Abstract
The H1 linker histone family is the most abundant group of eukaryotic chromatin-binding proteins. However, their contribution to chromosome structure and function remains incompletely understood. Here we use single-molecule fluorescence and force microscopy to directly visualize the behavior of H1 on various nucleic acid and nucleosome substrates. We observe that H1 coalesces around single-stranded DNA generated from tension-induced DNA duplex melting. Using a droplet fusion assay controlled by optical tweezers, we find that single-stranded nucleic acids mediate the formation of gel-like H1 droplets, whereas H1:dsDNA and H1:nucleosome droplets are more liquid-like. Molecular dynamics simulations reveal that multivalent and transient engagement of H1 with unpaired DNA strands drives their enhanced phase separation. Using eGFP-tagged H1, we demonstrate that inducing single-stranded DNA accumulation in cells causes an increase in H1 puncta that are able to fuse. We further show that H1 and RPA occupy separate nuclear regions, but that H1 colocalizes with the replication factor PCNA, particularly after DNA damage. Overall, our results provide a refined perspective on the diverse roles of H1 in genome organization and maintenance, and suggest its involvement at stalled replication forks.
Introduction
H1 proteins are a key component of eukaryotic chromatin, binding to the nucleosome core particle and linker DNA at the entry and exit sites of the nucleosome1–5. This binding configuration underlies the well-established role of H1 in local and higher-order chromatin compaction6–9. In addition, H1 functions in a variety of other DNA-templated processes including transcriptional regulation and DNA damage response10–12, the mechanisms for which are less understood. Dysregulation of H1 expression has been linked to human diseases including cancer13,14. Previous data showed that H1 is not stably associated with interphase chromosomes15 and is highly dynamic inside the nucleus16. These results indicate a more complex and nuanced regulatory repertoire for H1 than a purely architectural factor. Nonetheless, dynamic H1-DNA interactions in a reconstituted biochemical setting have not been directly observed.
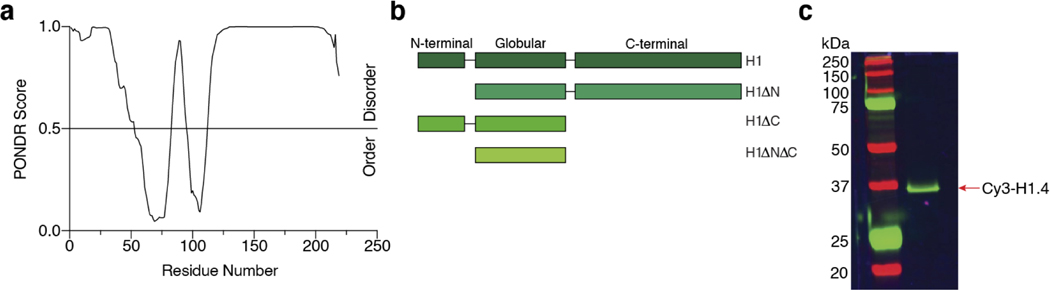
Higher eukaryotes contain multiple H1 subtypes, including eleven found in humans10. Each H1 protein contains a conserved globular domain flanked by a short disordered N-terminal domain (NTD) and a long unstructured C-terminal domain (CTD), with the lysine-rich CTD accounting for over 50% of the protein’s mass and two-thirds of its positive charge (Extended Data Fig. 1a, b). These intrinsically disordered regions (IDRs) indicate a potential for H1 to undergo liquid-liquid phase separation (LLPS), a phenomenon that has been implicated in myriad cellular processes17–19. Nucleic acids, mainly RNA, are known to modulate the viscoelastic properties of phase-separated protein droplets20,21. Differential phase separation with single-stranded (ss) and double-stranded (ds) DNA has also been observed for model peptides and proteins22–24. Indeed, recent studies reported that H1 can form condensates with DNA and nucleosome substrates under certain conditions25–28. However, the biophysical basis for H1 condensation remains unclear and no biological functions have been identified yet for these condensates. Here we present a comprehensive analysis demonstrating the differential phase separation of H1 with nucleic acid and chromatin substrates. We show that H1 specifically coalesces with single-stranded nucleic acids and readily forms phase-separated condensates exhibiting material properties distinct from those formed with dsDNA or nucleosomes. Finally, using both in vitro and cell-based assays, we provide evidence for potential involvement of H1:ssDNA interaction at stalled replication forks.
Results
H1 coalesces around slack but not taut ssDNA
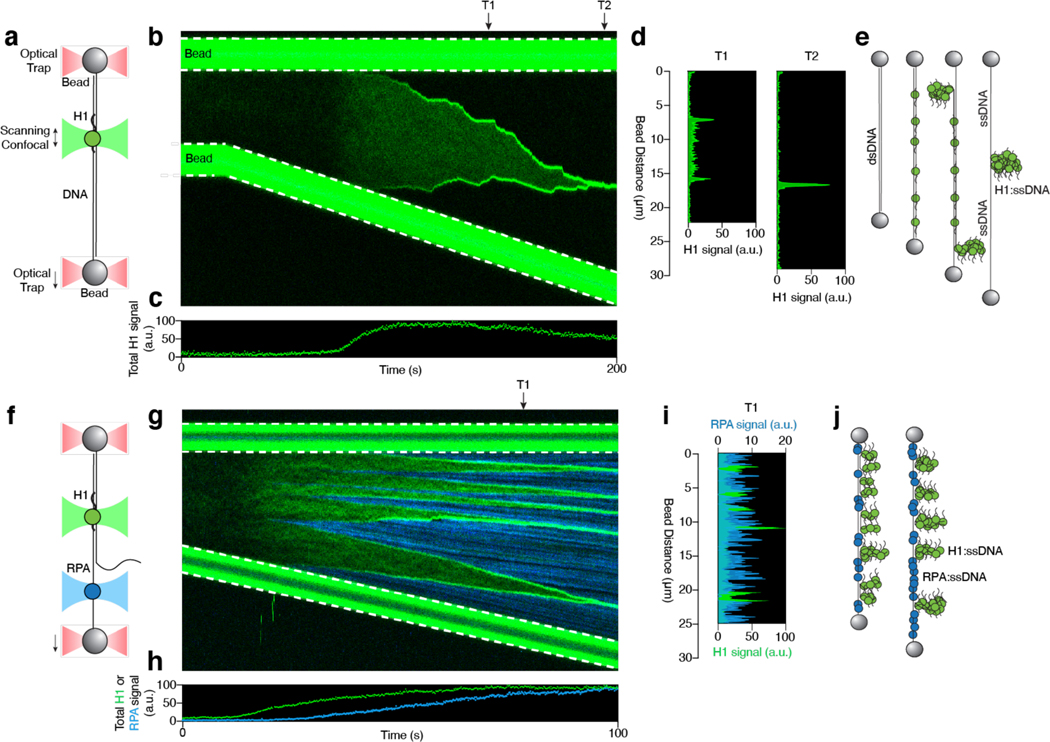
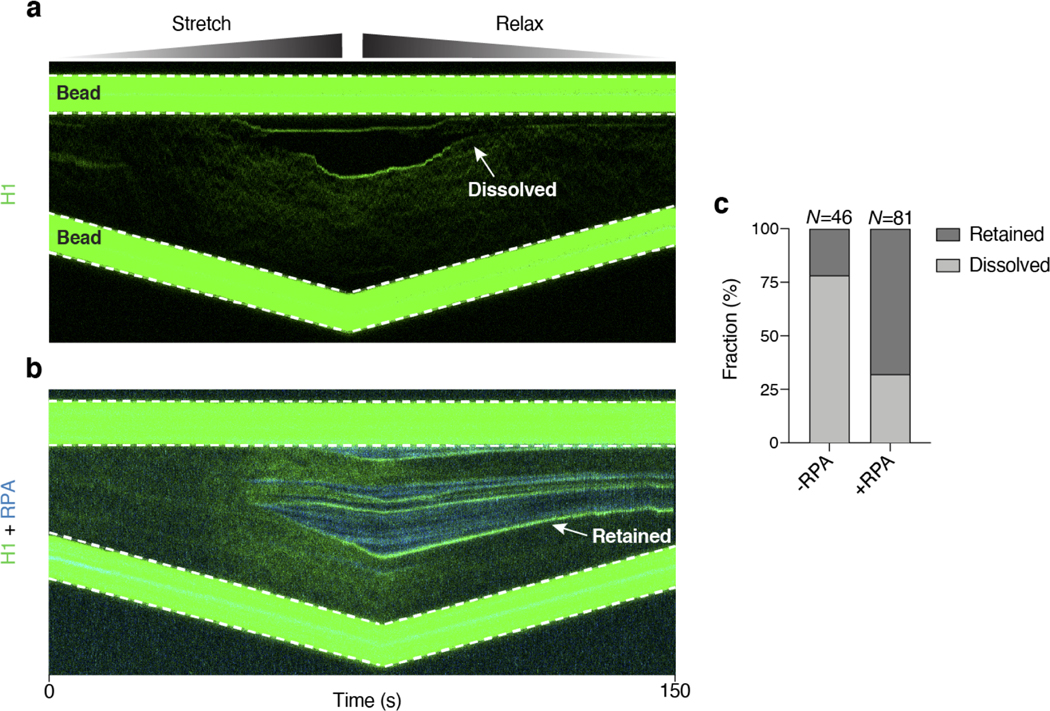
To directly visualize the behavior of H1 on DNA, we purified human H1.4 (hereafter referred to as H1), one of the major H1 subtypes in human cells, and labeled it with a Cy3 fluorophore (Extended Data Fig. 1c). We then added 15 nM of Cy3-H1 to a biotinylated phage λ genomic DNA [48.5 kilobasepair (kbp) in length] tethered between two laser-trapped beads. By moving the beads apart from each other, we applied an increasing force to the tether and monitored H1 binding along the DNA using scanning confocal microscopy (Fig. 1a). At forces where the dsDNA remained in B-form, we observed a low level of H1 binding (Fig. 1b). As the force approached the overstretching regime [> 60 picoNewton(pN)], H1 binding markedly intensified as evidenced by a dramatic increase in total Cy3 signal across the tether (Fig. 1c). Strikingly, H1 specifically accumulated at both ends of the tethered DNA as shown by the emergence of two fluorescent foci (“T1” in Fig. 1b, d). As the inter-bead distance continued to increase, the foci became brighter, migrating towards each other, and eventually merging into one singular spot (“T2” in Fig. 1b, d). Because the DNA was attached to the bead via the 3’ end of each strand, we posited that H1 preferentially binds to the untethered ssDNA created by force-induced unpeeling29 (Fig. 1e). Notably, H1 did not bind the other ssDNA strand that was attached to the bead (i.e. we detected no fluorescence signal between the H1 focus and its proximal bead) (Fig. 1b, e). Hence, under these experimental conditions, H1 does not bind ssDNA under tension. In other examples, we also observed H1 foci forming internally, presumably around relaxed ssDNA originating from the internal nicks naturally occurring within the DNA substrate (Extended Data Fig. 2a-c). In addition, we found this behavior to be unique to the linker histone, since core histones such as H2B exhibited force-insensitive binding and did not coalesce on ssDNA created by high tension (Extended Data Fig. 2d, e). This difference may be attributed to the fact that H1 proteins are structurally more disordered and have a higher net positive charge than the core histones10.
Fig. 1 |. H1 coalesces around nascent ssDNA.
a, Schematic of the combined single-molecule fluorescence and force microscopy. A biotinylated λ-DNA molecule (48.5 kbp) is tethered between two streptavidin-coated polystyrene beads. b, A representative kymograph of Cy3-H1 binding to DNA over time as the inter-bead distance was increased. c, Total H1 signal across the DNA as a function of time for the kymograph shown in b. d, Distribution of the H1 signal along the DNA at two specific time points (T1 and T2) as indicated by the arrows in b. e, Cartoon illustrating the distinct binding configurations of H1 on DNA under different tensions. ssDNA is created by force-induced unpeeling. f, Schematic of two-color imaging for simultaneous visualization of H1 and RPA binding to DNA. g, A representative kymograph of Cy3-H1 (green) and AlexaFluor488-RPA (blue) binding to DNA over time as the inter-bead distance was increased. h, Total H1 and RPA signals across the DNA as a function of time for the kymograph shown in g. i, Distribution of the H1 (green) and RPA (blue) signals along the DNA at a specific time point (T1) as indicated by the arrow in g. j, Cartoon illustrating that H1 and RPA occupy separate regions of the tethered DNA. H1 coalesces around relaxed ssDNA, whereas RPA binds to ssDNA under tension.
To confirm this interpretation, we used AlexaFluor488-labeled Replication Protein A (RPA), a well-studied eukaryotic ssDNA-binding protein that is capable of interacting with ssDNA under tension30 (Fig. 1f). Indeed, both H1 and RPA signals on DNA increased with tension (Fig. 1g, h), but with drastically distinct localization patterns. H1 foci were consistently observed on the flanks of RPA-bound regions of the tether in an anticorrelated manner (Fig. 1g, i). This finding suggests that H1 does not bind to ssDNA regions that are under tension, whereas RPA is excluded from H1 foci around the relaxed ssDNA (Fig. 1j). Next we examined the reversibility of H1 foci formation by sequentially stretching and relaxing the tether. Interestingly, we found that the majority of H1 foci dissolved upon tether relaxation (Extended Data Fig. 3a), presumably induced by the energetically favorable re-annealing of the two complementary ssDNA. In contrast, in the presence of RPA, most of the H1 foci persisted at low forces (Extended Data Fig. 3b, c), suggesting that RPA poses a barrier against the re-annealing of ssDNA and the dissolution of H1 foci. These results demonstrate that H1 coalesces with relaxed, untethered ssDNA, but not with ssDNA under tension.
