing relations. We submitted these findings to the GO consortium for further validation and received encouraging feedback. These indicate that our evidence-based approach can be utilized to uncover missing relations and erroneous existing relations in GO.
Keywords: Gene Ontology, ontology quality assurance, lexical patterns, missing relations, erroneous relations
Introduction
Ontologies are artifacts used to provide common controlled knowledge representation enabling knowledge sharing and reasoning in a particular domain. An ontology contains a set of classes (or concepts) that represent entities in a domain and a set of relations that define the semantic relations between the classes [1]. Ontologies have been extensively used in biomedical and health-related research and applications.
Gene Ontology (GO) is one such resource providing a computational representation of the current scientific knowledge on gene functions of different organisms [2]. The GO resource offers GO itself as well as GO annotations. The GO itself is the logical structure comprising terms for biological processes, molecular functions and cellular components as well as different types of relations that denote how each term is related to other terms (note that ‘class’, ‘concept’ and ‘term’ are interchangeably used in the context of GO). GO annotations link a specific gene product with a GO concept to describe its normal biological role [3, 4]. The 15 December 2021 release of GO, which is used in this paper, contains over 50 000 concepts. GO relationships include is-a, part of, has part, regulates, negatively regulates and positively regulates that link concepts with each other [5].
Modern biomedical ontologies such as GO can be large and complex. Although extreme care is always taken by human curators to make an ontology as accurate as possible, due to the size and complexity, introduction of unintentional errors or defects is difficult to avoid. Some identified defects are fixed as part of the ontology management life-cycle. However, systematic methods to uncover and fix quality defects in biomedical ontologies are still scarce. Manual efforts to audit biomedical ontologies are not really sustainable. Hence, automated or semi-automated auditing algorithms for ontology quality assurance are highly desirable.
Various approaches have been investigated to assess qualities of biomedical ontologies such as concept orientation, consistency, non-redundancy, soundness and comprehensive coverage [6, 7]. Amith et al. have categorized recent ontology quality assurance approaches to 10 categories including structure-based, lexical-based, semantic-based, abstraction-network-based and big data approaches [8].
Many lexical-based approaches leverage the ‘lexically suggest, logically define’ principle which states that the knowledge represented lexically in concept labels should be represented as axioms in the ontology [9]. For instance, van Damme et al. have proposed a method which clusters concepts by lexical regularities that their concept labels contain and extracts information from each cluster to suggest logical axioms for concepts in SNOMED CT [10]. Agrawal et al. have extensively investigated approaches where lexically similar concept sets are identified where inconsistent modeling may be prevalent [11–14]. Bodenreider has introduced a method to identify missing hierarchical relations in SNOMED CT by reasoning on logical definitions constructed by leveraging lexical features of concept labels [15].
With respect to GO, most of the quality assurance efforts have focused on enriching the ontology [16–18]. Other studies have tried to audit GO from different points of view. For instance, Ochs et al. have investigated two types of abstraction networks, called area taxonomy and partial-area taxonomy, to identify groups of anomalous concepts in the biological process subhierarchy of GO [19]. Abstraction networks are a form of compact summarizations of ontologies that have been extensively explored for ontology quality assurance [20–21]. Mougin has explored reasoning over relationships to detect redundant relations in GO, and identified missing necessary and sufficient conditions based on compositional structure of GO concept names [22]. Xing et al. developed a scalable approach combining the algorithmic ideas of dynamic programming and topological sort to exhaustively identify redundant hierarchical is-a relations in large ontologies including GO [23]. In previous works, we investigated a lexical-based inference approach [24] and a subsumption-based sub-term inference framework [25] to identify missing and erroneous hierarchical is-a relations in GO.
Relational defects such as missing or erroneous is-a relations in GO directly affect the quality of downstream research and applications that rely on the relational structure of GO. For instance, when retrieving genes and gene products annotated with a given GO concept, if the concept has a missing subtype, then the genes and gene products associated with this subtype will be excluded from the result; and if the concept has an erroneous subtype, then the genes and gene products associated with the subtype will be wrongly included in the result. More specifically, suppose we want to find all the genes and gene products associated with the GO concept ‘epithelial cell differentiation’ (with ID GO:0030855) using QuickGO [26]. Currently, QuickGO returns 45714 distinct gene products associated with GO:0030855. However, the current version (15 December 2021 release) of GO does not list the concept ‘melanocyte differentiation’ (GO:0030318), which is associated with 2357 distinct gene products, as a subtype of GO:0030855 (i.e. a missing is-a relation). Excluding the overlapping 158 gene products, the remaining 2199 gene products associated with ‘melanocyte differentiation’ (GO:0030318) will be missing from the search result. Therefore, it is imperative to ensure the quality of GO relations. In this paper, we introduce a novel evidence-based approach to uncovering missing relations and erroneous existing relations in GO (including but not limited to is-a relations).
Methods
The basic idea of our evidence-based approach is leveraging lexical patterns exhibited in related concept pairs (i.e. existing relations) in GO to identify potentially missing relations between unrelated concept pairs. We represent each GO concept’s name with a sequence of words along with part-of-speech tags. Such representation enables us to automatically generate lexical patterns from related concept pairs, serving as the first layer evidence to suggest potentially missing relations between unrelated concept pairs. For each suggested missing relation, we further identify a concept quadruple consisting of concepts in two existing relations as the second layer of evidence, which resembles the difference among the concept quadruple consisting of concepts in the missing relation and the existing relation based on which the missing relation is suggested.
Concept name representation
Given a concept  in GO, we represent its concept name as a sequence of words
in GO, we represent its concept name as a sequence of words  along with a sequence of part-of-speech tags
along with a sequence of part-of-speech tags 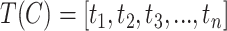 corresponding to each word, where
corresponding to each word, where 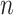 is the number of words in the concept name,
is the number of words in the concept name, 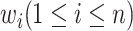 is the
is the 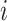 -th word in the concept name and
-th word in the concept name and 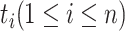 is the part-of-speech tag of
is the part-of-speech tag of 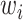 . For instance, GO concept
. For instance, GO concept  ‘nitric oxide biosynthetic process’ (GO:0006809) can be represented as
‘nitric oxide biosynthetic process’ (GO:0006809) can be represented as
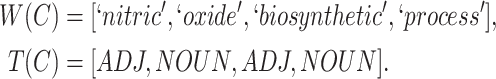 |
For the part-of-speech tagging, we used the English transformer pipeline of the open-source natural language processing library spaCy [27].
Computing related concept pairs
GO concepts are connected with various relationships including is-a, part-of, has-part, regulates, negatively-regulates and positively-regulates [5, 28]. A related concept pair is a pair of concepts that are directly or indirectly connected with a relationship. To obtain all the related concept pairs in GO, we first extract directly related concept pairs using the GOATOOLS python library [29], and then obtain indirectly related concept pairs by computing transitive closure leveraging the reasoning rules given in Table 1. For example, one of the reasoning rules is that if 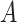 is-a
is-a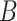 and
and 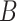 part-of
part-of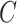 , then it can be inferred
, then it can be inferred 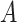 part-of
part-of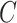 . Note that these inference rules may involve more complex cases combining multiple rules. For instance, if
. Note that these inference rules may involve more complex cases combining multiple rules. For instance, if 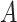 is-a
is-a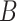 ,
, 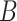 part-of
part-of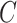 and
and 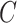 is-a
is-a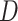 , then it can be inferred
, then it can be inferred 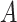 part-of
part-of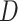 using the reasoning rules (2) and (7) in Table 1.
using the reasoning rules (2) and (7) in Table 1.
Table 1.
