Abstract
Background
Antidepressants are a first-line treatment for depression. However, only a third of individuals experience remission after the first treatment. Common genetic variation, in part, likely regulates antidepressant response, yet the success of previous genome-wide association studies has been limited by sample size. This study performs the largest genetic analysis of prospectively assessed antidepressant response in major depressive disorder to gain insight into the underlying biology and enable out-of-sample prediction.
Methods
Genome-wide analysis of remission (nremit = 1852, nnonremit = 3299) and percentage improvement (n = 5218) was performed. Single nucleotide polymorphism–based heritability was estimated using genome-wide complex trait analysis. Genetic covariance with eight mental health phenotypes was estimated using polygenic scores/AVENGEME. Out-of-sample prediction of antidepressant response polygenic scores was assessed. Gene-level association analysis was performed using MAGMA and transcriptome-wide association study. Tissue, pathway, and drug binding enrichment were estimated using MAGMA.
Results
Neither genome-wide association study identified genome-wide significant associations. Single nucleotide polymorphism–based heritability was significantly different from zero for remission (h2 = 0.132, SE = 0.056) but not for percentage improvement (h2 = −0.018, SE = 0.032). Better antidepressant response was negatively associated with genetic risk for schizophrenia and positively associated with genetic propensity for educational attainment. Leave-one-out validation of antidepressant response polygenic scores demonstrated significant evidence of out-of-sample prediction, though results varied in external cohorts. Gene-based analyses identified ETV4 and DHX8 as significantly associated with antidepressant response.
Conclusions
This study demonstrates that antidepressant response is influenced by common genetic variation, has a genetic overlap schizophrenia and educational attainment, and provides a useful resource for future research. Larger sample sizes are required to attain the potential of genetics for understanding and predicting antidepressant response.
Keywords: Antidepressant response, Depression, Genetics, GWAS, MDD, Polygenic score
Major depressive disorder (MDD) is the third leading cause of years lived with disability worldwide (1) and is a substantial risk factor for suicide (2). MDD confers a major personal, societal, and economic burden (3), partly because of the limited efficacy of treatment options.
In 2011 to 2014, 12.7% of individuals in the United States 12 years of age and over reported antidepressant medication use (4). The rate of antidepressant prescriptions is also increasing, with the number of prescriptions doubling in the United Kingdom in the decade prior to 2018 (5). Antidepressants are robustly linked to a reduction in depressive symptoms (6), but they are often ineffective: approximately 35% of patients remit after their primary treatment (7) and approximately 40% develop treatment-resistant depression (TRD), defined as not remitting after two or more antidepressants (8). For patients, the process of trialing antidepressants can be lengthy and demoralizing, delaying recovery and exposing patients to a range of potential side effects that reduce adherence and willingness to try new drugs (9). There is therefore great potential to improve treatment of depression through better understanding of the factors that control response to antidepressants and implementing this knowledge through individually tailored treatment.
Pharmacogenetic studies were expected to uncover loci with large effects on drug response and adverse events due to effects of pharmacokinetic or pharmacodynamic mechanisms. While associations between antidepressant plasma levels and drug-metabolizing enzymes CYP2D6 and CYP2C19 have been identified (10, 11, 12), previous research suggests that genes encoding these enzymes and other candidate genes account for a small proportion of variation in drug response (13,14). However, genotyping complexities for such candidate genes may contribute to limited findings.
Several genome-wide association studies (GWASs) have been performed to identify genetic predictors of antidepressant response. Although no robustly replicated associations have been detected to date (15, 16, 17, 18, 19), common single nucleotide polymorphisms (SNPs) are reported to explain 42% (SE = 18%; 95% confidence interval [CI], 7%–77%) of the variance (20). Pharmacogenetic studies are intensive to perform, requiring disease severity measures at baseline pretreatment and then longitudinally, with many studies being performed as part of a randomized controlled trial (15, 16, 17, 18). This clinically assessed approach provides high-quality data, though it has led to previous studies being limited in sample size, with <3000 patients with MDD in the largest GWAS to date. Further efforts to combine these individual cohorts to increase sample size for genetic studies are therefore required. Use of lighter phenotyping approaches such as electronic health record–derived TRD (21) may also provide novel insight, though it is unclear whether these different measures of antidepressant response have a common genetic basis.
In this study, we analyze genome-wide genetic data on clinically assessed antidepressant response from 5843 patients treated for MDD, combined from 13 international research studies. Using this novel data resource, we perform GWAS of remission and percentage improvement after receiving antidepressant medication, and undertake extensive post-GWAS analyses, made feasible through this increased sample size. This study aims to elucidate the genetic architecture of antidepressant response and use polygenic scores to establish the relationship between antidepressant response and mental health outcomes. We find, for the first time, a replicable polygenic signal of antidepressant response across studies.
Methods and Materials
Primary Samples and Measures
This study analyzed 13 cohorts (Table 1). Ten cohorts were of European ancestry and 3 were of East Asian ancestry (Supplement 1). All subjects provided written informed consent for pharmacogenetic analyses. These primary cohorts include individuals with a clinical diagnosis of MDD, who were assessed for depressive symptoms before and after treatment with antidepressants.
Table 1.
Cohorts of Individuals Diagnosed With Major Depressive Disorder and Assessed for Depressive Symptoms Before and After Treatment With Antidepressant Medication
| Study (Reference) | Country, Region | Study Design | Study Length, Weeks | Medication(s) | Measure | Median Age, Years | IQR for Age, Years | Female | Na | npercentage improvement | nremit | nnonremit |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| European Ancestry | ||||||||||||
| STAR∗D (52) | United States | Open label | 12 | Citalopram | QIDSC | 44 | 32–53 | 58% | 1163 | 1163 | 506 | 657 |
| GSRD (17) | Europe | Naturalistic | >4 | Various | MADRS | 52.5 | 43–61 | 66% | 1152 | 1152 | 189 | 963 |
| GENDEP (53) | Europe | Partially randomized RCT | 12 | Escitalopram, nortriptyline | MADRS | 43 | 33–51 | 63% | 783 | 783 | 291 | 365 |
| DAST (see Supplement 1) | Germany | Naturalistic inpatient | 6 | Various | HAMD-21 | 50 | 37–62 | 57% | 586 | 586 | 245 | 303 |
| PGRN-AMPS (54) | United States | Open label | 8 | Citalopram, escitalopram | QIDSC | 38.5 | 28–49 | 63% | 490 | 392 | 200 | 290 |
| GENPOD (18) | United Kingdom | Open label | 12 | Citalopram, reboxetine | BDI | 38 | 30–48 | 69% | 474 | 474 | 169 | 305 |
| PFZ (18) | United States | RCT | 6-8 | Sertraline, fluoxetine, paroxetine | HAMD-17 | 43 | 32–54 | 67% | 309 | 309 | 99 | 210 |
| Mayo (16) | United States | Open label | 8 | Citalopram, escitalopram | HAMD-17 | 37 | 29–51 | 62% | 156 | 156 | 80 | 76 |
| GSK (18) | United States | RCT | 8 | Escitalopram | HAMD-17 | 36 | 25.75–45 | 55% | 132 | 132 | 56 | 76 |
| GODS (18) | Switzerland | Open label | 8 | Paroxetine | MADRS | 37 | 29.5–43.5 | 52% | 71 | 71 | 17 | 54 |
| East Asian Ancestry | ||||||||||||
| Miaoli (16) | Taiwan | Open label | 8 | Escitalopram, paroxetine | HAMD-17 | 41 | 30–52 | 82% | 233 | 233 | 103 | 130 |
| Taipei (16) | Taiwan | Open label | 8 | Fluoxetine, citalopram | HAMD-17 | 46 | 34–59 | 55% | 174 | 174 | 45 | 129 |
| Japan (16) | Japan | RCT | 6 | Fluvoxamine, paroxetine | HAMD-17 | 44.5 | 32–56 | 47% | 120 | 120 | 78 | 42 |
| Total | 5843 | 5745 | 2078 | 3600 | ||||||||
BDI, Beck Depression Inventory; DAST, Depression and Sequence of Treatment; GENDEP, Genome Based Therapeutic Drugs for Depression; GENPOD, GENetic and clinical Predictors Of treatment response in Depression; GODS, Geneva Outpatient Depression Study; GSK, Glaxo Smith Kline; GSRD, Group for the Study of Resistant Depression; HAMD-17, 17-item Hamilton Depression Rating Scale; HAMD-21, 21-item Hamilton Depression Rating Scale; IQR, interquartile range; MADRS, Montgomery–Åsberg Depression Rating Scale; PFZ, Pfizer; PGRN-AMPS, Pharmacogenomics Research Network Antidepressant Medication Pharmacogenomic Study; QIDSC, Quick Inventory of Depressive Symptomatology; RCT, randomized controlled trial; STAR∗D, Sequenced Treatment Alternatives to Relieve Depression.
Number of participants included after quality control of genetic and clinical data.
Two measures of antidepressant response were defined: remission and percentage improvement. Remission is a binary measure attained when a patient’s depression symptom score decreases to a prespecified threshold for the rating scale (Supplement 1).
All analyses included covariates of the first 20 principal components of population structure, age, and gender. Analyses using the remission measure of response also included the baseline symptom score as a covariate, to control for depression severity.
Each cohort underwent standard quality control and 1000 Genomes Project phase 3 imputation using the RICOPILI pipeline on the LISA server (22) (Supplement 1 and Table S1 in Supplement 2).
Genome-wide Association Study
GWAS was performed using the RICOPILI pipeline (22) separately for studies with participants of European and of East Asian ancestry (Supplement 1). All other analyses were performed using only the European ancestry cohorts due to the limited sample size of the East Asian cohorts.
Gene-Level Association Analysis
Gene associations were estimated using MAGMA (23) and transcriptome-wide association study (TWAS) (24).
The MAGMA v1.06b SNP-wise mean model (±10-kb window) was used to perform gene-level association analysis based on the remission and percentage improvement GWAS p values. The analysis was based on genetic variants and linkage disequilibrium in the 1000 Genomes Project phase 3 dataset available on the MAGMA website (g1000_eur.bed/bim/fam). SNPs were assigned to genes using the MAGMA NCBI37.3.gene.loc file with a 10-kb window. False discovery rate (FDR) correction was used to control for multiple testing. See Supplement 1 for a description of gene set enrichment analysis using MAGMA.
