Abstract
Tn5 mutants of Sinorhizobium meliloti RMB7201 which swarmed 1.5 to 2.5 times faster than the parental strain in semisolid agar, moist sand, and viscous liquid were identified. These faster-swarming (FS) mutants outgrew the wild type 30- to 40-fold within 2 days in mixed swarm colonies. The FS mutants survived and grew as well as or better than the wild type under all of the circumstances tested, except in a soil matrix subjected to air drying. Exopolysaccharide (EPS) synthesis was reduced in each of the FS mutants when they were grown on defined succinate-nitrate medium, but the extent of reduction was different for each. It appears that FS behavior likely results from a modest, general derepression of motility involving an increased proportion of motile and flagellated cells and an increased average number of flagella per cell and increased average flagellar length. Spontaneous FS variants of RMB7201 were obtained at a frequency of about 1 per 10,000 to 20,000 cells by either enrichment from the periphery of swarm colonies or screening of colonies for reduced EPS synthesis on succinate-nitrate plates. The spontaneous FS variants and Tn5 FS mutants were symbiotically effective and competitive in alfalfa nodulation. Reversion of FS variants to wild-type behavior was sporadic, indicating that reversion is affected by unidentified environmental factors. Based on phenotypic and molecular differences between individual FS variants and mutants, it appears that there may be multiple genetic configurations that result in FS behavior in RMB7201. The facile isolation of spontaneous FS variants of Escherichia coli and Pseudomonas aeruginosa indicates that switching to FS behavior may be fairly common among bacterial species. The substantial growth advantage of FS mutants and variants wherever nutrient gradients exist suggests that switching to FS forms may be an important behavioral adaptation in natural environments.
Although flagellar motility and chemotaxis in bacteria have been studied intensively for more than 20 years (8), relatively little is known about how or when they actually operate in natural environments or how they are regulated to optimize growth and survival under diverse conditions. We are interested in the role of flagellar motility in the ability of Sinorhizobium meliloti to survive in soil, colonize roots, and symbiotically infect and nodulate its host, alfalfa. Strains of S. meliloti are motile and chemotactic. Nonmotile and nonchemotactic mutants of S. meliloti are substantially less competitive than the wild type at infecting and nodulating host roots (3, 12). Molecular aspects of motility and the regulation of behavior in S. meliloti differ in many respects from those seen in enteric bacteria (4, 19). Motility in S. meliloti is based on exclusive clockwise rotation of the bacterium’s two to eight peritrichous, complex flagella (17, 18, 23). Complex flagella are more rigid and more efficient for propulsion in viscous media than are plain flagella. S. meliloti cells are chemokinetic, swimming at higher speeds when exposed to higher attractant concentrations, with brief pauses or asynchronous flagellar rotation resulting in changes in the direction of swimming (4). The bacterium is attracted toward a variety of amino acids, dicarboxylic acids, and sugars (6), toward the nodulation gene-inducing flavonoids secreted by roots of its host (14), and toward unknown attractants secreted at localized sites on the host root (20).
Isolates of S. meliloti with greater motility or swarming have been obtained by serial enrichment for faster-moving cells from the periphery of soft agar swarm colonies (23) and by Tn5 mutagenesis (34). The faster-swarming (FS) isolate obtained by enrichment was reported to have greater flagellation than the wild type (23), but the genesis, stability, and consequences of this enhanced swarming behavior were not examined for either isolate. We thought that mutants with increased motility and taxis might be valuable tools for investigating the ecological consequences of altered behavioral activity and might also prove valuable in developing more competitive inoculants. The studies reported here examined several Tn5 mutants and spontaneous variants of S. meliloti RMB7201 which have FS behavior. Our studies revealed that the swarming behavior of S. meliloti is subject to spontaneous, relatively high-frequency switching between normal and FS activities and that FS cells differ quite significantly from wild-type cells in growth and survival under various conditions.
MATERIALS AND METHODS
Bacterial strains, plasmids, media, and buffers.
The bacterial strains and plasmids used in this study are listed in Table 1. Stock cultures were kept in 15% glycerol at −80°C. S. meliloti strains were routinely cultured in TY (containing 6 g of tryptone, 3 g of yeast extract, and 0.5 g of CaCl2 · 2H2O per liter) or in TY containing reduced concentrations of tryptone and yeast extract but maintaining the CaCl2 · 2H2O at 0.5 g/liter. S. meliloti was also cultured in a defined medium (NM) containing mineral salts and vitamins with 10 mM succinate and 5 mM nitrate as the carbon and nitrogen sources (32). Escherichia coli strains were grown in Luria-Bertani medium. All chemicals were analytical or reagent grade (Sigma Chemical Co. or Baker Chemical Co.). To improve reproducibility of behavior, all liquid cultures of S. meliloti were routinely started from glycerol stocks, grown at 28°C to late exponential or early stationary phase on a rotary shaker at 175 to 200 rpm, and then subcultured in fresh medium. Cells from the subcultures were harvested during early exponential phase (A590 = 0.15 to 0.30), the period of best motility for S. meliloti. Solid media were prepared by the addition of 1.5% Bacto Agar (Difco). Semisolid (swarm) agar contained 3 g of Bacto Agar per liter and either 1/20 strength TY or 1/10 strength NM medium unless otherwise specified. Motility, chemotaxis, and some swarm assays were carried out in chemotaxis buffer (CB). CB was found to be optimal for maintaining the structural integrity of complex flagella and, hence, motility in S. meliloti (32). This buffer consists of 10 mM HEPES, 0.1 mM CaCl2, and 0.01 mM sodium EDTA in high-performance liquid chromatography grade water, pH 7.0.
TABLE 1.