Gene Ontology reasoning rules for relationships is-a, part-of, has-part, regulates, negatively-regulates (n-regulates) and positively-regulates (p-regulates) [5, 28]
| Relationship | Reasoning rules | |
|---|---|---|
| is-a | (1) 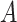 is-a is-a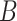 , , 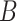 is-a is-a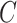
|
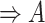 is-a
is-a
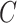
|
(2) 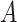 is-a is-a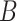 , , 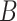 part-of part-of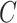
|
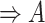 part-of
part-of
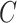
|
|
(3) 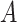 is-a is-a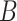 , , 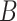 has-part has-part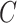
|
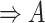 has-part
has-part
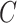
|
|
(4) 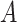 is-a is-a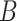 , , 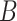 regulates regulates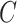
|
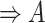 regulates
regulates
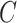
|
|
(5) 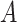 is-a is-a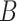 , , 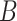 n-regulates n-regulates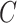
|
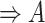 n-regulates
n-regulates
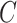
|
|
(6) 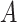 is-a is-a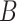 , , 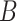 p-regulates p-regulates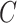
|
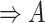 p-regulates
p-regulates
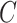
|
|
| part-of | (7) 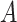 part-of part-of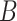 , , 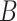 is-a is-a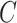
|
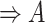 part-of
part-of
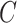
|
(8) 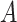 part-of part-of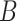 , , 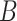 part-of part-of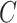
|
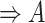 part-of
part-of
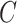
|
|
| has-part | (9) 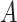 has-part has-part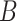 , , 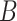 is-a is-a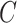
|
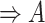 has-part
has-part
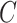
|
(10) 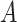 has-part has-part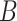 , , 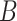 has-part has-part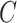
|
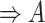 has-part
has-part
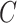
|
|
| regulates | (11) 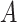 regulates regulates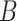 , , 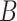 is-a is-a 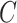
|
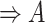 regulates
regulates
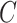
|
(12) 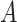 regulates regulates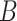 , , 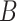 regulates regulates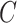
|
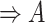 regulates
regulates
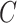
|
|
| n-regulates | (13) 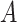 n-regulates n-regulates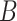 , , 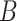 is-a is-a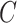
|
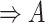 n-regulates
n-regulates
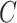
|
| p-regulates | (14) 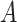 p-regulates p-regulates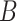 , , 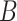 is-a is-a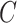
|
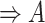 p-regulates
p-regulates
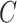
|
(15) 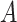 p-regulates p-regulates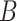 , , 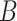 p-regulates p-regulates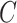
|
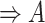 p-regulates
p-regulates
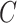
|
|
(16) 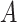 n-regulates n-regulates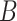 , , 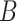 n-regulates n-regulates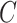
|
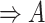 p-regulates
p-regulates
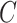
|
|

Given a concept 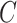 , Algorithm 1 presents our procedure to obtain all the concepts that
, Algorithm 1 presents our procedure to obtain all the concepts that 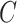 connects to via the is-a relationship; and Algorithm 2 demonstrates how to compute all the concepts that
connects to via the is-a relationship; and Algorithm 2 demonstrates how to compute all the concepts that 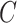 connects to via the part-of relationship. The concept pairs connected via other relationships can be similarly obtained. Note that such transitive closure for a given concept can be obtained through GOATOOLS (or other tools such as Owlready2 and OWL API) for the is-a relationship.
connects to via the part-of relationship. The concept pairs connected via other relationships can be similarly obtained. Note that such transitive closure for a given concept can be obtained through GOATOOLS (or other tools such as Owlready2 and OWL API) for the is-a relationship.
Extracting lexical patterns from concept pairs
We extract lexical patterns from pairs of concepts having at least one word in common. Given a pair of concepts (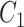 ,
, 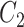 ) with
) with
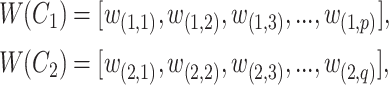 |
such that 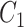 and
and 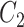 have a set of common words
have a set of common words 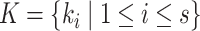 , where
, where 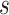 is the total number of commons words, we can generate a lexical pattern of (
is the total number of commons words, we can generate a lexical pattern of (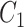 ,
, 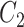 ):
):
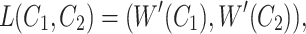 |
where 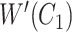 is obtained by replacing each common word
is obtained by replacing each common word 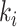 in
in 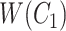 with an abstract label
with an abstract label 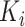 , and
, and 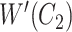 is obtained by replacing each common word
is obtained by replacing each common word 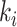 in
in 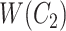 with
with 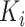 .
.
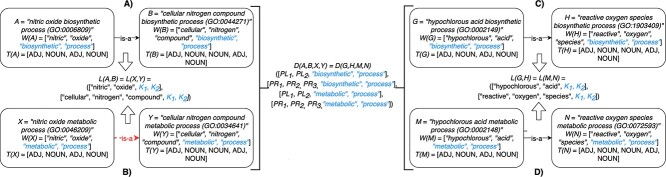
For instance, considering the following two concepts in Figure 1A:
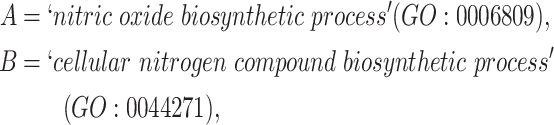 |
Figure 1.
(A) Existing is-a relation between concept 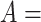 ‘nitric oxide biosynthetic process’ (GO:0006809) and concept B = ‘cellular nitrogen compound biosynthetic process’ (GO:0044271) that is leveraged to generate the lexical pattern
‘nitric oxide biosynthetic process’ (GO:0006809) and concept B = ‘cellular nitrogen compound biosynthetic process’ (GO:0044271) that is leveraged to generate the lexical pattern 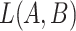 ; (B) Missing is-a relation (dashed arrow in red) between concept
; (B) Missing is-a relation (dashed arrow in red) between concept 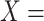 ‘nitric oxide metabolic process’ (GO:0046209) and concept
‘nitric oxide metabolic process’ (GO:0046209) and concept 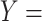 ‘cellular nitrogen compound metabolic process’ (GO:0034641) with the same lexical pattern; (C) and (D): Pair of existing is-a relations that resembles the difference between (A) and (B).
‘cellular nitrogen compound metabolic process’ (GO:0034641) with the same lexical pattern; (C) and (D): Pair of existing is-a relations that resembles the difference between (A) and (B).
they have two words in common, that is,  ‘biosynthetic’, ‘process’
‘biosynthetic’, ‘process’ . After replacing ‘biosynthetic’ with
. After replacing ‘biosynthetic’ with 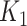 and ‘process’ with
and ‘process’ with 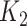 , the obtained lexical pattern is
, the obtained lexical pattern is
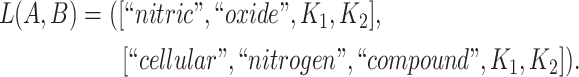 |
Similarly, in Figure 2B, for concepts
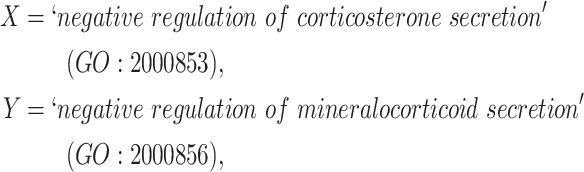 |
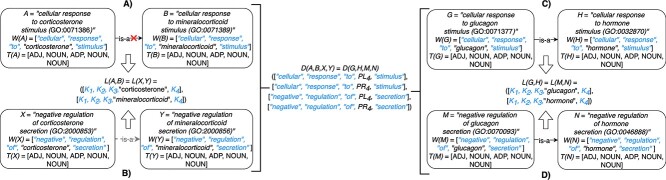
Figure 2.
(A) Erroneous existing is-a relation (red cross) between concept 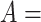 ‘cellular response to corticosterone stimulus’ (GO:0071386) and concept
‘cellular response to corticosterone stimulus’ (GO:0071386) and concept 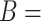 ‘cellular response to mineralocorticoid stimulus’ (GO:0071389) that is leveraged to generate the lexical pattern
‘cellular response to mineralocorticoid stimulus’ (GO:0071389) that is leveraged to generate the lexical pattern 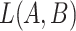 ; (B) Invalid missing is-a relation between GO:2000853 and GO:2000856 with the same lexical pattern; (C) and (D): Pair of existing is-a relations that resembles the difference between (A) and (B).
