Abstract
The microbial diversity of a deteriorated biological phosphorus removal reactor was investigated by methods not requiring direct cultivation. The reactor was fed with media containing acetate and high levels of phosphate (P/C weight ratio, 8:100) but failed to completely remove phosphate in the effluent and showed very limited biological phosphorus removal activity. Denaturing gradient gel electrophoresis (DGGE) of PCR-amplified 16S ribosomal DNA was used to investigate the bacterial diversity. Up to 11 DGGE bands representing at least 11 different sequence types were observed; DNA from the 6 most dominant of these bands was further isolated and sequenced. Comparative phylogenetic analysis of the partial 16S rRNA sequences suggested that one sequence type was affiliated with the alpha subclass of the Proteobacteria, one was associated with the Legionella group of the gamma subclass of the Proteobacteria, and the remaining four formed a novel group of the gamma subclass of the Proteobacteria with no close relationship to any previously described species. The novel group represented approximately 75% of the PCR-amplified DNA, based on the DGGE band intensities. Two oligonucleotide rRNA probes for this novel group were designed and used in a whole-cell hybridization analysis to investigate the abundance of this novel group in situ. The bacteria were coccoid and 3 to 4 μm in diameter and represented approximately 35% of the total population, suggesting a relatively close agreement with the results obtained by the PCR-based DGGE method. Further, based on electron microscopy and standard staining microscopic analysis, this novel group was able to accumulate granule inclusions, possibly consisting of polyhydroxyalkanoate, inside the cells.
The concept of modern activated sludge processes for industrial and human wastewater has expanded from the simple degradation of organic matter to the removal of nutrients such as nitrogen and phosphate. Biological phosphorus removal has become one of the most important processes, but little is known about the microbial groups participating in the reactions. In enhanced biological phosphorus removal (EBPR), groups of polyphosphate-accumulating bacteria are enriched in the activated sludge by recycling of the sludge in anaerobic and aerobic zones. In the anaerobic step, the polyphosphate-accumulating bacteria take up short-chain fatty acids and store them in granules as polyhydroxyalkanoates (PHA). The required energy and reducing equivalents for this process are thought to come from the degradation of polyphosphates and glycogen stored in the cells. In the aerobic step, the polyphosphate-accumulating bacteria utilize the stored PHA, take up available phosphates, and restore the pools of polyphosphates and glycogen (10, 15, 16, 23, 55). The polyphosphate-accumulating bacteria are thought to be enriched in the sludge, since strictly heterotrophic and aerobic bacteria are limited by the low concentrations of carbon source in the aerobic step.
Although the EBPR process provides a more economical solution than the chemical precipitation processes previously used, the process operation has not yet been fully optimized and therefore often fails. One of the difficulties in process optimization is the inability to isolate the responsible microorganisms and to verify the biochemical metabolism for the observed EBPR activity. For example, a number of researchers (8, 15, 29, 65) have repeatedly found Acinetobacter spp. to be the most common isolates and have demonstrated the ability of Acinetobacter spp. to accumulate polyphosphates during aerobic growth on acetate. However, Acinetobacter spp. have failed to exhibit the key biochemical transformation observed in EBPR sludge (20, 60). Not surprisingly, as in other natural systems, this bias is caused by the lack of culturability of the majority of the microorganisms in the activated sludge (61). When molecular techniques were implemented, Acinetobacter spp. were found to constitute only a small fraction of the total population (62, 63). A much higher microbial diversity was identified by molecular criteria (7, 28, 53), which showed that bacteria from the subclasses of the Proteobacteria accounted for up to 80% of the total sludge population (62, 63).
Another difficulty in the optimization of the EBPR process is the possible involvement of microbial competitors for substrate uptake under anaerobic conditions. Under these conditions, some bacteria can store carbon at the expense of other, previously stored compounds, such as glycogen (9, 25, 50). These bacteria can compete with the polyphosphate-accumulating bacteria for anaerobic uptake of the carbon source and, under certain conditions, can cause the failure of biological phosphorus removal (26). By gradually decreasing the P/C ratio in the feed from 20:100 to 2:100, Liu et al. observed a decrease in the sludge phosphate content along with a change in the microbial community from polyphosphate-accumulating to non-polyphosphate-accumulating bacteria (25, 26) and a parallel shift in the community DNA fingerprint (28). The shift in the microbial population was attributed to an inability of the polyphosphate-accumulating bacteria to compete for the C and P resources at this P/C ratio. Similar EBPR deterioration in the presence of a high-P-containing substrate (5, 16, 34, 43, 51) suggested that a variety of substrate-related changes in community structure or function may contribute to process failure. Descriptive information about the relevant populations is thus essential to better understanding and control of the EBPR process.
Molecular techniques based on the rRNA phylogenetic framework have demonstrated the ability to characterize community structure and activity in natural and engineered systems without prior cultivation and isolation (3, 41, 56). For example, the PCR-based 16S rRNA gene community fingerprinting methods have been extensively used to provide an estimate of the complexity of a microbial community or as a relative index of similarity to other communities (14, 27, 32, 38, 48, 59). Further, oligonucleotide probes specific for organisms of interest can be designed (2, 12, 56) from sequence data. Used in combination with membrane or in situ whole-cell hybridization, these can provide knowledge of the abundance, distribution, and activity of the organisms of interest under specific growth conditions (3, 12, 22, 36, 37, 44, 46). We applied these techniques to identify populations that may compete with polyphosphate-accumulating microorganisms in a deteriorated reactor system. We anticipate that the results will provide important information for defining populations associated with or contributing to impairment of reactor performance.
We used the denaturing gradient gel electrophoresis (DGGE) method to investigate the microbial diversity of sludge from a deteriorated EBPR reactor. DGGE utilizes the melting property of each unique DNA fragment in a denaturing gradient gel to separate different double-stranded DNA fragments with identical lengths, and its theory and application have been extensively described in previous reports (14, 38, 48, 59). 16S ribosomal DNA (rDNA) from the dominant sequence types was purified and sequenced, and the phylogenetic affiliations were determined. A novel group of the gamma subclass of the Proteobacteria was discovered, and nucleotide probes were designed for fluorescent in situ hybridization to identify and investigate the abundance of the novel group in the sludge directly. In addition, conventional microscopic and biochemical methods were applied to correlate the abundance of this group of bacteria with system performance.
MATERIALS AND METHODS
Reactor design and operation.
