Summary
Background
Previously, we uncovered a patient subgroup with highly malignant pancreatic cancer with serum markers CEA+/CA125+/CA19-9 ≥ 1000 U/mL (triple-positive, TP). However, the underlying immunosuppressive mechanism in the tumor immune microenvironment (TIME) of this subgroup is still unknown.
Methods
Human tissues were analyzed by flow cytometry, mass cytometry, and immunofluorescence staining. Mouse pancreatic ILC2s were expanded in vivo and used for RNA sequencing, chromatin immunoprecipitation (ChIP), and chemotaxis assays.
Findings
Through microarray data, we identified the accumulation of the hypoxia-induced factor-1α (HIF-1α) pathway in these TP patients. Via flow and mass cytometry, we discovered that a special subset of ILC2s were highly infiltrated in TP patients. Under the hypoxia microenvironment, ILC2s were found undergo a transition to a IL10+ regulatory phenotype, we named ILCregs which was correlated with pancreatic ductal adenocarcinoma (PDAC) progression. Further, neoadjuvant chemotherapy could ameliorate hypoxic tumor microenvironments so that significantly reverse the regulatory phenotype of ILCregs. Moreover, most tumor ILC2 were CD103−, which indicated its circulatory origin. The expression of Ccr2 was significantly upregulated on mouse ILCregs, and these cells selectively migrated to CCL2.
Interpretation
Our results indicate that the hypoxia microenvironment creates an immunosuppressive TIME by inducing ILCregs from a population of circulating group 2 ILCs in TP PDAC patients.
Funding
This study was jointly supported by the National Natural Science Foundation of China (U21A20374, 82173091, and 81701630)
Keywords: Group 2 innate lymphoid cells (ILC2s), Pancreatic ductal adenocarcinoma (PDAC), Hypoxia
Research in context.
Evidence before this study
PDAC is classified as an aggressive and fatal disease. We previously discovered a special patient subgroup characterized by serum markers CEA+/CA125+/CA19-9 ≥ 1000 U/mL (triple-positive, TP), which was associated with early metastasis, uncontrolled progressive and chemotherapy resistance. However, the tumor immune microenvironment (TIME) of this subgroup is yet to be explored.
Added-value of this study
Innate lymphoid cells (ILCs) have been proved a vital role in tumor microenvironment and promote high tumor plasticity. Previous study identified ILC2 as anti-cancer immune cells for PDAC. However, in our study, we revealed that hypoxia-treated ILC2s underwent a transition to IL10+ ILCregs in TP patients, which was correlated with pancreatic ductal adenocarcinoma (PDAC) progression.
Implications of all the available evidence
Our work indicated that substantial hypoxia promotes the formation of the immunosuppressive TIME through the induction of ILCregs from a population of hypoxic group 2 innate lymphoid cells (ILC2s) in TP patients, and knowledge of this mechanism may assist in increasing response rate to immunotherapy for treating PDAC.
Alt-text: Unlabelled box
Introduction
Despite the recent chemotherapeutic advancements in the treatment of pancreatic ductal adenocarcinoma (PDAC), the 5-year overall survival remains dismal and is <8%.1 Previously, we identified a subpopulation of patients with a preoperative serum signature of CEA+/CA125+/CA19-9 ≥ 1000 U/mL (triple-positive, TP) associated with early distant metastasis after radical pancreatectomy.2 However, its mechanism remained ambiguous. Therefore, a more in-depth investigation of the complexity within the tumor immune microenvironment (TIME) of this subgroup is needed to improve patients’ outcomes.
Hypoxia is a hallmark of the pancreatic cancer microenvironment, with the median pO2 being as low as 0–5.3 mmHg (0–0.7%) in tumors compared to the median pO2 of 24.3–92.7 mmHg (3.2–12.3%) in normal pancreas.3,4 Hypoxia-induced changes can not only induce cell cycle arrest, apoptosis, and glycolysis in cancer cells,5 but also have an important role in regulating immune cell effector functions.6, 7, 8
Innate lymphoid cells (ILCs) are innate immune cells with roles in parasitic infections, asthma, inflammation, and cancer. Moral et al. reported that IL33-activated group 2 ILCs (ILC2s) recruited T cells to boost anti-cancer immunity in PDAC tissues.9 However, Huang et al. demonstrated that IL33-treated ILC2s produced IL10 and played a protective role in islet transplantation.10 These results indicate that ILC2s are a highly dynamic cell type. And their phenotypes and functions are controlled by the microenvironment. Previous studies have treated mice with IL33, a canonical ILC2-activating cytokine. However, the effects of extremely low oxygen levels on ILC2s are still unclear, which may reveal the real status in PDAC tissue.
In the present study, we explored the adaptation of ILC2s to the hypoxic milieu in the TIME. We identified a unique immunosuppressive mechanism where hypoxia converts ILC2s to IL10+ ILCregs, helping to form a tolerogenic state in TP pancreatic cancer.
Methods
Ethics
The study was approved by the ethics committee of the Fudan University Shanghai Cancer Center (050432-4-1911D), and written informed consent was obtained from all patients. The animal protocol was approved by the Institutional Animal Care and Use Committee of the Fudan University Shanghai Cancer Center and carried out in accordance with the guidelines.
Human specimens
Human peripheral blood mononuclear cells (PBMCs) and tissues were obtained from the Fudan University Shanghai Cancer Center. Intra-tumor, peri-tumor, and non-tumor tissues, as well as peripheral blood, were obtained as shown in Fig. S1a from patients who underwent PDAC surgery. Normal pancreas and spleens were acquired from patients who received surgery for benign pancreatic cystic neoplasms. All samples were confirmed by postoperative pathology.
PBMCs were isolated by Ficoll gradient centrifugation as previously described.11 Human tissue specimens were minced into 1 mm fragments and digested with 0.1 mg/mL of Liberase TL (Roche Diagnostics, Indianapolis, IN, USA) and 10 U/mL of DNase I (Roche Diagnostics, Indianapolis, IN, USA) for 30 min. Cells were filtered and re-suspended in 40% Percoll for gradient centrifugation (Fig. S1a–d).
Mice
Five-week-old male C57BL/6 mice were purchased from Shanghai Jihui Co. (Shanghai, China). Mice were bred and housed under pathogen-free conditions at an animal facility. ILC2s (Lin−CD127+ST2+) were sorted by FACS from mice pancreas (Fig. S1e). The isolation of ILC2 was established as described previously, which is similar to human specimens. Briefly, mice tissue was minced and digested with 0.1 mg/mL of Liberase TL (Roche Diagnostics, Indianapolis, IN, USA) and 10 U/mL of DNase I (Roche Diagnostics, Indianapolis, IN, USA) for 30 min. Cells were filtered and re-suspended in 40% Percoll for gradient centrifugation.
For adoptive transfer, ILC2s (Lin−CD127+ST2+) were sorted from mice treated with IL-33 (400 ng, Thermo Fisher) every day for 7 days. For orthotopic PDAC tumors, the pancreas of each mouse was injected with 1 × 107 PANC-2 cells in 25 μL of phosphate-buffered saline (PBS, pH 7.4). For subcutaneous PDAC model, 1 × 106 PANC-2 cells were resuspended in sterile PBS and implanted subcutaneously. After three days, 1 × 105 ILC2s or PBS were injected every 3 days via the tail vein or intratumorally and sacrificed 2 weeks later. Tumor volumes (cm3) were calculated as 1/2 (length × width2).