TWAS integrates GWAS associations with external expression quantitative trait loci data to infer whether differential gene expression estimated from SNP data is associated with the GWAS phenotype. TWAS was performed using FUSION software (http://gusevlab.org/projects/fusion/) and precomputed multi-SNP predictors of gene expression based on data collected from multiple specific brain regions, thyroid tissue, pituitary gland, liver, and blood (Table S2 in Supplement 2). The transcriptome-wide significance threshold of p < 2.51 × 10−6 was estimated using a permutation procedure (25). To test whether the same causal SNP affects both the GWAS phenotype and gene expression, colocalization analysis was performed using the coloc package in R software (version 3.5.0; R Foundation for Statistical Computing) (26), as implemented by FUSION software.
Estimation of SNP-Based Heritability
The SNP-based heritability of remission and percentage improvement was estimated using individual-level data by genomic relatedness–based restricted maximum likelihood (GREML) in the software GCTA (genome-wide complex trait analysis) (27,28). The analysis was performed 1) across all cohorts, including a study covariate (mega-GREML); and 2) separately within each cohort and then inverse variance meta-analyzed (meta-GREML) (Supplement 1). Comparison of mega- and meta-GREML estimates can provide insight into the heterogeneity between cohorts, as only mega-GREML accounts for genetic covariances between cohorts. We converted SNP-based heritability estimates for remission to the liability scale using assuming a population prevalence of 0.357, reflecting the prevalence of remission across the cohorts in this study.
Leave-One-Out Polygenic Scoring
To determine whether polygenic scores derived from the remission and percentage improvement GWAS summary statistics predict antidepressant response in an independent sample, a leave-one-out polygenic scoring approach was used. This involves calculating polygenic scores within each cohort based on GWAS summary statistics derived using all other cohorts. Polygenic scores were calculated using PRSice V2 (29) (Supplement 1). One-sided p values were used to assess statistical significance, as we are testing the one-sided hypothesis that the polygenic score has a positive association with the outcome in the target sample.
Estimation of Genetic Overlap With Mental Health Phenotypes
We tested for evidence of genetic overlap between antidepressant response measures and seven mental health phenotypes: major depression (30), bipolar disorder (31), schizophrenia (32), attention-deficit/hyperactivity disorder (33), autism spectrum disorder (ASD) (34), anxiety (35), and problematic drinking (Alcohol Use Disorders Identification Test problem subscale) (36). Educational attainment (37) was also included, as it has strong correlations with the mental health disorders tested. Evidence of genetic overlap was assessed using polygenic scoring with AVENGEME (38), and linkage disequilibrium score regression (39). To avoid sample overlap between the major depression GWAS and the antidepressant response cohorts in this study, we used major depression GWAS summary statistics excluding overlapping cohorts (STAR∗D [Sequenced Treatment Alternatives to Relieve Depression], GENPOD [GENetic and clinical Predictors Of treatment response in Depression], GENDEP [Genome Based Therapeutic Drugs for Depression], PFZ [Pfizer]).
AVENGEME aggregates polygenic score association results across p-value thresholds to estimate genetic covariance between antidepressant response and the eight mental health phenotypes. AVENGEME parameters are provided in Table S3 in Supplement 2. Bonferroni correction was used to account for multiple testing for the eight discovery GWASs used.
Replication Cohorts and Analyses
Out-of-Sample Prediction
External validation of polygenic scores derived using the full GWAS results was also carried out. Five independent samples were used (Supplement 1). In brief, Janssen (N = 190, remission rate = 11.8%) (40), the Douglas Biomarker Study (N = 127, remission rate = 23.6%) (41), and the IRL-GREY (Incomplete Response in Late Life Depression: Getting to Remission) study (N = 307, remission rate = 52.4%) (42) prospectively assessed depressive symptoms, concordant with the discovery GWAS samples. In contrast, Generation Scotland (ntreatment resistant = 177, nnon–treatment resistant = 2455) (21) assessed electronic prescription data, and the AGDS (Australian Genetics of Depression Study) study (nresponders = 4368, nnonresponders = 6879) (43) collected retrospective self-report questionnaire data. Polygenic score association results were meta-analyzed across the prospectively assessed cohorts given their more comparable study design and antidepressant measures. One-sided p values were used to assess statistical significance.
Comparison of Genetic Covariance With Mental Health Phenotypes
Individual-level data were available for Generation Scotland enabling estimation of genetic covariance between TRD and mental health-related phenotypes using AVENGEME, as described above. Analyses in Generation Scotland were controlled for age, gender, and 20 principal components of population structure. When estimating genetic covariance between TRD and major depression, we used major depression GWAS summary statistics excluding Generation Scotland to avoid sample overlap.
Results
Descriptive statistics for the cohorts used in this study are available in Table 1 and in Figures S1 to S5 in Supplement 1.
GWAS of Antidepressant Response
Across the 10 European studies, 5151 individuals with remission data (1852 [36.0%] patients remitting) and 5218 participants with percentage improvement data were available. No variants were significantly associated with remission or percentage improvement (Figures S6 and S7 in Supplement 1, Tables S4 and S5 in Supplement 2). There was no evidence of confounding (Figures S8 and S9 in Supplement 1, Table S6 in Supplement 2)
No significant associations were identified in the East Asian GWASs (N = 527) (Figures S10 and S11 in Supplement 1). A comparison between East Asian and European GWAS results is shown in Supplement 1.
Gene-Level Association Results
MAGMA identified a significant association on chromosome 17 for ETV4 with both remission (pFDR = .016) and percentage improvement (pFDR = .016). Within the same region, DHX8 was also significantly associated with remission (pFDR = .046). The SNP associations within this region span multiple genes (Figure S12 in Supplement 1). Full MAGMA gene-based association results are shown in Tables S7 and S8 in Supplement 2.
TWAS identified no association achieving transcriptome-wide significance (p < 2.51 × 10−6). Further inspection of TWAS associations within the chromosome 17 region implicated by MAGMA highlighted SNP-associations with upregulation of BRCA1 (remission p = 1.96 × 10−4; percentage improvement p = 9.21 × 10−5; GTeX brain–caudate [basal ganglia]) and upregulation of TMEM106A (remission p = .0011; percentage improvement p = .0018; Young Finns Study [blood]). Colocalization analysis of these associations indicated shared casual variants for these genes’ differential expression and antidepressant response. Full TWAS results are given in Tables S9 and S10 in Supplement 2.
See Supplement 1 for gene set enrichment analysis results.
SNP-Based Heritability
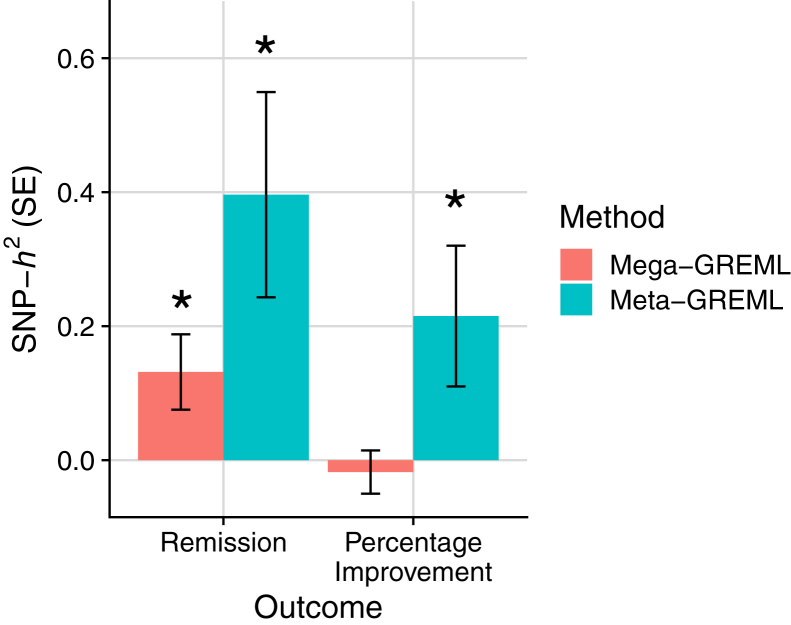
Analysis across all samples (mega-GREML) showed remission to have a significant nonzero SNP-based heritability (h2 = 0.132; SE = 0.056; 95% CI, 0.022 to 0.241; p = .009, liability scale assuming population prevalence of 0.357), whereas the SNP-based heritability for percentage improvement was not significantly different from zero (h2 = −0.018; SE = 0.032; 95% CI, −0.080 to 0.045; p = .303) (Figure 1).
Figure 1.
Single nucleotide polymorphism–based heritability (SNP-h2) estimates for remission and percentage improvement with SE bars. Figure shows across (mega-) and within (meta-) sample genomic relatedness–based restricted maximum likelihood (GREML) estimates. ∗Estimate is significantly different from zero, at p < .05.
The SNP-based heritability estimates from meta-analysis of within-sample estimates (meta-GREML) were significant for both remission (h2 = 0.396; SE = 0.153; 95% CI, 0.096 to 0.696; p = .010, liability scale assuming population prevalence of 0.357) and percentage improvement (h2 = 0.215; SE = 0.105; 95% CI, 0.009 to 0.421; p = .041) (Figure 1). See Figures S18 and S19 in Supplement 1 for meta-analysis forest plots.
See Supplement 1 for SNP-based heritability sensitivity analyses.
Out-of-Sample Prediction
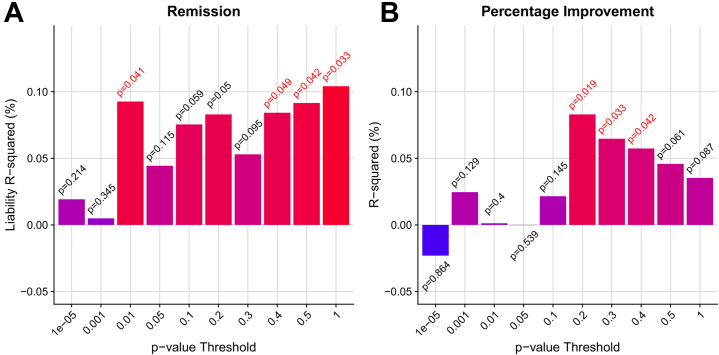
Leave-one-out polygenic score analysis provided evidence that polygenic scores derived using remission and percentage improvement GWAS results could both explain a statistically significant amount of variance out-of-sample (Figure 2). Both remission and percentage improvement explained ∼0.1% of the variance, with polygenic scores for multiple p-value thresholds associated at nominal significance.
Figure 2.
