ABSTRACT
Since echinocandins are recommended as first line therapy for invasive candidiasis, detection of resistance, mainly due to alteration in FKS protein, is of main interest. EUCAST AFST recommends testing both MIC of anidulafungin and micafungin, and breakpoints (BPs) have been proposed to detect echinocandin-resistant isolates. We analyzed MIC distribution for all three available echinocandins of 2,787 clinical yeast isolates corresponding to 5 common and 16 rare yeast species, using the standardized EUCAST method for anidulafungin and modified for caspofungin and micafungin (AM3-MIC). In our database, 64 isolates of common pathogenic species were resistant to anidulafungin, according to the EUCAST BP, and/or to caspofungin, using our previously published threshold (AM3-MIC ≥ 0.5 mg/L). Among these 64 isolates, 50 exhibited 21 different FKS mutations. We analyzed the capacity of caspofungin AM3-MIC and anidulafungin MIC determination in detecting isolates with FKS mutation. They were always identified using caspofungin AM3-MIC and the local threshold while some isolates were misclassified using anidulafungin MIC and EUCAST threshold. However, both methods misclassified four wild-type C. glabrata as resistant. Based on a large data set from a single center, the use of AM3-MIC testing for caspofungin looks promising in identifying non-wild-type C. albicans, C. tropicalis and P. kudiravzevii isolates, but additional multicenter comparison is mandatory to conclude on the possible superiority of AM3-MIC testing compared to the EUCAST method.
KEYWORDS: FKS mutation, MIC distribution, anidulafungin, antifungal resistance, caspofungin, common yeast, micafungin, rare yeast, yeasts
INTRODUCTION
Echinocandins inhibit cell wall synthesis by targeting the 1,3-β-d-glucan synthase encoded by FKS genes (1) and are the first-line recommended therapy of invasive candidiasis (2–4). Acquired resistance to echinocandins among yeasts remains rare (4–7). Some publications report a trend toward increasing the rate of echinocandin-resistant Candida glabrata, mostly in the USA but also recently in Germany (8–12), while others do not (13, 14). In any case, acquired echinocandin resistance has a major impact on patient management. It is therefore of utmost importance to reliably discriminate susceptible from resistant isolates in yeast species naturally susceptible to these drugs.
We previously showed that using caspofungin diluted in AM3 medium and a threshold of ≥ 0.5 mg/L is more stringent than RPMI to differentiate wild-type from non-wild-type isolates (i.e., with alteration in either Hot Spot (HS) 1 or 2 region of FKS protein) for Candida albicans, Candida tropicalis and Pichia kudriavzevii (synonym and current name for Candida krusei) (15–18). Between 2009 and 2014, we determined MIC of micafungin and caspofungin, using AM3 medium (AM3-MIC), and also MIC of anidulafungin using RPMI, according to the EUCAST procedure (https://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/AFST/Files/EUCAST_E_Def_7.3.2_Yeast_testing_definitive_revised_2020.pdf). We describe here the echinocandins MIC distribution for clinical isolates belonging to 5 frequent and 16 rare pathogenic yeast species (19). The isolates, mainly recovered during prospective, multicentric surveillance programs in France, were all studied using the same procedure at the French National Reference Center for Invasive Mycoses & Antifungals (NRCMA). In addition, the suitability of our threshold (AM3-MIC ≥0.5 mg/L for caspofungin) was compared with that of the EUCAST breakpoints (BPs; anidulafungin MIC > 0.032 mg/L for C. albicans and >0.064 mg/L for C. glabrata, C. tropicalis, and P. kudriavzevii to detect isolates harboring FKS mutations (15–17, 20).
RESULTS
Echinocandins susceptibility distribution.
A total of 2,787 clinical yeast isolates, mainly (88.1%) recovered from invasive human infections, were studied for their susceptibility to the three currently available echinocandins (Table 1; Table S1 in the supplemental material). The isolates belong to 21 different species (19 Ascomycetes corresponding to 11 genera, and 2 Basidiomycetes) with five species represented by more than 100 isolates (C. albicans, C. glabrata, C. tropicalis, Candida parapsilosis, and P. kudriavzevii).
TABLE 1.
Range of echinocandins MICs, MIC50, and MIC90 for the 2,787 yeast isolates received at the NRCMA between January 1 of 2009 and the December 31 of 2014, using EUCAST broth microdilution method
| Minimal inhibitory concentrations (MIC, mg/L) |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Caspofungina |
Micafungina |
Anidulafungin |
||||||||||
| Species | Synonym | n | Blood culture | MIC50 | MIC90 | Range | MIC50 | MIC90 | Range | MIC50 | MIC90 | Range |
| Candida albicans | 1198 | 1068 | 0.03 | 0.06 | ≤0.007–≥4 | 0.03 | 0.03 | ≤0.007–2 | ≤0.007 | ≤0.007 | ≤0.007–0.5 | |
| Candida glabrata | 466 | 398 | 0.06 | 0.125 | 0.03–≥4 | 0.03 | 0.03 | ≤0.007–2 | 0.03 | 0.06 | ≤0.007–2 | |
| Candida parapsilosis | 282 | 261 | 0.25 | 1 | 0.03–2 | 0.5 | 0.5 | 0.125–2 | 1 | 1 | 0.125–2 | |
| Candida orthopsilosis | 14 | 14 | 0.125 | 0.5 | 0.06–0.5 | 0.25 | 0.25 | 0.125–0.5 | 0.5 | 1 | 0.25–1 | |
| Candida metapsilosis | 14 | 12 | 0.06 | 0.25 | 0.06–0.5 | 0.125 | 0.25 | 0.06–0.5 | 0.25 | 0.25 | 0.06–0.25 | |
| Candida tropicalis | 238 | 214 | 0.03 | 0.06 | ≤0.007–≥4 | 0.03 | 0.03 | ≤0.007–2 | 0.015 | 0.03 | ≤0.007–1 | |
| Pichia kudriavzevii | Candida krusei | 137 | 105 | 0.125 | 0.25 | 0.06–4 | 0.06 | 0.125 | 0.03–2 | 0.06 | 0.06 | 0.015–0.5 |
| Clavispora lusitaniae | Candida lusitaniae | 85 | 74 | 0,06 | 0,125 | ≤0.007–4 | 0.06 | 0.125 | ≤0.007–4 | 0.06 | 0.125 | ≤0.007–1 |
| Saprochaete clavata | Geotrichum clavatum | 72 | 61 | ≥4 | ≥4 | 1–≥4 | ≥4 | ≥4 | 0.25–≥4 | 4 | 4 | 1–4 |
| Kluyveromyces marxianus | Candida kefyr | 63 | 57 | 0.03 | 0.03 | ≤0.007–0.5 | 0.06 | 0.06 | 0.03–0.5 | 0.03 | 0.06 | ≤0.007–0.5 |
| Candida dubliniensis | 35 | 31 | 0.03 | 0.03 | ≤0.007–0.06 | 0.03 | 0.03 | ≤0.007–0.03 | ≤0.007 | 0.015 | ≤0.007–0.015 | |
| Trichosporon asahii | 26 | 13 | 4 | ≥4 | 2–≥4 | 2 | ≥4 | 1–≥4 | 4 | 4 | 2–4 | |
| Meyerozyma guilliermondii | Candida guilliermondii | 23 | 19 | 0,125 | 0,125 | 0.03–0.25 | 0.25 | 0.25 | 0.06–0.5 | 0.5 | 1 | 0.25–2 |
| Magnusiomyces capitatus | Geotrichum capitatum | 22 | 16 | ≥4 | ≥4 | 4–≥4 | ≥4 | ≥4 | 1–≥4 | 2 | 4 | 0.25–4 |
| Galactomyces candidus | Geotrichum candidum | 20 | 1 | 1 | ≥4 | ≤0.007–≥4 | 0.5 | ≥4 | ≤0.007–≥4 | 2 | 4 | ≤0.007–4 |
| Candida inconspicua | 20 | 19 | 0,06 | 0,06 | 0.03–0.125 | 0.03 | 0.03 | ≤0.007–0.03 | ≤0.007 | 0.03 | ≤0.007–0.03 | |
| Saccharomyces cerevisiae | 19 | 44 | 0.125 | 0.25 | 0.125–0.25 | 0.125 | 0.25 | 0.06–0.25 | 0.06 | 0.25 | 0.03–0.5 | |
| Rhodotorula mucilaginosa | 18 | 17 | ≥4 | ≥4 | 4–≥4 | ≥4 | ≥4 | 4–≥4 | 4 | 4 | 2–4 | |
| Candida haemulonii | 13 | 8 | 0.03 | 0.06 | 0.03–0.25 | 0.06 | 0.125 | 0.06–0.125 | 0.06 | 0.125 | 0.03–0.125 | |
| Wickerhamomyces anomalus | Candida pelliculosa | 12 | 11 | 0,06 | 0.06 | 0.03–0.06 | 0.03 | 0.03 | ≤0.007–0.03 | 0.015 | 0.015 | ≤0.007–0.03 |
| Yarrowia lipolytica | Candida lipolytica | 10 | 4 | 0.125 | 0.5 | 0.06–4 | 0.25 | 0.25 | 0.25–0.5 | 0.25 | 0.5 | 0.125–4 |
Caspofungin and micafungin are diluted in AM3 medium.