Flow cytometry and mass cytometry staining
All antibodies were procured from BioLegend (San Diego, CA, USA), unless specified otherwise. The lineage cocktail for human cells included anti-CD16 (3G8), anti-CD11c (3.9), anti-CD123 (6H6), anti-CD19 (HIB19), anti-CD3 (UCHT1), anti-CD5 (L17F12), anti-CD94 (DX22), and anti-CD34 (581). Other antibodies for this purpose included anti-CRTH2 (BM16), anti-CD127 (A019D5), anti-CD117 (104D2), anti-ICOS (C398.4A), anti-PD1 (A17188B), anti-CD25 (BC96), anti-IL10 (JES3-9D7), anti-CTLA4 (BNI3), and anti-CCR2 (K036C2). Lineage antibodies for mice included anti-B220 (RA3-6B2), anti-CD3 (2C11), anti-TCRβ (H57), anti-TCRγδ (GL-3), anti-CD11b (M1/70), and anti-CD5 (53-7.3). Other antibodies for this purpose included anti-Thy1.2 (53-2.1), anti-CD25 (PC61.5), anti-ST2 (DJ8; MD Bioproducts, Oakdale, MN, USA), and anti-IL10 (JES5-2A5). Cell sorting was performed on a FACSAria II system (BD Biosciences, San Jose, CA, USA), whereas flow cytometric analysis was performed on a Canto-II system (BD Biosciences, San Jose, CA, USA). For cytokine staining, cells were incubated with the Leukocyte Activation Cocktail (BD Biosciences, San Jose, CA, USA) for 5 h and then stained with the Intracellular Cytokine Staining Kit (BD Pharmingen, San Jose, CA, USA). For transcriptional factor staining, the Fix/Perm Kit (eBioscience, San Diego, CA, USA) was used as previously described.11
Cytometry by time-of-flight (CyTOF) was performed by PLTTech Inc. (Hangzhou, China) as previously described.12 Briefly, human tumor tissue was digested and immune cells were extracted. Isolated human tumor immune cells were blocked and stained for 30 min with a surface marker. Then, the cells were stained with intracellular/nuclear antibodies after fixation and permeabilization. The cells were rinsed, and re-suspended. Then, the samples were run through the CyTOF instrument (Helios, Fluidigm, South San Francisco, CA, USA), and the signal data were collected.
Chromatin immunoprecipitation assay
To obtain sufficient numbers of ILC2s, mice were treated with 400 ng IL-33 for 7 consecutive days. Chromatin immunoprecipitation (ChIP) was performed as previously described.11,13 Anti-HIF-1α antibody (NB100, Novus) or IgG (negative control) were used. The primers were 5′-CATCAGATAAGACGAGATAACCCC-3′ (forward) and 5′-GAGTCCCCTATTGCCTTTCATT-3′ (reverse).
Luciferase reporter assays
To construct the Il10 promoter luciferase reporter plasmid, the HRE-like sequence (GGACGTGT) was ligated into the pGL3-basic plasmid (Promega, Madison, WI, USA). Luciferase reporter assays were performed as previously described.11
Cell culture
For hypoxia treatment, cells were exposed to hypoxia (1% O2) for the indicated time period. In reoxygenation group, RNA was collected from ILC2 after 24 h hypoxia culture (1% O2) + 24 h reoxygenation (20% O2). ILC2s were cultured in α-MEM supplemented with 20% FBS in the presence or absence of 10 ng/ml of IL-2, IL-7, or IL-33. Mouse CD3+ T cells were sorted by FACS and labeled with 0.5 µM CFSE in 1 ml of PBS for 15 min at 37 °C (Invitrogen, Carlsbad, CA, USA). CFSE-labeled CD3+ T cells were cultured with or without 24 h hypoxia-treated ILC2s (renewed daily) and examined 5 days later with Transwell (0.4 μm) with polyclonal-stimulated (10 μg/mL anti-CD3 and 10 μg/mL antiCD28). Human PBMC-derived ILC2s were cultured with 400 ng/ml CCL2 and examined 24 h later.
Human mast cell line (HMC-1) is a gift from Professor Hui Tang's Lab (Huashan Hospital, Fudan Univeristy, Shanghai, China). It is maintained in IMDM supplemented with 10% FBS and 5%L-glutamine.
Chemotaxis assay
Mouse pancreatic ILC2s (5 × 105) were added to the upper well (5 μm), and 400 ng/ml mouse CCL2 (R&D System, Minneapolis, MN, USA) was added to the lower chamber. Cells were cultured for 24 h, and the chemotactic index was calculated as the ratio of cells migrated with CCL2 to the cells migrated with the medium alone.
Immunofluorescent staining and immunohistochemistry
Paraffin-embedded 5 µm-thick pancreatic adenocarcinoma tumor tissue sections were deparaffinized and processed for immunofluorescent staining. Sections were incubated overnight with human anti-CD3 (SP162), anti-CD127 (Tyr449), or anti-CRTH2 (PA5-20332) at 4 °C. Sections were visualized under a fluorescence microscope (NIKON ECLIPSE C1, Japan), and images were acquired with a digital camera (NIKON DS-U3, Japan).
Immunohistochemistry (IHC) was performed as previously described to detect the expression of HIF-1α.14 According to the percentage of stained cells, staining intensity was scored as follows: 0 = 0–5% stained cells, 1 = 5–25%, 2 = 25–50%, 3 = 50–75%, and 4 = 75–100%. Based on the staining morphology, the following scores were assigned: 0 = negative, 1 = pallide-flavens grains, 2 = buffy grains, and 3 = brown-black grains. Both scores were multiplied to determine the score of each sample, with the minimum score being 0 and the maximum score being 12. Scores of 0–4, 5–8, and 9–12 were considered to indicate negative, positive, and strongly positive HIF-1α expression, respectively.
RNA sequencing
Total RNA from ILC2 or HMC-1 was extracted and sequenced as we previously described.15 RNA-seq data generated in this study have been deposited in the Gene Expression Omnibus (GEO) with the accession number GSE196812.
Gene set enrichment analysis (GSEA) and gene set variant analysis (GSVA)
GSEA and GSVA were conducted by “clusterProfiler” R package and “GSVA” R package. GSEA plots were based on unbiased analysis against all gene sets. The survminer R package was used to identify the optimal cutoff point for high and low expression.
Statistics and bioinformatics analysis
The Student t-test was used to compare the two groups, unless specified otherwise. Survival analysis was conducted using the Kaplan–Meier method and data were compared using the log-rank test. Data were expressed as means and SDs. P-values less than 0.05 (*p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001) were considered statistically significant.
Immune cells abundance in samples was estimated by ImmuCellAI16 and TIP (Tracking Tumor Immunophenotype).17 GEPIA (Gene Expression Profiling Interactive Analysis)18 was used to analyze the indicated genes expression level and survival association in TCGA data.
Role of the funding source
The funding agencies of this study had no role in the study design, data collection, analysis, interpretation of data, writing of paper, or in the decision to submit the paper for publication.