Polygenic prediction of antidepressant response from leave-one-out polygenic scoring for (A) remission and (B) percentage improvement. R2 estimates are signed to indicate positive or negative association. One-sided p values are shown above or below the bars, with p values < .05 highlighted in red.
Validation of polygenic scores based on the full antidepressant response GWAS summary statistics was carried out using five samples. Meta-analysis of polygenic score associations across the three prospectively assessed cohorts (Janssen, Douglas Biomarker Study, and IRL-GREY study) showed nominally significant evidence of association for the remission polygenic score (maximum liability R2 = 0.8%, p = .015) and a nonsignificant association for the percentage improvement score (maximum R2 = 0.2%, p = .091) (Figure S21 in Supplement 1). Results were highly variable across each prospectively assessed cohort. No association was found between polygenic scores in Generation Scotland or AGDS study cohorts. Full polygenic score replication results are in Tables S14 to S17 in Supplement 2.
Genetic Overlap With Mental Health Phenotypes
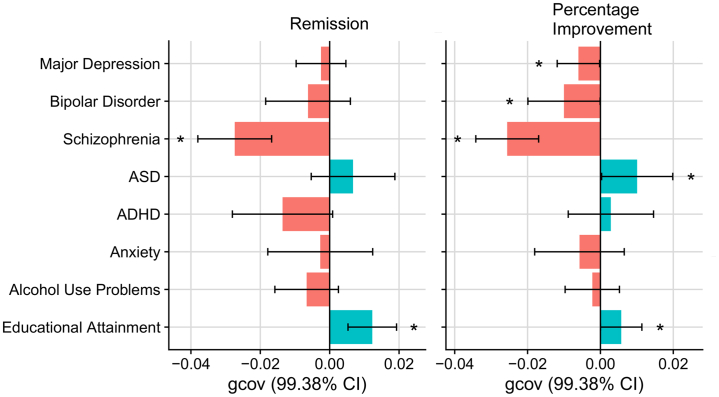
Both remission and percentage improvement showed a significant negative genetic covariance with schizophrenia, and significant positive genetic covariance with educational attainment (Figure 3; Tables S18 and S19 in Supplement 2). Percentage improvement also showed a significant negative covariance with major depression and bipolar disorder, and a significant positive genetic covariance with ASD. Linkage disequilibrium score regression genetic correlation estimates were broadly concordant, although they were nonsignificant (Figure S22 in Supplement 1). Subsequent conditional analysis, covarying for educational attainment polygenic scores, showed that the associations with psychiatric disorders were independent of the association with educational attainment (Figure S23 in Supplement 1).
Figure 3.
Genetic covariance (gcov) estimates between antidepressant response phenotypes and seven mental health phenotypes and educational attainment. Confidence intervals (CIs) were corrected for multiple testing. ADHD, attention-deficit/hyperactivity disorder; ASD, autism spectrum disorder.
Genetic overlap estimates between TRD in Generation Scotland and mental health phenotypes were congruent with results from primary samples, showing that genetic risk for schizophrenia was greater among individuals with TRD, and educational attainment genetic propensity was greater among individuals with non-TRD (Figure S24 in Supplement 1).
Discussion
Antidepressants are a common and effective strategy for treating MDD; however, remission rates are typically low, and factors affecting antidepressant response are poorly understood. This study is the largest genetic investigation of antidepressant response based on clinically defined cohorts. For the first time, we identify a polygenic profile for antidepressant response, which can predict across cohorts, and shows genetic correlations with traits that reflect clinical observations.
This study finds significant evidence that antidepressant response is influenced by common genetic variation. Meta-analysis of SNP-based heritability estimates within each cohort indicates that 20% to 40% of the variance in antidepressant response is attributable to common genetic variation, consistent with a previous analysis of a subset of these studies (20). However, the SNP-based heritability decreased substantially when estimating across cohorts simultaneously. Although the change in SNP-heritability was not statistically significant, these results suggest that antidepressant response in a broad context has a heritable component, but genetic differences can explain additional variability in antidepressant response within more specific contexts. Despite the apparent heterogeneity across individual cohorts, the sample sizes for antidepressant response are sufficiently large to detect a polygenic signal. Genetic studies for susceptibility to psychiatric disorders show that findings accrue after an inflection point in sample size is reached (30, 31, 32). This study’s findings for SNP-based heritability and out-of-sample polygenic prediction indicate that sample sizes for antidepressant response are reaching the inflection point and that larger studies will uncover more of the genetic component (44). Power calculations for detecting genome-wide significant variation, and the variance explained by corresponding polygenic scores, are provided in Figure S25 in Supplement 1. Interestingly, our findings suggest that the SNP-based heritability of remission is higher than for percentage improvement. The percentage improvement score might have lower heritability because of increased noise, in which this measure is more susceptible to random variation in depressive symptoms, is less comparable across the different depressive symptom scales used, or captures increases in depressive symptoms.
This study provides novel insight into the shared genetic basis between antidepressant response and mental health phenotypes. We show an association between high genetic liability of psychiatric disorders and poorer response, which mirrors conclusions of clinical studies (45). The schizophrenia polygenic risk score was negatively associated with antidepressant response, which is replicated in the TRD phenotype in Generation Scotland. Previous studies have shown that individuals with TRD may respond to antipsychotic medication (46). Our findings extend those reports by suggesting that individuals with antidepressant resistance also have a higher burden of schizophrenia genetic risk. We found some evidence that genetic liability to major depression is associated with poorer response to antidepressants. However, this association was only statistically significant for percentage improvement, and it requires replication. In addition, we report a novel finding that high ASD genetic liability increased the chance of remission. Another recent study reported that ASD genetic liability is associated with poorer response to cognitive behavioral therapy (47). If both these findings are replicated, it would suggest ASD genetic liability could serve as a differential predictor of response to antidepressants and cognitive behavioral therapy. We also identified a significant association between genetic propensity for educational attainment and improved antidepressant response as well as between genetic propensity for educational attainment and non-TRD. This may reflect the indirect measurement of socioeconomic status captured by educational attainment, which is supported by previous literature showing a positive association between antidepressant response and socioeconomic status (48). Future research should explore whether individuals with higher educational attainment have improved response due to factors such as adherence or joint psychological treatment.
Polygenic scores derived from the remission and percentage improvement GWASs both significantly predicted antidepressant response out of sample using a leave-one-out design. This is the first GWAS of antidepressant response able to predict significantly out of sample, representing an important advance in the field of antidepressant response genetics. Although the variance explained is low (R2 = 0.1%) and p values are close to the nominal significance threshold, this result is encouraging given the sample size of this study. For example, a recent GWAS of MDD explains only 1.9% of the variance in MDD, despite having a sample size 100 times greater than this study (30). Our finding suggests that a renewed effort to systematically collect new samples in which genetic associations with antidepressant response can be identified will improve the prediction of antidepressant response, helping to uncover its biological mechanisms and clinical associations, and eventually enable more accurate clinical predictors to be developed and applied.
This study provided limited insight into the biological underpinnings of antidepressant response implicating one locus on chromosome 17 surrounding ETV4 and DHX8. A previous study using neuronal cell lines and mouse models found that ETV4 mediates brain-derived neurotrophic factor (BDNF) induced hippocampal dendrite development and plasticity (49), congruent with the hypothesis that the mechanism of action for antidepressants is via hippocampal neuroplasticity (50). DHX8 has a less clear mechanistic link to antidepressant response with a broader function in messenger RNA splicing (51). Replication of the association at this locus is required before further experimental investigation.
In addition, no association was detected with genetic variation within classical pharmacokinetic candidate genes, such as CYP2D6 and CYP2C19, which have previously been robustly associated with antidepressant plasma levels (11). Although the enzymatic activity of CYP2D6 and CYP2C19 is largely regulated by common genetic variation, these variants include structural variants that are not well captured by GWAS arrays, and large effects on enzymatic activity are typically conferred by combinations of genetic variants (haplotypes), which GWAS does not assess. Therefore, the absence of an association at this point may be a false negative result. Furthermore, looking across individuals that have not been treated with a specific antidepressant or antidepressant class will reduce the likelihood of detecting pharmacokinetic effects.
Owing to a limited sample size, it was not possible to estimate genetic correlations between longitudinally assessed antidepressant response and TRD defined using electronic health records. However, comparison of shared genetic etiology with other mental health phenotypes indicated that these distinct measures of antidepressant response have a shared genetic basis. Further comparison and integration of these two approaches is warranted and may prove fruitful given the large gains in sample size that electronic health record–derived phenotypes can provide.
There are several limitations to this study that should be addressed in the future. First, large sample sizes are essential for robust identification of associated genetic variation and out-of-sample prediction. However, combining independently collected datasets inevitably introduces heterogeneity. Obtaining large homogeneous samples is particularly challenging for pharmacogenetic studies, as heterogeneity is driven not only by patient characteristics such as diagnosis and patient ascertainment, but also by differences in treatment such as the drug, dosage, duration, and co-pharmacotherapy. Although the cohorts within this study have many features in common, heterogeneity in antidepressant treatment is present. As sample sizes grow, analyses stratified by these factors will become more feasible, enabling detection of genetic effects relevant to each antidepressant, antidepressant class, or other treatment characteristics. Second, an important question to consider is whether the variance in depressive symptoms after treatment is due to antidepressant response or to other variables altering the course of depression. Although antidepressants have a significant effect on depressive symptoms, and their administration is the core feature of participants in this study, individuals may vary in depressive symptoms due to other factors affecting disease progression, such as clinical and sociodemographic variables and placebo response. This is a difficult issue to resolve but should be considered when interpreting the results. Future genetic studies incorporating the placebo arm of clinical trials may help identify genetic associations specific to antidepressant response. Third, this study has focused on changes in total depressive symptoms without considering symptom domain-specific changes or the presence of side effects. Given the wide range of depressive symptoms and the influence side effects can have on efficacy, consideration of these features may provide additional insights. Fourth, although this study included three cohorts of East Asian ancestry, further inclusion of cohorts with diverse ancestries is an important area. Genetic analysis within diverse populations helps to ensure that the findings are applicable to worldwide populations and can help fine-map causal variants underlying genetic associations.
In summary, this study identifies a polygenic profile for antidepressant response that predicts across studies and is negatively correlated with genetic susceptibility to schizophrenia, which could be used for prognostic purposes. While the current results have no clinical utility as a pharmacogenetic test, they indicate that studies with larger sample sizes could provide predictions explaining a substantial proportion of antidepressant response. We note that a prognostic test that enables even a modest increase in the proportion of patients that respond to antidepressants would have a substantial impact on recovery for many patients, given the high prevalence of depression. We hope that this study prompts both replication and extension to accelerate the development of pharmacogenetic testing for psychiatry.
