Abstract
The structure and post-translational processing of the SARS-CoV-2 spike glycoprotein (S) is intimately associated with the function of the virus and of sterilising vaccines. The surface of the S protein is extensively modified by glycans, and their biosynthesis is driven by both the wider cellular context, and importantly, the underlining protein structure and local glycan density. Comparison of virally derived S protein with both recombinantly derived and adenovirally induced proteins, reveal hotspots of protein-directed glycosylation that drive conserved glycosylation motifs. Molecular dynamics simulations revealed that, while the S surface is extensively shielded by N-glycans, it presents regions vulnerable to neutralising antibodies. Furthermore, glycans have been shown to influence the accessibility of the receptor binding domain and the binding to the cellular receptor. The emerging picture is one of unifying, principles of S protein glycosylation and an intimate role of glycosylation in immunogen structure and efficacy.
Graphical abstract
Introduction
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) spike (S) glycoprotein is important for the assembly and stability of the virus [1]. The entry of SARS-CoV-2 is primarily mediated by the interaction between S and the ACE2 receptor, expressed on the surface of the host cell [2,3]. The S glycoprotein comprises two subunits, S1 and S2. The receptor-binding domain (RBD) in S1 subunit interacts with the ACE2 receptor [4]. The S protein is extensively glycosylated, typically encoding 66 N-linked glycosylation sites per trimer and a lower number of O-linked glycans present at low occupancy [5∗∗, 6, 7]. The high density of viral glycosylation has prompted investigations into the functional role of glycans in a wide range of settings, from its impact on protein folding to its influence on immunogenicity.
The trimeric S protein is a key target for sterilising antibody responses [8,9] and a key component of all currently approved vaccines [10, 11, 12, 13, 14, 15]. S protein has also been used as reagent in serological testing [16,17], principally because of its primary role in viral infection and its exposure on the viral surface. There is a wide range of S protein constructs across the currently approved vaccines, which rarely utilise native sequences, with exception of the Sinovac vaccine construct [12] which corresponds to a deactivated virus with a native S protein sequence. Other formats are based on a range of non-native sequences and even the effective adenoviral-based ChAdOx1 nCoV-19 (AZD1222) vaccine [18] encoding native S protein sequence is codon optimised to improve expression level in human cell lines. Other design features used within the trimeric S protein formats include C-terminal truncations to remove the hydrophobic transmembrane regions required for solubilisation and the use of stabilising mutations, such as in the 2P, HexaPro, and S-closed formats [19, 20, 21]. For example, the RNA-based Pfizer/BioNTech vaccine encodes a codon-optimised S protein with the 2P mutations [10]. The proline mutations are incorporated in these formats to maintain the S protein in pre-fusion conformation as there is enhanced presentation of neutralising antibody (NAb) epitopes compared to the post-fusion conformation [22]. Notably, not all vaccine formats rely on trimeric S protein and the RBD-based constructs have shown promising immunogenicity [23] and safety [24].
The wide range of constructs at the basis of currently approved SARS-CoV-2 vaccines suggests that immunogenicity is not sensitive to precise glycan processing. In one extreme, robust immune responses have been generated from material derived from insects [25, 26, 27∗], plants [28,29], and mammalian expression systems [10,15]. Even in mammalian derived immunogens, wide range of cell-specific glycosylation is utilised from common recombinant expression systems such as HEK 293 to in vivo production by adenoviral and RNA-delivery systems [10,13]. In the latter case, it is anticipated that the bulk of production is located within phagocytic immune cells, which would be expected to impact cellular-specific glycosylation [30]. Previous studies in HIV-1 have shown differences in glycan composition at the basis of producer cells such as presence of lactosaminoglycans in macrophages whereas these glycan compositions are absent in peripheral blood mononuclear cells (PBMCs) [30]. Despite the apparent insensitivity of the immune response to glycan processing, terminal glycan structures could have implications for viral infectivity [31]. For example, S protein derived from pseudovirus expressed in lung epithelial cells have shown higher processing such as high sialylation content compared to protein expressed in HEK 293T cells and has shown to be associated with increased infectivity [31]. However, the fine processing of glycans does not play critical role in influencing immunogenicity, in fact, truncation of S protein glycosylation enhances the immune response [31]. Even the bulk change in glycosylation of S protein using chemical inhibitor, kifunensine, does not influence serological properties [32]. Despite these observations, at site-specific level, glycans on S are critical for maintaining the quaternary structure [33] and thus indirectly influences immunogenicity and antigenicity. Furthermore, in HIV-1, immunogen glycosylation is known to enhance the humoral immunity by influencing antigen trafficking to immune cells [34]. Overall, the glycosylation of SARS-CoV-2 S protein has a weaker impact on the immunological properties compared to more densely glycosylated protein such as HIV-1 [35].
Beyond the impact of glycans on the immune response to immunogens, there is growing evidence that viral glycosylation contributes to the inflammatory response to viral material [36]. Thus, understanding the fine structure of SARS-CoV-2 S protein glycan shield on the virus and of different immunogens can contribute to the understanding of the role of glycosylation in natural infection and vaccine development. Here, we review the parameters that are structurally affecting the glycosylation of the S protein and assess the extent to which recombinant S protein mimics that of the native virus (Figure 1 ).
Figure 1.
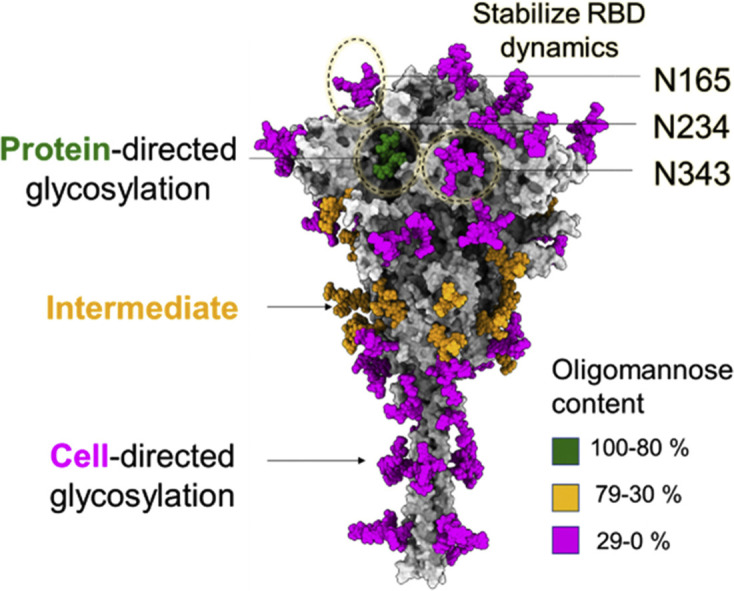
Glycosylation of viral-derived and recombinant spike protein. (a) Representation of cryo-electron tomograph (Cryo-ET) slice of SARS-CoV-2 virions reproduced from Yao et al. [37]. (b) Model of representative SARS-CoV-2 virus highlighting one S protein on the envelope, has been adapted to highlight a single spike from Yao et al. [37]. (c) Fine glycan processing of viral S representing content of oligomannose-type glycans and heat plot of percentage of oligomannose-type glycans of viral-derived S protein, using reported abundances [38,39]. (d) Glycan processing of recombinant S, values of which obtained from data produced earlier [38,39]. The glycan composition is categorised in three groups based on the abundance of oligomannose-type glycan content: green (100%–80%), orange (79%–30%) and magenta (29%–0%). The heat plot represents the percentage of oligomannose-type glycans at each site from a scale of 0% (white) to 100% (green).
