Abstract
The enzyme acetolactate decarboxylase (Ald) plays a key role in the regulation of the α-acetolactate pool in both pyruvate catabolism and the biosynthesis of the branched-chain amino acids, isoleucine, leucine, and valine (ILV). This dual role of Ald, due to allosteric activation by leucine, was used as a strategy for the isolation of Ald-deficient mutants of Lactococcus lactis subsp. lactis biovar diacetylactis. Such mutants can be selected as leucine-resistant mutants in ILV- or IV-prototrophic strains. Most dairy lactococcus strains are auxotrophic for the three amino acids. Therefore, the plasmid pMC004 containing the ilv genes (encoding the enzymes involved in the biosynthesis of IV) of L. lactis NCDO2118 was constructed. Introduction of pMC004 into ILV-auxotrophic dairy strains resulted in an isoleucine-prototrophic phenotype. By plating the strains on a chemically defined medium supplemented with leucine but not valine and isoleucine, spontaneous leucine-resistant mutants were obtained. These mutants were screened by Western blotting with Ald-specific antibodies for the presence of Ald. Selected mutants lacking Ald were subsequently cured of pMC004. Except for a defect in the expression of Ald, the resulting strain, MC010, was identical to the wild-type strain, as shown by Southern blotting and DNA fingerprinting. The mutation resulting in the lack of Ald in MC010 occurred spontaneously, and the strain does not contain foreign DNA; thus, it can be regarded as food grade. Nevertheless, its application in dairy products depends on the regulation of genetically modified organisms. These results establish a strategy to select spontaneous Ald-deficient mutants from transformable L. lactis strains.
Diacetyl is considered to be one of the most important compounds contributing to the final flavor and aroma in a range of fresh fermented dairy products, such as butter, buttermilk, cultured cream, and quark. Diacetyl is formed during the fermentation of milk by the gram-positive microaerophilic bacterium Lactococcus lactis subsp. lactis biovar diacetylactis and Leuconostoc spp. The rate of formation and the stability of diacetyl in fermented dairy products depend on the growth medium, temperature (3), and pH (19), and the presence or absence of oxygen (2, 5, 21). Many of these external factors are difficult to control in the production processes of fermented milk products due to established dairy technologies. However, the formation of diacetyl is to a large extent dependent on the properties of the applied bacterial strains. Therefore, the diacetyl level may be improved by using selected or tailor-made strains with altered metabolisms leading to improved diacetyl formation.
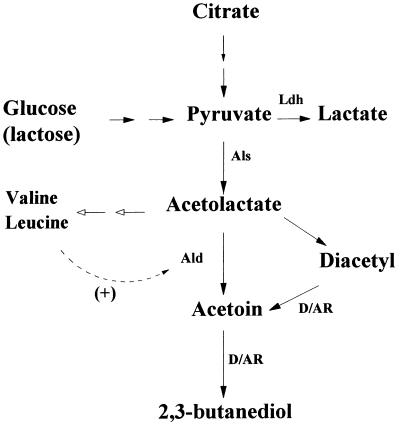
Recent studies of pyruvate and citrate metabolism in L. lactis and Leuconostoc spp. demonstrate that diacetyl is formed by a nonenzymatic oxidative decarboxylation of α-acetolactate (ALA) (Fig. 1) (18, 27). However, in L. lactis ALA is predominantly degraded to acetoin by the enzyme ALA decarboxylase (Ald). In this way the ALA pool available for diacetyl formation is actively and irreversibly diminished.
FIG. 1.
Pathway of diacetyl formation for L. lactis. Ldh, lactate dehydrogenase; Als, acetolactate synthase; Ald, acetolactate decarboxylase; D/AR, diacetyl reductase; open arrows, biosynthesis reactions; dotted line (+), allosteric activation.
A possible method for improving the diacetyl level would be to redirect and adjust the metabolic activities involved in the formation of ALA (for a review, see reference 9). The following approaches have been used: inactivation of ldh, encoding lactate dehydrogenase (Ldh) (11, 16, 26); overexpression of the ilvBN (4) or alsS genes (26), encoding the anabolic and catabolic acetolactate synthase (Als), respectively; inactivation of aldB, encoding Ald (14, 15, 34); and overexpression of nox, encoding NADH oxidase (8, 11). Also, some of these strategies have been combined (16, 26). In most of the strategies the pyruvate flux was redirected, resulting in significantly improved acetoin, but not diacetyl, production. One of the most promising solutions to enhance the formation of diacetyl is inactivation of the aldB gene, which results in an accumulation of the diacetyl precursor ALA (14, 15).
