Summary
Plastic waste imposes a serious problem to the environment and society. Hence, strategies for a circular plastic economy are demanded. One strategy is the engineering of polyester hydrolases toward higher activity for the biotechnological recycling of polyethylene terephthalate (PET). To provide tools for the rapid characterization of PET hydrolases and the detection of degradation products like terephthalic acid (TPA), we coupled a carboxylic acid reductase (CAR) and the luciferase LuxAB. CAR converted TPA into the corresponding aldehydes in Escherichia coli, which yielded bioluminescence that not only semiquantitatively reflected amounts of TPA in hydrolysis samples but is suitable as a high-throughput screening assay to assess PET hydrolase activity. Furthermore, the CAR-catalyzed synthesis of terephthalaldehyde was combined with a reductive amination cascade in a one-pot setup yielding the corresponding diamine, suggesting a new strategy for the transformation of TPA as a product obtained from PET biodegradation.
Subject areas: Sensor, Polymer chemistry, Enzyme engineering, Bioelectronics
Graphical abstract
Highlights
-
•
First bioreduction of terephthalic acid (TPA) by a carboxylic acid reductase in vivo
-
•
Real-time, high-throughput detection of TPA-derived aldehydes by luciferase LuxAB
-
•
Bioluminescence reflects TPA amounts, assessing (engineered) PET hydrolase activity
-
•
Transformation of TPA into the diamine through chemo-enzymatic one-pot cascade
Sensor; Polymer chemistry; Enzyme engineering; Bioelectronics
Introduction
The global production of plastics is rapidly increasing. More than 8% of the global petrochemical production – 4% as source for materials and 4% to cover energy demands – were consumed by plastic manufacturing industries (Hopewell et al., 2009). However, only a fraction of discarded plastic is recycled (Geyer et al., 2017). Consequently, efficient disposal and sustainable recycling strategies for plastic waste are urgently needed to reduce the risk of pollution imposed on ecosystems and human health (Eriksen et al., 2014; Rahimi and García, 2017; Vollmer et al., 2020; Wright and Kelly, 2017). Furthermore, to decrease both carbon dioxide (CO2) emissions and the dependence on fossil fuel-based resources, a circular plastic economy is regarded as the central – and vital – approach (Raoul et al., 2021; Sarah and Gloria, 2021; Simon et al., 2021; Wei et al., 2020).
Particularly, the biocatalysis-based recycling of polyethylene terephthalate (PET), which is extensively used to manufacture food packaging and beverage containers, has become a vivid field of research with the discovery of microbial PET-degrading enzymatic activities (Kawai et al., 2019, 2020; Tournier et al., 2020; Wei et al., 2020, 2022; Yoshida et al., 2016). So far, PET hydrolases from actinomycetes including different Thermobifida strains (Herrero Acero et al., 2011; Müller et al., 2005; Wei et al., 2014), from the bacterium Ideonella sakaiensis (Yoshida et al., 2016), and a commercial cutinase from the fungi Thermomyces insolens, formerly known as Humicola insolens, have been employed (Ronkvist et al., 2009). Recently, a variant of the compost metagenome-derived and highly thermostable leaf-branch compost cutinase (LCC) (Sulaiman et al., 2012) was engineered toward increased PET-hydrolyzing activity, which pushed the enzymatic depolymerization of PET from laboratory scales to industrially relevant metrics by degrading amorphized (i.e., pretreated) postconsumer PET bottles in only 10 h reaction time (Tournier et al., 2020). This and the fact that LCC as well as other PET hydrolases were found in public metagenome databases will certainly advance biotechnological plastic degradation and recycling in the near future (Bornscheuer, 2016; Danso et al., 2018; Wei et al., 2020, 2022).
Despite the many achievements in the last two decades, the activity of PET hydrolases is still assessed by simply measuring the weight loss of the residual bulk PET polymer after depolymerization (Wei et al., 2019a, 2019b; Yoshida et al., 2016) or the chromatographic analysis and quantification of degradation intermediates and/or products such as terephthalic acid (TPA) and its monoesters and diesters (Eberl et al., 2009; Herrero Acero et al., 2011; Palm et al., 2019). Recently, an isothermal titration calorimetry-based method has been established for directly assessing the enthalpy of ester hydrolysis, thus enabling a real-time monitoring of the enzymatic PET hydrolysis (Vogel et al., 2021). All these strategies suffer from laborious sample preparation and the only low to moderate sample throughput, impeding the characterization of novel biocatalysts – not only limited to polyester hydrolases – and the screening of large protein libraries (Markel et al., 2020; Wei et al., 2020, 2022; Yi et al., 2021). This obstacle was addressed by a Fenton chemistry-mediated fluorometric detection assay for TPA in a 96-well microtiter plate format, suitable for high-throughput (HT) screening applications (Pfaff et al., 2021; Wei et al., 2012). The assay is based on the formation of hydroxyl radicals mediated by an Fe(II)-ethylenediaminetetraacetic acid complex in the presence of molecular oxygen (O2) (Saran and Summer, 1999; Wei et al., 2012; Welch et al., 2002); hydroxyl radicals and TPA then react to the fluorescent 2-hydroxyterephthalate (λexcitation = 315 nm, λemission = 421 nm).
Complementary, genetically encoded biosensor systems have been used for the detection of small molecules and included transcription factors (TFs), riboswitches, or enzyme-coupled sensor devices (Bayer et al., 2021; Dietrich et al., 2010; Lehtinen et al., 2017; Liu et al., 2015, 2017; Yi et al., 2021). To date, only two biosensors have been reported to detect TPA in vivo. The first was assembled by Pardo et al. and comprised the TF TphR and its regulatory nucleotide sequences from Comamonas testosteroni and the superfolder green fluorescent protein (sfGFP) (Pardo et al., 2020). TphR is a transcriptional activator, which – upon binding of TPA – acts as the inducer of a gene cluster responsible for the conversion of TPA to protocatechuate in Comamonas strains (Kasai et al., 2010). Their TF-based biosensor system facilitated the screening of TPA transporter variants, in other words, the improved uptake of TPA from the environment in Acinetobacter baylyi ADP1 through fluorescence-activated cell sorting (Pardo et al., 2020). The second example featured sfGFP as the fluorescence reporter and the promiscuous TF XylS from Pseudomonas putida, which was engineered by Li and coworkers to bind TPA additionally to reported benzoic acid derivatives (Li et al., 2022). With the efficient detection of TPA in living cells, these sensing devices have yet to be tested for the directed evolution of PET hydrolases by the HT-assisted detection of TPA as PET degradation product.
Most recently, the luciferase LuxAB from Photorhabdus luminescens (P. luminescens) was introduced for the detection of structurally diverse aldehydes in Escherichia coli (E. coli) (Bayer et al., 2021). In the present work, the carboxylic acid reductase from Mycobacterium marinum (CARMm) was shown to transform TPA into the corresponding aldehydes 4-carboxybenzaldehyde (4-CBAL) and terephthalaldehyde (TAL) in vivo (Figure 1). The coupling of the enzymatic reduction to the LuxAB biosensor device yielded bioluminescence that semiquantitatively reflected increasing amounts of TPA in PET hydrolysis samples obtained through various hydrolases. The system not only provides a biosensor-based HT assay for TPA but the first biocatalytic route toward highly reactive TPA-derived aldehydes such as TAL, avoiding hazardous chemical procedures (Barnicki, 2017; Snell and Weissberger, 1940). Following the transformation of TAL in the same reaction vessel, the corresponding diamine was yielded and will allow for potential industrial applications (Brindell et al., 1976; Rohan et al., 2015; Suematsu et al., 1983; Wang et al., 2021).
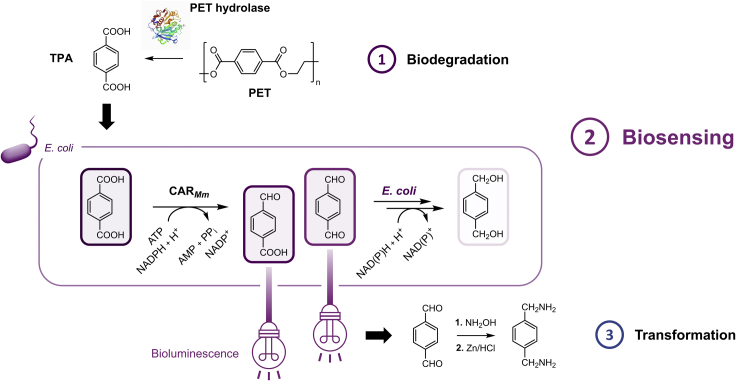
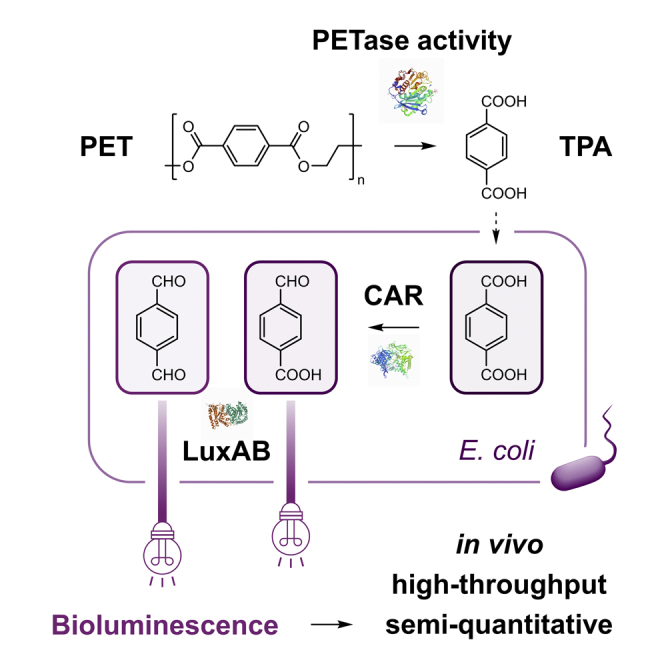
Figure 1.
Enzyme-coupled biosensor for the detection of TPA in E. coli
(1) The biocatalytic degradation of PET by hydrolases releases monomeric molecules including TPA and ethylene glycol (not shown). The PET hydrolase structure in the scheme was adapted from PDB: 6THT (Tournier et al., 2020). (2) TPA can be reduced to the corresponding dialdehydes and monoaldehydes by CARMm (accessory PPTNi not shown). These aldehydes are sensed by LuxAB, thereby emitting bioluminescence. Endogenous enzymes further reduce aldehydes to the corresponding primary alcohols. (3) The reactive TAL can be captured as aldoxime (not shown) and further converted to the diamine by reductive amination and basic work-up in a one-pot cascade, interconverting polymer precursors as future upcycling option after further optimization.