Role of glycans in protein folding and stability
The N-linked glycosylation of S protein is essential for folding and assembly in both recombinant formats and in viral settings. Glycosylation mediates interactions with the calnexin/calreticulin folding pathway and secretion checkpoint. Inhibitors targeting entry into the pathway, for example, inhibition of α-glucosidases by iminosugar-containing/based compounds, such as N-butyldeoxynojirimycin (NB-DNJ) and N-butyldeoxygalactonojirimicin (NB-DGJ), have led to a marked reduction in viral infectivity [40]. Beyond the capacity for glycans to mediate chaperone interactions, they can also impart physicochemical stability. Furthermore, the high degree of N-linked glycosylation of the SARS-CoV-2 S protein suggests a selective pressure on the virus that has driven the density of the glycan shield. Among the many different key functions driving the evolution of the shield, glycans can be aiding S folding and structural integrity [33,41], facilitate the interaction with cellular factors [42,43], or be shielding underlying epitopes [31∗∗, 33∗∗]. Because of this key functional role, the glycan shield evolved significantly along the phylogeny, and it is continually evolving [45], with the very recent loss of N370 glycosylation due to T372A mutation in SARS-CoV-2. Through molecular dynamics (MD) simulations, it has been observed that presence of both N234 and N370 results in tying the closed RBDs together, and likely hinders the RBD opening [45, 46, 47]. This possibly explains the absence of N370 glycan site in SARS-CoV-2. Similarly, we can speculate that SARS-CoV-2 glycosylation carries the signature of selective pressure from its host prior to zoonosis.
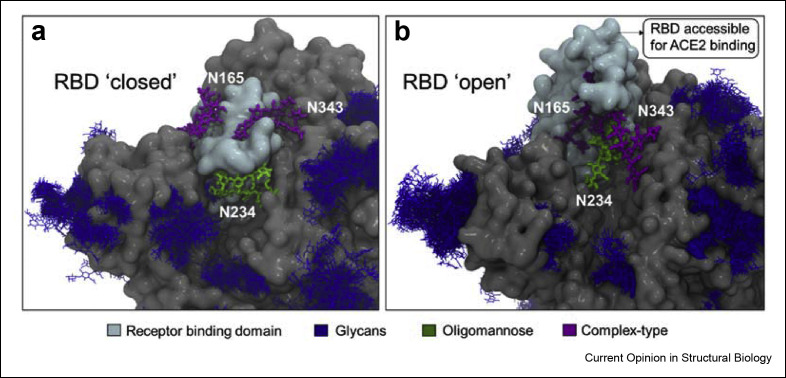
There is a direct relationship between the spatial accessibility of the glycosylation sites and the processing state [48,49]. Within the context of the S glycoprotein, MD simulations have shown that the N-glycan at position N234 is one of the least accessible, in both closed and open S conformations see Figure 2 , which explains the high abundance of large oligomannose-type glycans at this site [6,33]. The N234 glycan is also one the most ordered glycans observed in cryo-EM structures [19] and this glycan has also been shown by MD simulations to stabilise the RBD open conformation and its dynamics [33,45]. MD simulations experiments [33] have shown that the N234 glycan is able to access and effectively fill the cleft left vacant by the opening of the RBD and thus supporting its stability. This function is also supported by the N165 and N343 glycans, located in proximity (Figure 2). Indeed, deletion of these glycans through single point N234A, N165A and N343A mutations corresponds to a significantly reduced binding to the ACE2 receptor [33,41,50]. However, none of these single point mutations fully abrogate the binding suggesting that multiple residues are involved in RBD opening [33,41]. Further insight from MD and cryo-electron micrograph (cryo-EM) studies have shown that glycosylation at N343 is essential for gating the RBD opening and closing [41,45]. Overall, the interaction of the clustered N165, N234, and N343 glycosylation sites with the surrounding protein helps stabilise the orientation and dynamics of the open RBD conformation allowing the different possible orientations and thereby supporting receptor recognition [33,41,45] (Figure 2b).
Figure 2.
Role of glycans in stabilizing dynamics of RBD. Model representation of a) RBD ‘closed’ and b) RBD ‘open’ showing the interaction between neighbouring glycans, N165, N234 and N343. The RBD is accessible for ACE2 binding in ‘open’ state. The N343 glycan act as a “glycan gate” as it pushes the RBD from ‘down’ to the ‘up’ conformation. The N234 is modelled with Man9GlcNAc2 glycan represented in green, N165 and N343 are modelled with biantennary complex-type glycans [38,39]. The RBD is shown in cyan, and the remaining S protein highlighted in grey. These models are reproduced from previous studies [41,45].
Role of protein structure in glycan maturation
The glycosylation processing of viral glycoproteins is highly dependent on the accessibility of the glycan sites to the glycan processing enzymes in the ER and Golgi apparatus [51,52]. The glycan maturation is strongly influenced by the glycosylation site accessibility and can sterically be impaired by the local architecture of the protein [52]. In contrast to N-linked glycans which are added co-translationally in the ER, O-linked glycans are added in the Golgi apparatus and their presence is influenced by both the intrinsic susceptibility of the sequence to modification but also the local steric accessibility. Importantly, the proximity of potential O-glycosylation sites to the furin cleavage site, that generates the separate S1 and S2 subunits, means that their modification can impact viral maturation and infectivity [53].
