Abstract
The nitrogenase enzyme complex of Methanosarcina barkeri 227 was found to be more sensitive to NaCl than previously studied molybdenum nitrogenases are, with total inhibition of activity occurring at 190 mM NaCl, compared with >600 mM NaCl for Azotobacter vinelandii and Clostridium pasteurianum nitrogenases. Na+ and K+ had equivalent effects, whereas Mg2+ was more inhibitory than either monovalent cation, even on a per-charge basis. The anion Cl− was more inhibitory than acetate was. Because M. barkeri 227 is a facultative halophile, we examined the effects of external salt on growth and diazotrophy and found that inhibition of growth was not greater with N2 than with NH4+. Cells grown with N2 and cells grown with NH4+ produced equal concentrations of α-glutamate at low salt concentrations and equal concentrations of Nɛ-acetyl-β-lysine at NaCl concentrations greater than 500 mM. Despite the high energetic cost of fixing nitrogen for these osmolytes, we obtained no evidence that there is a shift towards nonnitrogenous osmolytes during diazotrophic growth. In vitro nitrogenase enzyme assays showed that at a low concentration (approximately 100 mM) potassium glutamate enhanced activity but at higher concentrations this compound inhibited activity; 50% inhibition occurred at a potassium glutamate concentration of approximately 400 mM.
Methanosarcina spp. have wide halotolerance ranges and have been found in osmotically diverse environments, such as freshwater sediments, sewage digestors, and seawater (2, 14). Strains which were originally considered to be freshwater have been found to adapt to more saline conditions, often losing their heteropolysaccharide (methanocondroitin) sacculus, thereby shifting from growth as clumps to growth as individual cells with an S-layer cell wall (35, 36). The strain used in this study, Methanosarcina barkeri 227, has been shown in physiological and biochemical studies to be a halotolerant organism, although most studies of its physiology and biochemistry have been conducted in nonmarine media (19, 20, 22, 23, 36).
Organisms adapt to the osmotic stress caused by high extracellular salt concentrations by producing increased levels of intracellular solutes called osmolytes, which are solutes that are more compatible with intracellular enzyme function than sodium chloride is. Methanosarcina spp. have been shown to accumulate primarily potassium α-glutamate at extracellular salt concentrations up to 500 mM and the unusual zwitterionic amino acid derivative Nɛ-acetyl-β-lysine at higher salt concentrations (30, 38). When the organisms are grown with yeast extract, glycine-betaine derived from the yeast extract is also found (31).
Methanosarcina spp. have been shown to be capable of nitrogen fixation (3, 22) (i.e., conversion of N2 to NH3). M. barkeri 227 has been shown to contain a typical two-component nitrogenase complex (20), although this complex has a low specific activity, and the genes encoding the nitrogenase components are part of a phylogenetic cluster which includes nitrogenase genes from Clostridium pasteurianum (7, 8, 33). Nitrogenases usually consist of two protein components, neither of which is individually active; the MoFe protein (dinitrogen reductase; component 1), an α2β2 tetramer (encoded by nifD and nifK), is the site of N2 binding and reduction, and the Fe protein (dinitrogen reductase; component 2) is a homodimer (encoded by nifH) which catalyzes the hydrolysis of ATP. The two components bind to one another to form a tight complex in which electrons are transferred from the Fe protein to the MoFe protein, after which the two proteins dissociate (27). Nitrogenase complex formation has been demonstrated to be a critical step in nitrogen reduction. Failure to form an active complex or failure to dissociate leads to a cessation of activity (10, 11). Salts have been shown to interfere with the ionic interactions involved in complex formation and, therefore, inhibit activity (5).
External salts have been shown to be inhibitory to nitrogen fixation by cells in some organisms, while other organisms have mechanisms to adapt to external salt concentrations. In this study, we examined the interactions between osmoregulation and nitrogen fixation in M. barkeri 227. We demonstrated that the M. barkeri nitrogenase is particularly sensitive to salt inhibition, whereas cells can grow diazotrophically in the presence of high external salt concentrations. In this paper we describe the effects of salt on diazotrophy in vivo and in vitro of various cations and anions. We also examined osmolyte accumulation in both diazotrophic and ammonium-grown cells to determine whether there was a shift towards nonnitrogenous osmolytes under diazotrophic conditions.
MATERIALS AND METHODS
Strains and growth conditions.
M. barkeri 227 (= ATCC 43241 = DSM 1538 = OCM 35) cultures were grown either in the disaggregating marine medium described by Sowers et al. (35) in the presence of 0 to 1.2 M NaCl or in the basal salts medium of and under the conditions described by Chien and Zinder (7, 35). Cysteine-HCl and vitamins, however, were omitted from the marine media (35). The basal medium was prepared anaerobically, and 50-ml portions were dispensed into a series of 126-ml bottles by the method of Lobo and Zinder (20) under an N2-CO2 (70:30) atmosphere (19). After autoclaving, each bottle was supplemented with 0.1 ml of 100% methanol, 0.1 ml of 20% (wt/vol) Na2S, and 0.1 ml of 10% (wt/vol) NaHCO3. Some bottles also received 0.05 ml of 5 M NH4Cl. All bottles were inoculated with 1 ml of M. barkeri grown in ammonia-free marine medium containing the relevant NaCl concentration and were incubated with agitation (200 rpm) at 37°C. Growth was determined principally by measuring methane production with a thermal conductivity gas chromatograph as described by Lobo and Zinder (19). Cells were harvested in the mid-log phase (when approximately 30 to 40 mmol of CH4 per liter was produced) for nitrogenase assays and osmolyte determinations.