H1 undergoes enhanced phase separation with ssDNA
Based on the single-molecule observations, we hypothesized that H1 forms phase-separated condensates with ssDNA. To test this hypothesis, we mixed full-length H1 with ssDNA of 75 nucleotides (nt) in length under physiological buffer conditions, which indeed yielded distinct phase-separated droplets (Supplementary Fig. 1a). We further validated the enrichment of H1 and ssDNA within the same droplets via fluorescence imaging using Cy3-labeled H1 and Cy5-labeled ssDNA (Fig. 2a). The phase separation propensity increased as a function of H1 and ssDNA concentrations, as shown by solution turbidity measurements (absorbance at 350 nm, A350) (Fig. 2b). Notably, the A350 value for H1 and ssDNA was greater than that of H1 and dsDNA across concentration regimes (Fig. 2b). We found that droplet formation required the presence of DNA but not other buffer components such as polyethylene glycol (PEG), which was used in low concentrations as a crowding agent to mimic the cellular environment (Supplementary Fig. 1a, b).
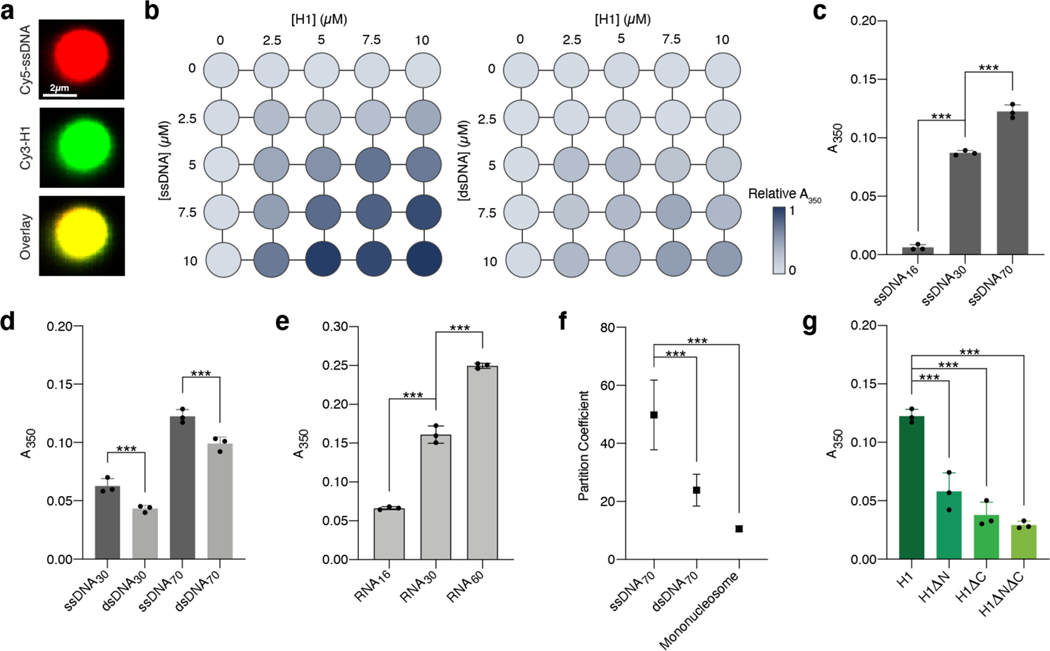
Fig. 2 |. H1 exhibits enhanced phase separation with single-stranded nucleic acids.
a, Fluorescence images of a representative droplet (among 20 independent droplets) formed by mixing 2.5 μM Cy3-H1 with 10 μM Cy5-ssDNA75 (10% labeled). b, Matrix diagram of solution turbidity values (A350) measured at different concentrations of H1 and either ssDNA75 (left) or dsDNA30 (right) (all unlabeled), normalized by the highest A350 value at 10 μM H1 and 10 μM ssDNA. c, A350 values for full-length H1 mixed with ssDNA of different lengths. ssDNA concentrations were normalized to yield the same total amount of nucleotides (44 μM ssDNA16, 23 μM ssDNA30, 10 μM ssDNA70). Significance calculated using a one-way ANOVA with Tukey’s test for multiple comparisons (***p<0.001). d, A350 values for H1 mixed with ssDNA or dsDNA of the same length and sequence. The concentrations of ss/ds DNA pairs were normalized to yield the same total amount of nucleotides (10 pM ssDNA30 / 5 μM dsDNA30; 10 pM ssDNA70 / 5 μM dsDNA70). Significance calculated using an unpaired t-test for each pair (***p<0.001). e, A350 values for H1 mixed with RNA of different lengths. RNA concentrations were normalized to yield the same total amount of nucleotides (44 μM RNA16, 23 μM RNA30, 12 μM RNA60). Significance calculated using a one-way ANOVA with Tukey’s test for multiple comparisons (***p<0.001). f, Partition coefficients determined for H1:Cy3-ssDNA70 (n=36 independent droplets), H1:Cy3-dsDNA70 (n=33 independent droplets), and H1:Cy3-H2A mononucleosome droplets (n=26 independent droplets). Data are presented as mean values ± SD. Significance calculated using a one-way ANOVA with Dunnett’s test for multiple comparisons (***p<0.001). g, A350 values for different H1 truncations mixed with 10 μM ssDNA70. Significance calculated using a one-way ANOVA with Dunnett’s test for multiple comparisons (***p<0.001). All experiments were performed with 2.5 μM H1 unless otherwise specified. All A350 experiments were performed with n=3 independent measurements. All data in bar charts are presented as mean values ± SD.
To examine how DNA length impacts the propensity of H1 :DNA phase separation, we used ssDNA ranging from 16 to 70 nucleotides (nt) and found that longer ssDNA substrates promote H1:ssDNA droplet formation (Fig. 2c). We then compared pairs of ssDNA and dsDNA of 30 or 70 nt/bp in length (ssDNA30 vs. dsDNA30; ssDNA70 vs. dsDNA70) after normalizing their molar concentrations for the total amount of nucleotides (Fig. 2d, Supplementary Fig. 1c). We found H1 to possess a significantly higher capacity to phase separate with ssDNA substrates than with dsDNA of the same length and sequence, in accordance with our single-molecule results. We also compared the propensity of H1 to phase separate with single-stranded RNA. Again, we observed that H1 has a greater tendency to phase separate with longer RNA molecules, a trend consistent with the DNA results (Fig. 2e). Moreover, H1 appears to phase separate more readily with RNA than with ssDNA of the same length (Fig. 2c, e).
We next examined how H1:ssDNA condensates differ from those formed between H1 and nucleosomal substrates, either mononucleosomes composed of a core histone octamer and 207 bp of DNA, or nucleosomal arrays. To confirm the previously reported H1-nucleosome interaction and phase separation26,27, we first utilized single-molecule fluorescence microscopy to observe H1:nucleosome colocalization and condensate formation on nucleosome-bound λ DNA (Extended Data Fig. 4a-d). We then performed solution turbidity measurements for H1 droplets formed with ssDNA, mononucleosomes, and 12-mer nucleosomal arrays. We found that bare ssDNA increased H1’s phase separation propensity compared to either of the nucleosomal substrates (Supplementary Fig. 1d). Additionally, we determined the partition coefficients of H1:ssDNA, H1:dsDNA, and H1:mononucleosome droplets and found that H1:ssDNA droplets yielded the highest value (Fig. 2f). These results suggest distinct properties of H1:ssDNA droplets compared with H1:dsDNA and H1:nucleosome droplets.
Given the known contributions of IDRs to H1 LLPS with dsDNA and mononucleosomes25,26, we truncated either NTD or CTD or both (the constructs were termed H1ΔN, H1ΔC, H1ΔNΔC, respectively; Extended Data Fig. 1b) and assessed the capacity of these truncated H1 proteins to phase separate with ssDNA. We found that H1:ssDNA phase separation was significantly impaired by deletion of the longer CTD and to a lesser extent, deletion of the NTD (Fig. 2g). When both tails were removed, the globular domain alone showed minimal phase separation with ssDNA (Fig. 2g). These results demonstrate that droplet formation between H1 and ssDNA is mediated by H1’s unstructured N- and C-terminal tails.
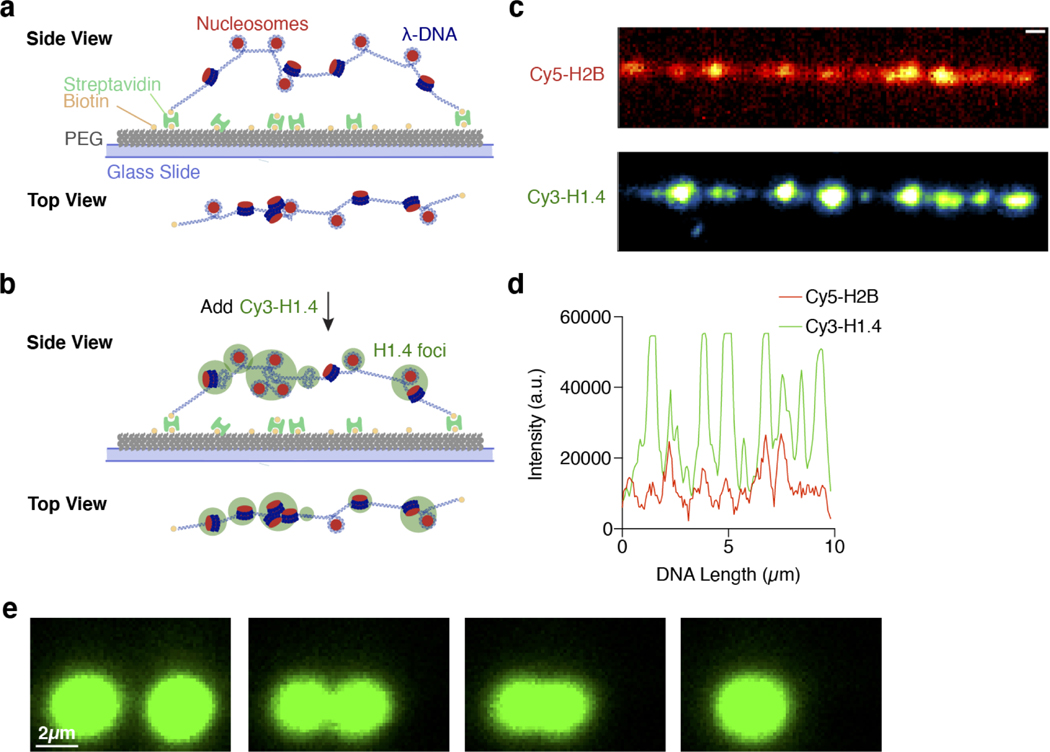
H1 droplets exhibit substrate-dependent material properties
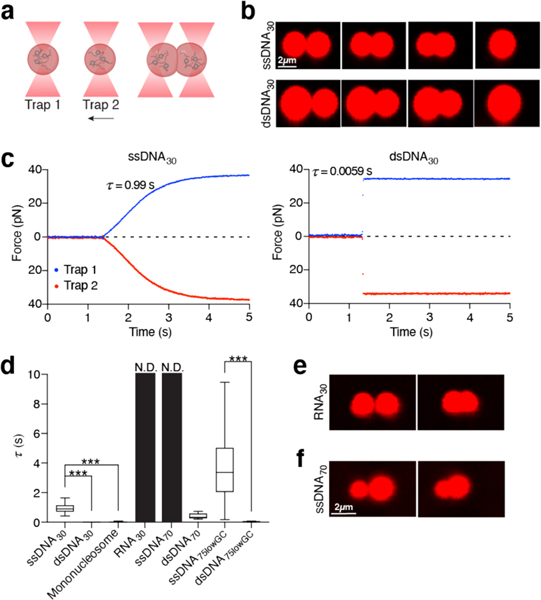
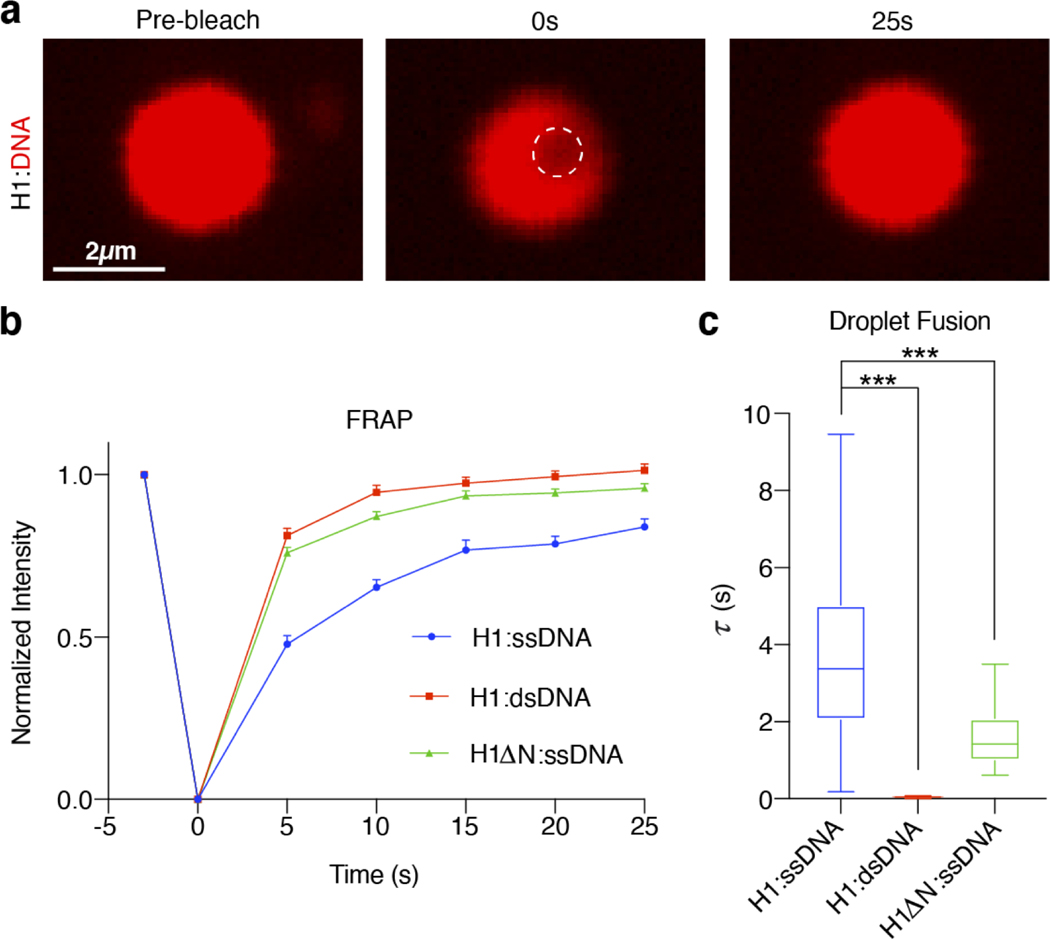
To further dissect the difference in physicochemical properties between H1:ssDNA droplets and those formed with either dsDNA or nucleosomes, we leveraged the ability of optical tweezers to trap and manipulate protein droplets due to their different refractive index relative to the surrounding medium31–33. We first generated H1:ssDNA droplets containing fluorescently labeled ssDNA for real-time visualization. Two micron-sized droplets were captured and brought into proximity by moving one trap towards the other (Fig. 3a). We observed that the two droplets eventually fused into a singular one after contact (Fig. 3b). Meanwhile, the force experienced by the droplets was recorded during the fusion process, displaying a gradual increase as the droplets were being pulled away from their respective trap center (Fig. 3c, Supplementary Fig. 2). When fusion was completed, the merged droplet was caught between two traps, yielding a steady-state force readout. This force measurement allowed us to quantify the timescale of the fusion process (τ), which lasted 0.96 ± 0.37 s for the H1:ssDNA30 droplets (Fig. 3d). We next performed the same droplet fusion experiment with H1:dsDNA30 droplets (Fig. 3b), and observed that they fused drastically faster (0.0046 ± 0.0013 s) than the ones containing ssDNA of the same length (Fig. 3c, d). Fusion of H1:mononucleosome droplets also occurred significantly faster (0.017 ± 0.0044 s) than H1:ssDNA droplets (Fig. 3d, Extended Data Fig. 4e). The marked differences in fusion kinetics suggest H1:ssDNA droplets are more viscous and gel-like, whereas H1:dsDNA and H1:mononucleosome droplets are more liquid-like. Notably, compared to ssDNA of the same length and sequence, H1:RNA droplets took much longer to fuse, if at all (Fig. 3d, e). These results reinforce our conclusion that H1 differentially coacervates with DNA, nucleosomes, and RNA. In addition, we conducted fluorescence recovery after photobleaching (FRAP) experiments, which also provide information about the fluidity of the droplets. We found that the recovery rate for H1:dsDNA droplets was faster than that for H1:ssDNA ones, again suggesting H1:dsDNA droplets are more dynamic and liquid-like (Extended Data Fig. 5).
