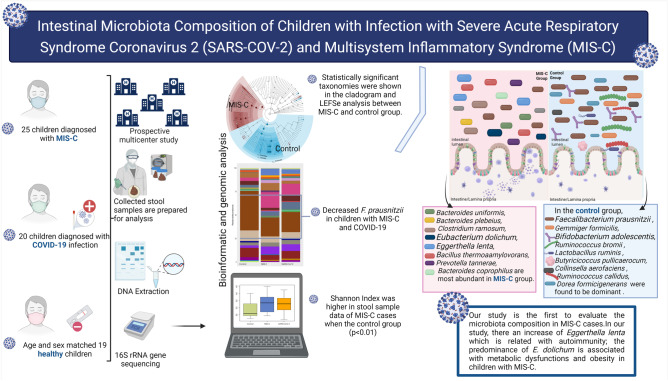
Abstract
Microbiota composition may play a role in the development, prognosis, or post-infection of COVID-19. There are studies evaluating the microbiota composition at the time of diagnosis and during the course of COVID-19, especially in adults, while studies in children are limited and no study available in children with multisystem inflammatory syndrome in children (MIS-C). This study was planned to compare intestinal microbiota composition in children diagnosed with MIS-C and acute COVID-19 infection with healthy children. In this prospective multicenter study, 25 children diagnosed with MIS-C, 20 with COVID-19 infection, and 19 healthy children were included. Intestinal microbiota composition was evaluated by 16 s rRNA gene sequencing. We observed changes of diversity, richness, and composition of intestinal microbiota in MIS-C cases compared to COVID-19 cases and in the healthy controls. The Shannon index was higher in the MIS-C group than the healthy controls (p < 0.01). At phylum level, in the MIS-C group, a significantly higher relative abundance of Bacteroidetes and lower abundance of Firmicutes was found compared to the control group. Intestinal microbiota composition changed in MIS-C cases compared to COVID-19 and healthy controls, and Faecalibacterium prausnitzii decreased; Bacteroides uniformis, Bacteroides plebeius, Clostridium ramosum, Eubacterium dolichum, Eggerthella lenta, Bacillus thermoamylovorans, Prevotella tannerae, and Bacteroides coprophilus were dominant in children with MIS-C. At species level, we observed decreased Faecalibacterium prausnitzii, and increased Eubacterium dolichum, Eggerthella lenta, and Bacillus thermoamylovorans in children with MIS-C and increased Bifidobacterium adolescentis and Dorea formicigenerasus in the COVID-19 group. Our study is the first to evaluate the microbiota composition in MIS-C cases. There is a substantial change in the composition of the gut microbiota: (1) reduction of F. prausnitzii in children with MIS-C and COVID-19; (2) an increase of Eggerthella lenta which is related with autoimmunity; and (3) the predominance of E. dolichum is associated with metabolic dysfunctions and obesity in children with MIS-C.
Conclusions: Alterations of the intestinal microbiota might be part of pathogenesis of predisposing factor for MIS-C. It would be beneficial to conduct more extensive studies on the cause-effect relationship of these changes in microbiota composition and their effects on long-term prognosis.
|
What is Known: • Microbiota composition may play a role in the development, prognosis, or post-infection of COVID-19. • However, the number of studies on children is limited, and no study on multisystem inflammatory syndrome in children is currently available (MIS-C). | |
|
What is New: • In individuals with MIS-C, the composition of the gut microbiota changed dramatically. • Decreased Faecalibacterium prausnitzii have been observed, increased Eggerthella lenta, which was previously linked to autoimmunity, and predominance of Eubacterium dolichum which was linked to metabolic dysfunction and obesity. |
Graphical abstract
Keywords: COVID-19, Multisystem inflammatory syndrome in children, MIS-C, Children, Microbiota, Microbiome
Introduction
By the end of February 2022, the COVID-19 pandemic had maintained its severity despite diverse strategies and increased vaccination rates worldwide [1]. Since the beginning of the pandemic, children and adolescents have accounted for a relatively small proportion of total COVID-19 cases compared with adults [2]. Disease burden and the age distribution of SARS-CoV-2 infection among children and adolescents has varied according to the variants in circulation, prevention strategies applied to combat the pandemic, and the vaccination rates in a community [2, 3]. Hospitalization, intensive care unit stays, and mortality rates have also been lowered in children, and the majority of patients have been shown to have an underlying illness [4, 5]. However, COVID-19 can cause long-term complications, such as multiple systemic inflammatory syndrome in children (MIS-C) and long-COVID [6–9]. MIS-C is a severe disease condition which characterized by fever, rash, conjunctivitis, gastrointestinal symptoms, and shock due to myocardial dysfunction in children who have SARS-COV-2 positivity or a history of exposure approximately 4–6 weeks before the onset of symptoms [2, 10]. Although genetic and immune system-related risk factors are thought to play a role in the pathogenesis of MIS-C, they have not been fully elucidated and the underlying etiology appears to be multifactorial. An autoimmune-mediated inflammatory process after infection, a cytokine storm initiated by a superantigen response, and a dysregulated immune response to exposure to SARS-CoV-2 viral antigens have been proposed as theories to explain its pathogenesis [11, 12]. Immunological mechanisms, such as humoral and cellular adaptive immunity and innate response, play roles in the development of systemic inflammatory syndrome [11]. Recent studies have also suggested that specific HLA haplotypes could be more frequently associated with the presence of this disease [13].
It has been shown that infections, especially of the gastrointestinal and respiratory systems, have effects on microbiota composition [14]. It is thought that microbiota composition may play a role in the development, prognosis, or post-infection of COVID-19 [15]. It has also been suggested that microbial interactions and competition, especially in the nasopharynx of children colonized by more viruses and bacteria compared with adults, may reduce the infectious potential of SARS-CoV-2 [16]. Some studies have evaluated microbiota composition at the time of diagnosis and during the course of COVID-19, especially in adults; however, few studies on microbiota composition in children have been conducted [17–19]. Moreover, no previous study has investigated microbiota composition in MIS-C. Therefore, this study aimed to compare intestinal microbiota composition in children diagnosed with MIS-C and acute COVID-19 infection with healthy children.
Materials and methods
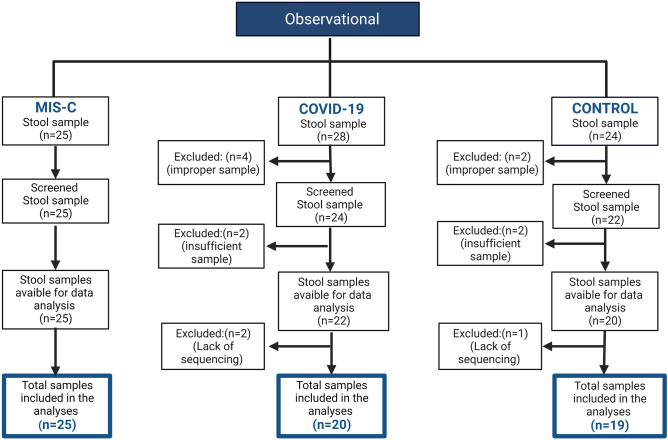
This prospective multicenter (i.e., five centers in four cities) study aimed to evaluate the microbiota composition in patients diagnosed with MIS-C or SARS-CoV-2, and a control sample of healthy children aged between 3 and 14 years. This clinical study was planned and performed in accordance with the Declaration of Helsinki and Good Clinical Practice guidelines, patient rights regulations, and ethical committees. Permission for the study was obtained from the Clinical Research Ethics Committee of Eskisehir Osmangazi University’s Faculty of Medicine based on Decision Number 12 on March 2, 2021. Permission for the clinical study was also obtained from the Ministry of Health in Turkey. Written informed consent was obtained from all parents and children prior to their inclusion in the study. The study was supported by Eskisehir Osmangazi University Scientific Research Project (TTU-2021–1750). The results of the study and flow chart are shown according to Strengthening the Organization and Reporting of Microbiome Studies (STORMS) [20] (Fig. 1).
Fig. 1.
Flow chart of the study groups
COVID-19 was diagnosed according to SARS-CoV-2 rt-PCR positivity in nasopharyngeal swab samples. To diagnose MIS-C, we used the Centers for Disease Control and Prevention (CDC) guidelines [21]. The exclusion criteria for the patient and control groups were as follows: the presence of chronic disease or routine medications; use of antibiotics/probiotics in the last 8 weeks; and treatment related to MIS-C or COVID-19 before the study samples were collected. The age, gender, height, body weight, and body mass index (BMI) of all cases at the time of admission were recorded, and anthropometric parameters were based on percentile scales according to the age group. Obesity was defined as a BMI at or above the 95th percentile and overweight at or above the 85th percentile on the CDC’s sex-specific age-for-age BMI growth charts [22]. Symptoms and signs were noted in all cases based on a detailed physical examination. SARS-CoV-2 rt-PCR and serological tests (SARS-CoV-2 IgM and/or IgG) were noted. At the time of diagnosis, white blood cell count, absolute neutrophil count, absolute lymphocyte count, platelet count, absolute eosinophil count, blood biochemistry (i.e., urea nitrogen, creatinine, transaminases, albumin, cardiac markers, and lactate dehydrogenase), inflammation biomarkers (i.e., serum procalcitonin, erythrocyte sedimentation rate, fibrinogen, ferritin, C-reactive protein, and IL-6 levels), and coagulation parameters (aPTT, PT, INR, and D-dimer) were extracted from medical records. We enrolled 24 aged-matched healthy children (matched with 25 children with MIS-C) served as the control group (Fig. 1). IgM/IgG rapid diagnosis kits (KIT-19 SARS-CoV-2 IgG/IgM Rapid Test Cassette, Devaju®, Turkey) were used to evaluate whether the healthy children to be included in the study had the disease previously; only children with negative test results were enrolled.
Stool samples were obtained from the participants at the time of their enrolment before initiation of any COVID-19/MIS-C-related therapy. The stool samples of at least 5 ml were taken in 50 cc Falcon tubes and then immediately frozen. The Falcon tubes were stored upright at − 80 °C without any treatment. All samples were delivered to the laboratory where the DNA analysis was conducted in accordance with cold chain requirements every 3 months.