Clostridium pasteurianum W5 was obtained from J. Chen, Virginia Polytechnic Institute and State University, and was grown anaerobically in the basal salts medium described by Lobo and Zinder except that 20 mM sucrose and 1 μg of d-biotin per liter were added (19). Azotobacter vinelandii CA was kindly provided by Paul Bishop (North Carolina State University) and was grown at 30°C under aerobic diazotrophic conditions in modified Burk medium supplemented with sucrose (20 g liter) and 0.8 mM NH4Cl (39). A. vinelandii and C. pasteurianum were harvested at an optical density at 660 nm of approximately 0.8.
Protein content determination.
Protein concentrations in cell extracts were determined by using the standard Bio-Rad (Richmond, Calif.) protein assay. A standard curve was constructed by using bovine serum albumin.
Osmolyte analysis.
The cells (10 ml) used for osmolyte analysis were harvested and washed by centrifugation at 15,000 × g for 20 min, resuspended in 75% ethanol–H2O, and lysed by using glass beads as described by Murray and Zinder (24). After five 1-min rounds of high-speed vortexing, more than 90% of the cells had lysed, as determined by light microscopy and protein content determination. Cell debris and glass particles were removed by microcentrifuging samples for approximately 5 min. The supernatant extract was then collected and evaporated to dryness. The dried solids were subsequently dissolved in high-performance liquid chromatography (HPLC) grade water and analyzed by thin-layer chromatography (TLC) or HPLC to determine the amino acid content.
A TLC analysis of intracellular carbohydrates was carried out by using a mobile phase consisting of n-propanol, ethyl acetate, H2O, and a 25% ammonia solution (50:10:30:10). Sugars were visualized by spraying the dried plate with p-anisaldehyde–H2SO4–ethanol–glacial acetic acid (3:3:54:0.6) and heating it at 100°C for approximately 5 min. A TLC analysis of amino acids was carried out by using a mobile phase consisting of n-butanol, acetic acid and H2O (60:20:20). After the plate was dried, the amino acids were visualized by spraying the plate with 0.3% (wt/vol) ninhydrin in acetone-glacial acetic acid (97:3). A gradient reverse-phase HPLC analysis of cell extracts was carried out by using a Bio-Rad BIO-SIL ODS-5S column after precolumn o-phthaldialdehyde derivatization with mobile phases consisting of (i) methanol, tetrahydrofuran, and sodium acetate (pH 7.6) (2:2:96) and (ii) methanol and water (80:20). M. Roberts kindly provided Nɛ-acetyl-β-lysine, which was used as a standard.
Nitrogenase assays.
The cell extracts used for nitrogenase assays were prepared from anaerobically harvested M. barkeri and C. pasteurianum cultures by lysing diazotrophic cells anaerobically with a French press as previously described by Lobo and Zinder (20). A. vinelandii cells were harvested aerobically but were lysed anaerobically with a French press. Nitrogenase activity was determined by the ethyne reduction assay in sealed, argon-flushed, 11-ml serum vials. A 1-ml portion of ethyne, produced by the method described by Lobo and Zinder (19), was added to each vial along with 0.5 ml of the reaction mixture and 0.25 ml of a 5 mM sodium dithionite solution flushed with argon (19). The argon-flushed reaction mixture contained 25 mM HEPES (pH 7.5), 10 mM ATP, 10 mM MgCl2, 50 mM creatine phosphate, and 100 μg of creatine kinase per ml. After the vials containing the reaction mixture had been incubated for 30 min at 30°C, approximately 1 mg of M. barkeri extract was added to each vial to initiate the assay, and the final aqueous volume was 1.0 ml. Salt solutions, generated by dissolving the relevant salts in 25 mM HEPES (pH 7.5) buffer and correcting the pH, were also flushed with argon and added to the vials with the reaction buffer at the appropriate concentrations. Potassium glutamate (pH 7.5) was prepared by dissolving free glutamic acid in HPLC grade water, adjusting the pH to 7.5 with KOH, and then flushing this solution with argon.
RESULTS
Effect of sodium chloride on nitrogenase activity in vitro.
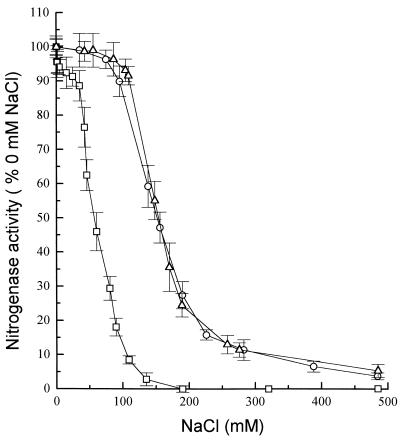
Nitrogenase complex formation has been demonstrated to be a critical step in nitrogen reduction, and salts have been shown to interfere with the ionic interactions involved in nitrogenase complex formation and thus inhibit activity (5, 9, 27). As part of an ongoing study of the physiology and biochemistry of diazotrophy in M. barkeri 227, we examined the effects of salts on diazotrophic activity in vitro. The M. barkeri nitrogenase was much more sensitive to NaCl inhibition than the two other eubacterial nitrogenases tested were. Total inhibition of the M. barkeri nitrogenase occurred in the presence of 190 mM NaCl, compared to >600 mM NaCl for the C. pasteurianum and A. vinelandii enzymes (Fig. 1); the latter results are similar to results obtained in previous studies (5, 9, 27). The inhibition curves in all cases were sigmoidal. It should be noted that the concentration of NaCl that completely inhibited nitrogenase activity (190 mM) was fivefold lower than the NaCl concentration required to inhibit the growth of diazotrophic cultures.
