ABSTRACT
The Mycobacterium tuberculosis genome contains an abundance of toxin-antitoxin (TA) systems, 50 of which belong to the VapBC family. The activity of VapC toxins is controlled by dynamic association with their cognate antitoxins—the toxin is inactive when complexed with VapB antitoxin but active when freed. Here, we determined the cellular target of two phylogenetically related VapC toxins and demonstrate how their properties can be harnessed for drug development. First, we used a specialized RNA sequencing (RNA-seq) approach, 5′ RNA-seq, to accurately identify the in vivo RNA target of M. tuberculosis VapC2 and VapC21 toxins. Both toxins exclusively disable initiator tRNAfMet through cleavage at a single, identical site within their anticodon loop. Consistent with the essential role and global requirement for initiator tRNAfMet in bacteria, expression of each VapC toxin resulted in potent translation inhibition followed by growth arrest and cell death. Guided by previous structural studies, we then mutated two conserved amino acids in the antitoxin (WR→AA) that resided in the toxin-antitoxin interface and were predicted to inhibit toxin activity. Both mutants were markedly less efficient in rescuing growth over time, suggesting that screens for high-affinity small-molecule inhibitors against this or other crucial VapB-VapC interaction sites could drive constitutive inactivation of tRNAfMet by these VapC toxins. Collectively, the properties of the VapBC2 and VapBC21 TA systems provide a framework for development of bactericidal antitubercular agents with high specificity for M. tuberculosis cells.
KEYWORDS: tRNA, RNA-seq, protein synthesis, translation, antibiotic, antitubercular, antimicrobial activity
INTRODUCTION
Among the ∼90 type II toxin-antitoxin (TA) systems in Mycobacterium tuberculosis, most belong to the 50-member VapBC family of RNases (designated VapC1 through VapC50) (1). All VapC toxins possess a PIN (PilT amino-terminal) catalytic domain containing 4-5 conserved acidic residues, all of which are responsible for coordinating one or more divalent cations in the catalytic center (2). It is not understood why the M. tuberculosis genome has the highest number of VapC TA systems among free-living bacteria; most bacteria have just one (3). Curiously, the few M. tuberculosis VapC toxins studied in detail have different targets that lead to dramatically different endpoints. The M. tuberculosis VapC20 and VapC26 toxins cleave 23S rRNA at a single site within the sarcin-ricin loop that is universally conserved within all domains of life. Cleavage of the sarcin-ricin loop by these two toxins leads to ribosome inactivation and translation inhibition (4, 5). In contrast, the M. tuberculosis VapC4 (4, 6, 7) and VapC11 (8) are isoacceptor-specific tRNases. The VapC4 toxin exclusively inactivates the sole tRNACys through cleavage at a single site within the anticodon sequence (6). Reduction in the levels of tRNACys mimics a state of Cys starvation that leads to the reprogramming of M. tuberculosis metabolism to specifically defend against oxidative and copper stresses (6).
Type II TA systems are composed of adjacent genes encoding two small (∼10-kDa) proteins, a toxin and its cognate antitoxin that inhibits toxin activity through formation of a stable TA protein-protein complex. In the prevailing model, stress conditions are proposed to play a major role in regulating the toxin-antitoxin balance by selectively activating antitoxin degradation (9, 10). Thus, specific stress conditions lead to a preponderance of free toxin, which exerts its growth-regulating and/or other functions within the bacterial cells (9, 10). To obtain a broader understanding of M. tuberculosis VapC toxin functions and how they might collaborate to survive the spectrum of stresses associated with the M. tuberculosis infection cycle, we are systematically identifying the RNA targets of each toxin and studying how they impact M. tuberculosis growth and physiology. Here, we identified tRNAfMet as a new target for the M. tuberculosis VapC family. We demonstrate that expression of either of the two toxins with this activity, VapC2 and VapC21, results in strong inhibition of protein synthesis and cell death. Finally, we highlight how their properties can be exploited for development of a shorter, more effective treatment regimen for tuberculosis (TB) infections with high specificity for M. tuberculosis cells while sparing beneficial resident bacteria.
RESULTS
Expression of phylogenetically related VapC toxins, VapC2 and VapC21, leads to growth inhibition.
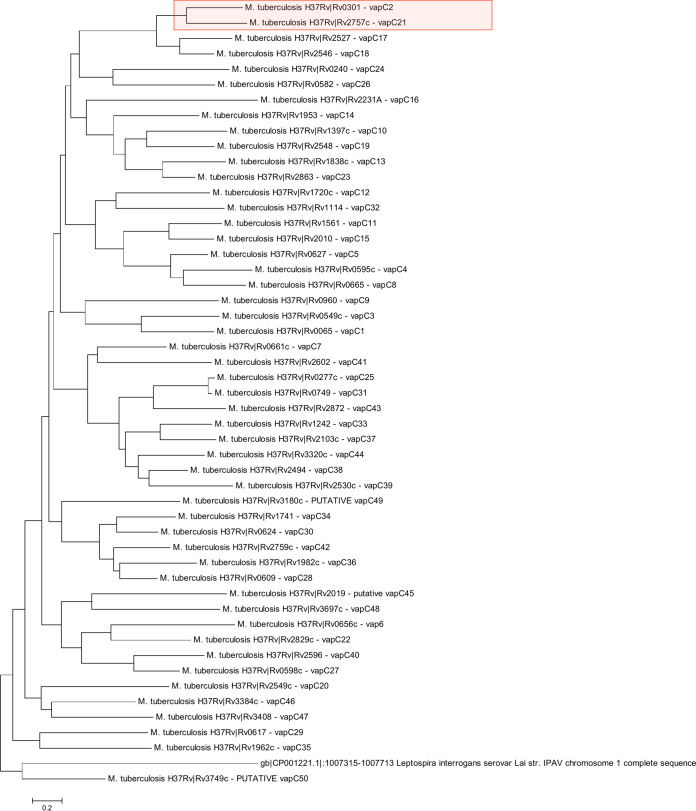
We created a phylogenetic tree with all 50 M. tuberculosis VapC toxins (Fig. 1). We identified two toxins, VapC2 and VapC21, together on one branch of the tree and posited that these two toxins likely have similar functions. Therefore, going forward we studied these two toxins in parallel.
FIG 1.
Phylogenetic tree of the 50 M. tuberculosis VapC toxins. VapC2 and VapC21 are highlighted in red.
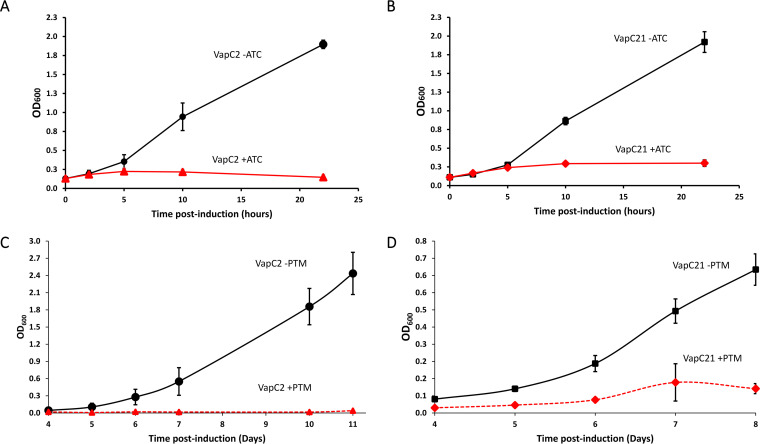
Following convention in the field, we individually expressed the two toxins of interest from an inducible expression plasmid in Mycobacterium smegmatis or M. tuberculosis. Tightly regulated inducible plasmids are used because the physiological triggers of nearly all bacterial TA systems are not known. Triggers that lead to expression of VapC2 and VapC21 have also not been rigorously established. Expression of either VapC2 or VapC21 toxin leads to strong growth arrest in both mycobacterial model organisms (Fig. 2). Growth begins to flatline approximately 5 h postinduction in M. smegmatis (Fig. 2A and B) and approximately 5 days postinduction in M. tuberculosis (Fig. 2C and D). This phenotype is in alignment with type II TA systems, where toxin expression characteristically leads to some degree of cell growth arrest regardless of toxin mechanism of action.
