Abstract
Most techniques used to assay the growth of microbes in natural communities provide no information on the relationship between microbial productivity and community structure. To identify actively growing bacteria, we adapted a technique from immunocytochemistry to detect and selectively isolate DNA from bacteria incorporating bromodeoxyuridine (BrdU), a thymidine analog. In addition, we developed an immunocytochemical protocol to visualize BrdU-labeled microbial cells. Cultured bacteria and natural populations of aquatic bacterioplankton were pulse-labeled with exogenously supplied BrdU. Incorporation of BrdU into microbial DNA was demonstrated in DNA dot blots probed with anti-BrdU monoclonal antibodies and either peroxidase- or Texas red-conjugated secondary antibodies. BrdU-containing DNA was physically separated from unlabeled DNA by using antibody-coated paramagnetic beads, and the identities of bacteria contributing to both purified, BrdU-containing fractions and unfractionated, starting-material DNAs were determined by length heterogeneity PCR (LH-PCR) analysis. BrdU-containing DNA purified from a mixture of DNAs from labeled and unlabeled cultures showed >90-fold enrichment for the labeled bacterial taxon. The LH-PCR profile for BrdU-containing DNA from a labeled, natural microbial community differed from the profile for the community as a whole, demonstrating that BrdU was incorporated by a taxonomic subset of the community. Immunocytochemical detection of cells with BrdU-labeled DNA was accomplished by in situ probing with anti-BrdU monoclonal antibodies and Texas red-labeled secondary antibodies. Using this suite of techniques, microbial cells incorporating BrdU into their newly synthesized DNA can be quantified and the identities of these actively growing cells can be compared to the composition of the microbial community as a whole. Since not all strains tested could incorporate BrdU, these methods may be most useful when used to gain an understanding of the activities of specific species in the context of their microbial community.
Methods used to study microbial communities typically either measure net rates of biochemical processes or employ molecular analyses to assess the diversity of community members. Few techniques link these methodological approaches by identifying community members responsible for biochemical transformations. One widely used technique for assessing community productivity is the measurement of microbial incorporation of radiolabeled thymidine (TdR) into newly synthesized DNA (8, 9). Thymidine incorporation measurements are used to estimate the number of cells added to microbial populations during pulse-labeling experiments, and these numbers are used to estimate carbon flux through the microbial compartments of complex ecosystems. In contrast, molecular genetic studies are used to identify the numerically dominant taxa resident in microbial communities. Phylogenetic analyses of 16S rRNA genes cloned from community DNA are commonly used to identify the microbes (10, 25). Neither of these techniques can discriminate the relative contributions of different microbial taxa to community productivity.
A variety of techniques have been used to quantify metabolically active bacteria in natural populations. In situ assays for cells containing nucleoid DNA (32) and ribosomes (13), autoradiographic detection of cells incorporating radioactive substrates (3, 22), redox dye detection of charged cell membranes (28), and cell enlargement assays for communities treated with the DNA replication inhibitor nalidixic acid (17) have all been applied to natural microbial communities. The number of active cells identified by these techniques varies widely. For instance, the number of nucleoid-containing cells (cells containing DNA as assessed by a modified 4′,6-diamidino-2-phenylindole [DAPI] staining protocol with an isopropanol wash) sometimes amounts to only 2% of the cells detected by the standard DAPI protocol (32). Yet, in replicate samples, an average of 49% of cells incorporated radiolabeled amino acids, 1% maintained a membrane redox potential, and 56% bound a universal rRNA probe, while only 29% contained visible nucleoids (15). To some extent, the discrepancies among the number of active cells detected by these techniques must reflect differences among community members in the activities of their metabolic processes, which could be related to phylogenetic diversity. None of these techniques is able to determine whether the metabolically most-active cells comprise a phylogenetic subset of the microbial community.
Techniques have also been developed to measure growth rates and productivity for particular taxa within natural communities. Growth rates for specific phylogenetic groups can be estimated from fluorescence intensities when fixed cells are hybridized to fluorescent, group-specific oligonucleotide probes (6). For culturable microorganisms, these fluorescence intensities can be correlated with growth rates (16). Taxon-specific productivity estimates can also be inferred from incorporation of radiolabeled tracer molecules into natural populations followed by immunochemical purification of cells binding to specific antibodies (2). This technique is most applicable to cultivable taxa, pure cultures of which can be used to produce antisera directed against cell surface antigens. Each of these methods enables the estimation of group-specific growth rates for microorganisms in natural communities. However, they cannot directly determine which taxa are most active in a microbial population.
Bromodeoxyuridine (BrdU) is structurally similar to thymidine and can be incorporated into newly synthesized DNA (31). In mammalian cell biology and medicine, immunocytochemical detection of BrdU-containing DNA has been used to define cell cycle parameters for cultured cells, to study the mechanisms of DNA repair, and to identify rapidly multiplying, cancerous foci in histological sections (7). Due to the widespread use of BrdU in medical research, both BrdU and high-quality anti-BrdU antibodies are commercially available at modest prices.