; (B) Invalid missing is-a relation between GO:2000853 and GO:2000856 with the same lexical pattern; (C) and (D): Pair of existing is-a relations that resembles the difference between (A) and (B).
they have four common words (i.e. 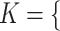 ‘negative’, ‘regulation’, ‘of’, ‘secretion’
‘negative’, ‘regulation’, ‘of’, ‘secretion’ ). After replacing ‘negative’ with
). After replacing ‘negative’ with 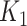 , ‘regulation’ with
, ‘regulation’ with 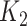 , ‘of’ with
, ‘of’ with 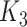 and ‘secretion’ with
and ‘secretion’ with 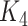 , the obtained lexical pattern is
, the obtained lexical pattern is
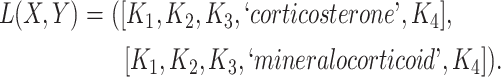 |
Generating difference patterns from concept quadruples
For two concept pairs with the same lexical pattern, we further generate a difference pattern to represent their different parts. More formally, given two concept pairs 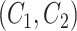 and
and 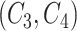 , we consider
, we consider 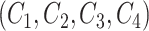 as a candidate concept quadruple if the following conditions are met:
as a candidate concept quadruple if the following conditions are met:
(1)
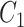 and
and 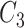 contain the same number of words, and have the same part-of-speech tags, i.e.
contain the same number of words, and have the same part-of-speech tags, i.e. 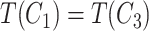 ;
;(2)
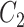 and
and 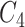 contain the same number of words, and have the same part-of-speech tags, i.e.
contain the same number of words, and have the same part-of-speech tags, i.e. 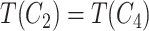 ; and
; and(3) the lexical pattern of concept pair
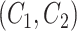 is the same as that of concept pair
is the same as that of concept pair 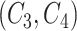 , i.e.
, i.e.  .
.
For a candidate concept quadruple 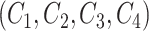 with
with
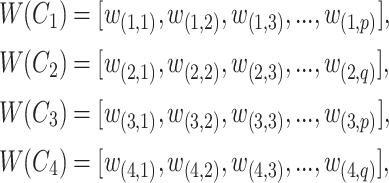 |
we can generate a difference pattern:
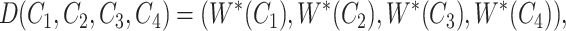 |
where 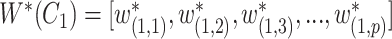 is defined as
is defined as
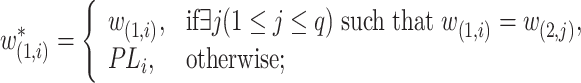 |
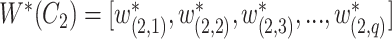 is defined as
is defined as
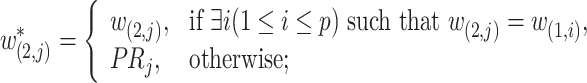 |
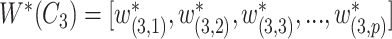 is defined as
is defined as
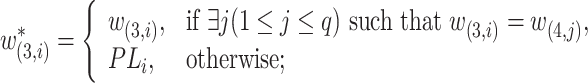 |
and 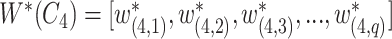 is defined as
is defined as
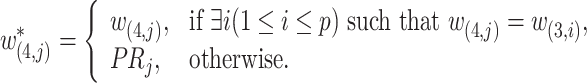 |
Here, 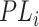
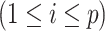 is an abstract label denoting that the corresponding word locates at the
is an abstract label denoting that the corresponding word locates at the 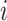 -th position of concept pair
-th position of concept pair 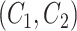 ’s left concept
’s left concept 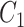 or concept pair
or concept pair 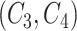 ’s left concept
’s left concept 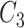 ; and
; and 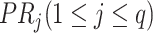 is an abstract label denoting that the corresponding word locates at the
is an abstract label denoting that the corresponding word locates at the 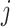 -th position of concept pair
-th position of concept pair 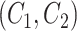 ’s right concept
’s right concept 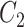 or concept pair
or concept pair 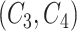 ’s right concept
’s right concept 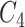 . Intuitively speaking,
. Intuitively speaking, 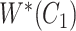 is obtained by replacing words in
is obtained by replacing words in 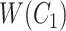 but not in
but not in 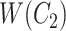 with abstract labels;
with abstract labels; 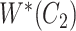 is obtained by replacing words in
is obtained by replacing words in 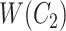 but not in
but not in 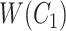 with abstract labels;
with abstract labels; 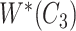 is obtained by replacing words in
is obtained by replacing words in 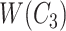 but not in
but not in 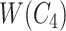 with abstract labels; and
with abstract labels; and 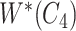 is obtained by replacing words in
is obtained by replacing words in 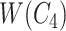 but not in
but not in 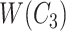 with abstract labels.
with abstract labels.
For example, consider the following four concepts in Figure 1C and Figure 1D:
 |
Concepts  and
and 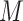 have the same number of words and the same part-of-speech tags. So does concepts
have the same number of words and the same part-of-speech tags. So does concepts 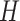 and
and 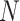 . In addition, concept pair
. In addition, concept pair 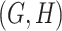 and concept pair
and concept pair 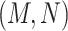 have the same lexical pattern:
have the same lexical pattern:
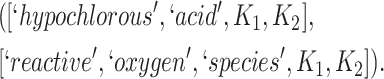 |
Therefore, 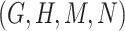 forms a candidate concept quadruple. For concept
forms a candidate concept quadruple. For concept  , since words ‘hypochlorous’ and ‘acid’ do not appear in
, since words ‘hypochlorous’ and ‘acid’ do not appear in 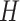 , they are replaced by labels
, they are replaced by labels 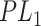 and
and 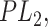 respectively, resulting in
respectively, resulting in 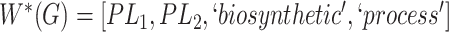 ; for concept
; for concept 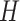 , since words ‘reactive’, ‘oxygen’ and ‘species’ does not appear in
, since words ‘reactive’, ‘oxygen’ and ‘species’ does not appear in 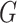 , they are replaced by labels
, they are replaced by labels 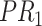 ,
, 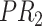 and
and 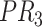 , respectively, resulting in
, respectively, resulting in 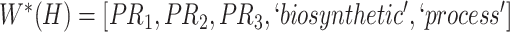 ; and similarly, we can obtain
; and similarly, we can obtain 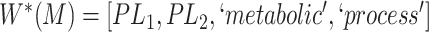 and
and 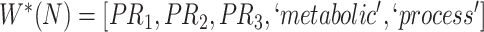 . Therefore, the difference pattern of
. Therefore, the difference pattern of 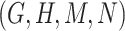 is
is
 |
Note that the difference pattern represents the difference between two pairs of concepts. In this example, we can see that the different parts are [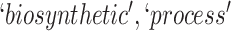 ] in concept pair
] in concept pair 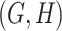 and [
and [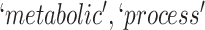 ] in concept pair
] in concept pair 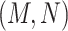 .
.
Evidence-based identification of relational defects
We focus on identifying relational defects regarding the following set of GO relationships: 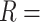 {is-a, part-of, has-part, regulates, negatively-regulates, positively-regulates}. For each relationship
{is-a, part-of, has-part, regulates, negatively-regulates, positively-regulates}. For each relationship 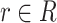 , we extract lexical patterns for all the related concept pairs connected via
, we extract lexical patterns for all the related concept pairs connected via 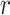 . Then we generate difference patterns for candidate concept quadruples
. Then we generate difference patterns for candidate concept quadruples 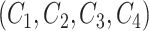 where
where 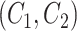 and
and 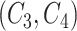 are related concept pairs connected via
are related concept pairs connected via  . We leverage these lexical patterns and difference patterns as two layers of evidence to identify potentially missing
. We leverage these lexical patterns and difference patterns as two layers of evidence to identify potentially missing 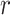 relations as follows.
relations as follows.
Given a pair of concepts 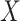 and
and 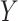 that are not related via any GO relationship, if
that are not related via any GO relationship, if
- (1) there exists a related concept pair
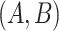 connected via
connected via 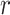 , such that
, such that
and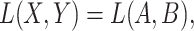
- (2) there exists a candidate concept quadruple
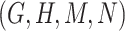 where
where 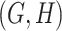 and
and 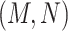 are related concept pairs connected via
are related concept pairs connected via 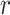 , such that
, such that 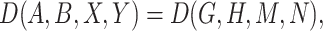
then we suggest a potentially missing 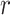 relation between concepts
relation between concepts 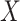 and
and 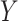 . Here, the related concept pair
. Here, the related concept pair 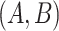 serves as the first layer of evidence, and the concept quadruple
serves as the first layer of evidence, and the concept quadruple 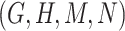 serves as the second layer of evidence. Note that a potentially missing relation may be derived by multiple first and second layers of evidence.
serves as the second layer of evidence. Note that a potentially missing relation may be derived by multiple first and second layers of evidence.