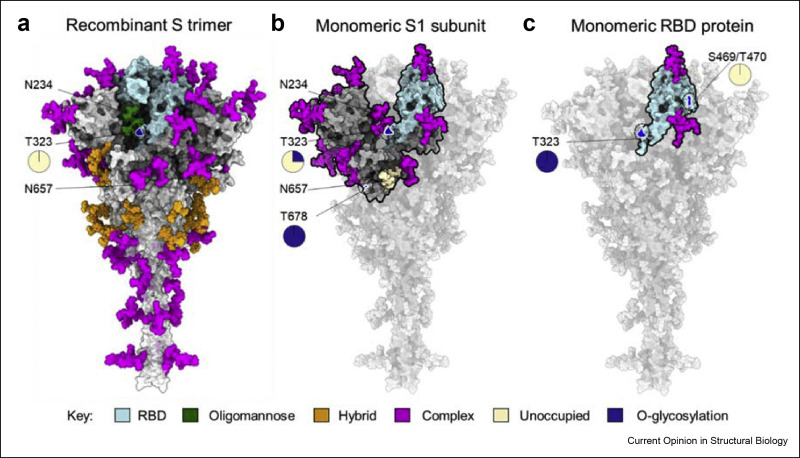
One extreme example of steric constraints limiting glycan maturation within the S protein is the N234 site where limited enzymatic access results in almost exclusively oligomannose-type glycans being presented at that site [6,33,38] (Figure 3 a). More broadly, comparison of S protein from wide variety of laboratories and expressed in different mammalian cell lines demonstrated consistent subpopulation of oligomannose-type glycans, indicating that this is an intrinsic property of the glycoprotein [38]. These mannose signatures were also observed in spike protein derived from infectious virus [38,39]. However, when only the S1 subunit is expressed, N234 is fully processed to complex-type glycosylation [39] consistent with the relaxation of steric restrictions to processing in the monomeric form [7,27,39] (Figure 3b).
Figure 3.
The influence of protein structure on glycan maturation. (a) Illustration of glycan composition of recombinant trimeric S protein of which values reproduced from Eldrid et al. [7]. The N-linked glycosylation takes place at specific sequon, Asn-X-Ser/Thr (X is any amino acid except proline) whereas O-linked glycosylation is not dictated by specific sequon and occurs on serine and threonine in exposed regions. The N-linked glycosylation is presented in three categories on basis of oligomannose content as described in Figure 1, oligomannose (green), hybrid (orange) and complex-type (magenta) glycans. The O-linked glycosylation at T323 site (see magnification) on trimeric S is present at low levels (0.2%) of which values obtained from Eldrid et al. [7]. (b) The glycan composition of recombinant monomeric S1 subunit of which values reproduced from Wang et al. [27] and Brun et al. [39]. Most of the N-glycan sites on S1 subunit is highly processed represented in magenta except N657 which is unoccupied, represented in wheat color. The O-glycosylation is present on S1 subunit at sites, T323 and T678 (see magnification). (e) The glycan composition of monomeric RBD protein (cyan) which binds to main host receptor (ACE2). The N-glycan sites of RBD are highly processed and are represented in magenta, values reproduced from Allen et al. [38]. The O-glycosylation was observed on monomeric RBD protein at sites T323 and S469/T470 (see magnification) [7].
Sialylation was another significant difference observed between S1 and trimeric S, the former expressing high glycan content of sialylated glycans with high branching compared to the latter which showed low sialylated glycan content including mostly mono-sialylated glycans [27]. Likewise, an increase in glycan content of O-glycosylation was observed in S1 recombinant protein compared to trimeric S. For example, T323 is more occupied in S1 recombinant protein, and T678 O-glycosylation was observed only on the isolated S1 subunit suggesting more accessibility to Golgi resident transferases [39,53] (Figure 3b). However, within the native trimeric sequence furin cleavage unmasks the T678 site which is modified around 25% with O-glycan structures [39]. Furthermore, when isolated RBD was expressed as a recombinant protein reveals high occupancy of O-glycosylation compared to trimeric S protein [7]. For example, T323 is fully occupied in monomeric RBD whereas this site shows low occupancy on trimeric S-protein [7]. In addition, the T470 O-glycosylation site was identified only on monomeric RBD, albeit at very low abundance [7] (Figure 3c). Similarly, the N-glycosylation sites, N331 and N343 of monomeric RBD revealed elevated branching and terminal processing when compared to trimeric S-protein [38]. These glycan characterisations of different viral protein formats highlight the impact of protein architecture on glycan structure. Although a wide variety of protein formats have shown to be effective vaccines, the changes in glycosylation may impact the immunogen efficacy, as has been observed in the case of HIV-1 [34].
Role of glycans in viral entry
The initial step for SARS-CoV-2 infection in the host cell is to bind with the host cell receptor and enter the target cells. While additional factors can enhance entry (see below), entry is principally mediated by S-ACE2 interaction [4,5,54]. ACE2 is ubiquitously expressed on epithelial, endothelial, and blood cells [55]. The viral S protein and ACE2 receptor both are extensively glycosylated, with ACE2 exhibiting 14 N-linked glycans across the dimer [5,6,19]. O-glycosylation was found at low levels on both ACE2 and S protein [5,7,39]. Previous studies have determined the effect of N and O-glycosylation processing of both S and ACE2 on S-ACE2 binding [56,57]. The glycan engineering of ACE2 did not significantly impact the ACE2-S binding [56,57], whereas that of S glycosylation had modest influence on binding [57]. In contrast to these recombinant settings, glycan processing of S protein can influence viral entry [57].
Beyond the impact of glycan maturation on receptor recognition, modification of the furin cleavage by O-linked glycosylation can impact viral entry. For example, P681H and P681R mutation found in highly transmissible alpha and delta variants, respectively, decreases O-glycosylation which potentially increases furin cleavage and may influence viral infectivity [53]. Together these results have highlighted the importance of S protein glycosylation on viral entry and abrogation of these glycans using inhibitors provides insight into intervention strategies to target the SARS-CoV-2 infection [58,59].
Although ACE2 is necessary for infection, there are carbohydrate-based attachment factors that can enhance infection particularly in cases where ACE2 expression is limiting [60]. This has the effect of broadening the viral tropism beyond that which is achieved with ACE2 expression alone. For example, cell surface heparan sulfate (HS) is recognised by SARS-CoV-2 S protein at a separate site from the site involved in ACE2 binding and enhances infection [61, 62, 63]. Specifically, HS binds to RBD and enhances the open conformation of RBD, thus exposing the RBD for ACE2 interaction and viral infection [61, 62, 63]. Furthermore, sialylated glycolipids, have shown interaction with RBD and facilitating in viral entry [64], which has also been exploited for diagnostic purposes [65].
Additionally, viral glycans can also be recognised by soluble and cellular lectins which play an important role in recognition. Nuclear magnetic resonance (NMR) studies and MD simulations have revealed the binding of human lectins such as galectins with N331 and N343 glycan sites of RBD [66]. Mannose-binding lectins (MBL) can recognise glycans on S protein, and have shown role in vitro in inhibiting SARS-CoV-2 infection by activation of lectin mediated complement pathway [67]. Furthermore, there are cellular proteases such as transmembrane serine protease 2 (TMPRSS2) which enhance viral infectivity by cleaving both S and ACE2 protein [54]. These attachment factors mediate the viral S protein interaction with ACE2 and their inhibition reduces viral infectivity [54,62,63,68].
