Summary
Molecular conformational engineering is to engineer flexible non-functional molecules into unique conformations to create novel functions just like natural proteins fold. Obviously, it is a grand challenge with tremendous opportunities. Based on the facts that natural proteins are only marginally stable with a net stabilizing energy roughly equivalent to the energy of two hydrogen bonds, and the energy barriers for the adatom diffusion of some metals are within a similar range, we propose that metal nanoparticles can serve as a general replacement of protein scaffolds to conformationally engineer protein fragments on the surface of nanoparticles. To prove this hypothesis, herein, we successfully restore the antigen-recognizing function of the flexible peptide fragment of a natural anti-lysozyme antibody on the surface of silver nanoparticles, creating a silver nanoparticle-base artificial antibody (Silverbody). A plausible mechanism is proposed, and some general principles for conformational engineering are summarized to guide future studies in this area.
Subject areas: Engineering, Nanoparticles, Diffusion potentials
Graphical abstract
Highlights
-
•
A silver NP-based artificial antibody is created by conformational engineering
-
•
Function emerges on NPs from non-functional peptide by mimicking the protein folding
-
•
A general mechanism is proposed for the conformational engineering on metal NPs
Engineering, Nanoparticles, Diffusion potentials
Introduction
Chemistry has long been considered as a mature discipline (Peplow, 2006), especially after artificial intelligence (AI) has been successfully used to plan chemical syntheses (Segler et al., 2018), and robots can be used to perform the syntheses (Burger et al., 2020). Yet, there is still a large area in chemistry seldom being explored to date. This novel area is molecular conformational engineering, i.e., controlling the angles of all rotatable chemical bonds within a molecule to arrange all its atoms in a specific conformation as a molecular machinery to perform sophisticated functions in a concerted manner, just like natural proteins fold (Yan et al., 2018). The natural protein folding can be regarded as a special case of molecular conformational engineering. However, even the special case of protein folding is still not fully understood (Cao, 2020a, 2020b; Dill and MacCallum, 2012; Narayan et al., 2000); the general task of molecular conformational engineering is apparently a grand challenge.
Generally, for most small molecules, when all their chemical bonds are set, their chemical properties are set, too. But for large molecules, especially those with many rotatable chemical bonds, their chemical properties are also determined by their conformations, i.e., the arrangements of all atoms in 3D space corresponding to each angle of those rotatable chemical bonds. The total number of conformations of a molecule increases exponentially with the number of its rotatable chemical bonds. Fundamentally, the molecular conformational engineering is intentionally controlling the rotation of all rotatable chemical bonds of a molecule.
Though it is still almost impossible for chemists to control the conformation of molecules with rotatable chemical bonds, nature has done it for billions of years. In fact, life emerged only after proteins mastered the technique of conformational engineering through billions of evolution. The protein structure is the best example of precisely controlling the rotation of chemical bonds: natural proteins can fold into their unique active conformation from their astronomical conformational spaces.
Apparently, intentionally manipulating molecules into specific conformations from their all possible enormous conformational spaces is far more complicated than the usual passive observation of conformational change. In certain sense, both the supramolecular chemistry and self-assembly are related to molecular conformation; however, they mainly focus on forming molecular complexes and aggregations, not controlling the active conformation of individual molecules. For example, most studies on the self-assembly of small molecules on gold surface intended to find certain beautiful patterns assembled by those molecules, but not controlling the conformation of individual molecules (Claridge et al., 2013; Kühnle et al., 2002; Liao et al., 2012; Love et al., 2005).
Protein-mimicking with nanoparticles (NPs) has been a long-pursuing goal for scientists (Kotov, 2010; Martinez-Veracoechea and Frenkel, 2011; Yang et al., 2013). Yet until now, instead of conformational engineering, most protein-mimicking studies just directly connected functional groups on NPs to enhance the already existing functions of those groups relying on multivalency effects (Huskens, 2006; Mammen et al., 1998; Wang et al., 2010; Weissleder et al., 2005) or synergy effects out of random combinatorial libraries (Bai et al., 2018; Okesola and Mata, 2018; Riccardj et al., 2017; Rufo et al., 2014; Yang and Liu, 2020; Zhang et al., 2014). In fact, the real challenge for mimicking proteins with NPs is not to endow NPs with protein-like functions but to endow NPs with protein-like functions in a protein-like way, i.e., creating novel functions on NPs by conformationally engineering non-functional groups.
Inspired by protein folding, after years of efforts, we managed to demonstrate for the first time that it is possible to create novel functions on NPs by the conformational engineering of non-functional groups (Yan et al., 2018). We have successfully restored the native conformation and function of the complementary-determining region (CDR) fragments of natural antibodies, which are unstructured and non-functional by themselves, on the surface of gold NPs (AuNPs), and created AuNP-based artificial antibodies, denoted as Goldbodies. The successful restoration of the native conformation and function of natural proteins’ flexible fragments on AuNPs implies that there must be some fundamental commonalities between AuNP and natural proteins, which keep the flexible fragments of proteins in the same conformation.
Interestingly, though numerous different interactions are involved, the net stabilizing energy for natural proteins is only about the energy of two O-H⋅⋅⋅O hydrogen bonds (around 10 kcal mol−1) (Taverna and Goldstein, 2002). By lucky coincidence, the energy barriers for the adatom diffusion of many metals including gold are also in the same range (Biener et al., 2007; Cossaro et al., 2008; Ehrlich and Stolt, 1980; Maksymovych et al., 2010; Martín et al., 1999; Müller and Ibach, 2006; Paez-Ornelas et al., 2020; Takano et al., 2003; Yu et al., 2006). Based on these two facts and the success of Goldbodies, we assume that the energy barrier for the metal adatom diffusion might serve as a general replacement for the long-range interactions in proteins to stabilize the native conformation of protein fragments. To prove this hypothesis as well as that the conformational engineering is not solely limited on AuNPs, herein, we select the widely studied silver NP (AgNP) (Arvizo et al., 2012; Chen et al., 2016; Flores et al., 2010; Paez-Ornelas et al., 2020; Wang et al., 2016) as a new scaffold for the conformational engineering of flexible protein fragments. As expected, the isolated flexible CDR3 peptide of cAB-lys3 (an anti-hen egg white lysozyme (HEWL) antibody) has been successfully engineered into its native conformation on silver NPs, producing a silver NP-based artificial antibody, denoted as Silverbody. Based on this study, we propose a plausible mechanism for both Goldbody and Silverbody, and discuss some general principles for molecular conformational engineering.