Fig. 3 |. H1 droplets exhibit distinct material properties depending on the nucleic acid/chromatin substrate.
a, Schematic of the controlled droplet fusion assay with optical tweezers (droplet created with BioRender). b, Snapshots of a representative fusion event for H1:Cy5-ssDNA30 (top) (among 11 independent fusion events) and H1:Cy5-dsDNA30 (bottom) (among 35 independent fusion events) droplets visualized by Cy5 fluorescence. c, Force profiles of the two traps during a representative fusion event for H1:ssDNA30 (left) and H1:dsDNA30 (right) droplets. τ represents the droplet fusion time. d, τ values for H1:ssDNA30 (n=11), H1:dsDNA30 (n=35), H1:mononucleosome (n=21), H1:dsDNA70 (n=11), H1:ssDNA75lowGC (n=56), H1:dsDNA75lowGC (n=24) droplets. The τ value for dsDNA30 is too small to be visible (0.0046 ± 0.0013 s). τ values for RNA30 and ssDNA70 were nondetermined (N.D.) because these droplets did not fuse during our observation window (at least 20 s after contact). The top and bottom edges of each box represent the 3rd and 1st quartiles of the data, and the middle line in each box represents the median value. The top and bottom whiskers represent the maximum and minimum values. Significance calculated between H1:ssDNA30/H1:dsDNA30/H1:mononucleosome using a one-way ANOVA with Dunnett’s test for multiple comparisons (***p<0.001). Significance calculated between H1:ssDNA75lowGc/H1:dsDNA75lowGc using an unpaired t-test (***p<0.001). e and f, Representative images of two H1:Cy5-RNA30 droplets (e) (among 10 independent experiments) or two H1:Cy5-ssDNA70 droplets (f) (among 10 independent experiments) that were unable to fuse. All experiments were performed with 2.5 μM H1 and 10 μM DNA/RNA (10% labeled) or 1.8 μM mononucleosomes.
Next we used the droplet fusion assay to evaluate how DNA length and base composition affect the material properties of H1 condensates. We found that a longer ssDNA70 with the same GC content as ssDNA30 renders the droplets unable to fuse over the observation window (Fig. 3d, f). Conversely, lowering the GC content of the ssDNA fluidifies the droplets and accelerates the fusion process (compare ssDNA70 and ssDNA75lowGC in Fig. 3d). Deletion of the H1 NTD also makes the droplets more liquid-like, as evidenced by a faster FRAP rate for HlΔN-containing droplets (Extended Data Fig. 5). Deletion of the H1 CTD greatly diminished droplet formation and thus droplet fusion experiments for this construct were infeasible.
Extensive H1:dsDNA contacts suppress phase separation
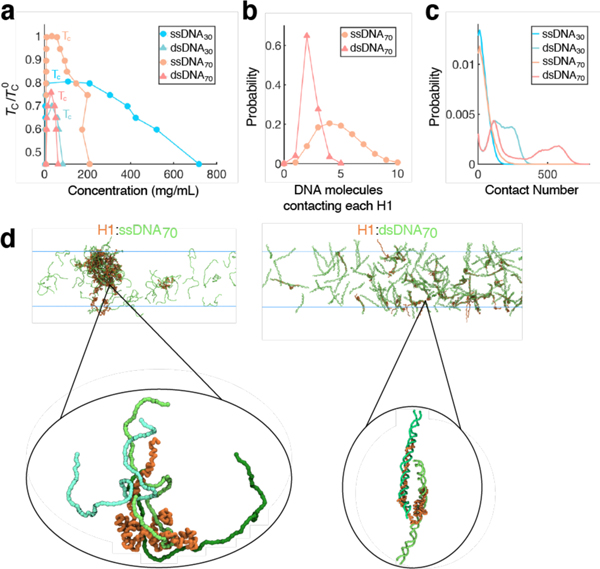
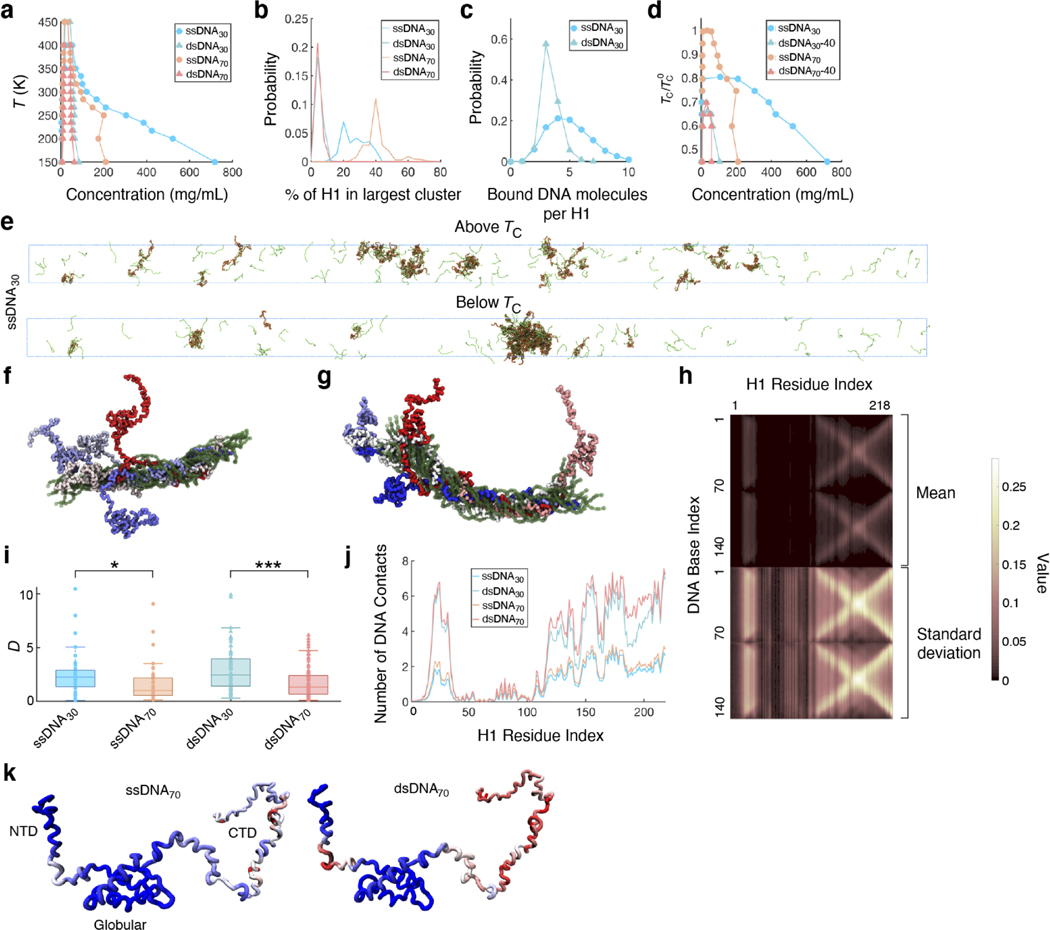
To obtain a detailed characterization of the internal organization of H1:DNA condensates, we carried out coarse-grained molecular dynamics (MD) simulations34,35. The force fields used in these simulations were fine-tuned to capture sequence-specific protein-protein interactions and the persistence lengths of ssDNA and dsDNA. The simulated critical temperatures (Tc) of phase separation in different systems match the A350 trends recorded experimentally (Fig. 4a, Extended Data Fig. 6a), supporting the usefulness of the in silico model for mechanistic exploration. Close examination of the simulated condensed phases reveals marked differences in their internal organization. H1 molecules coacervate with ssDNA into droplets at much higher densities than with dsDNA, corroborating our droplet fusion results; on the other hand, dsDNA molecules are more rigid, rendering them more resistant to dense packing (Fig. 4b-d, Extended Data Fig. 6b-e).
Fig. 4 |. Multivalency and interaction strength influence H1:DNA phase separation.
a,Computational phase diagrams in the temperature-concentration plane. For each system, we calculated the upper critical temperature for phase separation (TC). The displayed values are normalized by the TC with ssDNA70 (). b, Probability distribution of the number of DNA molecules contacting each H1 molecule. c, Probability distribution of the number of contacts formed between a single DNA molecule and a single H1 molecule. d, Example configurations of the condensates formed between H1 and ssDNA70 (left) or dsDNA70 (right). The insets highlight the different protein-DNA binding modes.
Simulation results suggest that H1 adopts distinct binding modes depending on the type and length of DNA. For example, consistent with their known high binding affinity25, H1 coils around dsDNA to form an extensive number of contacts (Fig. 4d). Notably, the H1:dsDNA complexes, while stable, remain highly dynamic, an observation that is in line with previous NMR measurements and results from the non-specific interactions between the two, similar to the H1:prothymosin-α complex36.It is noteworthy that H1 conformation varies significantly over time and across complexes (Extended Data Fig. 6f-h). The large number of contacts formed with individual dsDNA does limit the overall number of DNA molecules each H1 can bind. In contrast, contacts with individual ssDNA are sparser, and each H1 simultaneously engages more ssDNA than dsDNA molecules (Fig. 4b). Such multivalent interactions could explain ssDNA’s increased propensity to phase separate with H1 (ref. 37).Furthermore, longer DNA forms more contacts with H1 (Fig. 4c), yielding lower overall diffusion coefficients of H1 (Extended Data Fig. 6i). The relative affinity between binding partners is another known factor that dictates droplet properties38. Thus, the strength and multivalency of H1:DNA interactions collectively determine the observed phase separation trends. Lastly, in agreement with our experimental observations that the disordered tails of H1 control its phase separation, simulations also show that H1 interacts with DNA primarily through its CTD, although the NTD also contributes significantly (Extended Data Fig. 6j, k).
