Abstract
Despite its essential role in the (patho)physiology of several diseases, CB2R tissue expression profiles and signaling mechanisms are not yet fully understood. We report the development of a highly potent, fluorescent CB2R agonist probe employing structure-based reverse design. It commences with a highly potent, preclinically validated ligand, which is conjugated to a silicon-rhodamine fluorophore, enabling cell permeability. The probe is the first to preserve interspecies affinity and selectivity for both mouse and human CB2R. Extensive cross-validation (FACS, TR-FRET and confocal microscopy) set the stage for CB2R detection in endogenously expressing living cells along with zebrafish larvae. Together, these findings will benefit clinical translatability of CB2R based drugs.
Detection and visualization of the cannabinoid receptor type 2 by a cell-permeable high affinity fluorescent probe platform enables tracing receptor trafficking in live cells and in zebrafish.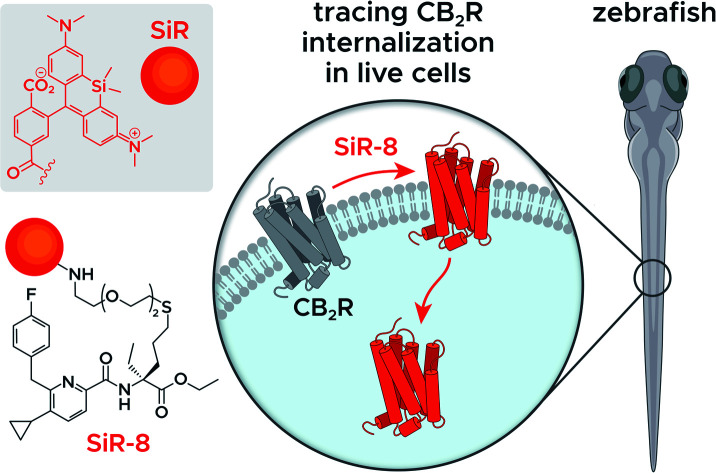
Introduction
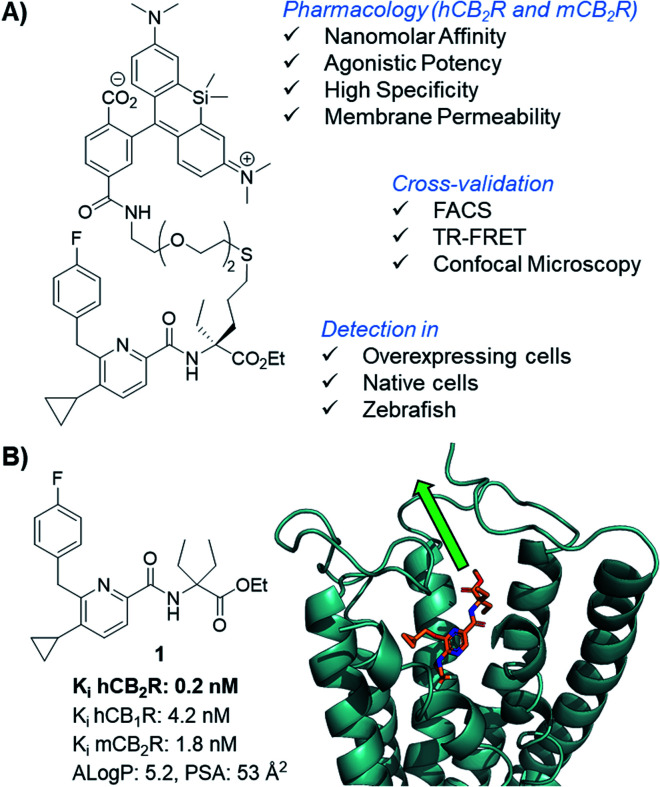
Cannabinoid type 1 and 2 receptors (CB1R and CB2R) are key transducers of extracellular stimuli in the endocannabinoid (eCB) system, a fundamental lipid signaling network in all vertebrates.1,2 Kidney, cardiovascular, gastrointestinal, lung, neurodegenerative and psychiatric disorders, along with pain and cancer are linked to eCB impairment.3–7 While CB1R is mainly expressed in the central nervous system and to a lesser extent in peripheral tissue, CB2R is found throughout the periphery and primarily expressed in immune cells.8,9 Tissue and cell-type specific receptor expression profiles remain largely uncharted because appropriate biological and chemical tools are lacking. This is aggravated by the absence of antibodies sufficiently specific for both human and rodent CB2R; which even if available would be precluded for use in translational imaging and incapable of intracellular permeation.10–12 The development of therapies involving novel CB2R agonists requires understanding of molecular and cellular mechanisms of action,13,14 thus, chemical probes targeting CB2R are needed. Herein we report the synthesis of a novel, selective, cell-permeable, high affinity CB2R-agonist fluorescent probe (Fig. 1A) along with its pharmacological validation.
Fig. 1. CB2R agonist 1 for the design of CB2R-selective fluorescent probe and docking experiments into the co-crystal structure of active state CB2R with agonist AM12033 (PDB 6KPC).15.
This is the first directly labeled small molecule probe to preserve binding affinity and agonistic efficacy at both human and mouse CB2R. The probe is applied and cross-validated using time-resolved fluorescence resonance energy transfer (TR-FRET) by flow cytometry and cellular trafficking studies. We showcase its application with super resolution live cell imaging in native cells and zebrafish.