Results
TP pancreatic cancer exhibit a distinct hypoxia tumor microenvironment with high ILC2s infiltrate
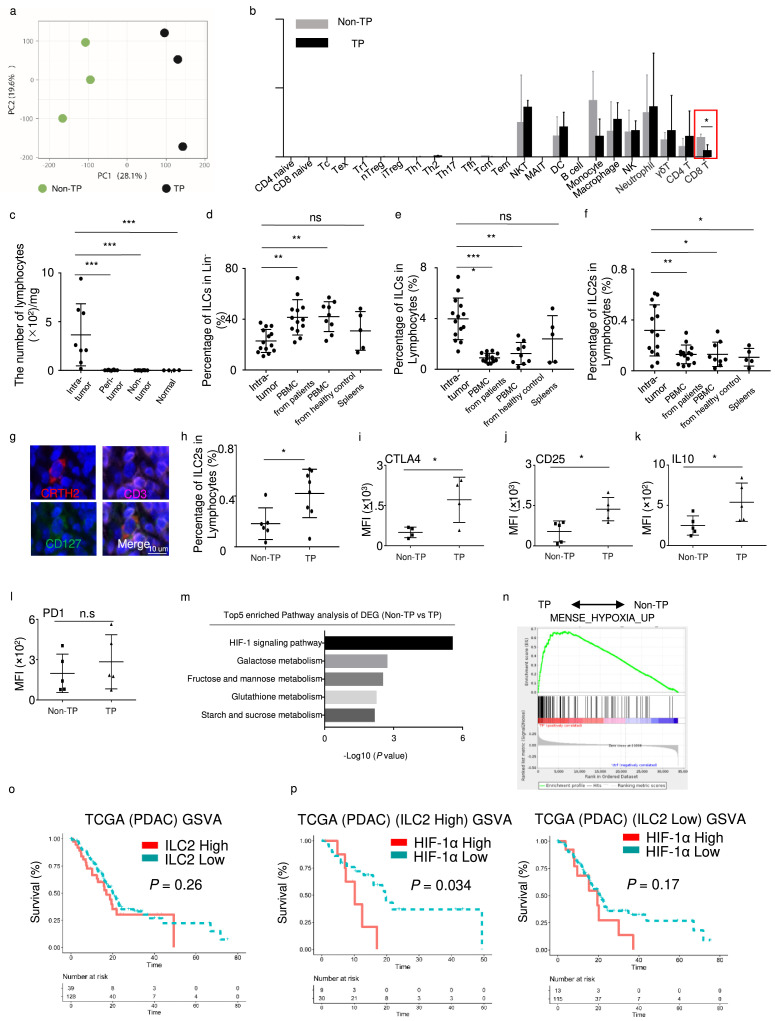
We analyzed the microarray data in patients with TP pancreatic cancer and patients who were non-CEA+/CA125+/CA19-9 ≥ 1000 U/mL (Non-TP).14 PCA (principal component analysis) showed that the two groups had distinct gene expression profiles (Figure 1a). To identify the immune cells components in the tumor microenvironment in PDAC, we estimated the abundance of major immune cells in TP and Non-TP patients using ImmuCellAI algorithm. The results showed that only CD8+ T cells were dramatically decreased in TP patients (Figure 1b). Previously, Moral et al.9 reported that ILC2 could prime and recruit CD8+ T cells into PDAC. That prompted us to investigate the status of ILC2 in TP patients. Therefore, we isolated lymphocytes from intra-tumor, peri-tumor, non-tumor, and normal tissues (Fig. S1a–d). Compared with intra-tumor tissues, very few lymphocytes were identified in the peri-tumor, non-tumor, and normal tissues (Figure 1c). Therefore, peri-tumor and non-tumor tissues were not included in further analysis due to the absence of lymphocytes. Although the percentage of ILCs (Lin−CD127+) in Lin− was significantly lower in tumor tissues than that in PBMC and spleen (Figure 1d), the proportions of ILCs (Lin−CD127+) and ILC2s (Lin−CD127+CRTH2+) in lymphocytes were significantly increased in intra-tumor tissues compared with PBMCs from patients, PBMCs from healthy controls, and spleens (Figure 1e, f). In immunofluorescence analysis, the number of ILC2s (CD3−CD127+CRTH2+) also significantly increased in tumor tissues (Figure. 1g, S1f). Furthermore, the ratio of ILC2s was increased in the TP patients compared to that in Non-TP patients (Figure 1h). And the levels of CTLA4, CD25, and IL10 in the TP ILC2s were much higher than those in the Non-TP-derived ILC2s (Figure. 1i–k, S1g). However, PD1 expression remained almost same (Figure 1l). To furtherly clarify the feature of tumor microenvironment in TP patients, we performed pathway analysis and gene set enrichment analysis (GSEA). The results revealed that the HIF-1 signaling pathway was significantly increased in TP patients (Figure 1m, n).
Figure 1.
Characterization of a novel ILC2 group in pancreatic cancer.
(a) Visualization of TP (n = 3) and Non-TP (n = 3) in PCA, computed on a gene expression matrix. (b) Major immune cells abundance in TP and Non-TP patients estimated by ImmuCellAI algorithm, using microarray data. (c) Lymphocyte numbers in intra-tumor (n = 8), peri-tumor (n = 8), non-tumor (n = 8), and normal tissues (n = 4), assessed with the Mann-Whitney U test. (d) Percentages of ILCs in Lin− in tumors (n = 14), PBMCs from patients (n = 13), PBMCs from healthy controls (n = 9), and spleens (n = 5). (e) Percentages of ILCs in lymphocytes from tumors (n = 14), PBMCs from patients (n = 13), PBMCs from healthy controls (n = 9), and spleens (n = 5). (f) Percentages of ILC2s in lymphocytes from tumors (n = 14), PBMCs from patients (n = 13), PBMCs from healthy controls (n = 9), and spleens (n = 5). (g) Representative immunofluorescence images (magnification, × 1500) of human PDAC tissue stained with antibodies against CRTH2 (red), CD3 (pink), and CD127 (green). Nuclei were stained with DAPI (blue). (h) Percentages of ILC2s in lymphocytes from Non-TP (n = 6) and TP (n = 8) patients. Statistical analysis of CTLA4 (n = 4) (i), CD25 (n = 5) (j), IL10 (n = 5) (k) and PD1 (n = 5) (l) in ILC2 from Non-TP and TP patients. (m) Pathway analysis of differentially expressed genes highly expressed in TP. (n) Gene set enrichment analysis (GSEA) of hypoxia pathway in TP and Non-TP patients. (o) Gene Set Variance Analysis (GSVA) using a 12-gene signature of ILC2s derived from single-cell sequencing analysis. Optimal cutoff point was determined by the survminer R package. Analysis of survival prediction within PDAC TCGA cohort. (p) Survival prediction of PDAC according to HIF-1α expression in “ILC2 high” or “ILC2 low” patients’ group. Data are expressed as means ± SD and compared using the unpaired t test (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
According to our previous ILC2 scRNA-seq study,15 we identified 12-gene signature of ILC2 compared to other cell types (IL13, HPGDS, KLRB1, GATA3, GADD45G, PLIN2, TNFSF10, STARD4, PHLDA1, ARL4A, KLRG1 and IGKC). Then, we employed gene set variation analysis (GSVA) to predict overall survival in PDAC TCGA data using this 12-gene signature. Optimal cutoff point was determined by the survminer R package. No difference in overall survival (OS) was observed between patients who were ILC2 high and ILC2 low in TCGA data (Figure 1o). Moreover, the ILC2-activating cytokines IL33 and TSLP in the TCGA database could not predict the prognosis (Fig. S1h, i). However, when we stratified ‘ILC2 high’ as HIF-1α high and HIF-1α low groups, the patients showed significantly lower OS in the HIF-1α high group (Figure 1p). This difference is nonsignificant in ‘ILC2 low’ group (Figure 1p). Therefore, the function of ILC2s needed to be further investigated under hypoxic microenvironment.
Hypoxia-treated ILC2s in tumor tissues correlate with PDAC progression
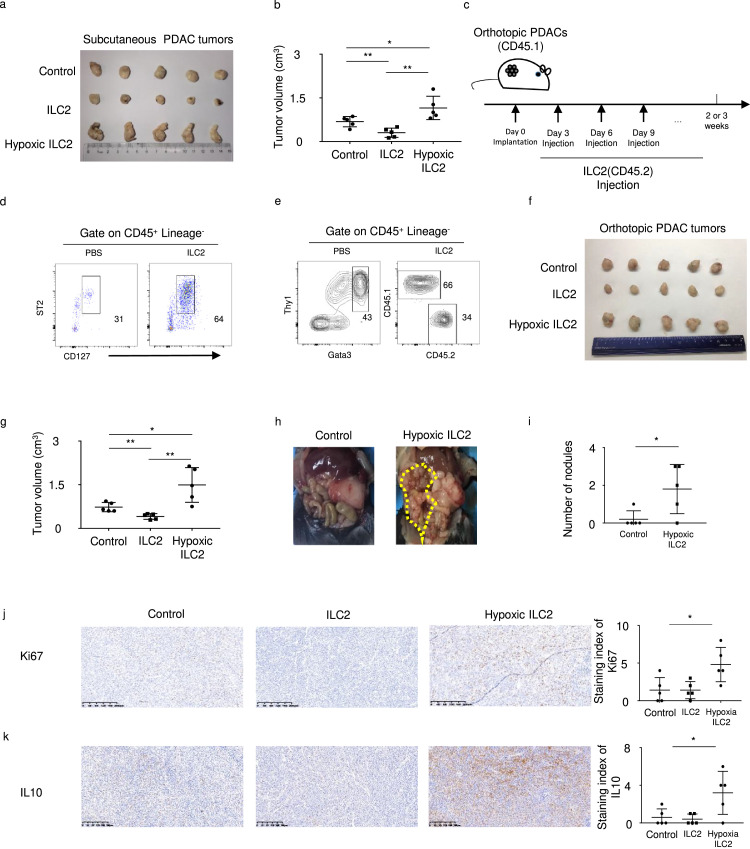
To further clarify the role of ILC2s in PDAC, we established a subcutaneous PDAC tumor model and transferred ILC2s or PBS intratumorally every 3 days. Consistent with the study of Moral et al.9 the tumor volume was significantly decreased in mice with IL33-activated ILC2 group. However, when we injected hypoxia-treated ILC2 cells, the tumor volume significantly increased (Figure 2a, b). Furthermore, we adoptively transferred CD45.2+ ILC2s into CD45.1 orthotopic tumor-bearing and control mice (Figure 2c), and confirmed the accumulation of ILC2s in tumor tissue (Figure 2d, e). The orthotopic PDAC tumors results showed that hypoxia treatment significantly reversed ILC2 function in PDAC (Figure 2f, g). Moreover, peritoneal metastasis was observed in mice of the hypoxia-treated ILC2 group (Figure 2h, i, Fig. S2). Furthermore, Ki67 and IL10 staining was significantly higher in the hypoxia-treated ILC2 group (Figure 2j, k).
