Abstract
The on-going Microbial Observatory Experiments on the International Space Station (ISS) revealed the presence of various microorganisms that may be affected by the distinct environment of the ISS. The low-nutrient environment combined with enhanced irradiation and microgravity may trigger changes in the molecular suite of microorganisms leading to increased virulence and resistance of microbes. Proteomic characterization of two Aspergillus fumigatus strains, ISSFT-021 and IF1SW-F4, isolated from HEPA filter debris and cupola surface of the ISS, respectively, is presented, along with a comparison to experimentally established clinical isolates Af293 and CEA10. In-depth analysis highlights variations in the proteome of both ISS-isolated strains when compared to the clinical strains. Proteins that showed increased abundance in ISS isolates were overall involved in stress responses, and carbohydrate and secondary metabolism. Among the most abundant proteins were Pst2 and ArtA involved in oxidative stress response, PdcA and AcuE responsible for ethanol fermentation and glyoxylate cycle, respectively, TpcA, TpcF, and TpcK that are part of trypacidin biosynthetic pathway, and a toxin Asp-hemolysin. This report provides insight into possible molecular adaptation of filamentous fungi to the unique ISS environment.
Keywords: Aspergillus fumigatus, proteome, the International Space Station
1. INTRODUCTION
The International Space Station (ISS) is a man-made closed habitat that functions as a platform to study the impact of the distinct space environment, which includes enhanced irradiation and microgravity on humans1–7, animals8,9, plants10–14 and microorganisms15–19. Most experiments conducted on board the ISS are precisely planned. Studied organisms are intentionally sent to the ISS to investigate the possible alterations in their physiology, using ground controls for comparison. However, one on-going ISS Microbial Observatory (ISS-MO) experiment focuses on studying hitchhikers that have followed humans and cargo aboard the ISS20. Thorough investigation of microbiological characteristics of closed habitats, like the ISS, are indispensable to National Aeronautics and Space Administration (NASA), as manned long-term space flight missions are within reach. A deeper understanding of microbes that coexist in closed habitats with humans remains imperative to astronauts’ health and the overall maintenance of closed systems.
Strict scrutiny of the microbiome and mycobiome of the ISS20–25, Mir26,27, and Skylab28, in the past, has revealed prevalence of fungal genera: Cladosporium, Penicillium, and Aspergillus in space environments. These fungi can be both beneficial and detrimental to mankind, as they produce a myriad of commercially useful bioactive compounds29–34, while also causing allergies35, infections36,37 and biodeterioration of habitats38–40. Aspergillus fumigatus, one of many fungal isolates identified in a recent ISS-MO study22, is a ubiquitous saprophytic fungus41. Its enormous adaptation capacity enables it to not only be omnipresent in the environment, but also to be a successful opportunistic pathogen42. A. fumigatus causes variety of health conditions spanning from allergies to potentially life-threatening invasive aspergillosis (IA) in immunocompromised individuals43,44. Initial characterization of two A. fumigatus ISS-isolates, ISSFT-021 and IF1SW-F4, showed no outstanding differences in their genomes and secondary metabolites profiles when compared to clinical isolates CEA10 and Af293, however both isolates were significantly more lethal in a larval zebra fish model of IA45. Considering that A. fumigatus becomes more virulent in space and therefore potentially more dangerous to astronauts’ health, it was pertinent to further investigate molecular changes of ISS-isolated strains.
Presented in this study are the unique differences observed in proteome of two ISS-isolated A. fumigatus strains, ISSFT-021 and IF1SW-F4, when compared to Af293 and CEA10. The goal of this study was to understand if the distinct environment of the ISS (low-nutrients, enhanced irradiation and microgravity) alters the proteome of A. fumigatus. Due to an existing gap in our understanding of how filamentous fungi molecularly adapt to space conditions, proteome investigation of these two ISS-isolated A. fumigatus strains was prudent.
2. METHODS
2.1. Isolation and identification of A. fumigatus.
Procedures to isolate and identify A. fumigatus collected from the ISS were described previously45. In brief, HEPA filter associated particulates were scraped, resuspended in sterile phosphate-buffered saline (PBS; pH 7.4) and spread onto potato dextrose agar (PDA) plates22. Cupola surfaces were sampled with sterile polyester wipes assembled at Jet Propulsion Laboratory (JPL) prior to space flight. After each sampling event on board the ISS, wipes were returned to JPL for subsequent processing. During that process, two of multiple isolates were identified via ITS region and subsequently whole genome sequencing (WGS) as A. fumigatus.
2.2. Growth conditions.
Af293, CEA10, ISSFT-021 and IF1SW-F4 were cultivated for 5 days at 30°C on glucose minimal medium (GMM) agar plates (6 g/L NaNO3, 0.52 g/L KCl, 0.52 g/L MgSO4·7H2O, 1.52 g/L KH2PO4, 10 g/L D-glucose, 15 g/L agar supplemented with 1 mL/L of Hutner’s trace elements) covered with cellophane membrane. Each Petri plate (D=10 cm) was inoculated with 10*106 spores/plate. For supplementary comparison, each strain was also cultured in potato-dextrose (PD, BD Difco, Franklin Lakes, NJ) and Czapek-dox (CD, BD Difco) liquid media for 2 days and 10 days, respectively.
2.3. Protein extraction.
Mycelia and spores from GMM agar plates were collected. The hyphae from the liquid media were collected on Whatman paper filter and washed with cell culture grade water. All samples were stored at −80 °C prior to protein extraction. The lysis buffer consisted of 100 mM triethylammonium bicarbonate (TEAB) with 1:100 Halt Protease Inhibitor Cocktail (Thermo Scientific, Rockford, IL) and 200 μg phenylmethylsulfonyl fluoride (Sigma-Aldrich, St. Louis, MO). The hyphae from the liquid media were first homogenized on ice using a Polytron (Kinematica AG, Bohemia, NY) with a speed setting of 5 for 3–5 times (1 min/time, 30 sec pause). Subsequently, the crude homogenates were subjected to a Precellys 24 homogenizer (Bertin, Rockville, MD) in which each sample was processed inside a 2 mL cryotube with 0.5 mm glass beads three times (at 4 °C, 6500 rpm, 1 min., repeated 3 times with 15 sec pauses in between). Mycelia from GMM were homogenized directly by bead beating due to their small volume. The lysed fungi were centrifuged at 17,000 g for 15 min. Protein concentrations in the supernatants were measured by Bradford assay (Bio-Rad Laboratories, Inc. Hercules, CA).
2.4. Tandem mass tag (TMT) labeling.
200 μg proteins from each group were TCA-precipitated. Obtained protein pellets were washed with ice-cold acetone, and resolubilized in 25 μL TEAB (100 mM) and 25 μL 2,2,2-trifluoroethanol (TFE). Subsequently, proteins were reduced with tris(2-carboxyethyl)phosphine (TCEP, 500 mM), alkylated with iodoacetamide (IAA) and digested overnight using mass spec grade trypsin/lysC (Promega, Madison, WI) at 37°C. The digested peptides were quantified using the Pierce Quantitative Colorimetric Peptide Assay (Thermo Scientific). 40 μg of peptides from each specific sample was labeled with the Thermo Scientific TMTsixplex Isobaric Mass Tagging Kit (Af293 with TMT6-126, CEA10 with TMT6-127, ISSFT-021 with TMT6-128, and IF1SW-F4 with TMT6-130) according to the manufacturer’s protocol. The TMT6-131 label was used as a reference that contained an equal amount of peptides from each of the samples. All labeled-peptide mixtures were combined into a single tube, mixed, and fractionated into eight fractions using the Thermo Scientific Pierce High pH Reversed-Phase Peptide Fractionation Kit. The fractionated samples were dried using a SpeedVac concentrator and re-suspended in 1% (v/v) formic acid prior to LC-MS/MS analysis.
2.5. LC-MS/MS analysis.
An Orbitrap Fusion Tribrid mass spectrometer with the Thermo EASY-nLC ion source, 75 μm × 2 cm Acclaim PepMap100 C18 trapping column, and 75 μm × 25 cm PepMap RSLC C18 analytical column was used to analyze the samples. Peptides were eluted at 45°C with a flow rate of 300 nL/min over a 110 min gradient, from 3–30% solvent B (100 min), 30–50% solvent B (3 min), 50–90% solvent B (2 min), and 90% solvent B (2 min). The solvent A was 0.1% formic acid in water and the solvent B was 0.1% formic acid in acetonitrile.