Results and discussion
Probe design, synthesis, and pharmacological characterization
Fluorescent imaging probes have emerged as sensitive tools with high degree of spatiotemporal resolution.16 Accordingly, we sought to develop a probe that would provide a well-validated translational path from preclinical pharmacological animal data to clinic. In earlier work,17,18 we described fluorescent probes based on a phytocannabinoid that was lacking desirable pharmacokinetics for translational applications. Accordingly, here we employ a drug-derived, reverse-design approach from a pharmacologically well-validated in vivo active compound class that would satisfy a number of desirable boundary conditions: affinity, potency and selectivity, chemical stability, water solubility and membrane permeability.19,20 A ligand that meets these criteria is 1,21–23 displaying picomolar selective binding affinity on human and mouse CB2R and full agonistic picomolar potency (Fig. 1B and ESI Table S1†). With this as a lead, a conjugation handle was required for fluorophore attachment.
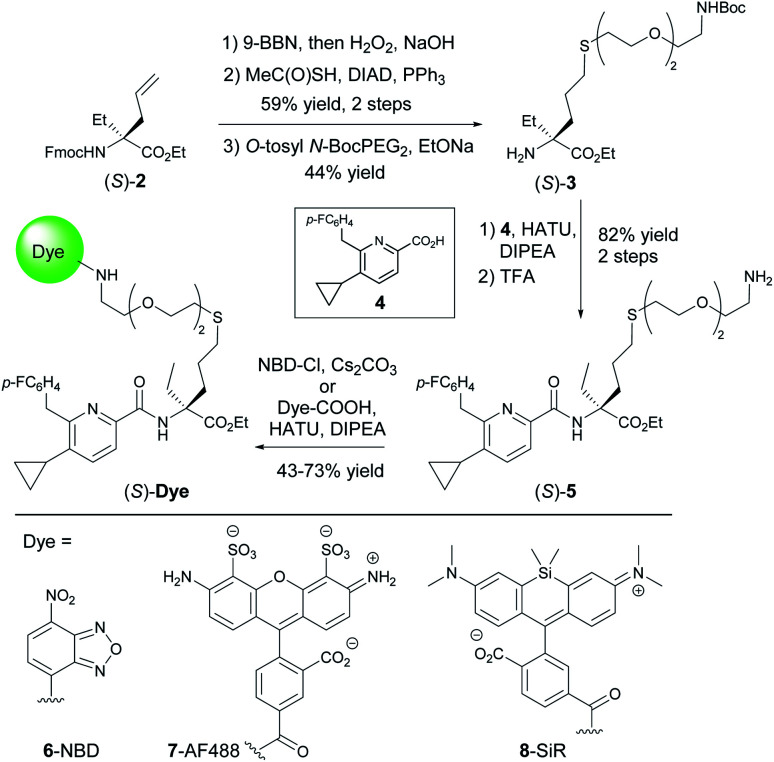
Prospective SAR studies supported by molecular modelling using docking poses of 1 suggested an exit vector (Fig. 1B, green arrow and ESI Tables S2 and 3†) for thioether linked spacers reaching out into the extracellular space, ultimately leading to the design of (S)-5 as a versatile lynchpin (Scheme 1). Its synthesis commenced with hydroboration of (S)-2, followed by thio-Mitsunobu reaction which allowed linker introduction. The absolute configuration of 2 was assigned by X-ray crystallography of a N-toluenesulfonyl-(S)-proline derivative (ESI-34, see ESI, S23 and S24†). Following deprotection and coupling, (S)-3 was isolated. Subsequent amide coupling of (S)-3 with acid 4 and deprotection furnished (S)-5. This compound was used to generate several different probes for study: 6-NBD, 7-AF488, and 8-SiR (Scheme 1). Their pharmacological investigation revealed that all probes displayed high potency, selectivity, and full agonistic properties showing high generality of the probe platform (Table 1 and ESI Table S1†). These CB2R-selective fluorescent probes are the first to preserve interspecies affinity and selectivity for both mouse and human CB2R. Interestingly, (S)-6-NBD displayed greater affinity than its enantiomer (Table 1) towards human and murine CB2R. A similar observation was made for fully labeled Alexa Fluor 488 probe pair 7 ((S)-7 hCB2R Ki: 44 nM vs. (R)-7 hCB2R Ki: 62 nM), where only 1.4-fold enantio-discrimination with regard to hCB2R binding affinity favoring the (S)-enantiomer was observed (ESI Table S1†). In particular, NBD probe (S)-6 and 8-SiR outperformed with regard to functional selectivity versus CB1R (hEC50 ratio CB1R/CB2R for (S)-6: >4545 and for 8: >149, Tables 1 and 1†). Consequently, we proceeded with the (S)-enantiomers for our studies. Probe optimization and dye selection were supported by careful evaluation of absorption and emission spectra in buffer (ESI Fig. S3 and Table S6†). In these studies, all probes were observed to be water soluble without any tendency to form aggregates. Broad lipophilicity range of Alog P values was observed, 5.3–10.6 for 7-AF488 and 8-SiR, respectively, suggesting good permeability for 8-SiR which is devoid of negative charges as per our design.
Scheme 1. Synthesis of probe candidates.