A sequencing batch reactor with a working volume of 4 liters was seeded with sludge from a local wastewater treatment plant. The reactor was operated with four 6-h anaerobic-aerobic cycles per day at room temperature (22°C). Each cycle consisted of an initial anaerobic period of 2.25 h, an aerobic period of 2.25 h, and a settling period of 1.5 h. In the beginning of the anaerobic stage, the reactor was fed with a growth substrate to maintain an organic loading of 1.2 g of carbon/day. The growth substrate was prepared daily from stock solutions and contained (per liter) 0.85 g of NaCH3COO · 3H2O, 50.4 mg of NH4Cl, 302 mg of NaH2PO4 · H2O, 360 mg of MgSO4 · 7H2O, 144 mg of KCl, 14 mg of CaCl2 · H2O, 1 mg of yeast extract, and 1.2 ml of mineral salt solution. The mineral salt solution (55) was composed of (per liter) 0.375 g of FeCl3 · 6H2O, 0.0375 g of H3BO3, 0.0075 g of CuSO4 · 5H2O, 0.045 g of KI, 0.03 g of MnCl2 · 4H2O, 0.015 g of Na2MoO4 · 2H2O, 0.03 g of ZnSO4 · 7H2O, 0.0375 g of CoCl2 · 6H2O, and 2.5 g of EDTA. The system was maintained with a solids retention time of 7 days and a hydraulic residence time of 12 h. The pH in the system was maintained at between 6.8 and 7.1 by use of a pH controller.
Sampling from the bioreactor.
The phosphate concentration in the biomass was measured by the ascorbic acid method after digestion with ammonium persulfate (4). Total organic carbon was measured by use of a Shimadzu TOC-5000 analyzer equipped with an ASI-5000 autosampler. At day 213, activated-sludge samples were taken from the end of the aerobic stage for the analysis of microbial community structure.
Bacteria.
Alicyclobacillus acidocaldarius ATCC 43030 was obtained from the American Type Culture Collection (Rockville, Md.) and was grown at 50°C as previously described (11). Nitrosomonas eutropha was obtained from Jodi Flax (Northwestern University).
Isolation and PCR amplification of DNA.
Total community DNA from the activated sludge was obtained after cell lysis, phenol-chloroform extraction, and ethanol precipitation by a protocol previously described (27). This DNA preparation was used as a DNA template in a PCR performed with 1× PCR buffer (Gibco BRL, Gaithersburg, Md.) containing a 200 μM concentration of each of the deoxynucleoside triphosphates, 1.5 mM MgCl2, a 0.1 μM concentration of each of the primers (Operon Technologies, Inc., Alameda, Calif.), and 2.5 U of Taq polymerase (Pharmacia Biotech Inc., Piscataway, N.J.) in a final volume of 100 μl. For amplification of 16 S rDNA for DGGE, the forward primer 968FGC (5′-AACGCGAAGAACCTTAC-3′) with a GC clamp (5′-CGCCCGGGGCGCGCCCCGGGCGGGGCGGGGGCACGGGGGG-3′) (54) and the reverse primer 1392R (5′-ACGGGCGGTGTGTAC-3′) (14) were used. The PCR was performed with a PTC-100 programmable thermal cycler (MJ Research Inc., Watertown, Mass.) and the following program: an initial denaturation at 94°C for 5 min; 30 cycles of denaturation (45 s at 94°C), annealing (45 s at 38°C), and extension (1 min at 72°C); and a final extension at 72°C for 5 min. Amplified DNA was verified by electrophoresis of 2 μl of the PCR product on a 1% agarose gel in 1× TAE buffer (20 mM Tris acetate [pH 7.4], 10 mM sodium acetate, 0.5 mM disodium EDTA).
DGGE.
DGGE was performed with the D-Gene system (Bio-Rad Laboratories, Hercules, Calif.) at 200 V, 60°C, and various electrophoresis times. Samples were loaded on a 6% (wt/vol) polyacrylamide gel (acrylamide:N,N′-methylenebisacrylamide ratio, 37.5:1 [Bio-Rad]) in 1× TAE buffer. The denaturing gradient in the gel was formed by mixing two stock solutions of 6% acrylamide containing 40% denaturing agent (2.8 M urea [Sigma Chemical Co., St. Louis, Mo.], 18.7% [vol/vol] formamide [Sigma] deionized with AG501-X8 mixed-bed resin [Bio-Rad]) and 60% denaturing agent (4.2 M urea, 24% formamide). The separated DNA was visualized with silver stain by the following procedure. The gel was rinsed briefly in water and shaken gently in fixing solution (10% ethanol, 0.5% acetic acid) for 2 h. The gel was transferred to a freshly made staining solution (1 g of AgNO3 per liter) and shaken gently for 20 min, followed by a brief rinse in water. The stained gel was developed in a freshly made developing solution (0.1 g of NaBH4 per liter, 4% formaldehyde, 1.5% [wt/vol] NaOH) until the desired exposure was achieved. The gel was fixed in 0.75% Na2CO3 and photographed with a charge-coupled device camera.
Isolation and sequencing of DGGE bands.
DNA bands in the DGGE gel were cut out with a razor blade, and the DNA of the excised bands was eluted overnight at 4°C in a 1.5-ml tube containing 100 μl of 1× TAE buffer. The DNA was amplified by PCR with primers for DGGE as described above and run on a second DGGE gel. This procedure was repeated two or three times to obtain clean DNA products for DNA sequencing. DNA sequences of excised bands were obtained from both 5′ and 3′ directions with infrared light-labeled primers 968F (forward primer without the GC clamp) and 1392R by use of a 4000L automated sequencer in accordance with the manufacturer’s instructions (Li-Cor, Lincoln, Nebr.).
Phylogenetic analysis and probe design.
The six partial sequences (∼390 bp) obtained were compared to available 16S rRNA sequences in GenBank by use of the NCBI Blast program. Bacterial sequences closely related to those six sequence types or representative sequences from different subclasses of the Proteobacteria were used in the phylogenetic analysis. These sequences were first aligned by use of a sequence alignment editor, the ARB program (provided by Oliver Strunk and Wolfgang Ludwig, Technical University of Munich, Munich, Germany). A distance matrix tree with bootstrapping was constructed by use of the neighbor-joining method of Saitou and Nei (49) provided in the ARB program. For distance correction, the algorithm of Jukes and Cantor (21) was used. A parsimony tree with bootstrapping was constructed by use of the DNAPARS method in the ARB program. Approximately 350 base positions that were identical in more than 50% of the aligned sequences were included in the analysis.
Oligonucleotide rRNA probes were designed from the retrieved sequences, and specificities were checked by use of the Check Probe program of the Ribosomal RNA Database Project (40). Two probes specific for the novel group of the gamma subclass of the Proteobacteria were designed. Gam1019 (Table 1), labeled with CY5, was specific for organisms represented by band 4, while Gam1278, labeled with CY3, was specific for organisms represented by bands 3, 5, and 6. Furthermore, a probe specific for the domain Bacteria (Eub338) and labeled with fluorescein isothiocyanate and another gamma subclass probe (Gam42a) labeled with CY3 were used. Probes were synthesized and labeled by Operon Technologies. Nucleotide sequences of probes Gam1019 and Gam1278 had one mismatch each to zero and five sequences that were available in GenBank, respectively. The specificity of probe Gam1019 was optimized by comparative hybridization to A. acidocaldarius, which has one mismatch to the target. This step could not be performed with Gam1278, since no known strain with one mismatch exists. The optimal conditions for the hybridization of Gam1019 and Gam1278 at 37°C were found to be formamide concentrations of 30 and 33%, respectively (Table 1). Probe Gam42a, specific for the gamma subclass of the Proteobacteria, was tested against a one-mismatch target, N. eutropha, from the beta subclass of the Proteobacteria, and was found to be specific at a formamide concentration of 45% at 37°C as previously reported (31).