L. lactis aldB mutants can be isolated by selection for growth in the presence of leucine, omitting the other branched-chain amino acids (BCAA), valine and isoleucine (14). The basis for this is the fact that the activity of Ald is subjected to allosteric regulation by leucine, which directly activates Ald, having in mind that ALA is a precursor for the amino acids leucine and valine (23). Leucine activates Ald, which then converts ALA to acetoin so that it is unavailable for the biosynthesis of leucine and valine (Fig. 1). Under conditions where valine is not supplemented the cells cannot grow. However, cells with a mutation in aldB accumulate ALA and can therefore synthesize valine and grow under such conditions (14).
Most of the dairy lactococcal strains have irreversibly lost the ability to synthesize BCAA (13, 29), which makes the selection method of limited use. In this study we show that the approach for isolating Ald mutants can be applied by introduction into dairy lactococcal strains of the genes coding for the enzymes involved in the biosynthesis of BCAA.
MATERIALS AND METHODS
Bacterial strains, plasmids, culture media, and reagents.
The bacterial strains and plasmids used are listed in Table 1. Escherichia coli JIM109 and TG1 and L. lactis subsp. lactis IL1403 were used as recipients during the construction of plasmids. L. lactis was routinely grown at 30°C in M17 broth (Oxoid, Basingstoke, United Kingdom) supplemented with either 0.5% (wt/vol) glucose or 5% (wt/vol) lactose or in chemically defined medium (CDM). The CDM contained (per liter) 5.0 g of glucose, 1 g of Na acetate, 0.6 g of NH4 citrate, 0.5 g of NH4Cl, 9.0 g of KH2PO4, 7.5 g of K2HPO4, 0.2 g of MgCl2 · 6H2O, 0.005 g of FeCl3 · 4H2O, 0.05 g of CaCl2 · 2H2O, 0.005 g of ZnSO4 · 7H2O, 0.0025 g of CoCl2 · 6H2O, 0.3 g of alanine, 0.2 g of arginine, 0.1 g of asparagine, 0.1 g of cysteine, 0.1 g of glutamine, 0.2 g of glycine, 0.05 g of histidine, 0.2 g of lysine, 0.1 g of methionine, 0.2 g of phenylalanine, 0.3 g of proline, 0.3 g of serine, 0.2 g of threonine, 0.1 g of tryptophan, 0.05 g of tyrosine, 10 mg of benzoic acid, 10 mg d-(+)-biotin, 1 mg of folic acid, 1 mg of nicotinic acid, 1 mg of pantothenic acid, 1 mg of riboflavin, 1 mg of thiamine, 2 mg of pyridoxine, 1 mg of cyanocobalamine, 5 mg of orotic acid, 5 mg of 2-deoxytimidine, 5 mg of inosine, 2.5 mg of dl-6,8-thioctic acid, and 5 mg of pyridoxamine. Where indicated, the medium was supplied with 0.05 g of leucine/liter, 0.05 g of isoleucine/liter, and 0.05 g of valine/liter. Anaerobic growth of L. lactis on agar plates was performed with Gas pack (Biomerieux, Marcy l’Etoile, France). E. coli JIM109 and TG1 were grown at 37°C in Luria-Bertani medium (32). When necessary, ampicillin (50 μg/ml) or erythromycin (100 μg/ml) was added for E. coli and erythromycin (5 μg/ml) was added for L. lactis. When required, o-nitrophenyl-β-d-galactopyranoside (ONPG) supplemented with 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) was used as described previously (32). All chemicals were purchased from Sigma (St. Louis, Mo.) or Merck (Darmstad, Germany).
TABLE 1.