Fecal DNA extraction, sequencing, and bioinformatics analysis
QuickGene (Kurabo, Japan) was used to extract DNA from the stool samples. First, 25 mg of each stool sample was transferred to a homogenization tube with 250 µl of tissue lysis (MDT) solution. To homogenize the solution, 15 mg of 0.1 mmø glass beads or 10 1.0 mmø zirconia beads were added to the tube and then homogenized for 2 × 120 s at 5,000 rpm. After the sample was homogenized, 25 µl of Proteinase K (EDT) solution was added and incubated at 56 °C for 60 min. The tube was then centrifuged at 15,000 g for 10 min at room temperature. After centrifugation, 200 µl of supernatant was transferred to a 1.5 ml microtube. Then 180 µl of cell lysis (LDT) solution was added and vortexed for 15 s. The microtube was left to incubate at 70 °C for 10 min. In the next step, 240 µl of 99% cold ethanol was added and vortexed for 15 s. The entire contents of the microtube were transferred to a QuickGene (Kurabo, Japan) filtered cassette, where washes and elutions were performed following the instrument protocol. Three washes were performed using 750 μl of wash buffer (WDT) solution. Based on the results of the extraction process, bacterial 16S ribosomal RNA (rRNA) gene target sequencing was performed from the materials obtained in the study. The resulting genomic DNA was amplified using 16S V3-V4 314F-860R primer sets, and library preparation was performed using a Nextera XT DNA library preparation kit and indices (Illumina, CA, USA). The amplicon library was cleaned by selecting large fragments (AMPure XP, Beckman Coulter). It was then normalized and aggregated. After the library was prepared, the NovoSeq 6000 (Illumina, CA, USA) instrument was used to run the sequencing.
Pair-ended Illumina reads (2 × 250) were transferred to the QIIME2 environment [23]. All samples had a sequence depth greater than 100 × , and no samples were omitted from the run. Quality clipping, chimera detection, and read cleaning were implemented using the QIIME2 Dada2 pipeline (via q2‐dada2) [24]. Amplicon sequence variants (ASVs) generated by Dada2 were mapped to the GreenGenes (http://greengenes.lbl.gov) database [25, 26]. The phyloseq object was created from qiime2 artifact files in the R 4.1 environment [27, 28]. Alpha diversity metrics (Chao-1 diversity and Simpson index) were calculated from the phyloseq object using the microbiome R package. Significant differences between groups were calculated using the Kruskal–Wallis rank sum test. Beta diversity was computed by phyloseq, including the Bray–Curtis, Jaccard, unweighted UniFrac, and weighted UniFrac distance metrics. Beta diversity statistical significance between groups was calculated using a PERMANOVA test via the Adonis function in the vegan R package. Intergroup p values were calculated using the Kruskal–Wallis test. Specific differences between groups were determined by differential abundance analysis using the Deseq2 R package [29]. Linear discriminant analysis effect size (LEFSe) analyses were performed between groups to determine statistically significant taxonomies [30].
Statistical analysis
The SPSS Statistical Package for the Social Sciences (Illinois, CA, USA) program was used for the statistical analysis. Descriptive statistical methods (i.e., median in percentage) Pearson’s chi-square, Kruskal–Wallis, and ANOVA tests were conducted to compare the qualitative data; p < 0.05 was accepted as statistically significant correlations between data. In this study, the R program was used to evaluate the microbiota analysis.
Results
A total of 64 children (27 boys, 37 girls) were included in the study (20 COVID-19 cases [10 boys, 10 girls], 25 MIS-C cases [16 boys, 9 girls], and 19 healthy children [11 boys, 8 girls]). There was no statistical difference between the study groups in terms of gender (p > 0.05). The median age was 7 years in the MIS-C group, 9.5 years in the COVID-19 group, and 8 years in the control group (Table 1). The age distribution of the MIS-C, COVID-19, and control groups was statistically similar (p < 0.05). In the MIS-C group, five children were obese (> 95th percentile), and nine children were overweight (85–95th percentile). It was observed that in the COVID-19 group, five subjects were obese (> 95th percentile) and seven subjects were overweight (85–95th percentile).
Table 1.
Demographic and clinical findings of the study groups
|
MIS-C (n = 25) |
COVID-19 (n = 20) |
Control (n = 19) |
|
|---|---|---|---|
| Age/years | 7 (5–10) | 9.5 (5.2–11) | 8 (7–10) |
| Body weight (kg) | 28 (20–43) | 41.5 (20.5–59.7) | 32 (26–37) |
| Height (cm) | 124 (115–145) | 137 (114–156) | 133 (120–140) |
| Body mass index (BMI) (kg/m2) | 17.8 (15.9–22.0) | 20.2 (15.7–24.1) | 18.9 (16.6–21.9) |
| Presence of overweight/obesity (n/%) | |||
| Overweight (85–95th p) | 9/25 (36) | 7/20 (3) | 0/19 (0) |
| Obese (> 95th p) | 5/25 (20) | 5/20 (25) | 0/19 (0) |
| Symptoms* (n/100) | |||
| Fever | 25/25 (100) | 15/20 (75) | |
| Cough | 4/25 (16) | 10/20 (50) | |
| Runny nose | 2/25 (8) | 5/20 (40) | |
| Diarrhea | 14/25 (56) | 5/20 (40) | |
| Tachypnea | 5/25 (20) | 5/20 (40) | |
| Myalgia | 12/25 (48) | 7/20 (35) | |
| Abdominal pain | 18/25 (72) | 6/20 (30) | |
| Headache | 9/25 (36) | 7/20 (35) | |
| System involvement** (n/%) | |||
| Hematological | 18/25 (72) | ||
| Gastrointestinal | 16/25 (64) | ||
| Cardiovascular | 10/25 (40) | ||
| Dermatological | 8/25 (32) | ||
| Respiratory | 5/25 (20) | ||
| Neurological | 2/25 (8) | ||
*Most patients had more than one symptom. **Most patients had more than one system involvement
Diagnoses were based on SARS-CoV-2 rt-PCR positivity in 95% of the COVID-19 cases and antigen positivity in one case. In the MIS-C cases, 24% (n = 6) were SARS-CoV-2 rt-PCR positive, 52% (n = 13) were SARS-CoV-2 IgG positive, 28% (n = 7) were both SARS-CoV-2 IgG and SARS-CoV-2 IgM positive, and antigen positivity was detected in one case. There was a history of contact in 10 cases (40%) in the MIS-C group.
In the COVID-19 cases, fever was the most common symptom (75%, n = 15), followed by cough (50%, n = 10). Other symptoms were diarrhea (40%, n = 5), tachypnea (40%, n = 5), runny nose (40%, n = 5), myalgia (35%, n = 7), headache (35%, n = 7), and abdominal pain (30%, n = 6). In the COVID-19 group, positive results were found in 13 cases (65%) of chest radiography and in 7 cases (35%) of computerized thorax tomography.
Fever was present in all MIS-C cases. The most common symptoms in the MIS-C patients were abdominal pain (72%, n = 18), diarrhea (56%, n = 14), and myalgia (48%, n = 12), followed by headache (36%, n = 9), tachypnea (20%, n = 5), cough (16%, n = 4), and runny nose (8%, n = 2). Most cases had more than one symptom. The most common sign was rash (44%). Other pathological findings were symptoms of conjunctivitis (24%), respiratory (20%), and neurological (8%) symptoms. Most patients in the study population had multisystem involvement. The most common system involvements were hematologic (72%) and gastrointestinal (64%). Other system involvements were cardiovascular (40%), dermatological (32%), respiratory (20%), and neurological (8%) (Table 1). The most common laboratory parameters in the MIS-C cases were elevated serum C-reactive protein levels in 16 cases (64%), elevated serum fibrinogen levels in 13 cases (52%), and elevated D-dimer levels in 12 cases (48%). Other markers were lymphopenia in 9 cases (36%), high ferritin in 8 cases (32%), high BNP in 8 cases (32%), high erythrocyte sedimentation rate in 4 cases (16%), high INR in 4 cases (16%), elevated serum IL-6 levels in 4 cases (16%), high levels of procalcitonin in 3 cases (12%), high levels of LDH in 1 case (4%), high levels of neutrophils in 2 cases (8%), and elevated liver function in two cases (8%).
Intestinal microbiota analysis
Alpha and beta diversity
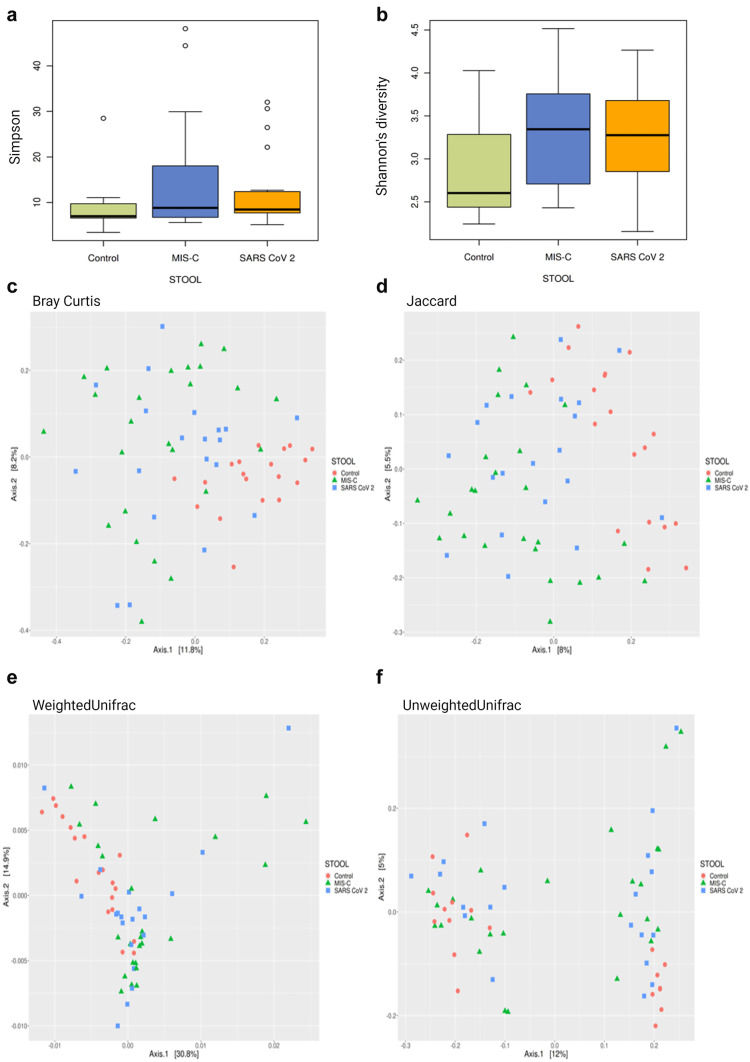
Although the median observed taxonomic units (OTUs) value observed in the stool samples taken from the MIS-C group was lower than those in the control and SARS-CoV-2 groups, no statistical difference was found in OTU levels, the Chao-1 index, and Simpson index (p > 0.05). Species dominance distribution between the 25th and 75th percentile range in both stool sample data was found to be greater in MIS-C cases compared to other groups (Fig. 2a). The Shannon index is a measure of richness and uniformity that considers the entropy used in the study. It was higher in the MIS-C cases compared with the control group (p < 0.01) (Fig. 2b). A statistical difference in the stool samples was found between the groups in the results of the Bray–Curtis (Fig. 2c), Jaccard (Fig. 2d), weighted UniFrac (Fig. 2e), and unweighted UniFrac (Fig. 2f) baseline coordinate analysis (PCoA) (p = 0.001).