FIG. 1.
Effect of NaCl on ethyne reduction rates as a measure of nitrogenase activity in extracts of M. barkeri, A. vinelandii, and C. pasteurianum. The results are means of values from three separate assays, and standard errors are indicated by bars. Symbols: □, M. barkeri 227; ▵, A. vinelandii; ○, C. pasteurianum. A 100% level of activity corresponded to 42.7, 1.1, and 49.7 nmol min−1 mg of protein−1 for A. vinelandii, M. barkeri 227, and C. pasteurianum, respectively.
Effect of NaCl concentration on M. barkeri 227 growth when N2 or NH4+ was the nitrogen source.
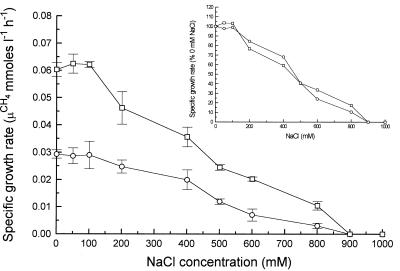
In light of the sensitivity of the M. barkeri 227 nitrogenase complex to salt inhibition, we examined whether this organism was capable of growing diazotrophically in the presence of high salt concentrations. Cultures of M. barkeri 227 which had previously been grown on low-osmotic-strength medium were adapted to grow in media containing a range of NaCl concentrations over a period of months by transferring cultures sequentially to media with higher salt concentrations once adequate growth was attained (i.e., once the amount of CH4 produced was >30 mmol per liter of culture). Like Sowers and Gunsalus (37), we observed that there was a lag in growth following inoculation of a culture into a higher-osmolarity medium but that a lag was not observed after subsequent transfers (37). Using cultures adapted in this way to different salt concentrations, we examined the effects of salt concentrations on growth rates, as measured by the exponential increase in methanogenesis, which has been shown to parallel growth in previous studies (19).
Diazotrophic cultures of M. barkeri 227 doubled approximately every 24 h in medium containing no NaCl, whereas ammonium-grown cultures doubled every 12 h (Fig. 2), which is consistent with previous results (19). The lower growth rate of the diazotrophic cells indicates that a large amount of energy was directed toward nitrogen fixation instead of growth. Adding up to 100 mM NaCl to the medium had little effect on the growth rate under both diazotrophic and ammonium-grown conditions. In the presence of NaCl concentrations greater than 100 mM, the growth rate under both conditions decreased, and neither culture was able to grow in the presence of NaCl concentrations greater than 800 mM. A comparison of the growth rates of diazotrophic and ammonium-grown cells in the presence of 0 to 800 mM NaCl (Fig. 2, insert) showed that these growth rates were roughly proportional to each other, indicating that NaCl did not inhibit nitrogen fixation more than it inhibited other cellular processes.
FIG. 2.
Effect of NaCl on growth rates of M. barkeri 227 grown in marine medium containing 50 mM methanol as the sole carbon source and either ammonium (□) or N2 (○) as the sole nitrogen source. The results are means of values for three separate cultures, and standard errors are indicated by bars. The cultures used as inocula were initially adapted to 0.8 M NaCl and then to the concentration used.
Effects of various anions and cations on nitrogenase activity in vitro.
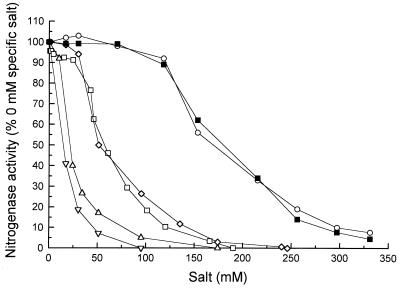
We examined the inhibition of M. barkeri nitrogenase by salts containing different cations and anions. All of the salts tested inhibited M. barkeri 227 nitrogenase and produced a characteristic sigmoidal inhibition curve. A comparison of the anions added with the monovalent cations Na+ and K+ clearly demonstrated that chloride was much more inhibitory to nitrogenase activity than acetate was (Fig. 3). A comparison of K+ and Na+ showed that the inhibitory activities of these ions were equivalent, whether chloride or acetate salts were used. Magnesium chloride was the most inhibitory chloride salt on a molar basis, but this compound is divalent, contains two chloride ions, and thus has a net higher salt concentration and greater total ionic strength than the other compounds tested. However, a comparison of the cations when acetate was the anion showed that magnesium diacetate was sixfold more inhibitory than either potassium acetate or sodium acetate. The difference between chloride inhibition and acetate inhibition when magnesium was the cation was not as marked, indicating that magnesium ions were the major inhibitor of nitrogenase activity in these salts.
