Abstract
Asparagine-linked glycosylation (N-glycosylation) of proteins in the cancer secretome has been gaining increasing attention as a potential biomarker for cancer detection and diagnosis. Small extracellular vesicles (sEVs) constitute a large part of the cancer secretome, yet little is known about whether their N-glycosylation status reflects known cancer characteristics. Here, we investigated the N-glycosylation of sEVs released from small-cell lung carcinoma (SCLC) and non–small-cell lung carcinoma (NSCLC) cells. We found that the N-glycans of SCLC-sEVs were characterized by the presence of structural units also found in the brain N-glycome, while NSCLC-sEVs were dominated by typical lung-type N-glycans with NSCLC-associated core fucosylation. In addition, lectin-assisted N-glycoproteomics of SCLC-sEVs and NSCLC-sEVs revealed that integrin αV was commonly expressed in sEVs of both cancer cell types, while the epithelium-specific integrin α6β4 heterodimer was selectively expressed in NSCLC-sEVs. Importantly, N-glycomics of the immunopurified integrin α6 from NSCLC-sEVs identified NSCLC-type N-glycans on this integrin subunit. Thus, we conclude that protein N-glycosylation in lung cancer sEVs may potentially reflect the histology of lung cancers.
Keywords: exosome (vesicle), extracellular vesicles, glycomics, glycosylation, integrins, lung cancers
Abbreviations: ALIX, ALG 2–interacting protein X; CD81, cluster of differentiation 81; ESI, electrospray ionization; FLOT-1, flotillin-1; LacdiNAc, N-acetylgalactosamine-GlcNAc; LC, liquid chromatography; LCC, large-cell lung carcinoma; LUAD, lung adenocarcinoma; MS, mass spectrometry; NSCLC, non–small-cell lung carcinoma; PA, 2-aminopyridine; PNGase F, peptide:N-glycanase F; PVDF, polyvinylidene difluoride; SCC, squamous-cell lung carcinoma; SCLC, small-cell lung carcinoma; sEV, small extracellular vesicle; SSA, Sambucus sieboldiana; WGA, wheat germ agglutinin
Lung cancer is a histologically complex disease that arises from different types of cells in the lungs. Small-cell lung carcinomas (SCLCs) and non–small-cell lung carcinomas (NSCLCs) are the two major lung cancer types classified based upon their histological properties (1). NSCLCs are further divided into three subtypes, lung adenocarcinomas (LUADs), squamous-cell lung carcinomas (SCCs), and large-cell lung carcinomas (LCCs), in descending order of frequency. Identification of the histological types of lung cancers, as well as the driver genes, is critical for clinical decisions to select the most effective treatments (2, 3, 4, 5). Thus, further understanding of the molecular basis that distinguishes lung cancer types may aid the diagnosis and discovery of druggable targets.
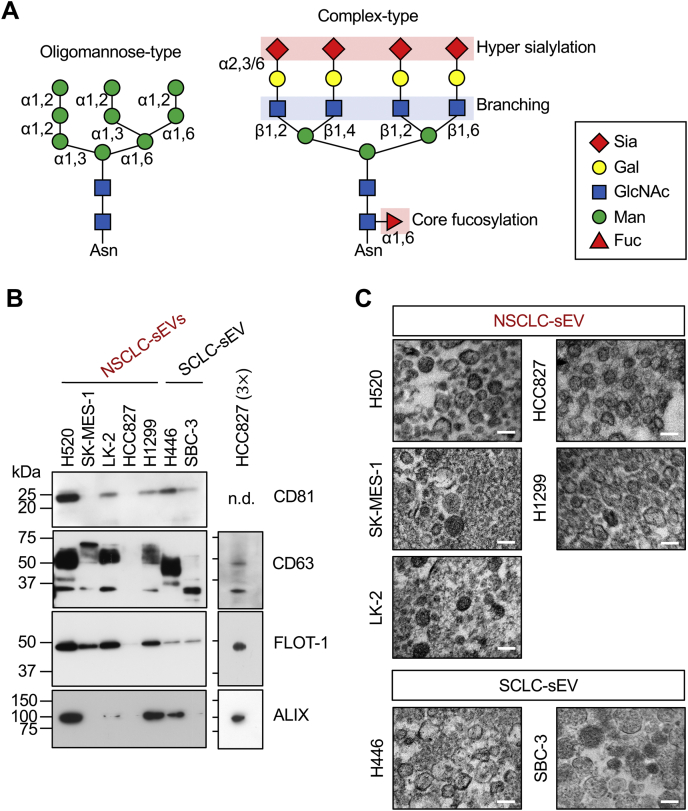
The surface of cells is covered with dense glycocalyx, which is covalently attached to proteins and lipids. Among several glycosylation types, asparagine-linked glycosylation (N-glycosylation) is a fundamental posttranslational modification of secretory and membrane proteins synthesized in the endoplasmic reticulum (6). In the endoplasmic reticulum, evolutionarily conserved oligomannose-type N-glycans are added to nascent polypeptides, and the glycans undergo structural remodeling to complex-type N-glycans during the maturation process of glycoproteins in the Golgi (Fig. 1A). The N-glycan remodeling process is tightly regulated by genetic, epigenetic, and environmental factors. These factors generate unique expression patterns of glycan-modifying enzymes in different cell contexts, thus creating a cell-type–specific N-glycome. For instance, the human brain N-glycome is abundant in unique structural motifs, such as bisecting GlcNAc, N-acetylgalactosamine-GlcNAc (LacdiNAc), and Lewis-type antennal fucose (7, 8, 9), while the human lung N-glycome is dominated by biantennary complex-type N-glycans (10). In contrast, dysregulation of the glycosylation-regulating mechanisms in malignant cells generates cancer-associated, functional N-glycan structures, for example, increased core fucosylation, more branching, and hypersialylation (Fig. 1A) (11, 12, 13). These structural changes on particular secretory and membrane glycoproteins have gained increasing attention as potential biomarkers for the detection and diagnosis of cancers, such as liver and prostate cancers (14, 15, 16).
Figure 1.
SCLC and NSCLC cells secrete qualitatively different sEVs.A, schematic representations of oligomannose-type and complex-type N-glycans. Sialic acids (Sia) are linked to galactose (Gal) via α2,3 or α2,6 linkage. β1,2- and β1,4-linked GlcNAc branching occurs on the α1,3-mannose (Man) arm of N-glycan. β1,2- and β1,6-linked GlcNAc branching occurs on the α1,6-Man arm of N-glycan. α1,6-core fucosylation occurs on the innermost GlcNAc residue. B, Western blot analysis of sEVs (5 μg of protein for left panels or 15 μg (3×) of protein for right panels) released from SCC (H520, SK-MES-1, and LK-2), LCC (H1299), LUAD (HCC827), and SCLC (H446 and SBC-3) cells. n.d., not detected. C, transmission electron microscopy (TEM) analysis of sEVs prepared in (B). The scale bars represent 200 nm. Western blots are representative images from three independent experiments. TEM analysis was performed once. LCC, large-cell lung carcinoma; LUAD, lung adenocarcinoma; NSCLC, non–small-cell lung carcinoma; SCC, squamous-cell lung carcinoma; SCLC, small-cell lung carcinoma; sEV, small extracellular vesicle.
A cancer secretome contains information that correlates with the cancer types and disease states (17, 18). Cancer cells secrete nanosized (50–200 nm), lipid bilayer–enclosed membrane vesicles that carry a wide range of macromolecules (e.g., messenger RNAs, micro-RNAs, membrane/soluble proteins, and glycans) (19, 20, 21, 22, 23, 24, 25). Extensive molecular profiling of these vesicles, termed small extracellular vesicles (sEVs), has revealed that the transcriptome and proteome of sEVs differ between healthy individuals and cancer patients, as well as between tumor and adjacent tissues and between cell lines (26, 27, 28, 29, 30, 31, 32). Interestingly, the expression profiles of membrane proteins, most of which are modified with N-glycans, in sEVs have been proposed to promote tumor progression, as well as to determine the metastatic sites of tumor cells. For instance, EV-associated integrins α6β1 and α6β4, which are laminin receptors, are linked to lung metastasis, while EV-associated integrin αVβ5, which binds to the Arg–Gly–Asp motif, promotes liver metastasis in mice (31). These membrane proteins, as well as several others (26, 27) that were released into culture medium can be detected in blood samples from cancer patients, highlighting the clinical relevance of sEVs as a potential biomarker for cancers. However, much remains unknown about whether the N-glycosylation profiles of cancer cell–derived sEVs are associated with the originating cell types and disease states.
In the present study, we performed a comparative N-glycan analysis of sEVs released from four histologically distinct lung cancer types (SCLC and NSCLC [LUAD, SCC, and LCC] cells) to examine whether the N-glycan signatures of the sEVs exist and reflect lung cancer types. The analysis revealed that SCLC-sEVs had no common N-glycan signatures, but they contained brain-associated structural motifs. In contrast, NSCLC-sEVs were dominated by typical lung-type N-glycans with NSCLC-associated core fucosylation. We identified several integrin subunits as N-glycoproteins in lung cancer sEVs and found that the αV subunit was expressed in both SCLC-sEVs and NSCLC-sEVs. However, the epithelium-specific integrin α6β4 heterodimer was selectively expressed in NSCLC-sEVs, and integrin α6 carried NSCLC-type N-glycans. Overall, our data reveal the molecular link between lung cancer types and the protein N-glycosylation of sEVs.
Results
SCLC and NSCLC cells secrete qualitatively different sEVs
We prepared sEVs from a conditioned medium of SCLC (H446 and SBC-3) and NSCLC (LUAD [HCC827], SCC [H520, SK-MES-1, and LK-2], and LCC [H1299]) cells. Equal amounts of the sEV preparations were analyzed for the expression of sEV marker proteins. We found that a cluster of differentiation 81 (CD81), CD63, flotillin-1 (FLOT-1), and ALG 2–interacting protein X (ALIX) were expressed at different levels in lung cancer sEVs and that HCC827-sEVs expressed little CD81 and very low amounts of CD63, FLOT-1, and ALIX as compared to the other sEVs (Fig. 1B). Transmission electron microscopy analysis of these sEV preparations revealed rounded shapes of the vesicles with sizes of 100 to 200 nm (Fig. 1C). These results indicate that SCLC and NSCLC cells secrete qualitatively different sEVs, but the expression pattern of the sEV markers does not correlate with the originating lung cancer types.
