Abstract
Several fungal isolates obtained from two cured meat products from Spain were identified as Penicillium nalgiovense by their morphological features and by DNA fingerprinting. All P. nalgiovense isolates showed antibiotic activity in agar diffusion assays, and their penicillin production in liquid complex medium ranged from 6 to 38 μg · ml−1. We constructed a restriction map of the penicillin gene cluster of P. nalgiovense and found that the organization of the penicillin biosynthetic genes (pcbAB, pcbC, and penDE) is the same as in Penicillium chrysogenum and Aspergillus nidulans. The pcbAB gene is located in an orientation opposite that of the pcbC and penDE genes in all three species. Significant amounts of penicillin were found in situ in the casing and the outer layer of salami meat during early stages of the curing process, coinciding with fungal colonization, but no penicillin was detected in the cured salami. The antibiotic produced in situ was sensitive to penicillinase.
Penicillium nalgiovense Laxa is one of two species in the Penicillium subgenus with white or pale green colonies (21, 23). P. nalgiovense was originally isolated from cheese (23) and cured meat products (10). In Spain, Italy, France, Switzerland, Hungary, Rumania, Germany, and some other European countries, dry sausages are usually ripened with molds. The development of molds on the surface of these sausages is required before they are considered to be cured and ready for consumption (26). P. nalgiovense gives a typical white appearance and characteristic odor and flavor to cured sausages and is available commercially as a starter culture for dry sausages (12). Fungal isolates from meat products have appeared to be very frequent producers of penicillin (2, 7). However, in situ penicillin production by fungi growing on meat products has not been demonstrated.
Genes involved in a particular biosynthetic pathway for secondary metabolites are often clustered in bacteria and fungi (11, 16). This phenomenon is especially common in the so-called dispensable pathways, i.e., pathways that are not required for normal growth or are required only under certain conditions. The genes involved in the biosynthesis of β-lactam antibiotics are located in clusters in filamentous fungi as well as in actinomycetes (1, 13, 15). In Penicillium chrysogenum, the penicillin biosynthetic genes pcbAB, pcbC, and penDE are clustered together in a 17-kb DNA fragment (6, 24). The long pcbAB gene (11.2 kb) is located in an orientation opposite that of pcbC and penDE.
Our objectives in this study were (i) to identify penicillin-producing isolates from cured sausages and (ii) to determine if those isolates have a penicillin gene cluster similar to that in P. chrysogenum and Aspergillus nidulans. We use these results to determine the role of penicillin in the control of undesirable bacteria during the curing process.
MATERIALS AND METHODS
Cured sausage types.
Filamentous fungi were isolated from the surface of salami (two different types, fuet artisan and fuet petit) and chorizo. Both contain minced pork meat, salt, and spices, and chorizo also contains paprika. Both products are fermented by lactic acid bacteria and then cured.
Isolation and identification of fungi.
The strains that we used are listed in Table 1. Isolates from salami and chorizo were obtained from the surface of the sausages with a sterile cotton swab impregnated with saline solution (0.9% NaCl) (19). Each sample was submerged in 1 ml of saline solution and homogenized by agitation. For colony isolation, 0.1 ml of the sample was plated on malt extract agar (MEA) (21) and incubated at 25°C for 7 days. Isolates obtained were identified by use of a taxonomic key based on morphological and pigmentation characteristics (21) and by DNA fingerprinting (18).
TABLE 1.
Origins of fungal strains used in this work
| Speciesa | Source |
|---|---|
| P. nalgiovense INBCC 100 | Commercial starter culture |
| P. nalgiovense INBCC 101 | Isolated from salami fuet artisan (Lérida, Spain) |
| P. nalgiovense INBCC 102 | Isolated from salami fuet petit (Barcelona, Spain) |
| P. nalgiovense INBCC 200 | Isolated from chorizo (Toro, Spain) |
| P. chrysogenum AS-P-78 | Antibióticos S.A. |
| Penicillium subgenus Penicillium strain INBCC 300 | Isolated from cured meat cecina (León, Spain) |
| P. nalgiovense NRRL 911 | NRRC |
| P. notatum ATCC 9478 | ATCC |
| P. viridicatum NRRL 963 | NRRC |
| P. commune NRRL 845 | NRRC |
| P. expansum NRRL 976 | NRRC |
| P. hirsutum NRRL 2032 | NRRC |
INBCC, Institute of Biotechnology (INBIOTEC) Culture Collection; NRRC, National Center for Agricultural Utilization Research; ATCC, American Type Culture Collection.