Results
Design and synthesis of the anti-lysozyme silverbody
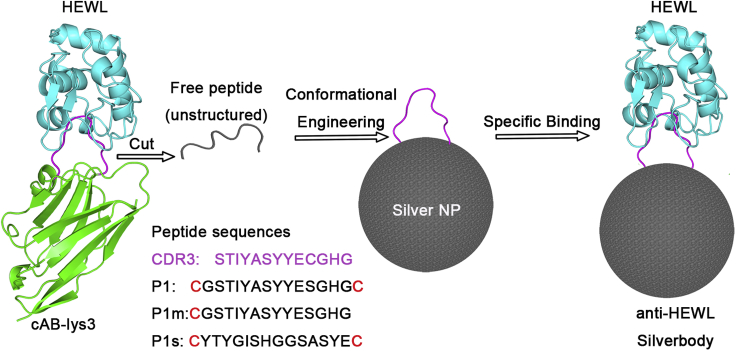
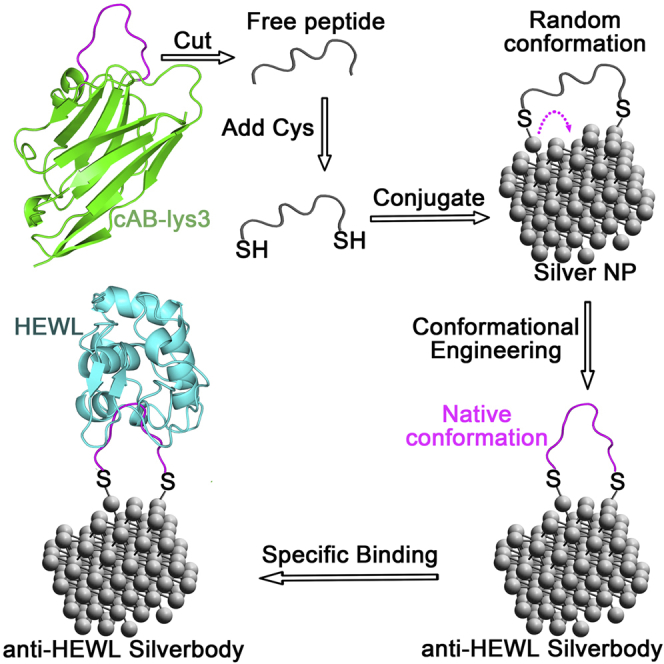
The common method for conjugating molecules onto NPs, which links the molecule to NPs with one covalent bond, lacks measures to manipulate the conformation of the conjugated molecule. As demonstrated by the design of Goldbodies (Yan et al., 2018), there has to be at least two linking positions between the conjugated molecule and the NP for conformational engineering. Figure 1 shows the rationale for conformationally engineering flexible peptides on AgNPs. The flexible CDR3 fragment of cAB-lys3 was modified as peptide P1 with one cysteine (Cys) residue added at each of its two terminals, so that each peptide could be conjugated onto AgNPs with two Ag-S bonds. These two Ag-S bonds were crucial for the conformational engineering, and the underlining mechanism will be discussed later.
Figure 1.
Conformationally engineering the flexible peptide on AgNPs
The peptide is adapted from the CDR3 loop (shown in magenta) of an anti-HEWL antibody (cAB-lys3, green), which is unstructured by itself. The restoration of the native conformation of the CDR3 on AgNP results in an anti-HEWL Silverbody. To make the scheme clear, only one peptide is shown on the AgNP.
To demonstrate the important role of the two Ag-S bonds, peptide P1m, which would form only one Ag-S bond with the AgNP (corresponding to the common conjugation method), was used as a negative control peptide. So, the difference between AgNP-P1 and AgNP-P1m was the net effect of conformational engineering. Another peptide, P1s, which has Cys at both terminals and the same composition as P1 but with a shuffled sequence, was set as a non-specific control peptide. Generally, if the conformational engineering works, the function (binding with HEWL) would be in following order: AgNP-P1 > AgNP-P1m > AgNP-P1s.
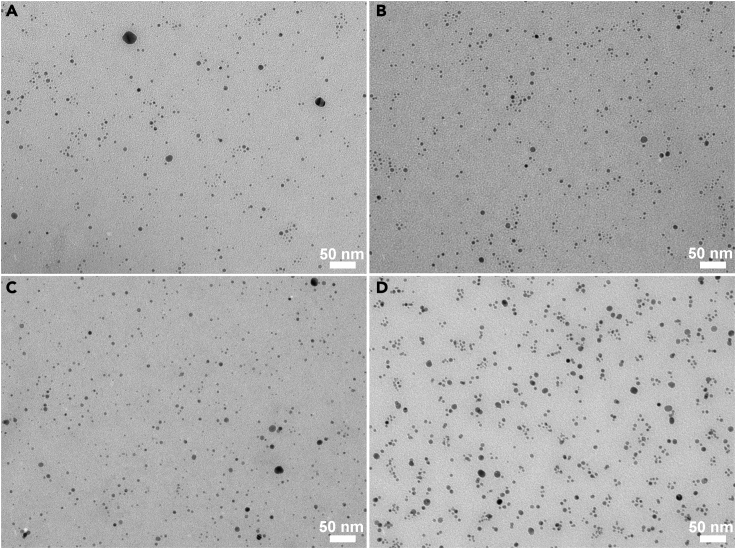
AgNPs with an average diameter of around 4.5 nm were synthesized by the common citrate-reduction method (Flores et al., 2010). The as-synthesized AgNPs were citrate-capped, negatively charged (Figure S1), and well dispersed with an average diameter of 4.43 ± 1.14 nm as determined by transmission electron microscope (TEM) (Figures 2 and S2). The average diameter of AgNPs in solutions increased slightly after conjugation with peptides (Figure S3). The mass concentration of the as-synthesized AgNPs was 30.4 ± 0.2 mg L−1 as determined using atomic absorption spectroscopy (AAS) (Table S1), and the corresponding molar concentration was thus estimated, supposing all AgNPs were spherical balls with a diameter of 4.5 nm. As well known, AgNPs were not very stable, and their size and dispersion might change with the storage time. So, after synthesis, AgNPs were immediately conjugated with peptides.
Figure 2.
TEM images of AgNPs and AgNP-peptide conjugates
(A) AgNPs, Related to Figures S2 and S3.
(B) AgNP-60P1 (Silverbody), Related to Figures S2 and S3.
(C) AgNP-60P1m, Related to Figures S2 and S3.
(D) AgNP-60P1s. Related to Figures S2 and S3.
The conjugation of peptides with AgNPs was efficient through Ag-S bonds (Luque and Santos, 2012; Poblete et al., 2016; Gondikas et al., 2012; Smith and Fickett, 1995). As detected by the fluorescence of peptide in solution, almost no free peptides could be detected after reaction with AgNPs (at least at the peptide:AgNP molar ratio up to 120:1. See Figure S4). After peptide conjugation, the AgNPs were more stable than the as-synthesized AgNPs.
Conformational engineering and characterization of the anti-lysozyme silverbody
As shown previously, there was an optimum peptide density on AuNPs for restoration of the native conformation and function of peptide P1 (Yan et al., 2018). Therefore, the same optimum peptide P1 density was directly adopted for the conjugation of P1 on AgNPs. On average, 60 P1 peptides were conjugated on one AgNP, and this AgNP-peptide conjugate is named as AgNP-60P1. Similarly, the two controls were prepared and named as AgNP-60P1m and AgNP-60P1s, respectively.