Selected pharmacological characteristics of fluoroprobes 6, 7, and 8a.
| Probe | Alog P | K i [nM] | cAMP EC50 [nM] (% efficacy) | ||||
|---|---|---|---|---|---|---|---|
| hCB2R | hCB1R | mCB2R | mCB1R | hCB2R | hCB1R | ||
| (S)-6-NBD | 6.3 | 9.1 | 617 | 33 | 691 | 2.2 (72) | >10 000 |
| (R)-6-NBD | 6.3 | 159 | 4925 | 622 | n.d. | 17 (84) | >10 000 |
| (S)-7-AF488 | 5.3 | 44 | 321 | 28 | >10 000 | 1.3 (100) | 86 (109) |
| (S)-8-SiR | 10.6 | 62 | 114 | 117 | 1892 | 67 (96) | >10 000 |
See ESI, S10 for detailed description of the assay protocols.
To experimentally probe permeability, the effective permeation coefficients of (R)- and (S)-6-NBD were measured in the parallel artificial membrane permeability assay (PAMPA). The fact that both passively permeated through membranes suggested the more lipophilic probe 8-SiR was likely to be cell-permeable24 thereby enabling studies of intracellular compartments. Conversely, negatively charged 7-AF488 was not capable of passive membrane permeation (see ESI, S43†), making it potentially useful in the investigation of extracellular receptor pools. To identify potential off-targets of our labeled ligands, 7-AF488 probe, having the highest potency, was screened against a customized panel of 50 representative receptors and enzymes. In this assay, ligand 7 exhibited a very clean selectivity profile (ESI Table S4†).
Validation of 8-SiR in overexpressing cells using complementary imaging techniques
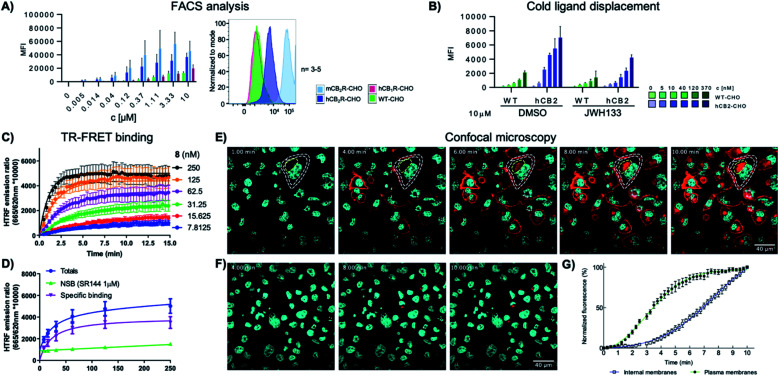
To study the specificity of 8-SiR for human and mouse CB2R, CHO cells overexpressing either hCB2R, mCB2R or hCB1R were incubated at various concentrations and analyzed in FACS experiments (Fig. 2A). In comparison to hCB1R overexpressing cells or WT-CHO control, 8-SiR was highly specific for CHO cells overexpressing either hCB2R or mCB2R. To confirm ligand specificity and exclude unspecific binding, we investigated whether these compounds can compete for the CB2R binding site with known cold CB2R ligand agonist JWH133 (ref. 25) (Fig. 2B). Following pre-incubation of WT-CHO or hCB2R-CHO cells with these, 8-SiR efficiently displaced CB2R-agonist JWH133 over a broad concentration range. This illustrates the high degree of target specificity of 8-SiR in a cellular setting targeting the active conformation of the receptors in its native cellular environment and thereby overruling the observed limited human CB1R/CB2R selectivity ratio in the binding assay by relevant cellular data.26 Similar high specificity and full displacement by JWH133 was also observed when 7-AF488 was analyzed in FACS (ESI Fig. S4†).
Fig. 2. (A) FACS analysis of the mean fluorescent intensity (MFI) of WT, hCB2R, mCB2R and hCB1R overexpressing CHO cells at different concentrations of 8-SiR. (B) FACS analysis of the MFI of WT and hCB2R-CHO cells pre-treated with JWH133 (10 μM) and stained with different concentrations of 8-SiR; see ESI† for details. (C and D) TR-FRET characterization of 8-SiR binding association (C) and saturation analysis (D) using HEK-hCB2R cell membranes. (E and F) Time-lapse confocal microscopy frames for hCB2R (E) and hCB1R (F). CHO cells co-stained with 8-SiR (red) and Hoechst 33342 (cyan, nucleus counter stain) at 1, 4, 6, 8 and 10 min; plasma and internal membranes are highlighted with white and yellow dashes, respectively. (G) Association curve of 0.4 μM 8-SiR on plasma membrane and internal membranes of hCB2R-CHO cells. See also ESI Videos S1 and S2.†.
As previously described by us,17 we assayed kinetic and equilibrium binding of 8-SiR via TR-FRET (Fig. 2C and D) and further confirmed its use as tracer for TR-FRET applications (see ESI, S16–S20†).