FIG 2.
Expression of VapC2 and VapC21 leads to growth inhibition. (A and B) Growth profile of M. smegmatis cells harboring anhydrotetracycline (ATC)-inducible plasmid expressing either VapC2 or VapC21 with (+ATC) or without (-ATC) the inducer. (C and D) Growth profile of M. tuberculosis cells harboring the pristinamycin (PTM)-inducible plasmid expressing either VapC2 or VapC21 with (+PTM) or without (-PTM) inducer.
VapC2 and VapC21 cleave multiple tRNAs in vitro.
Knowing that M. tuberculosis VapC toxins have been reported to cleave either tRNA or 23S rRNA (4, 5, 7, 8, 11), we first prepared recombinant VapC2 and VapC21 toxins and performed in vitro cleavage assays with a complete set of M. tuberculosis tRNAs. Several of the 45 tRNAs were either partially or completely cleaved when incubated with each toxin (shaded gray in Fig. S1A and B in the supplemental material). However, only tRNAfMet (designated “Met30” in Fig. S1A and B) was efficiently cleaved and generated stable cleavage products corresponding to two tRNA halves (shaded black in Fig. S1A and B). Although previous studies in our laboratory indicated that this in vitro cleavage assay typically suggests more tRNA targets than those we identify by 5′ RNA sequencing (RNA-seq) in vivo (6, 8), we have found that this in vitro assay is a reliable indicator of the class of RNA favored by an RNase toxin. Therefore, VapC2 and VapC21 are likely tRNases.
VapC2 and VapC21 cleave and inactivate only tRNAfMet in vivo.
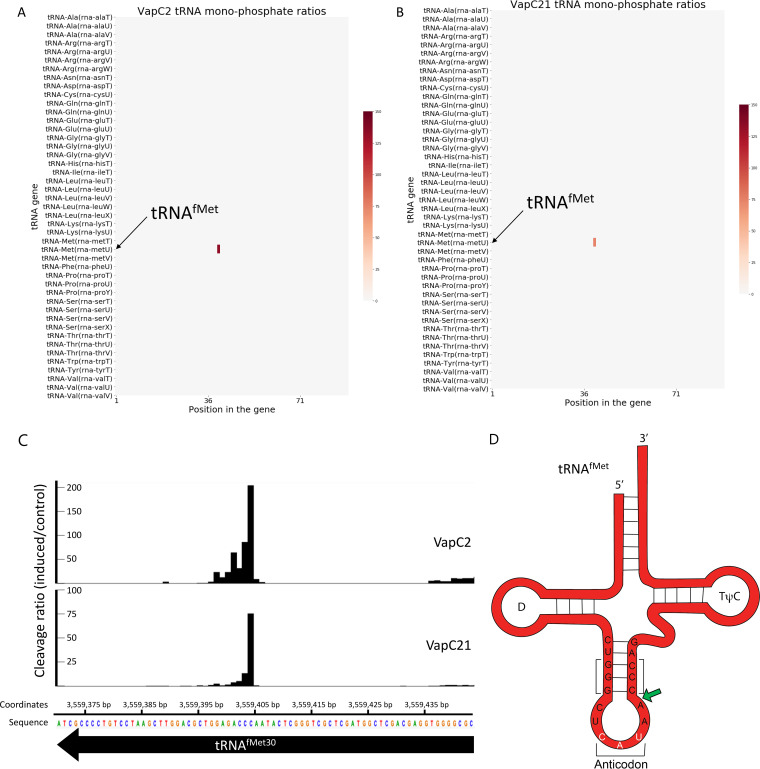
We have previously demonstrated that endoribonuclease toxins must be expressed in their native host for reliable identification of their true in vivo target(s) (6, 8). Therefore, we harvested total RNA from M. tuberculosis cells expressing VapC2 or VapC21 versus controls and performed 5′ RNA-seq to identify the RNA targets of each toxin in vivo. 5′ RNA-seq enables genome-wide RNA target identification coupled with high-resolution, single-nucleotide cleavage site mapping (12). For VapC toxins, 5′ RNA-seq amplifies RNAs carrying a 5′-monophosphate (5′-P) moiety generated upon cleavage by VapC. As suggested by in vitro cleavage assays, 5′ RNA-seq unequivocally identified tRNAfMet as the sole target of each toxin (Fig. 3A to C). Both toxins cleave only M. tuberculosis tRNAfMet within the anticodon loop between nucleotides (nt) 39 and 40 (Fig. 3D); they do not cleave the other two M. tuberculosis elongator tRNAMet species (metT and metV) shown near tRNAfMet (metU) in Fig. 3A and B heatmaps. The cleavage position is identical to that of the sole VapC toxin in Salmonella/Shigella that also targets initiator tRNAfMet (13) and adjacent to the site of tRNAfMet cleavage by two VapCs in Haemophilus influenzae (14). The VapC2 and VapC21 cleavage site is also in proximity to the three G-C pairs at the base of the stem that are crucial for initiation factor (IF)-3 discrimination of initiator tRNA versus elongator tRNAfMet (15) (Fig. 3D).
FIG 3.
VapC2 and VapC21 cleave and inactivate only tRNAfMet in vivo. (A and B) Heatmap representing fold change (induced/control) at each position for all 45 M. tuberculosis tRNAs was generated from 5′-P RNA-seq libraries after induction of either VapC2 (A) or VapC21 (B) in M. tuberculosis. (C) Histogram representing the ratio of cleavage by VapC2 and VapC21 toxin identified using 5′ RNA-seq at each nucleotide within the metU gene (tRNAfMet30) in M. tuberculosis after toxin induction. Genomic position and the negative-strand sequence are shown. (D) Representation of VapC2 and VapC21 target, tRNAfMet, showing the sequence of the anticodon stem-loop. The anticodon sequence (CAU) is highlighted in white; the toxin cleavage site is denoted with green arrow. The three G-C pairs at the base of the stem that are crucial for initiation factor (IF)-3 discrimination of initiator tRNA versus elongator tRNAfMet are bracketed (15).
VapC2 and VapC21 inhibit protein synthesis.