Strains and plasmids used
| Bacterial strain or plasmid | Characteristics | Source or reference |
|---|---|---|
| E. coli | ||
| JIM109 | JIM107 recA1 | 32 |
| TG1 | supE thiΔ(lac-pro) hsd(F′ traD proAB lacIqZM15) | 32 |
| L. lactis subsp. lactis | ||
| IL1403 | Plasmid free; r− m− | 6 |
| IL3222 | IL1403/pIL500 | 12 |
| L. lactis subsp. lactis biovar diacetylactis | ||
| DB0003 | ILV auxotroph | CHCCa |
| DB0033 | ILV auxotroph | CHCC |
| DB0410 | ILV auxotroph | CHCC |
| DB0427 | ILV auxotroph | CHCC |
| DB0444 | ILV auxotroph | CHCC |
| DB0449 | ILV auxotroph | CHCC |
| DB1341 | ILV auxotroph | CHCC |
| MC010 | DB0410 Ald− | This work |
| Plasmids | ||
| pIL500 | 18.5-kb XbaI fragment of L. lactis NCDO2118 in pIL 253 | 12 |
| pNEB193 | 2.7 kb; Ampr | New England Biolabs |
| pJIM2279 | 4.8kb; Ermr | 30 |
| pMC004 | 7.5-kb PacI fragment of pIL500 in pJIM2279; Ermr | This work |
CHCC, Chr. Hansen’s Culture Collection.
DNA preparation and manipulation.
Plasmid extraction of L. lactis and E. coli and pulsed-field gel electrophoresis (PFGE) were performed according to the published techniques (1, 32). Restriction endonucleases, T4 DNA ligase, calf intestine alkaline phosphatase, and T4 polymerase (Boehringer GmbH, Mannheim, Germany) were used as recommended by the supplier. Electroporation of L. lactis was performed as described by Holo and Nes (17). Transformation of E. coli was performed by the CaCl2 method as described previously (32).
Construction of pMC004.
A 7.5-kb PacI fragment, containing part of the leuD and ilv genes, of the plasmid pIL500 (12) originating from L. lactis NCDO2118 was cloned into the PacI-digested vector pNEB193 and transformed into E. coli TG1, selecting for Ampr. pNEB193-ilv was digested by KpnI, blunt ended by the use of T4 polymerase, and ligated into the plasmid vector pJIM2279 cut by EcoRV. The plasmid pJIM2279 is capable of carrying large DNA fragments in a stable manner and may also be switched from a high to a low copy number by the exclusion of the 22-bp linker flanked by two KpnI sites of the copF gene (30). The ligation was electroporated into E. coli TG1, selecting for Ermr. The hybrid pJIM2279–pNEB193-ilv isolated from the transformants was restricted by SalI, religated by T4 DNA ligase, and then electroporated in L. lactis IL1403. The 12.3-kb high-copy-number plasmid pJIM2279-ilv, designated pMC004, was isolated from the transformants and used later for the transformation of seven L. lactis subsp. lactis biovar diacetylactis strains (Table 1).
Protein analysis.
Cell-free extracts, protein separation by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and Western blot analysis with Ald-specific antibodies were performed as described by Goupil et al. (14).
Curing of the plasmid pMC004.
The clones in which the absence of Ald was confirmed by Western blotting were cured of the introduced plasmid by subcultivation in M17 medium without erythromycin. After three subcultivations inoculated with 0.1 ml of a 10−6 dilution of the previous culture in 10 ml of M17 medium, Erms clones were isolated by replica plating.
Southern hybridization.
Plasmids were extracted and separated by gel electrophoresis and blotted onto a nylon membrane (1, 32). Total cellular DNA was extracted according to the procedure of Hung and Bandziulis (20), digested by SmaI, and separated by PFGE (CHEF mapper XA system; Bio-Rad) with the following parameters: switch time, 3→8 s; run time, 24 h at 5.3 V/cm; ramp factor, −1.53. The fragments were then blotted onto a nylon membrane (GeneScreen Plus; Dupont, Wilmington, Del.). The two blots were hybridized to a radioactively labelled vector, and the blot of total cellular DNA was hybridized to the recombinant plasmid pMC004. Labelling was performed with a DNA labelling kit (MegaPrime; Amersham) as described by the supplier (Life Science, Little Chalfont, England). The hybridizations were performed by the formamide procedure (GeneScreen Plus; Dupont) as described by the suppliers.
Fermentations.
Product formation in L. lactis was investigated by cultivation in M17 broth supplemented with 5% (wt/vol) lactose and 0.2% (wt/vol) ammonium citrate. The medium was inoculated with a 2% (vol/vol) inoculum from an exponential culture, which was then grown under static conditions. The temperature was maintained at 30°C. Samples were taken at the time intervals indicated in Results.
Measurement of bacterial growth.