Next, we employed 8-SiR in confocal microscopy experiments. Currently available CB2R probes require long pre-incubation times (>30 min) in live-cell imaging experiments, contributing to unspecific staining.27–31 By contrast, we observed instantaneous labeling with 8-SiR. In accordance with the PAMPA assay (ESI Table S3†), probe 8-SiR is membrane permeable and successfully entered hCB2R-overexpressing cells, reaching internal receptor pools (Fig. 2E and ESI Video S2†). Besides the cell membrane, labeled hCB2R was detected in intracellular compartments, predominantly within perinuclear structures, reminiscent of Golgi complex and endoplasmic reticulum (see ESI Fig. S4† for higher magnification). Kinetic analysis of the staining of internal membranes with CB2R shows that after 10 minutes saturation had not occurred (Fig. 2G). Moreover, 8-SiR was nearly non-fluorescent in aqueous environments. This feature allows bright labeling of cellular membranes even in the continued presence of 8-SiR in culture media,24,32 which permits imaging over prolonged time periods. Importantly, in hCB1R-overexpressing CHO cells, 8-SiR did not label the membrane, demonstrating high CB2R-specificity and remarkably low unspecific binding (Fig. 2F and ESI Video S1†). For detection applications, cell-permeable agonist probes have the advantage of accumulating inside cells upon receptor internalization and following recycling events, which consecutively lead to signal amplification, improved image contrast and enhanced detection sensitivity.33
8-SiR enables CB2R visualization in live cells
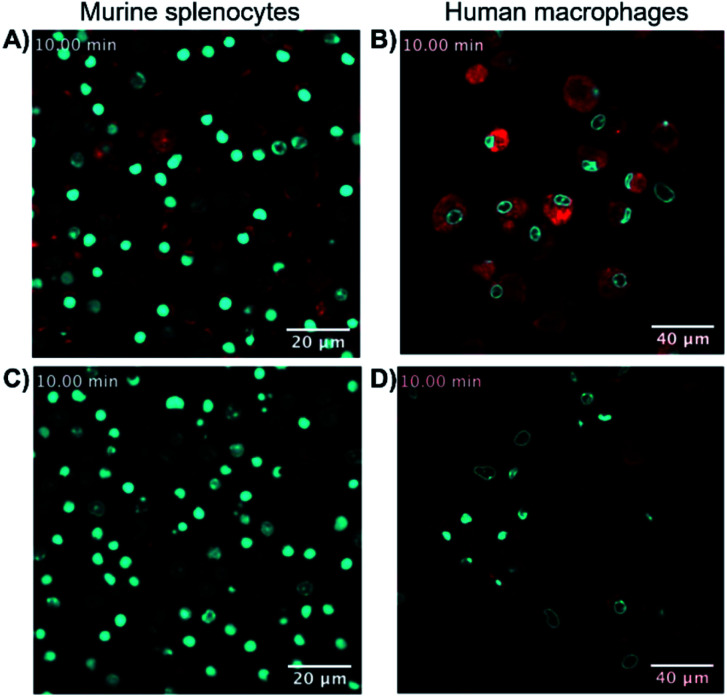
Finally, the ability of 8-SiR to visualize endogenous CB2R expression in non-transfected cells was tested (Fig. 3). A challenge in CB2R research is the very low expression level of the receptor in native cells, even when these are known to be responsive to CB2R activation.34,35 To this end, primary cultures of human macrophages derived from healthy donors and of murine splenocytes from healthy C57BL/6J wild-type mice were isolated. Incubation of primary cell populations with 8-SiR resulted in increased mean fluorescence (Fig. 3A, B, ESI Videos S3, S4, and S6†).
Fig. 3. Super resolution fluorescence microscopy in native cells using probe 8-SiR. Time-lapse confocal microscopy frames with 8-SiR in murine splenocytes (A and C) and human macrophages (B and D). Cells were pre-stained with Hoechst 33342 (cyan) to counter stain the nuclei and incubated for 10 min with 0.4 μM 8-SiR (A and B) or 0.6 μM 8-SiR and 4 μM JWH133 (C and D). See also ESI Videos S3 to S7.†.
Consistent with the results obtained previously with CHO-CB2R cells, 8-SiR produced robust, time-dependent membrane labeling and effected receptor internalization. Probe specificity towards human and mouse CB2R was evident by the abrupt reduction in observed fluorescence after co-incubating cells with 8-SiR with non-fluorescent JWH133, an agonist with high affinity for human and mouse CB2R25 (Fig. 3C, D and ESI Videos S5 and S7†). These competition studies demonstrate the capability of probe 8-SiR for real-time staining of CB2R at native levels with high specificity. Additionally, the excellent cell permeability of 8-SiR allows accurate tracing of subcellular CB2R receptor distribution and traffic dynamics as well as staining total cellular content of CB2R in real-time, live-cell imaging.
Imaging CB2R in vivo using 8-SiR
Having demonstrated the use of 8-SiR for detection of CB2R in native cells, we then studied the probes in live animals. Zebrafish embryos and larvae are an established model in biomedical research with high potential for clinical translation36–38 and show a remarkably high CB2R binding site sequence identity to the human receptor39,40 (ESI Fig. S2†). Probes 7-AF488 and 8-SiR were intravenously injected via the Duct of Cuvier at 3 days post fertilization (dpf), and their dynamics were analyzed using confocal microscopy (Fig. 4).