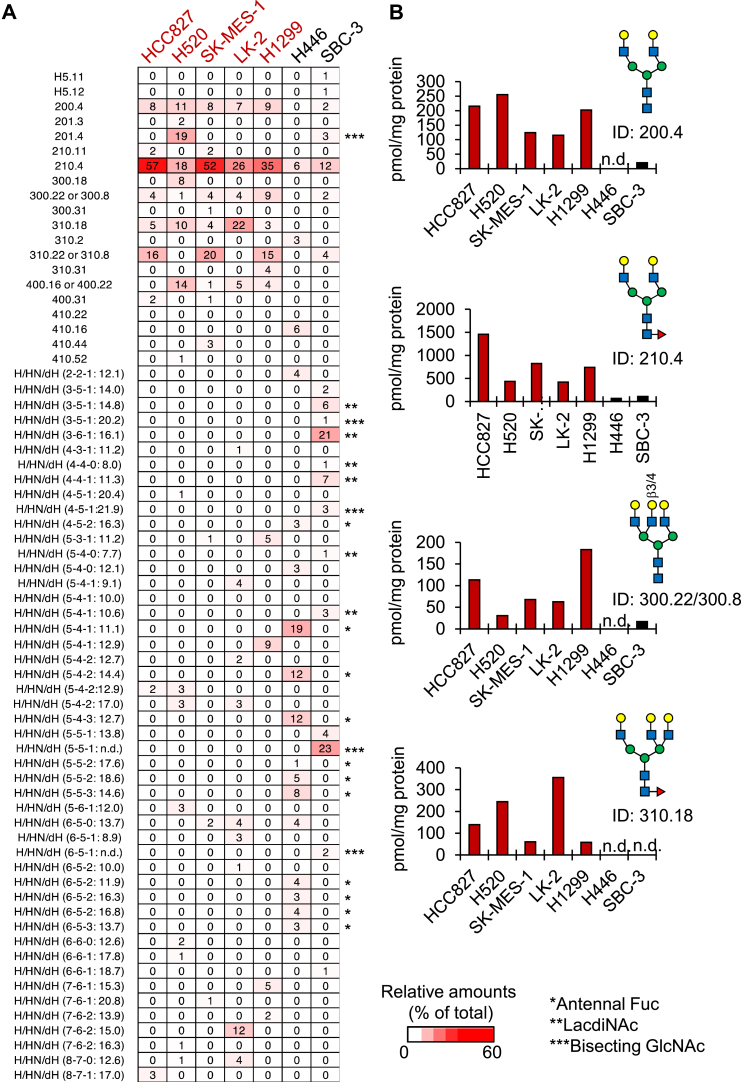
SCLC-sEVs and NSCLC-sEVs have different N-glycan profiles
We noticed that CD63 showed remarkable differences in molecular weight between lung cancer cell–derived sEVs (Fig. 1B). CD63 has three potential N-glycosylation sites (33), suggesting that the N-glycosylation status of sEVs differs between lung cancer types. Sialic acids are negatively charged monosaccharides typically located at the outermost part of N-glycans, and we found that N-glycans from SCLC-sEVs and NSCLC-sEVs were mainly monosialylated and disialylated at different levels (Fig. S1), indicating that the number of sialic acids of N-glycans on lung cancer cell–derived sEVs is not associated with lung cancer types.
Sialic acids were removed from the N-glycan preparations by sialidase treatment, and the core N-glycan structures were deduced by reversed-phase HPLC, followed by MALDI-TOF mass spectrometry (MS) or liquid chromatography (LC)-electrospray ionization (ESI)–MS/MS. Interestingly, the core N-glycan structures showed striking differences between SCLC-sEVs and NSCLC-sEVs (Figs. 2, A and B, S2–S5 and Data S1). NSCLC-sEVs were commonly enriched in biantennary and triantennary N-glycans, the core fucosylated forms being the major glycan structures (Figs. 2A, S2 and S3). In contrast, the N-glycan profiles of SCLC-sEVs showed considerable heterogeneity, with an enrichment of N-glycans with antennal fucose (73% of total) in H446-sEVs and those with LacdiNAc (40% of total) or bisecting GlcNAc (29% of total) in SBC-3-sEVs (Figs. 2A, S4 and S5). It should be noted that most N-glycan structures in SCLC-sEVs could not be deduced because the glycan database, GALAXY (http://www.glycoanalysis.info), used for structural analysis did not contain the corresponding reference structures. All of these results indicate that the N-glycans on SCLC-sEVs are structurally heterogeneous, while NSCLC-sEVs are characterized by the presence of core fucosylated, biantennary, and triantennary N-glycans.
Figure 2.
SCLC-sEVs and NSCLC-sEVs have different N-glycan profiles.A, the relative amounts of N-glycans in NSCLC-sEVs (H520, SK-MES-1, LK-2, H1299, and HCC827) and SCLC-sEVs (H446 and SBC-3) were shown as a heatmap. Glycan IDs for deduced N-glycan structures are based on the GALAXY database. Glycan IDs for undeduced N-glycan structures are indicated as the number of H/HN/dH (x-y-z), followed by their GU in reversed-phase HPLC. n.d., not determined. Asterisks indicate N-glycans with antennal fucose (∗), LacdiNAc (∗∗), and bisecting GlcNAc (∗∗∗). B, N-glycans that were enriched in NSCLC-sEVs were extracted from a heatmap in (A), and the amounts were expressed as pmol/mg protein of sEVs. n.d., not detected. See also Figs S2–S5 and Data S1. Three batches of sEVs were independently prepared, fluorescently labeled, and analyzed by ODS-HPLC after desialylation, and similar HPLC profiles were observed among three batches. Rigorous structural analysis by MS was performed once for the representative sample. LacdiNAc, N-acetylgalactosamine-GlcNAc; NSCLC, non–small-cell lung carcinoma; SCLC, small-cell lung carcinoma; sEV, small extracellular vesicle.
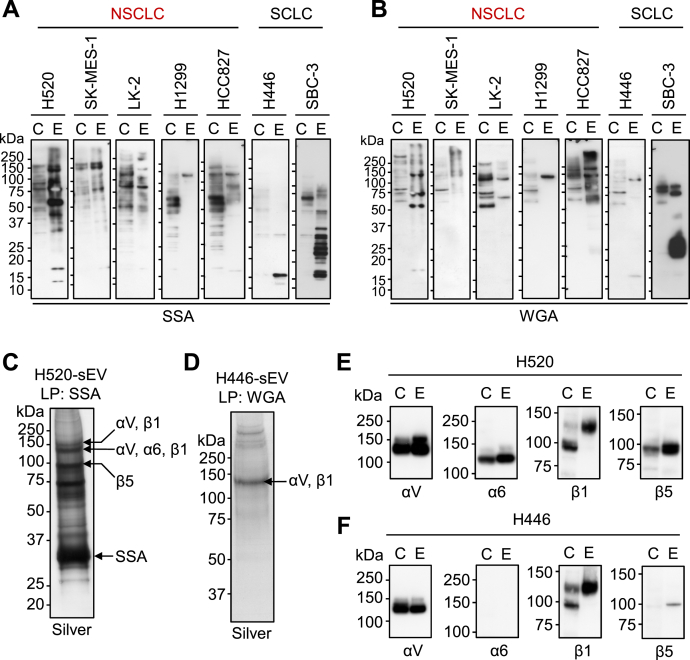
Some glycoproteins are enriched in SCLC-sEVs and NSCLC-sEVs
Our data clearly indicated that SCLC-sEVs and NSCLC-sEVs had different N-glycan profiles, but the carrier N-glycoproteins were not clear. We analyzed the glycoprotein profiles through lectin blotting using sialic acid–binding lectins, as sialic acids were present on the N-glycans of sEVs in all four lung cancer types (Fig. S1). The lectin from Sambucus sieboldiana (SSA) specifically binds to sialic acids linked to galactose via α2,6 (34). The SSA lectin reacted with multiple glycoproteins in SCLC-sEVs and NSCLC-sEVs (Fig. 3A), but H446-sEVs and H1299-sEVs showed relatively simple SSA-staining profiles. Most of the SSA-positive glycoproteins in SCLC-sEVs and NSCLC-sEVs had molecular weights greater than 50 kDa, but H446-sEVs carried 15 kDa SSA-positive glycoproteins. Wheat germ agglutinin (WGA) is a lectin that can recognize terminal GlcNAc residues, as well as sialic acid clusters, on multiantennary glycans (35). The WGA-staining pattern of sEVs was somewhat different from the SSA-staining pattern, and the analysis showed the presence of WGA-positive glycoproteins with molecular weights greater than 50 kDa in H446-sEVs (Fig. 3B). Comparison of SSA- and WGA-staining profiles between cells and the sEVs highlighted the enrichment of several glycoproteins in sEVs, indicating that some glycoproteins are enriched in SCLC-sEVs and NSCLC-sEVs.
Figure 3.
SCLC-sEVs and NSCLC-sEVs express several integrin subunits.A and B, lectin blot analysis of whole-cell lysates (C, 20 μg) and sEVs (E, 5 μg) using SSA (A) and WGA (B) lectins. C and D, lectin pull-down (LP) of glycoproteins from the detergent-solubilized sEVs using SSA-conjugated beads (for H520-sEVs) or WGA-conjugated beads (for H446-sEVs), followed by SDS-PAGE/silver staining. Arrows indicate integrin subunits detected in the corresponding bands in proteomics. E and F, validation of integrin expression by Western blot analysis. Whole-cell lysates (C, 20 μg) and sEVs (E, 5 μg) from H520 (E) and H446 (F) cells were analyzed. See also Data S2 and S3. Lectin blots (A and B) are representative images from three independent experiments. Experiments for (C−F) were performed once. NSCLC, non–small-cell lung carcinoma; SCLC, small-cell lung carcinoma; sEV, small extracellular vesicle; SSA, Sambucus sieboldiana; WGA, wheat germ agglutinin.