Bacterial growth was followed spectrophotometrically by measuring optical density at 600 nm with a Hitachi (Tokyo, Japan) U-1100 spectrophotometer. The correlation between cell dry weight and optical density was determined gravimetrically. A change of 1 absorbance unit at 600 nm corresponds to 0.36 g (dry weight) of cells per liter.
Measurement of fermentation products.
Lactate production was measured by high-pressure liquid chromatography (HPLC) with a Hitachi HPLC system equipped with a SHODEX SH1011 column. The mobile phase was 8 mM H2SO4 containing 0.3 mM EDTA. The column temperature was maintained at 60°C, and a flow rate of 1 ml/min was used. Samples were treated with perchloric acid prior to injection (1.7 ml of 0.1 M perchloric acid per 1 ml of sample, with subsequent centrifugation at 3,500 × g for 25 min). Concentrations of ALA, acetoin, and diacetyl were determined by headspace gas chromatography as described by Richelieu et al. (31). Specific product formation, expressed as millimolar product per gram (dry weight) of cells, was calculated for the values determined at the early stationary phase of growth.
RESULTS
Selection of Ald-deficient mutants.
A L. lactis strain, prototrophic for the BCAA, is unable to grow in a medium containing leucine if the medium is not simultaneously supplemented with valine (14). This is due to the allosteric activation of Ald by leucine (15, 25). As a consequence, the intracellular pool of ALA, the precursor for the biosynthesis of valine and leucine, is reduced, preventing further growth of the cell. Mutants that can grow in the presence of leucine alone can be selected. Such mutants have a mutation in the aldB gene encoding Ald (14, 15), which results in an increased intracellular pool of ALA and a regained ability to synthesize valine.
Seven strains of L. lactis subsp. lactis biovar diacetylactis (Table 1) were plated on CDM containing no leucine, isoleucine, or valine. All strains showed a growth requirement for these amino acids. They also were unable to grow in the presence of leucine and valine, which indicated that neither leu-ilv nor ilv genes were functionally expressed in these strains.
The constructed plasmid pMC004 (see Materials and Methods) contains the genes ilvA, -BN, -C and -D (12). All of the strains used were transformable by the vector pJIM2279; however, upon transformation of pMC004 into the seven BCAA-auxotrophic L. lactis strains, only transformants of two strains, DB0410 and DB0427, which had high frequencies of transformation (106/μg of DNA), were obtained. They were designated DB0410/pMC004 and DB0427/pMC004, respectively.
DB0410/pMC004 and DB0427/pMC004 were tested for growth on CDM supplemented with leucine and valine or with leucine alone. IL3222 (Table 1), harboring a leu-ilv fragment of L. lactis NCDO2118, was used as a positive control. DB0410/pMC004 and DB0427/pMC004 grew on CDM supplemented with leucine and valine but not on CDM supplemented with leucine alone, indicating that leucine caused a reduction in the ALA pool used for the biosynthesis of valine.
In order to select spontaneous leucine-resistant (LeuR) mutants, DB0410/pMC004 and DB0427/pMC004 (105 to 108 cells) were plated on CDM supplemented with leucine but not valine. After 3 days of incubation under anaerobic conditions, small colonies appeared against a transparent background of cell growth. Five LeuR mutants of DB0410/pMC004 and nine LeuR mutants of DB0427/pMC004 were selected for further study.
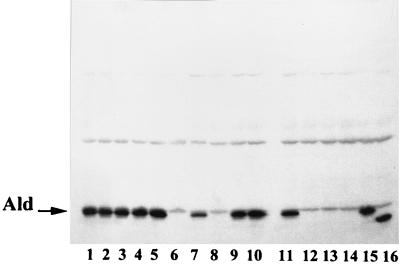
The presence or absence of Ald in the LeuR mutants of DB0410/pMC004 and DB0427/pMC004 was checked by Western blot analysis with Ald-specific antibodies (Fig. 2). Among the LeuR mutants in which Ald could not be detected (Ald−), one DB0410/pMC004 mutant (Fig. 2, lane 6), and five DB0427/pMC004 mutants (Fig. 2, lanes 8, 12, 13, 14, and 16) were chosen for further study. The other LeuR mutants that still expressed the Ald protein probably had a mutation in aldB which resulted in an inactive form of Ald.
FIG. 2.
