Abstract
In the quest of identifying newer molecular targets for the management of glucocorticoid-induced ocular hypertension (GC-OHT) and glaucoma (GCG), several microarray studies have attempted to investigate the genome-wide transcriptome profiling of primary human trabecular meshwork (TM) cells in response to dexamethasone (DEX). However, no studies are reported so far to demonstrate the temporal changes in the expression of genes in the cultured human TM cells in response to DEX treatment. Therefore, in the present study, the time-dependent changes in the genome-wide expression of genes in primary human TM cells after short (16 hours: 16 h) and long exposure (7 days: 7 d) of DEX was investigated using RNA sequencing. There were 199 (118 up-regulated; 81 down-regulated) and 525 (119 up-regulated; 406 down-regulated) DEGs in 16 h and 7 d treatment groups respectively. The unique genes identified in 16 h and 7 d treatment groups were 152 and 478 respectively. This study found a distinct gene signature and pathways between two treatment regimes. Longer exposure of DEX treatment showed a dys-regulation of Wnt and Rap1 signaling and so highlighted potential therapeutic targets for pharmacological management of GC-OHT/glaucoma.
Subject terms: Cell biology, Molecular biology, Medical research
Introduction
Glucocorticoids (GCs) use is widespread in ophthalmology and continues to be the mainstay of treatment for inflammatory eye diseases1. The prolonged use of GCs elevates intraocular pressure (IOP) in susceptible individuals2. It is reported that the increase in IOP happens in 40% of the individuals without glaucoma (steroid responder) and in up to 90% of primary open angle glaucoma2–4. The risk of IOP elevation, and the time-frame during which this occurs, is dependent on steroid potency, pharmacokinetics, duration of treatment, routes of administration, as well as individual differences in responsiveness5. The exact molecular mechanism for GC-induced elevated IOP is not clearly understood.
In the trabecular meshwork (TM), dexamethasone (DEX) is known to induce cell remodeling5–9 and extracellular matrix (ECM) remodeling which are implicated in ocular hypertension caused by GCs10. Several studies have examined global changes in gene expression in the cultured human TM cells treated with dexamethasone (DEX) using cDNA or oligonucleotide arrays9,11–20. However, the overall findings were not consistent across studies because of the different duration of treatment (varied from 24 h to 21 days) and different microarray platforms used in these studies. These studies were restricted due to a single/ limited time point.
Since the responses of the TM to DEX treatment are dose and duration dependent, no studies have been reported so far to demonstrate the temporal changes in the expression of genes in the cultured human TM cells. Therefore, in the present study, the time-dependent changes in the genome-wide expression of genes in primary human TM cells after short (16 hours: 16 h) and long exposure (7 days: 7 d) of 100 nM DEX were assessed using RNA-Sequencing (RNA-Seq). This is the first study report on the temporal changes in the expression of genes in the cultured human TM cells after two different DEX exposure time points. We hypothesized that the shorter DEX exposure may reveal acute and initiating alterations in gene expression which is responsible for the sustained ocular hypertensive phenotypes in the TM which persists with longer exposure of DEX. The results of the present study revealed a distinct gene signature observed between two treatment groups and found that long exposure of DEX treatment showed dys-regulation of two signaling pathways such as Wnt and Rap1 signaling. These pathways could serve as potential therapeutic targets for pharmacological management of GC-OHT/glaucoma.
Results
Quality of RNA-seq data
An average of 35 million reads were generated from each sample of 16 h treated group and obtained by de-multiplexing. Each base from mRNA-sequencing reads of 16 h and 7 d DEX-treated cells met the expected Phred score criteria, Q > 30.
In Principle Component Analysis, the samples got dispersed into different regions of the plot corresponding to the principal sources of variation within the experiment. Both control as well treated groups segregate on the first and second principal component axes, but there is a clear relationship between donor eyes, indicating the need to incorporate donor source as a factor in an expression analysis of this data.
Differential gene expression analysis
In total 16,335 and 17,499 genes with more than ten read counts were identified in 16 h (Group A) and 7 d (Group B) DEX-treated HTM cells respectively. There were 199 (118 up-regulated; 81 down-regulated) DEGs in Group A and 525 (119 up-regulated; 406 down-regulated) DEGs in Group B identified as differentially expressed with absolute fold change 2 and probability value lesser than 0.05. The relationship between statistical significance and log FC in normalized expression between the experimental groups are shown in the volcano plot (Fig. 1). The commonly expressed genes from Group C were found to be 47 and out of which ZBTB16, SAA1, OCA2, RGCC and FKBP5 were identified as the most up-regulated and KRT15, FST, and TP63 the most down-regulated genes. Unique genes identified in 16 h (Group D) and 7 d (Group E) treatment groups were 152 and 478 respectively (Fig. 2).
Figure 1.
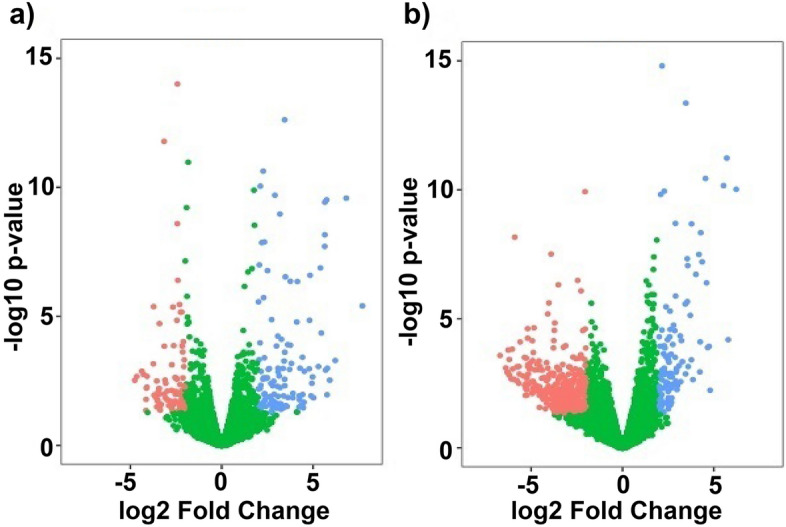
Volcano Plot Showing the Distribution of DEGs. The fold of change (log2) and p value (− log10) of genes from (a) 16 h DEX treatment (Group A), (b) 7 d DEX treatment (Group B) are represented.
Figure 2.
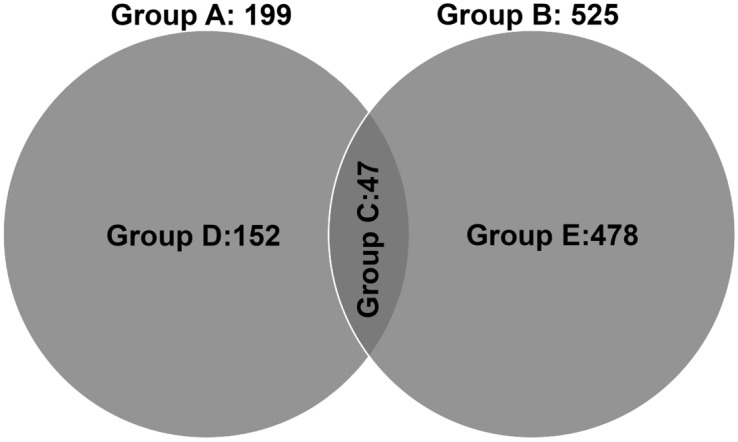
Venn diagram showing differentially expression groupings. DEGs of RNA seq data identified in different study groups are shown. Only genes with absolute fold change 2 and significant P value < 0.05 were included in these groupings. Group A: DEGs between DEX and ETH treated for short duration (16 h); Group B: DEGs between DEX and ETH treated for longer duration (7 d); Group C: DEGs that overlapping between Group A and Group B; Group D: Uniquely expressed DEGs of HTM cells exposed for 16 h (Group A minus Group C); Group E: Uniquely expressed DEGs of HTM cells exposed for 7 d (Group B minus Group C).