Bacterial strains and plasmids used in this study and their characteristics
| Bacterial strain or plasmid | Relevant characteristic(s) | Source or reference |
|---|---|---|
| S. meliloti | ||
| RMB7201 | Wild type; Smr Spcr, Ω insertion in chromosomal inositol gene | 9 |
| FS1 | Tn5 (Kmr) derivative of RMB7201, FS, reduced EPS in NM medium | This study |
| FS7 | Tn5 (Kmr) derivative of RMB7201, FS, reduced EPS in NM medium | This study |
| FS44 | Tn5 (Kmr) derivative of RMB7201, FS, reduced EPS in NM medium | This study |
| FS17 | Mini-Tn5 (Kmr) derivative of RMB7201, FS, reduced EPS in NM medium | This study |
| FS27 | Mini-Tn5 (Kmr) derivative of RMB7201, FS, reduced EPS in NM medium | This study |
| FS28 | Mini-Tn5 (Kmr) derivative of RMB7201, FS, reduced EPS in NM medium | This study |
| SV2 | Spontaneous FS, reduced-EPS variant of RMB7201 | This study |
| SV3 | Spontaneous intermediate-swarming, reduced-EPS variant of RMB7201 | This study |
| SV4 | Spontaneous FS, reduced-EPS variant of RMB7201 | This study |
| SV8 | Spontaneous FS, reduced-EPS variant of RMB7201 | This study |
| SV9 | Spontaneous FS, reduced-EPS variant of RMB7201 | This study |
| SV68 | Spontaneous intermediate-swarming, reduced-EPS variant of RMB7201 | This study |
| SV69 | Spontaneous intermediate-swarming, reduced-EPS variant of RMB7201 | This study |
| Rm1021 | Smr derivative of SU47 wild type | 26 |
| Rm7032 | exoA EPS nonproducer | 24 |
| Rm8468 | exoP EPS nonproducer | 25 |
| Rm8395 | exoR EPS overproduction | 15 |
| Rm8396 | exoS EPS overproduction | 15 |
| Rm7020 | exoC EPS overproduction | 16 |
| Rm7103 | exoX EPS overproduction | 40 |
| E. coli | ||
| WA803(pGS9) | Tn5 donor | 37 |
| CC118λpir(pUTmini-Tn5lac1) | Tn5 donor | 13 |
| P. aeruginosa PAO1 | Wild type | 36 |
| Plasmids | ||
| pM6 | exoR cosmid | 15 |
| pM13-1 | 5.3-kb subclone complements exoS | 15 |
| pD5 | Complements exoC | 24 |
Numbers of CFU of Tn5 mutant strains in mixtures with the parental strain were determined by subtraction of counts on TY medium containing streptomycin and spectinomycin at 100 μg/ml and kanamycin at 200 μg/ml from total counts on TY containing just streptomycin and spectinomycin. The ratio of mutant to parent bacteria was verified by testing representative colonies on swarm agar. A Spiral plater (model D; Spiral System, Inc., Cincinnati, Ohio) was used for all CFU determinations, and counts from duplicate or triplicate plates were averaged.
Transposon mutagenesis and Southern analysis.
S. meliloti RMB7201 was mutagenized with Tn5 through filter and plate mating with E. coli WA803(pGS9) (37) or with CC118λpir(pUTminiTn5-lacZ) (13). Genomic DNA preparation, restriction digestion, and Southern blotting were performed as previously described (35). DNA probes (the internal 3.3-kb HindIII fragment from Tn5 and the 5.0-kb EcoRI-BamHI fragment from pUTminiTn5-lacZ) were labeled with 32P (Amersham, Arlington Heights, Ill.) or biotin-14-dCTP (Life Technologies, Gaithersburg, Md.) in accordance with the supplier’s instructions. The blots were detected, respectively, with X-ray film or the BlueGene nonradioactive nucleic acid detection system (Life Technologies).
Motility and chemotaxis.
The percentage of motile cells was determined in a Petroff-Hauser counting chamber with a Zeiss IM 35 microscope as previously described (39). Motile-cell percentages were based on observation of 60 to 100 cells in two replicate suspensions and were generally reproducible within ± 10%. In some cases, video recorded data were analyzed to determine motile-cell percentages and analyze motile behavior. Chemotactic responses to attractants were assayed as described by Adler (1) and Palleroni (28) in chemotaxis chambers using quadruplicate 1-μl capillary tubes and 30-min assays. To ensure stringent comparison of chemotactic responsiveness to various attractants, carefully matched early log-phase cultures of FS mutant and wild-type strains were suspended in CB, the suspensions were mixed together and diluted to 107 cells of each strain per ml, and the number of cells of each strain entering capillaries was determined by differential plating. In a previous study (39), it was shown that entry of the wild-type strain into buffer-filled capillaries was not affected by the addition of a potent attractant to the bacterial suspension, indicating that attractants do not substantially enhance random motility and that buffer-filled capillaries are an adequate control for the random entry of bacteria.
Swarming in semisolid agar, sand, or viscous media.
Swarm colony development in semisolid agar typically involved inoculation of the center of a plate with 2.5 μl of a suspension of the mutant and/or the wild type. The plates were then sealed with Parafilm and incubated at 28°C. For analysis of swimming behavior and swarming in visous liquid medium, 0.4% high-viscosity carboxymethyl cellulose (CMC) was mixed with an equal volume of 1/10 strength TY. The rate of spreading of swarm colonies in CMC was determined by microscopic detection of cells at the periphery. To measure swarming in sand, washed quartz sand in a petri dish was moistened with 1/20 TY to field capacity, inoculated at the center of the plate with 2 μl of a bacterial suspension, and incubated for 1 to 3 days at 28°C. Spreading of the swarm colony in sand was determined by replication onto nutrient agar with a multiprong replicator. To measure the rate of swarm colony spreading in CB swarm agar, agar blocks were removed from points at various distances from the center 3 days after inoculation, homogenized, and plated.
Detection and quantification of EPS.
The amount of exopolysaccharide (EPS) produced by S. meliloti RMB7201 and FS mutants on solid NM succinate-nitrate medium was roughly estimated by direct observation of colonies on plates, usually after 2 or 3 days of incubation. To measure the quantity of EPS produced in liquid NM-succinate medium, cells were removed by centrifugation of stationary-phase cultures and EPS was precipitated from the supernatants with 3 volumes of ethanol, rinsed, and then dried. EPS was then either weighted directly or suspended in water by thorough sonication, and carbohydrate content was determined by the phenol-sulfuric acid method (33) with glucose as the standard.
Preparation of flagella and Western blotting.
Cells from mid-exponential-phase cultures were pelleted at low speed (2,000 × g), resuspended in CB, and mixed in a Waring blender (model 33BL79; Dynamics Corp. of America, New Hartford, Conn.) at full power for 20 s to shear flagella from the cells. Cells were removed by centrifugation at 15,000 × g for 15 min. The supernatant containing detached flagella was centrifuged at 100,000 × g for 2 h, and the pelleted flagella were suspended in CB. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western blot analysis of flagellin preparations were performed as described previously (38).
EM.
Flagella and flagellated cells were observed with a Philips 201 transmission electron microscope using the direct-staining, no-rinse method described previously (39). Cells for electron microscopy (EM) examination were harvested in early exponential phase and resuspended in CB containing 0.5 mM HEPES with a threefold dilution. Five-microliter aliquots of the cell suspensions were mixed with 1 μl of 2% uranyl acetate, and 2 μl of the mixture was transferred to a Formvar-coated grid and air dried. The percentage of cells with flagella was determined by examination of at least 100 representative cells on two or three independently prepared grids per sample. The average number and length of flagella were determined from electron micrographs of at least 50 cells per strain with a planimeter (Graphics Calculator; Numonics Co., Lansdale, Pa.).
Nodulation of alfalfa.
Nodulation tests in growth pouches were done as previously described (7). Nodule occupancy by mutant, revertant, or wild-type strains was determined on 30 to 70 nodules taken from the oldest part of the primary roots of 5 to 40 plants by plating homogenates of surface-sterilized nodules onto selective media and confirming the swarm phenotype of representative colonies on semisolid agar.
Soil drying tolerance.