Results
Optimization of whole-cell biotransformations and evaluation of HT assay conditions
In a previous study, the monooxygenase LuxAB from P. luminescens was expressed in E. coli K-12 MG1655 RARE (Kunjapur et al., 2014), herein referred to as E. coli RARE, and provided a reliable detection tool for aldehydes in living cells in a 96-well microplate format (Bayer et al., 2021), importantly, beyond the previously reported long-chain aliphatic aldehydes (Colepicolo et al., 1989). Furthermore, LuxAB was suitable to sense aldehydes, including aromatic products such as benzaldehyde, cuminaldehyde, and 2-phenylacetaldehyde that were enzymatically produced from carboxylic acid substrates by the co-expression of CARMm in the same cell (Bayer et al., 2021). Prompted by the structural relatedness of these aromatic aldehydes to TPA-derived aldehydes, the capabilities of (1) CARMm – to reduce one or both carboxylic acid functionalities of TPA to the aldehyde – and (2) LuxAB – to accept aldehyde products formed in situ, thereby yielding bioluminescence – were investigated.
Therefore, chemically competent E. coli BL21(DE3) cells were transformed with pACYCDuet-1/carMm:pptNi to co-express CARMm and a phosphopantetheinyl transferase from Nocardia iowensis (PPTNi) (Bayer et al., 2021). The PPT is required to posttranslationally modify apoCARs to yield the functional holo-CAR enzymes (Akhtar et al., 2013; Finnigan et al., 2017; Horvat and Winkler, 2020). Whereas TPA was not converted in resting cells (RCs) of untransformed E. coli, the detection of 4-(hydroxymethyl) benzaldehyde (4-HMBAL) and 1,4-benzenedimethanol (1,4-BDM; 32.7 ± 3.5% combined yields) by gas chromatography equipped with a flame ionization detector (GC/FID) indicated both the activity of CARMm toward TPA and the further reduction of aldehydes by endogenous host enzymes (Figure 1 and Table 1) (Bayer et al., 2017; Kunjapur et al., 2014; Kunjapur and Prather, 2015). However, biotransformation mixtures contained up to 75% unreacted TPA besides the over-reduced products after 24 h (Figure S1A). Although a similar conversion of TPA was achieved with RCs of E. coli RARE (Figure S1B), the utilization of E. coli BL21(DE3) Δlpp enhanced the bioreduction of TPA significantly (Figure 2A). RCs of the engineered strain harboring pACYCDuet-1/carMm:pptNi were prepared and biotransformations were carried out as outlined below. The resulting suspension contained a mixture of TPA (31.1 ± 5.9%), 4-CBAL (36.8 ± 9.9%), 4-HMBAL (6.5 ± 2.2%), and 1,4-BDM (13.7 ± 5.7%) according to GC/FID (Figure 2A); the highly reactive TAL could only be detected in traces. The nonessential lpp gene encodes one of the most abundant cellular proteins in terms of copy number (Li et al., 2014) and controls the (mechanical) properties of the inner and outer membrane (Asmar et al., 2017; Mathelié-Guinlet et al., 2020). Not only was its deletion suggested to affect the permeability of the cellular envelope for small molecules (Ni et al., 2007); it increased expression levels of CARMm according to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis (Figure S2). This may be explained by the reallocation of cellular resources (Li et al., 2014) and might provide a general approach to improve heterologous protein production.
Table 1.
List of compounds
| Compound (Abbreviation) | Retention time [min] | RRF |
|---|---|---|
| Terephthalic acid (TPA) | 6.90–7.00 | 0.243 |
| 4-Carboxybenzaldehyde (4-CBAL) | 3.70–3.80 | 0.297 |
| 4-(Hydroxymethyl) benzoic acid (4-HMBA) | 4.14 | 0.216 |
| Terephthalaldehyde (TAL) | 4.23 | 0.932 |
| 4-(Hydroxymethyl) benzaldehyde (4-HMBAL) | 5.22 | 1.132 |
| 1,4-Benzenedimethanol (1,4-BDM) | 5.51 | 1.168 |
| 1,4-bis-(Aminomethyl) benzene (1,4-bis-AMB) | 5.27 | 0.830 |
| Benzylamine (BAM) | 2.76 | 0.783 |
| Methyl benzoate (IS) | 3.28 | – |
The retention times for benzoic acid and 2-phenylacetic acid and their corresponding aldehydes and primary alcohols as well as GC/FID-based quantification were reported previously (Bayer et al., 2021). Relative response factors (RFFs) were used as mean values of independently prepared standard solutions (n ≥ 3) analyzed by GC/FID.
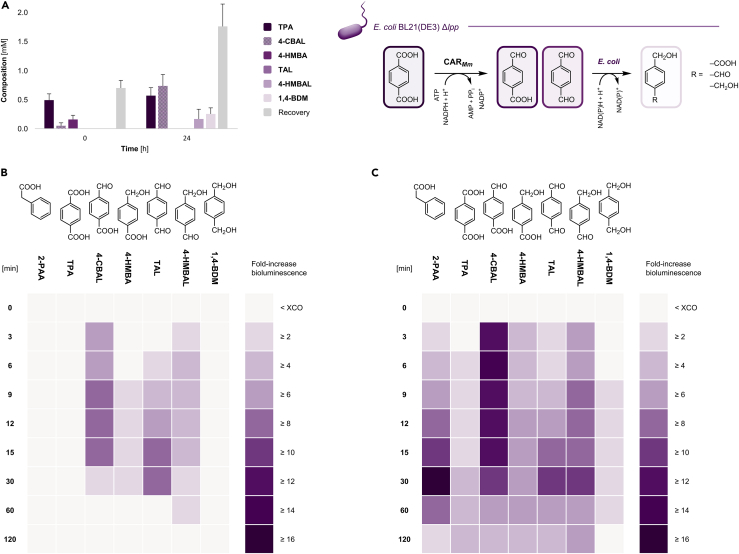
Figure 2.
Enzyme-coupled biosensor assembly in E. coli BL21(DE3) Δlpp
(A) CARMm reduces TPA to 4-CBAL and TAL, which are further reduced to 4-HMBA, 4-HMBAL, and 1,4-BDM by endogenous enzymes in vivo; PPTNi for posttranslational modification of CARMm is omitted for clarity. Experiments were performed in RCs of E. coli BL21(DE3) Δlpp (OD600 ≈ 10.0) co-expressing enzymes from pACYCDuet-1/carMm:pptNi (Bayer et al., 2021) in the presence of 2 mM TPA and 5% (ν/ν) DMSO as organic cosolvent. Sampling: 0 h (after the addition of TPA and mixing) and 24 h. Recoveries were reduced because of low solubility of TPA in resting cell medium (RCM) and the volatility of reaction compounds. GC yields are presented as mean values + standard deviation (SD) [mM] of biological replicates (n = 3); see also Figure S1.
(B) Direct detection of aldehydes (1 mM) by increasing bioluminescence over time in RCs of E. coli BL21(DE3) Δlpp expressing LuxAB from pLuxAB. (C) In situ production of aldehydes from carboxylates (1 mM) in RCs of E. coli BL21(DE3) Δlpp co-expressing LuxAB and CARMm/PPTNi; 2-phenyl acetic acid (2-PAA) was used as control. Experiments were performed in the presence of 1% (ν/ν) DMSO under HT assay conditions as described previously (Bayer et al., 2021); data presented as mean fold-increase bioluminescence obtained from biological replicates (n = 3). For results employing E. coli RARE, see Figure S3.
Subsequently, RCs of E. coli BL21(DE3) Δlpp as well as E. coli RARE were prepared either expressing only LuxAB or the luciferase together with CARMm/PPTNi. E. coli RARE exhibits reduced aromatic aldehyde-reducing activity (Kunjapur et al., 2014) and has been employed by various groups to increase the persistence of aldehydes for both their production in vivo (Bayer et al., 2017; Horvat and Winkler, 2020; Kunjapur et al., 2016) and their efficient detection (Bayer et al., 2021; Ressmann et al., 2019).
Satisfyingly, the previously established HT assay conditions yielded bioluminescence in the presence of TPA-derived aldehydes in both E. coli strains expressing LuxAB (Bayer et al., 2021). At 1 mM final concentration, the highest fold-increase in bioluminescence was observed in the presence of 4-CBAL and TAL, both elevating bioluminescence about 8-fold above background in RCs of E. coli BL21(DE3) Δlpp after 15 min, followed by 4-HMBAL (4-fold) (Figure 2B). As expected, TPA did not increase bioluminescence in RCs only expressing the biosensor, but signals increased more than 4-fold when co-expressing LuxAB and CARMm/PPTNi in the same cell (Figure 2C). Similar results were obtained with RCs of E. coli RARE upon the addition of TPA (Figure S3; 1 mM final concentration); to extenuate the cytotoxic effects of initially high aldehyde levels and in accordance with previous findings, 4-CBAL, TAL, and 4-HMBAL could be efficiently detected at 0.1 mM final concentration in E. coli RARE (Figure S3) (Bayer et al., 2021). The established reduction of 2-phenylacetic acid to 2-phenylacetaldehyde by CARMm was included as positive control for the HT assay because the latter is accepted by LuxAB (Figure 2B–2C). DMSO slightly increased background luminescence over time, which had also been shown for other cosolvents like ethanol and acetonitrile (Bayer et al., 2021). Supporting the results of the HT assay (Figure 2C), the activity of the CAR enzyme toward 4-CBAL and 4-HMBA could be confirmed by GC/FID analysis of extracts from biotransformations employing CARMm/PPTNi (Figures S1C–S1D).
Motivated by the functional CAR/luciferase biosensor couple for the detection of TPA, the assay was tested with hydrolysate samples obtained after the enzymatic degradation of PET.
Assaying TPA in PET hydrolysis samples under HT conditions
For the preparation of PET hydrolysates, the codon-optimized genes of LCC, the engineered variant LCC-ICCG (Tournier et al., 2020), and the polyester hydrolase-1 (PES-H1) (Zimmermann et al., 2019) were expressed from pET26b vectors in E. coli BL21(DE3) cultivated in auto-induction medium (AIM) supplemented with kanamycin and finally purified as described in this study.
