Abstract
We analyzed changes in bacterioplankton morphology and composition during enhanced protozoan grazing by image analysis and fluorescent in situ hybridization with group-specific rRNA-targeted oligonucleotide probes. Enclosure experiments were conducted in a small, fishless freshwater pond which was dominated by the cladoceran Daphnia magna. The removal of metazooplankton enhanced protozoan grazing pressure and triggered a microbial succession from fast-growing small bacteria to larger grazing-resistant morphotypes. These were mainly different types of filamentous bacteria which correlated in biomass with the population development of heterotrophic nanoflagellates (HNF). Small bacterial rods and cocci, which showed increased proportion after removal of Daphnia and doubling times of 6 to 11 h, belonged nearly exclusively to the beta subdivision of the class Proteobacteria and the Cytophaga-Flavobacterium cluster. The majority of this newly produced bacterial biomass was rapidly consumed by HNF. In contrast, the proportion of bacteria belonging to the gamma and alpha subdivisions of the Proteobacteria increased throughout the experiment. The alpha subdivision consisted mainly of rods that were 3 to 6 μm in length, which probably exceeded the size range of bacteria edible by protozoa. Initially, these organisms accounted for less than 1% of total bacteria, but after 72 h they became the predominant group of the bacterial assemblage. Other types of grazing-resistant, filamentous bacteria were also found within the beta subdivision of Proteobacteria and the Cytophaga-Flavobacterium cluster. We conclude that the predation regimen is a major structuring force for the bacterial community composition in this system. Protozoan grazing resulted in shifts of the morphological as well as the taxonomic composition of the bacterial assemblage. Grazing-resistant filamentous bacteria can develop within different phylogenetic groups of bacteria, and formerly underepresented taxa might become a dominant group when protozoan predation is the major selective pressure.
Planktonic bacteria are regulated by the availability of inorganic and organic nutrients (“bottom-up control”), by bacterivorous protists (“top-down control”), and by viral lysis. In addition, cladocerans, especially Daphnia spp., can replace protozoans as the major bacterial consumer in freshwater lakes (16, 28). An important issue in aquatic microbial ecology is elucidating the relative importance of resource limitation, grazing, and viral mortality of bacterioplankton communities. Most field studies approached these questions with methods which describe the average bacterial community response in terms such as abundance, biomass, production, and mortality rates. These studies resulted in good quantitative measurements of bacterial growth and loss rates and in convincing estimates of the limiting factors. However, they provided little information as to how the specific type of control results in qualitative changes of natural bacterial assemblages. This was largely due to methodological constraints and was also due to the main focus being on carbon and nutrient fluxes in most of these studies.
There is evidence that grazing is one of the major forces shaping the bacterial community structure (10). Predation balances bacterial production and therefore should be considered an important selective pressure, especially in more productive systems (20). So far, the grazing impact has been mainly studied with respect to the size structures of bacterial communities. Predator-prey interactions between bacteria and protozoans are known to affect the bacterial size structure in two ways: first, size-selective grazing, i.e., higher rates of encounter of and feeding on large bacteria (5, 7, 8), which results in a shift towards smaller cell sizes, and second, the development of bacteria which are too large to be ingested by protists, which results in the occurrence of grazing-resistant complex morphologies (9). The shift towards very small and very large bacteria, which both experience reduced grazing mortality, has been observed to occur in natural planktonic communities during increased protozoan grazing (10, 32).
Direct and indirect evidence implies that different mechanisms are involved when grazing-resistant bacteria appear in natural communities in response to enhanced protozoan grazing (reviewed in reference 20). However, the most obvious type of resistance, one that is accessible to a microscopic analysis, is manifested by bacterial cells or clusters of cells, such as filamentous, chain-forming bacteria or bacterial aggregates, which surpass small protozoans in size. Filamentous bacteria are widespread, at least in freshwater and coastal marine plankton, and it has been demonstrated that their occurrence is correlated with population maxima of protozoan grazers, especially heterotrophic nanoflagellates (HNF) (11, 14, 22, 32, 39).
The high phenotypic plasticity of many bacteria and the large diversity of natural bacterial assemblages favor a rapid response towards predation as a selective pressure (20). In experimental laboratory systems both potential mechanisms, i.e., taxonomic changes resulting in less-vulnerable species and the development of resistant phenotypes, have been demonstrated (9, 12, 31, 35, 37). In natural bacterial communities both mechanisms might occur simultaneously during the development of bacterial assemblages containing a high proportion of grazing-resistant cells. Molecular techniques for analyzing the bacterial community structure might reveal the relative degrees of importance of phenotypic and taxonomic changes in response to different predation regimens. These techniques also allow monitoring of changes in structure and function of bacterial assemblages during enhanced grazing pressure.
The goal of the present study was to analyze changes in morphology and composition of a natural planktonic bacterial assemblage in response to enhanced protozoan grazing after food web manipulation. In previous studies (17, 21) it became evident that Daphnia-dominated situations are especially suitable for such experiments because these filter-feeding cladocerans suppress the whole microbial food web, including most protists and large bacteria. The removal of Daphnia from a water sample generally triggers a microbial succession from fast-growing bacteria to phagotrophic protozoans and may initiate the development of bacterial forms with lower vulnerability to protozoans (17, 21). Therefore, such a system allows us to examine the potential of a bacterial community to develop grazing resistance and to analyze possible underlying mechanisms and resulting changes in bacterial taxonomic composition. Fluorescent in situ hybridization (FISH) with rRNA-targeted oligonucleotide probes enables a rapid and cultivation-independent analysis of the bacterial community structure (reviewed in reference 2) and has been used to examine the composition of freshwater bacterioplankton (1, 6, 29, 30, 42). Here, oligonucleotide probes for the major phylogenetic groups of planktonic bacteria were applied to study the bacterial composition during a predation-induced shift towards a community with a high proportion of grazing-resistant cells.
MATERIALS AND METHODS
Study site.
A small, fishless farm pond (15 m in diameter; approximately 1.2-m maximum depth) in eastern Schleswig-Holstein (Neudorf, Northern Germany) was used for the experiment because it is inhabited by a dense population of the cladoceran Daphnia magna throughout most of the year. The pond is only moderately eutrophied, as it does not receive input from fertilizers, but it is slightly dystrophic due to considerable input of organic matter from leaf fall and other surrounding vegetation. There is no growth of submerged macrophytes in the pond, and the phytoplankton biomass is extremely low (<5 μg of chlorophyll a liter−1) during periods of high densities of D. magna.
Bottle experiment.
Water samples were taken from various spots of the pond and used to fill a 50-liter container. From this container the experimental bottles (4.8-liter polycarbonate bottles; Nalgene) were filled. Three bottles were filled with unaltered water with the natural zooplankton density (referred to herein as the + DAPH bottles), and three bottles were filled with water which was screened through a 250-μm-pore-size mesh in order to remove the mesozooplankton (referred to herein as the − DAPH bottles). After fixation of the start samples the bottles were incubated in situ at approximately a 0.5-m depth and sampled twice per day (once in the morning and once in the evening) for a period of 1 week.
Enumeration of organisms.