The relative growth and survival of FS mutants and the parental strain during gradual drying in a soil matrix were examined in a sterilized silt-clay soil fraction. This silt-clay material was prepared by sieving fresh silt loam soil through 7-mesh screen, suspending 600 g of field-moist soil in 3 liters of tap water, and decanting it after 10 min of settling to remove floating debris and fine clay. Decantation was repeated twice before blending the suspension in a Waring blender in 500-ml portions for 30 s. The blended suspension was passed through an 80-mesh screen to remove large sand particles and then through a 325-mesh screen to remove particles larger than 45 μm. The silt-clay suspension was then washed three times by 20 min of settling and decantation to remove fine clay particles, autoclaved twice for 60 min, and then washed twice with sterile tap water before resuspension in a total volume of 600 ml. This suspension was dispensed by pipet into scintillation vials and dried at 100°C for 24 h. Mid-exponential-phase cultures of an FS mutant and the parental strain were washed and resuspended in CB containing 0.5 mM HEPES, mixed at a 1:1 ratio, and then inoculated into the dried silt-clay material to about 106 CFU per strain per g (dry weight) and a moisture level of about 75% of field capacity. The vials were kept open in the laboratory and allowed to dry at approximately 23°C and 50% relative humidity. In a subsequent experiment, the vials were placed in a plastic box with dishes containing saturated K2SO4 solution to maintain the relative humidity at 95%. Numbers of mutant and parental strain CFU were determined at various times after inoculation by extracting cells in 3 volumes of sterile water, vigorously vortexing them for 1 to 2 min, and then sonicating the suspension at 25% energy output in a Heat System-Ultrasonics, Inc., w-370 cup horn sonicator for 3 min. After allowing particles to settle for 3 min, the suspension was transferred to a screw-capped 50-ml tube and the sediment was extracted three more times. The cell recovery rate was 90 to 95%.
RESULTS
Isolation of FS mutants.
Screening of over 6,000 purified isolates from several Tn5 matings of S. meliloti RMB7201 with E. coli WA803(pGS9) on 1/20 TY semisolid agar plates for altered swarm colony development allowed identification of a variety of behavioral mutants. Nonswarming mutant isolates, with cells which were nonmotile or had severely impaired motility in liquid culture, were obtained at a frequency of about 3.0 × 10−3. Isolates with (i) reduced swarm rates, (ii) cells that appeared microscopically to have either impaired motility or a lower percentage of motile cells, or (iii) normal motility but impaired chemotaxis were obtained at a frequency of 3.6 × 10−3. Three mutants, FS1, FS7, and FS44, each from a different mating, swarmed at rates significantly faster than that of the parental strain. FS mutants were recovered at a frequency of 4.5 × 10−4.
Swarming behavior of FS mutants.
Table 2 shows the relative rates of swarm colony spreading for the wild type and the three FS mutants. Even in CB agar plates, where nutrient availability was very low and one might expect the wild type’s swarming capabilities to be fully derepressed, the FS mutants swarmed significantly faster than the parental strain. Cells of the FS mutants were present in greater numbers at all locations in these plates, from the center to the outer edges. The FS mutants also spread more rapidly through viscous liquid media and through a moist sand matrix, indicating that their FS behavior was not dependent on interactions with a semisolid agar lattice.
TABLE 2.
Swarming rates of S. meliloti RMB7201 and Tn5 FS mutants in various media
| Medium | Swarming rate (avg change in colony radius [mm/h] ± SD)a
|
|||
|---|---|---|---|---|
| RMB7201 | FS1 | FS7 | FS44 | |
| TY (1/20), 0.3% agar | 0.50 ± 0.019 | 1.0 ± 0.018 | 1.0 ± 0.018 | 1.0 ± 0.012 |
| Soil extract,b 0.3% agar | 0.35 ± 0.022 | 0.50 ± 0.00 | 0.50 ± 0.00 | NDc |
| CB, 0.3% agar | 0.29 ± 0.00 | 0.40 ± 0.036 | 0.40 ± 0.031 | 0.37 ± 0.036 |
| TY (1/20), moist sand | 0.64 ± 0.039 | 1.2 ± 0.076 | 1.1 ± 0.073 | 0.92 ± 0.14 |
| TY (1/20) in 0.2% CMC | 3.0 ± 0.43 | 4.6 ± 0.46 | 4.6 ± 0.29 | 3.6 ± 0.31 |
Values are averages taken from replicate plates in a representative experiment after 40 h, except for CMC-amended 1/20 TY, liquid medium, where rates were determined 6 to 9.5 h after inoculation.
Soil extract solution was obtained by adding 1 liter of tap water to 500 g of soil (Crosby silt loam), mixing it, and centrifuging it at 5,800 × g and 35,000 × g for 15 min each to remove particles. The soil solution was then concentrated ninefold in a rotary evaporator and filter sterilized through 0.2-μm-pore-size membranes.
ND, not determined.
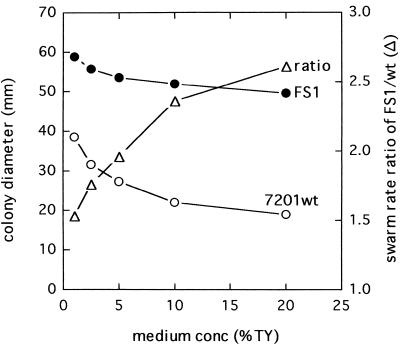
Nutrient concentration was found to affect the rate of spreading of both FS1 and the wild type (Fig. 1). However, the wild type was more sensitive to nutrient concentration than the mutant, so that the higher the nutrient concentration, the larger the difference in the rate of swarm colony growth between the mutant and parental strains (Fig. 1).
FIG. 1.
Effect of nutrient concentration on swarm rate. Swarm rates of S. meliloti RMB7201 (○) and FS1 (●) and the ratio of the FS1 swarm rate to the RMB7201 swarm rate (▵) are shown. The two strains were separately spotted into swarm agar containing 0.3% agar and 1/100, 1/40, 1/20, 1/10, or 1/5 strength TY. Swarm colony diameters were measured after 2 days of incubation at 28°C. Each datum point is the average of four replicates from a representative experiment. wt, wild type; Conc., concentration.
Swimming behavior and chemotactic responsiveness of FS mutants.
Microscopic analysis revealed that FS1 and FS7 had a pattern of swimming behavior not readily distinguishable from that of the RMB7201 parental strain, with fairly long, straight swims interrupted by abrupt changes in direction (39). The percentage of cells that showed active motility was consistently about 10% higher for the mutants than the parental strain, and the parental strain seemed to have a higher percentage of cells that changed direction at a high frequency. In contrast to the parental strain and the other FS mutants, FS44 was observed to change direction at high frequency in liquid media with no long swims. Interestingly, in viscous medium (0.2% CMC), FS44 swam long distances without frequent changes in direction.