Fig. 2.
Alpha and beta diversity plots to visualize the difference in microbiota structure between the MIS-C, SARS-CoV-2, and control groups. Simpson index comparison between the study groups. There was no significant difference in stool sample data between the groups in the study (p > 0.05). a Shannon index comparison between the study groups. Shannon index was higher in stool sample data of MIS-C cases when the control group (p < 0.01). b PCoA 2D plots of beta diversity analysis of the MIS-C, SARS-CoV-2, and control groups. Each dot represents a stool sample. Red circle, green triangle, and blue square represent control, MIS-C, and SARS-CoV-2 cases, respectively. c–f Between-sample dissimilarities were measured by Bray–Curtis distances (c), Jaccard distance (d), weighted UniFrac distances (e), and unweighted UniFrac distances (f). A statistical difference in the stool samples was found between the groups in the results of the Bray–Curtis (c), Jaccard (d), weighted UniFrac (e), and unweighted UniFrac (f) baseline coordinate analysis (PCoA) (p = 0.001)
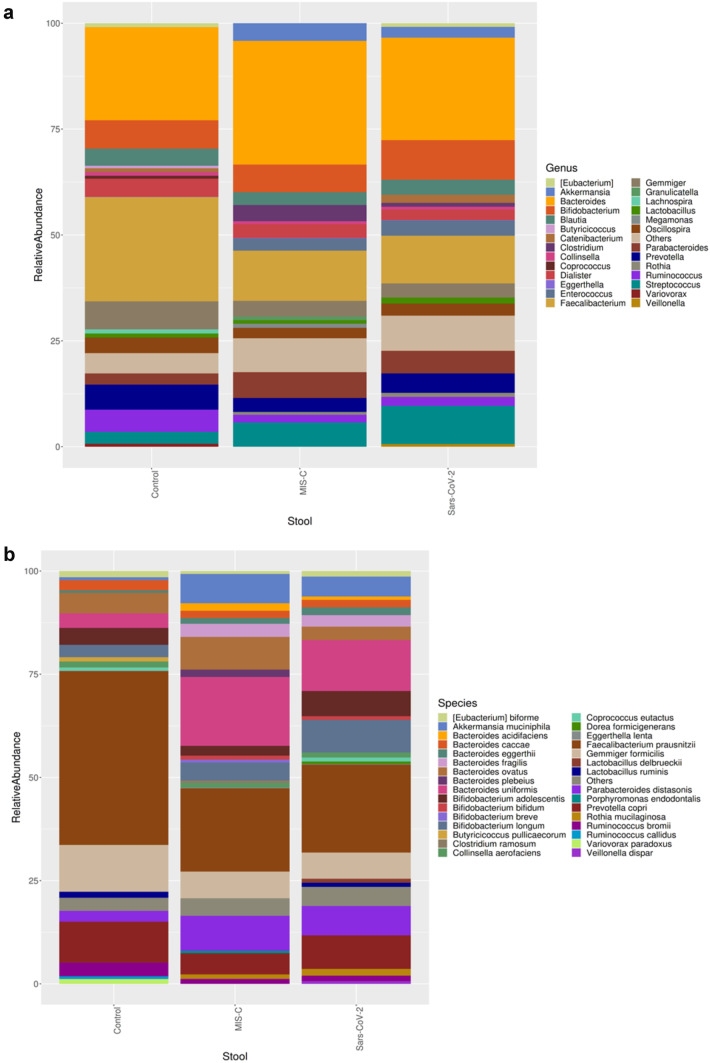
The MIS-C group showed the following phylum levels: Firmicutes (46%), Bacteroidetes (32%), Proteobacteria (11%), Actinobacteria (7%), and Verrucomicrobia (3%). In the control group, they were as follows: Firmicutes (64%), Bacteroidetes (23%), Actinobacteria (6%), and Proteobacteria (5%). In the SARS-CoV-2 group, the levels were as follows: Firmicutes (51%), Bacteroidetes (25%), Proteobacteria (12%), Actinobacteria (9%), and Verrucomicrobia (2%). Regarding the presence of Bacteroidetes, no difference was observed between the SARS-CoV-2 group and the control group; however, Bacteroidetes were higher in the MIS-C group compared with the other two groups.
Microbiota composition is shown according to genera and species levels in Fig. 3. At genera levels, the dominant genus in the healthy controls was Faecalibacterium (16%), which was decreased in the MIS-C (9%) group and the SARS-CoV-2 (8%) group. Bacteroides, Eggerthella, and Clostridium genera were significantly higher in the MIS-C group (21%, 10%, and 21%, respectively) than in the SARS-CoV-2 group (16%, 6%, and 2%, respectively) and the control group (15%, 2%, and 15%, respectively) (Fig. 3a).
Fig. 3.
Intestinal microbiota composition of the study groups at species and genus levels. Bacterial community relative abundance analysis at the genus (a) and species (b) levels (relative abundance > 1%; bacteria with relative abundances < 1% were pooled in the “others” category and sorted by total concentration)
At the species level, we observed abundances of Faecalibacterium prausnitzii, Gemmiger formicilis, Ruminococcus bromii, Bifidobacterium adolescentis, Lactobacillus ruminis, Butyricicoccus pullicaerocum, Collinsella aerofaciens, Ruminococcus callidledikans, and Dorea formicians in the healthy controls compared with the MIS-C group. The most abundant species in the MIS-C group were Bacteroides uniformis, Bacteroides plebeius, Clostridium ramosum, Eubacterium dolichum, Eggerthella lenta, Bacillus thermoamylovorans, Prevotella tannerae, and Bacteroides coprophilus. The SARS-CoV-2 group showed an increase in the relative abundance of Bacteroides coprophilus, Bifidobacterium adolescentis, Dorea formicigenerans, Ruminococcus albus, and Clostridium piliforme (Fig. 3b).
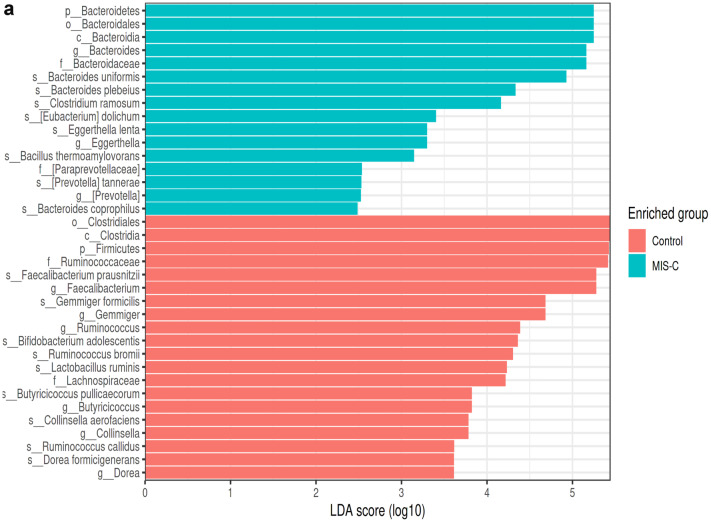
In this study, a LEfSe analysis (LDA threshold value > 2, p < 0.05) was used to determine significant bacterial compositions between groups. The results of the LEfSe analysis in the MIS-C group and the control group are shown in Fig. 4a. At the genus level, in the MIS-C group, Bacteroides, Eggerthella, and Prevotella were dominant, whereas Faecalibacterium, Gemmiger, Ruminococcus, Butyricicoccus, Collinsella, and Dorea were found to be dominant in the control group. At the species level, Bacteroides uniformis, Bacteroides plebeius, Clostridium ramosum, Eubacterium dolichum, Eggerthella lenta, Bacillus thermoaamylovorans, Prevotella tannerae, and Bacteroides coprophilus were most abundant in the MIS-C group. In the control group, Faecalibacterium prausnitzii, Gemmiger formicilis, Bifidobacterium adolescentis, Ruminococcus bromii, Lactobacillus ruminis, Butyricicoccus pullicaerocum, Collinsella aerofaciens, Ruminococcus callidus, and Dorea formicigenerans were found to be dominant (Fig. 4a).
Fig. 4.