TABLE 1.
Oligonucleotide probes used in this study
| Standardized probe namea | Abbreviation | Specificity | Sequence | FA (%)b | Reference or source |
|---|---|---|---|---|---|
| S-D-Bact-0338-a-A-18 | Eub338 | Bacteria | 5′-GCT GCC TCC CGT AGG AGT-3′ | 30 | 57 |
| L-Sc-gProt-0042-a-A-17 | Gam42a | Gamma subclass of Proteobacteria | 5′-GCC TTC CCA CAT CGT TT-3′ | 45 | 31 |
| S-S-gProt-1019-a-A-18 | Gam1019 | Band 4 | 5′-GGT TCC TTG CGG CAC CTC-3′ | 30 | This study |
| S-S-gProt-1278-a-A-20 | Gam1278 | Bands 3, 5, and 6 | 5′-ACG AGC GGC TTT TTG GGA TT-3′ | 33 | This study |
As described by Alm et al. (1).
Percentage of formamide (FA) in hybridization buffer during hybridization at 37°C.
In situ hybridization.
Samples for in situ hybridization were fixed in 3% formaldehyde as previously described (44). Cells were attached to poly-l-lysine-coated slides as described by Møller et al. (38) and dehydrated by sequential washes in 50, 70, and 96% ethanol (3 min each). Subsequently, 10 μl of hybridization solution (formamide as indicated in Table 1, 0.9 M NaCl, 100 mM Tris [pH 7.2], 0.1% sodium dodecyl sulfate [SDS]) containing 25 ng of probe was added to each hybridization well. Cells were incubated with hybridization solution for 16 h in a humid chamber at 37°C (57). For washing, slides were submersed in 80 ml of washing solution I (formamide as indicated in Table 1, 0.9 M NaCl, 100 mM Tris [pH 7.2], 0.1% SDS) at 37°C for 20 min and subsequently in washing solution II (0.9 M NaCl, 100 mM Tris [pH 7.2], 0.1% SDS) at 37°C for 20 min. Slides were briefly rinsed with Milli-Q water between each washing step.
Microscopy.
Hybridized samples were analyzed with a Carl Zeiss Axioplan epifluorescence microscope. The exitation source was a 100-W halogen bulb, and digital images were captured with a 12-bit cooled slow-scan charge-coupled device camera (CH250; Photometrics, Tucson, Ariz.). The camera was controlled by IPLap Spectrum 10 software (Signal Analytics, Vienna, Va.). Fluorescein, CY3, and CY5 were visualized by use of filter sets 10 and 15 (Carl Zeiss) and XF45 (Omega Opticals, Brattleboro, Vt.), respectively. Neisser staining of samples was performed as previously described (19), visualized under bright-field and phase-contrast illumination with an Olympus BH2 microscope, and photographed with an Olympus OM-4 camera on Kodak Ektachrome 400-ASA film. Image analysis was performed with Photoshop 4.0 software (Adobe, Mountain View, Calif.) and the NIH Image program (39a). Transmission electron microscopy was performed by a protocol previously described (17).
Nucleotide sequence accession numbers.
The sequences obtained in this study have been deposited in the GenBank, EMBL, and DDBJ nucleotide sequence databases under accession no. AF093776 (band 1), AF093779 (band 2), AF093777 (band 3), AF093778 (band 4), AF093780 (band 5), and AF093781 (band 6).
RESULTS AND DISCUSSION
Deterioration of EBPR processes has been reported in several studies (5, 16, 34, 43, 51). In the sludge from deteriorated processes, the dominance of certain microorganisms not performing EBPR has been observed (25). In these processes, the carbon substrate was completely taken up under anaerobic conditions, with an increase in the cellular PHA content (7, 26). It is suspected that, under certain conditions, these microorganisms can outcompete polyphosphate-accumulating bacteria for uptake of the growth substrate in the anaerobic step and thus contribute to process deterioration. Therefore, a better understanding of the diversity of these microbial competitors is a key element for optimizing the EBPR process.
Performance of the EBPR reactor.
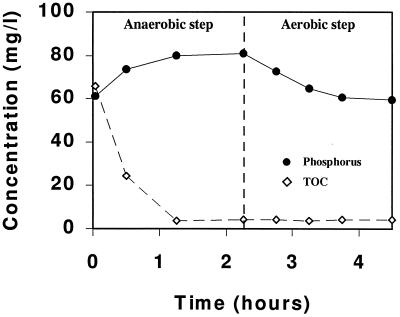
The EBPR reactor was fed with acetate as the main carbon source and operated under sequential anaerobic and aerobic conditions for more than 250 days. A high P/C weight ratio (8:100) was used to enrich for bacteria capable of removing phosphate, since bacteria involved in phosphate removal may store excessive amounts of phosphate as cellular polyphosphate. However, the reactor failed to completely remove the phosphate, and the EBPR process deteriorated throughout the course of the operation. The phosphate content of the sludge measured at the end of the aerobic stage over 250 days was, on average, 1.95% (dry weight), equivalent to the normal phosphate content in bacterial cells. This finding agrees with the observation that only little phosphate was released into the bulk solution under anaerobic conditions and taken up under subsequent aerobic conditions (Fig. 1). At the same time, acetate was rapidly taken up and accumulated in the cells under anaerobic conditions (Fig. 1). This finding suggests that the main fraction of the bacteria in the EBPR process did not use polyphosphate as an energy source for the uptake and storage of carbon reserves.
FIG. 1.
Profiles of the concentrations of phosphate and total organic carbon (TOC) in the sequencing batch reactor during an anaerobic-aerobic cycle after 213 days.
Microscopic analysis of sludge.
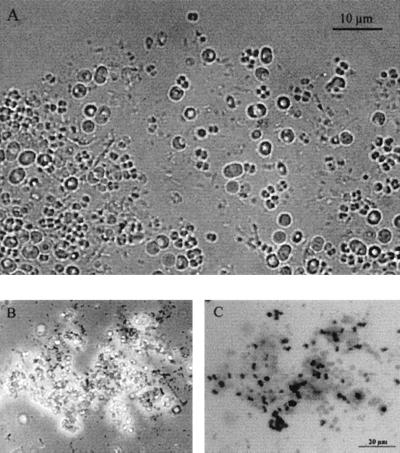
Phase-contrast microscopy (Fig. 2A) revealed the presence of several morphologically distinct bacteria in the sludge, including long rods, cells forming tetrads, dicoccoid cells, filamentous cells, and a large fraction of large coccoid cells. These morphotypes resembled those previously observed by Neisser and polyhydroxybutyrate (PHB) staining techniques (9, 25, 26). Large coccoid cells with a diameter of 3 to 4 μm were dominant in the sludge. Within this group, two slightly different morphologies were observed: bright cells with a high concentration of granules and darker cells with fewer granules.