SCLC-sEVs and NSCLC-sEVs express several integrin subunits
Our glycoprotein profiling of SCLC-sEVs and NSCLC-sEVs indicated the enrichment of glycoproteins larger than 50 kDa. We pulled down these glycoproteins from the detergent-solubilized sEVs with SSA-conjugated beads for H520-sEVs (Fig. 3C) and WGA-conjugated beads for H446-sEVs (Fig. 3D), and the protein bands were subjected to shotgun proteomics. The analysis revealed that several integrin subunits were enriched in the sEVs: αV, α6, β1, and β5 subunits in H520-sEVs (Fig. 3C and Data S2) and αV and β1 subunits in H446-sEVs (Fig. 3D and Data S3). We confirmed the expression of these integrin subunits by Western blot analysis (Fig. 3, E and F) and found that the integrin α6 subunit is expressed in H520-sEVs but not in H446-sEVs.
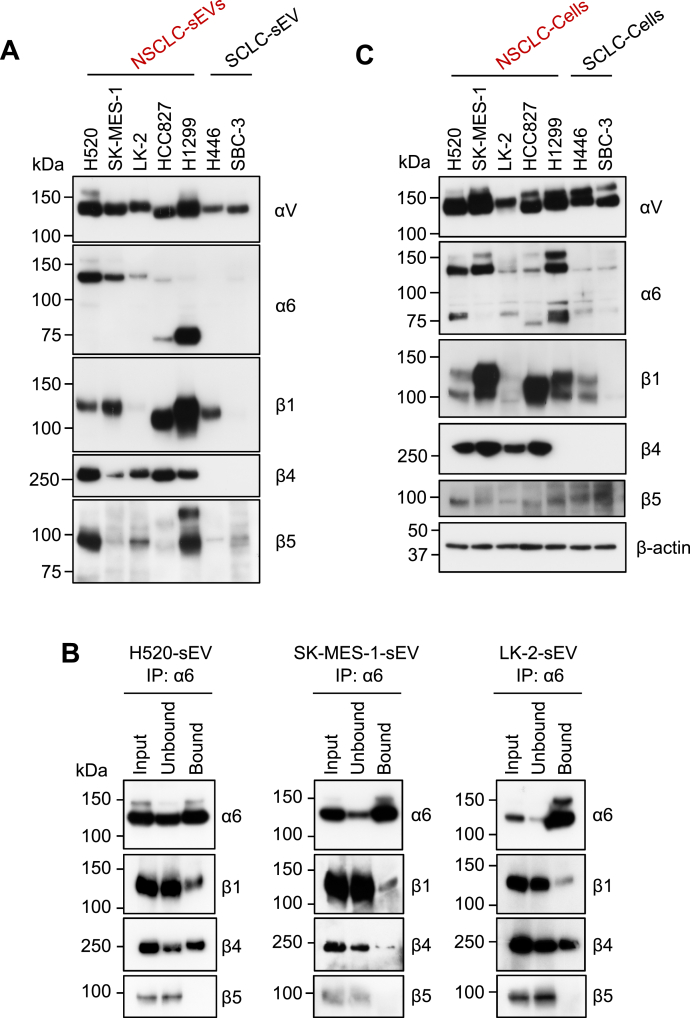
Integrin α6β4 expression is unique to NSCLC-sEVs
To test the hypothesis that integrin α6 expression may be unique to NSCLC-sEVs, we compared the protein-expression levels of integrin subunits between SCLC-sEVs (H446 and SBC-3) and NSCLC-sEVs (SCC [H520, SK-MES-1, and LK-2], LCC [H1299], and LUAD [HCC827]) and found that sEVs from all four lung cancer types contained significant amounts of integrin αV subunits, while integrin β1 and β5 subunits were expressed at different levels in these sEVs (Fig. 4A). In contrast, the integrin α6 subunit (pro [150 kDa], mature [130 kDa], and proteolytic [70 kDa] (36) forms) was detected almost exclusively in NSCLC-sEVs (Fig. 4A). The mature form of the integrin α6 subunit was detected in all NSCLC-sEVs, while the proteolytic 70 kDa form was highly enriched in LCC-sEVs and LUAD-sEVs. The integrin β4 subunit, which was not identified in our lectin-assisted glycoproteomics but is known to form a heterodimer with the integrin α6 subunit (37), was also detected exclusively in NSCLC-sEVs (Fig. 4A). Indeed, the integrin α6 subunit in detergent-solubilized SCC-sEVs was coimmunoprecipitated with the integrin β4 subunit but not with the β5 subunit (Fig. 4B). We also detected integrin α6β1 heterodimers in SCC-sEVs (Fig. 4B). The very low level of integrin α6 and β4 subunits in SCLC-sEVs was most likely because the SCLC cells rarely expressed these integrin subunits (Fig. 4C). Collectively, these results indicate that integrin α6β4 expression is unique to NSCLC-sEVs.
Figure 4.
Integrin α6β4 expression is unique to NSCLC-sEVs.A, Western blot analysis of sEVs (5 μg) prepared from SCC (H520, SK-MES-1, and LK-2), LCC (H1299), LUAD (HCC827), and SCLC (H446 and SBC-3) cells. B, immunoprecipitation (IP) of the integrin α6 subunit from the detergent-solubilized SCC-sEVs. Equal amounts of input, unbound, and bound fractions were analyzed by Western blot. C, Western blot analysis of whole-cell lysates (20 μg) prepared from SCC (H520, SK-MES-1, and LK-2), LCC (H1299), LUAD (HCC827), and SCLC (H446 and SBC-3) cells. Western blots (A and C) are representative images from three independent experiments. LCC, large-cell lung carcinoma; LUAD, lung adenocarcinoma; NSCLC, non–small-cell lung carcinoma; SCC, squamous-cell lung carcinoma; SCLC, small-cell lung carcinoma; sEV, small extracellular vesicle.
The integrin α6 subunit carries NSCLC-type N-glycans
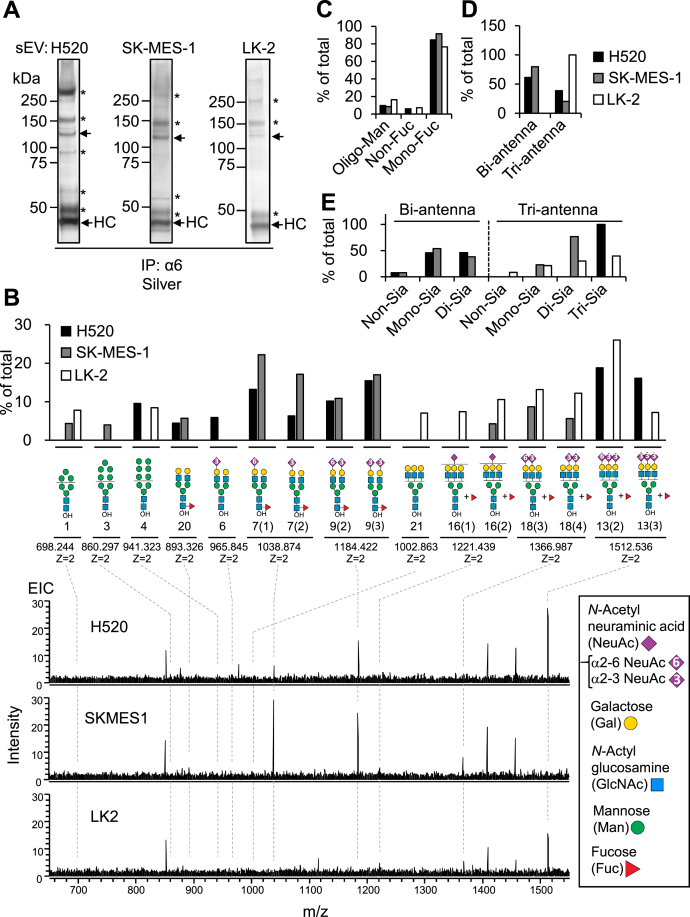
Finally, we examined whether the integrin α6 in NSCLC-sEVs was modified with NSCLC-type N-glycans. To this end, the integrin α6 subunit immunoprecipitated from the detergent-solubilized SCC-sEVs (Fig. 5A) was separated by SDS-PAGE and blotted onto a membrane, and the band corresponding to α6 was excised. N-glycans were then released from the band by peptide:N-glycanase F (PNGase F) digestion. We were able to identify only major structures of the N-glycans through LC-MS analysis due to the limited sample amounts, and we found that oligomannose-type and complex N-glycans were expressed on the integrin α6 subunit (Figs. 5B, S6 and Data S4). Most N-glycans of integrin α6 in all three SCC-sEVs were complex and monofucosylated (Fig. 5C and Data S4). A large majority of the complex-type N-glycans of the integrin α6 subunit in H520-sEVs and SK-MES-1-sEVs were biantennary and triantennary N-glycans, while almost all N-glycans of integrin α6 in LK-2-sEVs were triantennary (Fig. 5D and Data S4). The sialylation status of biantennary N-glycans on the integrin α6 subunit was very similar between H520-sEVs and SK-MES-1-sEVs, while that of triantennary N-glycans differed depending on the source of the sEVs (Fig. 5E and Data S4). Although the fucosylation sites of triantennary N-glycans on the integrin α6 subunit were not determined due to the lack of standard glycans, most of the biantennary N-glycans were modified with core fucose (Figs. 5B and S6). Collectively, these results indicate that the integrin α6 subunit in NSCLC-sEVs is modified with NSCLC-type N-glycans.
Figure 5.