DNA fingerprinting analysis.
Total DNA from all fungal strains was extracted by a small-scale procedure (8). Total DNA was digested with EcoRI and electrophoresed in a 0.7% agarose gel. Subsequent treatments for fixing the DNA to the gel were carried out as described previously (4). The probe used in the fingerprinting analysis was the oligonucleotide (GTG)5. Labeling of (GTG)5 with [γ-32P]ATP and hybridization were done essentially as described by Meyer et al. (18).
Determination of β-lactam antibiotic production in pure cultures.
For each strain, spores obtained from three petri dishes of potato dextrose agar medium (Difco, Detroit, Mich.) were inoculated into Erlenmeyer flasks with 100 ml of defined growth medium supplemented with yeast extract (containing, in grams per liter, the following: glucose, 40; NaNO3, 3; yeast extract, 2; KCl, 0.5; MgSO4 · 7H2O, 0.5; and FeSO4 · 7H2O, 0.01; pH adjusted to 6.0) (3) and incubated at 25°C with agitation (250 rpm) for 48 h. Ten milliliters of each culture was transferred to 100 ml of complex penicillin production medium (CPM) (containing, in grams per liter, the following: corn steep liquor, 20; lactose, 55; MgSO4 · 7H2O, 3; CaCO3, 10; KH2PO4, 7; and 64% potassium phenylacetate, 6.25 ml; pH adjusted to 6.8) (25) or liquid minimal medium (MM) (7). These cultures were incubated for 120 h at 25°C in an orbital shaker at 250 rpm. One-milliliter samples were taken at 24-h intervals from each culture. Each sample was centrifuged (10,000 × g for 15 min), and the supernatant was used for the quantitative determination of β-lactam antibiotic activity.
Micrococcus luteus ATCC 9341 was used as a bacterial indicator in agar diffusion assays with tryptic soy agar (TSA) medium (Difco) containing 1% agar. Solutions with increasing penicillin G (Antibióticos S.A., León, Spain) concentrations were used as controls. Simultaneously, control plates with penicillinase (2 μg from a Bacillus cereus UL1 penicillinase preparation [about 1 μg of protein] per ml of TSA medium) were inoculated with M. luteus to test if the antibiotic produced was degraded by penicillinase.
Penicillin production in situ in cured salami at different stages of ripening.
The production of penicillin in situ by P. nalgiovense was tested with three salami products obtained from the same manufacturer and inoculated with P. nalgiovense at different stages of ripening: (i) soft, unpalatable salami (not cured) with initial fungal colonization in which isolated P. nalgiovense colonies were observed; (ii) semidry (not fully cured) salami covered with a nonhomogeneous fungal mat; and (iii) cured, dried salami showing good organoleptic properties and a complete mat of fungal growth.
The production of penicillin was tested directly in the casing with 9-mm-diameter disks of salami casing, salami meat outer surface (0 to 4 mm deep), inner surface (4 to 8 mm deep), and inner core (>10 mm deep). The disks were assayed directly on a penicillin-sensitive M. luteus surface culture on 1% TSA medium. The antibiotic was allowed to diffuse in the plates in the cold (3 h at 5°C), and the cultures were incubated at 30°C for 24 h.
Alternatively, the penicillin was extracted with organic solvent as follows. Samples taken as described above from the casing, outer surface layer (0 to 4 mm deep), inner surface layer (4 to 8 mm deep), and inner core (>10 mm deep) of each type of salami were frozen in liquid nitrogen and ground in a mortar. The triturated material was suspended in 20 ml of saline solution (0.9% NaCl) (pH 7.0) and stirred at 5°C for 15 min. The solid material was removed by centrifugation at 3,000 × g and 5°C for 30 min. The aqueous solution was taken to pH 3.0 (with 1.0 N HCl) and extracted with an equal volume of ethyl acetate. The organic phase (free of the lipid interface) was collected, dried in a vacuum evaporator, and redissolved in 0.2 ml of distilled water at pH 8.0. Bioassays with 50-μl aliquots of the extracts were performed with M. luteus as indicated above. Similarly, bioassays were also run with 60-μl aliquots of the aqueous phase or the lipid interface.