In the aftermath of classic experiments establishing the semiconservative nature of DNA replication (20), researchers studying bacterial chromosome duplication routinely employed BrdU labeling to make newly synthesized DNA heavier than normal (19). The Escherichia coli and Bacillus subtilis strains used for these experiments would not incorporate exogenously supplied BrdU unless they had mutations affecting thymine synthesis (12). Because of these experiments, it has been widely assumed that wild-type bacteria do not take up BrdU (5). However, this assumption has not been tested. In light of the facts that many bacteria are capable of incorporating thymidine and that thymidine incorporation is routinely used to assess microbial productivity, it is worthwhile to determine whether natural populations and cultured bacteria of interest are indeed capable of incorporating this compound. If microorganisms important to natural populations can incorporate BrdU into their DNA, then a number of powerful techniques become available to microbial ecologists.
At present, it is not known whether biochemical transformations such as thymidine incorporation are performed equally by all taxa in microbial communities. However, it is likely that taxa differ in this regard due to innate physiological differences and in response to environmental conditions that promote or retard their growth. A technique to meld thymidine incorporation with molecular genetic analyses could be used to identify metabolically active taxa within microbial communities. We have developed a suite of techniques which can be used to detect microbial DNA synthesis in natural samples, count metabolically active cells, and also phylogenetically identify the active members of a microbial community. To this end, we have exploited the relatively inexpensive, commercially available reagents used by the biomedical community for BrdU labeling and immunocytochemistry.
MATERIALS AND METHODS
Bacterial strains, experimental growth conditions, and postincubation processing.
Clonal isolates of marine bacteria (Table 1) were grown in R2A medium (without agar) at 25°C (30). Experimental flasks were supplemented with 20 μM BrdU and 33 nM TdR, and control flasks were supplemented with 33 nM TdR only. For comparisons of BrdU incorporation among bacterial strains, cultures were incubated for two doublings of optical density at 660 nm (OD660), pelleted by centrifugation, resuspended in sucrose lysis buffer (750 mM sucrose, 400 mM NaCl, 50 mM Tris [pH 9.0], 20 mM EDTA), and frozen at −80°C. For immunocytochemistry, cultures were incubated for one doubling of OD660, fixed with 4% formaldehyde in culture medium at room temperature for 1 h, pelleted, washed and resuspended in phosphate-buffered saline (PBS), diluted 1:1 with 100% ethanol, and stored at −20°C. For purification of BrdU-containing DNA, cultures of Alteromonas sp. strain C250.5-4 were incubated for 1 h, during which the OD660 increased by a factor of 1.4 to 1.5, and Roseobacter sp. strain S34 was grown overnight without addition of BrdU or TdR. Cultures were pelleted and frozen in sucrose lysis buffer as described above.
TABLE 1.
Bacterial isolates used in these experiments
| Strain | Phylogenetic affiliation | Site of isolation | Isolator(s) | Reference |
|---|---|---|---|---|
| S34 | Roseobacter sp. (α-proteobacterium) | Sargasso Sea, surface | N. Adair and S. J. Giovannoni | |
| C250.5-4 | Alteromonas sp. (γ-proteobacterium) | Sargasso Sea, 250-m depth | B. Lanoil | |
| B250-17A | High-G+C gram-positive bacterium | Sargasso Sea, 250-m depth | B. Lanoil | |
| R2A-132 | Flavobacterium sp. | Oregon coast | M. Suzuki | 25 |
Field sampling.
Water was collected at Cronemiller Lake, a shallow, eutrophic impoundment at Oregon State University’s McDonald-Dunn Research Forest, at 11:30 a.m. on 13 February 1998. All containers used were acid-cleaned, sterile polypropylene. A sample was collected with a 20-liter carboy, which was opened just below the lake’s surface, and the lake water was immediately distributed through 10-μm-mesh Nytex fabric into 11 incubation bottles. With the exception of a control bottle that received no supplements, the bottles were supplemented with 33 nM TdR and 0, 0.02, 0.2, 2.0, or 20.0 μM BrdU. The bottles were enclosed in dark plastic and incubated in situ at 8°C for 1 h, with the exception of a 0 μM BrdU no-incubation control that was placed immediately on ice. After incubation, the bottles were placed on ice and transported back to the laboratory (ca. 30 min), and cells were pelleted by centrifugation. The cell pellets were resuspended in sucrose lysis buffer and stored at −80°C.
DNA isolation.
DNA was prepared from Roseobacter sp. strain S34, Alteromonas sp. strain C250.5-4, and Flavobacterium sp. strain R2A-132 by the sodium dodecyl sulfate-proteinase K-cetyltrimethylammonium bromide (CTAB) method (1). For paramagnetic-bead isolation of BrdU-containing DNA, Alteromonas sp. strain C250.5-4 DNA was further purified by equilibrium ultracentrifugation in cesium trifluoroacetate (1). DNA from Cronemiller Lake samples was prepared by the sodium dodecyl sulfate-proteinase K-CTAB method with the addition of a guanidinium isothiocyanate treatment (26). DNA from gram-positive strain B250-17A was isolated according to the guanidinium isothiocyanate protocol (26). All DNA preparations were treated with RNase A, and DNA concentrations were estimated by visual examination of ethidium bromide-stained agarose gels.