Bacteria and flagellates were preserved in formaldehyde (final concentration, 2%) and stored until processing (within 1 day of preservation) at 4°C. Then, 1-ml subsamples were filtered on black polycarbonate membranes (0.2-μm pore size, 25-mm diameter; Millipore) and stained with 4′,6-diamidino-2-phenylindole (DAPI) (final concentration, 100 μg ml−1) (33). DAPI preparations were stored at −20°C until bacteria and HNF were counted with an epifluorescence microscope (Axiophot II; Zeiss, Jena, Germany). At least 300 bacteria and 50 HNF were counted per sample (magnification, ×1,250). Filamentous bacteria, which were defined as elongated cells or chains >5 μm in length, were counted and sized (with an ocular grid) by examining strips across the filter. For volume calculations, filaments were considered to be cylinders with two hemispherical ends. Autotrophic flagellates were distinguished from heterotrophs by checking for chlorophyll a autofluorescence under blue light excitation. Ciliates were preserved with acid Lugol solution (final concentration, 1%) and counted in sedimenting chambers with an inverted microscope. Zooplankton >250 μm in length were preserved in sucrose-formaldehyde (final concentration, 4%) and counted and sized with a dissecting microscope equipped with a semi-automated image analyzer.
FISH.
For whole-cell in situ hybridization on membrane filters the procedure of Glöckner et al. (6) was used. Ten milliliters of sample collected on each sampling date was filtered on 0.2-μm-pore-size polycarbonate membrane filters (47-mm diameter; Nuclepore), air dried, and stored at −20°C until further processing. In situ hybridizations of sections from the filters were performed with the following oligonucleotide probes: EUB338, which is specific for Bacteria; BET42a, which is specific for the beta subdivision of the class Proteobacteria; ALF1b, which is specific for the alpha subdivision of Proteobacteria; GAM42a, which is specific for the gamma subdivision of Proteobacteria; and CF319a, which is specific for the Cytophaga-Flavobacterium cluster of the Cytophaga-Flavobacterium-Bacteroides phylum. For some selected sampling dates only, we used the probes BONE23a, for the β1 group of the beta subdivision of Proteobacteria, and HGC69a, for gram-positive bacteria with a high DNA G+C content. Oligonucleotide probes were synthesized with Cy3 fluorochrome at the 5′ end (Interactiva Biotechnologie GmbH, Ulm, Germany). Probe sequences, hybridization and washing buffers, formamide concentrations, and competitors (for probes BET42a, GAM42a, and BONE23a) were as described by Snaidr et al. (38). We checked the quality and specificity of all probes with positive control strains of the corresponding phylogenetic group and with negative controls. After hybridization, the filter sections were stained with DAPI (final concentration, 1 μg ml−1) and mounted on microscopic slides in glycerol medium (Citifluor AF 1; Citifluor, Ltd., Canterbury, United Kingdom). The slides were examined with an Axiophot II microscope (Zeiss) with the Zeiss filter set 01 for UV excitation (for detection of DAPI) and a Chroma HQ 41007 set (AF Analysentechnik, Tübingen, Germany) for green excitation (for detection of Cy3). For each probe and sample, from 10 to 30 fields or, for rare groups, strips across the whole filter were counted.
Image analysis.
For selected time points we measured the size distribution of hybridized bacterial cells (<10 μm in length) with an automated image analysis system as described by Pernthaler et al. (29). Briefly, the method consisted of recording images of the hybridized filter sections under UV and under green light excitation with a charge-coupled device camera. An image processing program extracts the fraction of the DAPI-stained cells that also show Cy3 fluorescence. After edge detection, automatic grey-level thresholding and binarization, the dimensions (pixel area and perimeter) of the DAPI-stained cells were measured and the cell volume was calculated according to standard formulas.
RESULTS
Microbial succession after elimination of zooplankton.
The filtration with the 250-μm-pore-size mesh removed more than 98% of the zooplankton, which was due to the fact that the plankton was dominated by large crustaceans. Mesozooplankton in the bottles containing unfiltered water consisted of D. magna (mean density, 40 animals liter−1; mean body length, 2.4 mm), Eudiaptomus spp. (adults and copepodites; mean density, 10 animals liter−1), and Cyclops spp. (adults and copepodites; mean density, 100 animals liter−1). In terms of zooplankton biomass (84% of total biomass) and potential rates of filtration from nanoplankton (>95% of estimated community filtration rates) D. magna was the predominating species of the zooplankton assemblage.
For the examination of the microbial succession as revealed in the DAPI preparations, we focused mainly on three operationally defined groups: (i) freely dispersed, single-cell bacteria (rods and cocci) which resemble the “normal” type of planktonic bacteria and which we considered to be edible by protozoa; (ii) filamentous bacteria (inedible by protozoa), which we defined as elongated bacteria (>5 μm in length), including such different types as straight filaments without visible septae, dividing cells which did not separate, and bacterial chains; and (iii) HNF, colorless flagellates, mainly in the 3- to 5-μm size range.
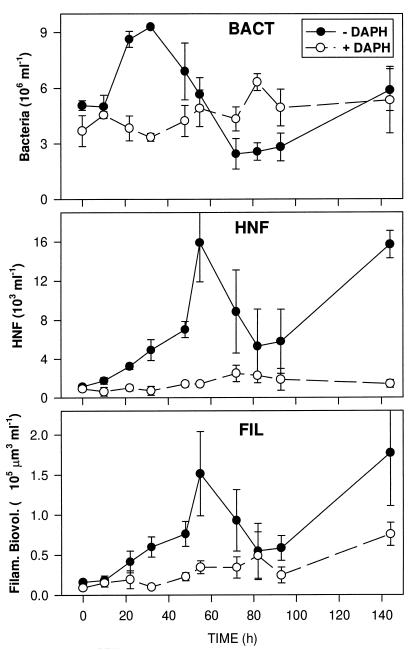
The situation at the start of the experiment was characterized by small numbers of protozoans (<1,000 HNF ml−1 and <100 ciliates liter−1), and the majority of the bacterial biomass consisted of freely dispersed small cells (approximately 5 × 106 cells ml−1; mean volume of 0.07 μm3). Besides normal-sized planktonic bacteria (0.4 to 1 μm in maximal dimensions), a large number of extremely small and faintly fluorescent particles were present; we assumed these to be viruses or bacterial remains released by grazers and did not enumerate them as bacteria (see Fig. 5). The development of bacteria, HNF, and bacterial filaments differed markedly between the treatments with and without zooplankton during the 6 days of incubation. A very pronounced microbial succession occurred in the − DAPH bottles after the zooplankton was removed (Fig. 1). After a lag phase of 10 h, the concentration of rod-shaped bacteria increased strongly for about 20 h and the increase nearly doubled the bacterial concentration. The subsequent decline in the concentration of bacteria was paralleled by an exponential increase of the HNF concentration. HNF consisted mainly of colorless chrysomonads, each with a spherical diameter of 3 to 5 μm (formalin-fixed and DAPI-stained cells) and reached, after 55 h, maximum concentrations of 11 × 103 ml−1 to 20 × 103 ml−1 in the three replicate bottles. This strong peak in HNF abundance was short-term and, except on the last sampling date, HNF otherwise remained at medium densities. Towards the end of the experiment larger flagellates and ciliates (mainly oligotrichous species) (maximum concentration, 7 cells ml−1) also appeared.