Southern blotting and hybridization.
Total DNA was extracted by the same small-scale procedure as that used for the DNA for fingerprinting (8). DNA fragments were separated by electrophoresis and blotted by standard techniques (22). The probe (a 7.2-kb NaeI fragment containing the penDE and pcbC genes and the 5′ end of pcbAB) was labeled by nick translation with [α-32P]dCTP as recommended by the manufacturer (Promega Corp., Madison, Wis.). This probe allowed us to check for the presence of the three penicillin genes and to map the restriction sites by comparing the band patterns and sizes. Prehybridization and hybridization were done with 30% formamide standard buffer (22) at 4°C. After hybridization, the membrane was washed once at room temperature for 20 min with 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–0.1% sodium dodecyl sulfate and twice at 42°C for 15 min each time with the same solution.
RESULTS
Identification of the fungal isolates from cured meat products.
Four different Penicillium strains were isolated from cured meat products (Table 1) and identified by the criteria of Pitt and Hocking (21). The strains isolated from fuet artisan and fuet petit salami were morphologically similar to the P. nalgiovense commercial starter culture INBCC 100. Colonies on MEA were flat (15 to 17 mm in diameter) and lacked exudate or soluble pigment. The reverse of the colony was strongly colored orange-brown. Colonies on Czapek yeast extract agar (CYA) (30 to 35 mm in diameter) were radially sulcate and had a clear exudate. The reverse of the colony was pale yellow. Colonies on MEA, CYA, and 25% glycerol–nitrate agar (21) were always white, even in old cultures.
Isolates from chorizo on MEA (25 to 30 mm in diameter) were white to yellow in the center and green at the borders. The reverse of the colony was yellow-brown but lighter than for P. nalgiovense NRRL 911. Colonies on CYA (35 to 40 mm in diameter) were white but had a pale brown center and were radially sulcate but less so than the colonies of the isolates from fuet. The reverse of the colony was pale yellow and very different from the dark brown for P. nalgiovense NRRL 911. Colonies of the isolates from chorizo were larger than those of the isolates from fuet and NRRL 911 on all culture media.
All isolates had a neutral to weakly acid reaction on creatine-sucrose neutral agar (20), except that strains isolated from fuet artisan salami showed a strongly acid reaction.
DNA fingerprinting.
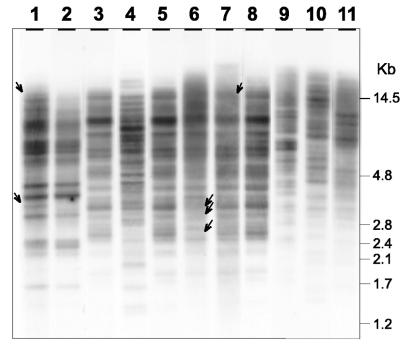
By fingerprinting of EcoRI-digested total DNAs of different isolates with the (GTG)5 probe, we were able to distinguish different species and in some cases to find minor differences between different isolates of the same species (Fig. 1). P. chrysogenum and Penicillium notatum (4), two species that have been classified as P. chrysogenum by Pitt and Hocking (21), had similar hybridization patterns. The P. nalgiovense isolates in Fig. 1, lanes 3, 5, 6, 7, and 8, had almost identical patterns. No differences were found between isolates from fuet petit, fuet artisan, and chorizo. Only a minor difference in the intensity of one band was observed between the commercial starter culture and the salami isolates. Strain NRRL 911 showed three differences in band patterns compared to the rest of P. nalgiovense isolates; these different bands (either absent or at a much lower intensity in NRRL 911) are indicated in Fig. 1. Differences in the intensities of DNA fingerprinting bands with the (GTG)5 probe have also been reported for some P. chrysogenum strains (4).
FIG. 1.
DNA fingerprinting of several isolates from cured sausages and control species of the genus Penicillium. Total DNAs were digested with EcoRI and probed with labeled oligonucleotide (GTG)5. Lanes: 1, P. notatum ATCC 9478; 2, P. chrysogenum AS-P-78; 3, P. nalgiovense INBCC 102 isolated from fuet petit; 4, P. viridicatum NRRL 963; 5, P. nalgiovense INBCC 101 isolated from fuet artisan; 6, P. nalgiovense NRRL 911; 7, P. nalgiovense INBCC 100 commercial starter culture; 8, P. nalgiovense INBCC 200 isolated from chorizo; 9, P. expansum NRRL 976; 10, P. hirsutum NRRL 2032; 11, P. commune NRRL 845. Bands that are different between P. nalgiovense strains and other strains are indicated by arrows (see the text for details).