Immunochemical detection of BrdU in genomic DNA dot blots.
BrdU-containing DNA was detected in dot blots by two methods, one employing a secondary antibody conjugated to a peroxidase enzyme and the other using a fluorescently labeled secondary antibody. Washes and incubations in both protocols were performed at 30°C in a Techne hybridization oven.
For the peroxidase method, 1.0 μg of DNA from each culture was spotted onto a Zetaprobe hybridization membrane as described previously (11). The membrane was treated with 25 ml of 1× digoxigenin blocking solution (0.1 mM maleic acid–0.15 M NaCl [pH 7.5] containing 1× blocking reagent; Boehringer) for 30 min and then with 6 ml of fresh blocking solution containing 12 μl of monoclonal mouse anti-BrdU immunoglobulin G (IgG) (3.9 mg of IgG/ml; Sigma) for 30 min, washed twice with 25 ml of maleic acid buffer (0.1 mM maleic acid, 0.15 M NaCl [pH 7.5]) for 15 min each time, incubated with 6 ml of maleic acid buffer containing 6 μl of secondary antibody (peroxidase-conjugated goat anti-mouse IgG [0.375 mg of IgG/ml; Sigma]) for 30 min, washed twice with 25 ml of maleic acid buffer for 15 min each time, and then washed twice with PBS containing 0.1% Tween 20 (PBS-Tween) for 5 min each time. Antibody binding was visualized on autoradiographic film by using Renaissance Western blot chemiluminescence detection reagents (New England Nuclear).
For the fluorescence method, 0.1-μg DNA samples from Cronemiller Lake bacterioplankton were spotted onto a Zetaprobe membrane. The membrane was treated with 25 ml of 5% nonfat dry milk in PBS-Tween for 30 min, washed with 25 ml of PBS-Tween for 5 min, incubated with 12 μl of monoclonal mouse anti-BrdU IgG in 6 ml of PBS-Tween for 30 min, washed twice with 25 ml of PBS-Tween for 5 min each time, incubated with 6 ml of PBS-Tween containing 50 μl of fluorescent secondary antibody (Texas red-conjugated goat anti-mouse IgG [2 mg IgG/ml; Molecular Probes]) for 30 min, and washed four times with 25 ml of PBS-Tween each time. Texas red fluorescence was electronically detected with an FMBIOII fluorescence scanner (Hitachi).
Immunocytochemical detection of BrdU-labeled bacteria.
BrdU-labeled and control cultures of Alteromonas sp. strain C250.5-4 were attached to slides and fixed as described previously (18). All incubations were performed at room temperature. Anti-BrdU monoclonal antibody (diluted 1:10 in PBS-Tween) was spotted onto the slides, which were then incubated in a humidified chamber for 2 to 3 h and washed twice with PBS-Tween for 15 min each time. A 10-μl volume of diluted (1:10 in PBS-Tween) Texas red-conjugated goat anti-mouse IgG was spotted onto the slides, which were subsequently incubated and washed as for the primary antibody. Cells were counterstained with a 1-μg/ml solution of DAPI (Sigma), covered with 1,4-diazabicyclo[2,2,2]octane (DAPCO; Sigma) solution, and sealed under coverslips. Slides were examined with a Leica model DMRB microscope equipped with a 75-W xenon vapor arc lamp. Images were captured with a Photometrics (Tucson, Ariz.) Star I cooled charge-coupled device camera, to which was attached a Photometrics Star I camera controller, and processed by using IP Labs Spectrum version 3.1 software (Signal Analytics Corporation, Vienna, Va.).
Immunochemical purification of BrdU-labeled DNA.
Immunoglobulin-coated paramagnetic beads were used to purify BrdU-containing DNA from experimental mixtures of DNA from labeled and unlabeled cultures and from environmental samples. For the culture experiment, the protocol was applied to mixed DNA from BrdU-labeled or unlabeled cultures of Alteromonas sp. strain C250.5-4 combined with approximately equal quantities of DNA from unlabeled Roseobacter sp. strain S34. For the field experiment, the protocol was applied to DNA from Cronemiller Lake bacterioplankton incubated with 20 μM BrdU and/or 33 nM TdR. All incubations were performed at room temperature. Herring sperm DNA (1.25 mg/ml in PBS) was boiled for 1 min, quickly frozen in dry ice-ethanol, thawed, mixed in a 9:1 ratio with monoclonal anti-BrdU antibodies (diluted 1:10 in PBS), and incubated for 30 min. DNA samples (1 μg [total] in 10 μl of PBS) were boiled for 1 min, frozen in dry ice-ethanol, thawed, mixed with 10 μl of the herring sperm DNA-antibody mixture, and incubated for 30 min. Goat anti-mouse IgG-coated paramagnetic beads (DYNAL Inc.) were washed once in PBS containing 1 mg of acetylated bovine serum albumin (Sigma) per ml (PBS-BSA), using a magnetic particle concentrator (DYNAL Inc.), and resuspended in PBS-BSA at their initial concentration. A 25-μl volume of beads was added to each sample, which was subsequently incubated for 30 min with constant agitation and then washed seven times with 0.5 ml of PBS-BSA each time. A BrdU-containing DNA fraction was eluted by adding 100 μl of 1.7 mM BrdU (in PBS-BSA) and incubating for 30 min with constant agitation. A 2-μl volume of glycogen (20 mg/ml; U.S. Biochemicals) was added to the bead supernatants, and DNA was isolated by two rounds of ethanol precipitation.