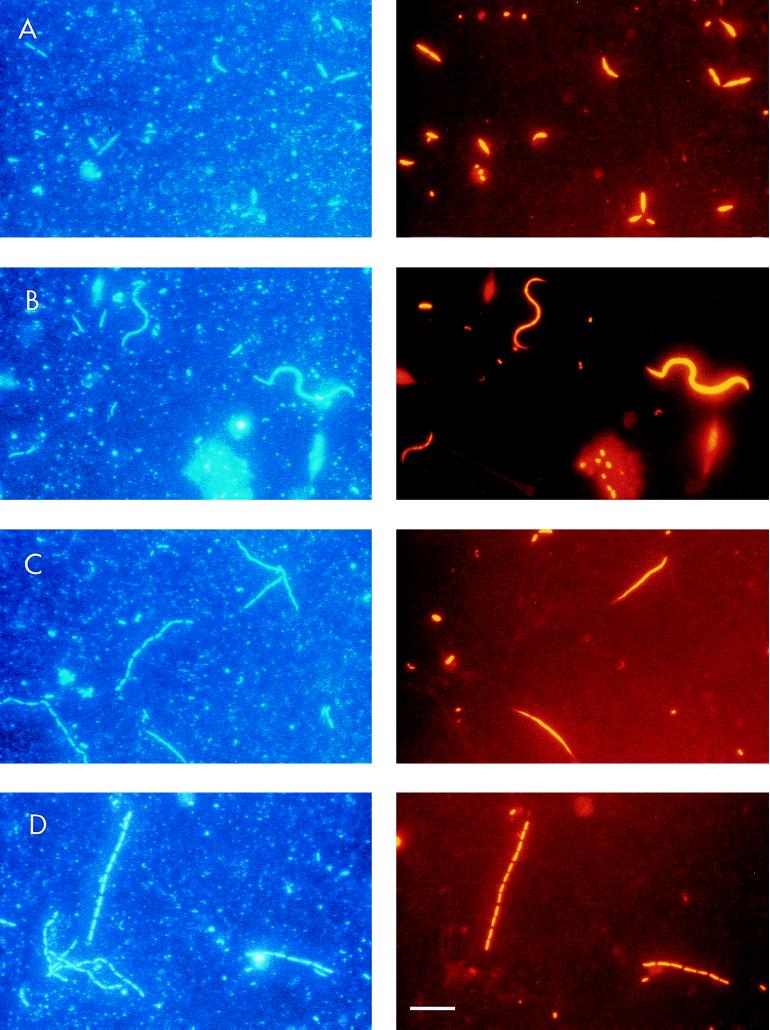
FIG. 5.
Epifluorescence photomicrographs of filamentous bacteria in − DAPH bottles after 72 h. The same microscopic fields are shown with UV excitation to visualize DAPI staining (left) and with Cy3 excitation to visualize probe-conferred fluorescence (right); the probes were ALF1b (A), BET42a (B and C), and CF319a (D). The bar in panel D corresponds to 10 μm and applies to all panels.
FIG. 1.
Development of total bacterioplankton (BACT) and HNF and biovolume of filaments (FIL) in the treatments with (open symbols) and without (closed symbols) metazooplankton. Values are means for three replicate treatments, and vertical bars show SDs.
A remarkable parallel trend was observed between population development of HNF and biovolume of filamentous bacteria (Fig. 1). Both abundance and average length of the filaments increased from the beginning of the experiment to maximum development, observed after 55 h; mean abundance increased from 2.1 × 104 ml−1 to 13.2 × 104 ml−1, and mean length increased from 7.8 μm to 11.8 μm. Also, the subsequent decline and the second increase at the end of the experiment closely matched the population development of HNF. In contrast to this succession, the microbial community in the bottles with zooplankton (+ DAPH) stayed relatively constant during the incubation. The concentration of bacteria fluctuated irregularly in the range from 3 × 106 ml−1 to 6 × 106 ml−1, and the concentrations of filaments and HNF increased slightly only in the second half of the experiment. The major portion of the metazooplankton in the bottles, especially the daphnids, were alive at the end of the experiment.
In situ hybridization and bacterial community composition.
In situ hybridizations with the probe EUB338 for Bacteria and the group-specific probes ALF1b, BET42a, GAM42a, and CF319 were performed for most sampling dates in order to cover the main sequences of the microbial succession in the − DAPH bottles. In addition, we tested other probes (BONE23a and HGC69a) for selected time points only. There were no picoalgae or other orange or red fluorescent particles in the samples, which facilitated the enumeration of Cy3-labeled hybridized bacterial cells. Bacteria on the filters used for hybridization were evenly distributed, with distribution comparable to that of normal DAPI preparations. To estimate the variability related to the hybridization procedure, we performed six parallel hybridizations from one filter (− DAPH, after 32 h) with the EUB probe. The coefficient of variation (CV; percent standard deviation [SD] of the mean) for EUB-positive cells was 4%, the CV for total bacteria was 10%, and hybridized cells accounted for 77 to 83% of the DAPI counts (CV was 3.5%). The greater variability of DAPI counts was probably due to the continuous size spectrum of DAPI-positive particles (see Fig. 5) and the use of a subjective threshold for counting DAPI-stained particles above a certain size as bacteria.
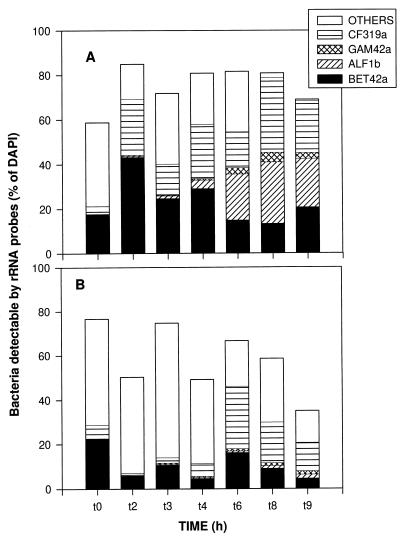
Hybridization efficiency, defined as the proportion of total bacteria which were detected with the EUB probe, varied during the course of the experiment between 35 and 85%, and higher values and less variability in the − DAPH bottles (mean efficiency, 74% ± 9%) than in the + DAPH bottles (mean efficiency, 59% ± 15%) were noted (Fig. 2). These numbers are restricted in that the exact number of total bacteria depended on the subjective threshold for counting DAPI-stained particles as bacteria. In general, only the smallest bacteria had no signal for hybridization with the EUB probe. At later periods in the experiment some filamentous chains which did not hybridize with the EUB probe also appeared in the − DAPH bottles. The proportion of bacteria which could be affiliated with the four group-specific probes for major phyla within the domain Bacteria was also higher in the − DAPH than in + DAPH bottles, where a major portion of EUB-positive cells remained unaffiliated with one of the specific groups (Fig. 2). This group, referred to as OTHERS in Fig. 2, comprised between 1 and 61% of the total bacteria, with mean values of 18% for the − DAPH bottles and 36% for the + DAPH bottles. An obvious decrease in the proportion of bacteria which hybridized with EUB but with none of the other probes occurred in − DAPH bottles throughout the course of the experiment and in + DAPH bottles towards the end of the experiment (Fig. 2). The major bacterial groups contributing to the count of bacteria detectable by rRNA probes were members of the beta subdivision of Proteobacteria and of the Cytophaga-Flavobacterium cluster. For − DAPH bottles members of the alpha subdivision of Proteobacteria also contributed significantly to the total count of bacteria detectable during the second half of the experiment (Fig. 2).