The integrin α6 subunit carries NSCLC-type N-glycans.A, IP of the integrin α6 subunit from detergent-solubilized SCC-sEVs (H520, SK-MES-1, and LK-2), followed by SDS-PAGE/silver staining. Heavy chain (HC) of immunoglobulin G used for immunoprecipitation. Arrows indicate the immunoprecipitated integrin α6 subunit. Asterisks indicate bands that were not detected with the anti-integrin α6 antibody in Western blot analysis. B, LC-ESI-MS analysis of major N-glycans detected in the immunoprecipitated integrin α6 subunit prepared in (A). The relative amounts (%, upper panel) of each glycan structure were calculated by setting the total peak intensities of all detected alditol N-glycans in each extracted-ion chromatogram (EIC) to 100%. Peak intensities of each alditol N-glycan were calculated based on the EIC in Fig. S6. Bottom panels show the average mass spectrum (28–55 min) in the base peak chromatogram of N-glycan alditols. The deduced structures of the major N-glycans are shown with the theoretical mass and the charge state. The structure number, observed mass, theoretical mass, peak intensities, and retention time are summarized in Data S4. C, relative amounts of oligomannose-type (Oligo-Man) and nonfucosylated (Non-Fuc) or monofucosylated (Mon-Fuc) complex-type N-glycans in total N-glycans. D, relative amounts of biantennary (Bi-antenna) and triantennary (Tri-antenna) N-glycans in the total complex-type N-glycans. E, sialylation profiles (Non-Sia, Mono-Sia, Di-Sia, and Tri-Sia) of biantennary and triantennary N-glycans. See also Fig. S6 and Data S4. All experiments were performed once. ESI, electrospray ionization; LC, liquid chromatography; MS, mass spectrometry; NSCLC, non–small-cell lung carcinoma; SCC, squamous-cell lung carcinoma; sEV, small extracellular vesicle.
Discussion
In the present study, we demonstrated that NSCLC-sEVs abundantly express core fucosylated, biantennary, and triantennary N-glycans. Furthermore, NSCLC-sEVs expressed the epithelium-specific integrin α6β4. Importantly, NSCLC-type N-glycans were present on the integrin α6 subunit in NSCLC-sEVs. In contrast, the N-glycans on SCLC-sEVs exhibited brain-type characteristics. These findings underscore the potential role of protein N-glycosylation of sEVs in identifying the originating lung cancer types.
SCLC and NSCLC are considered to arise from different lung cell types; SCLC cells have neuroendocrine properties, while NSCLC cells have epithelial origins (38). Our N-glycan analysis of lung cancer sEVs revealed that the N-glycan profiles of SCLC-sEVs (H446 and SCB-3) shared no common N-glycan features, potentially reflecting the heterogeneity in the originating cell’s properties (39, 40). Interestingly, a large majority of N-glycans in SCLC-sEVs contained characteristic structural motifs, that is, bisecting GlcNAc, LacdiNAc, or antennal fucose, which are known to be highly expressed in human brains (8, 9). Although the precise mechanism by which pulmonary neuroendocrine cells are generated is not fully understood, they are proposed to derive from epithelial cell lineage during lung development in mice (41) and SCLC cells express lineage-defining transcription factors that are critical for neuronal and neuroendocrine differentiation in humans (42, 43). These findings raise the possibility that this transcriptional regulation may contribute to the formation of brain-type N-glycome in SCLC cells and their sEVs. In contrast to SCLC-sEVs, NSCLC-sEVs (SCC [H520, SK-MES-1, and LK-2], LUAD [HCC827], and LCC [H1299]) had relatively homogeneous N-glycan repertories, depicting the characteristic homogeneity of their origin. Normal human lungs, wherein the pulmonary epithelial cells occupy a large majority of the cell types, reportedly express biantennary N-glycans with no core fucose as major N-glycoforms (10). Since most biantennary N-glycans had core fucose in NSCLC-sEVs, malignant transformation of the pulmonary epithelial cells may increase this glycan modification. Indeed, upregulation of the FUT8 gene, whose product FUT8 is responsible for the core fucosylation reaction (44), has been associated with the progression and poor prognosis of NSCLC (11, 45). Moreover, core fucosylation of the N-glycans on cell surface receptor proteins, for example, the transforming growth factor β1 receptor (46), E-cadherin (47), and integrins (48, 49), is known to influence the functions of these receptors in various disease settings, including emphysema and cancer. These findings raise the possibility that the core fucosylation levels of N-glycans displayed on NSCLC-sEVs may also be associated with disease progression.
We found that some fractions of the N-glycans in NSCLC-sEVs and SCLC-sEVs contained a β1,6-GlcNAc branch (e.g., GALAXY ID 310.18, 400.16/400.22, 400.31, 410.16, 410.44, and 410.52), which is formed by the action of β1,6-GlcNAc transferase 5 (MGAT5) (50, 51). It has been demonstrated that Mgat5 promotes tumor growth and metastasis in mouse mammary tumor models (52), and the high expression of MGAT5 is associated with poor prognosis in human hepatocellular carcinomas (53). The β1,6-GlcNAc branch can be further extended by poly-N-acetyllactosamine, which serves as a ligand for galectin molecules. The expression of the β1,6-GlcNAc branch on tumor endothelial cells is critical for galectin-1-mediated angiogenesis (54). Moreover, the β1,3-GlcNAc transferase 3 that initiates poly-N-acetyllactosamine extension is required for immunosuppression in triple-negative breast cancer cells through enhancement of the interaction between programmed cell death protein 1 and programmed cell death ligand 1. Therefore, it is tempting to speculate that the β1,6-GlcNAc branch on tumor cell–derived EVs plays a role in tumor progression by modulating the tumor microenvironment. However, further studies are necessary to validate this hypothesis.
Our lectin-assisted glycoproteomics of SCLC-sEVs and NSCLC-sEVs revealed the enrichment of several integrin α and β subunits. We identified the integrin αV subunit in sEVs from all lung cancer types tested, suggesting that this integrin subunit serves as pan–lung cancer sEV markers. In contrast, the expression of integrin α6 and β4 subunits was limited to NSCLC-sEVs. Importantly, our N-glycomics of the integrin α6 subunit purified from SCC-sEVs indicated that this integrin subunit carried NSCLC-type N-glycans, showing that integrin α6 expression and its N-glycan profiles in lung cancer cell–derived sEVs define the NSCLC origin. The integrin α6 subunit formed a heterodimer with the integrin β1 or β4 subunit in NSCLC-sEVs, confirming the formation of laminin receptors (37) on sEVs. Integrin α6β1 is known to be expressed in cells with various lineages (55). In contrast, integrin α6β4 is primarily expressed in hemidesmosomes in polarized epithelial cells and connects the cell to basement membranes (56), supporting our hypothesis that the integrin α6β4 expression in lung cancer cell–derived sEVs reflects the epithelial origin. Comprehensive proteomic comparison of sEVs that were prepared from human plasma and tissue explants (30) has revealed that integrin α6 and β4 subunits are rarely found in plasma-derived sEVs from tumor-bearing (α6, 4%; β4, 0%) or nontumor (α6, 21%; β4, 0%) donors. In contrast, these two integrins are more frequently found in sEVs released from tumor tissue explants derived from NSCLC patients (α6, 47%; β4, 29%) than in those released from nontumor tissue explants (α6, 27%; β4, 5%). These findings suggest that NSCLC-derived, sEV-associated integrin α6β4 is locally abundant in tumor lesions. The integrin α6β4 expression in sEVs is not limited to lung cancer cells (31) and may be distributed across various types of carcinoma-derived sEVs (30). Thus, further in-depth glycoproteomics, as well as glycomics, of cancer cell–derived sEVs are necessary to identify unique glycan–glycoprotein signatures that define individual cancer types.
Our N-glycomic analysis of the integrin α6 subunit in SCC-sEVs demonstrated the presence of oligomannose-type and complex N-glycans on this integrin subunit. Although the precise roles of N-glycosylation of the integrin α6 subunit in cancer progression remain elusive, N-glycosylation of some integrin subunits is known to have large impacts on receptor functions. For example, site-specific N-glycosylation of α5 and β1 subunits is critical for heterodimer formation, cell-surface trafficking, and ligand binding (57), and it also serves as a switching mechanism for α5β1 and α6β4 to be associated with the epidermal growth factor receptor (58). The integrin β4 subunit, an interaction partner of integrin α6, has an unusually large cytosolic domain involved in the regulation of hemidesmosome disassembly, migration, and invasion (59), raising the possibility that N-glycosylation of the integrin α6 subunit may also influence the metastatic potential of cancer cells by modulating the β4 functions.
In summary, we demonstrated that the integrin α6β4 expression and N-glycosylation status of the α6 subunit in NSCLC-sEVs are associated with the epithelial origin of lung cancer cells. We further showed that SCLC-sEVs are unique in that their N-glycans contain brain-related structural units. Despite the extensive proteomic profiling of cancer-derived and cancer cell–derived sEVs, the association of profiles with the originating cell types has not yet been fully elucidated. Thus, targeting the protein N-glycosylation of cancer sEVs may pave the way for detecting and distinguishing various cancer types and their origins, as well as developing therapeutic strategies.
Experimental procedures
Cell culture
Human LUAD (HCC827), SCC (H520, H446, and SK-MES-1), and LCC (H1299) cell lines were purchased from the American Type Culture Collection. Human SCLC cell lines (LK-2 and SBC-3) were purchased from the Japanese Collection of Research Bioresources Cell Bank and validated by short tandem repeat profiling (National Institute of Biomedical Innovation). All cell lines were routinely tested for Mycoplasma and confirmed to have no contamination using a LookOut Kit (MP0035; Sigma–Aldrich) according to the manufacturer’s instructions. H1299, HCC827, H520, H446, and LK-2 were maintained in RPMI1640 (187-02705 for H520 and H446, FUJIFILM Wako; R8758 for LK-2, Sigma–Aldrich). SK-MES-1 and SBC-3 were maintained in Eagle’s minimal essential medium (M4655; Sigma–Aldrich). All media contained 10% fetal bovine serum (10270106; Sigma–Aldrich) and 100 U/ml penicillin and 10 μg/ml streptomycin (P4333; Sigma–Aldrich). Cells were cultured at 37 °C in a 5% CO2 atmosphere.
