Abstract
Medulloblastoma has been categorized into four subgroups based on genetic, epigenetic, and transcriptional profiling. Radiation is used for treating medulloblastoma regardless of the subgroup. A better understanding of the molecular pathways determining radiotherapy response could help improve medulloblastoma treatment. Here, we investigated the role of the EZH2-dependent histone H3K27 trimethylation in radiotherapy response in medulloblastoma. The tumors in 47.2% of group 3 and 4 medulloblastoma patients displayed H3K27me3 deficiency. Loss of H3K27me3 was associated with a radioresistant phenotype, high relapse rates, and poor overall survival. In H3K27me3-deficient medulloblastoma cells, an epigenetic switch from H3K27me3 to H3K27ac occurred at specific genomic loci, altering the transcriptional profile. The resulting upregulation of EPHA2 stimulated excessive activation of the pro-survival AKT signaling pathway, leading to radiotherapy resistance. BET inhibition overcame radiation resistance in H3K27me3-deficient medulloblastoma cells by suppressing H3K27ac levels, blunting EPHA2 overexpression, and mitigating excessive AKT signaling. Additionally, BET inhibition sensitized medulloblastoma cells to radiation by enhancing the apoptotic response through suppression of Bcl-xL and upregulation of Bim. This work demonstrates a novel mechanism of radiation resistance in medulloblastoma and identifies an epigenetic marker predictive of radiotherapy response. Based on these findings, we propose an epigenetically guided treatment approach targeting radiotherapy resistance in medulloblastoma patients.
Keywords: Medulloblastoma, radiation therapy, epigenetics, DNA damage response, BET inhibitors
Introduction
Central nervous system tumors are the leading cause of cancer-related mortality in children. Medulloblastoma is the most common malignant brain tumor of the childhood (1). Medulloblastoma has been categorized into 4 molecular subgroups based on the patterns of genetic mutations as well as epigenetic and transcriptional profiles: WNT, SHH (sonic hedgehog), group 3 and group 4 (2,3). WNT medulloblastomas have the best prognosis with a 95% cure rate (4,5), whereas have an intermediate prognosis, with a 5-year survival rates of 70% (5). The non-WNT/SHH medulloblastomas consist of group 4 tumors that have an intermediate prognosis similar to the SHH subgroup (6,7) and group 3 tumors that have the worst outcomes of all four subgroups, with a 5 year survival rates of less than 50% (5,7). The poor prognosis of the latter subgroup is attributed to tumor resistance to radiation and chemotherapy resulting in high relapse rates (5,8).
The current treatment paradigm for medulloblastoma includes surgery, chemotherapy and radiotherapy regardless of the subgroup (9). Radiation is given to the entire craniospinal axis due to a high risk of the leptomeningeal spread of tumor cells to additional sites in the brain and spine (9) followed by a radiation boost to the tumor resection cavity (9,10). Clinical outcomes strongly depend on the efficacy of radiation treatment since recurrences are hardly salvageable and almost always lethal (8,11). Since relapses are attributed to tumor cells surviving radiotherapy (11,12) targeting radiation resistance in medulloblastoma would limit the risk of recurrence and improve clinical outcomes of this highly malignant disease. Thus, there is an unmet need to understand the mechanisms of radiation resistance driving treatment failure and tumor recurrence in medulloblastoma.
Radiation kills tumor cells by inducing DNA double-strand breaks (DSBs). Therefore, cellular DNA damage response (DDR) pathways are a crucial determinant of tumor response to radiation (13,14). The fate of a tumor cell following radiation treatment depends on signaling pathways regulating cell survival, DNA repair and cell death (14–16). Thus, an up-regulation of pro-survival signaling or DNA repair pathways or a blockade of programmed cell death can contribute to radiation resistance (14–16). Epigenetic alteration of chromatin by post-translational modification of histone proteins plays an important role in DDR (13,14). The histone methyl-transferase EZH2 (Enhancer Of Zeste 2 Polycomb Repressive Complex 2 Subunit) that is responsible for H3K27 tri-methylation (H3K27me3) was shown to promote both, cell survival (17) and tumor cell kill (18) following genotoxic treatment suggesting a tumor-specific role in DDR. Importantly, a strong variation of H3K27me3 levels was found within the group 3 and 4 medulloblastoma (19,20) suggesting that the H3K27me3 status may be an important determinant of radiation response in these tumors. However, the exact role of EZH2 and its product H3K27me3 in radiotherapy response in medulloblastoma has remained unknown.
Experimental models reflecting the genetic profile and clinical behavior of human tumors are critical to studying radiotherapy response in medulloblastoma. An intricate model of group 3 medulloblastoma has been developed by overexpressing MYC in murine CDKN2C−/−, p53−/− cerebellar granule neuronal progenitor cells (GNPs) sharing the developmental origin and the transcriptional profile of human group 3 medulloblastoma (21). Importantly, cells derived from murine tumors can be used as an in vitro model of group 3 medulloblastoma (20–22). We utilized this model to investigate the effect of the H3K27me3 status on radiation response in group 3 medulloblastoma. We show that loss of H3K27me3 occurring in 47.2% of group 3 and 4 tumor samples results in a radiation-resistant phenotype. We found that radiation resistance in H3K27me3-deficient cells is due to an epigenetic switch from H3K27me3 to H3K27ac resulting in increased expression of EPHA2 that stimulates excessive pro-survival AKT pathway signaling. Finally, we demonstrate that the Bromodomain and Extra-Terminal motif (BET) inhibitor JQ1 efficiently suppresses H3K27ac levels and restores radiation response in H3K27me3-deficient medulloblastoma cells. Collectively, our work elucidates the role of H3K27me3 in radiotherapy response in medulloblastoma and identifies a drug that can improve treatment outcomes in radioresistant tumors.
Materials and methods
Study approval
The study of human tumor tissue samples was approved by the Washington University School of Medicine Internal Review Board (IRB). A waiver of consent was approved by the IRB since the study retrospectively used human tumor tissues stored in the pathology department.
Cell Culture and cell treatment
Derivation of the murine group 3 medulloblastoma cell lines has been previously described (20,21). Murine medulloblastoma lines #51 and #68 and the human D425 line were cultured in Neurobasal media (Thermo Scientific) supplemented with N2, B27, 0.5mM L-glutamine, penicillin/streptomycin, 25μg/ml epidermal growth factor and 25μg/ml fibroblast growth factor at 37°C and 5% CO2. For adherent conditions used for experiments plates were coated with phosphate buffer saline (PBS) pH 7.4 with 0.5μg/ml of Matrigel (Corning, NY, USA CB40230A). CRISPR/Cas9-mediated knock-outs (KO) were performed in vitro following the derivation of cells from primary tumors. The D425 line was obtained from ATCC. The #51 and #68 murine lines were provided by Dr. Martine Roussel at St. Jude.
Antibodies, Primes and Chemical Inhibitors
Detailed information about Antibodies, Primes and Chemical Inhibitors is provided in Tables S1–3.
Patient sample selection and immunohistochemical staining
36 patients with non-WNT/SHH medulloblastoma that were treated with radiotherapy at Washington University School of Medicine between 2008 and 2019 were evaluated. The tumor tissue samples and data were accessed according to the guidelines of the Institutional Review Board of Washington University (IRB No. 201808120). Patients with non-WNT/SHH medulloblastoma were identified based on negative immunohistochemical staining for nuclear beta-catenin, YAP1 and GAB1 (23). Immunohistochemistry (IHC) of sectioned formalin-fixed paraffin-embedded tumor samples was performed using Ventana automated IHC staining device (Ventana Medical Systems, Tucson, AZ, USA). Sections were counterstained with hematoxylin for visualization of nuclei. An avidin-biotin horseradish peroxidase was applied to visualize antibodies using ImmunoCruz™ Rabbit LSAB Staining System (Santa-Cruz Biotechnology Inc.).
Cell irradiation
Cells were irradiated on an X-ray irradiator (RS-2000, RadSource Technologies).