The full MS survey scan (m/z 400–1500) was acquired at a resolution of 120,000 and an automatic gain control (AGC) target of 2×105 in the Orbitrap with the 50 ms maximum injection time for MS scans. Monoisotopic precursor ions were selected for fragmentation with charge states 2–7, within a ±10 ppm mass window, using a 70 s dynamic exclusion function. MS2 scans (m/z 400–2000) were performed using the linear ion trap with the 35% CID collision energy. The ion trap scan rate was set to “rapid”, with an AGC target of 4×103, and a maximum injection time of 150 ms. Subsequently, ten fragment ions from each MS2 experiment were subjected to an MS3 experiment. The MS3 scan (m/z 100–500) generated the TMT reporter ions in the linear ion trap using HCD at a 55% collision energy, a rapid scan rate and an AGC target of 5×103, and a maximum injection time of 250 ms.
2.6. Proteome data processing.
The Proteome Discoverer (version 2.1.0.81, Thermo Scientific) with searching engines Sequest-HT against an A. fumigatus Af293 protein database from NCBI containing 9845 non-redundant sequences was used to search all MS/MS spectra. The following parameters: 5 ppm tolerance for precursor ion masses and 0.6 Da tolerance for fragment ion masses were selected. The static modification settings included carbamidomethyl of cysteine residues, whereas dynamic modifications included oxidation of methionine, TMT6plex modification of lysine ε-amino groups and peptide N-termini, and acetyl modification of peptide N-terminus. A false discovery rate (FDR) of 1% for peptides and proteins was obtained using a target-decoy database search. The reporter ions integration tolerance was 0.5 Da while the co-isolation threshold was 75%. The average signal-to-noise threshold of all reporter peaks was greater than 10. The sum of all detected reporter ions of associated peptides from a protein was used to determine the total intensity of a reporter ion for that protein. The ratios between reporter and the reference reporter ions (TMT6–131) were used to estimate the abundance ratio of each protein. For the statistical analysis, the sum of reporter ion intensities for each protein was Log2 transformed and technical triplicate measurements for each protein were averaged. Only the proteins that were identified with at least one peptide detected in each technical replicate, and quantified in all three technical replicates, were considered for the analysis. One-way ANOVA was performed to identify proteins that are differentially expressed. Proteins with p-value ≤ 0.05 were further evaluated for up- and down-regulation using a cut-off value of ≥ ±1 fold (Log2) change.
3. RESULTS
3.1. Proteome analysis overview.
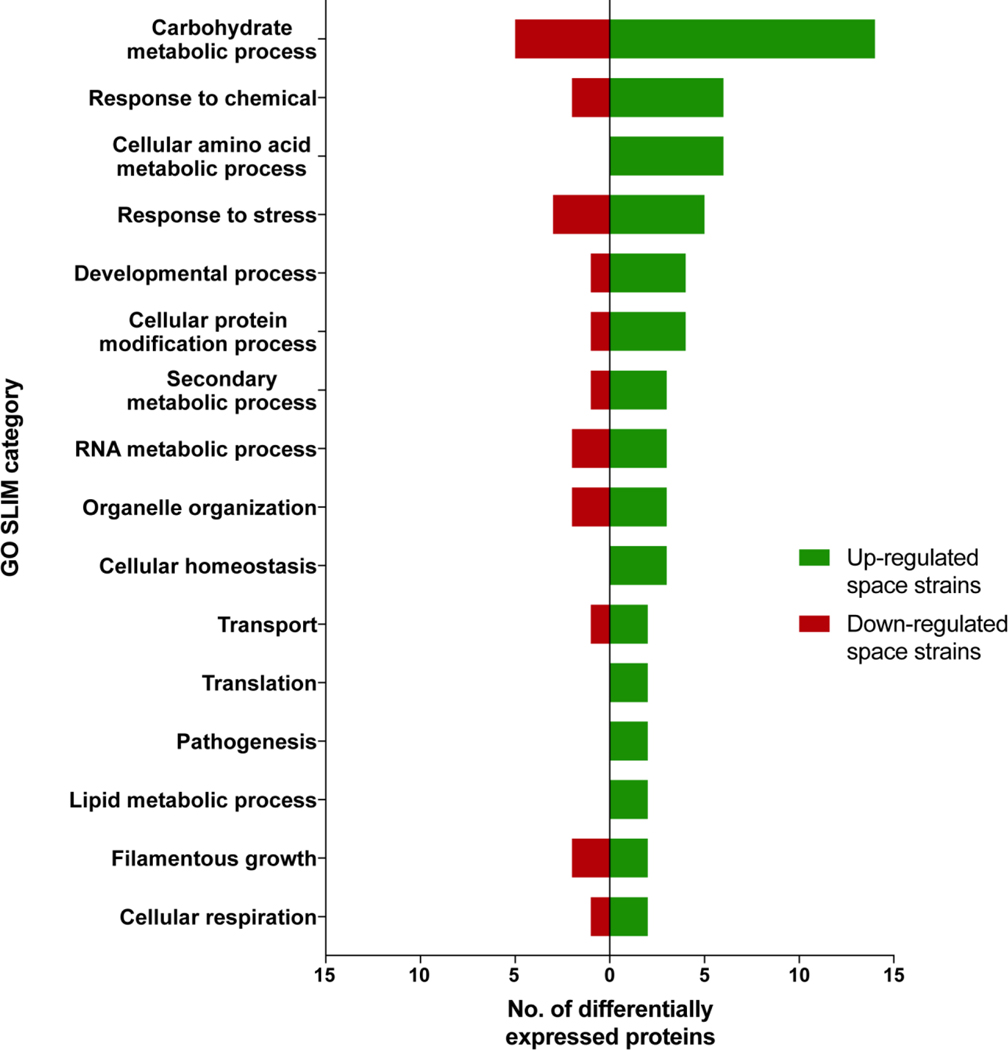
Proteins with altered abundance in ISS-isolated strains ISSFT-021 and IF1SW-F4, and clinical isolates Af293 and CEA10, were investigated upon extraction of total protein from each strain. Extracted proteins were digested into peptides and labeled using tandem mass tags (TMT), fractionated, and analyzed via LC-MS/MS followed by spectrum/sequence matching using A. fumigatus Af293 protein database (NCBI). The abundance ratios of all identified proteins were normalized to Af293, that enabled identification of 553, 464 and 626 increased and 314, 289 and 317 decreased in abundance proteins in CEA10, ISSFT-021 and IF1SW-F4 strains respectively (Table S-1, Figure S-1). When compared to both, Af293 and CEA10, 60 proteins showed increased and 32 decreased abundance in space strains only (fold change (FC) > |2|) (Table S-2). AspGD GO Slim terms46 were used to study the distribution of differentially abundant proteins in space strains. Analysis of proteins with increased abundance revealed involvement of 14 proteins in carbohydrate metabolic processes, eight in stress responses, five in secondary metabolism and toxins biosynthesis, and two in pathogenesis whereas five, three, one and zero proteins showed respectively decreased abundance in these categories. Proteins associated with cellular amino acid metabolic process (6), lipid cellular homeostasis (3), metabolic processes (2), pathogenesis (2), and translation (2) exhibited increased abundance in space strains only (Figure 1). FungiDB47 was used to carry out GO term enrichment analysis to gain a general understanding of biological processes possibly affected by unique environment of the ISS. The results revealed that significantly over-represented up-regulated biological processes included secondary metabolic processes (40% of all up-regulated proteins), carbohydrate metabolic processes (23%), and response to chemical (~15%), whereas significantly over-represented down-regulated processes included carbohydrate metabolic processes (15%), response to heat (6%), and mRNA metabolic processes (6%) (Table S-3).
F1. AspGD GO Slim terms of proteins differentially expressed in space strains when compared to Af293 and CEA10.
Differentially expressed proteins in ISSFT-021 and IF1SW-F4 are categorized into GO Slim categories using AspGD. Categories containing at least 2 up- or down-regulated proteins are presented.
