Summary
The different variants of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) have attracted most public concern because they caused “wave and wave” COVID-19 pandemic. The initial step of viral infection is mediated by the SARS-CoV-2 Spike (S) protein, which mediates the receptor recognition and membrane fusion between virus and host cells. Neutralizing antibodies (nAbs) targeting the S protein of SARS-CoV-2 have become promising candidates for clinical intervention strategy, while multiple studies have shown that different variants have enhanced infectivity and antibody resistance. Here, we explore the structure and function of STS165, a broadly inter-Spike bivalent nAb against SARS-CoV-2 variants and even SARS-CoV, contributing to further understanding of the working mechanism of nAbs.
Subject areas: Biochemistry applications, Molecular Structure, Structural biology, Virology
Graphical abstract
Highlights
-
•
STS165 broadly neutralizes different variants of SARS-CoV-2 and even SARS-CoV
-
•
STS165 exhibits inter-Spike bivalent binding characteristics
-
•
STS165 may work as an ideal partner to form therapeutic antibody cocktails
Biochemistry applications; Molecular Structure; Structural biology; Virology
Introduction
The ongoing coronavirus disease 2019 (COVID-19) pandemic, caused by different variants of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has highlighted the need for effective therapeutic interventions (Zhu et al., 2020). The SARS-CoV-2 is an enveloped, positive-strand RNA virus that belongs to the β-coronavirus family. Because of the selective pressure from natural and vaccine-induced immunity, SARS-CoV-2 is constantly evolving to escape the humoral immunity and creates another wave of infection, especially the Omicron variant of SARS-CoV-2. Omicron variant is a new variant of concern (VOC) identified in South Africa since November 2021 and has spread rapidly across the world.
One of the reasons for the prevalence of the Omicron variant is that over 30 mutations occur in the Spike protein on the virion surface, which is responsible for receptor recognition. During viral infection, the trimeric S protein binds to the host receptor and is cleaved into S1 and S2 subunits by host protease (Belouzard et al., 2009; Simmons et al., 2004, 2013; Song et al., 2018). S1 subunit contains the N-terminal domain (NTD) and receptor binding domain (RBD), which directly bind to the host receptor (Li et al., 2005; Yan et al., 2020). S2 subunit is responsible for membrane fusion between virus and host cells. The binding of S1 to the receptor and protease digestion trigger the conformational change of trimeric Spike and expose the fusion peptide of S2, enabling the virus entry into host cells (Belouzard et al., 2009; Millet and Whittaker, 2015; Simmons et al., 2005). Similar to the original SARS-CoV-2, the Omicron variant still exploits ACE2 for host infection (Cui et al., 2022).
Neutralizing antibodies (nAbs) against SARS-CoV-2 represent a promising clinical intervention. Many complex structures of nAbs targeting Spike protein have been determined, most of which used the Fab form rather than the full-length IgG form that contains two Fab fragments in a “Y” shape and indicates a putative bivalent binding effect. It has been reported that the bivalent binding of antibodies mediated a more potent function than Fab form in neutralizing viruses such as rhinovirus, Dengue virus, and even SARS-CoV-2 (Edeling et al., 2014; Hewat and Blaas, 1996; Yan et al., 2021). The structural and functional intra-Spike bivalent binding of nAbs has been revealed by our previous study and exhibited more potent neutralizing activity than monovalent Fab form against wild-type (WT) SARS-CoV-2 (Yan et al., 2021). However, whether there is another form of bivalent binding such as the single antibody that can bind to two Spike proteins (inter-Spike bivalent binding) remains unclear. Here, we report a neutralizing antibody STS165, which can neutralize the existing variants of SARS-CoV-2, including the Omicron variant. More importantly, the inter-Spike bivalent binding characteristics in SARS-CoV-2 was first revealed by the full-length IgG form of STS165.
Results
Identification of STS165 as a broadly neutralizing antibody
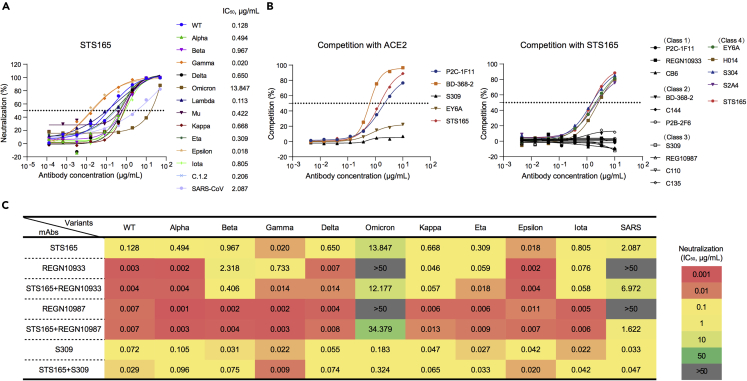
Briefly, STS165 is isolated from peripheral blood specific single memory B cells of a COVID-19 convalescent individual using SARS-CoV-2 RBD protein as bait. STS165 can neutralize 13 pseudoviruses of SARS-CoV-2 variants, including WT, Alpha, Beta, Gamma, Delta, Lambda, Mu, Kappa, Eta, Epsilon, Iota, C.1.2, and even Omicron, indicating its great potential. Especially, STS165 also neutralizes SARS-CoV with 50% inhibitory concentration (IC50) of 2.087 μg/mL (Figure 1A). Relatively high binding affinities to RBD proteins of SARS-CoV-2 variants and SARS-CoV in the nanomolar range or below contributed to the broadly neutralizing activities of STS165 (Figure S1). The binding affinity of STS165 to Omicron RBD was simply decreased by 3-fold than that to WT RBD, and its neutralizing activity was largely reduced by these mutations. Similar results were also observed in the binding and neutralizing activities of the other two nAbs, AZD8895, and AZD1061 (Cao et al., 2021), suggesting that the monomeric RBD protein could not fully represent the native conformational Spike trimer on the surface of virus membrane to evaluate the interaction between nAbs and virus, especially the neutralization. These data indicated that STS165 maintained the neutralization activity against Omicron and even SARS-CoV despite the significant reduction. These data also suggest that there are many key binding sites between STS165 and Omicron that deserve further exploration.
Figure 1.
Biochemical characteristics of a broadly neutralizing antibody STS165
(A) The broadly neutralizing antibody STS165 can neutralize 13 pseudoviruses of SARS-CoV-2 variants and SARS-CoV.
(B) The competition assay of STS165 with ACE2 (left) and other representative antibodies (right).
(C) The neutralization of 11 pseudoviruses by 4 antibodies (STS165, REGN10933, REGN10987, and S309) and antibody cocktails (STS165+REGN10933, STS165+REGN10987, and STS165+S309). The data represented here are means of at least two independent experiments. The curves are representatives of similar results.