Evaluation of cell survival by colony formation assay
Medulloblastoma cells were plated onto the matrigel-coated 6 well plates. 7 days post irradiation cell colonies were stained with crystal violet and colonies with more than 50 cells were counted.
Evaluation of cell survival by MTT assay
Medulloblastoma cells were plated onto the matrigel-coated 6 well plates. 7 days post irradiation cells were stained with 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) reagent (Fisher Scientific). MTT was dissolved in isopropanol and relative absorbance was quantified at 570nm in a plate reader (i3 SpectraMax, Molecular Devices). Percentage of viable cells was calculated as a quotient of relative absorbance of an irradiated sample and un-irradiated control sample.
Immunofluorescence
Cells were plated onto coverslips coated with Matrigel. Cells were washed with PBS, fixed with 4% paraformaldehyde for 20 minutes at room temperature (RT), permeabilized with 0.25% Triton-X 100 in PBS for 5 minutes at RT and washed with PBS. Cells were blocked with (3% BSA (bovine serum albumin) and 1% FCS (fetal calf serum) in PBS) and incubated with primary antibody at 4°C overnight. Cells were washed with PBS and incubated with goat anti-mouse Alexa Fluor 488 antibody for 1hr at RT in the dark. Coverslips were mounted on slides with ProLong Diamond anti-fade (Thermo Scientific, P36961). γH2AX foci from 50 nuclei for each time point were quantified using a fluorescent microscope (Axioplan2, Zeiss).
Annexin V labeling
Cells per well were seeded onto Matrigel coated 12-well plates. 48 hours post IR, cells were treated with Accutase, labeled with Annexin V per manufacturer’s protocol (Beckman Coulter) and Annexin V positive cell population was quantified using a flow cytometer (Miltenyi, MACS Quant) and analyzed by FlowJo software (BD Biosciences).
pH3 (S10) labeling
Cells were permeabilized with 0.25% Triton X 100 in PBS, and incubated with anti-pH3S10 AB for 2 hours at RT, washed with PBS and incubated with a secondary antibody labeled with Alexa Fluor 488 (Cell Signaling, A11034) for 30 minutes at RT in the dark. pH3S10-positive cell population was determined using a flow cytometer (Miltenyi, MACS Quant) and analyzed by FlowJo software (BD Biosciences).
Western blot
Whole cell protein lysates were prepared using cell lysis buffer containing NP40, protease and phosphatase inhibitors. Protein lysates were ultra-sonicated (Sonic Dismembrator, Fisher Scientific) thrice for 10 seconds. Western blot was performed as previously described (24).
RNA isolation, cDNA synthesis and qPCR
Total RNA was isolated using TRIZOL reagent according to the manufacturer’s instructions (Sigma, St Louis, MO). 1μg of RNA was subjected to reverse transcription into cDNA using High Capacity cDNA Reverse Transcriptase Kit (Applied Biosystems) according to the manufacturer’s instructions. qPCR was performed using SYBR Quantitative Real-Time (SYBR Select Master Mix, Applied Biosystem) on an ABI 7900HT Real time PCR platform (Applied Biosystem). Actb was used as the internal standard reference. Normalized expression was calculated using the comparative CT method and fold changes were derived from the 2−ΔΔCt values for each gene.
ChIP-Seq
Chromatin immunoprecipitation (ChIP) of H3K27me3 and H3K27ac followed by whole genome sequencing was previously described (20). Data was accessed (GEO: GSE84761) and downloaded from the SRA archives (SRP079643) in FASTQ format and aligned to the mouse reference genome (UCSC mm9) using BWA with default parameters. Duplicate reads were marked using Picard, and only nonduplicated reads were kept (Samtools [4] -q 1 -F 1024). The SPP R package was used to draw cross-correlation plots and estimate the smallest fragment size to use to extend each read. DeepTools was used to extend reads and scale bigwig files to 15M reads for visual comparison using IGV.
ChIP
ChIP assay was performed as previously described (24). Appropriate negative controls using normal rabbit IgG were performed for all ChIP experiments.
Generation of Lentiviral vectors and Particles for CRISPR/Cas9-mediated gene knock-out
The LentiCRISPR v2 plasmid was procured from Addgene (#52961) and gRNA sequences were synthesized by IDT Technologies. The sense oligo and the antisense oligo were annealed and ligated into the BsmBI-digested LentiCRISPR vector (Addgene). Cloned vector was transformed into XL1 blue competent cells (NEB). The cloned viral vector was co-transfected with packaging vectors psPAX2 and pMD2.G (both from Addgene) using polyethylenimine into 293T cells to generate lentiviral particles, culture medium was replaced with 5 mL of DMEM containing high glucose 24h later and viral supernatants were collected 48h later filtered and stored at −80°C.
Data Availability
ChIP-seq data (20) are available in the SRA archives (SRP079643), accession number (GEO: GSE84761).
Results
Loss of H3K27me3 is associated with increased relapse rates and reduced survival in group 3 and 4 medulloblastoma
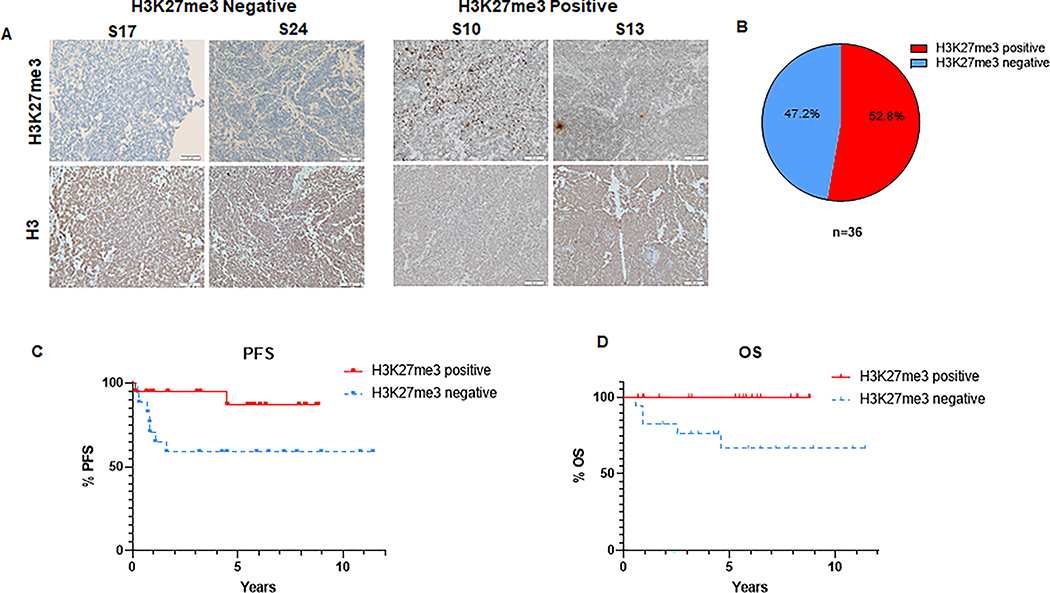
Group 3 and 4 medulloblastoma is characterized by a high frequency of genetic mutations as well as epigenetic dysregulation (5). In particular, H3K27me3 levels are affected by alterations in the function of the H3K27-specific histone methyl-transferase EZH2 or the corresponding histone de-methylase KDM6A in group 3 and 4 medulloblastoma (19). However, the extent of variation of the H3K27me3 levels in these tumors and the effect of the H3K27me3 status on treatment response remains to be elucidated. Thus, we determined the levels of H3K27me3 in tumor samples of group 3 and 4 medulloblastoma patients who underwent surgery followed by the standard-of-care radiotherapy. Importantly, current clinical practice does not routinely involve molecular profiling required to distinguish between group 3 and 4 medulloblastoma (2,7) but classifies tumors into WNT, SHH and non-WNT/SHH groups based on the immunohistochemical analysis of tumor markers that indicate an overactivation of the WNT or SHH pathways. Thus, we analyzed the non-WNT/SHH group that includes group 3 and 4 medulloblastoma. We found that 52.8% of tumors expressed H3K27me3 (Figure 1A and B) consistent with previous reports (19). However, we identified a significant subpopulation constituting 47.2% of analyzed tumors that was H3K27me3-deficient. Strikingly, H3K27me3 loss was associated with high rates of recurrence and poor overall survival compared to H3K27me3-proficient tumors (Figure 1 C and D). The HK27me3 status was not associated with the Myc amplification status, sex or age of the patients (Figure S1A–C). Our data indicates that H3K27me3 expression is frequently lost in group 3 and 4 medulloblastoma and lack of H3K27me3 is associated with a high risk of treatment failure following radiotherapy and poor survival of the patients.