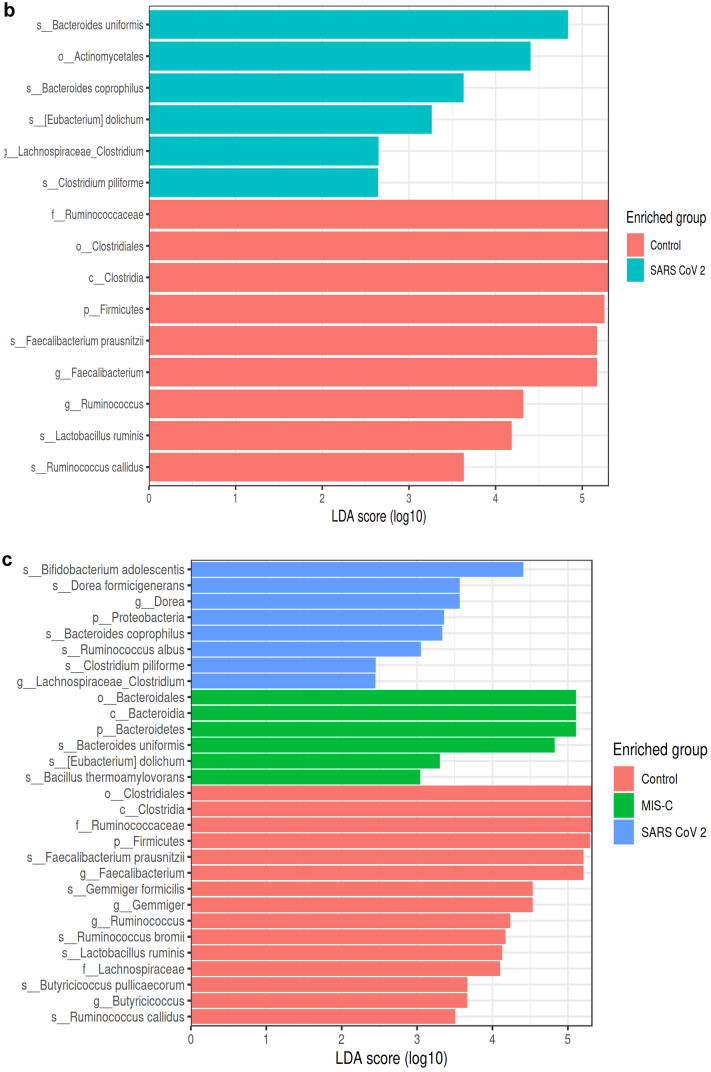
LEfSe analysis graphics showing bacterial taxa that were significantly different in abundance between the study groups. a–c LefSe analysis was performed to identify differentially abundant taxa for which the LDA scores are shown. Only species and functional modules with LDA effect size > 2 and FDR-corrected p value < 0.05 were plotted. Horizontal bars represent log.10 converted LDA score indicated by vertical dotted lines. Different colors represent different groups. p—phylum, c—class, o—order, f—family, g—genus, s—species. LEfSe analysis in stool samples for the MIS-C and control groups (control group (red), MIS-C (green)). a At the genus level, in the MIS-C group, Bacteroides (LDA score = 5.162, p = 0.018), Eggerthella (LDA score = 3.297, p = 0.017), and Prevotella (LDA score = 2.514, p = 0.038) were dominant, whereas Faecalibacterium (LDA score = 5.265, p = 0.0008), Gemmiger (LDA score = 4.666, p = 0.012), Ruminococcus (LDA score = 4.368, p = 0.000), Butyricicoccus (LDA score = 3.815, p = 0.002), Collinsella (LDA score = 3.752, p = 0.045), and Dorea (LDA score = 3.608, p = 0.045) were found to be dominant in the control group. At the species level, Bacteroides uniformis (LDA score = 4.926, p = 0.023), Bacteroides plebeius (LDA score = 4.332, p = 0.046), Clostridium ramosum (LDA score = 4.160, p = 0.046), Eubacterium dolichum (LDA score = 3.401, p = 0.023), Eggerthella lenta (LDA score = 3.296, p = 0.032), Bacillus thermoaamylovorans (LDA score = 3.146, p = 0.023), Prevotella tannerae (LDA score = 2.523, p = 0.038), and Bacteroides coprophilus (LDA score = 2.460, p = 0.041) are most abundant in the MIS-C group. In the control group, Faecalibacterium prausnitzii (LDA score = 5.265, p = 0.000), Gemmiger formicilis (LDA score = 4.666, p = 0.012), Bifidobacterium adolescentis (LDA score = 4.344, p = 0.008), Ruminococcus bromii (LDA score = 4.286, p = 0.000), Lactobacillus ruminis (LDA score = 4.231, p = 0.046), Butyricicoccus pullicaerocum (LDA score = 3.815, p = 0.002), Collinsella aerofaciens (LDA score = 3.752, p = 0.045), Ruminococcus callidus (LDA score = 3.609, p = 0.008), and Dorea formicigenerans (LDA score = 3.608, p = 0.000) were found to be dominant (a). LEfSe analysis in stool samples for the SARS-CoV-2 and control groups (control group (red), SARS-CoV-2 (blue)). b At the genus level, Lachnospiraceae_Clostridium (LDA score = 2.648, p = 0.039) was dominant in SARS-CoV-2 cases, while Faecalibacterium (LDA score = 5.168, p = 0.016) and Ruminococcus (LDA score = 4.316, p = 0.037) were dominant in the control group. At the species level, in the SARS-CoV-2 group, Bacteroides coprophilus (LDA score = 3.629, p = 0.002), Eubacterium dolichum (LDA score = 3.261, p = 0.012), Bacteroides uniformis (LDA score = 4.834, p = 0.011), and Clostridium piliforme (LDA score = 2.642, p = 0.039) were dominant, while Faecalibacterium prausnitzii (LDA score = 5.168, p = 0.016), Ruminococcus callidus (LDA score = 3.632, p = 0.003), and Lactobacillus ruminis (LDA score = 4.184, p = 0.013) were dominant in the control group (b). LEfSe analysis in stool samples for the MIS-C, SARS-CoV-2, and control groups (control group (red), MIS-C group (green), SARS-CoV-2 (blue)). c Bacteroides uniformis (LDA score = 4.820, p = 0.024), Bacillus thermoaamylovorans (LDA score = 3.040, p = 0.048), and Eubacterium dolichum (LDA score = 3.297, p = 0.002) were most abundant species in children with MIS-C at species level. Bacteroides coprophilus (LDA score = 3.331, p = 0.008), Bifidobacterium adolescentis (LDA score = 4.408, p = 0.016), Dorea formicigenerans (LDA score = 3.560, p = 0.000), Ruminococcus albus (LDA score = 3.049, p = 0.017), and Clostridium piliforme (LDA score = 2.450, p = 0.0130) were most abundant species in children with SARS-COV-2. In the control group, Faecalibacterium prausnitzii (LDA score = 5.201, p = 0.000), Gemmiger formicilis (LDA score = 4.524, p = 0.019), Ruminococcus bromii (LDA score = 4.169, p = 0.000), Lactobacillus ruminis (LDA score = 4.231, p = 0.046), Butyricicoccus pullicaerocum (LDA score = 3.660, p = 0.002), and Ruminococcus callidus (LDA score = 3.499, p = 0.000) were found to be dominant (c)
The results of the LEfSe analysis conducted to compare the SARS-CoV-2 group and the control group are shown in Fig. 4b. At the genus level, Lachnospiraceae_Clostridium was dominant in the SARS-CoV-2 group. Faecalibacterium and Ruminococcus were dominant in the control group. At the species level, in the SARS-CoV-2 group, Bacteroides coprophilus, Eubacterium dolichum, Bacteroides uniformis, and Clostridium piliforme were dominant. Faecalibacterium prausnitzii, Ruminococcus callidus, and Lactobacillus ruminis were dominant in the control group (Fig. 4b).
The results of the LEfSe analysis of three groups (MIS-C, SARS-CoV-2, and the controls) were also compared and Bacteroides uniformis, Bacillus thermoaamylovorans, and Eubacterium dolichum were the most abundant species in the MIS-C group at the species level. Bacteroides coprophilus, Bifidobacterium adolescentis, Dorea formicigenerans, Ruminococcus albus, and Clostridium piliforme were the most abundant species in the SARS-COV-2 group. In the control group, Faecalibacterium prausnitzii, Gemmiger formicilis, Ruminococcus bromii, Lactobacillus ruminis, Butyricicoccus pullicaerocum, and Ruminococcus callidus were dominant (Fig. 4c).
Discussion
In our study, we first showed that the diversity, richness, and composition of intestinal microbiota in the MIS-C group at the time of diagnosis differed from those in the healthy controls. These findings indicate that the composition of microbiota can deteriorate during the course of infection. Previous studies have evaluated microbiota composition at the time of diagnosis of COVID-19, during the course of the disease, and at post-disease follow-up, especially in adults. Chen et al. [31] showed that microbiota richness did not return to normal levels 6 months after recovery in adult patients with COVID-19; however, another study showed that dysbiosis improved rapidly after the disease, and there was no difference from the healthy control group [17]. Geographical and demographic differences have been observed in previous studies on the recovery of microbiota composition after COVID-19 [32]. Our study is the first to be conducted in children with MIS-C, and the fact that both alpha and beta diversity indicators and their relative abundance were different in the MIS-C cases compared with the healthy controls supported that microbiota composition changed in the MIS-C group.
In our study, the relative abundance of Bacteroidetes increased in the MIS-C group, and the Firmicutes:Bacteroides ratio decreased significantly in the MIS-C group compared with the healthy control group and the SARS-CoV-2 group. Endotoxins and metabolites of microorganisms causing inflammation in the lung have been shown to enter the bloodstream, causing an increase in the relative abundance of Bacteroidetes and a decrease in the relative abundance of Firmicutes in the intestinal composition [33]. In addition, previous findings showed that Firmicutes decreased and Bacteriodes increased in Kawasaki disease [34]. We also found an increased percentage of Proteobacteria (11%) in children with MIS-C compared to healthy controls (5%).
Compared with the healthy children in the MIS-C group, the dominant genera were Bacteroides, Eggerthella, and Prevotella, and the dominant species were Bacteroides uniformis, Bacteroides plebeius, Clostridium ramosum, Eubacterium dolichum, Eggerthella lenta, Bacillus thermoamylovorans, Prevotella tannerae, and Bacillus thermoamylovorans. The findings also showed that Faecalibacterium prausnitzii decreased in the MIS-C group compared with the healthy control group. F. prausnitzii, which is a notable member of intestinal microbiota, has positive effects on the immune system, as well as anti-inflammatory properties. Previous studies have shown that decreased F. prausnitzii could be considered a microbiological marker of gastrointestinal tract inflammation [35]. Decreased F. prausnitzii has been reported in cystic fibrosis [36], inflammatory bowel disease [37–39], hypertension [40], multiple sclerosis [41], obesity [42], type 2 diabetes [43, 44], rheumatoid arthritis [45], celiac disease [46], and in adults with COVID-19 [18, 47, 48]. In our study, we also found that F. prausnitzii decreased in the intestinal microbiota composition of the cases in the MIS-C group, which could be accepted as an indicator of gastrointestinal system involvement/inflammation in MIS-C cases.
Our findings also showed an increase at the genus level in the microbiota element Prevotella in the MIS-C group. Prevotella strains are anaerobic gram-negative bacteria of the Bacteroidetes phylum, and they are among the natural elements of microbiota composition. A previous study found a relationship between Prevotella strains and periodontitis, bacterial vaginitis, rheumatoid arthritis, and systemic and metabolic disorders [49]. In a study conducted on obese children in Mexico, the findings showed that Prevotella strains were increased [50]. It was concluded that there might be a relationship between Prevotella dominance and high zonulin levels in obese patients with colorectal cancer [51]. Although zonulin levels were not included in the present study, they were found to be high in MIS-C cases in a previous study [52]. In rheumatoid arthritis patients, it was shown that Prevotella activated the TLR-2 receptor of intestinal epithelial cells, stimulated the release of proinflammatory cytokines, such as IL-1β, IL-6, and IL-23, supported the activation of Th17 cells that lead to IL-17 production, and played a role in inflammation [53, 54]. In our study, we found that Prevotella at the genus level and Prevotella tannerae were significantly dominant in the intestine at the species level. Considering inflammation in MIS-C disease, further studies are needed on the genus and species of Prevotella in MIS-C cases.