Other Penicillium species used in this study had different hybridization patterns and were clearly distinguished from P. nalgiovense.
Antibiotic production.
Isolates from cured meat products, the commercial starter culture, and strain NRRL 911 were tested for their ability to suppress the growth of M. luteus.
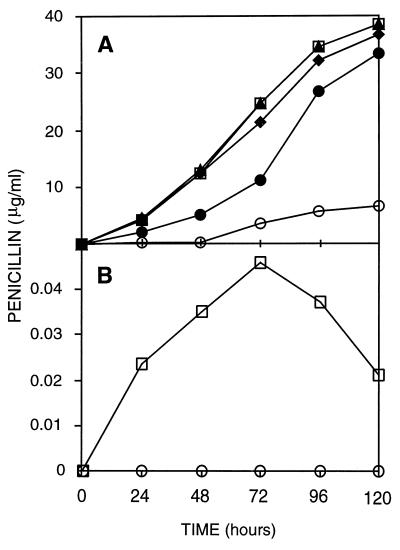
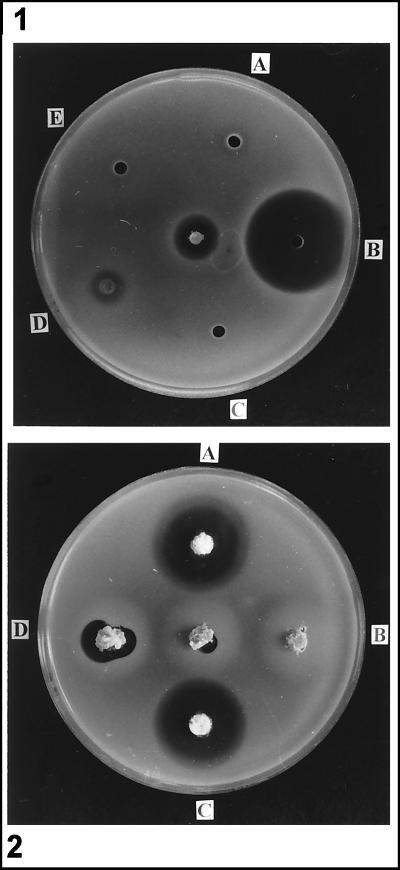
All the isolates examined produced antibiotics in CPM (Fig. 2A). The commercial starter culture produced significant levels of antibiotics in MM (Fig. 2B), while no inhibitory effect could be detected when P. nalgiovense NRRL 911 was grown in the same medium. The penicillin production levels of most P. nalgiovense isolates fell within the range for P. chrysogenum NRRL 1951 (wild type) under the same fermentation conditions (9), while the penicillin production level of P. nalgiovense NRRL 911 was similar to the average level observed for isolates of A. nidulans (17).
FIG. 2.
Penicillin production in flask cultures of different P. nalgiovense isolates in CPM (A) and MM (B). The isolates were obtained from fuet artisan (INBCC 101) (⧫), fuet petit (INBCC 102) (▴), chorizo (INBCC 200) (●), commercial starter culture INBCC 100 (□), and NRRL 911 (○). In MM, only the commercial starter culture and NRRL 911 were tested.
The antibiotic activity obtained after fermentation in CPM and MM was completely inactivated by the addition of penicillinase (see below) in agar diffusion assays with M. luteus, indicating that it corresponds to a penicillin.
Presence of the penicillin gene cluster in P. nalgiovense isolates.
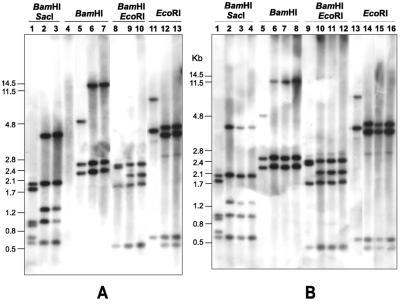
Hybridization signals with the 7.2-kb pcbC-penDE probe, corresponding to the penicillin genes, appeared in all P. nalgiovense isolates (all of them shown previously to be penicillin producers) but were absent in a Penicillium subgenus Penicillium isolate which did not produce the antibiotic (Fig. 3).