The enzymatic degradation of amorphous PET film (Gf-PET, purchased from Goodfellow Ltd.) by LCC, LCC-ICCG, and PES-H1 was adapted from Tournier et al. as outlined below (Tournier et al., 2020). Hydrolysates were processed as described in this study and analyzed by the CARMm/LuxAB biosensor system under HT conditions (Figure 3) as well as calibrated high-performance liquid chromatography (HPLC; Table S2).
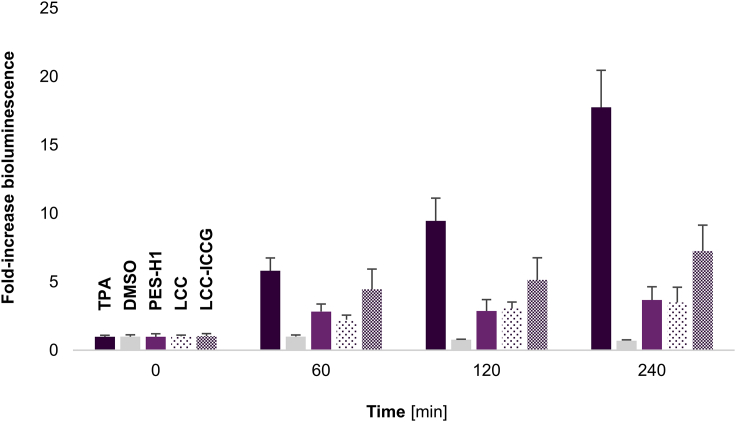
Figure 3.
PET hydrolysis samples analyzed under HT conditions in E. coli RARE
The enzyme-coupled biosensor system yielded bioluminescence in the presence of 1 mM TPA (positive control) and hydrolysates obtained by the enzymatic degradation of Gf-PET films by PES-H1, LCC, and LCC-ICCG; the bioluminescence did not increase in the presence of 1% (ν/ν) DMSO over monitoring time. Experiments were performed in RCs of E. coli RARE under HT assay conditions as described previously (Bayer et al., 2021); data presented as mean values of the fold-increase in bioluminescence + SD of biological replicates (n ≥ 3). For results employing E. coli BL21(DE3) Δlpp RCs, see Figure S4.
In the presence of 1 mM TPA, the bioluminescence increased about 4-fold and 5-fold in RCs of E. coli BL21(DE3) Δlpp and E. coli RARE, respectively, after 1 h. While the fold-increase in bioluminescence plateaued in E. coli BL21(DE3) Δlpp for 4 h (Figure S4), it increased more than 17-fold in E. coli RARE cells during the same reaction time (Figure 3). This difference can be explained by the distinct metabolic backgrounds of the two strains as highlighted earlier. The knockout of several alcohol dehydrogenases and aldo-keto reductases in E. coli RARE increases the persistence of (aromatic) aldehydes in vivo, including TPA-derived aldehydes (Kunjapur et al., 2014). In contrast, the activity of these endogenous enzymes in E. coli BL21(DE3) Δlpp continuously reduces reactive aldehydes to the corresponding primary alcohols such as 1,4-BDM (Figure S1), which is not a substrate for LuxAB (Figure 2B).
Whereas bioluminescence signals were elevated >3-fold with PET hydrolysates obtained by the wild-type enzymes PES-H1 and LCC, bioluminescence increased >7-fold in LCC-ICCG samples after 4 h (Figure 3). This may be attributed to higher concentrations of potassium terephthalate salts in PET hydrolysates obtained by the LCC-ICCG variant compared to LCC, for example. Based on the fold-increases, TPA concentrations in the supernatants of the three hydrolysates were calculated and suggested 38.3 ± 3.8 mM, 39.3 ± 1.3 mM, and 95.5 ± 10.7 mM for PES-H1, LCC, and LCC-ICCG, respectively. Similar TPA yields in the same concentration range (56 mM, 47 mM, and 111 mM, respectively) were determined by HPLC (Table S2).
Given that the biosensor system is operating in living cells, the cytotoxicity of both carboxylates and the corresponding aldehydes (Bayer et al., 2017, 2021; Kunjapur and Prather, 2015), as well as the transient nature of bioluminescence signals (Fleiss and Sarkisyan, 2019) may interfere with the quantitative determination, allowing for marginal deviations from HPLC data. Nonetheless, the analysis of PET hydrolysates under HT conditions employing RCs of E. coli RARE yielded a reproducible fold-increase in bioluminescence based on the enzymatic transformation of TPA into the corresponding aldehydes and their detection by LuxAB, ultimately, reflecting TPA concentrations in PET hydrolysate samples semiquantitatively.
Transformation of TAL by a chemo-enzymatic cascade in one pot
The chemical synthesis of (aromatic) aldehydes can be troublesome because of the high reactivity of the carbonyl group (Ferguson, 1946; Kunjapur and Prather, 2015). A promising alternative to specifically synthesize aldehydes are the well-established enzymatic reductions of carboxylates by CARs (Bayer et al., 2017, 2021; Butler and Kunjapur, 2020; Finnigan et al., 2017; Horvat and Winkler, 2020; Qu et al., 2018). CARMm readily accepts TPA as indicated by the bioluminescence signals in the LuxAB-based HT assay (Figures 2C and S2B) and confirmed by the detection of 4-CBAL and TAL as intermediates and the corresponding over-reduced compounds 4-HMBAL and 1,4-BDM according to GC/FID. 4-HMBAL and 1,4-BDM are exclusively formed by the endogenous activities of host enzymes (Bayer et al., 2017; Kunjapur et al., 2014) (Figures 2A and S1). To the best of our knowledge, the CAR-catalyzed reduction of TPA is the first reported biocatalytic route forming TPA-derived aldehydes such as TAL, substituting hazardous chemical procedures (Snell and Weissberger, 1940). Depending on the purity and downstream application of plastic monomers from biocatalytic degradations, not all TPA is suitable for the resynthesis of virgin PET. Therefore, (bio)chemical transformation strategies for the re-use (i.e., upcycling) of plastic precursors is of interest (Tiso et al., 2021). Recently, Sadler and Wallace synthesized vanillin from hydrolyzed waste PET by combining TPA-transforming enzymes from Comamonas sp. to yield intermediate catechol that was converted to the product by the activities of a CAR and an engineered catechol O-methyltransferase in E. coli RARE (Kunjapur and Prather, 2019; Sadler and Wallace, 2021).
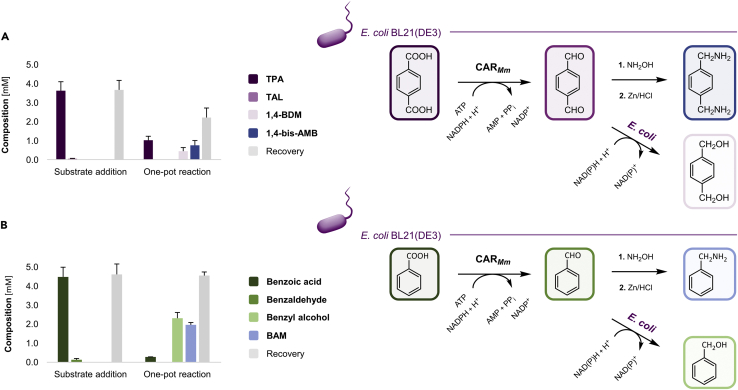
In the following proof-of-concept example, benzaldehyde and TAL were produced from benzoic acid and TPA, respectively, by CARMm/PPTNi in E. coli BL21(DE3) or E. coli RARE RCs. The aldehydes were quenched in the presence of an excess of hydroxylamine hydrochloride (NH2OH · HCl) to form the corresponding aldoximes. Subsequently, reductive amination was performed in one pot by the addition of zinc powder and acidification (Ayedi et al., 2013). After extraction under basic conditions, the expected primary amines – benzylamine (BAM; 35.3 ± 0.7%) and 1,4-bis-(aminomethyl) benzene (1,4-bis-AMB; 15.0 ± 5.0%) – could be detected by GC/FID (Figure 4); benzyl alcohol and 1,4-BDM, respectively, were the major byproducts. Structurally related diamines find applications in synthesis of polyurethanes and polyamides, for example (Wang et al., 2021). In addition, although not further investigated in this study, the formation of imines might contribute to the low yield and the poor recovery of material in reactions starting from TPA (Godoy-Alcántar et al., 2005; Simion et al., 2001).
Figure 4.
Chemo-enzymatic one-pot cascades
Carboxylates are reduced by CARMm in RCs of E. coli BL21(DE3) to the corresponding aldehydes; PPTNi is omitted for clarity. In the presence of NH2OH · HCl, the oximes are formed (not shown), which are reduced to the primary amines (shades of blue) after the addition of Zn/HCl to the same reaction vessel.
(A) The TAL intermediate yields the desired 1,4-bis-AMB, besides 1,4-BDM as the major byproduct. Recoveries were reduced due to low solubility of TPA in RCM containing 5% (ν/ν) DMSO as organic co-solvent, the volatility of reaction compounds, and the formation of yet to be identified byproducts such as imines (Godoy-Alcántar et al., 2005; Simion et al., 2001).
(B) Benzoic acid in the presence of 5% (ν/ν) ethanol was reduced to benzaldehyde, yielding the desired BAM after reductive amination and benzyl alcohol as the sole byproduct. Experiments were performed in RCs (OD600 ≈ 10.0) co-expressing enzymes from pACYCDuet-1/carMm:pptNi (Bayer et al., 2021). Sampling: (1) after the addition of NH2OH · HCl (2.2 and 1.1 equiv for TPA and benzoic acid, respectively) and carboxylic acid and mixing; (2) after performing the reductive amination in one-pot. GC yields are presented as mean values + SD [mM] of biological replicates (n = 3). Performance was similar with RCs of E. coli RARE producing 27.2 ± 6.6% BAM and 13.1 ± 8.0% 1,4-bis-AMB (n = 2).