FIG. 2.
Development of the relative compositions of the bacterial assemblages in − DAPH bottles (A) and + DAPH bottles (B). The total length of each bar represents the fraction of the count of DAPI-stained cells which was detectable with the probe EUB338. Values are means for three replicate treatments.
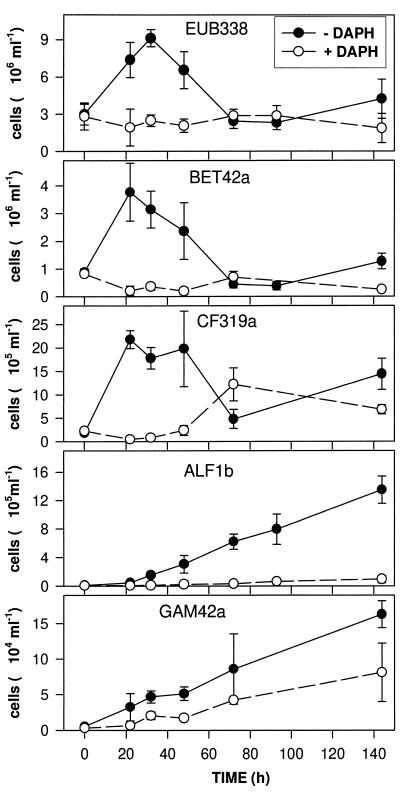
The population development of EUB-positive bacteria and of the four different bacterial subgroups is shown in Fig. 3. There was good correspondence between the three replicate treatments of − DAPH and + DAPH. The mean CV of all hybridizations was 26% in − DAPH bottles and 33% in + DAPH bottles; the values of CV for the different bacterial groups ranged from 22 to 29% in the former and from 23 to 44% in the latter. Concurrent with the morphological changes which were revealed in DAPI preparations, there was a bacterial community shift in − DAPH bottles, whereas cell numbers and overall composition stayed relatively constant in + DAPH bottles (Fig. 3). Numbers of EUB-positive cells reflect the development of total counts and show, in − DAPH bottles, the strong increase and subsequent decline to initial levels. The major portion of the rapid increase in the count of bacteria after the removal of zooplankton was comprised of cells belonging to the beta subdivision of Proteobacteria and to the Cytophaga-Flavobacterium cluster. Within the former group, the majority of the bacteria (75% ± 15%) at the peak observed after 22 h belonged to the beta 1 subgroup. Most cells that hybridized with BET42a and CF319a were freely suspended rods and cocci <1.5 μm in size. These cells declined in numbers during the increase of the count of HNF. Different dynamics were observed for the alpha and gamma subdivisions of Proteobacteria. Both groups continuously increased in numbers during the course of the experiment, irrespective of protozoan development. The alpha subdivision became a predominant group of the bacterial assemblage, with abundance higher by one order of magnitude than that of the gamma subdivision. The only significant change of bacterial community composition in + DAPH bottles was an increase in the number of cells that hybridized with probe CF319a during the end of the experiment. We did not detect positive signals with the probe for gram-positive bacteria (HGC69a) when we tested at several points in time during the bacterial succession in − DAPH bottles.
FIG. 3.
Development of different bacterial groups in the treatments with (open symbols) and without (closed symbols) Daphnia as determined by in situ hybridization with probes specific for Bacteria (EUB338), the beta subdivision of the class Proteobacteria (BET42a), the Cytophaga-Flavobacterium cluster (CF319a), the alpha subdivision of the class Proteobacteria (ALF1b), and the gamma subdivision of the class Proteobacteria (GAM42a). Values are means for three replicate treatments, and vertical bars show SDs. Note the different scales of the y axes.
Growth rates of bacteria and protists.
The removal of metazooplankton from − DAPH bottles relieved the microorganisms from grazing pressure, and the subsequent increase, first of the count of bacteria and later on the count of HNF, could be used to calculate net growth rates (19). This was done for all bacteria and for the different groups enumerated by hybridization. HNF developed with a delay of 1 day, and the growth rate was calculated for the period of their exponential increase. The values for the periods with the maximal increases for the different organisms are shown in Table 1. The highest growth rate, characterized by a doubling time of about 6 h, was achieved by bacteria that hybridized with CF319a. The other bacterial subgroups had fairly similar growth rates, characterized by doubling times of 11 to 13 h, which were comparable to the calculated growth rate of all bacterioplankton as calculated from the DAPI counts. The lower growth rates for EUB338-positive cells, with doubling times of around 20 h, might be explained by the fact that this group also included a substantial fraction of cells, which probably grew more slowly, not covered by the four group-specific probes.
TABLE 1.
Maximum growth rates of bacterioplankton, HNF, and different bacterial groups in the treatments without metazooplankton (− DAPH)
| Organism | Growth rate (h−1)a | Interval (h) |
|---|---|---|
| HNF | 0.053 ± 0.021 | 32–55 |
| Bacterioplankton | 0.046 ± 0.008 | 10–22 |
| EUB338-hybridized | 0.037 ± 0.009 | 0–33 |
| BET42a-hybridized | 0.065 ± 0.015 | 0–22 |
| CF319a-hybridized | 0.113 ± 0.007 | 0–22 |
| ALF1b-hybridized | 0.059 ± 0.006 | 0–72 |
| GAM42a-hybridized | 0.055 ± 0.018 | 0–72 |
Mean ± SD for three replicate treatments is shown. Rates were calculated for intervals with exponential increase.
Morphological structures of the different bacterial groups.
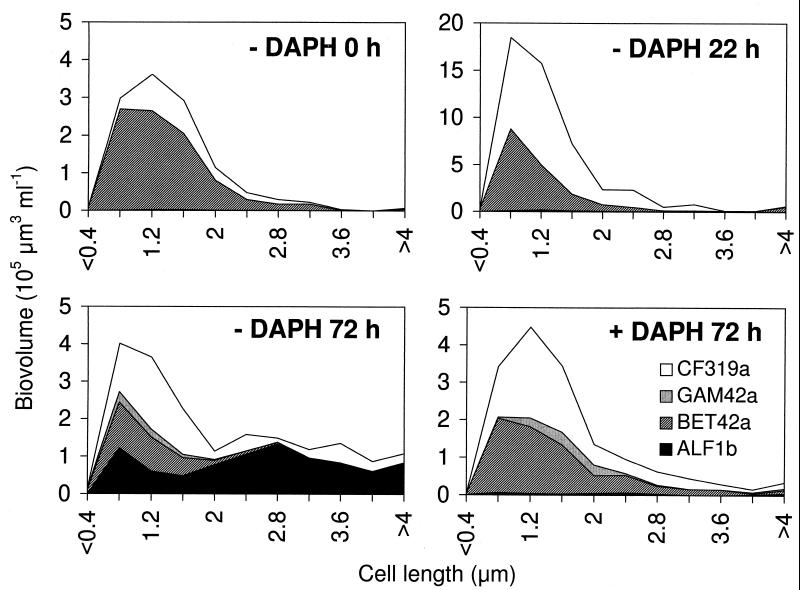
The shift in morphology of the bacterial assemblage (as visible in DAPI preparations) could be related to the shift in community composition (deduced from FISH results). For several samples taken on dates important in the succession, we analyzed the size structure of hybridized cells with image analysis and counted and separately sized filamentous bacteria within the different subgroups. The changes within the size structure of the freely dispersed single bacterial cells are shown for selected dates in Fig. 4. At the start of the experiment (− DAPH, 0 h) the majority of the bacterial biomass was in the cell length classes of 0.8 to 1.6 μm and mainly comprised of bacteria of the beta subdivision and, to a lesser extent, bacteria of the Cytophaga-Flavobacterium cluster. This remained virtually unchanged for 22 h after zooplankton removal, but the bacterial biomass was approximately three times greater (− DAPH, 22 h). After protozoan grazing had reduced the bacterial abundance to the initial level, the size distribution and phylogenetic composition was significantly changed (− DAPH, 72 h). A large proportion of the biovolume was comprised of cells 2 to 5 μm in length, and the majority of these larger bacteria hybridized with probe ALF1b. In treatments with Daphnia, the bacterial size distribution did not change after 72 h but the proportion of cells that hybridized with CF319a was larger (+ DAPH, 72 h).