Exploiting breaches within the glycan shield
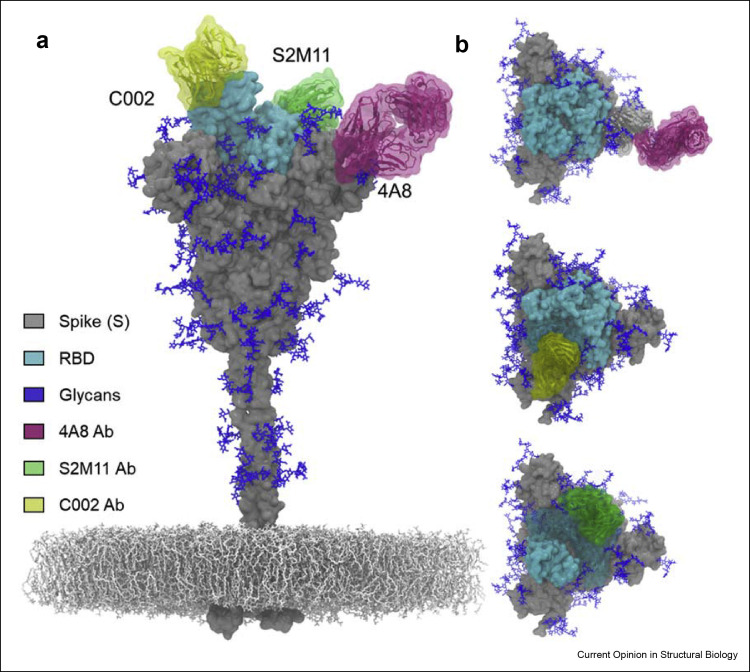
Despite the extensive glycosylation and dynamics of the S protein, antibodies can still recognise breaches within the glycan shield of S protein and can neutralise the virus [33,69]. Previous studies have demonstrated the glycan composition and accessible surface area (ASA) of these glycans using liquid chromatography mass spectrometry (LC-MS) and MD simulations respectively [6,33,38,69]. Despite the extensive coverage of glycans on the head domain, the glycans are sufficiently sparse for there to be still present extensive vulnerabilities to antibody binding [33,69] (Figure 4 a). In contrast, the glycans in the stalk region have shown effective glycan shielding in MD simulation studies suggesting less accessibility for antibodies to bind this region [33,69]. Recent studies have shown the isolation of antibodies against the head region including RBD, RBM (receptor binding motif), NTD (N-terminal domain) and S2 subunit from COVID-19 patients [70, 71, 72, 73]. The antibodies against RBD such as C002 and SM211 have shown recognition of quaternary epitope on the RBD. The C002 neutralising antibody (nAb) binds to a region which spans two RBDs, one in ‘down’ conformation adjacent to RBD in ‘up’ conformation (Figure 4b). The S2M11 antibody binds to two neighbouring RBDs and stabilise the close conformation of trimeric S protein (Figure 4b). Furthermore, Abs have also been elicited against NTD present on the head of the S protein [70,71,74]. One such antibody is 4A8 highlighted in Figure 4b, which recognises the NTD in an identical region despite the dynamics of RBD and have also shown to stabilise the NTD region. The N149 glycan site which is present in NTD might also be involved in NTD-4A8 interaction [70] (Figure 4b).
Figure 4.
Antibody recognition within the breaches of S protein glycan shield. (a) Representation of trimeric S protein with glycans (blue) present on it [33]. The RBD present on the head region of the S is highlighted in cyan. The stalk region is effectively shielded with glycans. The mesh network represents the virion membrane on which S protein is embedded. (b) Illustration of antibodies binding within the glycan shield of S protein obtained by structural alignment (www.pymol.org). The 4A8 nAb (pink, PDB 7C2L) targets the NTD region of the S protein [70]. The C002 neutralising antibody (nAb) (yellow, PBD 7K8T) is binding to RBD ‘up’ conformation [73]. The S2M11 (green, PDB 7K43) nAb recognises a quaternary epitope consisting of two neighbouring RBDs and stabilises the trimeric S in closed state [72].
In addition to the view of accessibility derived from static models, considering the dynamics of the system further enhances our understanding of the antigenic surface. Dynamic motions of the S protein can influence the glycan–glycan interaction and can bring the glycans of different domains in close contact. Using the network analysis approach, the glycans responsible for effective shielding were predicted [69,75,76]. The glycans, N234 and N165, involved in RBD dynamics demonstrated stronger glycan–glycan interaction in the open state compared to the closed state as shown in Figure 2b. The glycans at sites N603 and N616 have been shown to play a central role in connecting the upper head with the lower head as these glycans are responsible for the proper shielding of the S protein [69]. Neutralising antibodies exploit spaces between these shielded region where the glycan density is low or bind to epitopes containing glycans as has been observed in case of HIV-1 (Figure 4b) [75,77].
Further glycobiology perspectives in vaccine development and beyond
It has been established that underlying protein architecture has significant impact on glycosylation [52]. In one extreme trimeric viral spike exhibit oligomannose-type glycans whereas RBD domains are almost entirely devoid of such structures [6,33,38]. Given the known influence of glycan processing on immunogen targeting, it will be fruitful to understand the impact of protein formats on vaccine efficacy. Similarly, there is considerable opportunity to enhance the understanding of RNA-based [10] and adenoviral-based delivery system [13] by considering the range of vaccine materials derived from different cellular sources upon immunisation [39,78]. It is already established that protein modification can influence antigen stability, and it is likely that such modifications will also impact antigen distribution and cellular presentation [34,79,80]. Overall understanding the interplay between protein architecture, production routes, and glycan processing will illuminate how viral immunogens work and aid in their optimisation.
Conflict of interest statement
None declared.
Acknowledgement
This work was supported by the International AIDS Vaccine Initiative (IAVI) through the Collaboration for AIDS Vaccine Discovery (CAVD) grant, INV-008352/OPP1153692 funded by the Bill and Melinda Gates Foundation. The Science Foundation of Ireland (20/FFP-P/8809) is gratefully acknowledged for financial support. The opinions, findings and conclusions or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of the Science Foundation Ireland.