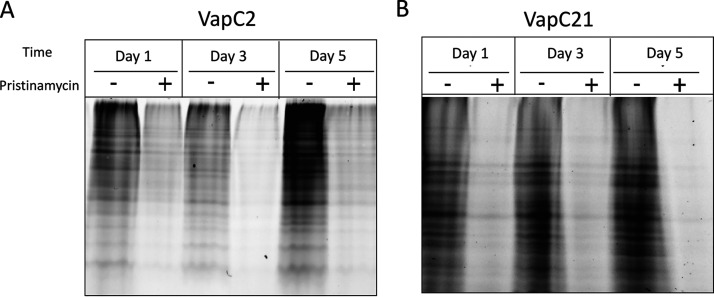
Since these two toxins disable a tRNA species critical for translation of all bacterial mRNAs, we next tested if their roles are to comprehensively shut down translation. We used a click chemistry approach for metabolic labeling to monitor de novo protein synthesis in M. tuberculosis cells with and without VapC2 or VapC21 toxin expression. Incorporation of the azide-containing Met mimetic azidohomoalanine (AHA) enabled fluorescent visualization of AHA-containing proteins upon coupling to the alkyne tetramethylrhodamine (TAMRA). As anticipated, translation was severely impaired at all intervals tested (days 1, 3 and 5 following toxin induction) (Fig. 4A and B).
FIG 4.
VapC2 and VapC21 inhibit protein synthesis. Newly synthesized proteins were labeled with the methionine mimetic AHA in induced and uninduced samples 1, 3, or 5 days post-induction of either VapC2 (A) or VapC21 (B) and visualized upon alkyne-TAMRA conjugation. Each lane was normalized to 10 μg of total protein.
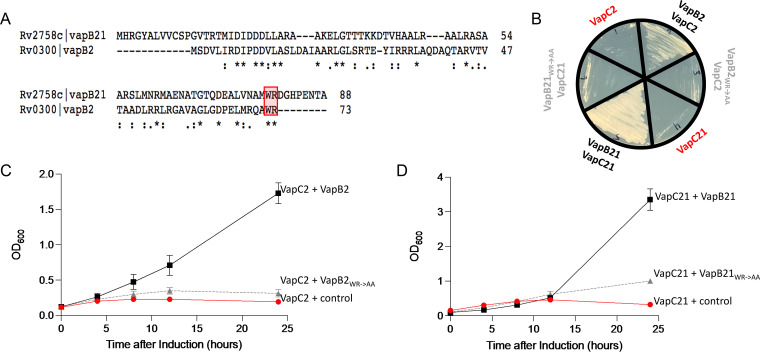
The C-terminal WR residues of VapB2 and VapB21 antitoxins contribute to inhibition of toxin activity.
M. tuberculosis VapB2 and VapB21 proteins are 18% identical and 28% similar (Fig. 5A). Clues to their precise mechanism of action were revealed by the X-ray crystal structure of the M. tuberculosis VapBC2 complex (16; note that the title of the reference mistakenly refers to this TA pair as VapBC-3). The VapB2 WR residues located at the carboxy terminus (boxed and highlighted in Fig. 5A) are critical for inhibition of VapC2 toxin enzymatic activity; they reside in the carboxy-terminal helix that obstructs the VapC2 RNA-binding groove and blocks access to the VapC2 active sites (16). More specifically, VapB2 W72 embeds into a crevice formed between the VapC toxin dimers; R73 disrupts the catalytic PIN domain of VapC2 by contacting three Mg2+-coordinating aspartates of its PIN domain (D9, D99, and D117, boxed in Fig. 6A) (16). The WR amino acid pair is also present in VapB21, but it is located 10 amino acids from the carboxy end (Fig. 5A).
FIG 5.
The C-terminal WR residues of VapB2 and VapB20 antitoxins are necessary for inhibition of the cognate VapC toxin. (A) VapB2 and VapB21 sequence alignments highlighting the position of the WR residues generated using Clustal Omega v. 1.2.4 (45). (B) Growth phenotypes of a colony used to inoculate each of the represented growth profiles in panels C and D; color coding matches detailed descriptions in the growth profiles. (C) Growth profiles of M. smegmatis expressing VapC2 toxin from the ATC-inducible pMC1s plasmid plus either the empty isovaleronitrile (IVN)-inducible pNIT plasmid (control, red circle), wild-type VapB2 antitoxin in pNIT (black square), or VapB2 W72A/R739778A mutant antitoxin in pNIT (WR→AA, gray triangle). (D) Growth profiles of M. smegmatis expressing VapC21 from the ATC-inducible pMC1s plasmid plus either the empty IVN-inducible pNIT plasmid (control, red circle), wild-type VapB21 antitoxin in pNIT (black square), or VapB21 (W79A/R80A) mutant antitoxin in pNIT (WR→AA, gray triangle). Growth curves performed in triplicate; standard deviation indicated.
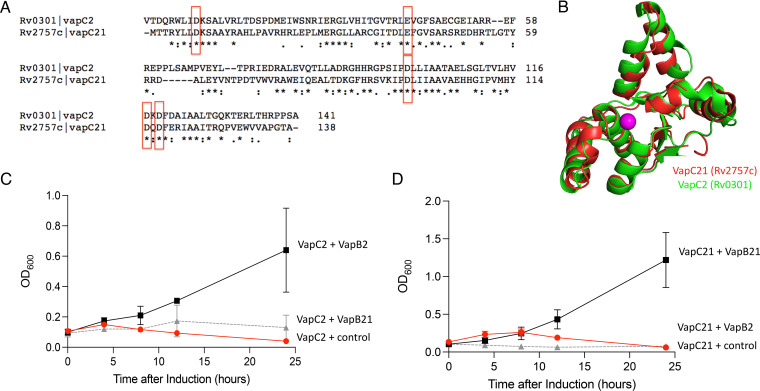
FIG 6.
VapC2 and VapC21 toxins are structurally similar, yet the VapB2 and VapB21 antitoxins are not interchangeable. (A) VapC2 and VapC21 sequence alignments were generated using Clustal Omega v 1.2.4 (45). The five conserved residues comprising the PIN domain are boxed in red. (B) Crystal structures of M. tuberculosis VapC2 (PDB 3H87) and VapC21 (PDB 5SV2) were superimposed using PyMOL. (C) Growth profiles of M. smegmatis carrying VapC2 toxin in pMC1s plasmid with either control pNIT vector (red circle), VapB2 antitoxin (black square), or VapB21 antitoxin (gray triangle). Growth curves performed in triplicate; standard deviation indicated. (D) Growth profiles of M. smegmatis carrying VapC21 toxin in pMC1s plasmid with either control pNIT vector (red circle), VapB21 antitoxin (black square), or VapB2 antitoxin (gray triangle).
We mutated the WR amino acid pair in VapB2 and VapB21 to alanines (WR→AA) and compared the abilities of mutant and wild-type antitoxins to rescue toxin-mediated reduction in growth rate in culture (Fig. 5C and D) and on an agar plate (Fig. 5B). Consistent with their critical position in the VapBC2 structure, both mutant WR→AA antitoxins were impaired in their ability to inhibit toxin activity (Fig. 5B to D). Therefore, these conserved WR residues appear to be excellent drug targets for inhibitors of the antitoxin-toxin interaction. We then measured cell viability from growth profiles by plating for CFU (see Fig. S2 in the supplemental material). Both mutant antitoxins were again impaired in their ability to rescue the toxin-mediated reduction in growth rate (Fig. S2A and B) but displayed only marginal reductions in recovery of viable cells with time (Fig. S2C and D). However, exclusive consideration of the plotted CFU-per-milliliter data does not fully reflect how the WR mutants impair growth rate. The same serial dilutions used for the CFU-per-milliliter data show that the mutant VapB2 antitoxin dramatically reduces cell growth rates on plates compared to wild-type VapB2 antitoxin (Fig. S2E to J). The colony sizes in the spotted aliquots of serial dilutions of cultures expressing VapB2 mutant antitoxin were markedly smaller 12 h postinduction (Fig. S2F and G). Finally, the CFU-per-milliliter plots in Fig. S2C and D with and without antitoxin show a 3-log difference in cell recovery, consistent with the clinical standard for a bactericidal effect, i.e., 99.9% killing or a ≥3-log10-CFU/mL reduction in colony count (17, 18). Therefore, expression of VapC2 or VapC21 toxins is bactericidal, not bacteriostatic.