3.2. Secondary metabolism and toxins.
The proteomic analysis revealed altered abundance of proteins involved in secondary metabolism (Table 1). Proteins involved in trypacidin biosynthetic pathway, including emodin O-methyltransferase TpcA (AFUA_4G14580), glutathione S-transferase TpcF (AFUA_4G14530), and dehydratase TpcK (AFUA_4G14470)48–50, were at least threefold more abundant in ISS-isolated strains than in clinical isolates. Arp1 (AFUA_2G17580), a scytalone dehydratase involved in conidial pigment biosynthesis51–53 was threefold more abundant, whereas Asp-hemolysin (Asp-HS; AFUA_3G00590)54 was eight times more abundant in ISS strains.
Table 1.
Proteins involved in secondary metabolism and toxin biosynthesis that revealed increased or decreased abundance
| Relative protein abundance* | ||||||
|---|---|---|---|---|---|---|
|
|
||||||
| ORF | Protein | Putative function/activity | CEA10 | ISSFT-021 | IF1SW-F4 | p-value |
|
| ||||||
| AFUA_4G14530 | TpcF/GstC | Glutathione S-transferase involved in trypacidin biosynthesis | 0.83 | 3.02 | 2.46 | 2.76E-06 |
| AFUA_3G00590 | AspHS | Asp-hemolysin; hemolytic toxin | 0.00 | 3.01 | 3.24 | 7.48E-09 |
| AFUA_4G14580 | TpcA | Emodin O-methyltransferase involved in trypacidin biosynthesis | −0.14 | 2.95 | 3.06 | 6.77E-07 |
| AFUA_4G14470 | TpcK | Dehydratase involved in trypacidin biosynthesis | 0.27 | 2.58 | 2.17 | 3.04E-03 |
| AFUA_2G17580 | Arp1 | Scytalone dehydratase involved in conidial pigment biosynthesis | 0.58 | 2.49 | 2.69 | 2.24E-07 |
| AFUA_4G00860 | DprA | Dehydrin-like protein / oxidative, osmotic and pH stress responses | −3.54 | −5.64 | −4.62 | 1.37E-07 |
Log2 fold change of CEA10, ISSFT-021, and IF1SW-F4 compared to Af293 (P < 0.05)
To confirm observed up-regulation of proteins involved in the trypacidin biosynthesis secondary metabolite profiles of the clinical and ISS isolates were acquired using high performance liquid chromatography-photodiode array detection-mass spectroscopy (HPL-DAD-MS) analysis. Examination of the production yields of monomethylsulochrin, the final intermediate in trypacidin biosynthesis, (trypacidin is not detectable until 7–8 days of growth50) showed ~600% and ~200% increased production in ISSFT-021 ad IF1SW-F4, respectively (Figure S-2). Additionally, proteome of clinical and ISS strains was examined in liquid CD and PD media and confirmed significant up-regulation of AspHS (Table S-4). Lastly, observed differences in sporulation capacity between the strains do not seem to be directly correlated with conidiaassociated protein expression levels (Figure S-3), suggesting that the observed variations are due to exposure to space environment rather than different growth rates.
3.3. Stress response.
Among proteins with altered abundance, 11 were involved in the stress response of A. fumigatus (Table 2). AFUA_5G11430, a quinone oxidoreductase, and Pst2 (AFUA_1G02820), an NADH-quinone oxidoreductase, involved in oxidative stress response were three and four times more abundant in space strains. Erythromycin esterase, AFUA_1G05850, and 3’ exoribonuclease AFUA_2G1598056 were at least twofold more abundant in ISS strains. Among proteins with decreased abundance several were heat shock proteins including Scf1 (AFUA_1G17370)57,58, and Awh11 (AFUA_6G12450)56. Other down-regulated protein was dehydrin-like protein DprA (AFUA_4G00860) that is known to play a role in oxidative stress response59.
Table 2.
Proteins involved in stress response that revealed increased or decreased abundance
| Relative protein abundance* | ||||||
|---|---|---|---|---|---|---|
|
|
||||||
| ORF | Protein | Putative function/activity | CEA10 | ISSFT-021 | IF1SW-F4 | p-value |
|
| ||||||
| AFUA_3G08470 | Glucose-6-phosphate 1- dehydrogenase Glutathione S-transferase |
1.41 | 3.06 | 3.33 | 8.66E-07 | |
| AFUA_4G14530 | TpcF/GstC | involved in trypacidin biosynthesis | 0.83 | 3.02 | 2.46 | 2.76E-06 |
| AFUA_4G11730 | GldB | Glycerol dehydrogenase | 1.29 | 2.83 | 3.70 | 5.34E-07 |
| AFUA_5G11430 | Quinone oxidoreductase | 0.95 | 2.55 | 2.51 | 1.45E-09 | |
| AFUA_2G03290 | ArtA | 14–3-3 family protein | 0.67 | 2.21 | 2.82 | 1.90E-07 |
| AFUA_1G02820 | Pst2 | NADH-quinone oxidoreductase | −0.37 | 2.10 | 2.55 | 9.33E-06 |
| AFUA_2G15980 | 3’ exoribonuclease family protein; pre-miRNA processing | 0.02 | 1.24 | 2.20 | 4.73E-03 | |
| AFUA_1G05850 | Erythromycin esterase | −0.16 | 1.22 | 2.48 | 6.61E-03 | |
| AFUA_1G17370 | Scf1 | Heat shock protein | −2.95 | −5.23 | −4.26 | 4.25E-07 |
| AFUA_6G12450 | Awh11 | Heat shock protein | −4.05 | −5.81 | −5.12 | 5.46E-05 |
| AFUA_4G00860 | DprA | Dehydrin-like protein / oxidative, osmotic and pH stress responses | −3.54 | −5.64 | −4.62 | 1.37E-07 |
Log2 fold change of CEA10, ISSFT-021, and IF1SW-F4 compared to Af293 (P < 0.05)
3.4. Carbohydrate metabolic processes.
Comparative analysis of proteomes of ISS-isolated strains, IF1SW-F4 and ISST-021, with clinical isolates CEA10 and Af293 revealed changes in abundance of proteins involved in carbohydrate metabolic processes (Table 3). Pyruvate decarboxylase PdcA (AFUA_3G11070), which catalyzes the first step in anaerobic conversion of pyruvate to ethanol60, was about three times more abundant in space strains when compared to clinical isolates. Proteins involved in glycerol metabolism, including glycerol dehydrogenase GldB (AFUA_4G11730), and glycerol kinase AFUA_4G1154055 were at least 2.5-fold more abundant in ISS-isolated strains. AFUA_5G10540 and AFUA_1G02140, which are homologues of A. niger An14g04190 and GdbA (An01g06120)61, respectively, and involved in glycogen biosynthesis and metabolism were at least twofold more abundant. Protein abundance of AFUA_3G08470, AFUA_4G08880, AFUA_7G_01830, and AFUA_1G14710 involved in glucose metabolism, increased twofold at minimum. Both, the 14–3-3 family protein ArtA (AFUA_2G03290)62 and malate synthase AcuE (AFUA_6G03540)63 were threefold more abundant when compared to clinical isolates. At minimum twofold increased abundance of phosphoketolase (AFUA_3G00370) and hexokinase HxkA (AFUA_2G05910)64 was observed. Among the decreased in abundance proteins were hydrolase Exg17 (AFUA_6G14490)65 and β−1,3-glucan modifying enzyme Sun1 (AFUA_7G05450)66.
Table 3.
