Abstract
Introduction:
Aerobic exercise maintains telomere length through increased human telomerase reverse transcriptase (hTERT) expression and telomerase enzyme activity. The impact of acute exercise on hTERT alternative splicing (AS) is unknown.
Purpose:
This study aimed to examine hTERT AS in response to acute treadmill running.
Methods:
A bacterial artificial chromosome mouse model containing the 54 kilobase hTERT gene locus inserted into its genome (hTERT-BAC) was utilized. The gastrocnemius, left ventricle, and brain were excised prior to (Pre), upon cessation (Post), and during recovery (1, 24, 48, and 72H; n = 5/time point) from treadmill running (30 min. at 60% max. speed). Full length (FL) hTERT and the “minus beta” (−β) AS variant (skips exons 7 and 8 and does not code for active telomerase) were measured by gel based and droplet digital RT-PCR methods. SF3B4 and SRSF2 protein expression were measured by western blotting.
Results:
Compared to Pre, FL hTERT increased at Post before decreasing during recovery in the gastrocnemius (48 and 72H; p ≤ 0.001) and left ventricle (24 h; p = 0.004). The percentage of FL hTERT in the gastrocnemius also increased during recovery (1 and 72H; p ≤ 0.017), whereas a decrease was observed in the left ventricle (1, 24, and 48 h; p ≤ 0.041). hTERT decreased in the brain (48 h), whereas FL hTERT percentage remained unaltered. SF3B4 protein expression decreased throughout recovery in the gastrocnemius and tended to be associated with FL hTERT (r = −0.348, p = 0.075) and −β in opposite directions (r = 0.345, p = 0.067).
Conclusions:
Endurance exercise increased hTERT gene expression and altered FL hTERT splicing contractile tissues and may maintain telomere length necessary to improve the function and health of the organism.
Keywords: hTERT, SF3B4, SRSF2, Alternative Splicing, Intronic Element
INTRODUCTION
Regular participation in endurance exercise is a powerful behavioral strategy that is known to promote healthy aging and has recently been shown to slow multiple aspects of the aging process at the cellular level (1). One well known cellular biomarker of the aging process is telomere length (1). Telomeres are 5′(TTAGGG)n3′ repeats located at the ends of linear chromosomes (2). In response to cellular replication, telomere lengths progressively shorten due to the inability of DNA polymerase to fully replicate telomeric DNA (3–5). This “end-replication-problem” is partially compensated by the ribonucleoprotein telomerase that generates new telomeric DNA repeats to maintain or prevent the excessive shortening of telomere lengths during cellular replication (6, 7). However, the telomerase enzyme is silenced or only present at extremely low levels in adult human somatic cells (8–9). Consequently, telomere length shortens with subsequent cellular replication cycles. When a few telomeres in a cell reach a critically shortened length, cells enter a state of replicative senescence that is associated with the development and progression of age-related disease pathology (10–12). Regular participation in endurance exercise (e.g., running) is known to maintain telomere lengths across the lifespan (13–15). Therefore, the identification of molecular mechanisms that activate telomerase and help to maintain telomere lengths are vital for understanding how exercise training impacts cellular aging and aids in improving the quality of life of an aging population.
One under explored area of investigation in the exercise and telomere field is the regulatory mechanisms of telomerase. Telomerase is a ribonucleoprotein consisting minimally of an RNA template (telomerase RNA component known as hTERC or hTR) and the protein catalytic component human telomerase reverse transcriptase (hTERT). hTERC is expressed constitutively across human cells and tissues (16, 17). In contrast, hTERT is tightly regulated, expressed at low levels, and is recognized as the rate-limiting component of the active telomerase enzyme in most cell types. hTERT is a 16-exon gene, and all 16 exons must be present in full and in sequence (i.e., full length [FL] hTERT) to encode for the catalytically active telomerase enzyme (16). In addition to FL hTERT, 21 alternatively spliced variants (ASVs) of hTERT have been identified that do not code for active telomerase enzyme activity (9). The most common ASV measured is termed minus beta (−β), which skips exons 7 and 8 of the reverse transcriptase (RT)-domain and places a premature termination codon in frame within exon 10 that could result in the transcript being degraded by the non-sense mediated (NMD) decay pathway (8). Utilizing primers spanning exons 5 through 9 of the RT-domain, Ulaner et al. (18) demonstrated that the FL hTERT transcript and −β ASV are co-expressed during early fetal kidney development. However, a shift in splicing away from the FL transcript was observed in favor of the −β ASV from weeks 17 to 18. Moreover, reduced FL hTERT occurred in parallel with the suppression of telomerase enzyme activity and shortening of telomere lengths (19). Additionally, the importance of alternative splicing of hTERT is underscored by the observation that the FL hTERT isoform is the minority (1–25%) of the transcriptional output and the ASVs make up the majority (99–75%) of the transcriptional output from the hTERT gene (16, 20, 21). These findings indicate that alternative splicing of hTERT is regulated, in part to help control the levels of active telomerase in cells and tissues.
Acute exercise is a physiological stressor sufficient to transiently increase hTERT mRNA expression and activate the telomerase enzyme (22–24). However, insights into the mechanisms of hTERT/telomerase regulation in response to acute exercise are largely limited to relative expression changes in circulating human immune cells, or in mouse tissues which constitutively express telomerase enzyme activity and do not rely upon alternative splicing as a mechanism of mTert regulation (25). More specifically, human TERT has intronic cis-elements in the RT-domain that are highly conserved in old world primates, but are not found in rodent species, and are critical for the alternative splicing pattern of hTERT (25–28). For example, intron 6 of hTERT pre-mRNA contains Block 6 (B6), a variable number tandem repeat (VNTR) composed of 38-nt repeats (also known as VNTR6-1st), and the 256-nt direct repeat 6 (DR6) cis-elements necessary for producing the −β ASV. In addition, the 258-nt DR8 cis-element located in intron 8 is necessary for the promotion or repression of FL hTERT when bound by specific trans-acting RNAbps (see Figure 1; 25–28). Therefore, studying how physiological stressors such as endurance exercise impact the alternative splicing regulatory mechanisms of mTert within traditional animal models would not provide meaningful insights to the human condition because they lack these important intronic elements that dictate alternative splicing choice. To overcome these species-specific differences, we utilized an hTERT-BAC (bacterial artificial chromosome) mouse model that has the entire hTERT gene locus inserted in its genome (29).
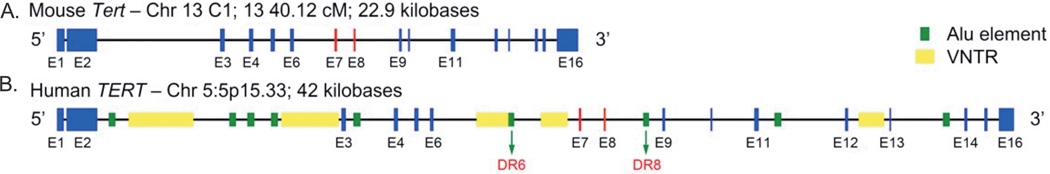
FIGURE 1–
Cartoon of mTert and hTERT genes. Both mTert and hTERT are 16 exon genes, and all 16 exons must be present in full to encode for the catalytically active telomerase enzyme. Although average exon lengths are similar (mTert: 269.56 ± 348.94 nucleotides; hTERT: 252.44 ± 326.38 nucleotides), average hTERT intron lengths are nearly twice as long compared to mTert (1241 ± 1324.82 vs 2524.2 ± 2772.03 nucleotides; see Table 1). In addition, hTERT introns contain numerous species specific cis-elements that are recognized by various trans-acting RNA binding proteins (RNAbps) and vital for the regulation of alternative splicing of hTERT pre-mRNA, including the −β ASV that excludes exon 7 and 8 of the RT-domain (in red).
The hTERT-BAC mouse model has previously been utilized to reveal species specific promoter cis-elements (29). However, hTERT alternative splicing has not been explored in the in vivo context in this mouse model. Furthermore, the mechanisms by which acute treadmill running impacts hTERT alternative splicing remain yet to be examined. The hTERT-BAC mouse model should allow for the confirmation that the splicing machinery of mice (trans-acting RNA binding proteins [RNA-bp]), which are highly conserved between mice and humans (30), recognizes the human specific TERT pre-mRNA cis-elements shown to regulate hTERT alternative splicing patterns (25–28). The hTERT-BAC mouse model will also allow us to test if exercise impacts hTERT alternative splicing patterns in tissues that are difficult to obtain from humans, such as whole skeletal muscle, heart, and brain. Likewise, the hTERT-BAC mouse will provide necessary understanding of the mechanisms by which exercise aids in the regulation of telomerase and potentially a mechanism as to how exercise slows the aging process.
Results from the present study confirm the recapitulation of human, tissue-specific splicing patterns (i.e., FL and −β) within hTERT-BAC mice. hTERT-BAC mice were then challenged with an acute bout of treadmill running and the response of hTERT gene expression and alternative splicing patterns were examined in skeletal muscle, heart, and brain tissues. Utilizing results from our previously published siRNA minigene screen against 515 potential RNAbps that manipulate hTERT splicing in HeLa cells (28), we examined changes in protein abundance of two trans-acting RNAbps predicted to regulate hTERT splicing choice, splicing factor 3b subunit 4 (SF3B4) and serine and arginine rich splicing factor 2 (SRSF2) (28, 31). We hypothesized that acute treadmill running would increase FL hTERT splicing and increase the expression levels of SF3B4 and SRSF2. We further hypothesized that FL hTERT expression levels would be positively correlated to SF3B4 and SRSF2 protein levels and inversely correlated to minus beta hTERT alternative splicing.