Figure 1: H3K27me3 status predicts clinical outcomes in group 3 and 4 medulloblastoma.
(A) Representative images of immunohistochemical (IHC) staining of H3K27me3 in tumor samples from non-WNT/SHH medulloblastoma patients (includes group 3 and 4 medulloblastoma). Representative images of H3K27me3-deficient tumors S17, S24 (left panel) and H3K27me3-proficient tumors S10, S13 (right panel) are shown. Lower panel shows the H3 staining of respective tumors (B) Pie chart demonstrating the proportion of H3K27me3-proficient versus H3K27me3-deficient tumors among the 36 analyzed samples from non-WNT/SHH medulloblastoma patients. (C) Kaplan-Meier graph showing the progression free survival (PFS) of non-WNT/SHH medulloblastoma patients following radiotherapy stratified by the H3K27me3 status (n=36), (p = 0.045). (D) Kaplan-Meier graph showing the overall survival (OS) of non-WNT/SHH medulloblastoma patients following radiotherapy stratified by the H3K27me3 status (n=36), (p = 0.014). (C-D) Median follow-up time was 5.8 and 6.8 years for H3K27me3-positive and H3K27me3-negative patients, respectively. Log-rank test was used for statistical analysis.
H3K27me3 deficiency results in a radioresistant phenotype in group 3 medulloblastoma
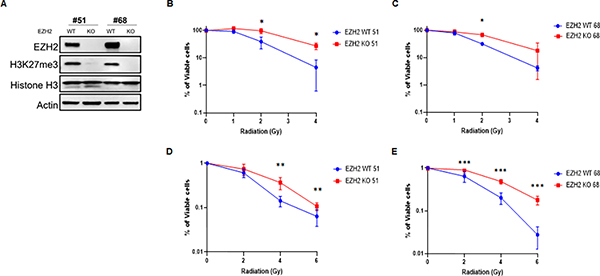
Understanding the molecular mechanisms whereby changes in the epigenetic profile affect radiation response is critical for therapeutic targeting of tumors (14). The role of the EZH2-dependent H3K27 tri-methylation in cellular response to radiation remains controversial with prior reports suggesting, both a pro-survival and an anti-survival role for EZH2 following DNA damage induction (17,18). After demonstrating that loss of H3K27me3 is associated with an early tumor recurrence after radiotherapy in group 3 and 4 medulloblastoma patients (Figure 1C and D) we sought to investigate the role of this histone modification in radiation response. For this purpose, we used cell lines derived from an in vivo group 3 medulloblastoma model established by overexpressing MYC in murine CDKN2C−/−, p53−/− cerebellar GNP cells and implanting them into mouse brains (20,21). The advantage of this model is that it resembles the genetic and transcriptional fingerprint of human group 3 medulloblastoma (21). To mimic an H3K27me3-deficient phenotype we use cell lines containing a CRISPR/Cas9-mediated knock-out (KO) of the EZH2 gene. As expected, EZH2 KO resulted in a loss of H3K27me3 (Figure 2A) allowing a direct comparison of isogenic cell lines that differ in the H3K27me3 status. Next, we examined the effect of the EZH2 KO on radiation response in group 3 medulloblastoma showing that loss of EZH2 renders tumor cells resistant to radiation (Figure 2B–E). Thus, we demonstrate that EZH2 loss results in a radiation resistant phenotype in group 3 medulloblastoma.
Figure 2. Loss of EZH2 results in a radiation resistant phenotype in group 3 medulloblastoma.
(A) Western blot (WB) analysis of indicated proteins in EZH2-WT (H3K27me3-proficient) and EZH2-KO (H3K27me3-deficient) group 3 medulloblastoma cells derived from orthotopic tumors that were established using murine Myc-expressing CDKN2C−/−, p53−/− cerebellar GNP. (B-C) MTT assay demonstrating survival in group 3 medulloblastoma cells ((B) clone #51, (C) clone #68). Cells were treated with indicated doses of ionizing radiation (IR) and percentage of viable cells was determined 7d later. (D-E) Colony formation assay demonstrating survival in EZH2-WT and EZH2-KO group 3 medulloblastoma cells ((D) clone #51, (E) clone #68). Cells were treated with indicated doses of ionizing radiation (IR) and survival was assessed by colony formation assay 7d later. (B-E) Data of three independent experiments are shown as mean +/− SD. Paired two-tailed T-test was used for statistical analysis (***p < 0.001; **p < 0.01; *p < 0.05).
H3K27me3 status does not affect DNA repair and cell death pathways after radiation
Next, we sought to elucidate the mechanism of radiation resistance in H3K27me3-deficent cells. First, we investigated if the H3K27me3 status affects DSB repair kinetics after radiation in medulloblastoma cells. Importantly, the initial number of DSBs induced by 1Gy radiation was similar in EZH2 wild-type (WT) and KO cells indicating that the structural chromatin changes due to the H3K27me3 loss do not affect the efficacy of DNA damage induction by radiation (Figure 3A and Figure S2A). Further, we demonstrate that the H3K27me3 status does not affect the kinetics of DSB repair (Figure 3A and Figure S2A) suggesting that radiation resistance in EZH2 KO cells is not due to enhanced DSB re-ligation.
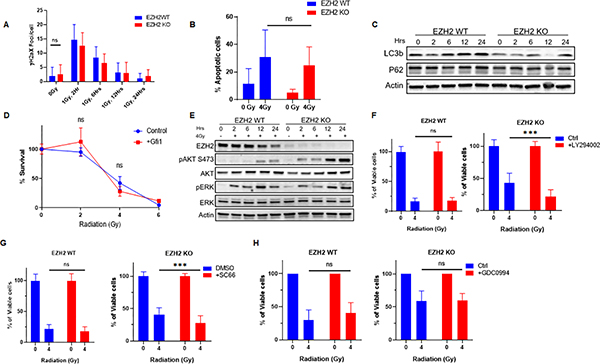
Figure 3. Overactivation of AKT signaling renders H3K27me3-deficient medulloblastoma cells resistant to radiation.
(A) Quantification of γH2AX foci in EZH2-WT and EZH2-KO #51 group 3 medulloblastoma cells. Cells were mock-irradiated or exposed to 1Gy IR and γH2AX foci were visualized by immunofluorescence and quantified at indicated time points following irradiation. (B) EZH2-WT and EZH2-KO #51 medulloblastoma cells were mock-irradiated or exposed 4Gy IR exposed and cell population undergoing apoptotic cell death was determined by Annexin V staining followed by flow cytometry analysis 48hrs after IR. (C) WB analysis of indicated proteins in EZH2-WT and EZH2-KO #51 medulloblastoma cells that were mock-irradiated or exposed to 4Gy IR. Cells were harvested for analysis at indicated time points following IR. (D) EZH2-WT #51 medulloblastoma cells stably overexpressing an empty control vector or GFI1 that were mock-irradiated or exposed to indicated doses of IR. Percentage of viable cells was determined by MTT 7d after IR. (E) WB analysis of indicated proteins in EZH2-WT and EZH2-KO #51 medulloblastoma cells that were mock-irradiated or exposed to 4Gy IR. Cells were harvested for analysis at indicated time points post-IR. (F-H) MTT assay showing percentage of viable cells in EZH2-WT and EZH2-KO #51 medulloblastoma cells. Cells were mock-treated or pre-treated with (F) 5μM PI3K inhibitor LY294002, (G) 5ng/ml AKT inhibitor SC66, or (H) 1μM ERK1 inhibitor GDC0994 for 24h and either mock-irradiated or exposed to 4Gy IR. Percentage of viable cells l was determined 7d after IR. (A, B, D, F-H) Data of three independent experiments are shown as mean +/− SD. Paired two-tailed T-test was used for statistical analysis (***p < 0.001).