Proteins involved in carbohydrate metabolism that revealed increased or decreased abundance
| Relative protein abundance* | ||||||
|---|---|---|---|---|---|---|
|
|
||||||
| ORF | Protein | Putative function/activity | CEA10 | ISSFT-021 | IF1SW-F4 | p-value |
|
| ||||||
| AFUA_3G11070 | PdcA | Pyruvate decarboxylase involved in ethanol fermentation pathway | 1.95 | 3.42 | 3.43 | 9.50E-08 |
| AFUA_4G11540 | Glycerol kinase | 1.52 | 3.18 | 2.94 | 5.99E-05 | |
| AFUA_3G08470 | Glucose-6-phosphate 1- dehydrogenase | 1.41 | 3.06 | 3.33 | 8.66E-07 | |
| AFUA_4G11730 | GldB | Glycerol dehydrogenase | 1.29 | 2.83 | 3.70 | 5.34E-07 |
| AFUA_1G14710 | Beta-glucosidase | 1.63 | 2.70 | 3.52 | 4.34E-05 | |
| AFUA_5G10540 | 1,4-alpha-glucan branching enzyme activity, glycogen biosynthesis | 0.91 | 2.23 | 3.06 | 1.39E-07 | |
| AFUA_2G03290 | ArtA | 14–3-3 family protein | 0.67 | 2.21 | 2.82 | 1.90E-07 |
| AFUA_6G03540 | AcuE | Malate synthase | 0.38 | 2.10 | 2.16 | 1.22E-07 |
| AFUA_1G02140 | Glycogen debranching enzyme | 0.87 | 1.98 | 3.18 | 1.08E-03 | |
| AFUA_3G00370 | Phosphoketolase | 0.51 | 1.88 | 2.84 | 3.50E-05 | |
| AFUA_4G04680 | FGGY-family carbohydrate kinase | 0.81 | 1.82 | 2.61 | 1.73E-05 | |
| AFUA_4G08880 | Glucose-6-phosphate 1-epimerase | −0.32 | 1.46 | 2.61 | 7.00E-05 | |
| AFUA_7G01830 | Ugp1 | UTP-glucose-1 -phosphate uridylyltransferase | 0.00 | 1.35 | 2.11 | 4.43E-05 |
| AFUA_2G05910 | HxkA | Hexokinase | −0.01 | 1.20 | 2.12 | 8.68E-07 |
| AFUA_7G05450 | Sun1 | Beta-1,3-glucan modifying enzyme | −0.61 | −1.87 | −2.15 | 4.13E-02 |
| AFUA_1G06910 | Arabinogalactan endo-1,4-beta- galactosidase activity | −0.43 | −1.53 | −2.04 | 9.05E-06 | |
| AFUA_6G05030 | Polysaccharide deacetylase | −0.09 | −1.21 | −1.23 | 1.53E-03 | |
| AFUA_6G14490 | Exg17 | O-glycosyl hydrolase | −0.21 | −1.41 | −1.33 | 4.68E-05 |
| AFUA_1G03140 | Glycosyl hydrolase | −0.31 | −1.31 | −1.44 | 1.39E-06 | |
Log2 fold change of CEA10, ISSFT-021, and IF1SW-F4 compared to Af293 (P < 0.05)
4. DISCUSSION
The proteome of A. fumigatus has been studied under various conditions, including short-67, and long-term hypoxia68, following exposure to antifungal agents like amphotericin B69, and voriconazole70, and during different developmental stages58. However, in this report we present unique proteome differences observed in the two ISS-isolated strains when compared to clinical isolates. To date, there is no report which elucidates the molecular response of filamentous fungi to the distinct environment of the ISS, despite previous reports of their presence on board of the ISS22 and Mir27,71. In-depth understanding of alterations triggered in the proteome of omnipresent A. fumigatus remains imperative for astronauts’ health, as it is an opportunistic pathogen that affects individuals with impaired immune system functions41.
Both ISS-isolated A. fumigatus strains displayed higher abundance of several proteins involved in trypacidin biosynthesis. Trypacidin is a potent mycotoxin produced by A. fumigatus conidia. It has been shown to have cytotoxic activity against A549 alveolar lung cells suggesting its importance during the infection76. Further, it has been reported that A. fumigatus conidia with disrupted production of trypacidin exhibit higher susceptibility to macrophage clearance49. Increased abundance of proteins involved in biosynthesis of trypacidin in both ISS–isolated strains may be possible cause of the reported increased virulence in the larval zebra fish model of both ISS-isolated strains when compared to clinical isolates45. Moreover, emodin and questin, precursors of trypacidin, are pigmented anthraquinones that have been reported to have a protective effect against UV in other organisms (Xanthoria elegans and Cetraria islandica) that produce these types of compounds50,77. It is therefore reasonable to assume that up-regulation of trypacidin production may be another adaptation to the enhanced irradiation environment of the ISS, however this hypothesis has to be validated by further experiments.
Further, the level of Asp-HS, which is a known toxin produced by A. fumigatus54,78 was highly increased in both ISS-isolates. While several studies have reported its hemolytic79 and cytotoxic80,81 activities in the past, a recent report showed lack of attenuated virulence when Asp-HS gene was deleted82. This discrepancy may be strain specific as the studies used different A. fumigatus strains to carry out the experiments or related to experimental models used in both studies, as at times results observed for in vitro analyses do not correlated with the outcomes observed in in vivo studies. The exact role of Asp-HS in the pathogenicity remains to be determined during future studies. Nonetheless, increased abundance of Asp-HS observed in both ISS-isolated strains appears to be a part of the A. fumigatus adaptation response to the unique ISS environment.
One of the increased in abundance proteins in ISS-isolated A. fumigatus strains was Arp1, which is one of the six enzymes involved in the DHN-melanin production51–53. Arp1 disruption resulted in production of reddish pink conidia with induced C3 binding that led to phagocytosis and killing of conidial spores during infection51,52. Increased abundance of Arp1 protein may, therefore, be another cause of the previously reported increased virulence in the larval zebra fish model45. Additionally, higher AlbA abundance, involved in DHN-melanin production72, was observed in JSC-093350089 A. niger isolated from the ISS73. Seemingly, both ISS-isolated species, A. fumigatus and A. niger, responded with increased melanin production to enhanced irradiation on board of the ISS. This observation is in agreement with previous reports of increased melanin production in fungi isolated from high-radiation environments such as the Chernobyl Power Plant accident sites or “Evolution Canyon”74,75. However, because none of the other enzymes involved in the melanin biosynthesis showed increased abundance when compared to both clinical isolates simultaneously, further assessment of melanin production is necessary to definitively conclude its increased production under ISS conditions.
Proteome analysis of both ISS-isolated A. fumigatus strains revealed altered levels of proteins involved in oxidative stress response. ArtA, a regulatory protein, was reported to be increased in abundance in response to incubation with H2O2, suggesting its importance in overcoming oxidative stress62. Pst2, which is also present in Candida albicans and Saccharomyces cerevisiae, was shown to be induced in response to oxidative stress83,84. Additionally, Pst2 was reported to be a post-translational modification target protein that is ubiquitinated under peroxide stress conditions in C. albicans85. Interestingly, such post-translational modifications play a role in several cellular processes in eukaryotes, including cell growth regulation and environmental adaptation, suggesting their importance in gaining survival advantage86. These data are in agreement with previous reports of induced oxidative stress response in humans, mice and yeast while in space87–91. Overall, increased abundance of proteins involved in oxidative stress response was also observed in Acinetobacter sp. Ver3 when exposed to UV irradiation92 and JSC-093350089 A. niger73. Although increased abundance of several oxidative stress-correlated proteins was observed in this study, it was previously reported that ISS-isolated strains were more resistant than Af293 to H2O2 exposure, but less resistant than CEA10, suggesting no adaptation45.
Among the more abundant proteins in both ISS-isolated strains several were involved in carbohydrate metabolism. Malate synthase AcuE (AFUA_6G03540) is one of the three key enzymes involved in glyoxylate cycle63, which has been shown to be crucial for fungal growth on C2 compounds and fatty acids as a sole carbon source93. In our earlier study it was documented that these two ISS-isolated strains significantly outgrew both clinical isolates45. Further, more abundant PdcA (AFUA_3G11070), a pyruvate decarboxylase, has been shown to be involved in ethanol fermentation60. These data therefore suggest possible adaptation of A. fumigatus to low-nutrient environment of the ISS20,22, which is in agreement with changes observed in proteome of A. fumigatus during starvation94. Similarly, up-regulation of proteins involved in starvation response was observed in ISS-isolated JSC-093350089 A. niger73.
This study presents comparative proteomic analysis of two ISS-isolated A. fumigatus strains, ISSFT-021 and IF1SW-F4 when compared to clinically established isolates Af293 and CEA10. Such comparison has enabled the identification of possible adaptation responses to the unique microgravity environment of the ISS, which includes increased abundance of stress response related proteins, and modulation of proteins involved in carbohydrate and secondary metabolism. To our knowledge this is the first report that focused on studying the proteomic changes in filamentous fungus isolated from on board of the ISS. Complex analyses of possible molecular alterations triggered by microgravity and enhanced irradiation will be pertinent to the future long-term manned space flights, as such an understanding is crucial for astronauts’ health and biodeterioration of the closed habitat.