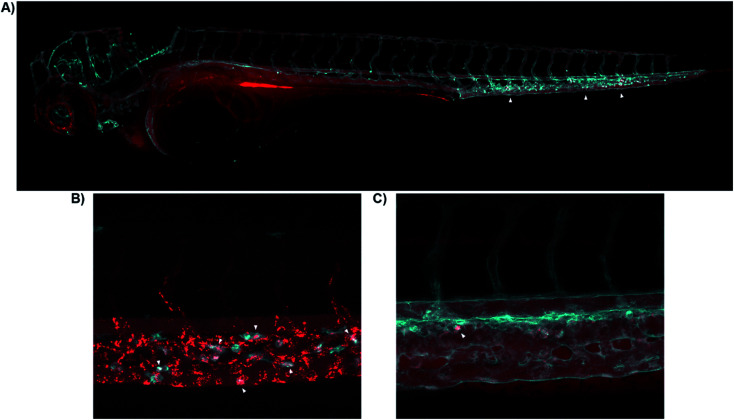
Fig. 4. Real-time in vivo visualization of CB2R in zebrafish embryos. (A) 8-SiR (red) injected in 3dpf Tg(fli1:EGFP) zebrafish larvae, which expresses EGFP fluorescent protein in the blood vasculature (cyan). 8-SiR was found freely circulating in the blood vasculature after injection, with increased intensity in the CHT region (white triangles). (B) 8-SiR injected in 3dpf Tg(mpeg1.1:mCherry-F) zebrafish larvae, which expresses mCherry-F fluorescent protein in macrophages (cyan), demonstrating its colocalization. (C) 7-AF488 (red) injected in 3dpf Tg(kdrl:mCherry) zebrafish larvae, which expresses mCherry fluorescent protein in the blood vasculature (cyan). See also ESI Videos S8 and S9.†.
Both probes were freely circulating as evidenced by fluorescence within the blood vasculature, which slowly decayed during the first hours post injection due to diffusion throughout the body (Fig. 4A). Over time, probes were extensively taken up at the Caudal Hematopoietic Tissue (CHT), showing as a strong punctate pattern (Fig. 4A). The CHT is a highly perfused tissue wherein hematopoietic stem cells and immune precursors such as macrophages and neutrophils develop during zebrafish embryogenesis.41 Given CB2R's implication in the immune system,42–44 we focused on CHT to study the possible uptake of the probes by the residing immune cells.45,46 However, the CHT is also the residing place for endothelial cells that remove macromolecular waste from the blood via scavenging receptors. Analysis revealed uptake of 8-SiR from motile cells (immune-related) and non-motile (endothelial) cells. Indeed, 8-SiR colocalized with local residing macrophages prior to its removal from blood circulation via endothelial cells (Fig. 4B, and ESI Videos S8 and S9†).
To avoid imaging of binding due to the unspecific scavenging function of endothelial cells, additional analyses were performed during the first 3 hours post injection. These demonstrated specific uptake of 7-AF488 and 8-SiR from motile cells both inside CHT as well as crawling into the adjacent fin, putatively endocytosed after contact with CB2R (Fig. 4C and ESI Video S9†). In contrast, injecting the fluorophores without the CB2R recognizing element resulted in unspecific fluorescence signal in all cellular membranes in contact47 (ESI Fig. S5 and Video S10†).
Conclusions
In conclusion, we have developed cell-permeable, fluorescent probe 8-SiR, based on a preclinical CB2R agonist 1. It displays high affinity, specificity, and agonistic potency. Moreover, the probe shows unprecedented, highly consistent interspecies affinity and potency for both human and mouse CB2R. Subsequent to validation in overexpressing systems, the probe was used for imaging in native cells and ultimately in live zebrafish. Specifically, in flow cytometry experiments we labelled CB2R overexpressing cells and demonstrated target specificity by competition experiments. In fluorescence-based TR-FRET assays, kinetic and equilibrium binding profiles for 8-SiR were determined. Fluoroprobe 8-SiR was employed to monitor intracellular CB2R distribution in real-time, live-cell imaging by super resolution confocal microscopy with overexpressing hCB2R-CHO cells and endogenously expressing native cells from human macrophages and mouse splenocytes. In zebrafish larvae, 8-SiR was found to be well tolerated after intravenous injection. It freely circulated inside the blood vasculature over prolonged times before colocalizing with CB2R expressing macrophages. This demonstrates the implementation of the probe in vivo and attendant aspects such as biosafety, biodistribution, and cellular uptake.
Probe 8-SiR provides researchers a unique and novel platform to: (1) overcome the large interspecies differences; (2) access a high specificity and low nanomolar affinity probe for CB2R with full agonist efficacy, and (3) perform an array of biologically and pharmacologically relevant experiments to detect CB2R and study its function. We anticipate that this novel highly translational agonist CB2R fluorescent probe will help to elucidate CB2R molecular and cellular mechanisms of action, as well as to unravel its expression levels in different disease states.
Data availability
Primary data for probe synthesis, characterization, photophysical measurements, and fluorescence imaging, as well as atomic coordinates are provided in the ESI.†
Author contributions
M. N., U. G., E. M. C., C. U. and J. F. conceived the research. M. N and U. G. acquired funding for this project. T. G., B. B., D. S., G. F.-C., W. G., A. C. R., N. G., M. H., J. F., K. G., J. R., C. J. H., P. J. MC., M. v. d. S., P. P., J. G., C. U., S. O., H. P. S., M. Ma., H. S., D. B. V., E. M. C., U. G. and M. N. designed the research approach. T. G., B. B, K. A., C. K., D. S., G. F.-C., Y. M., L. M., M. W., R. C. S., M. V. W., P. P., B. L. H., T. M., M. Me., N. R., E. J. K., W. G., A. A., E. A. K., S. H., C. R., E. A. Z., A. O., A. P., S. M., J. B., I. B.-C., F. D., G. W., D. H., T. v. d. W., H. M., Y. S., Z. V. V. G. F.-C, J. S., J. Br. and S. O. performed experiments, analyzed raw data, and worked on probe validation. T. G., C. K., D. S., G. F.-C, S. O., U. G., and M. N. analyzed all data generated, and wrote the manuscript. B. B., K. A., R. C. S., M. V. W., P. P., M. v. d. S., H. P. S., M. Ma., D. B. V., and E. M. C. provided useful comments and feedback for the manuscript. K. G. was not contactable during the submission process of the manuscript, but contributed to the work. All authors have given approval to the final version of the manuscript.