Eggerthella lenta is an anaerobic, non-spore-forming gram-positive bacillus that is among the natural members of the gut microbiota [55]. Previous studies have shown that Th17 activation may have effects on related inflammatory diseases, familial Mediterranean fever, some infections, and autoimmune diseases [56–61]. Crohn’s disease, ulcerative colitis, multiple sclerosis, and rheumatoid arthritis were found to be higher in the Behçet’s disease group than in the Eggerthella control group at the genus level [62, 63]. In our study, a significant increase was observed in the MIS-C group at both the genus (Eggerthella) and species levels (Eggerthella lenta). Considering autoimmunity and the Th17 pathway in the pathogenesis of MIS-C [64], our findings suggest that an increase in Eggerthella lenta may be associated with the pathogenesis. We also found that Eubacterium dolichum was dominant in both the MIS-C and COVID-19 groups in our study. The E. dolichum strain is predominant in obesity and metabolic dysfunctions, which have been defined as risk factors for MIS-C [65–68]. Similarly, the predominance of Clostridium ramosum, which was previously shown to increase in metabolic syndrome, obesity, and COVID-19 cases, in our study, it was thought to support the relationship between obesity and metabolic disorders and MIS-C [59, 69–73].
The clinical picture of MIS-C may be characterized by an exaggerated immune response and cytokine storm in children and adolescents, who may be genetically susceptible after exposure to SARS-CoV-2 [74]. Previous findings showed that genetic predisposition, environmental factors, and intestinal microbiota play roles in the development of abnormal immune responses and inflammation in many diseases [75–77]. It has also been shown that microbiota composition is impaired in many diseases (i.e., dysbiosis), but research has yet to determine whether these disorders were causes or effects. The findings of our study did not provide evidence for a cause–effect relationship between existing microbiota in MIS-C cases.
Previous studies on adults have reported a reduction in microbial diversity and a possible association between increased intestinal dysbiosis and disease severity in COVID-19-positive patients compared with healthy controls [78]. One hypothesis regarding the less severe course of COVID-19 infection in children compared with adults is that their oropharyngeal, nasopharyngeal, lung, and/or gastrointestinal microbiota are different from those of adults [16]. It has been suggested that, especially in the nasopharynx of children colonized with more viruses and bacteria compared with adults, microbial interactions and competition may reduce the infection potential of SARS-CoV-2 [16, 79]. In our study, different strains were determined by intestinal microbiota analysis at the time of diagnosis of COVID-19 in the children in that group, which had not been reported in other studies. In the COVID-19 group, the taxa Eubacterium, Faecalibacterium prausnitzii, Roseburia, and Lachnospiraceae, which are common in healthy children, were detected less frequently. The comparison of the COVID-19 group with the MIS-C group showed further differences. Previous studies have shown that Faecalibacterium species and bacterial diversity in general are reduced in patients infected with SARS-CoV-2 [72, 73, 78, 80]. A negative correlation was found between the presence/increase of Faecalibacterium prausnitzii, which has anti-inflammatory properties and is one of the important features of healthy gut microbiota, and disease severity [72, 81]. It has also been shown that microbiota change depending on inflammation in COVID-19 cases [18, 72, 82]. In another study, butyrate-producing F. prausnitzii, Clostridum butyricum, C. leptum, and Eubacterium rectale were significantly reduced during COVID-19 infection [81]. A reduction in short-chain fatty acids may downregulate ACE-2, one of the key receptors of the SARS-CoV-2 virus [83]. Experimental studies have shown that the depletion of ACE-2 promoted intestinal epithelial damage and increased sensitivity to inflammation [84]. In COVID-19 cases, microbiota may play a role in the pathogenesis of acute lung injury by various mechanisms, including the direct translocation of bacteria from the gut to the lung and the immune-modulating effects of microbe-related metabolites [85]. Increased levels of Enterobacteriaceae and Lachnospiraceae were observed in patients with severe ARDS [86]. These results suggest that microbiota can be used as a potential marker in the development and follow-up of ARDS in COVID-19 cases [87]. Previous research has shown that children with respiratory tract infections have changes in their gut microbiome [88]. Hasegawa et al. [89] discovered that infants with the Enterobacter/Veillonella-dominant profile had the lowest proportion of bronchiolitis and those with the Bacteroides-dominant profile had the highest. De Maio and colleagues recently conducted a comparative analysis in hospitalized infants younger than 6 months with acute bronchiolitis caused by RSV or non-RSV, and found no statistically significant differences in intestinal microbiota composition in children with bronchiolitis caused by RSV or non-RSV [90].
The long-term effects of SARS-CoV-2 infection in children are little understood [91]. To date, the majority of investigations have focused on adult populations. However, a subset of children appear to recover from their initial COVID-19 infection but have signs and symptoms and may be diagnosed with long-COVID [92]. Severe acute infection might be related with chronic symptoms; however, symptoms might be also related with chronic inflammation, autoimmunity, chronic endotheliopathy, microthrombosis, or viral persistence [91]. Microbiota composition changes might also play a role in long-COVID. In our study, we did not enroll children with long-COVID and potential further studies will clarify this interaction.
Our study has the following limitations. We collect a single fecal sample at the diagnosis, and it is not possible to evaluate the cause of consequence of. Our research was conducted during the period when the Alpha variant and the first part of the Delta variant were apparent in SARS-CoV-2 infection. Moreover, our results did not include the period when the Delta and Omicron variants were predominant. Because many factors affect microbiota composition in children, it was not possible to ensure that all the children in our study groups were homogeneously similar. Geography, diet, culture, traditions, physiological variations, and changes in lifestyle have been shown to affect microbiota composition. Most cases included in the SARS-CoV-2 group had mild to moderate symptoms and signs, and microbiota composition or prognoses could not be evaluated in severe cases. In addition, the children in the MIS-C and SARS-CoV-2 groups and in the healthy control group have been enrolled during the restrictions applied at the global level in response to the COVID-19 epidemic (e.g., curfews, school closures, increased hand hygiene, fewer social interactions, and travel restrictions).
Conclusion
Our study is the first in the literature to show that changes occurred in intestinal microbiota composition in MIS-C cases. It is not possible to conclude whether the changes we detected in the composition of the microbiota were causes or effects. Many previous studies have shown a decreased abundance of Faecalibacterium prausnitzii in various intestinal diseases, similar to the children in our MIS-C group. In our study, we also found an increase in the Eggerthella lenta strain in the MIS-C group compared with the healthy controls. The predominance of E. lenta in diseases associated with abnormal Th17 activation, such as rheumatoid arthritis, multiple sclerosis, and inflammatory bowel disease, shown in previous studies supports the association of E. lenta with autoimmunity. In addition, it is thought that the changes detected in microbiota composition in children may affect the immune, gastrointestinal system, metabolic, and brain–intestinal axis of the child at that time, as well as in adulthood [93]. Long-term observations of these changes, which we detected for the first time in our study, and their effects on prognoses should be conducted in a larger series.
Abbreviations
- ARDS
Acute respiratory distress syndrome
- BMI
Body mass index
- CDC
Centers for Disease Control and Prevention
- COVID-19
Coronavirus disease 2019
- IgM
Immunoglobulin M
- IgG
Immunoglobulin G
- IL-6
Interleukin 6
- MIS-C
Multisystem inflammatory syndrome in children
- LEFSe
Linear discriminant analysis effect size
- rt-PCR
Reverse transcription–polymerase chain reaction
- STORMS
Strengthening the Organization and Reporting of Microbiome Studies
Authors’ contributions
Dr Cansu Suskun conceptualized and designed the study, drafted the initial manuscript, and reviewed and revised the manuscript. Dr Sirin Guven, Dr Adem Karbuz, Dr Aslinur Ozkaya-Parlakay, Dr Yalcin Kara, Dr Ebru Kacmaz, Dr Aysın Boga, Dr Didem Inancli, Dr Belgin Gulhan, Dr Saliha Kanik Yuksek, Dr Eylem Kiral, Dr Gurkan Bozan, Dr Mehmet Ozgur Arslanoglu, Dr Mahmut Can Kizil, Dr Meltem Dinleyici, and Dr Tercan Us evaluated and recruited patients, recorded clinical and laboratory results, and collected the samples. Mucahit Kaya and Ahmet Varis designed the data collection instruments, carried out the initial analyses and sequencing, and performed laboratory and bioinformatic analysis. Dr Kilic, Dr Yilmaz-Ciftdogan, and Dr Vandenplas conceptualized and designed the study, and critically reviewed the manuscript for important intellectual content. EC Dinleyici conceptualized and designed the study, evaluated results, drafted the initial manuscript, and reviewed and revised the manuscript. All authors approved the final manuscript as submitted and agree to be accountable for all aspects of the work.
Funding
The study was supported by Eskisehir Osmangazi University Scientific Research Project (TTU-2021–1750).
Availability of data and material
Authors agree to participate all data, if requested.
Code availability
Not available.
Declarations
Ethics approval
Permission for the study was obtained from the Clinical Research Ethics Committee of Eskisehir Osmangazi University’s Faculty of Medicine based on Decision Number 12 on March 2, 2021. Permission for the clinical study was also obtained from the Ministry of Health in Turkey.
Consent to participate
Written informed consent was obtained from all parents and children prior to their inclusion in the study. This clinical study was planned and performed in accordance with the Declaration of Helsinki and Good Clinical Practice guidelines, patient rights regulations, and ethical committees.
Consent for publication
Not available.
Conflict of interest
The authors declare no competing interests.
Footnotes
The original online version of this article was revised: The co-author's name has been updated Saliha Kanik-Yuksek and cited as Kanik-Yuksek S.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
6/13/2022
A Correction to this paper has been published: 10.1007/s00431-022-04525-5
Contributor Information
Cansu Suskun, Email: cansuceyda93@gmail.com.
Omer Kilic, Email: omerkilic7@yahoo.com.
Dilek Yilmaz Ciftdogan, Email: drdilekyilmaz@hotmail.com.