Acknowledgments and Disclosures
The collection of the sample from the Group for the Study of Resistant Depression (GSRD) Consortium was supported by an unrestricted grant from Lundbeck for the GSRD. Lundbeck had no further role in the study design and the collection, analysis, and interpretation of data. The GENDEP (Genome Based Therapeutic Drugs for Depression) study was funded by a European Commission Framework 6 grant (EC Contract Ref. No. LSHB-CT-2003-503428). H. Lundbeck provided nortriptyline and escitalopram for the GENDEP study. GlaxoSmithKline and the UK National Institute for Health Research of the Department of Health contributed to the funding of the sample collection at the Institute of Psychiatry, London. GENDEP Illumina array genotyping was funded in part by a joint grant from the UK Medical Research Council and GlaxoSmithKline (Grant No. G0701420). The GENPOD (GENetic and clinical Predictors Of treatment response in Depression) trial was funded by the UK Medical Research Council and supported by the UK Mental Health Research Network. The genotyping of GENPOD samples was supported by the Innovative Medicine Initiative Joint Undertaking under Grant No. 115008, of which resources are composed of European Union and the European Federation of Pharmaceutical Industries and Associations (EFPIA) in-kind contribution and financial contribution from the European Union’s Seventh Framework Programme (Grant No. FP7/2007-2013). EFPIA members Pfizer, GlaxoSmithKline, and F. Hoffmann-La Roche have contributed work and samples to the project presented here. The PFZ (Pfizer), GSK (GlaxoSmithKline), and GODS were supported by the Innovative Medicine Initiative Joint Undertaking (IMI-JU) under Grant No. 115008 of which resources are composed of European Union and EFPIA) in-kind contribution and financial contribution from the European Union’s Seventh Framework Programme (FP7/2007-2013). EFPIA members Pfizer, GlaxoSmithKline, and F. Hoffmann La-Roche have contributed work and samples to the project presented here. The PGRN-AMPS (Pharmacogenomics Research Network Antidepressant Medication Pharmacogenomic Study) study data were obtained via the database of Genotypes and Phenotypes (dbGAP) (Accession No. phs000670.v1.p1). Funding support for the PGRN-AMPS study was provided by the National Institute of General Medical Sciences, National Institutes of Health, through the PGRN grant to Principal Investigators R. Weinshilboum and L. Wang (Grant No. U19 GM61388). D. Mrazek served as the Principal Investigator for the PGRN-AMPS study within the Mayo Clinic PGRN program. Genome-wide genotyping was performed at the RIKEN Center for Genomic Medicine, with funding provided by RIKEN. The datasets used for the analyses described in this manuscript were obtained from the dbGaP at http://www.ncbi.nlm.nih.gov/gap/. Generation Scotland is supported by the Wellcome Trust (Reference Nos. 104036/Z/14/Z and 216767/Z/19/Z), the Chief Scientist Office of the Scottish Government Health Department (Grant No. CZD/16/6), and the Scottish Funding Council (Grant No. HR03006). Data linkage and analysis in Generation Scotland is supported by the Medical Research Council (Grant No. MC_PC_17209). For the Douglas Biomarker Study, the genotyping of the samples was funded and generated by Janssen Research & Development, LLC. Major funding for the Psychiatric Genetics Consortium (PGC) is from the U.S. National Institutes of Health (Grant Nos. U01 MH109528 and U01 MH109532). OP and CML were funded by the National Institute for Health Research (NIHR) Biomedical Research Centre at South London and Maudsley NHS Foundation Trust and King’s College London. The authors acknowledge use of the research computing facility at King’s College London, Rosalind (https://rosalind.kcl.ac.uk), which is delivered in partnership with the NIHR Maudsley BRC, and part-funded by capital equipment grants from the Maudsley Charity (Award No. 980) and Guy's & St. Thomas' Charity (Grant Nos. TR130505). The views expressed are those of the authors and not necessarily those of the NHS, the NIHR, or the Department of Health and Social Care.
We are deeply indebted to the investigators who comprise the PGC, and to the hundreds of thousands of subjects who have shared their life experiences with PGC investigators. Statistical analyses for the PGC were carried out on the NL Genetic Cluster Computer (http://www.geneticcluster.org) hosted by SURFsara. 23andMe, Inc. used GWAS summary statistics for major depression including 23andMe participants. We thank the research participants and employees of 23andMe for making the work regarding major depression possible.
A previous version of this article was published as a preprint on medRxiv: https://www.medrxiv.org/content/10.1101/2020.12.11.20245035v1.
The PGC’s policy is to make genome-wide summary results public. Summary statistics for remission and percentage improvement GWAS are available on the PGC website (https://www.med.unc.edu/pgc).
CML has served on the scientific advisory board for Myriad Neuroscience. KJA has received two research grants in the last 2 years from Janssen Inc., Canada (fellowship grants for trainees), and provided consultancy services in the last three years for Otsuka Canada Pharmaceutical Inc., Lundbeck Canada, and HLS Therapeutics. AS is or has been consultant/speaker for: Abbott, AbbVie, Angelini, AstraZeneca, Clinical Data, Boehringer, Bristol Myers Squibb, Eli Lilly, GlaxoSmithKline, InnovaPharma, Italfarmaco, Janssen, Lundbeck, Naurex, Pfizer, Polifarma, Sanofi, and Servier. AMM has received research support from the Sackler Trust and speakers fees from Janssen and Illumina. MK has received grant funding from the Japan Society for the Promotion of Science, SENSHIN Medical Research Foundation, and Japan Research Foundation for Clinical Pharmacology; and speaker honoraria from Dainippon-Sumitomo Pharma, Otsuka, Meiji-Seika Pharma, Eli Lilly, MSD K.K., GlaxoSmithKline, Pfizer, Janssen Pharmaceutical, Shionogi, Mitsubishi Tanabe Pharma, Takeda Pharmaceutical, Lundbeck, and Ono Pharmaceutical. QSL is an employee of Janssen Research & Development, LLC, and a shareholder in Johnson & Johnson, the parent company of the Janssen companies and declares that, except for income received from her primary employer, no financial support or compensation has been received from any individual or corporate entity over the past 3 years for research or professional service, and there is no personal financial holding that could be perceived as constituting a potential conflict of interest. DS has received grant/research support from GlaxoSmithKline and Lundbeck; and served as a consultant or on advisory boards for AstraZeneca, Bristol Myers Squibb, Eli Lilly, Janssen, and Lundbeck. Stuart Montgomery has served as a consultant or on advisory boards for AstraZeneca, Bristol Myers Squibb, Forest, Johnson & Johnson, Leo, Lundbeck, Medelink, Neurim, Pierre Fabre, and Richter. Siegfried Kasper has received grants/research support, consulting fees, and/or honoraria within the last 3 years from Angelini, AOP Orphan Pharmaceuticals AG, Celgene GmbH, Eli Lilly, Janssen-Cilag Pharma GmbH, KRKA-Pharma, Lundbeck A/S, Mundipharma, Neuraxpharm, Pfizer, Sanofi, Schwabe, Servier, Shire, Sumitomo Dainippon Pharma Co. Ltd., and Takeda. Joseph Zohar has received grant/research support from Lundbeck, Servier, Brainsway, and Pfizer; served as a consultant or on advisory boards for Servier, Pfizer, Abbott, Lilly, Actelion, AstraZeneca, and Roche; and served on speakers bureaus for Lundbeck, Roche, Lilly, Servier, Pfizer, and Abbott. Julien Mendlewicz has served on the Board of the Lundbeck International Neuroscience Foundation and on the advisory board for Servier. Patrick F. Sullivan has served on the scientific advisory board for Pfizer, Inc., and on the advisory committee for Lundbeck. All other authors report no biomedical financial interests or potential conflicts of interest.