Accession number
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE95 partner repository with the dataset identifier PXD008517.
Supplementary Material
Figure S-1. AspGD GO Slim terms of proteins normalized to Af293. Differentially expressed proteins in CEA10, ISSFT-021 and IF1SW-F4 are categorized into GO Slim categories using AspGD. Categories containing at least 5 up- or down-regulated proteins are presented.
Figure S-2. Monomethylsulochrin yields in clinical and ISS-isolated strains. A) Extracted ion count (EIC) traces of monomethylsulochrin in a positive mode. B) Quantification of of monomethylsulochrin production using positive mode EIC when compared to Af293. Significance was determined using Welch’s t-test.
Figure S-3. Sporulation capacity of clinical and ISS-isolated strains. Average values of conidial counts from 3 biological replicates are plotted after 3, 4 and 5 days of growth. Statistical significance was determined using multiple t-test with Holm-Sidak correction.
Table S-1. Up- and down-regulated proteins in CEA10, ISSFT-021 and IF1SW-F4 when normalized to Af293
Table S-2. Up- and down-regulated proteins in space strains when compared to Af293 and CEA10
Table S-3. GO enrichment analysis of proteins in space strains only
Table S-4. Differentially expressed proteins in CEA10, ISSFT-021 and IF1SW-F4 when grown on solid GMM, and liquid PD and CD media when normalized to Af293
Highlights:
Proteome characterization of Aspergillus fumigatus isolated from the International Space Station (ISS).
The distinct ISS environment (enhanced radiation, microgravity and scarce nutrients) alters A. fumigatus proteome.
When compared to clinical isolates CEA10 and Af293 the majority of up-regulated proteins are involved in carbohydrate and secondary metabolism, and oxidative stress response.
Acknowledgements
Part of the research described in this publication was carried out at the Jet Propulsion Laboratory (JPL), California Institute of Technology, under a contract with NASA. This research was funded by NASA’s Space Biology grants # NNN13D111T (19-12829-26) and NNN13D111T (19-12829-27) to K.V. and funded a student fellowship to AB. The City of Hope Mass Spectrometry and Proteomics core facility used by AC and MK was supported in parts by National Cancer Institute of the National Institutes of Health under award number P30 CA33572. We thank Jay Perry, Marshall Space Flight Center, for providing the HEPA filter and the implementation team of the Microbial Observatory (Microbial Tracking) project at NASA Ames Research Center. We appreciate the technical help of Alexandra Checinska Sielaff in isolating the fungal cultures.
Footnotes
Competing interests
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- (1).Benjamin CL; Stowe RP; St. John L; Sams CF; Mehta SK; Crucian BE; Pierson DL; Komanduri KV Decreases in Thymopoiesis of Astronauts Returning from Space Flight. JCI Insight 1 (12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Crucian B; Stowe R; Mehta S; Uchakin P; Quiriarte H; Pierson D; Sams C. Immune System Dysregulation Occurs during Short Duration Spaceflight on Board the Space Shuttle. J. Clin. Immunol 2013, 33 (2), 456–465. [DOI] [PubMed] [Google Scholar]
- (3).Cucinotta FA Space Radiation Risks for Astronauts on Multiple International Space Station Missions. PLOS ONE 2014, 9 (4), e96099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Mehta SK; Laudenslager ML; Stowe RP; Crucian BE; Feiveson AH; Sams CF; Pierson DL Latent Virus Reactivation in Astronauts on the International Space Station. NPJ Microgravity 2017, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Mehta SK; Cohrs RJ; Forghani B; Zerbe G; Gilden DH; Pierson DL Stress-Induced Subclinical Reactivation of Varicella Zoster Virus in Astronauts. J. Med. Virol 2004, 72 (1), 174–179. [DOI] [PubMed] [Google Scholar]
- (6).Ombergen AV; Demertzi A; Tomilovskaya E; Jeurissen B; Sijbers J; Kozlovskaya IB; Parizel PM; Heyning P. H. V. de; Sunaert S; Laureys S; et al. The Effect of Spaceflight and Microgravity on the Human Brain. J. Neurol 2017, 264 (1), 18–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Williams D; Kuipers A; Mukai C; Thirsk R. Acclimation during Space Flight: Effects on Human Physiology. Can. Med. Assoc. J 2009, 180 (13), 1317–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Ijiri K. Life-Cycle Experiments of Medaka Fish Aboard the International Space Station. Adv. Space Biol. Med 2003, 9, 201–216. [DOI] [PubMed] [Google Scholar]
- (9).Tavella S; Ruggiu A; Giuliani A; Brun F; Canciani B; Manescu A; Marozzi K; Cilli M; Costa D; Liu Y; et al. Bone Turnover in Wild Type and Pleiotrophin-Transgenic Mice Housed for Three Months in the International Space Station (ISS). PLOS ONE 2012, 7 (3), e33179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Driss-Ecole D; Legué V; Carnero-Diaz E; Perbal G. Gravisensitivity and Automorphogenesis of Lentil Seedling Roots Grown on Board the International Space Station. Physiol. Plant 2008, 134 (1), 191–201. [DOI] [PubMed] [Google Scholar]
- (11).Kittang A-I; Iversen T-H; Fossum KR; Mazars C; Carnero-Diaz E; BoucheronDubuisson E; Le Disquet I; Legué V; Herranz R; Pereda-Loth V; et al. Exploration of Plant Growth and Development Using the European Modular Cultivation System Facility on the International Space Station. Plant Biol. 2014, 16 (3), 528–538. [DOI] [PubMed] [Google Scholar]
- (12).Link BM; Durst SJ; Zhou W; Stankovic B. Seed-to-Seed Growth of Arabidopsis Thaliana on the International Space Station. Adv. Space Res 2003, 31 (10), 2237–2243. [DOI] [PubMed] [Google Scholar]
- (13).Sychev VN; Levinskikh MA; Gostimsky SA; Bingham GE; Podolsky IG Spaceflight Effects on Consecutive Generations of Peas Grown Onboard the Russian Segment of the International Space Station. Acta Astronaut. 2007, 60 (4), 426–432. [Google Scholar]
- (14).Tepfer D; Zalar A; Leach S. Survival of Plant Seeds, Their UV Screens, and NptII DNA for 18 Months Outside the International Space Station. Astrobiology 2012, 12 (5), 517–528. [DOI] [PubMed] [Google Scholar]
- (15).Benoit MR; Li W; Stodieck LS; Lam KS; Winther CL; Roane TM; Klaus DM Microbial Antibiotic Production Aboard the International Space Station. Appl. Microbiol. Biotechnol 2006, 70 (4), 403–411. [DOI] [PubMed] [Google Scholar]
- (16).Leys N; Baatout S; Rosier C; Dams A; s’Heeren C; Wattiez R; Mergeay M. The Response of Cupriavidus Metallidurans CH34 to Spaceflight in the International Space Station. Antonie Van Leeuwenhoek 2009, 96 (2), 227. [DOI] [PubMed] [Google Scholar]
- (17).Rabbow E; Rettberg P; Baumstark-Khan C; Horneck G. The SOS-LUX-LACFLUORO-Toxicity-Test on the International Space Station (ISS). Adv. Space Res 2003, 31 (6), 1513–1524. [DOI] [PubMed] [Google Scholar]
- (18).Timmery S; Hu X; Mahillon J. Characterization of Bacilli Isolated from the Confined Environments of the Antarctic Concordia Station and the International Space Station. Astrobiology 2011, 11 (4), 323–334. [DOI] [PubMed] [Google Scholar]