More specifically, for concepts 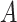 and
and 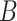 , given that they have common words and are related via
, given that they have common words and are related via 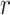 , we assume that the different words between
, we assume that the different words between 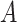 and
and 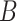 are highly likely to make their
are highly likely to make their 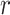 relation hold, which is leveraged as the first layer of evidence for suggesting an
relation hold, which is leveraged as the first layer of evidence for suggesting an 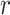 relation between concepts
relation between concepts 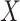 and
and 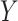 , because
, because 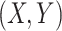 have the same lexical pattern as
have the same lexical pattern as 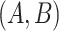 (i.e. the different words between
(i.e. the different words between 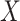 and
and 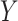 are the same as the different words between
are the same as the different words between 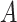 and
and 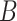 ). For instance, for concept
). For instance, for concept 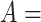 ‘nitric oxide biosynthetic process’ (GO:0006809) and concept
‘nitric oxide biosynthetic process’ (GO:0006809) and concept 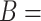 ‘cellular nitrogen compound biosynthetic process’ (GO:0044271) in Figure 1A related by is-a, we assume that ‘nitric oxide’ in
‘cellular nitrogen compound biosynthetic process’ (GO:0044271) in Figure 1A related by is-a, we assume that ‘nitric oxide’ in 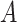 and ‘cellular nitrogen compound’ in
and ‘cellular nitrogen compound’ in 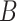 are highly likely to make the is-a relation hold; and this serves as the first layer of evidence for us to suggest a potentially missing is-a relation between concept
are highly likely to make the is-a relation hold; and this serves as the first layer of evidence for us to suggest a potentially missing is-a relation between concept 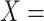 ‘nitric oxide metabolic process’ (GO:0046209) and concept
‘nitric oxide metabolic process’ (GO:0046209) and concept 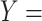 ‘cellular nitrogen compound metabolic process’ (GO:0034641) in Figure 1B.
‘cellular nitrogen compound metabolic process’ (GO:0034641) in Figure 1B.
Although concept pair 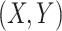 have the same lexical pattern with concept pair
have the same lexical pattern with concept pair 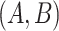 , the common words of
, the common words of 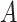 and
and 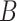 are distinct from that of
are distinct from that of 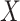 and
and 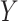 . Therefore, we seek further evidence of such distinction among other related
. Therefore, we seek further evidence of such distinction among other related 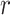 concept pairs in candidate concept quadruples (i.e. difference pattern). For the above example
concept pairs in candidate concept quadruples (i.e. difference pattern). For the above example 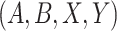 in Figure 1A and Figure 1B, the difference pattern is ‘biosynthetic process’ versus ‘metabolic process’; and there exists a candidate concept quadruple
in Figure 1A and Figure 1B, the difference pattern is ‘biosynthetic process’ versus ‘metabolic process’; and there exists a candidate concept quadruple 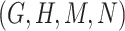 where
where  and
and  are related is-a concept pairs (see Figure 1C and Figure 1D), such that
are related is-a concept pairs (see Figure 1C and Figure 1D), such that 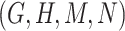 have the same difference pattern (‘biosynthetic process’ versus ‘metabolic process’), the second layer of evidence.
have the same difference pattern (‘biosynthetic process’ versus ‘metabolic process’), the second layer of evidence.
Note that in some instances, the same lexical pattern could be obtained through different relationship types. We discard such patterns as they would suggest multiple types of missing relations among the same two concepts (e.g. 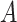 is-a
is-a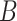 and
and 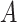 part-of
part-of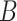 both being suggested), which is unlikely to be true.
both being suggested), which is unlikely to be true.
In addition, it is possible that a suggested missing relation can be inferred by other suggested missing relations and existing GO relations using the reasoning rules in Table 1. To identify such cases, we check whether each suggested missing relation is included in the transitive closure computed with all the other suggested missing relations and existing relations in GO. Such suggestions are redundant and hence removed. For example, consider the following two suggestions for missing relationships: (1) regulates relation between concepts ‘regulation of NK T cell differentiation’ (GO:0051136) and ‘NK T cell activation’ (GO:0051132); (2) is-a relation between the concepts ‘regulation of NK T cell differentiation’ (GO:0051136) and ‘regulation of NK T cell activation’ (GO:0051133). However, GO currently has the regulates relation between concepts: ‘regulation of NK T cell activation’ (GO:0051133) and ‘NK T cell activation’ (GO:0051132) which together with (2) infers (1) through reasoning rule (4) in Table 1.
For the potentially missing relations automatically suggested by our approach, manual review by domain experts is required to assess their validity. If a suggested missing 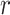 relation between concepts
relation between concepts 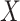 and
and 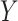 is agreed by domain experts, then it is considered a valid missing relation (e.g. is-a relation between ‘nitric oxide metabolic process’ and ‘cellular nitrogen compound metabolic process’ in Figure 1B). However, if a suggested missing
is agreed by domain experts, then it is considered a valid missing relation (e.g. is-a relation between ‘nitric oxide metabolic process’ and ‘cellular nitrogen compound metabolic process’ in Figure 1B). However, if a suggested missing 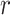 relation between concepts
relation between concepts 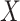 and
and 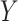 is disagreed by domain experts, then the concept pair
is disagreed by domain experts, then the concept pair 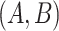 that is leveraged as the first layer of evidence to suggest the missing relation is further examined as follows: if the
that is leveraged as the first layer of evidence to suggest the missing relation is further examined as follows: if the 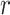 relation between concepts
relation between concepts 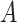 and
and 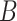 is agreed by domain experts, then we consider the suggested missing
is agreed by domain experts, then we consider the suggested missing 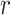 relation between
relation between 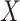 and
and 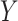 is a false positive suggested by our approach; but if the
is a false positive suggested by our approach; but if the 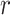 relation between concepts
relation between concepts 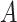 and
and 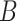 is disagreed by domain experts, then it is considered as a valid erroneous existing relation.
is disagreed by domain experts, then it is considered as a valid erroneous existing relation.
For instance, Figure 2B shows a potentially missing is-a relation between concepts 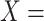 ‘negative regulation of corticosterone secretion’ (GO:2000853) and
‘negative regulation of corticosterone secretion’ (GO:2000853) and 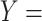 ‘negative regulation of mineralocorticoid secretion’ (GO:2000856) suggested by our approach by leveraging an existing is-a relation between concepts
‘negative regulation of mineralocorticoid secretion’ (GO:2000856) suggested by our approach by leveraging an existing is-a relation between concepts 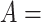 ‘cellular response to corticosterone stimulus’ (GO:0071386) and
‘cellular response to corticosterone stimulus’ (GO:0071386) and 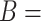 ‘cellular response to mineralocorticoid stimulus’ (GO:0071389) as shown in Figure 2A. However, the suggested is-a relation between ‘negative regulation of corticosterone secretion’ and ‘negative regulation of mineralocorticoid secretion’ is disagreed by domain experts, since mineralocorticoid is considered a subtype of corticosterone (not the other way around) [30]. Further, the is-a relation between the evidence concept pair ‘cellular response to corticosterone stimulus’ and ‘cellular response to mineralocorticoid stimulus’ is also disagreed by domain experts, and thus an erroneous existing is-a relation.
‘cellular response to mineralocorticoid stimulus’ (GO:0071389) as shown in Figure 2A. However, the suggested is-a relation between ‘negative regulation of corticosterone secretion’ and ‘negative regulation of mineralocorticoid secretion’ is disagreed by domain experts, since mineralocorticoid is considered a subtype of corticosterone (not the other way around) [30]. Further, the is-a relation between the evidence concept pair ‘cellular response to corticosterone stimulus’ and ‘cellular response to mineralocorticoid stimulus’ is also disagreed by domain experts, and thus an erroneous existing is-a relation.