LH-PCR.
Bacterial 16S rRNA gene fragments in experimental DNA mixtures, in lake water populations, and eluted from antibody-coated paramagnetic beads, were PCR amplified with primer 338R (E. coli positions 338 to 355) and fluorescently labeled primer EUBB-FAM (positions 8 to 28). Naturally occurring length heterogeneities which distinguish bacterial phylogenetic groups were assayed by length heterogeneity PCR (LH-PCR) (29). PCR product concentrations were kept below 10 μM to avoid amplification biases (29).
RESULTS
Growth of bacterial cultures in the presence of BrdU.
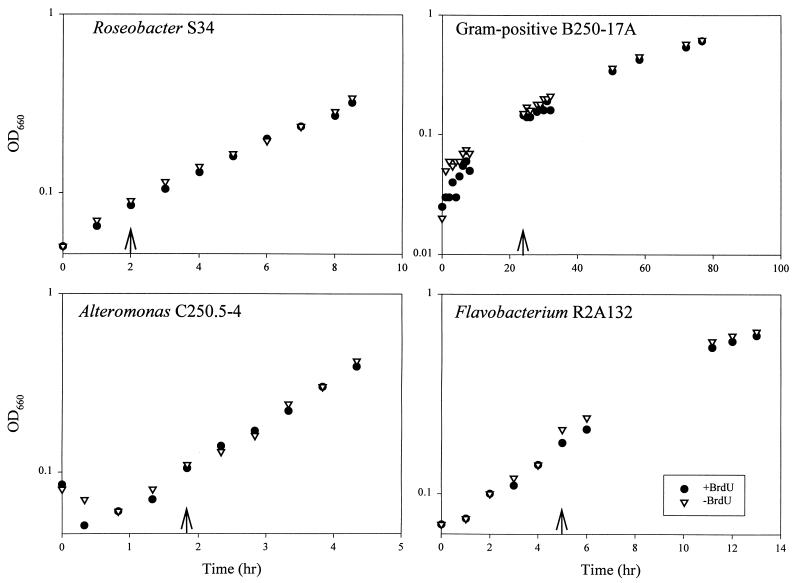
Growth kinetics for bacterial cultures treated with BrdU and TdR were similar to those of controls treated with TdR alone, indicating a low level of toxicity for BrdU under these experimental conditions (Fig. 1). Previous studies have established that the concentration of TdR added to the culture medium (33 nM) is sufficient to inhibit the activity of thymidylate synthase, an enzyme required for de novo synthesis of thymidine monophosphate, in bacterial strains capable of importing TdR (23). Inhibition of thymidylate synthase activity forces dependence on imported TdR, resulting in increased incorporation of exogenously supplied [3H]TdR. Although we have not established that thymidylate synthase activity was inhibited during our experiments, TdR was included in our labeling protocol with the intention of inhibiting thymidylate synthase and thereby maximizing BrdU incorporation. The fact that the growth kinetics for BrdU-labeled and control cultures were identical indicates that BrdU had a low toxicity level in the presence of thymidylate synthase-inhibiting concentrations of TdR.
FIG. 1.
Growth kinetics for bacterial cultures with and without BrdU. Arrows indicate times at which cultures were supplemented with BrdU and TdR (+BrdU) or TdR alone (−BrdU). Cultures were harvested at the final time points and used to prepare DNA for dot blots.
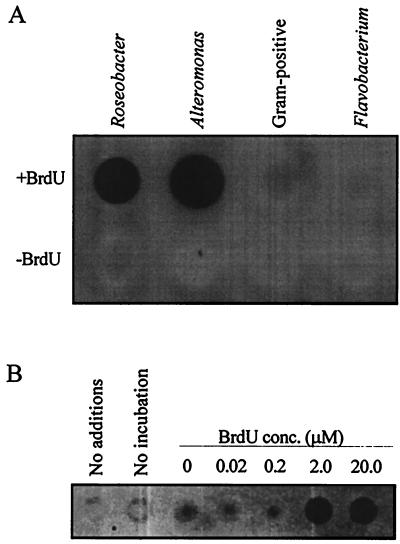
Dot blot detection of BrdU-labeled DNA.
DNA from some BrdU-labeled cultures and lake water gave positive signals, while unlabeled control DNAs did not react with the antibody probes (Fig. 2). Although an extensive phylogenetic survey was beyond the scope of this study, we tested four bacterial strains, representing a broad sample of bacterial diversity, for their capacity to incorporate exogenously supplied BrdU into DNA during growth. Two strains, one a member of the α subclass of the class Proteobacteria and the other a γ-proteobacterium, assimilated BrdU into DNA, but two strains, one a flavobacterium and the other a gram-positive bacterium, failed to assimilate BrdU during growth.