Conflicts of interest
The authors have no conflicts to declare.
Supplementary Material
Acknowledgments
M. N., T. G., B. B., Y. M., L. M. M. K.-W. and M. W. would like to thank Edgar Specker, Sandra Miksche and the NMR core facility of the FMP for their excellent support on compound characterization. We are grateful to Barth van Rossum for preparing graphical artwork. We greatly acknowledge the chiral separation of key building block 2 by Daniel Zimmerli and Erik Hunziker. Some aspects of developing TR-FRET assay were supported by the Swiss National Science Foundation grant 159748 to D. B. V., E. J. K. was funded by the IBSA Foundation for Scientific Research. M. Ma. and S. O. thank Lucia Scipioni and Antonio Totaro for cell culture and technical support, and Daunia Laurenti for her technical assistance in live imaging. They are also grateful to the Italian Ministry of Education, University and Research (MIUR) for partial financial support under the competitive grant PRIN 2017. The determination of solubility data by Catherine Karrer and log D values by Aynur Ekiciler is greatly acknowledged. We furthermore thank Björn Wagner, Virginie Micallef and Joelle Muller for the generation of PAMPA data. We thank Mathias Christmann, Rainer Haag and PD Daniel Häussinger for helpful discussions. This work was partially supported by the Sino-German Research Project grant (GZ 1271) to M. N. K. G. Z. V. V. and P. P. are thankful for the support of the Intramural Research Program of NIAAA, NIH. E. M. C. is grateful to ETH Zürich for financial support. R. C. S. and P. P. are awardees of the Scholarship Fund of the Swiss Chemical Industry (SSCI).
Electronic supplementary information (ESI) available: Supplementary figures and tables; general synthetic methods; compound synthesis and characterization; molecular docking; in vitro pharmacology; fluorescence spectroscopy; TR-FRET kinetic CB2R binding assay; FACS analysis; time-lapse confocal imaging; in vivo zebrafish imaging; NMR spectra; and N-terminal SNAP-hCB2R sequence. CCDC 1923120. For ESI and crystallographic data in CIF or other electronic format see DOI: 10.1039/d1sc06659e
References
- Munro S. Thomas K. L. Abu-Shaar M. Nature. 1993;365:61–65. doi: 10.1038/365061a0. [DOI] [PubMed] [Google Scholar]
- Devane W. A. Dysarz F. A. Johnson M. R. Melvin L. S. Howlett A. C. Mol. Pharmacol. 1988;34:605–613. [PubMed] [Google Scholar]
- Pacher P. Mechoulam R. Prog. Lipid Res. 2011;50:193–211. doi: 10.1016/j.plipres.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon J. Hohmann A. G. Br. J. Pharmacol. 2008;153:319–334. doi: 10.1038/sj.bjp.0707531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picone R. P. Kendall D. A. Mol. Endocrinol. 2015;29:801–813. doi: 10.1210/me.2015-1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malfitano A. M. Basu S. Maresz K. Bifulco M. Dittel B. N. Semin. Immunol. 2014;26:369–379. doi: 10.1016/j.smim.2014.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertwee R. G. Philos. Trans. R. Soc. 2012;367:3353–3363. doi: 10.1098/rstb.2011.0381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turcotte C. Blanchet M. R. Laviolette M. Flamand N. Cell. Mol. Life Sci. 2016;73:4449–4470. doi: 10.1007/s00018-016-2300-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galiegue S. Mary S. Marchand J. Dussossoy D. Carriere D. Carayon P. Bouaboula M. Shire D. Le Fur G. Casellas P. Eur. J. Biochem. 1995;232:54–61. doi: 10.1111/j.1432-1033.1995.tb20780.x. [DOI] [PubMed] [Google Scholar]
- Zhang H. Y. Shen H. Jordan C. J. Liu Q. R. Gardner E. L. Bonci A. Xi Z. X. Acta Pharmacol. Sin. 2019;40:398–409. doi: 10.1038/s41401-018-0037-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cecyre B. Thomas S. Ptito M. Casanova C. Bouchard J. F. Naunyn Schmiedebergs Arch. Pharmacol. 2014;387:175–184. doi: 10.1007/s00210-013-0930-8. [DOI] [PubMed] [Google Scholar]