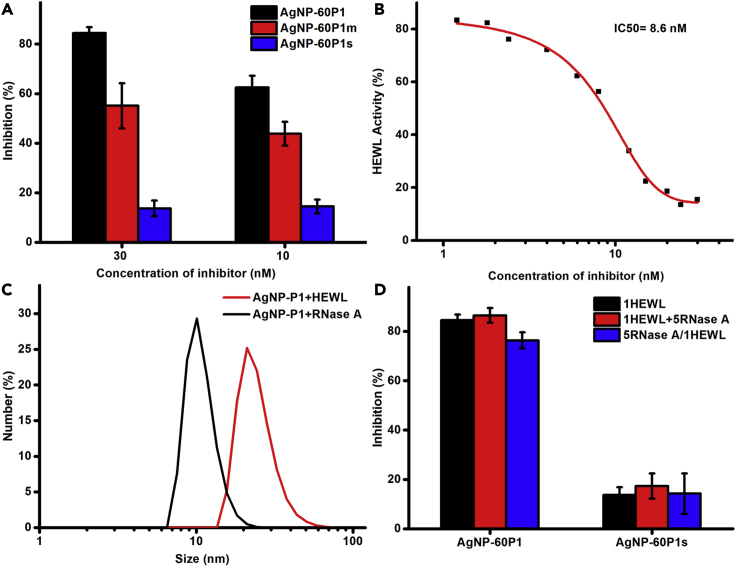
If the conformational engineering technique, which was originally developed for Goldbody on AuNPs (Liu et al., 2022; Luo et al., 2020; Yan et al., 2018), would also effectively work on AgNPs, then AgNP-60P1 would directly bind into the active cleft of HEWL, inhibiting the enzymatic activity of HEWL. Fortunately, AgNP-60P1 did inhibit the activity of HEWL (Figure 3A), demonstrating that the conformational engineering technique worked well on AgNPs too. A more convincing fact is that both of the two controls (AgNP-60P1m and AgNP-60P1s) showed much less inhibitive effects on HEWL, indicating that there was a specific interaction between AgNP-60P1 and HEWL. Therefore, AgNP-60P1 was indeed an artificial anti-HEWL antibody, which is referred to as Silverbody hereafter. Actually, the inhibition of the anti-HEWL Silverbody on HEWL (the half inhibitory concentration (IC50) value was around 8.6 nM for 30 nM HEWL (Figure 3B)) was comparable to that of the anti-HEWL Goldbody (the IC50 value was around 7.9 nM for 30 nM HEWL (Yan et al., 2018)).
Figure 3.
Interactions between HEWL and AgNP-peptide conjugates
(A) The different inhibitions of the enzymatic activity of 30 nM HEWL by 30 nM (left) or 10 nM (right) AgNP-60P1 (the Silverbody, black), AgNP-60P1m (red), and AgNP-60P1s (blue). Error bars indicate SDs (n = 3).
(B) IC50 measurement of the Silverbody for 30 nM HEWL.
(C) Sizes of the Silverbody after mixed with RNase A (black) or HEWL (red) as measured by DLS.
(D) Little influence of RNase A on the inhibition of enzymatic activity of HEWL by the Silverbody (AgNP-60P1, left) and the control AgNP-60P1s with different incubation procedures. Black columns: Conjugates were incubated with pure HEWL; red columns: Conjugates were incubated with the mixture of HEWL and RNase A (in a 1:5 M ratio); blue columns: Conjugates were pre-incubated with RNase A, and then HEWL was added into the mixture. Error bars indicate SDs (n = 3).
The specific interaction between the anti-HEWL Silverbody and HEWL was demonstrated not only by the different inhibition of HEWL by Silverbody, AgNP-60P1m, and AgNP-60P1s (Figure 3A) but also by the Silverbody’s selective binding with HEWL from other proteins, including RNase A (RNase A), a protein very similar to HEWL in many aspects (Yan et al., 2018). As shown in Figure 3C, the DLS (dynamic light scattering) data of the mixture of the Silverbody and HEWL show significantly increased size, indicating the formation of the Silverbody/HEWL complex, but DLS could not detect the presence of the Silverbody/RNase A complex from their mixture. Furthermore, when HEWL was mixed with 5-fold more RNase A, or even after the Silverbody had been pre-incubated with RNase A, the specific inhibition of HEWL by the Silverbody was almost not affected (Figure 3D).
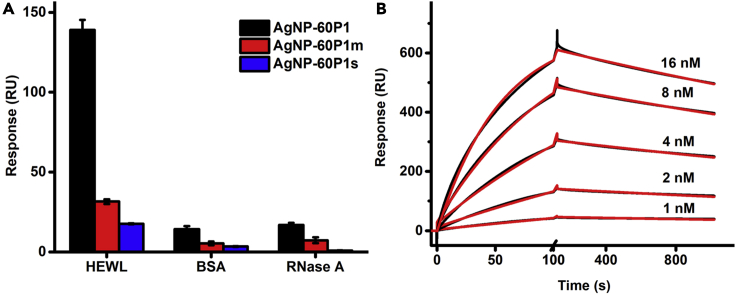
The specific interaction between the anti-HEWL Silverbody and HEWL was further characterized by surface plasmon resonance (SPR) technique on a Biacore T200 (Cytiva), with HEWL and two control proteins (RNase A and BSA, the most abundant protein in serum) immobilized on a CM5 chip (Figure 4A). Once again, the SPR binding data showed that the anti-HEWL Silverbody bound strongly to HEWL, but only bound weakly (non-specific) to RNase A and BSA. And as expected, both of the control peptide conjugations, AgNP-P1m and AgNP-P1s, only bound weakly (non-specific) with all three proteins including HEWL.
Figure 4.
SPR binding between HEWL and AgNP-peptide conjugates
(A) SPR binding of AgNP-60P1 (the Silverbody, black), AgNP-60P1m (red), and AgNP-60P1s (blue) onto the immobilized HEWL, BSA, and RNase A. Error bars indicate SDs (n = 3).
(B) SPR binding kinetic curves (black) of the Silverbody at different concentrations (indicated) onto the immobilized HEWL. Red curves are the fitting lines. Related to Figures S5.
To quantify the affinity between the anti-HEWL Silverbody and HEWL, the binding kinetics was measured by SPR with HEWL immobilized on the CM5 chip (Figures 4B and S5). The binding kinetics data could be fitted with the simple 1:1 model, and we obtained an apparent affinity (KD) of 2.0 × 10−10 M (kon = 1.2 × 106 M−1 s−1 and koff = 2.3 × 10−4 s−1). The apparent affinity was comparable to that of the previous anti-HEWL Goldbody (KD = 1.5 × 10−10 M) (Yan et al., 2018), but much higher than that of the antibody cAB-lys3 (Desmyter et al., 1996). It has to be pointed out that the multivalent effect contributed a lot to the apparent affinity (Yan et al., 2018), because the Silverbody, like the previous Goldbody (Yan et al., 2018), had many binding sites for HEWL on one particle.
Discussion
Mechanism of conformational engineering on metal NPs
All the above experiments have demonstrated that the native conformation and the antigen-recognition function of the CDR3 fragment of cAB-lys3 were successfully restored on AgNPs, thus proving that, like AuNPs, AgNPs can also be used as scaffolds to restore (conformationally engineer) the native conformation of flexible protein fragments. It has to be pointed out that the CDR3 peptide (peptide P1) was unstructured and non-functional by itself. As the protein folding problem is still not fully understood (despite the recent breakthrough on protein structure prediction made by AlphaFold2 (Jumper et al., 2021), which is basically just a numerical approximation of a partial problem of protein folding), it is amazing how simple NPs such as AuNPs and AgNPs could replace the complicated protein scaffold to restore the native conformation and function of the flexible protein fragments.