FIG. 2.
Microscopic observations of activated sludge from the sequencing batch reactor. (A) Phase-contrast image showing the microbial diversity of the sludge. (B) Phase-contrast image of Neisser-stained sludge. (C) Bright-field image of the Neisser-stained sludge in panel B; black indicates phosphate accumulation by the bacteria. The scale bar on panel C (20 μm) also applies to panel B.
Microscopic investigation of the sludge stained with the Neisser stain revealed that the deteriorated EBPR process was dominated by organisms that did not accumulate polyphosphate. Bacteria capable of accumulating polyphosphate (positive Neisser stain) were present only in low numbers (Fig. 2B and C). This finding is consistent with the relatively low phosphate content in the sludge. The large coccoid bacteria clearly visible as large white cells in Fig. 2B also did not stain, suggesting that these bacteria did not accumulate polyphosphate under the reactor operating conditions.
Analysis of microbial diversity.
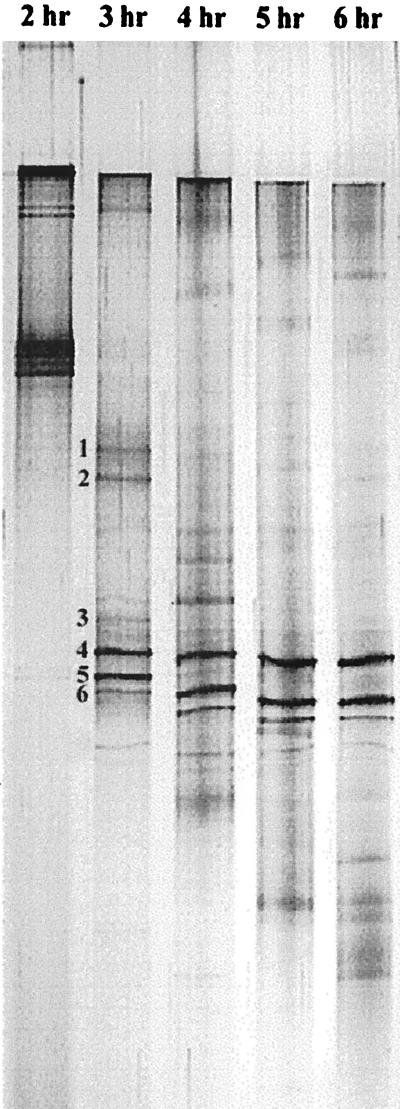
The microbial diversity of deteriorated EBPR systems was analyzed by DGGE of PCR-amplified 16S rDNA (Fig. 3). Electrophoresis for 1 to 6 h was used to optimize the separation of the amplified 16S rDNA. A running time of 3 to 4 h gave the best resolution of the 16S rDNA, with up to 11 bands or sequence types observed. A running time of 3.5 h was chosen for subsequent analyses. The migration of several bands was observed even after extended running times, emphasizing the importance of proper selection of running conditions. Although only a few dominant morphotypes were observed, a complex microbial diversity with at least 11 different sequence types was documented in the DGGE analysis of amplified rDNA. This observation was consistent with previous findings obtained with either clone libraries (7) or the restriction fragment length polymorphism approach (28).
FIG. 3.
Silver-stained DGGE pattern of PCR-amplified 16S rDNA from the total DNA extracted from the deteriorated EBPR reactor. Electrophoresis was done for 2 to 6 h with a 1-h interval to optimize the running time. Up to 11 bands were clearly observed in the original gel. The six most dominant bands, which were further isolated and sequenced, are indicated next to the 3-h lane.
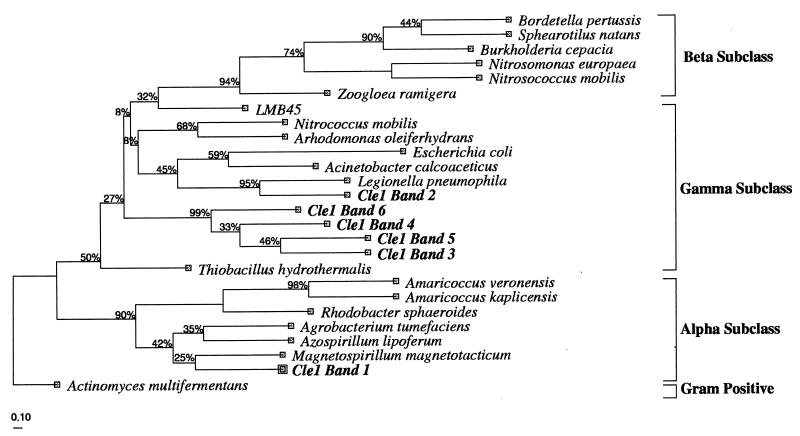
The six most dominant fragments were isolated (Fig. 3) and reamplified by PCR. Before DNA sequencing, the recovered DGGE bands were run on a DGGE gel to confirm their positions relative to the original sample. This step was repeated at least two times to obtain a pure DNA product for sequencing, since reamplification of a (presumably) single fragment often resulted in the formation of multiple amplicons from the adjacent bands. Partial 16S rDNA sequences of approximately 430 nucleotides were obtained from the DNA from the six dominant bands. These sequences were compared with the sequences available in GenBank, and the phylogenetic affiliation of the sequences was further analyzed by the neighbor-joining method. It was previously shown that relatively short sequences, such as those obtained from DGGE analysis, are sufficient for an approximate phylogenetic identification (52, 53, 64). The resulting phylogenic tree (Fig. 4) indicates that organisms represented by band 1 belonged to the alpha subclass of the Proteobacteria, most closely related to Magnetospirillum magnetotacticum (87% similarity). Bacteria of the genus Amaricoccus, tetrad-forming bacteria in the alpha subclass of the Proteobacteria, were previously isolated from deteriorated EBPR sludge (6, 9, 33). However, this group of bacteria was not observed in the present study. The organisms represented by band 2 belonged to the gamma subclass of the Proteobacteria, most closely affiliated with Legionella pneumophila. A similar observation was previously reported for EBPR-activated sludge, in which this group of bacteria constituted approximately 2% of the total number of clones (7).
FIG. 4.