- (19).Vaishampayan PA; Rabbow E; Horneck G; Venkateswaran KJ Survival of Bacillus Pumilus Spores for a Prolonged Period of Time in Real Space Conditions. Astrobiology 2012, 12 (5), 487–497. [DOI] [PubMed] [Google Scholar]
- (20).Venkateswaran K. Microbial Characteristics of ISS Environmental Surfaces. 2017. [Google Scholar]
- (21).Venkateswaran K; Vaishampayan P; Cisneros J; Pierson DL; Rogers SO; Perry J. International Space Station Environmental Microbiome — Microbial Inventories of ISS Filter Debris. Appl. Microbiol. Biotechnol 2014, 98 (14), 6453–6466. [DOI] [PubMed] [Google Scholar]
- (22).Checinska A; Probst AJ; Vaishampayan P; White JR; Kumar D; Stepanov VG; Fox GE; Nilsson HR; Pierson DL; Perry J; et al. Microbiomes of the Dust Particles Collected from the International Space Station and Spacecraft Assembly Facilities. Microbiome 2015, 3 (1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Vesper SJ; Wong W; Kuo CM; Pierson DL Mold Species in Dust from the International Space Station Identified and Quantified by Mold-Specific Quantitative PCR. Res. Microbiol 2008, 159 (6), 432–435. [DOI] [PubMed] [Google Scholar]
- (24).Satoh K; Nishiyama Y; Yamazaki T; Sugita T; Tsukii Y; Takatori K; Benno Y; Makimura K. Microbe-I: Fungal Biota Analyses of the Japanese Experimental Module KIBO of the International Space Station before Launch and after Being in Orbit for about 460 Days. Microbiol. Immunol 2011, 55 (12), 823–829. [DOI] [PubMed] [Google Scholar]
- (25).Satoh K; Yamazaki T; Nakayama T; Umeda Y; Alshahni MM; Makimura M; Makimura K. Characterization of Fungi Isolated from the Equipment Used in the International Space Station or Space Shuttle. Microbiol. Immunol 2016, 60 (5), 295–302. [DOI] [PubMed] [Google Scholar]
- (26).Kawamura Y; Li Y; Liu H; Huang X; Li Z; Ezaki T. Bacterial Population in Russian Space Station “Mir.” Microbiol. Immunol 2001, 45 (12), 819–828. [DOI] [PubMed] [Google Scholar]
- (27).Makimura K; Hanazawa R; Takatori K; Tamura Y; Fujisaki R; Nishiyama Y; Abe S; Uchida K; Kawamura Y; Ezaki T; et al. Fungal Flora on Board the Mir-Space Station, Identification by Morphological Features and Ribosomal DNA Sequences. Microbiol. Immunol 2001, 45 (5), 357–363. [DOI] [PubMed] [Google Scholar]
- (28).Brockett RM; Ferguson JK; Henney MR Prevalence of Fungi during Skylab Missions. Appl. Environ. Microbiol 1978, 36 (2), 243–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Fleming A. On the Antibacterial Action of Cultures of a Penicillium, with Special Reference to Their Use in the Isolation of B. Influenzæ. Br. J. Exp. Pathol 1929, 10 (3), 226–236. [Google Scholar]
- (30).Chain E; Florey HW; Gardner AD; Heatley NG; Jennings MA; Orr-Ewing J; Sanders AG PENICILLIN AS A CHEMOTHERAPEUTIC AGENT. The Lancet 1940, 236 (6104), 226–228. [Google Scholar]
- (31).Alberts AW Discovery, Biochemistry and Biology of Lovastatin. Am. J. Cardiol 1988, 62 (15), J10–J15. [DOI] [PubMed] [Google Scholar]
- (32).Frishman WH; Rapier RC Lovastatin: An HMG-CoA Reductase Inhibitor for Lowering Cholesterol. Med. Clin. North Am 1989, 73 (2), 437–448. [DOI] [PubMed] [Google Scholar]
- (33).Max B; Salgado JM; Rodríguez N; Cortés S; Converti A; Domínguez JM Biotechnological Production of Citric Acid. Braz. J. Microbiol 2010, 41 (4), 862–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Ikram-ul-Haq; Khurshid S; Ali S; Ashraf H; Qadeer MA; Rajoka MI Mutation of Aspergillus Niger for Hyperproduction of Citric Acid from Black Strap Molasses. World J. Microbiol. Biotechnol 2001, 17 (1), 35–37. [Google Scholar]
- (35).Simon-Nobbe B; Denk U; Pöll V; Rid R; Breitenbach M. The Spectrum of Fungal Allergy. Int. Arch. Allergy Immunol 2008, 145 (1), 58–86. [DOI] [PubMed] [Google Scholar]
- (36).Sharpe RA; Bearman N; Thornton CR; Husk K; Osborne NJ Indoor Fungal Diversity and Asthma: A Meta-Analysis and Systematic Review of Risk Factors. J. Allergy Clin. Immunol 2015, 135 (1), 110–122. [DOI] [PubMed] [Google Scholar]
- (37).Segal BH Aspergillosis. N. Engl. J. Med 2009, 360 (18), 1870–1884. [DOI] [PubMed] [Google Scholar]
- (38).Andersen B; Frisvad JC; Søndergaard I; Rasmussen IS; Larsen LS Associations between Fungal Species and Water-Damaged Building Materials ▿. Appl. Environ. Microbiol 2011, 77 (12), 4180–4188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Sterflinger K. Fungi: Their Role in Deterioration of Cultural Heritage. Fungal Biol. Rev 2010, 24 (1), 47–55. [Google Scholar]
- (40).Bashir U; Hafeez R. DETERIORATION OF PAINTED WALL SURFACE BY FUNGAL SAPROBES: ISOLATION AND IDENTIFICATION. Pak. J. Phytopathol 2016, 28 (1), 09–13. [Google Scholar]
- (41).Tekaia F; Latgé J-P Aspergillus Fumigatus: Saprophyte or Pathogen? Curr. Opin. Microbiol 2005, 8 (4), 385–392. [DOI] [PubMed] [Google Scholar]
- (42).Kwon-Chung KJ; Sugui JA Aspergillus Fumigatus—What Makes the Species a Ubiquitous Human Fungal Pathogen? PLOS Pathog. 2013, 9 (12), e1003743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Dagenais TRT; Keller NP Pathogenesis of Aspergillus Fumigatus in Invasive Aspergillosis. Clin. Microbiol. Rev 2009, 22 (3), 447–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Taccone FS; Van den Abeele A-M; Bulpa P; Misset B; Meersseman W; Cardoso T; Paiva J-A; Blasco-Navalpotro M; De Laere E; Dimopoulos G; et al. Epidemiology of Invasive Aspergillosis in Critically Ill Patients: Clinical Presentation, Underlying Conditions, and Outcomes. Crit. Care Lond. Engl 2015, 19, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Knox BP; Blachowicz A; Palmer JM; Romsdahl J; Huttenlocher A; Wang CCC; Keller NP; Venkateswaran K. Characterization of Aspergillus Fumigatus Isolates from Air and Surfaces of the International Space Station. mSphere 2016, 1 (5), e00227–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Cerqueira GC; Arnaud MB; Inglis DO; Skrzypek MS; Binkley G; Simison M; Miyasato SR; Binkley J; Orvis J; Shah P; et al. The Aspergillus Genome Database: Multispecies Curation and Incorporation of RNA-Seq Data to Improve Structural Gene Annotations. Nucleic Acids Res. 2014, 42 (D1), D705–D710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Stajich JE; Harris T; Brunk BP; Brestelli J; Fischer S; Harb OS; Kissinger JC; Li W; Nayak V; Pinney DF; et al. FungiDB: An Integrated Functional Genomics Database for Fungi. Nucleic Acids Res. 2012, 40 (Database issue), D675–D681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Burns C; Geraghty R; Neville C; Murphy A; Kavanagh K; Doyle S. Identification, Cloning, and Functional Expression of Three Glutathione Transferase Genes from Aspergillus Fumigatus. Fungal Genet. Biol. FG B 2005, 42 (4), 319–327. [DOI] [PubMed] [Google Scholar]
- (49).Mattern DJ; Schoeler H; Weber J; Novohradská S; Kraibooj K; Dahse H-M; Hillmann F; Valiante V; Figge MT; Brakhage AA Identification of the Antiphagocytic Trypacidin Gene Cluster in the Human-Pathogenic Fungus Aspergillus Fumigatus. Appl. Microbiol. Biotechnol 2015, 99 (23), 10151–10161. [DOI] [PubMed] [Google Scholar]