Taken together, these results demonstrate that the antitoxin WR residues contribute to, but are not necessarily the sole mediators of, a stable toxin-antitoxin complex. Nevertheless, the WR and amino acids in proximity to the WR residues in the VapB2 tertiary structure remain an important therapeutic target because of their clear functional significance. A more comprehensive interrogation of the VapB2-VapC2 interaction sites (∼40% of VapB2 residues contact VapC2 [16]) will likely lead to identification of more drug target sites. Although the structure of VapBC21 has not been reported, the antitoxin may interact with the toxin in a similar manner since VapB21 also contains the WR motif.
VapC2 and VapC21 toxins are structurally similar, yet the VapB2 and VapB21 antitoxins are not interchangeable.
Finally, we aligned the two toxin sequences (37% identity and 49% overall similarity) (Fig. 6A) and modeled their tertiary structures (Fig. 6B). In spite of their similarity, when we coexpressed these toxins with their noncognate antitoxins, we determined that they were not able to functionally interact, i.e., the VapC2-VapB21 and VapC21-VapB2 pairs still exhibited toxicity as assessed by growth arrest compared to the cognate TA complexes (Fig. 6C and D). Therefore, although there are reports of cross talk between noncognate toxins and antitoxins in M. tuberculosis (19), this was not the case for VapBC2 and VapBC21.
DISCUSSION
To date, M. tuberculosis VapC toxins are now known to specifically cleave and inactivate two classes of RNAs, tRNAs and 23S rRNA, leading to two distinct outcomes. tRNA-cleaving toxins can target specific isoacceptors for mRNA codons or, as demonstrated here, tRNAfMet. We have recently reported that VapC toxins that target tRNA isoacceptors used during elongation of newly synthesized proteins have a more subtle effect: they remodel the M. tuberculosis transcriptome and proteome toward an outcome beneficial to this pathogen (6). In contrast, bacterial toxins that cleave and inactivate initiator tRNAfMet (13, 14) or the 23S rRNA sarcin-ricin loop (4, 5) cause inhibition of protein synthesis. Therefore, prolonged inactivation of ribosomes through cleavage at the extremely conserved sarcin-ricin loop—present in 23S rRNA of ribosomes in all domains of life—should be deadly to bacteria and harmful to humans as well. In contrast, prolonged inactivation of the initiator tRNAfMet, leading to global inhibition of translation and cell death (see Fig. S2 in the supplemental material), is expected to be deadly to only bacteria and not human cells since tRNAfMet is absent in eukaryotes. This distinction suggests the use of the VapBC2 and VapBC21 TA systems as novel targets for development of more effective antitubercular agents that are less susceptible to resistance.
The drugs currently used for TB treatment were developed >50 years ago by the pharmaceutical industry (20). At that time, the prescribed drug regimens led to a significant decline in TB in the industrialized world, so there was little incentive to continue its drug discovery and development programs. This gap in development has left us with a limited arsenal for treatment options with very few drugs that are also effective against M. tuberculosis persister cells present during latent TB. Though difficult to prove formally, it is believed that most bacilli in latent M. tuberculosis infection are not actively replicating but remain viable. Thus, there is an urgent need for the development of new TB drugs that (i) clear M. tuberculosis infection faster because they also kill persisters, (ii) are effective against multidrug-resistant (MDR) and extremely drug-resistant (XDR) TB, (iii) are less susceptible to being disarmed through the development of cellular resistance mechanisms, and (iv) are not contraindicated in combination with HIV antiretroviral therapy.
The ability of VapC2 and VapC21 to specifically target tRNAfMet suggests that the development of small-molecule inhibitors (or peptide inhibitors) of the VapBC2 and/or VapBC21 complexes would be a worthy path to pursue in follow-on studies. This proposed strategy for antimicrobial development relies on tricking cells into committing suicide using a small molecule or peptide to break up the T-A complex (preferably by selectively binding the antitoxin only as to not alter toxin function) and prevent its interaction with the toxin (illustrated in Fig. 7). This approach should result in an excess of free toxin with specificity to initiator tRNAfMet, leading to widespread translation inhibition and cell death (Fig. 7). In addition to the antitoxin WR residues that contribute to stable TA complex formation, there are many other interaction interfaces. In the case of VapB2, nearly 40% of its residues are known to participate in dimerization with VapC2 (16). The critical attributes that underlie our hypothesis that VapB2 or VapB21 antitoxins are viable antibiotic targets are reviewed below.
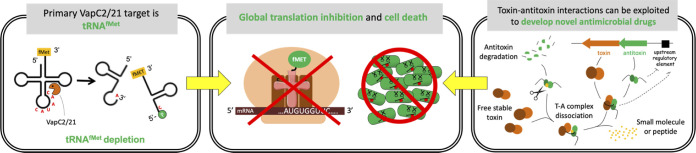
FIG 7.
Summary of proposed VapC2 and VapC21 mechanism of action and application as targets for antitubercular development. VapC2 and VapC21 specifically cleave and inactivate initiator tRNAfMet (left panel). Sustained toxin expression leads to depletion of this tRNA which is essential for translation of M. tuberculosis mRNAs, resulting in cell death (center panel). These properties can be harnessed for drug development as illustrated in the right panel. TA modules typically reside in an autoregulated operon (repressed by antitoxin alone or the TA complex); the antitoxin gene precedes the toxin gene (top right of right panel). After their transcription and translation, the antitoxin protein is intrinsically unfolded (and thus more susceptible to proteolytic cleavage) until it assembles with its cognate antitoxin. Identification of small-molecular inhibitors (yellow) of the T-A interaction is predicted to drive this dynamic system toward excess, free toxin and cell death because free antitoxin with bound inhibitor will be susceptible to degradation by M. tuberculosis proteases (scissors).
First, free VapB antitoxin is intrinsically unstable in vivo. When the antitoxin is not associated with its cognate toxin, it is readily degraded by one or more cellular proteases (Fig. 7, top left of right panel). Early biological observations of antitoxin instability in vivo (21–23) are now supported by elegant structural and biophysical data. Several high-resolution structures of VapBC protein complexes compared to VapB apo (antitoxin-only) structures have been reported (24–26). In general, the antitoxins have a two-domain structure in which the carboxy-terminal domain is intrinsically disordered and folds only when associated with cognate toxins. Therefore, if a drug prevented an M. tuberculosis antitoxin from associating with its cognate toxin, the antitoxin should be degraded by M. tuberculosis proteases (Fig. 7, top left of right panel). Although M. tuberculosis lacks Lon and HsIUV proteases (27), it does have ClpCP/XP proteases (28). TA systems are substrates of ClpCP proteases in M. tuberculosis (28).
Second, free VapC toxins should lead to cell death in actively dividing cells as well as those in the nonreplicating persistent state characteristic of latent TB. It is well established that TA systems initiate the cell suicide program after enduring an extended state of growth arrest, once they have sustained irreparable damage (29, 30). Therefore, the normal progression for TA toxin initiation of the suicide program is from growth-arrested cells, not from actively growing cells. Since M. tuberculosis cells in the latent state of the disease are also not actively replicating but viable, the cells are somewhat analogous to those that are in the growth-arrested, viable state due to TA toxin activation. These parallels suggest that TA toxins can mediate the suicide of both actively growing and latent-state M. tuberculosis cells. However, the efficacy of this approach for treatment of latent tuberculosis is dependent on the presence of functional toxin and antitoxin proteins in these M. tuberculosis cells, which has not been established.