Next, we investigated whether a disruption in cell death signaling is responsible for radioresistance in EZH2 KO cells. We observed a moderate induction of apoptotic cell death in medulloblastoma cells following irradiation and the EZH2 status did not affect the rates of apoptosis (Figure 3B). Consistent, with this finding there was no difference in the expression of pro- and anti-apoptotic factors between the EZH2 WT and KO cells (Figure S2B). We also investigated whether autophagic cell death plays a role in radiation resistance in the EZH2 KO cells but found no significant conversion of LC3-I to LC3-II or change in p62 levels in medulloblastoma cells regardless of the EZH2 status (Figure 3C). Finally, we demonstrate a transient G2/M arrest following irradiation that was not affected by the EZH2 status (Figure S2C and D) (25) suggesting that an altered cell cycle checkpoint signaling is not responsible for the radiation resistant phenotype in EZH2 KO cells.
Radiation resistance in H3K27me3-deficient cells is due to overactivation of AKT signaling
After excluding alterations in DNA repair and cell death signaling as the cause of radiation resistance in H3K27me3-deficient EZH2 KO cells we turned our attention to signaling pathways that promote cell survival following DNA damage. Importantly, increased expression of the transcription factor GFI1 was shown to accelerate cell proliferation and promote an aggressive phenotype in H3K27me3-deficient medulloblastoma (20). However, an overexpression of GFI1 did not induce radiation resistance in H3K27me3-proficient EZH2 WT cells (Figure 3D). Since multiple reports have indicated a potential role of the PI3K/AKT signaling pathway in radiation resistance (15,16) we determined whether AKT is activated in our system following exposure to irradiation. Strikingly, we observed a rapid increase of pAKT (Ser473) levels in EZH2 KO cells following radiation that significantly exceeded pAKT (Ser473) levels in EZH2 WT cells (Figure 3E). Consistent with this finding, inhibition of the AKT signaling pathway using a small molecule inhibitor of PI3K that is upstream of AKT (Figure 3F) or a direct inhibition of AKT (Figure 3G) restored radiation response in the H3K27me3-deficient medulloblastoma cells. Notably, PI3K or AKT inhibition did not have a significant effect on radiation sensitivity in EZH2 WT cells (Figure 3F and G). These data suggest that excessive AKT signaling is responsible for radiation resistance in medulloblastoma cells lacking H3K27me3.
Additionally, we investigated the role of the pro-survival MAPK signaling pathway in radiation response in medulloblastoma. However, we did not observe an effect of the EZH2 status on MAPK signaling following radiation (Figure 3E). Consistently, chemical inhibition of ERK did not affect radiation response in medulloblastoma cells (Figure 3H).
Epigenetic switch stimulates AKT signaling through up-regulation of EPHA2 in H3K27me3-deficient medulloblastoma cells
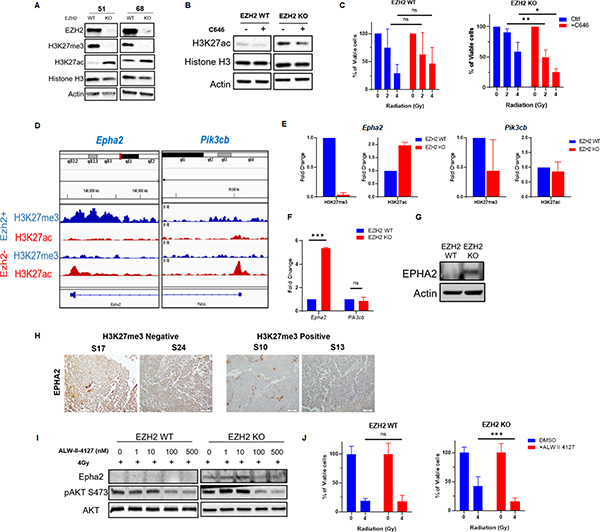
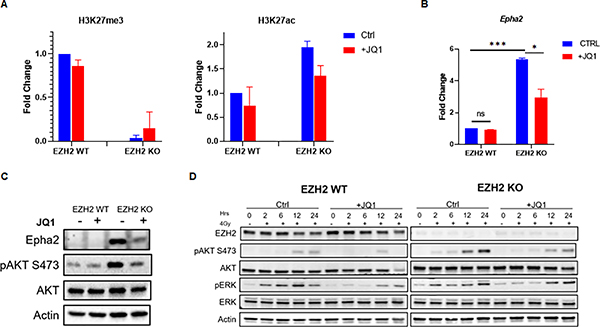
In certain tumor types a decrease in H3K27me3 levels results in a reciprocal elevation in H3K27ac (26). Increase in H3K27ac can affect the tumor phenotype by altering the transcriptional profile of cancer cells and driving an overexpression of oncogenes (26). We show that H3K27me3 loss results in a reciprocal increase of H3K27ac levels in EZH2 KO medulloblastoma cells (Figure 4A). Thus, we investigated whether an epigenetic shift from H3K27me3 to H3K27ac is responsible for radiation resistance in H3K27me3-deficient medulloblastoma. Indeed, chemical inhibition of the CPB/p300 histone acetyl-transferase that is responsible for H3K27 acetylation reduced the elevated levels of H3K27ac (Figure 4B) and restored radiation response in EZH2 KO cells without affecting radiation sensitivity in EZH2 WT cells (Figure 4C). Since H3K27ac is a histone modification that increases transcriptional activity (27,28) we hypothesized that a rise in H3K27ac levels at specific genetic loci alters the expression of genes regulating the AKT pathway. This leads to enhanced AKT signaling and radiation resistance in H3K27me3-deficient cells. Using ChIP-Seq we analyzed the H3K27me3 and H3K27ac levels within the genes encoding for AKT pathway members and regulators in EZH2 WT versus KO cells (Figure S3). We found that loss of H3K27me3 in the promotor regions of the Pik3cb and Epha2 genes was associated with a local increase in H3K27ac (Figure 4D). However, we did not detect this epigenetic switch within the promoters or the gene body of other genes that were screened (Figure S3). Next, we used ChIP-qPCR to confirm the local epigenetic changes within the Pik3cb and Epha2 promotor regions in EZH2 WT versus EZH2 KO cells that were detected by ChIP-Seq (Figure 4E). As expected, we observed reduced levels of H3K27me3 within the Epha2 and Pik3cb promotors (Figure 4E). However, an increase in H3K27ac was only found within the Epha2 but not the Pik3cb promotor (Figure 4E). We show that this epigenetic shift is associated with an increased transcription of the Epha2 gene in EZH2 KO cells resulting in elevated EPHA2 protein levels (Figure 4F and G). Consistent with a lack of the reciprocal H3K27ac increase within the Pik3cb gene promotor its transcription was not affected by the EZH2 status (Figure 4F). Importantly, the expression of PTEN, an important regulator of the AKT pathway, was not affected by the EZH2 status (Figure S4). Finally, we demonstrate that loss of H3K27me3 correlates with increased EPHA2 expression levels in tumor samples from medulloblastoma patients (Figure 4H) supporting the clinical relevance of our finding.
Figure 4. EZH2 loss stimulates the AKT pathway through an epigenetic switch that up-regulates EPHA2.