- (50).Throckmorton K; Lim FY; Kontoyiannis DP; Zheng W; Keller NP Redundant Synthesis of a Conidial Polyketide by Two Distinct Secondary Metabolite Clusters in Aspergillus Fumigatus. Environ. Microbiol 2016, 18 (1), 246–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Tsai HF; Washburn RG; Chang YC; Kwon-Chung KJ Aspergillus Fumigatus Arp1 Modulates Conidial Pigmentation and Complement Deposition. Mol. Microbiol 1997, 26 (1), 175–183. [DOI] [PubMed] [Google Scholar]
- (52).Tsai H-F; Wheeler MH; Chang YC; Kwon-Chung KJ A Developmentally Regulated Gene Cluster Involved in Conidial Pigment Biosynthesis in Aspergillus Fumigatus. J. Bacteriol 1999, 181 (20), 6469–6477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Pihet M; Vandeputte P; Tronchin G; Renier G; Saulnier P; Georgeault S; Mallet R; Chabasse D; Symoens F; Bouchara J-P Melanin Is an Essential Component for the Integrity of the Cell Wall of Aspergillus Fumigatus Conidia. BMC Microbiol. 2009, 9, 177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Yokota K; Shimada H; Kamaguchi A; Sakaguchi O. Studies on the Toxin of Aspergillus Fumigatus. VII. Purification and Some Properities of Hemolytic Toxin (Asp-Hemolysin) from Culture Filtrates and Mycelia. Microbiol. Immunol 1977, 21 (1), 11–22. [DOI] [PubMed] [Google Scholar]
- (55).Teutschbein J; Albrecht D; Pötsch M; Guthke R; Aimanianda V; Clavaud C; Latgé J-P; Brakhage AA; Kniemeyer O. Proteome Profiling and Functional Classification of Intracellular Proteins from Conidia of the Human-Pathogenic Mold Aspergillus Fumigatus. J. Proteome Res 2010, 9 (7), 3427–3442. [DOI] [PubMed] [Google Scholar]
- (56).Nierman WC; Pain A; Anderson MJ; Wortman JR; Kim HS; Arroyo J; Berriman M; Abe K; Archer DB; Bermejo C; et al. Genomic Sequence of the Pathogenic and Allergenic Filamentous Fungus Aspergillus Fumigatus. Nature 2005, 438 (7071), 1151–1156. [DOI] [PubMed] [Google Scholar]
- (57).Soriani FM; Malavazi I; Savoldi M; Espeso E; Dinamarco TM; Bernardes LAS; Ferreira MES; Goldman MHS; Goldman GH Identification of Possible Targets of the Aspergillus Fumigatus CRZ1 Homologue, CrzA. BMC Microbiol. 2010, 10, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Suh M-J; Fedorova ND; Cagas SE; Hastings S; Fleischmann RD; Peterson SN; Perlin DS; Nierman WC; Pieper R; Momany M. Development Stage-Specific Proteomic Profiling Uncovers Small, Lineage Specific Proteins Most Abundant in the Aspergillus Fumigatus Conidial Proteome. Proteome Sci. 2012, 10 (1), 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Hoi JWS; Lamarre C; Beau R; Meneau I; Berepiki A; Barre A; Mellado E; Read ND; Latgé J-P A Novel Family of Dehydrin-like Proteins Is Involved in Stress Response in the Human Fungal Pathogen Aspergillus Fumigatus. Mol. Biol. Cell 2011, 22 (11), 1896–1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Grahl N; Puttikamonkul S; Macdonald JM; Gamcsik MP; Ngo LY; Hohl TM; Cramer RA In Vivo Hypoxia and a Fungal Alcohol Dehydrogenase Influence the Pathogenesis of Invasive Pulmonary Aspergillosis. PLoS Pathog. 2011, 7 (7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Yuan X-L; Kaaij RM van der; Hondel CAMJJ van den; Punt PJ; Maarel, van der MJEC; Dijkhuizen L; Ram AFJ Aspergillus Niger Genome-Wide Analysis Reveals a Large Number of Novel Alpha-Glucan Acting Enzymes with Unexpected Expression Profiles. Mol. Genet. Genomics 2008, 279 (6), 545–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Lessing F; Kniemeyer O; Wozniok I; Loeffler J; Kurzai O; Haertl A; Brakhage AA The Aspergillus Fumigatus Transcriptional Regulator AfYap1 Represents the Major Regulator for Defense against Reactive Oxygen Intermediates but Is Dispensable for Pathogenicity in an Intranasal Mouse Infection Model. Eukaryot. Cell 2007, 6 (12), 2290–2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Olivas I; Royuela M; Romero B; Monteiro MC; Mínguez JM; Laborda F; Lucas JRD Ability to Grow on Lipids Accounts for the Fully Virulent Phenotype in Neutropenic Mice of Aspergillus Fumigatus Null Mutants in the Key Glyoxylate Cycle Enzymes. Fungal Genet. Biol 2008, 45 (1), 45–60. [DOI] [PubMed] [Google Scholar]
- (64).Albrecht D; Guthke R; Brakhage AA; Kniemeyer O. Integrative Analysis of the Heat Shock Response in Aspergillus Fumigatus. BMC Genomics 2010, 11, 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Mouyna I; Hartl L; Latgé J-P β−1,3-Glucan Modifying Enzymes in Aspergillus Fumigatus. Front. Microbiol 2013, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (66).Willger SD; Puttikamonkul S; Kim K-H; Burritt JB; Grahl N; Metzler LJ; Barbuch R; Bard M; Lawrence CB; Jr RAC A Sterol-Regulatory Element Binding Protein Is Required for Cell Polarity, Hypoxia Adaptation, Azole Drug Resistance, and Virulence in Aspergillus Fumigatus. PLOS Pathog. 2008, 4 (11), e1000200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (67).Vödisch M; Scherlach K; Winkler R; Hertweck C; Braun H-P; Roth M; Haas H; Werner ER; Brakhage AA; Kniemeyer O. Analysis of the Aspergillus Fumigatus Proteome Reveals Metabolic Changes and the Activation of the Pseurotin A Biosynthesis Gene Cluster in Response to Hypoxia. J. Proteome Res 2011, 10 (5), 2508–2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (68).Barker BM; Kroll K; Vödisch M; Mazurie A; Kniemeyer O; Cramer RA Transcriptomic and Proteomic Analyses of the Aspergillus Fumigatus Hypoxia Response Using an Oxygen-Controlled Fermenter. BMC Genomics 2012, 13 (1), 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (69).Gautam P; Shankar J; Madan T; Sirdeshmukh R; Sundaram CS; Gade WN; Basir SF; Sarma PU Proteomic and Transcriptomic Analysis of Aspergillus Fumigatus on Exposure to Amphotericin B. Antimicrob. Agents Chemother 2008, 52 (12), 4220–4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (70).Amarsaikhan N; Albrecht-Eckardt D; Sasse C; Braus GH; Ogel ZB; Kniemeyer O. Proteomic Profiling of the Antifungal Drug Response of Aspergillus Fumigatus to Voriconazole. Int. J. Med. Microbiol. IJMM 2017, 307 (7), 398–408. [DOI] [PubMed] [Google Scholar]
- (71).Alekhova TA; Aleksandrova AA; Novozhilova TI; Lysak LV; Zagustina NA; Bezborodov AM [Monitoring of microbial degraders in manned space stations]. Prikl. Biokhim. Mikrobiol 2005, 41 (4), 435–443. [PubMed] [Google Scholar]