FIG. 3.
Effects of various cations and anions on ethyne reduction rates as a measure of nitrogenase activity in cell extracts of M. barkeri 227. The results are means of values from three separate assays, and the standard deviations were less than 10%. Symbols: ○, potassium acetate; ■, sodium acetate; □, NaCl; ◊, KCl; ▵, magnesium acetate; ▿, MgCl2. A 100% level of activity corresponded to 52.4 nmol h−1 mg of protein−1.
Osmolytes and osmotic regulation of M. barkeri 227 under diazotrophic and ammonium-grown conditions.
We examined the internal solutes (osmolytes) present in M. barkeri cells grown in media supplemented with different amounts of NaCl. Analysis of the amino acids present in ammonium-grown cells (Table 1) revealed a nearly 10-fold increase in the level of α-glutamate in cells grown in medium supplemented with 500 mM NaCl compared to the level in cells grown without added NaCl, while cells grown in the presence of 800 mM NaCl showed no further increase in the level of α-glutamate. No Nɛ-acetyl-β-lysine was detected in cells grown in medium to which NaCl was not added, low amounts of this compound were detected in cells grown in the presence of 500 mM NaCl, and the amount of Nɛ-acetyl-β-lysine was nearly equal to the amount of α-glutamate in cells grown in the presence of 800 mM NaCl. There were also slight increases in the amounts of alanine and valine in the presence of elevated NaCl concentrations. These results are in agreement with other results obtained for Methanosarcina spp. (16, 30, 37, 38).
TABLE 1.
Determination of major osmolytes in 75% ethanol extracts of M. barkeri 227 cells grown in the presence of 0, 500, or 800 mM NaCl
| NaCl concn in medium (mM) | Nitrogen source | Intracellular concn (nmol mg of protein−1) ofa:
|
||||
|---|---|---|---|---|---|---|
| α-Glutamate | Nɛ-Acetyl-β-lysine | Alanine | Valine | Glycine | ||
| 0 | N2 | 205.7 (19.2) | 0.0 (0.0) | 67.8 (4.5) | 12.1 (2.4) | 18.3 (3.2) |
| NH4+ | 197.8 (18.2) | 0.0 (0.0) | 68.9 (5.9) | 30.6 (7.6) | 19.9 (1.2) | |
| 500 | N2 | 1,763 (125.2) | 189.3 (27.6) | 117.8 (8.9) | 126.6 (15.6) | 55.2 (4.6) |
| NH4+ | 1,933 (108.7) | 204.6 (14.6) | 60.9 (5.6) | 35.3 (3.8) | 51.0 (4.2) | |
| 800 | N2 | 1,875 (105.7) | 2,004 (167.9) | 142.3 (12.5) | 154.2 (12.4) | 61.2 (2.5) |
| NH4+ | 1,883 (156.8) | 2,103 (98.9) | 104.6 (4.5) | 108.9 (11.6) | 63.2 (3.9) | |
The values are means (standard errors) based on three replicates.
Examination of diazotrophically grown cells showed that the levels of α-glutamate and Nɛ-acetyl-β-lysine that were present were essentially identical to the levels in ammonium-grown cells (Table 1). Other amino acids were present at much lower concentrations in high-salt medium (Table 1), and neither β-glutamate nor β-glutamine, osmolytes detected in other methanogenic archaea (18, 29, 30), was detected in any of the cell extracts from either culture. Examination of cell extracts by TLC did not reveal any detectable carbohydrate osmolytes, such as trehalose, under any growth conditions.
Effect of potassium glutamate on nitrogenase activity.
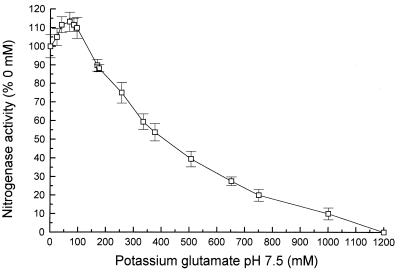
Since α-glutamate is apparently the major osmolyte in M. barkeri cells grown in the presence of NaCl concentrations up to 500 mM and since previous work has shown that potassium ions are used as counterions to glutamate in a number of archaea when they are grown under osmotic stress conditions (18, 37), we examined the effects of potassium α-glutamate on M. barkeri nitrogenase in vitro (Fig. 4). Addition of 100 to 200 mM potassium glutamate led to an initial increase in activity. At higher concentrations, the activity decreased, and complete inhibition occurred at a potassium glutamate concentration of approximately 1.2 M.
FIG. 4.
Effect of potassium glutamate on ethyne reduction rates as a measure of nitrogenase activity in cell extracts of M. barkeri 227. The results are means of values from three separate assays, standard errors are indicated by bars. A 100% level of activity corresponded to 60.4 nmol h−1 mg of protein−1.