Evaluation
To evaluate the effectiveness of our approach, all the potential missing relations obtained are manually reviewed by our local domain experts (authors YY, MB and WJZ who have expertise in systems biology and genomics). Any disagreements among the experts are resolved through discussion. For each potentially missing relation, the domain experts are provided with the concept names and web links (in QuickGO [26]) of the two concepts involved in the relation. If a potentially missing relation is confirmed as valid by domain experts, then we consider it as a true missing relation; otherwise, domain experts are further provided with the concept pair that was leveraged as the first layer of evidence to suggest the missing relation. If the evidence concept pair is confirmed to have a valid relation by domain experts, then we consider the original missing relation as a false positive; however, if the evidence concept pair is confirmed to be an invalid relation, then we consider the evidence concept pair as an erroneous existing relation.
Results
In this work, we used the 15 December 2021 release of GO with 50 757 concepts. We focused on auditing the following GO relationships: is-a, part-of, has-part, regulates, negatively-regulates and positively-regulates.
The distribution of each relationship in terms of the number of direct relations, number of direct and indirect relations and number of extracted lexical patterns can be found in Table 2. Take the is-a relationship as an example, there were 70 759 direct is-a relations, a total of 496 502 direct and indirect is-a relations and 290 849 lexical patterns extracted.
Table 2.
The numbers of relations, lexical patterns and potentially missing relations for each relationship
| No. of direct | No. of direct & | No. of lexical | No. of potentially | |
|---|---|---|---|---|
| relations | indirect relations | patterns | missing relations | |
| is-a | 70 759 | 496 502 | 290 849 | 702 |
| part-of | 8118 | 204 180 | 38 099 | 144 |
| regulates | 3550 | 162 927 | 23 901 | 19 |
| has-part | 808 | 17 349 | 3516 | 1 |
| Total | 83 235 | 880 958 | 356 365 | 866 |
In total, our approach suggested 2722 cases of potentially missing relations in GO, among which 1856 relations can be inferred by others (these redundant relations can be found in the Supplementary file ‘Redundant relations.xlsx’). Removal of such redundant relations resulted in 866 potentially missing relations. The number of potentially missing relations suggested for each relationship can also be found in Table 2. For instance, 702 potentially missing is-a relations were suggested. Note that the approach suggested only two negatively-regulates potential missing relations which were both found to be redundant. The method did not suggest any positively-regulates potential missing relations.
The 866 potentially missing relations were suggested by 764 unique lexical patterns. Out of these, 688 lexical patterns suggested only one potentially missing relation, while 76 suggested more than one potentially missing relation. Table 3 shows 10 examples of lexical patterns and the number of potentially missing relations each pattern suggested. For instance, lexical pattern 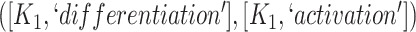 was leveraged to suggest five potentially missing is-a relations.
was leveraged to suggest five potentially missing is-a relations.
Table 3.
Ten examples of lexical patterns suggesting the most potentially missing relations and the number of potentially missing relations suggested by each pattern.
| Lexical pattern | Relationship | No. of potentially |
|---|---|---|
| missing relations suggested | ||
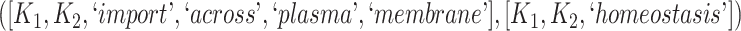
|
is-a | 6 |
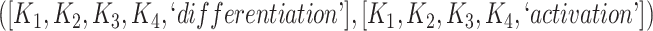
|
is-a | 6 |
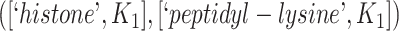
|
is-a | 5 |
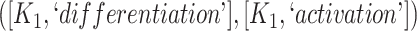
|
is-a | 5 |
|
is-a | 4 |
|
is-a | 4 |
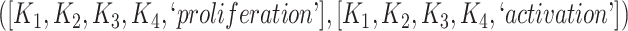
|
is-a | 4 |
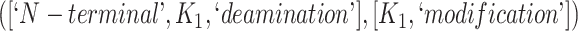
|
is-a | 4 |
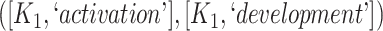
|
is-a | 3 |
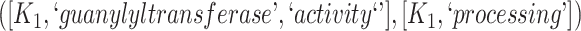
|
is-a | 3 |
Evaluation results
The entire set of 866 potentially missing relations suggested by this approach was evaluated by local domain experts. Table 4 shows the number of potentially missing relations suggested by our approach, number of valid missing relations according to local domain experts and number of valid erroneous existing relations according to local domain experts, for each relationship. For instance, there were 702 potentially missing is-a relations suggested by our approach, of which 661 were identified by local domain experts to be valid missing is-a relations and 41 revealed valid erroneous existing is-a relations. Out of 866 potentially missing relations suggested by our approach, 821 were identified by local domain experts to be valid missing relations and 45 revealed valid erroneous existing relations (see Supplementary files ‘Missing relations.xlsx’ and ‘Erroneous relations.xlsx’ for details).
Table 4.
The numbers of potentially missing relations suggested by our approach, valid missing relations according to local domain experts and valid erroneous existing relations according to local experts.
| No. of potentially | No. of valid | No. of valid | |
|---|---|---|---|
| missing relations | missing relations | erroneous relations | |
| is-a | 702 | 661 | 41 |
| part-of | 144 | 143 | 1 |
| has-part | 1 | 1 | 0 |
| regulates | 19 | 16 | 3 |
| Total | 866 | 821 | 45 |
Table 5 lists 10 examples of valid relational defects in the random sample, including a missing part-of relation between ‘cardiac right atrium formation’ (GO:0003217) and ‘heart formation’ (GO:0060914), and an erroneous is-a relation between ‘hypochlorous acid metabolic process’ (GO:0002148) and ‘organic acid metabolic process’ (GO:0006082).
Table 5.
Ten examples of valid missing relations (M) or erroneous existing relations (E) according to local domain experts.
| Source concept | Relationship | Target concept | Type |
|---|---|---|---|
| ganglion formation (GO:0061554) | is-a | animal organ formation (GO:0048645) | M |
| positive regulation of RIG-I signaling pathway (GO:1900246) | is-a | positive regulation of defense response (GO:0031349) | M |
| geranyl diphosphate biosynthetic process (GO:0033384) | is-a | cellular lipid biosynthetic process (GO:0097384) | M |
| hypochlorous acid metabolic process (GO:0002148) | is-a | organic acid metabolic process (GO:0006082) | E |
| negative regulation of cell septum assembly (GO:1901892) | is-a | negative regulation of cytokinesis (GO:0032466) | E |
| cardiac right atrium formation (GO:0003217) | part-of | heart formation (GO:0060914) | M |
| endocardial cushion fusion (GO:0003274) | part-of | endocardial cushion formation (GO:0003272) | M |
| regulation of cellotriose catabolic process (GO:2000936) | regulates | polysaccharide catabolic process (GO:0000272) | M |
| regulation of glycogen catabolic process (GO:0005981) | regulates | glucose catabolic process (GO:0006007) | M |
| polyadenylation-dependent ncRNA catabolic process (GO:0043634) | has-part | ncRNA processing (GO:0034470) | M |
Time complexity and running time
We analyze the time complexity of our approach as follows. Given an ontology, let 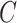 be the number of concepts in the ontology,
be the number of concepts in the ontology, 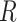 be the number of relations (direct and indirect) in the ontology and
be the number of relations (direct and indirect) in the ontology and 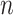 be the maximum number of words contained in concepts. Then, the time complexity for generating lexical patterns from related concept pairs is
be the maximum number of words contained in concepts. Then, the time complexity for generating lexical patterns from related concept pairs is 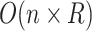 . For generating difference patterns from existing relations, the time complexity is
. For generating difference patterns from existing relations, the time complexity is 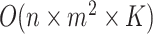 , where
, where 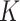 is the number of generated lexical patterns and
is the number of generated lexical patterns and 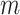 is the maximum number of relations exhibiting a lexical pattern. For the last step to identify potential missing relations, the time complexity is
is the maximum number of relations exhibiting a lexical pattern. For the last step to identify potential missing relations, the time complexity is 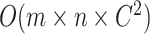 . Therefore, the time complexity for the overall approach is
. Therefore, the time complexity for the overall approach is 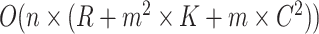 . Note that in the 15 December 2021 release of GO used in this work, the maximum number of words contained in concepts
. Note that in the 15 December 2021 release of GO used in this work, the maximum number of words contained in concepts 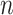 is 27, while the average number is 4.54. On the other hand, the maximum number of relations exhibiting a lexical pattern
is 27, while the average number is 4.54. On the other hand, the maximum number of relations exhibiting a lexical pattern 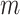 is 566, while the average number is 1.17.
is 566, while the average number is 1.17.