- Marchalant Y. Brownjohn P. W. Bonnet A. Kleffmann T. Ashton J. C. J. Histochem. Cytochem. 2014;62:395–404. doi: 10.1369/0022155414530995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler D. S. Vappiani J. Caneque T. Lam E. Y. N. Ward A. Gilan O. Chan Y. C. Hienzsch A. Rutkowska A. Werner T. Wagner A. J. Lugo D. Gregory R. Ramirez Molina C. Garton N. Wellaway C. R. Jackson S. MacPherson L. Figueiredo M. Stolzenburg S. Bell C. C. House C. Dawson S. J. Hawkins E. D. Drewes G. Prinjha R. K. Rodriguez R. Grandi P. Dawson M. A. Science. 2017;356:1397–1401. doi: 10.1126/science.aal2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon G. M. Niphakis M. J. Cravatt B. F. Nat. Chem. Biol. 2013;9:200–205. doi: 10.1038/nchembio.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua T. Li X. Wu L. Iliopoulos-Tsoutsouvas C. Wang Y. Wu M. Shen L. Johnston C. A. Nikas S. P. Song F. Song X. Yuan S. Sun Q. Wu Y. Jiang S. Grim T. W. Benchama O. Stahl E. L. Zvonok N. Zhao S. Bohn L. M. Makriyannis A. Liu Z. J. Cell. 2020;180:655–665. doi: 10.1016/j.cell.2020.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoddart L. A. Kilpatrick L. E. Briddon S. J. Hill S. J. Neuropharmacology. 2015;98:48–57. doi: 10.1016/j.neuropharm.2015.04.033. [DOI] [PubMed] [Google Scholar]
- Sarott R. C. Westphal M. Pfaff P. Korn C. Sykes D. A. Gazzi T. Brennecke B. Atz K. Weise M. Mostinski Y. Hompluem P. Koers E. Miljus T. Roth N. J. Asmelash H. Vong M. C. Piovesan J. Guba W. Rufer A. Kusznir E. A. Huber S. Raposo C. Zirwes E. A. Osterwald A. Pavlovic A. Moes S. Beck J. Benito-Cuesta I. Grande T. Ruiz de Martin S. Yeliseev A. A. Drawnel F. Widmer G. Holzer D. van der Wel T. Mandhair H. Yuan C.-Y. Drobyski W. Saroz Y. Grimsey N. L. Honer M. Fingerle J. Gawrisch K. Romero J. Hillard C. Varga Z. van der Stelt M. Pacher P. Gertsch J. McCormick P. Ullmer C. Oddi S. Maccarrone M. Veprintsev D. Nazaré M. Grether U. Carreira E. M. J. Am. Chem. Soc. 2020;142:16953–16964. doi: 10.1021/jacs.0c05587. [DOI] [PubMed] [Google Scholar]
- Westphal M. V. Sarott R. C. Zirwes E. A. Osterwald A. Guba W. Ullmer C. Grether U. Carreira E. M. Chem.–Eur. J. 2020;26:1380–1387. doi: 10.1002/chem.201904584. [DOI] [PubMed] [Google Scholar]
- Blagg J. Workman P. Cancer Cell. 2017;32:9–25. doi: 10.1016/j.ccell.2017.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Workman P. Collins I. Chem. Biol. 2010;17:561–577. doi: 10.1016/j.chembiol.2010.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bissantz C., Grether U., Hebeisen P., Kimbara A., Liu Q., Nettekoven M., Prunotto M., Roever S., Rogers-Evans M., Schulz-Gasch T., Ullmer C., Wang Z. and Yang W., WO2012168350A1, 2012
- Haider A. Gobbi L. Kretz J. Ullmer C. Brink A. Honer M. Woltering T. J. Muri D. Iding H. Burkler M. Binder M. Bartelmus C. Knuesel I. Pacher P. Herde A. M. Spinelli F. Ahmed H. Atz K. Keller C. Weber M. Schibli R. Mu L. Grether U. Ametamey S. M. J. Med. Chem. 2020;63:10287–10306. doi: 10.1021/acs.jmedchem.0c00778. [DOI] [PubMed] [Google Scholar]
- Haider A. Kretz J. Gobbi L. Ahmed H. Atz K. Burkler M. Bartelmus C. Fingerle J. Guba W. Ullmer C. Honer M. Knuesel I. Weber M. Brink A. Herde A. M. Keller C. Schibli R. Mu L. Grether U. Ametamey S. M. J. Med. Chem. 2019;62:11165–11181. doi: 10.1021/acs.jmedchem.9b01280. [DOI] [PubMed] [Google Scholar]
- Lukinavičius G. Umezawa K. Olivier N. Honigmann A. Yang G. Plass T. Mueller V. Reymond L. Corrêa Jr I. R. Luo Z. G. Schultz C. Lemke E. A. Heppenstall P. Eggeling C. Manley S. Johnsson K. Nat. Chem. 2013;5:132–139. doi: 10.1038/nchem.1546. [DOI] [PubMed] [Google Scholar]
- Soethoudt M. Grether U. Fingerle J. Grim T. W. Fezza F. de Petrocellis L. Ullmer C. Rothenhausler B. Perret C. van Gils N. Finlay D. MacDonald C. Chicca A. Gens M. D. Stuart J. de Vries H. Mastrangelo N. Xia L. Alachouzos G. Baggelaar M. P. Martella A. Mock E. D. Deng H. Heitman L. H. Connor M. Di Marzo V. Gertsch J. Lichtman A. H. Maccarrone M. Pacher P. Glass M. van der Stelt M. Nat. Commun. 2017;8:13958–13972. doi: 10.1038/ncomms13958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The binding data are generated from previously frozen membranes, while flow cytometry experiments are performed with viable cells. We hypothesize that membrane lysate preparation affects CB2R and CB1R structure differentially, with detrimental effects on the observed selectivity. In the cAMP assay, which is also performed on living cells, the behaviour is consistently similar to that in flow cytometry, confocal imaging. Therefore, in the subsequent much more relevant multiple whole cell evaluations targeting the agonist binding site in its native conformation we observed consistently a very high selectivity.