In Group A, the genes such as CCAT1 (FC = 5.9), CIDEC (FC = 5.7), NDST4 (FC = 5.7), FAM105A (FC = 5.6) and APOB (FC = 4.2) genes were found to be highly up-regulated, while LRRN1 (FC = − 4.1), FGF16 (FC = − 4.1) and SERPINB2 (FC = − 3.7) were down-regulated and these genes were uniquely expressed in Group D whereas, in Group B, MYOC (FC = 5.7), APOD (FC = 4.7), PRODH (FC = 4.5), LSP1 (FC = 4.2) and IGF2 (FC = 3.7) genes were identified as highly up-regulated, while NPY (FC = − 6.7), PRSS22 (FC = − 6.3), ACPP (FC = − 6.2) and RLN1 (FC = − 6.1) were down-regulated genes and were found to be uniquely expressed in Group E (Supplementary Table S3a–e). Interestingly, a greater number of significantly DEGs were identified in 7 d treatment group as compared to 16 h treatment group which indicates that prolonged exposure of DEX significantly alters the gene expression profile in HTM cells.
Pathways enrichment analysis
The pathway enrichment analysis of DEGs from each group revealed some unique significantly altered pathways (Table 1). They were clustered into several functional categories. Interestingly, Drug metabolism—cytochrome P450 and tyrosine metabolism pathways were commonly expressed as up-regulated pathways in Group A (16 h) and Group B (7 d). The up-regulation of cytokine-cytokine receptor interaction pathway was found only in 16 h group (Group D; unique 16 h) whereas Rap1 signaling pathway was observed only in 7 d group (Group E; unique 7 d). The down-regulated pathways found in Group D were hematopoietic cell lineage, neuroactive ligand-receptor interaction and calcium signaling pathway and in Group E, the pathways such as Wnt signaling pathway, pathways in cancer, cell adhesion molecules and proteoglycans in cancer pathways were found (Table 1).
Table 1.
List of enriched up/down-regulated pathways.
| Group | Pathway | Fold enrichment | P value | Genes |
|---|---|---|---|---|
| Group A | Tyrosine metabolism | 20.7 | 8.3E−04 | PNMT, MAOA, ADH1B, AOX1 |
| Cytokine-cytokine receptor interaction | 3.7 | 4.0E−02 | LEP, CCL3L3, IL1R2, IL18, IFNLR1 | |
| Amphetamine addiction | 8.2 | 4.9E−02 | GRIA1, MAOA, PPP1R1B | |
| Drug metabolism—cytochrome P450 | 8.0 | 5.1E−02 | MAOA, ADH1B, AOX1 | |
| Neuroactive ligand-receptor interaction | − 5.3 | 4.0E−03 | GRIN2A, GABRA4, TACR1, GRPR, MCHR1, NTSR1 | |
| Cytokine-cytokine receptor interaction | − 5.1 | 1.4E−02 | IL1A, CSF2, IL1B, TNFRSF9, CXCR4 | |
| Calcium signaling pathway | − 5.5 | 3.2E−02 | GRIN2A, TACR1, GRPR, NTSR1 | |
| Salmonella infection | − 8.9 | 4.2E−02 | IL1A, CSF2, IL1B | |
| Hematopoietic cell lineage | − 8.5 | 4.5E−02 | IL1A, CSF2, IL1B | |
| Rheumatoid arthritis | − 8.4 | 4.6E−02 | IL1A, CSF2, IL1B | |
| Group B | Drug metabolism—cytochrome P450 | 15.2 | 3.6E−05 | ADH4, MAOA, ADH1B, ADH1A, AOX1, FMO2 |
| Tyrosine metabolism | 24.6 | 4.1E−05 | ADH4, MAOA, ADH1B, ADH1A, AOX1 | |
| Retinol metabolism | 10.7 | 5.5E−03 | ADH4, ADH1B, ADH1A, AOX1 | |
| Fatty acid degradation | 12.3 | 2.3E−02 | ADH4, ADH1B, ADH1A | |
| Glycolysis/gluconeogenesis | 7.7 | 5.5E−02 | ADH4, ADH1B, ADH1A | |
| Chemical carcinogenesis | 6.4 | 7.5E−02 | ADH4, ADH1B, ADH1A | |
| Cell adhesion molecules (CAMs) | − 4.6 | 8.6E−06 | ICAM2, SELE, SELP, HLA-DMB, CDH3, CLDN3, CDH1, PECAM1, HLA-DRA, HLA-DOA, HLA-DQA2, HLA-DQA1, HLA-DRB1, HLA-DQB1 | |
| Allograft rejection | − 8.9 | 1.1E−04 | HLA-DMB, HLA-DRA, HLA-DOA, HLA-DQA2, HLA-DRB1, HLA-DQA1, HLA-DQB1 | |
| Basal cell carcinoma | − 6.9 | 1.3E−04 | WNT10B, SHH, WNT10A, WNT7B, PTCH2, FZD10, WNT2, WNT4 | |
| Type I diabetes mellitus | − 7.8 | 2.3E−04 | HLA-DMB, HLA-DRA, HLA-DOA, HLA-DQA2, HLA-DRB1, HLA-DQA1, HLA-DQB1 | |
| Inflammatory bowel disease (IBD) | − 5.8 | 3.8E−04 | HLA-DMB, RORC, HLA-DRA, HLA-DOA, HLA-DQA2, HLA-DRB1, HLA-DQA1, HLA-DQB1 | |
| Autoimmune thyroid disease | − 6.3 | 7.5E−04 | HLA-DMB, HLA-DRA, HLA-DOA, HLA-DQA2, HLA-DRB1, HLA-DQA1, HLA-DQB1 | |
| Pancreatic secretion | − 4.0 | 3.5E−03 | CPA3, CPB1, FXYD2, KCNQ1, PLA2G2A, CLCA2, CD38, ATP1A2 | |
| cAMP signaling pathway | − 2.6 | 9.2E−03 | GRIA2, CHRM1, NPY, FXYD2, PDE3B, CNGA1, ADRB1, ATP1A2, SSTR1, SSTR2, GRIN1 | |
| Hippo signaling pathway | − 2.8 | 1.5E−02 | WNT10B, WNT10A, CDH1, WNT7B, RASSF6, FZD10, WNT2, BMP7, WNT4 | |
| Wnt signaling pathway | − 2.7 | 2.7E−02 | WNT10B, WNT10A, SFRP2, MMP7, WNT7B, FZD10, WNT2, WNT4 | |
| Signaling pathways regulating pluripotency of stem cells | − 2.7 | 2.9E−02 | WNT10B, WNT10A, WNT7B, IGF1, FZD10, WNT2, ISL1, WNT4 | |