In this work, we ran this approach 10 times on an iMac with an M1 processor and 16GB of RAM. The average time taken was 94 min.
Discussion
In this paper, we introduced an evidence-based approach leveraging automatically extracted lexical patterns to facilitate identification of two types of relational defects in GO: missing relations and erroneous existing relations. A vast majority of potentially missing relations suggested by our approach are is-a relations. This is expected as the majority of relations in GO are is-a relations. According to local domain experts, 94.8% of potentially missing relations (821 out of 866) are valid missing relations and 5.2% of them (45 out of 866) revealed valid erroneous existing relations. This indicates the effectiveness of our approach that leverages lexical patterns and difference patterns derived from existing GO relations as two layers of evidence.
For the erroneous existing relations identified, considering the is-a relation between ‘hypochlorous acid metabolic process’ (GO:0002148) and ‘organic acid metabolic process’ (GO:0006082), this is invalid since hypochlorous acid is not an organic acid as it does not contain a carbon. Among the 45 erroneous existing relations identified, seven were is-a relations with ‘hypochlorous acid metabolic process’ (GO:0002148) as the parent. Local domain experts suggested that some erroneous existing relations may be better represented using a different relationship. For instance, the concepts ‘negative regulation of cohesin loading’ (GO:0071923) and ‘negative regulation of sister chromatid cohesion’ (GO:0045875) may be better connected through a part-of relation than the existing is-a relation. There were 16 such cases among the erroneous existing relations identified.
Part-of-speech tagging tool selection
Note that we chose spaCy for performing part-of-speech tagging of concept names. We also experimented with two other NLP libraries: NLTK [31] and StanfordNLP [32], and compared their results of potentially missing relations with spaCy’s. The comparison showed that a vast majority of cases identified by NLTK and StanfordNLP (83.74% and 81.22% respectively) were also identified by spaCy. On the other hand, a considerable number of cases identified by spaCy were not identified by NLTK and StanfordNLP (40.6% and 41.84%, respectively).
Concept distance in missing relations
We define a distance measure to quantify the closeness of concepts involved in the missing relations. Given a missing relation between source concept 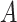 and target concept
and target concept 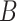 , the distance between
, the distance between 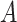 and
and 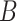 is defined as
is defined as
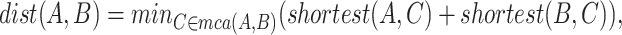 |
where 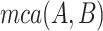 denotes the set of minimal common ancestors of concepts
denotes the set of minimal common ancestors of concepts 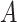 and
and 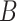 , and
, and 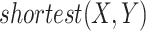 denotes the length of the shortest path between concepts
denotes the length of the shortest path between concepts 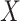 and
and 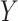 . For instance, considering the missing relation between source concept ‘ganglion formation’ (GO:0061554) and target concept ‘animal organ formation’ (GO:0048645) in Table 5, they have one minimal common ancestor ‘anatomical structure formation involved in morphogenesis’ (GO:0048646), which is their direct parent. Therefore, the distance between these two concepts is 2.
. For instance, considering the missing relation between source concept ‘ganglion formation’ (GO:0061554) and target concept ‘animal organ formation’ (GO:0048645) in Table 5, they have one minimal common ancestor ‘anatomical structure formation involved in morphogenesis’ (GO:0048646), which is their direct parent. Therefore, the distance between these two concepts is 2.
Figure 3 shows a distribution plot of the distances between concept pairs involved in the 821 missing relations assessed by local domain experts. It can be seen that most of the missing relations are observed among concepts closed to each other. Especially, for is-a, part-of and has-part, a majority of missing relations are observed among concept-pairs with a distance of 3 (i.e. uncle–nephew pairs). On the other hand, regulates relations are generally observed among rather distant concept-pairs (distances between 9 and 12). This may indicate that it is more likely to find missing relations by analyzing local subgraphs of an ontology, such as uncle–nephew subgraphs [33] and non-lattice subgraphs [34–37].
Figure 3.
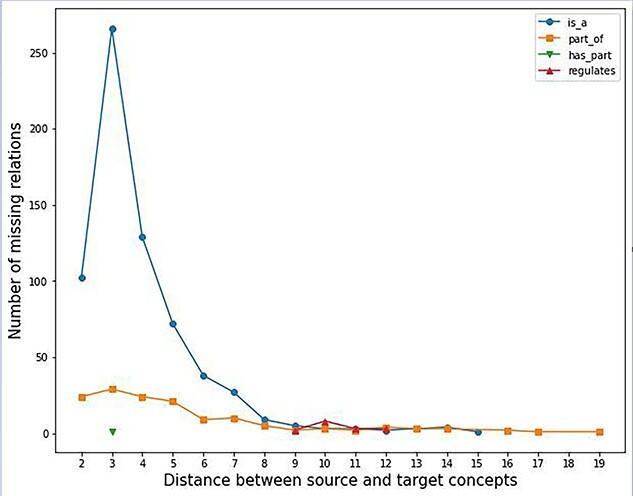
Distribution plot of the distances between concept pairs in missing relations validated by domain experts.
Comparison with related work
A major difference between this work and other lexical pattern-based work to audit GO is that the lexical patterns are generated automatically rather than being manually crafted. For instance, in a previous study, we used three conditional rules (monotonicity, intersection and sub-concept rules) that were manually defined to uncover missing and erroneous is-a relations in GO [25]. Such manual creation of lexical patterns may take extensive exploration of existing concepts and relations of an ontology which is very time-consuming and may require thorough domain knowledge about the ontology. Therefore, automated generation of such patterns from existing relations in the ontology is a considerable improvement in lexical-pattern-based ontological auditing. In addition, only is-a relations were investigated in [25], while this work covers a variety of relationships including is-a, part-of, has-part, regulates, negatively-regulates and positively-regulates. It should also be noted that a vast majority (85.8%) of relational defects identified by this approach is not identifiable by the manually curated rules in [25]. Additionally, the local domain expert evaluation in this work is much more rigorous because the entire set of 866 potentially missing relations suggested by our approach has been assessed. In the previous work [25], only a random subset of 210 samples was assessed.
Ontology auditing approaches are discovery oriented in their nature and different approaches are intended to address different types of issues. This makes it harder to compare different approaches in terms of their performance, as there is a lack of gold standard for quality issues in an ontology. However, purely based on the percentage of valid quality issues assessed by local domain experts, our approach in this work outperforms the previous approach using manually crafted lexical patterns in [25], where the monotonicity, intersection and subconcept rules revealed only 60.61%, 60.49% and 46.03% valid quality issues, respectively, based on local domain experts’ evaluation of 210 instances.
Another advantage of our work over approaches like the one employed by Agrawal et al. [11] is that the manual effort needed to uncover quality defects is considerably less in our approach. Agrawal et al. approach requires an extensive manual evaluation of the problematic areas of the ontology to locate the exact quality issues. However, this work directly provides the two concepts where a missing relation may exist and the experts only need to validate whether it is accurate.
GO consortium feedback
We have reached out to the GO consortium and submitted our suggested changes as a whole (821 missing relations and 45 erroneous existing relations) for further validation and incorporation to GO. The initial review by the GO editorial team indicated that most of the missing relations and erroneous existing relations we identified seem correct. And they have also independently identified some of the issues we found, and are already working on addressing them, including adding missing axioms for some GO terms, working with external ontology teams (e.g. Chemical Entities of Biological Interest [ChEBI] [38]) and restructuring specific parts of the ontology.