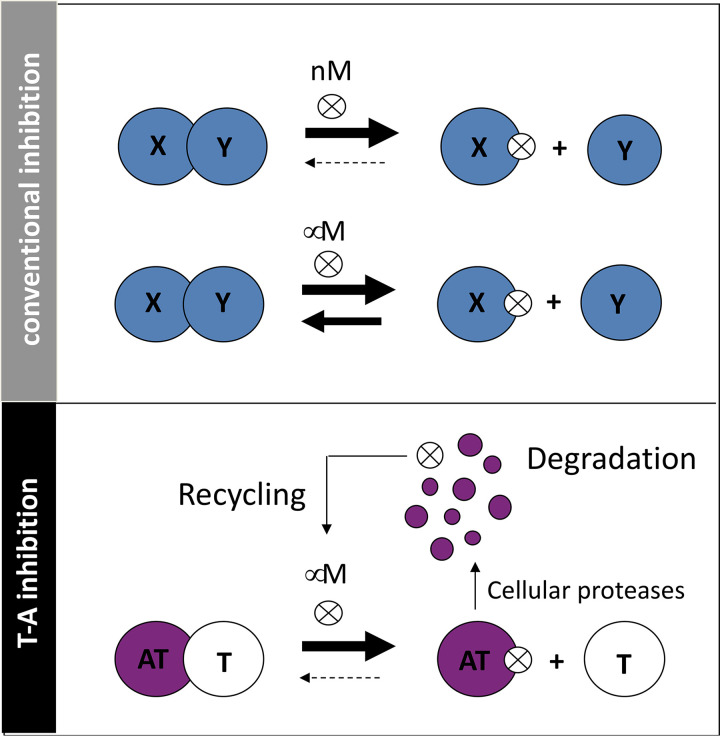
Third, characteristic autoregulation of the TA operon should potentiate the effects of drugs that prevent T-A association (31). Autoregulation (when the presence of excess operon gene products represses its transcription) serves to potentiate the efficacy of the drug since the levels of both putative repressors of the operon—the antitoxin alone and the TA complex—are minimized or eliminated. This is predicted to lead to runaway transcription of the TA module and overproduction of both the toxin (good because this further accelerates suicide) and antitoxin (not a problem because newly synthesized antitoxin should be bound by inhibitor and degraded and the inhibitor should be recycled). This feature is important because it also pushes the equilibrium toward free toxin when a small molecule/peptide binds to the antitoxin. Since the equilibrium is predicted to favor free toxin when there is concomitant degradation of the antitoxin and recycling of the inhibitor, nanomolar concentrations (typically the gold standard for small molecule inhibitors) may not be necessary (Fig. 8).
FIG 8.
Small-molecular inhibitors of T-A association are predicted to be effective at lower affinities than conventional protein complexes. (Top) In conventional inhibition of protein-protein interaction with small molecules or peptides (⊗), it is most desirable to select those that bind at nanomolar affinities to favor maintenance of the dissociated complex; micromolar affinities are often not potent enough to keep the proteins dissociated. (Below) In contrast, for TA complexes the intrinsic instability of the antitoxin coupled with inhibitor recycling upon antitoxin degradation is predicted to potentiate the efficacy of inhibitors; micromolar affinities may be sufficient to favor stable dissociation of the two proteins.
Fourth, the presence of two known tRNAfMet family members in M. tuberculosis may also prevent the acquisition of drug resistance. Since we demonstrated that the carboxy-terminal WR residues of the VapB2 and VapB21 antitoxins are essential for association with their cognate toxins (recall that noncognate binding of VapB2-VapC21 and VapB21-VapB2 does not occur [Fig. 6C and D]), this WR domain is a likely region for inhibitors to bind. Target redundancy could safeguard against acquisition of resistance by mutation of either antitoxin (e.g., which cannot bind to the inhibitor) or the toxin (e.g., a mutation of any one of the residues comprising the PIN domain that abrogates enzymatic activity [32]). The probability that a single mutation event occurs in one protein of the VapB2-VapC2 pair is high given the selective pressure. However, the likelihood that a second independent drug-inactivating mutation appears in one member of the VapB21-VapC21 pair is lower, making acquisition of resistance less likely.
Inhibition of protein-protein interactions through small-molecule inhibitors enables the manipulation of key intracellular processes that dictate bacterial growth, cancer progression, and viral replication. Although it was once viewed as a more challenging strategy relative to small-molecule inhibition of the enzymatic function of a single protein, the number of success stories continues to grow (33–36). Some of the highest-visibility examples include disruption of p53/MDM2 to induce apoptosis and inhibit tumor growth (37), disruption of ToxT transcriptional activator dimerization to inhibit transcription of the cholera toxin gene (38), inhibition of the interaction of the CCR5 receptor with gp-120 to block entry of HIV into macrophages and T cells expressing CD4 receptors (39), and inhibition of the antiapoptotic activity of Bcl family proteins by blocking their interaction with SH3 domain-containing antiapoptotic proteins for cancer treatment (40). In fact, there are multiple examples of protein-protein interaction inhibitors that have reached clinical development (36).
Finally, this approach should overcome another Achilles’ heel of conventional antibiotic therapy—cells must be actively growing to be killed. Because all strains of pathogenic M. tuberculosis carry these two TA modules, this approach should kill not only actively growing M. tuberculosis, including multidrug-resistant strains, but M. tuberculosis persisters as well. Also, VapBC TA systems are typically present only in genomes of bacterial pathogens and archaea. Therefore, antitubercular agents targeting VapBC2 and -21 should be selective for killing of M. tuberculosis by minimizing widespread killing of all beneficial resident flora that can lead to detrimental colonization by other pathogens (such as Clostridium difficile). In conclusion, the attributable benefits of the development of efficacious, novel VapBC2- and VapBC21-based antibiotics from test tube to the clinic are significant. This class of novel antitubercular agents has the potential to dramatically reduce the duration of treatment required for eradication of all viable M. tuberculosis cells from infected individuals.
MATERIALS AND METHODS
Strains, plasmids, and reagents.
VapC2 (Rv0301) and VapC21 (Rv2757c) were amplified by PCR from M. tuberculosis H37Rv genomic DNA. The amplified gene was cloned adjacent to an anhydrotetracycline (ATC)-inducible promoter in the pMC1s plasmid (41) or a pristinamycin (PTM)-inducible promoter in the pNW2046 (pMYT769-derived [42]) plasmid. Induction of VapC toxins was obtained by adding ATC to the media to a final concentration of 200 ng/mL, except for the experiments shown in Fig. 5 and 6, which used 50 ng/mL; ATC was replenished in cultures every 48 h. PTM was added to the media to a final concentration of 1 μg/mL.
All M. tuberculosis experiments were performed using M. tuberculosis attenuated strain mc2 6206 (ΔpanCD ΔleuCD, generously provided by the William Jacobs laboratory, Albert Einstein College of Medicine). M. tuberculosis cells were grown under constant shaking at 200 rpm at 37°C in 7H9 Middlebrook medium containing 1× oleic acid-albumin-dextrose-catalase (OADC) supplement (BD Biosciences), 0.05% tyloxapol (Sigma), and appropriate antibiotic (kanamycin at 25 μg/mL or spectinomycin at 100 μg/mL) for plasmid selection. The medium was also supplemented with 50 μg/mL of pantothenic acid and 100 μg/mL of leucine for growth of M. tuberculosis mc2 6206.