DISCUSSION
In this study, M. barkeri 227 was moderately halotolerant; it was able to grow at salt concentrations up to 0.8 M. It never made the transition to being halophilic as described by Sowers and Gunsalus (37) for M. barkeri 227 and other Methanosarcina spp., who found that optimal growth occurred in the presence of NaCl concentrations near 0.4 M and growth occurred in medium containing up to 1.2 M NaCl. Attempts to effect this transition by duplicating the growth conditions and media used by Sowers and Gunsalus (37), including adding trimethylamine (1), repeatedly failed. A culture of NaCl-adapted M. barkeri 227, kindly provided by K. Sowers, grew optimally in the presence of 0.4 M NaCl in our growth medium and was able to grow in the presence of NaCl concentrations up to 1.2 M as previously described (data not shown). Microscopic observations of the cultures grown in the presence of 0.4 M NaCl showed that the M. barkeri 227 culture used by Sowers and Gunsalus (37) formed essentially single cells, while our culture formed small clumps, indicating that the cells maintained a methanocondroitin outer layer. The culture of M. barkeri 227 in our laboratory has been transferred repeatedly in nonmarine medium for more than 14 years, and it is possible that some mutation has occurred which does not allow the organism to make the transition to a more halophilic state.
M. barkeri 227 appeared to be equally halotolerant whether the nitrogen source present in the medium was N2 or ammonium. Few studies have been carried out to determine the effect of external salt on the growth rate with different nitrogen sources. Diazotrophic growth of Rhodobacter capsulatus was inhibited by NaCl concentrations of more than 0.1 M, while other nitrogen sources (with the exception of nitrate) allowed growth under these conditions (13). Osmoregulation in the diazotrophic bacteria Azotobacter sp. and Klebsiella pneumoniae has been shown to involve first accumulation of α-glutamate and potassium ions and then production of proline and trehalose; however, in this study the cultures were not studied under diazotrophic conditions and were not assayed for nitrogenase activity (21). In a study of the salt-tolerant organism Rhizobium leguminosarum biovar viciae C1204b, Chien et al. (6) demonstrated that there was an increase in the intracellular glutamate concentration in response to salt stress under diazotrophic conditions.
The nitrogenase complex of M. barkeri was readily inhibited by relatively low salt concentrations in vitro. Salt inhibition studies have been utilized in the past to gain insight into nitrogenase component interactions and complex formation in a wide variety of free-living diazotrophs (9). According to the model of Deits and Howard (9), NaCl inhibits nitrogenase activity in two ways. First, NaCl reduces the affinity of the Fe protein for Mg-ATP. Second, it conceals the charged residues involved in component interactions, thereby inhibiting the formation of the iron protein-molybdenum iron protein complex. A sigmoidal inhibition curve, such as the curves obtained for salt inhibition of M. barkeri 227 nitrogenase, indicates that such inhibition occurs and suggests that there are multiple sites of interaction.
In our study, complete nitrogenase inhibition occurred at NaCl concentrations of <200 mM, which are much lower than the NaCl concentrations that inhibit the molybdenum nitrogenases of A. vinelandii and K. pneumoniae (typically 600 mM NaCl) (5, 9) and the molybdenum nitrogenase of C. pasteurianum in our studies. The sigmoidal inhibition curves obtained for salts indicate that the two nitrogenase components of M. barkeri 227 interact in a cooperative binding mechanism which involves a number of charged amino acid residues on the surfaces of the two components. The inhibition by low salt concentrations suggests that the M. barkeri 227 nitrogenase components do not have as high an affinity for one another as the nitrogenase components of other organisms have, which may be a manifestation of the low specific activities which we have observed for M. barkeri 227 (19).
Our study also demonstrated that certain ions are much more inhibitory to in vitro nitrogenase activity in M. barkeri 227 than other ions are. Chloride was the most inhibitory anion. This inhibitory effect is thought to be due to masking of the surface charge, particularly the arginine residues clearly demonstrated in Azotobacter strains to be important in component interactions (9, 41). Na+ and K+ inhibited M. barkeri nitrogenase equally, whereas Mg2+ was the most inhibitory cation, as it was also for A. vinelandii, K. pneumoniae, and Rhodospirillum rubrum nitrogenases (9, 32, 40). It has been suggested that Mg2+ has two modes of inhibitory activity. It may reduce the affinity of component 2 for Mg-ATP, and it may interfere with component interactions; the latter is considered more important (5, 9). From our data it appears that salt inhibition is directly related to the total charge of the ion and its relative charge density. Interestingly, the least inhibitory salt tested was potassium glutamate, one of the primary osmolytes in M. barkeri.
Like Sowers et al. (37, 38), we found that the predominant osmolyte in M. barkeri 227 cells growing in medium containing 500 mM NaCl was α-glutamate; in the studies of Sowers et al. α-glutamate had K+ as a counterion, while at higher external salt concentrations the zwitterion Nɛ-acetyl-β-lysine became significant. Our enzyme studies showed that potassium α-glutamate at a concentration of 400 mM, approximately the maximum concentration found in the cells (38), caused approximately 50% inhibition of M. barkeri 227 nitrogenase in vitro. A large number of organisms produce potassium glutamate as the initial response to osmotic stress but employ zwitterionic or neutral osmoprotectants (Nɛ-acetyl-β-lysine, proline, glycine-betaine, or trehalose) at high osmotic strengths to minimize the cation concentration. Unfortunately, not enough pure Nɛ-acetyl-β-lysine was available to test its effect on M. barkeri 227 nitrogenase.