(A) WB analyzing the expression of indicated proteins in EZH2-WT and EZH2-KO #51 and #68 medulloblastoma cells. (B) WB analysis of indicated proteins in EZH2-WT and EZH2-KO #51 medulloblastoma cells that were mock-treated or treated with 10μM CBP/p300 inhibitor C646 for 24h. (C) MTT assay showing percentage of viable cells in EZH2-WT and EZH2-KO #51 medulloblastoma cells. Cells were mock-treated or pre-treated with 10μM CBP/p300 inhibitor C646 for 24h and either mock-irradiated or exposed to indicated doses of IR. Percentage of viable cells was determined 7d after IR. (D) ChIP-Seq analysis of the H3K27me3 and H3K27ac levels within the indicated genomic loci in EZH2-WT and EZH2-KO #51 medulloblastoma cells. (E) ChIP analysis of H3K27me3 and H3K27ac levels within the Epha2 (left panel) and Pik3cb (right panel) gene promotors in EZH2-WT and EZH2-KO medulloblastoma cells. (F) qRT-PCR analysis showing the transcriptional activity of indicated genes in EZH2-WT and EZH2-KO #51 medulloblastoma cells. (G) WB analysis of indicated proteins in EZH2-WT and EZH2-KO #51 medulloblastoma cells. (H) Images of IHC staining of EPHA2 in H3K27me3-deficient S17, S24 (left panel) and H3K27me3-proficient S10, S13 (right panel) tumor samples from non-WNT/SHH medulloblastoma patients. (I) WB analysis of indicated proteins in EZH2-WT and EZH2-KO #51 medulloblastoma cells that were mock-treated or pre-treated with indicated concentrations of the EPHA2 inhibitor ALW-II-4127 for 24h followed by exposure to 4Gy IR. Cells were harvested for WB analysis 2h following IR. (J) MTT assay showing percentage of viable cells in EZH2-WT and EZH2-KO #51 medulloblastoma cells. Cells were pre-treated with 100nM EPHA2 inhibitor ALW-II-4127 for 24h and either mock-irradiated or exposed to 4Gy IR. Percentage of viable cells was determined 7d after IR. (C, F, J) Data of three independent experiments are shown as mean +/− SD. Paired two-tailed T-test was used for statistical analysis (***p < 0.001; **p < 0.01; *p < 0.05). (E) Mean of two independent experiments is shown.
Based on these findings we focused on the role of EPHA2 as a potential mediator of AKT-dependent radiation resistance in H3K27me3-deficient medulloblastoma cells. EPHA2 is a tyrosine kinase that can function as a regulator of the AKT signaling pathway. Overexpression of EPHA2 or EPHA2 stimulation with progranulin enhances the AKT kinase activity (29,30). Thus, we tested whether excessive AKT signaling in EZH2 KO medulloblastoma cells is due to EPHA2 overexpression. Indeed, blocking EPHA2 activity normalized AKT signaling following radiation and restored radiation response in EZH2 KO cells (Figure 4I and J). Together, our data indicates that an overexpression of EPHA2 drives an overactivation of the AKT signaling pathway and radiation resistance in H3K27me3-deficient medulloblastoma.
BET inhibition targets radiation resistance in H3K27me3-deficient medulloblastoma
Radiotherapy resistance is a major challenge in group 3 medulloblastoma resulting in high rates of disease recurrence (5,8). Thus, we sought to establish a therapeutic strategy targeting radiation resistance in H3K27me3-deficient group 3 medulloblastoma. Multiple targeted therapies have been proposed for group 3 and 4 medulloblastoma including CDK4/6 (31,32) and HER2/neu (33,34) inhibitors. However, while moderately improving radiation response in EZH2 WT cells CDK4/6 inhibition failed to mitigate radiation resistance in EZH2 KO cells (Figure S5A). Further, targeting HER2/neu had no effect on radiation response of medulloblastoma cells (Figure S5B).
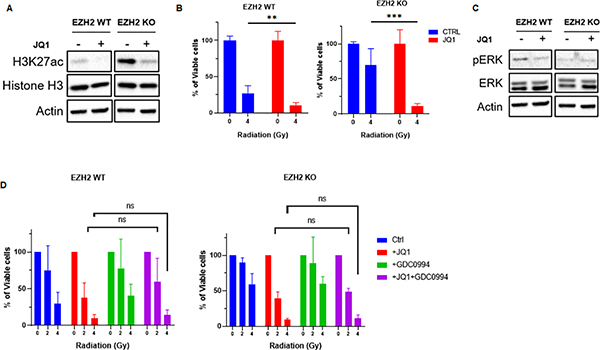
Thus, we decided to directly target the epigenetic shift causing radiation resistance in the H3K27me3-deficient cells. Our data shows that CPB/p300 inhibition partially reduces the elevated H3K27ac levels and improves radiation response in EZH2 KO cells (Figure 4B and C). However, we sought a more effective approach of targeting the elevated H3K27ac levels. Notably, bromodomain proteins mediate the p300-dependent histone acetylation (35) and BET inhibition can suppress H3K27ac levels (26,35). Thus, we tested whether BET inhibition by a small molecule inhibitor JQ1 restores radiation response in H3K27me3-deficient cells. We found that BET inhibition is more efficient in suppressing H3K27ac levels in EZH2 KO cells than CBP/p300 inhibition (Figure 5A and 4B). Consequently, BET inhibition by JQ1 was highly efficient in restoring radiation response in multiple EZH2 KO cell lines (Figure 5B and Figure S5C). Importantly, JQ1 sensitized EZH2 KO cells to radiation beyond restoring their radiation response to the level of EZH2 WT cells (Figure 5B). Further, we found that JQ1 suppressed the baseline levels of H3K27ac in EZH2 WT cells (Figure 5A and Figure S5D) and sensitized them to radiation (Figure 5B). Collectively, our data indicate that BET inhibition effectively targets radiation resistance in H3K27me3-deficient medulloblastoma cells by suppressing H3K27ac levels.
Figure 5. BET inhibition targets radiation resistance in H3K27me3-deficient medulloblastoma cells.
(A) WB analysis of indicated proteins in EZH2-WT and EZH2-KO #51 medulloblastoma cells that were mock-treated or treated with 0.1μM BET inhibitor JQ1 for 24h. (B) MTT assay showing percentage of viable cells l in EZH2-WT and EZH2-KO #51 medulloblastoma cells. Cells were mock-treated or pre-treated with 0.1μM BET inhibitor JQ1 for 24h and either mock-irradiated or exposed to indicated doses of IR. Percentage of viable cells l was determined 7d after IR. (C) WB analysis of indicated proteins in EZH2-WT and EZH2-KO #51 medulloblastoma cells that were mock-treated or pre-treated with 0.1μM BET inhibitor JQ1 for 24h and either mock-irradiated or exposed to 4Gy IR. Cells were harvested for WB analysis 24h following IR. Same cell lysates and, therefore same actin loading control was used as in Figure 5A. (D) MTT assay showing percentage of viable cells in EZH2-WT and EZH2-KO #51 medulloblastoma cells. Cells were mock-treated or pre-treated with 0.1μM BET inhibitor JQ1, 1μM ERK1 inhibitor GDC0994 or a combination of both for 24h and either mock-irradiated or exposed to indicated doses of IR. Percentage of viable cells was determined 7d after IR. (B, D) Data of three independent experiments are shown as mean +/− SD. Paired two-tailed T-test was used for statistical analysis (***p < 0.001; **p < 0.01; *p < 0.05).
Notably, BET inhibition has been shown to upregulate ERK1 signaling in several EZH2-deficient cancer cell lines, which can promote cell survival and attenuate the effect of BET inhibition on tumor cell kill (26). However, we did not observe an effect of BET inhibition on ERK1 signaling in medulloblastoma cells (Figure 5C). Consistently, we found no synergistic effect on radiosensitivity when combining BET and ERK1 inhibitors (Figure 5D).
BET inhibition suppresses excessive AKT signaling and alters the transcriptional profile in favor of pro-apoptotic factors
Next, we investigated the mechanism whereby BET inhibition affects radiation response in medulloblastoma. Consistent with a global reduction in H3K27ac levels (Figure 5A) BET inhibition suppressed the elevated levels of H3K27ac within the Epha2 gene promotor in H3K27me3-deficient medulloblastoma cells (Figure 6A). This reduced the Epha2 gene transcription and normalized EPHA2 expression levels (Figure 6B and C). Consequently, BET inhibition mitigated excessive AKT signaling in EZH2 KO cells following radiation (Figure 6D) consistent with its ability to restore radiation response in these medulloblastoma cells (Figure 5B). To validate these findings we have established a human H3K27me3-deficient group 3 medulloblastoma model by knocking out EZH2 in p53-mutant Myc-amplified patient-derived D425 medulloblastoma cells (36) (Figure S6A). Using this model we show that JQ1 effectively suppresses excessive AKT signaling in EZH2 KO cells and sensitizes them to radiation (Figure S6B and C).