The MIC90 for the three echinocandins was ≤ 0.06 mg/L for C. albicans, C. dubliniensis, and C. tropicalis while it was ≥ 0.5 mg/L for C. parapsilosis. Candida parapsilosis sensu stricto had higher MIC50 and MIC90 than Candida metapsilosis and Candida orthopsilosis, with C. metapsilosis exhibiting the lowest MIC values. Candida glabrata and P. kudriavzevii displayed caspofungin and anidulafungin MIC50 and MIC90 values higher than those obtained with C. albicans. Micafungin MICs were similar to those of C. albicans for C. glabrata but higher for P. kudriavzevii. Clavispora lusitaniae had MIC90 comparable to P. kudriavzevii for the three echinocandins (Table 1).
Among the rare species of Ascomycetes, Candida inconspicua and Wickerhamomyces anomalus behaved as C. albicans did while Kluyveromyces marxianus exhibited higher anidulafungin MIC90, and Candida haemulonii higher micafungin and anidulafungin MIC90. Saccharomyces cerevisiae, Meyerozyma guilliermondii, and Yarrowia lipolytica had MIC50 and MIC90 higher than those of C. albicans with anidulafungin MIC50 and MIC90 ≥ 1 mg/L for M. guilliermondii. Finally, Saprochaete clavata, Magnusiomyces capitatus, Galactomyces candidus, Rhodotorula mucilaginosa, and Trichosporon asahii had MIC90 ≥ 4 mg/L for the three echinocandins.
Strains ATCC22019 and ATCC6258 used as internal quality control exhibited stable anidulafungin, caspofungin, and micafungin MIC values over the study period. These MIC values were consistent with target values described by the EUCAST for anidulafungin (Fig. S1).
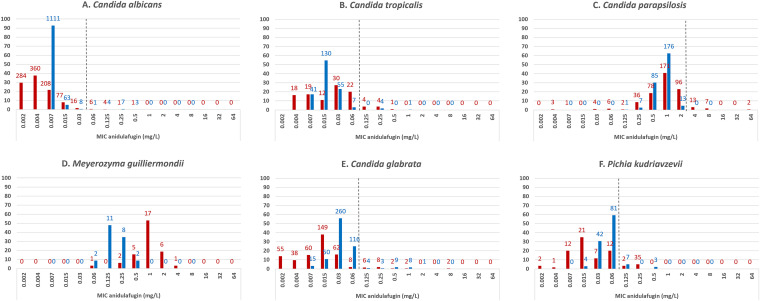
To compare our data with those recorded in the EUCAST database in terms of distribution and range of anidulafungin MICs, we used the online available EUCAST data (https://mic.eucast.org/Eucast2/SearchController/search.jsp?action=performSearch&BeginIndex=0&Micdif=mic&NumberIndex=50&Antib=716&Specium=-1). Both data sets (NRCMA and EUCAST) included similar number of isolates for the most common species. For C. albicans, C. tropicalis, C. parapsilosis, M. guilliermondii, C. glabrata, and P. kudriavzevii isolates tested at the NRCMA, MIC distribution of anidulafungin was similar to that found online (Fig. 1A to 1F). Based on the EUCAST BP (R > 0.03 mg/L), 1.3% (15/1,198) of the NRCMA C. albicans isolates were resistant. Furthermore, 2.1% (5/238) of C. tropicalis isolates, 5.4% of C. glabrata (25/466), and 7.3% of P. kudriavzevii (10/137) were resistant to anidulafungin (R > 0.06 mg/L), while none of the C. parapsilosis was (R > 4 mg/L).
FIG 1.
MIC distributions of anidulafungin for (A) Candida albicans, (B) Candida tropicalis, (C) Candida parapsilosis, (D) Meyerozyma guilliermondii, (E) Candida glabrata, (F) Pichia kudriavzevii. NRCMA data in blue, EUCAST data in red. The number of isolates is indicated for each MIC value. The dotted line corresponds to the EUCAST BP.
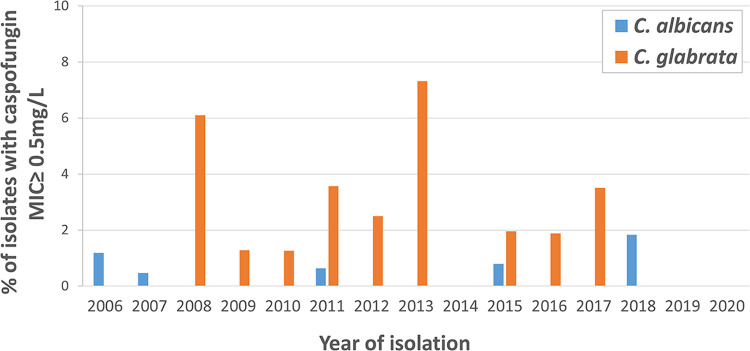
Local cutoff values were thus determined for isolates recovered in the YEASTS surveillance (14) program, i.e., without bias in isolates' selection and for species with more than 30 MIC values available (Table S2). Compared to the T-ECOFF (tentative epidemiological cutoff) values available on the anidulafungin rationale document (European Committee on Antimicrobial Susceptibility Testing, Anidulafungin: Rationale for the Clinical Breakpoints, version 3.0, 2020; http://www.eucast.org), the local cutoffs for anidulafungin were higher (1 dilution) for C. glabrata, C. tropicalis, and (2 dilutions) P. kudriavzevii, and lower (1 dilution) for C. albicans and C. parapsilosis. ECOFF values and local cutoffs were not comparable for caspofungin and micafungin given that AM3 medium was used locally for dilution instead of RPMI. To assess whether the proportion of C. albicans and C. glabrata isolates with caspofungin AM3-MIC ≥ 0.5 mg/L increased over time, we analyzed the data according to the year of isolation up to 2020 since the YEASTS program is still ongoing (Fig. 2). We observed variations in the proportion of isolates with caspofungin AM3-MICs ≥ 0.5 mg/L (between 0 and 1.8% for C. albicans, and between 0 and 7.3% for C. glabrata), but no trends toward an increasing proportion of resistant isolates over time.
FIG 2.
Percentages of C. albicans and C. glabrata isolates with caspofungin MIC ≥ 0.5 mg/L, according to the year of isolation. Candida albicans (blue) and C. glabrata (orange) isolates were recovered during the YEASTS surveillance program between 2006 and 2020. Linear trends curves were determined.
Analysis of FKS mutation.
We sequenced FKS genes for the 64 isolates considered resistant to anidulafungin based on the EUCAST BP (15 C. albicans, 25 C. glabrata, 5 C. tropicalis, and 10 P. kudriavzevii) and those above the caspofungin threshold (20 C. albicans, 25 C. glabrata, 4 C. tropicalis, 5 P. kudriavzevii, and 1 K. marxianus). Of these 64 isolates, 46 were considered resistant using both criteria (15 C. albicans, 23 C. glabrata, 4 C. tropicalis, and 4 P kudriavzevii). Overall, 50/64 exhibited alterations in FKS protein: 20/20 C. albicans, 3/5 C. tropicalis, 21/27 C. glabrata, 5/11 P. kudriavzevii, and 1/1 K. marxianus (Table 2, Fig. 3).
TABLE 2.
