Abstract
Approximately 8% of the world population and 35 to 45% of East Asians are carriers of the hereditary disorder, aldehyde dehydrogenase 2 (ALDH2) deficiency. ALDH2 plays a central role in the liver to metabolize ethanol. With the common E487K variant, there is a deficiency of ALDH2 function; when ethanol is consumed, there is a systemic accumulation of acetaldehyde, an intermediate product in ethanol metabolism. In ALDH2 deficient individuals, ethanol consumption acutely causes the “Alcohol Flushing Syndrome” with facial flushing, tachycardia, nausea and headaches. With chronic alcohol consumption, ALDH2 deficiency is associated with a variety of disorders, including a remarkably high risk for aerodigestive tract cancers. Acetaldehyde is a known carcinogen. The epidemiologic data relating to the association of ALDH2 deficiency and cancer risk are striking: ALDH2 homozygotes that are moderate to heavy consumers of ethanol have a 7- to 12-fold increased risk for esophageal cancer, making ALDH2 deficiency the most common hereditary disorder associated with an increased cancer risk. In this review, we summarize the genetics and biochemistry of ALDH2, the epidemiology of cancer risk associated with ALDH2 deficiency, the metabolic consequences of ethanol consumption associated with ALDH2 deficiency and gene therapy strategies to correct ALDH2 deficiency and its associated cancer risk. With the goal of reducing the risk of aerodigestive tract cancers, in the context that ALDH2 is a hereditary disorder and ALDH2 functions primarily in the liver, ALDH2 deficiency is an ideal target for the application of adeno-associated virus-mediated liver directed gene therapy to prevent cancer.
Introduction
Aldehyde dehydrogenase 2 (ALDH2) deficiency, an autosomal recessive disorder involving ethanol metabolism, is one of the most common hereditary disorders, affecting 560 million people, representing 8% of the world population1, 2. ALDH2 deficiency is very common in the East Asian population, with an allele frequency of 35–45%3, 4. Ethanol metabolism involves two major NAD-dependent enzymes, alcohol dehydrogenase (ADH) and ALDH25. With ethanol consumption, ADH converts ethanol to acetaldehyde and then ALDH2 metabolizes acetaldehyde to acetate6 (Figure 1). Mutations in the ALDH2 gene (GenBank Nt_029419), most commonly E487K (referred to as the ALDH2*2 allele, the normal allele is ALDH2*1), result in a deficiency of ALDH2 function with subsequent systemic accumulation of acetaldehyde following ethanol consumption.
Figure 1.
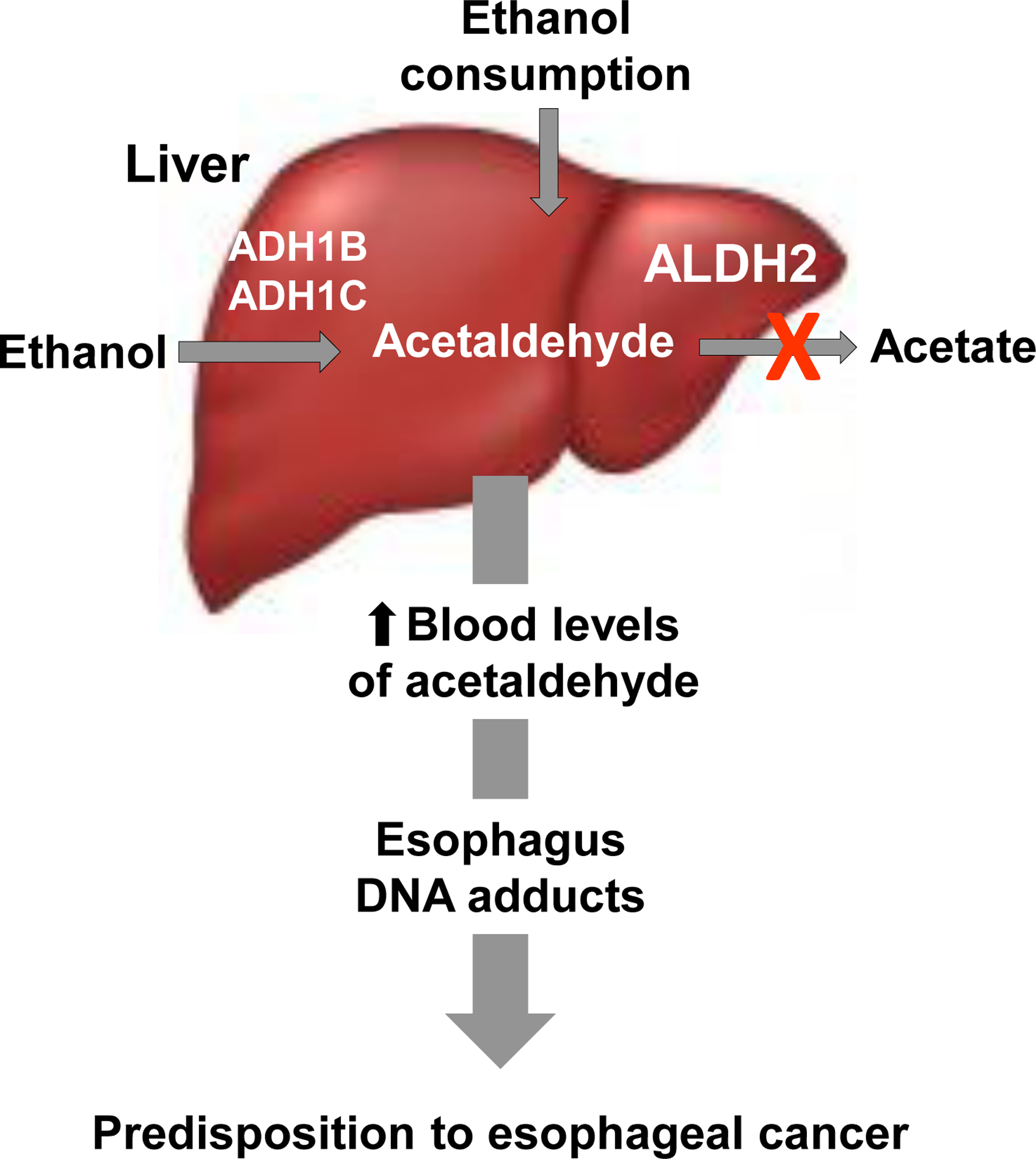
The high risk for esophageal cancer in ALDH2 deficiency results from a combination of the ALDH2 deficiency state and ethanol consumption. The liver is the major site of ethanol metabolism. Alcohol dehydrogenases (ADH1B, 1C) convert ethanol to acetaldehyde, which is metabolized by ALDH2 to acetate. With mutations in ALDH2, there is a deficiency in liver ALDH2 enzymatic activity and with ethanol intake acetaldehyde accumulates. Acutely, this causes the “Alcohol Flushing Syndrome,” but with chronic alcohol consumption, chronic elevated levels of acetaldehyde are associated with DNA damage and the accumulation of DNA adducts in the esophagus, predisposing to esophageal cancer.
The consequences of ethanol consumption in individuals with ALDH2 deficiency are significant. Acutely, ethanol consumption by ALDH2 deficient individuals causes the “Alcohol Flushing Syndrome,” with facial flushing, tachycardia, nausea and headaches1. With chronic ethanol ingestion, ALDH2 deficiency is associated with a variety of neurologic, endocrine, cardiovascular and dermatologic disorders, aberrant drug metabolism, and the subject of this review, a marked increased risk of upper aerodigestive tract cancer of the oral cavity pharynx, larynx and esophagus (reviewed in Chen et al2).
The increased risk of ALDH2 deficiency for upper digestive tract cancers, particularly esophageal cancer, is one of the highest risk factors for cancer. Of those who chronically drink alcohol, both ALDH2 homozygotes and heterozygotes are at risk of developing esophageal cancer1, 7 and the risk increases with the extent of drinking8, 9. Strikingly, ALDH2*2/*2 homozygotes that drink alcohol and also smoke cigarettes have a 50-fold risk of esophageal cancer10, 11. The focus of this review is the opportunity to use gene therapy to reduce the high risk for aerodigestive cancer associated with ALDH2 deficiency.
Aldehyde Dehydrogenase 2
The human ALDH group of enzymes includes 19 isozymes responsible for metabolizing alcohol byproducts. Of this group, there are two major ALDH isoforms, the cytosolic ALDH1 and mitochondrial ALDH26. Mitochondrial ALDH2 is the most efficient in metabolizing alcohol-derived acetaldehyde because of its extremely low Km (~0.2 μM) for acetaldehyde, 900-fold lower compared to cytosolic ALDH112.