The chemotactic responsiveness of FS1 to glutamine, succinate, glucose, and alfalfa seed exudate was similar to that of the wild type (Table 3). Further assays were conducted to determine chemotactic responses toward other attractants, including phenylalanine, tryptophan, aspartic acid, sucrose, mannitol, cycloleucine, itaconic acid, 4,7-dihydroxyflavone, and p-hydroxybenzoic acid. The wild type, FS1, FS7, and FS44 did not differ significantly in responsiveness to these compounds at a 95% confidence level (data not shown).
TABLE 3.
Chemotactic responsiveness of wild-type S. meliloti RMB7201 and mutant FS1
| Attractant (conc) | Avg chemotaxis ratioa ± SD
|
|
|---|---|---|
| RMB7201 | FS1 | |
| Glutamine (10 mM) | 37 ± 5.8 | 43 ± 5.2 |
| Glucose (30 mM) | 1.6 ± 0.28 | 1.8 ± 0.2 |
| Succinate (30 mM) | 3.2 ± 0.8 | 3.7 ± 0.5 |
| Alfalfa seed exudate (1/4) | 93 ± 21 | 125 ± 17 |
| Alfalfa seed exudate (1/16) | 53 ± 16 | 66 ± 4.7 |
| Alfalfa seed exudate (1/64) | 18 ± 5.8 | 22 ± 1.9 |
The chemotaxis ratio is the average number of cells entering attractant-filled capillaries divided by the average number of cells (ca. 3,000/30 min) entering buffer-filled control capillaries. Ratios are based on average numbers of CFU from four replicate capillary tubes and are from a single experiment that was repeated with similar results. Differences in the chemotaxis ratio between the wild type and FS1 were not significant at the 95% confidence level by t-test analysis.
Growth and survival of FS mutants in vitro.
Various media were inoculated with a 1:1 mixture of FS1 and the wild type cultured as indicated below, and numbers of CFU were determined by selective plating, with strain identity confirmed by swarm rate testing of 50 to 100 colonies. In a liquid shake culture on 1/10 strength TY, where motility can provide no significant advantage, growth rates of the wild type and the FS mutant were essentially identical (Table 4). However, in still cultures on the same medium, the number of FS1 cells was 1.8-fold higher than that of wild-type cells at stationary phase. The FS mutants had an even greater advantage over the parental strain in semisolid agar, outgrowing the parental strain by 30- to 40-fold in 2 to 3 days.
TABLE 4.
Competitive growth of a 1:1 mixed inoculum of the RMB7201 wild type with an FS mutant or variant in semisolid agar and shake and still cultures.
| Culture conditions | Strain mixed with wild type | Ratio of FS to wild-type cells ± SDa |
|---|---|---|
| Shake cultureb in 1/10 TY | FS1 | 1.0 ± 0.12 |
| FS17 | 1.0 ± 0.03 | |
| SV8 | 1.0 ± 0.04 | |
| Still culturec in 1/10 TY | FS1 | 1.8 ± 0.37 |
| FS17 | 1.6 ± 0.10 | |
| SV8 | 1.6 ± 0.09 | |
| Agar (0.3%), 1/20 TYd | FS1 | 33 ± 9.6 |
| FS17 | 30 ± 4.0 | |
| SV8 | 35 ± 2.7 | |
| SV68 | 27 ± 4.0 | |
| SV69 | 20 ± 1.7 | |
| SV3 | 36e ± 3.3 | |
| Agar (0.3%), CB | FS1 | 10 ± 1.2 |
These values are from three replicate cultures in a representative experiment after correction for the ratio of the two strains in the inoculum. The ratio of wild-type to FS isolate colonies was determined by differential antibiotic sensitivity, colony morphology, and swarm tests for FS17 and by colony morphology and swarm tests on at least 100 colonies for SV3, SV8, SV68, and SV69.
Replicate cultures with 15 ml of medium were inoculated with about 107 cells each of the FS strain and the wild type, cultured at 200 rpm at 28°C for 2 days to stationary phase, and then plated to determine relative CFU.
Replicate cultures in 6 ml of medium in culture tubes (18 by 150 mm) were inoculated with about 106 cells each of the FS strain and the wild type, cultured at 28°C undisturbed for 5 days to stationary phase, and then plated to determine relative CFU.
Replicate swarm agar plates were inoculated with about 106 cells of the FS strain and the wild type and cultured for 3 days, and then the agar was vortexed vigorously for 2 min and the suspension was plated for determination of relative CFU.
The swarm rate of colonies of SV3 cells from the periphery of the swarm was about 1.8-fold that of the wild type during tests to determine the ratio of the two strains in the 3-day swarm colony, faster than the swarm rate of the original isolate, ca. 1.4-fold faster than the wild type.
When FS1 and the wild type were incubated separately in still cultures of CB or unfiltered pond water (Mirror Lake, Ohio State University campus), the survival rates of the two strains were very similar. The CFU counts of both the parental strain and FS1 after 3 months of incubation in the CB starvation buffer were only two- to threefold lower than the CFU counts recovered immediately after inoculation. The CFU counts recovered from unfiltered pond water after 4 months were 104-fold lower than those recovered immediately after inoculation, but the ratio of wild-type to FS1 bacteria was essentially unchanged.
To date, the only circumstance under which the wild-type isolate has shown a significant growth or survival advantage over the FS1 mutant has been in a sterile silt-clay soil that was allowed to dry extensively. Both FS1 and FS7 died out several times faster than the wild type (Table 5). In a similar experiment in which drying was slower, the populations of both the parental strain and FS1 increased more than 100-fold during the first 2 weeks and that of the parental strain increased more extensively than that of the mutant. The populations then declined and stabilized at densities of about 107 cells/g (dry weight), with FS1 populations seven or eight times smaller than the parental population (data not shown).
TABLE 5.
Survival of S. meliloti RMB7201, FS1, and FS7 during drying in a silt-clay matrixa
| Strains mixed | Avg no. of CFU (% survival)
|
|||
|---|---|---|---|---|
| Day 1b | Day 3 | Day 5 | Day 11 | |
| Wild type plus FS1 | ||||
| Wild type | 1.1 × 107 (83) | 4.9 × 104 (0.40) | 3.5 × 104 (0.30) | 4.9 × 103 (0.04) |
| FS1 | 7.3 × 106 (59) | 1.4 × 103 (0.13) | 6.8 × 103 (0.063) | 5.3 × 102 (0.005) |
| Wild type plus FS7 | ||||
| Wild type | 2.3 × 106 (16) | 5.3 × 104 (0.37) | 2.0 × 104 (0.14) | 1.6 × 103 (0.011) |
| FS7 | 2.0 × 106 (15) | 1.5 × 103 (0.12) | 2.0 × 103 (0.02) | 1.1 × 102 (0.001) |
Values are averages from duplicate samples in a single experiment. CFU per vial = CFU/1.6 g of dry silt-clay matrix.
The moisture content of the silt-clay matrix was 8.75% by weight on day 1 and below 0.8% on days 3, 5, and 11.
Flagellation and motility of FS mutants.