| Pathways in cancer | − 1.8 | 3.9E−02 | WNT10B, WNT10A, WNT7B, PTCH2, CBLC, KLK3, FZD10, IGF1, MMP9, SHH, GNG4, CDH1, WNT2, NKX3-1, WNT4 | |
| cGMP-PKG signaling pathway | − 2.4 | 5.1E−02 | NOS3, FXYD2, PDE2A, PDE3B, CNGA1, ADRB1, ATP1A2, ADRA2A | |
| Adrenergic signaling in cardiomyocytes | − 2.4 | 7.3E−02 | MYL4, TNNT2, FXYD2, KCNQ1, ADRB1, ATP1A2, CACNG4 | |
| Cardiac muscle contraction | − 3.1 | 7.5E−02 | MYL4, TNNT2, FXYD2, ATP1A2, CACNG4 | |
| Group C | Tyrosine metabolism | 45.4 | 2E−3 | MAOA, ADH1B, AOX1 |
| Drug metabolism—cytochrome P450 | 23.3 | 6E−3 | MAOA, ADH1B, AOX1 | |
| Tryptophan metabolism | 26.5 | 6.8E−2 | MAOA, AOX1 | |
| Group D | Cytokine-cytokine receptor interaction | 4.5 | 5.1E−2 | CCL3L3, IL1R2, IL18, IFNLR1 |
| Neuroactive ligand-receptor interaction | − 5.4 | 1.1E−2 | GRIN2A, GABRA4, TACR1, GRPR, NTSR1 | |
| Calcium signaling pathway | − 6.7 | 1.9E−2 | GRIN2A, TACR1, GRPR, NTSR1 | |
| Salmonella infection | − 10.8 | 2.8E−2 | IL1A, CSF2, IL1B | |
| Hematopoietic cell lineage | − 10.3 | 3.1E−2 | IL1A, CSF2, IL1B | |
| Rheumatoid arthritis | − 10.2 | 3.2E−2 | IL1A, CSF2, IL1B | |
| Amoebiasis | − 8.5 | 4.4E−2 | CSF2, SERPINB2, IL1B | |
| Measles | − 6.7 | 6.7E−2 | IL1A, IL1B, TACR1 | |
| Circadian rhythm | − 19.3 | 9.5E−2 | PER3, NR1D1 | |
| Group E | Drug metabolism—cytochrome P450 | 10.5 | 3.1E−2 | ADH4, ADH1A, FMO2 |
| Rap1 signaling pathway | 4.5 | 5.2E−2 | FPR1, KDR, FGFR4, RAPGEF5 | |
| Cell adhesion molecules (CAMs) | − 4.8 | 0.000 | ICAM2, SELE, SELP, HLA-DMB, CDH3, CLDN3, CDH1, PECAM1, HLA-DRA, HLA-DOA, HLA-DQA2, HLA-DQA1, HLA-DRB1, HLA-DQB1 | |
| Type I diabetes mellitus | − 8.2 | 0.000 | HLA-DMB, HLA-DRA, HLA-DOA, HLA-DQA2, HLA-DRB1, HLA-DQA1, HLA-DQB1 | |
| Antigen processing and presentation | − 5.2 | 1E−3 | CD74, HLA-DMB, HLA-DRA, HLA-DOA, HLA-DQA2, HLA-DRB1, HLA-DQA1, HLA-DQB1 | |
| cAMP signaling pathway | − 2.7 | 7E-3 | GRIA2, CHRM1, NPY, FXYD2, PDE3B, CNGA1, ADRB1, ATP1A2, SSTR1, SSTR2, GRIN1 | |
| Proteoglycans in cancer | − 2.7 | 7E−3 | WNT10B, WNT10A, HPSE2, WNT7B, CBLC, IGF1, FZD10, WNT2, HOXD10, MMP9, WNT4 | |
| Wnt signaling pathway | − 2.8 | 2.1E−2 | WNT10B, WNT10A, SFRP2, MMP7, WNT7B, FZD10, WNT2, WNT4 | |
| Signaling pathways regulating pluripotency of stem cells | − 2.8 | 2.3E−2 | WNT10B, WNT10A, WNT7B, IGF1, FZD10, WNT2, ISL1, WNT4 | |
| Pathways in cancer | − 1.9 | 2.7E−2 | WNT10B, WNT10A, WNT7B, PTCH2, CBLC, KLK3, FZD10, IGF1, MMP9, SHH, GNG4, CDH1, WNT2, NKX3-1, WNT4 | |
| Phagosome | − 2.6 | 3.2E−2 | HLA-DMB, SFTPA2, HLA-DRA, HLA-DOA, HLA-DQA2, HLA-DRB1, HLA-DQA1, HLA-DQB1 | |
| Hippo signaling pathway | − 2.6 | 3.3E−2 | WNT10B, WNT10A, CDH1, WNT7B, FZD10, WNT2, BMP7, WNT4 | |
| cGMP-PKG signaling pathway | − 2.5 | 4.1E−2 | NOS3, FXYD2, PDE2A, PDE3B, CNGA1, ADRB1, ATP1A2, ADRA2A | |
| Adrenergic signaling in cardiomyocytes | − 2.5 | 6E−2 | MYL4, TNNT2, FXYD2, KCNQ1, ADRB1, ATP1A2, CACNG4 | |
| Cardiac muscle contraction | − 3.3 | 6.5E−2 | MYL4, TNNT2, FXYD2, ATP1A2, CACNG4 | |
| Dilated cardiomyopathy | − 2.9 | 9E−2 | DES, TNNT2, ADRB1, IGF1, CACNG4 |
Group A: DEGs between DEX and ETH treated for short duration (16 h); Group B: DEGs between DEX and ETH treated for longer duration (7 d); Group C: DEGs that overlapping between Group A and Group B; Group D: Uniquely expressed DEGs of HTM cells exposed for 16 h (Group A minus Group C); Group E: Uniquely expressed DEGs of HTM cells exposed for 7 d (Group B minus Group C).
Validation RNA seq data by RT-PCR
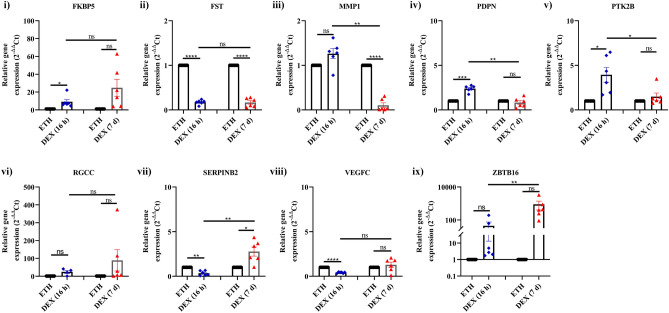
To validate results obtained from the RNA-seq analysis, 9 candidate genes were selected for q-PCR analysis based on their statistical significance in the RNA-seq data set and on the basis of published literature providing evidence of their involvement in glaucoma pathophysiology. Eight of the nine candidate genes demonstrated consistent trends with RNA seq data (Fig. 3, Table 2). Though MMP-1 expression was found to be down-regulated in RNA-seq data, it was found to be up-regulated in qPCR analysis but the difference was not statistically significant.
Figure 3.
Validation of DEGs by qPCR. Expression profile of selected genes identified from RNA-seq validated by qPCR is shown. Primary HTM cells were treated with 100 nM DEX or 0.1% ETH for 16 h and 7 d. Gene expressions were normalized to GAPDH (16 h group) and ACTB (7 d group), and analyzed using the 2−ΔΔCT method. The data is represented as mean ± SEM. *P < 0.01; **P < 0.005; ***P < 0.001; ****P < 0.0001. Paired 2-tailed Student’s t test.
Table 2.