The human ALDH2 gene located on chromosome 12q24 codes for a 517-amino acid polypeptide13. The ALDH2 protein is a 56 kDa tetramer with four identical subunits that each contain a catalytic domain, a coenzyme or NAD+ binding domain, and an oligomerization domain14. Together, the tetrameric structure contributes to normal functioning of enzymatic ALDH2. The most common ALDH2*2 mutation representing 33 to 51% of all ALDH2 mutations associated with ALDH2 deficiency, results in dysfunction of the ALDH2 tetramer15. With the ALDH2*2/*2 genotype, the E487K mutation disturbs hydrogen bond stabilization and modifies the αG helix and the arginine 475 loop. Disruptions in these interactions impair the structure of the NAD+ binding site and rearrange residues in the catalytic domain16–18. Because of these modifications, the Km for NAD+ increases by 200-fold such that ALDH2 is unable to combine with NAD+ or coenzymes and is limited in enzymatic activity16.
The amino acid configuration in the tetrameric structure (E487 or K487) affects functionality of the ALDH2 enzyme. Enzymatic activity is 100% when all 4 subunits have the E487 amino acid but decreases if the subunit sequence is modified. If the tetramer has a single subunit with the K487 variant (1/4) and three normal E487 remaining subunits, the tetramer has 48% reduced ALDH2 activity. The presence of the K487 variant in two subunits (2/4) and three subunits (3/4) reduces ALDH2 activity to 12 and 5%, respectively. When all four subunits (4/4) have the K487 variant, ALDH2 activity is 4%19. With ethanol consumption, the consequence of reduced ALDH2 activity is the systemic accumulation of acetaldehyde, resulting in the clinical manifestations of the deficiency state.
Prevalence
The ALDH2*2 allele has a high prevalence in East Asians20. Among these population groups, the ALDH2*2 allele is observed in 25 to 45% of Chinese, Korean and Taiwanese origin and ~50% of Japanese, while nearly absent in other population groups such as Caucasians and African Americans20, 21 (Table I). For example, the ALDH2*2 allele frequency is 0.0004 in Latino/Admixed American, 0.0003 in South Asian, 0.0002 in African/African American, 0.0001 in European (Finnish), 0.00002 in European (non-Finnish) and is absent in the Ashkenazi Jewish population3.
Table I.
Prevalence of ALDH2*2 Allele in Global Populations
| Population allele frequencies (AF)1 | |||
|---|---|---|---|
| Population | gnomAd | 1000 Genomes | NCBI ALFA |
| East Asian | 0.2554 | 0.174 | 0.244 |
| Latino/Admixed American | 0.0004094 | 0.003 | 0 |
| South Asian | 0.0002698 | 0 | 0.001 |
| African/African-American | 0.0002073 | 0.002/0 | 0.009/0 |
| Finnish | 0.00008095 | 0 | * |
| European | 0.00002404 | 0 | 0.0000822 |
| Ashkenazi Jewish | 0.000 | 0 | * |
Population allele frequencies for the rs671 single nucleotide polymorphism (ALDH2*2 E487K mutation) reported in National Center for Biotechnology (NCBI) databases including gnomAD, 1000 Genomes Project Phase 3, and NCBI Allele Frequency Aggregatory (ALFA).
Allele frequency not reported
Because other population groups also experience alcohol flushing syndrome in response to alcohol consumption, studies have looked at other variants in the ALDH2 gene that cause the ALDH2 deficiency state. Healthy donors from a blood bank in Mexico City, identified two novel ALDH2 variants (P92T and V304M). The Exome Aggregation Consortium (ExAC) database (formerly ExAC, now gnomAD database) identified five missense variants (P92T, V304M, I41V, T244M and R338W) present at a frequency greater than 0.5%. These variants are at low allele frequencies (~2.5–2.6%) among Latinos compared to the high frequency of the E487K classic ALDH2*2 variant among East Asians15. In all genome-wide association studies for alcohol use disorders, the ALDH2*2 polymorphism is observed at a noticeably greater significance compared to any other variant22.
Metabolic Consequences of the Major ALDH2 Deficiency Variant with Ethanol Consumption
ALDH2*2 carriers have a dysfunctional ALDH2 enzyme, which limits the capacity of the liver to eliminate acetaldehyde and results in the accumulation of acetaldehyde in the blood following alcohol consumption23, 24. Because of the reduced ALDH2 activity, ALDH2*2/*2 and ALDH2*1/*2 individuals have blood acetaldehyde levels that are markedly higher than ALDH2*1/*1 individuals following alcohol consumption25. In normal individuals, blood acetaldehyde levels range from 2 to 20 μM24. In ALDH2*2/*2 homozygotes who consume low to moderate levels of ethanol, blood acetaldehyde levels peak at 23 to 81 μM and in ALDH2*1/*2 heterozygotes, 8 to 26 μM26.
Acetaldehyde is highly reactive and can directly interact with DNA, introducing point mutations27, 28. Since acetaldehyde can directly bind to DNA, it can interfere with DNA replication, leading to double-stranded breaks24. Some of these chromosomal aberrations are repaired by proteins of the homologous recombination repair pathway including Rad51 and phosphorylated H2A histone family member X (γH2AX), which are both used clinically as biomarkers for the detection of DNA damage24.
Acetaldehyde forms acetaldehyde-DNA adducts that are mutagenic and potentially carcinogenic29. Several of the common mutagenic DNA adducts include N2-ethylidene-2’-deoxyguanosine (N2-ethylidene-dG; often quantified by its reduced, stable form as N2-ethyl-2’-deoxyguanosine [N2-Et-dG]30), N2-(2,6-dimethyl-1,3-dioxan-4-yl)-deoxyguanosine and α-methyl-γ-OH-propano-deoxyguanosine (Cr-PdG)31. Of these, N2-ethylidene-dG, the most abundant DNA adduct derived from exposure to acetaldehyde, is a common biomarker used to detect acetaldehyde-derived DNA damage32–34. N2-Et-dG is detectable in leukocytes of human heavy ethanol drinkers with the ALDH2*2 genotype35–37 38. Cr-PdG has the capacity to generate secondary lesions that arise as DNA-protein crosslinks or DNA inter-strand crosslinks27. Because these lesions impair DNA replication and promote cell death, Cr-PdG is considered genotoxic and highly mutagenic27, 39. Together, N2-Et-dG and Cr-PdG initiate replication errors and mutations in oncogenes or onco-suppressor genes and promote carcinogenesis27, 39.
In addition to DNA adducts, acetaldehyde protein adducts can be generated by the interaction of acetaldehyde with either lysine residues or the alpha amino group of N-terminal amino acids27. These adducts then alter the structure and function of the protein and, in the cases of enzymes, disrupt enzymatic activity27. For example, acetaldehyde forms adducts with the DNA repair mechanism enzyme, O6-methylguanine methyltransferase27. Because the formation of this protein adduct disables the DNA repair function of the enzyme, carcinogenesis may be induced27.
Several lines of evidence link ALDH2 deficiency, alcohol consumption, systemic elevation in acetaldehyde levels, DNA damage and the increased risk for esophageal cancer. Yukawa et al40 identified significantly higher levels of DNA adducts (N2-Et-dG, Cr-PdG, and N2-ethylidene-dG) in ALDH2-deficient Japanese alcoholics. In animal studies, Matsuda et al32 observed elevated levels of N2-ethylidene-dG in the liver of heterozygous (Aldh2+/−) and homozygous ALDH2 knockout (Aldh2−/−) mice consuming 20% ethanol for 5 weeks32, 34. Yukawa et al37 observed significantly elevated esophageal N2-ethylidene-dG levels in Aldh2−/− mice compared to control mice after intraperitoneal administration of ethanol, consistent with the concept that circulating ethanol-derived acetaldehyde levels contribute to induction of esophageal DNA damage. Amanuma et al34 studied whether DNA damage was induced in the esophagus of ALDH2 wildtype (Aldh2+/+) and Aldh2−/− mice following chronic consumption of 10% ethanol for 8 weeks. Elevated levels of γ-H2AX and N2-ethylidene-dG were observed in the esophagus of Aldh2−/− mice following ethanol consumption indicating a significant degree of DNA damage compared with Aldh2+/+ mice34. These findings of elevated N2-ethylidene-dG are similar to those observed in ALDH2-deficient Japanese alcoholics40.
Consistent with the data indicating that systemic acetaldehyde generated by chronic ethanol ingestion will induce cancer-related changes in the esophagus, elevated serum acetaldehyde levels and esophageal damage and adducts were observed in Aldh2−/− knockout and E487 knockin (Aldh2E487K+/+) mice chronically administered alcohol in the drinking water41–43. Aldh2−/− and Aldh2E487K+/+ mice had significantly higher serum acetaldehyde levels compared to wild type mice after 12 wk ethanol consumption. ALDH2 also metabolizes other aldehydes including malondialdehyde (MDA), a reactive aldehyde derived from oxidative stress with implications in DNA adduct formation and mutagenesis. MDA levels in the liver were significantly higher in Aldh2−/− and Aldh2E487K+/+ mice drinking ethanol for 12 weeks compared to wild type mice43.
Risk for Esophageal Cancer Associated in Humans with ALDH2 Deficiency and Ethanol Consumption
The combination of ALDH2 deficiency and ethanol consumption, the focus of this review, are major risk factors for the development of aerodigestive cancers1, 5, 7–11, 25, 28, 29, 34, 37, 44–76. In addition to aerodigestive cancers, there is extensive literature detailing the association of ALDH2 deficiency and ethanol consumption with an increased risk for many other disorders, including other cancers and non-cancer related diseases such as cardiovascular disease, diabetes, and neurodegenerative disease. For details, there are several reviews2, 10, 49, 53, 57–59, 65, 73, 75, 77, 78.