FIG. 2.
Immunochemical detection of BrdU in DNA dot blots. (A) DNA from cultured bacteria grown in the presence (+) or absence (−) of BrdU, probed with anti-BrdU monoclonal antibodies and peroxidase-conjugated secondary antibodies. (B) DNA from Cronemiller Lake bacterioplankton, incubated with or without BrdU, probed with anti-BrdU monoclonal antibodies and Texas red-conjugated secondary antibodies.
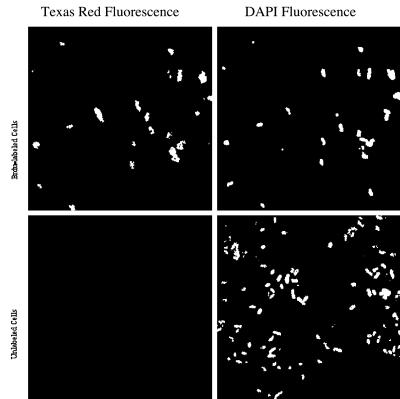
Immunocytochemical detection of BrdU in labeled cells.
Most cells in the BrdU-labeled culture fluoresced red under 560-nm excitation, while cells in the unlabeled culture showed no Texas red fluorescence (Fig. 3). The fraction of BrdU-positive cells exhibiting Texas red fluorescence (85%) may represent the proportion of cells undergoing DNA replication during the BrdU pulse-labeling, which persisted for one cell doubling cycle as judged by changes in the OD660. DAPI fluorescence in the immunocytochemistry preparations was distinguishable from background fluorescence but was reduced relative to that of routine preparations made for bacterial-cell counting. Reduced DAPI fluorescence can be attributed to the fact that DNA in the target cells must be denatured to allow for anti-BrdU antibody binding.
FIG. 3.
Immunocytochemical detection of BrdU-labeled Alteromonas sp. strain C250.5-4. The panels on the left show fluorescence from Texas red-conjugated secondary antibodies bound to anti-BrdU monoclonal antibodies; the panels on the right show DAPI fluorescence. The upper panels show BrdU-labeled cultures; the lower panels show unlabeled cultures.
Immunochemical purification of BrdU-labeled DNA from mixed DNAs of labeled and unlabeled cultures.
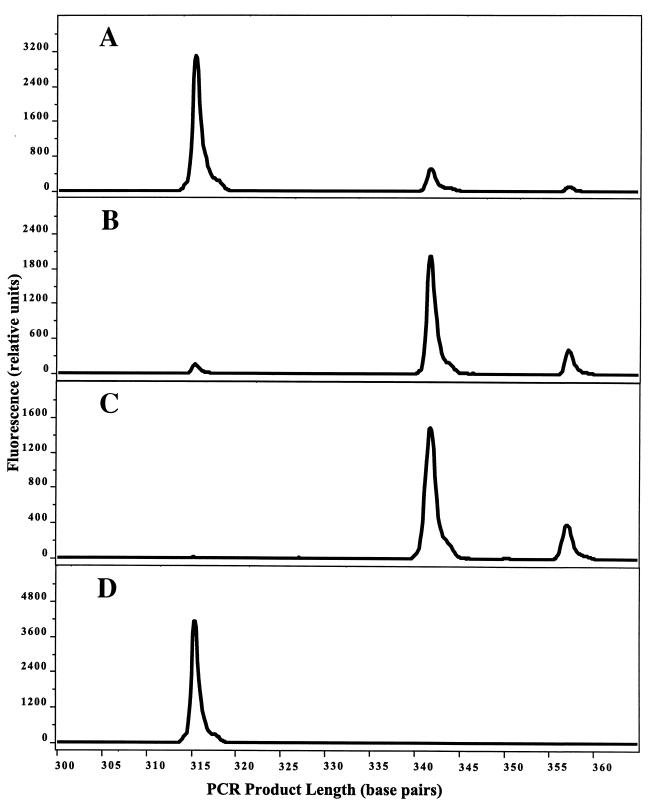
LH-PCR analysis indicated a significant enrichment of Alteromonas DNA in the eluate from a labeled Alteromonas-unlabeled Roseobacter DNA mixture, relative to that of the starting mixture (Fig. 4). Roseobacter and Alteromonas DNAs can be distinguished by LH-PCR, which detects naturally occurring differences in the number of nucleotides between positions 27 and 355 (E. coli numbering) in their 16S rRNA genes. The LH-PCR gene fragment from Alteromonas sp. strain C250.5-4 is 342 nucleotides long and the fragment from Roseobacter sp. strain S34 is 315 nucleotides in length, as judged from electropherograms. Comparison of peak areas for the 342- and 315-nucleotide fragments indicates a 91-fold enrichment of Alteromonas DNA when purified from a mixture containing BrdU-labeled Alteromonas DNA (Fig. 4) and a 2.8-fold enrichment when purified from a control mixture of unlabeled DNAs (data not shown). Enrichment with antibody-coated paramagnetic beads proved to be an effective method of separating BrdU-labeled DNA from a background of unlabeled DNA in a form suitable for taxonomic analysis by LH-PCR or other molecular methods.