Meanwhile, we have put 20 sample issues (15 missing relations and five erroneous existing relations) in the GO-ontology tracking system on GitHub [39]. As of 10 March 2022, seven issues have received feedback, where six of them were agreed by the GO editorial team and revealed different remediation solutions (see Table 6). For instance, the missing part-of relation between ‘bone growth’ (GO:0098868) and ‘bone development’ (GO:0060348) has been directly added to GO; and the erroneous existing is-a relation between ‘purine nucleobase biosynthetic process’ (GO:0009113) and ‘pigment biosynthetic process’ (GO:0046148) has been directly removed from GO. In the case of the missing is-a relation between ‘xylan catabolic process’ (GO:0045493) and ‘hemicellulose catabolic process’ (GO:2000895), the issue was found to be a missing is-a relation between concepts ‘xylan’ (CHEBI:37166) and ‘hemicellulose’ (CHEBI:61266) in the external ontology ChEBI that GO reuses. We have reported this missing is-a relation to ChEBI (which has been added), and thus the former relation can be inferred in GO.
Table 6.
Six valid missing relations (M) or erroneous existing relations (E), which were further validated by the GO editorial team and incorporated into GO.
| Relation | Type | Solution |
|---|---|---|
| bone growth (GO:0098868) | M | Relation added |
| part-of | ||
| bone development (GO:0060348) | ||
| xylan catabolic process (GO:0045493) | M | External ontology changed |
| is-a | ||
| hemicellulose catabolic process (GO:2000895) | ||
| positive regulation of establishment of turgor in appressorium (GO:0075041) | M | GO:0075041 obsoleted |
| is-a | ||
| positive regulation of appressorium maturation (GO:0075037) | ||
| purine nucleobase biosynthetic process (GO:0009113) | E | Relation removed |
| is-a | ||
| pigment biosynthetic process (GO:0046148) | ||
| rhizobactin 1021 biosynthetic process (GO:0019289) | E | Relation removed |
| is-a | ||
| catechol-containing compound biosynthetic process (GO:0009713) | ||
| positive regulation of prosthetic group metabolic process (GO:0051200) | E | GO:0051200 obsoleted |
| is-a | ||
| positive regulation of cellular protein metabolic process (GO:0032270) |
Note that certain issues uncovered by our approach have helped with identification of additional issues in GO. For instance, while reviewing the missing is-a relation between ‘positive regulation of establishment of turgor in appressorium’ (GO:0075041) and ‘positive regulation of appressorium maturation’ (GO:0075037), the GO editorial team has decided to obsolete not only GO:0075041, but also eight additional concepts including ‘regulation of establishment of turgor in appressorium’ (GO:0075040) and ‘negative regulation of establishment of turgor in appressorium’ (GO:0075042).
However, the GO editorial team did not agree with a missing is-a relation between ‘histone methylation’ (GO:0016571) and ‘peptidyl-lysine methylation’ (GO:0018022). The first layer of evidence leveraged by our approach to suggest this relation is an existing is-a relation between ‘histone acetylation’ (GO:0016573) and ‘peptidyl-lysine acetylation’ (GO:0018394). According to the GO editorial team, histones can also be methylated on residues other than lysine, while it looks like that aetlyation is only on lysines [44].
Limitations and future directions
When generating lexical patterns for concept pairs, we require that the two concepts in a concept pair need to share at least one common word. Therefore, the suggested missing relations are among such concept pairs with common words. Since over 99% of concepts in GO have at least one unrelated concept with common words, almost all the GO concepts were considered for missing relation identification by this approach. However, there might be other missing relations among concept pairs that do not share any common words that this approach misses. In the future, we plan to explore whether leveraging ancestors’ lexical features could help identify relational defects for concept pairs without common words.
In addition, certain lexical patterns generated by our approach may be similar and could be further grouped or generalized. For instance, the following two lexical patterns (see Table 3) are similar:
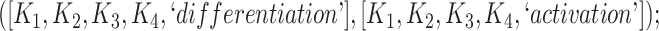 |
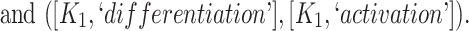 |
These two lexical patterns could be grouped and generalized to a single lexical pattern:
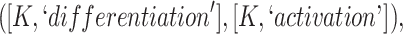 |
where  represents one or more common words between the two concepts. Such generalization may uncover additional potentially missing relations as the pattern does not require a specific number of common words.
represents one or more common words between the two concepts. Such generalization may uncover additional potentially missing relations as the pattern does not require a specific number of common words.
Since a lexical pattern can be generated by a pair of concepts with an indirect relation (through reasoning rules in Table 1), an identified missing relation using this lexical pattern may also be indirect. That is, there may be an intermediate missing relation (which is more specific) from which the former missing relation can be inferred. Given the significant amount of manual effort needed to uncover such intermediate missing relations, it is highly desirable to develop automated or semi-automated methods that can identify such root cause issues that lead to the indirect missing relations.
Although our approach is capable of automatically suggesting potentially missing relations based on two layers of evidence, the manual evaluation by domain experts showed that a few cases revealed erroneous existing relations. It remains a challenge to automatically identify such erroneous existing relations to further reduce manual effort by domain experts.
Additionally, although we have submitted all the findings evaluated by local domain experts to the GO consortium, it requires GO editorial team’s further adjudication and diligence to come up with specific remediation measures (e.g. directly adding a missing relation, directly removing an erroneous existing relation, obsoleting a concept, adding another missing relation in an external ontology that GO reuses) and perform GO content modification.
Since our approach only requires the concept names and relational structures of an ontology, which are fundamental to biomedical ontologies, it is generally applicable to audit relations in other biomedical ontologies. We plan to apply it to other biomedical ontologies like SNOMED CT and National Cancer Institute thesaurus, and evaluate the effectiveness of this approach for other ontologies.
Conclusions
In this work, we presented an evidence-based approach to identify relational defects regarding is-a, part-of, has-part, regulates, negatively regulates and positively regulates relationships in GO. We were able to automatically extract lexical patterns from concept pairs and difference patterns from concept quadruples as two layers of evidence to suggest potentially missing relations. Both local domain experts’ evaluation and GO consortium’s encouraging feedback indicated the effectiveness of our evidence-based approach, which can be utilized to uncover missing relations and erroneous existing relations in GO.
Key Points
Biomedical ontology quality assurance is a critical component of ontology management to ensure that an ontology provides accurate knowledge representation to downstream applications that rely on them.
We developed a two-layered, evidence-based approach to extract lexical patterns from existing relations and automatically suggest potentially missing relations in Gene Ontology.
Local domain experts’ evaluation and GO consortium’s feedback indicate that our evidence-based approach can be utilized to uncover missing relations and erroneous existing relations in GO.
Supplementary Material
Funding
This work was supported by the National Institutes of Health (R01LM013335 and R01NS116287 to L.C.; 1UL1TR003167 to W.J.Z.); the Cancer Prevention and Research Institute of Texas (RP170668 to W.J.Z.); and the National Science Foundation (2047001 to L.C.).
Author Biographies
Rashmie Abeysinghe is a Research Scientist at The University of Texas Health Science Center at Houston, Houston, TX, USA. His research interests include biomedical ontologies, machine learning, information extraction and big data analytics.
Yuntao Yang is a PhD student in the School of Biomedical Informatics at The University of Texas Health Science Center at Houston, Houston, TX, USA. His research interests include data science and translational bioinformatics.
Mason Bartels is an MS student in the School of Biomedical Informatics at The University of Texas Health Science Center at Houston, Houston, TX, USA. His research interests include genomics and translational bioinformatics.
W. Jim Zheng is a Professor in the School of Biomedical Informatics at The University of Texas Health Science Center at Houston, Houston, TX, USA. His research interests include large scale data integration and mining, translational bioinformatics and precision medicine.
Licong Cui is an Assistant Professor in the School of Biomedical Informatics at The University of Texas Health Science Center at Houston, Houston, TX, USA. Her research interests include biomedical ontologies and neuroinformatics.
Author contributions statement
L.C. and R.A. conceived this study. R.A. designed and implemented the algorithms, generated and analyzed the results and prepared the evaluation sample. Y.Y., M.B. and W.J.Z. performed the evaluation. R.A. and L.C. analyzed the evaluation results. R.A. and L.C. wrote the manuscript. All authors have read and approved the final manuscript.