FIG. 3.
Southern hybridization of total DNAs from different P. nalgiovense isolates and P. chrysogenum AS-P-78 with a probe internal to the penicillin biosynthetic gene cluster. The probe used was a 7.2-kb NaeI fragment containing the pcbC and penDE genes and the 5′ region of the pcbAB gene (see Fig. 4). The enzymes used for the DNA digestions are indicated above each panel. DNA size markers (in kilobases) are indicated at the left. (A) Lanes 1, 5, 8, and 11, P. chrysogenum AS-P-78; lanes 2, 6, 9, and 12, P. nalgiovense NRRL 911; lanes 3, 7, 10, and 13, P. nalgiovense INBCC 100 (commercial starter culture); lane 4, Penicillium sp. strain INBCC 300 isolated from cecina (nonproducer of penicillin; note that there is no hybridization with the penicillin probe). (B) Lanes 1, 5, 9, and 13, P. chrysogenum AS-P-78; lanes 2, 6, 10, and 14, P. nalgiovense INBCC 200 isolated from chorizo; lanes 3, 7, 11, and 15, P. nalgiovense INBCC 102 isolated from fuet petit; lanes 4, 8, 12, and 16, P. nalgiovense INBCC 101 isolated from fuet artisan.
Organization of the penicillin biosynthesis gene cluster in P. nalgiovense isolates.
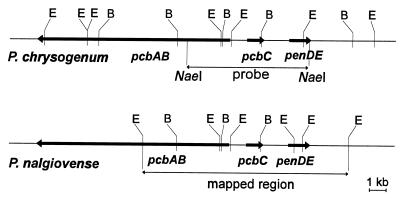
Hybridization signals common to P. nalgiovense and P. chrysogenum in BamHI and EcoRI fragments indicated that the three penicillin biosynthetic genes are clustered in P. nalgiovense and that many restriction sites are conserved within the cluster in P. nalgiovense compared to P. chrysogenum. Taking into account the hybridization patterns of both species, we constructed a restriction map of the P. nalgiovense cluster (Fig. 4). Most BamHI and EcoRI sites were conserved in the mapped region with respect to P. chrysogenum, while a new EcoRI site appeared in the P. nalgiovense penDE gene.
FIG. 4.
Restriction maps of the P. chrysogenum and P. nalgiovense gene clusters for penicillin biosynthesis. The orientation of the pcbAB, pcbC, and penDE genes is conserved in both species. The P. chrysogenum NaeI probe used in the Southern hybridizations (Fig. 3) is indicated on the P. chrysogenum map.
Relevance of in situ penicillin production by P. nalgiovense in meat products.
Results of bioassays of solvent-extracted antibiotic production by P. nalgiovense growing in situ on the cured and semicured sausages are shown in Fig. 5. As shown in panel 1 of Fig. 5, there was moderate penicillin production in solvent extracts of the casing and in the lipidic interface from semicured salami. No detectable levels of penicillin were found either in the casing or in the meat of cured salami. Similarly, using direct bioassays of penicillin production in the casing or the salami meat, we found significant levels of penicillin in the soft salami, namely, 1.25 μg/cm2 of casing, 0.66 μg/g of meat in the outer surface layer, and 0.08 μg/g of meat in the inner surface layer. The antibiotic produced was completely degraded by penicillinase and was extracted with ethyl acetate, indicating that the side chain in the penicillin molecules was not polar.
FIG. 5.
Bioassays of antibiotic produced in situ by P. nalgiovense growing in salami. (Panel 1) Solvent-extracted antibiotic from cured or semicured salami. A, Casing from dry cured salami; B, casing from semicured salami (0.013 μg/cm2 of casing); C, lipidic interface from the solvent extraction of cured salami; D, aqueous phase from the extraction of semicured salami; E, aqueous phase from the extraction of cured salami; center, lipidic interface from semicured salami. (Panel 2) Direct assays of soft salami (not cured). A, Salami casing; B, inner core of salami; C, casing (region different from that in A); D, outer salami layer; center, inner salami layer.