METHODS
Animals
All animal experiments were approved by the University of Michigan Institutional Animal Care and Use Committee (IACUC) and conducted as per the institutional guidelines. C57BL/6-Tg(TERT)C10Hode/J (~ 8-weeks old) were purchased from The Jackson Laboratory (Bar Harbor, ME, USA). The transgenic hTERT+ founder mice were housed at 25°C on a 12:12-h light-dark cycle, fed ad libitum laboratory mouse chow (Prolab RMH 3000, 5P00, LabDiet by Purina, Nestlé, Vevey, Switzerland), and given free access to water. Following a 2-week acclimation period, breeding pairs were established for the hTERT-BAC mouse colony that maintain the C57BL/6 background and express the hTERT gene (29). Following each litter, pups were weaned after three weeks, and PCR genotyping was carried out using tail DNA (KAPA Biosystems, Wilmington, MA, USA). hTERT-BAC mice that were hTERT+ were confirmed from extracted tail DNA using transgene hTERT (F: 5’-GGAAGGCAGGAGGCTCTTGG-3’; R: 5’-TGCACACACTTGTGCCCTTGC-3’; 431-bp) and control primer pairs (F: 5’-CAAATGTTGCTTGTCTGGTG-3’; R: 5’-GTCAGTCGAGTGCACAGTTT-3’; 200-bp) in EmeraldAmp MAX PCR Master Mix (Takara Bio, Mountain View, CA, USA) on a C1000 Touch Thermal Cycler (Bio-Rad Laboratories, Hercules, CA, USA) under the following conditions: heat activation at 98°C for 5 minutes, followed by 35 cycles of denaturation at 95°C for 10 seconds, annealing at 60°C for 30 seconds, and an extension at 72°C for 1 minute. A final extension step at 72°C for 5 minutes was also performed. The results were confirmed on a 1.2% agarose gel (see SDC 1, Supplemental Figure 1, hTERT-BAC mouse genotyping).
Treadmill Acclimation and Incremental Exercise Testing Protocol
Confirmed hTERT+ transgenic male and female hTERT-BAC mice were aged to 8-weeks before undergoing a 10-day treadmill acclimation protocol as previously described (Figure 2A; 32, 33). In brief, the treadmill was set at a 7% incline for all treadmill sessions and mice were introduced to gradual increases in speed and shock stimuli during each session on the treadmill. Forty-eight hours after the last acclimation session, each mouse completed an incremental exercise test to determine their peak treadmill running speed. The peak treadmill running speed test was initiated with each mouse being placed on a motionless treadmill for 2 minutes before increasing the belt speed to 5 meters per minute for minutes 2 through 5. At minute 5, the treadmill speed was increased at a consistent pace of 1 meter per minute until the running ability of each mouse was impaired (determined by the animal’s refusal to run even when provoked for ≥ 30 seconds). The final speed at which the mouse refused to run was recorded as the peak treadmill running speed.
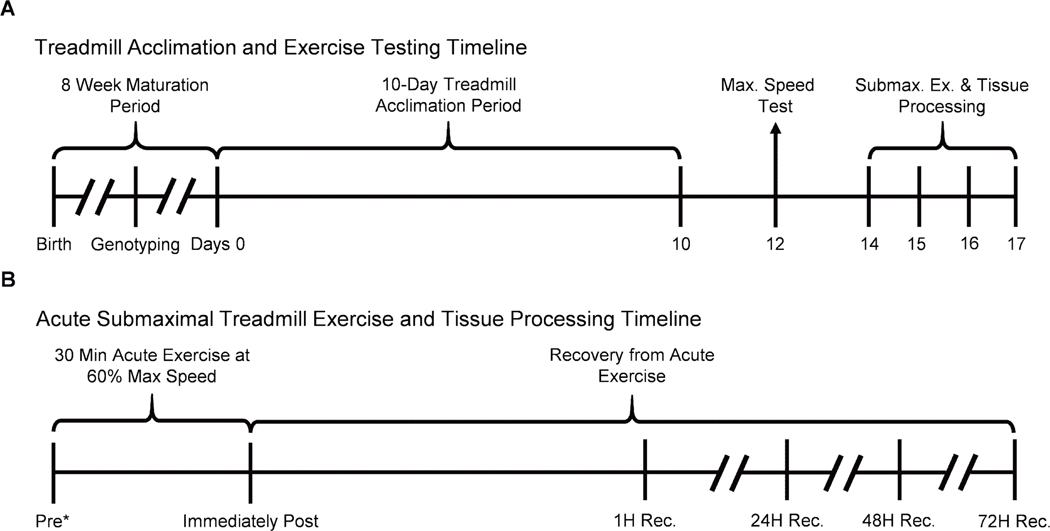
FIGURE 2–
Timelines of treadmill acclimation, acute submaximal treadmill exercise testing, and tissue processing in hTERT-BAC mice. A, Upon birth, mice were weaned after 3 wk, genotyped to determine the presence of the hTERT transgene, and age to 8 wk old. Eight-week-old hTERT-BAC mice were then subjected to a progressive treadmill acclimation protocol over the course of 10 d (days 0–10). After a 48-h rest period, hTERT-BAC mice participated in a progressive speed test to determine maximal running speed on the treadmill (day 12). Finally, after another 48-h period of rest, mice participated in an acute, submaximal bout of treadmill exercise for 30 min at 60% of maximal running speed and tissues were processed (days 14–17; panel A). B, A total of 30 hTERT-BAC mice were included in the acute submaximal treadmill exercise protocol and tissues were processed before (Pre), immediately after, and at 1, 24, 48, and 72 h into recovery (n = 5 per time point; panel B). *Preexercise group were exposed to a motionless treadmill for 30min before to tissue processing.
Acute Treadmill Exercise Protocol
Forty-eight hours after peak treadmill running speed test, the mice were exposed to 30 minutes of submaximal treadmill running at 60% of their peak speed (Figures 2A and B). Mice were separated into six experimental groups (n = 5 per group; 2–3 males and females per group). The pre-exercise group (Pre) were exposed to a motionless treadmill for 30 minutes before being euthanized for tissue processing. This is important as the rodents might have developed a stress response associated with the treadmill resulting from the maximal speed test only 48-hours prior, and the exposure to the motionless treadmill would serve to control for any potential physiological responses that would otherwise confound subsequent measures. The remaining 25 mice underwent a 30-min treadmill exercise bout and were euthanized either immediately following acute treadmill exercise (Post) or at 1, 24, 48, or 72 hours into recovery from the exercise bout (1H, 24H, 48H, and 72H).
Cell and Tissue Processing
Bone marrow cells were collected from the femur of both legs of three adult male (6-months old) hTERT-BAC mice in DMEM supplemented with 4% FBS (Fisher Scientific, Hampton, NH, USA). BM cells were flushed out by centrifugation for 15 seconds at 10,000 x g, treated with red blood cell lysis buffer for 1 minute (Sigma Aldrich, St. Louis, MO, USA), and filtered through a 70 μM mesh strainer. Collected cells were then washed and pelleted by centrifugation for 5 minutes at 2,000 rpm, counted, and divided into 1 mL aliquots for storage at −80°C (24 hours) before transfer to liquid nitrogen until processing and analyzed for gene expression. In addition, gastrocnemius, heart (left ventricle), whole brain, liver, gonadal white adipose tissue, and testes were dissected and flash frozen in liquid nitrogen for future analysis from baseline and exercised hTERT-BAC mice. Prior to analysis for specific biomolecules (i.e., RNA and protein), tissues were powdered with a mortar and pestle in liquid nitrogen. The powdered samples were either processed immediately or stored at −80°C until further analysis.
Gel-based and Droplet Digital reverse transcription PCR
Total RNA from bone marrow was isolated using the RNeasy Plus Mini Kit (Qiagen) and tissue RNA was extracted using QIAzol and RNeasy® Universal Mini Kit (Qiagen, Hilden, Germany). Isolated RNA was quantified by spectrophotometry using a NanoDrop 2000 and cDNA was synthesized using SuperScript IV First-Strand cDNA Synthesis System primed with oligodT and random hexamers at a 1:1 ratio (Invitrogen, Carlsbad, CA). All cDNAs were diluted 1:4 using nuclease-free water and stored at −20 °C until analysis of hTERT expression levels qualitatively and quantitatively by gel-based RT-PCR and RT-droplet digital PCR (ddPCR), respectively. For hTERT splice variant analyses, diluted cDNAs were used within 48 hours of production. NIH-3T3 (mouse) and HeLa (human) cells were utilized as species specific positive/negative controls to demonstrate primer specificity.
For qualitative gel-based RT-PCR assays, species specific primers spanning exons 5 through 9 of the RT-domain were utilized (mTert [F: 5’-CGGACAAAACATCCTCACCT-3’; R: 5’-CGAAACACAGACTGCAGAGC-3’]; hTERT [F: 5’-GCCTGAGCTGTACTTTGTCAA-3’; R: 5’-AGGCTGCAGAGCAGCGTGGAGAGG-3’]). PCR was performed using a C1000 Touch Thermal Cycler in 2x EmeraldAmp MAX PCR Master Mix along with 2 μL of cDNA (tissue specific input concentrations specified in each figure caption). Amplification conditions: initial denaturation at 98° for 5 minutes, followed by 27–33 cycles (species and tissue specific cycle numbers specified in each figure caption) of denaturation at 95°C for 10 seconds, annealing at 60°C for 30 seconds, and extension at 72°C for 1 minute. A final extension step at 72°C for 5 minutes was also performed. PCR products were resolved by polyacrylamide gel electrophoresis and stained with GelRed® (Biotium, Hayward, CA, USA) to qualitatively demonstrate alternative splicing patterns for mTert and hTERT in tissues excised from hTERT-BAC mice.