FIG. 4.
Immunochemical purification of BrdU-containing DNA, analyzed by LH-PCR. (A) Experimental mixture containing DNA from a BrdU-labeled Alteromonas cultures and an unlabeled Roseobacter culture. (B) DNA immunochemically purified from the mixture by using anti-BrdU monoclonal antibodies and paramagnetic beads. (C) BrdU-labeled Alteromonas DNA. (D) Unlabeled Roseobacter DNA. The electropherogram for unlabeled Alteromonas DNA was identical to that shown in panel C.
Discrimination of bacterial taxa incorporating BrdU in a lake water microbial community.
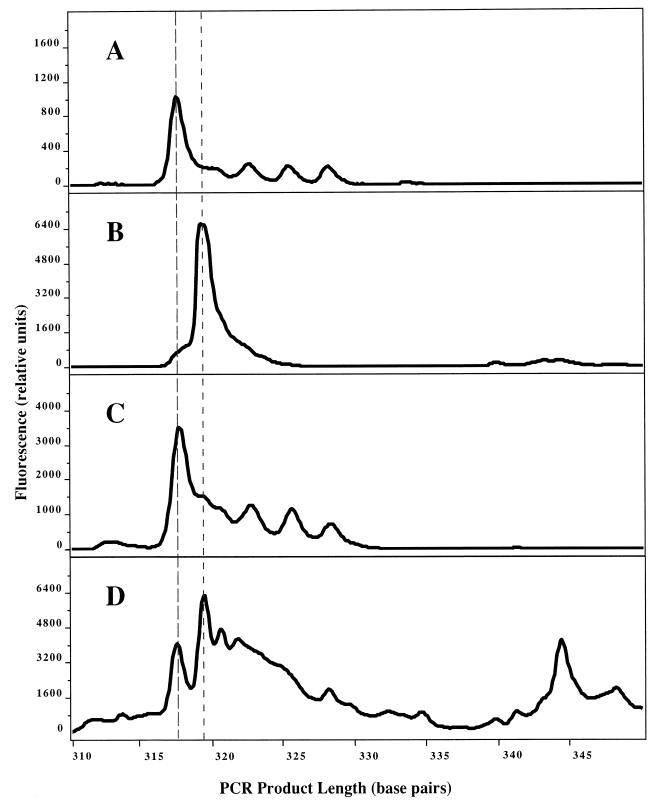
Paramagnetic-bead purification of BrdU-containing DNA was used to distinguish a taxonomic subset of bacteria incorporating BrdU in the Cronemiller Lake community (Fig. 5). LH-PCR of unfractionated DNA from both BrdU-labeled and unlabeled communities revealed a taxonomically complex community structure dominated by bacteria with an LH-PCR fragment length of 317 nucleotides. In contrast, BrdU-containing DNA purified from the labeled community contained a different spectrum of taxa, dominated by bacteria with a fragment length of 319 nucleotides. DNA from control preparations of immunochemically treated, unlabeled lake water DNA gave low yields of PCR amplification products (ca. 10% of the amount obtained from BrdU-containing DNA [data not shown]) which nonetheless exhibited a complex LH-PCR pattern. Actively growing bacteria incorporating BrdU can therefore be discriminated from the majority of bacterioplankton in a natural community, which may not be synthesizing DNA.
FIG. 5.
LH-PCR analysis of active microbial taxa in Cronemiller Lake. (A) BrdU-labeled lake water DNA. (B) Active fraction isolated from labeled lake water DNA by using anti-BrdU monoclonal antibodies and paramagnetic beads. (C) Control (unlabeled lake water DNA). (D) Control fraction isolated from unlabeled lake water DNA by using anti-BrdU monoclonal antibodies and paramagnetic beads. Dashed lines indicate the positions of the major peaks at 317 and 319 nucleotides.
DISCUSSION
Immunochemical and immunocytochemical manipulation of microbial DNA labeled with BrdU is a new, nonradioactive technology which can be used to link molecular methods of community structure analysis to biogeochemical measurements of community productivity. In this report, we have demonstrated methods to (i) detect BrdU incorporated into DNA during pulse-labeling experiments, (ii) distinguish BrdU-incorporating cells from unlabeled cells, and (iii) purify BrdU-containing DNA from unlabeled DNA for molecular genetic analysis. In addition, we have demonstrated the ability of bacterial cultures and a natural microbial community to incorporate BrdU into newly synthesized DNA, and we have discriminated the active, BrdU-incorporating taxa in a lake water microbial community. Application of these techniques in conjunction with higher-resolution methods of population analysis, such as denaturing gradient gel electrophoresis, cloning and sequencing, or hybridization to group-specific oligonucleotide probes, has the potential to resolve debates over the level of metabolic activity of currently unculturable taxa in their natural environment (4). Also, BrdU techniques may be useful for studying the mechanics of microbial community dynamics: actively growing bacteria identified by BrdU incorporation may either be increasing their representation in the community or be subject to above-average mortality. Most importantly, future experiments with BrdU-labeled microbial communities will provide an understanding of which microbial taxa are responsible for TdR uptake in biogeochemical studies of complex ecosystems.