DISCUSSION
Molds grow on a variety of food products. In many cases, they are deleterious, producing undesirable alterations of the food or feed products, e.g., aflatoxin production in corn or peanuts. However, in some cases (e.g., cheese ripening), the proteolytic and lipolytic activities of fungi are clearly beneficial, contributing decisively to the organoleptic characteristics of traditional foods. Molds may also produce antibacterial agents that help to preserve foods.
Dry sausages are ripened with molds in many countries, but the exact role of fungi in the salami casing is unknown. Our results clearly indicate that there is a significant level of penicillin production by P. nalgiovense isolates in cured meat products. The conserved arrangement of the penicillin biosynthetic genes among different species indicates that the penicillin gene cluster is derived from a common ancestor of P. nalgiovense, P. chrysogenum, and A. nidulans. Clustering also may be important in the regulation of expression of the three genes (14).
We found that penicillin is produced during fungal colonization of the salami casing in the early stages of the curing process, in agreement with previous reports (2, 7). This is one of the first examples of the production of an antibiotic in a natural substrate and provides support for the role of antibiotics as antagonists in nature (5). The penicillin in the salami casing diffuses to the outer layer of the meat and becomes undetectable in the salami core. Cured salami (as available in the market) had no detectable levels of penicillin. The reason for the decay of penicillin during the curing process is unclear, although it may be due to inactivation by exposure of the antibiotic to an acid pH for prolonged periods of time.
Colonization by P. nalgiovense of salami during the early stages of the curing process may be beneficial by preventing the growth of undesirable bacteria on the surface of the soft sausage. Cured salami does not contain detectable levels of penicillin, excluding the possible induction of undesirable cross-resistance to β-lactam antibiotics from penicillin present in the meat. Disruption of the penicillin gene might be useful for obtaining starter strains unable to produce penicillin, although the ability to colonize the salami also must be maintained in the disarmed strains.
ACKNOWLEDGMENTS
This research was supported by a grant from the Diputación de León (León, Spain). F. Laich was supported by a MUTIS fellowship from the Agencia Española de Cooperación Internacional (AECI).
We thank B. Martín and J. Merino for technical assistance.
REFERENCES
- 1.Aharonowitz Y, Cohen G, Martín J F. Penicillin and cephalosporin biosynthetic genes: structure, organization, regulation and evolution. Annu Rev Microbiol. 1992;46:461–495. doi: 10.1146/annurev.mi.46.100192.002333. [DOI] [PubMed] [Google Scholar]
- 2.Andersen S J, Frisvad J C. Penicillin production by Penicillium nalgiovense. Lett Appl Microbiol. 1994;19:486–488. doi: 10.1111/j.1472-765x.1994.tb00988.x. [DOI] [PubMed] [Google Scholar]
- 3.Anné J. Somatic hybridization between Penicillium species after induced fusion of their protoplasts. Agricultura. 1977;25:1–117. [Google Scholar]
- 4.Cantoral J M, Gutiérrez S, Fierro F, Gil-Espinosa S, van Liempt H, Martín J F. Biochemical characterization and molecular genetics of nine mutants of Penicillium chrysogenum impaired in penicillin biosynthesis. J Biol Chem. 1993;268:737–744. [PubMed] [Google Scholar]
- 5.Demain A L. Do antibiotics function in nature? Search. 1980;11:148. [Google Scholar]
- 6.Díez B, Gutiérrez S, Barredo J L, van Solingen P, van der Voort L H M, Martín J F. The cluster of penicillin biosynthetic genes. Identification and characterization of the pcbAB gene encoding the α-aminoadipyl-cysteinyl-valine synthetase and linkage to the pcbC and penDE genes. J Biol Chem. 1990;265:16358–16365. [PubMed] [Google Scholar]
- 7.Färber P, Geisen R. Antagonistic activity of the food-related filamentous fungus Penicillium nalgiovense by the production of penicillin. Appl Environ Microbiol. 1994;60:3401–3404. doi: 10.1128/aem.60.9.3401-3404.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fierro F, Montenegro E, Gutiérrez S, Martín J F. Mutants blocked in penicillin biosynthesis show a deletion of the entire penicillin gene cluster at a specific site within a conserved hexanucleotide sequence. Appl Microbiol Biotechnol. 1996;44:597–604. doi: 10.1007/BF00172491. [DOI] [PubMed] [Google Scholar]
- 9.Fierro, F., and J. F. Martín. Unpublished data.