Figure 2.
Hypoxia-treated ILC2s in tumors correlate with PDAC progression.
(a) Left panel shows resected tumors and (b) right panel shows tumor volume in a subcutaneous PDAC mouse model with or without injection of ILC2s (n = 5). (c) Adoptive transfer of CD45.1-ILC2s into CD45.2 mice (d) Representative flow cytometry data for pancreas ILC2 in mice with or without ILC2s injection. (e) Reconstitution of mice PDAC ILC2 cells. (f) Orthotopic PDAC mouse model with or without injection of ILC2s (n = 5). (g) Tumor volume in orthotopic PDAC mouse model with or without injection of ILC2s (n = 5). (h) The left panel shows peritoneal metastasis in the orthotopic PDAC mouse model, and (i) the right panel shows statistical analysis of the number of nodules (n = 5). (j) The left panel shows Ki67 staining in control, ILC2, or hypoxia-treated ILC2 groups (magnification, × 100) and the right panel shows statistical analysis of staining index of Ki67 (n = 5). (k) The left panel shows IL10 staining in control, ILC2, or hypoxia-treated ILC2 groups (magnification, × 100) and the right panel shows statistical analysis of staining index of IL10 (n = 5). Data are expressed as means ± SD and compared using the unpaired t test (*P < 0.05; **, P < 0.01; ***, P < 0.001).
Hypoxia induced ILC2 regulatory conversion
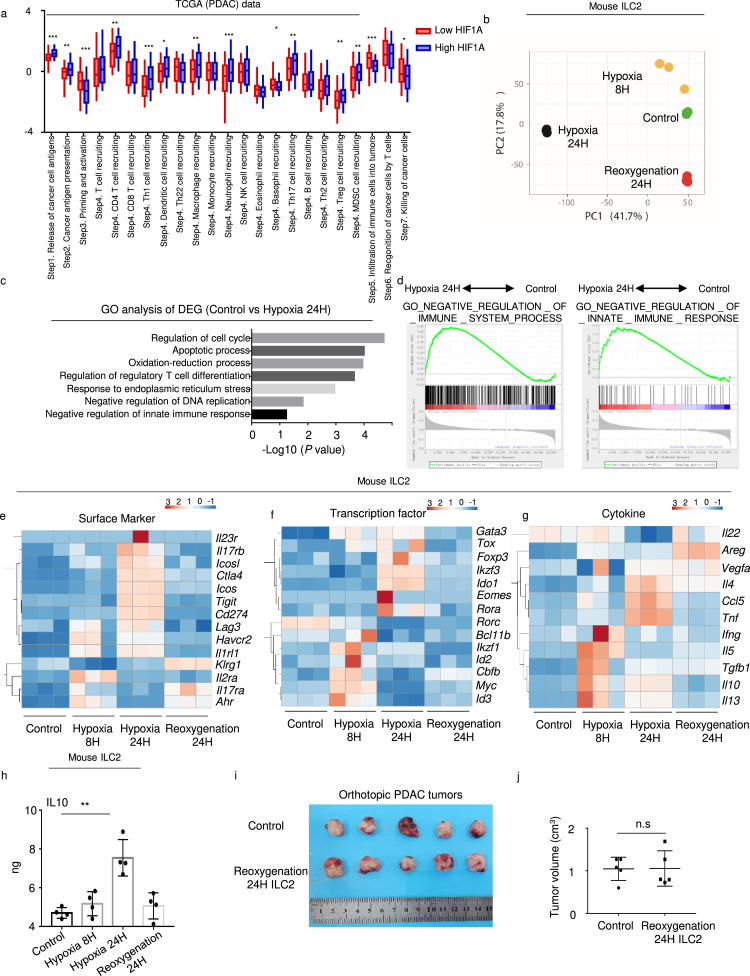
In accordance with the results of our previous study,4 HIF1A expression significantly increased in PDAC tumor tissues (Fig. S3a). Furthermore, the TIP tool was employed for analyzing the anti-cancer immune response. In HIF1A high group, priming and activation (Step 3), infiltration of immune cells into tumors (Step5) and killing of cancer cells (Step7) were found to be significantly downregulated (Figure 3a). In the meantime, macrophage recruiting (Step4), Treg cell recruiting (Step4) and MDSC recruiting (Step4) were significantly increased (Figure 3a). Thus, hypoxia may induce tumor immunosuppression in PDAC. To investigate the dynamic change of ILC2s under hypoxic culture, we sorted and cultured hypoxic mouse ILC2s at different time points for RNA-seq. PCA verified the different gene profiles in these groups (Figure 3b). Differentially expressed genes in 24 h treated and untreated ILC2s were enriched for the negative regulation of the immune response (Figure 3c). GSEA revealed that the expression of negatively regulated genes were increased in the 24 h hypoxia ILC2s group (Figure 3d). Surface markers Ctla4 and Il2ra, transcription factors Gata3 and Ido1, and cytokines Il10, Tgfb1, and Vegf gradually increased in the ILC2s after hypoxia treated, and decreased after reoxygenation 24 h later (Figure 3e–g). Given that IL-10 is the feature cytokine of Treg cell-mediated suppression, we confirmed the increase in IL-10 induced by hypoxia and decrease after reoxygenation using ELISA in hypoxic mouse ILC2s (Figure 3h). Therefore, we named these hypoxic ILC2s as ILCregs. Furthermore, tumor volume remained almost same after reoxygenation 24 h ILC2 injection compared to control group in orthotopic PDAC tumors (Figure 3i, j). Collectively, these results indicate that hypoxia reversibly induces the conversion of ILC2s to ILCregs, a regulatory phenotype.
Figure 3.
Hypoxia promotes ILC2 transition to ILCregs in PDAC.
(a) Differences in the various steps of the anti-cancer immunity between high- and low-HIF1α groups in TCGA (PDAC) data. RNA-seq was performed with mouse ILC2s with or without hypoxia treatment. (b) PCA of the control, 8 h hypoxia, 24 h hypoxia, and 24 h reoxygenation ILC2s groups (n = 3). (c) GO analysis of differentially expressed genes between control and 24 h hypoxia ILC2s. (d) Gene set enrichment analyses and (e) (f) (g) heatmap of representative genes in the ILC2 RNA-seq. (h) ELISA detection of IL10 expression in mouse ILC2s under hypoxia treatments (n = 4). (i) Orthotopic PDAC mouse model with or without injection of reoxygenation 24 h ILC2s (n = 5). (j) Tumor volume in orthotopic PDAC mouse model with or without injection of reoxygenation 24h ILC2s (n = 5). Data are expressed as means ± SD and compared using the unpaired t test (*,P < 0.05; **, P < 0.01; ***, P < 0.001).