The utility of BrdU incorporation studies will be subject to limitations similar to those that apply to TdR incorporation. Both techniques are limited to the analysis of taxa capable of incorporating exogenously provided nucleotide precursors into their DNA (14, 27). In this regard, it is important to note that the two strains which incorporated BrdU in our experimental cultures are representatives of the α- and γ-proteobacterial groups, which dominate the marine bacterioplankton populations (24). It is also believed that flavobacteria and gram-positive bacteria, which did not incorporate BrdU in our experiments, do not predominate in marine or freshwater systems (21). Additional culture studies are required to better characterize the capacity for BrdU incorporation among diverse taxa and to provide comparisons to incorporation of [3H]TdR. The lake water community experiment demonstrated BrdU incorporation by microbial populations in a natural setting.
In contrast to TdR incorporation experiments, it will be possible to identify taxa incorporating label in BrdU field studies. This will be useful for demonstrating the applicability of this method to particular environmental situations. Manipulation of environmental conditions by means of nutrient amendment or other methods may be used in conjunction with BrdU labeling to demonstrate that apparently inactive taxa are nonetheless physiologically capable of incorporating BrdU. In such situations, nucleotide precursor uptake studies will accurately reflect the activity of the microbial community.
Our observation that some bacteria fail to assimilate BrdU shows that results from natural ecosystems, such as those presented in Fig. 5, must be interpreted with caution. While BrdU incorporation can be used to prove that specific populations of bacteria in a natural ecosystem are growing, this method cannot be used conversely to prove that a population is not growing, unless it is also demonstrated that the species in question can assimilate BrdU. Similar caveats apply to thymidine assimilation, which is nonetheless used widely to provide estimates of net production by bacterial communities.
ACKNOWLEDGMENTS
We thank B. Lanoil, C. Mathews, E. Sherr, B. Sherr, and A. Vella for helpful discussions; the McDonald-Dunn Research Forest personnel for permission to sample Cronemiller Lake; and L. Young for assistance with bacterial cultures.
This work was supported by National Science Foundation grant DEB9709012.
REFERENCES
- 1.Ausubel F A, Brent R, Kingston R E, Moore D D, Smith J A, Seidman J G, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley and Sons; 1988. [Google Scholar]
- 2.Bard D G, Ward B B. A species-specific bacterial productivity method using immunomagnetic separation and radiotracer experiments. J Microbiol Methods. 1997;28:207–219. [Google Scholar]
- 3.Brock T D. Bacterial growth rate in the sea: direct analysis by thymidine autoradiography. Science. 1967;155:81–83. doi: 10.1126/science.155.3758.81. [DOI] [PubMed] [Google Scholar]
- 4.Choi J W, Sherr E B, Sherr B F. Relation between presence-absence of a visible nucleoid and metabolic activity in bacterioplankton cells. Limnol Oceanogr. 1996;41:1161–1168. [Google Scholar]
- 5.Coote J G, Binnie C. Tolerance to bromodeoxyuridine in a thymidine-requiring strain of Bacillus subtilis. J Gen Microbiol. 1986;132:481–492. doi: 10.1099/00221287-132-2-481. [DOI] [PubMed] [Google Scholar]
- 6.DeLong E F, Wickham G S, Pace N R. Phylogenetic stains: ribosomal RNA-based probes for the identification of single cells. Science. 1989;243:1360–1363. doi: 10.1126/science.2466341. [DOI] [PubMed] [Google Scholar]
- 7.Dolbeare F. Bromodeoxyuridine: a diagnostic tool in biology and medicine, part I: Historical perspectives, histochemical methods and cell kinetics. Histochem J. 1995;27:339–369. [PubMed] [Google Scholar]
- 8.Fuhrman J A, Azam F. Bacterioplankton secondary production estimates for coastal waters of British Columbia, Antarctica, and California. Appl Environ Microbiol. 1980;39:1085–1095. doi: 10.1128/aem.39.6.1085-1095.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fuhrman J A, Azam F. Thymidine incorporation as a measure of heterotrophic bacterioplankton production in marine surface waters: evaluation and field results. Mar Biol. 1982;66:109–120. [Google Scholar]
- 10.Giovannoni S J, Britschgi T B, Moyer C L, Field K G. Genetic diversity in Sargasso Sea bacterioplankton. Nature. 1990;345:60–62. doi: 10.1038/345060a0. [DOI] [PubMed] [Google Scholar]