Species specific primers were utilized for total mTert and hTERT gene quantification by RT-ddPCR. Absolute quantification of mTert gene expression levels were determined using primers and probes specific for exon 2 (F: 5’-ACCCCATCAGGCAAAT-3’; R: 5’-GAGGTGCCTACTGCAGAGAAA-3’; universal probe #15 [Roche; Basal, Switzerland]; 25). Total hTERT gene expression levels were determined by summing the absolute quantification of the potential FL, exon 7–8 containing transcripts (determined by PCR of exons 7–8 – Forward primer exon 7: 5’-ACAGTTCGTGGCTCACCTG-3’; Reverse primer exon 8: 5’-GCGTAGGAAGACGTCGAAGA-3’; universal probe #52 [Roche; Basal, Switzerland] in exon 8) plus the −β, exon 7–8 skipping ASV (determined by PCR of exon 6–9 containing transcripts – Forward primer exon 6/9 junction: 5’-CAAGAGCCACGTCCTACGTC-3’; Reverse primer exon 10: 5’-CAAGAAATCATCCACCAAACG-3’; universal probe #58 [Roche; Basal, Switzerland] in exon 9; 28). The summation of exon 7–8 containing and exon 7–8 skipping transcripts has been shown to account for ≥ 95% of total hTERT, suggesting that this strategy should provide a good estimate of total cellular hTERT transcripts (21). It is possible that exon 7–8 containing transcripts, such as the minus α (skipping of the first 36 nucleotides at the 5̍ end of exon 6) and minus γ (exon 11 skipping) transcripts that yields hTERT catalytically inactive, could be included in this analysis. However, these transcripts account for < 5% of total hTERT (21). A 2 μL sample of cDNA (tissue specific input concentrations specified in each figure caption) was combined with species- and transcript-specific primers and probe sets, and 2x ddPCR Supermix for Probes (Bio-Rad Laboratories) in a 20 μL final volume mix. Droplets were generated using 70 μL of Droplet Generation Oil for Probes using the QX200 Droplet Generator (Bio-Rad Laboratories). Generated droplets were loaded into a 96-well plate and amplified under the following thermocycling conditions: initial denaturation at 95° for 10 minutes, followed by 40 cycles of denaturation at 94°C for 30 seconds and annealing at either 55°C (mTert) or 60°C (hTERT) for 1 minute. A final enzyme denaturation step at 98°C for 5 minutes was also performed. Droplets were analyzed using the QX200 Droplet Reader and QuantaSoft software systems, respectively (Bio-Rad Laboratories). Full length and alternatively spliced hTERT percentages were determined as previously described (25) and according to the following formulas: Percent Potential FL = Exon 7–8 containing transcript / Total transcript (total transcripts = exon 7–8 containing transcripts + exon 7–8 lacking transcripts); Percent alternatively spliced = 100 – Percent Potential FL.
Candidate RNA Binding Protein Selection
Our laboratory has recently performed an siRNA minigene screen against 515 potential RNAbps in HeLa cells, resulting in the identification of 93 genes observed to promote FL hTERT splicing and 17 genes identified to reduce FL hTERT splicing in favor of an increase in the −β ASV (28). Experimentally, we have confirmed that two RNAbps, neuro-oncological ventral antigen 1 (NOVA1) and polypyrimidine tract-binding protein 1 (PTBP1), bind directly to the DR8 cis-element within hTERT pre-mRNA to promote FL splicing in non-small cell lung cancer cells (25, 28). However, NOVA1 and PTBP1 expression within skeletal and cardiac tissue are low to undetectable according to data on The Human Protein Atlas (HPA) (34). SF3B4 and SRSF2 splicing factors were among the most robust promoters of FL hTERT from our minigene screen analysis (28) and in silico analysis indicates that both could potentially interact directly with hTERT sequences. SF3B4 is vital for spliceosome assembly and involved in branch point recognition (35) and the SF3B4 motif is located in hTERT intron 8 (3’ end of the direct repeat 8 [DR8] cis-element; 35, 36). In addition, SRSF2 regulates spliceosome assembly and recognizes exonic splicing enhancer motifs to facilitate exon inclusion and promote FL hTERT splicing through its predicted interaction at the intron 6/exon 7 boundary of hTERT pre-mRNA (31, 37).
Western Blot Analysis
To examine the presence of the SF3B4 and SRSF2 splicing factors, total protein lysates were generated from the gastrocnemius and left ventricle using T-per® tissue protein extraction reagent containing Halt® protease and phosphatase inhibitor cocktail (ThermoFisher Scientific, Waltham, MA, USA), and protein concentrations were determined using Pierce 660nm Protein Assay Kit (ThermoFisher Scientific). Protein samples were prepared using 2x Laemmli buffer with β-mercaptoethenol (Bio-Rad Laboratories) and boiled for 5 minutes at 95°C. 15 μg of protein lysate were resolved by SDS-polyacrylamide gel electrophoresis, transferred to polyvinylidene fluoride membranes, and total protein loading was determined with ponceau stain (Sigma Aldrich). Membranes were subsequently destained in 0.1 M NaOH, washed in phosphate-buffered saline with Tween (PBST), incubated for 1 hour in 5% non-fat dry milk, then incubated overnight at 4°C with a rabbit polyclonal antibody for SF3B4 (Cat. No. ab157117; Abcam, Cambridge, UK) or rabbit monoclonal antibody for SRSF2 (Cat. No. ab204916; Abcam) diluted to 1:2000 and 1:4000, respectively, in 5% non-fat dry milk. The following morning, membranes were washed in PBST, incubated for 1 hour with anti-rabbit IgG, HRP-linked secondary antibody (Cat. No. 7074; Cell Signaling Technology, Danvers, MA, USA), and detected in SuperSignal West Femto Maximum Sensitivity Substrate (ThermoFisher Scientific) using a ChemiDoc XRS+ Imaging System (Bio-Rad Laboratories). Protein expression was normalized to total protein using ImageJ.
Statistical Analyses
Unless otherwise noted, a one-way analysis of variance (ANOVA) was utilized to determine the impact of acute exercise on total tissue specific mTert and hTERT gene expression and alternative splicing changes. Pearson’s correlations were utilized to examine the relationship of SF3B4 and SRSF2 protein expression with FL hTERT and −β splicing percentages in the gastrocnemius and left ventricle. Of note, no significant sex differences were observed for any outcome variable under investigation. Statistical significance was defined as a p-value ≤ 0.05 ± standard error of mean (SEM).
RESULTS
hTERT is Alternatively Spliced within the hTERT-BAC Mouse Model
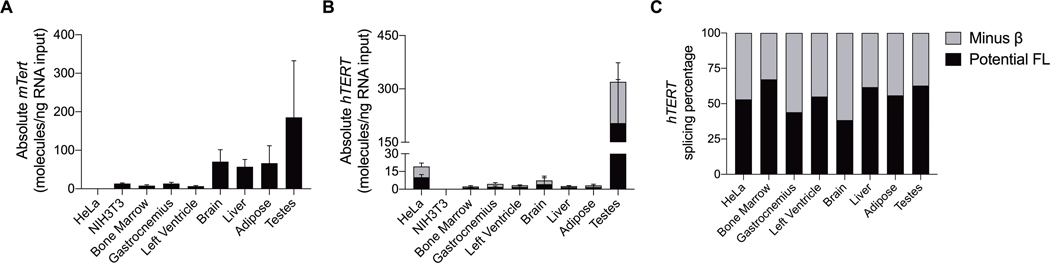
The hTERT-BAC mouse contains the full 54-kb hTERT gene encompassing an 11-kb upstream region, all 16 exonic and 15 intronic sequences, and a 1.2-kb downstream region (29). This hTERT-BAC mouse model allows for the measurement of the hTERT transgene along with the endogenous mTert gene and exhibits hTERT gene expression levels within mouse cells and tissue types at similar expression levels observed in human tissue and cell samples (29). To confirm these results and confirm that our RT-PCR assays are specific for mTert versus hTERT, we measured gene expression within the hTERT RT-domain by RT-PCR (primers spanning exons 5–9) and RT-ddPCR using primers for both the mTert (exon 2) and hTERT (FL: exon 7–8 included; −β ASV: exon 7–8 excluded) genes in bone marrow cells and multiple tissue types (testes are a known telomerase positive tissue in both rodents and humans) obtained from male hTERT-BAC mice (n = 3; 6 months old). Immortal mouse fibroblasts (NIH3T3; mTert and telomerase positive) and HeLa cells (hTERT and telomerase positive) were utilized as reference lines for mTert and hTERT gene expression, respectively. As expected, mTert was expressed as a single band (536 bp) in all tissue/cells collected, as well as the NIH3T3 cells, demonstrating that only the potential FL mTert transcript is present. mTert was not detected in HeLa cells (human cervical carcinoma) indicating that our mTert primers do not detect hTERT. Two bands representing potential FL with all five intact exons (5–6–7–8–9; 423 bp) and the −β ASV that skips exons 7 and 8 (5–6–9; 246 bp; see SDC 2, Supplemental Figure 2, Qualitative alternative splicing expression of the hTERT gene across tissues collected from hTERT-BAC mice) were observed in all tissue/cells collected, as well as HeLa cells but not NIH3T3 cells. These data indicate that our hTERT primers are specific for human sequences and do not detect mouse sequences. Similar primer specificity was observed following RT-ddPCR analysis for mTert and hTERT (Figures 3A and B). The absolute quantification of hTERT expression was low across tissue/cell samples except for in the testes, which is expected (Figure 3B; 29), and the percentage of FL hTERT splicing relative to total hTERT ranged from ~ 35% in brain to ~ 65% in the gastrocnemius (Figure 3C).