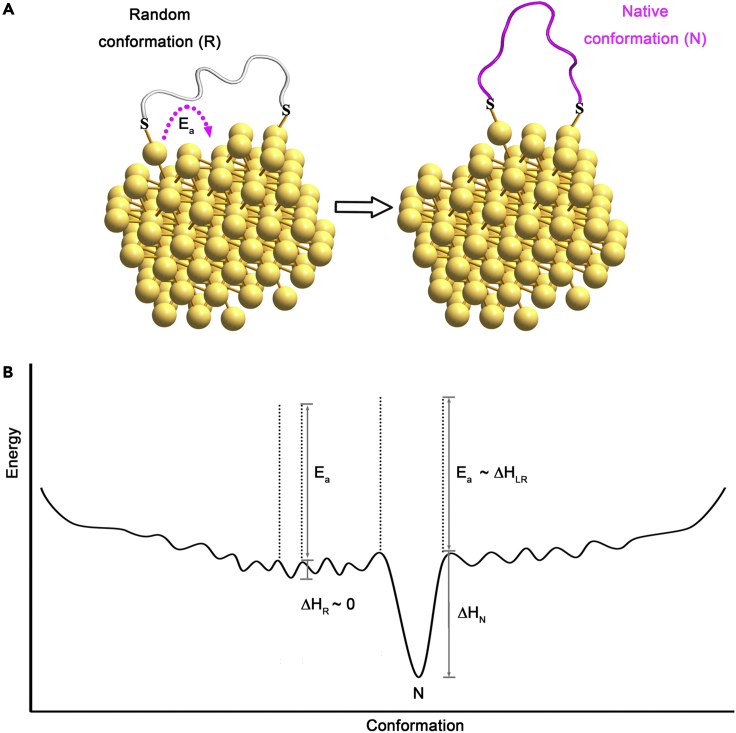
To answer this fundamental question, we proposed the “Confined Lowest Energy Structure Fragment” (CLESF) hypothesis (Cao, 2020a) (later unified as the CLEF hypothesis (Cao, 2020b)) for the protein folding problem. The main idea of the CLEF hypothesis is that the whole peptide chain of a protein can be divided into many CLEFs. Each CLEF is a semi-independent folding unit, whose native conformation is at least one of the energy minima (not necessarily the lowest one) as schematically shown in Figure 5B. But the favorable enthalpy (ΔHN) of the CLEF is usually not big enough to counterbalance the large entropy of the enormous non-native conformations, so the isolated protein fragments are usually unstructured (random coil or a huge number of conformations). In the protein, there are a small number of key residues to hold all the CLEFs together with strong long-range interactions, such as disulfide bond and hydrophobic packing. The CLEF hypothesis provides a simple solution for the protein folding problem, and a simple explanation for the success of conformational engineering on AuNPs and AgNPs: The Au-S and Ag-S bonds are satisfactory replacements of the strong long-range interactions within the original antibody to stable the native conformation of the CDR peptide.
Figure 5.
Mechanism of the restoration of the native conformation of peptide fragments of natural proteins
(A) Model for restoring the native conformation of peptide fragments conjugated on the surface of metal NPs through M-S bonds.
(B) Typical energy landscape of a CLEF, whose native conformation has favorable enthalpy (ΔHN), while the random conformation does not (ΔHR ∼0). When the peptide (CLEF) is conjugated onto AuNP or AgNP, the activation energy for the adatom diffusion (Ea) will increase the energy well, and then the native conformation is stabilized.
Figure 5 illustrates a general mechanism for the conformational engineering of those initially unstructured peptides into their native conformations on NPs like AuNPs and AgNPs. As mentioned previously, a distinguishable feature of the current conformational engineering is the two anchoring positions for each peptide on NPs. The first advantage of the two anchors is to greatly reduce the entropy of the non-native state of the peptide by excluding a large number of conformations (Liu et al., 2022). As the free peptide fragment (e.g. P1) is unstructured, when it forms two metal-S (M-S) bonds with the adatoms of one NP, there would still be a large number of possible conformations for the conjugated peptide, corresponding to different distances between the two S atoms (Figure 5A). As the native conformation of the peptide requires a specific distance between the two S atoms, the conformational engineering therefore requires the initially formed distance between the two S atoms being changed, either by the movement of S atoms (breaking of M-S bonds or sliding of the S atoms on the NP surface) or by the movement of the adatoms (together with S atoms). Because the M-S bond is weaker than most covalent bonds, it is possible for the peptide to break one M-S bond and hop to another adatom (hoping mode). In addition, it has been reported that S atoms could form Au-S-Au bridging bonds and the S atoms can move (slide) from one Au adatom to another, and the mobility of Au adatoms has been also generally observed (Biener et al., 2007; Cossaro et al., 2008; Maksymovych et al., 2010; Yu et al., 2006).
All three modes (hoping and sliding of S, and diffusion of M adatom) for the change of the span of the peptide on NPs are possible, but which one dominates depends on their activation energy (Ea) and experimental conditions (Figure 5A). Because the Ea for breaking M-S bonds is the largest one, the M adatom diffusion and the S atom sliding are likely the major modes for metal NPs at the experimental conditions used here. In principle, no matter which mode is adopted, there is generally little net energy gain from the movement of M-S bond on the surface of NPs (neglecting the energy difference between the adatoms at the starting and ending locations on the metal surface). However, when the adatom (and also the M-S) moves to the right position corresponding to the right distance between the two S atoms for the native conformation of the peptide, the native conformation of the peptide will be stabilized. In other words, the reason that those initially unstructured peptides can fold to their native conformation on NPs is the coupling of the favorable enthalpy (ΔHN) of the native conformation of the peptide and the stability (the activation energy for the mobility, Ea) of S-M bonds. That is, the coupling will set a high energy barrel (ΔHN + Ea) for the unfolding of the native conformation of the peptide to overcome. Basically, the Ea plays a similar role in stabilizing the native conformation of the peptide as that of the key long-range interactions (ΔHLR) of the original protein for the peptide fragment (Figure 5B) (Cao, 2020a, 2020b).
Principles for molecular conformational engineering
Together with previous Goldbodies (Liu et al., 2022; Luo et al., 2020; Yan et al., 2018), the current success of Silverbody highlights the general possibility of conformational engineering on NPs. Though molecular conformational engineering is still in its infancy, some principles might be concluded from the successes of Goldbody and Silverbody, as well as from the theoretical consideration.