It is widely known that RBD-specific nAbs could be divided into four classes according to the competition with ACE2 and the accessibility of epitopes recognized on the RBD in up or down conformation (Barnes et al., 2020). Using competition ELISA, we predicted the binding epitope of STS165 based on its competition with ACE2. STS165 exhibited high capacity in competing with the ACE2 receptor, which is similar to the representative Class 1 antibody P2C-1F11 and Class 2 antibody BD-368-2 (Figure 1B, left panel). To identify a more precise epitope recognized by STS165, we further measured its competition with other 14 monoclonal antibodies whose epitopes have been well studied via structural biology technology. The 14 antibodies included are representative ones falling in the four classical classes that have been identified: Class 1: P2C-1F11, REGN10933, CB6; Class 2: BD-368-2, C144, P2B-2F6; Class 3: S309, REGN10987, C110, C135; Class 4: EY6A, H014, S304, S2A4 (Ju et al., 2020; Li et al., 2022; Piccoli et al., 2020). As shown in Figure 1B (right panel), the STS165 antibody could compete with four Class 4 antibodies, indicating that STS165 belongs to the Class 4 antibody whose recognition site was far away from the receptor-binding motif (RBM).
Many diverse SARS-CoV-2 variants have emerged and spread around the world. These variants are resistant to specific monoclonal antibodies that have been authorized or approved for clinical use. Especially, the Omicron has escaped most of the authorized nAbs except S309 (Cao et al., 2021). Thus, it is urgent to rescue the lost neutralization of these nAbs against Omicron and ongoing variants. As STS165 can neutralize 13 variants including Beta, Delta, and Omicron, it may be an ideal partner to form a therapeutic antibody cocktail. This cocktail is not only an effective treatment but may also protect against antibody resistance caused by virus escape mutants that could arise in response to selective pressure from single antibody treatments. We evaluated the escape of the 11 pseudoviruses from neutralization by the STS165 in combination with other 3 non-competition nAbs approved for clinical use (REGN10933, REGN10987, and S309). As shown in Figures 1C and S2, the level of REGN10933 against Beta was lower than that against WT, displaying a significant decrease in IC50 (2.318 μg/mL). Importantly, the inhibition values of REGN10933 and REGN10987 against Omicron and SARS-CoV were below 50% when tested at the highest concentration (50 μg/mL). However, the approved antibodies REGN10933 and REGN10987 in combination with STS165, respectively, restored their neutralizing activities against Omicron and SARS-CoV to some extent. Notably, for other variants, STS165 cocktail therapy with REGN10933, REGN10987, and S309 antibodies, respectively, also maintained their neutralizing activities against pseudoviruses. In summary, these results indicate that STS165 is not only a broadly neutralizing antibody but also can be used as a partner with different approved antibodies to resist constantly emerging SARS-CoV-2 variants.
Structure determination of Omicron S-6p bound with STS165
To characterize the binding epitope recognized by STS165, we aimed to determine the cryo-EM structure of the Omicron variant of SARS-CoV-2 S-6p bound with STS165. The S-6p protein of the Omicron variant was purified by Ni-NTA affinity resin. To test whether this protein is functional, we determined the structure of Omicron S-6p bound with the peptidase domain (PD) of ACE2. This structure is almost identical to the previously reported structures (Figures S3, S4, and S5 and Table S1) (Cui et al., 2022). The IgG form of STS165 and Omicron S-6p were mixed and incubated at a molar ratio of ∼3.6 to 1 for 1 h and applied to size exclusion chromatography (SEC) to remove excess antibodies (Figure S6A). The peak fractions containing the complex were concentrated for further cryo-EM sample preparation.
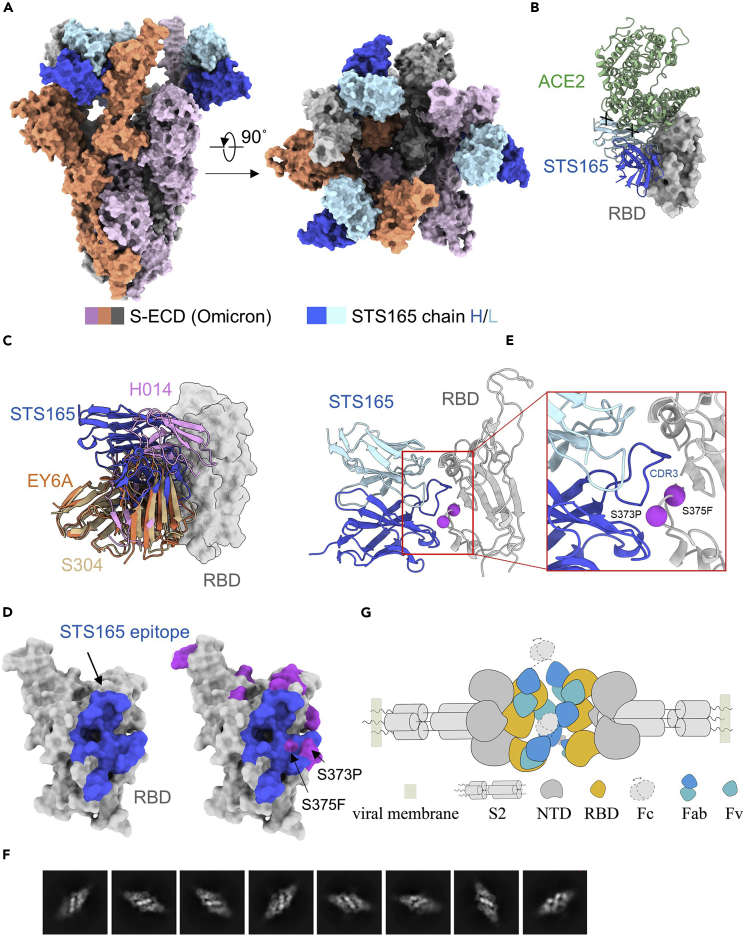
The cryo-EM structure of Omicron S-6p bound with STS165 was determined at an overall resolution of 3.3 Å (Figures 2A, S5, S6, and S7 and Table S1). However, the resolution at the interface between RBD and STS165 was not high enough to reveal a detailed interaction network because of the flexibility of this region even with focused refinement applied. To characterize the putative epitope, we docked an Alphafold predicted STS165 into the density for analysis. When aligned with the ACE2-binding site on the RBD, we can find the overlap between ACE2 and STS165, consistent with our competition assays (Figure 2B).
Figure 2.
Cryo-EM structure of S-ECD (Omicron) in complex with STS165
(A) The domain-colored cryo-EM structure of S-ECD (Omicron) in complex with STS165 is shown in two perpendicular views. STS165 binds to three “up” RBDs, which the heavy and light chains of it are colored blue and cyan, respectively. Each of the Spike trimer is colored pink, orange, and gray, respectively.
(B) Structural comparison between ACE2 (green) and STS165 bound to RBD, and the spatial overlap of them is represented by a black x.
(C) Structural classification of some Class 4 representative nAbs, which are similar to the STS165 epitope. H014 (PDB: 7CAH), EY6A (PDB: 6ZFO), and S304 (PDB: 7R6X).
(D) The binding epitope of STS165 bound to the RBD is colored blue on the left, which overlaps with the mutated residues (purple) relative to the RBD (Wuhan-Hu-1) on S373 and S375.
(E) The binding interface between STS165 and RBD (Omicron). The mutated residues relative to the RBD (Wuhan-Hu-1) are shown as purple spheres.
(F) The representative bivalent binding 2D classification of Omicron S-6p bound to STS165.
(G) The proposed inter-Spike bivalent binding model of STS165 based on the structures.