Phylogenetic affiliation of the 16S rDNA isolated from the sequencing batch reactor. The parsimony phylogenetic tree was calculated by use of the DNAPARS method with bootstrapping. The bold names correspond to the sequence types isolated from the DGGE analysis, while the other sequences used in the analysis were obtained from GenBank. The tree was rooted with the 16S rDNA sequence of a gram-positive bacterium, Actinomyces multifermentans, as an outgroup. The scale bar corresponds to 0.10 substitution per nucleotide position. Cle1, Clemson sample 1.
The remaining four of the most dominant sequence types recovered by DGGE fractionation from the sludge belonged to the gamma subclass of the Proteobacteria and formed a highly related group with no close relationship to any previously characterized bacteria. The bacterial species most closely related to this novel group was an environmental clone, LBM45, isolated from the deep waters of Lake Michigan (B. MacGregor, Northwestern University). This finding suggests that the bacterial group represented by bands 3 to 6 is very likely a novel group of organisms in the gamma subclass of the Proteobacteria. Based on the intensity of the DGGE bands, the novel group of the four related organisms accounted for approximately 75% of the PCR-amplified DNA in the sample (data not shown), an amount about two times higher than the actual abundance of the bacteria found by in situ hybridization (see below). This finding is not consistent with that of a previous study in which the gamma subclass group was found to constitute only a minority of the population in a deteriorated EBPR reactor (7). This discrepancy may have been caused by a difference in the enrichment conditions or biases associated with DNA isolation and amplification. In the previous study, settled sewage was used as a substrate, as opposed to the acetate used in this study. Moreover, a shorter enrichment process was used by Bond et al. (7), and a slower substrate uptake rate was observed under anaerobic conditions in their study than in the present study. However, a comparison is also complicated because the previously identified partial sequences (7) only partly overlapped the region of the 16S rRNA gene sequence determined in this study. Longer 16S rDNA fragments are required for a better determination of the phylogenetic positions of novel sequence types. This information must be obtained by independent cloning and sequencing methods, since larger DNA fragments are poorly resolved by DGGE.
In situ identification of the novel group of bacteria.
In situ hybridization with oligonucleotide probes targeting rRNA is a valuable method for the identification and monitoring of specific organisms in natural or engineered systems. Oligonucleotide probes were designed to monitor the four organisms in the novel group of the gamma subclass in the sludge. From the sequence information obtained from the DGGE analysis, it was not possible to design a probe specific for the entire group. Therefore, two oligonucleotide probes, Gam1019 and Gam1278, targeting the organisms represented by band 4 and by bands 3, 5, and 6, respectively, were designed to target the novel group. In order to identify closely related strains, sets of hierarchical probes with increasing specificities can be used (3, 35). In the present study, two more general probes were used, a probe specific for the entire gamma subclass of the Proteobacteria (Gam42a) and a probe targeting the domain Bacteria (Eub338).
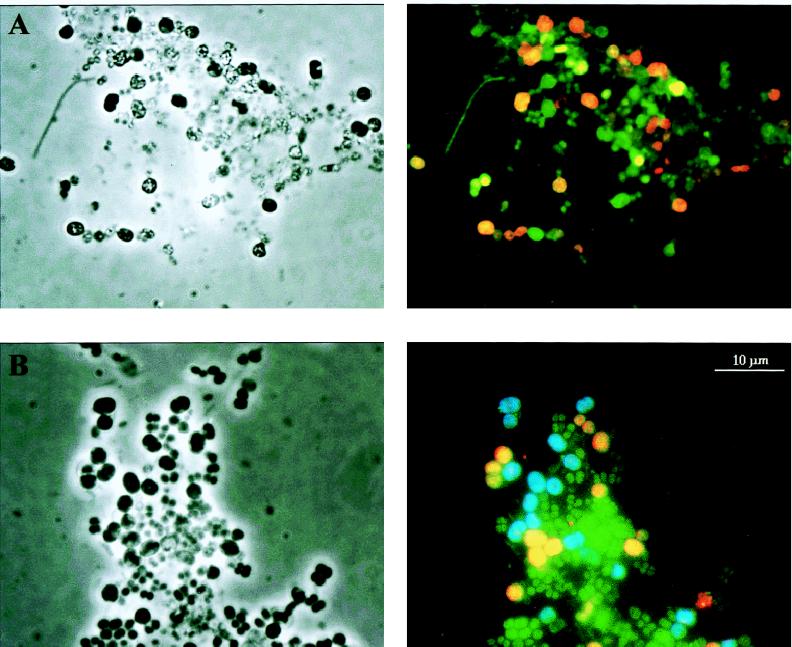
As shown in Fig. 5, all cells showed clear signals with the eubacterial probe, indicating that adequate amounts of rRNA for detection by in situ hybridization were present in the cells. The two group-specific probes, Gam1019 and Gam1278, separately hybridized with two different types of large coccoid cells in the sludge (Fig. 5B), together constituting approximately 35% of the total population. The large coccoid cells were found mainly in small clusters in the sludge, and dispersed cells were observed mainly when mild sonication was imposed on the sludge. Electron microscopy showed that a thin membrane-like boundary of unknown origin surrounded these clusters (Fig. 6). The two types of coccoid cells, with a diameter of approximately 3 to 4 μm, were morphologically similar but still distinguishable by phase-contrast microscopy, since the cells targeted by Gam1019 were darker and more dense than the cells targeted by Gam1278. These two probes hybridized only to the large coccoid cells in the sample, suggesting that a high degree of specificity for the target bacteria in the sludge had been obtained. Although the Gam1278 probe targeted three different sequence types (bands 3, 5, and 6), only one morphotype was observed after whole-cell hybridization. It is likely that the closely related strains have similar morphologies. Another possibility, although unlikely, is that a single strain may contain different copy numbers of the 16S rRNA (39) which, after PCR amplification, may give rise to multiple fragments. The Gam1019 probe, specific for the bacteria represented by band 4, hybridized to a similar type of large coccoid cells, further suggesting that the closely related phylotypes might have almost identical morphologies.
FIG. 5.
In situ hybridization of activated sludge from the deteriorated EBPR reactor. The left side shows phase-contrast images, and the right side shows epifluorescence micrographs of the corresponding areas. (A) In situ hybridization with probes specific for the domain Bacteria (green) and the gamma subclass of the Proteobacteria (red). (B) In situ hybridization with probes specific for the domain Bacteria (green) and for the novel subgroup of the gamma subclass of the Proteobacteria; the probe specific for the bacteria from band 4 is shown in blue, and the probe specific for the bacteria from bands 3, 5, and 6 is shown in yellow. The scale bar applies to all of the images.
FIG. 6.