Comparison between RNA sequencing findings and qPCR.
| S. no. | Name of the gene | Log2 fold change | |||
|---|---|---|---|---|---|
| Group A | Group B | ||||
| RNA seq | qPCR | RNA seq | qPCR | ||
| 1 | FKBP5 | 2.91 | 2.97 | 4.47 | 3.94 |
| 2 | FST | − 2.45 | − 2.61 | − 2.04 | − 2.88 |
| 3 | MMP1 | − 1.97 | 0.30 | − 1.47 | − 4.53 |
| 4 | PDPN | 1.35 | 1.24 | − 0.001 | − 0.60 |
| 5 | PTK2B | 2.34 | 1.79 | 1.8 | 0.38 |
| 6 | RGCC | 4.79 | 3.54 | 3.59 | 4.48 |
| 7 | SERPINB2 | − 3.73 | − 2.10 | − 0.41 | 1.32 |
| 8 | VEGFC | − 1.32 | − 1.30 | − 0.82 | − 0.10 |
| 9 | ZBTB16 | 7.70 | 3.19 | 6.47 | 8.90 |
Group A: DEGs between DEX and ETH treated 16 h; Group B: DEGs between DEX and ETH treated for 7 d.
FKBP5 FK506 binding protein 5, FST follistatin, MMP1 matrix metalloproteinase-1, PDPN podoplanin; protein tyrosine kinase 2 beta, RGCC regulator of cell cycle, SERPINB2 serpin family B member 2, VEGFC vascular endothelial growth factor-C, ZBTB16 zinc finger and BTB domain containing 16.
Discussion
The present study utilized RNA-seq to explore genome-wide, time-dependent changes in the expression of genes in primary HTM cells in response to DEX treatment for a shorter (16 h) or longer duration (7 d) of exposure. Several previous studies identified the global changes in the expression of DEGs in response to DEX treatment in cultured HTM cells using cDNA and oligonucleotide arrays11–20. However, no studies are available to-date to demonstrate the temporal changes in the expression of genes in cultured HTM cells in response to DEX treatment. Therefore, in the present study two different DEX exposure time points were chosen: short duration (16 h) and long duration (7 d) of exposure. A shorter DEX exposure time may reveal acute and initiating alterations in gene expression which is responsible for the sustained ocular hypertensive phenotypes in the TM cells. The long exposure time of 7 d was chosen to investigate genes altered by chronic GC exposure to further identify the molecular changes in the HTM cells resulting in GC-induced OHT and glaucoma and also to identify novel therapeutic targets.
Our study revealed a distinct gene signature and their associated pathways that were identified for both the time points studied. Interestingly, the 7 d post DEX treatment showed a higher number of significantly altered genes (DEGs: 525–119, up-regulated and 406, down-regulated) as compared to 16 h treatment (DEGs: 199–118, up-regulated and 81, down-regulated) which indicates that prolonged exposure of DEX significantly alters the gene expression profile in HTM cells. A common number of genes that were up-regulated in the previous studies were identified in the present study in both the groups12,13,15,17,18,21,22. However, the expression of these genes varied in fold change with time points studied (16 h versus 7 d) which confirms that DEX-induced mRNA expression is time dependent.
The present study also revealed a novel set of genes which are not reported earlier12,13,15,17,18,21–23. This could be due to differences in the GC responsiveness of the human donor eyes used in the present study and in fact RNA-Seq technology offers significant benefits over previous microarray technologies in terms of offering a broader dynamic range and the sequencing coverage depth allows easier detection of rare and low-abundance transcripts.
Myocilin (MYOC), a known DEX-inducible gene was significantly altered in 7 d post-treatment group (log2FC: 5.79) as compared to 16 h group (log2FC:1.3)21. It has been identified as one of the four major pathogenic genes for POAG and JOAG22. ZBTB16 (Zinc Finger and BTB Domain Containing 16), a transcriptional repressor involved in cell cycle control and suppress the expression of several genes that regulate cell proliferation were highly expressed in both groups. However, the role of ZBTB16 in HTM cells and steroid-induced glaucoma still remains unclear23. The oculo-cutaneous albinism 2 (OCA2) which is prominently associated with pigmentation and acts as a determinant of brown or blue eye color was identified as one of the highly expressed genes in both short and long-term treatment groups (log2FC:6.8 (16 h); log2FC:5.7 (7 d). FK506 binding protein 5 (FKBP5), a co-chaperone of heat shock protein 90 (hsp90) which regulates GC receptor sensitivity was up-regulated at both time points. Loss of DNA methylation in FKBP5 by GC, increases FKBP5 transcription12.
The serum amyloid A proteins (SAA1 and SAA2), which are acute phase apo-lipoproteins reported to play an important role in inflammation, infection and tissue repair were found to be up-regulated in both time-points studied19,20. However, the intensity of gene expression was higher in 7 d DEX-treated group as compared to 16 h group. Moreover, another isoform of serum amyloid A protein 4, SAA4 was additionally found only in 16 h group. The Angiopoietin-like 7 (ANGPTL7) protein, a member of the ANGPTL family was one of the most up-regulated in DEX-treated HTM cells at two different time points studied. The overexpression of ANGPTL7 in human TM and aqueous humor was found to alter the expression of collagen and fibronectin, myocilin and MMP124,25. Rare protein-altering variants in ANGPTL7 lower IOP and protect against glaucoma26.
Among the most commonly down-regulated genes identified between two groups, follistatin (FST) is known to be induced by DEX treatment in TM cells18. FST is a stress responsive protein exerting protective effects by neutralizing TGF-β2 signaling by antagonizing BMP and activin27. FST is expressed in the normal and glaucomatous TM28,29 and this expression is increased in a dose-dependent manner with TGF-β2 treatment29. In this study, FST was significantly down-regulated in both groups (log2FC: 16 h: − 2.4; 7 d: − 2.0) and it was corroborated by qPCR assay. The biological activities and underlying mechanisms for FST involvement in the TM is not understood clearly but it is proposed that FST reduces ROS production in TM cells by interacting with members of the NADPH oxidase inhibitor (NOX) family30. Interestingly, FST can be regulated at both transcriptional and post-transcriptional levels31,32, which allows an opportunity to manipulate FST expression therapeutically.
Out of 47 commonly found DEGs between two treatment groups, the expression of complement factor 7 (C7), solute carrier organic anion transporter family member 2A1 (SLCO2A1) and Ras Association Domain Family Member 6 (RASSF6) were differentially altered. For example, the C7 was up-regulated in 16 h treatment group (log2FC: 5.0) whereas in the 7 d treatment group, the expression was down-regulated (log 2FC: − 2.5). C7 is a terminal component of the complement cascade and can form a membrane attack complex with other complement components (C5b, C6, C8 and C9) which constitutes a part of the innate immune system. The expression of C7 on the cell membrane acts as a regulator of an excessive pro-inflammatory reaction. In some tumors, C7 acts as a tumor suppressor and found to show a decreased mRNA expression level correlated with an increased aggressiveness of the tumors33. Likewise, RASSF6 was differentially expressed in response to DEX treatment and is suppressed in human cancers and its low expression level is associated with poor prognosis34. However, the association of C7, RASSF6 and GC-induced glaucoma is not known and warrants further investigation.
Solute carrier organic anion transporter family member 2A1 (SLCO2A1) gene is known to encode the prostaglandin transporter PGT35 and is involved in the process of uptake and clearance of prostaglandins36. The mRNA expression of this transporter is present in a variety of human ocular tissues especially in the ciliary epithelium and choroid/RPE complex37. Glucocorticoids are known to inhibit the production of prostaglandins in many cell types and tissues38. In the TM, it has been demonstrated that the DEX treatment reduced the production of prostaglandin by 60%39. The present study revealed that longer exposure of DEX significantly down-regulated this transporter as compared to short exposure and hence the regulation of genes upon DEX treatment is time-dependent.