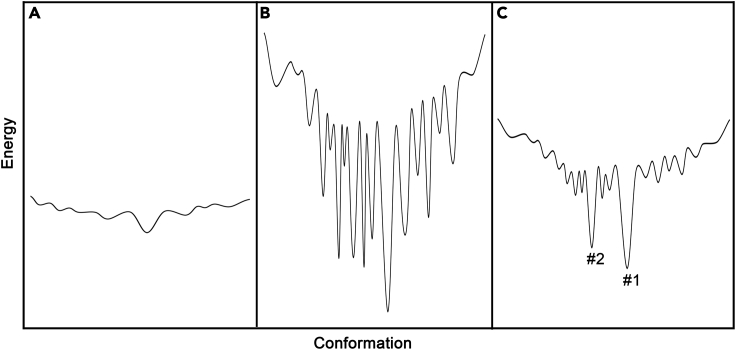
The first principle is about the molecules to be conformationally engineered. Apparently, not every molecule could be engineered into a unique conformation, just as not any arbitrary peptide could be folded. In fact, the foldable peptides including all natural proteins are only a very tiny portion in the astronomical amino acid sequence space. Figure 6 schematically shows three types of energy landscapes for molecules with rotatable chemical bonds. From the statistical thermodynamics point of view, the molecules with a relatively flat energy landscape (Figure 6A) are not suitable to be conformationally engineered, because they do not have energetically favorable conformations, or the favorable enthalpy for the lowest energy conformations is too small to stabilize the conformation. As for those molecules whose energy landscapes have many deep wells (Figure 6B), they are also difficult to be conformationally engineered, because they are prone to be kinetically trapped in those unwanted conformations that also have large favorable enthalpies. Therefore, suitable molecules should have a typical energy landscape like that of CLEFs as shown in Figures 5B and 6C, i.e., the molecules with one or a few conformations that have reasonably favorable enthalpies (largely equivalent to the energy of two hydrogen bonds). Even for these molecules, they can only be conformationally engineered into those favorable conformations (energy minima). It has to be pointed out that the goal for conformational engineering is not necessarily the one with the lowest energy, so both conformations #1 and #2 in Figure 6C are the possible conformations, just like that the same peptide fragment could adopt different native conformations in different proteins or at different conditions (Liu et al., 2022; Luo et al., 2020).
Figure 6.
Typical energy landscapes of molecules with large conformation space
(A) Molecules with an energy landscape too shallow.
(B) Molecules with an energy landscape too rough.
(C) Molecules with an energy landscape suitable for conformational engineering.
Naturally, all CLEFs from natural proteins are possible to be conformationally restored on suitable NPs. As CLEFs have some tolerance on the peptide terminals, peptide fragments that consist of the majority of CLEFs are all possible to be restored on suitable NPs. Usually, after the proper peptide fragment is selected from natural proteins, it has to be properly modified, such as the mutation of strong binding residues (e.g. Cys, Lysine, and Arginine) to prevent the strong interaction with NPs at unwanted positions, and the incorporation of Cys residues at suitable positions in order to form at least two M-S bonds with NPs (Liu et al., 2022; Luo et al., 2020; Yan et al., 2018).
Besides, peptides screened from phage display (Smith, 2019) are also excellent candidates for conformational engineering. In principle, non-natural peptides and non-peptidyl molecules could also be designed to have suitable energy landscapes like that showed in Figure 6C for further conformational engineering to achieve better properties.
The second principle is about the technical measures to manipulate the conformation of the selected molecules. According to the mechanism illustrated in Figure 5, the basic requirement for the technical measures is to provide a stabilizing energy for the target conformation of the molecule. The technical measures include choosing proper anchoring positions in the target molecules and suitable anchoring methods (for example the positions mutated to Cys in peptide P1 for forming M-S bonds), and choosing suitable scaffolds to stabilize the conformation of the target molecules. Apparently, there should be at least two anchoring positions for each target molecule, so they can reduce the flexibility of the molecules and exclude a major portion of the conformational space to achieve a large favorable entropy effect. And the scaffolds basically provide extra enthalpy benefit and should accommodate the right orientation of the anchoring positions. Generally, the favorable scaffolds are those that cause little or no extra energy difference for molecular conformations corresponding to different distances between the anchoring positions, such as AuNPs and AgNPs.
Besides NPs, organic molecules, including peptides, are also possible to be used as scaffolds. As the CLEF hypothesis points out, natural proteins could be regarded as peptides (CLEFs) assembled together, helping each other to stabilize their favorable conformations. This might be how natural proteins evolved at the first place as proposed by the “stone age” or “CLEF age” hypothesis (Cao, 2020a, 2020b).
It is also possible to conformationally engineer different molecules on the same NPs, for example to produce Goldbodies or Silverbodies with multiple specificities. More challenging and more marvelous are to arrange different molecules onto one “particle” to carry out novel functions, just like how natural proteins evolved from assembling different CLEFs (Cao, 2020a, 2020b). As the human-manipulated evolution can produce much better enzymes or antibodies than natural ones (Arnold, 2019; Smith, 2019; Winter, 2019), the conformational engineering approach may also produce unprecedented molecules/particles with unimaginable properties.
In summary, a silver NP-based artificial antibody, denoted as Silverbody, has been successfully synthesized by conformationally engineering non-functional flexible peptides on the surface of AgNPs. The results show that besides AuNPs, other NPs, such as AgNPs could also be used as scaffolds to restore the native conformation and function of fragments of proteins. And a general mechanism is proposed for the conformational engineering on NPs. Accordingly, a new class of artificial proteins could be made, with both the excellent stability of NPs and the high specificity of proteins, demonstrating enormous opportunities in the new area of conformational engineering. It is foreseeable that as life emerged after natural proteins “mastered conformational engineering” through billion years’ evolution, wonderful things will happen when chemists master the skills of molecular conformational engineering.
Limitations of the study
Although the restoration of original antigen-recognizing function is unambiguously demonstrated by enzymatic inhibition and binding affinity measurements with different control experiments, the restoration of the native conformation of the peptide on AgNPs is only a conclusion deduced on the basis of structure-determined function. Because there is currently no available technique to determine the structure of the conformationally engineered loop peptides on AgNPs, computational simulations with semi-empirical/force field would be helpful to provide the direct evidence of the engineered conformation of the peptides in the future.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and virus strains | ||
| Micrococcus lysodeikticus cells | Sangon Biotech | A008736 |
| Chemicals, peptides, and recombinant proteins | ||
| Silver nitrate | Sinopharm Chemical Reagent | CAS: 7761-88-8 |
| Sodium tetrahydroborate | Sinopharm Chemical Reagent | CAS: 16940-66-2 |
| Trisodium citrate dehydrate | Sinopharm Chemical Reagent | CAS: 6132-04-3 |
| Sodium dihydrogen phosphate | Sinopharm Chemical Reagent | CAS: 13472-35-0 |
| Disodium hydrogen phosphate | Sinopharm Chemical Reagent | CAS: 7558-79-4 |
| Hydrochloric acid | Sinopharm Chemical Reagent | CAS: 7647-01-0 |
| Nitric acid | Sinopharm Chemical Reagent | CAS: 7697-37-2 |
| Sodium hydroxide | Sinopharm Chemical Reagent | CAS: 1310-73-2 |
| Lysozyme from chicken egg white | Sigma-Aldrich | L4919; CAS: 12650-88-3 |
| Bovine serum albumin | Sigma-Aldrich | B2064; CAS: 9048-46-8 |
| Ribonuclease A | Sangon Biotech | B694345; CAS: 9001-99-4 |
| Acetate 4.5 | Cytiva | BR100350 |
| 10×HBS-EP Buffer | Cytiva | BR100669 |
| Peptide-Pep1 (P1) | Shanghai Science Peptide Biological Technology Co., Ltd. | N/A |
| Peptide-Pep1m (P1m) | Shanghai Science Peptide Biological Technology Co., Ltd. | N/A |
| Peptide-Pep1s (P1s) | Shanghai Science Peptide Biological Technology Co., Ltd. | N/A |
| Critical commercial assays | ||
| Amine coupling kit | Cytiva | BR100050 |
| Deposited data | ||
| Raw and analyzed data | This paper | https://doi.org/10.17632/22vwv5yk6s.1 |
| Software and algorithms | ||
| ProtParam tool | SIB Swiss Institute of Bioinformatics | https://web.expasy.org/protparam/#userconsent# |
| Nano Measurer 1.2.5 | Jie Xu, Fudan Univ. | shine6832@163.com |
| Biacore T200 Software v3.0 | Cytiva | https://www.cytivalifesciences.com.cn/zh/cn/shop/protein-analysis/spr-label-free-analysis/software |
| Other | ||
| Surface plasmon resonance CM5 sensor chip | Cytiva | 29149603 |
Resource availability
Lead contact
Further information and requests for resources should be directed to and will be fulfilled by the lead contact, Aoneng Cao (ancao@shu.edu.cn).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
None.