As shown in Table 6, the percentage of flagellated and motile cells was slightly (1.1- to 1.2-fold) higher for the FS mutants than for the parental strain. Based on EM comparisons, the FS mutants had a 1.6-fold greater average number of flagella per cell and a 1.6-fold greater average flagellar length. Thus, overall, EM analysis indicated that the amount of flagellin attached to FS mutant cells was about 1.15 × 1.6 × 1.6 = 2.9 times greater than the amount of flagellin attached to wild-type cells. Western blot analysis confirmed that approximately two to three times as much flagellin was attached to the FS mutant cells and free in the culture medium as in the parental strain culture (data not shown). Growth of the bacteria at low rates of shaking (50 rpm) had no discernible effect on the percentage of motile or flagellated cells or on the average length of the flagella attached to cells (data not shown), indicating that shearing of flagella during growth or sample preparation was not a problem affecting the assessment of differences in flagellation. After exposure of the FS mutants to semistarvation conditions in CB for several hours, both motility and flagellation were reduced, with motility reaching zero in about 24 h. In terms of motility, the FS mutants and the parental strain were affected to comparable extents. Motility diminished more quickly and extensively than flagellation (Table 6), indicating rapid arrest of flagellar motors in response to nutrient deprivation (39).
TABLE 6.
Motility and flagellation of FS mutants
| Strain | Time after resuspension in CBa
|
|||||
|---|---|---|---|---|---|---|
| 0 h
|
4 h
|
|||||
| % Motileb | % Flagellatedc | No. of Flagella/celld | Avg flageller lengthe (μm) | % Motileb | % Flagellatedc | |
| RMB7201 | 42 | 47 | 1.9 | 2.4 | 33 | 44 |
| FS1 | 55 | 57 | 2.4 | 3.8 | 43 | 55 |
| FS7 | 53 | 46 | NDf | ND | 35 | 44 |
| FS44 | 53 | 51 | ND | ND | 35 | 46 |
Cells from log-phase 1/10 TY cultures were washed and gently resuspended in CB.
Motile-cell percentages are averages of counts from several fields and two independent cultures from a representative experiment. Averages from replicate aliquots normally varied by ±5%.
Flagellated-cell percentages are averages of counts from about 60 to 100 cells on two separate grids from the same two independent cultures used for motile-cell percentage assays. Averages from replicate grids normally varied by less than 5%.
Average number of flagella per flagellated cell, determined from electron micrographs of at least 100 cells per strain on replicate grids. Values were reproducible within ± 10% between grids.
Average length of flagella per flagellated cell, determined by planimetry from micrographs of at least 50 cells per strain on replicate grids.
ND, not determined.
EPS production.
Colonies of the RMB7201 wild type were large and mucoid on both TY agar and NM-succinate-nitrate minimal agar. However, on NM-succinate-nitrate agar, colonies of FS1 and FS44 were considerably smaller than those of the wild type and relatively dry in appearance, indicating that these mutants might be defective in EPS synthesis on this medium. Based on two experiments with duplicate samples, mutants FS1, FS7, and FS44 produced 41, 68, and 18% of the EPS produced by the wild type in liquid NM-succinate-nitrate medium, respectively, although they produced essentially the same amount of EPS as the wild type in TY liquid medium (with a delay of 1 day for FS1 and FS44). On plates containing Calcofluor white M2R, colonies of the three FS mutants fluoresced strongly under UV light, indicating that they all produced EPS-I (24).
To further explore possible links between regulation of EPS synthesis and regulation of swarming motility in S. meliloti, the rates of swarming of several well-characterized EPS-overproducing and -nonproducing mutants of S. meliloti 1021 were determined. Strains Rm7095, Rm7096, and Rm7020 are EPS-I overproducers caused by insertional mutations in the regulatory genes exoR, exoS, and exoC, respectively (15, 16, 31). These strains were found to be essentially nonswarming in 1/10 NM semisolid agar for the first 2 days after inoculation. After 2 days, Rm7095 swarmed at a 50% rate in 1/10 NM agar while Rm7096 swarmed at a 50% rate in 1/20 TY semisolid agar. When complementing plasmids pM6, pM13-1, and pD5 (15, 24, 31) were introduced into Rm7095, Rm7096, and Rm7020, respectively, the strains swarmed normally. Strain Rm7103, an exoX mutant which also overproduces EPS-I (40), swarmed at a rate about 20% lower than that of the wild type. Both of the EPS-nonproducing mutants tested (Table 1) swarmed at the same rate as the parental strain.
Isolation of spontaneous FS variants and additional Tn5 FS mutants.
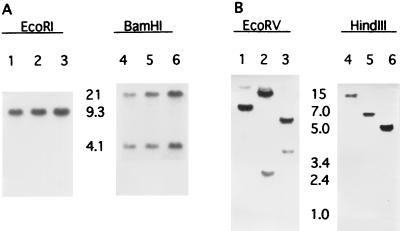
Southern analysis of FS1, FS7, and FS44 revealed that all three mutants had identical restriction patterns, indicating that the three insertions were located at essentially the same site (Fig. 2A). However, homologous recombination of the Tn5-containing sequence from FS1 back into the parental strain did not yield isolates with either the FS phenotype or altered EPS synthesis, even though the recombinants appeared to have the Tn5 inserted in the same location as FS1 (39a). Therefore, in order to more rigorously test the correlation between FS behavior and Tn5 insertions at this locus, as well as to identify other possibly critical sequences, additional FS mutants were isolated by two cycles of enrichment of a mini-Tn5 mating mixture for cells from the periphery of 6-day-old swarm colonies on NM semisolid agar.
FIG. 2.
Southern analysis of six independent FS mutants of S. meliloti RMB7201. Panel A shows FS1 (lanes 1 and 4), FS7 (lanes 2 and 5), and FS44 (lanes 3 and 6) probed with the 3.2-kb HindIII fragment from wild-type Tn5. Panel B shows FS17 (lanes 1 and 4), FS27 (lanes 2 and 5), and FS28 (lanes 3 and 6) probed with the 5.0-kb EcoRI-BamHI fragment from mini-Tn5-lacZ1. RMB7201 DNA has no detectable sequence homology to wild-type Tn5 but shows a faint band when probed with the mini-Tn5 sequence. The values between the lanes are approximate molecular sizes in kilobases.
Three new mini-Tn5 mutants, FS17, FS27, and FS28, each having the same FS–reduced-EPS phenotype as FS1, were isolated after two cycles of enrichment of the mating mixture. Whereas the restriction patterns for the original mutants, FS1, FS7, and FS44, were identical to each other for each of the four enzymes tested, the patterns for FS17, FS27, and FS28 differed from each other (Fig. 2B).