Sirin Guven, Email: sirin2006@gmail.com.
Adem Karbuz, Email: karbuzadem@hotmail.com.
Aslinur Ozkaya Parlakay, Email: aslinur.o@gmail.com.
Yalcın Kara, Email: dryalcinkara@hotmail.com.
Ebru Kacmaz, Email: elvinduru3446@gmail.com.
Aslihan Sahin, Email: aslihansahn@gmail.com.
Aysun Boga, Email: aysunboga@gmail.com.
Didem Kizmaz Isancli, Email: didemkizmaz@gmail.com.
Belgin Gulhan, Email: docbelgin@yahoo.com.
Saliha Kanik-Yuksek, Email: salihakanik@gmail.com.
Eylem Kiral, Email: dr_eylem@hotmail.com.
Gurkan Bozan, Email: drgurkanbozan@gmail.com.
Mehmet Ozgür Arslanoglu, Email: ozgurarslanoglu@yahoo.com.tr.
Mahmut Can Kizil, Email: mcankizil@hotmail.com.
Meltem Dinleyici, Email: meltemayata@hotmail.com.
Tercan Us, Email: tercanus@ogu.edu.tr.
Ahmet Varis, Email: a.varis@diagen.com.tr.
Mucahit Kaya, Email: m.kaya@diagen.com.tr.
Yvan Vandenplas, Email: yvan.vandenplas@uzbrussel.be.
Ener Cagri Dinleyici, Email: timboothtr@yahoo.com.
References
- 1.World Health Organization (WHO) WHO coronavirus (COVID-19) dashboard Available from: https://covid19.who.int/. Accessed 26 Feb 2022.
- 2.Blanchard-Rohner G, Didierlaurent A, Tilmanne A, Smeesters P, Marchant A. Pediatric COVID-19: immunopathogenesis, transmission and prevention. Vaccines (Basel)9. 2021 doi: 10.3390/vaccines9091002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xue FX, Shen KL. COVID-19 in children and the importance of COVID-19 vaccination. World J Pediatr. 2021;17:462–466. doi: 10.1007/s12519-021-00466-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kompaniyets L, Agathis NT, Nelson JM, Preston LE, Ko JY, Belay B, Pennington AF, Danielson ML, DeSisto CL, Chevinsky JR, Schieber LZ, Yusuf H, Baggs J, Mac Kenzie WR, Wong KK, Boehmer TK, Gundlapalli AV, Goodman AB. Underlying medical conditions associated with severe COVID-19 illness among children. JAMA Netw Open. 2021;4:e2111182. doi: 10.1001/jamanetworkopen.2021.11182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Feldstein LR, Tenforde MW, Friedman KG, Newhams M, Rose EB, Dapul H, Soma VL, et al. Characteristics and outcomes of US children and adolescents with multisystem inflammatory syndrome in children (MIS-C) compared with severe acute COVID-19. JAMA. 2021;325:1074–1087. doi: 10.1001/jama.2021.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yilmaz Ciftdogan D, Ekemen Keles Y, Cetin BS, Dalgic Karabulut N, Emiroglu M, Bagci Z, Buyukcam A et al (2022) COVID-19 associated multisystemic inflammatory syndrome in 614 children with and without overlap with Kawasaki disease-Turk MIS-C study group. Eur J Pediatr:1–13. 10.1007/s00431-022-04390-2 [DOI] [PMC free article] [PubMed]
- 7.Soriano JB, Murthy S, Marshall JC, Relan P, Diaz JV. A clinical case definition of post-COVID-19 condition by a Delphi consensus. Lancet Infect Dis. 2021 doi: 10.1016/S1473-3099(21)00703-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fainardi V, Meoli A, Chiopris G, Motta M, Skenderaj K, Grandinetti R, Bergomi A, Antodaro F, Zona S, Esposito S. Long COVID in children and adolescents. Life (Basel)12. 2022 doi: 10.3390/life12020285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jiang L, Tang K, Levin M, Irfan O, Morris SK, Wilson K, Klein JD, Bhutta ZA. COVID-19 and multisystem inflammatory syndrome in children and adolescents. Lancet Infect Dis. 2020;20:e276–e288. doi: 10.1016/S1473-3099(20)30651-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yilmaz Ciftdogan D, Ekemen Keles Y, Karbuz A, Cetin BS, Elmas Bozdemir S, Kepenekli Kadayifci E, Metin Akcan O, et al. Multisystem inflammatory syndrome in children associated with COVID-19 in 101 cases from Turkey (Turk-MISC study) J Paediatr Child Health. 2022 doi: 10.1111/jpc.15913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mazer MB, Bulut Y, Brodsky NN, Lam FW, Sturgill JL, Miles SM, Shein SL, Carroll CL, Remy KE. Multisystem inflammatory syndrome in children: host immunologic responses. Pediatr Crit Care Med. 2022 doi: 10.1097/PCC.0000000000002897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ramaswamy A, Brodsky NN, Sumida TS, Comi M, Asashima H, Hoehn KB, Li N, et al. Immune dysregulation and autoreactivity correlate with disease severity in SARS-CoV-2-associated multisystem inflammatory syndrome in children. Immunity. 2021;54:1083–1095.e1087. doi: 10.1016/j.immuni.2021.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Porritt RA, Paschold L, Rivas MN, Cheng MH, Yonker LM, Chandnani H, Lopez M, Simnica D, Schultheiß C, Santiskulvong C, Van Eyk J, McCormick JK, Fasano A, Bahar I, Binder M, Arditi M. HLA class I-associated expansion of TRBV11-2 T cells in multisystem inflammatory syndrome in children. J Clin Invest131. 2021 doi: 10.1172/JCI146614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Enaud R, Prevel R, Ciarlo E, Beaufils F, Wieërs G, Guery B, Delhaes L. The gut-lung axis in health and respiratory diseases: a place for inter-organ and inter-kingdom crosstalks. Front Cell Infect Microbiol. 2020;10:9. doi: 10.3389/fcimb.2020.00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Antunes AEC, Vinderola G, Xavier-Santos D, Sivieri K. Potential contribution of beneficial microbes to face the COVID-19 pandemic. Food Res Int. 2020;136:109577. doi: 10.1016/j.foodres.2020.109577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zimmermann P, Curtis N. Why is COVID-19 less severe in children? A review of the proposed mechanisms underlying the age-related difference in severity of SARS-CoV-2 infections. Arch Dis Child. 2020 doi: 10.1136/archdischild-2020-320338. [DOI] [PubMed] [Google Scholar]
- 17.Newsome RC, Gauthier J, Hernandez MC, Abraham GE, Robinson TO, Williams HB, Sloan M, Owings A, Laird H, Christian T, Pride Y, Wilson KJ, Hasan M, Parker A, Senitko M, Glover SC, Gharaibeh RZ, Jobin C. The gut microbiome of COVID-19 recovered patients returns to uninfected status in a minority-dominated United States cohort. Gut Microbes. 2021;13:1–15. doi: 10.1080/19490976.2021.1926840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yeoh YK, Zuo T, Lui GC, Zhang F, Liu Q, Li AY, Chung AC, Cheung CP, Tso EY, Fung KS, Chan V, Ling L, Joynt G, Hui DS, Chow KM, Ng SSS, Li TC, Ng RW, Yip TC, Wong GL, Chan FK, Wong CK, Chan PK, Ng SC. Gut microbiota composition reflects disease severity and dysfunctional immune responses in patients with COVID-19. Gut. 2021;70:698–706. doi: 10.1136/gutjnl-2020-323020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu R, Liu P, Zhang T, Wu Q, Zeng M, Ma Y, Jin X, Xu J, Zhang Z, Zhang C. Progressive deterioration of the upper respiratory tract and the gut microbiomes in children during the early infection stages of COVID-19. J Genet Genomics. 2021;48:803–814. doi: 10.1016/j.jgg.2021.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mirzayi C, Renson A, Zohra F, Elsafoury S, Geistlinger L, Kasselman LJ, Eckenrode K, et al. Reporting guidelines for human microbiome research: the STORMS checklist. Nat Med. 2021;27:1885–1892. doi: 10.1038/s41591-021-01552-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Centers for Disease Control and Prevention (2020) Multisystem inflammatory syndrome in children (MIS-C) associated with coronavirus disease 2019 (COVID-19). https://emergency.cdc.gov/han/2020/han00432.asp. Accessed 10 Oct 2021.
- 22.Centers for Disease Control and Prevention. Division of nutrition, physical activity, and obesity. Use and interpretation of the WHO and CDC growth charts for children from birth to 20 years in the United States. Lasted Updated: May 2013. https://www.cdc.gov/nccdphp/dnpao/growthcharts/resources/growthchart-508.pdf. Accessed 1 Jan 2022.
- 23.Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA, Alexander H, et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol. 2019;37:852–857. doi: 10.1038/s41587-019-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJ, Holmes SP. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods. 2016;13:581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schloss PD (2021) Amplicon sequence variants artificially split bacterial genomes into separate clusters. mSphere 6:e0019121. 10.1128/mSphere.00191-21. [DOI] [PMC free article] [PubMed]
- 26.Werner JJ, Koren O, Hugenholtz P, DeSantis TZ, Walters WA, Caporaso JG, Angenent LT, Knight R, Ley RE. Impact of training sets on classification of high-throughput bacterial 16s rRNA gene surveys. Isme j. 2012;6:94–103. doi: 10.1038/ismej.2011.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McMurdie PJ, Holmes S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE. 2013;8:e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.R Core Team (2021) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria, https://www.R-project.org/.