Method details
Synthesis of AgNPs
AgNPs with an average diameter of around 4.5 nm were synthesized by the common citrate-reduction method (Flores et al., 2010) with a little modification. Before synthesis, all glassware was cleaned with aqua regia and rinsed thoroughly with ultrapure water. The reaction temperature was controlled at 4°C. Briefly, 1 mL of 17 mM sodium citrate was diluted with 15 mL of water, and then 1 mL of 5 mM AgNO3 solution was added. After stirred for 1 min, 100 μL of 100 mM freshly prepared sodium tetrahydroborate was added into the mixture with a syringe pump. The reaction mixture was stirred for 105 min to complete the reaction. The as-prepared AgNPs were stored at 4°C in the dark.
Determination of the concentration of the AgNPs
The as-synthesized AgNP solution was first ultracentrifuged with an Amicon Ultra 15 filter (MWCO: 30 kD) at 1500 g for 15 min to remove the unreacted Ag ions. The purified AgNPs were digested in aqua regia, and the mass concentration of Ag element (CAg) was determined by atomic absorption spectroscopy (PinAAcle 900T, Perkin Elmer).
Preparation of AgNP-peptide conjugates
The pH of the AgNPs solution was adjusted to 7.4 using 0.2 M trisodium citrate. The peptide dissolved in phosphate buffer (PB) solution (0.01 M, pH 7.4) was added dropwise to the AgNPs solution, and the mixture was stirred for 12 h at room temperature. The conjugation efficiency was determined by measuring the fluorescence of the total peptides before the reaction and the unreacted peptides in the filtrate after ultracentrifugation with an Amicon Ultra 15 filter (MWCO: 30 kD) at 1500 g for 15 min. Since the conjugation was very efficient, the peptide density or the average number of peptide molecules per AgNP was simply achieved by mixing the peptide and the AgNP solution in stoichiometric proportions for 12 h at room temperature.
Characterization of AgNPs and AgNP-peptide conjugates
The size and morphology of AgNPs and AgNP-peptide conjugates were characterized by TEM (200CX; JEOL) and UV-Vis spectroscopy (U-3010; Hitachi). The hydrodynamic diameters and zeta-potentials were measured with a Nanosizer ZS90 (DLS; Malvern) at 25°C.
Enzymatic activity assay of lysozyme
The enzymatic activity of HEWL was assayed using the method reported previously (Luo et al., 2020; Yan et al., 2018), by monitoring changes of the absorbance at 450 nm, using Micrococcus lysodeikticus (M. lysodeikticus) as the substrate. The enzymatic digestion of M. lysodeikticus within first 90 s was recorded on the U-3010 UV-Vis spectrophotometer (Hitachi). The stock suspension of M. lysodeikticus (1/3 g·L-1) was freshly prepared in PB buffer (0.1 M, pH 6.2), and the HEWL stock solution (1.5 × 10−7 M) was prepared in PB buffer (0.01 M, pH 7.4). In a typical assay, 0.5 mL of the HEWL stock solution was added to 1 mL of 75 nM AgNP-peptide conjugates and mixed well for 1 min. Then 1 mL of the stock suspension of M. lysodeikticus was added to the mixture. After short vigorous mixing, the mixture was quickly transferred to a cuvette for the absorbance measurement. All samples were pre-incubated at 25°C and all assays were conducted at 25°C.
IC50 determination
The IC50 of AgNP-60P1 to inhibit HEWL was determined by measuring the enzymatic activity of 30 nM (final concentration) HEWL in the presence of 0, 1.2, 1.8, 2.4, 4.0, 6.0, 8.0, 12, 15, 20, 24, or 30 nM (final concentration) AgNP-60P1, following the above assay procedure.
SPR experiments
SPR experiments were carried out on a Biacore T200 instrument (Cytiva) at 25°C. The standard HBS EP buffer was used as the running buffer. HEWL, RNase A, and BSA were coupled to different channels of CM5 chips of series S following the standard amine coupling procedure.
To investigate the binding specificity of the Silverbody, the running buffer-diluted 2 nM AgNP-60P1, 2 nM AgNP-60P1m, or 2 nM AgNP-60P1s was injected into the HEWL-, BSA-, and RNase A-immobilized channels, at a flow rate of 30 μL·min-1.
For binding kinetics measurements, HEWL was coupled to a CM5 chip at a much lower level than the above binding specificity experiments to prevent the possible binding of one AgNP-60P1 particle with more than one HEWL, so that the binding kinetics could be fitted with the 1:1 model. Then, different concentrations of AgNP-60P1 (diluted in the running buffer) were injected into the HEWL-immobilized channel at a flow rate of 30 μL·min-1.
Quantification and statistical analysis
The molar concentration of the as-synthesized AgNP solution ([AgNPs]) was estimated from the mass concentration of Ag measured by AAS, assuming that all AgNPs were spherical with the same diameter (the average diameter measured by TEM). The equation for estimating [AgNPs] is:
where, NA is the Avogadro constant, ρ is the density of Ag (10.492 g·cm-3) (Smith and Fickett, 1995), and r is the average radius of AgNPs determined by TEM.
Using the average radius of 2.25 nm, the calculated molar concentration of the as-synthesized AgNPs was 100.7 nM. Considering AgNPs were not perfectly spherical with a distribution of size, the calculated concentration was only a rough estimation, therefore, the decimal was omitted for easy dilution (the as-synthesized AgNP solutions were used as the stock solutions).
The slopes of the recorded absorbance changes at 450 nm were calculated as the activity of HEWL. The relative activities of HEWL in the presence of different additives were calculated as the ratio of the corresponding slopes to the slope of free HEWL, and the inhibition rates were calculated as the percentage of relative activity loss.
The activity data vs the logarithm of inhibitor concentrations were plotted and fitted to the logistic function to obtain the IC50 value.
The SPR kinetic data of the binding of the Silverbody with HEWL were fitted with the simple 1:1 model using the Biacore T200 Software v3.0.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (Nos. 31871007, 32071404, 22071145, and 31771105) and the National Key Research and Development Plan of China (No.2016YFA0201602).
Author contributions
A.C. conceived the project; A.C. and H.W. supervised the project. J.X., T.G., L.S., Y.W., and C.L. conducted the experiments. All authors analyzed the data. A.C., J.X., and H.W. co-wrote manuscript with contribution from all authors.
Declaration of interests
The authors declare no competing interests.
Published: June 17, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.104324.
Contributor Information
Haifang Wang, Email: hwang@shu.edu.cn.
Aoneng Cao, Email: ancao@shu.edu.cn.