- 11.Gordon D A, Giovannoni S J. Detection of stratified microbial populations related to Chlorobium and Fibrobacter species in the Atlantic and Pacific oceans. Appl Environ Microbiol. 1996;62:1171–1177. doi: 10.1128/aem.62.4.1171-1177.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hewitt R, Suit J C, Billen D. Utilization of 5-bromouracil by thymineless bacteria. J Bacteriol. 1967;93:86–89. doi: 10.1128/jb.93.1.86-89.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hicks R E, Amann R I, Stahl D A. Dual staining of natural bacterioplankton with 4′,6-diamidino-2-phenylindole and fluorescent oligonucleotide probes targeting kingdom-level 16S rRNA sequences. Appl Environ Microbiol. 1992;58:2158–2163. doi: 10.1128/aem.58.7.2158-2163.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jeffrey W H, Paul J H. Thymidine uptake, thymidine incorporation, and thymidine kinase activity in marine bacterium isolates. Appl Environ Microbiol. 1990;56:1367–1372. doi: 10.1128/aem.56.5.1367-1372.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Karner M, Fuhrman J A. Determination of active marine bacterioplankton: a comparison of universal 16S rRNA probes, autoradiography, and nucleoid staining. Appl Environ Microbiol. 1997;63:1208–1213. doi: 10.1128/aem.63.4.1208-1213.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kerkhof L, Ward B B. Comparison of nucleic acid hybridization and fluorometry for measurement of the relationship between RNA/DNA ratio and growth rate in a marine bacterium. Appl Environ Microbiol. 1993;59:1303–1309. doi: 10.1128/aem.59.5.1303-1309.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kogura K, Simidu U. A tentative direct microscopic method for counting living marine bacteria. Can J Microbiol. 1979;25:415–420. doi: 10.1139/m79-063. [DOI] [PubMed] [Google Scholar]
- 18.Lanoil B D, Giovannoni S J. Identification of bacterial cells by chromosomal painting. Appl Environ Microbiol. 1997;63:1118–1123. doi: 10.1128/aem.63.3.1118-1123.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lark K G. Regulation of chromosome replication and segregation in bacteria. Bacteriol Rev. 1966;30:3–32. doi: 10.1128/br.30.1.3-32.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meselson M, Stahl F W. The replication of DNA in Escherichia coli. Proc Natl Acad Sci USA. 1958;44:671–682. doi: 10.1073/pnas.44.7.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Methé B A, Hiorns W D, Zehr J P. Contrasts between marine and freshwater bacterial community composition: analyses of communities in Lake George and six other Adirondack lakes. Limnol Oceanogr. 1998;43:368–374. [Google Scholar]
- 22.Meyer-Reil L-A. Autoradiography and epifluorescence microscopy combined for the determination of number and spectrum of actively metabolizing bacteria in natural waters. Appl Environ Microbiol. 1978;36:506–512. doi: 10.1128/aem.36.3.506-512.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moriarty D J W. Measurement of bacterial growth rates in aquatic systems from rates of nucleic acid synthesis. Adv Microb Ecol. 1986;9:246–292. [Google Scholar]
- 24.Mullins T D, Britschgi T B, Krest R L, Giovannoni S J. Genetic comparisons reveal the same unknown bacterial lineages in Atlantic and Pacific bacterioplankton communities. Limnol Oceanogr. 1995;40:148–158. [Google Scholar]
- 25.Pace N R. A molecular view of microbial diversity and the biosphere. Science. 1997;276:734–740. doi: 10.1126/science.276.5313.734. [DOI] [PubMed] [Google Scholar]
- 26.Pitcher D G, Saunders N A, Owen R J. Rapid extraction of bacterial genomic DNA with guanidium thiocyanate. Lett Appl Microbiol. 1989;8:151–156. [Google Scholar]
- 27.Pollard P C, Moriarty D J W. Validity of the tritiated thymidine method for estimating bacterial growth rates: measurement of isotope dilution during DNA synthesis. Appl Environ Microbiol. 1984;48:1076–1083. doi: 10.1128/aem.48.6.1076-1083.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rodriguez G G, Phipps D, Ishiguro K, Ridgway H F. Use of a fluorescent redox probe for direct visualization of actively respiring bacteria. Appl Environ Microbiol. 1992;58:1801–1808. doi: 10.1128/aem.58.6.1801-1808.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Suzuki M T, Giovannoni S J. Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl Environ Microbiol. 1996;62:625–630. doi: 10.1128/aem.62.2.625-630.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Suzuki M T, Rappé M S, Haimberger Z W, Winfield H, Adair N, Ströbel J, Giovannoni S J. Bacterial diversity among small-subunit rRNA gene clones and cellular isolates from the same seawater sample. Appl Environ Microbiol. 1997;63:983–989. doi: 10.1128/aem.63.3.983-989.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weygand F, Wacker A, Dellweg H. Stoffwechseluntersuchungen bei Mikroorganism mit Hilfe radioaktiver Isotope. II. Kompetitive und nicht-kompetitive Enthemmung von 5-82Br-Uracil. Z Naturforsch. 1952;7b:19–25. [Google Scholar]
- 32.Zweifel U L, Hagström A. Total counts of marine bacteria include a large fraction of non-nucleoid-containing bacteria (ghosts) Appl Environ Microbiol. 1995;61:2180–2185. doi: 10.1128/aem.61.6.2180-2185.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]