We found that the levels of α-glutamate and Nɛ-acetyl-β-lysine, which contain one and two atoms of nitrogen per molecule, respectively, were essentially identical in diazotrophic and ammonium-grown cells, and we failed to detect osmolytes lacking N, such as trehalose. These findings are of interest since nitrogen fixation is an energetically costly process for a cell, thought to require 8 to 16 mol of ATP per mol of N2 fixed (28). This is clearly the case for M. barkeri 227, since diazotrophic growth caused significant reductions in the cell growth rate (Fig. 1) (19) and the cell yield (19). The nitrogen-free disaccharide trehalose has been detected in the archaeon Sulfolobus sulfataricus as an osmolyte. Studies of the diazotrophic phototroph Ectothiorhodospira halochloris also failed to show any switch from nitrogen-containing osmolytes to trehalose under nitrogen limitation conditions (12). Calculations assuming that about 10% of the M. barkeri 227 cell dry weight is N (22) and that the cells contain approximately 70% water (25) indicated that the N content of the cells should increase approximately 13% when the culture contains 0.4 M potassium α-glutamate and 26% when the culture contains 0.4 M Nɛ-acetyl-β-lysine, so the effects of producing these osmolytes on the cellular N budget should be modest in the presence of all but the highest salt concentrations. In this study we also failed to detect any other osmolytes, such as β-glutamate, which is a common osmolyte in marine methanogens (38). It has been suggested that beta amino acids, such as Nɛ-acetyl-β-lysine and β-glutamate, are good osmolytes as they are not substrates for α-amino acid-utilizing enzymes and presumably do not interfere with cellular metabolism (30, 38).
Here we demonstrated that under osmotic stress conditions M. barkeri 227 fixes nitrogen in the presence of high levels of free cytosolic amino acids, particularly α-glutamate. Glutamate and its related metabolites α-ketoglutarate and glutamine have been demonstrated to play important roles in regulation of gene expression and enzyme activity in diverse eubacteria. Little is known about nitrogen state regulation in the archaea, but homologues of genes encoding some nitrogen state-sensing proteins have been found in the genome sequences of members of the Euryarchaeota (4, 17, 34). One such gene is glnB, encoding the PII protein, which is part of a regulatory network that senses α-ketoglutarate and glutamine (15, 26). It therefore appears that sensing of the nitrogen state within M. barkeri is not related to the free amino acid pool or, more specifically, α-glutamate. Work is under way to examine these compounds and to relate them to nitrogenase activity and gene expression.
ACKNOWLEDGMENTS
We thank M. Roberts for providing a cellular extract of Methanococcus thermolithotrophicus to aid in our osmolyte studies and K. Sowers for providing an NaCl-adapted culture of M. barkeri 227. We thank Bob Sherwood for help in identifying the amino acids discussed in this paper.
This research was supported by grant DE-FG02-85ER13370 from the U.S. Department of Energy.
REFERENCES
- 1.Ahring B K, Alatriste-Mondragon F, Westermann P, Mah R A. Effects of cations on Methanosarcina thermophila TM-1 growing in moderate concentrations of acetate: production of single cells. Appl Microbiol Biotechnol. 1991;35:686–689. [Google Scholar]
- 2.Balch W E, Fox G E, Magrum L J, Woese C R, Wolfe R S. Methanogens: re-evaluation of a unique biological group. Microbiol Rev. 1979;43:260–296. doi: 10.1128/mr.43.2.260-296.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Belay N, Sparling R, Daniels L. Dinitrogen fixation by a thermophilic methanogenic bacterium. Nature (London) 1984;312:286–288. doi: 10.1038/312286a0. [DOI] [PubMed] [Google Scholar]
- 4.Bult C J, White O, Olsen G J, Zhou L, Fleischmann R D, Sutton G G, Blake J A, FitzGerald L M, Clayton R A, Gocayne J D, Kerlavage A R, Dougherty B A, Tomb J-F, Adams M D, Reich C I, Overbeek R, Kirkness E F, Weinstock K G, Merrick J M, Glodek A, Scott J L, Geoghagen N S M, Weidman J F, Fuhrmann J L, Nguyen D, Utterback T R, Kelley J M, Peterson J D, Sadow P W, Hanna M C, Cotton M D, Roberts K M, Hurst M A, Kaine B P, Borodovsky M, Klenk H-P, Fraser C M, Smith H O, Woese C R, Venter J C. Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science. 1996;273:1058–1073. doi: 10.1126/science.273.5278.1058. [DOI] [PubMed] [Google Scholar]
- 5.Burns A, Watt C D, Wang Z C. Salt inhibition of nitrogenase catalysis and salt effects on the separate protein components. Biochemistry. 1985;24:3932–3936. [Google Scholar]
- 6.Chien C-T, Manundu J, Cavaness J, Dandurand L-M, Orser C S. Characterization of salt-tolerant and salt-sensitive mutants of Rhizobium leguminosarum biovar viciae strain C1204b. FEMS Microbiol Lett. 1992;90:135–140. doi: 10.1016/0378-1097(92)90617-w. [DOI] [PubMed] [Google Scholar]