Mutations in FKS for isolates of C. albicans, C. glabrata, P. kudriavzevii, C. tropicalis, and K. marxianus having caspofungin MIC ≥ 0.5 mg/L or resistant to anidulafungin according to the EUCAST BP
| Range of MICs (mg/L) |
|||||||
|---|---|---|---|---|---|---|---|
| Species (nbr of isolates sequenced) | Nbr of isolates non-WT | Mutation | Localization | Caspo | Mica | Anidula | Mutations described |
| Candida albicans (n = 20) | 1 | F641S | HS1 | 1 | 0.25 | 0.25 | 37 |
| 9 | S645P | HS1 | 4–>4 | 0.5 –>4 | 0.25–1 | 36 | |
| 8 | S645S/P | HS1 | 0.5–2 | 0.25–1 | 0.015–1 | 36 | |
| 1 | R647G | HS1 | 0.5 | 0.5 | 0.03 | 16 | |
| 1 | R1361G | HS2 | 0.5 | 0.25 | 0.25 | 16 | |
| Candida glabrata (n = 27) | 1 | F625S | FKS1HS1 | >4 | 0.5 | 2 | 11 |
| 4 | DelF659 | FKS2HS1 | 4–>4 | 0.25–2 | 1–2 | 11 | |
| 1 | F659S | FKS2HS1 | 0.5 | 0.06 | 0.5 | 43 | |
| 1 | F659V | FKS2HS1 | 4 | 0.06 | 1 | 39 | |
| 7 | S663P | FKS2HS1 | 1–>4 | 0.125–2 | 0.25–4 | 11 | |
| 1 | S663F | FKS2HS1 | 1 | 0.25 | 1 | 43 | |
| 1 | F659S +L664V | FKS2HS1 | 0.5 | 0.06 | 1 | 16 | |
| 1 | S663P +M1439R | FKS2HS1 + FKS2 non-hotspot alteration | 4 | 0.5 | 1 | This study | |
| 3 | R1378S | FKS2HS2 | 1–4 | 0.06–0.125 | 0.5–1 | 53 | |
| 1 | DelF659 + K335N | FKS2HS1 + FKS3 non-hotspot alteration | >4 | 1 | 2 | This study | |
| Pichia kudriavzevii (n = 11) | 1 | F655L | HS1 | 2 | 1 | 1 | 44 |
| 2 | S659F | HS1 | 4 | 2 | 0.5 | 38 | |
| 2 | S659S/P | HS1 | 0.5–1 | 0.125 | 0.06–0.125 | 38 | |
| Candida tropicalis (n = 5) | 2 | S645P | HS1 | 4 | 0.5 | 0.5 | 54 |
| 1 | F641L + I1368S | HS1 +HS2 | >4 | 2 | 2 | This study | |
| Kluyveromyces marxianus (n = 1) | 1 | F651F/S | HS1 | 0.5 | 0.5 | 1 | This study |
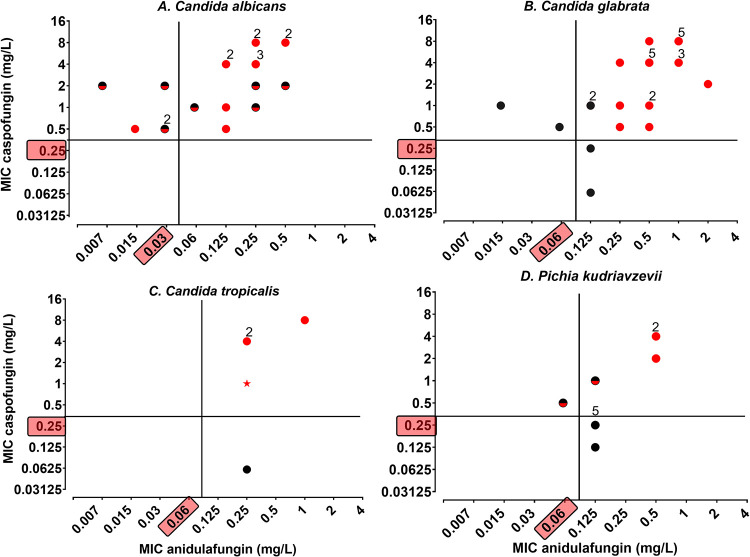
FIG 3.
Schematic classification of isolates according to anidulafungin EUCAST BPs or NRCMA criteria (caspofungin MIC ≥ 0.5 mg/L), and FKS sequence. Isolates were defined as wild type (WT) when the FKS protein sequence was similar to that of the reference strain (black dot), and as non-wild-type (NWT) if it exhibited amino acid mutation in the HS1 or HS2 region (red dot for homozygous mutation; red and black dot for heterozygous mutation). The anidulafungin BP defined by the EUCAST AFST subcommittee and the CNRMA caspofungin threshold (MIC ≥ 0.5 mg/L) are highlighted in the y and x axis, respectively. The number of isolates is indicated above the dot when superior to 1. (A) Twenty Candida albicans isolates resistant to anidulafungin (R > 0.03 mg/L, n = 15) or caspofungin (n = 20). (B) Twenty-seven Candida glabrata isolates resistant to anidulafungin (R > 0.06 mg/L, n = 25) or caspofungin (n = 25). (C) Five Candida tropicalis isolates resistant to anidulafungin (R > 0.06 mg/L, n = 5) or caspofungin (n = 4). The star symbol corresponds to the isolate for which HS1 was not sequenced. (D) Eleven Pichia kudriavzevii isolates resistant to anidulafungin (R > 0.06 mg/L, n = 10) or caspofungin (n = 5).
The 9 isolates considered resistant using EUCAST BP and “susceptible” using the NRCMA criteria (1 C. tropicalis, 2 C. glabrata, and 6 P. kudriavzevii) had a wild-type FKS sequence (Fig. 3B to D). Among the 8 isolates considered susceptible using EUCAST BP and “resistant” using the NRCMA criteria, 6 had FKS mutation (5 C. albicans and 1 P. kudriavzevii, Fig. 3A and D), and two had a wild-type FKS sequence (2 C. glabrata isolates, Fig. 3C).
Overall, 21 different mutations were identified, and the majority (17/21) were localized in the HS1 region. Five different mutations or combination of mutations were found for C. albicans, 10 for C. glabrata, 3 for P. kudriavzevii, 2 for C. tropicalis, and 1 for K. marxianus (Table 2).
For C. albicans, modification at S645 was the most prominent amino acid substitution (17/20) in homozygous and heterozygous forms (9/17 and 8/17, respectively; Table 2).
In C. glabrata, amino acid modification in S663 (S663P [n = 7]; S663F [n = 1]) and F659 (DelF659 [n = 4]; F659V [n = 1]; F659S [n = 1]) in HS1 of FKS2 were the most frequent mutations. An unreported mutation localized upstream of the HS2 region of FKS3 was also found. For three isolates, combination of mutations including one known mutation in HS1 of FKS2 and another mutation localized downstream HS2 of FKS2 or upstream HS1 of FKS3 were found. The 4 isolates with deletion in position F659 had high MICs of the three echinocandins, whereas the five isolates with single or combined F659S or F659V substitution had low micafungin MIC (AM3-MIC = 0.06 mg/L). Four isolates with caspofungin AM3-MIC ≥ 0.5 mg/L had no mutation in HS1 nor HS2 for FKS1 2 and 3. Among those four isolates, two were resistant to anidulafungin (MIC = 0.125 mg/L) and two were susceptible (MIC ≤ 0.06 mg/L).
For C. tropicalis, two isolates had mutations S645P in HS1, one isolate had two mutations in HS1 and HS2, and for one isolate, amplification of the HS1 region was technically not possible. One isolate of K. marxianus with elevated MIC for the three echinocandins has an undescribed heterozygous mutation F651S.
Sensitivity (SE), specificity (SP), prevalence (P), positive and negative predictive values (PPV and NPV, respectively) were calculated on the subset of isolates that were sequenced to assess the efficacy of caspofungin or anidulafungin MIC determination and the EUCAST BPs/local thresholds in detecting isolates with FKS mutation (Table 3). The data set included 62 isolates belonging to species for which anidulafungin EUCAST BP were defined (C. albicans, C. glabrata, C. tropicalis, and P. kudriavzevii). PPV and NPV were 92% and 100% for caspofungin, and 80% and 25% for anidulafungin, respectively. The proportion of isolates with FKS mutation correctly identified (i.e., sensitivity) was 100% for caspofungin (NRCMA threshold) and 88% for anidulafungin (EUCAST BP), while the proportion of wild-type isolates correctly identified (i.e., specificity) was 69% for caspofungin (4 C. glabrata nonmutated isolates had caspofungin AM3-MIC ≥ 0.5 mg/L) and 15% for anidulafungin (11 isolates considered resistant to anidulafungin with no FKS mutation uncovered, including 4 C. glabrata, 6 P. kudriavzevii, and 1 C. tropicalis).
TABLE 3.
Calculation of positive predictive value (PPV) and negative predictive value (NPV) of the caspofungin local threshold and anidulafungin BP for isolates of C. albicans (n = 20), C. glabrata (n = 27), C. tropicalis (n = 4), and P. kudriavzevii (n = 11)a
| FKS protein |
||||
|---|---|---|---|---|
| Categorization | Non-WT | WT | PPV | NPV |
| Caspofungin MIC ≥ 0.5 mg/L | TP = 49 | FP = 4 | 92% | |
| Caspofungin MIC < 0.5 mg/L | FN = 0 | TN = 9 | 100% | |
| Anidulafungin R | TP = 43 | FP = 11 | 80% | |
| Anidulafungin S | FN = 6 | TN = 2 | 25% | |
TP, true positives; FP, false positives; FN, false negatives; TN, true negatives. R and S, resistant or susceptible to anidulafungin according to the BP EUCAST; WT, wild-type FKS protein sequence.