FIGURE 3–
Quantitative RT-droplet digital PCR mTert and hTERT gene expression across hTERT-BAC mice tissues. The absolute number of mTert and hTERT molecules per nanograms of RNA input were determined across tissue/cell types isolated from adult male hTERT+ transgenic hTERT mice (n = 3; 6 months old). RNA from cDNAs were amplified with species specific primers–mTert (exon 2); hTERT (exon 7–8 containing [FL] and excluding [−β]). Testes are a known telomerase positive tissue in mice and humans, thereby serving as a positive control tissue. Mouse immortal fibroblasts (NIH-3T3) and human HeLa cell lines were utilized as positive and negative controls for each species-specific primer pair. Data are presented as means ± SD. Total cDNA input–bone marrow: 5 ng; gastrocnemius, left ventricle, brain, liver, adipose, testes, HeLa, and NIH3T3: 12.5 ng.
To understand potential mechanisms for the regulatory differences of alternative splicing between mice and humans for the TERT gene, we next compared to genomic sequences of mice and humans. Alternative splicing is controlled by the combination of cis (sequence) and trans (protein)-elements (38). By comparing the sequences between humans and mice we might reveal cis-elements important in the regulation of hTERT alternative splicing patterns not known to occur within mice. Compared to the mTert gene locus, the hTERT gene locus is nearly twice as long (22.9 vs. 42.0 kilobases; Figure 1). Although genomic sequence analysis revealed that the average exon length of the 16 exons is similar (mTert: 269.56 ± 348.94 nucleotides; hTERT: 252.44 ± 326.38 nucleotides), average intron length is over twice as long in hTERT compared to mTert (1,241 ± 1,324.82 vs. 2,524.2 ± 2,772.03 nucleotides; Figure 1; Table 1). In addition to the differences in mTert/hTERT promoter regions that result in the repression of hTERT transcription in fibroblasts (29), Roy et al. (39) demonstrated that larger intron length contributes to proteomic and organismal diversity through alternative RNA splicing. This could indicate that the regulatory information for the hTERT alternative splicing pattern is contained in the much larger intronic non-coding sequences of the hTERT gene compared to the mTert gene (26, 28). Indeed, hTERT introns 2, 6, 8, 11, 12 and 13 all contain elements that are unique to the human gene and are known potential regulators of alternative splicing (i.e., Alu elements; 21). Therefore, differences in mTert/hTERT gene expression and telomerase enzyme activity between mice and human also likely result from alternative RNA splicing facilitated by conserved trans-acting RNA binding proteins (RNAbps) recognizing intronic cis-elements (e.g., B6, DR6, and DR8 and other yet to be determined empirically) present in the long intronic regions of hTERT, but not mTert, pre-mRNA.
Table 1.
mTert and hTERT Genomic Sequence Length
| Intron | Intron Length (bp) | Features of hTERT | |
|---|---|---|---|
| mTert | hTERT | ||
|
| |||
| 2 | 5,477 | 10,688 | Alu, VNTR2.1, 2.2 |
| 3 | 1,072 | 2,090 | Alu |
| 6 | 1,791 | 6,360 | Alu, VNTR6.1, 6.2, DR6 |
| 8 | 1,904 | 2,485 | Alu, DR8 |
| 11 | 1,743 | 3,803 | Alu |
| 12 | 469 | 1,814 | VNTR12 |
| 13 | 1,108 | 3,186 | Alu |
|
| |||
| All Exons | 4,313 | 4,039 | Highly Conserved |
| All Introns | 18,615 | 37,863 | Evolutionarily Divergent |
Abbreviations: DR: Direct Repeat; VNTR: Variable Number Tandem Repeat
Acute Treadmill Exercise Modulates hTERT Expression and Alternative Splicing Patterns
Total hTERT Gene Expression
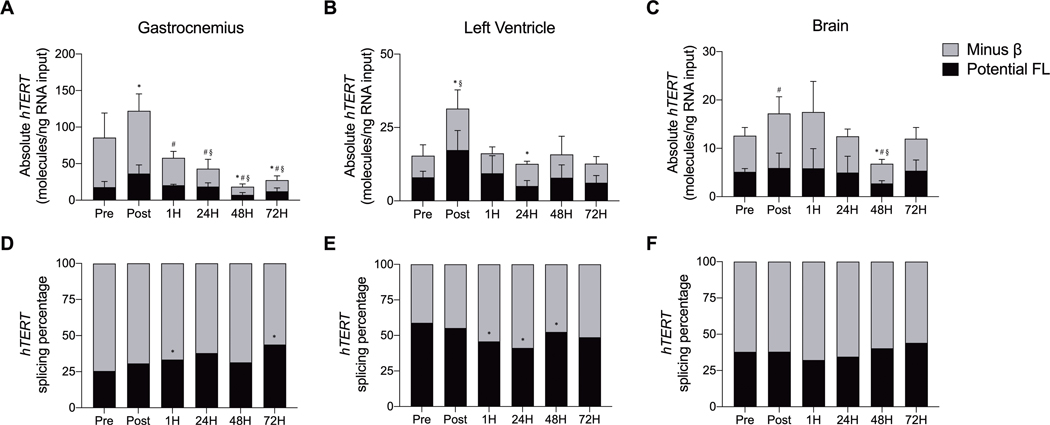
Next, the impact of acute submaximal treadmill running on hTERT gene expression and alternative RNA splicing were examined in the gastrocnemius, left ventricle, and brain (Figures 4A-C; see SDC 3, Supplemental Figure 3, Qualitative mTert gene (exons 5–9) across tissues collected from hTERT-BAC mice prior to, immediately following, and throughout recovery from acute submaximal treadmill exercise). Compared to Pre, total hTERT gene expression (FL hTERT plus the −β ASV) significantly decreased in gastrocnemius at 24, 48, and 72H into recovery from acute treadmill exercise (F [5, 21] = 13.812, p ≤ 0.004). In the left ventricle, total hTERT expression significantly increased at Post before returning to Pre exercise levels throughout recovery (F [5, 22] = 3.662, p = 0.015). Finally, total hTERT expression was significantly decreased in the brain at 48H into recovery (p = 0.007), but an overall response was not observed (F [5, 22] = 2.167, p = 0.095).
FIGURE 4–
Quantitative hTERT gene expression and alternative splicing across tissues collected from hTERT-BAC mice by droplet digital PCR before, immediately after, and throughout recovery from acute submaximal treadmill exercise in the gastrocnemius (panels A and D), left ventricle (panels B and E), and brain (panels C and F). * indicates a significant change in total FL (exon 7–8 including) hTERT transcripts (panels A-C) and FL alternative splicing percentage (panels D-F; relative to Pre; P ≤ 0.05); # indicates a significant change in total −β (7–8 excluding) hTERT transcripts (relative to Pre; P ≤ 0.05); § indicates a significant change in total (FL plus −β) hTERT transcripts (relative to Pre; P ≤ 0.05). Total cDNA input–gastrocnemius and left ventricle: 12 ng; brain: 25 ng. Data are presented as means ± SD. Splicing percentage data are regraphed of the absolute data (A and D panels, B and E panels, C and F panels).
FL hTERT Expression
Acute submaximal treadmill exercise transiently increased FL hTERT at Post in the gastrocnemius and left ventricle before a significant decrease was observed during recovery in the gastrocnemius (48 and 72H; F [5, 22] = 11.035, p ≤ 0.001), left ventricle (24H; F [5, 22] = 4.840, p = 0.004), and brain (48H; F [5, 23] = 0.918, p = 0.487).
Expression of the −β ASV
In the gastrocnemius, acute submaximal treadmill exercise significantly decreased expression of the −β ASV throughout recovery (1, 24, 48, and 72H; F [5, 22] = 12.431, p ≤ 0.001). No statistically significant changes were observed in the left ventricle (F [5, 20] = 2.236, p = 0.091). A significant increase in the −β ASV was observed in the brain at Post followed by a decrease 48H into recovery (F [5, 22] = 3.657, p = 0.015).
Alternative hTERT Splicing Percentages
Acute submaximal treadmill exercise did not alter the overall percentage of hTERT splicing in the gastrocnemius (F [5, 22] = 1.170, p = 0.355), left ventricle (F [5, 24] = 1.176, p = 0.350), or the brain (F [5, 24] = 1.681, p = 0.154; Figures 4D-F; see SDC 3, Supplemental Figure 3, Qualitative mTert gene (exons 5–9) across tissues collected from hTERT-BAC mice prior to, immediately following, and throughout recovery from acute submaximal treadmill exercise). When individual time points were compared to Pre, a pattern emerged suggesting that hTERT alternative splicing patterns were differentially impacted within both contractile tissues. More specifically, a significant increase in the percentage of FL hTERT was observed in the gastrocnemius at 1 and 72H (p ≤ 0.017), whereas a decrease was observed in the left ventricle at 1, 24, and 48H (p ≤ 0.042).