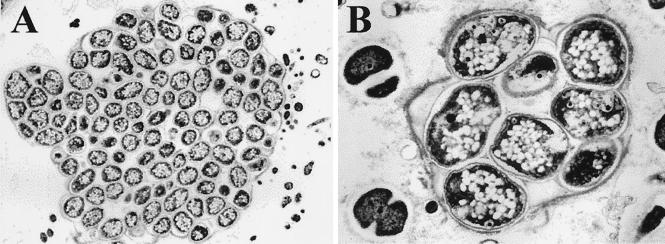
Transmission electron microscopy of the dominant coccoid bacteria, which have morphological traits similar to those of the novel group of bacteria from the gamma subclass of the Proteobacteria found in the activated sludge. (A) Bacterial cluster with a fine layer of membrane surrounding the bacteria. (B) Large coccoid bacteria from the novel group of the gamma subclass of the Proteobacteria with an accumulation of granules, possibly consisting of PHA, inside the cells.
The gamma subclass probe hybridized only to the large coccoid bacteria in the sludge (Fig. 5A), indicating that the correct specificity of the probes was obtained. Surprisingly, the probe specific for the gamma subclass did not hybridize with the bacteria corresponding to band 4 (Fig. 5), which belong to the gamma subclass. This observation provides additional evidence that the Gam1278 and Gam1019 probes detected different bacterial strains. It further suggests that the actual abundance of organisms of the gamma subclass of the Proteobacteria in natural and engineered systems may not be correctly identified with a gamma subclass-specific probe (Gam42a) during membrane or whole-cell rRNA hybridizations. This probe has been extensively used in the quantification of organisms of the gamma subclass of the Proteobacteria (31, 62, 63). It was not possible to check the specificity of the probe in the Ribosomal Database Project database (30) due to the rather limited numbers of published 23S rDNA sequences. It is possible that more specific probes can be designed when larger numbers of 16S or 23S rDNA sequences are known.
From the in situ hybridizations, it was estimated that the novel group of large coccoid cells constituted approximately 35% of the total number of cells in the sludge. This finding is different from the results of the DGGE analysis, which showed that the 16S rDNA sequences from the novel group constituted approximately 75% of the total amount of PCR-amplified rDNA. This difference is likely due to a bias associated with the DNA extraction and PCR amplification steps (13, 18, 24, 42, 45, 47, 58, 66) but is rather insignificant in comparison to the extent of bias reported in the aforementioned studies. Nevertheless, it shows the importance of directly determining the actual presence and abundance of the populations of interest by rRNA-based techniques after characterization of the microbial diversity by DNA-based methods.
Bacteria resembling the novel group of coccoid bacteria described above in morphology showed no response to Neisser staining (Fig. 2B and C) and therefore did not seem to store polyphosphates in granules. Bacteria with a similar morphology have been observed before in deteriorated sludge, and it was suggested that the bacteria accumulated glycogen instead of polyphosphates (25). They were suspected of deriving the energy required for anaerobic substrate uptake from the glycolysis of glycogen, thereby enabling them to compete with polyphosphate-accumulating bacteria. Electron microscopy revealed that the large coccoid bacteria contained many large bright granules (Fig. 6), possibly PHA, inside the cells. This result further indicates that these bacteria might be able to compete with polyphosphate-accumulating bacteria in sludge.
Nucleic acid methods were used here to identify a novel group of as-yet-uncultured bacteria which were present in high concentrations in the sludge. In situ hybridization with probes targeting rRNA served to better estimate the abundance of these bacteria in the sludge and provided a method for investigating the relationship of these bacteria to the deterioration of the EBPR process. We are now evaluating the use of these probes to screen enrichment cultures designed for the eventual isolation of these bacteria. Only pure cultures will provide the basis for a more complete understanding of the role of these bacteria in activated-sludge processes.
ACKNOWLEDGMENTS
We thank Tom Geisbert, United States Army Medical Research Institute of Infectious Diseases, Fort Detrick, Md., for performing electron microscopy analysis and Barbara MacGregor, Northwestern University, Evanston, Ill., for kind help with phylogenetic analysis.
REFERENCES
- 1.Alm E W, Oerther D B, Larsen N, Stahl D A, Raskin L. The oligonucleotide probe database. Appl Environ Microbiol. 1996;62:3557–3559. doi: 10.1128/aem.62.10.3557-3559.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amann R I, Krumholz L, Stahl D A. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J Bacteriol. 1990;172:762–770. doi: 10.1128/jb.172.2.762-770.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amann R I, Ludwig W, Schleifer K H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.American Public Health Association. Standard methods for the examination of water and wastewater. 17th ed. Washington, D.C: American Public Health Association; 1989. [Google Scholar]
- 5.Barnard J L, Stevens G M, Leslie P J. Design strategies for nutrient removal plants. Water Sci Technol. 1985;17:233–242. [Google Scholar]
- 6.Blackall L L, Rossetti S, Christensson C, Cunningham M, Hartman P, Hugenholtz P, Tandoi V. The characterization and description of representatives of ‘G’ bacteria from activated sludge plants. Lett Appl Microbiol. 1997;25:63–69. doi: 10.1046/j.1472-765x.1997.00176.x. [DOI] [PubMed] [Google Scholar]
- 7.Bond P L, Hugenholtz P, Keller J, Blackall L L. Bacterial community structures of phosphate-removing and non-phosphate-removing activated sludge from sequencing batch reactors. Appl Environ Microbiol. 1995;61:1910–1916. doi: 10.1128/aem.61.5.1910-1916.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brodisch K E U. Interaction of different groups of micro-organisms in biological phosphate removal. Water Sci Technol. 1985;17:89–97. [Google Scholar]
- 9.Cech J S, Hartman P. Competition between polyphosphate and polysaccharide accumulating bacteria in enhanced biological phosphate removal systems. Water Res. 1993;27:1219–1225. [Google Scholar]
- 10.Comeau Y, Hall K J, Hancock R E W, Oldham W K. Biochemical model for enhanced biological phosphorus removal. Water Res. 1986;20:1511–1521. [Google Scholar]
- 11.Darland G, Brock T D. Bacillus acidocaldarius sp. nov., an acidophilic thermophilic spore-forming bacterium. J Gen Microbiol. 1971;67:9–15. [Google Scholar]
- 12.DeLong E F, Wickham G S, Pace N R. Phylogenetic stains: ribosomal RNA-based probes for identification of single cells. Science. 1989;243:1360–1362. doi: 10.1126/science.2466341. [DOI] [PubMed] [Google Scholar]