Neoadjuvant chemotherapy reverses the ILC2 regulatory phenotype
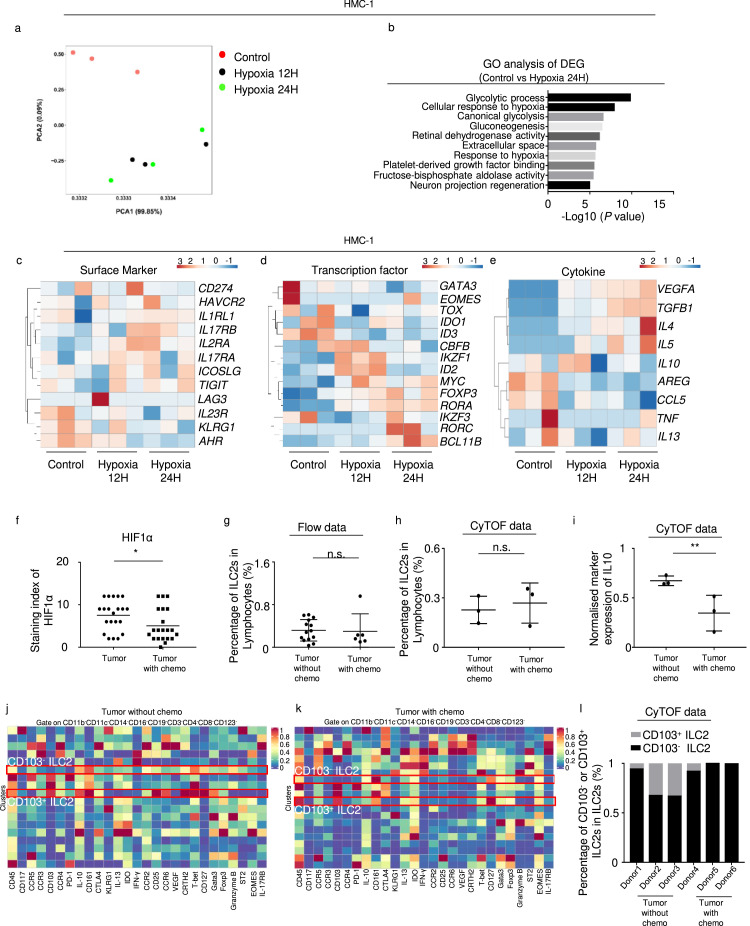
In order to investigate whether this effect is a trait confined to ILC2s or not, we cultured human mast cell line (HMC-1) under hypoxia condition for indicated time period. PCA (principal component analysis) showed the distinct gene expression profiles (Figure 4a). GO analysis between control and 24 h hypoxia HMC-1 group was mainly enriched in metabolite process (Figure 4b). In addition, surface markers CD274 and IL2RA, transcription factors GATA3 and ID2, and cytokines IL10 remained almost same under hypoxia culture (Figure 4c–e). Therefore, unlike ILC2s, mast cells did not undergo regulatory conversion after hypoxia culture.
Figure 4.
Neoadjuvant chemotherapy reverses the ILC2 regulatory phenotype.
PCA plot (a) and GO analysis of differentially expressed genes (b) in control vs Hypoxia 24H group in HMC-1 (n = 3). (c) (d) (e) Heatmap of representative genes in the HMC-1 RNA-seq. (f) Statistical analysis of HIF-1α in tumor samples with or without chemotherapy (n = 21). (g) Percentage of ILC2s in lymphocytes from tumors with (n = 6) or without chemotherapy (n = 14) (flow data). (h) Percentages of ILC2s in lymphocytes from tumors with or without chemotherapy (CyTOF data) (n = 3). (i) Values of IL10 in ILC2s with or without chemotherapy (n = 3). Heatmap of ILC2s from tumors without (j) or with (k) chemotherapy pre-gated on CD45+ lineage− cells (CD11b, CD11c, CD14, CD16, CD19, CD3, CD4, CD8) in CyTOF, indicated by the rectangle drawn with the red border. (l) Percentage of CD103− and CD103+ ILC2s in ILC2s, each bar indicated one patient. Data are expressed as means ± SD and compared using the unpaired t test (*P < 0.05; **, P < 0.01; ***, P < 0.001).
Since nab-paclitaxel has been reported to induce a marked decrease in cancer stroma,19 we investigated whether neoadjuvant chemotherapy could ameliorate hypoxic tumor environments and affect ILC2s. We obtained samples from patients who received neoadjuvant AG (nab-paclitaxel + gemcitabine) prior to surgery. Immunohistochemistry assay confirmed that the expression of HIF-1α significantly decreased after neoadjuvant chemotherapy (Figure. 4f and S3b). Although the percentage of ILC2s in lymphocytes remained almost the same both in flow (Figure 4g) and CyTOF data (Figure 4h), the IL10 levels of ILC2s significantly decreased after chemotherapy (Figure 4i), indicating that chemotherapy reversed the ILCregs group. Moreover, via mass cytometry, we identified two different subsets of ILC2s both in tumor without and with chemotherapy, in which one was CD103+ and the other was CD103− (Figure 4j, k). Interestingly, the majority of ILC2 in PDAC was CD103− (Figure 4l).
Tumor infiltrated ILC2 migrated via CCL2/CCR2
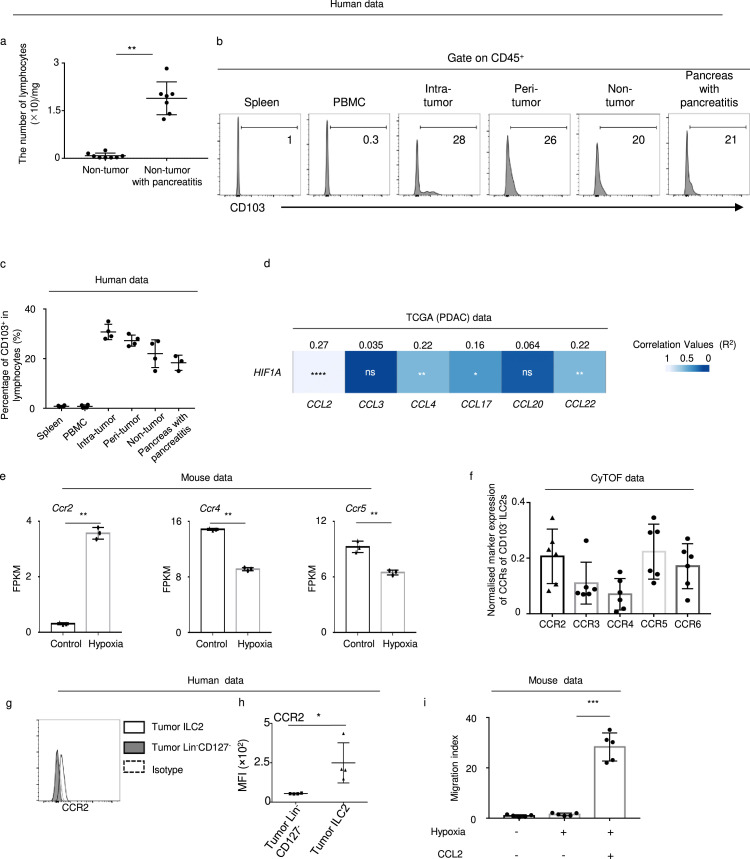
The origin of lymphocytes in the intra-tumor PDAC tissues was unclear. There were very few lymphocytes in the non-tumor pancreas; however, it increased significantly in pancreas tissue with pancreatitis (Figure 5a), in which only 20% lymphocytes were CD103+ and slightly elevated in intra-tumor tissues (Figure 5b, c). These results indicate that the pancreatic tumor related lymphocytes like ILC2 might be migrated from the circulation.
Figure 5.
PDAC ILC2s are derived from the circulation and correlate with CCL2.
(a) Number of lymphocytes in non-tumor tissues with (n = 7) or without pancreatitis (n = 8). (b) Representative flow plot of CD103 in lymphocytes in the indicated tissues. (c) Percentage of CD103+ in lymphocytes from spleen (n = 4), PBMC (n = 4), intra-tumor (n = 4), peri-tumor (n = 4), and non-tumor tissues (n = 4) as well as pancreas with pancreatitis (n = 3). (d) Correlation between the indicated chemokine and HIF1A in the TCGA (PDAC) database. (e) FPKM of Ccr2, Ccr4, and Ccr5 in control and 24h hypoxia mouse ILC2 groups. (f) Expression of CCR2, CCR3, CCR4 CCR5, and CCR6 in CD103− ILC2 (CyTOF data) (n = 6). (g) Representative flow plot and (h) statistical analysis of CCR2 in human tumor-derived ILC2s and Lin−CD127− (n = 4). (i) Mouse ILC2s migrate in response to CCL2 under hypoxia condition (n = 5). Data are expressed as means ± SD and compared using the unpaired t test (*,P < 0.05; **, P < 0.01; ***, P < 0.001).