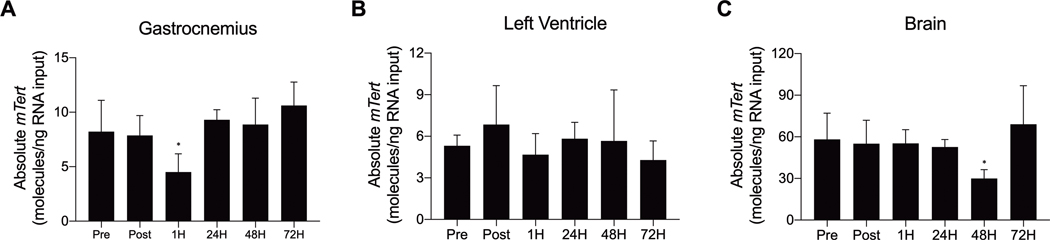
Total mTert Gene Expression
We also explored endogenous mouse Tert (mTert) gene expression in our model. In response to acute submaximal treadmill exercise, mTert mRNA expression significantly decreased in the gastrocnemius (F [5, 24] = 4.964, p = 0.003) and the brain (F [5, 24] = 3.114, p = 0.026) 1 and 48H into recovery, respectively (Figures 5A and C; see SDC 4, Supplemental Figure 4, Qualitative hTERT gene (exons 5–9) across tissues collected from hTERT-BAC mice prior to, immediately following, and throughout recovery from acute submaximal treadmill exercise). No mTert expression changes were observed in the left ventricle (Figures 5B).
FIGURE 5–
Quantitative mTert gene expression across tissues collected from hTERT-BAC mice by droplet digital PCR before, immediately after, and throughout recovery from acute submaximal treadmill exercise. Significant decrease in mTert gene expression were observed in the gastrocnemius and whole brain 1 and 48 h into recovery, respectively (panels A and C). No changes were observed in the left ventricle (panel B). * indicates a significant change (relative to Pre; P ≤ 0.05). Data are presented as means ± SD. Total cDNA input–gastrocnemius and left ventricle: 12 ng; brain: 25 ng.
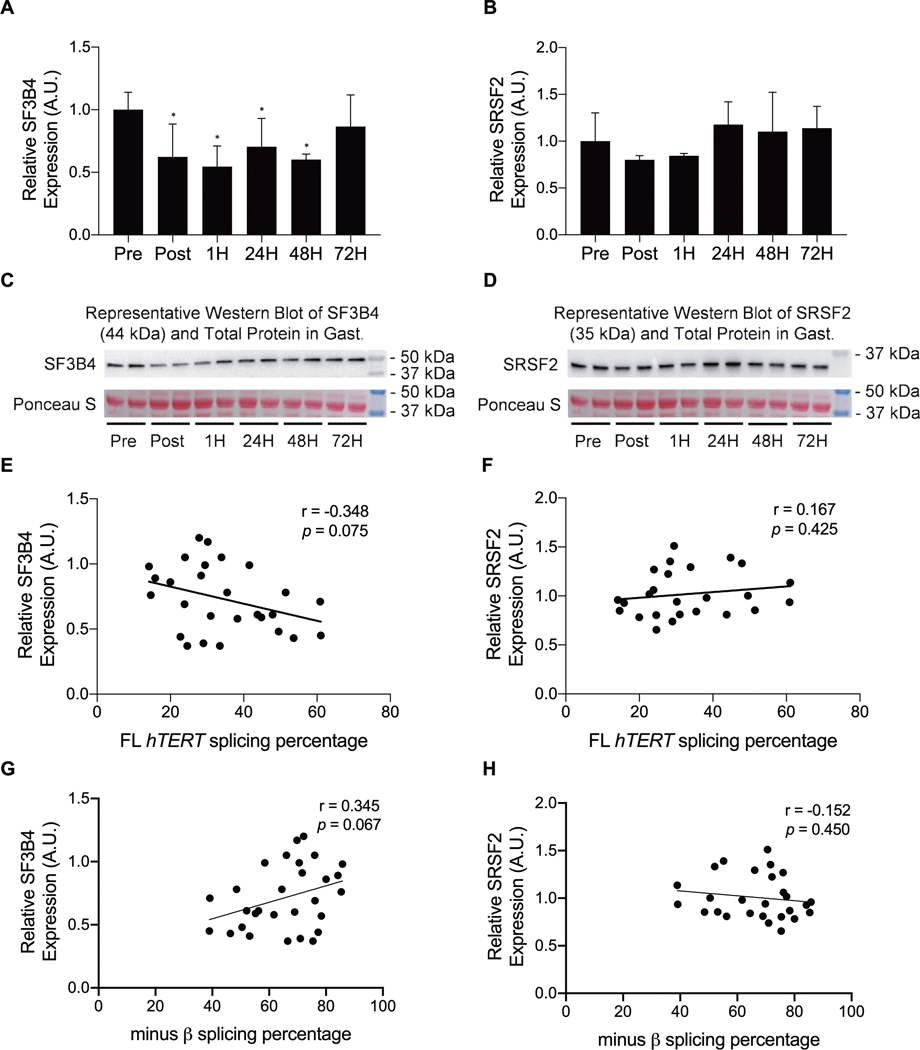
Acute Treadmill Exercise Differentially Regulates the SF3B4 and SRSF2 RNAbps
Finally, potential trans-acting RNAbps involved in the regulation of FL hTERT splicing were examined. In response to acute submaximal treadmill exercise, SF3B4 protein expression decreased immediately post and throughout recovery in the gastrocnemius (1, 24, and 48H; F [5, 28] = 2.814, p = 0.040; Figures 6A and C), whereas no significant changes in SRSF2 protein expression were observed (F [5, 26] = 2.567, p = 0.058; Figures 6B and D). The relationships of hTERT splicing isoform percentages with the SF3B4 and SRSF2 protein expression were also examined in response to acute submaximal treadmill running. FL hTERT and −β splicing percentages in the gastrocnemius tended to be associated with SF3B4 (Pearson’s correlation r = −0.348, p = 0.075; r = 0.345, p = 0.067, respectively; Figures 6E and G). No associations were observed between FL hTERT and −β splicing percentage and SRSF2 protein expression in the gastrocnemius (Pearson’s correlation r = 0.167, p = 0.425; r = −0.152, p = 0.450, respectively; Figures 6F and H).
Figure 6.
SF3B4 and SRSF2 protein expression in the gastrocnemius in response to acute submaximal treadmill exercise. SF3B4 protein expression significantly decreased immediately following, and throughout recovery (panels A and C), whereas SRSF2 protein expression tended to decrease (panels B and D). In addition, although SF3B4 tended toward a negative association with FL hTERT minus β splicing percentage in response to acute submaximal treadmill exercise (panels E) and G, no associations were observed between SRSF2 protein expression and hTERT splicing (panels F and H). * indicates a significant change (relative to Pre; p ≤ 0.05). Data are presented as means ± S.D.
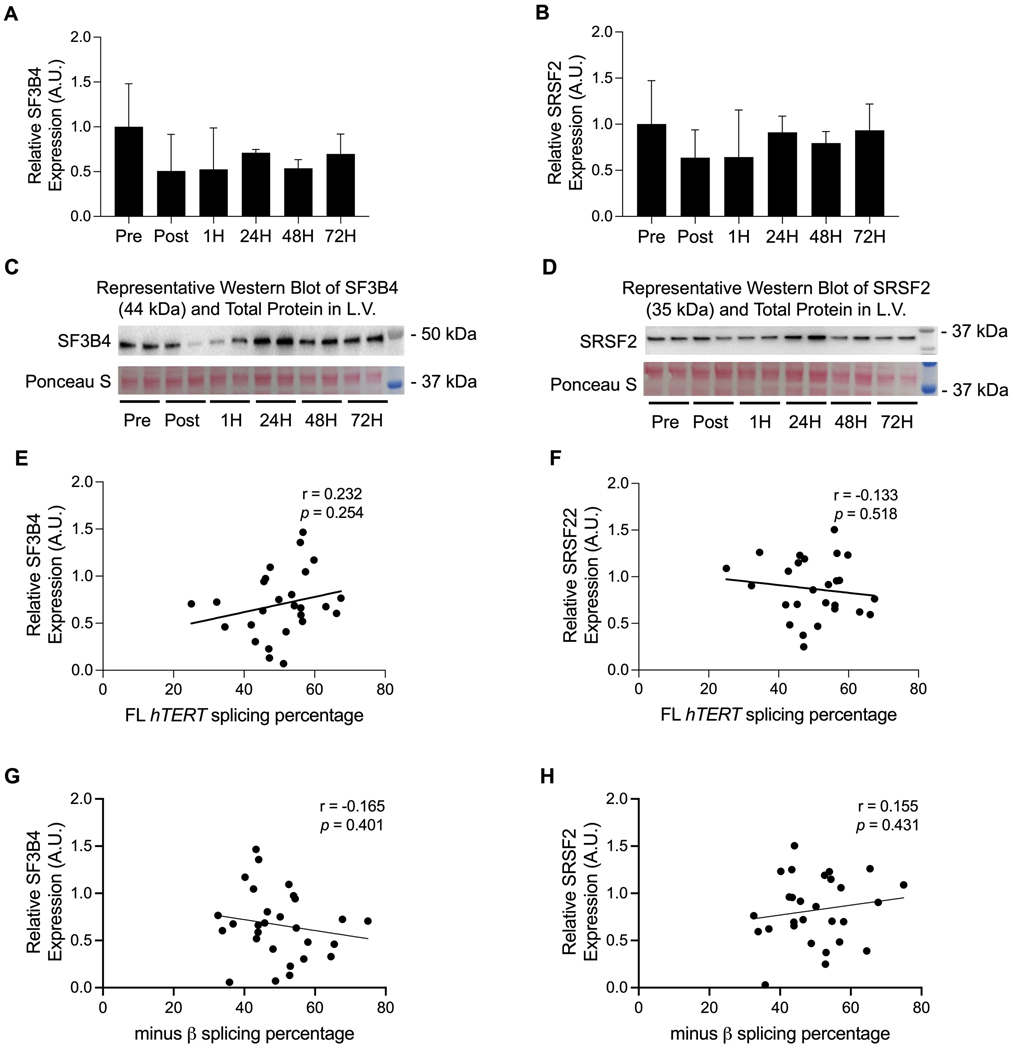
Finally, overall SF3B4 and SRSF2 protein expression in the left ventricle remained unchanged in response to acute submaximal treadmill exercise (F [5, 27] = 1.420, p = 0.256; F [5, 28] = 0.982, p = 0.450; Figures 7A-D). Likewise, no relationships were observed between FL hTERT or −β splicing percentage and SF3B4 (Pearson’s correlation r = 0.232, p = 0.254; r = −0.165, p = 0.401, respectively; Figures 7E and G) or SRSF2 protein expression in the left ventricle (Pearson’s correlation r = −0.133, p = 0.518; r = 0.155, p = 0.431, respectively; Figures 7F and H). However, the same general pattern between skeletal muscle and cardiac tissues was observed. Of note, no associations among total, FL, or −β absolute transcripts with the SF3B4 and SRSF2 RNAbps were observed in the gastrocnemius and left ventricle at baseline or in response to acute treadmill running.