Figure 6. BET inhibition suppresses excessive AKT signaling.
(A) ChIP analysis of H3K27me3 and H3K27ac levels within the Epha2 gene promotor in EZH2-WT and EZH2-KO #51 medulloblastoma cells. Cells were mock-treated or pre-treated with 0.1μM BET inhibitor JQ1 for 24h. (B) qRT-PCR analysis showing the transcript levels of the Epha2 gene in EZH2-WT and EZH2-KO #51 medulloblastoma cells that were mock-treated or pre-treated with 0.1μM BET inhibitor JQ1 for 24h. (C) WB analysis of indicated proteins in EZH2-WT and EZH2-KO #51 medulloblastoma cells that were mock-treated or pre-treated with 0.1μM BET inhibitor JQ1 for 24h. (D) WB analysis of indicated proteins in EZH2-WT and EZH2-KO #51 medulloblastoma cells that were mock-treated or pre-treated with 0.1μM BET inhibitor JQ1 for 24h and irradiated with 4Gy. Cells were harvested for analysis at indicated time points post-IR. (A) Data of two independent experiments are shown as mean. (B) Data of three independent experiments are shown as mean +/− SD. Paired two-tailed T-test was used for statistical analysis (***p < 0.001; **p < 0.01; *p < 0.05).
Notably, BET inhibition had only a mild effect on ERK activation after radiation in EZH2 WT and KO cells (Figure 6D). Together with our finding that ERK signaling does not affect radiation response in medulloblastoma cells (Figure 5C and D) this data suggests that the MAPK pathway is not involved in mediating radiosensitization by BET inhibition.
BET inhibition alters the transcriptional profile in favor of pro-apoptotic factors
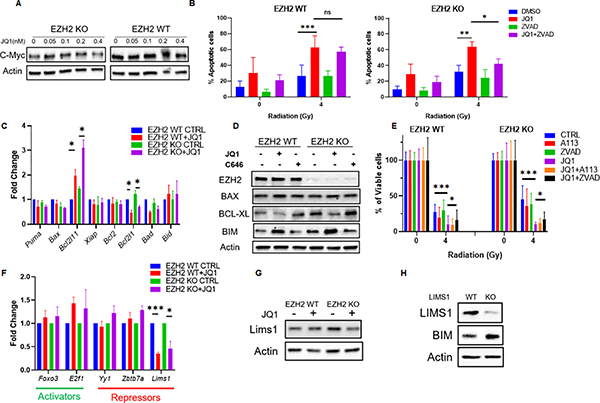
The question remained how BET inhibition facilitates radiosensitization of H3K27me3-deficient EZH2 KO cells beyond restoring radiation response. The additional degree of radiosensitization in the EZH2 KO cells correlated with the ability of JQ1 to sensitize the H3K27me3-proficient EZH2 WT cells to radiation. Thus, we hypothesized that this is due to transcriptional alteration resulting from a nearly complete suppression of the H3K27ac levels by BET inhibition. BET inhibitors are known to affect the expression of c-Myc (37,38). However, we found that JQ1 does not affect Myc expression in the group 3 medulloblastoma model used in this study (Figure 7A). Interestingly, we observed that JQ1 increases the rates of apoptotic cell death after radiation independent of the H3K27me3 status (Figure 7B). Thus, we investigated the effect of JQ1 on transcription of multiple pro- and anti-apoptotic genes. We demonstrate that BET inhibition suppresses the transcription of the Bcl2l1 gene resulting in reduced Bcl-XL protein levels in both, EZH2 WT and KO cells, Figure 7C and D). This correlates with the ability of JQ1 to effectively suppress H3K27ac levels in both, EZH2 WT and KO cells (Figure 5A and Figure S5C). In contrast, p300/CBP inhibitor C646 did not affect Bcl-XL levels (Figure 7D) consistent with an attenuated H3K27ac suppression by C646 (Figure 4B). Notably, inhibition of Bcl-XL did not enhance the radiosensitization induced by JQ1 (Figure 7E). Further, blocking caspase activation alleviated the radiosensitization induced by BET inhibition (Figure 7E). These findings suggest that an enhancement of apoptotic response due to a down-regulation of Bcl-XL contributes to the radiosensitizing effect of BET inhibition in medulloblastoma cells independent of the H3K27me3-status.
Figure 7. BET inhibition alters the transcriptional profile in favor of pro-apoptotic factors.
A) WB analysis of indicated proteins in EZH2-WT and EZH2-KO #51 medulloblastoma cells that were mock-treated or treated with indicated concentrations of the BET inhibitor JQ1 for 24h. (B) EZH2-WT and EZH2-KO #51 medulloblastoma cells were mock-treated or pre-treated with 0.1μM BET inhibitor JQ1 for 24h and were mock-irradiated or exposed 4Gy IR. Cell population undergoing apoptotic cell death was determined by Annexin V staining followed by flow cytometry analysis 48h after IR. (C) qRT-PCR analysis showing the transcript levels of indicated genes in EZH2-WT and EZH2-KO #51 medulloblastoma cells that mock-treated or treated with 0.1μM BET inhibitor JQ1 for 24h. (D) WB analysis of indicated proteins in EZH2-WT and EZH2-KO #51 medulloblastoma cells that were mock-treated or treated with 10μM CBP/p300 inhibitor C646 or 0.1μM BET inhibitor JQ1 24h. (E) MTT assay showing percentage of viable cells in EZH2-WT and EZH2-KO #51 medulloblastoma cells. Cells were mock-treated or pre-treated with 1μM Bcl-XL inhibitor A1331852, 0.1μM BET inhibitor JQ1, 25 μM pan-caspase inhibitor z-VAD-FMK or indicated combinations thereof for 24h and either mock-irradiated or exposed to 4Gy IR. Percentage of viable cells was determined 7d after IR. Data of three independent experiments are shown as mean +/− SD. Paired one-tailed T-test was used for statistical analysis (***p < 0.001; *p < 0.05). (F) qRT-PCR analysis showing the transcript levels of indicated genes in EZH2-WT and EZH2-KO #51 medulloblastoma cells that mock-treated or treated with 0.1μM BET inhibitor JQ1 for 24h. (G) WB analysis of indicated proteins in EZH2-WT and EZH2-KO #51 medulloblastoma cells that were mock-treated or treated with 0.1μM BET inhibitor JQ1 for 24h. (H) WB analysis of indicated proteins in EZH2-WT #51 medulloblastoma cells 4. (B, C, F) Data of three independent experiments are shown as mean +/− SD. Paired two-tailed T-test was used for statistical analysis (***p < 0.001; **p < 0.01; *p < 0.05).
Surprisingly, BET inhibition significantly increased the transcription of the Bcl2l11 gene that encodes for the pro-apoptotic factor Bim in EZH2 WT and KO cells (Figure 7C and D). We observed a moderate reduction of the H3K27ac levels within the Bcl2l11 promotor following JQ1 treatment (Figure S7). Since a decrease in H3K27ac levels results in a transcriptionally repressive chromatin state (26,39) we, therefore, assumed that up-regulation of BIM expression by BET inhibition occurs indirectly. Therefore, we explored the effect of BET inhibition on a panel of transcriptional factors that regulate the Bcl2l11 gene. As expected, JQ1 did not affect the transcription of genes encoding for activators of the Bcl2l11 gene (Figure 7F). However, we found that JQ1 attenuates the expression of the Lims1 gene (Figure 7F and G) encoding for a transcriptional repressor of the Bcl2l11 gene. A KO of LIMS1 mimicked the effect of BET inhibition on Bim expression raising BIM levels in medulloblastoma cells (Figure 7H). Collectively, our results indicate that in addition to restoring radiation response in H3K27me3-deficient cells (Figure S8A) BET inhibition induces radiosensitization in an H3K27me3-independent manner by stimulating apoptosis (Figure S8B). Notably, we have displayed all individual data points for cell viability assays in Figures S9 and S10.