The STS165 binding Omicron S-6p exhibits a conformation with three “up” RBDs, each of which is bound by an STS165 fragment (Figure 2A). For an overall view, STS165 targets the lateral side of RBD, which is a highly conserved epitope between the SARS-CoV and SARS-CoV-2 (Lv et al., 2020; Zhou et al., 2020a). STS165 can be classified into Class 4, which also includes H014, EY6A, and S304 (Figure 2C). We mapped the epitope of STS165 on the RBD of Omicron variant and found that the S373P and S375F might be included (Figures 2D and 2E). We propose these mutations might alter the neutralizing activities of STS165 against the Omicron variant by changing the local environment. We also mapped the epitope of STS165 on the RBD of other SARS-CoV-2 variants, including Alpha, Beta, Gamma, Delta, Lambda, Mu, Kappa, Eta, Epsilon, Iota, and C.1.2 and found that no mutated residues were involved in this region, suggesting the conservation of this epitope (Figures S8 and S9). The Class 4 antibodies have been reported to have largely lost the neutralizing ability to the Omicron variant (Ju et al., 2022a), while STS165 still keeps its neutralizing ability. This might be attributed to the detailed epitope variation among these antibodies of Class 4.
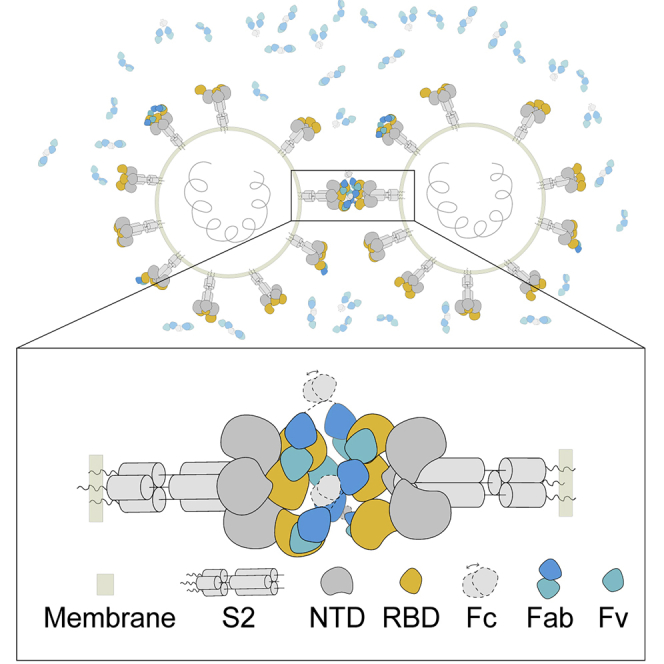
Intriguingly, we found a special binding pattern of STS165 in our 2D classification, of which the “head to head Spike” is revealed for the first time (Figure 2F). This pattern was not observed in the Fab-form of STS165 incubated with the Omicron variant (data not shown). This binding pattern indicates that the IgG form of STS165 can bind to two distinct Spikes simultaneously, revealing the inter-Spike bivalent binding characteristics. However, we could not solve the structure of the two-Spike binding pattern because of the preferred orientation of protein particles. Based on the structure of STS165 and Omicron S-6p complex, we proposed a bivalent binding model of STS165 in which at least two STS165 in IgG form can bind to the “up” RBD of distinct Spikes and limit them in a straight line (Figure 2G). This unique bivalent binding manner may enhance the neutralizing ability of STS165. Indeed, the neutralizing assay of STS165 in IgG form and Fab form also confirmed this hypothesis. Compared with the IgG form, the Fab form of STS165 nearly or totally lost the neutralizing activities against WT, Beta, Delta, and Omicron (Figure S10). To further explore the potential mechanism, we measured the binding affinities of the Fab form of STS165 to WT and Omicron RBD proteins, respectively (Figure S11). Compared with the IgG form (Figure S1), the binding affinity of STS165-Fab to WT or Omicron RBD was largely decreased by more than 50-fold, which was partially responsible for the serious reduction in neutralizing activity.
Discussion
The COVID-19 pandemic has lasted for more than two years around the world (Zhou et al., 2020b). As a variety of variants continue to appear, SARS-CoV-2 largely escaped the antibody immunity elicited by the natural virus infection and vaccine immunization (Dejnirattisai et al., 2021; McCallum et al., 2021; Planas et al., 2021; Wang et al., 2021). Before the Omicron emerged, any VOC or variant of interest (VOI) carried no more than three mutations in the RBD region. These mutations mainly decreased the neutralizing activities of Class 1, 2, and 3 nAbs to some extent (Cheng et al., 2021; Liu et al., 2021a; Yuan et al., 2021). None of key escape mutations was found to be located in the binding epitope of Class 4 nAbs, which were once considered as the most broad-spectrum nAbs (Barnes et al., 2020; Lv et al., 2020; Zhou et al., 2020a). However, the RBD of Omicron contained an alarming number of 15 mutations, some of which were first identified in SARS-CoV-2 variants and conferred the greater antibody resistance of Omicron (Cameroni et al., 2021; Cao et al., 2021; Dejnirattisai et al., 2022; Liu et al., 2021b). In particular, S371L appeared in the binding epitope of Class 4 nAbs and mediated the resistance of Omicron to most of these nAbs (Ju et al., 2022a; Liu et al., 2021b). Fortunately, we isolated a Class 4 nAb, named STS165, from a COVID-19 convalescent individual, retaining a degree of neutralizing activity against Omicron. Particularly, STS165 is characterized by the relatively high binding affinity to the RBD protein and the stable complex with the Spike trimer of Omicron, which facilitates the determination of the real cryo-EM structure of this class of nAbs bound with Omicron variant. The future research might well modify these Class 4 nAbs to improve their neutralizing activities based on the structural interaction. To our knowledge, this study for the first time directly observed an inter-Spike bivalent binding mode of a SARS-CoV-2 nAb through the application of the full IgG-form antibody to determine the structure. This kind of “head to head Spike” binding pattern in that two distinct Spike trimers are recognized and limited in a straight line by IgGs is unique. Our findings also demonstrated that this bivalent IgG form of STS165 had a higher binding affinity and neutralizing activity than its monovalent form of Fab. In addition, the bivalent binding of STS165 to distinct Spike proteins may increase the engulfing capacity and antigen-presenting function of macrophages against SARS-CoV-2 in vivo, which is needed to be further explored in the future. Overall, our functional and structural studies reveal a broadly nAb, STS165, targeting a conserved epitope on the SARS-CoV-2 RBD. The inter-Spike binding property of STS165 can enhance the neutralization activities and might mitigate the risk of viral escape. These results provide clues for antibody-based cocktail therapy interventions against COVID-19.