H1 forms phase-separated puncta in vivo
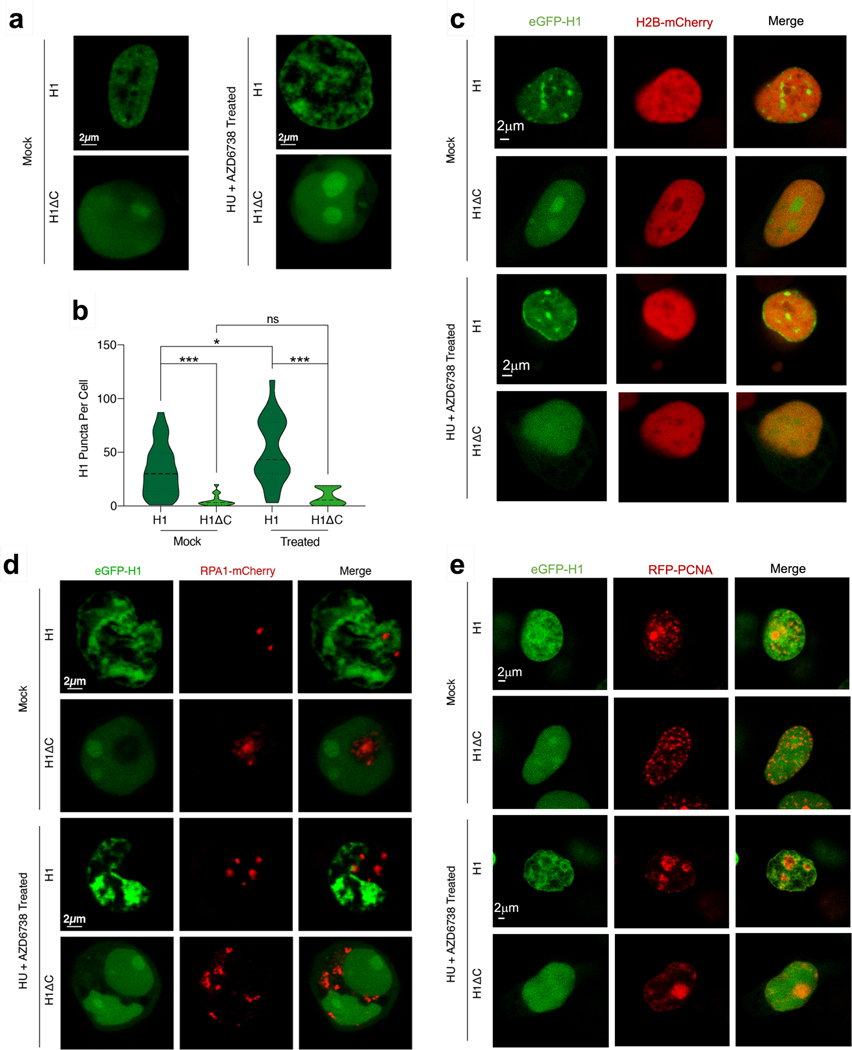
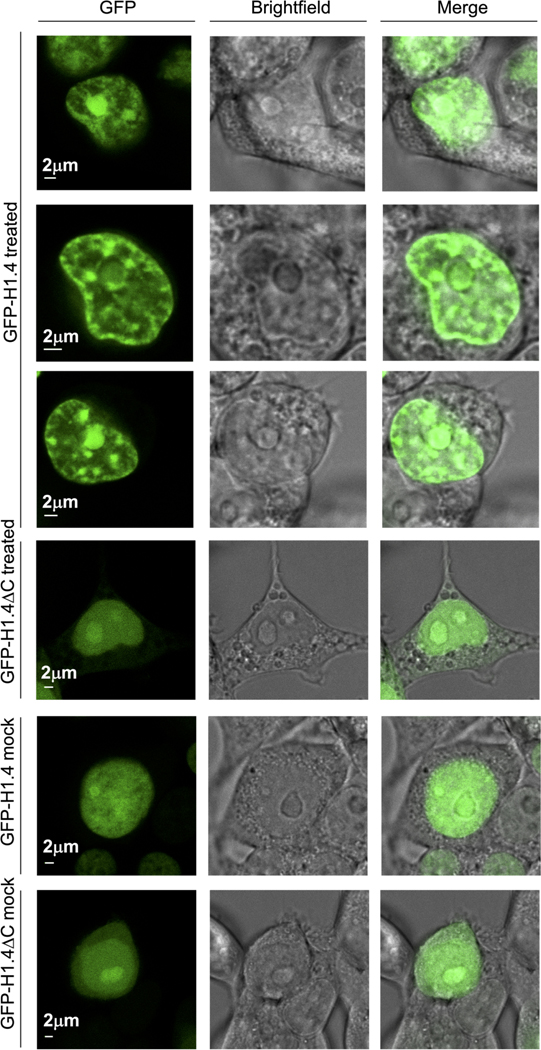
To observe the H1 localization pattern in a cellular context, we expressed H1.4 N-terminally tagged with eGFP (eGFP-H1)39,40 in Human Embryonic Kidney 293T (HEK293T) cells and performed live-cell confocal imaging. We confirmed that recombinant eGFP-tagged H1 can still phase separate with ssDNA and eGFP-H1:ssDNA droplets can fuse in vitro (Supplementary Fig. 3). Cells expressing full-length eGFP-H1 displayed multiple nuclear puncta (Fig. 5a), consistent with previously observed patterns27. In contrast, cells expressing eGFP-tagged CTD-deleted H1 (eGFP-H1ΔC) displayed a striking deficiency in puncta formation, instead exhibiting a diffused signal throughout the nucleus (Fig. 5a, b). Further, we co-transfected HEK293T cells with eGFP-H1 and H2B tagged with mCherry (H2B-mCherry) to demonstrate chromatin localization for our H1 construct. We found that the overall background of diffuse H1 and H2B signals co-existed over the entire nucleus. However, we observed additional bright eGFP-H1 puncta that were not reflected in the H2B-mCherry distribution (Fig. 5c, Supplementary Fig. 4, 5). This result suggests that although H1 is largely present on intact chromatin as expected, some puncta may be forming within unchromatinized regions of the DNA. We also overlaid the fluorescence signal from eGFP-H1 and eGFP-H1ΔC cells with brightfield images to determine whether the puncta corresponded to identifiable nuclear features. While each nucleus typically had a large region of accumulated H1 in the nucleolus, additional puncta were also observed in the rest of the nucleus (Extended Data Fig. 7).
Fig. 5 |. H1 puncta in the nucleus are CTD-dependent and show a distinct localization pattern.
a, Representative confocal fluorescence images of HEK293T cells transfected with either eGFP-H1 or eGFP-HlAC and treated with either mock or 2 mM HU + 20 μM AZD6738 for 12 h. eGFP-H1, mock (n=29 independent nuclei); eGFP-H1ΔC, mock (n=16); eGFP-H1, treated (n=22); eGFP-H1ΔC, treated (n=12). b, Violin plot showing the distribution of number of H1 puncta per cell from a for eGFP-H1 and eGFP-HlAC cells after mock or HU + AZD6738 treatment. Significance calculated using Welch’s t-test (*p<0.05, ***p<0.001). c-e, Representative confocal fluorescence images of HEK293T cells co-transfected with eGFP-H1/eGFP-H1ΔC and H2B-mCherry (c), RPA-mCherry (d), or RFP-PCNA (e) and treated with either mock or 2 mM HU + 20 μM AZD6738 (18-h DNA damage treatment for c and e, 12-h treatment for d). For c, eGFP-H1, mock (n=16 independent nuclei); eGFP-H1ΔC, mock (n=13); eGFP-H1, treated (n=20); eGFP-H1ΔC, treated (n=12). For d, eGFP-H1, mock (n=16); eGFP-H1ΔC, mock (n=12); eGFP-H1, treated (n=15); eGFP-H1ΔC, treated (n=12). For e, eGFP-H1, mock (n=11); eGFP-H1ΔC, mock (n=9); eGFP-H1, treated (n=14); eGFP-H1ΔC, treated (n=12).
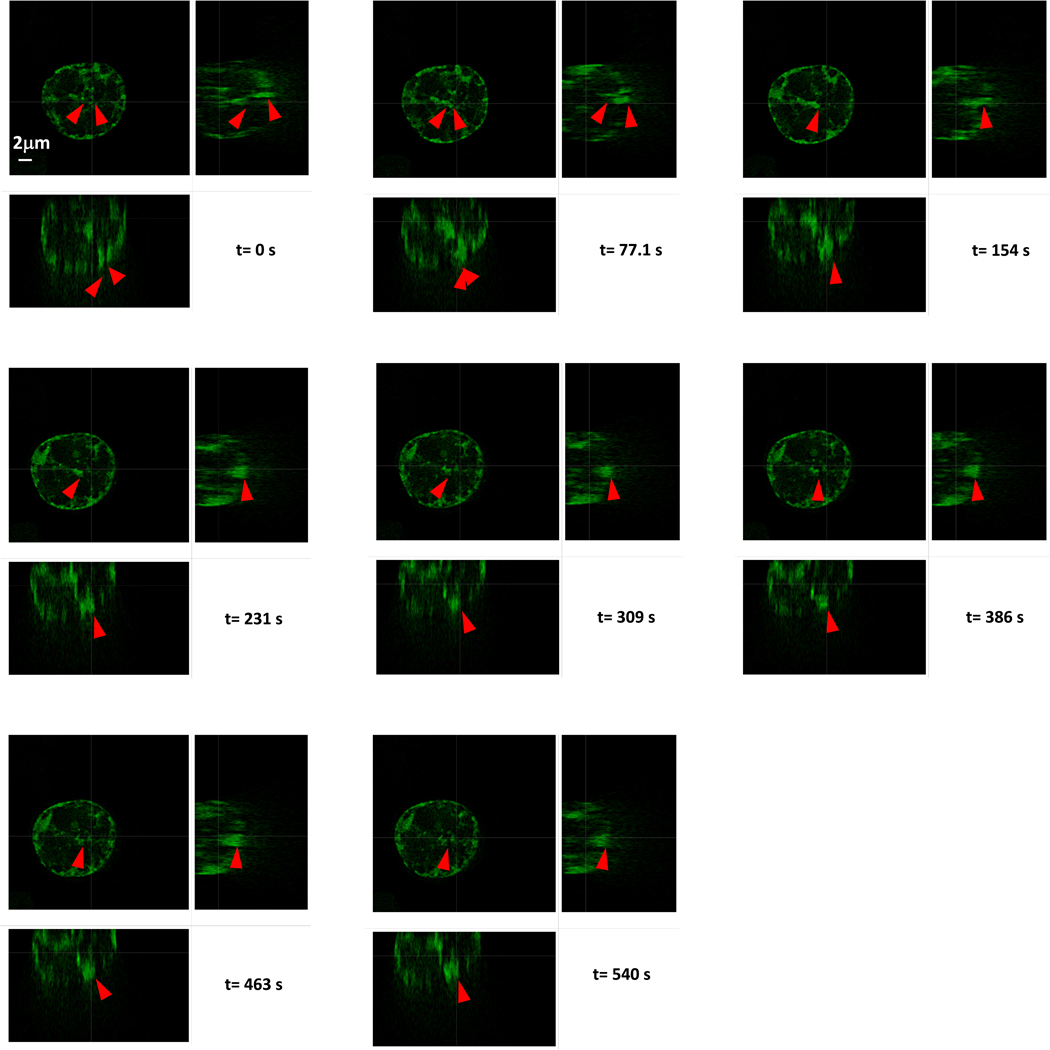
We next sought to verify whether these puncta represented H1-mediated LLPS. To this end, we treated eGFP-H1 and eGFP-H1ΔC transfected cells with both hydroxyurea (HU) and the ATR kinase inhibitor ceralasertib (AZD6738). HU induces replication stress and the accumulation of ssDNA in the cell, while AZD6738 impairs the DNA damage response to allow the ssDNA to persist for longer41,42. We then examined whether there was an increase in H1 coacervation in response to the treatment. After treatment with HU and AZD6738, we observed a significant increase in puncta formation in the eGFP-H1 cells only (Fig. 5a, b). Importantly, this increase was not observed in the eGFP-H1ΔC cells, supporting our in vitro observation that H1:ssDNA coalescence depends on the H1 CTD. Finally, to directly demonstrate LLPS in cells, we took continuous Z-stack images of the nuclei over 10-minute windows and observed frequent fusion events where two puncta merged in several planes and persisted as one for the remainder of the imaging window (Extended Data Fig. 8, Supplementary Fig. 6).