FIG. 4.
Biovolume size distributions of freely dispersed rods and cocci within the different bacterial subclasses as determined by whole-cell in situ hybridization and image analysis. Data for treatments without Daphnia (− DAPH) at the start of the experiment, after 22 h, and after 72 h and data for treatments with Daphnia (+ DAPH) after 72 h are shown. Note the different y-axis scaling for − DAPH 22 h.
Different types of filamentous bacteria were visible in + DAPH and − DAPH bottles, but only in the latter treatment did they develop in larger numbers. According to the morphology visible in epifluorescence microscopy, we distinguished five different types of filaments (Table 2), to which we assigned all cells with a length of >5 μm. The filament types 1, 2, and 4 were found only within one bacterial subgroup, whereas types 3 and 5 occurred, with relatively similar morphology, among several bacterial subgroups. Examples of the most important filament morphologies are shown in the photomicrographs presented in Fig. 5. We analyzed the filament biovolume and group affiliation in more detail at 72 h, which was shortly after the maximum development of filaments. Filamentous bacteria of the different subgroups were sized based on Cy3 fluorescence, as this was generally much more visible than DAPI fluorescence (Fig. 5).
TABLE 2.
Morphological diversity of filamentous bacteria as revealed by fluorescent oligonucleotide probing
| Filament type | General filament morphology | Length (μm) | Probe(s) | Example |
|---|---|---|---|---|
| 1 | Long rods; dividing stages | 5–10 | ALF1b | Fig. 5A |
| 2 | Strongly curved and S-shaped filaments with pointed ends | 7–25 | BET42a | Fig. 5B |
| 3 | Long linear threads; septae not visible | 5–55 | BET42a, CF319a | |
| 4 | Slender, slightly curved filaments with pointed ends; septae not visible | 5–35 | BET42a | Fig. 5C |
| 5 | Small to medium-sized rods, arranged in chains | 5–100 | CF319a, BET42a; EUB338-negative | Fig. 5D |
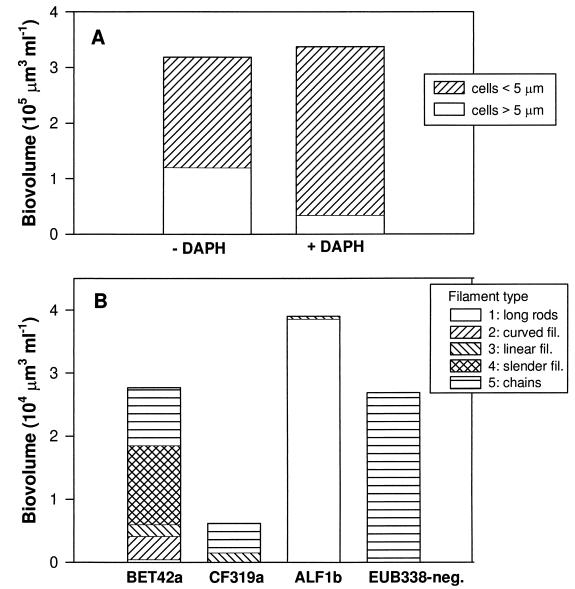
After 72 h, the total bacterial biomasses were approximately the same in − DAPH and + DAPH bottles but the proportions of filamentous and nonfilamentous bacteria differed strongly (Fig. 6A). In − DAPH bottles the filamentous biovolume was 54%, whereas in + DAPH bottles it was 11%, of the nonfilamentous biovolume. Highly diverse filament types, belonging to different phylogenetic groups of bacteria, were present in − DAPH bottles (Fig. 6B). Bacteria belonging to the alpha subdivision of Proteobacteria were the major contributors to total filament biovolume. They nearly exclusively showed one type of filament: long rods, many in the dividing stages, 5 to 10 μm in length (Fig. 5A). They looked similar to and are probably a continuation of the long rods, ranging from 2 to 5 μm in length, which are included in the size distribution presented in Fig. 4. Bacteria belonging to the beta subdivision of Proteobacteria also substantially contributed to the total biovolume of filaments and expressed a high diversity of filament types. S-shaped and curved cells were the most conspicuous (Fig. 5B), but long threads (Fig. 5C) and chains of small rods also occurred within this subdivision. Bacteria of the Cytophaga-Flavobacterium cluster also contributed to filamentous forms, mainly as chains of rods with a total length of up to 100 μm (Fig. 5D). Filaments within the gamma subdivision of Proteobacteria were extremely rare, comprising less than 1% of total cells in this subdivision, and therefore not included in this analysis. Another significant group of filamentous bacteria, forming chains of various lengths but with a consistently similar cell morphology, did not show a signal indicating hybridization with the probe EUB338 (Fig. 6).
FIG. 6.
(A) Biovolume distribution between freely dispersed rods and cocci <5 μm in length and filaments >5 μm in length after 72 h in + DAPH and − DAPH bottles. Means for three replicate treatments (mean CV was 20%) are shown. (B) distribution of biovolume after 72 h among different filamentous morphotypes in − DAPH bottles for the different bacterial groups as revealed by in situ hybridization. Filament types 1 through 5 are as characterized in Table 2. EUB338-negative refers to filaments which did not hybridize with the probe EUB338. fil., filaments.
DISCUSSION
Our enclosure experiment clearly demonstrated that bacterivorous protozoans not only affect the overall bacterial abundance but also can strongly change the morphological and phylogenetic composition of a planktonic bacterial community. Intense grazing pressure in the treatments without metazooplankton, exerted mainly by HNF, promoted the development of grazing-resistant bacterial filaments and elongated cell forms. These developed in different phylogenetic groups, but only one of them, the alpha subdivision of Proteobacteria, became a dominant part of the bacterial assemblage.