This review comes from a themed issue on Protein-Carbohydrate complexes and glycosylation
Edited by Sylvie Ricard-Blum and Joseph Zaia
References
- 1.Santopolo S., Riccio A., Santoro M.G. The biogenesis of SARS-CoV-2 spike glycoprotein: multiple targets for host-directed antiviral therapy. Biochem Biophys Res Commun. 2021;538:80–87. doi: 10.1016/j.bbrc.2020.10.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shang J., Ye G., Shi K., Wan Y., Luo C., Aihara H., Geng Q., Auerbach A., Li F. Structural basis of receptor recognition by SARS-CoV-2. Nat. 2020;581:221–224. doi: 10.1038/s41586-020-2179-y. 5817807 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang Q., Zhang Y., Wu L., Niu S., Song C., Zhang Z., Lu G., Qiao C., Hu Y., Yuen K.Y., et al. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell. 2020;181:894–904.e9. doi: 10.1016/j.cell.2020.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- Zhao P., Praissman J.L., Grant O.C., Cai Y., Xiao T., Rosenbalm K.E., Aoki K., Kellman B.P., Bridger R., Barouch D.H., et al. Virus-receptor interactions of glycosylated SARS-CoV-2 spike and human ACE2 receptor. Cell Host Microbe. 2020;28:586–601.e6. doi: 10.1016/j.chom.2020.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]; Comprehensive study of spike and ACE2 receptor using mass spectrometry and molecular dynamics simulation, determined the essential roles for glycosylation.
- 6.Watanabe Y., Allen J.D., Wrapp D., McLellan J.S., Crispin M. Site-specific glycan analysis of the SARS-CoV-2 spike. Science. 2020;369:330–333. doi: 10.1126/science.abb9983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eldrid C.F.S., Allen J.D., Newby M.L., Crispin M. Suppression of O-linked glycosylation of the SARS-CoV-2 spike by quaternary structural restraints. Anal Chem. 2021;93:14392–14400. doi: 10.1021/acs.analchem.1c01772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brouwer P.J.M.M., Caniels T.G., van der Straten K., Snitselaar J.L., Aldon Y., Bangaru S., Torres J.L., Okba N.M.A.A., Claireaux M., Kerster G., et al. Potent neutralizing antibodies from COVID-19 patients define multiple targets of vulnerability. Science. 2020;369:643–650. doi: 10.1126/science.abc5902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ju B., Zhang Q., Ge J., Wang R., Sun J., Ge X., Yu J., Shan S., Zhou B., Song S., et al. Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nat. 2020;584:115–119. doi: 10.1038/s41586-020-2380-z. 5847819 2020. [DOI] [PubMed] [Google Scholar]
- 10.Vogel A.B., Kanevsky I., Che Y., Swanson K.A., Muik A., Vormehr M., Kranz L.M., Walzer K.C., Hein S., Güler A., et al. BNT162b vaccines protect rhesus macaques from SARS-CoV-2. Nature. 2021;592:283–289. doi: 10.1038/s41586-021-03275-y. [DOI] [PubMed] [Google Scholar]
- 11.Corbett K.S., Flynn B., Foulds K.E., Francica J.R., Boyoglu-Barnum S., Werner A.P., Flach B., O'Connell S., Bock K.W., Minai M., et al. vol. 383. 2020. pp. 1544–1555. (Evaluation of the mRNA-1273 vaccine against SARS-CoV-2 in nonhuman primates). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gao Q., Bao L., Mao H., Wang L., Xu K., Yang M., Li Y., Zhu L., Wang N., Lv Z., et al. Development of an inactivated vaccine candidate for SARS-CoV-2. Science. 2020 doi: 10.1126/science.abc1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Folegatti P.M., Ewer K.J., Aley P.K., Angus B., Becker S., Belij-Rammerstorfer S., Bellamy D., Bibi S., Bittaye M., Clutterbuck E.A., et al. Safety and immunogenicity of the ChAdOx1 nCoV-19 vaccine against SARS-CoV-2: a preliminary report of a phase 1/2, single-blind, randomised controlled trial. Lancet. 2020;396:467–478. doi: 10.1016/S0140-6736(20)31604-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mercado N.B., Zahn R., Wegmann F., Loos C., Chandrashekar A., Yu J., Liu J., Peter L., McMahan K., Tostanoski L.H., et al. Single-shot Ad26 vaccine protects against SARS-CoV-2 in rhesus macaques. Nat. 2020;586:583–588. doi: 10.1038/s41586-020-2607-z. 5867830 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bos R., Rutten L., van der Lubbe J.E.M., Bakkers M.J.G., Hardenberg G., Wegmann F., Zuijdgeest D., de Wilde A.H., Koornneef A., Verwilligen A., et al. Ad26 vector-based COVID-19 vaccine encoding a prefusion-stabilized SARS-CoV-2 Spike immunogen induces potent humoral and cellular immune responses. npj Vaccines. 2020 doi: 10.1038/s41541-020-00243-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Amanat F., Stadlbauer D., Strohmeier S., Nguyen T.H.O.O., Chromikova V., McMahon M., Jiang K., Arunkumar G.A., Jurczyszak D., Polanco J., et al. A serological assay to detect SARS-CoV-2 seroconversion in humans. Nat Med. 2020;26:1033–1036. doi: 10.1038/s41591-020-0913-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Faustini S.E., Jossi S.E., Perez-Toledo M., Shields A.M., Allen J.D., Watanabe Y., Newby M.L., Cook A., Willcox C.R., Salim M., et al. Development of a high-sensitivity ELISA detecting IgG, IgA and IgM antibodies to the SARS-CoV-2 spike glycoprotein in serum and saliva. Immunology. 2021:13349. doi: 10.1111/imm.13349. 00:imm. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van Doremalen N., Lambe T., Spencer A., Belij-Rammerstorfer S., Purushotham J.N., Port J.R., Avanzato V.A., Bushmaker T., Flaxman A., Ulaszewska M., et al. ChAdOx1 nCoV-19 vaccine prevents SARS-CoV-2 pneumonia in rhesus macaques. Nature. 2020;586:578–582. doi: 10.1038/s41586-020-2608-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.-L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hsieh C.-L., Goldsmith J.A., Schaub J.M., DiVenere A.M., Kuo H.-C., Javanmardi K., Le K.C., Wrapp D., Lee A.G., Liu Y., et al. Structure-based design of prefusion-stabilized SARS-CoV-2 spikes. Science. 2020;369:1501–1505. doi: 10.1126/science.abd0826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Juraszek J., Rutten L., Blokland S., Bouchier P., Voorzaat R., Ritschel T., Bakkers M.J.G., Renault L.L.R., Langedijk J.P.M. Stabilizing the closed SARS-CoV-2 spike trimer. Nat Commun. 