In addition to the known DEX-induced genes reported in cultured HTM cells, our study revealed some unique genes identified in each treatment group (16 h and 7 d) (Supplementary Table S3d,e). For example, plasminogen activator inhibior2 (PAI-2) or serpineB2 (SERPINB2), a serine protease inhibitor of the serpin superfamily induced during inflammatory processes and infections in many cell types such as macrophages, fibroblasts, endothelial cells and differentiating keratinocytes40. In the present study, it is interesting to note that SERPINB2 was significantly down-regulated in 16 h treatment group (log2FC: − 3.7) whereas in 7 d treatment group, the other form of SERPINB11was significantly down-regulated (log2FC: − 4.9). Validation of SERPINB2 decreased expression could not be achieved by q-PCR (Fig. 3). Significant glucocorticoid down-regulation of SERPINB2 expression in the TM was also reported after 6–8 days of DEX treatment15. SERPINE1 (PAI-1), a phylogenetically distinct but regulated coagulation factor is induced in the TM when treated with TGFβ2 which is proposed to inhibit ECM degradation via MMP regulation. TGFβ2 induced PAI-1 increased substrate elasticity and contribute aberrant ECM deposition in glaucoma41. However, the role of SERPINB2 and SERPINB11in the TM is not clearly understood. A detailed investigation in this line may give insight into the involvement of SERPINB2 and other members of this family in the pathogenesis of GC-induced glaucoma.
Similarly, neuropeptide (NPY) was significantly down-regulated only in 7 d treatment group (log2FC: − 6.7) and not found in 16 h post treatment. This peptide is known to be involved in various physiological and homeostatic processes in both central and peripheral nervous systems. The expression of NPY was documented in the retina of both mammalian and non-mammalian species where it is reported to prevent neuronal cell death induced by excitotoxic insults42,43. It is also expressed in drainage angle of mammalian eyes44. However, the physiological role of NPY in trabecular meshwork cells is not clearly understood which warrants a detailed investigation to understand its role in GC-OHT and glaucoma.
In this study, several functional pathways were significantly altered in both the groups after DEX treatment. The significant down-regulation of WNT signaling was observed only in 7 d DEX treated HTM cells with decreased expression of WNT10B (logFC = − 2.8), WNT10A (logFC = − 2.8), WNT7B (logFC = − 2.6), WNT2 (logFC = − 3.5) and WNT4 (logFC = − 2.0). The WNT signaling antagonist SFRP2 (logFC = − 3.7) was also identified as down-regulated only in 7 d treatment group. These findings suggest that the prolonged exposure of DEX in HTM cells resulted in the inhibition of WNT signaling. Previously, it was shown that there is an existence of a cross-inhibition between the TGFβ/Smad and canonical Wnt pathways in the TM and such cross-inhibition was mediated by Smad4 and β-catenin45. In a metanalysis of human microarray DEX expression datasets and bovine RNA -seq these two pathways were shown to be in TM cells in response to DEX treatment46. Among the 11 pathways that were altered between human and bovine TM cells in response to DEX treatment, only cell adhesion and Wnt signaling pathways were found similar in the present study.
In a recent study, it is shown that GR and WNT signaling inhibit each other in the TM, and that activation of Wnt signaling could prevent the adverse effect of glucocorticoids in the eye47. In the above study, elevated levels of Dickkopf-related protein1 (Dkk1) which is a canonical Wnt signaling inhibitor was found in the aqueous humor and TM of glaucoma patients. Dkk1 enhanced GR signaling and knockdown of Dkk1 or Wnt signaling activators decreased GR signaling in human TM cells. In contrast, the inhibition of canonical Wnt signaling by β-catenin knockdown increased GC-induced up-regulation of ECM proteins. In addition, it has been demonstrated that adenovirus mediated Wn3a expression decreased GC-induced OHT in mouse eyes. These observations strongly suggest that Wnt activation may be of potential strategy for the future treatment for GC-induced OHT and glaucoma.
The Rap1 signaling pathway was also significantly up-regulated only in 7 d treatment group. Rap1 regulates diverse cellular functions including focal adhesions, cell junction formation and cell polarity. A recent study by Zhu et al.48 identified rottlerin as a potential therapeutic agent for POAG through bio-informatics analysis of microarray data sets derived from POAG TMs and found that rottlerin could reverse the glaucomatous phenotype in both in vitro and steroid-induced experimental mouse model. Target prediction identified that Rap1 signaling as a part of a potential pathway underlying the effects of rottlerin. However, the role of Rap1 signaling has not been well studied in glaucoma. Further investigation into the role of the Rap1 signaling pathway in glaucoma including steroid glaucoma is warranted.
Interestingly, the cytokine-cytokine receptor interaction pathway was significantly enriched only in 16 h group as compared to 7 d group. This is in agreement with the observation by Peng et al.49 that this was the pathway which was significantly enriched in the aqueous humor of patients with POAG and Sturge–Weber syndrome induced secondary glaucoma. Cytokine-cytokine receptor interactions may play a role in immune-pathogenesis of glaucoma.
The findings of the present study may have a clinical relevance that DEX implants are increasingly being used for retinal vascular diseases due to increase in prevalence in our aging society. Long-term use of these implants results in elevated IOP which represents a major clinical challenge. This study opens up the avenues to design a steroid response susceptibility test to screen patients at risk before initiating steroid therapy for any eye inflammatory conditions. Some molecular targets with the potential of developing into drug therapy for the management of steroid glaucoma and POAG were identified in the present study.
Our study has some limitations. The history of steroid responsiveness of the donor eyes used in the present study was not known. Adaptation of an alternative method to better screen the steroid responsiveness of the donor eyes may provide a more disease appropriate cohort for examining global gene expression and aid in potential novel target identification. Due to limited availability of human donor eyes, HTM cells derived from two different ethnicities and RNA seq experiments conducted at two different centers were utilized in the present study which may contribute to variations in the gene expression profile. However, utmost care was taken to avoid the experimental variations by including appropriate vehicle controls for each DEX-treated HTM cells from the same donor.
In conclusion, this is the first report on the time-dependent effect of DEX on gene expression of primary human TM cells using RNA seq technology. Interestingly, both 16 h and 7 d DEX treatment groups demonstrated a distinct gene signature and their associated pathways in addition to common genes and pathways that were reported previously. Longer exposure of DEX in HTM cells showed down-regulation of Wnt signaling and up-regulation of Rap1 signaling. The cross-talk of these pathways with GR signaling warrants further experimental validation. Manipulation of these pathways have the potential to develop novel therapies for the management of GC-OHT/glaucoma.
Materials and methods
Human trabecular meshwork (HTM) cell culture
Human cadaveric eyes used for 16 h and 7 d duration were provided by the Liverpool Research Eye Bank, University of Liverpool, United Kingdom and Rotary Aravind International Eye Bank, Aravind Eye Hospital, Madurai, India, respectively. The characteristics of donor eyes used for this study are given in Supplementary Table S1. All eyes were examined under the dissecting microscope for any gross ocular pathological changes and only macroscopically normal eyes were used for the experiments. In Liverpool the medical and surgical history was available and all donors had no previous eye surgery or glaucoma. For eyes used in 7 d group, no medical and ocular history were available; however, the status of phakic/aphakic and history and duration of diabetes were available for those eye. Sixteen hours (16 h) (short DEX exposure time) and 7 days (7 d) long DEX exposure time) experiments were conducted at the University of Liverpool, UK and Aravind Medical Research Foundation, India, respectively.