For mutation studies, VapB2 (Rv0300) and VapB21 (Rv2758c) were PCR amplified from M. tuberculosis H37Rv genomic DNA and cloned into an isovaleronitrile (IVN)-inducible pNIT (43) plasmid. Mutations were created using PCR mutagenesis, and the sequences of the resulting products were confirmed. M. smegmatis mc2155 used for mutation studies was grown in 7H9 Middlebrook medium containing 1× ADN supplement (0.5% bovine albumin, 0.2% dextrose, 0.085% NaCl), 0.05% Tween 80, and the appropriate antibiotic (zeocin at 25 μg/mL). M. smegmatis cells were grown under constant shaking at 200 rpm at 37°C. For coexpression, cells were induced at an optical density at 600 nm (OD600) of 0.1 by adding ATC and IVN to a final concentration of 50 ng/mL and 1 μM, respectively. CFU per milliliter were assessed in triplicate by decimal serial dilutions in phosphate-buffered saline containing 0.05% Tween 80 (PBST). Five-microliter aliquots of each dilution were carefully transferred onto the surface of 7H11 Middlebrook agar containing 1× ADN, kanamycin, and zeocin. Colonies were counted after 3 days of incubation at 37°C.
Purification of VapC2 and VapC21 toxins.
Escherichia coli BL21(DE3) cells transformed with either pET28a-vapC2 or pET28a-vapC21 were grown to exponential phase in M9 minimal medium supplemented with 0.1% glycerol and 50 μg/mL kanamycin at 37°C. Induction of the protein was achieved by adding 1 mM isopropyl-1-thio-d-galactopyranoside (IPTG). After 3 h, cells were harvested by centrifugation and resuspended in lysis buffer (50 mM NaH2PO4 [pH 8.0], 500 mM NaCl, 20 mM imidazole, 10 mM β-mercaptoethanol, 1 mM phenylmethylsulfonyl fluoride [PMSF], 1 mg/mL lysozyme). Cell pellets were then lysed by sonication, and lysates were applied to a nickel-nitrilotriacetic acid (Ni-NTA) resin (Qiagen) to purify the protein as described previously (32).
In vitro synthesis of tRNA.
M. tuberculosis tRNAs were synthesized in vitro using a synthetic DNA oligonucleotide containing the T7 RNA polymerase promoter and the 5′ end of the tRNA gene of interest and a second oligonucleotide corresponding to the 3′ end of the tRNA gene (44). Oligonucleotides were designed to have a region of overlap, thus providing a template for PCR. The annealed oligonucleotides were then extended using Taq DNA polymerase to create a double-stranded DNA (dsDNA) containing the entire tRNA gene preceded by the T7 promoter (tDNA). The product was then resolved on a 2% agarose gel to confirm its size and purified using the QIAquick gel extraction kit (Qiagen). Two hundred nanograms of the tDNA was then transcribed in vitro using the RiboMAX large-scale RNA production system (Promega) as recommended by the manufacturer. The transcription reaction mixture was separated on a 9% polyacrylamide, 7 M urea gel and visualized by staining with SYBR Gold to confirm the size and purity of the transcribed tRNA. The tRNA transcript was then excised from the gel and incubated for 18 h at 37°C in elution buffer (1 mM EDTA, 0.5 M ammonium acetate, 10 mM magnesium acetate, 0.1% SDS). The eluate was ethanol precipitated and resuspended in nuclease-free water.
In vitro tRNA cleavage assay.
VapC2 and VapC21 cleavage reactions were performed as described in the work of Cruz et al. (7) with minimal modification. Briefly, ∼50 pmol of purified VapC toxin was individually incubated with 2 pmol of each of the 45 in vitro-synthesized tRNAs from the M. tuberculosis H37Rv genome for 3 h at 37°C in 10 mM HEPES (pH 7.5), 15 mM KCl, 3 mM MgCl2, and 10% glycerol. Cleavage reaction mixtures were resolved on a 15% urea-PAGE gel, stained with SYBR Gold, and visualized in a UV transilluminator.
RNA isolation.
Mycobacterial cells were grown in the absence or presence of inducer for 72 h. Cells were centrifuged at 2,000 × g for 10 min at 4°C, and supernatants were removed. The cell pellets were resuspended in 1 mL of TRIzol and transferred to lysing tubes (Bertin Corp.) containing 0.1-mm glass beads. Cells were lysed with 3 cycles of 30 s each at 10,000 rpm using the Precellys Evolution homogenizer (Bertin Corp.) with 2-min cooldown periods on ice in between each cycle. The lysate was centrifuged at 14,000 rpm at 4°C, and the supernatant was used to extract total RNA using the Direct-zol RNA MiniPrep Plus kit (Zymo Research). RNA was treated with an additional step of genomic DNA removal in a Turbo DNase (ThermoFisher) digestion reaction for 30 min at 37°C and repurified using the Zymo RNA Clean and Concentrator kit. The extracted RNA was quantified using a μCuvette in a BioSpectrometer (Eppendorf).
5′ monophosphate RNA-seq.
Preparation of 5′ monophosphate libraries was performed as described in the work of Barth et al. (6). In brief, the Illumina small RNA 5′ adapter (5′-GUUCAGAGUUCUACAGUCCGACGAUCNNNNNN-3′) was ligated to 3 μg of total RNA using T4 RNA ligase 1 (New England Biolabs) at 4°C overnight. The ligated reaction mixture was electrophoresed on a 6% (wt/vol) polyacrylamide-7 M urea gel, and excess nonligated adapters were excluded upon gel excision and purification. The purified RNA was used in a Superscript IV reverse transcription reaction (ThermoFisher) using the primer 5′-GCCTTGGCACCCGAGAATTCCANNNNNNNNN-3′, and the resulting cDNA was electrophoresed on a 10% (wt/vol) polyacrylamide-7 M urea gel and size selected for fragments from 80 to 500 nt. The cDNA libraries were amplified in 12 cycles of PCR using Phusion HF DNA polymerase (ThermoFisher). The oligonucleotides used for PCR amplification were RP1 (5′-AATGATACGGCGACCACCGAGATCTACACGTTCAGAGTTCTACAGTCCGA-3′) and RPIX (5′-CAAGCAGAAGACGGCATACGAGATNNNNNNGTGACTGGAGTTCCTTGGCACCCGAGAATTCCA-3′), where the N’s represent the individual Illumina barcodes for each library. After electrophoresis on a 10% (wt/vol) polyacrylamide gel, amplified DNA between the sizes of 150 and 450 bp was isolated by gel excision and purified. The libraries were sequenced using the Illumina HiSeq 4000 platform at NYU Langone’s Genome Technology Center. Data analysis was performed as described in the work of Barth et al. (11). Reads were considered if they had (i) at least 1 read per million of mapped reads for mRNAs, (ii) 5 reads per million for tRNAs in the induced sample, and (iii) a fold change of at least 20.
Labeling of newly synthesized protein.