Footnotes
Major Depressive Disorder Working Group of the Psychiatric Genomics Consortium. Primary contact: Cathryn Lewis. Naomi R. Wray,1,2 Stephan Ripke,3,4,5 Manuel Mattheisen,6,7,8 Maciej Trzaskowski,1 Enda M. Byrne,1 Abdel Abdellaoui,9 Mark J. Adams,10 Esben Agerbo,11,12,13 Tracy M. Air,14 Till F.M. Andlauer,15,16 Silviu-Alin Bacanu,17 Marie Bækvad-Hansen,13,18 Aartjan T.F. Beekman,19 Tim B. Bigdeli,17,20 Elisabeth B. Binder,15,21 Julien Bryois,22 Henriette N. Buttenschøn,13,23,24 Jonas Bybjerg-Grauholm,13,18 Na Cai,25,26 Enrique Castelao,27 Jane Hvarregaard Christensen,8,13,24 Toni-Kim Clarke,10 Jonathan R.I. Coleman,28 Lucía Colodro-Conde,29 Baptiste Couvy-Duchesne,2,30 Nick Craddock,31 Gregory E. Crawford,32,33 Gail Davies,34 Ian J. Deary,34 Franziska Degenhardt,35 Eske M. Derks,29 Nese Direk,36,37 Conor V. Dolan,9 Erin C. Dunn,38,39,40 Thalia C. Eley,28 Valentina Escott-Price,41 Farnush Farhadi Hassan Kiadeh,42 Hilary K. Finucane,43,44 Jerome C. Foo,45 Andreas J. Forstner,35,46,47,48 Josef Frank,45 Héléna A. Gaspar,28 Michael Gill,49 Fernando S. Goes,50 Scott D. Gordon,29 Jakob Grove,8,13,24,51 Lynsey S. Hall,10,52 Christine Søholm Hansen,13,18 Thomas F. Hansen,53,54,55 Stefan Herms,35,47 Ian B. Hickie,56 Per Hoffmann,35,47 Georg Homuth,57 Carsten Horn,58 Jouke-Jan Hottenga,9 David M. Hougaard,13,18 David M. Howard,10,28 Marcus Ising,59 Rick Jansen,19 Ian Jones,60 Lisa A. Jones,61 Eric Jorgenson,62 James A. Knowles,63 Isaac S. Kohane,64,65,66 Julia Kraft,4 Warren W. Kretzschmar,67 Zoltán Kutalik,68,69 Yihan Li,67 Penelope A. Lind,29 Donald J. MacIntyre,70,71 Dean F. MacKinnon,50 Robert M. Maier,2 Wolfgang Maier,72 Jonathan Marchini,73 Hamdi Mbarek,9 Patrick McGrath,74 Peter McGuffin,28 Sarah E. Medland,29 Divya Mehta,2,75 Christel M. Middeldorp,9,76,77 Evelin Mihailov,78 Yuri Milaneschi,19 Lili Milani,78 Francis M. Mondimore,50 Grant W. Montgomery,1 Sara Mostafavi,79,80 Niamh Mullins,28 Matthias Nauck,81,82 Bernard Ng,80 Michel G. Nivard,9 Dale R. Nyholt,83 Paul F. O’Reilly,28 Hogni Oskarsson,84 Michael J. Owen,60 Jodie N. Painter,29 Carsten Bøcker Pedersen,11,12,13 Marianne Giørtz Pedersen,11,12,13 Roseann E. Peterson,17,85 Wouter J. Peyrot,19 Giorgio Pistis,27 Danielle Posthuma,86,87 Jorge A. Quiroz,88 Per Qvist,8,13,24 John P. Rice,89 Brien P. Riley,17 Margarita Rivera,28,90 Saira Saeed Mirza,36 Robert Schoevers,91 Eva C. Schulte,92,93 Ling Shen,62 Jianxin Shi,94 Stanley I. Shyn,95 Engilbert Sigurdsson,96 Grant C.B. Sinnamon,97 Johannes H. Smit,19 Daniel J. Smith,98 Hreinn Stefansson,99 Stacy Steinberg,99 Fabian Streit,45 Jana Strohmaier,45 Katherine E. Tansey,100 Henning Teismann,101 Alexander Teumer,102 Wesley Thompson,13,54,103,104 Pippa A. Thomson,105 Thorgeir E. Thorgeirsson,99 Matthew Traylor,106 Jens Treutlein,45 Vassily Trubetskoy,4 André G. Uitterlinden,107 Daniel Umbricht,108 Sandra Van der Auwera,109 Albert M. van Hemert,110 Alexander Viktorin,22 Peter M. Visscher,1,2 Yunpeng Wang,13,54,104 Bradley T. Webb,111 Shantel Marie Weinsheimer,13,54 Jürgen Wellmann,101 Gonneke Willemsen,9 Stephanie H. Witt,45 Yang Wu,1 Hualin S. Xi,112 Jian Yang,2,113 Futao Zhang,1 Volker Arolt,114 Bernhard T. Baune,114,115,116 Klaus Berger,101 Dorret I. Boomsma,9 Sven Cichon,35,47,117,118 Udo Dannlowski,114 E.J.C. de Geus,9,119 J. Raymond DePaulo,50 Enrico Domenici,120 Katharina Domschke,121,122 Tõnu Esko,5,78 Hans J. Grabe,109 Steven P. Hamilton,123 Caroline Hayward,124 Andrew C. Heath,89 Kenneth S. Kendler,17 Stefan Kloiber,59,125,126 Glyn Lewis,127 Qingqin S. Li,128 Susanne Lucae,59 Pamela A.F. Madden,89 Patrik K. Magnusson,22 Nicholas G. Martin,29 Andrew M. McIntosh,10,34 Andres Metspalu,78,129 Ole Mors,13,130 Preben Bo Mortensen,11,12,13,24 Bertram Müller-Myhsok,15,131,132 Merete Nordentoft,13,133 Markus M. Nöthen,35 Michael C. O’Donovan,60 Sara A. Paciga,134 Nancy L. Pedersen,22 Brenda W.J.H. Penninx,19 Roy H. Perlis,38,135 David J. Porteous,105 James B. Potash,136 Martin Preisig,27 Marcella Rietschel,45 Catherine Schaefer,62 Thomas G. Schulze,45,93,137,138,139 Jordan W. Smoller,38,39,40 Kari Stefansson,99,140 Henning Tiemeier,36,141,142 Rudolf Uher,143 Henry Völzke,102 Myrna M. Weissman,74,144 Thomas Werge,13,54,145 Cathryn M. Lewis,28,146 Douglas F. Levinson,147 Gerome Breen,28,148 Anders D. Børglum,8,13,24 Patrick F. Sullivan.22,149,150
1Institute for Molecular Bioscience, The University of Queensland, Brisbane, QLD, AU; 2Queensland Brain Institute, The University of Queensland, Brisbane, QLD, AU; 3Analytic and Translational Genetics Unit, Massachusetts General Hospital, Boston, MA, US; 4Department of Psychiatry and Psychotherapy, Universitätsmedizin Berlin Campus Charité Mitte, Berlin, DE; 5Medical and Population Genetics, Broad Institute, Cambridge, MA, US; 6Department of Psychiatry, Psychosomatics and Psychotherapy, University of Wurzburg, Wurzburg, DE; 7Centre for Psychiatry Research, Department of Clinical Neuroscience, Karolinska Institutet, Stockholm, SE; 8Department of Biomedicine, Aarhus University, Aarhus, DK; 9Dept of Biological Psychology & EMGO+ Institute for Health and Care Research, Vrije Universiteit Amsterdam, Amsterdam, NL; 10Division of Psychiatry, University of Edinburgh, Edinburgh, GB; 11Centre for Integrated Register-based Research, Aarhus University, Aarhus, DK; 12National Centre for Register-Based Research, Aarhus University, Aarhus, DK; 13iPSYCH, The Lundbeck Foundation Initiative for Integrative Psychiatric Research, DK; 14Discipline of Psychiatry, University of Adelaide, Adelaide, SA, AU; 15Department of Translational Research in Psychiatry, Max Planck Institute of Psychiatry, Munich, DE; 16Department of Neurology, Klinikum rechts der Isar, Technical University of Munich, Munich, DE; 17Department of Psychiatry, Virginia Commonwealth University, Richmond, VA, US; 18Center for Neonatal Screening, Department for Congenital Disorders, Statens Serum Institut, Copenhagen, DK; 19Department of Psychiatry, Vrije Universiteit Medical Center and GGZ inGeest, Amsterdam, NL; 20Virginia Institute for Psychiatric and Behavior Genetics, Richmond, VA, US; 21Department of Psychiatry and Behavioral Sciences, Emory University School of Medicine, Atlanta, GA, US; 22Department of Medical Epidemiology and Biostatistics, Karolinska Institutet, Stockholm, SE; 23Department of Clinical Medicine, Translational Neuropsychiatry Unit, Aarhus University, Aarhus, DK; 24iSEQ, Centre for Integrative Sequencing, Aarhus University, Aarhus, DK; 25Human Genetics, Wellcome Trust Sanger Institute, Cambridge, GB; 26Statistical genomics and systems genetics, European Bioinformatics Institute (EMBL-EBI), Cambridge, GB; 27Department of Psychiatry, Lausanne University Hospital and University of Lausanne, Lausanne, CH; 28Social, Genetic and Developmental Psychiatry Centre, King’s College London, London, GB; 29Genetics and Computational Biology, QIMR Berghofer Medical Research Institute, Brisbane, QLD, AU; 30Centre for Advanced Imaging, The University of Queensland, Brisbane, QLD, AU; 31Psychological Medicine, Cardiff University, Cardiff, GB; 32Center for Genomic and Computational Biology, Duke University, Durham, NC, US; 33Department of Pediatrics, Division of Medical Genetics, Duke University, Durham, NC, US; 34Centre for Cognitive Ageing and Cognitive Epidemiology, University of Edinburgh, Edinburgh, GB; 35Institute of Human Genetics, University of Bonn, School of Medicine & University Hospital Bonn, Bonn, DE; 36Epidemiology, Erasmus MC, Rotterdam, Zuid-Holland, NL; 37Psychiatry, Dokuz Eylul University School Of Medicine, Izmir, TR; 38Department of Psychiatry, Massachusetts General Hospital, Boston, MA, US; 39Psychiatric and Neurodevelopmental Genetics Unit (PNGU), Massachusetts General Hospital, Boston, MA, US; 40Stanley Center for Psychiatric Research, Broad Institute, Cambridge, MA, US; 41Neuroscience and Mental Health, Cardiff University, Cardiff, GB; 42Bioinformatics, University of British Columbia, Vancouver, BC, CA; 43Department of Epidemiology, Harvard T.H. Chan School of Public Health, Boston, MA, US; 44Department of Mathematics, Massachusetts Institute of Technology, Cambridge, MA, US; 45Department of Genetic Epidemiology in Psychiatry, Central Institute of Mental Health, Medical Faculty Mannheim, Heidelberg University, Mannheim, Baden-Württemberg, DE; 46Department of Psychiatry (UPK), University of Basel, Basel, CH; 47Department of Biomedicine, University of Basel, Basel, CH; 48Centre for Human Genetics, University of Marburg, Marburg, DE; 49Department of Psychiatry, Trinity College Dublin, Dublin, IE; 50Psychiatry & Behavioral Sciences, Johns Hopkins University, Baltimore, MD, US; 51Bioinformatics Research Centre, Aarhus University, Aarhus, DK; 52Institute of Genetic Medicine, Newcastle University, Newcastle upon Tyne, GB; 53Danish Headache Centre, Department of Neurology, Rigshospitalet, Glostrup, DK; 54Institute of Biological Psychiatry, Mental Health Center Sct. Hans, Mental Health Services Capital Region of Denmark, Copenhagen, DK; 55iPSYCH, The Lundbeck Foundation Initiative for Psychiatric Research, Copenhagen, DK; 56Brain and Mind Centre, University of Sydney, Sydney, NSW, AU; 57Interfaculty Institute for Genetics and Functional Genomics, Department of Functional Genomics, University Medicine and Ernst Moritz Arndt University Greifswald, Greifswald, Mecklenburg-Vorpommern, DE; 58Roche Pharmaceutical Research and Early Development, Pharmaceutical Sciences, Roche Innovation Center Basel, F. Hoffmann-La Roche Ltd, Basel, CH; 59Max