Preparation of sEVs
Cells were seeded onto 10 cm dishes at concentrations of 1 × 106/cm2 (for H1299), 2 × 106/cm2 (for HCC827), 5 × 106/cm2 (for H520), 2 × 106/cm2 (for LK-2), 2 × 106/cm2 (for SK-MES-1), 2 × 106/cm2 (for H446), and 1 × 106/cm2 (for SBC-3) and cultured for 4 days. On day 4, the cells were washed twice with PBS (D8537; Sigma–Aldrich) and cultured in 10 ml of fetal bovine serum–free medium for 36 h. The medium was centrifuged at 160g for 10 min at 25 °C, and the supernatant was again centrifuged at 20,000g for 20 min at 4 °C (P28S rotor, Himac 80WX ultracentrifuge, Eppendorf Himac) to remove large EVs. The supernatant thus obtained was ultracentrifuged at 100,000g for 70 min at 4 °C (P50AT2 rotor, Himac CP80WX ultracentrifuge, Eppendorf Himac). The pellet fraction, which contained sEVs, was resuspended in 20 ml of PBS and pelleted by ultracentrifugation at 100,000g for 70 min at 4 °C using the same rotor. The sEVs were resuspended in 1 ml of PBS and pelleted by ultracentrifugation at 100,000g for 45 min (S55A2 rotor, Himac CS100FNX ultracentrifuge, Eppendorf Himac). The resultant sEV pellet was resuspended in small volumes of PBS and used for assays without freezing.
Preparation of whole-cell lysates
Cells were lysed for 15 min at 0 °C in a lysis buffer containing 50 mM tris(hydroxymethyl)aminomethane (Tris)–HCl (pH 7.4), 150 mM NaCl, 1% Triton X-100, 1 mM EDTA, and a protease inhibitor cocktail (04693124001; Sigma–Aldrich). The cell lysate was centrifuged at 20,000g for 10 min at 4 °C, and the supernatant was recovered as whole-cell lysates.
Fluorescent labeling of N-glycans
N-glycans were released from sEVs and whole-cell lysates (50 μg of protein) with 1000 U of PNGase F (P0704; New England Biolabs) according to the manufacturer’s instruction. The released glycans were purified with BlotGlyco (BS-45403; Sumitomo Bakelite) and fluorescently labeled with 2-aminopyridine (PA, 011-14181; FUJIFILM Wako) according to the manufacturer’s instruction.
Desialylation of PA-labeled glycans
PA-labeled glycans were incubated with 10 mU of neuraminidase from Arthrobacter ureafaciens (Sigma–Aldrich) in 20 μl of 10 mM sodium acetate buffer (pH 5.5) for 20 h at 37 °C. The reaction was terminated by the addition of 60 μl of ethanol, followed by incubation for 15 min at 0 °C. The reaction mixture was centrifuged at 20,000g for 10 min at 4 °C, and the supernatant was evaporated to dryness.
Western blot and lectin blot analyses
sEVs (5 μg of protein) and a whole-cell lysate (20 μg of protein) were denatured in a Laemmli sample buffer for 3 min at 100 °C. The denatured proteins were separated by SDS-PAGE and transferred to a polyvinylidene difluoride (PVDF) membrane (IPVH07850; Merck). Antibodies used for blotting were CD81 (1:10,000, B-11; Santa Cruz Biotechnology), integrin β1 (1:1,000, 4706S; Cell Signaling Technology), integrin β4 (1:1,000, 4707S; Cell Signaling Technology), integrin β5 (1:1,000, 4708S; Cell Signaling Technology), integrin αV (1:5,000, AF1219; R&D Systems), and integrin α6 (1:5,000, AF1350; R&D Systems). For lectin blotting, denatured sEVs (5 μg of protein) and whole-cell lysates (20 μg of protein) were separated by SDS-PAGE and transferred to a nitrocellulose membrane (10600002; General Electric Healthcare). The membrane was blocked with PBS containing 0.1% (v/v) Tween 20 and incubated with a biotinylated lectin (agglutinin) from SSA (J218; J-Oil Mills) or Triticum vulgare (WGA, J220; J-Oil Mills). The biotinylated lectins were detected using a VECTASTAIN ABC Kit (PK-4000; Vector Laboratories).
Lectin pull-down assay
sEVs that were prepared from 200 ml of a conditioned medium of H520 and H446 were lysed in 100 μl of PBS containing 0.5% SDS for 10 min at 25 °C and diluted with 1 ml of PBS containing 1% (v/v) Triton X-100. After centrifugation at 20,000g for 10 min at 4 °C, the diluted lysates (1 ml) were incubated with 100 μl of agarose beads conjugated with SSA (for H520) or WGA (for H446) lectin for 2 h at 4 °C with gentle agitation. The beads were washed five times with 1 ml of PBS containing 0.1% Triton X-100, and bound proteins were eluted by heating with 100 μl of 2× Laemmli sample buffer for 15 min at 70 °C. The eluted proteins were separated by SDS-PAGE, and the gel was stained with silver using a silver staining kit (299-58901; FUJIFILM Wako) according to the manufacturer’s instruction.
Immunoprecipitation
sEVs that were prepared from 1 l of a conditioned medium of H520, SK-MES-1, and LK-2 were lysed in 500 μl of PBS containing 1% NP-40 (2111; BioVision). After stirring for 5 s, the homogenate was centrifuged at 20,000g for 5 min at 4 °C, and the cleared supernatant (500 μl) was incubated with 40 μl of protein A/G-immobilized agarose beads (sc-2003; Santa Cruz Biotechnology) prior to incubation with 8 μg of anti-integrin α6 antibodies (ab105669; Abcam) for 12 h at 4 °C with gentle agitation. The beads were then washed three times with PBS containing 0.1% NP-40 and denatured in 40 μl of Laemmli sample buffer for 3 min at 100 °C.
HPLC
PA-labeled glycans were analyzed via HPLC using an LC-2030C system (Shimadzu Corporation) equipped with an RF-20Axs detector (Shimadzu Corporation). Anion-exchange HPLC was carried out using a TSKgel DEAE-5PW column (10 μm, 750 × 7.5 mm i.d., Tosoh Bioscience) (61). The elution was performed using a two-solvent gradient system as follows: eluent A (10% [v/v] acetonitrile + 0.01% [v/v] triethylamine) and eluent B (10% [v/v] acetonitrile + 7.4% [v/v] triethylamine + 3% [v/v] acetic acid). The gradient program was set at a flow rate of 1 ml/min (expressed as the percentage of eluent B): 0 to 40 min, 0 to 20%; 40 to 50 min, 100%; 50 to 60 min, 0%. PA-glycans were detected by measuring the fluorescence (excitation wavelength, 310 nm; emission wavelength, 380 nm). Reversed-phase HPLC was carried out using a Shim-pack HRC-ODS column (5 μm, 150 × 6 mm i.d., Shimadzu Corporation) (61). The elution was performed using a two-solvent gradient system as follows: eluent C (10 mM sodium phosphate buffer [pH 3.8]) and eluent D (eluent C + 0.5% [v/v] n-butanol). The gradient program was set at a flow rate of 1 ml/min (expressed as the percentage of eluent D): 0 to 60 min, 20 to 50%; 60 to 65 min, 100%; 65 to 85 min, 20%. PA-glycans were detected by measuring the fluorescence (excitation wavelength, 320 nm; emission wavelength, 400 nm).
MS
Desialylated, PA-labeled N-glycans were fractionated by reversed-phase HPLC and analyzed via either MALDI-TOF-MS or LC-ESI-MS. For MALDI-TOF-MS, the fractionated glycans (1 μl) were mixed with 1 μl of 2,5-dihydroxybenzoic acid (10 mg/ml in 50% acetonitrile/0.1% TFA) on a target plate. The spots were air-dried, and spectra were obtained in the positive mode using an Autoflex mass spectrometer (Bruker Daltonics) operated in the reflector mode. LC-ESI-MS of the fractionated PA-glycans was performed on an LTQ XL ion trap mass spectrometer (Thermo Fisher Scientific) connected to an LC-20A HPLC system (Shimadzu Corporation). The glycans were desalted on a graphitized carbon (Hypercarb, 5 μm; Thermo Fisher Scientific) packed in a cartridge column (0.3 × 5 mm; CERI) at a flow rate of 0.06 ml/min. The cartridge was equilibrated with aqueous formic acid (0.3%, v/v) and held for 5 min after sample injection. The glycans were eluted by steeply increasing the ratio of acetonitrile with formic acid (0.5%, v/v) to 95% in 2 min and then held for 1 min prior to reequilibration for 11 min. The ESI-MS settings used were as follows: spray voltage, 4 kV; source heater, 250 °C; sheath gas, 30 units; auxiliary gas, 5 units; capillary temperature, 175 °C; tube lens voltage, 80 V. MS2 experiments were performed by collision-induced dissociation in a data-dependent mode or an isolation of the selected precursor ion. MS data were recorded and analyzed using Xcalibur 4.0 software (Thermo Scientific). Structural identification was based on the elution position on the HPLC column and MS data in the GALAXY database (62).
Proteomics analysis
The protein bands in SDS-PAGE were excised, reduced with DTT, and alkylated with acrylamide. The proteins were digested in gel with trypsin (tosyl phenylalanyl chloromethyl ketone treated; Worthington Biochemical Co) at 37 °C overnight. An aliquot of each digestion mixture was separated using a nano-ESI spray column (NTCC-360, 0.075 mm internal diameter × 105 mm length, 3 μm, Nikkyo Technos Co) at a flow rate of 300 nl/min and applied online to a Q-Exactive Mass Spectrometer (Thermo Fisher Scientific) with a nanospray ion source. MS and MS/MS data were acquired using the data-dependent Top 10 method. The obtained MS/MS data were searched against the NCBInr 20160711 database with Mascot Version 2.5 software (Matrix Science) using the following parameters: taxonomy, Homo sapiens (human) (326,427 sequences); type of search, MS/MS ion; enzyme, trypsin; fixed modification, none; variable modifications, acetyl (protein N-term), deamidated (NQ), Gln->pyro-Glu (N-term Q), oxidation (M), propionamide (C); mass values, monoisotopic; peptide mass tolerance, ± 15 ppm; fragment mass tolerance, ± 20 mmu; max missed cleavages, 3; instrument type, ESI-TRAP. Proteome Discoverer 2.2 was used to generate peak list and the expectation value for accepting individual spectra in Mascot search was < 0.05.