DISCUSSION
Thanks to the missions of the NRCMA, we have been able to generate over time antifungal susceptibility data concerning isolates of yeasts responsible for invasive infections. We here analyzed the echinocandins susceptibility of 2,787 isolates belonging to 21 yeast species, including the 5 most frequent ones in France, but also emerging and rare Ascomycetous and Basidiomycetous yeast species. The susceptibility profiles that we observed matched those usually reported for common and rare species (5, 19, 21–28).
We then assessed whether the EUCAST BPs for anidulafungin and our local threshold for caspofungin using our AM3-MIC testing lead to similar categorization of wild-type and mutated isolates. The anidulafungin EUCAST BPs were liable to categorize isolates without FKS mutation as resistant (1 C. tropicalis, 6 P. kudriavzevii, 4 C. glabrata) (12), but they also categorize isolates with mutation as susceptible (2 C. albicans and 1 P. kudriavzevii). However, for C. glabrata this threshold was also liable to categorize wild-type isolates as caspofungin resistant (n = 4), two of which were also considered as anidulafungin resistant according to the EUCAST BP. The absence of mutation associated with high MIC value for echinocandins is already described (12, 13, 29). One explanation is that these isolates corresponded to a mixture of susceptible and resistant isolates that we did not uncover despite testing single colonies. Another hypothesis is that the presence of a mutation localized in another part of the gene or in another gene was involved in the acquired resistance that would require whole genome sequencing to uncover. Furthermore, the concept of area of technical uncertainty (ATU) already described in the EUCAST standardized method, could be applied. In fact, C. glabrata isolates susceptible to anidulafungin and exhibiting caspofungin AM3-MIC of 0.5 or 1 mg/L do not harbor FKS protein alteration. Of note, given the mode of MIC distribution for anidulafungin observed in our center, an elevation of the anidulafungin BP value for C. glabrata by one dilution (MIC > 0.125 mg/L) could be an option for us to correctly categorize the mutated isolates.
The EUCAST-AFST subcommittee has ruled out the use of caspofungin MICs to categorize isolates, because of inter and intralaboratory variations of MICs (8, 30, 31), and the same lack of laboratory-to-laboratory reproducibility was shown for CLSI testing (31). Thus, it considers that isolates of frequent species resistant to anidulafungin and micafungin should be considered resistant to caspofungin without further assessment (32). Nevertheless, we and another team already pointed out that determination of caspofungin MICs seems more reliable for detecting FKS mutated isolates than anidulafungin (16, 18, 33). We thus wonder whether reintroducing caspofungin in the panel of echinocandin drugs used to look for FKS-mutated isolates is not worth it.
Similarly, the EUCAST-AFST subcommittee does not recommend AM3 medium because of the possible variations related to manufacturers and even batches, the medium being more complex than RPMI (34). We did not observe variations in the caspofungin AM3-MIC for the quality control strains over the 6 years of the study (Fig. S1). We acknowledge that this lack of variation could rely on the single center evaluation. More specifically, plates and AM3 were purchased from unique manufacturers even if several batches were used over time, and inoculum determination and OD measurement were performed with the same readers, and by a single laboratory technician team. Finally, testing caspofungin in AM3 could modify the susceptibility profiles of the yeasts preventing its routine use, which was not the case.
To support our results, we looked for possible biases in our conclusions. We wondered whether the mutations observed were those usually described. Overall, of the 64 isolates sequenced, 50 harbored 21 different alterations in the FKS proteins. Resistance to echinocandins has been associated with amino acid substitutions in HS1 and/or HS2 regions on FKS protein for the most frequent species (C. albicans, C. glabrata, C. tropicalis, P. kudriavzevii) (8, 11, 20, 35–39) but also for less frequent species such as C. lusitaniae, C. dubliniensis, and K. marxianus (20, 40–42). FKS alterations are most commonly substitutions in the HS1 region, but deletions and stop codons have also been reported in C. glabrata (8). In fact, FKS mutations are more frequently described for C. glabrata than for any other yeast species (5, 8, 16). Mutations found here for C. albicans and C. glabrata corresponded mostly to the mutations frequently described (43–45). We also found 2 previously unknown putative mutations localized near the HS1 or HS2 regions for C. glabrata isolates, 1 combination of mutation in HS1 and HS2 for C. tropicalis, and 1 unknown mutation in the HS1 region for K. marxianus. For C. albicans, S645P mutation was found in homozygous and heterozygous forms leading to higher caspofungin MICs for homozygous (AM3-MIC > 2 mg/L) than heterozygous (AM3-MIC between 0.5 and 2 mg/L), suggesting the presence of wild-type and mutant enzymes as already reported (1). Even if MIC determination, DNA extraction, and FKS sequencing were performed on single colonies for isolates with heterozygous mutation, it does not exclude that a mixture of wild-type and resistant population was present in the original sample leading to low MIC value and detection of heterozygous form of enzyme. Influence of the amino-acid alteration on the MIC value has been previously reported with mutation in F641, S645, and R1361 associated with pronounced MIC elevations while other mutations such as R647 were associated with less elevated MIC values (46). This was not confirmed here. For C. glabrata, isolates with deletion in position F659 were “resistant” to three echinocandins, whereas isolates with F659S or F659V substitution have low AM3-MIC for micafungin (43). In the same way, isolates with a combination of mutations localized in positions 659 and 664 in the HS1 region had high MIC of caspofungin and anidulafungin but low MIC of micafungin (AM3-MIC = 0.06 mg/L). Among the 11 isolates of P. kudriavzevii studied, 5 had mutations in HS1 and also L701M heterozygous or homozygous mutation localized near the HS1 region. This mutation has already been described in echinocandin-susceptible isolates, and its implication in resistance is unclear (17, 44, 47). These results suggest that we did not introduce a bias due to a particular mutation when using our caspofungin threshold.
In conclusion, based on a large data set on isolates collected through prospective microbiological surveillance networks in France, we showed that detection of resistant isolates in common yeast species is not always trivial despite published BPs, and that our single center experience looks promising. Whether our results are convincing enough to trigger additional studies or whether alternative methods such as mass spectrometry using MALDI-TOF, or real-time PCR, can be more reliably used to detect resistant isolates remains to be determined (45, 48–51).
MATERIALS AND METHODS
Isolates.
The French National Reference Center for Invasive Mycoses and Antifungals (NRCMA) provides expertise on isolates involved in invasive fungal infections from >200 hospitals in France. Between the January 1 of 2009 and the December 1 of 2014, all yeast isolates (n = 3,337) sent to the NRCMA were identified and tested for antifungal susceptibility. Isolates were sent mainly in the context of the epidemiological surveillance (72.5%, 2,418/3,337 including 1,963 isolates from the YEASTS program in the Paris area [6, 7]), or for specific expertise on species identification/typing or antifungal susceptibility testing (27.5%, 918/3,337). Isolates of Cryptococcus neoformans and Cryptococcus gattii species complex were excluded from this study. Species with 10 or more isolates were analyzed in the present study (n = 2,787). Isolates were identified at the species levels as previously described (52).
The dual source of isolates prevented calculation of echinocandin resistance incidence. Nevertheless, we used the exhaustive YEASTS surveillance program on candidemia in the Paris area to determine the percentage of C. albicans and C. glabrata isolates with caspofungin AM3-MIC ≥ 0.5 mg/L according to the year of isolation between 2006 and 2020.
Antifungal susceptibility.
MICs were determined for anidulafungin by using the standardized broth microdilution method from the European Committee on Antimicrobial Susceptibility Testing (EUCAST) (sterile tissue culture plates, 96-well flat bottom in clear polystyrene, TPP Techno Plastic Products AG, Switzerland, Reference 92096). For caspofungin and micafungin, AM3 medium (BD Difco, USA, Reference 224320) was used for dilution (15–18). All MICs were determined on the same day using the same inoculum. Quality control strains (ATCC22019, ATCC6258) were included in each set. The concentrations corresponding to the MIC that inhibited 50% (MIC50) and 90% (MIC90) of the isolates were determined for species with 10 or more isolates studied (Table 1 MICs). ECOFFs (epidemiological cutoff values) are defined by EUCAST as “the highest MIC for organisms devoid of phenotypically detectable acquired resistance mechanisms” and correspond to the upper end of the wild-type MIC distribution. Given that our data come from a unique center, we could only determine local cutoff values. We thus calculate 99.9% local cutoff values for anidulafungin, micafungin, and caspofungin, for species considered as susceptible to echinocandins (excluding S. clavata) with 30 or more isolates tested, following the EUCAST SOP10 recommendation (MIC distributions and epidemiological cutoff value [ECOFF] setting, EUCAST SOP 10.0, 2017. http://www.eucast.org) and using the ECOFFfinder program v2.0. To avoid bias of selection, isolates not recovered during the YEASTS program (active surveillance program of fungemia in the Paris area [19]) were excluded. The clinical BPs or T-ECOFF values determined by EUCAST for some species and some antifungal agents were used to calculate percentage of resistant (R) isolates (https://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/AFST/Clinical_breakpoints/AFST_BP_v10.0_200204_updatd_links_200924.pdf; https://mic.eucast.org/Eucast2/SearchController/).