2021 doi: 10.1038/s41467-020-20321-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sanders R.W., Moore J.P. Virus vaccines: proteins prefer prolines. Cell Host Microbe. 2021;29:327–333. doi: 10.1016/j.chom.2021.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Valdes-Balbin Y., Santana-Mederos D., Paquet F., Fernandez S., Climent Y., Chiodo F., Rodríguez L., Sanchez Ramirez B., Leon K., Hernandez T., et al. Molecular aspects concerning the use of the SARS-CoV-2 receptor binding domain as a target for preventive vaccines. ACS Cent Sci. 2021;7:757–767. doi: 10.1021/acscentsci.1c00216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Toledo-Romani M.E., Garcia-Carmenate M., Silva C.V., Baldoquin-Rodriguez W., Pérez M.M., Gonzalez M.C.R., Moreno B.P., Hernández I.C.M., Romero R.G.-M., Tabio O.S., et al. Efficacy and safety of SOBERANA 02, a COVID-19 conjugate vaccine in heterologous three-dose combination. medRxiv. 2021 doi: 10.1101/2021.10.31.21265703. [DOI] [Google Scholar]
- 25.Li T., Zheng Q., Yu H., Wu D., Xue W., Xiong H., Huang X., Nie M., Yue M., Rong R., et al. SARS-CoV-2 spike produced in insect cells elicits high neutralization titres in non-human primates. Emerg Microb Infect. 2020;9:2076–2090. doi: 10.1080/22221751.2020.1821583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shinde V., Bhikha S., Hoosain Z., Archary M., Bhorat Q., Fairlie L., Lalloo U., Masilela M.S.L., Moodley D., Hanley S., et al. Efficacy of NVX-CoV2373 covid-19 vaccine against the B.1.351 variant. N Engl J Med. 2021;384:1899–1909. doi: 10.1056/NEJMoa2103055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Baudys J., Bundy J.L., Solano M., Keppel T., Barr J.R. Comprehensive analysis of the glycan complement of SARS-CoV-2 spike proteins using signature ions-triggered electron-transfer/higher-energy collisional dissociation (EThcD) mass spectrometry. Anal Chem. 2020;92:14730–14739. doi: 10.1021/acs.analchem.0c03301. [DOI] [PMC free article] [PubMed] [Google Scholar]; Extensive analysis of trimeric S and S1 subunit using mass spectrometry.
- 28.Ward B.J., Gobeil P., Séguin A., Atkins J., Boulay I., Charbonneau P.Y., Couture M., D'Aoust M.A., Dhaliwall J., Finkle C., et al. Phase 1 randomized trial of a plant-derived virus-like particle vaccine for COVID-19. Nat Med. 2021;276:1071–1078. doi: 10.1038/s41591-021-01370-1. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Makatsa M.S., Tincho M.B., Wendoh J.M., Ismail S.D., Nesamari R., Pera F., de Beer S., David A., Jugwanth S., Gededzha M.P., et al. SARS-CoV-2 antigens expressed in plants detect antibody responses in COVID-19 patients. Front Plant Sci. 2021;12:550. doi: 10.3389/fpls.2021.589940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Willey R.L., Shibata R., Freed E.O., Cho M.W., Martin M.A. Differential glycosylation, virion incorporation, and sensitivity to neutralizing antibodies of human immunodeficiency virus type 1 envelope produced from infected primary T-lymphocyte and macrophage cultures. J Virol. 1996;70:6431–6436. doi: 10.1128/jvi.70.9.6431-6436.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H.-Y., Liao H.-Y., Chen X., Wang S.-W., Cheng C.-W., Shahed-Al-Mahmud M., Liu Y.-M., Mohapatra A., Chen T.-H., Lo J.M., et al. Vaccination with SARS-CoV-2 spike protein lacking glycan shields elicits enhanced protective responses in animal models. Sci Transl Med. 2022 doi: 10.1126/scitranslmed.abm0899. [DOI] [PMC free article] [PubMed] [Google Scholar]; Extensive analysis of glycosylation of S protein expressed in different cell lines and also revealed that truncation of glycans on S protein induces enhance protective response.
- 32.Chawla H., Jossi S.E., Faustini S.E., Samsudin F., Allen J.D., Watanabe Y., Newby M.L., Marcial-Juárez E., Lamerton R.E., McLellan J.S., et al. Glycosylation and serological reactivity of an expression-enhanced SARS-CoV-2 viral spike mimetic. J Mol Biol. 2022;434:167332. doi: 10.1016/j.jmb.2021.167332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casalino L., Gaieb Z., Goldsmith J.A., Hjorth C.K., Dommer A.C., Harbison A.M., Fogarty C.A., Barros E.P., Taylor B.C., McLellan J.S., et al. Beyond shielding: the roles of glycans in the SARS-CoV-2 spike protein. ACS Cent Sci. 2020;6:1722–1734. doi: 10.1021/acscentsci.0c01056. [DOI] [PMC free article] [PubMed] [Google Scholar]; Detailed molecular dynamics studies of glycans of SARS-CoV-2, determined glycan sites critical for RBD dynamics.
- 34.Tokatlian T., Read B.J., Jones C.A., Kulp D.W., Menis S., Chang J.Y.H., Steichen J.M., Kumari S., Allen J.D., Dane E.L., et al. Innate immune recognition of glycans targets HIV nanoparticle immunogens to germinal centers. Science. 2019;363:649–654. doi: 10.1126/science.aat9120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mccoy L.E., Falkowska E., Doores K.J., Le K., Sok D., Van Gils M.J., Euler Z. Incomplete neutralization and deviation from sigmoidal neutralization curves for HIV broadly neutralizing monoclonal antibodies. PLoS Pathog. 2015;11:1005110. doi: 10.1371/journal.ppat.1005110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Watanabe Y., Bowden T.A., Wilson I.A., Crispin M. Exploitation of glycosylation in enveloped virus pathobiology. Biochim Biophys Acta Gen Subj. 2019;1863:1480–1497. doi: 10.1016/j.bbagen.2019.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yao H., Song Y., Chen Y., Wu N., Xu J., Sun C., Zhang J., Weng T., Zhang Z., Wu Z., et al. Molecular architecture of the SARS-CoV-2 virus. Cell. 2020;183:730–738.e13. doi: 10.1016/j.cell.2020.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen J.D., Chawla H., Samsudin F., Zuzic L., Shivgan A.T., Watanabe Y., He W., Callaghan S., Song G., Yong P., et al. Site-specific steric control of SARS-CoV-2 spike glycosylation. Biochemistry. 2021;60:2153–2169. doi: 10.1021/acs.biochem.1c00279. [DOI] [PMC free article] [PubMed] [Google Scholar]; Extensive glycan analysis of viral S protein and recombinant S protein produced in different laboratories.