Upper aero digestive tract cancers, including esophageal, oral and laryngeal cancer, together constitute 3.5 to 4.0% of all malignancies71. Individuals homozygous for ALDH2*2 that drink alcohol have a 7- to 12-fold increased risk of upper aerodigestive tract cancers1, 7, 71. Consistent with the knowledge that the ALDH2 tetramer with two K487 subunits has only 12% normal activity19, the increased risk for esophageal cancer includes not only ALDH2*2 homozygotes, but also ALDH2*2 heterozygotes. High hazard ratios are found in ALDH2*2 heterozygote Japanese men who consumed more than 23 g of ethanol on occasion and drink more than 5 times per week74. Heterozygote heavy drinkers, or those that consumed more than 46 g of ethanol on occasion and drank 5 days or more per week also have a high odds ratio74. Association studies between risk for esophageal cancer and amount of alcohol consumption demonstrate a strong association by a significant odds ratio with the rs671 polymorphism in ALDH2*2 homozygotes or heterozygotes in both moderate drinkers and heavy drinkers44, 55, 56, 62, 68, 69, 76.
The risk for esophageal cancer in association with ALDH2 deficiency is dependent on the extent of alcohol consumption. ALDH2*2 homozygous individuals that avoid alcohol consumption are protected from the high risk associated with esophageal cancer61. The risk of ALDH2 deficient individuals developing esophageal cancer increases 3-fold from those who never consumed alcohol to moderate drinkers and 2-fold from moderate drinkers to heavy drinkers54, 70, 72. Meta-analysis of the association between ALDH2-deficient individuals and esophageal cancer in Japan and China60 reported an odds ratio of 1.28 in never drinkers, 3.12 in moderate drinkers, and 7.12 in heavy drinkers60. The ALDH2*1/*2 genotype is associated with a high risk of esophageal cancer in Taiwanese, Chinese, and Japanese moderate drinkers (odds ratio 4.74–6.21) and heavy drinkers (9.21–9.75). This risk is lower in regions of Mainland China moderate drinkers (1.98) or heavy drinkers (1.31)60. The highest risk of esophageal cancer is observed in ALDH2*2 homozygote cigarette smokers that consume alcohol who have an odds ratio 50, with a 25-year earlier onset of esophageal carcinoma10, 11.
Consequences of Excess ALDH2 Activity Drug Resistance
While ALDH2 deficiency and alcohol consumption is associated with increased aerodigestive cancer risk, excess ALDH2 activity is linked to drug resistance79–81. Various studies report a correlation between ALDH2 overexpression and multi-drug resistance against common cancer drugs such as antimetabolic agents and microtubule inhibitors79–81. In microtubule-inhibitor resistant head and neck cancer cells, upregulated ALDH2 expression is reduced by treatment with disulfiram (Antabuse), an ALDH2 inhibitor, and copper, a combination treatment used to override drug resistance, causing apoptosis of the cancer cells82. When ALDH2 is silenced with small interfering RNA (siRNA), cytotoxicity of anticancer drugs, such as Taxol, is enhanced and drug resistance is inhibited in lung and head and neck cancer cell lines82.
Prior Therapeutic Strategies Relevant to ALDH2
Attempts at therapies to enhance ALDH2 enzymatic activity have used enzymatic activators or substances to mitigate acetaldehyde elevation. Alda-1, an ALDH2 activator, enhances ALDH2 enzymatic activity and inhibits alcohol-induced oxidative stress83, 84. In human ALDH2*2 knockin mice, Alda-1 significantly increased liver ALDH2 levels and reduced esophageal DNA damage levels after ethanol consumption85. In rats, ALDH2*2 recombinant homotetramers treated with Alda-1 had an 11-fold increase in ALDH2 enzymatic activity71, 83. Although no ALDH2 activators have moved forward to clinical trials, together, these studies indicate that Alda-1-mediated activation of ALDH2 theoretically could serve as a therapeutic intervention in reducing the toxic effects of acetaldehyde83.
“Essential AD2” is a nutritional supplement reported to increase ALDH2 activity and reduce acetaldehyde levels, thereby reducing the flushing reaction experienced by ALDH2 deficient individuals following alcohol consumption. Subjects with ALDH2 deficiency receiving essential AD2 daily for 28 days had decreased blood acetaldehyde levels following alcohol use86.
ADH inhibitors impede the metabolism of alcohol to acetaldehyde. For example, 4-methylpyrazol (4-MP) competitively inhibits the oxidation of ethanol to acetaldehyde by ADH and therefore reduces the ethanol elimination rate. When ALDH2 deficient individuals and individuals with normal ALDH2 ingested 4-MP orally, after 2 hours, both the ALDH2 deficient and normals groups showed a reduction (38% and 46%, respectively) in ethanol elimination rate, with a rise in blood ethanol levels and a decrease in acetaldehyde levels. The flushing response was suppressed in both groups following treatment87.
In a mimic of the naturally occurring ALDH2 deficiency state, a variety of strategies have focused on reducing alcohol intake by inhibiting ALDH2 activity, including the FDA approved drug disulfiram (Antabuse)88, daidzein89, kudzu extract90, puerarin91, bitter herbs (gentian, tangerine peel)92 and decinol93. These remedies increase acetaldehyde and induce symptoms similar to the ALDH2 deficiency state after alcohol intake with the goal of reducing alcohol use. The effect of these compounds on cancer risk have not been evaluated.
Gene Therapy to Prevent Cancer Risk Associated with ALDH2 Deficiency
Since ALDH2 deficiency is a hereditary disorder that primarily manifests in the liver, it should be amenable to adeno-associated virus (AAV)-mediated gene therapy to augment the inactive mitochondrial ALDH2 enzyme with a normal ALDH2*1 allele. By doing so, ALDH2 enzymatic activity could be restored and acetaldehyde generated following ethanol consumption metabolized to non-toxic acetate, theoretically reducing the risk for esophageal cancer.
Matsumura et al43, 94 used two murine models to assess the feasibility of using gene therapy to prevent esophageal cancer associated with ethanol consumption in the ALDH2 deficiency state. The Aldh2 knockout mouse (Aldh2−/−) has undetectable ALDH2 protein or enzymatic activity41. The Aldh2 E487K knockin mouse (Aldh2E487K+/+, also called ALDH2*2 knockin) has the ALDH2*2 (E487K) lysine mutation found in ALDH2-deficient individuals inserted in the mouse Aldh2 gene, resulting in significantly reduced levels of ALDH2 enzymatic activity42.
To demonstrate that AAV-mediated gene therapy can correct the ALDH2 deficiency state and prevent the accumulation of acetaldehyde after ethanol consumption and the resultant acetaldehyde-induced esophageal DNA abnormalities, an AAVrh.10 serotype gene transfer vector coding for normal human ALDH2 was generated (Figure 2A). Two studies were carried out, one to assess the acute effects of ethanol94 and the other the chronic effects of ethanol43. Both studies used the Aldh2−/− and Aldh2E487K+/+ models. The following describes the results with the Aldh2E487K+/+ model; for details, see Matsumura et al43, 94. By delivering an AAVrh.10hALDH2 to murine hepatocytes, wild-type liver enzymatic activity was normalized, and serum acetaldehyde levels were significantly lower after acute alcohol consumption (Figure 2B, C). When Aldh2E487K+/+ mice were treated with an AAVrh.10hALDH2, there was a significant reduction in γH2AX positive cells in the esophageal epithelium and significant reduction in the number of N2-Et-dG adducts formed in the esophageal DNA when compared with AAVrh.10control-treated Aldh2E487K+/+ mice (Figure 3). DNA adducts and DNA damage are precursors to the formation of esophageal cancer, thus prevention of their formation by treatment with AAVrh.10hALDH2 suggests that restoration of ALDH2 expression and function in the liver could help mitigate the risk of esophageal cancer in ALDH2 deficient individuals.
Figure 2.