Transmission electron microscopy
sEVs were fixed for 30 min at 4 °C in a 2% osmium tetroxide solution (Nisshin EM) and postfixed for 30 min at room temperature in half-strength Karnovsky fixative containing 2.5% glutaraldehyde, 2% paraformaldehyde, and 100 mM phosphate buffer (pH 7.4). The fixed specimens were dehydrated, embedded in epoxy resin EPON-812 (TAAB Laboratories Equipment Ltd), and subjected to ultrathin sectioning. The sections were mounted on collodion-coated copper grids, stained with uranyl acetate and lead citrate, and observed under an electron microscope (Hitachi H-7650, Hitachi High-Technologies).
Glycomics analysis
Integrin α6 that was immunoprecipitated from H520-sEVs, SK-MES-1-sEVs, or LK-2-sEVs was separated by SDS-PAGE, and proteins on the gel were transferred to a PVDF membrane. The PVDF membrane was washed once with ethanol for 1 min, followed by three washes with water for 1 min each prior to staining for 5 min with Direct Blue 71 (800 μl of solution A: 0.1% (w/v) Direct Blue 71 [Sigma–Aldrich] in 10 ml of solution B: acetic acid:ethanol:water = 1:4:5). After destaining the membrane with solution B for 1 min, it was dried at 25 °C for 18 h. The band corresponding to integrin α6, as well as the corresponding position of the lane that had the anti-integrin α6 antibody alone, was excised from the membrane. The excised bands were placed into a 96-well plate separately, covered with 100 μl of 1% (w/v) poly(vinylpyrrolidone) 40,000 in 50% (v/v) methanol, agitated for 20 min, and washed with water (100 μl × five times). N-glycans were released from the membrane by incubation with a reaction mixture containing 2 μl of PNGase F (2 U/membrane; Roche) and 8 μl of water. The free N-glycans were reduced to alditol N-glycans and desalted on a cation-exchange column as described previously (63), with some modifications (64). The alditol N-glycans were analyzed by LC-ESI-MS and MS/MS as described previously (64). Briefly, alditol N-glycans were separated on a carbon column (5 μm HyperCarb, 1 mm I.D. × 100 mm, Thermo Fisher Scientific) using an Accela HPLC pump (flow rate, 50 μl/min; column oven, 40 °C) under the following gradient conditions involving a sequence of isocratic and two segmented linear gradients: 0 to 8 min, 10 mM NH4HCO3; 8 to 38 min, 6.75 to 15.75% (v/v) acetonitrile in 10 mM NH4HCO3; 38 to 73 min, 15.75 to 40.5% (v/v) acetonitrile in 10 mM NH4HCO3, increasing to 81% (v/v) acetonitrile in 10 mM N NH4HCO3 for 7 min. They were then reequilibrated with 10 mM NH4HCO3 for 15 min. The eluate was continuously introduced into an ESI source (Thermo Fisher Scientific). With regard to the mass spectrometer (LTQ Orbitrap XL, Hybrid Linear Ion Trap-Orbitrap mass spectrometer, Thermo Fisher Scientific), the voltage of the capillary source was set at 3 kV, and the temperature of the transfer capillary was maintained at 300 °C. The capillary voltage and tube lens voltage were set at −18.0 V and −112.8 V, respectively. MS spectra were obtained in the negative polarity using an Orbitrap (mass range, m/z 500–2500; resolution, 15,000; mass accuracy, 5 ppm), and MS/MS spectra were obtained using an Ion Trap (data-dependent top 3, collision-induced dissociation). Monoisotopic masses of glycans observed via MS were analyzed to find possible monosaccharide compositions using the GlycoMod tool available on the ExPASy server (http://au.expasy.org/tools/glycomod; mass tolerance for precursor ions, ±0.01 Da), and the proposed glycan structures were selected from the GlyConnect database (https://glyconnect.expasy.org/) linked via GlycoMod and according to the separation pattern on a carbon column as reported previously (65). The peak intensities of each alditol N-glycan were calculated on the extracted-ion chromatogram using Xcalibur software version 2.2 (Thermo Fisher Scientific). The relative abundances (%) of each glycan structure were calculated by setting the total peak intensities of all detected alditol N-glycans in each extracted-ion chromatogram to 100%.
Data availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Yoichiro Harada (yoharada3@mc.pref.osaka.jp).
Material availability
This study did not generate new unique reagents.
Data and code availability
Glycomic raw data and the identification result file for glycan-structure analysis via LC-ESI-MS have been deposited in GlycoPOST (announced ID: GPST000188). The URL is https://glycopost.glycosmos.org/preview/111108106860af1dd73867e (PIN: 7784). The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (60) partner repository with the dataset identifier PXD030865 and 10.6019/PXD030865.
Supporting information
This article contains supporting information (66, 67, 68).
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We wish to thank our laboratory members and Dr Junichi Takagi (Osaka University) for fruitful discussions, and Dr Yasuhiko Kizuka (Gifu University) for technical advice.
Author contributions
Y. H. and H. I. conceptualization; K. K., M. N., T. S., T. F., K. H., H. Y., K. T., Keiko Mizuno, and Kentaro Machida investigation; K. K., Y. H., M. N., T. S., T. F., K. H., and N. D. data curation; K. K., Y. H., M. N., T. S., T. F., K. H., and H. I. writing–original draft; K. K. and H. I. writing–reviewing and editing; Y. H., Y. M., N. T., K. K., T. K., N. D., I. M., and H. I. supervision.
Funding and addditional information
This work was partly supported by grants from the SENSHIN Medical Research Foundation (Y. H.), the Kodama Memorial Fund for Medical Research (Y. H.), JSPS KAKENHI (Grant Number JP19K06546 to Y. H. and JP17K09661 and JP21K08160 to H. I.), the Japan Agency of Medical Research and Development (AMED) (JP20ek0410052 to H. I.), and the Takeda Science Foundation (Y. H.).
Edited by Gerald Hart
Contributor Information
Yoichiro Harada, Email: yoharada3@mc.pref.osaka.jp.
Hiromasa Inoue, Email: inoue@m2.kufm.kagoshima-u.ac.jp.
Supporting information
References
- 1.Travis W.D., Brambilla E., Nicholson A.G., Yatabe Y., Austin J.H.M., Beasley M.B., Chirieac L.R., Dacic S., Duhig E., Flieder D.B., Geisinger K., Hirsch F.R., Ishikawa Y., Kerr K.M., Noguchi M., et al. The 2015 world health organization classification of lung tumors: Impact of genetic, clinical and radiologic advances since the 2004 classification. J. Thorac. Oncol. 2015;10:1243–1260. doi: 10.1097/JTO.0000000000000630. [DOI] [PubMed] [Google Scholar]
- 2.Rusch V., Klimstra D., Venkatraman E., Pisters P.W., Langenfeld J., Dmitrovsky E. Overexpression of the epidermal growth factor receptor and its ligand transforming growth factor alpha is frequent in resectable non-small cell lung cancer but does not predict tumor progression. Clin. Cancer Res. 1997;3:515–522. [PubMed] [Google Scholar]
- 3.Brabender J., Danenberg K.D., Metzger R., Schneider P.M., Park J., Salonga D., Holscher A.H., Danenberg P.V. Epidermal growth factor receptor and HER2-neu mRNA expression in non-small cell lung cancer Is correlated with survival. Clin. Cancer Res. 2001;7:1850–1855. [PubMed] [Google Scholar]
- 4.Soda M., Choi Y.L., Enomoto M., Takada S., Yamashita Y., Ishikawa S., Fujiwara S., Watanabe H., Kurashina K., Hatanaka H., Bando M., Ohno S., Ishikawa Y., Aburatani H., Niki T., et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448:561–566. doi: 10.1038/nature05945. [DOI] [PubMed] [Google Scholar]
- 5.Rikova K., Guo A., Zeng Q., Possemato A., Yu J., Haack H., Nardone J., Lee K., Reeves C., Li Y., Hu Y., Tan Z., Stokes M., Sullivan L., Mitchell J., et al. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell. 2007;131:1190–1203. doi: 10.1016/j.cell.2007.11.025. [DOI] [PubMed] [Google Scholar]
- 6.Harada Y., Ohkawa Y., Kizuka Y., Taniguchi N. Oligosaccharyltransferase: A gatekeeper of health and tumor progression. Int. J. Mol. Sci. 2019;20:6074. doi: 10.3390/ijms20236074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Capela A., Temple S. LeX/ssea-1 is expressed by adult mouse CNS stem cells, identifying them as nonependymal. Neuron. 2002;35:865–875. doi: 10.1016/s0896-6273(02)00835-8. [DOI] [PubMed] [Google Scholar]
- 8.Yanagisawa M., Yu R.K. The expression and functions of glycoconjugates in neural stem cells. Glycobiology. 2007;17:57R–74R. doi: 10.1093/glycob/cwm018. [DOI] [PubMed] [Google Scholar]