Autoradiogram of Western blot analysis showing presence or absence of the Ald protein for the leucine-resistant derivatives of DB0410/pMC004 and DB0427/pMC004. Lanes 1 and 7, wild-type strains DB0410 and DB0427; lanes 2 to 6, derivatives of DB0410/pMC004; lanes 8 to 16, derivatives of DB0427/pMC004. The band corresponding to Ald is indicated by an arrow. Lanes 6, 8, and 12 to 14, absence of Ald; lane 16, truncated form of Ald. The other bands originate from nonspecific binding of the secondary antibodies used.
Plasmid curing of the Ald mutants.
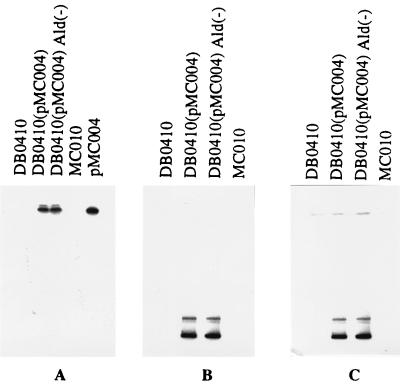
The selected Ald− mutants were then cured of pMC004 by screening for the loss of Ermr. Fifteen Erms clones of each strain were checked for isoleucine auxotrophy by plating them on CDM supplemented with leucine and valine. Four strains, one derived from DB0410/pMC004 and three derived from DB0427/pMC004, were selected, and their plasmids were purified. In all four strains pMC004 could not be detected. However, the three strains derived from DB0427/pMC004 had lost one or more of their original plasmids in addition to pMC004 (data not shown). Only one Ald− mutant of DB0410, designated MC010, contained all of the original plasmids. DB0410, MC010, and their intermediate derivatives, DB0410/pMC004 and DB0410/pMC004 Ald−, were tested subsequently for the presence of the vector (pJIM2279) and pMC004. Total cellular DNA of these four strains was prepared, digested with SmaI, and separated by PFGE (results not shown). In the intermediate derivatives one band with high electrophoretic mobility was detected, corresponding to pMC004. Except for this band, no differences were observed, demonstrating that no major genomic rearrangements had occurred in MC010 compared to DB0410. Figure 3 presents the results of Southern blot hybridization of the vector to plasmid preparation (Fig. 3A), and to total cellular DNA (Fig. 3B) and hybridization of pMC004 to total cellular DNA (Fig. 3C), showing that vector DNA was present only in DB0410/pMC004 and in the derived DB0410/pMC004 Ald− and not in the resulting MC010 and that the presence of pMC004 is seen only in the intermediate DB0410/pMC004 and the derived DB0410/pMC004 Ald−. The results of the Southern blot analysis clearly show that MC010 contains no residual sequences from either JIM2279 or pMC004.
FIG. 3.
Autoradiograms of Southern blot hybridization of the radioactively labelled vector pJIM2279 (A and B) or pMC004 (C) to plasmid preparation of DB0410, MC010, and the intermediate derivatives DB0410/pMC004 and DB0410/pMC004 Ald−. (A) Plasmids were digested by EcoRI, separated by gel electrophoresis, and blotted onto nylon membrane. (B and C) Total cellular DNA was digested by SmaI, separated by PFGE, and blotted onto nylon membrane.
Fermentation products of MC010.