Limitations of study
Despite the exceptional findings for the STS165 targeting the different variants of SARS-CoV-2, there are two limitations in our study. First, the resolution of the interface between STS165 and the Spike protein is not high enough for model building, so we cannot clearly define the interaction map in this region. Second, the molecular mechanism for the inter-Spike bivalent binding mode of IgG-form STS165 remains unclear because of the preferred orientation of protein particles. We will unveil the relevant mechanisms underlying the inter-Spike bivalent binding by STS165 in the future.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| P2C-1F11 | Ge et al., 2021 | PDB code 7CDI |
| P2B-2F6 | Ju et al., 2020 | PDB code 7BWJ |
| REGN10933 | Hansen et al., 2020 | PDB code 6XDG |
| REGN10987 | Hansen et al., 2020 | PDB code 6XDG |
| CB6 | Shi et al., 2020 | PDB code 7C01 |
| BD-368-2 | Du et al., 2020 | PDB code 7CHH |
| C144 | Barnes et al., 2020 | PDB code 7K90 |
| C110 | Barnes et al., 2020 | PDB code 7K8V |
| C135 | Barnes et al., 2020 | PDB code 7K8Z |
| S309 | Pinto et al., 2020 | PDB code 6WPS |
| EY6A | Zhou et al., 2020a | PDB code 6ZCZ |
| H014 | Lv et al., 2020 | PDB code 7CAI |
| S304 | Piccoli et al., 2020 | PDB code 7JW0 |
| S2A4 | Piccoli et al., 2020 | PDB code 7JVC |
| CD3-Pacific Blue | BD Pharmingen | Cat#558117; RRID: AB_397038 |
| CD8-Pacific Blue | BD Pharmingen | Cat#558207; RRID: AB_397058 |
| CD14-Pacific Blue | BD Pharmingen | Cat#558121; RRID: AB_397041 |
| CD19-PE/Cy7 | BD Pharmingen | Cat#560911; RRID: AB_396893 |
| CD27-APC/H7 | BD Pharmingen | Cat#560222; RRID: AB_1645474 |
| FITC anti-IgG | BD Pharmingen | Cat#555786; RRID: AB_396121 |
| APC Anti-6×His | Abcam | Cat#ab72579; RRID: AB_1267597 |
| PE Anti-6×His | Abcam | Cat#ab72467; RRID: AB_1267596 |
| Bacterial and virus strains | ||
| E. coli DH5α | Invitrogen | Cat# 18265017 |
| SARS-CoV-2/wild-type (WT) | Ju et al., 2022a | N/A |
| SARS-CoV-2/Alpha | Ju et al., 2022a | N/A |
| SARS-CoV-2/Beta | Ju et al., 2022a | N/A |
| SARS-CoV-2/Gamma | Ju et al., 2022a | N/A |
| SARS-CoV-2/Delta | Ju et al., 2022a | N/A |
| SARS-CoV-2/Omicron | Ju et al., 2022a | N/A |
| SARS-CoV-2/Lambda | Ju et al., 2022b | N/A |
| SARS-CoV-2/Mu | Ju et al., 2022a | N/A |
| SARS-CoV-2/Kappa | Ju et al., 2022a | N/A |
| SARS-CoV-2/Eta | Ju et al., 2022a | N/A |
| SARS-CoV-2/Epsilon | Ju et al., 2022a | N/A |
| SARS-CoV-2/Iota | Ju et al., 2022a | N/A |
| SARS-CoV-2/C.1.2 | Ju et al., 2022a | N/A |
| SARS-CoV/wild-type (WT) | Ju et al., 2022a | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| SMM 293T-II medium | Sino Biological Inc. | Cat#SMM 293-T2 |
| SARS-CoV-2 WT RBD | Ju et al., 2022a | N/A |
| SARS-CoV-2 Mu RBD | Ju et al., 2022a | N/A |
| SARS-CoV-2 C.1.2 RBD | Ju et al., 2022a | N/A |
| SARS-CoV-2 Omicron RBD | Sino Biological Inc. | Cat#40592-V08H121 |
| SARS-CoV-2 Lambda RBD | Sino Biological Inc. | Cat# 40592-V08H113 |
| SARS-CoV RBD | Sino Biological Inc. | Cat#40150-V08B2 |
| anti-Flag M2 affinity resin | Sigma-Aldrich | Cat#A2220 |
| Ni-NTA agarose | QIAGEN | Cat#30230 |
| Superdex 200, 10/300 GL | GE Healthcare | Cat#17-5175-01 |
| Superose 6, 10/300 GL | GE Healthcare | Cat#17-5172-01 |
| Penicillin-Streptomycin (10,000 U/mL) | Gibco | Cat# 15140163 |
| Ampicillin | Amresco | Cat# 69-52-3 |
| Imidazole | Sigma-Aldrich | Cat# I5513 |
| Trizma base | Sigma-Aldrich | Cat# 77-86-1 |
| Polyethylenimines (PEIs) 25K | PolySciences | Cat# 23966 |
| Trypsin | Gibco | Cat#25200-072 |
| Fetal bovine serum | Gibco | Cat#10099-141C |
| Dulbecco's Modified Eagle Medium | Gibco | Cat#11965-092 |
| Papain | Sigma Aldrich | Cat#P4762-1G |
| HEPES(1M) Buffer Solution | Gibco | Cat#15630-080 |
| Opti-MEM Reduced Serum Medium | Gibco | Cat#51985034 |
| DEAE-Dextran hydrochloride | Sigma Aldrich | Cat#D9885-10G |
| Critical commercial assays | ||
| LIVE/DEAD™ Fixable Dead Cell Stain Kits | Invitrogen | Cat#L34964 |
| Bright-Lite Luciferase Assay System | Vazyme Biotech | Cat#DD1204-03 |
| TMB substrate | Sangon Biotech | Cat#E661007-0100 |
| HRP Conjugation Kit | Abcam | Cat#ab102890 |
| Gold Hi EndoFree Plasmid Maxi Kit | CWBIO | Cat#CW2104M |
| Deposited data | ||
| Coordinates of Spike of Omicron and PD of ACE2 | This paper | PDB: 7XID EMDB: EMD-33203 |
| Coordinates of Spike of Omicron and STS165 | This paper | PDB: 7XIC EMDB: EMD-33202 |
| Experimental models: Cell lines | ||
| HEK293F | Gibco | Cat# R79007 |
| Human: HEK293T | ATCC | Cat#CRL-3216 |
| HEK293T expressing human ACE2 | YEASEN Biotech | Cat#41107ES03 |
| Recombinant DNA | ||
| ECD of SARS-CoV-2 Spike (Omicron 6p) cloned onto a modified pCAG vector (His tag) | This paper | N/A |
| PD of human ACE2 cloned onto a modified pCAG vector (Flag tag) | This paper | N/A |
| pNL4-3.Luc.R-E- | NIH AIDS Reagent Program | N/A |
| Software and algorithms | ||
| MotionCorr | Li et al., 2013 | http://cryoem.ucsf.edu/software/driftcorr.html |
| MotionCor2 | Li et al., 2013 | http://cryoem.ucsf.edu/software/driftcorr.html |
| Gctf | Zhang, 2016 | http://www.mrc-lmb.cam.ac.uk/kzhang/Gctf/ |
| RELION 2.0 | Kimanius et al., 2016 | http://www2.mrc-lmb.cam.ac.uk/relion |
| RELION 3.0 | Zivanov et al., 2018 | https://www3.mrc-lmb.cam.ac.uk/relion/ |
| CRYOSPARC | Punjani et al., 2017 | https://www.cryosparc.com/ |
| COOT | Emsley et al., 2010 | http://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot |
| UCSF Chimera | Pettersen et al., 2004 | http://www.cgl.ucsf.edu/chimera |
| PHENIX | Adams et al., 2010 | https://www.phenix-online.org |
| PyMOL | PyMOL | http://www.pymol.org |
| Graphpad Prism 8 | GraphPad | https://www.graphpad.com/ |
| Biacore Evaluation software 3.0 | GE Healthcare | https://www.gehealthcare.com |
| STAR Software | Star Software Technologies | http://www.starsoftwarein.com/ |
| Other | ||
| R1.2/1.3 300 mesh Au grids | Quantifoil | Q17080 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Zheng Zhang (zhangzheng1975@aliyun.com).