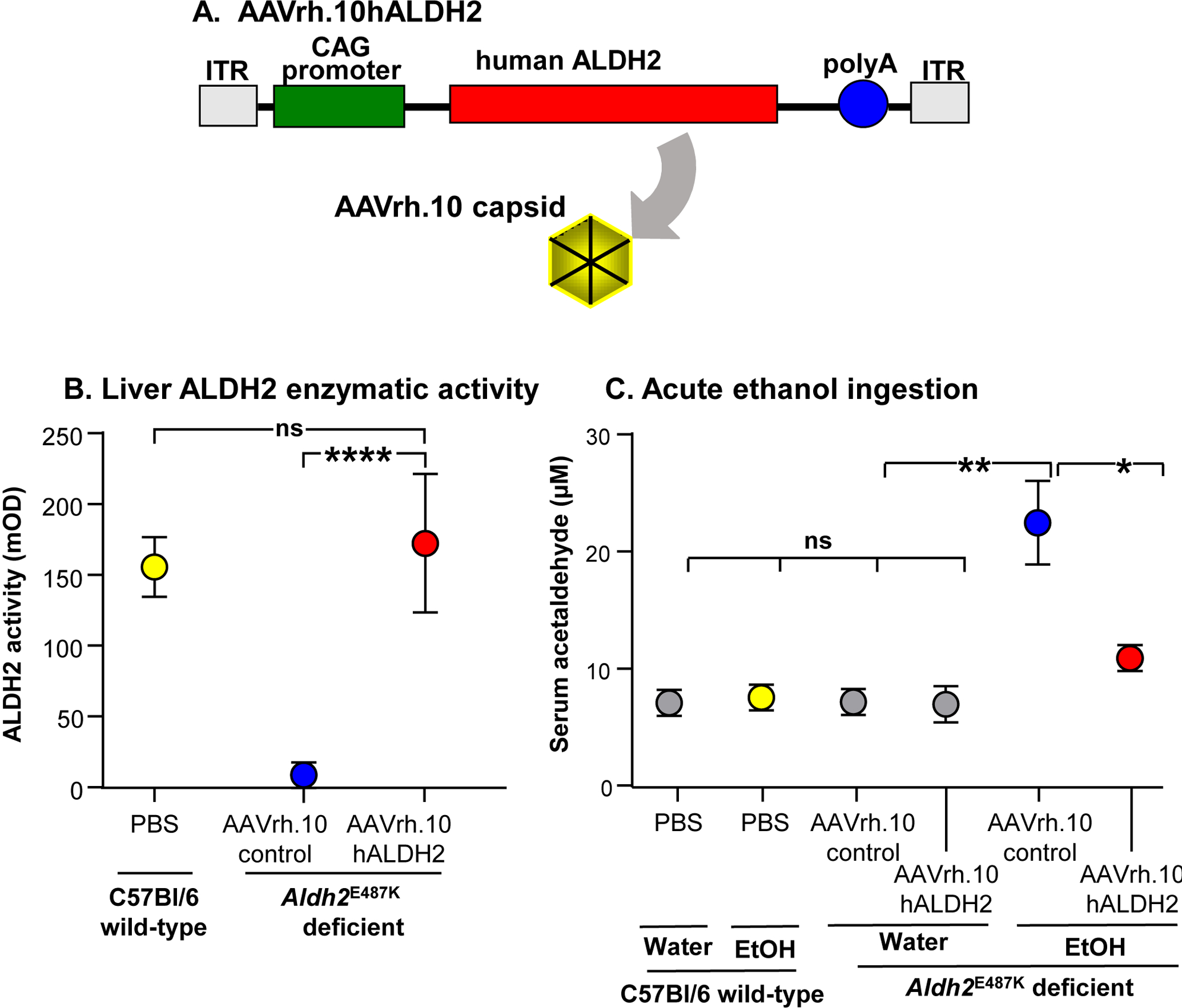
AAVrh.10hALDH2 correction of ALDH2 deficiency in the Aldh2E487K+/+ murine model. A. AAVrh.10hALDH2 construct. The expression cassette including the AAV2 inverted terminal repeats (ITR), CAG promoter, and the human ALDH2 coding sequence (hALDH2) packaged in the AAVrh.10 capsid. B. AAVrh.10hALDH2-mediated liver human ALDH2-driven enzymatic activity in Aldh2E487K+/+ mice. Four wk following intravenous administration, Aldh2E487K+/+ deficient mice had ALDH2 enzyme activity similar to that of wild-type mice (1011 genome copies). C. AAVrh.10hALDH2-mediated prevention of serum acetaldehyde accumulation in Aldh2E487K+/+ mice administered ethanol acutely. Four wk post-administration of AAVrh.10hALDH2, mice were given water or ethanol (4 g/kg body weight) by intragastric gavage and serum was collected 6 hr later for acetaldehyde quantitation. Values are presented as means ± SEM. *p<0.05, ** p<0.01, *** p<0.001, **** p<0.0001. Data from Matsumura et al94 with permission.
Figure 3.
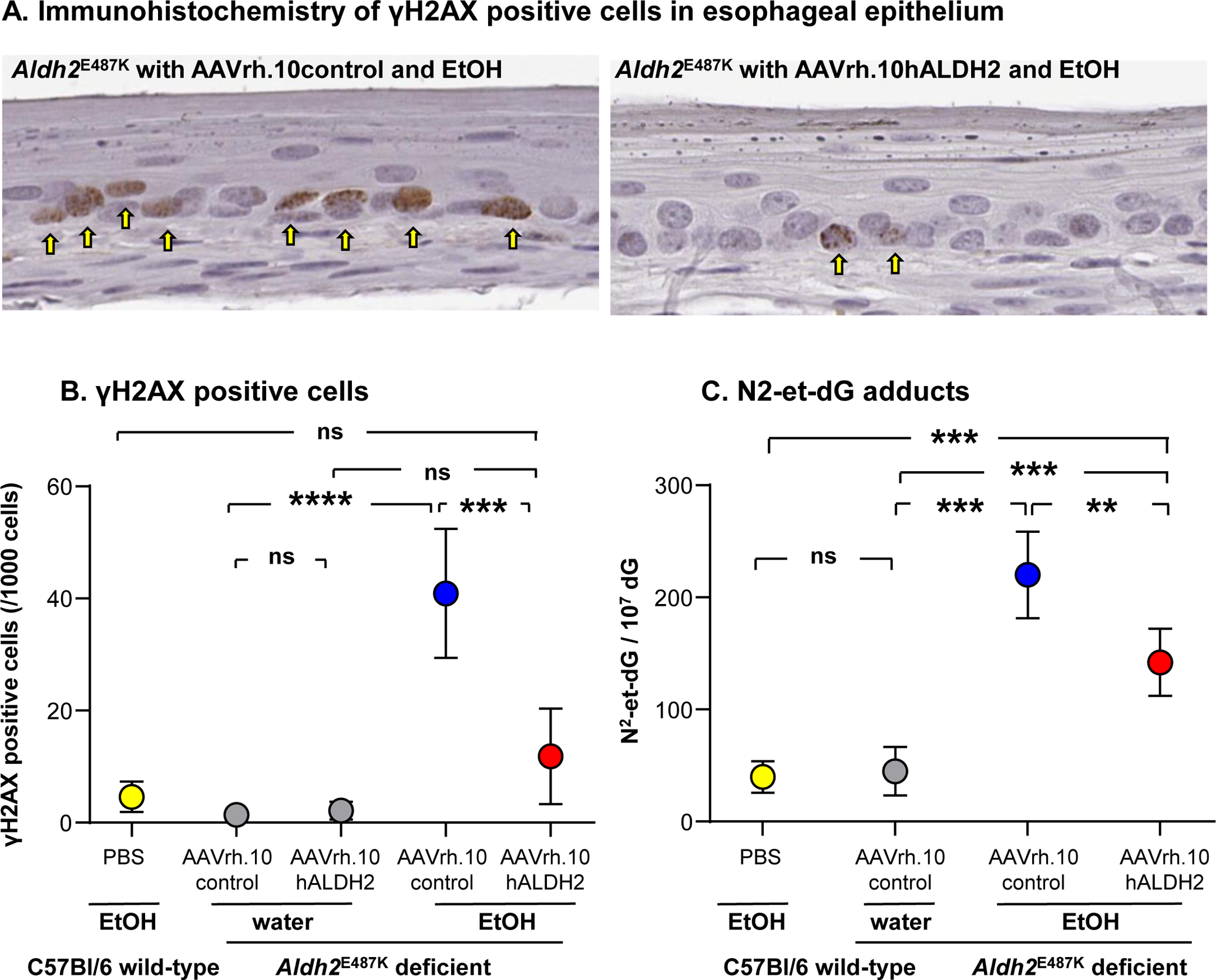
Effect of AAVrh.10hALDH2 therapy on DNA damage and adducts in the esophageal epithelium after 12 wk chronic ethanol exposure. Aldh2E487K+/+ mice were intravenously administered AAVrh.10hALDH2 or AAVrh.10control (1011 genome copies), and C57Bl/6 mice were administered PBS. Four wk after vector administration, mice were challenged with water or ethanol in water for 12 wk. A. Representative immunohistochemical staining of γH2AX positive cells in esophageal epithelium of Aldh2E487K+/+ mice. B. Quantitation of γH2AX positive cells in esophageal epithelium Aldh2E487K+/+ after chronic ethanol exposure. C. DNA was extracted from whole esophagus and N2-et-dG adducts were quantified by LC-MS/MS. Values are presented as means ± SEM. ** p<0.01, *** p<0.001, **** p<0.0001. Data from Matsumura et al43, with permission.
Alternative Gene Therapy Strategies
Other gene therapies have focused on ALDH2 with therapies designed to reduce chronic ethanol intake and binge drinking. Instead of correction of the ALDH2 deficiency state, these therapies were developed to inhibit the ALDH2 enzyme function to induce the common Alcohol Flushing Syndrome associated with ALDH2 deficiency, with the goal of deterring alcohol intake.