- Singh S. Oyagawa C. R. M. Macdonald C. Grimsey N. L. Glass M. Vernall A. J. ACS Med. Chem. Lett. 2019;10:209–214. doi: 10.1021/acsmedchemlett.8b00597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spinelli F. Giampietro R. Stefanachi A. Riganti C. Kopecka J. Abatematteo F. S. Leonetti F. Colabufo N. A. Mangiatordi G. F. Nicolotti O. Perrone M. G. Brea J. Loza M. I. Infantino V. Abate C. Contino M. Eur. J. Med. Chem. 2020;188:112037. doi: 10.1016/j.ejmech.2020.112037. [DOI] [PubMed] [Google Scholar]
- Zhang S. Shao P. Bai M. Bioconjugate Chem. 2013;24:1907–1916. doi: 10.1021/bc400328m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling X. Zhang S. Shao P. Li W. Yang L. Ding Y. Xu C. Stella N. Bai M. Biomaterials. 2015;57:169–178. doi: 10.1016/j.biomaterials.2015.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrov R. R. Ferrini M. E. Jaffar Z. Thompson C. M. Roberts K. Diaz P. Bioorg. Med. Chem. Lett. 2011;21:5859–5862. doi: 10.1016/j.bmcl.2011.07.099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L. Frei M. S. Salim A. Johnsson K. J. Am. Chem. Soc. 2019;141:2770–2781. doi: 10.1021/jacs.8b11134. [DOI] [PubMed] [Google Scholar]
- Reynolds F. Kelly K. A. Mol. Imaging. 2011;10:407–419. doi: 10.2310/7290.2011.00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper A. Singh S. Hook S. Tyndall J. D. A. Vernall A. J. Pharmacol. Rev. 2017;69:316–353. doi: 10.1124/pr.116.013243. [DOI] [PubMed] [Google Scholar]
- Grimsey N. L. Goodfellow C. E. Dragunow M. Glass M. Biochim. Biophys. Acta. 2011;1813:1554–1560. doi: 10.1016/j.bbamcr.2011.05.010. [DOI] [PubMed] [Google Scholar]
- Gomes M. C. Mostowy S. Trends Microbiol. 2020;28:10–18. doi: 10.1016/j.tim.2019.08.005. [DOI] [PubMed] [Google Scholar]
- Patton E. E. Tobin D. M. Dis. Models Mech. 2019;12:dmm039370. doi: 10.1242/dmm.039370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patton E. E. Zon L. I. Langenau D. M. Nat. Rev. Drug Discovery. 2021;20:611–628. doi: 10.1038/s41573-021-00210-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krug II R. G. Clark K. J. Gene. 2015;570:168–179. doi: 10.1016/j.gene.2015.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oltrabella F. Melgoza A. Nguyen B. Guo S. Dev., Growth Differ. 2017;59:194–210. doi: 10.1111/dgd.12351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murayama E. Kissa K. Zapata A. Mordelet E. Briolat V. Lin H.-F. Handin R. I. Herbomel P. Immunity. 2006;25:963–975. doi: 10.1016/j.immuni.2006.10.015. [DOI] [PubMed] [Google Scholar]
- Esain V. Kwan W. Carroll K. J. Cortes M. Liu S. Y. Frechette G. M. Sheward L. M. Nissim S. Goessling W. North T. E. Stem Cells. 2015;33:2596–2612. doi: 10.1002/stem.2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y. J. Fan H. B. Jin Y. Ren C. G. Jia X. E. Wang L. Chen Y. Dong M. Zhu K. Y. Dong Z. W. Ye B. X. Zhong Z. Deng M. Liu T. X. Ren R. J. Biol. Chem. 2013;288:13551–13562. doi: 10.1074/jbc.M113.453811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Martin I. Herrero-Turrion M. J. Marron Fdez de Velasco E. Gonzalez-Sarmiento R. Rodriguez R. E. Gene. 2007;389:36–44. doi: 10.1016/j.gene.2006.09.016. [DOI] [PubMed] [Google Scholar]
- Arias-Alpizar G. Koch B. Hamelmann N. M. Neustrup M. A. Paulusse J. M. J. Jiskoot W. Kros A. Bussmann J. Nanomedicine. 2021;34:102395. doi: 10.1016/j.nano.2021.102395. [DOI] [PubMed] [Google Scholar]
- Campbell F. Bos F. L. Sieber S. Arias-Alpizar G. Koch B. E. Huwyler J. Kros A. Bussmann J. ACS Nano. 2018;12:2138–2150. doi: 10.1021/acsnano.7b06995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarin M. Faelens R. Giusti A. De Croze N. Leonard M. Cabooter D. Annaert P. de Witte P. Ny A. Sci. Rep. 2021;11:12229. doi: 10.1038/s41598-021-91612-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Primary data for probe synthesis, characterization, photophysical measurements, and fluorescence imaging, as well as atomic coordinates are provided in the ESI.†