- Brun J., Vasiljevic S., Gangadharan B., Hensen M., V Chandran A., Hill M.L., Kiappes J.L., Dwek R.A., Alonzi D.S., Struwe W.B., et al. Assessing antigen structural integrity through glycosylation analysis of the SARS-CoV-2 viral spike. ACS Cent Sci. 2021;7:586–593. doi: 10.1021/acscentsci.1c00058. [DOI] [PMC free article] [PubMed] [Google Scholar]; Detailed analysis of viral glycan composition, adenovirally associated vaccine (ChAdOx) and S1 subunit using mass spectrometery.
- 40.Casas-Sanchez A., Romero-Ramirez A., Hargreaves E., Ellis C.C., Grajeda B.I., Estevao I.L., Patterson E.I., Hughes G.L., Almeida I.C., Zech T., et al. Inhibition of protein N- glycosylation blocks SARS-CoV-2 infection. MBio. 2022 doi: 10.1128/mbio.03718-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sztain T., Ahn S.H., Bogetti A.T., Casalino L., Goldsmith J.A., Seitz E., McCool R.S., Kearns F.L., Acosta-Reyes F., Maji S., et al. A glycan gate controls opening of the SARS-CoV-2 spike protein. Nat Chem. 2021;1310:963–968. doi: 10.1038/s41557-021-00758-3. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]; Cryo electron micrograph and molecula dynamics studies depicting the role of N343 in stabilizing RBD dynamics.
- 42.Li B., Wang L., Ge H., Zhang X., Ren P., Guo Y., Chen W., Li J., Zhu W., Chen W., et al. Identification of potential binding sites of sialic acids on the RBD domain of SARS-CoV-2 spike protein. Front Chem. 2021:9. doi: 10.3389/fchem.2021.659764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hoffmann D., Mereiter S., Jin Oh Y., Monteil V., Elder E., Zhu R., Canena D., Hain L., Laurent E., Grünwald-Gruber C., et al. Identification of lectin receptors for conserved SARS-CoV-2 glycosylation sites. EMBO J. 2021:40. doi: 10.15252/embj.2021108375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Harbison A.M., Fogarty C.A., Phung T.K., Satheesan A., Schulz B.L., Fadda E. Fine-tuning the spike: role of the nature and topology of the glycan shield in the structure and dynamics of the SARS-CoV-2 S. Chem Sci. 2022 doi: 10.1039/D1SC04832E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kang L., He G., Sharp A.K., Wang X., Brown A.M., Michalak P., Weger-Lucarelli J. A selective sweep in the Spike gene has driven SARS-CoV-2 human adaptation. Cell. 2021;184:4392–4400.e4. doi: 10.1016/j.cell.2021.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang S., Liang Q., He X., Zhao C., Ren W., Yang Z., Wang Z., Ding Q., Deng H., Wang T., et al. Loss of Spike N370 glycosylation as an important evolutionary event for the enhanced infectivity of SARS-CoV-2. Cell Res. 2022 doi: 10.1038/s41422-021-00600-y. 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zacchi L.F., Schulz B.L. N-glycoprotein macroheterogeneity: biological implications and proteomic characterization. Glycoconj J. 2015;33:359–376. doi: 10.1007/s10719-015-9641-3. 333 2015. [DOI] [PubMed] [Google Scholar]
- 49.Mathew C., Weiß R.G., Giese C., Lin C.W., Losfeld M.E., Glockshuber R., Riniker S., Aebi M. Glycan–protein interactions determine kinetics of N-glycan remodeling. RSC Chem Biol. 2021;2:917–931. doi: 10.1039/d1cb00019e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Henderson R., Edwards R.J., Mansouri K., Janowska K., Stalls V., Kopp M., Haynes B.F., Acharya P. Glycans on the SARS-CoV-2 spike control the receptor binding domain conformation. bioRxiv Prepr Serv Biol. 2020 doi: 10.1101/2020.06.26.173765. [DOI] [Google Scholar]
- 51.Hang I., Lin C., Grant O.C., Fleurkens S., Villiger T.K., Soos M., Morbidelli M., Woods R.J., Gauss R., Aebi M. Analysis of site-specific N -glycan remodeling in the endoplasmic reticulum and the Golgi. Glycobiology. 2015;25:1335–1349. doi: 10.1093/glycob/cwv058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rudd P.M., Dwek R.A. Glycosylation: heterogeneity and the 3D structure of proteins. Crit Rev Biochem Mol Biol. 1997;32:1–100. doi: 10.3109/10409239709085144. [DOI] [PubMed] [Google Scholar]
- Zhang L., Mann M., Syed Z.A., Reynolds H.M., Tian E., Samara N.L., Zeldin D.C., Tabak L.A., Ten Hagen K.G. Furin cleavage of the SARS-CoV-2 spike is modulated by O-glycosylation. Proc Natl Acad Sci U S A. 2021;118:2109905118. doi: 10.1073/pnas.2109905118. [DOI] [PMC free article] [PubMed] [Google Scholar]; Detailed enzyme assays provided the evidence for role of O-glycosylation in furin cleavage and thus viral infection.