FKS sequencing.
According to previous studies, Hot Spot (HS) 1 and 2 regions of the FKS gene were sequenced for C. albicans, C. glabrata, C. tropicalis, P. kudriavzevii, and K. marxianus isolates having AM3-MIC caspofungin ≥ 0.5 mg/L, with primers listed in Table 4. Of note, for C. glabrata HS1 and HS2 regions were sequenced for FKS1, FKS2, and FKS3 genes. Reference strains (C. albicans CBS 562, C. glabrata ATCC 2001, C. tropicalis ATCC 750, P. kudriavzevii ATCC 6258, K. marxianus CBS 712) were used as positive control for PCR amplification and as wild-type reference for FKS protein sequences. For K. marxianus, numbering was based on the protein sequence of the 1-3beta glucan synthase (GenBank accession number BAO40851.1). For isolates resistant to anidulafungin and/or with AM3-MIC caspofungin ≥ 0.5 mg/L and having a wild-type sequence, at least five colonies were checked for MIC and FKS sequences. For the 12 isolates with heterozygous mutation in FKS (8 C. albicans, 2 P. kudriavzevii, and 1 K. marxianus), two single colonies were isolated. For these single colonies, DNA extraction, FKS sequencing of the HS regions, and MIC determination were performed. Among the 12 isolates analyzed, two P. kudriavzevii (CNRMA13.91, CNRMA12.1267) were identified as a possible mixture of wild-type and mutated isolates, but only mutated isolates were recovered, and one C. albicans (CNRMA9.37) was identified as a mixture of wild type and resistant isolates with wild-type FKS and mutated (heterozygous) FKS, respectively.
TABLE 4.
Primers used for amplification and sequencing of HS1 and HS2 regions of FKS genes
| Species | Region | Primer | Sequence 5′ 3′ | Ref |
|---|---|---|---|---|
| Candida albicans | HS1 | GSC1f | GAAATCGGCATATGCTGTGTC | 36 |
| GSC1r | AATGAACGACCAATGGAGAAG | |||
| HS2 | CAS2f | ACCACCAAGATTGGTGCTG | 17 | |
| CAS2r | TATCTAGCACCACCAACGG | |||
| Pichia kudriavzevii | HS1 | CKS1f | ACTGCATCGTTTGCTCCTCT | 17 |
| CKS1r | GAACATGATCAATTGCCAAC | |||
| HS2 | CKS2f | CCGGTATGGGAGAACAAATG | ||
| CKS2r | CACCACCAATGGAAACATCA | |||
| Candida tropicalis | HS1 | CTS1-1f | ATGGTTCAGTATAGGTGGATG | 17 |
| CTS1-1r | AAGGAACGACCAATGGAGAAG | |||
| HS2 | CTS1-2f | ACTACCAAGATTGGTGCTG | ||
| CTS1-2r | TATCTAGCACCACCAACAG | |||
| Candida glabrata | FKS2-HS1 | FKS2F | GGCCACTGTTTTATTCTTCTCG | 35 |
| FKS2R | GTAAATGTTCTCTGTACATGGA | |||
| FKS2-HS2 | CG2f | ACAACTAAGATTGGTGCAG | 54 | |
| CG2r | TAACGAGCACCACCCACA | |||
| FKS1-HS1 | FKS1f | GTTGCAGTCGCTACATTGCTA | 35 | |
| FKS1r | TAGCGTTCCAGACTTGGGAA | |||
| FKS1-HS2 | FKS1HS2f | GGTATTTCAAAGGCTCAAAAGGG | 39 | |
| FKS1HS2r | ATGGAGAGAACAGCAGGGCG | |||
| FKS3-HS1 | FKS3f | TGGAGCCCAGCACTTAACAA | 35 | |
| FKS3r | GTCCATCTCGGATGTTGCTA | |||
| FKS3-HS2 | CG3-HS2f | TTATGCAGAGGAACCTGCTC | 54 | |
| CG3-HS2r | GTGCCATCGACAGTAAGTGA | |||
| Kluyveromyces marxianus | HS1 | CKHS1f | GGTGGTTTATTCACTTCCTACA | 42 |
| CKHS1r | GCGTAGCCAAAGATTGAGCA | |||
| HS2 | CKHS2f | AAGATTGGTGCYGGTATGGG | ||
| CKHS2r | RGTDGCAAAACCTCTAGCAGT |
Positive and negative predictive values.
Positive and negative predictive values (PPV and NPV, respectively) correspond to the proportion of positive and negative results in a test that are true positive and true negative results, respectively. In the present study, PPV indicates the proportion of non-wild-type isolates for isolates resistant to anidulafungin (based on EUCAST BPs) or for isolates with caspofungin AM3-MIC ≥ 0.5 mg/L. The ideal value of PPV and NPV is 1 (100%).
Sensitivity (SE), specificity (SP), and prevalence (P) were determined to calculate PPV and NPV, using the number of wild-type and non-wild-type isolates according to the MIC value for caspofungin or anidulafungin. The sensitivity (SE) measures the proportion of positives correctly identified (i.e., isolates with FKS mutation resistant to anidulafungin or with caspofungin AM3-MIC ≥ 0.5 mg/L). The specificity (SP) measures the proportion of negatives correctly identified (i.e., wild-type isolates susceptible to anidulafungin or with caspofungin AM3-MIC < 0.5 mg/L). SE, SP, and P were calculated as follows:
PPV and NPV were calculated as follows:
ACKNOWLEDGMENTS
The authors express their deep acknowledgments to Maiken Arendrup (Statens Serum Institut, University of Copenhagen, Rigshospitalet, Denmark) for helpful discussion.