DNA damage-induced H1 puncta colocalize with PCNA
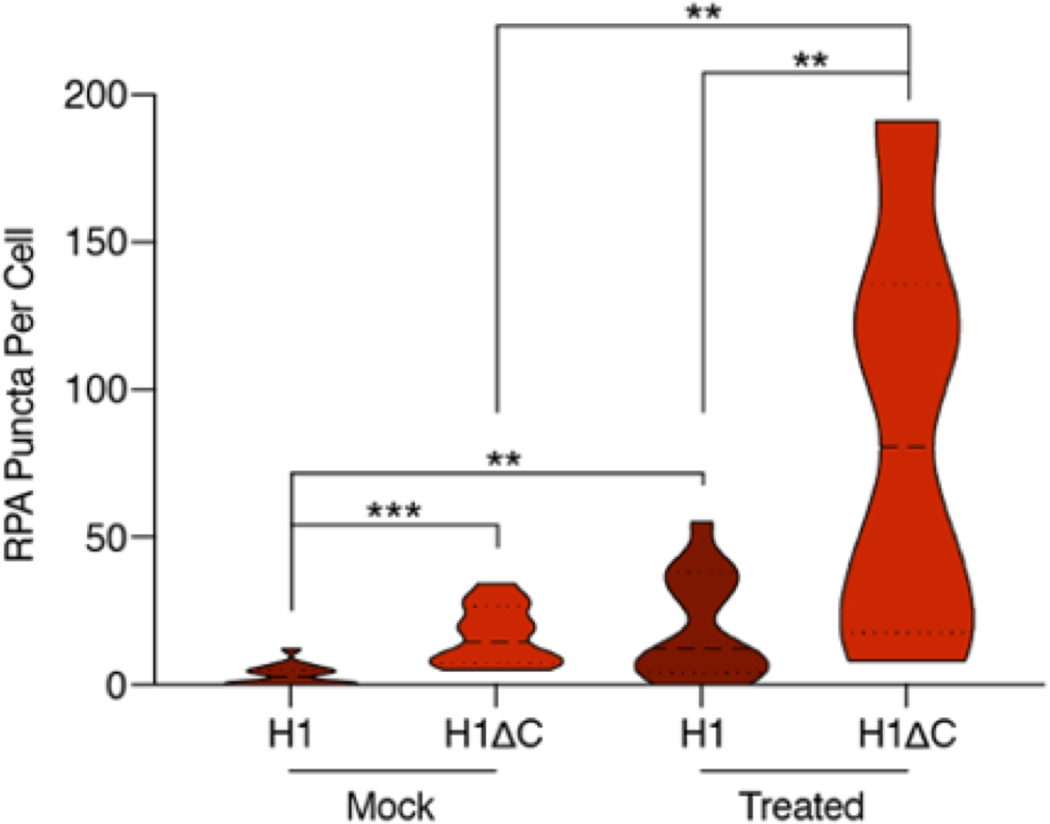
To better understand the contribution of ssDNA to H1 nuclear puncta, we co-transfected HEK293T cells with eGFP-H1 and RPA1 tagged with mCherry (RPA-mCherry). In agreement with our in vitro single-molecule results, H1 and RPA appeared to be localized to distinct nuclear regions (Fig. 5d). Notably, eGFP-H1ΔC cells contained significantly more RPA puncta than eGFP-H1 cells (Extended Data Fig. 9), indicating that the absence of H1:ssDNA condensates frees up additional ssDNA sites for RPA to bind. As expected, following treatment with HU and AZD6738, we detected a significant increase in the number of RPA puncta in both eGFP-H1 and eGFP-H1ΔC cells (Extended Data Fig. 9). The number of H1 puncta only increased in eGFP-H1 cells.
These results indicate that at least some of the puncta may be forming at the stalled replication forks caused by HU and AZD6738 treatment. To test this hypothesis, we co-transfected HEK293T cells with eGFP-H1 or eGFP-H1ΔC and RFP-tagged proliferating cell nuclear antigen (PCNA), an essential component of the replication machinery43. Our confocal microscopy data indeed revealed colocalization between H1 and PCNA (Fig. 5e, Supplementary Fig. 7, 8, 9). This result led us to examine H1 binding to a forked DNA substrate in vitro. We tethered the forked DNA substrate at low tension (~1 pN) in our single-molecule optical tweezers setup (Extended Data Fig. 10a). We observed strong Cy3-H1 signal at the fork junction and only minimal binding at the rest of the dsDNA tether (Extended Data Fig. 10b). Interestingly, when we stretched this substrate containing H1 at the fork, we observed no fork movement (Extended Data Fig. 10b). In contrast, the fork junction was unpeeled by force in the absence of H1 as indicated by the binding of AlexaFluor488-RPA (Extended Data Fig. 10c). Two-color experiments further supported that H1 shields the DNA fork against force-induced duplex separation (Extended Data Fig. 10d). These results demonstrate that H1 can accumulate on relaxed ssDNA proximal to DNA forks and suggest that H1:ssDNA condensation may stabilize the fork junction.
Discussion
In this work, we report that H1, a prominent and highly abundant group of proteins in eukaryotic cells, exhibits disparate propensities to phase separate with ssDNA, dsDNA, and chromatinized DNA. Single-stranded nucleic acids mediate enhanced H1 condensation, rendering the resulting droplets gel-like and slow to fuse. Our experimental results are corroborated by in silico modeling, which indicates more multivalent binding of H1 to ssDNA compared to dsDNA is what leads to the observed differences in the droplet properties. While affinity above a certain threshold is required for LLPS, very stable contacts— in which two polymers can only interact in a one-to-one fashion— turn out to suppress the multivalent interactions needed for phase separation44. As such, the condensation behavior of H1 is likely dependent on the relative abundance of certain DNA/chromatin species in the nucleus.
The ability of H1 to distinguish between different forms of nucleic acid and chromatin substrates present in the nucleus implies an expanded role for H1 in genome organization and maintenance beyond its known function in chromatin compaction. In this study, we observed that CTD-dependent H1 puncta became more prevalent with induced accumulation of ssDNA in cells. Based on our single-molecule data, while H1 occupies the majority of ssDNA under slack, only RPA can interact with ssDNA under tension, which could be exerted by force-generating motor proteins. These results, combined with our in vivo data demonstrating that H1 and RPA occupy separate nuclear regions, indicate that they serve distinct functions in genome maintenance. Moreover, we found that H1 accumulates around replication forks and colocalizes with PCNA. These data suggest H1 may protect replication forks from force-induced peeling and partition these sites from the rest of the genome. Given H1 is highly abundant, mobile, and known to contribute to LLPS in the nucleus, it may possess broader cellular functions including a role in DNA damage response mediated by ssDNA coacervation—a possibility illuminated by our results which merits future investigation. In accordance with this view, H1 has been shown to inhibit homologous recombination, and its depletion leads to altered sensitivity to DNA damage45,46.
Additionally, our results show that H1 readily phase separates with RNA, and H1:RNA condensates are distinguished by their gel- or solid-like behavior. We also observed accumulation of H1 in the nucleoli in addition to puncta formation. Given the prevalence of both coding and non-coding RNAs in the cell, H1:RNA condensates may serve important biological functions. For example, it is conceivable that the nucleolar localization of certain H1 subtypes is directed by their RNA interaction and condensation, which could be further modulated by cell-cycle-dependent posttranslational modifications such as phosphorylation that alters the protein’s charge47.
Finally, the human H1 subtypes diverge in homology primarily in their CTD composition. While these variants exhibit distinct abilities to bind and compact chromatin48, the extent of this functional variability remains controversial. In accordance with previous studies reporting the importance of the CTD in H1 LLPS with dsDNA and nucleosomes, our results provide unambiguous evidence that the CTD is also a key mediator of H1 condensation with single-stranded nucleic acids. It is therefore possible that the divergent CTDs of H1 subtypes may enhance or attenuate their ability to coalesce with and partition single-stranded nucleic acid substrates, resulting in distinct roles for each subtype in genome maintenance and damage response. The experimental and computational platforms developed in this study pave the way for a systematic interrogation of H1 subtypes, posttranslational modifications, and disease-associated mutations, which together constitute a highly regulated and multifaceted network of linker histones49.
Methods
Protein purification and labeling
All recombinant H1 proteins and mutants were purified as previously described with minor modifications50. Briefly, Rosetta DE3 cells expressing His-SUMO-H1.4-GyrA-His were induced with IPTG and expressed for 12 h before harvesting. After rod sonication, lysate was incubated with Ni-NTA beads (Bio-Rad) and eluted. The eluent was treated with recombinant Ulp-1 (1:100 w/v) and 500 mM β-mercaptoethanol, followed by the addition of solid urea to a final concentration of 6 M and the pH was adjusted to 9.0. This was then loaded onto a HiTrap SP cation exchange column (Cytiva) and subjected to a gradient of 100% H1 purification buffer A (6 M urea, 20 mM Tris pH 9.0, 200 mM NaCl) to 100% buffer B (6 M urea, 20 mM Tris pH 9.0, 1 M NaCl) of an AKTA FPLC system (GE Healthcare). Fractions containing full-length H1 were pooled and purified on a semi-preparative C18 HPLC column on a gradient of 0–70% buffer B and freeze-dried until use. H1 proteins were resuspended in H1 buffer (20 mM Tris pH 7.5, 200 mM NaCl) before use.
Recombinant H1.4A4C was purified as described above. Fluorescent labeling was performed based on a previously published protocol51. Briefly, H1.4A4C was dissolved in 4 mL of histone labeling buffer (6 M Guanidine, 20 mM Tris pH 7.5, 0.2 mM TCEP). Three molar equivalents of Cy3-maleimide (ApexBio) in DMF were added and mixed gently at room temperature followed by the addition of 1 mM β-mercaptoethanol to quench the reaction. The resultant mixture was purified on a semi-preparative reverse-phase C18 HPLC column on a gradient of 0—70% buffer B, and freeze-dried before resuspension in H1 buffer. Recombinant human H2BT49C was purified and labeled as previously described52. AlexaFluor488-RPA was prepared as previously described53.
Recombinant human histone octamers were purified and labelled as previously described52. In brief, core histones and their mutants were individually expressed in BL21 (DE3) cells, extracted from inclusion bodies, and purified under denaturing conditions using Q and SP ion exchange columns (GE Healthcare). H2AK12C and H4L50C were labeled with Cy3 maleimide (GE Healthcare) or LD655 maleimide (Lumidyne Technologies) respectively under denaturing conditions54. Octamers were reconstituted with equal ratio of each histone and purified by gel filtration as described previously55. Mononucleosomes and nucleosome arrays were assembled by salt gradient dialysis as described previously56.
Correlative single-molecule fluorescence and force assay of H1-DNA interactions
λ DNA preparation. To create a terminally biotinylated dsDNA template, the 12-base 5’ overhang on each end of genomic DNA from bacteriophage λ (48,502 bp; Roche) was filled in with a mixture of natural and biotinylated nucleotides by the exonuclease-deficient DNA polymerase I Klenow fragment (New England BioLabs). Reaction was conducted by incubating 10 nM λ DNA, 33 μM each of dGTP/dATP/biotin-11-dUTP/biotin-14-dCTP (Thermo Fisher), and 5 U Klenow in 1× NEB2 buffer at 37 °C for 45 min, followed by heat inactivation for 20 min at 75 °C. DNA was then ethanol precipitated overnight at −20 °C in 2.5× volume cold ethanol and 300 mM sodium acetate pH 5.2. Precipitated DNA was recovered by centrifugation at 20,000× g for 15 min at 4 °C. After removing the supernatant, the pellet was air-dried, resuspended in TE buffer (10 mM Tris-HCl pH 8.0, 1 mM EDTA) and stored at 4°C.
Forked substrate construction.
The λ DNA replication fork substrate containing a 5’-biotinylated fork at one end and a 5’-biotinylated duplex at the other was prepared essentially as described previously57. The replication fork structure was composed of synthetic oligonucleotides (IDT) and attached to λ DNA (New England BioLabs) by annealing λ DNA, ForkLead 172-mer, and ForkLag 95-mer with 5’ dual-biotins at a molar ratio of 1:10:60 in TE buffer plus 37.5 mM NaCl. The reaction was heated to 65 °C for 30 min then cooled slowly to room temperature over 2 h. Duplex DNA for ligation to the opposite end of λ was prepared by annealing FarLag 41-mer and FarLead 29-mer with 5’ dual-biotins at a molar ratio of 1:1.5 by the same process. Ligation was performed on λ-ForkLead-ForkLag with T4 DNA Ligase in 1× T4 DNA Ligase Buffer (New England BioLabs) at room temperature. After one hour of ligation of the replication fork to one end of λ DNA, the annealed FarLag-FarLead duplex was added to the reaction at a molar ratio of 1:100 (λ:FarLag) and ligation was continued for 3 h at room temperature. The reaction was stopped by adding 23 mM EDTA and ligase was heat inactivated by incubation at 65 °C for 20 min. The ligated λ DNA construct was then purified by gel filtration to separate excess oligos and un-ligated forks from λ DNA. The DNA mix was brought to >100 mM NaCl and loaded onto a 5-mL Sepharose 4B (GE Healthcare) column which was equilibrated and eluted in 40 mM Tris-HCl pH 8.0, 100 mM NaCl, and 1 mM EDTA. Elution fractions were analyzed by 1% agarose gel electrophoresis and fractions containing the desired construct were pooled and stored at 4 °C.
Oligonucleotide sequences (5’P = 5’ phosphate; * = phosphorothioate bond; 5’BB = 5’ dual biotin): ForkLead: 5’P-GGGCGGCGACCTACCGATGTGGTAGGAAGTGAGAATTGGAGAGTGTGTTTTTtttttttttttttttttttttttttttttttttttgaggaaagaatgttggtgagggttgggaaGTGGAAGGATGGGCTCGAGAGGTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTT*T*T*TForkLag: 5’BB-TTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTCACACTCTCCAATTCTCACTTCCTACCACATCGGTFarLead: 5’BB-AGAGACCTCAGTGCAATGGACGTAGGAAGFarLag: 5’P-AGGTCGCCGCCCCTTCCTACGTCCATTGCACTGAGGTCTCT
Single-molecule experiments.