The great majority of the isolates obtained from mini-Tn5 mating enrichments and examined by Southern blot analysis had the FS–reduced-EPS phenotype of FS1 but did not contain mini-Tn5 insertions (data not shown). It seemed possible that such FS isolates might be spontaneous FS variants of the wild type. To test this possibility, evidence of spontaneous FS variant formation was sought by both enrichment from the periphery of wild-type swarm colonies and by screening of the wild type for reduced-EPS variants on NM-succinate-nitrate agar. Spontaneous variants of the wild type showing FS behavior were obtained in several experiments, although not in every experiment (Table 7). Some of the spontaneous FS variants (e.g., SV3, SV68, and SV69) swarmed at rates intermediate between those of the wild type and FS1. While most of the spontaneous FS variants were stable during purification, storage, and subculture, some were not. Overall, spontaneous FS-reduced EPS variants of the wild type appeared at a frequency of about 1 in 10,000 to 20,000 in both broth cultures and swarm colonies.
TABLE 7.
Spontaneous FS variants obtained by serial enrichment or direct screening of the RMB7201 wild type
| Isolation method | Phenotype | Representative isolate(s) | % of total |
|---|---|---|---|
| Serial enrichmenta | Normal swarm rate, normal EPS | SV1 | 25–50 |
| Intermediate swarm rate (>1.4–<1.6 × wild type); reduced EPS synthesis | SV3, SV5, SV6 | 30–50 | |
| Increased swarm rate (1.8 × wild type); reduced EPS synthesis | SV2, SV4, SV8 | 30–70 | |
| Increased swarm rate, normal EPS synthesis | SV19 | <1 | |
| Direct screeningb | Normal swarm rate, reduced EPS | 93 | |
| Increased swarm rate (>1.6 × wild-type rate), reduced EPS | SV8 | 2 | |
| Intermediate swarm rate (>1.4–<1.6 × wild-type rate), reduced EPS | SV68, SV69 | 1 | |
| Normal swarm rate, reduced EPS | SV2-5 | 4 |
Isolates were obtained from three independent experiments after serial enrichment as described in the text.
Isolates were identified after screening of approximately 254,000 colonies in two separate experiments.
Reversion of the FS, reduced-EPS variants and mutants to normal swarming and EPS synthesis was observed in several experiments but was not consistent or predictable. Reversion, detected by mucoid colony formation on NM-succinate-nitrate agar, was observed in three of nine separate experiments with SV8. Reversion to wild-type behavior occurred at frequencies ranging from about 0.01% to about 5% in the three positive trials (data not shown). Reversion during host plant infection and nodule formation was also examined. When FS1, FS7, FS44, FS17, FS27, FS28, SV3, SV8, SV9, SV68, and SV69 were inoculated onto alfalfa roots in growth pouches, they all formed normal numbers of normal nodules. This establishes that these isolates are derivatives of S. meliloti, not some contaminating species, and that these FS mutants and variants are symbiotically infective and effective. Randomly selected nodules generated by various FS isolates were crushed to determine the swarming and EPS phenotypes of the nodule occupants. Revertants of several of the strains were observed in the first experiment, but there were none in the second, larger experiment (data not shown).
Growth rate of FS isolates obtained by enrichment or indirect screening.
As shown in Table 4, both FS17, a mini-Tn5 mutant, and SV8, a spontaneous FS variant, behaved like FS1 with respect to growth relative to the wild type in shake, still, and swarm agar cultures. FS variants SV68 and SV69, with intermediate swarm rates (ca. 1.4 times faster than that of the wild type), had less of a growth advantage (20- to 27-fold) in swarm agar than did faster-swarming strains FS1, FS17, and SV8. SV3, another FS isolate with an intermediate swarm rate, had a large (36-fold) growth advantage in swarm agar, like that of FS1. Cells from the periphery of these colonies swarmed at rates equal to that of FS1 rather than the initial, intermediate rate characteristic of SV3, indicating that SV3 spontaneously switched from intermediate to fast swarming.
DISCUSSION
Spontaneous FS variants of S. meliloti RMB7201 were readily isolated from the periphery of growing swarm colonies, and their swarming behavior has generally proven stable during subsequent subculture and testing. In addition, FS variants were reproducibly obtained by screening of individual colonies for diminished EPS synthesis on NM agar, a trait associated with FS behavior in almost all of the FS isolates of RMB7201. The ability to identify FS variants based on the EPS phenotype is important because it demonstrates that the formation of FS variants in a population of RMB7201 cells occurs independently of growth in a swarm colony or the presence of sustained nutrient gradients. The formation of FS variants occurred spontaneously at a frequency of about 1 in 10,000 to 20,000 cells. This frequency seems too high to be readily explained by random mutation. Nonetheless, FS behavior is clearly heritable. Further studies are in progress to determine the genetic mechanisms underlying FS variant formation and reversion.
Several observations indicate that there may be multiple genetic configurations that result in FS behavior. (i) The restriction patterns of FS17, FS27, and FS28 are different, although they have the same FS-EPS phenotype (Fig. 2B). (ii) The restriction patterns and swarming behavior of FS1, FS7, and FS44 are the same, but these mutants differ in EPS regulation or swimming behavior. (iii) Spontaneous FS variants (e.g., SV3 and SV69) have been isolated with intermediate swarm rates. These genetic and phenotypic differences between FS isolates emphasize the potential complexity of FS switching. They indicate a need for caution in extrapolating the behavior of one FS isolate to others and suggest that there could be a number of cryptic traits (like reduced EPS synthesis on defined media) associated with FS behavior that remain to be identified.
Given that cells of S. meliloti do spontaneously switch between normal and increased swarming behavior, it is of interest to make a tentative assessment of the likely costs and benefits of FS behavior to populations of these bacteria in natural environments. FS1, FS17, and SV8 were found to grow at the same rate as the wild type in mixed shake cultures on moderately rich media (Table 4). This indicates that the metabolic costs of FS behavior, relative to normal swarming behavior, are modest. The parental strain and FS1 showed very similar abilities to survive for long periods of in vitro storage under conditions of very low nutrient availability. This also suggests that FS behavior is not much more costly in terms of metabolic resources than normal swarming behavior, perhaps because FS switching does not affect the extensive downregulation of motility under starvation conditions (Table 6 and reference 39).
The FS mutants had a consistent and significant growth advantage over the parental strain under all in vitro conditions in which appreciable nutrient gradients developed, including still cultures, moist sand, and swarm agar. FS variants SV68 and SV69, with intermediate swarm rates (ca. 1.3- to 1.5-fold greater than that of the wild type), appeared to have intermediate (20- to 27-fold) growth advantages in swarm agar, indicating that the competitive growth advantage of FS behavior is proportional to the rate of swarming. The faster growth of the FS mutants and variants in swarm colonies should probably be attributed to better access to available nutrients near the periphery. The periphery of swarm colonies generated by equal numbers of mutant and wild-type cells was 100-fold enriched for FS mutant cells.