Primary cultures of HTM cells were established and characterized as described previously50–53. Confluent cultures of HTM cells were treated with either 100 nM DEX or 0.1% ethanol (ETH) as a vehicle control for 16 h (n = 6) and 7 d (n = 8). The medium was exchanged every other day for 7 d treatment group. At the end of the respective treatments, RNA was extracted for gene expression profiling by RNA sequencing. Human TM cells between passages 3–7 were used for the experiments.
RNA extraction
Total RNA was isolated from cultured TM cells after 16 h or 7 d of treatment using the commercial Qiagen Universal All Prep (Qiagen, UK) kit and TRIZOL reagent (Sigma, MO, USA) as per manufacturer’s specifications respectively. Total RNA was quantified initially on the Nanodrop-1000 (Thermofisher Scientific, DE, UK) and quality was determined by the Bioanalyser 2100 (Agilent, UK). HTM cells with an RIN > 9.5 and a concentration of 100 ng/μl or higher were used for RNA sequencing54.
Expression profiling by RNA sequencing
RNA sequencing for 16 h and 7 d DEX–treated HTM cells were conducted at Exiqon Services, Denmark and Sandoor Life sciences, Hyderabad, India respectively as described previously15,23.
RNA seq data analysis
The paired-end raw reads of both 16 h and 7 d DEX-treated HTM cells strains were subjected to bioinformatic analysis to identify differentially expressed genes (DEG). Briefly, the quality of each base was assessed by FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/), and low-quality reads, PCR duplicates and adapter sequences were eliminated using Cutadapt3. The processed high quality paired end reads from each HTM cells were mapped with human reference genome hg38/GRCh38 using HISAT255 and then expression of genes in read counts was obtained by quantification using FeatureCounts56. Genes with less than 10 read counts were excluded before global level normalization by Quintile strategy.
Differential gene expression analysis using raw counts was carried out using EdgeR from Bioconductor package in R programme57, with the DEX treated group compared to vehicle treated group at the 16 h and 7 d time points. Models of differential expression at a series of group comparisons were constructed using the limma package and a Benjamini–Hochberg (BH) correction was applied. Genes with absolute fold change (log2) value 2, and BH P value ≤ 0.05 were considered as DEGs and taken for further analysis. For comparison, the DEGs were segregated into five groups: Group A: DEGs between DEX and ETH treated for short duration (16 h); Group B: DEGs between DEX and ETH treated for longer duration (7 d); Group C: DEGs overlapping between Group A and Group B; Group D: Uniquely expressed DEGs of HTM cells exposed for 16 h (Group A minus Group C); Group E: Uniquely expressed DEGs of HTM cells exposed for 7 d (Group B minus Group C).
Pathway enrichment analysis
Pathways associated with DEGs identified from RNA-seq were identified by Database for Annotation, Visualization and Integrated Discovery (DAVID) v6.8 (10.1186/gb-2003-4-9-r60)58,59. Pathways with significant P value (less than 0.05) and gene count greater than three were considered).
Validation of RNA seq by qPCR
DEGs identified in the RNA seq data of 16 h and 7 d treatment groups were further validated by qPCR. Nine candidate genes were selected for q-PCR analysis based on their statistical significance in the RNA-seq data set and on the basis of published literature providing evidence of their involvement in glaucoma pathophysiology12,15,17,20,23,27,40,51. The list of primers for the selected genes for validation by qPCR is shown in the Supplementary Table S2. The expression of significantly altered differentially expressed genes identified in 16 h RNA seq data was validated by qPCR as described previously51. For 7 d treatment group, qPCR was performed using ABI-QuantStudio 5 (Applied Biosystems, MA, USA).
All mRNA were measured at CT threshold levels and normalized with the average CT values of a reference gene; GAPDH (for 16 h treatment). Values were expressed as fold increase over the corresponding values for control by the 2−ΔΔCT method. The expression of genes in DEX treated HTM cells in logFC ratio was calculated by normalizing with reference control (ACTB; 7 d treatment) and vehicle control.
Statistical analysis
All statistical analysis of expression of genes were carried out by using Graph Pad Prism (version.8.0.2) (Graph Pad software, CA, USA). Statistical significance between groups was analyzed by using un-paired two-tailed Student-t-test. All data are represented in mean ± Standard Error Mean (SEM) or otherwise specified. P value < 0.05 was considered as statistically significant.
Institutional review board statement
This study was approved by the standing Human Ethics Committee of Aravind Medical Research Foundation, Madurai, Tamil Nadu, India (ID NO. RES2017006BAS-7 d treatment group) and University of Liverpool ethics review board (RETH000833-16 h treatment group) handled in accordance to the tenets of the Declaration of Helsinki.
Supplementary Information
Acknowledgements
The authors acknowledge the Rotary Aravind International Eye Bank, Aravind Eye Hospital, Madurai, India and the Liverpool Research Eye Bank, University of Liverpool, United Kingdom for providing human donor eyes for 16 h and 7 d treatment groups respectively for the present study.
Author contributions
Conceptualization: S.S. & C.E.W. Data Curation: S.S., C.E.W. & D.B. Formal Analysis: K.K., K.L., R.H., S.S., C.E.W. & D.B. Investigation: S.S., C.E.W. & D.B. Methodology: K.K., K.L. & R.H. Project administration: S.S. & C.E.W. Resources: S.S., C.E.W., D.B., D.A.S. Supervision: S.S. & C.E.W. Validation: S.S. & C.E.W. Software: D.B., K.K., K.L. Writing—Original Draft: K.K., K.L., R.H., S.S. & C.E.W. Writing—Review and Editing: D.B., R.K., V.M., B.L., D.A.S., K.G. & C.S. Funding acquisition: S.S., C.S. & C.E.W.
Funding
This study was supported by the Department of Biotechnology (DBT)-Wellcome Trust/India Alliance fellowship ([Grant Number: IA/I/16/2/502694] awarded to Dr. Senthilkumari Srinivasan) and Glaucoma UK; Fight for Sight (UK) [Grant awarded to Professor Colin E Willoughby]; NC3Rs PhD studentship (UK) [Grant awarded to Dr Carl Sheridan and Professor Colin E Willoughby].
Data availability
The RNA-sequencing data of the present study are publicly available in NCBI (http://www.ncbi.nlm.nih.gov/sra) under the accession numbers: PRJNA781014 (16 h) and PRJNA729873 (7 d).
Competing interests
The authors declare no competing interests.
Footnotes
The original online version of this Article was revised: The original version of this Article contained an error in the spelling of the author Karen Lester, which was incorrectly given as Lester Karen. The Author contribution statement now reads: “Conceptualization: S.S. & C.E.W. Data Curation: S.S., C.E.W. & D.B. Formal Analysis: K.K., K.L., R.H., S.S., C.E.W. & D.B. Investigation: S.S., C.E.W. & D.B. Methodology: K.K., K.L. & R.H. Project administration: S.S. & C.E.W. Resources: S.S., C.E.W., D.B., D.A.S. Supervision: S.S. & C.E.W. Validation: S.S. & C.E.W. Software: D.B., K.K., K.L. Writing—Original Draft: K.K., K.L., R.H., S.S. & C.E.W. Writing—Review and Editing: D.B., R.K., V.M., B.L., D.A.S., K.G. & C.S. Funding acquisition: S.S., C.S. & C.E.W.”
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Kandasamy Kathirvel and Karen Lester.