Members of the French Mycoses Study Group who contributed to the data are, in alphabetical order of the cities, all the French microbiologists and mycologists who sent isolates for characterization of unusual antifungal susceptibility profiles or to contribute to the ongoing surveillance program on the epidemiology of invasive fungal infections in France (YEASTS and RESSIF programs): N. Brieu (CH Aix); T. Chouaki, C. Damiani, A. Totet (CHU Amiens); J. P. Bouchara, D. Chabasse, M. Pihet (CHU Angers); S. Bland (CH Annecy); V. Blanc (CH Antibes); S. Branger (CH Avignon); A. P. Bellanger, L. Millon (CHU Besançon); C. Plassart (CH Beauvais); I. Poilane (Hôpital Jean Verdier, Bondy); I. Accoceberry, L. Delhaes, B. Couprie, F. Gabriel (CH Bordeaux); J. Dunand, A. L. Roux, V. Sivadon-Tardy (Hôpital Ambroise Paré, Boulogne Billancourt); F. Laurent (CH, Bourg en Bresse); S. Legal, E. Moalic, G. Nevez, D. Quinio (CHU Brest); M. Cariou (CH Bretagne Sud); J. Bonhomme, C. Duhamel (CHU, Caen); B. Podac (CH, Chalon sur Saône); S. Lechatch (CH, Charleville-Mézières); C. Soler (Hopital d’Instruction des armées, Clamart); M. Cambon, C. Nourrisson, P. Poirier, D. Pons (CHU, Clermont Ferrand); O. Augereau, I. Grawey (CH, Colmar); N. Fauchet (CHIC, Créteil); A. Bonnin, F. Dalle (CHU, Dijon); P. Cahen, P. Honderlick (CMC, Foch); N. Desbois, C. Miossec (CHU, Fort de France); J. L. Hermann (Hôpital Raymond Poincaré, Garches); M. Cornet, R. Grillot, B. Lebeau, D. Maubon, H. Pelloux (CHU, Grenoble); M. Nicolas (CHU, Guadeloupe); C. Aznar, D. Blanchet, J. F. Carod, M. Demar, (CHU, Guyane); A. Angoulvant (Hôpital Bicêtre, le Kremlin Bicêtre); C. Ciupek (CH, Le Mans); A. Gigandon (Hôpital Marie Lannelongue, Le Plessis Robinson); B. Bouteille (CH Limoges); E. Frealle, D. Poulain, B. Sendid (CHU Lille); D. Dupont, J. Menotti, F. Persat, M.-A. Piens, M. Wallon (CHU, Lyon); C. Cassagne, S. Ranque (CHU, Marseille); T. Benoit-Cattin, L. Collet (CH Mayotte); A. Fiacre (CH Meaux); N. Bourgeois, L. Lachaud, P. Rispail, Y. Sterkers (CHU, Montpellier); M. Machouart (CHU, Nancy); F. Gay-Andrieu, P. Lepape, F. Morio (CHU, Nantes); O. Moquet (CH, Nevers); S. Lefrançois (Hôpital Américain, Neuilly); M. Sasso (CHU, Nimes); F. Reibel (GH, Nord-Essone); M. Gari-Toussaint, L. Hasseine (CHU Nice); L. Bret, D. Poisson (CHR Orléans); S. Brun (Hôpital Avicenne, Paris); C. Bonnal, C. Chochillon, S. Houzé (Hôpital Bichat, Paris); A. Paugam (Hôpital Cochin, Paris); N. Ait-Ammar, F. Botterel, R. Chouk (CHU Henri Mondor, Paris); M. E. Bougnoux, E. Sitterle (Hôpital Necker, Paris); A. Fekkar, R. Piarroux (Hôpital Pitié Salpêtrière, Paris); J. Guitard, C. Hennequin, J.-L. Poirot (Hôpital St Antoine, Paris); M. Gits-Muselli, S. Hamane, C. Lacroix (Hôpital Saint Louis, Paris); S. Bonacorsi, P. Mariani (Hôpital Robert Debré, Paris); D. Moissenet (Hôpital Trousseau, Paris); C. Kauffmann-Lacroix, A. Minoza, E. Perraud, M. H. Rodier (CHU Poitiers); G. Colonna (CH, Porto Vecchio); A. Huguenin, D. Toubas (CHU Reims); S. Chevrier, J. P. Gangneux, F. Robert-Gangneux, C. Guigen (CHU Rennes); O. Belmonte, G. Hoarau, M. C. Jaffar Bandjee, J. Jaubert, S. Picot, N. Traversier (CHU Réunion); L. Favennec, G. Gargala (CHU, Rouen); N. Godineau, C. Tournus (CH, St Denis); C. Mahinc, H. Raberin (CHU, St Etienne); V. Letscher Bru (CHU, Strasbourg); S. Cassaing (CHU, Toulouse); P. Patoz (CH Tourcoing); E. Bailly, J. Chandenier, G. Desoubeaux (CHU Tours); F. Moreau (CH Troyes); P. Munier (CH Valence); E. Mazars (CH Valenciennes); O. Eloy (CH Versailles); E. Chachaty (Institut Gustave Roussy, Villejuif); and members of the NRCMA (Institut Pasteur, Paris): A. Bertho, C. Blanc, A. Boullié, C. Gautier, V. Geolier, D. Hoinard, and D. Raoux-Barbot for technical help, and K. Boukris-Sitbon, F. Lanternier, A. Alanio, and D. Garcia-Hermoso for their expertise and contribution to the surveillance programs.
This work was supported by Santé Publique France and Institut Pasteur. The funders had no role in study design, data collection, and interpretation, or the decision to submit the work for publication.
Footnotes
Supplemental material is available online only.
Contributor Information
Marie Desnos-Ollivier, Email: mdesnos@pasteur.fr.
the French Mycoses Study Group:
N. Brieu, T. Chouaki, C. Damiani, A. Totet, J. P. Bouchara, D. Chabasse, M. Pihet, S. Bland, V. Blanc, S. Branger, A. P. Bellanger, L. Millon, C. Plassart, I. Poilane, I. Accoceberry, L. Delhaes, B. Couprie, F. Gabriel, J. Dunand, A. L. Roux, V. Sivadon-Tardy, F. Laurent, S. Legal, E. Moalic, G. Nevez, D. Quinio, M. Cariou, J. Bonhomme, C. Duhamel, B. Podac, S. Lechatch, C. Soler, M. Cambon, C. Nourrisson, P. Poirier, D. Pons, O. Augereau, I. Grawey, N. Fauchet, A. Bonnin, F. Dalle, P. Cahen, P. Honderlick, N. Desbois, C. Miossec, J. L. Hermann, M. Cornet, R. Grillot, B. Lebeau, D. Maubon, H. Pelloux, M. Nicolas, C. Aznar, D. Blanchet, J. F. Carod, M. Demar, A. Angoulvant, C. Ciupek, A. Gigandon, B. Bouteille, E. Frealle, D. Poulain, B. Sendid, D. Dupont, J. Menotti, F. Persat, M.-A. Piens, M. Wallon, C. Cassagne, S. Ranque, T. Benoit-Cattin, L. Collet, A. Fiacre, N. Bourgeois, L. Lachaud, P. Rispail, Y. Sterkers, M. Machouart, F. Gay-Andrieu, P. Lepape, F. Morio, O. Moquet, S. Lefrançois, M. Sasso, F. Reibel, M. Gari-Toussaint, L. Hasseine, L. Bret, D. Poisson, S. Brun, C. Bonnal, C. Chochillon, S. Houzé, A. Paugam, N. Ait-Ammar, F. Botterel, R. Chouk, M. E. Bougnoux, E. Sitterle, A. Fekkar, R. Piarroux, J. Guitard, C. Hennequin, J.-L. Poirot, M. Gits-Muselli, S. Hamane, C. Lacroix, S. Bonacorsi, P. Mariani, D. Moissenet, C. Kauffmann-Lacroix, A. Minoza, E. Perraud, M. H. Rodier, G. Colonna, A. Huguenin, D. Toubas, S. Chevrier, J. P. Gangneux, F. Robert-Gangneux, C. Guigen, O. Belmonte, G. Hoarau, M. C. Jaffar Bandjee, J. Jaubert, S. Picot, N. Traversier, L. Favennec, G. Gargala, N. Godineau, C. Tournus, C. Mahinc, H. Raberin, V. Letscher Bru, S. Cassaing, P. Patoz, E. Bailly, J. Chandenier, G. Desoubeaux, F. Moreau, P. Munier, E. Mazars, O. Eloy, and E. Chachaty
REFERENCES
- 1.Perlin DS. 2007. Resistance to echinocandin-class antifungal drugs. Drug Resist Updat 10:121–130. 10.1016/j.drup.2007.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ullmann AJ, Cornely OA, Donnelly JP, Akova M, Arendrup MC, Arikan-Akdagli S, Bassetti M, Bille J, Calandra T, Castagnola E, Garbino J, Groll AH, Herbrecht R, Hope WW, Jensen HE, Kullberg BJ, Lass-Florl C, Lortholary O, Meersseman W, Petrikkos G, Richardson MD, Roilides E, Verweij PE, Viscoli C, Cuenca-Estrella M, Group EFIS, ESCMID Fungal Infection Study Group. 2012. ESCMID* guideline for the diagnosis and management of Candida diseases 2012: developing European guidelines in clinical microbiology and infectious diseases. Clin Microbiol Infect 18(Suppl 7):1–8. 10.1111/1469-0691.12037. [DOI] [PubMed] [Google Scholar]
- 3.Pappas PG, Kauffman CA, Andes DR, Clancy CJ, Marr KA, Ostrosky-Zeichner L, Reboli AC, Schuster MG, Vazquez JA, Walsh TJ, Zaoutis TE, Sobel JD. 2016. Clinical practice guideline for the management of candidiasis: 2016 update by the Infectious Diseases Society of America. Clin Infect Dis 62:e1–e50. 10.1093/cid/civ933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bretagne S, Desnos-Ollivier M, Sitbon K, Lortholary O, Che D, Dromer F, Participants of the Yeasts. 2021. No impact of fluconazole to echinocandins replacement as first-line therapy on the epidemiology of yeast fungemia (hospital-driven active surveillance, 2004–2017, Paris, France). Front Med (Lausanne) 8:641965. 10.3389/fmed.2021.641965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Castanheira M, Deshpande LM, Davis AP, Rhomberg PR, Pfaller MA. 2017. Monitoring antifungal resistance in a global collection of invasive yeasts and molds: application of CLSI epidemiological cutoff values and whole-genome sequencing analysis for detection of azole resistance in Candida albicans. Antimicrob Agents Chemother 61:e00906-17. 10.1128/AAC.00906-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lortholary O, Renaudat C, Sitbon K, Desnos-Ollivier M, Bretagne S, Dromer F, French Mycoses Study G, The French Mycoses Study Group. 