Single-molecule experiments were performed at room temperature on a LUMICKS C-Trap instrument53. A computer-controlled stage enabled rapid movement of the optical traps within a microfluidic flow cell. Laminar flow separated channels 1–3, which were used to form DNA tethers between 3.23-μm streptavidin-coated polystyrene beads (Spherotech) held in traps with a stiffness of 0.6 pN/nm. Under constant flow, a single bead was caught in each trap in channel 1. The traps were then quickly moved to channel 2 containing the biotinylated DNA. By moving one trap against the direction of flow but toward the other trap, and vice versa, a DNA tether could be formed and detected via a change in the force-extension (F-x) curve. The traps were then moved to channel 3 containing only buffer, and the presence of a single DNA was verified by the F-x curve. Orthogonal channel 4 served as a protein loading and imaging channel. Flow was turned off during data acquisition. Force data were collected at 100 kHz. AlexaFluor488 and Cy3 fluorophores were excited by 488-nm and 532-nm laser lines, respectively. Kymographs were generated via a confocal line scan through the center of the two beads.
For H1 and RPA binding experiments, single DNA tethers were moved to channel 4 where they were incubated with 15 nM Cy3-H1. In all experiments using Cy3-H1, the inter-bead distance began at ~14 μm, and the DNA was stretched between the beads by moving the right trap at a constant velocity (0.1 μm/s) in the x-direction. In experiments with H1 and RPA, 10 nM AlexaFluor488-RPA was added simultaneously to channel 4. In experiments using a forked DNA substrate, Cy3-H1 was incubated with the tethered substrate in channel 4 and, when RPA was also used in the experiment, the tether with bound H1 was moved to channel 5 containing AlexaFluor488-RPA followed by tether stretching. In experiments with H2B, single DNA tethers were incubated with 15 nM Cy3-H2B and imaged as described for H1.
Data analysis.
Bead photon counts were removed from the kymographs using either manual removal or a local search method to maximize the sum over the size of the beads’ autofluorescence. The distance and confocal photon count measurements between these removed regions were then used to plot line scans of the kymographs. A photon count threshold value was used to define the foci points in the kymograph traces (110% of the maximum photon count from the dsDNA region of the kymograph). To analyze the reversibility of H1 foci formation, the lumicks.pylake Python package’s greedy line tracking algorithm was applied to define line traces in the regions where the DNA tether was being relaxed58,59. Line traces present in the first 20% of the region were analyzed. A trace was counted as dissolved if the trace ended before the last 20% of the region. Traces that were still present in the last 20% of the region were counted as retained.
TIRF experiments.
Imaging was conducted on a total-internal-reflection fluorescent microscope (Olympus IX83 cellTIRF). PEG slides were prepared as previously described52. The assembled flow chamber was infused with 20 μL of 0.2 mg/mL streptavidin (Thermo Fisher Scientific), incubated for 5 minutes, and washed with 250 μL of 50 mM Tris, 150 mM NaCl, and 0.0075% Tween pH 7.5 (T150 buffer). Biotinylated λ DNA (LUMICKS) was immobilized by slowly injecting a 10–20 pM solution at a volume of 40–80 μL over the course of 2 minutes followed by T150 buffer to wash away molecules that were not immobilized on the streptavidin surface. For the nucleosome experiments, we adopted a previously described protocol60. In brief, in situ nucleosome formation was achieved by flowing labeled Cy3-H2B histone octamer with NAP1 into the chamber followed by a 5-min incubation and wash step with T150 buffer. Next, a solution containing 150 pM of Cy3-H1.4, imaging buffer (T150 buffer, 4% (w/v) glucose, 1.5 mg/mL glucose oxidase, 0.072 mg/mL catalase, 2 mM Trolox), and 30 nM TOTO-3 was flowed into the microfluidic chamber for imaging.
Solution turbidity measurements
Phase separation experiments were performed in 20-μL volumes of 20 mM Tris-HCl pH 7.5, 200 mM NaCl, and 10% PEG8000, at a DNA concentration of 10 μM and an H1 concentration of 2.5 μM unless otherwise specified. Absorbance measurements were taken at 350 nm on a NanoDrop spectrophotometer (Thermo Scientific) after 10-min incubation at room temperature.
Droplet imaging and manipulation
Droplet imaging.
H1:nucleic acid droplets were formed as described above for the solution turbidity assay and injected into a home-made flow cell. Droplets were imaged on the C-Trap instrument either by brightfield microscopy or fluorescence microscopy using a 532-nm or 639-nm laser. For fluorescence recovery after photobleaching (FRAP) experiments, droplets were partially bleached (100% laser power) for 2 s, followed by 2D confocal scanning (10% laser power) every 5 s using a custom script titled “FRAP droplet imaging” (https://harbor.lumicks.com/single-script/3a796fac-dbb3-4fe1-8ce7-8b0cf8c25ad9). Images were uploaded to FIJI as a stack and analyzed by the FRAP Profiler plugin.
Partition coefficient determination.
Confocal image data were extracted from the .h5 files via the lumicks.pylake python library, and droplet masks were generated using the scipy python library. Briefly, Otsu thresholding of the image was followed by scipy.morphology.closing, scipy.morphology.remove_small_objects (min_size=6 pixels), and finally skimage.measure.label to define the mask for droplet regions. The partition coefficient was then calculated by extracting the mean intensity of each droplet region divided by the mean intensity of the background region of the image.
Controlled droplet fusion experiments.
Two droplets were captured by the 1064-nm infrared laser in the dual traps of C-Trap (5% laser power) and visualized by 2D confocal scanning every 5 s. One trap was manually stepped towards the other trap in 200-nm intervals after every image scan until the fusion process started. The force for each trap was recorded concurrently.
Data analysis.
The droplet fusion times (τ) were calculated by fitting sigmoidal curves to the force-time data over the time windows of droplet fusion. The fit equation used is , where F is the normalized magnitude of high-frequency force data (78 kHz) from the stationary optical trap, t is the time value of each force data point, a and b are the generalized fit parameters, and t1/2 is another fit parameter that approximates the time of half-maximum force. The fits were then used to calculate τ as defined by the time lag between 30% and 80% of the normalized force values.
Force and fluorescence data from the .h5 files generated from C-trap experiments were analyzed using tools in the lumicks.pylake Python library supplemented with other Python modules (Numpy, Matplotlib, Pandas)61 in a custom GUI Python script titled “C-Trap .h5 File Visualization GUI” (https://harbor.lumicks.com/single-script/c5b103a4-0804-4b06-95d3-20a08d65768f). This script was used to extract confocal images and fusion traces from droplet formation, fusion, and FRAP experiments.
MD simulations
Force field.
Coarse-grained, implicit-solvent molecular dynamics simulations were performed with one bead per protein residue or DNA base using the maximum entropy optimized force field (MOFF) for proteins62 and the molecular renormalization group coarse-graining model (MRG-CG) for DNA63. The MOFF force field was complemented with structure-based modeling potentials in the ordered domain of H1 to stabilize its tertiary structure64. The strength of the structure-based potentials was tuned to reproduce the root-mean-square fluctuations from all-atom simulations. The original DNA model was parameterized with explicit ions. We rescaled its bonded, angle, and fan interactions by a factor of 0.9 to reproduce the persistence lengths of ssDNA and dsDNA in simulations with implicit ions. Electrostatic interactions within and between protein and DNA molecules were described using the Debye-Huckel potential and a distance-dependent dielectric constant62. A salt concentration of 150 mM was used in simulations to account for the uneven partition of salts in complex coacervation, with lower values in the condensed phase65. In addition to electrostatic interactions, an excluded volume term of was introduced to prevent overlap between protein and DNA molecules, where r is the distance between two beads and ϵ0 = 1.6264 × 10−3 kJ mol−1 nm12. All simulations were performed using the GROMACS simulation package66.
Slab simulation methodology.
Simulations with a ratio of 4DNA:1H1 were performed following the slab methodology to determine critical temperatures62,67. The ssDNA simulations included 160 DNA molecules and 40 H1 molecules, while those with dsDNA were performed with 320 DNA molecules and 80 H1 molecules. To check for finite-size effects, we also performed simulations with 160 DNA molecules and 40 H1 molecules for dsDNA and obtained similar results. For each setup, the molecules were initially placed in a large simulation box of size 100 nm × 100 nm × 100 nm. We then performed the steepest descent energy minimization, followed by a 0.1 μs long NPT simulation at 150 K and 1 bar using a Parrinello-Rahman isotropic barostat and time coupling constant of 1 ps. The NPT simulation collapsed the molecules into a dense phase. The z-dimension of the simulation box was then expanded by ~20 times the original size, resulting in a droplet with a dilute phase on either side. We then performed an NVT simulation for 0.1 μs with a time coupling constant of 100 ps. During the simulation, we raised the temperature from 150 K to the desired value. The resulting equilibrated system was run for 2 μs in the NVT ensemble. Configurations collected at every 1-ns steps from the second half of the trajectories were used for analysis.
Data analysis.
For each simulated configuration, we first computed a molecular contact matrix. Contacts between two molecules, protein or DNA, were determined if any of their particles are within 1 nm.Using a depth-first search algorithm over the network constructed from the contact matrix, we identified the largest cluster. The high and low densities at a given temperature were determined using molecules whose centers of mass were in regions within 2.5 nm of the largest cluster or 150 nm or more away from it. These densities were used to obtain the critical temperature (TC) by fitting to the analytical expression , where (β = 0.325 is the universality class of a 3D Ising model. Unlike the analytical Ising model, which assumes a homogeneous system, our system is highly heterogeneous, with strong H1:DNA interactions and substantially weaker H1:H1 and DNA:DNA interactions. As such, we note that there is additional uncertainty in TC arising from the sensitivity in density estimation. In particular, while TC was predicted to be less than 300 K for some systems, large clusters with a significant percentage of H1 are prevalent. Therefore, we used simulation data obtained at 300 K to analyze protein-DNA binding and diffusion coefficients. MDAnalysis was used to help with analysis68.
Live-cell imaging
General.
Live-cell imaging was conducted on a Leica SP8 confocal microscope with an inverted stand. HEK293T cells (ATCC, CRL-3216) were cultured in Dulbecco’s modified Eagle’s medium with 10% fetal bovine serum and 4 mM glutamine at 37 °C in 5% CO2. For experiments investigating replication stress, cells were subjected to either mock (1% PBS and 0.1% DMSO, v/v) or treatment conditions. Treatment involved exposure to 2 mM hydroxyurea (Sigma-Aldrich) and 20 μM ceralasertib (MedChemExpress) for 12–18 h before imaging. At least ten cells were measured for each experimental condition. Puncta were counted using the 3D Object Counter plugin in FIJI version 2.0.0 (ref. 69).Images were manually thresholded with a size filter minimum of 10 voxels.
Colocalization experiments.
HEK293T cells were transfected with either N-terminally eGFP-tagged H1.4 (eGFP-H1) or the N-terminally eGFP-tagged H1.4 C-terminal deletion mutant (eGFP-H1ΔC) alone or in combination with C-terminally mCherry-tagged 70-kDa subunit of the heterotrimeric RPA (RPA-mCherry), N-terminally mtagRFP-tagged PCNA (RFP-PCNA), or C-terminally mCherry-tagged H2B (H2B-mCherry) using Lipofectamine 2000 (Invitrogen) according to manufacturer’s protocols. Cells were transfected 18 h prior to DNA damage or mock treatment. Puncta were counted using the 3D Object Counter plugin in FIJI version 2.0.0. Images were manually thresholded with a size filter minimum of 10 voxels. Colocalization coefficients were measured using the Coloc2 plugin in FIJI version 2.0.0.
Z-stack timelapse imaging.
HEK293T cells were transfected with either N-terminally eGFP-tagged H1.4 (eGFP-H1) or the N-terminally eGFP-tagged H1.4 C-terminal deletion mutant (eGFP-H1ΔC), and treated for 18 h before imaging. Continuous Z-stack images through single nuclei were taken over the course of 10 min. Data were visualized in Imaris version 9.7 and slices in a single X, Y and Z plane were then taken to give the timelapse images.
Data availability
Statistical source data for Figures 1–5 and Extended Data Figures 2-6, 9 and the unprocessed gel image for Extended Data Figure 1c are provided with this paper. Other data are available upon reasonable request.
Code availability
All specified scripts used to run C-Trap experiments or analyze their results can be accessed on LUMICKS Harbor (harbor.lumicks.com). All custom-written codes will be made available upon request.
Statistics and reproducibility
Statistical tests and p values are reported in figure legends. All experiments were independently repeated at least three times with similar results. Representative results are shown in figures.
Extended Data
Extended Data Fig. 1. H1 purification and labeling.
a, Predictor of natural disordered regions (PONDR) score for the H1.4 amino acid sequence (www.pondr.com). A score of >0.5 is considered intrinsically disordered. b, Schematic of the domain structures of full-length H1 and NTD/CTD-truncated H1 constructs. c, Representative SDS-PAGE gel scanned for fluorescence showing purified Cy3-labeled H1.4 (among 5 independent preparations).
Extended Data Fig. 2. H1 and core histones respond differently to force applied to the DNA tether.