As shown in CyTOF data, PDAC-derived ILC2s expressed CCR2, CCR3, CCR4, CCR5, and CCR6 (Figure 4j, k). According to a study by Weston et al.20 PBMC-derived ILC2s do not express CCR3. Therefore, we investigated the ligands of CCR2, CCR4, CCR5, and CCR6 and observed that the levels of CCL2, CCL3, CCL4, CCL17, CCL20, and CCL22 were significantly upregulated in tumors (Fig. S4). CCL2 (CCR2), CCL4 (CCR4), and CCL22 (CCR5) had the highest correlation values with HIF1A (Figure 5d). Of these, RNA-Seq data from hypoxic mouse ILC2s showed that the Ccr2 level, rather than Ccr4 and Ccr5, was upregulated in hypoxic conditions (Figure 5e). In addition, CyTOF data showed that CD103− ILC2 expressed high level of CCR2 (Figure 5f). Flow data confirmed that PDAC-derived ILC2s expressed higher CCR2 than Lin−CD127− cells (Figure 5g, h). Moreover, CCL2 could induce the migration of mouse ILC2s under in vitro hypoxic conditions for 24 h (Figure 5i). Taken together, these results indicate that circulatory ILC2s may selectively respond to CCL2 and migrate into tumors.
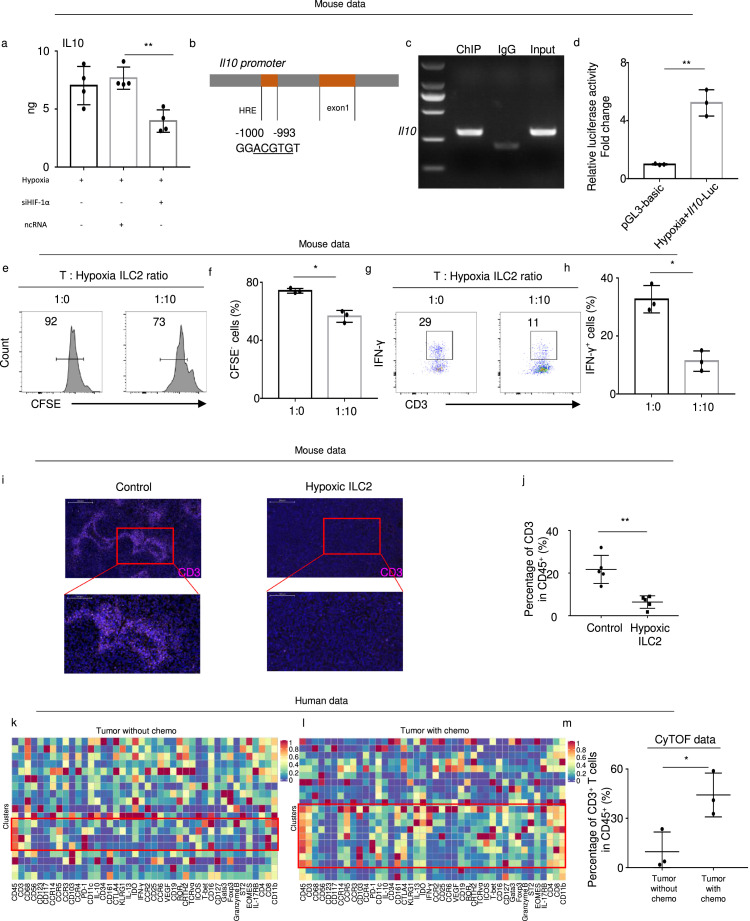
Hypoxia-induced IL10+ ILCregs suppress the proliferation of CD3+ T cells
Knockdown of Hif-1α in ILC2 cells significantly decreased the IL10 level (Figure 6a). To investigate the interaction between Hif-1α and Il10 in ILC2s, we searched for HREs in the promoter regions of Il10 and identified a high-scoring HRE-like binding site of ILC2s (Figure 6b). Chromatin immunoprecipitation verified that HIF-1α directly bound to the -1000 to -993 bp (GGACGTGT) promoter of Il10 in mouse ILC2s (Figure 6c). Subsequently, we transfected the Il10 luciferase reporter. Compared with the control, the Il10 reporter was increased under hypoxic conditions, indicating that an HRE motif existed in the promoter fragment of the Il10 gene (Figure 6d).
Figure 6.
Hypoxia-induced IL10+ ILCregs suppress the proliferation of CD4+ T cells and ILC2s.
(a) Expression of IL10 in mouse ILC2s under hypoxic conditions for 24 h after Hif-1α knockdown by siRNA compared with controls (as detected by ELISA) (n = 4). (b) Schematic depiction of the Il10 promoter suggested a putative HRE sequence. (c) ChIP analysis confirming the existence of the HRE in the promoter of the Il10 gene using anti-HIF-1α. (d) Luciferase activities of Il10 under hypoxia (n = 3). (e) The percentage of CFSE− T cells after 5-days culture with or without ILC2s and (f) statistical analysis of the percentage of CFSE− T cells (n = 3). (g) The percentage of IFN-γ+ cells after 5-day culture with or without ILC2s and (h) statistical analysis of the percentage of IFN-γ+ cells (n = 3). (i) Representative immunofluorescence images (magnification, × 400) of mouse PDAC tissue stained for CD3. Nuclei were stained with DAPI (blue). The lower panel presents magnified views (× 1500) of the area within the red boxes in the left panel. (j) The percentage of CD3+ cells in CD45+ cells identified by flow cytometry in mice orthotopic PDAC (n = 5). (k) (l) Heatmap of CD3+ T cells in TP tumors with or without chemotherapy in CyTOF, indicated by the rectangle drawn with the red border. (m) Percentages of CD3+ T cells in lymphocytes from tumors with or without chemotherapy (CyTOF data) (n = 3). Data are expressed as means ± SD and compared using the unpaired t test (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
To examine immunosuppression function of ILCregs, we co-cultured hypoxia-treated and untreated mouse ILC2s with mouse CD3+ T cells. As expected, CD3+ T cell proliferation was inhibited in the ILCregs (Figure 6e, f). In addition, the ILCregs significantly decreased secretion of IFN-γ in the CD3+ T cells (Figure 6g, h). Moreover, when we injected ILCregs into mice, CD3+ T cells in tumor were significantly decreased (Figure 6i, j). Notably, CyTOF data showed that chemotherapy reversed this effect and increased CD3+ T cells in PDAC tissues (Figure 6k–m). These results therefore highlight that the immunosuppressive function conversion of ILC2 to ILCreg may inhibit T cells function in PDAC tumor microenvironment.
Discussion
For decades, gemcitabine was the only chemotherapy choice for patients with PDAC. Recent advancements in adjuvant therapies, such as FOLFIRINOX and AG, have been reported to improve limited survival but with increased toxicity, especially in TP patients.2 Patient responses to immune checkpoint inhibitors, such as anti-programmed cell death protein 1 (PD-1), anti-programmed death-ligand 1 (PD-L1), and anti-cytotoxic T lymphocyte-associated antigen 4 (CTLA-4), alone or in combination, are quite disappointing.21,22 Previous studies have reported that PDAC has an infiltrated-excluded TIME, and lymphocytes localized along the border of the tumor mass are in the invasive margin.23,24 Therefore, PDAC has been classified as a “cold” tumor,23 which may explain why immunotherapy has been largely unsuccessful. However, in this article, we found a substantial population of lymphocytes in intra-tumor tissues and few lymphocytes in peri-tumor and non-tumor tissues, which prompted us to speculate that the low response rate to immunotherapy might be due to the immunosuppressive mechanisms in the TIME of PDAC patients.
Hypoxia is known as a characteristic feature of the PDAC microenvironment.25 To date, large bodies of work on hypoxia have focused on the cancer cells, and have taken little note of immune cells. This is the first study to report that hypoxia induced a regulatory ILC2 phenotype. In response to hypoxia, ILC2s expressed Ido1, Ctla4, Pdcd1, Il2ra, and Il10. Moreover, we identified a functional HRE in the promoter of Il10. Hypoxic ILC2s also inhibited T-cell proliferation compared with the control. These findings indicate that ILCregs can be generated from ILC2s under hypoxic conditions.