References
- 1. Rubin DL, Shah NH, Noy NF. Biomedical ontologies: a functional perspective. Brief Bioinform 2008;9(1):75–90. [DOI] [PubMed] [Google Scholar]
- 2. The Gene Ontology Consortium . The Gene Ontology resource: 20 years and still GOing strong. Nucleic Acids Res 2019;47(D1):D330–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. The Gene Ontology Consortium . About the GO. http://geneontology.org/docs/introduction-to-go-resource/ (25 January 2022, date last accessed).
- 4. Francis RW. GOLink: finding cooccurring terms across Gene Ontology namespaces. Int. J Genomics 2013;2013:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. The Gene Ontology Consortium . Relations in the Gene Ontology. http://geneontology.org/docs/ontology-relations/ (25 January 2022, date last accessed).
- 6. Geller J, Perl Y, Cui L, et al. Quality assurance of biomedical terminologies and ontologies. J Biomed Inform 2018;86:106–8. [DOI] [PubMed] [Google Scholar]
- 7. Zhu X, Fan JW, Baorto DM, et al. A review of auditing methods applied to the content of controlled biomedical terminologies. J Biomed Inform 2009;42(3):413–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Amith M, He Z, Bian J, et al. Assessing the practice of biomedical ontology evaluation: Gaps and opportunities. J Biomed Inform 2018;80:1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rector A, Iannone L. Lexically suggest, logically define: quality assurance of the use of qualifiers and expected results of post-coordination in SNOMED CT. J Biomed Inform 2012;45(2):199–209. [DOI] [PubMed] [Google Scholar]
- 10. Damme P, Quesada-Martínez M, Cornet R, et al. From lexical regularities to axiomatic patterns for the quality assurance of biomedical terminologies and ontologies. J Biomed Inform 2018;84:59–74. [DOI] [PubMed] [Google Scholar]
- 11. Agrawal A, Perl Y, Ochs C, et al. Algorithmic detection of inconsistent modeling among SNOMED CT concepts by combining lexical and structural indicators. In: Jun Huan (ed) 2015 IEEE international conference on bioinformatics and biomedicine (BIBM). Piscataway, New Jersey: IEEE, 2015, 476–83. [Google Scholar]
- 12. Agrawal A, Revelo P. Analysis of the consistency in the structural modeling of SNOMED CT and CORE problem list concepts. In: Xiaohua Hu (ed) 2017 IEEE International Conference on Bioinformatics and Biomedicine (BIBM). Piscataway, New Jersey: IEEE, 2017, 292–6. [Google Scholar]
- 13. Agrawal A. Evaluating lexical similarity and modeling discrepancies in the procedure hierarchy of SNOMED CT. BMC Med Inform Decis Mak 2018;18(4):27–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Agrawal A, Qazi K. Detecting modeling inconsistencies in SNOMED CT using a machine learning technique. Methods 2020;179:111–8. [DOI] [PubMed] [Google Scholar]
- 15. Bodenreider O. Identifying Missing Hierarchical Relations in SNOMED CT from Logical Definitions Based on the Lexical Features of Concept Names. 2016. http://ceur-ws.org/Vol-1747/IT601_ICBO2016.pdf (25 January 2022 date last accessed). [PMC free article] [PubMed] [Google Scholar]
- 16. Dutkowski J, Kramer M, Surma MA, et al. A Gene Ontology inferred from molecular networks. Nat Biotechnol 2013;31(1):38–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Liu W, Liu J, Rajapakse JC. Gene Ontology enrichment improves performances of functional similarity of genes. Sci Rep 2018;8(1):1–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Peng J, Wang T, Wang J, et al. Extending Gene Ontology with gene association networks. Bioinformatics 2016;32(8):1185–94. [DOI] [PubMed] [Google Scholar]
- 19. Ochs C, Perl Y, Halper M, et al. Quality assurance of the Gene Ontology using abstraction networks. J Bioinform Comput Biol 2016;14(03):1642001. [DOI] [PubMed] [Google Scholar]
- 20. Halper M, Gu H, Perl Y, et al. Abstraction networks for terminologies: supporting management of “big knowledge”. Artif Intell Med 2015;64(1):1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ochs C, Geller J, Perl Y, et al. A tribal abstraction network for SNOMED CT target hierarchies without attribute relationships. J Am Med Inform Assoc 2015;22(3):628–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mougin F. Identifying redundant and missing relations in the Gene Ontology. Stud Health Technol Inform 2015;210:195–9. [PubMed] [Google Scholar]
- 23. Xing G, Zhang GQ, Cui L. FEDRR: fast, exhaustive detection of redundant hierarchical relations for quality improvement of large biomedical ontologies. BioData Min 2016;9:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Abeysinghe R, Hinderer EW, Moseley HN, et al. Auditing subtype inconsistencies among Gene Ontology concepts. In: Xiaohua Hu (ed) 2017 IEEE International Conference on Bioinformatics and Biomedicine (BIBM). Piscataway, New Jersey: IEEE, 2017, 1242–5. [Google Scholar]
- 25. Abeysinghe R, Hinderer EW, Moseley HNB, et al. SSIF: Subsumption-based Sub-term Inference Framework to audit Gene Ontology. Bioinformatics 2020;36(10):3207–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Binns D, Dimmer E, Huntley R, et al. QuickGO: a web-based tool for Gene Ontology searching. Bioinformatics 2009;25(22):3045–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Explosion AI. spaCy: Industrial-Strength Natural Language Processing in Python. https://spacy.io/ (25 January 2022, date last accessed).
- 28. The OBO Foundry. Relations Ontology . http://www.obofoundry.org/ontology/ro.html (25 January 2022, date last accessed).
- 29. Klopfenstein DV, Zhang L, Pedersen BS, et al. GOATOOLS: A Python library for Gene Ontology analyses. Sci Rep 2018;8(1):10872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Raff H. CORT, Cort, B, Corticosterone, and now Cortistatin: Enough Already! Endocrinology 2016;157(9):3307–8. [DOI] [PubMed] [Google Scholar]
- 31. Loper E, Bird S. In: Chris Brew, and Michael Rosner (eds) Proceedings of the ACL-02 Workshop on Effective tools and methodologies for teaching natural language processing and computational linguistics. Association for Computational Linguistics, Stroudsburg, Pennsylvania 2002, 63–70.
- 32. Manning CD, Surdeanu M, Bauer J, et al. The Stanford CoreNLP natural language processing toolkit. In: Kalina Bontcheva, Jingbo Zhu (eds) 52nd annual meeting of the association for computational linguistics: system demonstrations, Stroudsburg, Pennsylvania: Association for Computational Linguistics, 2014, 55–60.
- 33. Luo L, Feng J, Yu H, et al. Automatic Structuring of Ontology Terms Based on Lexical Granularity and Machine Learning: Algorithm Development and Validation. JMIR Med Inform 2020;8(11):e22333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cui L, Zhu W, Tao S, et al. Mining non-lattice subgraphs for detecting missing hierarchical relations and concepts in SNOMED CT. J Am Med Inform Assoc 2017;24(4):788–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cui L, Bodenreider O, Shi J, et al. Auditing SNOMED CT hierarchical relations based on lexical features of concepts in non-lattice subgraphs. J Biomed Inform 2018;78:177–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Abeysinghe R, Brooks MA, Talbert J, et al. Quality assurance of NCI Thesaurus by mining structural-lexical patterns. In: AMIA annual symposium proceedings 2017, p. 364. American Medical Informatics Association. 2017:364. [PMC free article] [PubMed] [Google Scholar]
- 37. Abeysinghe R, Brooks MA, Cui L. Leveraging non-lattice subgraphs to audit hierarchical relations in NCI Thesaurus. In: AMIA annual symposium proceedings 2019, p. 982. American Medical Informatics Association. [PMC free article] [PubMed] [Google Scholar]
- 38. Degtyarenko K, De Matos P, Ennis M, et al. ChEBI: a database and ontology for chemical entities of biological interest. Nucleic Acids Res 2007;36(suppl_1):D344–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. The Gene Ontology Consortium . GO-ontology tracking system. https://github.com/geneontology/go-ontology/issues (23 January 2022, date last accessed).
- 40. Lawrence M, Daujat S, Schneider R. Lateral thinking: how histone modifications regulate gene expression. Trends Genet 2016;32(1):42–56. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.