Planck Institute of Psychiatry, Munich, DE; 60MRC Centre for Neuropsychiatric Genetics and Genomics, Cardiff University, Cardiff, GB; 61Department of Psychological Medicine, University of Worcester, Worcester, GB; 62Division of Research, Kaiser Permanente Northern California, Oakland, CA, US; 63Psychiatry & The Behavioral Sciences, University of Southern California, Los Angeles, CA, US; 64Department of Biomedical Informatics, Harvard Medical School, Boston, MA, US; 65Department of Medicine, Brigham and Women’s Hospital, Boston, MA, US; 66Informatics Program, Boston Children’s Hospital, Boston, MA, US; 67Wellcome Trust Centre for Human Genetics, University of Oxford, Oxford, GB; 68Institute of Social and Preventive Medicine (IUMSP), Lausanne University Hospital and University of Lausanne, Lausanne, VD, CH; 69Swiss Institute of Bioinformatics, Lausanne, VD, CH; 70Division of Psychiatry, Centre for Clinical Brain Sciences, University of Edinburgh, Edinburgh, GB; 71Mental Health, NHS 24, Glasgow, GB; 72Department of Psychiatry and Psychotherapy, University of Bonn, Bonn, DE; 73Statistics, University of Oxford, Oxford, GB; 74Psychiatry, Columbia University College of Physicians and Surgeons, New York, NY, US; 75School of Psychology and Counseling, Queensland University of Technology, Brisbane, QLD, AU; 76Child and Youth Mental Health Service, Children’s Health Queensland Hospital and Health Service, South Brisbane, QLD, AU; 77Child Health Research Centre, University of Queensland, Brisbane, QLD, AU; 78Estonian Genome Center, University of Tartu, Tartu, EE; 79Medical Genetics, University of British Columbia, Vancouver, BC, CA; 80Statistics, University of British Columbia, Vancouver, BC, CA; 81DZHK (German Centre for Cardiovascular Research), Partner Site Greifswald, University Medicine, University Medicine Greifswald, Greifswald, Mecklenburg-Vorpommern, DE; 82Institute of Clinical Chemistry and Laboratory Medicine, University Medicine Greifswald, Greifswald, Mecklenburg-Vorpommern, DE; 83Institute of Health and Biomedical Innovation, Queensland University of Technology, Brisbane, QLD, AU; 84Humus, Reykjavik, IS; 85Virginia Institute for Psychiatric & Behavioral Genetics, Virginia Commonwealth University, Richmond, VA, US; 86Clinical Genetics, Vrije Universiteit Medical Center, Amsterdam, NL; 87Complex Trait Genetics, Vrije Universiteit Amsterdam, Amsterdam, NL; 88Solid Biosciences, Boston, MA, US; 89Department of Psychiatry, Washington University in Saint Louis School of Medicine, Saint Louis, MO, US; 90Department of Biochemistry and Molecular Biology II, Institute of Neurosciences, Biomedical Research Center (CIBM), University of Granada, Granada, ES; 91Department of Psychiatry, University of Groningen, University Medical Center Groningen, Groningen, NL; 92Department of Psychiatry and Psychotherapy, University Hospital, Ludwig Maximilian University Munich, Munich, DE; 93Institute of Psychiatric Phenomics and Genomics (IPPG), University Hospital, Ludwig Maximilian University Munich, Munich, DE; 94Division of Cancer Epidemiology and Genetics, National Cancer Institute, Bethesda, MD, US; 95Behavioral Health Services, Kaiser Permanente Washington, Seattle, WA, US; 96Faculty of Medicine, Department of Psychiatry, University of Iceland, Reykjavik, IS; 97School of Medicine and Dentistry, James Cook University, Townsville, QLD, AU; 98Institute of Health and Wellbeing, University of Glasgow, Glasgow, GB; 99deCODE Genetics / Amgen, Reykjavik, IS; 100College of Biomedical and Life Sciences, Cardiff University, Cardiff, GB; 101Institute of Epidemiology and Social Medicine, University of Münster, Münster, Nordrhein-Westfalen, DE; 102Institute for Community Medicine, University Medicine Greifswald, Greifswald, Mecklenburg-Vorpommern, DE; 103Department of Psychiatry, University of California, San Diego, San Diego, CA, US; 104KG Jebsen Centre for Psychosis Research, Norway Division of Mental Health and Addiction, Oslo University Hospital, Oslo, NO; 105Medical Genetics Section, CGEM, IGMM, University of Edinburgh, Edinburgh, GB; 106Clinical Neurosciences, University of Cambridge, Cambridge, GB; 107Internal Medicine, Erasmus MC, Rotterdam, Zuid-Holland, NL; 108Roche Pharmaceutical Research and Early Development, Neuroscience, Ophthalmology and Rare Diseases Discovery & Translational Medicine Area, Roche Innovation Center Basel, F. Hoffmann-La Roche Ltd, Basel, CH; 109Department of Psychiatry and Psychotherapy, University Medicine Greifswald, Greifswald, Mecklenburg-Vorpommern, DE; 110Department of Psychiatry, Leiden University Medical Center, Leiden, NL; 111Virginia Institute for Psychiatric & Behavioral Genetics, Virginia Commonwealth University, Richmond, VA, US; 112Computational Sciences Center of Emphasis, Pfizer Global Research and Development, Cambridge, MA, US; 113Institute for Molecular Bioscience; Queensland Brain Institute, The University of Queensland, Brisbane, QLD, AU; 114Department of Psychiatry, University of Münster, Münster, Nordrhein-Westfalen, DE; 115Department of Psychiatry, Melbourne Medical School, University of Melbourne, Melbourne, AU; 116Florey Institute for Neuroscience and Mental Health, University of Melbourne, Melbourne, AU; 117Institute of Medical Genetics and Pathology, University Hospital Basel, University of Basel, Basel, CH; 118Institute of Neuroscience and Medicine (INM-1), Research Center Juelich, Juelich, DE; 119Amsterdam Public Health Institute, Vrije Universiteit Medical Center, Amsterdam, NL; 120Centre for Integrative Biology, Università degli Studi di Trento, Trento, Trentino-Alto Adige, IT; 121Department of Psychiatry and Psychotherapy, Medical Center - University of Freiburg, Faculty of Medicine, University of Freiburg, Freiburg, DE; 122Center for NeuroModulation, Faculty of Medicine, University of Freiburg, Freiburg, DE; 123Psychiatry, Kaiser Permanente Northern California, San Francisco, CA, US; 124Medical Research Council Human Genetics Unit, Institute of Genetics and Molecular Medicine, University of Edinburgh, Edinburgh, GB; 125Department of Psychiatry, University of Toronto, Toronto, ON, CA; 126Centre for Addiction and Mental Health, Toronto, ON, CA; 127Division of Psychiatry, University College London, London, GB; 128Neuroscience Therapeutic Area, Janssen Research and Development, LLC, Titusville, NJ, US; 129Institute of Molecular and Cell Biology, University of Tartu, Tartu, EE; 130Psychosis Research Unit, Aarhus University Hospital, Risskov, Aarhus, DK; 131Munich Cluster for Systems Neurology (SyNergy), Munich, DE; 132University of Liverpool, Liverpool, GB; 133Mental Health Center Copenhagen, Copenhagen Universtity Hospital, Copenhagen, DK; 134Human Genetics and Computational Biomedicine, Pfizer Global Research and Development, Groton, CT, US; 135Psychiatry, Harvard Medical School, Boston, MA, US; 136Psychiatry, University of Iowa, Iowa City, IA, US; 137Department of Psychiatry and Behavioral Sciences, Johns Hopkins University, Baltimore, MD, US; 138Department of Psychiatry and Psychotherapy, University Medical Center Göttingen, Goettingen, Niedersachsen, DE; 139Human Genetics Branch, NIMH Division of Intramural Research Programs, Bethesda, MD, US; 140Faculty of Medicine, University of Iceland, Reykjavik, IS; 141Child and Adolescent Psychiatry, Erasmus MC, Rotterdam, Zuid-Holland, NL; 142Psychiatry, Erasmus MC, Rotterdam, Zuid-Holland, NL; 143Psychiatry, Dalhousie University, Halifax, NS, CA; 144Division of Translational Epidemiology, New York State Psychiatric Institute, New York, NY, US; 145Department of Clinical Medicine, University of Copenhagen, Copenhagen, DK; 146Department of Medical & Molecular Genetics, King’s College London, London, GB; 147Psychiatry & Behavioral Sciences, Stanford University, Stanford, CA, US; 148NIHR Maudsley Biomedical Research Centre, King’s College London, London, GB; 149Genetics, University of North Carolina at Chapel Hill, Chapel Hill, NC, US; 150Psychiatry, University of North Carolina at Chapel Hill, Chapel Hill, NC, US.
Naomi R. Wray, Stephan Ripke, Manuel Mattheisen, Cathryn M. Lewis, Douglas F. Levinson, Gerome Breen, Anders D. Børglum, and Patrick F. Sullivan contributed equally to the Major Depressive Disorder Working Group of the Psychiatric Genomics Consortium.
European Group for the Study of Resistant Depression (GSRD) Consortium. Primary contact: Alessandro Serretti. Siegfried Kasper,1 Joseph Zohar,2 Daniel Souery,3 Stuart Montgomery,4 Diego Albani,5 Gianluigi Forloni,5 Panagiotis Ferentinos,6 Dan Rujescu,7 Julien Mendlewicz.8
1Department of Psychiatry and Psychotherapy, Medical University Vienna, Austria; 2Department of Psychiatry, Sheba Medical Center, Tel Hashomer, and Sackler School of Medicine, Tel Aviv University, Israel; 3Laboratoire de Psychologie Médicale, Université Libre de Bruxelles and Psy Pluriel, Centre Européen de Psychologie Médicale, Brussels; 4Imperial College School of Medicine, London, UK; 5Laboratory of Biology of Neurodegenerative Disorders, Neuroscience Department, Istituto di Ricerche Farmacologiche Mario Negri IRCCS, Milan, Italy; 6Department of Psychiatry, Athens University Medical School, Athens, Greece; 7University Clinic for Psychiatry, Psychotherapy and Psychosomatic, Martin-Luther-University Halle-Wittenberg, Germany; 8Université Libre de Bruxelles.
Supplementary material cited in this article is available online at https://doi.org/10.1016/j.bpsgos.2021.07.008.
Contributor Information
Oliver Pain, Email: oliver.pain@kcl.ac.uk.