- 54.Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.-H., Nitsche A., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li W., Moore M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A., Somasundaran M., Sullivan J.L., Luzuriaga K., Greenough T.C., et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Allen J.D., Watanabe Y., Chawla H., Newby M.L., Crispin M. Subtle influence of ACE2 glycan processing on SARS-CoV-2 recognition. J Mol Biol. 2021;433:166762. doi: 10.1016/j.jmb.2020.166762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yang Q., Hughes T.A., Kelkar A., Yu X., Cheng K., Park S.J., Huang W.C., Lovell J.F., Neelamegham S. Inhibition of SARS-CoV-2 viral entry upon blocking N-and O-glycan elaboration. Elife. 2020;9:1–44. doi: 10.7554/eLife.61552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Huang H.C., Lai Y.J., Liao C.C., Yang W.F., Huang K Bin, Lee I.J., Chou W.C., Wang S.H., Wang L.H., Hsu J.M., et al. Targeting conserved N-glycosylation blocks SARS-CoV-2 variant infection in vitro. EBioMedicine. 2021;74:103712. doi: 10.1016/j.ebiom.2021.103712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shang J., Wan Y., Luo C., Ye G., Geng Q., Auerbach A., Li F. Cell entry mechanisms of SARS-CoV-2. Proc Natl Acad Sci U S A. 2020;117:11727–11734. doi: 10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hamming I., Timens W., Bulthuis M.L.C., Lely A.T., Navis G.J., van Goor H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J Pathol. 2004;203:631–637. doi: 10.1002/path.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Clausen T.M., Sandoval D.R., Spliid C.B., Pihl J., Perrett H.R., Painter C.D., Narayanan A., Majowicz S.A., Kwong E.M., McVicar R.N., et al. SARS-CoV-2 infection depends on cellular heparan sulfate and ACE2. Cell. 2020;183:1043–1057.e15. doi: 10.1016/j.cell.2020.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mycroft-West C.J., Su D., Pagani I., Rudd T.R., Elli S., Gandhi N.S., Guimond S.E., Miller G.J., Meneghetti M.C.Z., Nader H.B., et al. Heparin inhibits cellular invasion by SARS-CoV-2: structural dependence of the interaction of the spike S1 receptor-binding domain with heparin. Thromb Haemostasis. 2020;120:1700–1715. doi: 10.1055/s-0040-1721319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kwon P.S., Oh H., Kwon S.J., Jin W., Zhang F., Fraser K., Hong J.J., Linhardt R.J., Dordick J.S. Sulfated polysaccharides effectively inhibit SARS-CoV-2 in vitro. Cell Discov. 2020;61:1–4. doi: 10.1038/s41421-020-00192-8. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nguyen L., McCord K.A., Bui D.T., Bouwman K.M., Kitova E.N., Elaish M., Kumawat D., Daskhan G.C., Tomris I., Han L., et al. Sialic acid-containing glycolipids mediate binding and viral entry of SARS-CoV-2. Nat Chem Biol. 2021;181:81–90. doi: 10.1038/s41589-021-00924-1. 2021. [DOI] [PubMed] [Google Scholar]
- 65.Baker A.N., Richards S.J., Guy C.S., Congdon T.R., Hasan M., Zwetsloot A.J., Gallo A., Lewandowski J.R., Stansfeld P.J., Straube A., et al. The SARS-COV-2 spike protein binds sialic acids and enables rapid detection in a lateral flow point of care diagnostic device. ACS Cent Sci. 2020;6:2046–2052. doi: 10.1021/acscentsci.0c00855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lenza M.P., Oyenarte I., Diercks T., Quintana J.I., Gimeno A., Coelho H., Diniz A., Peccati F., Delgado S., Bosch A., et al. Structural characterization of N-linked glycans in the receptor binding domain of the SARS-CoV-2 spike protein and their interactions with human lectins. Angew Chem Int Ed. 2020;59:23763–23771. doi: 10.1002/anie.202011015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Stravalaci M., Pagani I., Paraboschi E.M., Pedotti M., Doni A., Scavello F., Mapelli S.N., Sironi M., Perucchini C., Varani L., et al. Recognition and inhibition of SARS-CoV-2 by humoral innate immunity pattern recognition molecules. Nat Immunol. 2022;23:275–286. doi: 10.1038/s41590-021-01114-w. [DOI] [PubMed] [Google Scholar]
- 68.Paiardi G., Richter S., Oreste P., Urbinati C., Rusnati M., Wade R.C. The binding of heparin to spike glycoprotein inhibits SARS-CoV-2 infection by three mechanisms. J Biol Chem. 2022;298:101507. doi: 10.1016/j.jbc.2021.101507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ghorbani M., Brooks B.R., Klauda J.B. Exploring dynamics and network analysis of spike glycoprotein of SARS-COV-2. Biophys J. 2021;120:2902–2913. doi: 10.1016/j.bpj.2021.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chi X., Yan R., Zhang J., Zhang G., Zhang Y., Hao M., Zhang Z., Fan P., Dong Y., Yang Y., et al. A neutralizing human antibody binds to the N-terminal domain of the Spike protein of SARS-CoV-2. Science. 2020;369:650–655. doi: 10.1126/science.abc6952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cerutti G., Guo Y., Zhou T., Gorman J., Lee M., Rapp M., Reddem E.R., Yu J., Bahna F., Bimela J., et al. Potent SARS-CoV-2 neutralizing antibodies directed against spike N-terminal domain target a single supersite. Cell Host Microbe. 2021;29:819–833.e7. doi: 10.1016/j.chom.2021.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tortorici M.A., Beltramello M., Lempp F.A., Pinto D., Dang H.V., Rosen L.E., McCallum M., Bowen J., Minola A., Jaconi S., et al. Ultrapotent human antibodies protect against SARS-CoV-2 challenge via multiple mechanisms. Science. 2020;370:950–957. doi: 10.1126/science.abe3354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Barnes C.O., Jette C.A., Abernathy M.E., Dam K.-M.A., Esswein S.R., Gristick H.B., Malyutin A.G., Sharaf N.G., Huey-Tubman K.E., Lee Y.E., et al. SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies. Nature. 2020;588:682–687. doi: 10.1038/s41586-020-2852-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Liu L., Wang P., Nair M.S., Yu J., Rapp M., Wang Q., Luo Y., Chan J.F.W., Sahi V., Figueroa A., et al. Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike. Nature. 2020 doi: 10.1038/s41586-020-2571-7. [DOI] [PubMed] [Google Scholar]
- 75.Lemmin T., Soto C., Stuckey J., Kwong P.D. Microsecond dynamics and network analysis of the HIV-1 SOSIP env trimer reveal collective behavior and conserved microdomains of the glycan shield. Structure. 2017;25:1631–1639.e2. doi: 10.1016/j.str.2017.07.018. [DOI] [PubMed] [Google Scholar]
- 76.Berndsen Z.T., Chakraborty S., Wang X., Cottrell C.A., Torres J.L., Diedrich J.K., López C.A., Yates J.R., van Gils M.J., Paulson J.C., et al. Visualization of the HIV-1 Env glycan shield across scales. Proc Natl Acad Sci U S A. 2020;117:28014–28025. doi: 10.1073/pnas.2000260117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Stewart-Jones G.B.E., Soto C., Lemmin T., Chuang G.Y., Druz A., Kong R., Thomas P.V., Wagh K., Zhou T., Behrens A.J., et al. Trimeric HIV-1-Env structures define glycan shields from clades A, B, and G. Cell. 2016;165:813–826. doi: 10.1016/j.cell.2016.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Watanabe Y., Mendonça L., Allen E.R., Howe A., Lee M., Allen J.D., Chawla H., Pulido D., Donnellan F., Davies H., et al. Native-like SARS-CoV-2 spike glycoprotein expressed by ChAdOx1 nCoV-19/AZD1222 vaccine. ACS Cent Sci. 2021;7:594–602. doi: 10.1021/acscentsci.1c00080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Thai R., Moine G., Desmadril M., Servent D., Tarride J.L., Ménez A., Léonetti M. Antigen stability controls antigen presentation. J Biol Chem. 2004;279:50257–50266. doi: 10.1074/jbc.M405738200. [DOI] [PubMed] [Google Scholar]
- 80.Doyle H.A., Mamula M.J. Post-translational protein modifications in antigen recognition and autoimmunity. Trends Immunol. 2001;22:443–449. doi: 10.1016/s1471-4906(01)01976-7. [DOI] [PubMed] [Google Scholar]