- 9.Lee J., Ha S., Kim M., Kim S.W., Yun J., Ozcan S., Hwang H., Ji I.J., Yin D., Webster M.J., Shannon Weickert C., Kim J.H., Yoo J.S., Grimm R., Bahn S., et al. Spatial and temporal diversity of glycome expression in mammalian brain. Proc. Natl. Acad. Sci. U. S. A. 2020;117:28743–28753. doi: 10.1073/pnas.2014207117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jia N., Byrd-Leotis L., Matsumoto Y., Gao C., Wein A.N., Lobby J.L., Kohlmeier J.E., Steinhauer D.A., Cummings R.D. The human lung glycome reveals novel glycan ligands for influenza A virus. Sci. Rep. 2020;10:5320. doi: 10.1038/s41598-020-62074-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen C.Y., Jan Y.H., Juan Y.H., Yang C.J., Huang M.S., Yu C.J., Yang P.C., Hsiao M., Hsu T.L., Wong C.H. Fucosyltransferase 8 as a functional regulator of nonsmall cell lung cancer. Proc. Natl. Acad. Sci. U. S. A. 2013;110:630–635. doi: 10.1073/pnas.1220425110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pinho S.S., Reis C.A. Glycosylation in cancer: Mechanisms and clinical implications. Nat. Rev. Cancer. 2015;15:540–555. doi: 10.1038/nrc3982. [DOI] [PubMed] [Google Scholar]
- 13.Jia L., Zhang J., Ma T., Guo Y., Yu Y., Cui J. The function of fucosylation in progression of lung cancer. Front. Oncol. 2018;8:565. doi: 10.3389/fonc.2018.00565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haga Y., Uemura M., Baba S., Inamura K., Takeuchi K., Nonomura N., Ueda K. Identification of multisialylated LacdiNAc structures as highly prostate cancer specific glycan signatures on PSA. Anal. Chem. 2019;91:2247–2254. doi: 10.1021/acs.analchem.8b04829. [DOI] [PubMed] [Google Scholar]
- 15.Aoyagi Y., Suzuki Y., Igarashi K., Saitoh A., Oguro M., Yokota T., Mori S., Nomoto M., Isemura M., Asakura H. The usefulness of simultaneous determinations of glucosaminylation and fucosylation indices of alpha-fetoprotein in the differential diagnosis of neoplastic diseases of the liver. Cancer. 1991;67:2390–2394. doi: 10.1002/1097-0142(19910501)67:9<2390::aid-cncr2820670928>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 16.Sato Y., Nakata K., Kato Y., Shima M., Ishii N., Koji T., Taketa K., Endo Y., Nagataki S. Early recognition of hepatocellular carcinoma based on altered profiles of alpha-fetoprotein. N. Engl. J. Med. 1993;328:1802–1806. doi: 10.1056/NEJM199306243282502. [DOI] [PubMed] [Google Scholar]
- 17.Xue H., Lu B., Lai M. The cancer secretome: A reservoir of biomarkers. J. Transl Med. 2008;6:52. doi: 10.1186/1479-5876-6-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Robinson J.L., Feizi A., Uhlen M., Nielsen J. A systematic investigation of the malignant functions and diagnostic potential of the cancer secretome. Cell Rep. 2019;26:2622–2635.e5. doi: 10.1016/j.celrep.2019.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Becker A., Thakur B.K., Weiss J.M., Kim H.S., Peinado H., Lyden D. Extracellular vesicles in cancer: Cell-to-Cell mediators of metastasis. Cancer Cell. 2016;30:836–848. doi: 10.1016/j.ccell.2016.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Raposo G., Stoorvogel W. Extracellular vesicles: Exosomes, microvesicles, and friends. J. Cell Biol. 2013;200:373–383. doi: 10.1083/jcb.201211138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kalluri R., LeBleu V.S. The biology, function, and biomedical applications of exosomes. Science. 2020;367 doi: 10.1126/science.aau6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harada Y., Nakajima K., Suzuki T., Fukushige T., Kondo K., Seino J., Ohkawa Y., Suzuki T., Inoue H., Kanekura T., Dohmae N., Taniguchi N., Maruyama I. Glycometabolic regulation of the biogenesis of small extracellular vesicles. Cell Rep. 2020;33:108261. doi: 10.1016/j.celrep.2020.108261. [DOI] [PubMed] [Google Scholar]
- 23.Yamamoto M., Harada Y., Suzuki T., Fukushige T., Yamakuchi M., Kanekura T., Dohmae N., Hori K., Maruyama I. Application of high-mannose-type glycan-specific lectin from Oscillatoria Agardhii for affinity isolation of tumor-derived extracellular vesicles. Anal. Biochem. 2019;580:21–29. doi: 10.1016/j.ab.2019.06.001. [DOI] [PubMed] [Google Scholar]
- 24.Harada Y., Kizuka Y., Tokoro Y., Kondo K., Yagi H., Kato K., Inoue H., Taniguchi N., Maruyama I. N-glycome inheritance from cells to extracellular vesicles in B16 melanomas. FEBS Lett. 2019;593:942–951. doi: 10.1002/1873-3468.13377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kosaka N., Yoshioka Y., Fujita Y., Ochiya T. Versatile roles of extracellular vesicles in cancer. J. Clin. Invest. 2016;126:1163–1172. doi: 10.1172/JCI81130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Melo S.A., Luecke L.B., Kahlert C., Fernandez A.F., Gammon S.T., Kaye J., LeBleu V.S., Mittendorf E.A., Weitz J., Rahbari N., Reissfelder C., Pilarsky C., Fraga M.F., Piwnica-Worms D., Kalluri R. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature. 2015;523:177–182. doi: 10.1038/nature14581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peinado H., Aleckovic M., Lavotshkin S., Matei I., Costa-Silva B., Moreno-Bueno G., Hergueta-Redondo M., Williams C., Garcia-Santos G., Ghajar C., Nitadori-Hoshino A., Hoffman C., Badal K., Garcia B.A., Callahan M.K., et al. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat. Med. 2012;18:883–891. doi: 10.1038/nm.2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Costa-Silva B., Aiello N.M., Ocean A.J., Singh S., Zhang H., Thakur B.K., Becker A., Hoshino A., Mark M.T., Molina H., Xiang J., Zhang T., Theilen T.M., Garcia-Santos G., Williams C., et al. Pancreatic cancer exosomes initiate pre-metastatic niche formation in the liver. Nat. Cell Biol. 2015;17:816–826. doi: 10.1038/ncb3169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Asano N., Matsuzaki J., Ichikawa M., Kawauchi J., Takizawa S., Aoki Y., Sakamoto H., Yoshida A., Kobayashi E., Tanzawa Y., Nakayama R., Morioka H., Matsumoto M., Nakamura M., Kondo T., et al. A serum microRNA classifier for the diagnosis of sarcomas of various histological subtypes. Nat. Commun. 2019;10:1299. doi: 10.1038/s41467-019-09143-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hoshino A., Kim H.S., Bojmar L., Gyan K.E., Cioffi M., Hernandez J., Zambirinis C.P., Rodrigues G., Molina H., Heissel S., Mark M.T., Steiner L., Benito-Martin A., Lucotti S., Di Giannatale A., et al. Extracellular vesicle and particle biomarkers define multiple human cancers. Cell. 2020;182:1044–1061.e18. doi: 10.1016/j.cell.2020.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hoshino A., Costa-Silva B., Shen T.L., Rodrigues G., Hashimoto A., Tesic Mark M., Molina H., Kohsaka S., Di Giannatale A., Ceder S., Singh S., Williams C., Soplop N., Uryu K., Pharmer L., et al. Tumour exosome integrins determine organotropic metastasis. Nature. 2015;527:329–335. doi: 10.1038/nature15756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kosaka N., Kogure A., Yamamoto T., Urabe F., Usuba W., Prieto-Vila M., Ochiya T. Exploiting the message from cancer: The diagnostic value of extracellular vesicles for clinical applications. Exp. Mol. Med. 2019;51:1–9. doi: 10.1038/s12276-019-0219-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Metzelaar M.J., Wijngaard P.L., Peters P.J., Sixma J.J., Nieuwenhuis H.K., Clevers H.C. CD63 antigen. A novel lysosomal membrane glycoprotein, cloned by a screening procedure for intracellular antigens in eukaryotic cells. J. Biol. Chem. 1991;266:3239–3245. [PubMed] [Google Scholar]
- 34.Shibuya N., Tazaki K., Song Z.W., Tarr G.E., Goldstein I.J., Peumans W.J. A comparative study of bark lectins from three elderberry (Sambucus) species. J. Biochem. 1989;106:1098–1103. doi: 10.1093/oxfordjournals.jbchem.a122972. [DOI] [PubMed] [Google Scholar]
- 35.Itakura Y., Nakamura-Tsuruta S., Kominami J., Tateno H., Hirabayashi J. Sugar-binding profiles of chitin-binding lectins from the hevein family: A comprehensive study. Int. J. Mol. Sci. 2017;18:1160. doi: 10.3390/ijms18061160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Davis T.L., Rabinovitz I., Futscher B.W., Schnolzer M., Burger F., Liu Y., Kulesz-Martin M., Cress A.E. Identification of a novel structural variant of the alpha 6 integrin. J. Biol. Chem. 2001;276:26099–26106. doi: 10.1074/jbc.M102811200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Takada Y., Ye X., Simon S. The integrins. Genome Biol. 2007;8:215. doi: 10.1186/gb-2007-8-5-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Swanton C., Govindan R. Clinical implications of genomic discoveries in lung cancer. N. Engl. J. Med. 2016;374:1864–1873. doi: 10.1056/NEJMra1504688. [DOI] [PubMed] [Google Scholar]
- 39.Carney D.N., Gazdar A.F., Bepler G., Guccion J.G., Marangos P.J., Moody T.W., Zweig M.H., Minna J.D. Establishment and identification of small cell lung cancer cell lines having classic and variant features. Cancer Res. 1985;45:2913–2923. [PubMed] [Google Scholar]
- 40.Miyamoto H. Establishment and characterization of an adriamycin-resistant subline of human small cell lung cancer cells. Acta Med. Okayama. 1986;40:65–73. doi: 10.18926/AMO/31923. [DOI] [PubMed] [Google Scholar]