Field observations in a variety of lakes revealed that grazing-resistant bacterial morphotypes, such as filaments and aggregates, frequently constitute an important portion of the bacterial biomass, especially during periods of intense protozoan grazing (11, 22, 39). If a large proportion of planktonic bacterial biomass is constituted of grazing-resistant bacteria, this has important ecological consequences; for example, a decrease in trophic transfer efficiency and a reduction in nutrient regeneration might lower the overall system productivity (20). However, besides the fact that the appearance of these resistant morphotypes is related to certain food web constellations, we know very little about the underlying regulating mechanisms, ecology, and taxonomic affiliation of these bacteria.
Meso- and microcosm studies with intact natural communities and defined and controlled manipulations are a compromise between purely descriptive field observations and laboratory studies conducted under very reduced conditions. Situations in which large filter-feeding zooplankton predominate in the plankton community offer an interesting and suitable food web constellation in which to analyze grazing-mediated changes in the bacterial community structure. This is especially the case when Daphnia species predominate because they achieve a high total biomass and very high community filtration rates (24) which reduce all protists to very low levels (16). We assume that bacterial communities in Daphnia-predominated systems express few adaptations towards protozoan grazing. Therefore, the development of grazing resistance and the impact of protozoan grazing on bacterial community composition can be followed and analyzed after elimination of zooplankton. In previous experiments (17) it became obvious that the removal of Daphnia spp. in such situations initiates a heterotrophic microbial succession with the following sequence: (i) increase of the number of freely dispersed single bacterial cells, (ii) increase of the number of nanoflagellates and decline of that of bacteria, and (iii) development of grazing-resistant bacteria and later of larger protozoans. Grazing-resistant bacteria which develop in the third phase are not necessarily filamentous forms but can also be aggregates (18), other complex morphologies, or bacteria with unknown escape mechanisms (20). This conceptual outline of the microbial succession is similar to the one generally observed after substrate enrichment in decomposition studies (20) and was also evident in the present experiment.
We have to restrict our analysis of the dynamics of protozoan-resistant bacteria and those that are edible by protozoa to morphological criteria because we still do not possess the appropriate methods to quantify other potential mechanisms of grazing resistance in bacteria (20). In our microcosm experiment, we defined bacteria that are >5 μm in length as filaments and as inedible for the majority of protozoans. This approximation is suggested by the size of the flagellates, which are the principal bacterivores and mostly <5 μm in diameter. According to this definition, nearly 40% of the total bacterial biomass was protozoan grazing resistant after 72 h in − DAPH bottles, in contrast to about 10% of that in + DAPH bottles (Fig. 6). This value is probably an underestimation for − DAPH bottles, as particles within the size range of 2 to 5 μm are already ingested at substantially reduced rates by HNF compared to bacteria 1 μm in size (22a). This is also indicated by the shift in size distribution of nonfilamentous bacteria, mainly members of the alpha subdivision of Proteobacteria, towards larger cell sizes in − DAPH bottles (Fig. 4). S̆imek et al. (37) estimated from in situ hybridizations of bacteria inside protozoan food vacuoles that a cell length of about 3 μm was the maximum ingestible particle size for the nanoflagellate Bodo saltans. In an oligomesotrophic lake the abundance of bacterial cells that were >2.4 μm correlated strongly with the abundance of HNF, therefore suggesting the existence of a size refuge at this cell length (32). A comparable bacterial size limit for the nanoflagellates might have been the case in our experiment. We have to be aware that there is some controversy regarding the exact size estimations of bacteria derived from staining with different fluorochromes (40) and that DAPI staining may underestimate bacterial size. For larger rods and filaments, cell length measurements yielded 20 to 30% higher values from Cy3 fluorescence than from DAPI fluorescence of the same cells, but no such differences were apparent for small cells (32a). It has been shown in laboratory experiments that actively growing bacteria, which generally have an increased cell length or are at a dividing stage, are subject to higher rates of feeding by nanoflagellates and ciliates (8, 34), presumably due to higher rates of encounter with the grazers (36). However, increased grazing mortality probably changes to grazing resistance with small changes in cell elongation. This was probably the reason for the strong increase in the number of large rods of the alpha subdivision of Proteobacteria, which became a dominant group in the bacterial community due to the protozoan impact.
The development of a bacterial community with a high proportion of grazing-resistant forms was not unexpected. In enclosure experiments designed similarly to ours (17), nearly 90% of the bacterial biomass consisted of grazing-resistant bacterial morphotypes 96 h after removal of zooplankton. Filamentous bacteria in eutrophic lakes can achieve a high biomass during protozoan maxima that is comparable to the peaks observed in our experiment (22, 39). In addition to the data acquired in previous fractionation experiments, in which the bacterial community shift was monitored only from the morphological aspect as revealed in DAPI preparations (19, 23), we obtained a better insight into this succession by using group-specific oligonucleotide probes. Information on the composition of freshwater bacterioplankton is still very limited. However, recent studies using 16S rRNA sequences (15, 27) or FISH (1, 6, 30, 42) indicate that the beta subdivision of Proteobacteria seems to be globally distributed and abundant in freshwater environments. The next most abundant phylogenetic groups identified in these studies were members of the Cytophaga-Flavobacterium cluster and of the alpha subdivision of the class Proteobacteria. This general view of the phylogenetic composition of freshwater bacterial communities is also confirmed in the present study. Bacteria that hybridized with probes BET42a and CF319a represented a substantial portion of the bacterial community in this pond.
The dynamics of both groups closely resembled that of total bacterioplankton, and their abundance increased strongly after removal of zooplankton and declined to below the initial levels after development of HNF (Fig. 3). Interestingly, cells that hybridized with probe ALF1b did not follow this pattern but seemed to be rather unaffected by the large flagellate population, and their number continued to increase throughout the experiment. The large rods and cells at dividing stages, which made up the major portion of the biovolume in this bacterial subgroup (Fig. 4 and Fig. 5A) had probably just reached the cell length which protected them against ingestion by nanoflagellates. This represents an example in which grazing-resistant morphotypes, belonging to an initially minor bacterial taxon, become a dominant part of the bacterial assemblage due to high grazing pressure.
Principally two different mechanisms can be involved in the formation of grazing-resistant bacteria. First, predation-resistant morphotypes can appear due to phenotypic alterations of normal morphotypes, as shown for isolated strains (12, 35). This might be triggered by chemical release by the predators (release of kairomones), which is a widespread mechanism in planktonic food webs (reviewed in reference 25) but has not yet been shown for bacterium-protozoan interactions. Cell elongation can also be indirectly mediated by predators due to an increase in specific growth rate due to grazing (12). Second, changed selection conditions, i.e., towards higher predation pressure and less substrate competition, might increase the abundance of previously uncommon taxa with permanently low grazer vulnerability. The second mechanism would result, without any phenotypic plasticity, in a shift in the overall bacterial community composition. However, a combination of both mechanisms is possible, i.e., an increase in abundance of a phenotypically altered species, thereby changing the community composition as well (12, 13). Different mechanisms for the development of grazing resistance are likely to occur simultaneously in natural complex bacterial communities. In our experiment, the continuous size range from small to large elongated rods, the high net growth rates, and the many dividing stages of the bacteria that hybridized with probe ALF1b suggest that a mechanism similar to that demonstrated for several bacterial isolates (12, 13) might have been active: the reduction of bacterial abundance by grazers increases the substrate flow per cell and increases the cell-specific growth rate. This in turn results in larger, more-elongated cell forms and thus shifts the size distribution into the size range of grazing-resistant bacteria. However, we need more specific probes to follow the population dynamics of individual species within the whole community in order to reveal the exact underlying mechanisms.