Discussion
The expanding number of new PET hydrolases from natural resources including metagenomes as well as protein engineering endeavors calls for tools for their rapid characterization (Wei et al., 2022; Wiltschi et al., 2020). Furthermore, the functional assessment of these enzymes depends – with very few exceptions (Pfaff et al., 2021; Vogel et al., 2021) – on chromatographic methods characterized by only modest sample throughputs (Markel et al., 2020; Wei et al., 2020; Yi et al., 2021). To address this issue, this work coupled the activity of CARMm to reduce TPA in different E. coli strains to the corresponding aldehydes (4-CBAL, TAL, and 4-HMBAL) with the genetically encoded biosensor LuxAB from P. luminescens. The latter emits detectable bioluminescence in the presence of TPA-derived aldehydes (Figures 1, 2 and S3). As TPA is a building block of PET, the CAR/LuxAB couple was employed to detect terephthalates in hydrolysate samples obtained from PET degradation, catalyzed by the wild-type polyester hydrolases PES-H1 and LCC and the engineered variant LCC-ICCG. Not only was TPA reliably detected by reproducible fold-increase in bioluminescence values in independently carried out assay set-ups under HT conditions (Figures 2C and 3); samples containing terephthalate from PET hydrolysis by PES-H1, LCC, and LCC-ICCG exhibited steady fold-increases over 4 h under HT assay conditions in E. coli RARE, which was in agreement with HPLC data. This sufficed to distinguish between wild-type enzymes and a variant with increased PET degradation activity and offers a semiquantitative screening tool for PET hydrolase libraries in the future.
Lastly, with the biocatalytic production of TAL from TPA, we accessed a highly reactive aldehyde intermediate that could be transformed into the corresponding primary diamine, for example, in aqueous reaction media (Figure 4A). The chemo-enzymatic three-step cascade also yielded >30% BAM from benzoic acid via the aldehyde and aldoxime intermediates (Figure 4B).
In conclusion, the presented work featured a complementary biosensor tool for the HT detection of TPA in living cells and suggested new routes for the bio-based interconversion of polymer building blocks, supporting efforts toward a circular plastic economy, the reduction of CO2 emissions, and the stewardship of resources.
Limitations of the study
Although the utilization of E. coli BL21(DE3) Δlpp significantly increased the CAR-catalyzed conversion of TPA in vivo, it was not superior to the established E. coli RARE strain for the LuxAB-based detection of aldehydes over longer reaction times because of their different metabolic backgrounds. However, the reproducible detection of TPA by the CARMm/LuxAB-coupled biosensor under HT assay conditions in E. coli RARE enabled the semiquantitative assessment of terephthalate salts in the supernatants obtained from the biocatalytic degradation by various PET hydrolases. Even though calculated yields were in the same concentration range according to calibrated HPLC, discrepancies arise from operating the biosensor system in whole-cells of E. coli because of the cytotoxicity of TPA and the corresponding aldehydes, for example. Accordingly, the bioluminescence yielded by the LuxAB-catalyzed reaction is transient and influenced by the metabolic background, the viability and physiological state of cells including aeration; because LuxAB is a monooxygenase, the generation of bioluminescence depends on the aldehyde substrate and O2. In addition, expression levels of enzymes, intracellular cofactor availability, and the background luminescence in living cells can add to variations but are easily addressed by appropriate (negative) controls, and the normalization of bioluminescence signals as discussed in detail previously (Bayer et al., 2021).
The interconversion of TPA into 1,4-bis-AMB through a three-step chemo-enzymatic cascade operating in one-pot only yielded only 15.0 ± 5.0% of the diamine and could not be improved by employing E. coli RARE, for example. The poor recovery of material (<50%) can be explained by the low solubility of TPA in aqueous solutions and the volatility of reaction intermediates. Furthermore, the formation of imines from aldehyde and amine precursors in aqueous solutions has been reported (Godoy-Alcántar et al., 2005; Simion et al., 2001) and will be investigated as a contributing factor in the future. Nonetheless, the reductive amination could be achieved in an aqueous buffer system, which advances the original protocol (Ayedi et al., 2013) and puts it in the context of transforming PET-derived TPA.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and virus strains | ||
| E. coli BL21(DE3) | Thermo Scientific™ | Cat#EC0114 |
| E. coli BL21(DE3) Δlpp | This paper | N/A |
| E. coli DH5α | Thermo Scientific™ | Cat#18265017 |
| E. coli K-12 MG1655 RARE | Prof. K.L.J. Prather (Kunjapur et al., 2014) | Addgene Bacterial strain #61440 |
| Chemicals, peptides, and recombinant proteins | ||
| PET film | Goodfellow GmbH | Cat#ES301445 |
| Terephthalic acid (TPA; CAS: 100-21-0) | Sigma-Aldrich | Cat#185361 |
| 4-Carboxybenzaldehyde (4-CBAL; CAS: 619-66-9) | Acros | Cat#154580050 |
| 4-(Hydroxymethyl) benzoic acid (4-HMBA; CAS: 3006-96-0) | Sigma-Aldrich | Cat#382639 |
| Terephthalaldehyde (TAL; CAS: 623-27-8) | Alfa Aesar | Cat#A14930 |
| 4-(Hydroxymethyl) benzaldehyde (4-HMBAL; CAS: 52010-97-6) | Carbosynth Ltd | Cat#FH140138 |
| 1,4-Benzenedimethanol (1,4-BDM; CAS: 589-29-7) | TCI | Cat#D0605 |
| 1,4-bis-(Aminomethyl) benzene (1,4-bis-AMB; CAS: 539-48-0) | Sigma-Aldrich | Cat#8.41656 |
| Benzoic acid (CAS: 65-85-0) | Sigma-Aldrich | Cat#242381 |
| Benzaldehyde (CAS: 100-52-7) | Acros | Cat#378361000 |
| Benzyl alcohol (CAS: 100-51-6) | Fluka | Cat#77013 |
| Benzylamine (BAM; CAS: 100-46-9) | Sigma-Aldrich | Cat#185701 |
| Methyl benzoate (CAS: 93-58-3) | Sigma-Aldrich | Cat#M29908 |
| 2-Phenylacetic acid (2-PAA; CAS: 103-82-2) | Fluka | Cat#78490 |
| 2-Phenylacetaldehyde (2-PAAL; CAS: 122-78-1) | Acros | Cat#37091 |
| 2-Phenylethanol (CAS: 60-12-8) | Fluka | Cat#77861 |
| Lysonase™ Bioprocessing Reagent | Merck-Millipore | Cat#71230 |
| ROTI®Garose-His/Co Beads | Carl Roth | Cat#1235.1 |
| Recombinant protein (C-term. 6xHis, purified): leaf-branch compost cutinase (LCC) | This study | G9BY57 |
| Recombinant protein (C-term. 6xHis, purified): leaf-branch compost cutinase variant (LCC-ICCG) | This study | PDB: 6THT |
| Recombinant protein (C-term. 6xHis, purified): polyester hydrolase-1 (PES-H1) | This study | PDB: 7CUV |
| Q5® polymerase | NEB | Cat#M0491S |
| Q5® mutagenesis kit | NEB | Cat#E0554S |
| Deposited data | ||
| Raw and analyzed data | This paper | N/A |
| Experimental models: Organisms/strains | ||
| E. coli strains, see above: Bacterial and virus strains | This paper | N/A |
| Oligonucleotides | ||
| lpp-up_F, primer for strain engineering, see Table S1 | This paper (Thermo Scientific™) | N/A |
| lpp-up_R, primer for strain engineering, see Table S1 | This paper (Thermo Scientific™) | N/A |
| lpp-down_F, primer for strain engineering, see Table S1 | This paper (Thermo Scientific™) | N/A |
| lpp-down_R, primer for strain engineering, see Table S1 | This paper (Thermo Scientific™) | N/A |
| pTarget_F, primer for strain engineering, see Table S1 | This paper (Thermo Scientific™) | N/A |
| pTarget_R, primer for strain engineering, see Table S1 | This paper (Thermo Scientific™) | N/A |
| Δlpp-gRNA_F, primer for strain engineering, see Table S1 | This paper (Thermo Scientific™) | N/A |
| Δlpp-gRNA_R, primer for strain engineering, see Table S1 | This paper (Thermo Scientific™) | N/A |
| Recombinant DNA | ||
| Plasmid: pCDFduo/luxAB | Bayer et al., 2021 | NCBI: WP_088373098 (luxA); NCBI: P19840 (luxB) |
| Plasmid: pACYCDuet-1/carMm:pptNi | Bayer et al., 2021 | NCBI: WP_012393886 (carMm); NCBI: ABI83656 (pptNi) |
| Plasmid: pET26b/lcc | This paper (BioCat GmbH); (Tournier et al., 2020) | NCBI: G9BY57 |
| Plasmid: pET26b/lcc-ICCG | This paper (BioCat GmbH); (Tournier et al., 2020) | PDB: 6THT |
| Plasmid: pET26b/pes-H1 | This paper (BioCat GmbH); (Zimmermann et al., 2019) | PDB: 7CUV |
| Plasmid: pCas | Addgene (Jiang et al., 2015) | Addgene Plasmid #62225 |
| Plasmid: pTarget | Addgene (Jiang et al., 2015) | Addgene Plasmid #62226 |
| Plasmid: pTarget-Δlpp | This paper | N/A |
| Software and algorithms | ||
| Geneious Prime® 2022.0.2 | Biomatters Ltd | www.geneious.com |
| OligoEvaluator™ | Sigma-Aldrich | http://www.oligoevaluator.com/LoginServlet |
| Microsoft Office 16.0 | Microsoft Corporation | www.microsoft.com |
| Other | ||
| 96-well plate (flat bottom, black polystyrene) | Greiner Bio-One | Cat#655079 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Dr. Thomas Bayer (thomas.bayer@uni-greifswald.de).
Materials availability
-
•
For the assembly of the pTarget-Δlpp plasmid in this study, the templates pCas (#62225) and pTarget (#62226) were purchased from Addgene (Watertown, USA). Subsequently, pTarget-Δlpp and pCas were used to knock-out the lpp gene from the genome of E. coli BL21(DE3). The genes encoding the leaf-branch compost cutinase (LCC) and the LCC-ICCG variant (Tournier et al., 2020) and the polyester hydrolase-1 (PES-H1) (Zimmermann et al., 2019) were codon-optimized for the expression in E. coli, synthesized, and cloned in frame with the C-terminal 6xHis tag present in pET26b by the BioCat GmbH (Heidelberg, Germany). Accession numbers of proteins are provided in the Key resources table.
-
•
E. coli BL21(DE3) (#EC0114) and DH5α (#18265017) were initially purchased from Thermo Scientific™ (Darmstadt, Germany) and propagated as described below. E. coli BL21(DE3) Δlpp is available from the lead contact upon request. E. coli RARE was acquired from the Prather group (Kunjapur et al., 2014) but is also available from Addgene (#61440).
-
•
There are restrictions to the availability of the previously constructed pCDFduo/luxAB, herein referred to as pLuxAB, and pACYCDuet-1/carMm:pptNi plasmids (Bayer et al., 2021) due to material transfer agreements (MTAs). Further information is available from the lead contact upon request.