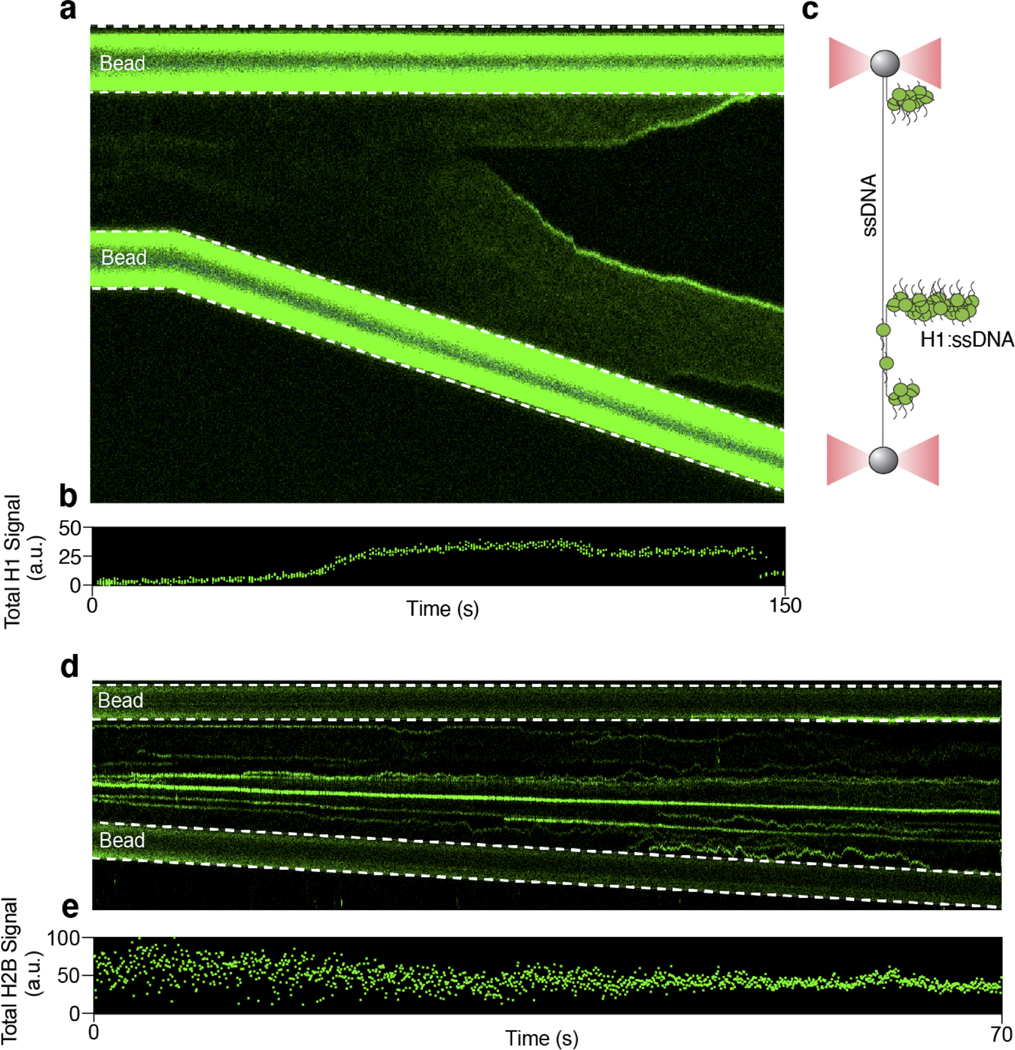
a, A kymograph showing Cy3-H1 binding to λ-DNA over time as the inter-bead distance was increased. b, Total Cy3 intensity across the DNA tether over time for the kymograph shown in a. c, Schematic of the final H1 binding configuration for the example shown in a. d, A representative kymograph showing Cy3-H2B binding to λ-DNA over time as the inter-bead distance was increased. e, Total Cy3 intensity across the DNA tether over time for the kymograph shown in d.
Extended Data Fig. 3. Reversibility of H1:ssDNA foci formed on tethered DNA.
a, A representative kymograph showing reversible formation and dissolution of Cy3-H1 foci during DNA tether stretching and relaxation. b, A representative kymograph showing the persistence of Cy3-H1 foci after tether relaxation in the presence of AlexaFluor488-RPA. c, Fraction of H1 foci dissolved versus retained in the absence or presence of RPA.
Extended Data Fig. 4. H1 colocalizes and forms condensates with nucleosomes.
a, Schematic of the total-internal-reflection fluorescence (TIRF) microscopy assay using surface-immobilized λ DNA loaded with Cy5-H2B nucleosomes. b, Schematic of the experimental setup in a after Cy3-H1 is added to the flow chamber. c, Representative fluorescence images (among 3 independentexperiments) of Cy5-H2B nucleosomes (top) and Cy3-H1 (bottom) on λ DNA. Scale bar: 0.5 μm. d, Fluorescence intensity profiles of Cy5-H2B and Cy3-H1 over the DNA length for the images in c. e, Snapshots of a representative fusion event for H1:Cy3-H2A mononucleosome droplets visualized by Cy3 fluorescence (among 21 independent fusion events).
Extended Data Fig. 5. Additional analyses of the biophysical properties of H1:DNA droplets.
a, A representative series of images (among 28 independent experiments) during the photobleaching and fluorescence recovery of an H1:Cy5-ssDNA75 droplet. b, Kinetics of fluorescence recovery for H1:Cy5-ssDNA75 (blue) (n=28), H1:Cy5-dsDNA75 (red) (n=15), and H1AN:ssDNA75 (green) (n=17) droplets. Data are presented as mean values ± SEM. c, Droplet fusion time (τ) for H1:Cy5-ssDNA75 (n=56), H1:Cy5-dsDNA75 (n=24), and H1AN:ssDNA75 (n=15) droplets. The top and bottom edges of each box represent the 3rd and 1st quartiles of the data, and the middle line in each box represents the median value. The top and bottom whiskers represent the maximum and minimum values. Significance calculated using a one-way ANOVA with Dunnett’s test for multiple comparisons (***p<0.001).
Extended Data Fig. 6. Computational examination of H1:DNA phase separation.
a, Phase behavior of the four simulated systems (H1:ssDNA30, H1:dsDNA30, H1:ssDNA70, H1:dsDNA70) as a function of temperature. b, Probability distribution of the percentage of H1 molecules found in the largest cluster at a temperature of 300 K. c, Probability distribution of the number of DNA molecules bound to each H1 molecule. d, Test of finite size effects on the computational phase diagrams in the temperature-concentration plane. For dsDNA, smaller systems with 40 H1 and 160 DNA molecules were used, but the trends shown in Fig. 3a are conserved. e, Representative configurations for the ssDNA30 system above and below TC. f, A single H1:dsDNA70 pair at different time points in our simulation. g, Different pairs ofH1:dsDNA70 at one time point. h, Contact map of bound H1:DNA pairs. Data include all H1:DNA pairs that have at least one residue in contact. The top half of the matrix represents the mean values of the fraction of time each residue-nucleotide pair is in contact, given that the H1 and DNA molecules are in contact. The bottom half represents the standard deviation of this matrix across time and ensemble. The significantly larger standard deviations relative to the mean values support the conformational heterogeneity of H1:dsDNA complexes. i, Diffusion coefficient (D) of H1 in each system at 300 K. All values were normalized by the median D for ssDNA70. Each data point represents the D value for an individual H1 molecule over the course of our simulation (n=40 for ssDNA, n=80 for dsDNA). Significance determined using a two-sample t-test (*p<0.05, ***p<0.001). The top and bottom edges of each box represent the 3rd and 1st quartiles of the data respectively, and the middle line indicates the median value. Whiskers extend to the minimum and maximum values of the data set that are within 2.7σ, where σ is the standard deviation. j, Average number of DNA residues in contact with any given H1 residue as a function of the H1 residue index. k, Average number of DNA residues in contact with any given H1 residue projected onto a structural model of H1. Data range from most contacts (red) to fewest contacts (blue).
Extended Data Fig. 7. Additional live-cell images of eGFP-H1 and eGFP-H1DC.
Representative confocal fluorescence images of HEK293T cells transfected with either eGFP-H1 or eGFP-H1ΔC and treated with either mock or 2 mM HU + 20 μM AZD6738 for 18h, and their corresponding brightfield images (among 10 independent nuclei imaged for each condition).
Extended Data Fig. 8. Timepoints from continuous Z-stack monitoring for merging.
Representative slices from continuous Z-stack imaging over 10 minutes of HEK293T cells transfected with eGFP-H1 and treated with 2 mM HU + 20 μM AZD6738 for 18h(among 6 imaging acquisitions on independent nuclei). For each timepoint, top left=x plane, top right=y plane, bottom left=z plane.
Extended Data Fig. 9. Quantification of RPA puncta in the nucleus.
Violin plot showing the distribution of number of RPA puncta per cell for eGFP-H1 and eGFP-H1ΔC transfected cells after mock or HU + AZD6738 treatment. Significance calculated using Welch’s t-test (**p<0.01, ***p<0.001). eGFP-H1, mock (n=16 independent nuclei), eGFP-H1ΔC, mock (n=12), eGFP-H1, treated (n=15), eGFP-H1ΔC, treated (n=12).
Extended Data Fig. 10. H1 interaction with forked DNA in vitro.
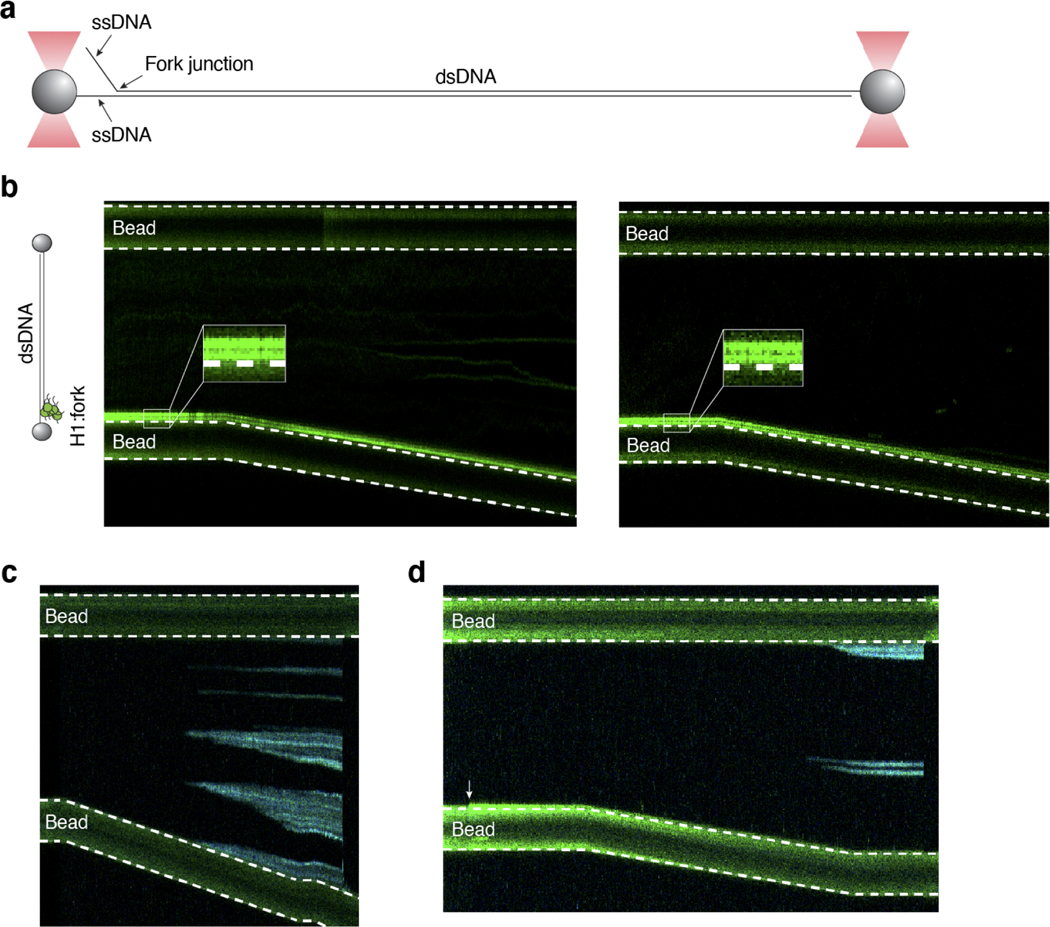
a, Schematic of a tethered DNA substrate containing a fork junction near one end of the tether. b, Two representative kymographs showing Cy3-H1 coalescing with relaxed ssDNA towards the fork junction near the bead. A schematic of the H1 binding configuration is shown on the left. Imaging was performed with green laser on. c, A representative kymograph showing AlexaFluor488-RPA binding to ssDNA regions formed by unpeeling from tethered ends and stochastically occurring internal nicks or by melting of dsDNA as the inter-bead distance was increased. Imaging was performed with blue laser on. d, A representative kymograph obtained with Cy3-H1 and AlexaFluor488-RPA showing that H1:ssDNA condensate (white arrow) prevents further force-induced ssDNA unpeeling at the fork junction, which would result in an expansion of RPA-bound ssDNA near the fork region. Imaging was performed with both green and blue lasers on.
Supplementary Material
Acknowledgements
We thank the O’Donnell laboratory (Rockefeller University) for RPA, Alexey Soshnev (Rockefeller) for discussions, and Murray Tipping and Yevgeniy Romin (MSK Molecular Cytology core facility) for advice with live-cell imaging and analysis. A.O. and A.L. are supported by the National Science Foundation Graduate Research Fellowship. T.N. is supported by a Medical Scientist Training Program grant from the National Institute of General Medical Sciences of the National Institutes of Health (NIH) under award number T32GM007739 to the Weill Cornell/Rockefeller/Sloan Kettering Tri-Institutional MD/PhD Program. B.Z. is supported by the National Institutes of Health grant R35GM133580. Y.D. is supported by NIH grant R35GM138386, CCSG core grant P30CA008748, the Mr. William H. Goodwin and Mrs. Alice Goodwin and the Commonwealth Foundation for Cancer Research and the Center for Experimental Therapeutics at MSKCC, the Parker Institute for Cancer Immunotherapy and the Anna Fuller Cancer Research Foundation. S.L. is supported by the Robertson Foundation, the Alfred P. Sloan 327 Foundation, and an NIH Director’s New Innovator Award (DP2HG010510). Y.D. and S.L. also acknowledge support from the Pershing Square Sohn Cancer Research Alliance. Y.D. is a Josie Robertson Young Investigator.