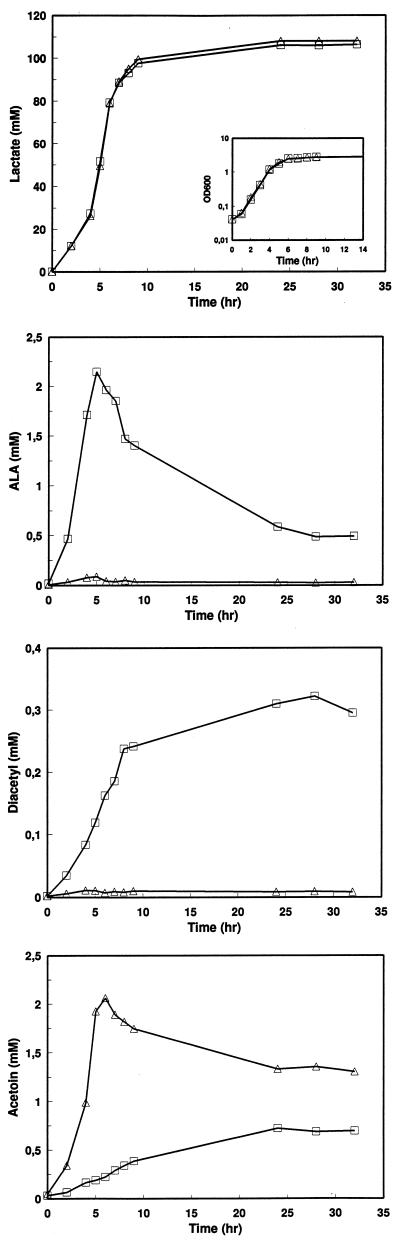
The effect of the Ald impairment was investigated by analyzing the product formation in the isogenic pair DB0410 and the Ald-deficient mutant MC010. The strains were batch cultivated in M17 supplemented with citrate under static conditions. The formation of ALA, acetoin, and diacetyl were followed during the fermentation. Growth rates, final optical densities, and acidification rates did not differ for the isogenic strains (Fig. 4). In the MC010 culture, ALA accumulated (1.64 mM/g [dry weight] of cells) and less acetoin was produced (0.45 mM/g [dry weight] of cells) compared to DB0410, which produced almost no ALA (0.04 mM/g [dry weight] of cells) and more acetoin (2.06 mM/g [dry weight] of cells). MC010 produced significantly more diacetyl (0.27 mM/g [dry weight] of cells) than DB0410 (0.01 mM/g [dry weight] of cells).
FIG. 4.
Comparison of growth rates (inset graph) and the formation of lactate, ALA, acetoin, and diacetyl by the wild-type DB0410 (▵) and by MC010 (□). The strains were cultivated in M17 supplemented with lactose and citrate under static conditions. Time zero indicates the time of inoculation.
DISCUSSION
It has previously been reported that spontaneous Ald-deficient mutants of L. lactis can be isolated from isoleucine-valine (IV)- or isoleucine-leucine-valine (ILV)-prototrophic strains (14). Here we have shown that Ald-deficient mutants can be selected from ILV-auxotrophic L. lactis strains if the ilv genes have been previously introduced into these strains. The introduction of the plasmid pMC004 containing the ilv genes into two dairy strains of L. lactis, DB0410 and DB0427, restores the isoleucine prototrophy. Furthermore, the resulting strains are unable to grow if only leucine, but not valine, is supplemented in the medium, indicating that the allosteric property of Ald, i.e., activation by leucine, is conserved in the dairy strains. This enabled the isolation of the spontaneous Ald-deficient mutants of DB0410/pMC004 and DB0427/pMC004 and establishes a strategy for not only ILV-prototrophic strains but also ILV-auxotrophic strains.
The Ald deficiency of the isolated strain MC010 resulted in a substantially changed ALA metabolism and improved diacetyl formation (Fig. 4). Since Ald has been shown to be the only enzyme involved in conversion of ALA to acetoin (14), it is presumed that in MC010, acetoin is formed by enzymatic reduction of diacetyl and partially by nonenzymatic decarboxylation of ALA. The increased diacetyl level is therefore due to the conversion of ALA to diacetyl combined with the slower enzymatic conversion of diacetyl to acetoin. The effect of Ald deficiency on ALA accumulation and on an increased capacity for diacetyl formation was as observed in other Ald-deficient strains, such as D1 (24), Ru4 (26, 33), and several strains which were recently selected by Goupil et al. (15), suggesting that with respect to increased diacetyl formation, inactivation of Ald is one of the most promising strategies.
The mutation which caused the deficiency of Ald in MC010 occurred spontaneously, and prior introduction of the plasmid pMC004 enabled the selection of the mutant. According to the results obtained by PFGE it appears that no major changes in the genome occurred. It was also shown that, after being cured of the plasmid, the Ald mutant does not contain any residual DNA of pMC004. Such a strain can be considered food grade and might be applied as a component of starter cultures for dairy fermentations. Trials in which the Ald-deficient MC010 was used as a component of a mixed starter culture and applied to milk fermentation show promise for an improved potential for diacetyl formation (7). According to current European Economic Community regulations (10), the Ald mutant MC010 must be approved prior to trials on a laboratory scale. However, in the United States, such a strain is ready and acceptable for food application.
ACKNOWLEDGMENTS
This work was partially financed by the Danish Academy for Technical Sciences under contract EF 572/95-0072-ATV.
We thank Anette Wind for providing DNA fingerprints and Marianne Maigaard and Fergal Patrick Rattray for proofreading the manuscript.