-
•
Otherwise, this study did not generate new unique reagents.
Experimental model and subject details
E. coli BL21(DE3), E. coli BL21(DE3) Δlpp, E. coli DH5α, and E. coli RARE were propagated in 4–5 mL lysogeny broth (LB) medium (25 g L−1; Sigma-Aldrich, Buchs, Switzerland) in Infors HT Multitron incubator shakers (Bottmingen, Switzerland) at 37°C with shaking (150–180 rpm) for 12–16 h. If not stated otherwise, chemically competent E. coli cells were produced by using 0.1 M CaCl2 and transformed with plasmid DNA (25–100 ng) by heat-shock at 42°C for 45 s as previously described (Bayer et al., 2021). For the efficient transformation of E. coli RARE, plasmids were passed through E. coli DH5α (Bayer et al., 2021). E. coli transformants harboring pLuxAB and pACYCDuet-1/carMm:pptNi were propagated in LB medium supplemented with streptomycin (25 μg·mL-1) and chloramphenicol (34 μg·mL-1), respectively. Only half the concentration of antibiotics was used for the selection and subsequent propagation of strains harboring both plasmids. For the selection and propagation on plates, LB containing 1.5% (ω/ν) agar (Carl Roth, Karlsruhe, Germany) and supplemented with antibiotics – if applicable – was used.
Method details
Strain engineering
The non-essential lpp gene encodes a cellular ‘bulk’ protein (Li et al., 2014), which controls the (mechanical) properties of the inner and outer membrane and the width of the periplasmic space (Asmar et al., 2017; Mathelié-Guinlet et al., 2020). The deletion of the lpp gene from the E. coli genome has been suggested to affect the permeability of the cellular envelope for small molecules (Ni et al., 2007) that might also influence the uptake of TPA and derivatives. Furthermore, expression levels of CARMm/PPTNi were increased in E. coli BL21(DE3) Δlpp according to SDS-PAGE analysis (Figure S2). This may be due to the re-allocation of cellular resources (Li et al., 2014).
E. coli BL21(DE3) Δlpp was constructed by using a previously developed two-plasmid-based CRISPR/Cas9 system (Jiang et al., 2015). The two key plasmids, pCas (#62225) and pTarget (#62226), were purchased from Addgene (Watertown, USA). The pTarget-Δlpp plasmid was constructed by first engineering the flanking sequence of the lpp gene by the assembly of three DNA fragments using a sequence- and ligation-independent cloning extract-based protocol (Zhang et al., 2012): (1) the lpp-up fragment (amplified from the E. coli genome employing the primers lpp-up_F/R), (2) the lpp-down fragment (amplified from the genome of E. coli using the primers lpp-down_F/R), and (3) the pTarget fragment. The latter was amplified by using the primers pTarget_F/R. Next, the guide RNA (gRNA) was introduced employing the Q5® mutagenesis kit (New England Biolabs, Frankfurt/Main, Germany) with the primer pair Δlpp-gRNA_F/R. The resulting pTarget-Δlpp was Sanger sequenced (Eurofins Genomics, Ebersberg, Germany) to confirm the insertion of gRNA and the flanking sequences of the lpp gene. Subsequently, E. coli BL21(DE3) Δlpp strain was constructed by using pCas and pTarget-Δlpp according to Jiang and co-workers (Jiang et al., 2015). Briefly, competent E. coli BL21(DE3) cells were transformed with pCas as described above. In the resulting pCas transformants, the λ-Red system was induced with 0.2% (ω/ν) arabinose and electrocompetent cells were prepared. Next, the pTarget-Δlpp plasmid was introduced by electroporation; transformants were selected on LB agar plates supplemented with kanamycin and streptomycin. Colony polymerase chain reaction (PCR) was performed for genotyping the colonies. The plasmids pTarget-Δlpp and pCas were cured sequentially in the presence of 0.5 mM isopropyl-β-D-thiogalactopyranoside (IPTG) and by culturing at 37°C, respectively. Finally, the successful construction of E. coli BL21(DE3) Δlpp was confirmed by PCR amplification and DNA sequencing of the genome sequence flanking the knocked-out lpp gene.
Desalted DNA oligonucleotides were ordered from Invitrogen/Thermo Fisher Scientific and dissolved in nuclease-free water (Invitrogen, Darmstadt, Germany). Primer sequences are given in Table S1. PCRs were performed on a Biometra TAdvanced thermal cycler (Analytik Jena, Jena, Germany) employing Q5® polymerase as suggested by the supplier (New England Biolabs).
Enzyme production and resting cell preparation
Production of LuxAB and CARMm/PPTNi was performed in E. coli transformants harboring pLuxAB and pACYCDuet-1/carMm:pptNi, respectively. For cultivation, auto-induction medium (AIM; 2.5% (ω/ν) lysogeny broth medium, 1 mM MgSO4, 25 mM (NH4)2SO4, 50 mM KH2PO4, 50 mM Na2HPO4, 5% (ω/ν) glycerol, 0.5% (ω/ν) glucose, 2% (ω/ν) α-lactose) supplemented with chloramphenicol (34 μg·mL-1) and streptomycin (25 μg·mL-1), respectively, was used. Only half the concentration of antibiotics was used for the selection and subsequent cultivation of strains harboring both plasmids. The AIM was adapted from Studier (Studier, 2005).
Briefly, a single colony of the desired strain was grown in LB medium supplemented with the appropriate antibiotic(s) at 37°C (180 rpm) for 12–16 h. AIM containing antibiotic(s) was inoculated with 0.2% (ν/ν) preculture in baffled flasks and incubated in Infors HT Multitron incubator shakers (Bottmingen, Switzerland) at 37°C (180 rpm) for 4–6 h (6 h for co-transformants, 5 h for cells harboring pLuxAB, and 4 h for cells harboring pACYCDuet-1/carMm:pptNi). Enzyme production was performed at 20°C (150 rpm) for 16–20 h. The optical density at 600 nm (OD600) of cultures was determined with a UV-1280 spectrophotometer (Shimadzu, Kyoto, Japan). Cells were harvested by centrifugation (4,000 ✕ g, 4°C) for 15–20 min using a Heraeus Fresco 17 centrifuge or a Heraeus Labofuge 400R (Thermo Fisher Scientific) (Bayer et al., 2021).
RCs of E. coli were prepared after cultivation by re-suspension in RCM (22 mM KH2PO4, 42 mM Na2HPO4, 8.56 mM NaCl, 1 mM MgSO4, 1 mM CaCl2 and 1% (ω/ν) glucose) to a final OD600 ≈ 10.0 as previously described (Bayer et al., 2021).
Similarly, LCC, LCC-ICCG, and PES-H1 were produced in E. coli BL21(DE3) transformants by cultivation in AIM supplemented with kanamycin (50 μg·mL-1) at 21°C (150 rpm) for 23 h (Tournier et al., 2020). To produce PES-H1, the time of cultivation was reduced from 23 h to 20 h. Cells were harvested, re-suspended in lysis buffer (7 mL per g wet cell pellet; 50 mM Na2HPO4, 300 mM NaCl, pH 8), and processed according to Tournier and co-workers (Tournier et al., 2020). After cell disruption by freezing/thawing, 1 μL LysonaseTM Bioprocessing Reagent (#71230; Merck-Millipore, Darmstadt, Germany) per mL cell suspension was added and incubated at 28°C (220 rpm) for 1 h. They lysate was clarified by centrifugation (6,000 ✕ g, 4°C) for 45 min. Subsequently, enzymes were purified from the supernatant through their C-terminal 6xHis tags by cobalt affinity chromatography (ROTI® Garose-His/Co Beads, Carl Roth, Karlsruhe, Germany). The three hydrolases were eluted with elution buffer (50 mM Na2HPO4, 300 mM NaCl, pH 8.0) supplemented with 250 mM (LCC and LCC-ICCG) or 100 mM imidazole (PES-H1). The target proteins were desalted with 50 mM Na2HPO4 buffer (pH 8.0) and used for the hydrolysis of Gf-PET film (Goodfellow, Hamburg, Germany) as describe below.
Protein expression was confirmed by 12.5% (ω/ν) SDS-PAGE of denatured whole-cell samples normalized to OD600 = 7.0 (Figure S2) or purified enzymes, using the Mini-PROTEAN electrophoresis system (Bio-Rad, Feldkirchen, Germany) and following standard protocols; gels were stained with InstantBlueTM Protein Stain (Expedeon, Heidelberg, Germany) for at least 30 min (Bayer et al., 2021).
PET hydrolysis
The enzymatic degradation of amorphous PET film (Goodfellow, Hamburg, Germany) was performed in 1 M potassium phosphate buffer (pH 8.0) at 72°C with shaking (1 000 rpm; ThermoMixer C, Eppendorf, Hamburg, Germany) for 24 h; the enzyme concentration of 1 mg per g PET in a total reaction volume of 1.5 mL was used as published before (Tournier et al., 2020). The clarified supernatants were analyzed according to Palm et al. by reverse-phase HPLC on a VWR Hitachi LaChrom Elite system (VWR International, Radnor, USA), equipped with a Kinetex® column (5 μM EVO C18 100 Å, 150 x 4.6 mm; Phenomenex®, Aschaffenburg, Germany), with a gradient of acetonitrile and 0.1% (ν/ν) formic acid in water at 30°C after injection of 10 μL sample. Within 12 min, acetonitrile was increased from 5 to 44% and then to 70% after 15 min. The ratio remained constant for another 3 min. TPA was detected at 240 nm and quantification was facilitated by standard calibration using commercial reference compounds (Palm et al., 2019). Samples were prepared from independent PET hydrolysis experiments with LCC, LCC-ICCG, and PES-H1 and subsequent HPLC measurments and quantification (n ≥ 2; Table S2).
LuxAB-based detection of TPA-derived aldehydes in vivo (96-well plate format)
RCs of the desired E. coli strain expressing either LuxAB or LuxAB together with CARMm/PPTNi were prepared as before in biological replicates (n ≥ 3). To 198 μL RCs (OD600 ≈ 10.0) per well, 2 μL of the target compound, dissolved in DMSO, were added to a final concentration of 1 mM concentration, if not stated otherwise, in a total volume of 200 μL containing 1% (ν/ν) DMSO as co-solvent per well (flat bottom, black polystyrene 96-well plate, #655079; Greiner Bio-One, Frickenhausen, Germany). It was mixed gently. The bioluminescence was measured immediately on a VarioskanTM LUX multimode plate reader (Thermo Fisher Scientific). The change in bioluminescence was monitored at 25°C for up to 1 h and the fold-increase in bioluminescence above background in the presence of directly added or enzymatically produced aldehydes calculated as described in detail previously (Bayer et al., 2021). Data were generated from biological replicates and presented as mean values + SD (n ≥ 3). These results are shown in Figures 2B–2C and Figure S3 for RCs of E. coli BL21(DE3) Δlpp and E. coli RARE, respectively.