Discussion
Radiation resistance is a major challenge in treatment of medulloblastoma resulting in tumor relapse and poor clinical outcomes. Our work reveals that the epigenetic modification H3K27me3 is a predictive marker of radiation response in non-WNT/SHH medulloblastoma and establishes a strategy for targeting radiation resistance in H3K27me3-deficient tumors. We show that loss of H3K27me3 leads to an epigenetic shift from H3K27me3 to H3K27ac. This alters the transcriptional profile in medulloblastoma cells resulting in a radiation resistant phenotype due to an overactivation of the AKT signaling pathway (schematic model shown in Figure S8). Further, we demonstrate that BET inhibition is an effective therapeutic approach for targeting radiation resistance in H3K27me3-deficient tumors. Thus, our results provide a foundation for clinical use of BET inhibitors to improve outcomes in a defined group of medulloblastoma patients.
Our work establishes a novel role of the epigenetic switch from H3K27 tri-methylation to acetylation in radiation response in medulloblastoma. H3K27me3 plays a critical part in transcriptional regulation by maintaining a transcriptionally repressive chromatin state (40,41). We demonstrate that loss of H3K27me3 results in a reciprocal increase of the transcriptionally activating histone mark H3K27ac (26–28). This epigenetic shift is associated with an overexpression of EPHA2, which enhances the DNA damage triggered activation of the AKT signaling pathway and promotes radiation resistance of tumor cells. These findings are consistent with prior reports demonstrating that the AKT pathway is an important driver of radiation resistance in cancer (15,16). Interestingly, an epigenetic switch from H3K27me3 to H3K27ac has been reported in pediatric midline glioma (42) and posterior fossa A type ependymoma (43) suggesting that it may be a potential therapeutic target in these highly aggressive pediatric malignancies.
Our findings demonstrate an important role of EPHA2 in radiation resistance in medulloblastoma. EPHA2 is a tyrosine kinase that has been implicated in regulation of the PI3K/AKT signaling pathway (29,30,44). Interestingly, EPHA2 can exhibit opposite effects on AKT activity. Whereas EPHA2 stimulation with Ephrin-A inhibits AKT activity (44) overexpression of EPHA2 or stimulation with progranulin enhances AKT signaling in an Ephrin-independent manner (29,30). The latter mechanism appears to regulate the AKT signaling pathway in group 3 medulloblastoma where an overexpression of EPHA2 drives excessive AKT signaling in H3K27me3-deficient cells. The exact biochemical mechanism whereby EPHA2 stimulates AKT signaling in medulloblastoma remains to be elucidated. It is likely, however, that EPHA2 expression levels and ligand selectivity determine its effect on the AKT pathway (29,30,44,45).
Our work has important clinical implications for medulloblastoma patients. We propose including the immunohistochemical analysis of the H3K27me3 status in the diagnostic evaluation of non-WNT/SHH medulloblastoma. The H3K27me3 status can serve as a predictive marker of radiotherapy response and help identifying patients with radioresistant tumors that are at an increased risk of tumor recurrence. Further, we propose combining BET inhibitors with radiotherapy to improve outcomes in H3K27me3-deficient group 3 medulloblastoma patients, which will need to be tested in in vivo models followed by clinical trials. Collectively, our work elucidates a new role of the EZH2-dependent histone modification H3K27me3 in radiotherapy response in medulloblastoma and provides a clinically suitable approach for targeting radiation resistance in this aggressive pediatric brain tumor.
Interestingly, our discovery of EPHA2 up-regulation in H3K27me3-deficient medulloblastoma has an important implication for the emerging field of CAR T-cell immunotherapy. Whereas CAR T-cell therapy has been successfully used in treatment of hematologic malignancies its application to solid tumors, especially tumors of the CNS has constituted a major challenge (46). A recent work has demonstrated a development of CAR T-cells targeting EPHA2 in a mouse model of group 3 medulloblastoma (47). However, a successful translation of this treatment approach into clinic requires an identification of patients with EPHA2-expressing tumors. In this context, our findings suggest that the H3K27me3 status can be used as a marker to select group 3 medulloblastoma patients that can benefit from EPHA2-targeted CAR T-cell therapy. Together, our work bears significant clinical implications for the fields of radiotherapy and immunotherapy.
Supplementary Material
Statement of Significance.
Our work demonstrates a novel epigenetic mechanism of radiation resistance in medulloblastoma and identifies a therapeutic approach to improve outcomes in these patients.
Acknowledgements
We thank Dr. Martine Roussel (St. Jude Children’s Research Hospital, Memphis, TN) for generously providing the EZH2-WT, EZH2-KO and GFI1-overexpressing murine group 3 medulloblastoma cell lines.
Grant Support
This work was supported by a grant from the NIH (1K08CA256170-01A1) and the Childhood Cancer Research Grant by The Andrew McDonough B+ Foundation to M. Goldstein.
Footnotes
The authors declare no potential conflicts of interest.
References
- 1.Pui CH, Gajjar AJ, Kane JR, Qaddoumi IA, Pappo AS. Challenging issues in pediatric oncology. Nat Rev Clin Oncol 2011;8:540–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Northcott PA, Jones DT, Kool M, Robinson GW, Gilbertson RJ, Cho YJ, et al. Medulloblastomics: the end of the beginning. Nat Rev Cancer 2012;12:818–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Juraschka K, Taylor MD. Medulloblastoma in the age of molecular subgroups: a review. J Neurosurg Pediatr 2019;24:353–63 [DOI] [PubMed] [Google Scholar]
- 4.Ellison DW, Onilude OE, Lindsey JC, Lusher ME, Weston CL, Taylor RE, et al. beta-Catenin status predicts a favorable outcome in childhood medulloblastoma: the United Kingdom Children’s Cancer Study Group Brain Tumour Committee. J Clin Oncol 2005;23:7951–7 [DOI] [PubMed] [Google Scholar]
- 5.Kool M, Korshunov A, Remke M, Jones DT, Schlanstein M, Northcott PA, et al. Molecular subgroups of medulloblastoma: an international meta-analysis of transcriptome, genetic aberrations, and clinical data of WNT, SHH, Group 3, and Group 4 medulloblastomas. Acta Neuropathol 2012;123:473–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cho YJ, Tsherniak A, Tamayo P, Santagata S, Ligon A, Greulich H, et al. Integrative genomic analysis of medulloblastoma identifies a molecular subgroup that drives poor clinical outcome. J Clin Oncol 2011;29:1424–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Northcott PA, Korshunov A, Witt H, Hielscher T, Eberhart CG, Mack S, et al. Medulloblastoma comprises four distinct molecular variants. J Clin Oncol 2011;29:1408–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koschmann C, Bloom K, Upadhyaya S, Geyer JR, Leary SE. Survival After Relapse of Medulloblastoma. J Pediatr Hematol Oncol 2016;38:269–73 [DOI] [PubMed] [Google Scholar]
- 9.Laprie A, Hu Y, Alapetite C, Carrie C, Habrand JL, Bolle S, et al. Paediatric brain tumours: A review of radiotherapy, state of the art and challenges for the future regarding protontherapy and carbontherapy. Cancer Radiother 2015;19:775–89 [DOI] [PubMed] [Google Scholar]
- 10.Gajjar A, Chintagumpala M, Ashley D, Kellie S, Kun LE, Merchant TE, et al. Risk-adapted craniospinal radiotherapy followed by high-dose chemotherapy and stem-cell rescue in children with newly diagnosed medulloblastoma (St Jude Medulloblastoma-96): long-term results from a prospective, multicentre trial. Lancet Oncol 2006;7:813–20 [DOI] [PubMed] [Google Scholar]