2017. The risk and clinical outcome of candidemia depending on underlying malignancy. Intensive Care Med 43:652–662. 10.1007/s00134-017-4743-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lortholary O, Renaudat C, Sitbon K, Madec Y, Denoeud-Ndam L, Wolff M, Fontanet A, Bretagne S, Dromer F, The French Mycosis Study Group. 2014. Worrisome trends in incidence and mortality of candidemia in intensive care units (Paris area, 2002–2010.). Intensive Care Med 40:1303–1312. 10.1007/s00134-014-3408-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arendrup MC, Perlin DS. 2014. Echinocandin resistance: an emerging clinical problem? Curr Opin Infect Dis 27:484–492. 10.1097/QCO.0000000000000111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shields RK, Nguyen MH, Press EG, Kwa AL, Cheng S, Du C, Clancy CJ. 2012. The presence of an FKS mutation rather than MIC is an independent risk factor for failure of echinocandin therapy among patients with invasive candidiasis due to Candida glabrata. Antimicrob Agents Chemother 56:4862–4869. 10.1128/AAC.00027-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fekkar A, Dannaoui E, Meyer I, Imbert S, Brossas JY, Uzunov M, Mellon G, Nguyen S, Guiller E, Caumes E, Leblond V, Mazier D, Fievet MH, Datry A. 2014. Emergence of echinocandin-resistant Candida spp. in a hospital setting: a consequence of 10 years of increasing use of antifungal therapy? Eur J Clin Microbiol Infect Dis 33:1489–1496. 10.1007/s10096-014-2096-9. [DOI] [PubMed] [Google Scholar]
- 11.Alexander BD, Johnson MD, Pfeiffer CD, Jimenez-Ortigosa C, Catania J, Booker R, Castanheira M, Messer SA, Perlin DS, Pfaller MA. 2013. Increasing echinocandin resistance in Candida glabrata: clinical failure correlates with presence of FKS mutations and elevated minimum inhibitory concentrations. Clin Infect Dis 56:1724–1732. 10.1093/cid/cit136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aldejohann AM, Herz M, Martin R, Walther G, Kurzai O. 2021. Emergence of resistant Candida glabrata in Germany. JAC Antimicrob Resist 3:dlab122. 10.1093/jacamr/dlab122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McCarty TP, Lockhart SR, Moser SA, Whiddon J, Zurko J, Pham CD, Pappas PG. 2018. Echinocandin resistance among Candida isolates at an academic medical centre 2005–15: analysis of trends and outcomes. J Antimicrob Chemother 73:1677–1680. 10.1093/jac/dky059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lortholary O, Desnos-Ollivier M, Sitbon K, Fontanet A, Bretagne S, Dromer F, French Mycosis Study G. 2011. Recent exposure to caspofungin or fluconazole influences the epidemiology of candidemia: a prospective multicenter study involving 2,441 patients. Antimicrob Agents Chemother 55:532–538. 10.1128/AAC.01128-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baixench MT, Aoun N, Desnos-Ollivier M, Garcia-Hermoso D, Bretagne S, Ramires S, Piketty C, Dannaoui E. 2007. Acquired resistance to echinocandins in Candida albicans: case report and review. J Antimicrob Chemother 59:1076–1083. 10.1093/jac/dkm095. [DOI] [PubMed] [Google Scholar]
- 16.Dannaoui E, Desnos-Ollivier M, Garcia-Hermoso D, Grenouillet F, Cassaing S, Baixench MT, Bretagne S, Dromer F, Lortholary O, French Mycoses Study G. 2012. Candida spp. with acquired echinocandin resistance, France, 2004–2010. Emerg Infect Dis 18:86–90. 10.3201/eid1801.110556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Desnos-Ollivier M, Bretagne S, Raoux D, Hoinard D, Dromer F, Dannaoui E. 2008. Mutations in the fks1 gene in Candida albicans, C. tropicalis, and C. krusei correlate with elevated caspofungin MICs uncovered in AM3 medium using the method of the European Committee on Antibiotic Susceptibility Testing. Antimicrob Agents Chemother 52:3092–3098. 10.1128/AAC.00088-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Desnos-Ollivier M, Dromer F, Dannaoui E. 2008. Detection of caspofungin resistance in Candida spp. by Etest. J Clin Microbiol 46:2389–2392. 10.1128/JCM.00053-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bretagne S, Renaudat C, Desnos-Ollivier M, Sitbon K, Lortholary O, Dromer F, French Mycosis Study Group. 2017. Predisposing factors and outcome of uncommon yeast species-related fungaemia based on an exhaustive surveillance programme (2002–14). J Antimicrob Chemother 72:1784–1793. 10.1093/jac/dkx045. [DOI] [PubMed] [Google Scholar]
- 20.Desnos-Ollivier M, Moquet O, Chouaki T, Guerin AM, Dromer F. 2011. Development of echinocandin resistance in Clavispora lusitaniae during caspofungin treatment. J Clin Microbiol 49:2304–2306. 10.1128/JCM.00325-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Desnos-Ollivier M, Robert V, Raoux-Barbot D, Groenewald M, Dromer F. 2012. Antifungal susceptibility profiles of 1698 yeast reference strains revealing potential emerging human pathogens. PLoS One 7:e32278. 10.1371/journal.pone.0032278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pfaller MA, Castanheira M, Diekema DJ, Messer SA, Moet GJ, Jones RN. 2010. Comparison of European Committee on Antimicrobial Susceptibility Testing (EUCAST) and Etest methods with the CLSI broth microdilution method for echinocandin susceptibility testing of Candida species. J Clin Microbiol 48:1592–1599. 10.1128/JCM.02445-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meletiadis J, Curfs-Breuker I, Meis JF, Mouton JW. 2017. In vitro antifungal susceptibility testing of Candida isolates with the EUCAST methodology, a new method for ECOFF determination. Antimicrob Agents Chemother 61:e02372-16. 10.1128/AAC.02372-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bonfietti LX, Martins MDA, Szeszs MW, Pukiskas SBS, Purisco SU, Pimentel FC, Pereira GH, Silva DC, Oliveira L, Melhem M. d S C. 2012. Prevalence, distribution and antifungal susceptibility profiles of Candida parapsilosis, Candida orthopsilosis and Candida metapsilosis bloodstream isolates. J Med Microbiol 61:1003–1008. 10.1099/jmm.0.037812-0. [DOI] [PubMed] [Google Scholar]
- 25.Canton E, Peman J, Quindos G, Eraso E, Miranda-Zapico I, Alvarez M, Merino P, Campos-Herrero I, Marco F, de la Pedrosa EG, Yague G, Guna R, Rubio C, Miranda C, Pazos C, Velasco D, FUNGEMYCA Study Group. 2011. Prospective multicenter study of the epidemiology, molecular identification, and antifungal susceptibility of Candida parapsilosis, Candida orthopsilosis, and Candida metapsilosis isolated from patients with candidemia. Antimicrob Agents Chemother 55:5590–5596. 10.1128/AAC.00466-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lovero G, Borghi E, Balbino S, Cirasola D, De Giglio O, Perdoni F, Caggiano G, Morace G, Montagna MT. 2016. Molecular identification and echinocandin susceptibility of Candida parapsilosis complex bloodstream isolates in Italy, 2007–2014. PLoS One 11:e0150218. 10.1371/journal.pone.0150218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cuenca-Estrella M, Gomez-Lopez A, Mellado E, Monzon A, Buitrago MJ, Rodriguez-Tudela JL. 2009. Activity profile in vitro of micafungin against Spanish clinical isolates of common and emerging species of yeasts and molds. Antimicrob Agents Chemother 53:2192–2195. 10.1128/AAC.01543-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stavrou AA, Perez-Hansen A, Lackner M, Lass-Florl C, Boekhout T. 2020. Elevated minimum inhibitory concentrations to antifungal drugs prevail in 14 rare species of candidemia-causing Saccharomycotina yeasts. Med Mycol 58:987–995. 10.1093/mmy/myaa005. [DOI] [PubMed] [Google Scholar]