Sanchez et al95 delivered an ALDH2 short hairpin RNA (shRNA) packaged in AAV serotype 2 under the control of the U6 promoter in vitro to HEK-293T cells and HepG2 cells. The goal was to inhibit mitochondrial ALDH2 enzymatic activity in vitro using a self-complimentary AAV encoding a shRNA specific to the human ALDH2 transcript95. To observe the effect of the ALDH2 vector on acetaldehyde levels, human cell lines were exposed to ethanol, resulting in increasing acetaldehyde levels. If applied to humans, this inhibitory shRNA approach would induce the ALDH2 deficiency state. While this may discourage alcohol consumption because it would establish the Alcohol Flushing Syndrome, it theoretically would also result in an increased risk for the ALDH2 deficiency associated disorders, including esophageal cancer.
In another gene therapy, a lentivirus vector augmenting ALDH2 expression resulted in reduction of ethanol intake. Karahanian et al96 administered a lentiviral vector coding for rat ALDH2 directly into the ventral tegmental area of 3- to 4- month-old female rats. Following the single injection, ethanol intake was greatly reduced for a 45-day period96. A similar result in rats has been observed with the ALDH2 activator Alda-197.
Translation of AAV-based ALDH2 Gene Therapy to the Clinic
Based on the preclinical data that a single intravenous administration of AAVrh.10-based delivery of the human ALDH2 coding sequence to ALDH2 deficient mice prevents chronic alcohol ingestion from inducing systemic acetaldehyde accumulation and consequent accumulation of DNA adducts and damage43, it is relevant to consider how these preclinical observations could be translated to humans with ALDH2 deficiency. In this context, we propose a two-step clinical design.
First, we suggest that the phase I clinical study to demonstrate safety and biochemical efficacy of AAV-based gene therapy enroll ALDH2-deficient homozygotes with a history of daily ethanol consumption who smoke cigarettes and have esophageal biopsy evidence of pre-cancerous lesions (moderate to severe dysplasia)98, 99. This population has an extraordinarily high risk for development of esophageal cancer. In addition to the marked increased risk of esophageal cancer in ALDH2*2 homozygotes who continue to drink1, 11, 65, 100–106, the presence of moderate to severe esophageal dysplasia is associated with a 15.8-fold increased risk of developing esophageal cancer over 3.5 yr107. Prior to gene therapy, each subject will be assessed for the biochemical response to oral administration of a standard amount of ethanol, with subsequent assessment of serum levels of acetaldehyde and acetate, quantification of breath ethanol levels and facial flushing. The following day, the subject will be administered the gene therapy, with doses determined by the results of a toxicology study in experimental animals. Four wk after administration, the subject will be rechallenged with the same standard ethanol dose, with assessment for the same parameters as prior to therapy. We expect that pre-therapy, the ethanol challenge will result in elevated ethanol and acetaldehyde levels and low acetate levels. However, post-therapy, the ethanol challenge should lead to elevated ethanol and acetate levels, but low or no increase in acetaldehyde levels. Pre-therapy facial flushing will be increased with ethanol challenge, but post-therapy facial flushing should be minimal.
Second, successful demonstration of safety and biochemical efficacy in the Phase I trial will provide support for initiating a Phase II clinical trial focused on prevention of esophageal cancer. The specific design of the Phase II trial will depend on the results of the Phase I trial, but because the risk for esophageal cancer is so high, the optimal risk-benefit group would be cigarette smoking ALDH2*2 homozygotes that continue to drink alcohol and had esophageal biopsy evidence of moderate to severe dysplasia. Compared to a placebo group, the primary outcome would be reduction in the development of esophageal cancer over the study period.
Conclusion
In this review, we have outlined the risk of developing esophageal cancer in those with ALDH2 deficiency that consume alcohol. Extensive epidemiologic studies have linked alcohol consumption in ALDH2-deficient individuals with an increased risk of esophageal cancer. The risk of cancer is largely due to the accumulation of acetaldehyde in ALDH2 deficiency because these individuals are unable to metabolize acetaldehyde to non-toxic acetate. As systemic acetaldehyde accumulates following alcohol consumption, the aldehyde acts as a genetic modifier by binding DNA and disrupting cellular repair mechanisms. Acetaldehyde-DNA and acetaldehyde-protein adducts form that are potentially carcinogenic. In the context that ALDH2 deficiency is a hereditary disorder and that the liver is the dominant site of ethanol metabolism, AAV gene therapy to augment normal ALDH2 enzymatic function in the liver provides an opportunity to use gene therapy to prevent the risk of esophageal cancer in ALDH2 deficient individuals who continue to consume alcohol.
Acknowledgments.
We thank N. Mohamed for editorial assistance. These studies were supported, in part, by NIH R41AA027739 and R41AA028465.
Footnotes
Conflict of interest: RGC is consultant and holds equity in LEXEO Therapeutics; LEXEO has an option with Weill Cornell Medical College to license the ALDH2 gene therapy program. RGC is also a co-inventor on a patent application related to this topic (US application number 16/321,023).
References
- 1.Brooks PJ, Enoch MA, Goldman D, Li TK, Yokoyama A. The alcohol flushing response: an unrecognized risk factor for esophageal cancer from alcohol consumption. PLoS Med 2009; 6(3): e50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen CH, Ferreira JC, Gross ER, Mochly-Rosen D. Targeting aldehyde dehydrogenase 2: new therapeutic opportunities. Physiol Rev 2014; 94(1): 1–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alföldi J, Wang Q et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 2020; 581(7809): 434–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Phan L, Jin Y, Zhang H, Qiang W, Shekhtman E, Shao D et al. ALFA: Allele Frequency Aggregator. In. www.ncbi.nlm.nih.gov/snp/docs/gsr/alfa/: National Center for Biotechnology Information, U.S. National Library of Medicine,, 2020. [Google Scholar]
- 5.Crous-Bou M, Rennert G, Cuadras D, Salazar R, Cordero D, Saltz Rennert H et al. Polymorphisms in alcohol metabolism genes ADH1B and ALDH2, alcohol consumption and colorectal cancer. PLoS One 2013; 8(11): e80158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Klyosov AA, Rashkovetsky LG, Tahir MK, Keung WM. Possible role of liver cytosolic and mitochondrial aldehyde dehydrogenases in acetaldehyde metabolism. Biochemistry 1996; 35(14): 4445–56. [DOI] [PubMed] [Google Scholar]
- 7.Yokoyama A, Muramatsu T, Ohmori T, Higuchi S, Hayashida M, Ishii H. Esophageal cancer and aldehyde dehydrogenase-2 genotypes in Japanese males. Cancer Epidemiol Biomarkers Prev 1996; 5(2): 99–102. [PubMed] [Google Scholar]
- 8.Hiraki A, Matsuo K, Wakai K, Suzuki T, Hasegawa Y, Tajima K. Gene-gene and gene-environment interactions between alcohol drinking habit and polymorphisms in alcohol-metabolizing enzyme genes and the risk of head and neck cancer in Japan. Cancer Sci 2007; 98(7): 1087–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee CH, Lee JM, Wu DC, Goan YG, Chou SH, Wu IC et al. Carcinogenetic impact of ADH1B and ALDH2 genes on squamous cell carcinoma risk of the esophagus with regard to the consumption of alcohol, tobacco and betel quid. Int J Cancer 2008; 122(6): 1347–56. [DOI] [PubMed] [Google Scholar]
- 10.Morita M, Kumashiro R, Kubo N, Nakashima Y, Yoshida R, Yoshinaga K et al. Alcohol drinking, cigarette smoking, and the development of squamous cell carcinoma of the esophagus: epidemiology, clinical findings, and prevention. Int J Clin Oncol 2010; 15(2): 126–34. [DOI] [PubMed] [Google Scholar]