Grazing-resistant morphotypes in our enclosure experiment were not restricted to the alpha subdivision of Proteobacteria, although this was quantitatively the most important group. Different types of filamentous bacteria, belonging to the beta subdivision of Proteobacteria, the Cytophaga-Flavobacterium cluster, and an as yet unaffiliated member of the bacterioplankton (which is EUB338 negative), increased in abundance and accounted for a significant fraction of bacterial biomass after the protozoan population maxima occurred (Fig. 6). Filament formation seems to be a phylogenetically widespread mechanism for resisting protozoan predation. In activated sludge, an environment with extremely high protozoan grazing pressure, there can be found many filamentous bacterial species which belong to all the bacterial subclasses which were quantified in this study with the group-specific probes (41). Besides complex bacterial morphologies, growth in detritus aggregates occurred in − DAPH bottles and the abundance of these aggregates increased during the experiment. However, as attached bacteria were estimated to comprise not more than 10% of total bacterial biomass, we did not include these in our analysis. It was obvious, however, that bacteria from all subgroups detected by the group-specific probes were present in these aggregates (data not shown), which probably present a temporary predation refuge (4, 18).
The application of group-specific rRNA-targeted oligonucleotides is a relatively rapid and easy way to obtain an idea of the overall bacterial community composition and of the major shifts which occur due to changes in biotic or abiotic conditions. However, with respect to our analysis of the predator-mediated bacterial succession in the microcosm experiment, we have to be aware of the methodological limitations. Group-specific probes can reveal only shifts between and not changes within the groups tested for. Phylogenetic groups, such as the alpha, beta, and gamma subdivisions of the class Proteobacteria and the Cytophaga-Flavobacterium cluster, include bacteria with very diverse morphologies and physiologies. The other restriction of FISH is the fact that in some planktonic habitats a high percentage of total bacteria do not hybridize with oligonucleotide probes (6, 23). This was not a major problem here as the hybridization efficiency was generally high. It was more problematic for the analysis of bacterial community changes that a rather large proportion of EUB338-positive cells could not be assigned by one of the four group-specific probes (Fig. 2). There are insufficient data to judge whether this is a general phenomenon in freshwater bacterioplankton. In a high mountain lake this proportion was also large (20 to 60%) (1), whereas it was generally small in a study on lake snow particles (42). Although sequencing of amplified and cloned 16S rRNA-encoding genes from bacterioplankton revealed that members of the phyla Proteobacteria and Cytophaga-Flavobacterium-Bacteroides are major phylogenetic groups in freshwater plankton (3, 15), some other bacterial clusters for which no oligonucleotide probes have been designed yet, such as Actinomycetales and Verrucomicrobiales, and low G+C content gram-positive bacteria were found (15). Another possible reason for the low detectability with the group-specific probes is that some of them, such as CF319a, do not hybridize with all known species within that group (26). Therefore, for these types of ecological studies there is a continued need for more oligonucleotide probes that are specific on various phylogenetic levels.
An important question which cannot be answered at the moment is whether, besides the filaments, bacteria with different predation resistance strategies developed. Even during peak abundance of protists, a substantial portion of bacteria in the edible size range remained in the bottles in the experiments; for example, the abundance of members of the gamma subdivision of Proteobacteria increased continuously although they consisted nearly exclusively of small rods. This may be due to other, nonmorphological, resistance mechanisms as suggested before (20) or, alternatively, to high growth rates of these bacteria which compensate for grazing losses (31). Our knowledge of the composition and seasonal succession of freshwater bacterioplankton is still very limited, and the same is true for our knowledge of the properties of bacteria which influence their susceptibility to predators. However, with the help of the rapidly evolving methodology for analyzing the bacterial community composition, we can expect many new insights into how grazers impact the phenotypic appearance and population dynamics of natural bacterial assemblages.
ACKNOWLEDGMENTS
We thank Sybille Liedtke for excellent technical assistance, Martin Hahn for comments on an early version of the manuscript, and Nancy Zehrbach for linguistic improvements.
REFERENCES
- 1.Alfreider A, Pernthaler J, Amann R, Sattler B, Glöckner F-O, Wille A, Psenner R. Community analysis of the bacterial assemblages in the winter cover and pelagic layers of a high mountain lake by in situ hybridization. Appl Environ Microbiol. 1996;62:2138–2144. doi: 10.1128/aem.62.6.2138-2144.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amann R I, Ludwig W, Schleifer K-H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bahr M, Hobbie J E, Sogin M L. Bacterial diversity in an arctic lake: a freshwater SAR11 cluster. Aquat Microb Ecol. 1996;11:271–277. [Google Scholar]
- 4.Brettar I, Höfle M. Influence of ecosystematic factors on survival of Escherichia coli after large-scale release into lake water mesocosms. Appl Environ Microbiol. 1992;58:2201–2210. doi: 10.1128/aem.58.7.2201-2210.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chrzanowski T H, S̆imek K. Prey-size selection by freshwater flagellated protozoa. Limnol Oceanogr. 1990;35:1429–1436. [Google Scholar]
- 6.Glöckner F O, Amann R, Alfreider A, Pernthaler J, Psenner R, Trebesius K, Schleifer K H. An in situ hybridization protocol for detection and identification of planktonic bacteria. Syst Appl Microbiol. 1996;19:403–406. [Google Scholar]
- 7.González J M, Sherr E B, Sherr B F. Size-selective grazing on bacteria by natural assemblages of estuarine flagellates and ciliates. Appl Environ Microbiol. 1990;56:583–589. doi: 10.1128/aem.56.3.583-589.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.González J M, Sherr E B, Sherr B F. Differential feeding by marine flagellates on growing versus starving, and on motile versus nonmotile, bacterial prey. Mar Ecol Prog Ser. 1993;102:257–267. [Google Scholar]
- 9.Güde H. Grazing by protozoa as selection factor for activated sludge bacteria. Microb Ecol. 1979;5:225–237. doi: 10.1007/BF02013529. [DOI] [PubMed] [Google Scholar]
- 10.Güde H. The role of grazing on bacteria in plankton succession. In: Sommer U, editor. Plankton ecology. Succession in plankton communities. Berlin, Germany: Springer Verlag; 1989. pp. 337–364. [Google Scholar]
- 11.Güde H, Haibel B, Müller H. Development of planktonic bacterial populations in a water column of Lake Constance (Bodensee-Obersee) Arch Hydrobiol. 1985;105:59–77. [Google Scholar]
- 12.Hahn M W, Höfle M G. Grazing pressure by a bacterivorous flagellate reverses the relative abundance of Comamonas acidovorans PX54 and Vibrio strain CB5 in chemostat cocultures. Appl Environ Microbiol. 1998;64:1910–1918. doi: 10.1128/aem.64.5.1910-1918.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hahn M W, Moore E R B, Höfle M G. Bacterial filament formation, a defense mechanism against flagellate grazing, is growth rate controlled in bacteria of different phyla. Appl Environ Microbiol. 1999;65:25–35. doi: 10.1128/aem.65.1.25-35.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Havskum H, Hansen A S. Importance of pigmented and colourless nano-sized protists as grazers on nanoplankton in a phosphate-depleted Norwegian fjord and in enclosures. Aquat Microb Ecol. 1997;12:139–151. [Google Scholar]
- 15.Hiorns W D, Methé B A, Nierzwicki-Bauer S A, Zehr J P. Bacterial diversity in Adirondack mountain lakes as revealed by 16S rRNA gene sequences. Appl Environ Microbiol. 1997;63:2957–2960. doi: 10.1128/aem.63.7.2957-2960.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jürgens K. The impact of Daphnia on microbial food webs—a review. Mar Microb Food Webs. 1994;8:295–324. [Google Scholar]
- 17.Jürgens K, Arndt H, Rothhaupt K O. Zooplankton-mediated changes of bacterial community structure. Microb Ecol. 1994;27:27–42. doi: 10.1007/BF00170112. [DOI] [PubMed] [Google Scholar]
- 18.Jürgens K, Arndt H, Zimmermann H. Impact of metazoan and protozoan grazers on bacterial biomass distribution in microcosm experiments. Aquat Microb Ecol. 1997;12:131–138. [Google Scholar]
- 19.Jürgens K, Gasol J M, Massana R, Pedrós-Alió C. Control of heterotrophic bacteria and protozoans by Daphnia pulex in the epilimnion of Lake Ciso. Arch Hydrobiol. 1994;131:55–78. [Google Scholar]
- 20.Jürgens K, Güde H. The potential importance of grazing-resistant bacteria in planktonic systems. Mar Ecol Prog Ser. 1994;112:169–188. [Google Scholar]
- 21.Jürgens K, Jeppesen E. Cascading effects on microbial food web structure in a dense macrophyte bed. In: Jeppesen E, Søndergaard M, Søndergaard M, Christoffersen K, editors. The structuring role of submerged macrophytes in lakes. New York, N.Y: Springer-Verlag; 1998. pp. 262–273. [Google Scholar]
- 22.Jürgens K, Stolpe G. Seasonal dynamics of crustacean zooplankton, heterotrophic nanoflagellates and bacteria in a shallow, eutrophic lake. Freshw Biol. 1995;33:27–38. [Google Scholar]
- 22a.Jürgens, K. Unpublished results.