The disadvantages of FS behavior are not so apparent. The only circumstance in which FS isolates were found to have an appreciably lower growth or survival rate is in a sterile soil matrix subjected to air drying (Table 5). Even here it is important to recognize that the poor survival of the FS mutants relative to the parental strain during such drying might be due to associated changes in EPS synthesis or some other cryptic, coordinately regulated trait rather than FS behavior per se. EPS synthesis is mentioned specifically because it is downregulated in almost all of the FS mutants and variants and because EPS synthesis is known to be important to drying tolerance (30). However, FS7, which had only a 30% reduction in EPS synthesis on NM-succinate-nitrate medium, survived air drying of the matrix just as poorly as FS1, which had a 60% reduction in EPS synthesis (Table 5). This suggests that FS behavior, rather than the differences in regulation of EPS synthesis is the most important determinant of differential survival in a drying soil. If this is so, the reason and mechanism are unclear. All considered, there is still no satisfactory explanation for the persistence of the majority of RMB7201 cells and isolates in the normal genetic configuration rather than an FS configuration.
The cellular-biochemical mechanisms responsible for the 1.4- to 2.5-fold faster spreading rates of the FS mutants and variants are not yet clearly established. From Table 3, it appears that FS behavior does not involve changes like the two- to sixfold increase in general chemotactic responsiveness seen in cells of RMB7201 after transfer to starvation conditions (39). The longer and more numerous flagella on the FS mutants could account for much of the observed twofold faster spreading rate, although a linear dependence of swarm rate on flagellation is not certain. Since S. meliloti has four independently transcribed flagellin genes (5, 29), it is also possible that the subunit composition of flagella on FS cells differs from that of flagella on wild-type cells. Based on the upregulation of swarming caused by Tn5 insertions, normally a loss-of-function event, and on the increased swarming rate of the wild type, approaching that of the FS mutants, when the concentration of added nutrients approached zero (Fig. 1), it seems that the increased spreading rate of FS mutants and variants may best be explained by a modest, general derepression of flagellar synthesis.
The frequent association of altered swarming behavior with altered EPS synthesis in FS mutants and variants suggests that some relatively strong, common regulatory factor(s) is involved, similar, perhaps, to phenotypic switching in Pseudomonas solanacearum, where both EPS and motility are coordinately affected by spontaneous mutations in phcA (10, 11). Mutations in EPS regulatory genes exoR, exoC, exoX, and exoS of S. meliloti result in elevated EPS synthesis (15, 31). Our finding that these mutations also result in reduced swarming suggests that changes in the expression of such genes could be involved in at least some instances of FS switching in S. meliloti.
We suspect that switching to FS behavior is a fairly common, although unappreciated, behavioral capability of bacteria. In the literature, it is apparent that many workers (e.g., 2, 21–23, 27) have routinely enriched their strains for more motile isolates to use in motility and chemotaxis assays, following the early suggestion of Adler (1). It seems likely that many of the isolates selected in this way were spontaneous FS variants with reasonable genetic stability. If so, the behavior of the wild-type forms of these strains remains to be characterized. In preliminary tests, it has proven rather easy to isolate FS variants of E. coli CC118λpir and Pseudomonas aeruginosa PAO1 after two to four cycles of swarm plate enrichment. The FS variants we obtained of these two model species were stable on storage and plating, swarmed in 0.35% agar at rates 1.5 to 2 times faster than those of the stock cultures, had swimming behavior similar to that of the wild type under the microscope, and outgrew the parental forms by 37- and 83-fold, respectively, after 2 days in 1/10 TY swarm agar (data not shown). If FS switching and behavior are indeed common phenomena, then further studies to examine the ecological consequences of FS behavior and the genetic mechanisms of FS variant formation and reversion in a number of representative species are clearly warranted.
ACKNOWLEDGMENTS
We appreciate the assistance of Elke Kretschmar and Robert Whitmoyer in the use of the electron microscope and thank John Leigh and Graham Walker for providing strains and plasmids. Helpful comments on the research and manuscript were provided by Gustavo Caetano-Anolles, David Coplin, Jyan-Chyun Jang, Jayne Robinson, and John Streeter. Their ideas and criticisms are much appreciated.
Partial support for salaries, supplies, and publication costs was provided by state and federal funds appropriated to the Ohio Agricultural Research and Development Center, Ohio State University.
Footnotes
Ohio Agricultural Research and Development Center contribution number 98-22.
REFERENCES
- 1.Adler J. A method for measuring chemotaxis and use of the method to determine optimum conditions for chemotaxis by E. coli. J Gen Microbiol. 1973;74:77–91. doi: 10.1099/00221287-74-1-77. [DOI] [PubMed] [Google Scholar]
- 2.Aguilar J M M, Ashby A M, Richards A J M, Loake G J, Watson M D, Shaw C H. Chemotaxis of R. leguminosarum biovar phaseoli towards flavonoid inducers of the symbiotic nodulation genes. J Gen Microbiol. 1988;134:2741–2746. [Google Scholar]
- 3.Ames P, Bergman K. Competitive advantage provided by bacterial motility in formation of nodules by Rhizobium meliloti. J Bacteriol. 1981;148:728–929. doi: 10.1128/jb.148.2.728-729.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Armitage J P, Schmitt R. Bacterial chemotaxis: Rhodobacter sphaeroides and Sinorhizobium meliloti—variations on a theme? Microbiology. 1997;143:3671–3682. doi: 10.1099/00221287-143-12-3671. [DOI] [PubMed] [Google Scholar]
- 5.Bergman K, Nulty E, Su L. Mutations in the two flagellin genes of Rhizobium meliloti. J Bacteriol. 1991;173:3716–3723. doi: 10.1128/jb.173.12.3716-3723.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bergman K, Gulash-Hoffee M, Hovestadt R E, Larosiliere R C, Ronco II P G, Su L. Physiology of behavioral mutants of Rhizobium meliloti: evidence for a dual chemotaxis pathway. J Bacteriol. 1988;170:3249–3254. doi: 10.1128/jb.170.7.3249-3254.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bhuvaneswari T V, Bhagwat A A, Bauer W D. Transient susceptibility of root cells in four common legumes to nodulation by rhizobia. Plant Physiol. 1981;68:1144–1149. doi: 10.1104/pp.68.5.1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blair D F. How bacteria sense and swim. Annu Rev Microbiol. 1995;49:489–522. doi: 10.1146/annurev.mi.49.100195.002421. [DOI] [PubMed] [Google Scholar]