Materials availability
All unique/stable reagents generated in this study are available from the lead contact with a completed Materials Transfer Agreement.
Experimental model and subject details
The recombinant protein was expressed using the HEK293F cells at 37°C under 5% CO2 in a Multitron-Pro shaker (Infors, 130 rpm). When the cell density reached 2.0 ×106 cells/mL, the plasmid was transiently transfected into the cells.
Method details
Protein expression and purification
The extracellular domain (ECD) (1-1208 a.a) of S protein of SARS-CoV-2 Omicron variant was cloned into the pCAG vector (Invitrogen) with six proline substitutions at residues 817, 892, 899, 942, 986 and 987 and a C-terminal T4 fibritin trimerization motif followed by 10×His tag. A “GSAS” mutation at residues 682 to 685 was introduced into ECD to prevent the host furin protease digestion. This construct was hereafter referred to as S-ECD (Omicron). The peptidase domain (PD) (19-615 a.a) of human ACE2 was also cloned into the pCAG vector (Invitrogen) with an N-terminal signal peptide of secreted luciferase and a C-terminal Flag tag. The mutants were generated with a standard two-step PCR-based strategy. All the plasmids used to transfect cells were prepared by Gold Hi EndoFree Plasmid Maxi Kit (CWBIO).
To transfect one liter of cell culture, about 1.5 mg of the plasmid was premixed with 3 mg of polyethylenimines (PEIs) (Polysciences) in 50 mL of fresh medium for 15 min before adding to cell culture. Cells were removed and medium was collected by centrifugation at 4000×g for 15 min after 60 h post-transfection.
The secreted S-ECD (Omicron) was purified by Ni-NTA affinity resin (Qiagen). The nickel resin loaded was rinsed with the wash buffer 1 containing 25 mM HEPES (pH 7.0), 500 mM NaCl, and washed with wash buffer 2 containing 25 mM HEPES (pH 7.0), 150 mM NaCl and 30 mM imidazole. Protein was eluted by wash buffer 2 plus 270 mM imidazole. Then the Ni-NTA eluent was subjected to size-exclusion chromatography (Superose 6 Increase 10/300 GL, GE Healthcare) in buffer containing 25 mM HEPES (pH 7.0), 150 mM NaCl. The peak fractions were collected and stored at −80°C.
The secreted PD was purified by anti-FLAG M2 affinity resin (Sigma Aldrich). After loading two times, the anti-FLAG M2 resin was washed with the wash buffer 3 containing 25 mM HEPES (pH 7.0), 150 mM NaCl. The protein was eluted with the wash buffer 3 plus 0.2 mg/mL flag peptide. The eluent of PD was then concentrated and subjected to size-exclusion chromatography (Superdex 200 Increase 10/300 GL, GE Healthcare) in buffer containing 25 mM HEPES (pH 7.0), 150 mM NaCl.
The S-ECD (Omicron) was incubated with PD or STS165 at a molar ratio of about 1:6 or 1:3.6 for 1 h. To remove excessive PD or STS165, the mixture was subjected to size-exclusion chromatography (Superose 6 Increase 10/300 GL, GE Healthcare) in buffer containing 25 mM HEPES (pH 7.0), 150 mM NaCl. The peak fractions containing protein complexes were collected for EM analysis.
Recombinant antibody production
Paired immunoglobulin heavy and light chain genes of STS165 antibody were obtained from Smart-seq2 Genomics V(D)J sequencing, and analysis was submitted to recombinant monoclonal antibody synthesis. Fastqc was used to perform quality control on the sequenced raw data. Reads were quality filtered with the cutoff threshold for average base quality score set at 20. The filtered reads were mapped against human reference genome (version: GRCh38) using STAR software. We use Trinity software to de novo assemble the transcripts of RNA-seq data. TransDecoder was used to identify the open reading frame and annotate the BCR sequence of the assembled contig. We extracted the annotated BCR sequence from all assembled mRNA transcripts. All the BCR sequences were manually corrected. Variable genes of published nAbs were downloaded from the National Center of Biotechnology Information (NCBI) and synthesized by Sangon Biotech and GenScript. Protein data bank (PDB) codes were P2C-1F11 (7CDI) (Ge et al., 2021), P2B-2F6 (7BWJ) (Ju et al., 2020), REGN10933 (6XDG) and REGN10987 (6XDG) (Hansen et al., 2020), CB6 (7C01) (Shi et al., 2020), BD-368-2 (7CHH) (Du et al., 2020), C144 (7K90), C110 (7K8V), and C135 (7K8Z) (Barnes et al., 2020), S309 (6WPS) (Pinto et al., 2020), EY6A (6ZCZ) (Zhou et al., 2020a), H014 (7CAI) (Lv et al., 2020), S304 (7JW0) and S2A4 (7JVC) (Piccoli et al., 2020), respectively. All variable genes of nAbs were cloned into IgG1 expression vectors containing heavy and light chain constant regions and then instantaneously co-transfected into 293F cells by using polyethylenimine (PEI, Polyscience). Seven days after transfection, antibodies were purified from the culture supernatants by protein A affinity chromatography.
Competitive enzyme-linked immunosorbent assay
96-well plates were coated with SARS-CoV-2 WT RBD proteins (Sino Biological) at 2 μg/mL in PBS at 4°C overnight The plates were washed with PBST (0.05% Tween-20 in PBS) buffer and blocked with 5% skim milk and 2% bovine albumin in PBS at RT for 1 h. Serially 3-fold diluted monoclonal antibodies were mixed with HRP (Abcam) conjugated human ACE2 protein or STS165 antibody in equal volume and then added to the plates and incubated at 37°C for 1 h. Finally, the TMB substrate (Sangon Biotech) was added to wells and incubated at RT for 20 min and the reaction was stopped by 2 M H2SO4. OD450 and OD630 were measured by ELISA microplate reader. The competitive effect is determined by comparing the ratio of different antibodies to control antibodies.
Binding analysis by surface plasmon resonance (SPR)
The binding assays of monoclonal antibodies to the RBD proteins of SARS-CoV-2 variants and SARS-CoV were performed using the Biacore 8K system (GE Healthcare). Specifically, one flow cell of the CM5 sensor chips were covalently coated with the RBD proteins (Sino Biological) in 10 mM sodium acetate buffer (pH 5.0) for a final RU (response units) of around 250, whereas the other flow cell was left uncoated and blocked as a control. All the assays were run at a flow rate of 30 μL/min in HBS-EP buffer (10 mM HEPES pH 7.4, 150 mM NaCl, 3 mM EDTA, and 0.05% Tween-20). Serially diluted antibodies were injected for 60 s respectively and the resulting data were fit in a 1:1 binding model with Biacore Evaluation software (GE Healthcare). Every measurement was performed three times and the individual values were used to produce the mean affinity constant and standard deviation.