The effect of VapC toxins on protein synthesis was determined using a methionine mimetic, azidohomoalanine (AHA). M. tuberculosis cells harboring VapC toxin were grown to an OD600 of 0.1. and the toxin was induced by adding PTM. AHA was added to the sample cultures at 1, 3, and 5 days following induction with PTM to a 50 μM final concentration and incubated for 24 h. Cells were pelleted, resuspended in lysis buffer (2% CHAPS {3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate}, 8 M urea), and lysed using the Precellys Evolution homogenizer as described above for RNA isolation. The lysate was centrifuged, and the proteins from the supernatant were linked to an alkyne-containing fluorophore (TAMRA) using the Click-IT protein reaction buffer kit (ThermoFisher) following manufacturer’s protocol. Ten micrograms of protein from each sample was resolved on a 9% SDS-PAGE gel and imaged with a Typhoon FLA 9500 (GE Healthcare) image system.
Data availability.
RNA-seq data have been deposited in the NCBI Sequence Read Archive under BioProject ID PRJNA764776.
ACKNOWLEDGMENTS
We thank the William Jacobs laboratory, Albert Einstein College of Medicine, for providing the M. tuberculosis attenuated strain mc2 6206 and Bruno Abbadi for providing expertise on the bactericidal effects of VapC2 and VapC21.
The New York University Genome Technology Center (which performed Illumina sequencing) was partially supported by the Cancer Center Support Grant P30CA016087 at the Laura and Isaac Perlmutter Cancer Center. This work was funded in part by a New Jersey Commission on Cancer Research predoctoral fellowship DFHS18PPC045 to U.C., a fellowship from CAPES – Brazilian Federal Agency for Support and Evaluation of Graduate Education within the Ministry of Education of Brazil to V.C.B., and National Institutes of Health grant RO1 AI154464 to N.A.W.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Sala A, Bordes P, Genevaux P. 2014. Multiple toxin-antitoxin systems in Mycobacterium tuberculosis. Toxins (Basel) 6:1002–1020. 10.3390/toxins6031002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Senissar M, Manav MC, Brodersen DE. 2017. Structural conservation of the PIN domain active site across all domains of life. Protein Sci 26:1474–1492. 10.1002/pro.3193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pandey DP, Gerdes K. 2005. Toxin-antitoxin loci are highly abundant in free-living but lost from host-associated prokaryotes. Nucleic Acids Res 33:966–976. 10.1093/nar/gki201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Winther K, Tree JJ, Tollervey D, Gerdes K. 2016. VapCs of Mycobacterium tuberculosis cleave RNAs essential for translation. Nucleic Acids Res 44:9860–9871. 10.1093/nar/gkw781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Winther KS, Brodersen DE, Brown AK, Gerdes K. 2013. VapC20 of Mycobacterium tuberculosis cleaves the sarcin-ricin loop of 23S rRNA. Nat Commun 4:2796. 10.1038/ncomms3796. [DOI] [PubMed] [Google Scholar]
- 6.Barth VC, Chauhan U, Zeng J, Su X, Zheng H, Husson RN, Woychik NA. 2021. Mycobacterium tuberculosis VapC4 toxin engages small ORFs to initiate an integrated oxidative and copper stress response. Proc Natl Acad Sci USA 118:e2022136118. 10.1073/pnas.2022136118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cruz JW, Sharp JD, Hoffer ED, Maehigashi T, Vvedenskaya IO, Konkimalla A, Husson RN, Nickels BE, Dunham CM, Woychik NA. 2015. Growth-regulating Mycobacterium tuberculosis VapC-mt4 toxin is an isoacceptor-specific tRNase. Nat Commun 6:7480. 10.1038/ncomms8480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cintron M, Zeng JM, Barth VC, Cruz JW, Husson RN, Woychik NA. 2019. Accurate target identification for Mycobacterium tuberculosis endoribonuclease toxins requires expression in their native host. Sci Rep 9:5949. 10.1038/s41598-019-41548-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harms A, Brodersen DE, Mitarai N, Gerdes K. 2018. Toxins, targets, and triggers: an overview of toxin-antitoxin biology. Mol Cell 70:768–784. 10.1016/j.molcel.2018.01.003. [DOI] [PubMed] [Google Scholar]
- 10.Masuda H, Inouye M. 2017. Toxins of prokaryotic toxin-antitoxin systems with sequence-specific endoribonuclease activity. Toxins 9:140. 10.3390/toxins9040140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barth VC, Zeng JM, Vvedenskaya IO, Ouyang M, Husson RN, Woychik NA. 2019. Toxin-mediated ribosome stalling reprograms the Mycobacterium tuberculosis proteome. Nat Commun 10:3035. 10.1038/s41467-019-10869-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schifano JM, Vvedenskaya IO, Knoblauch JG, Ouyang M, Nickels BE, Woychik NA. 2014. An RNA-seq method for defining endoribonuclease cleavage specificity identifies dual rRNA substrates for toxin MazF-mt3. Nat Commun 5:3538. 10.1038/ncomms4538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Winther KS, Gerdes K. 2011. Enteric virulence associated protein VapC inhibits translation by cleavage of initiator tRNA. Proc Natl Acad Sci USA 108:7403–7407. 10.1073/pnas.1019587108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Walling LR, Butler JS. 2018. Homologous VapC toxins inhibit translation and cell growth by sequence-specific cleavage of tRNA(fMet). J Bacteriol 200:e00582-17. 10.1128/JB.00582-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barraud P, Schmitt E, Mechulam Y, Dardel F, Tisne C. 2008. A unique conformation of the anticodon stem-loop is associated with the capacity of tRNAfMet to initiate protein synthesis. Nucleic Acids Res 36:4894–4901. 10.1093/nar/gkn462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Min AB, Miallau L, Sawaya MR, Habel J, Cascio D, Eisenberg D. 2012. The crystal structure of the Rv0301-Rv0300 VapBC-3 toxin-antitoxin complex from M. tuberculosis reveals a Mg(2)(+) ion in the active site and a putative RNA-binding site. Protein Sci 21:1754–1767. 10.1002/pro.2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Levison ME. 2004. Pharmacodynamics of antimicrobial drugs. Infect Dis Clin North Am 18:451–465. 10.1016/j.idc.2004.04.012. [DOI] [PubMed] [Google Scholar]
- 18.Pankey GA, Sabath LD. 2004. Clinical relevance of bacteriostatic versus bactericidal mechanisms of action in the treatment of Gram-positive bacterial infections. Clin Infect Dis 38:864–870. 10.1086/381972. [DOI] [PubMed] [Google Scholar]
- 19.Zhu L, Sharp JD, Kobayashi H, Woychik NA, Inouye M. 2010. Noncognate Mycobacterium tuberculosis toxin-antitoxins can physically and functionally interact. J Biol Chem 285:39732–39738. 10.1074/jbc.M110.163105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Williams KJ, Duncan K. 2007. Current strategies for identifying and validating targets for new treatment-shortening drugs for TB. Curr Mol Med 7:297–307. 10.2174/156652407780598575. [DOI] [PubMed] [Google Scholar]