- 11.Lee CH, Wu DC, Wu IC, Goan YG, Lee JM, Chou SH et al. Genetic modulation of ADH1B and ALDH2 polymorphisms with regard to alcohol and tobacco consumption for younger aged esophageal squamous cell carcinoma diagnosis. Int J Cancer 2009; 125(5): 1134–42. [DOI] [PubMed] [Google Scholar]
- 12.Eriksson CJ, Sippel HW. The distribution and metabolism of acetaldehyde in rats during ethanol oxidation-I. The distribution of acetaldehyde in liver, brain, blood and breath. Biochem Pharmacol 1977; 26(3): 241–7. [DOI] [PubMed] [Google Scholar]
- 13.Raghunathan L, Hsu LC, Klisak I, Sparkes RS, Yoshida A, Mohandas T. Regional localization of the human genes for aldehyde dehydrogenase-1 and aldehyde dehydrogenase-2. Genomics 1988; 2(3): 267–9. [DOI] [PubMed] [Google Scholar]
- 14.Ehrig T, Bosron WF, Li TK. Alcohol and aldehyde dehydrogenase. Alcohol Alcohol 1990; 25(2–3): 105–16. [DOI] [PubMed] [Google Scholar]
- 15.Chen CH, Ferreira JCB, Joshi AU, Stevens MC, Li SJ, Hsu JH et al. Novel and prevalent non-East Asian ALDH2 variants; Implications for global susceptibility to aldehydes’ toxicity. EBioMedicine 2020; 55: 102753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Larson HN, Weiner H, Hurley TD. Disruption of the coenzyme binding site and dimer interface revealed in the crystal structure of mitochondrial aldehyde dehydrogenase “Asian” variant. J Biol Chem 2005; 280(34): 30550–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Larson HN, Zhou J, Chen Z, Stamler JS, Weiner H, Hurley TD. Structural and functional consequences of coenzyme binding to the inactive asian variant of mitochondrial aldehyde dehydrogenase: roles of residues 475 and 487. J Biol Chem 2007; 282(17): 12940–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matsumoto S, Araki M, Isaka Y, Ono F, Hirohashi K, Ohashi S et al. E487K-Induced Disorder in Functionally Relevant Dynamics of Mitochondrial Aldehyde Dehydrogenase 2. Biophysical journal 2020; 119(3): 628–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weiner H, Wei B, Zhou J. Subunit communication in tetrameric class 2 human liver aldehyde dehydrogenase as the basis for half-of-the-site reactivity and the dominance of the oriental subunit in a heterotetramer. Chem Biol Interact 2001; 130–132(1–3): 47–56. [DOI] [PubMed] [Google Scholar]
- 20.Dandré F, Cassaigne A, Iron A. The frequency of the mitochondrial aldehyde dehydrogenase I2 (atypical) allele in Caucasian, Oriental and African black populations determined by the restriction profile of PCR-amplified DNA. Molecular and cellular probes 1995; 9(3): 189–93. [DOI] [PubMed] [Google Scholar]
- 21.Goedde HW, Agarwal DP, Fritze G, Meier-Tackmann D, Singh S, Beckmann G et al. Distribution of ADH2 and ALDH2 genotypes in different populations. Human genetics 1992; 88(3): 344–6. [DOI] [PubMed] [Google Scholar]
- 22.Kranzler HR, Zhou H, Kember RL, Vickers Smith R, Justice AC, Damrauer S et al. Genome-wide association study of alcohol consumption and use disorder in 274,424 individuals from multiple populations. Nature communications 2019; 10(1): 1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maejima R, Iijima K, Kaihovaara P, Hatta W, Koike T, Imatani A et al. Effects of ALDH2 genotype, PPI treatment and L-cysteine on carcinogenic acetaldehyde in gastric juice and saliva after intragastric alcohol administration. PLoS One 2015; 10(4): e0120397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kotova N, Vare D, Schultz N, Gradecka Meesters D, Stepnik M, Grawe J et al. Genotoxicity of alcohol is linked to DNA replication-associated damage and homologous recombination repair. Carcinogenesis 2013; 34(2): 325–30. [DOI] [PubMed] [Google Scholar]
- 25.Yang SJ, Wang HY, Li XQ, Du HZ, Zheng CJ, Chen HG et al. Genetic polymorphisms of ADH2 and ALDH2 association with esophageal cancer risk in southwest China. World J Gastroenterol 2007; 13(43): 5760–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Enomoto N, Takase S, Yasuhara M, Takada A. Acetaldehyde metabolism in different aldehyde dehydrogenase-2 genotypes. Alcohol Clin Exp Res 1991; 15(1): 141–4. [DOI] [PubMed] [Google Scholar]
- 27.Setshedi M, Wands JR, Monte SM. Acetaldehyde adducts in alcoholic liver disease. Oxidative medicine and cellular longevity 2010; 3(3): 178–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Seitz HK, Stickel F. Acetaldehyde as an underestimated risk factor for cancer development: role of genetics in ethanol metabolism. Genes Nutr 2010; 5(2): 121–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu HS, Oyama T, Matsuda T, Isse T, Yamaguchi T, Tanaka M et al. The effect of ethanol on the formation of N2-ethylidene-dG adducts in mice: implications for alcohol-related carcinogenicity of the oral cavity and esophagus. Biomarkers : biochemical indicators of exposure, response, and susceptibility to chemicals 2012; 17(3): 269–74. [DOI] [PubMed] [Google Scholar]
- 30.Wang M, Yu N, Chen L, Villalta PW, Hochalter JB, Hecht SS. Identification of an acetaldehyde adduct in human liver DNA and quantitation as N2-ethyldeoxyguanosine. Chem Res Toxicol 2006; 19(2): 319–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang M, McIntee EJ, Cheng G, Shi Y, Villalta PW, Hecht SS. Identification of DNA adducts of acetaldehyde. Chem Res Toxicol 2000; 13(11): 1149–57. [DOI] [PubMed] [Google Scholar]
- 32.Matsuda T, Matsumoto A, Uchida M, Kanaly RA, Misaki K, Shibutani S et al. Increased formation of hepatic N2-ethylidene-2’-deoxyguanosine DNA adducts in aldehyde dehydrogenase 2-knockout mice treated with ethanol. Carcinogenesis 2007; 28(11): 2363–6. [DOI] [PubMed] [Google Scholar]
- 33.Nagayoshi H, Matsumoto A, Nishi R, Kawamoto T, Ichiba M, Matsuda T. Increased formation of gastric N(2)-ethylidene-2’-deoxyguanosine DNA adducts in aldehyde dehydrogenase-2 knockout mice treated with ethanol. Mutat Res 2009; 673(1): 74–7. [DOI] [PubMed] [Google Scholar]
- 34.Amanuma Y, Ohashi S, Itatani Y, Tsurumaki M, Matsuda S, Kikuchi O et al. Protective role of ALDH2 against acetaldehyde-derived DNA damage in oesophageal squamous epithelium. Scientific reports 2015; 5: 14142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Matsuda T, Yabushita H, Kanaly RA, Shibutani S, Yokoyama A. Increased DNA damage in ALDH2-deficient alcoholics. Chem Res Toxicol 2006; 19(10): 1374–8. [DOI] [PubMed] [Google Scholar]
- 36.Balbo S, Hashibe M, Gundy S, Brennan P, Canova C, Simonato L et al. N2-ethyldeoxyguanosine as a potential biomarker for assessing effects of alcohol consumption on DNA. Cancer Epidemiol Biomarkers Prev 2008; 17(11): 3026–32. [DOI] [PubMed] [Google Scholar]
- 37.Yukawa Y, Ohashi S, Amanuma Y, Nakai Y, Tsurumaki M, Kikuchi O et al. Impairment of aldehyde dehydrogenase 2 increases accumulation of acetaldehyde-derived DNA damage in the esophagus after ethanol ingestion. Am J Cancer Res 2014; 4(3): 279–84. [PMC free article] [PubMed] [Google Scholar]
- 38.Fang JL, Vaca CE. Detection of DNA adducts of acetaldehyde in peripheral white blood cells of alcohol abusers. Carcinogenesis 1997; 18(4): 627–32. [DOI] [PubMed] [Google Scholar]
- 39.Brooks PJ, Theruvathu JA. DNA adducts from acetaldehyde: implications for alcohol-related carcinogenesis. Alcohol 2005; 35(3): 187–93. [DOI] [PubMed] [Google Scholar]
- 40.Yukawa Y, Muto M, Hori K, Nagayoshi H, Yokoyama A, Chiba T et al. Combination of ADH1B*2/ALDH2*2 polymorphisms alters acetaldehyde-derived DNA damage in the blood of Japanese alcoholics. Cancer Sci 2012; 103(9): 1651–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kitagawa K, Kawamoto T, Kunugita N, Tsukiyama T, Okamoto K, Yoshida A et al. Aldehyde dehydrogenase (ALDH) 2 associates with oxidation of methoxyacetaldehyde; in vitro analysis with liver subcellular fraction derived from human and Aldh2 gene targeting mouse. FEBS Lett 2000; 476(3): 306–11. [DOI] [PubMed] [Google Scholar]