FIGURE 7–
SF3B4 and SRSF2 protein expression in the left ventricle did not significantly change in response to acute submaximal treadmill exercise (A-D). Likewise, no associations were observed among SF3B4 and SRSF2 protein expression with FL hTERT and −β splicing percentage in response to acute submaximal treadmill exercise (E-H). Data are presented as means ± SD.
DISCUSSION
This investigation utilized the hTERT-BAC mouse model to recapitulate human hTERT mRNA expression and splicing patterns across various tissues at rest. The hTERT-BAC mouse model also allowed for the first documentation of the timecourse for hTERT mRNA expression and alternative splicing following acute submaximal treadmill running exercise in skeletal muscle, heart, and brain. Significant increases in hTERT gene expression immediately following exercise followed by splicing isoform ratio changes during recovery from exercise were observed in skeletal muscle and heart (Figure 5). In the brain, only hTERT gene expression changes were observed 48 hrs into recovery (Figure 5). Collectively, endurance exercise increases hTERT gene expression and FL hTERT splicing in contractile tissues following exercise, indicating that both transcriptional and post-transcriptional mechanisms are important in the adaptation of muscle tissues to endurance type exercise.
Typically, hTERT expression is difficult to detect in human tissues due to its low abundance, which has contributed to the assumption that hTERT is transcriptionally silenced (18). However, recent evidence indicates that this may not be the case and that certain physiological stressors may result in transient increases in hTERT expression (22–24). Lacking in the literature is an in-depth characterization of the tissue-specific response and regulation of the hTERT gene to a potent and healthy physiological stressor, such as an acute bout of endurance exercise. While ideally this study would be done in human volunteers, obtaining repeat biopsies from skeletal muscle, cardiac tissue, and the brain is not feasible. Thus, we utilized a transgenic mouse model containing the complete hTERT gene in the form of an integrated bacterial artificial chromosome to complete the present study. The present results most closely mimic the response of an untrained, young-adult human population to a similar acute bout of endurance exercise. Extrapolating our findings to humans, the present results indicate that acute endurance exercise rapidly initiates a signaling cascade leading to the activation of transcription factors and RNAbps that elicit transient changes to hTERT. Importantly, each of the contractile tissue may have undergone a slightly different regulatory response to match the specificity of each tissue.
In skeletal muscle, the acute response of hTERT gene expression or splicing to exercise in untrained humans is unknown. Our results lend to the hypothesis that the increased expression of hTERT mRNA observed immediately post exercise (relative to Pre; Figure 5) is likely due to gene transcription factors activated by the acute stress response to endurance exercise and closely related to hTERT transactivation. The biological relevance of the early increase in hTERT gene expression could be a stress response of the active muscles (i.e., gastrocnemius) to form TERT protein necessary to protect the genome from genotoxic stresses (i.e., increase oxidative stress during higher intensity unaccustomed exercise causing DNA damage) during recovery from exercise (40). Alternatively, this early increase hTERT expression could be to form mitochondrial protective FL hTERT during exercise, which has been observed to associate with the respiratory complex in mitochondrial and reduce the production of reactive oxygen species (41). During recovery from exercise, we observed significant shift towards the FL hTERT splicing isoform (Figure 5), which could serve to tightly regulate the amount of FL hTERT to maintain telomeres in activated satellite cells or other proliferating cell types that are aiding in muscle repair and regeneration following exercise (42). Although the cell source of altered hTERT gene expression within skeletal muscle is unknown, the ability to measure hTERT in a more concentrated sample of skeletal muscle containing all hTERT-expressing cell populations (e.g., immune and stem cells) is likely driving the observed changes within the gastrocnemius of the hTERT-BAC mouse. In contrast to our results, a previous study did not observe hTERT expression changes in highly trained endurance athletes following several marathon races completed in succession (43). Importantly, Laye et al. (43) performed explant primary cultures of proliferating myoblasts prior to measuring hTERT gene expression, and it is possible that the duration of time and the treatment of the cells/tissue between the cessation of exercise and subsequent hTERT measurements were sufficiently long to result in loss of the immediate increase of hTERT caused by the exercise stimulus. Thus, both training status and timing of downstream measures may play important roles in being able to observe changes in hTERT in skeletal muscle tissues.
Similar to skeletal muscle, the increase in total hTERT gene expression observed within the heart immediately post exercise could be a response to form TERT proteins during recovery from exercise to promote genome stability and reduced mitochondrial stress (44, 45). In contrast to skeletal muscle, −β ASV isoform expression increased in the heart during recovery from acute exercise (Figure 5E). The function of the −β ASV is currently debated. For example, the exclusion of exons 7 and 8 and inclusion of a premature termination codon within exon 10 could cause degradation of the hTERT transcript by NMD pathways and prevent telomerase enzyme activity (8). To test if the −β variant was translated into a protein in cells, several groups have performed ribosome profiling and have detected the −β ASV to be associated with polysome fractions in greater abundance compared to monosome fractions. These findings suggest that endogenous −β hTERT is translated into protein beyond the pioneering round of translation necessary for NMD (20, 46). Additionally, Listerman et al. (20) overexpressed the −β ASV and documented that the −β isoform is associated with the RNA template of telomerase (hTERC) and reduced telomerase activity, thereby acting as a dominant-negative telomerase. In two publications, the −β ASV was expressed in cells during treatment with genotoxic drugs, and it was observed that −β protected the cells from apoptosis even in the absence of canonical telomerase activity (20, 47). These findings indicate that some of the −β ASVs are escaping degradation and being translated into a truncated and functional protein in human cancer and stem cells to confer protection from genotoxic insults and promote cellular survival (20, 46, 47). As such, the tissue-specific regulation of hTERT alternative splicing might serve to maintain tissue and cellular homeostasis of cardiac muscle in response to various levels of physiological stress independent of telomerase enzyme activity and telomere length maintenance. Therefore, hTERT-BAC mouse model utilization might provide novel insights into hTERT alternative splicing regulation as a mechanism for telomere-dependent and independent functions of telomerase at rest and in response to acute endurance exercise.
The exact cell type responsible for the observed hTERT mRNA expression changes in the heart and brain following exercise remains unknown and is thus far unmeasurable in human populations. Single cell RNA sequencing from The Human Protein Atlas suggests that hTERT is expressed at low concentrations across cardiomyocytes, endothelial cells, fibroblasts, and immune cells within the heart and within the basal ganglia, midbrain, and cerebral cortex regions of the brain (34), and it is likely that all cell types are contributing to changes in hTERT mRNA expression within the present study. Interestingly, hTERT/telomerase within the brain appears to be regulated differently compared to other human tissues and the precise mechanisms have yet to be determined during development and throughout the aging process (29). As such, the mechanisms responsible for reduced hTERT gene expression observed 48 hours into recovery remain unknown. Given the challenges of investigating tissue-specific hTERT regulation in humans, future investigations should take careful consideration to isolate specific cell-types and organ regions within hTERT-BAC mice to further identify the specific source of hTERT mRNA expression and its change in response to physiological stress. Such findings are necessary to better understand the role of hTERT as a potential mechanism involved in the regulation of tissue-specific adaptations to endurance exercise in human populations.
The present study is also the first to demonstrate that hTERT alternative splicing patterns are tissue-specific in hTERT-BAC mice at rest and differentially regulated in contractile tissues following acute submaximal treadmill running. Alternative RNA splicing is an evolutionarily conserved mechanism that promotes proteome and organismal diversity in a manner dependent upon larger intron lengths (39, 48). Our laboratory has recently confirmed by long-read RNA-sequencing that FL hTERT containing all 16 exons accounts for only 40% of all transcripts in human cervical cancer (HeLa) cells (25). These results support data from The Cancer Genome Atlas splice variant database that hTERT is alternatively spliced most frequently to other, non-catalytic isoforms across 31 cancers (21), and suggest that alternative splicing of hTERT is a critical regulatory mechanism of telomerase in aging and cancer cells.
hTERT pre-mRNA contains multiple cis-elements that are evolutionarily conserved in humans and old-world primates, but are not present in rodents or other mammals (Figure 1 and Table 1; 26, 27). In contrast, the mTert gene contains shorter introns that lack the DR6, B6, and DR8 cis-elements (Figure 2; 26), and is therefore primarily expressed as the FL transcript (≥ 85%, and data presented here in SDC 2, Supplemental Figure 2; 25). Alternative splicing is also regulated by trans-acting RNAbps that bind to the intronic cis-elements, and together, comprise the “splicing code” responsible for the construction of the final mRNA transcript. Although the specific RNAbps that manipulate hTERT splicing choice remain yet to be fully identified in specific cell and tissue types and under various physiological and pathological states, the human steady-state alternative splicing patterns expressed within the hTERT-BAC mouse model supports one of two hypotheses. Firstly, the development of longer intron lengths containing specialized cis-elements within hTERT pre-mRNA are likely vital for controlling hTERT splicing choice, fine-tuning telomerase enzyme activity, and regulating telomere length-dependent replicative capacity of cells across the lifespan. Secondly, intronic cis-elements, including B6, DR6 and DR8 that are conserved among old-world primates, but not in mice (26, 27), are nonetheless recognized by the conserved trans-factors in mice and result in the human TERT alternative splicing pattern. To identify trans-acting RNAbps that regulate FL hTERT splicing, protein expression of the SF3B4 and SRSF2 splicing factors was measured prior to and following acute submaximal treadmill exercise. However, the lack of SF3B4 or SRSF2 association with changes in FL and −β in the gastrocnemius or left ventricle observed in the present study suggests that additional research is necessary to identify potential RNAbps that are transiently and intermittently regulated by endurance exercise to regulate hTERT alternative splicing.