Psesudovirus neutralization assay
Pseudovirus were generated by co-transfection of 20 μg of human immunodeficiency virus backbones expressing firefly luciferase (pNL4-3.Luc.R-E-) and 10 μg of expression vectors encoding the respective spike proteins into HEK-293T cells. After a 48 h transfection, cell culture containing virus supernatants was collected by centrifugation. The neutralization activity of the antibody was evaluated by incubating the serially diluted antibodies with an equal volume of pseudovirus at 37°C for 1 h. The virus-antibody mixture was then transferred to an all-white 96-well cell culture plate, and HEK-293T-hACE2 cells were subsequently added to the plates. 48 h post-incubation, the culture medium was removed and 100 μL of the Bright-Lite Luciferase reagent (Vazyme Biotech) was added to the plates. After a 2-min shock incubation at RT, the cell plates were measured by luminescence using the Varioskan™LUX multimode microplate reader (Thermo Fisher Scientific). Half-maximal inhibitory concentrations (IC50) were calculated using GraphPad Prism 8.0 software by log (inhibitor) vs. normalized response - Variable slope (four parameters) model.
Cryo-EM sample preparation and data acquisition
S-ECD (Omicron)-PD complex and S-ECD (Omicron)-STS165 complex was concentrated to ∼1.5 mg/mL and applied to the grids. Aliquots (3.3 μL) of the protein were placed on glo-discharged holey carbon grids (Quantifoil Au R1.2/1.3). The grids were blotted for 3.0 s or 3.5 s and flash-frozen in liquid ethane cooled by liquid nitrogen with Vitrobot (Mark IV, Thermo Fisher Scientific). The prepared grids were transferred to a Titan Krios operating at 300 kV equipped with Gatan K3 detector and GIF Quantum energy filter. Movie stacks were automatically collected using AutoEMation (Lei and Frank, 2005), with a slit width of 20 eV on the energy filter and a defocus range from −1.4 μm to −1.8 μm in super-resolution mode at a nominal magnification of 81,000×. Each stack was exposed for 2.56 s with an exposure time of 0.08 s per frame, resulting in a total of 32 frames per stack. The total dose rate was approximately 50 e−/Å2 for each stack. The stacks were motion corrected with MotionCor2 (Zheng et al., 2017) and binned 2-fold, resulting in a pixel size of 1.087 Å/pixel. Meanwhile, dose weighting was performed (Grant and Grigorieff, 2015). The defocus values were estimated with Gctf (Zhang, 2016).
Data processing
Particles for S-ECD (Omicron) in complex with STS165 and PD of ACE2 were automatically picked using Relion 3.0.6 (Kimanius et al., 2016; Scheres, 2012a, b; Zivanov et al., 2018) manually selected micrographs. After 2D classification with Relion, good particles were selected and subject to multiple cycles of heterogeneous refinement without symmetry using cryoSPARC (Punjani et al., 2017). The good particles were selected and subjected to Non-uniform Refinement (beta), Local CTF Refinement, and Local Refinement with C1 symmetry, resulting in the 3D reconstruction of the whole structures. For the interface between RBD of S-ECD (Omicron) and PD, the dataset was subject to focused refinement with an adapted mask and reference on the RBD-PD sub-complex to improve the map quality. The re-extracted dataset was 3D classified with heterogeneous refinement focused on the RBD-PD sub-complex. Then the good particles were selected and subject to Local Refinement, resulting in the 3D reconstruction of better quality on the RBD-PD sub-complex.
The resolution was estimated with the gold-standard Fourier shell correlation 0.143 criterion (Rosenthal and Henderson, 2003) with high-resolution noise substitution (Chen et al., 2013). Refer to Figures S4, S5, and S6 and Table S1 for details of data collection and processing.
Model building and structure refinement
For the model building of the complex of S-ECD (Omicron) in complex with STS165 and PD of ACE2, the atomic model of the 2 PD-bound S (p) (PDB ID: 7DX8) were used as templates, which were molecular dynamics flexible fitted (Trabuco et al., 2008) into the whole cryo-EM map of the complex and the focused-refined cryo-EM map of the RBD-PD sub-complex, respectively. A predicted model of STS165 was first obtained using alphafold2.1 (Jumper et al., 2021), which was further manually adjusted based on the cryo-EM map with Coot (Emsley et al., 2010). Each residue was manually checked with the chemical properties taken into consideration during model building. Several segments, whose corresponding densities were invisible, were not modeled. Structural refinement was performed in Phenix (Adams et al., 2010) with secondary structure and geometry restraints to prevent overfitting. To monitor the potential overfitting, the model was refined against one of the two independent half maps from the gold-standard 3D refinement approach. Then, the refined model was tested against the other map. Statistics associated with data collection, 3D reconstruction and model building were summarized in Table S1.
Quantification and statistical analysis
The independent experiment replicates were indicated in the figure legends. Half-maximal inhibitory concentrations (IC50) of mAbs were calculated using GraphPad Prism 8.0 software by log (inhibitor) vs. normalized response - Variable slope (four parameters) model. The values of binding affinity (KD) of mAbs were calculated using Biacore Evaluation software 3.0 (GE Healthcare) by Multi-cycle kinetics/affinity model. We did not perform any statistical analysis in this study.
Acknowledgments
We thank the Cryo-EM Facility and Supercomputer Center of Westlake University for providing cryo-EM and computation support, respectively. This work was funded by the National Science Fund for Distinguished Young Scholars (82025022), the National Natural Science Foundation of China (92169204, 82002140, 82171752, 82101861), the National Key Plan for Scientific Research and Development of China (2021YFC0864500), the Guangdong Basic and Applied Basic Research Foundation (2021B1515020034, 2021A1515011009), the Shenzhen Science and Technology Program (RCYX20200714114700046), the Science and Technology Innovation Committee of Shenzhen Municipality (JSGG20200207155251653, JSGG20200807171401008, JSGG20210901145200002, KQTD20200909113758004), and the start-up funds from the Southern University of Science and Technology (To R.Y.).
Author contributions
Z.Z., B.J., and R.Y. conceived the project. R.Y., B.J., Y.L., and Q.F. designed the experiments. All the authors did the experiments and contributed to data analysis. Z.Z., B.J., and R.Y. wrote the manuscript. All authors read and approved this version of manuscript.
Declaration of interests
Patent application has been filed on the monoclonal antibody STS165 (patent application number: 202210489241.1; patent applicants: National Clinical Research Center for Infectious Disease and Shenzhen Third People’s Hospital). Q.F., B.Z., F.Q., B.J., and Z.Z. are inventors.
Published: June 17, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.104431.
Contributor Information
Renhong Yan, Email: yanrh@sustech.edu.cn.
Bin Ju, Email: jubin2013@163.com.
Zheng Zhang, Email: zhangzheng1975@aliyun.com.