- (72).Chiang Y-M; Meyer KM; Praseuth M; Baker SE; Bruno KS; Wang CCC Characterization of a Polyketide Synthase in Aspergillus Niger Whose Product Is a Precursor for Both Dihydroxynaphthalene (DHN) Melanin and Naphtho-γ-Pyrone. Fungal Genet. Biol 2011, 48 (4), 430–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (73).Romsdahl J; Blachowicz A; Chiang AJ; Singh N; Stajich JE; Kalkum M; Venkateswaran K; Wang CCC Characterization of Aspergillus Niger Isolated from the International Space Station. mSystems 2018, 3 (5), e00112–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (74).Dadachova E; Casadevall A. Ionizing Radiation: How Fungi Cope, Adapt, and Exploit with the Help of Melanin. Curr. Opin. Microbiol 2008, 11 (6), 525–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (75).Singaravelan N; Grishkan I; Beharav A; Wakamatsu K; Ito S; Nevo E. Adaptive Melanin Response of the Soil Fungus Aspergillus Niger to UV Radiation Stress at “Evolution Canyon”, Mount Carmel, Israel. PLoS ONE 2008, 3 (8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (76).Gauthier T; Wang X; Sifuentes Dos Santos J; Fysikopoulos A; Tadrist S; Canlet C; Artigot MP; Loiseau N; Oswald IP; Puel O. Trypacidin, a Spore-Borne Toxin from Aspergillus Fumigatus, Is Cytotoxic to Lung Cells. PloS One 2012, 7 (2), e29906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (77).Nybakken L; Solhaug KA; Bilger W; Gauslaa Y. The Lichens Xanthoria Elegans and Cetraria Islandica Maintain a High Protection against UV-B Radiation in Arctic Habitats. Oecologia 2004, 140 (2), 211–216. [DOI] [PubMed] [Google Scholar]
- (78).Maličev E; Chowdhury HH; Maček P; Sepčić K. Effect of Ostreolysin, an Asp-Hemolysin Isoform, on Human Chondrocytes and Osteoblasts, and Possible Role of Asp-Hemolysin in Pathogenesis. Med. Mycol 2007, 45 (2), 123–130. [DOI] [PubMed] [Google Scholar]
- (79).Ebina K; Ichinowatari S; Yokota K. Studies on Toxin of Aspergillus Fumigatus. XXII. Fashion of Binding of Asp-Hemolysin to Human Erythrocytes and Asp-HemolysinBinding Proteins of Erythrocyte Membranes. Microbiol. Immunol 1985, 29 (2) , 91–101. [DOI] [PubMed] [Google Scholar]
- (80).Kumagai T; Nagata T; Kudo Y; Fukuchi Y; Ebina K; Yokota K. Cytotoxic Activity and Cytokine Gene Induction of Asp-Hemolysin to Murine Macrophages. Nihon Ishinkin Gakkai Zasshi Jpn. J. Med. Mycol 1999, 40 (4), 217–222. [DOI] [PubMed] [Google Scholar]
- (81).Kumagai T; Nagata T; Kudo Y; Fukuchi Y; Ebina K; Yokota K. [Cytotoxic activity and cytokine gene induction of Asp-hemolysin to vascular endothelial cells]. Yakugaku Zasshi 2001, 121 (4), 271–275. [DOI] [PubMed] [Google Scholar]
- (82).Wartenberg D; Lapp K; Jacobsen ID; Dahse H-M; Kniemeyer O; Heinekamp T; Brakhage AA Secretome Analysis of Aspergillus Fumigatus Reveals Asp-Hemolysin as a Major Secreted Protein. Int. J. Med. Microbiol. IJMM 2011, 301 (7), 602–611. [DOI] [PubMed] [Google Scholar]
- (83).Li L; Naseem S; Sharma S; Konopka JB Flavodoxin-Like Proteins Protect Candida Albicans from Oxidative Stress and Promote Virulence. PLOS Pathog. 2015, 11 (9), e1005147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (84).Pedroso N; Gomes-Alves P; Marinho HS; Brito VB; Boada C; Antunes F; Herrero E; Penque D; Cyrne L. The Plasma Membrane-Enriched Fraction Proteome Response during Adaptation to Hydrogen Peroxide in Saccharomyces Cerevisiae. Free Radic. Res 2012, 46 (10), 1267–1279. [DOI] [PubMed] [Google Scholar]
- (85).Leach MD; Stead DA; Argo E; MacCallum DM; Brown AJP Molecular and Proteomic Analyses Highlight the Importance of Ubiquitination for the Stress Resistance, Metabolic Adaptation, Morphogenetic Regulation and Virulence of Candida Albicans. Mol. Microbiol 2011, 79 (6), 1574–1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (86).Leach MD; Brown AJP Posttranslational Modifications of Proteins in the Pathobiology of Medically Relevant Fungi. Eukaryot. Cell 2012, 11 (2), 98–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (87).Baqai FP; Gridley DS; Slater JM; Luo-Owen X; Stodieck LS; Ferguson V; Chapes SK; Pecaut MJ Effects of Spaceflight on Innate Immune Function and Antioxidant Gene Expression. J. Appl. Physiol. Bethesda Md 1985 2009, 106 (6), 1935–1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (88).Crabbé A; Nielsen-Preiss SM; Woolley CM; Barrila J; Buchanan K; McCracken J; Inglis DO; Searles SC; Nelman-Gonzalez MA; Ott CM; et al. Spaceflight Enhances Cell Aggregation and Random Budding in Candida Albicans. PloS One 2013, 8 (12), e80677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (89).Rizzo AM; Corsetto PA; Montorfano G; Milani S; Zava S; Tavella S; Cancedda R; Berra B. Effects of Long-Term Space Flight on Erythrocytes and Oxidative Stress of Rodents. PLOS ONE 2012, 7 (3), e32361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (90).Stein TP Space Flight and Oxidative Stress. Nutr. Burbank Los Angel. Cty. Calif 2002, 18 (10), 867–871. [DOI] [PubMed] [Google Scholar]
- (91).Versari S; Longinotti G; Barenghi L; Maier JAM; Bradamante S. The Challenging Environment on Board the International Space Station Affects Endothelial Cell Function by Triggering Oxidative Stress through Thioredoxin Interacting Protein Overexpression: The ESA-SPHINX Experiment. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol 2013, 27 (11), 4466–4475. [DOI] [PubMed] [Google Scholar]
- (92).Kurth D; Belfiore C; Gorriti MF; Cortez N; Farias ME; Albarracín VH Genomic and Proteomic Evidences Unravel the UV-Resistome of the Poly-Extremophile Acinetobacter Sp. Ver3. Front. Microbiol 2015, 6, 328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (93).De Lucas JR; Domínguez AI; Valenciano S; Turner G; Laborda F. The AcuH Gene of Aspergillus Nidulans, Required for Growth on Acetate and Long-Chain Fatty Acids, Encodes a Putative Homologue of the Mammalian Carnitine/Acylcarnitine Carrier. Arch. Microbiol 1999, 171 (6), 386–396. [DOI] [PubMed] [Google Scholar]
- (94).Anjo SI; Figueiredo F; Fernandes R; Manadas B; Oliveira M. A Proteomic and Ultrastructural Characterization of Aspergillus Fumigatus’ Conidia Adaptation at Different Culture Ages. J. Proteomics 2017, 161 (Supplement C), 47–56. [DOI] [PubMed] [Google Scholar]
- (95).Vizcaíno JA; Csordas A; del-Toro N; Dianes JA; Griss J; Lavidas I; Mayer G; Perez-Riverol Y; Reisinger F; Ternent T; et al. 2016 Update of the PRIDE Database and Its Related Tools. Nucleic Acids Res. 2016, 44 (D1), D447–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S-1. AspGD GO Slim terms of proteins normalized to Af293. Differentially expressed proteins in CEA10, ISSFT-021 and IF1SW-F4 are categorized into GO Slim categories using AspGD. Categories containing at least 5 up- or down-regulated proteins are presented.
Figure S-2. Monomethylsulochrin yields in clinical and ISS-isolated strains. A) Extracted ion count (EIC) traces of monomethylsulochrin in a positive mode. B) Quantification of of monomethylsulochrin production using positive mode EIC when compared to Af293. Significance was determined using Welch’s t-test.
Figure S-3. Sporulation capacity of clinical and ISS-isolated strains. Average values of conidial counts from 3 biological replicates are plotted after 3, 4 and 5 days of growth. Statistical significance was determined using multiple t-test with Holm-Sidak correction.
Table S-1. Up- and down-regulated proteins in CEA10, ISSFT-021 and IF1SW-F4 when normalized to Af293
Table S-2. Up- and down-regulated proteins in space strains when compared to Af293 and CEA10
Table S-3. GO enrichment analysis of proteins in space strains only
Table S-4. Differentially expressed proteins in CEA10, ISSFT-021 and IF1SW-F4 when grown on solid GMM, and liquid PD and CD media when normalized to Af293