- 23.Karner M, Fuhrman J A. Determination of active marine bacterioplankton: a comparison of universal 16S rRNA probes, autoradiography, and nucleoid staining. Appl Environ Microbiol. 1997;63:1208–1213. doi: 10.1128/aem.63.4.1208-1213.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lampert W. The relationship between zooplankton biomass and grazing: a review. Limnologica. 1988;19:11–20. [Google Scholar]
- 25.Larsson P, Dodson S. Invited review—chemical communication in planktonic animals. Arch Hydrobiol. 1993;129:129–155. [Google Scholar]
- 26.Manz W, Amann R, Ludwig W, Vancanneyt M, Schleifer K H. Application of a suite of 16S rRNA-specific oligonucleotide probes designed to investigate bacteria of the phylum Cytophaga-Flavobacter-Bacteroides in the natural environment. Microbiology. 1996;142:1097–1106. doi: 10.1099/13500872-142-5-1097. [DOI] [PubMed] [Google Scholar]
- 27.Methe B A, Hiorns W D, Zehr J P. Contrasts between marine and freshwater bacterial community composition: analyses of communities in Lake George and six other Adirondack lakes. Limnol Oceanogr. 1998;43:368–374. [Google Scholar]
- 28.Pace M L, McManus G B, Findlay S E G. Planktonic community structure determines the fate of bacterial production in a temperate lake. Limnol Oceanogr. 1990;35:795–808. [Google Scholar]
- 29.Pernthaler J, Alfreider A, Posch T, Andreatta S, Psenner R. In situ classification and image cytometry of pelagic bacteria from a high mountain lake (Gossenköllersee, Austria) Appl Environ Microbiol. 1997;63:4778–4783. doi: 10.1128/aem.63.12.4778-4783.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pernthaler J, Glöckner F-O, Unterholzner S, Alfreider A, Psenner R, Amann R. Seasonal community and population dynamics of pelagic bacteria and archaea in a high mountain lake. Appl Environ Microbiol. 1998;64:4299–4306. doi: 10.1128/aem.64.11.4299-4306.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pernthaler J, Posch T, S̆imek K, Vrba J, Amann R, Psenner R. Contrasting bacterial strategies to coexist with a flagellate predator in an experimental microbial assemblage. Appl Environ Microbiol. 1997;63:596–601. doi: 10.1128/aem.63.2.596-601.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pernthaler J, Sattler B, S̆imek K, Schwarzenbacher A, Psenner R. Top-down effects on the size-biomass distribution of a freshwater bacterioplankton community. Aquat Microb Ecol. 1996;10:255–263. [Google Scholar]
- 32a.Pernthaler, J. Unpublished data.
- 33.Porter K G, Feig Y S. The use of DAPI for identifying and counting aquatic microflora. Limnol Oceanogr. 1980;25:943–947. [Google Scholar]
- 34.Sherr B F, Sherr E B, McDaniel J. Effect of protistan grazing on the frequency of dividing cells in bacterioplankton assemblages. Appl Environ Microbiol. 1992;58:2381–2385. doi: 10.1128/aem.58.8.2381-2385.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shikano S, Luckinbill L S, Kurihara Y. Changes of traits in a bacterial population associated with protozoan predation. Microb Ecol. 1990;20:75–84. doi: 10.1007/BF02543868. [DOI] [PubMed] [Google Scholar]
- 36.Shimeta J. Diffusional encounter of submicrometer particles and small cells by suspension feeders. Limnol Oceanogr. 1993;38:456–465. [Google Scholar]
- 37.S̆imek K, Vrba J, Pernthaler J, Posch T, Hartman P, Nedoma J, Psenner R. Morphological and compositional shifts in an experimental bacterial community influenced by protists with contrasting feeding modes. Appl Environ Microbiol. 1997;63:587–595. doi: 10.1128/aem.63.2.587-595.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Snaidr J, Amann R, Huber I, Ludwig W, Schleifer K H. Phylogenetic analysis and in situ identification of bacteria in activated sludge. Appl Environ Microbiol. 1997;63:2884–2896. doi: 10.1128/aem.63.7.2884-2896.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sommaruga R, Psenner R. Permanent presence of grazing-resistant bacteria in a hypertrophic lake. Appl Environ Microbiol. 1995;61:3457–3459. doi: 10.1128/aem.61.9.3457-3459.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Suzuki M T, Sherr E B, Sherr B F. DAPI direct counting underestimates bacterial abundances and average cell size compared to AO direct counting. Limnol Oceanogr. 1993;38:1566–1570. [Google Scholar]
- 41.Wagner M, Amann R, Kämpfer P, Assmus B, Hartmann A, Hutzler P, Springer N, Schleifer K H. Identification and in situ detection of gram-negative filamentous bacteria in activated sludge. Syst Appl Microbiol. 1994;17:405–417. [Google Scholar]
- 42.Weiss P, Schweitzer B, Amann R, Simon M. Identification in situ and dynamics of bacteria on limnetic organic aggregates (lake snow) Appl Environ Microbiol. 1996;62:1998–2005. doi: 10.1128/aem.62.6.1998-2005.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]