Change history
7/28/2022
A Correction to this paper has been published: 10.1038/s41598-022-17427-1
Contributor Information
Colin E. Willoughby, Email: c.willoughby@ulster.ac.uk
Srinivasan Senthilkumari, Email: ss_kumari@aravind.org.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-022-12443-7.
References
- 1.Gordon DM. Prednisone and prednisolone in ocular disease* *From the Department of Surgery (Ophthalmology), New York Hospital, Cornell University Medical Center. Am. J. Ophthalmol. 1956;41:593–600. [PubMed] [Google Scholar]
- 2.Becker B, Mills DW. Corticosteroids and intraocular pressure. Arch. Ophthalmol. 1963;70:500–507. doi: 10.1001/archopht.1963.00960050502012. [DOI] [PubMed] [Google Scholar]
- 3.Bernstein HN, Mills DW, Becker B. Steroid-induced elevation of intraocular pressure. Arch. Ophthalmol. 1963;70:15–18. doi: 10.1001/archopht.1963.00960050017005. [DOI] [PubMed] [Google Scholar]
- 4.Jones R, Rhee DJ. Corticosteroid-induced ocular hypertension and glaucoma: A brief review and update of the literature. Curr. Opin. Ophthalmol. 2006;17:163–167. doi: 10.1097/01.icu.0000193079.55240.18. [DOI] [PubMed] [Google Scholar]
- 5.Clark AF, et al. Glucocorticoid-induced formation of cross-linked actin networks in cultured human trabecular meshwork cells. Investig. Ophthalmol. Vis. Sci. 1994;35:281–294. [PubMed] [Google Scholar]
- 6.Matsumoto Y, Johnson DH. Dexamethasone decreases phagocytosis by human trabecular meshwork cells in situ. Investig. Ophthalmol. Vis. Sci. 1997;38:1902–1907. [PubMed] [Google Scholar]
- 7.Clark AF, et al. Dexamethasone alters F-actin architecture and promotes cross-linked actin network formation in human trabecular meshwork tissue. Cell Motil. Cytoskelet. 2005;60:83–95. doi: 10.1002/cm.20049. [DOI] [PubMed] [Google Scholar]
- 8.Raghunathan VK, et al. Dexamethasone stiffens trabecular meshwork, trabecular meshwork cells, and matrix. Investig. Ophthalmol. Vis. Sci. 2015;56:4447–4459. doi: 10.1167/iovs.15-16739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Faralli JA, et al. Genomic/proteomic analyses of dexamethasone-treated human trabecular meshwork cells reveal a role for GULP1 and ABR in phagocytosis. Mol. Vis. 2019;25:237–254. [PMC free article] [PubMed] [Google Scholar]
- 10.Yemanyi F, Vranka J, Raghunathan VK. Glucocorticoid-induced cell-derived matrix modulates transforming growth factor β2 signaling in human trabecular meshwork cells. Sci. Rep. 2020;10:1–18. doi: 10.1038/s41598-020-72779-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tripathi RC. Ultrastructure of the exit pathway of the aqueous in lower mammals. A preliminary report on the ‘angular aqueous plexus’. Exp. Eye Res. 1971;12:311–314. doi: 10.1016/0014-4835(71)90155-2. [DOI] [PubMed] [Google Scholar]
- 12.Matsuda A, et al. DNA methylation analysis of human trabecular meshwork cells during dexamethasone stimulation. Investig. Opthalmol Vis. Sci. 2015;56:3801. doi: 10.1167/iovs.14-16008. [DOI] [PubMed] [Google Scholar]
- 13.Bermudez JY, et al. A comparison of gene expression profiles between glucocorticoid responder and non-responder bovine trabecular meshwork cells using RNA sequencing. PLoS ONE. 2017;12:e0169671. doi: 10.1371/journal.pone.0169671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ishibashi T, et al. cDNA microarray analysis of gene expression changes induced by dexamethasone in cultured human trabecular meshwork cells. Investig. Ophthalmol. Vis. Sci. 2002;43:3691–3697. [PubMed] [Google Scholar]
- 15.Lo WR, et al. Tissue differential microarray analysis of dexamethasone induction reveals potential mechanisms of steroid glaucoma. Investig. Opthalmol. Vis. Sci. 2003;44:473. doi: 10.1167/iovs.02-0444. [DOI] [PubMed] [Google Scholar]
- 16.Leung YF, et al. The dual role of dexamethasone on anti-inflammation and outflow resistance demonstrated in cultured human trabecular meshwork cells. Mol. Vis. 2003;9:425–439. [PubMed] [Google Scholar]
- 17.Rozsa FW, et al. Gene expression profile of human trabecular meshwork cells in response to long-term dexamethasone exposure. Mol. Vis. 2006;12:125–141. [PubMed] [Google Scholar]
- 18.Hoheisel JD. Microarray technology: Beyond transcript profiling and genotype analysis. Nat. Rev. Genet. 2006;7:200–210. doi: 10.1038/nrg1809. [DOI] [PubMed] [Google Scholar]
- 19.Fan BJ, Wang DY, Tham CCY, Lam DSC, Pang CP. Gene expression profiles of human trabecular meshwork cells induced by triamcinolone and dexamethasone. Investig. Opthalmol. Vis. Sci. 2008;49:1886. doi: 10.1167/iovs.07-0414. [DOI] [PubMed] [Google Scholar]
- 20.Nehmé A, Lobenhofer EK, Stamer WD, Edelman JL. Glucocorticoids with different chemical structures but similar glucocorticoid receptor potency regulate subsets of common and unique genes in human trabecular meshwork cells. BMC Med. Genomics. 2009;2:58. doi: 10.1186/1755-8794-2-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Borrás T. The effects of myocilin expression on functionally relevant trabecular meshwork genes: A mini-review. J. Ocul. Pharmacol. Ther. 2014;30:202–212. doi: 10.1089/jop.2013.0218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang H, et al. Research progress on human genes involved in the pathogenesis of glaucoma (review) Mol. Med. Rep. 2018;18:656–674. doi: 10.3892/mmr.2018.9071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wasim M, et al. PLZF/ZBTB16, a glucocorticoid response gene in acute lymphoblastic leukemia, interferes with glucocorticoid-induced apoptosis. J. Steroid Biochem. Mol. Biol. 2010;120:218–227. doi: 10.1016/j.jsbmb.2010.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuchtey J, et al. Angiopoietin-like 7 secretion is induced by glaucoma stimuli and its concentration is elevated in glaucomatous aqueous humor. Investig. Opthalmol. Vis. Sci. 2008;49:3438. doi: 10.1167/iovs.07-1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Comes N, Buie LK, Borrás T. Evidence for a role of angiopoietin-like 7 (ANGPTL7) in extracellular matrix formation of the human trabecular meshwork: Implications for glaucoma. Genes Cells. 2011;16:243–259. doi: 10.1111/j.1365-2443.2010.01483.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tanigawa Y, et al. Rare protein-altering variants in ANGPTL7 lower intraocular pressure and protect against glaucoma. PLoS Genet. 2020;16:e1008682. doi: 10.1371/journal.pgen.1008682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang L, Liu K, Han B, Xu Z, Gao X. The emerging role of follistatin under stresses and its implications in diseases. Gene. 2018;639:111–116. doi: 10.1016/j.gene.2017.10.017. [DOI] [PubMed] [Google Scholar]
- 28.Wordinger RJ, et al. Expression of bone morphogenetic proteins (BMP), BMP receptors, and BMP associated proteins in human trabecular meshwork and optic nerve head cells and tissues. Mol. Vis. 2002;8:241–250. [PubMed] [Google Scholar]