Supplemental information
Data and code availability
-
•
Structural data for the Spike of Omicron-PD of ACE2 complex and Spike of Omicron-STS165 complex have been deposited at the Protein Data Bank (http://www.rcsb.org) and the Electron Microscopy Data Bank (http://www.emdataresource.org) and are publicly available as of the date of publication. Accession numbers are listed in the key resources table.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Adams P.D., Afonine P.V., Bunkoczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W., et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/s0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes C.O., Jette C.A., Abernathy M.E., Dam K.M.A., Esswein S.R., Gristick H.B., Malyutin A.G., Sharaf N.G., Huey-Tubman K.E., Lee Y.E., et al. SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies. Nature. 2020;588:682–687. doi: 10.1038/s41586-020-2852-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belouzard S., Chu V.C., Whittaker G.R. Activation of the SARS coronavirus spike protein via sequential proteolytic cleavage at two distinct sites. Proc. Natl. Acad. Sci. U S A. 2009;106:5871–5876. doi: 10.1073/pnas.0809524106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameroni E., Bowen J.E., Rosen L.E., Saliba C., Zepeda S.K., Culap K., Pinto D., VanBlargan L.A., De Marco A., di Iulio J., et al. Broadly neutralizing antibodies overcome SARS-CoV-2 Omicron antigenic shift. Nature. 2021;602:664–670. doi: 10.1038/d41586-021-03825-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y., Wang J., Jian F., Xiao T., Song W., Yisimayi A., Huang W., Li Q., Wang P., An R., et al. Omicron escapes the majority of existing SARS-CoV-2 neutralizing antibodies. Nature. 2021;602:657–663. doi: 10.1038/d41586-021-03796-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S., McMullan G., Faruqi A.R., Murshudov G.N., Short J.M., Scheres S.H., Henderson R. High-resolution noise substitution to measure overfitting and validate resolution in 3D structure determination by single particle electron cryomicroscopy. Ultramicroscopy. 2013;135:24–35. doi: 10.1016/j.ultramic.2013.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L., Song S., Fan Q., Shen S., Wang H., Zhou B., Ge X., Ju B., Zhang Z. Cross-neutralization of SARS-CoV-2 Kappa and Delta variants by inactivated vaccine-elicited serum and monoclonal antibodies. Cell Discov. 2021;7:112. doi: 10.1038/s41421-021-00347-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Z., Liu P., Wang N., Wang L., Fan K., Zhu Q., Wang K., Chen R., Feng R., Jia Z., et al. Structural and functional characterizations of altered infectivity and immune evasion of SARS-CoV-2 Omicron variant. bioRxiv. 2022 doi: 10.1101/2021.12.29.474402. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejnirattisai W., Huo J., Zhou D., Zahradnik J., Supasa P., Liu C., Duyvesteyn H.M.E., Ginn H.M., Mentzer A.J., Tuekprakhon A., et al. OPTIC Consortium. ISARIC4C Consortium SARS-CoV-2 Omicron-B.1.1.529 leads to widespread escape from neutralizing antibody responses. Cell. 2022;185:467–484.e15. doi: 10.1016/j.cell.2021.12.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejnirattisai W., Zhou D., Supasa P., Liu C., Mentzer A.J., Ginn H.M., Zhao Y., Duyvesteyn H.M.E., Tuekprakhon A., Nutalai R., et al. Antibody evasion by the P.1 strain of SARS-CoV-2. Cell. 2021;184:2939–2954.e9. doi: 10.1016/j.cell.2021.03.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du S., Cao Y., Zhu Q., Yu P., Qi F., Wang G., Du X., Bao L., Deng W., Zhu H., et al. Structurally resolved SARS-CoV-2 antibody shows high efficacy in severely infected hamsters and provides a potent cocktail pairing strategy. Cell. 2020;183:1013–1023.e13. doi: 10.1016/j.cell.2020.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edeling M.A., Austin S.K., Shrestha B., Dowd K.A., Mukherjee S., Nelson C.A., Johnson S., Mabila M.N., Christian E.A., Rucker J., et al. Potent dengue virus neutralization by a therapeutic antibody with low monovalent affinity requires bivalent engagement. PLoS Pathog. 2014;10:e1004072. doi: 10.1371/journal.ppat.1004072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P., Lohkamp B., Scott W.G., Cowtan K. Features and development of Coot. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge J., Wang R., Ju B., Zhang Q., Sun J., Chen P., Zhang S., Tian Y., Shan S., Cheng L., et al. Antibody neutralization of SARS-CoV-2 through ACE2 receptor mimicry. Nat. Commun. 2021;12:250. doi: 10.1038/s41467-020-20501-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant T., Grigorieff N. Measuring the optimal exposure for single particle cryo-EM using a 2.6 A reconstruction of rotavirus VP6. Elife. 2015;4:e06980. doi: 10.7554/elife.06980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen J., Baum A., Pascal K.E., Russo V., Giordano S., Wloga E., Fulton B.O., Yan Y., Koon K., Patel K., et al. Studies in humanized mice and convalescent humans yield a SARS-CoV-2 antibody cocktail. Science. 2020;369:1010–1014. doi: 10.1126/science.abd0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewat E.A., Blaas D. Structure of a neutralizing antibody bound bivalently to human rhinovirus 2. EMBO J. 1996;15:1515–1523. doi: 10.1002/j.1460-2075.1996.tb00495.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju B., Zhang Q., Ge J.W., Wang R.K., Sun J., Ge X.Y., Yu J.Z., Shan S.S., Zhou B., Song S., et al. Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature. 2020;584:115–119. doi: 10.1038/s41586-020-2380-z. [DOI] [PubMed] [Google Scholar]
- Ju B., Zheng Q., Guo H., Fan Q., Li T., Song S., Sun H., Shen S., Zhou X., Xue W., et al. Immune escape by SARS-CoV-2 Omicron variant and structural basis of its effective neutralization by a broad neutralizing human antibody VacW-209. Cell Res. 2022;32:491–494. doi: 10.1038/s41422-022-00638-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju B., Zhou B., Song S., Fan Q., Ge X., Wang H., Cheng L., Guo H., Shu D., Liu L., Zhang Z. Potent antibody immunity to SARS-CoV-2 variants elicited by a third dose of inactivated vaccine. Clin. Transl. Med. 2022;12:e732. doi: 10.1002/ctm2.732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., Tunyasuvunakool K., Bates R., Zidek A., Potapenko A., et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimanius D., Forsberg B.O., Scheres S.H., Lindahl E. Accelerated cryo-EM structure determination with parallelisation using GPUs in RELION-2. Elife. 2016;5:e18722. doi: 10.7554/elife.18722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei J., Frank J. Automated acquisition of cryo-electron micrographs for single particle reconstruction on an FEI Tecnai electron microscope. J. Struct. Biol. 2005;150:69–80. doi: 10.1016/j.jsb.2005.01.002. [DOI] [PubMed] [Google Scholar]
- Li X., Mooney P., Zheng S., Booth C.R., Braunfeld M.B., Gubbens S., Agard D.A., Cheng Y. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat. Methods. 2013;10:584–590. doi: 10.1038/nmeth.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D., Sempowski G.D., Saunders K.O., Acharya P., Haynes B.F. SARS-CoV-2 neutralizing antibodies for COVID-19 prevention and treatment. Annu. Rev. Med. 2022;73:1–16. doi: 10.1146/annurev-med-042420-113838. [DOI] [PubMed] [Google Scholar]