It is important to consider that the rate of telomere shortening, not absolute telomere length per se, is considered a more important factor related to the risk of cellular senescence during the aging process (1). Cells maintain robust mechanisms which protect telomeres from stress and/or replication induced shortening, such as chromatin configuration and the shelterin protein complex (49). As such, active telomerase might not always be critically necessary to maintain telomeres during or in response to external stimuli. The differential regulation of alternative splicing within the gastrocnemius and left ventricle, despite similar response patterns in hTERT mRNA expression, suggest a difference in the mechanisms that tightly regulate hTERT isoform selection. For example, while transcriptional regulation of hTERT might serve as an initial response to physiological stimuli, the pre-mRNA splicing machinery might serve as a secondary system to fine tune the abundance of catalytically active FL hTERT to prevent excess telomerase enzyme activity and aberrant telomere elongation within skeletal and/or cardiac muscle. The elucidation of the hTERT “splicing code” could help to identify novel molecular targets useful in both aging and cancer associated research. For example, the targeting the interactions of specific RNAbps with intronic cis-elements of hTERT provides therapeutic potential to promote (or repress) FL hTERT splicing, telomerase enzyme activity, and alter telomere length as a means to control cellular proliferation. These types of therapies would be useful in regenerative medicine approaches (promoting FL hTERT) and anti-cancer therapeutics (repressing FL hTERT).
In conclusion, this study provides necessary insight into the intronic cis-elements that, together with trans-acting RNAbps, regulate hTERT splicing choice in cell and tissue types which are difficult obtain from human populations. These data also highlight the importance for utilizing human gene BAC transgenic animals when studying alternative splicing for genes that have human specific (not conserved in mice) alternative splicing patterns. Our study is not without limitations, such as the small sample size and lack of robust comparison among male and female mice. Another limitation to this study is that hTERT and mouse telomerase RNA are enzymatically incompatible, not able to generate active telomerase. Thus, we are unable to precisely determine the contribution of the altered hTERT transgene expression on telomerase enzyme activity and telomere length maintenance within the hTERT-BAC mouse model due to species specific differences in the RNA component of telomerase (50). The mice used in this study are therefore unlikely to have hTERT-derived “extra” telomerase activity, which may cause unexpected cellular outcomes. Nonetheless, results from the present investigation provide a promising method to understand hTERT expression and alternative splicing regulation as a fundamental mechanism linking telomerase enzyme activity and telomere length maintenance in response to various physiologically relevant states. Future studies utilizing the hTERT-BAC model are needed to determine the tissue specific RNAbps that are impacted by exercise and result in the alternative splicing pattern following exercise. Such investigations are of particular interest to understanding the capacity of hTERT alternative splicing to regulate the cellular aging process in response acute and chronic endurance exercise across all stages of the lifespan.
Supplementary Material
SDC 1: Supplemental 1.docx - hTERT-BAC mouse genotyping.
SDC 3: Supplemental 3.docx - Qualitative mTert gene (exons 5–9) across tissues collected from hTERT -BAC mice prior to, immediately following, and throughout recovery from acute submaximal treadmill exercise.
SDC 2: Supplemental 2_revised.docx - Qualitative alternative splicing expression of the hTERT gene across tissues collected from hTERT -BAC mice.
SDC 4: Supplemental 4_revised.docx -Qualitative hTERT gene (exons 5–9) across tissues collected from hTERT -BAC mice prior to, immediately following, and throughout recovery from acute submaximal treadmill exercise.
Acknowledgments
This research was supported by the National Institutes of Health and National Cancer Institute under award number 5R00CA197672-04, along with the University of Michigan’s School of Kinesiology Marie Hartwig Research Fund. This research utilized the HeLa cell line. Henrietta Lacks, and the HeLa cell line that was established from her tumor cells without her knowledge or consent in 1951, have made significant contributions to scientific progress and advances in human health. We are grateful to Henrietta Lacks, now deceased, and to her surviving family members for their contributions to biomedical research.
Funding Source:
This research was supported by the National Institutes of Health and National Cancer Institute under award number 5R00CA197672-04, along with the University of Michigan’s School of Kinesiology Marie Hartwig Research Fund.
Footnotes
Conflict of Interest
The authors have no commercial or financial conflicts of interest to declare. The results are presented clearly, honestly, and without fabrication, falsification, or inappropriate data manipulation. The results of the present study do not constitute endorsement by the American College of Sports Medicine.
REFERENCES
- 1.Rebelo-Marques A, De Sousa Lages A, Andrade R, Ribeiro CF, Mota-Pinto A, Carrilho F, Espregueira-Mendes J. Aging Hallmarks: The Benefits of Physical Exercise. Front Endocrinol (Lausanne). 2018;9:258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moyzis RK, Buckingham JM, Cram LS, Dani M, Deaven LL, Jones MD, Meyne J, Ratliff RL, Wu JR. A highly conserved repetitive DNA sequence, (TTAGGG)n, present at the telomeres of human chromosomes. Proc Natl Acad Sci U S A. 1988;85(18):6622–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harley CB, Futcher AB, Greider CW. Telomeres shorten during ageing of human fibroblasts. Nature. 1990;345(6274):458–60. [DOI] [PubMed] [Google Scholar]
- 4.Levy MZ, Allsopp RC, Futcher AB, Greider CW, Harley CB. Telomere end-replication problem and cell aging. J Mol Biol. 1992;225(4):951–60. [DOI] [PubMed] [Google Scholar]
- 5.Lormand JD, Buncher N, Murphy CT, Kaur P, Lee MY, Burgers P, Wang H, Kunkel TA, Opresko PL. DNA polymerase δ stalls on telomeric lagging strand templates independently from G-quadruplex formation. Nucleic Acids Res. 2013;41(22):10323–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Greider CW, Blackburn EH. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell. 1985;43(2 Pt 1):405–13. [DOI] [PubMed] [Google Scholar]
- 7.Morin GB. The human telomere terminal transferase enzyme is a ribonucleoprotein that synthesizes TTAGGG repeats. Cell. 1989;59(3):521–9. [DOI] [PubMed] [Google Scholar]
- 8.Kilian A, Bowtell DD, Abud HE, Hime GR, Venter DJ, Keese PK, Duncan EL, Reddel RR, Jefferson RA. Isolation of a candidate human telomerase catalytic subunit gene, which reveals complex splicing patterns in different cell types. Hum Mol Genet. 1997;6(12):2011–9. [DOI] [PubMed] [Google Scholar]
- 9.Hrdlicková R, Nehyba J, Bose HR Jr. Alternatively spliced telomerase reverse transcriptase variants lacking telomerase activity stimulate cell proliferation. Mol Cell Biol. 2012;32(21):4283–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hayflick L, Moorhead PS. The serial cultivation of human diploid cell strains. Exp Cell Res. 1961;25:585–621. [DOI] [PubMed] [Google Scholar]
- 11.Zou Y, Sfeir A, Gryaznov SM, Shay JW, Wright WE. Does a sentinel or a subset of short telomeres determine replicative senescence? Mol Biol Cell. 2004;15(8):3709–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Childs BG, Durik M, Baker DJ, van Deursen JM. Cellular senescence in aging and age-related disease: from mechanisms to therapy. Nat Med. 2015;21(12):1424–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ludlow AT, Zimmerman JB, Witkowski S, Hearn JW, Hatfield BD, Roth SM. Relationship between physical activity level, telomere length, and telomerase activity. Med Sci Sports Exerc. 2008;40(10):1764–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ludlow AT, Witkowski S, Marshall MR, Wang J, Lima LC, Guth LM, Spangenburg EE, Roth SM. Chronic exercise modifies age-related telomere dynamics in a tissue-specific fashion. J Gerontol A Biol Sci Med Sci. 2012a;67(9):911–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Puterman E, Weiss J, Lin J, Schilf S, Slusher AL, Johansen KL, Epel ES. Aerobic exercise lengthens telomeres and reduces stress in family caregivers: A randomized controlled trial - Curt Richter Award Paper 2018. Psychoneuroendocrinology. 2018;98:245–252. [DOI] [PubMed] [Google Scholar]
- 16.Yi X, Shay JW, Wright WE. Quantitation of telomerase components and hTERT mRNA splicing patterns in immortal human cells. Nucleic Acids Res. 2001;29(23):4818–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Avilion AA, Piatyszek MA, Gupta J, Shay JW, Bacchetti S, Greider CW. Human telomerase RNA and telomerase activity in immortal cell lines and tumor tissues. Cancer Res. 1996;56(3):645–650. [PubMed] [Google Scholar]