Supplemental information
Data and code availability
Data generated in this study have deposited in Mendeley Data (https://doi.org/10.17632/22vwv5yk6s.1). Key words: anti-lysozyme silverbody.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Arnold F.H. Innovation by evolution: bringing new chemistry to life (Nobel Lecture) Angew. Chem. Int. Ed. 2019;58:14420–14426. doi: 10.1002/anie.201907729. [DOI] [PubMed] [Google Scholar]
- Arvizo R.R., Bhattacharyya S., Kudgus R.A., Giri K., Bhattacharya R., Mukherjee P. Intrinsic therapeutic applications of noble metal nanoparticles: past, present and future. Chem. Soc. Rev. 2012;41:2943–2970. doi: 10.1039/c2cs15355f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Y., Chotera A., Taran O., Liang C., Ashkenasy G., Lynn D.G. Achieving biopolymer synergy in systems chemistry. Chem. Soc. Rev. 2018;47:5444–5456. doi: 10.1039/c8cs00174j. [DOI] [PubMed] [Google Scholar]
- Biener M.M., Biener J., Friend C.M. Sulfur-induced mobilization of Au surface atoms on Au(111) studied by real-time STM. Surf. Sci. 2007;601:1659–1667. doi: 10.1016/j.susc.2007.01.041. [DOI] [Google Scholar]
- Burger B., Maffettone P.M., Gusev V.V., Aitchison C.M., Bai Y., Wang X., Li X., Alston B.M., Li B., Clowes R., et al. A mobile robotic chemist. Nature. 2020;583:237–241. doi: 10.1038/s41586-020-2442-2. [DOI] [PubMed] [Google Scholar]
- Cao A. The last secret of protein folding: the real relationship between long-range interactions and local structures. Protein J. 2020;39:422–433. doi: 10.1007/s10930-020-09925-w. [DOI] [PubMed] [Google Scholar]
- Cao A. “Confined lowest energy structure fragments (CLESFs)” hypothesis for protein structure and the “stone age” of protein prebiotic evolution. Acta Phys. Chim. Sin. 2020;36:1907002. doi: 10.3866/pku.whxb201907002. [DOI] [Google Scholar]
- Chen N., Song Z.-M., Tang H., Xi W.-S., Cao A., Liu Y., Wang H. Toxicological effects of Caco-2 cells following short-term and long-term exposure to Ag nanoparticles. Int. J. Mol. Sci. 2016;17:974. doi: 10.3390/ijms17060974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claridge S.A., Liao W.-S., Thomas J.C., Zhao Y., Cao H.H., Cheunkar S., Serino A.C., Andrews A.M., Weiss P.S. From the bottom up: dimensional control and characterization in molecular monolayers. Chem. Soc. Rev. 2013;42:2725–2745. doi: 10.1039/c2cs35365b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cossaro A., Mazzarello R., Rousseau R., Casalis L., Verdini A., Kohlmeyer A., Floreano L., Scandolo S., Morgante A., Klein M.L., Scoles G. X-ray diffraction and computation yield the structure of alkanethiols on Gold (111) Science. 2008;321:943–946. doi: 10.1126/science.1158532. [DOI] [PubMed] [Google Scholar]
- Desmyter A., Transue T.R., Ghahroudi M.A., Dao Thi M.H., Poortmans F., Hamers R., Muyldermans S., Wyns L. Crystal structure of a camel single-domain VH antibody fragment in complex with lysozyme. Nat. Struct. Mol. Biol. 1996;3:803–811. doi: 10.1038/nsb0996-803. [DOI] [PubMed] [Google Scholar]
- Dill K.A., MacCallum J.L. The Protein-folding problem, 50 years on. Science. 2012;338:1042–1046. doi: 10.1126/science.1219021. [DOI] [PubMed] [Google Scholar]
- Ehrlich G., Stolt K. Surf ace diffusion. Ann. Rev. Phys. Chem. 1980;31:603–637. doi: 10.1146/annurev.pc.31.100180.003131. [DOI] [Google Scholar]
- Flores C.Y., Diaz C., Rubert A., Benítez G.A., Moreno M.S., Fernández Lorenzo de Mele M., Salvarezza R.C., Schilardi P.L., Vericat C. Spontaneous adsorption of silver nanoparticles on Ti/TiO2 surfaces. Antibacterial effect on Pseudomonas Aeruginosa. J. Colloid Interf. Sci. 2010;350:402–408. doi: 10.1016/j.jcis.2010.06.052. [DOI] [PubMed] [Google Scholar]
- Gondikas A.P., Morris A., Reinsch B.C., Marinakos S.M., Lowry G.V., Hsu-Kim H. Cysteine-induced modifications of zero-valent silver nanomaterials: implications for particle surface chemistry, aggregation, dissolution, and silver speciation. Environ. Sci. Technol. 2012;46:7037–7045. doi: 10.1021/es3001757. [DOI] [PubMed] [Google Scholar]
- Huskens J. Multivalent interactions at interfaces. Curr. Opin. Chem. Biol. 2006;10:537–543. doi: 10.1016/j.cbpa.2006.09.007. [DOI] [PubMed] [Google Scholar]
- Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., Tunyasuvunakool K., Bates R., Žídek A., Potapenko A., et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotov N.A. Inorganic nanoparticles as protein mimics. Science. 2010;330:188–189. doi: 10.1126/science.1190094. [DOI] [PubMed] [Google Scholar]
- Kühnle A., Linderoth T.R., Hammer B., Besenbacher F. Chiral recognition in dimerization of adsorbed cysteine observed by scanning tunnelling microscopy. Nature. 2002;415:891–893. doi: 10.1038/415891a. [DOI] [PubMed] [Google Scholar]
- Liao W.-S., Cheunkar S., Cao H.H., Bednar H.R., Weiss P.S., Andrews A.M. Subtractive patterning via chemical lift-off lithography. Science. 2012;337:1517–1521. doi: 10.1126/science.1221774. [DOI] [PubMed] [Google Scholar]
- Liu Q., Sheng L., Liu Y.-Y., Gao T., Wang H., Liu Y., Cao A. A potential MDM2 inhibitor formed by restoring the native conformation of the p53 α-helical peptide on gold nanoparticles. ChemMedChem. 2022;17 doi: 10.1002/cmdc.202100623. [DOI] [PubMed] [Google Scholar]
- Love J.C., Estroff L.A., Kriebel J.K., Nuzzo R.G., Whitesides G.M. Self-assembled monolayers of thiolates on metals as a form of nanotechnology. Chem. Rev. 2005;105:1103–1169. doi: 10.1002/chin.200532281. [DOI] [PubMed] [Google Scholar]
- Luo L., Liu Y.-Y., Gao T., Wang X., Chen J., Wang H., Liu Y., Cao A. Characterization of the specific interactions between nanoparticles and proteins at residue-resolution by alanine scanning mutagenesis. ACS Appl. Mater. Inter. 2020;12:34514–34523. doi: 10.1021/acsami.0c05994. [DOI] [PubMed] [Google Scholar]
- Luque N.B., Santos E. Ab initio studies of Ag-S bond formation during the adsorption of L-cysteine on Ag(111) Langmuir. 2012;28:11472–11480. doi: 10.1021/la302376w. [DOI] [PubMed] [Google Scholar]
- Maksymovych P., Voznyy O., Dougherty D.B., Sorescu D.C., Yates J.T. Gold adatom as a key structural component in self-assembled monolayers of organosulfur molecules on Au (111) Prog. Surf. Sci. 2010;85:206–240. doi: 10.1016/j.progsurf.2010.05.001. [DOI] [Google Scholar]