- 7.Chien Y-T, Zinder S H. Cloning, DNA sequencing, and characterization of a nifD-homologous gene from the archaeon Methanosarcina barkeri 227 which resembles nifD1 from the eubacterium Clostridium pasteurianum. J Bacteriol. 1994;176:6590–6598. doi: 10.1128/jb.176.21.6590-6598.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chien Y T, Zinder S H. Cloning, functional organization, transcript studies, and phylogenetic analysis of the complete nitrogenase structural genes (nifHDK2) and associated genes in the archaeon Methanosarcina barkeri 227. J Bacteriol. 1996;178:143–148. doi: 10.1128/jb.178.1.143-148.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deits T L, Howard J B. Effects of salts on Azotobacter vinelandii nitrogenase activities. J Biol Chem. 1990;265:3859–3867. [PubMed] [Google Scholar]
- 10.Emerich D W, Burris R H. Complementary functioning of the component proteins of nitrogenase from several bacteria. J Bacteriol. 1978;134:936–943. doi: 10.1128/jb.134.3.936-943.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Emerich D W, Ljones T, Burris R H. Nitrogenase: properties of the catalytically inactive complex between the Azotobacter vinelandii MoFe protein and the Clostridium pasteurianum Fe protein. Biochim Biophys Acta. 1978;527:359–369. doi: 10.1016/0005-2744(78)90350-9. [DOI] [PubMed] [Google Scholar]
- 12.Galinski E A, Herzog R M. The role of trehalose as a substitute for nitrogen-containing compatible solutes (Ectothiorhospira halochloris) Arch Microbiol. 1990;153:607–613. [Google Scholar]
- 13.Igeno M I, Del Moral C G, Castillo F, Caballero F J. Halotolerance of the phototrophic bacterium Rhodobacter capsulatus E1F1 is dependent on the nitrogen source. Appl Environ Microbiol. 1995;61:2970–2975. doi: 10.1128/aem.61.8.2970-2975.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jones W J, Nagle D P, Jr, Whitman W B. Methanogens and the diversity of archaebacteria. Microbiol Rev. 1987;51:135–177. doi: 10.1128/mr.51.1.135-177.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kamberov E S, Atkinson M R, Ninfa A J. The Escherichia coli PII signal transduction protein is activated upon binding 2-ketoglutarate and ATP. J Biol Chem. 1995;270:17797–17807. doi: 10.1074/jbc.270.30.17797. [DOI] [PubMed] [Google Scholar]
- 16.Kenealy W R, Thompson T E, Schubert K R, Zeikus J G. Ammonia assimilation and synthesis of alanine, aspartate, and glutamate in Methanosarcina barkeri and Methanobacterium thermoautotrophicum. J Bacteriol. 1982;150:1357–1365. doi: 10.1128/jb.150.3.1357-1365.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klenk H P, Clayton R A, Tomb J F, White O, Nelson K E, Ketchum K A, Dodson R J, Gwinn M, Hickey E K, Peterson J D, Richardson D L, Kerlavage A R, Graham D E, Kyrpides N C, Fleischmann R D, Quackenbush J, Lee N H, Sutton G G, Gill S, Kirkness E F, Dougherty B A, McKenny K, Adams M D, Loftus B, Peterson S, Reich C, McNeil K, Badger J H, Glodek A, Zhou L, Overbeek R, Gocayne J D, Weidman F, McDonald L, Utterback T, Cotton M D, Spriggs T, Artiach P, Kaine B, Sykes S M, Sadow P W, D’Andrea K P, Bowman C, Fujii C, Garland S, Mason T M, Olsen G J, Fraser C M, Smith H O, Woese C R, Venter J C. The complete genome sequence of the hyperthermophilic, sulphate-reducing archaeon Archaeoglobus fulgidus. Nature (London) 1997;390:364–370. doi: 10.1038/37052. [DOI] [PubMed] [Google Scholar]
- 18.Lai M-C, Sowers K R, Robertson D E, Roberts M F, Gunsalus R P. Distribution of compatible solutes in the halophilic methanogenic archaebacteria. J Bacteriol. 1991;173:5352–5358. doi: 10.1128/jb.173.17.5352-5358.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lobo A L, Zinder S H. Diazotrophy and nitrogenase activity in the archaebacterium Methanosarcina barkeri 227. Appl Environ Microbiol. 1988;54:1656–1661. doi: 10.1128/aem.54.7.1656-1661.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lobo A L, Zinder S H. Nitrogenase in the methanogenic archaebacterium Methanosarcina barkeri strain 227. J Bacteriol. 1990;172:6789–6796. doi: 10.1128/jb.172.12.6789-6796.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Madkour M A, Smith L T, Smith G M. Preferential osmolyte accumulation: a mechanism of osmotic stress adaptation in diazotrophic bacteria. Appl Environ Microbiol. 1990;56:2876–2881. doi: 10.1128/aem.56.9.2876-2881.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murray P A, Zinder S H. Nitrogen fixation by a methanogenic archaebacterium. Nature (London) 1984;312:284–286. [Google Scholar]