REFERENCES
- 1.Anderson D G, McKay L L. Simple and rapid method for isolating large plasmid DNA from lactic streptococci. Appl Environ Microbiol. 1983;46:549–552. doi: 10.1128/aem.46.3.549-552.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bassit N, Boquien C-Y, Picque D, Corrieu G. Effect of initial oxygen concentration on diacetyl and acetoin production by Lactococcus lactis subsp. lactis biovar diacetylactis. Appl Environ Microbiol. 1993;59:1893–1897. doi: 10.1128/aem.59.6.1893-1897.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bassit N, Boquien C-Y, Picque D, Corrieu G. Effect of temperature on diacetyl and acetoin production by Lactococcus lactis subsp. lactis biovar. diacetylactis CNRZ 483. J Dairy Res. 1995;62:123–129. doi: 10.1128/aem.59.6.1893-1897.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benson K H, Godon J-J, Renault P, Griffin H G, Gasson M J. Effect of ilvBN encoded α-acetolactate synthase expression on diacetyl production in Lactococcus lactis. Appl Microbiol Biotechnol. 1996;45:107–111. [Google Scholar]
- 5.Boumerdassi H, Desmazeaud M, Monnet C, Boquein C, Corrieu G. Improvement of diacetyl production by Lactococcus lactis ssp. lactis CNRZ 483 through oxygen control. J Dairy Sci. 1996;79:775–781. [Google Scholar]
- 6.Chopin A, Chopin M-C, Moillo-Batt A, Langella P. Two plasmid-determined restriction and modification systems in Streptococcus lactis. Plasmid. 1984;11:260–263. doi: 10.1016/0147-619x(84)90033-7. [DOI] [PubMed] [Google Scholar]
- 7.Curic, M., B. Jelle, and D. Nilsson. Personal communication.
- 8.De Filipe F L, van de Zanden W, Marrug J, Hugenholtz J. Abstracts of the 5th Symposium on Lactic Acid Bacteria. 1996. Metabolic engineering of NADH-oxidation in Lactococcus lactis, abstr. G48. [Google Scholar]
- 9.De Voss W M. Metabolic engineering of sugar catabolism in lactic acid bacteria. Antonie Leeuwenhoek. 1996;70:223–242. doi: 10.1007/BF00395934. [DOI] [PubMed] [Google Scholar]
- 10.European Economic Community. Official Journal of the European Communities; 8.5.90; Council Directive of 23 April 1990 on the deliberate release into the environment of genetically modified organisms (90/220/EEC). Brussels, Belgium: ELC Secretariat; 1990. [Google Scholar]
- 11.Gasson M J, Benson K, Swindell S, Griffin H. Metabolic engineering of the Lactococcus lactis diacetyl pathway. Lait. 1996;76:33–40. [Google Scholar]
- 12.Godon J-J, Chopin M-C, Ehrlich S D. Branched-chain amino acid biosynthesis genes in Lactococcus lactis. J Bacteriol. 1992;174:6580–6589. doi: 10.1128/jb.174.20.6580-6589.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Godon J-J, Delorme C, Bardowski J, Chopin M-C, Ehrlich S D, Renault P. Gene inactivation in Lactococcus lactis: branched-chain amino acid biosynthesis. J Bacteriol. 1993;175:4383–4390. doi: 10.1128/jb.175.14.4383-4390.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goupil N, Cortier G, Ehrlich S D, Renault P. Imbalance of leucine flux in Lactococcus lactis and its use for the isolation of diacetyl-overproducing strains. Appl Environ Microbiol. 1996;62:2636–2640. doi: 10.1128/aem.62.7.2636-2640.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goupil-Feuillerat N, Cocaign-Bousquet M, Godon J-J, Ehrlich S D, Renault P. Dual role of α-acetolactate decarboxylase in Lactococcus lactis subsp. lactis. J Bacteriol. 1997;179:6285–6293. doi: 10.1128/jb.179.20.6285-6293.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Henriksen, C. M., and D. Nilsson. Personal communication.