- 21.Aizenman E, Engelberg-Kulka H, Glaser G. 1996. An Escherichia coli chromosomal “addiction module” regulated by guanosine [corrected] 3′,5′-bispyrophosphate: a model for programmed bacterial cell death. Proc Natl Acad Sci USA 93:6059–6063. 10.1073/pnas.93.12.6059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cherny I, Gazit E. 2004. The YefM antitoxin defines a family of natively unfolded proteins: implications as a novel antibacterial target. J Biol Chem 279:8252–8261. 10.1074/jbc.M308263200. [DOI] [PubMed] [Google Scholar]
- 23.Prysak MH, Mozdzierz CJ, Cook AM, Zhu L, Zhang Y, Inouye M, Woychik NA. 2009. Bacterial toxin YafQ is an endoribonuclease that associates with the ribosome and blocks translation elongation through sequence-specific and frame-dependent mRNA cleavage. Mol Microbiol 71:1071–1087. 10.1111/j.1365-2958.2008.06572.x. [DOI] [PubMed] [Google Scholar]
- 24.Kang SM, Jin C, Kim DH, Lee Y, Lee BJ. 2020. Structural and functional study of the Klebsiella pneumoniae VapBC toxin-antitoxin system, including the development of an inhibitor that activates VapC. J Med Chem 63:13669–13679. 10.1021/acs.jmedchem.0c01118. [DOI] [PubMed] [Google Scholar]
- 25.Kang S-M, Kim D-H, Lee K-Y, Park SJ, Yoon H-J, Lee SJ, Im H, Lee B-J. 2017. Functional details of the Mycobacterium tuberculosis VapBC26 toxin-antitoxin system based on a structural study: insights into unique binding and antibiotic peptides. Nucleic Acids Res 45:8564–8580. 10.1093/nar/gkx489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee IG, Lee SJ, Chae S, Lee KY, Kim JH, Lee BJ. 2015. Structural and functional studies of the Mycobacterium tuberculosis VapBC30 toxin-antitoxin system: implications for the design of novel antimicrobial peptides. Nucleic Acids Res 43:7624–7637. 10.1093/nar/gkv689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Darwin KH, Ehrt S, Gutierrez-Ramos JC, Weich N, Nathan CF. 2003. The proteasome of Mycobacterium tuberculosis is required for resistance to nitric oxide. Science 302:1963–1966. 10.1126/science.1091176. [DOI] [PubMed] [Google Scholar]
- 28.Ziemski M, Leodolter J, Taylor G, Kerschenmeyer A, Weber-Ban E. 2021. Genome-wide interaction screen for Mycobacterium tuberculosis ClpCP protease reveals toxin-antitoxin systems as a major substrate class. FEBS J 288:111–126. 10.1111/febs.15335. [DOI] [PubMed] [Google Scholar]
- 29.Amitai S, Yassin Y, Engelberg-Kulka H. 2004. MazF-mediated cell death in Escherichia coli: a point of no return. J Bacteriol 186:8295–8300. 10.1128/JB.186.24.8295-8300.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pedersen K, Christensen SK, Gerdes K. 2002. Rapid induction and reversal of a bacteriostatic condition by controlled expression of toxins and antitoxins. Mol Microbiol 45:501–510. 10.1046/j.1365-2958.2002.03027.x. [DOI] [PubMed] [Google Scholar]
- 31.Fraikin N, Goormaghtigh F, Van Melderen L. 2020. Type II toxin-antitoxin systems: evolution and revolutions. J Bacteriol 202:e00763-19. 10.1128/JB.00763-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sharp JD, Cruz JW, Raman S, Inouye M, Husson RN, Woychik NA. 2012. Growth and translation inhibition through sequence-specific RNA binding by Mycobacterium tuberculosis VapC toxin. J Biol Chem 287:12835–12847. 10.1074/jbc.M112.340109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Arkin MR, Tang Y, Wells JA. 2014. Small-molecule inhibitors of protein-protein interactions: progressing toward the reality. Chem Biol 21:1102–1114. 10.1016/j.chembiol.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bojadzic D, Buchwald P. 2018. Toward small-molecule inhibition of protein-protein interactions: general aspects and recent progress in targeting costimulatory and coinhibitory (immune checkpoint) interactions. Curr Top Med Chem 18:674–699. 10.2174/1568026618666180531092503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jin L, Wang W, Fang G. 2014. Targeting protein-protein interaction by small molecules. Annu Rev Pharmacol Toxicol 54:435–456. 10.1146/annurev-pharmtox-011613-140028. [DOI] [PubMed] [Google Scholar]
- 36.Scott DE, Bayly AR, Abell C, Skidmore J. 2016. Small molecules, big targets: drug discovery faces the protein-protein interaction challenge. Nat Rev Drug Discov 15:533–550. 10.1038/nrd.2016.29. [DOI] [PubMed] [Google Scholar]
- 37.Vassilev LT, Vu BT, Graves B, Carvajal D, Podlaski F, Filipovic Z, Kong N, Kammlott U, Lukacs C, Klein C, Fotouhi N, Liu EA. 2004. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science 303:844–848. 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- 38.Hung DT, Shakhnovich EA, Pierson E, Mekalanos JJ. 2005. Small-molecule inhibitor of Vibrio cholerae virulence and intestinal colonization. Science 310:670–674. 10.1126/science.1116739. [DOI] [PubMed] [Google Scholar]
- 39.Dorr P, Westby M, Dobbs S, Griffin P, Irvine B, Macartney M, Mori J, Rickett G, Smith-Burchnell C, Napier C, Webster R, Armour D, Price D, Stammen B, Wood A, Perros M. 2005. Maraviroc (UK-427,857), a potent, orally bioavailable, and selective small-molecule inhibitor of chemokine receptor CCR5 with broad-spectrum anti-human immunodeficiency virus type 1 activity. Antimicrob Agents Chemother 49:4721–4732. 10.1128/AAC.49.11.4721-4732.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oltersdorf T, Elmore SW, Shoemaker AR, Armstrong RC, Augeri DJ, Belli BA, Bruncko M, Deckwerth TL, Dinges J, Hajduk PJ, Joseph MK, Kitada S, Korsmeyer SJ, Kunzer AR, Letai A, Li C, Mitten MJ, Nettesheim DG, Ng S, Nimmer PM, O’Connor JM, Oleksijew A, Petros AM, Reed JC, Shen W, Tahir SK, Thompson CB, Tomaselli KJ, Wang B, Wendt MD, Zhang H, Fesik SW, Rosenberg SH. 2005. An inhibitor of Bcl-2 family proteins induces regression of solid tumours. Nature 435:677–681. 10.1038/nature03579. [DOI] [PubMed] [Google Scholar]
- 41.Ehrt S, Guo XV, Hickey CM, Ryou M, Monteleone M, Riley LW, Schnappinger D. 2005. Controlling gene expression in mycobacteria with anhydrotetracycline and Tet repressor. Nucleic Acids Res 33:e21. 10.1093/nar/gni013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Forti F, Crosta A, Ghisotti D. 2009. Pristinamycin-inducible gene regulation in mycobacteria. J Biotechnol 140:270–277. 10.1016/j.jbiotec.2009.02.001. [DOI] [PubMed] [Google Scholar]
- 43.Pandey AK, Raman S, Proff R, Joshi S, Kang C-M, Rubin EJ, Husson RN, Sassetti CM. 2009. Nitrile-inducible gene expression in mycobacteria. Tuberculosis (Edinb) 89:12–16. 10.1016/j.tube.2008.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sisido M, Ninomiya K, Ohtsuki T, Hohsaka T. 2005. Four-base codon/anticodon strategy and non-enzymatic aminoacylation for protein engineering with non-natural amino acids. Methods 36:270–8. 10.1016/j.ymeth.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 45.Sievers F, Higgins DG. 2018. Clustal omega for making accurate alignments of many protein sequences. Protein Sci 27:135–145. 10.1002/pro.3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 and S2. Download aac.01896-21-s0001.pdf, PDF file, 1.8 MB (691.3KB, pdf)
Data Availability Statement
RNA-seq data have been deposited in the NCBI Sequence Read Archive under BioProject ID PRJNA764776.