GSRD Consortium:
Siegfried Kasper, Joseph Zohar, Daniel Souery, Stuart Montgomery, Diego Albani, Gianluigi Forloni, Panagiotis Ferentinos, Dan Rujescu, and Julien Mendlewicz
Major Depressive Disorder Working Group of the Psychiatric Genomics Consortium:
Naomi R. Wray, Stephan Ripke, Manuel Mattheisen, Maciej Trzaskowski, Enda M. Byrne, Abdel Abdellaoui, Mark J. Adams, Esben Agerbo, Tracy M. Air, Till F.M. Andlauer, Silviu-Alin Bacanu, Marie Bækvad-Hansen, Aartjan T.F. Beekman, Tim B. Bigdeli, Elisabeth B. Binder, Julien Bryois, Henriette N. Buttenschøn, Jonas Bybjerg-Grauholm, Na Cai, Enrique Castelao, Jane Hvarregaard Christensen, Toni-Kim Clarke, Jonathan R.I. Coleman, Lucía Colodro-Conde, Baptiste Couvy-Duchesne, Nick Craddock, Gregory E. Crawford, Gail Davies, Ian J. Deary, Franziska Degenhardt, Eske M. Derks, Nese Direk, Conor V. Dolan, Erin C. Dunn, Thalia C. Eley, Valentina Escott-Price, Farnush Farhadi Hassan Kiadeh, Hilary K. Finucane, Jerome C. Foo, Andreas J. Forstner, Josef Frank, Héléna A. Gaspar, Michael Gill, Fernando S. Goes, Scott D. Gordon, Jakob Grove, Lynsey S. Hall, Christine Søholm Hansen, Thomas F. Hansen, Stefan Herms, Ian B. Hickie, Per Hoffmann, Georg Homuth, Carsten Horn, Jouke-Jan Hottenga, David M. Hougaard, David M. Howard, Marcus Ising, Rick Jansen, Ian Jones, Lisa A. Jones, Eric Jorgenson, James A. Knowles, Isaac S. Kohane, Julia Kraft, Warren W. Kretzschmar, Zoltán Kutalik, Yihan Li, Penelope A. Lind, Donald J. MacIntyre, Dean F. MacKinnon, Robert M. Maier, Wolfgang Maier, Jonathan Marchini, Hamdi Mbarek, Patrick McGrath, Peter McGuffin, Sarah E. Medland, Divya Mehta, Christel M. Middeldorp, Evelin Mihailov, Yuri Milaneschi, Lili Milani, Francis M. Mondimore, Grant W. Montgomery, Sara Mostafavi, Niamh Mullins, Matthias Nauck, Bernard Ng, Michel G. Nivard, Dale R. Nyholt, Paul F. O’Reilly, Hogni Oskarsson, Michael J. Owen, Jodie N. Painter, Carsten Bøcker Pedersen, Marianne Giørtz Pedersen, Roseann E. Peterson, Wouter J. Peyrot, Giorgio Pistis, Danielle Posthuma, Jorge A. Quiroz, Per Qvist, John P. Rice, Brien P. Riley, Margarita Rivera, Saira Saeed Mirza, Robert Schoevers, Eva C. Schulte, Ling Shen, Jianxin Shi, Stanley I. Shyn, Engilbert Sigurdsson, Grant C.B. Sinnamon, Johannes H. Smit, Daniel J. Smith, Hreinn Stefansson, Stacy Steinberg, Fabian Streit, Jana Strohmaier, Katherine E. Tansey, Henning Teismann, Alexander Teumer, Wesley Thompson, Pippa A. Thomson, Thorgeir E. Thorgeirsson, Matthew Traylor, Jens Treutlein, Vassily Trubetskoy, André G. Uitterlinden, Daniel Umbricht, Sandra Van der Auwera, Albert M. van Hemert, Alexander Viktorin, Peter M. Visscher, Yunpeng Wang, Bradley T. Webb, Shantel Marie Weinsheimer, Jürgen Wellmann, Gonneke Willemsen, Stephanie H. Witt, Yang Wu, Hualin S. Xi, Jian Yang, Futao Zhang, Volker Arolt, Bernhard T. Baune, Klaus Berger, Dorret I. Boomsma, Sven Cichon, Udo Dannlowski, E.J.C. de Geus, J. Raymond DePaulo, Enrico Domenici, Katharina Domschke, Tõnu Esko, Hans J. Grabe, Steven P. Hamilton, Caroline Hayward, Andrew C. Heath, Kenneth S. Kendler, Stefan Kloiber, Glyn Lewis, Qingqin S. Li, Susanne Lucae, Pamela A.F. Madden, Patrik K. Magnusson, Nicholas G. Martin, Andrew M. McIntosh, Andres Metspalu, Ole Mors, Preben Bo Mortensen, Bertram Müller-Myhsok, Merete Nordentoft, Markus M. Nöthen, Michael C. O’Donovan, Sara A. Paciga, Nancy L. Pedersen, Brenda W.J.H. Penninx, Roy H. Perlis, David J. Porteous, James B. Potash, Martin Preisig, Marcella Rietschel, Catherine Schaefer, Thomas G. Schulze, Jordan W. Smoller, Kari Stefansson, Henning Tiemeier, Rudolf Uher, Henry Völzke, Myrna M. Weissman, Thomas Werge, Cathryn M. Lewis, Douglas F. Levinson, Gerome Breen, Anders D. Børglum, and Patrick F. Sullivan
Supplementary Material
References
- 1.Vos T., Allen C., Arora M., Barber R.M., Bhutta Z.A., Brown A., et al. Global, regional, and national incidence, prevalence, and years lived with disability for 310 diseases and injuries, 1990–2015: A systematic analysis for the Global Burden of Disease Study 2015. Lancet. 2016;388:1545–1602. doi: 10.1016/S0140-6736(16)31678-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brådvik L., Mattisson C., Bogren M., Nettelbladt P. Long-term suicide risk of depression in the Lundby cohort 1947–1997—Severity and gender. Acta Psychiatr Scand. 2008;117:185–191. doi: 10.1111/j.1600-0447.2007.01136.x. [DOI] [PubMed] [Google Scholar]
- 3.Sobocki P., Jönsson B., Angst J., Rehnberg C. Cost of depression in Europe. J Ment Health Policy Econ. 2006;9:87–98. [PubMed] [Google Scholar]
- 4.Pratt L.A., Brody D.J., Gu Q. Antidepressant use among persons aged 12 and over: United States, 2011-2014. NCHS Data Brief. 2017;283:1–8. [PubMed] [Google Scholar]
- 5.Iacobucci G. NHS prescribed record number of antidepressants last year. BMJ. 2019;364:l1508. doi: 10.1136/bmj.l1508. [DOI] [PubMed] [Google Scholar]
- 6.Cipriani A., Furukawa T.A., Salanti G., Chaimani A., Atkinson L.Z., Ogawa Y., et al. Comparative efficacy and acceptability of 21 antidepressant drugs for the acute treatment of adults with major depressive disorder: A systematic review and network meta-analysis. Focus (Am Psychiatr Pub) 2018;16:420–429. doi: 10.1176/appi.focus.16407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Trivedi M.H., Rush A.J., Wisniewski S.R., Nierenberg A.A., Warden D., Ritz L., et al. Evaluation of outcomes with citalopram for depression using measurement-based care in STAR∗ D: Implications for clinical practice. Am J Psychiatry. 2006;163:28–40. doi: 10.1176/appi.ajp.163.1.28. [DOI] [PubMed] [Google Scholar]
- 8.Souery D., Serretti A., Calati R., Oswald P., Massat I., Konstantinidis A., et al. Switching antidepressant class does not improve response or remission in treatment-resistant depression. J Clin Psychopharmacol. 2011;31:512–516. doi: 10.1097/JCP.0b013e3182228619. [DOI] [PubMed] [Google Scholar]
- 9.Wang S.-M., Han C., Bahk W.-M., Lee S.-J., Patkar A.A., Masand P.S., Pae C.-U. Addressing the side effects of contemporary antidepressant drugs: A comprehensive review. Chonnam Med J. 2018;54:101–112. doi: 10.4068/cmj.2018.54.2.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grasmäder K., Verwohlt P.L., Rietschel M., Dragicevic A., Müller M., Hiemke C., et al. Impact of polymorphisms of cytochrome-P450 isoenzymes 2C9, 2C19 and 2D6 on plasma concentrations and clinical effects of antidepressants in a naturalistic clinical setting. Eur J Clin Pharmacol. 2004;60:329–336. doi: 10.1007/s00228-004-0766-8. [DOI] [PubMed] [Google Scholar]
- 11.McAlpine D.E., Biernacka J.M., Mrazek D.A., O’Kane D.J., Stevens S.R., Langman L.J., et al. Effect of cytochrome P450 enzyme polymorphisms on pharmacokinetics of venlafaxine. Ther Drug Monit. 2011;33:14–20. doi: 10.1097/FTD.0b013e3181fcf94d. [DOI] [PubMed] [Google Scholar]
- 12.Huezo-Diaz P., Perroud N., Spencer E.P., Smith R., Sim S., Virding S., et al. CYP2C19 genotype predicts steady state escitalopram concentration in GENDEP. J Psychopharmacol. 2012;26:398–407. doi: 10.1177/0269881111414451. [DOI] [PubMed] [Google Scholar]
- 13.van Westrhenen R., Aitchison K.J., Ingelman-Sundberg M., Jukić M.M. Pharmacogenomics of antidepressant and antipsychotic treatment: How far have we got and where are we going? Front Psychiatry. 2020;11:94. doi: 10.3389/fpsyt.2020.00094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Solomon H.V., Cates K.W., Li K.J. Does obtaining CYP2D6 and CYP2C19 pharmacogenetic testing predict antidepressant response or adverse drug reactions? Psychiatry Res. 2019;271:604–613. doi: 10.1016/j.psychres.2018.12.053. [DOI] [PubMed] [Google Scholar]
- 15.GENDEP Investigators. MARS Investigators. STAR∗D Investigators Common genetic variation and antidepressant efficacy in major depressive disorder: A meta-analysis of three genome-wide pharmacogenetic studies. Am J Psychiatry. 2013;170:207–217. doi: 10.1176/appi.ajp.2012.12020237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Biernacka J.M., Sangkuhl K., Jenkins G., Whaley R.M., Barman P., Batzler A., et al. The International SSRI Pharmacogenomics Consortium (ISPC): A genome-wide association study of antidepressant treatment response. Transl Psychiatry. 2015;5:e553. doi: 10.1038/tp.2015.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fabbri C., Kasper S., Kautzky A., Bartova L., Dold M., Zohar J., et al. Genome-wide association study of treatment-resistance in depression and meta-analysis of three independent samples, 2018/11/23. Br J Psychiatry. 2019;214:36–41. doi: 10.1192/bjp.2018.256. [DOI] [PubMed] [Google Scholar]
- 18.Tansey K.E., Guipponi M., Perroud N., Bondolfi G., Domenici E., Evans D., et al. Genetic predictors of response to serotonergic and noradrenergic antidepressants in major depressive disorder: A genome-wide analysis of individual-level data and a meta-analysis. PLoS Med. 2012;9 doi: 10.1371/journal.pmed.1001326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fabbri C., Tansey K.E., Perlis R.H., Hauser J., Henigsberg N., Maier W., et al. New insights into the pharmacogenomics of antidepressant response from the GENDEP and STAR∗ D studies: Rare variant analysis and high-density imputation. Pharmacogenomics J. 2018;18:413–421. doi: 10.1038/tpj.2017.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tansey K.E., Guipponi M., Hu X., Domenici E., Lewis G., Malafosse A., et al. Contribution of common genetic variants to antidepressant response. Biol Psychiatry. 2013;73:679–682. doi: 10.1016/j.biopsych.2012.10.030. [DOI] [PubMed] [Google Scholar]