- 29.Pham CD, Iqbal N, Bolden CB, Kuykendall RJ, Harrison LH, Farley MM, Schaffner W, Beldavs ZG, Chiller TM, Park BJ, Cleveland AA, Lockhart SR. 2014. Role of FKS mutations in Candida glabrata: MIC values, echinocandin resistance, and multidrug resistance. Antimicrob Agents Chemother 58:4690–4696. 10.1128/AAC.03255-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Arendrup MC, Garcia-Effron G, Lass-Flörl C, Lopez AG, Rodriguez-Tudela J-L, Cuenca-Estrella M, Perlin DS. 2010. Echinocandin susceptibility testing of Candida species: comparison of EUCAST EDef 7.1, CLSI M27-A3, Etest, disk diffusion, and agar dilution methods with RPMI and IsoSensitest media. Antimicrob Agents Chemother 54:426–439. 10.1128/AAC.01256-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Espinel-Ingroff A, Arendrup MC, Pfaller MA, Bonfietti LX, Bustamante B, Canton E, Chryssanthou E, Cuenca-Estrella M, Dannaoui E, Fothergill A, Fuller J, Gaustad P, Gonzalez GM, Guarro J, Lass-Florl C, Lockhart SR, Meis JF, Moore CB, Ostrosky-Zeichner L, Pelaez T, Pukinskas SR, St-Germain G, Szeszs MW, Turnidge J. 2013. Interlaboratory variability of Caspofungin MICs for Candida spp. using CLSI and EUCAST methods: should the clinical laboratory be testing this agent? Antimicrob Agents Chemother 57:5836–5842. 10.1128/AAC.01519-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Arendrup MC, Cuenca-Estrella M, Lass-Florl C, Hope W, EUCAST-AFST. 2012. EUCAST technical note on the EUCAST definitive document EDef 7.2: method for the determination of broth dilution minimum inhibitory concentrations of antifungal agents for yeasts EDef 7.2 (EUCAST-AFST). Clin Microbiol Infect 18:E246–E247. 10.1111/j.1469-0691.2012.03880.x. [DOI] [PubMed] [Google Scholar]
- 33.Castanheira M, Woosley LN, Diekema DJ, Messer SA, Jones RN, Pfaller MA. 2010. Low prevalence of FKS1 hot spot 1 mutations in a worldwide collection of Candida strains. Antimicrob Agents Chemother 54:2655–2659. 10.1128/AAC.01711-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lozano-Chiu M, Nelson PW, Lancaster M, Pfaller MA, Rex JH. 1997. Lot-to-lot variability of antibiotic medium 3 used for testing susceptibility of Candida isolates to amphotericin B. J Clin Microbiol 35:270–272. 10.1128/jcm.35.1.270-272.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Katiyar S, Pfaller M, Edlind T. 2006. Candida albicans and Candida glabrata clinical isolates exhibiting reduced echinocandin susceptibility. Antimicrob Agents Chemother 50:2892–2894. 10.1128/AAC.00349-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Park S, Kelly R, Kahn JN, Robles J, Hsu MJ, Register E, Li W, Vyas V, Fan H, Abruzzo G, Flattery A, Gill C, Chrebet G, Parent SA, Kurtz M, Teppler H, Douglas CM, Perlin DS. 2005. Specific substitutions in the echinocandin target Fks1p account for reduced susceptibility of rare laboratory and clinical Candida sp. isolates. Antimicrob Agents Chemother 49:3264–3273. 10.1128/AAC.49.8.3264-3273.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Balashov SV, Park S, Perlin DS. 2006. Assessing resistance to the echinocandin antifungal drug caspofungin in Candida albicans by profiling mutations in FKS1. Antimicrob Agents Chemother 50:2058–2063. 10.1128/AAC.01653-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tavernier E, Desnos-Ollivier M, Honeyman F, Srour M, Fayard A, Cornillon J, Augeul-Meunier K, Guyotat D, Raberin H. 2015. Development of echinocandin resistance in Candida krusei isolates following exposure to micafungin and caspofungin in a BM transplant unit. Bone Marrow Transplant 50:158–160. 10.1038/bmt.2014.230. [DOI] [PubMed] [Google Scholar]
- 39.Thompson GR, Wiederhold NP, Vallor AC, Villareal NC, Lewis JS, Patterson TF. 2008. Development of caspofungin resistance following prolonged therapy for invasive candidiasis secondary to Candida glabrata infection. Antimicrob Agents Chemother 52:3783–3785. 10.1128/AAC.00473-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Asner SA, Giulieri S, Diezi M, Marchetti O, Sanglard D. 2015. Acquired multidrug antifungal resistance in Candida lusitaniae during therapy. Antimicrob Agents Chemother 59:7715–7722. 10.1128/AAC.02204-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Prigent G, Ait-Ammar N, Levesque E, Fekkar A, Costa JM, El Anbassi S, Foulet F, Duvoux C, Merle JC, Dannaoui E, Botterel F. 2017. Echinocandin resistance in Candida species isolates from liver transplant recipients. Antimicrob Agents Chemother 61:e01229-16. 10.1128/AAC.01229-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fekkar A, Meyer I, Brossas JY, Dannaoui E, Palous M, Uzunov M, Nguyen S, Leblond V, Mazier D, Datry A. 2013. Rapid emergence of echinocandin resistance during Candida kefyr fungemia treatment with caspofungin. Antimicrob Agents Chemother 57:2380–2382. 10.1128/AAC.02037-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Arendrup MC, Perlin DS, Jensen RH, Howard SJ, Goodwin J, Hope W. 2012. Differential in vivo activities of anidulafungin, caspofungin, and micafungin against Candida glabrata isolates with and without FKS resistance mutations. Antimicrob Agents Chemother 56:2435–2442. 10.1128/AAC.06369-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Forastiero A, Garcia-Gil V, Rivero-Menendez O, Garcia-Rubio R, Monteiro MC, Alastruey-Izquierdo A, Jordan R, Agorio I, Mellado E. 2015. Rapid development of Candida krusei echinocandin resistance during caspofungin therapy. Antimicrob Agents Chemother 59:6975–6982. 10.1128/AAC.01005-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Perlin DS, Wiederhold NP. 2017. Culture-independent molecular methods for detection of antifungal resistance mechanisms and fungal identification. J Infect Dis 216:S458–S465. 10.1093/infdis/jix121. [DOI] [PubMed] [Google Scholar]
- 46.Lackner M, Tscherner M, Schaller M, Kuchler K, Mair C, Sartori B, Istel F, Arendrup MC, Lass-Florl C. 2014. Positions and numbers of FKS mutations in Candida albicans selectively influence in vitro and in vivo susceptibilities to echinocandin treatment. Antimicrob Agents Chemother 58:3626–3635. 10.1128/AAC.00123-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Prigitano A, Esposito MC, Cogliati M, Pitzurra L, Santamaria C, Tortorano AM. 2014. Acquired echinocandin resistance in a Candida krusei blood isolate confirmed by mutations in the fks1 gene. New Microbiol 37:237–240. [PubMed] [Google Scholar]
- 48.Vatanshenassan M, Arastehfar A, Boekhout T, Berman J, Lass-Florl C, Sparbier K, Kostrzewa M. 2019. Anidulafungin susceptibility testing of Candida glabrata isolates from blood cultures by the MALDI Biotyper antibiotic (antifungal) susceptibility test rapid assay. Antimicrob Agents Chemother 63:e00554-19. 10.1128/AAC.00554-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vella A, De Carolis E, Mello E, Perlin DS, Sanglard D, Sanguinetti M, Posteraro B. 2017. Potential use of MALDI-ToF mass spectrometry for rapid detection of antifungal resistance in the human pathogen Candida glabrata. Sci Rep 7:9099. 10.1038/s41598-017-09329-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vatanshenassan M, Boekhout T, Lass-Florl C, Lackner M, Schubert S, Kostrzewa M, Sparbier K. 2018. Proof of concept for MBT ASTRA, a rapid matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS)-based method to detect caspofungin resistance in Candida albicans and Candida glabrata. J Clin Microbiol 56:e00420-18. 10.1128/JCM.00420-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Delavy M, Cerutti L, Croxatto A, Prod'hom G, Sanglard D, Greub G, Coste AT. 2019. Machine learning approach for Candida albicans fluconazole resistance detection using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Front Microbiol 10:3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Desnos-Ollivier M, Bretagne S, Boullie A, Gautier C, Dromer F, Lortholary O, French Mycoses Study Group. 2019. Isavuconazole MIC distribution of 29 yeast species responsible for invasive infections (2015–2017). Clin Microbiol Infect 25:634.e1–634.e4. 10.1016/j.cmi.2019.02.007. [DOI] [PubMed] [Google Scholar]
- 53.Locke JB, Almaguer AL, Zuill DE, Bartizal K. 2016. Characterization of in vitro resistance development to the novel echinocandin CD101 in Candida species. Antimicrob Agents Chemother 60:6100–6107. 10.1128/AAC.00620-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Grosset M, Desnos-Ollivier M, Godet C, Kauffmann-Lacroix C, Cazenave-Roblot F. 2016. Recurrent episodes of Candidemia due to Candida glabrata, Candida tropicalis and Candida albicans with acquired echinocandin resistance. Med Mycol Case Rep 14:20–23. 10.1016/j.mmcr.2016.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tables S1 and S2 and Fig. S1. Download aac.01725-21-s0001.pdf, PDF file, 0.3 MB (274.6KB, pdf)