Morita and his colleagues reported that RA (retinoic acid) induced IL-10 secretion in ILC2s in asthma patients.26 However, the TCGA database did not show any differences in the levels of ALDH1A1 and ALDH1A2 in PDAC (Fig. S5a), which encode RALDH1 and RALDH2 and covert retinol to RA. Although ALDH1A3 expression was higher in tumor tissues, its level was very low compared with the other two genes (Fig. S5b). On the other hand, Wang et al. reported that ILCregs were present in mouse and human intestines in both the inflammatory and steady state.27 In the present study, we cannot detect ILCregs in the steady state in normal pancreas, and the expression of Id3 decreased in mouse hypoxic ILC2s, which was found to be essential for the development of intestinal ILCregs. Therefore, these results indicated that hypoxia-induced ILCregs in PDAC might be different with that in the intestine and lung.
A previous study characterized ILC2s as anti-cancer immune cells in PDAC.9 Consistent with the study by Moral et al.9 the injection of ILC2s into mice could decrease tumor volume. However, the injection of hypoxic ILC2s increased the tumor volume and induced peritoneal metastasis, indicating that hypoxia-treated ILC2s promoted PDAC progression. The discrepancy between our results and those of Moral et al. might be due to differences in hypoxia treatment of cells, which might reflect the real status in a tumor microenvironment. Moreover, we did not find an association between IL33, the ILC2-activating cytokine, and better prognosis in TCGA, as well as another ILC2-activating cytokine, TSLP. Our CyTOF data also demonstrated that PDAC-ILC2s expressed low levels of IL33R.
Homma et al.28 Murakami et al.29 and Tsuchikawa et al.30 have conclusively established that the number of CD8+ T cells increased and the number of FOXP3+ cells decreased in response to gemcitabine-based chemotherapy. However, whether the AG regimen could immunologically alter TIME still remained unclear. It was believed that tumors treated with nab-paclitaxel contained less abundant CAFs. As expected, the present study found that the AG regimen significantly reduced the hypoxic area in tumors. We employed a combination of flow cytometry and CyTOF data to show that the percentage of PDAC-derived ILC2s did not differ with or without chemotherapy. However, the regulatory phenotype of ILC2s was reversed after chemotherapy. Combined with in vitro data, these results might indicate that re-oxygenation of the hypoxic tumor area can reverse the phenotype of ILCregs.
High expression level of CEA+/CA125+/CA19-9 indicated abundant mucins presented in TIME of TP patients. Therefore, there might exist other mechanisms independent of immune modulation in TP patients. Thomas et al. reported that CA125/MUC16 activated AKT and GSK3β pathways through ErbB receptors enhancing PDAC progression.31 Muniyan et al. revealed the interaction of CA125/MUC16 and FAK in PDAC.32 Recently, Engle et al. established a genetically engineered mouse models (GEMM) that expressed CA19-9.33 CA19-9 expression in mice could not only promote rapid and severe pancreatitis, but also accelerate pancreatic cancer progression. Therefore, this maybe a suitable mice model for the investigation of both immune and nonimmune mechanisms of TP patients in future research.
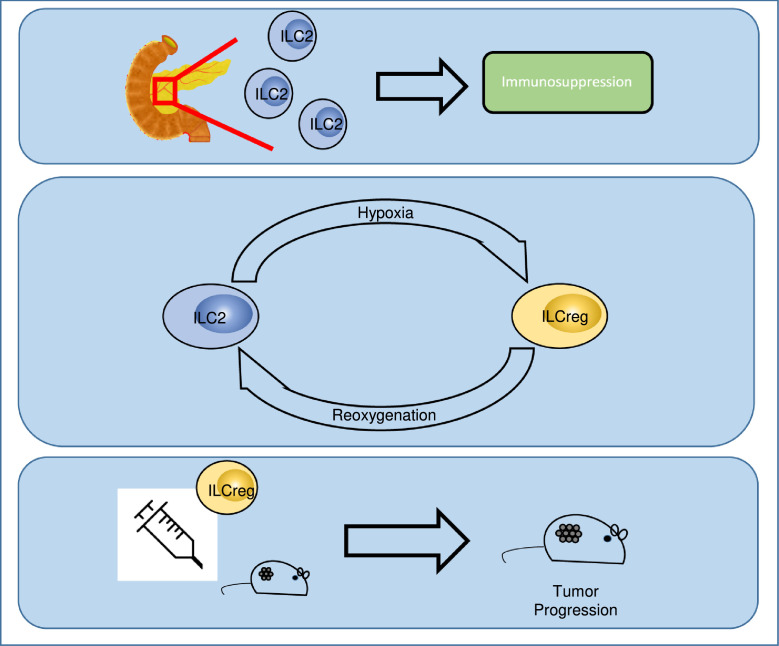
In conclusion, we showed that inhibitory ILC2s are enriched in TP PDAC patients and identified a new regulatory pathway by which ILC2s are converted to ILCregs under hypoxic conditions. This is the first study to report that hypoxia induced a regulatory ILC2 phenotype. Our findings support a novel mechanism to explain how ILCs adapt to hypoxia and promote PDAC progression (Figure 7).
Figure 7.
Schematic model depicting hypoxia-induced ILCreg from ILC2 in PDAC tumour microenvironment.
Substantial hypoxia exists in PDAC tumour microenvironment. When the circulating ILC2s are recruited to the tumour, hypoxia-treated ILC2s underwent a reversible transition to ILCregs. This subsets of ILC2 counteract effective T cell responses and promote tumor progression.
Contributors
LY, KJ and ZL: Conceptualization, data curation, formal analysis, investigation, methodology, supervision, validation, visualization, writing – original draft and editing. LY, ZX, and HX: Data curation and formal analysis. XL: Visualization and writing – original draft. HL: Formal analysis and resources. TL, WZ, XH: Data curation, formal analysis and investigation. WW (Wenquan Wang): Methodology, supervision, and validation. HG: Writing – original draft and editing, LL: Data curation, formal analysis and investigation. WW (Weiding Wu) and XY: Conceptualization, supervision, validation. All of the authors (LY, KJ, ZL, ZX, HX, XL, HL, TL, WZ, XH, WW (Wenquan Wang), HG, LL, WW (Weiding Wu), and XY) discussed the results and approved the manuscript.
Data sharing statement
The data used in this study can be obtained by request to LY(yelongyun@fudanpci.org) upon reasonable request. RNA-seq raw data were deposited to GEO database (accession code GSE196812).
Declaration of interests
The authors declare no potential conflicts of interest.
Acknowledgments
This study was jointly supported by the National Natural Science Foundation of China (U21A20374, 82173091, and 81701630), Shanghai Municipal Science and Technology Major Project (21JC1401500), Scientific Innovation Project of Shanghai Education Committee (2019-01-07-00-07-E00057), Clinical Research Plan of Shanghai Hospital Development Center (SHDC2020CR1006A), Xuhui District Artificial Intelligence Medical Hospital Cooperation Project (2021-011), Research Project of Shanghai Municipal Health Commission (20214Y0396, 20194Y0375), Shanghai Pujiang Program (21PJD014) and China Scholarship Council scholarship (No. 201808330152). The funding agencies had no role in study design, data collection, and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2022.104016.
Contributor Information
Liang Liu, Email: liuliang@fudanpci.org.
Weiding Wu, Email: wuweiding@fudanpci.org.
Xianjun Yu, Email: yuxianjun@fudanpci.org, yuxianjun@fudan.edu.cn.