- 9.Bosworth A H, Williams M K, Albrecht K A, Hankinson T R, Kwiatkowski R, Beynon J, Ronson C W, Cannon F, Wacek T J, Triplett E W. Alfalfa yield response to inoculation with recombinant strains of Rhizobium meliloti carrying an extra copy of dctA and/or modified nifA expression. Appl Environ Microbiol. 1994;60:3815–3832. doi: 10.1128/aem.60.10.3815-3832.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brumbley S M, Denny T P. Cloning of wild-type Pseudomonas solanacearum phcA, a gene that when mutated alters expression of multiple traits that contribute to virulence. J Bacteriol. 1990;172:5677–5685. doi: 10.1128/jb.172.10.5677-5685.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brumbley S M, Carney B F, Denny T P. Phenotype conversion in Pseudomonas solanacearum due to spontaneous inactivation of PhcA, a putative LysR transcriptional regulator. J Bacteriol. 1993;175:5477–5487. doi: 10.1128/jb.175.17.5477-5487.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Caetano-Anolles G, Wall L G, De Micheli A T, Macchi E M, Bauer W D, Favelukes G. Role of motility and chemotaxis in efficiency of nodulation by Rhizobium meliloti. Plant Physiol. 1988;86:1228–1235. doi: 10.1104/pp.86.4.1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Lorenzo V, Herrero M, Jakubzik U, Timmis K P. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J Bacteriol. 1990;172:6568–6572. doi: 10.1128/jb.172.11.6568-6572.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dharmatilake A J, Bauer W D. Chemotaxis of Rhizobium meliloti towards nodulation gene-inducing compounds from alfalfa roots. Appl Environ Microbiol. 1992;58:1153–1158. doi: 10.1128/aem.58.4.1153-1158.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Doherty D, Leigh J A, Glazebrook J, Walker G C. Rhizobium meliloti mutants that overproduce the R. meliloti acidic Calcofluor-binding exopolysaccharide. J Bacteriol. 1988;170:4249–4256. doi: 10.1128/jb.170.9.4249-4256.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Finan T M, Kunkel B, De Vos G F, Signer E R. Second symbiotic megaplasmid in Rhizobium meliloti carrying exopolysaccharide and thiamine synthesis genes. J Bacteriol. 1986;167:66–72. doi: 10.1128/jb.167.1.66-72.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Götz R, Limmer N, Ober K, Schmitt R. Motility and chemotaxis in two strains of Rhizobium with complex flagella. J Gen Microbiol. 1982;128:789–798. [Google Scholar]
- 18.Götz R, Schmitt R. Rhizobium meliloti swims by unidirectional, intermittent rotation of right-handed flagellar helices. J Bacteriol. 1987;169:3146–3150. doi: 10.1128/jb.169.7.3146-3150.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Greck M, Platzer J, Sourjik V, Schmitt R. Analysis of a chemotaxis operon in R. meliloti. Mol Microbiol. 1995;15:989–1000. doi: 10.1111/j.1365-2958.1995.tb02274.x. [DOI] [PubMed] [Google Scholar]
- 20.Gulash M, Ames P, LaRosiliere R C, Bergman K. Rhizobia are attracted to localized sites on legume roots. Appl Environ Microbiol. 1984;48:149–152. doi: 10.1128/aem.48.1.149-152.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaempf C, Greenberg E P. Negative chemotaxis in Spirochaeta aurantia. Curr Microbiol. 1992;21:187–192. [Google Scholar]
- 22.Kape R, Parniske M, Werner D. Chemotaxis and nod gene activity of Bradyrhizobium japonicum in response to hydroxycinnamic acids and isoflavonoids. Appl Environ Microbiol. 1991;57:316–319. doi: 10.1128/aem.57.1.316-319.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krupski G, Götz R, Ober K, Pleier E, Schmitt R. Structure of complex flagellar filament in Rhizobium meliloti. J Bacteriol. 1985;162:361–366. doi: 10.1128/jb.162.1.361-366.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leigh J A, Signer E R, Walker G C. Exopolysaccharide-defecient mutants of Rhizobium meliloti that form ineffective nodules. Proc Natl Acad Sci USA. 1985;82:6231–6235. doi: 10.1073/pnas.82.18.6231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Long S, Reed J W, Himawan J, Walker G C. Genetic analysis of a cluster of genes required for synthesis of the Calcofluor-binding exopolysaccharide of Rhizobium meliloti. J Bacteriol. 1988;170:4239–4248. doi: 10.1128/jb.170.9.4239-4248.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meade H M, Long S R, Ruvkun G B, Brown S E, Ausubel F M. Physical and genetic characterization of symbiotic and auxotrophic mutants of Rhizobium meliloti induced by transposon Tn5 mutagenesis. J Bacteriol. 1982;149:114–122. doi: 10.1128/jb.149.1.114-122.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nikata T, Sumida K, Kato J, Ohtake H. Rapid method for analyzing bacterial behaviorial responses to chemical stimuli. Appl Environ Microbiol. 1992;58:2250–2254. doi: 10.1128/aem.58.7.2250-2254.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Palleroni N J. Chamber for bacterial chemotaxis experiments. Appl Environ Microbiol. 1976;32:729–730. doi: 10.1128/aem.32.5.729-730.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pleier E, Schmitt R. Expression of two Rhizobium meliloti flagellin genes and their contribution to the complex filament structure. J Bacteriol. 1991;173:2077–2085. doi: 10.1128/jb.173.6.2077-2085.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Potts M. Desiccation tolerance of prokaryotes. Microbiol Rev. 1994;58:755–805. doi: 10.1128/mr.58.4.755-805.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reuber T L, Long S, Walker G C. Regulation of Rhizobium meliloti exo genes in free-living cells and in planta examined by using TnphoA fusions. J Bacteriol. 1991;173:426–434. doi: 10.1128/jb.173.2.426-434.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Robinson J B, Tuovinen O H, Bauer W D. Role of divalent cations in the subunit association of complex flagella from Rhizobium meliloti. J Bacteriol. 1992;174:3896–3902. doi: 10.1128/jb.174.12.3896-3902.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Robyt J F, White B J. Biochemical techniques, theory and practice. Prospect Heights, Ill: Waveland Press, Inc.; 1987. [Google Scholar]
- 34.Ronco P G., II . Tn5 mutagenesis of the motility and chemotaxis genes. M. S. thesis. Boston, Mass: Northeastern University; 1988. [Google Scholar]
- 35.Sambrook J E, Fritsch F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 36.Seed P C, Passador L, Iglewski B H. Activation of the Pseudomonas aeruginosa lasI gene by LasR and the Pseudomonas autoinducer PAI: an autoinduction regulatory hierarchy. J Bacteriol. 1995;177:654–659. doi: 10.1128/jb.177.3.654-659.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Selvaraj G, Iyer V N. Suicide plasmid vehicles for insertion mutagenesis in Rhizobium meliloti and related bacteria. J Bacteriol. 1983;156:1292–1300. doi: 10.1128/jb.156.3.1292-1300.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tuhela L, Robinson J B, Tuovinen O H. Characterization of chemotactic responses and flagella of Hyphomicrobium strain W1-1B. J Bacteriol. 1998;180:3003–3006. doi: 10.1128/jb.180.11.3003-3006.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wei X, Bauer W D. Starvation-induced changes in motility, chemotaxis, and flagellation of Rhizobium meliloti. Appl Environ Microbiol. 1998;64:1708–1714. doi: 10.1128/aem.64.5.1708-1714.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39a.Wei, X., and W. D. Bauer. Unpublished data.
- 40.Zhan H, Leigh J A. Two genes that regulate exopolysaccharide production in Rhizobium meliloti. J Bacteriol. 1990;172:5254–5259. doi: 10.1128/jb.172.9.5254-5259.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]