- 23.Murray P A, Zinder S H. Nutritional requirements of Methanosarcina thermophila strain TM-1. Appl Environ Microbiol. 1985;50:49–55. doi: 10.1128/aem.50.1.49-55.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murray P A, Zinder S H. Polysaccharide reserve material in the acetotrophic methanogen Methanosarcina thermophila strain TM-1: accumulation and mobilization. Arch Microbiol. 1987;147:109–116. [Google Scholar]
- 25.Neidhardt F C, Ingraham J L, Schaechter M. Physiology of the bacterial cell. Sunderland, Mass: Sinauer Associates; 1990. [Google Scholar]
- 26.Ninfa A J, Atkinson M R, Kamberov E S, Feng J, Ninfa E J. Control of nitrogen assimilation by the NRI-NRII two-component system of enteric bacteria. In: Hoch J A, Silhavy T J, editors. Two-component signal transduction. Washington, D.C: ASM Press; 1995. pp. 67–88. [Google Scholar]
- 27.Peters J W, Fisher K, Dean D R. Identification of a nitrogenase protein-protein interaction site defined by residues 59 and 67 within the Azotobacter vinelandii Fe protein. J Biol Chem. 1993;269:28076–28083. [PubMed] [Google Scholar]
- 28.Peters J W, Fisher K, Dean D R. Nitrogenase structure and function: a biochemical-genetic perspective. Annu Rev Microbiol. 1995;49:335–366. doi: 10.1146/annurev.mi.49.100195.002003. [DOI] [PubMed] [Google Scholar]
- 29.Robertson D E, Lai M-C, Gunsalus R P, Roberts M F. Composition, variation, and dynamics of major osmotic solutes in Methanohalophilus strain FDF1. Appl Environ Microbiol. 1992;58:2438–2443. doi: 10.1128/aem.58.8.2438-2443.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Robertson D E, Noll D, Roberts M F. Free amino acid dynamics in marine methanogens. β-Amino acids as compatible solutes. J Biol Chem. 1992;267:14893–14901. [PubMed] [Google Scholar]
- 31.Robertson D E, Noll D, Roberts M F, Menaia J A G F, Boone D R. Detection of the osmoregulator betaine in methanogens. Appl Environ Microbiol. 1990;56:563–565. doi: 10.1128/aem.56.2.563-565.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shah V K, Davis L C, Brill W J. Nitrogenase. I. Repression and derepression of the iron-molybdenum protein and iron proteins of nitrogenase in Azotobacter vinelandii. Biochim Biophys Acta. 1972;256:498–511. doi: 10.1016/0005-2728(72)90078-3. [DOI] [PubMed] [Google Scholar]
- 33.Sibold L, Henriquet M, Possot O, Aubert J-P. Nucleotide sequence of nifH regions from Methanobacterium ivanovii and Methanosarcina barkeri and characterization of glnB-like genes. Res Microbiol. 1991;142:5–12. doi: 10.1016/0923-2508(91)90091-n. [DOI] [PubMed] [Google Scholar]
- 34.Smith D R, Doucette-Stamm L A, Deloughery C, Lee H, Dubois J, Aldredge T, Bashirzadeh R, Blakely D, Cook R, Gilbert K, Harrison D, Hoang L, Keagle P, Lumm W, Pothier B, Qiu D, Spadafora R, Vicaire R, Wang Y, Wierzbowski J, Gibson R, Jiwani N, Caruso A, Bush D, Safer H, Patwell D, Prabhakar S, McDougall S, Shimer G, Goyal A, Pietrokovski S, Church G M, Daniels C J, Mao J I, Rice P, Nolling J, Reeve J N. Complete genome sequence of Methanobacterium thermoautotrophicum Delta-H: functional analysis and comparative genomics. J Bacteriol. 1997;179:7135–7155. doi: 10.1128/jb.179.22.7135-7155.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sowers K R, Boone J E, Gunsalus R P. Disaggregation of Methanosarcina spp. and growth as single cells at elevated osmolarity. Appl Environ Microbiol. 1993;59:3832–3839. doi: 10.1128/aem.59.11.3832-3839.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sowers K R, Gunsalus R P. Adaptation for growth at various saline concentrations by the archaebacterium Methanosarcina thermophila. J Bacteriol. 1988;170:998–1002. doi: 10.1128/jb.170.2.998-1002.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sowers K R, Gunsalus R P. Halotolerance in Methanosarcina spp.: role of Nɛ-acetyl-β-lysine, α-glutamate, glycine betaine, and K+ as compatible solutes for osmotic adaptation. Appl Environ Microbiol. 1995;61:4382–4388. doi: 10.1128/aem.61.12.4382-4388.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sowers K R, Robertson D E, Noll D, Gunsalus R P, Roberts M F. N-Acetyl-β-lysine: an osmolyte synthesized by methanogenic archaebacteria. Proc Natl Acad Sci USA. 1990;87:9083–9087. doi: 10.1073/pnas.87.23.9083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Strandberg G W, Wilson P W. Formation of a nitrogen-fixing enzyme system in Azotobacter vinelandii. Can J Microbiol. 1968;14:25–31. doi: 10.1139/m68-005. [DOI] [PubMed] [Google Scholar]
- 40.Thorneley R N F, Willison K R. Nitrogenase of Klebsiella pneumoniae. Inhibition of acetylene reduction by magnesium ion explained by the formation of an inactive dimagnesium-adenosine triphosphate complex. Biochem J. 1974;139:211–214. doi: 10.1042/bj1390211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wolle D, Kim C, Dean D, Howard J B. Ionic interactions in the nitrogenase complex. Properties of Fe-protein containing substitutions for Arg-100. J Biol Chem. 1992;267:3667–3673. [PubMed] [Google Scholar]