- 13.Farrelly V, Rainey F A, Stackebrandt E. Effect of genome size and rrn gene copy number on PCR amplification of 16S rRNA genes from a mixture of bacterial species. Appl Environ Microbiol. 1995;61:2798–2801. doi: 10.1128/aem.61.7.2798-2801.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ferris M J, Muyzer G, Ward D M. Denaturing gradient gel electrophoresis profiles of 16S rRNA-defined populations inhabiting a hot spring microbial mat community. Appl Environ Microbiol. 1996;62:340–346. doi: 10.1128/aem.62.2.340-346.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fuh G W, Chen M. Microbiological basis of phosphate removal in the activated sludge process for the treatment of wastewater. Microb Ecol. 1975;2:119–138. doi: 10.1007/BF02010434. [DOI] [PubMed] [Google Scholar]
- 16.Fukase T, Shibata M, Miyaji Y. The role of an anaerobic stage on the biological phosphorus removal. Water Sci Technol. 1985;17:69–80. [Google Scholar]
- 17.Geisbert T W, Jaax N K. Marburg hemorrhagic fever: report of a case studied by immunohistochemistry and electron microscopy. Ultrastruct Pathol. 1998;22:3–17. doi: 10.3109/01913129809032253. [DOI] [PubMed] [Google Scholar]
- 18.Hansen M C, Nielsen T T, Givskov M, Molin S. Biased 16S rDNA PCR amplification caused by interference from DNA flanking the template region. FEMS Microbiol Ecol. 1998;26:141–149. [Google Scholar]
- 19.Jenkins D, Richard M G, Diagger G T. Manual on the causes and control of activated sludge bulking and foaming. Pretoria, Republic of South Africa: Water Research Commission; 1986. [Google Scholar]
- 20.Jenkins D, Tandoi V. The applied microbiology of enhanced biological phosphate removal accomplishments and needs. Water Res. 1991;25:1471–1478. [Google Scholar]
- 21.Jukes T H, Cantor C R. Evolution of protein molecules. In: Munro H M, editor. Mammalian protein metabolism. New York, N.Y: Academic Press, Inc.; 1969. pp. 21–132. [Google Scholar]
- 22.Kerkhof L, Ward B B. Comparison of nucleic acid hybridization and fluorometry for measurement of the relationship between RNA/DNA ratio and growth rate in a marine bacterium. Appl Environ Microbiol. 1993;59:1303–1309. doi: 10.1128/aem.59.5.1303-1309.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kortstee G J J, Appeldoorn K J, Bontning C F C, van Niel E W J, van Veen H W. Biology of polyphosphate-accumulating bacteria involved in enhanced biological phosphorus removal. FEMS Microbiol Rev. 1994;15:137–153. doi: 10.1111/j.1574-6976.1994.tb00131.x. [DOI] [PubMed] [Google Scholar]
- 24.Liesack W, Weyland H, Stackebrandt E. Potential risk of gene amplification by PCR as determined by 16S rDNA analysis of a mixed-culture of strict barophilic bacteria. Microb Ecol. 1991;21:191–198. doi: 10.1007/BF02539153. [DOI] [PubMed] [Google Scholar]
- 25.Liu W, Mino T, Nakamura K, Matsuo T. Glycogen accumulating population and its anaerobic substrate uptake in anaerobic-aerobic activated sludge without biological phosphorus removal. Water Res. 1996;30:75–82. [Google Scholar]
- 26.Liu W, Nakamura K, Matsuo T, Mino T. Internal energy-based competition between polyphosphate- and glycogen-accumulating bacteria in biological phosphorus removal reactors—effect of P/C feeding ratio. Water Res. 1997;31:1430–1438. [Google Scholar]
- 27.Liu W, Marsh T L, Cheng H, Forney L J. Characterization of microbial diversity by determining terminal restriction fragment length polymorphism of 16S ribosomal DNA. Appl Environ Microbiol. 1997;63:4516–4522. doi: 10.1128/aem.63.11.4516-4522.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu W, Marsh T L, Forney L J. Determination of the microbial diversity of anaerobic-aerobic activated sludge by a novel molecular biological technique. Water Sci Technol. 1997;37:417–422. [Google Scholar]
- 29.Lötter L H. The role of bacterial phosphate metabolism in enhanced phosphorus removal from the activated sludge process. Water Sci Technol. 1985;17:127–138. [Google Scholar]
- 30.Maidak B L, Olsen G J, Larsen N, Overbeek R, McCaughey M J, Woese C R. The Ribosomal Database Project (RDP) Nucleic Acids Res. 1996;24:82–85. doi: 10.1093/nar/24.1.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Manz W, Amann R I, Ludwig W, Wagner M, Schleifer K H. Phylogenetic oligodeoxynucleotide probes for the major subclasses of Proteobacteria: problems and solutions. Syst Appl Microbiol. 1992;15:593–600. [Google Scholar]
- 32.Martinez-Murcia A J, Acinas S G, Rodriguez-Valera F. Evaluation of prokaryotic diversity by restrictase digestion of 16S rDNA directly amplified from hypersaline environments. FEMS Microbiol Ecol. 1995;17:247–256. [Google Scholar]
- 33.Maszenan A M, Seviour R J, Patel B K C, Rees G N, McDougall B M. Amaricoccus gen. nov., a gram-negative coccus occurring in regular packages or tetrads, isolated from activated sludge biomass, and descriptions of Amaricoccus veronensis sp. nov., Amaricoccus tamworthensis sp. nov., Amaricoccus macauensis sp. nov., and Amaricoccus kaplicensis sp. nov. Int J Syst Bacteriol. 1997;47:727–734. doi: 10.1099/00207713-47-3-727. [DOI] [PubMed] [Google Scholar]
- 34.Matsuo Y. Effect of the anaerobic solids retention time on enhanced biological phosphorus removal. Water Sci Technol. 1994;30:193–202. [Google Scholar]
- 35.Mau M, Timmis K N. Use of subtractive hybridization to design habitat-based oligonucleotide probes for investigation of natural bacterial communities. Appl Environ Microbiol. 1998;64:185–191. doi: 10.1128/aem.64.1.185-191.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Møller S, Kristensen C S, Poulsen L K, Carstensen J M, Molin S. Bacterial growth on surfaces: automated image analysis for quantification of growth rate-related parameters. Appl Environ Microbiol. 1995;61:741–748. doi: 10.1128/aem.61.2.741-748.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Møller S, Pedersen A R, Poulsen L K, Arvin E, Molin S. Activity and three-dimensional distribution of toluene-degrading Pseudomonas putida in a multispecies biofilm assessed by quantitative in situ hybridization and scanning confocal laser microscopy. Appl Environ Microbiol. 1996;62:4632–4640. doi: 10.1128/aem.62.12.4632-4640.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Muyzer G, De Wall E C, Uitterlinden A G. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol. 1993;59:695–700. doi: 10.1128/aem.59.3.695-700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mylvaganam S, Dennis P P. Sequence heterogeneity between the two genes encoding 16S rRNA from the halophilic archaebacterium Haloarcula marismotui. Genetics. 1992;130:399–410. doi: 10.1093/genetics/130.3.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39a.National Institutes of Health. 22 June 1998, posting date. [Online.] NIH Image program. ftp:zippy.nimh.nih.gov. [13 August 1998, date last accessed.]