Appendix. Supplementary materials
References
- 1.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70(1):7–30. doi: 10.3322/caac.21590. [DOI] [PubMed] [Google Scholar]
- 2.Liu L., Xu H., Wang W., et al. A preoperative serum signature of CEA+/CA125+/CA19-9 >/= 1000 U/mL indicates poor outcome to pancreatectomy for pancreatic cancer. Int J Cancer. 2015;136(9):2216–2227. doi: 10.1002/ijc.29242. [DOI] [PubMed] [Google Scholar]
- 3.Koong A.C., Mehta V.K., Le Q.T., et al. Pancreatic tumors show high levels of hypoxia. Int J Radiat Oncol Biol Phys. 2000;48(4):919–922. doi: 10.1016/s0360-3016(00)00803-8. [DOI] [PubMed] [Google Scholar]
- 4.Ye L.Y., Zhang Q., Bai X.L., Pankaj P., Hu Q.D., Liang T.B. Hypoxia-inducible factor 1alpha expression and its clinical significance in pancreatic cancer: a meta-analysis. Pancreatology. 2014;14(5):391–397. doi: 10.1016/j.pan.2014.06.008. [DOI] [PubMed] [Google Scholar]
- 5.Daniel S.K., Sullivan K.M., Labadie K.P., Pillarisetty V.G. Hypoxia as a barrier to immunotherapy in pancreatic adenocarcinoma. Clin Transl Med. 2019;8(1):10. doi: 10.1186/s40169-019-0226-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hsu T.S., Lai M.Z. Hypoxia-inducible factor 1alpha plays a predominantly negative role in regulatory T cell functions. J Leukocyte Biol. 2018;104(5):911–918. doi: 10.1002/JLB.MR1217-481R. [DOI] [PubMed] [Google Scholar]
- 7.Schwartz M., Zhang Y., Rosenblatt J.D. B cell regulation of the anti-tumor response and role in carcinogenesis. J Immunother Cancer. 2016;4:40. doi: 10.1186/s40425-016-0145-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Henze A.T., Mazzone M. The impact of hypoxia on tumor-associated macrophages. J Clin Invest. 2016;126(10):3672–3679. doi: 10.1172/JCI84427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moral J.A., Leung J., Rojas L.A., et al. ILC2s amplify PD-1 blockade by activating tissue-specific cancer immunity. Nature. 2020;579(7797):130–135. doi: 10.1038/s41586-020-2015-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang Q., Ma X., Wang Y., et al. IL-10 producing type 2 innate lymphoid cells prolong islet allograft survival. EMBO Mol Med. 2020;12(11):e12305. doi: 10.15252/emmm.202012305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ye L.Y., Chen W., Bai X.L., et al. Hypoxia-induced epithelial-to-mesenchymal transition in hepatocellular carcinoma induces an immunosuppressive tumor microenvironment to promote metastasis. Cancer Res. 2016;76(4):818–830. doi: 10.1158/0008-5472.CAN-15-0977. [DOI] [PubMed] [Google Scholar]
- 12.Xu X., Ye L., Zhang Q., et al. Group-2 innate lymphoid cells promote hepatocellular carcinoma progression via CXCL2-neutrophil induced immunosuppression. Hepatology. 2021 doi: 10.1002/hep.31855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ye L., Pan J., Liang M., et al. A critical role for c-Myc in group 2 innate lymphoid cell activation. Allergy. 2020;75(4):841–852. doi: 10.1111/all.14149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ji S., Zhang B., Liu J., et al. ALDOA functions as an oncogene in the highly metastatic pancreatic cancer. Cancer Lett. 2016;374(1):127–135. doi: 10.1016/j.canlet.2016.01.054. [DOI] [PubMed] [Google Scholar]
- 15.Xu X., Ye L., Zhang Q., et al. Group-2 innate lymphoid cells promote HCC progression through CXCL2-neutrophil-induced immunosuppression. Hepatology. 2021;74(5):2526–2543. doi: 10.1002/hep.31855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miao Y.R., Zhang Q., Lei Q., et al. ImmuCellAI: a unique method for comprehensive T-cell subsets abundance prediction and its application in cancer immunotherapy. Adv Sci. 2020;7(7) doi: 10.1002/advs.201902880. (Weinh) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu L., Deng C., Pang B., et al. TIP: a web server for resolving tumor immunophenotype profiling. Cancer Res. 2018;78(23):6575–6580. doi: 10.1158/0008-5472.CAN-18-0689. [DOI] [PubMed] [Google Scholar]
- 18.Tang Z., Kang B., Li C., Chen T., Zhang Z. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019;47(W1):W556–WW60. doi: 10.1093/nar/gkz430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alvarez R., Musteanu M., Garcia-Garcia E., et al. Stromal disrupting effects of nab-paclitaxel in pancreatic cancer. Br J Cancer. 2013;109(4):926–933. doi: 10.1038/bjc.2013.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weston C.A., Rana B.M.J., Cousins D.J. Differential expression of functional chemokine receptors on human blood and lung group 2 innate lymphoid cells. J Allergy Clin Immunol. 2019;143(1):410–413. doi: 10.1016/j.jaci.2018.08.030. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li T.J., Wang W.Q., Yu X.J., Liu L. Killing the “BAD”: Challenges for immunotherapy in pancreatic cancer. Biochim Biophys Acta Rev Cancer. 2020;1874(1) doi: 10.1016/j.bbcan.2020.188384. [DOI] [PubMed] [Google Scholar]
- 22.Ahn D.H., Ramanathan R.K., Bekaii-Saab T. Emerging therapies and future directions in targeting the tumor stroma and immune system in the treatment of pancreatic adenocarcinoma. Cancers. 2018;10(6) doi: 10.3390/cancers10060193. (Basel) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Binnewies M., Roberts E.W., Kersten K., et al. Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat Med. 2018;24(5):541–550. doi: 10.1038/s41591-018-0014-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van den Ende T., van den Boorn H.G., Hoonhout N.M., et al. Priming the tumor immune microenvironment with chemo(radio)therapy: a systematic review across tumor types. Biochim Biophys Acta Rev Cancer. 2020;1874(1) doi: 10.1016/j.bbcan.2020.188386. [DOI] [PubMed] [Google Scholar]
- 25.Sarcar B., Li X., Fleming J.B. Hypoxia-induced autophagy degrades stromal lumican into tumor microenvironment of pancreatic ductal adenocarcinoma: a mini-review. J Cancer Treat Diagn. 2019;3(1):22–27. doi: 10.29245/2578-2967/2019/1.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morita H., Kubo T., Ruckert B., et al. Induction of human regulatory innate lymphoid cells from group 2 innate lymphoid cells by retinoic acid. J Allergy Clin Immunol. 2019;143(6):2190–2201. doi: 10.1016/j.jaci.2018.12.1018. e9. [DOI] [PubMed] [Google Scholar]
- 27.Wang S., Xia P., Chen Y., et al. Regulatory innate lymphoid cells control innate intestinal inflammation. Cell. 2017;171(1):201–216. doi: 10.1016/j.cell.2017.07.027. e18. [DOI] [PubMed] [Google Scholar]
- 28.Homma Y., Taniguchi K., Murakami T., et al. Immunological impact of neoadjuvant chemoradiotherapy in patients with borderline resectable pancreatic ductal adenocarcinoma. Ann Surg Oncol. 2014;21(2):670–676. doi: 10.1245/s10434-013-3390-y. [DOI] [PubMed] [Google Scholar]
- 29.Murakami T., Homma Y., Matsuyama R., et al. Neoadjuvant chemoradiotherapy of pancreatic cancer induces a favorable immunogenic tumor microenvironment associated with increased major histocompatibility complex class I-related chain A/B expression. J Surg Oncol. 2017;116(3):416–426. doi: 10.1002/jso.24681. [DOI] [PubMed] [Google Scholar]
- 30.Tsuchikawa T., Hirano S., Tanaka E., et al. Novel aspects of preoperative chemoradiation therapy improving anti-tumor immunity in pancreatic cancer. Cancer Sci. 2013;104(5):531–535. doi: 10.1111/cas.12119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thomas D., Sagar S., Liu X., et al. Isoforms of MUC16 activate oncogenic signaling through EGF receptors to enhance the progression of pancreatic cancer. Mol Ther. 2021;29(4):1557–1571. doi: 10.1016/j.ymthe.2020.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Muniyan S., Haridas D., Chugh S., et al. MUC16 contributes to the metastasis of pancreatic ductal adenocarcinoma through focal adhesion mediated signaling mechanism. Genes Cancer. 2016;7(3-4):110–124. doi: 10.18632/genesandcancer.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Engle D.D., Tiriac H., Rivera K.D., et al. The glycan CA19-9 promotes pancreatitis and pancreatic cancer in mice. Science. 2019;364(6446):1156–1162. doi: 10.1126/science.aaw3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.