- 42.Zambelli VO, Gross ER, Chen CH, Gutierrez VP, Cury Y, Mochly-Rosen D. Aldehyde dehydrogenase-2 regulates nociception in rodent models of acute inflammatory pain. Sci Transl Med 2014; 6(251): 251ra118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Matsumura Y, Li N, Alwaseem H, Pagovich OE, Crystal RG, Greenblatt MB et al. Systemic Adeno-Associated Virus-Mediated Gene Therapy Prevents the Multiorgan Disorders Associated with Aldehyde Dehydrogenase 2 Deficiency and Chronic Ethanol Ingestion. Hum Gene Ther 2020; 31(3–4): 163–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Katoh T, Kaneko S, Kohshi K, Munaka M, Kitagawa K, Kunugita N et al. Genetic polymorphisms of tobacco- and alcohol-related metabolizing enzymes and oral cavity cancer. Int J Cancer 1999; 83(5): 606–9. [DOI] [PubMed] [Google Scholar]
- 45.Nomura T, Noma H, Shibahara T, Yokoyama A, Muramatusu T, Ohmori T. Aldehyde dehydrogenase 2 and glutathione S-transferase M 1 polymorphisms in relation to the risk for oral cancer in Japanese drinkers. Oral oncology 2000; 36(1): 42–6. [DOI] [PubMed] [Google Scholar]
- 46.Matsuo K, Hamajima N, Shinoda M, Hatooka S, Inoue M, Takezaki T et al. Gene-environment interaction between an aldehyde dehydrogenase-2 (ALDH2) polymorphism and alcohol consumption for the risk of esophageal cancer. Carcinogenesis 2001; 22(6): 913–6. [DOI] [PubMed] [Google Scholar]
- 47.Boonyaphiphat P, Thongsuksai P, Sriplung H, Puttawibul P. Lifestyle habits and genetic susceptibility and the risk of esophageal cancer in the Thai population. Cancer letters 2002; 186(2): 193–9. [DOI] [PubMed] [Google Scholar]
- 48.Yokoyama A, Kato H, Yokoyama T, Tsujinaka T, Muto M, Omori T et al. Genetic polymorphisms of alcohol and aldehyde dehydrogenases and glutathione S-transferase M1 and drinking, smoking, and diet in Japanese men with esophageal squamous cell carcinoma. Carcinogenesis 2002; 23(11): 1851–9. [DOI] [PubMed] [Google Scholar]
- 49.Enzinger PC, Mayer RJ. Esophageal cancer. N Engl J Med 2003; 349(23): 2241–52. [DOI] [PubMed] [Google Scholar]
- 50.Yang CX, Matsuo K, Ito H, Hirose K, Wakai K, Saito T et al. Esophageal cancer risk by ALDH2 and ADH2 polymorphisms and alcohol consumption: exploration of gene-environment and gene-gene interactions. Asian Pacific journal of cancer prevention : APJCP 2005; 6(3): 256–62. [PubMed] [Google Scholar]
- 51.Chen YJ, Chen C, Wu DC, Lee CH, Wu CI, Lee JM et al. Interactive effects of lifetime alcohol consumption and alcohol and aldehyde dehydrogenase polymorphisms on esophageal cancer risks. Int J Cancer 2006; 119(12): 2827–31. [DOI] [PubMed] [Google Scholar]
- 52.Guo YM, Wang Q, Liu YZ, Chen HM, Qi Z, Guo QH. Genetic polymorphisms in cytochrome P4502E1, alcohol and aldehyde dehydrogenases and the risk of esophageal squamous cell carcinoma in Gansu Chinese males. World J Gastroenterol 2008; 14(9): 1444–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hiyama T, Yoshihara M, Tanaka S, Chayama K. Genetic polymorphisms and head and neck cancer risk (Review). International journal of oncology 2008; 32(5): 945–73. [PubMed] [Google Scholar]
- 54.Boccia S, Hashibe M, Gallì P, De Feo E, Asakage T, Hashimoto T et al. Aldehyde dehydrogenase 2 and head and neck cancer: a meta-analysis implementing a Mendelian randomization approach. Cancer Epidemiol Biomarkers Prev 2009; 18(1): 248–54. [DOI] [PubMed] [Google Scholar]
- 55.Cui R, Kamatani Y, Takahashi A, Usami M, Hosono N, Kawaguchi T et al. Functional variants in ADH1B and ALDH2 coupled with alcohol and smoking synergistically enhance esophageal cancer risk. Gastroenterology 2009; 137(5): 1768–75. [DOI] [PubMed] [Google Scholar]
- 56.Oze I, Matsuo K, Hosono S, Ito H, Kawase T, Watanabe M et al. Comparison between self-reported facial flushing after alcohol consumption and ALDH2 Glu504Lys polymorphism for risk of upper aerodigestive tract cancer in a Japanese population. Cancer Sci 2010; 101(8): 1875–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Seitz HK, Becker P. Alcohol metabolism and cancer risk. Alcohol Res Health 2007; 30(1): 38–41, 44–7. [PMC free article] [PubMed] [Google Scholar]
- 58.Seitz HK, Meier P. The role of acetaldehyde in upper digestive tract cancer in alcoholics. Transl Res 2007; 149(6): 293–7. [DOI] [PubMed] [Google Scholar]
- 59.Seitz HK, Stickel F. Molecular mechanisms of alcohol-mediated carcinogenesis. Nature reviews. Cancer 2007; 7(8): 599–612. [DOI] [PubMed] [Google Scholar]
- 60.Yang SJ, Yokoyama A, Yokoyama T, Huang YC, Wu SY, Shao Y et al. Relationship between genetic polymorphisms of ALDH2 and ADH1B and esophageal cancer risk: a meta-analysis. World J Gastroenterol 2010; 16(33): 4210–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fang P, Jiao S, Zhang X, Liu Z, Wang H, Gao Y et al. Meta-analysis of ALDH2 variants and esophageal cancer in Asians. Asian Pacific journal of cancer prevention : APJCP 2011; 12(10): 2623–7. [PubMed] [Google Scholar]
- 62.Ji YB, Tae K, Ahn TH, Lee SH, Kim KR, Park CW et al. ADH1B and ALDH2 polymorphisms and their associations with increased risk of squamous cell carcinoma of the head and neck in the Korean population. Oral oncology 2011; 47(7): 583–7. [DOI] [PubMed] [Google Scholar]
- 63.Li QD, Li H, Wang MS, Diao TY, Zhou ZY, Fang QX et al. Multi-susceptibility genes associated with the risk of the development stages of esophageal squamous cell cancer in Feicheng County. BMC gastroenterology 2011; 11: 74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang Y, Ji R, Wei X, Gu L, Chen L, Rong Y et al. Esophageal squamous cell carcinoma and ALDH2 and ADH1B polymorphisms in Chinese females. Asian Pacific journal of cancer prevention : APJCP 2011; 12(8): 2065–8. [PubMed] [Google Scholar]
- 65.Cadoni G, Boccia S, Petrelli L, Di Giannantonio P, Arzani D, Giorgio A et al. A review of genetic epidemiology of head and neck cancer related to polymorphisms in metabolic genes, cell cycle control and alcohol metabolism. Acta otorhinolaryngologica Italica : organo ufficiale della Societa italiana di otorinolaringologia e chirurgia cervico-facciale 2012; 32(1): 1–11. [PMC free article] [PubMed] [Google Scholar]
- 66.Gu H, Gong D, Ding G, Zhang W, Liu C, Jiang P et al. A variant allele of ADH1B and ALDH2, is associated with the risk of esophageal cancer. Exp Ther Med 2012; 4(1): 135–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Matsuo K, Rossi M, Negri E, Oze I, Hosono S, Ito H et al. Folate, alcohol, and aldehyde dehydrogenase 2 polymorphism and the risk of oral and pharyngeal cancer in Japanese. European journal of cancer prevention : the official journal of the European Cancer Prevention Organisation (ECP) 2012; 21(2): 193–8. [DOI] [PubMed] [Google Scholar]
- 68.Gao Y, He Y, Xu J, Xu L, Du J, Zhu C et al. Genetic variants at 4q21, 4q23 and 12q24 are associated with esophageal squamous cell carcinoma risk in a Chinese population. Human genetics 2013; 132(6): 649–56. [DOI] [PubMed] [Google Scholar]
- 69.Wu M, Chang SC, Kampman E, Yang J, Wang XS, Gu XP et al. Single nucleotide polymorphisms of ADH1B, ADH1C and ALDH2 genes and esophageal cancer: a population-based case-control study in China. Int J Cancer 2013; 132(8): 1868–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tsai ST, Wong TY, Ou CY, Fang SY, Chen KC, Hsiao JR et al. The interplay between alcohol consumption, oral hygiene, ALDH2 and ADH1B in the risk of head and neck cancer. Int J Cancer 2014; 135(10): 2424–36. [DOI] [PubMed] [Google Scholar]
- 71.Li R, Zhao Z, Sun M, Luo J, Xiao Y. ALDH2 gene polymorphism in different types of cancers and its clinical significance. Life sciences 2016; 147: 59–66. [DOI] [PubMed] [Google Scholar]
- 72.Lachenmeier DW, Salaspuro M. ALDH2-deficiency as genetic epidemiologic and biochemical model for the carcinogenicity of acetaldehyde. Regulatory toxicology and pharmacology : RTP 2017; 86: 128–136. [DOI] [PubMed] [Google Scholar]
- 73.Matejcic M, Gunter MJ, Ferrari P. Alcohol metabolism and oesophageal cancer: a systematic review of the evidence. Carcinogenesis 2017; 38(9): 859–872. [DOI] [PubMed] [Google Scholar]