- 11.Sethi RV, Giantsoudi D, Raiford M, Malhi I, Niemierko A, Rapalino O, et al. Patterns of failure after proton therapy in medulloblastoma; linear energy transfer distributions and relative biological effectiveness associations for relapses. Int J Radiat Oncol Biol Phys 2014;88:655–63 [DOI] [PubMed] [Google Scholar]
- 12.Ramaswamy V, Remke M, Bouffet E, Faria CC, Perreault S, Cho YJ, et al. Recurrence patterns across medulloblastoma subgroups: an integrated clinical and molecular analysis. Lancet Oncol 2013;14:1200–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Mol Cell 2010;40:179–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goldstein M, Kastan MB. The DNA damage response: implications for tumor responses to radiation and chemotherapy. Annu Rev Med 2015;66:129–43 [DOI] [PubMed] [Google Scholar]
- 15.Li HF, Kim JS, Waldman T. Radiation-induced Akt activation modulates radioresistance in human glioblastoma cells. Radiat Oncol 2009;4:43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schuurbiers OC, Kaanders JH, van der Heijden HF, Dekhuijzen RP, Oyen WJ, Bussink J. The PI3-K/AKT-pathway and radiation resistance mechanisms in non-small cell lung cancer. J Thorac Oncol 2009;4:761–7 [DOI] [PubMed] [Google Scholar]
- 17.Xia H, Yu CH, Zhang Y, Yu J, Li J, Zhang W, et al. EZH2 silencing with RNAi enhances irradiation-induced inhibition of human lung cancer growth in vitro and in vivo. Oncol Lett 2012;4:135–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zeidler M, Varambally S, Cao Q, Chinnaiyan AM, Ferguson DO, Merajver SD, et al. The Polycomb group protein EZH2 impairs DNA repair in breast epithelial cells. Neoplasia 2005;7:1011–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Robinson G, Parker M, Kranenburg TA, Lu C, Chen X, Ding L, et al. Novel mutations target distinct subgroups of medulloblastoma. Nature 2012;488:43–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vo BT, Li C, Morgan MA, Theurillat I, Finkelstein D, Wright S, et al. Inactivation of Ezh2 Upregulates Gfi1 and Drives Aggressive Myc-Driven Group 3 Medulloblastoma. Cell Rep 2017;18:2907–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kawauchi D, Robinson G, Uziel T, Gibson P, Rehg J, Gao C, et al. A mouse model of the most aggressive subgroup of human medulloblastoma. Cancer Cell 2012;21:168–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang ZJ, Ellis T, Markant SL, Read TA, Kessler JD, Bourboulas M, et al. Medulloblastoma can be initiated by deletion of Patched in lineage-restricted progenitors or stem cells. Cancer Cell 2008;14:135–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Orr BA. Pathology, diagnostics, and classification of medulloblastoma. Brain Pathol 2020;30:664–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goldstein M, Derheimer FA, Tait-Mulder J, Kastan MB. Nucleolin mediates nucleosome disruption critical for DNA double-strand break repair. Proc Natl Acad Sci U S A 2013;110:16874–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu B, Kim ST, Lim DS, Kastan MB. Two molecularly distinct G(2)/M checkpoints are induced by ionizing irradiation. Mol Cell Biol 2002;22:1049–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang X, Yan J, Zhang M, Wang Y, Chen Y, Fu X, et al. Targeting Epigenetic Crosstalk as a Therapeutic Strategy for EZH2-Aberrant Solid Tumors. Cell 2018;175:186–99 e19 [DOI] [PubMed] [Google Scholar]
- 27.Holmqvist PH, Mannervik M. Genomic occupancy of the transcriptional co-activators p300 and CBP. Transcription 2013;4:18–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Raisner R, Kharbanda S, Jin L, Jeng E, Chan E, Merchant M, et al. Enhancer Activity Requires CBP/P300 Bromodomain-Dependent Histone H3K27 Acetylation. Cell Rep 2018;24:1722–9 [DOI] [PubMed] [Google Scholar]
- 29.Wang Y, Liu Y, Li G, Su Z, Ren S, Tan P, et al. Ephrin typeA receptor 2 regulates sensitivity to paclitaxel in nasopharyngeal carcinoma via the phosphoinositide 3kinase/Akt signalling pathway. Mol Med Rep 2015;11:924–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Neill T, Buraschi S, Goyal A, Sharpe C, Natkanski E, Schaefer L, et al. EphA2 is a functional receptor for the growth factor progranulin. J Cell Biol 2016;215:687–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cook Sangar ML, Genovesi LA, Nakamoto MW, Davis MJ, Knobluagh SE, Ji P, et al. Inhibition of CDK4/6 by Palbociclib Significantly Extends Survival in Medulloblastoma Patient-Derived Xenograft Mouse Models. Clin Cancer Res 2017;23:5802–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Whittaker S, Madani D, Joshi S, Chung SA, Johns T, Day B, et al. Combination of palbociclib and radiotherapy for glioblastoma. Cell Death Discov 2017;3:17033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nellan A, Rota C, Majzner R, Lester-McCully CM, Griesinger AM, Mulcahy Levy JM, et al. Durable regression of Medulloblastoma after regional and intravenous delivery of anti-HER2 chimeric antigen receptor T cells. J Immunother Cancer 2018;6:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fink MY, Chipuk JE. Survival of HER2-Positive Breast Cancer Cells: Receptor Signaling to Apoptotic Control Centers. Genes Cancer 2013;4:187–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu T, Kamikawa YF, Donohoe ME. Brd4’s Bromodomains Mediate Histone H3 Acetylation and Chromatin Remodeling in Pluripotent Cells through P300 and Brg1. Cell Rep 2018;25:1756–71 [DOI] [PubMed] [Google Scholar]
- 36.Ivanov DP, Coyle B, Walker DA, Grabowska AM. In vitro models of medulloblastoma: Choosing the right tool for the job. J Biotechnol 2016;236:10–25 [DOI] [PubMed] [Google Scholar]
- 37.Xu L, Chen Y, Mayakonda A, Koh L, Chong YK, Buckley DL, et al. Targetable BET proteins- and E2F1-dependent transcriptional program maintains the malignancy of glioblastoma. Proc Natl Acad Sci U S A 2018;115:E5086–E95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell 2011;146:904–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci U S A 2010;107:21931–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim KH, Roberts CW. Targeting EZH2 in cancer. Nat Med 2016;22:128–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yamaguchi H, Hung MC. Regulation and Role of EZH2 in Cancer. Cancer Res Treat 2014;46:209–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Piunti A, Hashizume R, Morgan MA, Bartom ET, Horbinski CM, Marshall SA, et al. Therapeutic targeting of polycomb and BET bromodomain proteins in diffuse intrinsic pontine gliomas. Nat Med 2017;23:493–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Michealraj KA, Kumar SA, Kim LJY, Cavalli FMG, Przelicki D, Wojcik JB, et al. Metabolic Regulation of the Epigenome Drives Lethal Infantile Ependymoma. Cell 2020;181:1329–45 e24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miao H, Li DQ, Mukherjee A, Guo H, Petty A, Cutter J, et al. EphA2 mediates ligand-dependent inhibition and ligand-independent promotion of cell migration and invasion via a reciprocal regulatory loop with Akt. Cancer Cell 2009;16:9–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang NY, Fernandez C, Richter M, Xiao Z, Valencia F, Tice DA, et al. Crosstalk of the EphA2 receptor with a serine/threonine phosphatase suppresses the Akt-mTORC1 pathway in cancer cells. Cell Signal 2011;23:201–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wagner J, Wickman E, DeRenzo C, Gottschalk S. CAR T Cell Therapy for Solid Tumors: Bright Future or Dark Reality? Mol Ther 2020;28:2320–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Donovan LK, Delaidelli A, Joseph SK, Bielamowicz K, Fousek K, Holgado BL, et al. Locoregional delivery of CAR T cells to the cerebrospinal fluid for treatment of metastatic medulloblastoma and ependymoma. Nat Med 2020;26:720–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
ChIP-seq data (20) are available in the SRA archives (SRP079643), accession number (GEO: GSE84761).