- 21.Wigmore E.M., Hafferty J.D., Hall L.S., Howard D.M., Clarke T.-K., Fabbri C., et al. Genome-wide association study of antidepressant treatment resistance in a population-based cohort using health service prescription data and meta-analysis with GENDEP. Pharmacogenomics J. 2020;202:329–341. doi: 10.1038/s41397-019-0067-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lam M., Awasthi S., Watson H.J., Goldstein J., Panagiotaropoulou G., Trubetskoy V., et al. RICOPILI: Rapid Imputation for COnsortias PIpeLIne. Bioinformatics. 2020;36:930–933. doi: 10.1093/bioinformatics/btz633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.de Leeuw C.A., Mooij J.M., Heskes T., Posthuma D. MAGMA: Generalized gene-set analysis of GWAS data. PLoS Comput Biol. 2015;11 doi: 10.1371/journal.pcbi.1004219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gusev A., Ko A., Shi H., Bhatia G., Chung W., Penninx B.W.J.H., et al. Integrative approaches for large-scale transcriptome-wide association studies. Nat Genet. 2016;48:245–252. doi: 10.1038/ng.3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pain O., Pocklington A.J., Holmans P.A., Bray N.J., O’Brien H.E., Hall L.S., et al. Novel insight into the aetiology of autism spectrum disorder gained by integrating expression data with genome-wide association statistics. Biol Psychiatry. 2019;86:265–273. doi: 10.1016/j.biopsych.2019.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Giambartolomei C., Vukcevic D., Schadt E.E., Franke L., Hingorani A.D., Wallace C., Plagnol V. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet. 2014;10 doi: 10.1371/journal.pgen.1004383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang J., Lee S.H., Goddard M.E., Visscher P.M. GCTA: A tool for genome-wide complex trait analysis. Am J Hum Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee S.H., Yang J., Chen G.-B., Ripke S., Stahl E.A., Hultman C.M., et al. Estimation of SNP heritability from dense genotype data. Am J Hum Genet. 2013;93:1151–1155. doi: 10.1016/j.ajhg.2013.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Choi S.W., O’Reilly P.F. PRSice-2: Polygenic Risk Score software for biobank-scale data. Gigascience. 2019;8:giz082. doi: 10.1093/gigascience/giz082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wray N.R., Ripke S., Mattheisen M., Trzaskowski M., Byrne E.M., Abdellaoui A., et al. Genome-wide association analyses identify 44 risk variants and refine the genetic architecture of major depression. Nat Genet. 2018;50:668–681. doi: 10.1038/s41588-018-0090-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stahl E.A., Breen G., Forstner A.J., McQuillin A., Ripke S., Trubetskoy V., et al. Genome-wide association study identifies 30 loci associated with bipolar disorder. Nat Genet. 2019;51:793–803. doi: 10.1038/s41588-019-0397-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pardiñas A.F., Holmans P., Pocklington A.J., Escott-Price V., Ripke S., Carrera N., et al. Common schizophrenia alleles are enriched in mutation-intolerant genes and in regions under strong background selection. Nat Genet. 2018;50:381–389. doi: 10.1038/s41588-018-0059-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Demontis D., Walters R.K., Martin J., Mattheisen M., Als T.D., Agerbo E., et al. Discovery of the first genome-wide significant risk loci for attention deficit/hyperactivity disorder. Nat Genet. 2019;51:63–75. doi: 10.1038/s41588-018-0269-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grove J., Ripke S., Als T.D., Mattheisen M., Walters R.K., Won H., et al. Identification of common genetic risk variants for autism spectrum disorder. Nat Genet. 2019;51:431–444. doi: 10.1038/s41588-019-0344-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Purves K.L., Coleman J.R.I., Meier S.M., Rayner C., Davis K.A.S., Cheesman R., et al. A major role for common genetic variation in anxiety disorders. Mol Psychiatry. 2020;25:3292–3303. doi: 10.1038/s41380-019-0559-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sanchez-Roige S., Palmer A.A., Fontanillas P., Elson S.L., 23andMe Research Team, the Substance Use Disorder Working Group of the Psychiatric Genetics Consortium, et al. Genome-wide association study meta-analysis of the Alcohol Use Disorders Identification Test (AUDIT) in two population-based cohorts. Am J Psychiatry. 2018;176:107–118. doi: 10.1176/appi.ajp.2018.18040369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee J.J., Wedow R., Okbay A., Kong E., Maghzian O., Zacher M., et al. Gene discovery and polygenic prediction from a genome-wide association study of educational attainment in 1.1 million individuals. Nat Genet. 2018;50:1112–1121. doi: 10.1038/s41588-018-0147-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Palla L., Dudbridge F. A fast method that uses polygenic scores to estimate the variance explained by genome-wide marker panels and the proportion of variants affecting a trait. Am J Hum Genet. 2015;97:250–259. doi: 10.1016/j.ajhg.2015.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bulik-Sullivan B., Finucane H.K., Anttila V., Day F.R., ReproGen Consortium, Psychiatric Genetics Consortium, et al. An atlas of genetic correlations across human diseases and traits. Nat Genet. 2015;47:1236–1241. doi: 10.1038/ng.3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rapaport M.H., Gharabawi G.M., Canuso C.M., Mahmoud R.A., Keller M.B., Bossie C.A., et al. Effects of risperidone augmentation in patients with treatment-resistant depression: Results of open-label treatment followed by double-blind continuation. Neuropsychopharmacology. 2006;31:2505–2513. doi: 10.1038/sj.npp.1301113. [DOI] [PubMed] [Google Scholar]
- 41.Ju C., Fiori L.M., Belzeaux R., Theroux J.-F., Chen G.G., Aouabed Z., et al. Integrated genome-wide methylation and expression analyses reveal functional predictors of response to antidepressants. Transl Psychiatry. 2019;9:254. doi: 10.1038/s41398-019-0589-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lenze E.J., Mulsant B.H., Blumberger D.M., Karp J.F., Newcomer J.W., Anderson S.J., et al. Efficacy, safety, and tolerability of augmentation pharmacotherapy with aripiprazole for treatment-resistant depression in late life: A randomised, double-blind, placebo-controlled trial. Lancet. 2015;386:2404–2412. doi: 10.1016/S0140-6736(15)00308-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Byrne E.M., Kirk K.M., Medland S.E., McGrath J.J., Colodro-Conde L., Parker R., et al. Cohort profile: The Australian genetics of depression study. BMJ Open. 2020;10(5):e032580. doi: 10.1136/bmjopen-2019-032580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim Y., Zerwas S., Trace S.E., Sullivan P.F. Schizophrenia genetics: Where next? Schizophr Bull. 2011;37:456–463. doi: 10.1093/schbul/sbr031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Perlman K., Benrimoh D., Israel S., Rollins C., Brown E., Tunteng J.-F., et al. A systematic meta-review of predictors of antidepressant treatment outcome in major depressive disorder. J Affect Disord. 2019;243:503–515. doi: 10.1016/j.jad.2018.09.067. [DOI] [PubMed] [Google Scholar]
- 46.Zhou X., Keitner G.I., Qin B., Ravindran A.V., Bauer M., Del Giovane C., et al. Atypical antipsychotic augmentation for treatment-resistant depression: A systematic review and network meta-analysis. Int J Neuropsychopharmacol. 2015;18:pyv060. doi: 10.1093/ijnp/pyv060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Andersson E., Crowley J.J., Lindefors N., Ljótsson B., Hedman-Lagerlöf E., Boberg J., et al. Genetics of response to cognitive behavior therapy in adults with major depression: A preliminary report. Mol Psychiatry. 2019;24:484–490. doi: 10.1038/s41380-018-0289-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cohen A., Gilman S.E., Houck P.R., Szanto K., Reynolds C.F. Socioeconomic status and anxiety as predictors of antidepressant treatment response and suicidal ideation in older adults. Soc Psychiatry Psychiatr Epidemiol. 2009;44:272–277. doi: 10.1007/s00127-008-0436-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fontanet P.A., Ríos A.S., Alsina F.C., Paratcha G., Ledda F. Pea3 transcription factors, Etv4 and Etv5, are required for proper hippocampal dendrite development and plasticity. Cereb Cortex. 2018;28:236–249. doi: 10.1093/cercor/bhw372. [DOI] [PubMed] [Google Scholar]
- 50.Liu W., Ge T., Leng Y., Pan Z., Fan J., Yang W., Cui R. The role of neural plasticity in depression: From hippocampus to prefrontal cortex. Neural Plast. 2017;2017:6871089. doi: 10.1155/2017/6871089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jurica M.S., Licklider L.J., Gygi S.R., Grigorieff N., Moore M.J. Purification and characterization of native spliceosomes suitable for three-dimensional structural analysis. RNA. 2002;8:426–439. doi: 10.1017/s1355838202021088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Garriock H.A., Kraft J.B., Shyn S.I., Peters E.J., Yokoyama J.S., Jenkins G.D., et al. A genomewide association study of citalopram response in major depressive disorder. Biol Psychiatry. 2010;67:133–138. doi: 10.1016/j.biopsych.2009.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Uher R., Perroud N., Ng M.Y.M., Hauser J., Henigsberg N., Maier W., et al. Genome-wide pharmacogenetics of antidepressant response in the GENDEP project. Am J Psychiatry. 2010;167:555–564. doi: 10.1176/appi.ajp.2009.09070932. [DOI] [PubMed] [Google Scholar]
- 54.Mrazek D.A., Biernacka J.M., McAlpine D.E., Benitez J., Karpyak V.M., Williams M.D., et al. Treatment outcomes of depression: The pharmacogenomic research network antidepressant medication pharmacogenomic study. J Clin Psychopharmacol. 2014;34:313–317. doi: 10.1097/JCP.0000000000000099. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.