For the screening of PET hydrolysates, samples were clarified by centrifugation (13,000 ✕ g, 1 min) using a Heraeus Labofuge 400R (Thermo Fisher Scientific). The supernatant was diluted 1:10 by mixing 100 μL of the hydrolysis sample with 200 μL DMSO and 700 μL RCM. Subsequently, 10 μL of the resulting dilution were added to 190 μL RCs (OD600 ≈ 10.0) per well. The 96-well plate was processed as before and the bioluminescence was measured up to 4 h. Data were generated from biological replicates and presented as mean values + SD (n ≥ 3). These results are shown in Figures 3 and S4 for RCs of E. coli RARE and E. coli BL21(DE3) Δlpp, respectively.
Whole-cell biotransformations and chemo-enzymatic cascade one-pot reaction
RCs (OD600 ≈ 10.0) expressing CARMm/PPTNi were prepared as before. Whole-cell biotransformations were performed in glass vials with screwcaps (4 mL) at 2–5 mM TPA concentration and 5% (ν/ν) DMSO as co-solvent (Vtotal = 0.5 mL) in Infors HT Multitron incubator shakers (Bottmingen, Switzerland) at 25°C (230–250 rpm) for 0–24 h. For GC analysis, samples (100 μL) of the biotransformation mixtures were taken immediately after the addition of substrate and mixing (t ≈ 0 h) and again after 24 h. Subsequently, samples were acidified with 2 M HCl (10 μL) and extracted two times with ethyl acetate (200 μL) containing 1 mM methyl benzoate as internal standard (IS) by vortexing for 1 min. For phase separation, samples were centrifuged (13,000 ✕ g, 4°C) for 1 min. The combined organic phases were desiccated over Na2SO4 and transferred into a GC vial with insert, capped, and submitted to GC analysis. Compound identification was performed by the comparisons of retention times of commercial standards (Table 1), unless stated otherwise; quantification and calculation of GC yields were performed by employing relative response factors (RRFs) as described in detail previously (Bayer et al., 2021) and below. Data were generated from biological replicates and presented as mean values + SD (n ≥ 3). When employing RCs of E. coli BL21(DE3) or E. coli RARE, reaction mixtures contained mainly unreacted TPA after 24 h (58.2 ± 16.0% for E. coli BL21(DE3) and 51.0 ± 12.1% for E. coli RARE as shown in Figures S1A and S1B, respectively). Conversions of TPA could be improved by the utilization of E. coli BL21(DE3) Δlpp. After 24 h reaction time, biotransformation mixtures only contained 31.1 ± 5.9% TPA, besides TPA-derived aldehydes including 4-CBAL and the over-reduced 4-HMBAL and 1,4-BDM (Figure 2A). Additionally, 4-CBAL and 4-(hydroxymethyl) benzoic acid (4-HMBA) were identified as substrates for CARMm as shown in Figures S1C and S1D, respectively. These results are in agreement with the LuxAB-based detection of corresponding aldehydes employing either RCs of E. coli BL21(DE3) Δlpp (Figure 2) or E. coli RARE (Figure S4) in the HT assay. Furthermore, whole-cell biotransformations suggest the activity of endogenous enzymes (e.g., aldehyde dehydrogenases) that oxidize aldehydes to the corresponding carboxylic acids (Figures S1C–S1E). This is in accordance with previous findings (Bayer et al., 2017).
For the chemo-enzymatic reaction in one-pot, TPA was reduced by CARMm in whole-cell biotransformations under the conditions given above (Vtotal = 0.1 mL) in the presence of 2.2–2.5 eq NH2OH · HCl in biological replicates (n ≥ 2). After 12–16 h of reaction time, 5.5 eq zinc powder were added and the cell suspension acidified with 10 M HCl (20 μL). After incubation with shaking at room temperature for 4 h, 30% (ν/ν) ammonia solution (10 μL) and 5 M NaOH (10 μL) were added (Ayedi et al., 2013). It was mixed for 15 min before extracting two times with ethyl acetate (200 μL) containing 1 mM IS as before. Combined organic phases were dried over Na2SO4 and submitted to GC analysis using a GC-2010 Plus (Shimadzu) equipped with a flame ionization detector (FID; Shimadzu) and a ZB-5MSi column (length: 30 m; inner diameter: 0.25 mm; film thickness: 0.25 μm) from Phenomenex (Torrance, USA). GC/FID method (hydrogen, 0.96 mL·min-1 flow rate; injector and detector: 320°C): 100°C, hold 1 min, 20°C per min to 250°C, hold 5 min; total time: 13.5 min.
Quantification and statistical analysis
Quantification was performed for (1) biocatalytic reactions performed in whole cells of E. coli (i.e., RCs) in biological replicates (n ≥ 3) and in combination with reductive amination reactions in one-pot (n ≥ 2) and (2) the enzymatic degradation of PET for independent hydrolysis reactions (n ≥ 2). Compound identification was realized by the comparison of the retention times of commercial standards by (1) GC/FID and (2) HPLC analysis. From the corresponding peak areas, yields were calculated (1) by employing RFFs for each compound of interest (Table 1) and (2) by linear standard calibration for TPA (slope = 49,134.00; axis intercept = 58,547.00; R2 > 0.99).
For the semi-quantitative assessment of amounts of TPA in PET hydrolysis samples, dilutions were analyzed by the CAR/LuxAB-based HT assay employing biological replicates of RCs (n ≥ 3). The fold-increase in bioluminescence was proportional to the concentration of TPA as described in the main text and could be calculated from TPA samples with known concentration (1 mM).
Statistical analysis included the calculations of mean values, SDs, and the determination coefficient (R2) by the integrated functions of the standard spreadsheet software Microsoft Excel (version 16.0).
GC yields are presented as bars representing mean values + SD in Figures 2A, 4, and S1. Both experimental and statistical details can also be found in the corresponding figure legends and in the main text.
HPLC yields are given as mean values ± SD in Table S2. Experimental details can be found in the main text and statistical details in the legend of Table S2.
The mean fold-increase in bioluminescence + SD is depicted as bars in Figures 3 and S4. Both experimental and statistical details can also be found in the corresponding figure legends and in the main text.
Additional resources
The OligoEvaluatorTM (Sigma-Aldrich; http://www.oligoevaluator.com/LoginServlet) was used to predict secondary structures and dimer formation of DNA oligonucleotides (Table S1).
Acknowledgments
T.B. was funded through the Erwin Schrödinger Fellowship (project no.: J4231-B21) granted by the Austrian Science Fund (FWF). R.W., L.P., Y.B., and U.T.B. acknowledge funding received from the European Union’s Horizon 2020 research and innovation program under grant agreement no. 870294 (MIX-UP project) and no. 953214 (upPE-T project). S.W. was supported by a Humboldt research fellowship. The authors would like to thank Prof. K.L.J. Prather (Massachusetts Institute of Technology, USA) for donating E. coli RARE. We thank Dr. C.W. Grathwol (University of Greifswald, Institute of Pharmacy, Germany) for providing reference compounds and I. Menyes (University of Greifswald, Institute of Biochemistry, Germany) for technical support regarding analytics. The Symrise AG (Holzminden, Germany) kindly provided the genes encoding the enzymes CARMm and PPTNi. Research was supported by the Federal Ministry of Food and Agriculture (BMEL; FKZ 22001617), Germany.
Author contributions
Conceptualization, T.B. and R.W.; Methodology, T.B. and R.W.; Investigation, T.B., A.B., Y.B., L.P., and S.W.; Resources, U.T.B. and R.W.; Data Curation, T.B. and L.P.; Writing – Original Draft, T.B.; Writing – Reviewing & Editing, A.B., U.T.B., Y.B., L.P., R.W., and S.W.; Visualization, T.B. and A.B.; Supervision, T.B., U.T.B., and R.W.; Project Administration, T.B., U.T.B., and R.W.; Funding Acquisition, T.B., U.T.B., R.W., and S.W.
Declaration of interests
The authors declare no competing interests.
Published: May 20, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.104326.
Contributor Information
Thomas Bayer, Email: thomas.bayer@uni-greifswald.de.
Ren Wei, Email: ren.wei@uni-greifswald.de.
Supplemental information
Data and code availability
-
•
The genome of E. coli BL21(DE3) and associated metadata were retrieved from the National Center for Biotechnology Information (NCBI; GenBank: CP001509.3). The accession numbers of protein sequences are listed in the Key resources table.
-
•
This paper does not report original code.
-
•
Any additional information required to re-analyze the data reported in this paper is available from the lead contact upon request.