- 18.Ulaner GA, Hu JF, Vu TH, Giudice LC, Hoffman AR. Telomerase activity in human development is regulated by human telomerase reverse transcriptase (hTERT) transcription and by alternate splicing of hTERT transcripts. Cancer Res. 1998;58(18):4168–72. [PubMed] [Google Scholar]
- 19.Ulaner GA, Hu JF, Vu TH, Giudice LC, Hoffman AR. Tissue-specific alternate splicing of human telomerase reverse transcriptase (hTERT) influences telomere lengths during human development. Int J Cancer. 2001;91(5):644–9. [PubMed] [Google Scholar]
- 20.Listerman I, Sun J, Gazzaniga FS, Lukas JL, Blackburn EH. The major reverse transcriptase-incompetent splice variant of the human telomerase protein inhibits telomerase activity but protects from apoptosis. Cancer Res. 2013;73(9):2817–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Slusher AL, Kim JJ, Ludlow AT. The Role of Alternative RNA Splicing in the Regulation of hTERT, Telomerase, and Telomeres: Implications for Cancer Therapeutics. Cancers (Basel). 2020;12(6):1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chilton WL, Marques FZ, West J, Kannourakis G, Berzins SP, O’Brien BJ, Charchar FJ. Acute exercise leads to regulation of telomere-associated genes and microRNA expression in immune cells. PLoS One. 2014;9(4):e92088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Denham J, O’Brien BJ, Prestes PR, Brown NJ, Charchar FJ. Increased expression of telomere-regulating genes in endurance athletes with long leukocyte telomeres. J Appl Physiol (1985). 2016;120(2):148–58. [DOI] [PubMed] [Google Scholar]
- 24.Werner CM, Hecksteden A, Morsch A, Zundler J, Wegmann M, Kratzsch J, Thiery J, Hohl M, Bittenbring JT, Neumann F, Böhm M, Meyer T, Laufs U. Differential effects of endurance, interval, and resistance training on telomerase activity and telomere length in a randomized, controlled study. Eur Heart J. 2019;40(1):34–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sayed ME, Yuan L, Robin JD, Tedone E, Batten K, Dahlson N, Wright WE, Shay JW, Ludlow AT. NOVA1 directs PTBP1 to hTERT pre-mRNA and promotes telomerase activity in cancer cells. Oncogene. 2019;38(16):2937–2952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wong MS, Chen L, Foster C, Kainthla R, Shay JW, Wright WE. Regulation of telomerase alternative splicing: a target for chemotherapy. Cell Rep. 2013;3(4):1028–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wong MS, Shay JW, Wright WE. Regulation of human telomerase splicing by RNA:RNA pairing. Nat Commun. 2014;5:3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ludlow AT, Wong MS, Robin JD, Batten K, Yuan L, Lai TP, Dahlson N, Zhang L, Mender I, Tedone E, Sayed ME, Wright WE, Shay JW. NOVA1 regulates hTERT splicing and cell growth in non-small cell lung cancer. Nat Commun. 2018;9(1):3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Horikawa I, Chiang YJ, Patterson T, Feigenbaum L, Leem SH, Michishita E, Larionov V, Hodes RJ, Barrett JC. Differential cis-regulation of human versus mouse TERT gene expression in vivo: identification of a human-specific repressive element. Proc Natl Acad Sci U S A. 2005;102(51):18437–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gerstberger S, Hafner M, Ascano M, Tuschl T. Evolutionary conservation and expression of human RNA-binding proteins and their role in human genetic disease. Adv Exp Med Biol. 2014a;825:1–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang F, Cheng Y, Zhang C, Chang G, Geng X. A novel antisense oligonucleotide anchored on the intronic splicing enhancer of hTERT pre-mRNA inhibits telomerase activity and induces apoptosis in glioma cells. J Neurooncol. 2019;143(1):57–68. [DOI] [PubMed] [Google Scholar]
- 32.Ludlow AT, Lima LC, Wang J, Hanson ED, Guth LM, Spangenburg EE, Roth SM. Exercise alters mRNA expression of telomere-repeat binding factor 1 in skeletal muscle via p38 MAPK. J Appl Physiol (1985). 2012b;113(11):1737–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ludlow AT, Gratidão L, Ludlow LW, Spangenburg EE, Roth SM. Acute exercise activates p38 MAPK and increases the expression of telomere-protective genes in cardiac muscle. Exp Physiol. 2017;102(4):397–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Uhlén M, Fagerberg L, Hallström BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson Å, Kampf C, Sjöstedt E, Asplund A, Olsson I, Edlund K, Lundberg E, Navani S, Szigyarto CA, Odeberg J, Djureinovic D, Takanen JO, Hober S, Alm T, Edqvist PH, Berling H, Tegel H, Mulder J, Rockberg J, Nilsson P, Schwenk JM, Hamsten M, von Feilitzen K, Forsberg M, Persson L, Johansson F, Zwahlen M, von Heijne G, Nielsen J, Pontén F. Proteomics. Tissue-based map of the human proteome. Science. 2015;347(6220):1260419. [DOI] [PubMed] [Google Scholar]
- 35.Golas MM, Sander B, Will CL, Lührmann R, Stark H. Molecular architecture of the multiprotein splicing factor SF3b. Science. 2003;300(5621):980–4. [DOI] [PubMed] [Google Scholar]
- 36.Tanaka Y, Ohta A, Terashima K, Sakamoto H. Polycistronic expression and RNA-binding specificity of the C. elegans homologue of the spliceosome-associated protein SAP49. J Biochem. 1997;121(4):739–45. [DOI] [PubMed] [Google Scholar]
- 37.Liu HX, Chew SL, Cartegni L, Zhang MQ, Krainer AR. Exonic splicing enhancer motif recognized by human SC35 under splicing conditions. Mol Cell Biol. 2000;20(3):1063–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang Y, Liu J, Huang BO, Xu YM, Li J, Huang LF, Lin J, Zhang J, Min QH, Yang WM, Wang XZ. Mechanism of alternative splicing and its regulation. Biomed Rep. 2015;3(2):152–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roy M, Kim N, Xing Y, Lee C. The effect of intron length on exon creation ratios during the evolution of mammalian genomes. RNA. 2008;14(11):2261–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rosen J, Jakobs P, Ale-Agha N, Altschmied J, Haendeler J. Non-canonical functions of Telomerase Reverse Transcriptase - Impact on redox homeostasis. Redox Biol. 2020;34:101543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ahmed S, Passos JF, Birket MJ, Beckmann T, Brings S, Peters H, Birch-Machin MA, von Zglinicki T, Saretzki G. Telomerase does not counteract telomere shortening but protects mitochondrial function under oxidative stress. J Cell Sci. 2008;121(Pt 7):1046–53. [DOI] [PubMed] [Google Scholar]
- 42.Ramunas J, Yakubov E, Brady JJ, Corbel SY, Holbrook C, Brandt M, Stein J, Santiago JG, Cooke JP, Blau HM. Transient delivery of modified mRNA encoding TERT rapidly extends telomeres in human cells. FASEB J. 2015;29(5):1930–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Laye M, Solomon TJP, Karstoft K, Pedersen KK, Nielsen SD, Pedersen BK. Increased shelterin mRNA expression in peripheral blood mononuclear cells and skeletal muscle following an ultra-long-distance running event. J Appl Physiol (1985). 2021;112(5):773–81. [DOI] [PubMed] [Google Scholar]
- 44.Ait-Aissa K, Heisner JS, Norwood Toro LE, Bruemmer D, Doyon G, Harmann L, Geurts A, Camara AKS, Beyer AM. Telomerase Deficiency Predisposes to Heart Failure and Ischemia-Reperfusion Injury. Front Cardiovasc Med. 2019;6:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Haendeler J, Dröse S, Büchner N, Jakob S, Altschmied J, Goy C, Spyridopoulos I, Zeiher AM, Brandt U, Dimmeler S. Mitochondrial telomerase reverse transcriptase binds to and protects mitochondrial DNA and function from damage. Arterioscler Thromb Vasc Biol. 2009;29(6):929–35. [DOI] [PubMed] [Google Scholar]
- 46.Radan L, Hughes CS, Teichroeb JH, Vieira Zamora FM, Jewer M, Postovit LM, Betts DH. Microenvironmental regulation of telomerase isoforms in human embryonic stem cells. Stem Cells Dev. 2014;23(17):2046–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fleisig HB, Hukezalie KR, Thompson CA, Au-Yeung TT, Ludlow AT, Zhao CR, Wong JM. Telomerase reverse transcriptase expression protects transformed human cells against DNA-damaging agents, and increases tolerance to chromosomal instability. Oncogene. 2016;35(2):218–27. [DOI] [PubMed] [Google Scholar]
- 48.Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet. 2008;40(12):1413–5. [DOI] [PubMed] [Google Scholar]
- 49.Kim W, Ludlow AT, Min J, Robin JD, Stadler G, Mender I, Lai TP, Zhang N, Wright WE, Shay JW. Regulation of the Human Telomerase Gene TERT by Telomere Position Effect-Over Long Distances (TPE-OLD): Implications for Aging and Cancer. PLoS Biol. 2016;14(12):e2000016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen JL, Greider CW. Determinants in mammalian telomerase RNA that mediate enzyme processivity and cross-species incompatibility. EMBO J. 2003;22(2):304–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
SDC 1: Supplemental 1.docx - hTERT-BAC mouse genotyping.
SDC 3: Supplemental 3.docx - Qualitative mTert gene (exons 5–9) across tissues collected from hTERT -BAC mice prior to, immediately following, and throughout recovery from acute submaximal treadmill exercise.
SDC 2: Supplemental 2_revised.docx - Qualitative alternative splicing expression of the hTERT gene across tissues collected from hTERT -BAC mice.
SDC 4: Supplemental 4_revised.docx -Qualitative hTERT gene (exons 5–9) across tissues collected from hTERT -BAC mice prior to, immediately following, and throughout recovery from acute submaximal treadmill exercise.