- 17.Holo H, Nes I F. High-frequency transformation, by electroporation, of Lactococcus lactis subsp. cremoris grown with glycine in osmotically stabilized media. Appl Environ Microbiol. 1989;55:3119–3123. doi: 10.1128/aem.55.12.3119-3123.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hugenholtz J, Starrenburg M. Diacetyl production by different strains of Lactococcus lactis subsp. lactis biovar. diacetylactis and Leuconostoc spp. Appl Microbiol Biotechnol. 1992;38:17–22. [Google Scholar]
- 19.Hugenholtz J. Citrate metabolism in lactic acid bacteria. FEMS Microbiol Rev. 1993;12:165–178. [Google Scholar]
- 20.Hung L, Bandziulis R. Megabase DNA analysis: chromosomal DNA perparation, restriction, and pulsed-field electorphoresis. Promega Notes. 1990;24:1–3. [Google Scholar]
- 21.Kaneko T, Takahashi M, Suzuki H. Acetoin fermentation by citrate-positive Lactococcus lactis subsp. lactis 3022 grown aerobically in the presence of hemin or Cu2+ Appl Environ Microbiol. 1990;56:2644–2649. doi: 10.1128/aem.56.9.2644-2649.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marugg J D, Goelling D, Stahl U, Ledeboer A M, Toonen M Y, Verhue W M, Verrips C T. Identification and characterization of the α-acetolactate synthase gene from Lactococcus lactis subsp. lactis biovar diacetylactis. Appl Environ Microbiol. 1994;60:1390–1394. doi: 10.1128/aem.60.4.1390-1394.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Monnet C, Phalip V, Schmitt P, Divies C. Comparison of α-acetolactate decarboxylase in Lactococcus spp. and Leuconostoc spp. Biotechnol Lett. 1994;16:257–262. [Google Scholar]
- 24.Monnet C, Schmitt P, Divies C. Diacetyl production in milk by an α-acetolactic accumulating strain of Lactococcus lactis ssp. lactis biovar. diacetylactis. J Dairy Sci. 1994;77:2916–2924. [Google Scholar]
- 25.Phalip V, Monnet C, Schmitt P, Renault P, Godon J-J, Divies C. Purification and properties of α-acetolactate decarboxylase from Lactococcus lactis subsp. lactis NCDO 2118. FEBS Lett. 1994;351:95–99. doi: 10.1016/0014-5793(94)00820-5. [DOI] [PubMed] [Google Scholar]
- 26.Platteeuw C, Hugenholtz J, Starrenburg M, van Alen-Boerrigter I, de Vos W M. Metabolic engineering of Lactococcus lactis: influence of the overproduction of α-acetolactate synthase in strains deficient in lactate dehydrogenase as a function of culture conditions. Appl Environ Microbiol. 1995;61:3967–3971. doi: 10.1128/aem.61.11.3967-3971.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ramos A, Jordan N K, Cogan T, Santos H. 13C nuclear magnetic resonance studies of citrate and glucose cometabolism in Lactococcus lactis. Appl Environ Microbiol. 1994;60:1739–1748. doi: 10.1128/aem.60.6.1739-1748.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ramos A, Lolkema J S, Konings W N, Santos H. Enzyme basis for pH regulation of citrate and pyruvate metabolism by Leuconostoc oenos. Appl Environ Microbiol. 1995;61:1303–1310. doi: 10.1128/aem.61.4.1303-1310.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Renault P, Godon J-J, Goupil N, Delorme C, Cortier G, Ehrlich S D. Metabolic operons in Lactococci. Genetics of Streptococci, Enterococci and Lactococci. Dev Biol Stand. 1995;85:431–441. [PubMed] [Google Scholar]
- 30.Renault P, Corthier G, Goupil N, Delorme C, Ehrlich S D. Plasmid vectors for Gram-positive bacteria switching from high to low copy number. Gene. 1996;183:175–182. doi: 10.1016/s0378-1119(96)00554-9. [DOI] [PubMed] [Google Scholar]
- 31.Richelieu M, Houlberg U, Nielsen J C. Determination of α-acetolactic acid and volatile compounds by headspace gas chromatography. J Dairy Sci. 1997;80:1918–1925. [Google Scholar]
- 32.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 33.Starrenburg M, Hugenholtz J. Citrate fermentation by Lactococcus and Leuconostoc spp. Appl Environ Microbiol. 1991;57:3535–3540. doi: 10.1128/aem.57.12.3535-3540.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Swindell S R, Benson K H, Griffin H G, Renault P, Ehrlich S D, Gasson M. Genetic manipulation of the pathway for diacetyl metabolism in Lactococcus lactis. Appl Environ Microbiol. 1996;62:2641–2643. doi: 10.1128/aem.62.7.2641-2643.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]