- 74.Iwasaki M, Budhathoki S, Yamaji T, Tanaka-Mizuno S, Kuchiba A, Sawada N et al. Inclusion of a gene-environment interaction between alcohol consumption and the aldehyde dehydrogenase 2 genotype in a risk prediction model for upper aerodigestive tract cancer in Japanese men. Cancer Sci 2020; 111(10): 3835–3844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Salaspuro M Local Acetaldehyde: Its Key Role in Alcohol-Related Oropharyngeal Cancer. Visceral medicine 2020; 36(3): 167–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Choi CK, Yang J, Kweon SS, Cho SH, Kim HY, Myung E et al. Association between ALDH2 polymorphism and esophageal cancer risk in South Koreans: a case-control study. BMC cancer 2021; 21(1): 254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chang JS, Hsiao JR, Chen CH. ALDH2 polymorphism and alcohol-related cancers in Asians: a public health perspective. J Biomed Sci 2017; 24(1): 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Joo Kang S, Shin CM, Sung J, Kim N. Association Between ALDH2 Polymorphism and Gastric Cancer Risk in Terms of Alcohol Consumption: A Meta-Analysis. Alcohol Clin Exp Res 2021; 45(1): 6–14. [DOI] [PubMed] [Google Scholar]
- 79.Zhang H, Fu L. The role of ALDH2 in tumorigenesis and tumor progression: Targeting ALDH2 as a potential cancer treatment. Acta pharmaceutica Sinica. B 2021; 11(6): 1400–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Moreb JS, Ucar D, Han S, Amory JK, Goldstein AS, Ostmark B et al. The enzymatic activity of human aldehyde dehydrogenases 1A2 and 2 (ALDH1A2 and ALDH2) is detected by Aldefluor, inhibited by diethylaminobenzaldehyde and has significant effects on cell proliferation and drug resistance. Chem Biol Interact 2012; 195(1): 52–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gao YH, Wu ZX, Xie LQ, Li CX, Mao YQ, Duan YT et al. VHL deficiency augments anthracycline sensitivity of clear cell renal cell carcinomas by down-regulating ALDH2. Nature communications 2017; 8: 15337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang NN, Wang LH, Li Y, Fu SY, Xue X, Jia LN et al. Targeting ALDH2 with disulfiram/copper reverses the resistance of cancer cells to microtubule inhibitors. Experimental cell research 2018; 362(1): 72–82. [DOI] [PubMed] [Google Scholar]
- 83.Chen C-H, Budas GR, Churchill EN, Disatnik M-H, Hurley TD, Mochly-Rosen D. Activation of aldehyde dehydrogenase-2 reduces ischemic damage to the heart. Science 2008; 321(5895): 1493–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhang T, Zhao Q, Ye F, Huang CY, Chen WM, Huang WQ. Alda-1, an ALDH2 activator, protects against hepatic ischemia/reperfusion injury in rats via inhibition of oxidative stress. Free radical research 2018; 52(6): 629–638. [DOI] [PubMed] [Google Scholar]
- 85.Hirohashi K, Ohashi S, Amanuma Y, Nakai Y, Ida T, Baba K et al. Protective effects of Alda-1, an ALDH2 activator, on alcohol-derived DNA damage in the esophagus of human ALDH2*2 (Glu504Lys) knock-in mice. Carcinogenesis 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Fujioka K, Gordon S. Effects of “Essential AD2” Supplement on Blood Acetaldehyde Levels in Individuals Who Have Aldehyde Dehydrogenase (ALDH2) Deficiency. Am J Ther 2018. [DOI] [PubMed] [Google Scholar]
- 87.Vakevainen S, Tillonen J, Salaspuro M. 4-Methylpyrazole decreases salivary acetaldehyde levels in aldh2-deficient subjects but not in subjects with normal aldh2. Alcohol Clin Exp Res 2001; 25(6): 829–34. [PubMed] [Google Scholar]
- 88.Lipsky JJ, Shen ML, Naylor S. In vivo inhibition of aldehyde dehydrogenase by disulfiram. Chem Biol Interact 2001; 130–132(1–3): 93–102. [DOI] [PubMed] [Google Scholar]
- 89.Keung WM, Vallee BL. Daidzin: a potent, selective inhibitor of human mitochondrial aldehyde dehydrogenase. Proc Natl Acad Sci U S A 1993; 90(4): 1247–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lukas SE, Penetar D, Su Z, Geaghan T, Maywalt M, Tracy M et al. A standardized kudzu extract (NPI-031) reduces alcohol consumption in nontreatment-seeking male heavy drinkers. Psychopharmacology 2013; 226(1): 65–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Penetar DM, Toto LH, Farmer SL, Lee DY, Ma Z, Liu Y et al. The isoflavone puerarin reduces alcohol intake in heavy drinkers: a pilot study. Drug and alcohol dependence 2012; 126(1–2): 251–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Rajput HJA, Medicine I. Treatment of Chronic Alcoholism: An Integrated Approach. 2014; 3: 1–4. [Google Scholar]
- 93.Kushner S, Han D, Oscar-Berman M, William Downs B, Madigan MA, Giordano J et al. Declinol, a Complex Containing Kudzu, Bitter Herbs (Gentian, Tangerine Peel) and Bupleurum, Significantly Reduced Alcohol Use Disorders Identification Test (AUDIT) Scores in Moderate to Heavy Drinkers: A Pilot Study. J Addict Res Ther 2013; 4(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Matsumura Y, Stiles KM, Reid J, Frenk E, Cronin S, Pagovich OE et al. Gene Therapy Correction of Aldehyde Dehydrogenase 2 Deficiency. Mol Ther Methods Clin Dev 2019; 15: 72–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sanchez AC, Li C, Andrews B, Asenjo JA, Samulski RJ. AAV Gene Therapy for Alcoholism: Inhibition of Mitochondrial Aldehyde Dehydrogenase Enzyme Expression in Hepatoma Cells. Hum Gene Ther 2017; 28(9): 717–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Karahanian E, Rivera-Meza M, Tampier L, Quintanilla ME, Herrera-Marschitz M, Israel Y. Long-term inhibition of ethanol intake by the administration of an aldehyde dehydrogenase-2 (ALDH2)-coding lentiviral vector into the ventral tegmental area of rats. Addict Biol 2015; 20(2): 336–44. [DOI] [PubMed] [Google Scholar]
- 97.Rivera-Meza M, Vásquez D, Quintanilla ME, Lagos D, Rojas B, Herrera-Marschitz M et al. Activation of mitochondrial aldehyde dehydrogenase (ALDH2) by ALDA-1 reduces both the acquisition and maintenance of ethanol intake in rats: A dual mechanism? Neuropharmacology 2019; 146: 175–183. [DOI] [PubMed] [Google Scholar]
- 98.Pastorino R, Iuliano L, Vecchioni A, Arzani D, Milic M, Annunziata F et al. Effect of alcohol dehydrogenase-1B and −7 polymorphisms on blood ethanol and acetaldehyde concentrations in healthy subjects with a history of moderate alcohol consumption. Drug Test Anal 2018; 10(3): 488–495. [DOI] [PubMed] [Google Scholar]
- 99.(NIAAA) NIoAAaA. Drinking Levels Defined. In. National Institute on Alcohol Abuse and Alcoholism (NIAAA) |: https://www.niaaa.nih.gov/alcohol-health/overview-alcohol-consumption/moderate-binge-drinking, 2020. [Google Scholar]
- 100.Bolli R, Jeroudi MO, Patel BS, DuBose CM, Lai EK, Roberts R et al. Direct evidence that oxygen-derived free radicals contribute to postischemic myocardial dysfunction in the intact dog. Proc Natl Acad Sci U S A 1989; 86(12): 4695–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Yokoyama M, Yokoyama A, Yokoyama T, Hamana G, Funazu K, Kondo S et al. Mean corpuscular volume and the aldehyde dehydrogenase-2 genotype in male Japanese workers. Alcohol Clin Exp Res 2003; 27(9): 1395–401. [DOI] [PubMed] [Google Scholar]
- 102.Muto M, Takahashi M, Ohtsu A, Ebihara S, Yoshida S, Esumi H. Risk of multiple squamous cell carcinomas both in the esophagus and the head and neck region. Carcinogenesis 2005; 26(5): 1008–12. [DOI] [PubMed] [Google Scholar]
- 103.Asakage T, Yokoyama A, Haneda T, Yamazaki M, Muto M, Yokoyama T et al. Genetic polymorphisms of alcohol and aldehyde dehydrogenases, and drinking, smoking and diet in Japanese men with oral and pharyngeal squamous cell carcinoma. Carcinogenesis 2007; 28(4): 865–74. [DOI] [PubMed] [Google Scholar]
- 104.Ding JH, Li SP, Cao HX, Wu JZ, Gao CM, Su P et al. Polymorphisms of alcohol dehydrogenase-2 and aldehyde dehydrogenase-2 and esophageal cancer risk in Southeast Chinese males. World J Gastroenterol 2009; 15(19): 2395–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Weinshenker D Cocaine sobers up. Nat Med 2010; 16(9): 969–70. [DOI] [PubMed] [Google Scholar]
- 106.Yokoyama A, Omori T, Yokoyama T. Alcohol and aldehyde dehydrogenase polymorphisms and a new strategy for prevention and screening for cancer in the upper aerodigestive tract in East Asians. Keio J Med 2010; 59(4): 115–30. [DOI] [PubMed] [Google Scholar]
- 107.Dawsey SM, Lewin KJ, Wang GQ, Liu FS, Nieberg RK, Yu Y et al. Squamous esophageal histology and subsequent risk of squamous cell carcinoma of the esophagus. A prospective follow-up study from Linxian, China. Cancer 1994; 74(6): 1686–92. [DOI] [PubMed] [Google Scholar]