References
- Akhtar M.K., Turner N.J., Jones P.R. Carboxylic acid reductase is a versatile enzyme for the conversion of fatty acids into fuels and chemical commodities. Proc. Natl. Acad. Sci. U S A. 2013;110:87–92. doi: 10.1073/pnas.1216516110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asmar A.T., Ferreira J.L., Cohen E.J., Cho S.-H., Beeby M., Hughes K.T., Collet J.-F. Communication across the bacterial cell envelope depends on the size of the periplasm. PLoS Biol. 2017;15:e2004303. doi: 10.1371/journal.pbio.2004303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayedi M.A., Le Bigot Y., Ammar H., Abid S., Gharbi R.E., Delmas M. Synthesis of primary amines by one-pot reductive amination of aldehydes. Synth. Commun. 2013;43:2127–2133. doi: 10.1080/00397911.2012.714830. [DOI] [Google Scholar]
- Barnicki S.D. In: Handbook of Industrial Chemistry and Biotechnology. Kent J.A., Bommaraju T.V., Barnicki S.D., editors. Springer International Publishing; 2017. Synthetic organic chemicals; pp. 423–530. [DOI] [Google Scholar]
- Bayer T., Becker A., Terholsen H., Kim I.J., Menyes I., Buchwald S., Balke K., Santala S., Almo S.C., Bornscheuer U.T. LuxAB-based microbial cell factories for the sensing, manufacturing and transformation of industrial aldehydes. Catalysts. 2021;11:953–1017. doi: 10.3390/catal11080953. [DOI] [Google Scholar]
- Bayer T., Milker S., Wiesinger T., Winkler M., Mihovilovic M.D., Rudroff F. In vivo synthesis of polyhydroxylated compounds from a “hidden reservoir” of toxic aldehyde species. ChemCatChem. 2017;9:2919–2923. doi: 10.1002/cctc.201700469. [DOI] [Google Scholar]
- Bornscheuer U.T. Feeding on plastic. Science. 2016;351:1154–1155. doi: 10.1126/science.aaf2853. [DOI] [PubMed] [Google Scholar]
- Brindell G.D., Lillwitz L.D., Wuskell J.P., Dunlop A.P. Polymer applications of some terephthalaldehyde derivatives. Ind. Eng. Chem. Prod. Res. Dev. 1976;15:83–88. doi: 10.1021/i360057a017. [DOI] [Google Scholar]
- Butler N., Kunjapur A.M. Carboxylic acid reductases in metabolic engineering. J. Biotechnol. 2020;307:1–14. doi: 10.1016/j.jbiotec.2019.10.002. [DOI] [PubMed] [Google Scholar]
- Colepicolo P., Cho K.W., Poinar G.O., Hastings J.W. Growth and luminescence of the bacterium Xenorhabdus luminescens from a human wound. Appl. Environ. Microbiol. 1989;55:2601–2606. doi: 10.1128/aem.55.10.2601-2606.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danso D., Schmeisser C., Chow J., Zimmermann W., Wei R., Leggewie C., Li X., Hazen T., Parales R.E., Streit W.R. New insights into the function and global distribution of polyethylene terephthalate (PET)-degrading bacteria and enzymes in marine and terrestrial metagenomes. Appl. Environ. Microbiol. 2018;84 doi: 10.1128/AEM.02773-17. e02773–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich J.A., McKee A.E., Keasling J.D. High-throughput metabolic engineering: advances in small-molecule screening and selection. Annu. Rev. Biochem. 2010;79:563–590. doi: 10.1146/annurev-biochem-062608-095938. [DOI] [PubMed] [Google Scholar]
- Eberl A., Heumann S., Brückner T., Araujo R., Cavaco-Paulo A., Kaufmann F., Kroutil W., Guebitz G.M. Enzymatic surface hydrolysis of poly(ethylene terephthalate) and bis(benzoyloxyethyl) terephthalate by lipase and cutinase in the presence of surface active molecules. J. Biotechnol. 2009;143:207–212. doi: 10.1016/j.jbiotec.2009.07.008. [DOI] [PubMed] [Google Scholar]
- Eriksen M., Lebreton L.C.M., Carson H.S., Thiel M., Moore C.J., Borerro J.C., Galgani F., Ryan P.G., Reisser J. Plastic pollution in the world’s oceans: more than 5 trillion plastic pieces weighing over 250,000 tons afloat at sea. PLoS One. 2014;9 doi: 10.1371/journal.pone.0111913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson L.N. The synthesis of aromatic aldehydes. Chem. Rev. 1946;38:227–254. doi: 10.1021/cr60120a002. [DOI] [PubMed] [Google Scholar]
- Finnigan W., Thomas A., Cromar H., Gough B., Snajdrova R., Adams J.P., Littlechild J.A., Harmer N.J. Characterization of carboxylic acid reductases as enzymes in the toolbox for synthetic chemistry. ChemCatChem. 2017;9:1005–1017. doi: 10.1002/cctc.201601249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleiss A., Sarkisyan K.S. A brief review of bioluminescent systems (2019) Curr. Genet. 2019;65:877–882. doi: 10.1007/s00294-019-00951-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geyer R., Jambeck J.R., Law K.L. Production, use, and fate of all plastics ever made. Sci. Adv. 2017;3:e1700782. doi: 10.1126/sciadv.1700782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godoy-Alcántar C., Yatsimirsky A.K., Lehn J.-M. Structure-stability correlations for imine formation in aqueous solution. J. Phys. Org. Chem. 2005;18:979–985. doi: 10.1002/poc.941. [DOI] [Google Scholar]
- Herrero Acero E., Ribitsch D., Steinkellner G., Gruber K., Greimel K., Eiteljoerg I., Trotscha E., Wei R., Zimmermann W., Zinn M., et al. Enzymatic surface hydrolysis of PET: effect of structural diversity on kinetic properties of cutinases from Thermobifida. Macromolecules. 2011;44:4632–4640. doi: 10.1021/ma200949p. [DOI] [Google Scholar]
- Hopewell J., Dvorak R., Kosior E. Plastics recycling: challenges and opportunities. Philos. Trans. R. Soc. B Biol. Sci. 2009;364:2115–2126. doi: 10.1098/rstb.2008.0311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvat M., Winkler M. In vivo reduction of medium- to long-chain fatty acids by carboxylic acid reductase (CAR) enzymes: limitations and solutions. ChemCatChem. 2020;12:5076–5090. doi: 10.1002/cctc.202000895. [DOI] [Google Scholar]
- Jiang Y., Chen B., Duan C., Sun B., Yang J., Yang S. Multigene editing in the Escherichia coli genome via the CRISPR-Cas9 system. Appl. Environ. Microbiol. 2015;81:2506–2514. doi: 10.1128/AEM.04023-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasai D., Kitajima M., Fukuda M., Masai E. Transcriptional regulation of the terephthalate catabolism operon in Comamonas sp. strain E6. Appl. Environ. Microbiol. 2010;76:6047–6055. doi: 10.1128/AEM.00742-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai F., Kawabata T., Oda M. Current state and perspectives related to the polyethylene terephthalate hydrolases available for biorecycling. ACS Sustain. Chem. Eng. 2020;8:8894–8908. doi: 10.1021/acssuschemeng.0c01638. [DOI] [Google Scholar]
- Kawai F., Kawabata T., Oda M. Current knowledge on enzymatic PET degradation and its possible application to waste stream management and other fields. Appl. Microbiol. Biotechnol. 2019;103:4253–4268. doi: 10.1007/s00253-019-09717-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunjapur A.M., Hyun J.C., Prather K.L.J. Deregulation of S-adenosylmethionine biosynthesis and regeneration improves methylation in the E. coli de novo vanillin biosynthesis pathway. Microb. Cell Factories. 2016;15:61. doi: 10.1186/s12934-016-0459-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunjapur A.M., Prather K.L.J. Development of a vanillate biosensor for the vanillin biosynthesis pathway in E. coli. ACS Synth. Biol. 2019;8:1958–1967. doi: 10.1021/acssynbio.9b00071. [DOI] [PubMed] [Google Scholar]
- Kunjapur A.M., Prather K.L.J. Microbial engineering for aldehyde synthesis. Appl. Environ. Microbiol. 2015;81:1892–1901. doi: 10.1128/AEM.03319-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunjapur A.M., Tarasova Y., Prather K.L.J. Synthesis and accumulation of aromatic aldehydes in an engineered strain of Escherichia coli. J. Am. Chem. Soc. 2014;136:11644–11654. doi: 10.1021/ja506664a. [DOI] [PubMed] [Google Scholar]
- Lehtinen T., Santala V., Santala S. Twin-layer biosensor for real-time monitoring of alkane metabolism. FEMS. Microbiol. Lett. 2017;364:1–7. doi: 10.1093/femsle/fnx053. [DOI] [PubMed] [Google Scholar]
- Li G.-W., Burkhardt D., Gross C., Weissman J.S. Quantifying absolute protein synthesis rates reveals principles underlying allocation of cellular resources. Cell. 2014;157:624–635. doi: 10.1016/j.cell.2014.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Nina M.R.H., Zhang X., Bai Y. Engineering transcription factor XylS for sensing phthalic acid and terephthalic acid: an application for enzyme evolution. ACS Synth. Biol. 2022;11:1106–1113. doi: 10.1021/acssynbio.1c00275. [DOI] [PubMed] [Google Scholar]
- Liu D., Evans T., Zhang F. Applications and advances of metabolite biosensors for metabolic engineering. Metab. Eng. 2015;31:35–43. doi: 10.1016/j.ymben.2015.06.008. [DOI] [PubMed] [Google Scholar]
- Liu Y., Zhuang Y., Ding D., Xu Y., Sun J., Zhang D. Biosensor-based evolution and elucidation of a biosynthetic pathway in Escherichia coli. ACS Synth. Biol. 2017;6:837–848. doi: 10.1021/acssynbio.6b00328. [DOI] [PubMed] [Google Scholar]
- Markel U., Essani K.D., Besirlioglu V., Schiffels J., Streit W.R., Schwaneberg U. Advances in ultrahigh-throughput screening for directed enzyme evolution. Chem. Soc. Rev. 2020;49:233–262. doi: 10.1039/C8CS00981C. [DOI] [PubMed] [Google Scholar]
- Mathelié-Guinlet M., Asmar A.T., Collet J.-F., Dufrêne Y.F. Lipoprotein Lpp regulates the mechanical properties of the E. coli cell envelope. Nat. Commun. 2020;11:1789. doi: 10.1038/s41467-020-15489-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller R.-J., Schrader H., Profe J., Dresler K., Deckwer W.-D. Enzymatic Degradation of poly(ethylene terephthalate): rapid hydrolyse using a hydrolase from T. fusca. Macromol. Rapid Commun. 2005;26:1400–1405. doi: 10.1002/marc.200500410. [DOI] [Google Scholar]
- Ni Y., Reye J., Chen R.R. Lpp deletion as a permeabilization method. Biotechnol. Bioeng. 2007;97:1347–1356. doi: 10.1002/bit.21375. [DOI] [PubMed] [Google Scholar]