- 40.Olsen G J, Larsen N, Woese C R. The Ribosomal RNA Database Project. Nucleic Acids Res. 1991;19(Suppl.):2017–2021. doi: 10.1093/nar/19.suppl.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pace N R, Stahl D A, Lane D J, Olsen G J. The analysis of natural microbial populations by ribosomal RNA sequences. Adv Microb Ecol. 1986;9:1–55. [Google Scholar]
- 42.Picard C, Ponsonnet C, Paget E, Nesme X, Simonet P. Detection and enumeration of bacteria in soil by direct DNA extraction and polymerase chain reaction. Appl Environ Microbiol. 1992;58:2717–2722. doi: 10.1128/aem.58.9.2717-2722.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pitman A R, Trim B C, van Dalsen L. Operating experience with biological nutrient removal at the Johannesburg Bushkoppie works. Water Sci Technol. 1988;20:51–61. [Google Scholar]
- 44.Poulsen L K, Ballard G, Stahl D A. Use of rRNA fluorescence in situ hybridization for measuring the activity of single cells in young and established biofilms. Appl Environ Microbiol. 1993;59:1354–1360. doi: 10.1128/aem.59.5.1354-1360.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rainey F A, Ward N, Sly L I, Stackebrandt E. Dependence on the taxon composition of clone libraries for PCR amplified, naturally occurring 16S rDNA, on the primer pair and the cloning system used. Experientia. 1994;50:796–797. [Google Scholar]
- 46.Raskin L, Poulsen L K, Noguera D R, Rittmann B E, Stahl D A. Quantification of methanogenic groups in anaerobic biological reactors by oligonucleotide probe hybridization. Appl Environ Microbiol. 1994;60:1241–1248. doi: 10.1128/aem.60.4.1241-1248.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reysenbach A, Giver L J, Wickham G S, Pace N R. Differential amplification of rRNA genes by polymerase chain reaction. Appl Environ Microbiol. 1992;58:3417–3418. doi: 10.1128/aem.58.10.3417-3418.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rölleke S, Muyzer G, Wawer C, Wanner G, Lubitz W. Identification of bacteria in a biodegraded wall painting by denaturing gradient gel electrophoresis of PCR-amplified gene fragments coding for 16S rRNA. Appl Environ Microbiol. 1996;62:2059–2065. doi: 10.1128/aem.62.6.2059-2065.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 50.Satoh H, Mino T, Matsuo T. Uptake of organic substrates and accumulation of polyhydroxyalkanoates linked with glycolysis of intracellular carbohydrates under anaerobic conditions in the biological excess phosphate removal process. Water Sci Technol. 1992;26:933–942. [Google Scholar]
- 51.Satoh H, Mino T, Matsuo T. Deterioration of enhanced biological phosphorus removal by the domination of microorganisms without polyphosphate accumulation. Water Sci Technol. 1994;30:203–211. [Google Scholar]
- 52.Schmidt T M, DeLong E F, Pace N R. Analysis of a marine picoplankton community by 16S rRNA gene cloning and sequencing. J Bacteriol. 1991;173:4371–4378. doi: 10.1128/jb.173.14.4371-4378.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schuppler M, Mertens F, Schön G, Göbel U B. Molecular characterization of nocardioform actinomycetes in activated sludge by 16S rRNA analysis. Microbiology. 1995;141:513–521. doi: 10.1099/13500872-141-2-513. [DOI] [PubMed] [Google Scholar]
- 54.Smalla K, Wachtendorf U, Heuer H, Liu W-T, Forney L J. Analysis of Biolog-GN substrate utilization patterns by microbial communities. Appl Environ Microbiol. 1998;64:1220–1225. doi: 10.1128/aem.64.4.1220-1225.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Smolders G J F, van der Meij J, van Loosdrecht M C M, Heijnen J J. Model of the anaerobic metabolism of the biological phosphorus removal process: stoichiometry and pH influence. Biotechnol Bioeng. 1994;43:461–470. doi: 10.1002/bit.260430605. [DOI] [PubMed] [Google Scholar]
- 56.Stahl D A, Flesher B, Mansfield H R, Montgomery L. Use of phylogenetically based hybridization probes for studies of ruminal microbial ecology. Appl Environ Microbiol. 1988;54:1079–1084. doi: 10.1128/aem.54.5.1079-1084.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stahl D A, Amann R I. Development and application of nucleic acid probes. In: Stackebrandt E, Goodfellow M, editors. Nucleic acid techniques in bacterial systematics. New York, N.Y: John Wiley & Sons, Inc.; 1991. pp. 205–248. [Google Scholar]
- 58.Tebbe C C, Vahjen W. Interference of humic acids and DNA extracted directly from soil in detection and transformation of recombinant DNA from bacteria and a yeast. Appl Environ Microbiol. 1993;59:2657–2665. doi: 10.1128/aem.59.8.2657-2665.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Teske A, Wawer C, Muyzer G, Ramsing N B. Distribution of sulfate-reducing bacteria in a stratified fjord (Mariager Fjord, Denmark) as evaluated by most-probable-number counts and denaturing gradient gel electrophoresis of PCR-amplified ribosomal DNA fragments. Appl Environ Microbiol. 1996;62:1405–1415. doi: 10.1128/aem.62.4.1405-1415.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Van Loosdrecht M C M, Smolders G J, Kuba T, Heijnen J J. Metabolism of micro-organisms responsible for enhanced biological phosphorus removal from wastewater. Antonie Leeuwenhoek. 1997;71:109–116. doi: 10.1023/a:1000150523030. [DOI] [PubMed] [Google Scholar]
- 61.Wagner M, Amann R, Lemmer H, Schleifer K H. Probing activated sludge with oligonucleotides specific for proteobacteria: inadequacy of culture-dependent methods for describing microbial community structure. Appl Environ Microbiol. 1993;59:1520–1525. doi: 10.1128/aem.59.5.1520-1525.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wagner M, Erhart R, Manz W, Amann R I, Lemmer H, Wedi D, Schleifer K H. Development of an rRNA-targeted oligonucleotide probe specific for the genus Acinetobacter and its application for in situ monitoring in activated sludge. Appl Environ Microbiol. 1994;60:792–800. doi: 10.1128/aem.60.3.792-800.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wallner G, Erhart R, Amann R I. Flow cytometric analysis of activated sludge with rRNA-targeted probes. Appl Environ Microbiol. 1995;61:1859–1866. doi: 10.1128/aem.61.5.1859-1866.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ward D M, Weller R, Bateson M M. 16S rRNA sequences reveal numerous uncultured microorganisms in a natural community. Nature. 1990;345:63–65. doi: 10.1038/345063a0. [DOI] [PubMed] [Google Scholar]
- 65.Wentzel M C, Loewenthal R E, Ekama G A, Marais G V R. Enhanced polyphosphate organism cultures in activated sludge systems. Part 1. Enhanced culture development. Water SA. 1988;14:81–92. [Google Scholar]
- 66.Wilson I G. Inhibition and facilitation of nucleic acid amplification. Appl Environ Microbiol. 1997;63:3741–3751. doi: 10.1128/aem.63.10.3741-3751.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]