- Li F., Li W., Farzan M., Harrison S.C. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science. 2005;309:1864–1868. doi: 10.1126/science.1116480. [DOI] [PubMed] [Google Scholar]
- Liu C., Ginn H.M., Dejnirattisai W., Supasa P., Wang B., Tuekprakhon A., Nutalai R., Zhou D., Mentzer A.J., Zhao Y., et al. Reduced neutralization of SARS-CoV-2 B.1.617 by vaccine and convalescent serum. Cell. 2021;184:4220–4236.e13. doi: 10.1016/j.cell.2021.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Iketani S., Guo Y., Chan J.F.W., Wang M., Liu L., Luo Y., Chu H., Huang Y., Nair M.S., et al. Striking antibody evasion manifested by the Omicron variant of SARS-CoV-2. Nature. 2021;602:676–681. doi: 10.1038/d41586-021-03826-3. [DOI] [PubMed] [Google Scholar]
- Lv Z., Deng Y.Q., Ye Q., Cao L., Sun C.Y., Fan C., Huang W., Sun S., Sun Y., Zhu L., et al. Structural basis for neutralization of SARS-CoV-2 and SARS-CoV by a potent therapeutic antibody. Science. 2020;369:1505–1509. doi: 10.1126/science.abc5881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCallum M., Bassi J., De Marco A., Chen A., Walls A.C., Di Iulio J., Tortorici M.A., Navarro M.J., Silacci-Fregni C., Saliba C., et al. SARS-CoV-2 immune evasion by the B.1.427/B.1.429 variant of concern. Science. 2021;373:648–654. doi: 10.1126/science.abi7994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millet J.K., Whittaker G.R. Host cell proteases: critical determinants of coronavirus tropism and pathogenesis. Virus Res. 2015;202:120–134. doi: 10.1016/j.virusres.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Piccoli L., Park Y.J., Tortorici M.A., Czudnochowski N., Walls A.C., Beltramello M., Silacci-Fregni C., Pinto D., Rosen L.E., Bowen J.E., et al. Mapping neutralizing and immunodominant sites on the SARS-CoV-2 spike receptor-binding domain by structure-guided high-resolution serology. Cell. 2020;183:1024–1042.e21. doi: 10.1016/j.cell.2020.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto D., Park Y.J., Beltramello M., Walls A.C., Tortorici M.A., Bianchi S., Jaconi S., Culap K., Zatta F., De Marco A., et al. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature. 2020;583:290–295. doi: 10.1038/s41586-020-2349-y. [DOI] [PubMed] [Google Scholar]
- Planas D., Veyer D., Baidaliuk A., Staropoli I., Guivel-Benhassine F., Rajah M.M., Planchais C., Porrot F., Robillard N., Puech J., et al. Reduced sensitivity of SARS-CoV-2 variant Delta to antibody neutralization. Nature. 2021;596:276–280. doi: 10.1038/s41586-021-03777-9. [DOI] [PubMed] [Google Scholar]
- Punjani A., Rubinstein J.L., Fleet D.J., Brubaker M.A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods. 2017;14:290–296. doi: 10.1038/nmeth.4169. [DOI] [PubMed] [Google Scholar]
- Rosenthal P.B., Henderson R. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J. Mol. Biol. 2003;333:721–745. doi: 10.1016/j.jmb.2003.07.013. [DOI] [PubMed] [Google Scholar]
- Scheres S.H.W. A Bayesian view on cryo-EM structure determination. J. Mol. Biol. 2012;415:406–418. doi: 10.1109/isbi.2012.6235807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres S.H. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi R., Shan C., Duan X., Chen Z., Liu P., Song J., Song T., Bi X., Han C., Wu L., et al. A human neutralizing antibody targets the receptor binding site of SARS-CoV-2. Nature. 2020;584:120–124. doi: 10.1038/s41586-020-2381-y. [DOI] [PubMed] [Google Scholar]
- Simmons G., Gosalia D.N., Rennekamp A.J., Reeves J.D., Diamond S.L., Bates P. Inhibitors of cathepsin L prevent severe acute respiratory syndrome coronavirus entry. Proc. Natl. Acad. Sci. U S A. 2005;102:11876–11881. doi: 10.1073/pnas.0505577102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons G., Reeves J.D., Rennekamp A.J., Amberg S.M., Piefer A.J., Bates P. Characterization of severe acute respiratory syndrome-associated coronavirus (SARS-CoV) spike glycoprotein-mediated viral entry. Proc. Natl. Acad. Sci. U S A. 2004;101:4240–4245. doi: 10.1073/pnas.0306446101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons G., Zmora P., Gierer S., Heurich A., Pohlmann S. Proteolytic activation of the SARS-coronavirus spike protein: cutting enzymes at the cutting edge of antiviral research. Antiviral Res. 2013;100:605–614. doi: 10.1016/j.antiviral.2013.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W., Gui M., Wang X., Xiang Y. Cryo-EM structure of the SARS coronavirus spike glycoprotein in complex with its host cell receptor ACE2. PLoS Pathog. 2018;14:e1007236. doi: 10.1371/journal.ppat.1007236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trabuco L.G., Villa E., Mitra K., Frank J., Schulten K. Flexible fitting of atomic structures into electron microscopy maps using molecular dynamics. Structure. 2008;16:673–683. doi: 10.1016/j.str.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Nair M.S., Liu L., Iketani S., Luo Y., Guo Y., Wang M., Yu J., Zhang B., Kwong P.D., et al. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature. 2021;593:130–135. doi: 10.1038/s41586-021-03398-2. [DOI] [PubMed] [Google Scholar]
- Yan R., Wang R., Ju B., Yu J., Zhang Y., Liu N., Wang J., Zhang Q., Chen P., Zhou B., et al. Structural basis for bivalent binding and inhibition of SARS-CoV-2 infection by human potent neutralizing antibodies. Cell Res. 2021;31:517–525. doi: 10.1038/s41422-021-00487-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan R., Zhang Y., Li Y., Xia L., Guo Y., Zhou Q. Structural basis for the recognition of the SARS-CoV-2 by full-length human ACE2. Science. 2020;367:1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M., Huang D., Lee C.C.D., Wu N.C., Jackson A.M., Zhu X., Liu H., Peng L., van Gils M.J., Sanders R.W., et al. Structural and functional ramifications of antigenic drift in recent SARS-CoV-2 variants. Science. 2021;373:818–823. doi: 10.1126/science.abh1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K. Gctf: real-time CTF determination and correction. J. Struct. Biol. 2016;193:1–12. doi: 10.1016/j.jsb.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng S.Q., Palovcak E., Armache J.P., Verba K.A., Cheng Y., Agard D.A. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods. 2017;14:331–332. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D., Duyvesteyn H.M.E., Chen C.P., Huang C.G., Chen T.H., Shih S.R., Lin Y.C., Cheng C.Y., Cheng S.H., Huang Y.C., et al. Structural basis for the neutralization of SARS-CoV-2 by an antibody from a convalescent patient. Nat. Struct. Mol. Biol. 2020;27:950–958. doi: 10.1038/s41594-020-0480-y. [DOI] [PubMed] [Google Scholar]
- Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., et al. China Novel Coronavirus Investigating and Research Team A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zivanov J., Nakane T., Forsberg B.O., Kimanius D., Hagen W.J., Lindahl E., Scheres S.H. New tools for automated high-resolution cryo-EM structure determination in RELION-3. Elife. 2018;7:e42166. doi: 10.7554/elife.42166. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
Structural data for the Spike of Omicron-PD of ACE2 complex and Spike of Omicron-STS165 complex have been deposited at the Protein Data Bank (http://www.rcsb.org) and the Electron Microscopy Data Bank (http://www.emdataresource.org) and are publicly available as of the date of publication. Accession numbers are listed in the key resources table.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.