- 10.Fink-Gremmels J, El-Banna A, Leistner L. Developing mould starter cultures for meat products. Fleischwirtschaft. 1988;68:1292–1294. [Google Scholar]
- 11.Keller N P, Hohn T M. Metabolic pathway gene clusters in filamentous fungi. Fung Gen Biol. 1997;21:17–29. [PubMed] [Google Scholar]
- 12.Leistner L. Mould-fermented foods: recent developments. Food Biotechnol. 1990;4:433–441. [Google Scholar]
- 13.Martín J F. New aspects of genes and enzymes for β-lactam antibiotic biosynthesis. Appl Microbiol Biotechnol. 1998;50:1–15. doi: 10.1007/s002530051249. [DOI] [PubMed] [Google Scholar]
- 14.Martín J F, Gutiérrez S. Genes for β-lactam antibiotic biosynthesis. Antonie Leeuwenhoek. 1995;67:181–200. doi: 10.1007/BF00871213. [DOI] [PubMed] [Google Scholar]
- 15.Martín J F, Liras P. Organization and expression of genes involved in the biosynthesis of antibiotics and other secondary metabolites. Annu Rev Microbiol. 1989;43:173–206. doi: 10.1146/annurev.mi.43.100189.001133. [DOI] [PubMed] [Google Scholar]
- 16.Martín J F, Gutiérrez S, Coque J J R, Montenegro E, Fernández F J, Velasco J, Fierro F, Gil S, Marcos A T, Arenós C, Rodríguez A, Cardoza R E, Llarena F J, Enguita F, Lumbreras M A, Liras P. Organization and expression of genes for biosynthesis of β-lactam antibiotics in filamentous fungi and actinomycetes. In: Alberghina L, Frontali L, Sensi P, editors. Progress in biotechnology. Vol. 9. Amsterdam, The Netherlands: Elsevier Science B. V.; 1994. pp. 737–744. [Google Scholar]
- 17.Merrick M J, Caten C E. The inheritance of penicillin titre in wild type isolates of Aspergillus nidulans. J Gen Microbiol. 1975;86:283–293. doi: 10.1099/00221287-86-2-283. [DOI] [PubMed] [Google Scholar]
- 18.Meyer W, Koch A, Niemann C, Beyermann B, Epplen J, Börner T. Differentiation of species and strains among filamentous fungi by DNA fingerprinting. Curr Genet. 1991;19:239–242. doi: 10.1007/BF00336493. [DOI] [PubMed] [Google Scholar]
- 19.Motilva Casado M J, Díaz Borrás M A, Vila Aguilar M. Fungal flora present on the surface of cured Spanish ham. Methodological study for its isolation and identification. Fleischwirtschaft. 1991;71:1300–1302. [Google Scholar]
- 20.Pitt J I. A modified creatine sucrose medium for differentiation of species in Penicillium subgenus Penicillium. J Appl Bacteriol. 1993;75:559–563. doi: 10.1111/j.1365-2672.1993.tb01595.x. [DOI] [PubMed] [Google Scholar]
- 21.Pitt J I, Hocking A D. Fungi and food spoilage. London, United Kingdom: Blackie Academic & Professional; 1997. p. 593. [Google Scholar]
- 22.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 23.Samson R, Hoekstra E, Frisvad J, Filtenborg O. Introduction to food-borne fungi. Baarn, The Netherlands: Centraalbureau voor Schimmelcultures; 1995. p. 322. [Google Scholar]
- 24.Smith D J, Burnham M K R, Bull J H, Hodgson J E, Ward J M, Browne P, Brown J, Barton B, Earl A J, Turner G. β-Lactam antibiotic biosynthetic genes have been conserved in clusters in prokaryotes and eukaryotes. EMBO J. 1990;9:741–747. doi: 10.1002/j.1460-2075.1990.tb08168.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Somerson N L, Demain A L, Nunheimer T D. Reversal of lysine inhibition of penicillin production by α-aminoadipic acid. Arch Biochem. 1961;93:238–241. [Google Scholar]
- 26.van der Riet W B. Studies on the mycoflora of biltong. S Afr Food Rev. 1976;3:105. , 107, 109, 111. [Google Scholar]