- 41.Kuo C.S., Krasnow M.A. formation of a neurosensory organ by epithelial cell slithering. Cell. 2015;163:394–405. doi: 10.1016/j.cell.2015.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Borromeo M.D., Savage T.K., Kollipara R.K., He M., Augustyn A., Osborne J.K., Girard L., Minna J.D., Gazdar A.F., Cobb M.H., Johnson J.E. ASCL1 and NEUROD1 reveal heterogeneity in pulmonary neuroendocrine tumors and regulate distinct genetic programs. Cell Rep. 2016;16:1259–1272. doi: 10.1016/j.celrep.2016.06.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pozo K., Kollipara R.K., Kelenis D.P., Rodarte K.E., Ullrich M.S., Zhang X., Minna J.D., Johnson J.E. ASCL1, NKX2-1, and PROX1 co-regulate subtype-specific genes in small-cell lung cancer. iScience. 2021;24:102953. doi: 10.1016/j.isci.2021.102953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Uozumi N., Yanagidani S., Miyoshi E., Ihara Y., Sakuma T., Gao C., Teshima T., Fujii S., Shiba T., Taniguchi N. Purification and cDNA cloning of porcine brain GDP-L-Fuc:N-Acetyl-β-D-Glucosaminide α1→6Fucosyltransferase. J. Biol. Chem. 1996;271:27810–27817. doi: 10.1074/jbc.271.44.27810. [DOI] [PubMed] [Google Scholar]
- 45.Honma R., Kinoshita I., Miyoshi E., Tomaru U., Matsuno Y., Shimizu Y., Takeuchi S., Kobayashi Y., Kaga K., Taniguchi N., Dosaka-Akita H. Expression of fucosyltransferase 8 is associated with an unfavorable clinical outcome in non-small cell lung cancers. Oncology. 2015;88:298–308. doi: 10.1159/000369495. [DOI] [PubMed] [Google Scholar]
- 46.Wang X., Inoue S., Gu J., Miyoshi E., Noda K., Li W., Mizuno-Horikawa Y., Nakano M., Asahi M., Takahashi M., Uozumi N., Ihara S., Lee S.H., Ikeda Y., Yamaguchi Y., et al. Dysregulation of TGF-beta1 receptor activation leads to abnormal lung development and emphysema-like phenotype in core fucose-deficient mice. Proc. Natl. Acad. Sci. U. S. A. 2005;102:15791–15796. doi: 10.1073/pnas.0507375102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Osumi D., Takahashi M., Miyoshi E., Yokoe S., Lee S.H., Noda K., Nakamori S., Gu J., Ikeda Y., Kuroki Y., Sengoku K., Ishikawa M., Taniguchi N. Core fucosylation of E-cadherin enhances cell-cell adhesion in human colon carcinoma WiDr cells. Cancer Sci. 2009;100:888–895. doi: 10.1111/j.1349-7006.2009.01125.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhao Y., Sato Y., Isaji T., Fukuda T., Matsumoto A., Miyoshi E., Gu J., Taniguchi N. Branched N-glycans regulate the biological functions of integrins and cadherins. FEBS J. 2008;275:1939–1948. doi: 10.1111/j.1742-4658.2008.06346.x. [DOI] [PubMed] [Google Scholar]
- 49.Zhao Y., Itoh S., Wang X., Isaji T., Miyoshi E., Kariya Y., Miyazaki K., Kawasaki N., Taniguchi N., Gu J. Deletion of core fucosylation on alpha3beta1 integrin down-regulates its functions. J. Biol. Chem. 2006;281:38343–38350. doi: 10.1074/jbc.M608764200. [DOI] [PubMed] [Google Scholar]
- 50.Shoreibah M.G., Hindsgaul O., Pierce M. Purification and characterization of rat kidney UDP-N-acetylglucosamine: alpha-6-D-mannoside beta-1,6-N-acetylglucosaminyltransferase. J. Biol. Chem. 1992;267:2920–2927. [PubMed] [Google Scholar]
- 51.Gu J., Nishikawa A., Tsuruoka N., Ohno M., Yamaguchi N., Kangawa K., Taniguchi N. Purification and characterization of UDP-N-acetylglucosamine: alpha-6-D-mannoside beta 1-6N-acetylglucosaminyltransferase (N-acetylglucosaminyltransferase V) from a human lung cancer cell line. J. Biochem. 1993;113:614–619. doi: 10.1093/oxfordjournals.jbchem.a124091. [DOI] [PubMed] [Google Scholar]
- 52.Granovsky M., Fata J., Pawling J., Muller W.J., Khokha R., Dennis J.W. Suppression of tumor growth and metastasis in Mgat5-deficient mice. Nat. Med. 2000;6:306–312. doi: 10.1038/73163. [DOI] [PubMed] [Google Scholar]
- 53.Liu H., Wu Q., Liu Y., Liu W., Zhang W., Pan D., Xu J. Prognostic significance of beta1,6-N-acetylglucosaminyltransferase V expression in patients with hepatocellular carcinoma. Jpn. J. Clin. Oncol. 2015;45:844–853. doi: 10.1093/jjco/hyv080. [DOI] [PubMed] [Google Scholar]
- 54.Croci D.O., Cerliani J.P., Dalotto-Moreno T., Mendez-Huergo S.P., Mascanfroni I.D., Dergan-Dylon S., Toscano M.A., Caramelo J.J., Garcia-Vallejo J.J., Ouyang J., Mesri E.A., Junttila M.R., Bais C., Shipp M.A., Salatino M., et al. Glycosylation-dependent lectin-receptor interactions preserve angiogenesis in anti-VEGF refractory tumors. Cell. 2014;156:744–758. doi: 10.1016/j.cell.2014.01.043. [DOI] [PubMed] [Google Scholar]
- 55.Luo B.H., Carman C.V., Springer T.A. Structural basis of integrin regulation and signaling. Annu. Rev. Immunol. 2007;25:619–647. doi: 10.1146/annurev.immunol.25.022106.141618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mercurio A.M., Rabinovitz I., Shaw L.M. The alpha 6 beta 4 integrin and epithelial cell migration. Curr. Opin. Cell Biol. 2001;13:541–545. doi: 10.1016/s0955-0674(00)00249-0. [DOI] [PubMed] [Google Scholar]
- 57.Gu J., Taniguchi N. Regulation of integrin functions by N-glycans. Glycoconj J. 2004;21:9–15. doi: 10.1023/B:GLYC.0000043741.47559.30. [DOI] [PubMed] [Google Scholar]
- 58.Hang Q., Isaji T., Hou S., Zhou Y., Fukuda T., Gu J. N-Glycosylation of integrin alpha5 acts as a switch for EGFR-mediated complex formation of integrin alpha5beta1 to alpha6beta4. Sci. Rep. 2016;6:33507. doi: 10.1038/srep33507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stewart R.L., O'Connor K.L. Clinical significance of the integrin alpha6beta4 in human malignancies. Lab. Invest. 2015;95:976–986. doi: 10.1038/labinvest.2015.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Perez-Riverol Y., Csordas A., Bai J., Bernal-Llinares M., Hewapathirana S., Kundu D.J., Inuganti A., Griss J., Mayer G., Eisenacher M., Perez E., Uszkoreit J., Pfeuffer J., Sachsenberg T., Yilmaz S., et al. The PRIDE database and related tools and resources in 2019: Improving support for quantification data. Nucl. Acids Res. 2019;47:D442–D450. doi: 10.1093/nar/gky1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tomiya N., Awaya J., Kurono M., Endo S., Arata Y., Takahashi N. Analyses of N-linked oligosaccharides using a two-dimensional mapping technique. Anal. Biochem. 1988;171:73–90. doi: 10.1016/0003-2697(88)90126-1. [DOI] [PubMed] [Google Scholar]
- 62.Takahashi N., Kato K. GALAXY(Glycoanalysis by the three axes of MS and chromatography): A web application that assists structural analyses of N-glycans. Trends Glycosci. Glycotechnol. 2003;15:235–251. [Google Scholar]
- 63.Wilson N.L., Schulz B.L., Karlsson N.G., Packer N.H. Sequential analysis of N- and O-linked glycosylation of 2D-PAGE separated glycoproteins. J. Proteome Res. 2002;1:521–529. doi: 10.1021/pr025538d. [DOI] [PubMed] [Google Scholar]
- 64.Nakano M., Saldanha R., Gobel A., Kavallaris M., Packer N.H. Identification of glycan structure alterations on cell membrane proteins in desoxyepothilone B resistant leukemia cells. Mol. Cell Proteomics. 2011;10 doi: 10.1074/mcp.M111.009001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pabst M., Bondili J.S., Stadlmann J., Mach L., Altmann F. Mass + retention time = structure: A strategy for the analysis of N-glycans by carbon LC-ESI-MS and its application to fibrin N-glycans. Anal. Chem. 2007;79:5051–5057. doi: 10.1021/ac070363i. [DOI] [PubMed] [Google Scholar]
- 66.Wuhrer M., Koeleman C.A., Hokke C.H., Deelder A.M. Mass spectrometry of proton adducts of fucosylated N-glycans: Fucose transfer between antennae gives rise to misleading fragments. Rapid Commun. Mass Spectrom. 2006;20:1747–1754. doi: 10.1002/rcm.2509. [DOI] [PubMed] [Google Scholar]
- 67.Green E.D., Adelt G., Baenziger J.U., Wilson S., Van Halbeek H. The asparagine-linked oligosaccharides on bovine fetuin. Structural analysis of N-glycanase-released oligosaccharides by 500-megahertz 1H NMR spectroscopy. J. Biol. Chem. 1988;263:18253–18268. [PubMed] [Google Scholar]
- 68.Townsend R.R., Hardy M.R., Cumming D.A., Carver J.P., Bendiak B. Separation of branched sialylated oligosaccharides using high-pH anion-exchange chromatography with pulsed amperometric detection. Anal. Biochem. 1989;182:1–8. doi: 10.1016/0003-2697(89)90708-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Yoichiro Harada (yoharada3@mc.pref.osaka.jp).
Material availability
This study did not generate new unique reagents.
Data and code availability
Glycomic raw data and the identification result file for glycan-structure analysis via LC-ESI-MS have been deposited in GlycoPOST (announced ID: GPST000188). The URL is https://glycopost.glycosmos.org/preview/111108106860af1dd73867e (PIN: 7784). The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (60) partner repository with the dataset identifier PXD030865 and 10.6019/PXD030865.
Glycomic raw data and the identification result file for glycan-structure analysis via LC-ESI-MS have been deposited in GlycoPOST (announced ID: GPST000188). The URL is https://glycopost.glycosmos.org/preview/111108106860af1dd73867e (PIN: 7784). The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (60) partner repository with the dataset identifier PXD030865 and 10.6019/PXD030865.