- Mammen M., Choi S.-K., Whitesides G.M. Polyvalent interactions in biological systems: implications for design and use of multivalent ligands and inhibitors. Angew. Chem. Int. Ed. 1998;37:2754–2794. doi: 10.1002/(sici)1521-3773(19981102)37:20<2754::aid-anie2754>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- Martín H., Carro P., Hernández Creus A., González S., Salvarezza R.C., Arvia A.J. Kinetics and mechanism of gold dendrite electroformation on C(0001). Activation energy for gold adatom surface diffusion. J. Phys. Chem. B. 1999;103:3900–3907. doi: 10.1021/jp982695k. [DOI] [Google Scholar]
- Martinez-Veracoechea F.J., Frenkel D. Designing super selectivity in multivalent nano-particle binding. Proc. Natl. Acad. Sci. U.S.A. 2011;108:10963–10968. doi: 10.1073/pnas.1105351108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller J.E., Ibach H. Migration of point defects at charged Cu, Ag, and Au (100) surfaces. Phys. Rev. B. 2006;74:085408. doi: 10.1103/physrevb.74.085408. [DOI] [Google Scholar]
- Narayan M., Welker E., Wedemeyer W.J., Scheraga H.A. Oxidative folding of proteins. Acc. Chem. Res. 2000;33:805–812. doi: 10.1021/ar000063m. [DOI] [PubMed] [Google Scholar]
- Okesola B.O., Mata A. Multicomponent self-assembly as a tool to harness new properties from peptides and proteins in material design. Chem. Soc. Rev. 2018;47:3721–3736. doi: 10.1039/c8cs00121a. [DOI] [PubMed] [Google Scholar]
- Paez-Ornelas J.I., Fernández-Escamilla H.N., Takeuchi N., Guerrero-Sánchez J. Adsorption and diffusion mechanisms of silver ad-atoms on Ag and Cu(110) surfaces: a first principles study. Mater. Today Commun. 2020;25:101461. doi: 10.1016/j.mtcomm.2020.101461. [DOI] [Google Scholar]
- Peplow M. Chemists get out begging bowl to avert closure. Nature. 2006;441:12–13. doi: 10.1038/441012a. [DOI] [PubMed] [Google Scholar]
- Poblete H., Agarwal A., Thomas S.S., Bohne C., Ravichandran R., Phopase J., Comer J., Alarcon E.I. New insights into peptide-silver nanoparticle interaction: deciphering the role of cysteine and lysine in the peptide sequence. Langmuir. 2016;32:265–273. doi: 10.1021/acs.langmuir.5b03601. [DOI] [PubMed] [Google Scholar]
- Riccardi L., Gabrielli L., Sun X., De Biasi F., Rastrelli F., Mancin F., De Vivo M. Nanoparticle-based receptors mimic protein-ligand recognition. Chem. 2017;3:92–109. doi: 10.1016/j.chempr.2017.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rufo C.M., Moroz Y.S., Moroz O.V., Stöhr J., Smith T.A., Hu X., Degrado W.F., Korendovych I.V. Short peptides self-assemble to produce catalytic amyloids. Nat. Chem. 2014;6:303–309. doi: 10.1038/nchem.1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segler M.H.S., Preuss M., Waller M.P. Planning chemical syntheses with deep neural networks and symbolic AI. Nature. 2018;555:604–610. doi: 10.1038/nature25978. [DOI] [PubMed] [Google Scholar]
- Smith D.R., Fickett F.R. Low-temperature properties of silver. J. Res. Natl. Inst. Stand. Technol. 1995;100:119–171. doi: 10.6028/jres.100.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith G.P. Phage display: simple evolution in a petri dish (Nobel Lecture) Angew. Chem. Int. Ed. 2019;58:14428–14437. doi: 10.1002/anie.201908308. [DOI] [PubMed] [Google Scholar]
- Takano J., Doyama M., Kogure Y. Motion and conversion energies of adatom and adatom clusters on gold(001) surface. Thin Solid Films. 2003;424:45–49. doi: 10.1016/s0040-6090(02)00905-7. [DOI] [Google Scholar]
- Taverna D.M., Goldstein R.A. Why are proteins marginally stable? Proteins: Struct. Funct. Genet. 2002;46:105–109. doi: 10.1002/prot.10016. [DOI] [PubMed] [Google Scholar]
- Wang J., Tian S., Petros R.A., Napier M.E., Desimone J.M. The complex role of multivalency in nanoparticles targeting the transferrin receptor for cancer therapies. J. Am. Chem. Soc. 2010;132:11306–11313. doi: 10.1021/ja1043177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.-W., Tang H., Wu D., Liu D., Liu Y., Cao A., Wang H. Enhanced bactericidal toxicity of silver nanoparticles by the antibiotic gentamicin. Environ. Sci. Nano. 2016;3:788–798. doi: 10.1039/c6en00031b. [DOI] [Google Scholar]
- Weissleder R., Kelly K., Sun E.Y., Shtatland T., Josephson L. Cell-specific targeting of nanoparticles by multivalent attachment of small molecules. Nat. Biotechnol. 2005;23:1418–1423. doi: 10.1038/nbt1159. [DOI] [PubMed] [Google Scholar]
- Winter G. Harnessing evolution to make medicines (Nobel Lecture) Angew. Chem. Int. Ed. 2019;58:14438–14445. doi: 10.1002/anie.201909343. [DOI] [PubMed] [Google Scholar]
- Yan G.-H., Wang K., Shao Z., Luo L., Song Z.-M., Chen J., Jin R., Deng X., Wang H., Cao Z., et al. Artificial antibody created by conformational reconstruction of the complementary-determining region on gold nanoparticles. Proc. Natl. Acad. Sci. U.S.A. 2018;115:E34–E43. doi: 10.1073/pnas.1713526115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S.-T., Liu Y., Wang Y.-W., Cao A. Biosafety and bioapplication of nanomaterials by designing protein-nanoparticle interactions. Small. 2013;9:1635–1653. doi: 10.1002/smll.201201492. [DOI] [PubMed] [Google Scholar]
- Yang Y., Liu Z. Chemiluminescent nanosystems for imaging cancer chemodynamic therapy. Chem. 2020;6:2127–2129. doi: 10.1016/j.chempr.2020.08.013. [DOI] [Google Scholar]
- Yu M., Bovet N., Satterley C.J., Bengió S., Lovelock K.R.J., Milligan P.K., Jones R.G., Woodruff D.P., Dhanak V. True nature of an archetypal self-assembly system: mobile Au-thiolate species on Au(111) Phys. Rev. Lett. 2006;97:166102. doi: 10.1103/physrevlett.97.166102. [DOI] [PubMed] [Google Scholar]
- Zhang C., Xue X., Luo Q., Li Y., Yang K., Zhuang X., Jiang Y., Zhang J., Liu J., Zou G., Liang X.J. Self-assembled peptide nanofibers designed as biological enzymes for catalyzing ester hydrolysis. ACS Nano. 2014;8:11715–11723. doi: 10.1021/nn5051344. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data generated in this study have deposited in Mendeley Data (https://doi.org/10.17632/22vwv5yk6s.1). Key words: anti-lysozyme silverbody.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.