- 29.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, Huttenhower C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen Y, Gu S, Chen Y, Lu H, Shi D, Guo J, Wu WR, Yang Y, Li Y, Xu KJ, Ding C, Luo R, Huang C, Yu L, Xu M, Yi P, Liu J, Tao JJ, Zhang H, Lv L, Wang B, Sheng J, Li L. Six-month follow-up of gut microbiota richness in patients with COVID-19. Gut. 2022;71:222–225. doi: 10.1136/gutjnl-2021-324090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hung YP, Lee CC, Lee JC, Tsai PJ, Ko WC. Gut dysbiosis during COVID-19 and potential effect of probiotics. Microorganisms. 2021;9(8):1605. doi: 10.3390/microorganisms9081605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jabczyk M, Nowak J, Hudzik B, Zubelewicz-Szkodzińska B. Microbiota and its impact on the immune system in COVID-19—a narrative review. J Clin Med. 2021;10(19):4537. doi: 10.3390/jcm10194537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen J, Yue Y, Wang L, Deng Z, Yuan Y, Zhao M, Yuan Z, Tan C, Cao Y. Altered gut microbiota correlated with systemic inflammation in children with Kawasaki disease. Sci Rep. 2020;10:14525. doi: 10.1038/s41598-020-71371-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martín R, Miquel S, Benevides L, Bridonneau C, Robert V, Hudault S, Chain F, Berteau O, Azevedo V, Chatel JM, Sokol H, Bermúdez-Humarán LG, Thomas M, Langella P (2017) Functional characterization of novel Faecalibacterium prausnitzii strains isolated from healthy volunteers: a step forward in the use of F. prausnitzii as a next-generation probiotic. Front Microbiol 8:1226. 10.3389/fmicb.2017.01226 [DOI] [PMC free article] [PubMed]
- 36.Bruzzese E, Callegari ML, Raia V, Viscovo S, Scotto R, Ferrari S, Morelli L, Buccigrossi V, Lo Vecchio A, Ruberto E, Guarino A. Disrupted intestinal microbiota and intestinal inflammation in children with cystic fibrosis and its restoration with Lactobacillus GG: a randomised clinical trial. PLoS ONE. 2014;9:e87796. doi: 10.1371/journal.pone.0087796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sokol H, Pigneur B, Watterlot L, Lakhdari O, Bermúdez-Humarán LG, Gratadoux JJ, Blugeon S, Bridonneau C, Furet JP, Corthier G, Grangette C, Vasquez N, Pochart P, Trugnan G, Thomas G, Blottière HM, Doré J, Marteau P, Seksik P, Langella P. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci U S A. 2008;105:16731–16736. doi: 10.1073/pnas.0804812105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Machiels K, Joossens M, Sabino J, De Preter V, Arijs I, Eeckhaut V, Ballet V, Claes K, Van Immerseel F, Verbeke K, Ferrante M, Verhaegen J, Rutgeerts P, Vermeire S. A decrease of the butyrate-producing species Roseburia hominis and Faecalibacterium prausnitzii defines dysbiosis in patients with ulcerative colitis. Gut. 2014;63:1275–1283. doi: 10.1136/gutjnl-2013-304833. [DOI] [PubMed] [Google Scholar]
- 39.Lavelle A, Sokol H. Gut microbiota-derived metabolites as key actors in inflammatory bowel disease. Nat Rev Gastroenterol Hepatol. 2020;17:223–237. doi: 10.1038/s41575-019-0258-z. [DOI] [PubMed] [Google Scholar]
- 40.Mushtaq N, Hussain S, Zhang S, Yuan L, Li H, Ullah S, Wang Y, Xu J. Molecular characterization of alterations in the intestinal microbiota of patients with grade 3 hypertension. Int J Mol Med. 2019;44:513–522. doi: 10.3892/ijmm.2019.4235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Clemente JC, Manasson J, Scher JU. The role of the gut microbiome in systemic inflammatory disease. BMJ. 2018;360:j5145. doi: 10.1136/bmj.j5145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maioli TU, Borras-Nogues E, Torres L, Barbosa SC, Martins VD, Langella P, Azevedo VA, Chatel JM. Possible benefits of Faecalibacterium prausnitzii for obesity-associated gut disorders. Front Pharmacol. 2021;12:740636. doi: 10.3389/fphar.2021.740636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Furet JP, Kong LC, Tap J, Poitou C, Basdevant A, Bouillot JL, Mariat D, Corthier G, Doré J, Henegar C, Rizkalla S, Clément K. Differential adaptation of human gut microbiota to bariatric surgery-induced weight loss: links with metabolic and low-grade inflammation markers. Diabetes. 2010;59:3049–3057. doi: 10.2337/db10-0253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang X, Shen D, Fang Z, Jie Z, Qiu X, Zhang C, Chen Y, Ji L. Human gut microbiota changes reveal the progression of glucose intolerance. PLoS ONE. 2013;8:e71108. doi: 10.1371/journal.pone.0071108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chu XJ, Cao NW, Zhou HY, Meng X, Guo B, Zhang HY, Li BZ. The oral and gut microbiome in rheumatoid arthritis patients: a systematic review. Rheumatology (Oxford) 2021;60:1054–1066. doi: 10.1093/rheumatology/keaa835. [DOI] [PubMed] [Google Scholar]
- 46.De Palma G, Nadal I, Medina M, Donat E, Ribes-Koninckx C, Calabuig M, Sanz Y. Intestinal dysbiosis and reduced immunoglobulin-coated bacteria associated with coeliac disease in children. BMC Microbiol. 2010;10:63. doi: 10.1186/1471-2180-10-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sun Z, Song ZG, Liu C, Tan S, Lin S, Zhu J, Dai FH, Gao J, She JL, Mei Z, Lou T, Zheng JJ, Liu Y, He J, Zheng Y, Ding C, Qian F, Zheng Y, Chen YM. Gut microbiome alterations and gut barrier dysfunction are associated with host immune homeostasis in COVID-19 patients. BMC Med. 2022;20:24. doi: 10.1186/s12916-021-02212-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu Q, Mak JWY, Su Q, Yeoh YK, Lui GC, Ng SSS, Zhang F, Li AYL, Lu W, Hui DS, Chan PK, Chan FKL, Ng SC. Gut microbiota dynamics in a prospective cohort of patients with post-acute COVID-19 syndrome. Gut. 2022;71:544–552. doi: 10.1136/gutjnl-2021-325989. [DOI] [PubMed] [Google Scholar]
- 49.Larsen JM. The immune response to Prevotella bacteria in chronic inflammatory disease. Immunology. 2017;151:363–374. doi: 10.1111/imm.12760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nirmalkar K, Murugesan S, Pizano-Zárate ML, Villalobos-Flores LE, García-González C, Morales-Hernández RM, Nuñez-Hernández JA, Hernández-Quiroz F, Romero-Figueroa MDS, Hernández-Guerrero C, Hoyo-Vadillo C, García-Mena J. Gut microbiota and endothelial dysfunction markers in obese mexican children and adolescents. Nutrients10. 2018 doi: 10.3390/nu10122009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sánchez-Alcoholado L, Ordóñez R, Otero A, Plaza-Andrade I, Laborda-Illanes A, Medina JA, Ramos-Molina B, Gómez-Millán J, Queipo-Ortuño MI. Gut microbiota-mediated inflammation and gut permeability in patients with obesity and colorectal cancer. Int J Mol Sci21. 2020 doi: 10.3390/ijms21186782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yonker LM, Gilboa T, Ogata AF, Senussi Y, Lazarovits R, Boribong BP, Bartsch YC, et al. Multisystem inflammatory syndrome in children is driven by zonulin-dependent loss of gut mucosal barrier. J Clin Invest131. 2021 doi: 10.1172/JCI149633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Paul AK, Paul A, Jahan R, Jannat K, Bondhon TA, Hasan A, Nissapatorn V, Pereira ML, Wilairatana P, Rahmatullah M. Probiotics and amelioration of rheumatoid arthritis: significant roles of Lactobacillus casei and Lactobacillus acidophilus. Microorganisms9. 2021 doi: 10.3390/microorganisms9051070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tsai YW, Dong JL, Jian YJ, Fu SH, Chien MW, Liu YW, Hsu CY, Sytwu HK. Gut microbiota-modulated metabolomic profiling shapes the etiology and pathogenesis of autoimmune diseases. Microorganisms9. 2021 doi: 10.3390/microorganisms9091930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kageyama A, Benno Y, Nakase T (1999) Phylogenetic evidence for the transfer of Eubacterium lentum to the genus Eggerthella as Eggerthella lenta gen. nov., comb. nov. Int J Syst Bacteriol 49 Pt 4:1725–1732. 10.1099/00207713-49-4-1725 [DOI] [PubMed]
- 56.Qin J, Li Y, Cai Z, Li S, Zhu J, Zhang F, Liang S, et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature. 2012;490:55–60. doi: 10.1038/nature11450. [DOI] [PubMed] [Google Scholar]
- 57.Miyake S, Kim S, Suda W, Oshima K, Nakamura M, Matsuoka T, Chihara N, Tomita A, Sato W, Kim SW, Morita H, Hattori M, Yamamura T. Dysbiosis in the gut microbiota of patients with multiple sclerosis, with a striking depletion of species belonging to Clostridia XIVa and IV clusters. PLoS ONE. 2015;10:e0137429. doi: 10.1371/journal.pone.0137429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen J, Wright K, Davis JM, Jeraldo P, Marietta EV, Murray J, Nelson H, Matteson EL, Taneja V. An expansion of rare lineage intestinal microbes characterizes rheumatoid arthritis. Genome Med. 2016;8:43. doi: 10.1186/s13073-016-0299-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cao J, Wang C, Zhang Y, Lei G, Xu K, Zhao N, Lu J, Meng F, Yu L, Yan J, Bai C, Zhang S, Zhang N, Gong Y, Bi Y, Shi Y, Chen Z, Dai L, Wang J, Yang P. Integrated gut virome and bacteriome dynamics in COVID-19 patients. Gut Microbes. 2021;13:1–21. doi: 10.1080/19490976.2021.1887722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jiang S, E J, Wang D, Zou Y, Liu X, Xiao H, Wen Y, Chen Z, Eggerthella lenta bacteremia successfully treated with ceftizoxime: case report and review of the literature. Eur J Med Res. 2021;26:111. doi: 10.1186/s40001-021-00582-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bilge NY, Brocal VP, Kaşifoğlu T, Bilge U, Kasifoglu N, Moya A, Dinleyici E (2020) AB1035 intestinal microbiota composition of adult patients with familial Mediterranean fever and healthy controls (the Rheuma-BIOTA study). BMJ Publishing Group Ltd
- 62.Yasar Bilge NS, Pérez Brocal V, Kasifoglu T, Bilge U, Kasifoglu N, Moya A, Dinleyici EC. Intestinal microbiota composition of patients with Behçet’s disease: differences between eye, mucocutaneous and vascular involvement. The Rheuma-BIOTA study Clin Exp Rheumatol. 2020;38(Suppl 127):60–68. [PubMed] [Google Scholar]