- Palm G.J., Reisky L., Böttcher D., Müller H., Michels E.A.P., Walczak M.C., Berndt L., Weiss M.S., Bornscheuer U.T., Weber G. Structure of the plastic-degrading Ideonella sakaiensis MHETase bound to a substrate. Nat. Commun. 2019;10:1717–1810. doi: 10.1038/s41467-019-09326-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardo I., Jha R.K., Bermel R.E., Bratti F., Gaddis M., McIntyre E., Michener W., Neidle E.L., Dale T., Beckham G.T., Johnson C.W. Gene amplification, laboratory evolution, and biosensor screening reveal MucK as a terephthalic acid transporter in Acinetobacter baylyi ADP1. Metab. Eng. 2020;62:260–274. doi: 10.1016/j.ymben.2020.09.009. [DOI] [PubMed] [Google Scholar]
- Pfaff L., Breite D., Badenhorst C.P.S., Bornscheuer U.T., Wei R. In: Methods in Enzymology. Weber G., Bornscheuer U.T., Wei R., editors. Academic Press; 2021. Fluorimetric high-throughput screening method for polyester hydrolase activity using polyethylene terephthalate nanoparticles; pp. 253–270. [DOI] [PubMed] [Google Scholar]
- Qu G., Guo J., Yang D., Sun Z. Biocatalysis of carboxylic acid reductases: phylogenesis, catalytic mechanism and potential applications. Green Chem. 2018;20:777–792. doi: 10.1039/C7GC03046K. [DOI] [Google Scholar]
- Rahimi A., García J.M. Chemical recycling of waste plastics for new materials production. Nat. Rev. Chem. 2017;1 doi: 10.1038/s41570-017-0046. [DOI] [Google Scholar]
- Meys R., Kätelhön A., Bachmann M., Winter B., Zibunas C., Suh S., Bardow A. Achieving net-zero greenhouse gas emission plastics by a circular carbon economy. Science. 2021;374:71–76. doi: 10.1126/science.abg9853. [DOI] [PubMed] [Google Scholar]
- Ressmann A.K., Schwendenwein D., Leonhartsberger S., Mihovilovic M.D., Bornscheuer U.T., Winkler M., Rudroff F. Substrate-independent high-throughput assay for the quantification of aldehydes. Adv. Synth. Catal. 2019;361:2538–2543. doi: 10.1002/adsc.201900154. [DOI] [Google Scholar]
- Rohan R., Pareek K., Cai W., Zhang Y., Xu G., Chen Z., Gao Z., Dan Z., Cheng H. Melamine-terephthalaldehyde-lithium complex: a porous organic network based single ion electrolyte for lithium-ion batteries. J. Mater. Chem. A. 2015;3:5132–5139. doi: 10.1039/C4TA06855F. [DOI] [Google Scholar]
- Ronkvist Å.M., Xie W., Lu W., Gross R.A. Cutinase-catalyzed hydrolysis of poly(ethylene terephthalate) Macromolecules. 2009;42:5128–5138. doi: 10.1021/ma9005318. [DOI] [Google Scholar]
- Sadler J.C., Wallace S. Microbial synthesis of vanillin from waste poly(ethylene terephthalate) Green Chem. 2021;23:4665–4672. doi: 10.1039/D1GC00931A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakadellis S., Rosetto G. Achieving a circular bioeconomy for plastics. Science. 2021;373:49–50. doi: 10.1126/science.abj3476. [DOI] [PubMed] [Google Scholar]
- Saran M., Summer K.H. Assaying for hydroxyl radicals: hydroxylated terephthalate is a superior fluorescence marker than hydroxylated benzoate. Free Radic. Res. 1999;31:429–436. doi: 10.1080/10715769900300991. [DOI] [PubMed] [Google Scholar]
- Simion A., Simion C., Kanda T., Nagashima S., Mitoma Y., Yamada T., Mimura K., Tashiro M. Synthesis of imines, diimines and macrocyclic diimines as possible ligands, in aqueous solution. J. Chem. Soc. Perkin Trans. 2001;1:2071–2078. doi: 10.1039/B102749M. [DOI] [Google Scholar]
- Simon N., Raubenheimer K., Urho N., Unger S., Azoulay D., Farrelly T., Sousa J., van Asselt H., Carlini G., Sekomo C., et al. A binding global agreement to address the life cycle of plastics. Science. 2021;373:43–47. doi: 10.1126/science.abi9010. [DOI] [PubMed] [Google Scholar]
- Snell J.M., Weissberger A. Synthesis of terephthalaldehyde. Org. Synth. 1940;20:92. doi: 10.15227/orgsyn.020.0092. [DOI] [Google Scholar]
- Studier F.W. Protein production by auto-induction in high-density shaking cultures. Protein Expr. Purif. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- Suematsu K., Nakamura K., Takeda J. Synthesis of aromatic polyimines by the condensation of aromatic dialdehyde and diamine. Colloid Polym. Sci. 1983;261:493–501. doi: 10.1007/BF01419833. [DOI] [Google Scholar]
- Sulaiman S., Yamato S., Kanaya E., Kim J.-J., Koga Y., Takano K., Kanaya S. Isolation of a novel cutinase homolog with polyethylene terephthalate-degrading activity from leaf-branch compost by using a metagenomic approach. Appl. Environ. Microbiol. 2012;78:1556–1562. doi: 10.1128/AEM.06725-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiso T., Narancic T., Wei R., Pollet E., Beagan N., Schröder K., Honak A., Jiang M., Kenny S.T., Wierckx N., et al. Towards bio-upcycling of polyethylene terephthalate. Metab. Eng. 2021;66:167–178. doi: 10.1016/j.ymben.2021.03.011. [DOI] [PubMed] [Google Scholar]
- Tournier V., Topham C.M., Gilles A., David B., Folgoas C., Moya-Leclair E., Kamionka E., Desrousseaux M.-L., Texier H., Gavalda S., et al. An engineered PET depolymerase to break down and recycle plastic bottles. Nature. 2020;580:216–219. doi: 10.1038/s41586-020-2149-4. [DOI] [PubMed] [Google Scholar]
- Vogel K., Wei R., Pfaff L., Breite D., Al-Fathi H., Ortmann C., Estrela-Lopis I., Venus T., Schulze A., Harms H., et al. Enzymatic degradation of polyethylene terephthalate nanoplastics analyzed in real time by isothermal titration calorimetry. Sci. Total Environ. 2021;773:145111. doi: 10.1016/j.scitotenv.2021.145111. [DOI] [PubMed] [Google Scholar]
- Vollmer I., Jenks M.J.F., Roelands M.C.P., White R.J., van Harmelen T., de Wild P., van der Laan G.P., Meirer F., Keurentjes J.T.F., Weckhuysen B.M. Beyond mechanical recycling: giving new life to plastic waste. Angew. Chem. Int. Ed. 2020;59:15402–15423. doi: 10.1002/anie.201915651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Gao S., Wang J., Xu S., Li H., Chen K., Ouyang P. The production of biobased diamines from renewable carbon sources: Current advances and perspectives. Chin. J. Chem. Eng. 2021;30:4–13. doi: 10.1016/j.cjche.2020.12.009. [DOI] [Google Scholar]
- Wei R., Oeser T., Billig S., Zimmermann W. A high-throughput assay for enzymatic polyester hydrolysis activity by fluorimetric detection. Biotechnol. J. 2012;7:1517–1521. doi: 10.1002/biot.201200119. [DOI] [PubMed] [Google Scholar]
- Wei R., Oeser T., Then J., Kühn N., Barth M., Schmidt J., Zimmermann W. Functional characterization and structural modeling of synthetic polyester-degrading hydrolases from Thermomonospora curvata. AMB Express. 2014;4:44. doi: 10.1186/s13568-014-0044-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei R., Breite D., Song C., Gräsing D., Ploss T., Hille P., Schwerdtfeger R., Matysik J., Schulze A., Zimmermann W. Biocatalytic degradation efficiency of postconsumer polyethylene terephthalate packaging determined by their polymer microstructures. Adv. Sci. 2019;6:1900491. doi: 10.1002/advs.201900491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei R., Song C., Gräsing D., Schneider T., Bielytskyi P., Böttcher D., Matysik J., Bornscheuer U.T., Zimmermann W. Conformational fitting of a flexible oligomeric substrate does not explain the enzymatic PET degradation. Nat. Commun. 2019;10:5581. doi: 10.1038/s41467-019-13492-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei R., Tiso T., Bertling J., O’Connor K., Blank L.M., Bornscheuer U.T. Possibilities and limitations of biotechnological plastic degradation and recycling. Nat. Catal. 2020;3:867–871. doi: 10.1038/s41929-020-00521-w. [DOI] [Google Scholar]
- Wei R., von Haugwitz G., Pfaff L., Mican J., Badenhorst C.P.S., Liu W., Weber G., Austin H.P., Bednar D., Damborsky J., Bornscheuer U.T. Mechanism-based design of efficient PET hydrolases. ACS Catal. 2022;12:3382–3396. doi: 10.1021/acscatal.1c05856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch K.D., Davis T.Z., Aust S.D. Iron autoxidation and free radical generation: effects of buffers, ligands, and chelators. Arch. Biochem. Biophys. 2002;397:360–369. doi: 10.1006/abbi.2001.2694. [DOI] [PubMed] [Google Scholar]
- Wiltschi B., Cernava T., Dennig A., Galindo Casas M., Geier M., Gruber S., Haberbauer M., Heidinger P., Herrero Acero E., Kratzer R., et al. Enzymes revolutionize the bioproduction of value-added compounds: from enzyme discovery to special applications. Biotechnol. Adv. 2020;40:107520. doi: 10.1016/j.biotechadv.2020.107520. [DOI] [PubMed] [Google Scholar]
- Wright S.L., Kelly F.J. Plastic and human health: a micro issue? Environ. Sci. Technol. 2017;51:6634–6647. doi: 10.1021/acs.est.7b00423. [DOI] [PubMed] [Google Scholar]
- Yi D., Bayer T., Badenhorst C.P.S., Wu S., Doerr M., Höhne M., Bornscheuer U.T. Recent trends in biocatalysis. Chem. Soc. Rev. 2021;50:8003–8049. doi: 10.1039/D0CS01575J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida S., Hiraga K., Takehana T., Taniguchi I., Yamaji H., Maeda Y., Toyohara K., Miyamoto K., Kimura Y., Oda K. A bacterium that degrades and assimilates poly(ethylene terephthalate) Science. 2016;351:1196–1199. doi: 10.1126/science.aad6359. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Werling U., Edelmann W. SLiCE: a novel bacterial cell extract-based DNA cloning method. Nucleic Acids Res. 2012;40:e55. doi: 10.1093/nar/gkr1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann W., Wei R., Hille P., Oeser T., Schmidt J. 2019. EP3517608A1: New Polypeptides Having a Polyester Degrading Activity and Uses Thereof. https://patents.google.com/patent/EP3517608A1/en. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
The genome of E. coli BL21(DE3) and associated metadata were retrieved from the National Center for Biotechnology Information (NCBI; GenBank: CP001509.3). The accession numbers of protein sequences are listed in the Key resources table.
-
•
This paper does not report original code.
-
•
Any additional information required to re-analyze the data reported in this paper is available from the lead contact upon request.