Footnotes
Competing interests statement
The authors declare no competing interests.
References
- 1.Osunsade A. et al. A Robust Method for the Purification and Characterization of Recombinant Human Histone H1 Variants. Biochemistry 58, 171–176 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fierz B. et al. Histone H2B ubiquitylation disrupts local and higher-order chromatin compaction. Nat Chem Biol 7, 113–9 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li S, Zheng EB, Zhao L. & Liu S. Nonreciprocal and Conditional Cooperativity Directs the Pioneer Activity of Pluripotency Transcription Factors. Cell Rep 28, 2689–2703 e4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wasserman MR, Schauer GD, O’Donnell ME & Liu S. Replication Fork Activation Is Enabled by a Single-Stranded DNA Gate in CMG Helicase. Cell 178, 600–611 e16 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harada BT et al. Stepwise nucleosome translocation by RSC remodeling complexes. Elife 5(2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Luger K, Rechsteiner TJ & Richmond TJ Preparation of nucleosome core particle from recombinant histones. Methods Enzymol 304, 3–19 (1999). [DOI] [PubMed] [Google Scholar]
- 7.Lee KM & Narlikar G. Assembly of nucleosomal templates by salt dialysis. Curr Protoc Mol Biol Chapter 21, Unit 21 6 (2001). [DOI] [PubMed]
- 8.Schauer G, Finkelstein J. & O’Donnell M. In vitro Assays for Eukaryotic Leading/Lagging Strand DNA Replication. Bio Protoc 7(2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sbalzarini IF & Koumoutsakos P. Feature point tracking and trajectory analysis for video imaging in cell biology. J Struct Biol 151, 182–95 (2005). [DOI] [PubMed] [Google Scholar]
- 10.Mangeol P, Prevo B. & Peterman EJ KymographClear and KymographDirect: two tools for the automated quantitative analysis of molecular and cellular dynamics using kymographs. Mol Biol Cell 27, 1948–57 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crickard JB, Moevus CJ, Kwon Y, Sung P. & Greene EC Rad54 Drives ATP Hydrolysis-Dependent DNA Sequence Alignment during Homologous Recombination. Cell 181, 1380–1394 e18 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harris CR et al. Array programming with NumPy. Nature 585, 357–362 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Latham AP & Zhang B. Consistent Force Field Captures Homologue-Resolved HP1 Phase Separation. J Chem Theory Comput (2021). [DOI] [PMC free article] [PubMed]
- 14.Savelyev A. & Papoian GA Chemically accurate coarse graining of double-stranded DNA. Proc Natl Acad Sci U S A 107, 20340–5 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Woodcock CL, Skoultchi AI & Fan Y. Role of linker histone in chromatin structure and function: H1 stoichiometry and nucleosome repeat length. Chromosome Res 14, 17–25 (2006). [DOI] [PubMed] [Google Scholar]
- 16.Song F. et al. Cryo-EM study of the chromatin fiber reveals a double helix twisted by tetranucleosomal units. Science 344, 376–80 (2014). [DOI] [PubMed] [Google Scholar]
- 17.Bednar J. et al. Structure and Dynamics of a 197 bp Nucleosome in Complex with Linker Histone H1. Mol Cell 66, 384–397 e8 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou BR et al. Distinct Structures and Dynamics of Chromatosomes with Different Human Linker Histone Isoforms. Mol Cell 81, 166–182 e6 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rudnizky S. et al. Extended and dynamic linker histone-DNA Interactions control chromatosome compaction. Mol Cell 81, 3410–3421 e4 (2021). [DOI] [PubMed] [Google Scholar]
- 20.Xiao B, Freedman BS, Miller KE, Heald R. & Marko JF Histone H1 compacts DNA under force and during chromatin assembly. Mol Biol Cell 23, 4864–71 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.White AE, Hieb AR & Luger K. A quantitative investigation of linker histone interactions with nucleosomes and chromatin. Sci Rep 6, 19122 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Perisic O, Portillo-Ledesma S. & Schlick T. Sensitive effect of linker histone binding mode and subtype on chromatin condensation. Nucleic Acids Res 47, 4948–4957 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Willcockson MA et al. H1 histones control the epigenetic landscape by local chromatin compaction. Nature 589, 293–298 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hergeth SP & Schneider R. The H1 linker histones: multifunctional proteins beyond the nucleosomal core particle. EMBO Rep 16, 1439–53 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fyodorov DV, Zhou BR, Skoultchi AI & Bai Y. Emerging roles of linker histones in regulating chromatin structure and function. Nat Rev Mol Cell Biol 19, 192–206 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fan Y. et al. Histone H1 depletion in mammals alters global chromatin structure but causes specific changes in gene regulation. Cell 123, 1199–212 (2005). [DOI] [PubMed] [Google Scholar]
- 27.Torres CM et al. The linker histone H1.0 generates epigenetic and functional intratumor heterogeneity. Science 353(2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yusufova N. et al. Histone H1 loss drives lymphoma by disrupting 3D chromatin architecture. Nature 589, 299–305 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Arimura Y, Shih RM, Froom R. & Funabiki H. Structural features of nucleosomes in interphase and metaphase chromosomes. Mol Cell (2021). [DOI] [PMC free article] [PubMed]
- 30.Catez F, Ueda T. & Bustin M. Determinants of histone H1 mobility and chromatin binding in living cells. Nat Struct Mol Biol 13, 305–10 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Banani SF, Lee HO, Hyman AA & Rosen MK Biomolecular condensates: organizers of cellular biochemistry. Nat Rev Mol Cell Biol 18, 285–298 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alberti S, Gladfelter A. & Mittag T. Considerations and Challenges in Studying Liquid-Liquid Phase Separation and Biomolecular Condensates. Cell 176, 419–434 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Larson AG & Narlikar GJ The Role of Phase Separation in Heterochromatin Formation, Function, and Regulation. Biochemistry 57, 2540–2548 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Elbaum-Garfinkle S. et al. The disordered P granule protein LAF-1 drives phase separation into droplets with tunable viscosity and dynamics. Proc Natl Acad Sci U S A 112, 7189–94 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang H. et al. RNA Controls PolyQ Protein Phase Transitions. Mol Cell 60, 220–30 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nott TJ, Craggs TD & Baldwin AJ Membraneless organelles can melt nucleic acid duplexes and act as biomolecular filters. Nat Chem 8, 569–75 (2016). [DOI] [PubMed] [Google Scholar]
- 37.Vieregg JR et al. Oligonucleotide-Peptide Complexes: Phase Control by Hybridization. J Am Chem Soc 140, 1632–1638 (2018). [DOI] [PubMed] [Google Scholar]
- 38.Shakya A. & King JT DNA Local-Flexibility-Dependent Assembly of Phase-Separated Liquid Droplets. Biophys J 115, 1840–1847 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Turner AL et al. Highly disordered histone H1-DNA model complexes and their condensates. Proc Natl Acad Sci U S A 115, 11964–11969 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gibson BA et al. Organization of Chromatin by Intrinsic and Regulated Phase Separation. Cell 179, 470–484 e21 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shakya A, Park S, Rana N. & King JT Liquid-Liquid Phase Separation of Histone Proteins in Cells: Role in Chromatin Organization. Biophys J 118, 753–764 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Muzzopappa F, Hertzog M. & Erdel F. DNA length tunes the fluidity of DNA-based condensates. Biophys J (2021). [DOI] [PMC free article] [PubMed]
- 43.Smith SB, Cui Y. & Bustamante C. Overstretching B-DNA: the elastic response of individual double-stranded and single-stranded DNA molecules. Science 271, 795–9 (1996). [DOI] [PubMed] [Google Scholar]
- 44.van Mameren J. et al. Unraveling the structure of DNA during overstretching by using multicolor, single-molecule fluorescence imaging. Proc Natl Acad Sci U S A 106, 18231–6 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Alshareedah I. et al. Interplay between Short-Range Attraction and Long-Range Repulsion Controls Reentrant Liquid Condensation of Ribonucleoprotein-RNA Complexes. J Am Chem Soc 141, 14593–14602 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rhine K. et al. ALS/FTLD-Linked Mutations in FUS Glycine Residues Cause Accelerated Gelation and Reduced Interactions with Wild-Type FUS. Mol Cell 80, 666–681 e8 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ghosh A. & Zhou HX Determinants for Fusion Speed of Biomolecular Droplets. Angew Chem Int Ed Engl 59, 20837–20840 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Savelyev A. & Papoian GA Chemically accurate coarse graining of double-stranded DNA. Proc Natl Acad Sci U S A 107, 20340–5 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Latham AP & Zhang B. Consistent Force Field Captures Homologue-Resolved HP1 Phase Separation. J Chem Theory Comput (2021). [DOI] [PMC free article] [PubMed]
- 50.Borgia A. et al. Extreme disorder in an ultrahigh-affinity protein complex. Nature 555, 61–66 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Espinosa JR et al. Liquid network connectivity regulates the stability and composition of biomolecular condensates with many components. Proc Natl Acad Sci U S A 117, 13238–13247 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dignon GL, Zheng W, Best RB, Kim YC & Mittal J. Relation between single-molecule properties and phase behavior of intrinsically disordered proteins. Proc Natl Acad Sci U S A 115, 9929–9934 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Misteli T, Gunjan A, Hock R, Bustin M. & Brown DT Dynamic binding of histone H1 to chromatin in living cells. Nature 408, 877–881 (2000). [DOI] [PubMed] [Google Scholar]
- 54.Hendzel MJ, Lever MA, Crawford E. & Th’ng JP The C-terminal domain is the primary determinant of histone H1 binding to chromatin in vivo. J Biol Chem 279, 20028–34 (2004). [DOI] [PubMed] [Google Scholar]
- 55.Koc A, Wheeler LJ, Mathews CK & Merrill GF Hydroxyurea arrests DNA replication by a mechanism that preserves basal dNTP pools. J Biol Chem 279, 223–30 (2004). [DOI] [PubMed] [Google Scholar]
- 56.Foote KM et al. Discovery and Characterization of AZD6738, a Potent Inhibitor of Ataxia Telangiectasia Mutated and Rad3 Related (ATR) Kinase with Application as an Anticancer Agent. J Med Chem 61, 9889–9907 (2018). [DOI] [PubMed] [Google Scholar]
- 57.Thakar T. et al. Ubiquitinated-PCNA protects replication forks from DNA2-mediated degradation by regulating Okazaki fragment maturation and chromatin assembly. Nat Commun 11, 2147 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang Y, Xu B, Weiner BG, Meir Y. & Wingreen NS Decoding the physical principles of two-component biomolecular phase separation. Elife 10(2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Downs JA, Kosmidou E, Morgan A. & Jackson SP Suppression of homologous recombination by the Saccharomyces cerevisiae linker histone. Mol Cell 11, 1685–92 (2003). [DOI] [PubMed] [Google Scholar]
- 60.Hashimoto H. et al. Histone H1 variant, H1R is involved in DNA damage response. DNA Repair (Amst) 6, 1584–95 (2007). [DOI] [PubMed] [Google Scholar]
- 61.Zheng Y. et al. Histone H1 phosphorylation is associated with transcription by RNA polymerases I and II. J Cell Biol 189, 407–15 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Osunsade A. et al. A Robust Method for the Purification and Characterization of Recombinant Human Histone H1 Variants. Biochemistry 58, 171–176 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Izzo A. & Schneider R. The role of linker histone H1 modifications in the regulation of gene expression and chromatin dynamics. Biochim Biophys Acta 1859, 486–95 (2016). [DOI] [PubMed] [Google Scholar]
- 64.Clementi C, Nymeyer H. & Onuchic JN Topological and energetic factors: what determines the structural details of the transition state ensemble and “en-route” intermediates for protein folding? An investigation for small globular proteins. J Mol Biol 298, 937–53 (2000). [DOI] [PubMed] [Google Scholar]
- 65.Sing CE & Perry SL Recent progress in the science of complex coacervation. Soft Matter 16, 2885–2914 (2020). [DOI] [PubMed] [Google Scholar]
- 66.Berendsen HJC, van der Spoel D. & van Drunen R. GROMACS: A message-passing parallel molecular dynamics implementation. Comput Phys Commun 91, 43–56 (1995). [Google Scholar]
- 67.Dignon GL, Zheng W, Kim YC, Best RB & Mittal J. Sequence determinants of protein phase behavior from a coarse-grained model. PLoS Comput Biol 14, e1005941 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Michaud-Agrawal N, Denning EJ, Woolf TB & Beckstein O. MDAnalysis: a toolkit for the analysis of molecular dynamics simulations. J Comput Chem 32, 2319–27 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bolte S. & Cordelieres FP A guided tour into subcellular colocalization analysis in light microscopy. J Microsc 224, 213–32 (2006). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Statistical source data for Figures 1–5 and Extended Data Figures 2-6, 9 and the unprocessed gel image for Extended Data Figure 1c are provided with this paper. Other data are available upon reasonable request.