- 29.Fitzgerald AM, Benz C, Clark AF, Wordinger RJ. The effects of transforming growth factor-β2 on the expression of follistatin and activin A in normal and glaucomatous human trabecular meshwork cells and tissues. Investig. Ophthalmol. Vis. Sci. 2012;53:7358. doi: 10.1167/iovs.12-10292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lim R, et al. Activin and NADPH-oxidase in preeclampsia: Insights from in vitro and murine studies. Am. J. Obstet. Gynecol. 2015;212:e1–e12. doi: 10.1016/j.ajog.2014.07.021. [DOI] [PubMed] [Google Scholar]
- 31.Chen CYA, Shyu A-B. AU-rich elements: Characterization and importance in mRNA degradation. Trends Biochem. Sci. 1995;20:465–470. doi: 10.1016/s0968-0004(00)89102-1. [DOI] [PubMed] [Google Scholar]
- 32.Lin C, et al. Transcriptional activation of follistatin by Nrf2 protects pulmonary epithelial cells against silica nanoparticle-induced oxidative stress. Sci. Rep. 2016;6:1–11. doi: 10.1038/srep21133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ying L, et al. Complement component 7 (C7), a potential tumor suppressor, is correlated with tumor progression and prognosis. Oncotarget. 2016;7:86536. doi: 10.18632/oncotarget.13294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hossain S, et al. The RASSF6 tumor suppressor protein regulates apoptosis and cell cycle progression via retinoblastoma protein. Mol. Cell. Biol. 2018 doi: 10.1128/MCB.00046-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kanai N, et al. Identification and characterization of a prostaglandin transporter. Science. 1995;268:866–869. doi: 10.1126/science.7754369. [DOI] [PubMed] [Google Scholar]
- 36.Schuster VL, Chi Y, Lu R. The prostaglandin transporter: Eicosanoid reuptake, control of signaling, and development of high-affinity inhibitors as drug candidates. Trans. Am. Clin. Climatol. Assoc. 2015;126:248–257. [PMC free article] [PubMed] [Google Scholar]
- 37.Kraft ME, et al. The prostaglandin transporter OATP2A1 is expressed in human ocular tissues and transports the antiglaucoma prostanoid latanoprost. Investig. Ophthalmol. Vis. Sci. 2010;51:2504–2511. doi: 10.1167/iovs.09-4290. [DOI] [PubMed] [Google Scholar]
- 38.Weidenfeld J, Lysy J, Shohami E. Effect of dexamethasone on prostaglandin synthesis in various areas of the rat brain. J. Neurochem. 1987;48:1351–1354. doi: 10.1111/j.1471-4159.1987.tb05670.x. [DOI] [PubMed] [Google Scholar]
- 39.Partridge CA, Weinstein BI, Southren AL, Gerritsen ME. Dexamethasone induces specific proteins in human trabecular meshwork cells. Investig. Ophthalmol. Vis. Sci. 1989;30:1843–1847. [PubMed] [Google Scholar]
- 40.Stojadinovic O, et al. Novel genomic effects of glucocorticoids in epidermal keratinocytes: Inhibition of apoptosis, interferon-gamma pathway, and wound healing along with promotion of terminal differentiation. J. Biol. Chem. 2007;282:4021–4034. doi: 10.1074/jbc.M606262200. [DOI] [PubMed] [Google Scholar]
- 41.Fuchshofer R, Stephan DA, Russell P, Tamm ER. Gene expression profiling of TGFβ2- and/or BMP7-treated trabecular meshwork cells: Identification of Smad7 as a critical inhibitor of TGF-β2 signaling. Exp. Eye Res. 2009;88:1020. doi: 10.1016/j.exer.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ohuchi T, et al. Neuropeptide-induced [Ca2+]i transients in cultured bovine trabecular cells. Investig. Ophthalmol. Vis. Sci. 1992;33:1676–1684. [PubMed] [Google Scholar]
- 43.Santos-Carvalho A, Elvas F, Álvaro AR, Ambrósio AF, Cavadas C. Neuropeptide Y receptors activation protects rat retinal neural cells against necrotic and apoptotic cell death induced by glutamate. Cell Death Dis. 2013;4:e636. doi: 10.1038/cddis.2013.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stone RA, Laties AM, Emson PC. Neuropeptide Y and the ocular innervation of rat, guinea pig, cat and monkey. Neuroscience. 1986;17:1207–1216. doi: 10.1016/0306-4522(86)90088-6. [DOI] [PubMed] [Google Scholar]
- 45.Webber HC, Bermudez JY, Sethi A, Clark AF, Mao W. Crosstalk between TGFβ and Wnt signaling pathways in the human trabecular meshwork. Exp. Eye Res. 2016;148:97. doi: 10.1016/j.exer.2016.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liesenborghs I, et al. The molecular processes in the trabecular meshwork after exposure to corticosteroids and in corticosteroid-induced ocular hypertension. Investig. Opthalmol. Vis. Sci. 2020;61:24. doi: 10.1167/iovs.61.4.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sugali CK, et al. The canonical Wnt signaling pathway inhibits the glucocorticoid receptor signaling pathway in the trabecular meshwork. Am. J. Pathol. 2021;191:1020–1035. doi: 10.1016/j.ajpath.2021.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhu J, Wang Y, Hu Q, Yuan R, Ye J. Rottlerin acts as a therapeutic in primary open-angle glaucoma by targeting the trabecular meshwork via activation of Rap1 signaling. Pharmacol. Res. 2020;159:104780. doi: 10.1016/j.phrs.2020.104780. [DOI] [PubMed] [Google Scholar]
- 49.Peng C, et al. Characteristic cytokine profiles of aqueous humor in glaucoma secondary to sturge-weber syndrome. Front. Immunol. 2020;11:4. doi: 10.3389/fimmu.2020.00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stamer WD, Seftor REB, Williams SK, Samaha HAM, Snyder RW. Isolation and culture of human trabecular meshwork cells by extracellular matrix digestion. Curr. Eye Res. 1995;14:611–617. doi: 10.3109/02713689508998409. [DOI] [PubMed] [Google Scholar]
- 51.Lester, K. L. Reverse Engineering Glaucoma. Scholarly Journal. PhD. Thesis, The University of Liverpool. (2018). https://livrepository.liverpool.ac.uk/3022786/. Accessed 16 September 2021.
- 52.Ashwinbalaji S, et al. SB772077B, A new rho kinase inhibitor enhances aqueous humour outflow facility in human eyes. Sci. Rep. 2018;8:15472. doi: 10.1038/s41598-018-33932-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Keller KE, et al. Consensus recommendations for trabecular meshwork cell isolation, characterization and culture. Exp. Eye Res. 2018;171:164–173. doi: 10.1016/j.exer.2018.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kathirvel K, et al. A comparative genome-wide transcriptome analysis of glucocorticoid responder and non-responder primary human trabecular meshwork cells. BioRxiv. 2021 doi: 10.1101/2021.09.21.461315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kim D, Paggi JM, Park C, Bennett C, Salzberg SL. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019;37:907–915. doi: 10.1038/s41587-019-0201-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liao Y, Smyth GK, Shi W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 57.Robinson MD, McCarthy DJ, Smyth GK. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2008;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-sequencing data of the present study are publicly available in NCBI (http://www.ncbi.nlm.nih.gov/sra) under the accession numbers: PRJNA781014 (16 h) and PRJNA729873 (7 d).