- 63.Forbes JD, Chen CY, Knox NC, Marrie RA, El-Gabalawy H, de Kievit T, Alfa M, Bernstein CN, Van Domselaar G. A comparative study of the gut microbiota in immune-mediated inflammatory diseases-does a common dysbiosis exist? Microbiome. 2018;6:221. doi: 10.1186/s40168-018-0603-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Brodin P. SARS-CoV-2 infections in children: understanding diverse outcomes. Immunity. 2022;55:201–209. doi: 10.1016/j.immuni.2022.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pallister T, Jackson MA, Martin TC, Glastonbury CA, Jennings A, Beaumont M, Mohney RP, Small KS, MacGregor A, Steves CJ, Cassidy A, Spector TD, Menni C, Valdes AM. Untangling the relationship between diet and visceral fat mass through blood metabolomics and gut microbiome profiling. Int J Obes (Lond) 2017;41:1106–1113. doi: 10.1038/ijo.2017.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Turnbaugh PJ, Bäckhed F, Fulton L, Gordon JI. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe. 2008;3:213–223. doi: 10.1016/j.chom.2008.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Martin B, DeWitt PE, Russell S, Anand A, Bradwell KR, Bremer C, Gabriel D, Girvin AT, Hajagos JG, McMurry JA, Neumann AJ, Pfaff ER, Walden A, Wooldridge JT, Yoo YJ, Saltz J, Gersing KR, Chute CG, Haendel MA, Moffitt R, Bennett TD. Characteristics, outcomes, and severity risk factors associated with SARS-CoV-2 infection among children in the US National COVID Cohort Collaborative. JAMA Netw Open. 2022;5:e2143151. doi: 10.1001/jamanetworkopen.2021.43151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Muscogiuri G, Cantone E, Cassarano S, Tuccinardi D, Barrea L, Savastano S, Colao A. Gut microbiota: a new path to treat obesity. Int J Obes Suppl. 2019;9:10–19. doi: 10.1038/s41367-019-0011-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Woting A, Pfeiffer N, Loh G, Klaus S, Blaut M (2014) Clostridium ramosum promotes high-fat diet-induced obesity in gnotobiotic mouse models. mBio 5:e01530–01514. 10.1128/mBio.01530-14. [DOI] [PMC free article] [PubMed]
- 70.Le Chatelier E, Nielsen T, Qin J, Prifti E, Hildebrand F, Falony G, Almeida M, et al. Richness of human gut microbiome correlates with metabolic markers. Nature. 2013;500:541–546. doi: 10.1038/nature12506. [DOI] [PubMed] [Google Scholar]
- 71.Woting A, Blaut M. The intestinal microbiota in metabolic disease. Nutrients. 2016;8:202. doi: 10.3390/nu8040202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zuo T, Zhang F, Lui GCY, Yeoh YK, Li AYL, Zhan H, Wan Y, Chung ACK, Cheung CP, Chen N, Lai CKC, Chen Z, Tso EYK, Fung KSC, Chan V, Ling L, Joynt G, Hui DSC, Chan FKL, Chan PKS, Ng SC. Alterations in gut microbiota of patients with COVID-19 during time of hospitalization. Gastroenterology. 2020;159:944–955.e948. doi: 10.1053/j.gastro.2020.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li S, Yang S, Zhou Y, Disoma C, Dong Z, Du A, Zhang Y, et al. Microbiome profiling using shotgun metagenomic sequencing identified unique microorganisms in COVID-19 patients with altered gut microbiota. Front Microbiol. 2021;12:712081. doi: 10.3389/fmicb.2021.712081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sharma C, Ganigara M, Galeotti C, Burns J, Berganza FM, Hayes DA, Singh-Grewal D, Bharath S, Sajjan S, Bayry J. Multisystem inflammatory syndrome in children and Kawasaki disease: a critical comparison. Nat Rev Rheumatol. 2021;17:731–748. doi: 10.1038/s41584-021-00709-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kowalska-Duplaga K, Gosiewski T, Kapusta P, Sroka-Oleksiak A, Wędrychowicz A, Pieczarkowski S, Ludwig-Słomczyńska AH, Wołkow PP, Fyderek K. Differences in the intestinal microbiome of healthy children and patients with newly diagnosed Crohn’s disease. Sci Rep. 2019;9:18880. doi: 10.1038/s41598-019-55290-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wu HJ, Wu E. The role of gut microbiota in immune homeostasis and autoimmunity. Gut Microbes. 2012;3:4–14. doi: 10.4161/gmic.19320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dehner C, Fine R, Kriegel MA. The microbiome in systemic autoimmune disease: mechanistic insights from recent studies. Curr Opin Rheumatol. 2019;31:201–207. doi: 10.1097/BOR.0000000000000574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Liu TFD, Philippou E, Kolokotroni O, Siakallis G, Rahima K, Constantinou C (2021) Gut and airway microbiota and their role in COVID-19 infection and pathogenesis: a scoping review. Infection:1–33. 10.1007/s15010-021-01715-5. [DOI] [PMC free article] [PubMed]
- 79.Nickbakhsh S, Mair C, Matthews L, Reeve R, Johnson PCD, Thorburn F, von Wissmann B, Reynolds A, McMenamin J, Gunson RN, Murcia PR. Virus-virus interactions impact the population dynamics of influenza and the common cold. Proc Natl Acad Sci U S A. 2019;116:27142–27150. doi: 10.1073/pnas.1911083116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tao W, Zhang G, Wang X, Guo M, Zeng W, Xu Z, Cao D, Pan A, Wang Y, Zhang K, Ma X, Chen Z, Jin T, Liu L, Weng J, Zhu S. Analysis of the intestinal microbiota in COVID-19 patients and its correlation with the inflammatory factor IL-18. Med Microecol. 2020;5:100023. doi: 10.1016/j.medmic.2020.100023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tang L, Gu S, Gong Y, Li B, Lu H, Li Q, Zhang R, Gao X, Wu Z, Zhang J, Zhang Y, Li L. Clinical significance of the correlation between changes in the major intestinal bacteria species and COVID-19 severity. Engineering (Beijing) 2020;6:1178–1184. doi: 10.1016/j.eng.2020.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gopal AB, Chakraborty S, Padhan PK, Barik A, Dixit P, Chakraborty D, Poirah I, Samal S, Sarkar A, Bhattacharyya A. Silent hypoxia in COVID-19: a gut microbiota connection. Curr Opin Physiol. 2021;23:100456. doi: 10.1016/j.cophys.2021.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Venegas-Borsellino C, Sankararaman S, Roche K, Burns J, Landis RM. Impact of COVID-19 on the intestinal microbiome. Curr Nutr Rep. 2021;10:300–306. doi: 10.1007/s13668-021-00375-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hashimoto T, Perlot T, Rehman A, Trichereau J, Ishiguro H, Paolino M, Sigl V, Hanada T, Hanada R, Lipinski S, Wild B, Camargo SM, Singer D, Richter A, Kuba K, Fukamizu A, Schreiber S, Clevers H, Verrey F, Rosenstiel P, Penninger JM. ACE2 links amino acid malnutrition to microbial ecology and intestinal inflammation. Nature. 2012;487:477–481. doi: 10.1038/nature11228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ferreira C, Viana SD, Reis F. Is gut microbiota dysbiosis a predictor of increased susceptibility to poor outcome of COVID-19 patients? An Update Microorganisms. 2020;9(1):53. doi: 10.3390/microorganisms9010053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Dickson RP, Schultz MJ, van der Poll T, Schouten LR, Falkowski NR, Luth JE, Sjoding MW, Brown CA, Chanderraj R, Huffnagle GB, Bos LDJ (2020) Lung microbiota predict clinical outcomes in critically ill patients. Am J Respir Crit Care Med 201:555–563. 10.1164/rccm.201907-1487OC. [DOI] [PMC free article] [PubMed]
- 87.He Y, Wang J, Li F, Shi Y. Main clinical features of COVID-19 and potential prognostic and therapeutic value of the microbiota in SARS-CoV-2 infections. Front Microbiol. 2020;11:1302. doi: 10.3389/fmicb.2020.01302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Harper A, Vijayakumar V, Ouwehand AC, Ter Haar J, Obis D, Espadaler J, Binda S, Desiraju S, Day R. Viral infections, the microbiome, and probiotics. Front Cell Infect Microbiol. 2021;10:596166. doi: 10.3389/fcimb.2020.596166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hasegawa K, Linnemann RW, Mansbach JM, Ajami NJ, Espinola JA, Petrosino JF, Piedra PA, Stevenson MD, Sullivan AF, Thompson AD, Camargo CA., Jr The fecal microbiota profile and bronchiolitis in infants. Pediatrics. 2016;138(1):e20160218. doi: 10.1542/peds.2016-0218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Osmanov IM, Spiridonova E, Bobkova P, Gamirova A, Shikhaleva A, Andreeva M, Blyuss O, El-Taravi Y, DunnGalvin A, Comberiati P, Peroni DG, Apfelbacher C, Genuneit J, Mazankova L, Miroshina A, Chistyakova E, Samitova E, Borzakova S, Bondarenko E, Korsunskiy AA, Konova I, Hanson SW, Carson G, Sigfrid L, Scott JT, Greenhawt M, Whittaker EA, Garralda E, Swann OV, Buonsenso D, Nicholls DE, Simpson F, Jones C, Semple MG, Warner JO, Vos T, Olliaro P, Munblit D; and the Sechenov StopCOVID Research Team Risk factors for post-COVID-19 condition in previously hospitalised children using the ISARIC Global follow-up protocol: a prospective cohort study. Eur Respir J. 2022;59(2):2101341. doi: 10.1183/13993003.01341-2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Buonsenso D, Di Gennaro L, De Rose C, Morello R, D’Ilario F, Zampino G, Piazza M, Boner AL, Iraci C, O’Connell S, Cohen VB, Esposito S, Munblit D, Reena J, Sigfrid L, Valentini P. Long-term outcomes of pediatric infections: from traditional infectious diseases to long COVID. Future Microbiol. 2022;17:551–571. doi: 10.2217/fmb-2022-0031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.De Maio F, Buonsenso D, Bianco DM, Giaimo M, Fosso B, Monzo FR, Sali M, Posteraro B, Valentini P, Sanguinetti M. Comparative fecal microbiota analysis of infants with acute bronchiolitis caused or not caused by respiratory syncytial virus. Front Cell Infect Microbiol. 2022;12:815715. doi: 10.3389/fcimb.2022.815715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Derrien M, Alvarez AS, de Vos WM. The gut microbiota in the first decade of life. Trends Microbiol. 2019;27:997–1010. doi: 10.1016/j.tim.2019.08.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Authors agree to participate all data, if requested.
Not available.